- 1Anhui Provincial Key Laboratory of Microbial Control, Engineering Research Center of Fungal Biotechnology, Ministry of Education, School of Forestry and Landscape Architecture, Anhui Agricultural University, Hefei, China
- 2Graduate School of Bio-Applications and Systems Engineering, Tokyo University of Agriculture and Technology, Tokyo, Japan
- 3Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou, China
- 4Anhui Academy of Science and Technology, Hefei, China
Insects’ olfactory receptor plays a central role in detecting chemosensory information from the environment. Odorant receptors (ORs) and ionotropic receptors (IRs) are two types of olfactory receptors, and they are essential for the recognition of ligands at peripheral neurons. Apriona germari (Hope) (Coleoptera: Cerambycidae) is one of the most serious insect pests that cause damage to economic trees and landscaping trees, resulting in massive environmental damages and economic losses. Olfactory-based management strategy has been suggested as a promising strategy to control this wood-boring beetle. However, the olfactory perception mechanism in A. germari is now almost unknown. In the present study, RNA sequencing analysis was used to determine the transcriptomes of adult A. germari antennae. Among 36,834 unigenes derived from the antennal assembly, we identified 42 AgerORs and three AgerIRs. Based on the tissue expression pattern analysis, 27 AgerORs displayed a female-biased expression. Notably, AgerOR3, 5, 13, 33, and 40 showed a significant female-biased expression and were clustered with the pheromone receptors of Megacyllene caryae in the phylogenetic tree, suggesting that these AgerORs could be potential pheromone receptors for sensing male-produced sex pheromones in A. germari. The AgerIRs expression profile demonstrated that AgerIR2 had high expression levels in male labial palps, suggesting that this receptor may function to detect female-deposited trail-sex pheromone blend of A. germari. In addition, the phylogenetic tree showed that the Orco gene of five cerambycidae species was highly conservative. These results provide a foundation for further studies on the molecular mechanisms of olfactory chemoreception in A. germari apart from suggesting novel targets for the control of this pest in the future.
Introduction
Insects have developed a set of highly specialized and sensitive olfactory system that can accurately identify miniscule and highly specific odor substances in a complex natural environment and perform adaptive behaviors by long-term selective evolution (Jacquin-Joly and Merlin, 2005; Martin et al., 2011). The accurate recognition of chemical signals in insects is mainly accomplished by the bristles of sensory taste and smell neurons on their cuticles, among which the olfactory sensilla are mainly distributed in the antennae and the maxillary palp of insects. It is thought that olfactory recognition in insects is a complex process involving multiple chemosensory-associated proteins, including chemosensory proteins, odorant receptors (ORs), odorant-binding proteins, odorant-degrading enzymes, ionotropic receptors (IRs), and sensory neuron membrane proteins (Sato and Touhara, 2009; Leal, 2013). The initial steps in odor detection involve the binding of an odor to the OR displayed on the dendrites of olfactory sensory neurons, which leads to signal transduction (chemical signals become electrical signals), before they are transmitted to the central nervous system for complete odor perception (Vogt and Riddiford, 1981; Durand et al., 2011; Bohbot and Dickens, 2012). Therefore, olfactory receptors are one of the key components of the olfactory system (Gong et al., 2008; Zhan et al., 2018). Since the discovery of olfactory receptors in Drosophila melanogaster in 1999, the olfactory receptor system has been a key link in understanding the molecular recognition mechanism of insects’ chemical signals. This also provides the basis for the specific control of agricultural and forestry pests through insect smell.
The olfactory receptors of insects include ORs and IRs. ORs are composed of about 400 amino acids and has seven transmembrane domains, with the N-terminal on the inner side of the cell and C terminus on the outer side of the cell, in contrast to the G-protein coupled receptor family (Benton et al., 2006; Lundin et al., 2007). The OR of insects can be divided into two categories: one is typical odor receptor, which includes common odor receptors and sex pheromone receptors representing the majority of ORs, which are lowly conserved between subspecies; the other is atypical odor receptor (Orco) which comprises a small number of proteins, highly conserved between subspecies, but is not directly involved in the perception of odor molecules (Nichols and Luetje, 2010; Nichols et al., 2011; Wang et al., 2018). ORs mainly recognize odorants like volatiles and less volatile matter, including plant volatiles, fragrance material, herbivore-induced plant volatiles, sex pheromones, and intraspecific volatiles such as alarm pheromones and aggregation pheromones (Dweck et al., 2013; Cattaneo et al., 2017; Zhang et al., 2017; Wu et al., 2018). IRs are olfactory receptors identified by Benton in the antennae of fruit flies, belonging to the ionotropic glutamate receptor family (Benton et al., 2009). IRs and ORs play a complementary role in odor recognition (Abuin et al., 2011), although IRs and ORs generally recognize different odor substances. ORN-expressing IRs are less sensitive to odor substances than those of ORN-expressing ORs, and the electrophysiological response rate of IR to odor substances is also slower than that of OR (Yao et al., 2005; Getahun et al., 2012). Functional studies of IRs in D. melanogaster have shown that they play an important role in the perception of the environment, such as smell, taste, humidity, and temperature (Hussain et al., 2016; Chen and Amrein, 2017; Knecht et al., 2017; Prieto-Godino et al., 2017; Budelli et al., 2019).
The brown mulberry longhorn beetle, Apriona germari (Hope) (Coleoptera: Cerambyciade, Lamiinae), native to China, Japan, North Korea, Thailand, and India, is one of the most serious insect pests that cause damage to economic trees and landscaping trees (Huang, 1999; Bi et al., 2017). It is a polyphagous xylophage that mainly damages Populus spp., Morus alba L., Broussonetia papyrifera L., Ulmus pumila L., Malus pumila Mill., Cerasusus pseudocerasus G., Pyrus spp., and Citrus reticulata (Huang, 1999; Zhang et al., 2011). In recent years, many provinces in China have vigorously promoted the program of returning farmland to forests, and as a result, areas of Populus spp. and Morus alba L. have expanded rapidly. However, damage caused by wood-boring beetles occurs almost every year. Among them, A. germari has been listed as a “second-level” harmful forest pest, with an affected area of over 666.67 km2, threatening the consolidation of afforestation efforts, economic benefits, and ecological effects (Lu et al., 2001; Tang et al., 2013). Given the devastating damage of A. germari to forests, an efficient and sustainable control tool still remains a challenge. Olfactory-based management strategy has been considered as a promising strategy to control this wood-boring beetle. However, olfactory perception mechanisms in A. germari are currently almost unknown. In order to increase our understanding of the olfactory receptor genes involved in this beetle, we carried out our studies by focusing on (1) analyzing the transcriptome data of adult A. germari antennae using bioinformatics, followed by screening of and identifying olfactory receptor genes, (2) examining the expression of olfactory receptor genes of both male and female adults using quantitative real-time polymerase chain reaction (qRT-PCR), and (3) providing valuable information for analyzing the role of olfactory receptor genes involved in the molecular chemoreception mechanisms of olfaction in A. germari.
Materials and Methods
Insect and Tissue Collections
The newly emerged adults of A. germari were collected from a poplar plantation in Bengbu Forest Farm, Anhui Province, China, in June 2018. The branches of B. papyrifera were collected in a centrifugal tube (50 ml) with a temperature of 25 ± 1°C and humidity of 65 ± 5%. Vigorous male and female adults were selected, and their antennae, maxillary palps, labial palps, and abdominal parts were immediately snap-frozen in liquid nitrogen and stored separately at −80°C until use.
RNA Extraction, cDNA Library Construction, and Next-Generation Sequencing
Total RNA was extracted from A. germari adults’ antennae (both male and female) by TRIzol method. The RNA samples were sent to Novogene Co., Ltd. (Beijing, China) for non-reference transcriptome sequencing. cDNA library construction and next-generation sequencing were performed as previously described (Sun et al., 2018). In brief, RNA purity was checked using a NanoPhotometer spectrophotometer (IMPLEN, CA, United States). NEBNext® UltraTM RNA Library Prep Kit (NEB, United States) was used to generate sequencing libraries, and index codes were added to attribute sequences to each sample. The cBot Cluster Generation System was used to generate a cluster of the index-coded samples with TruSeq PE Cluster Kit v3-cBot-HS (Illumia, San Diego, CA, United States), following the manufacturer’s instructions. After cluster generation, an Illumina Hiseq platform was used to sequence the library preparations and generate paired-end reads.
Assembly and Functional Annotation
Transcriptome assembly was accomplished based on clean reads using Trinity (V2.4.0) to generate transcripts (Grabherr et al., 2011). The clean reads obtained by sequencing were spliced to obtain transcripts, and the longest transcript of each gene was selected as Unigene. The transcript sequence was compared with protein databases NCBI (non-redundant, Nr), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Clusters of Orthologous Groups of proteins (E-value < 10–5) by Blastx and nucleic acid databases Nt (E-value < 10–5) by Blastn. The transcript was annotated as the protein with the highest consistency. Open reading frame (ORF) finder1 was used to find ORFs for related genes. The transmembrane prediction of receptor genes is based on TMHMM Server v. 2.02 online software. All nucleic acid sequences were translated into amino acid sequences by Primer Premier 5 software.
Phylogenetic Analysis
The amino acid sequences of candidate receptor genes were aligned by BioEdit with similar reported species. Then, MEGA5 was used to make phylogenetic trees by neighbor-joining method and bootstrap with 1,000 replicates (Saitou and Nei, 1987). The evolutionary tree of candidate ORs was constructed from the protein sequences of Anoplophora chinensis (Sun et al., 2018), Anoplophora glabripennis (Mitchell et al., 2017), Agrilus planipennis (Mitchell et al., 2020), Dendroctonus ponderosae (Andersson et al., 2013), Tribolium castaneum (Engsontia et al., 2008), Anomala corpulenta (Li et al., 2015), Monochamus alternates (Wang et al., 2014), Megacyllene caryae (Mitchell et al., 2012), and Tenebrio molitor (Liu et al., 2015). The evolutionary tree of candidate IRs was built with the aligned protein sequences from A. chinensis, I. typographus, A. glabripennis, D. ponderosae, D. melanogaster (Benton et al., 2009), Phyllotreta striolata (Wu et al., 2016), T. castaneum (Croset et al., 2010), A. corpulenta, M. alternates, and T. molitor.
qRT-PCR Validation of ORs and IRs
Total RNA was isolated from 30 antennae, 80 maxillary palps, 80 labial palps, and 30 body ends from each sex. Isolated RNA was reverse-transcribed into cDNA using PrimeScriptTMRT reagent kit with gDNA Eraser (Perfect Real Time, TaKaRa, Beijing, China). qRT-PCR validation was carried out as described in Sun et al. (2018) but using different primers (Supplementary Table S1). Three biological replications were carried out for each sample and measured in three technique replications. The variability of each gene expression in different tissues was confirmed by using Q-Gene method (Simon, 2003). Graphical plot mapping was done by GraphPad prism v5.0 Software (GraphPad Software Inc., CA, United States). The relative expression of mRNA of each gene (mean ± SD) was analyzed using one-way ANOVA (SPSS22.0 for Windows, IBM, United States), followed by Duncan’s new multiple-range test (a = 0.05).
Results
Transcriptome Sequencing and Homology Assembly
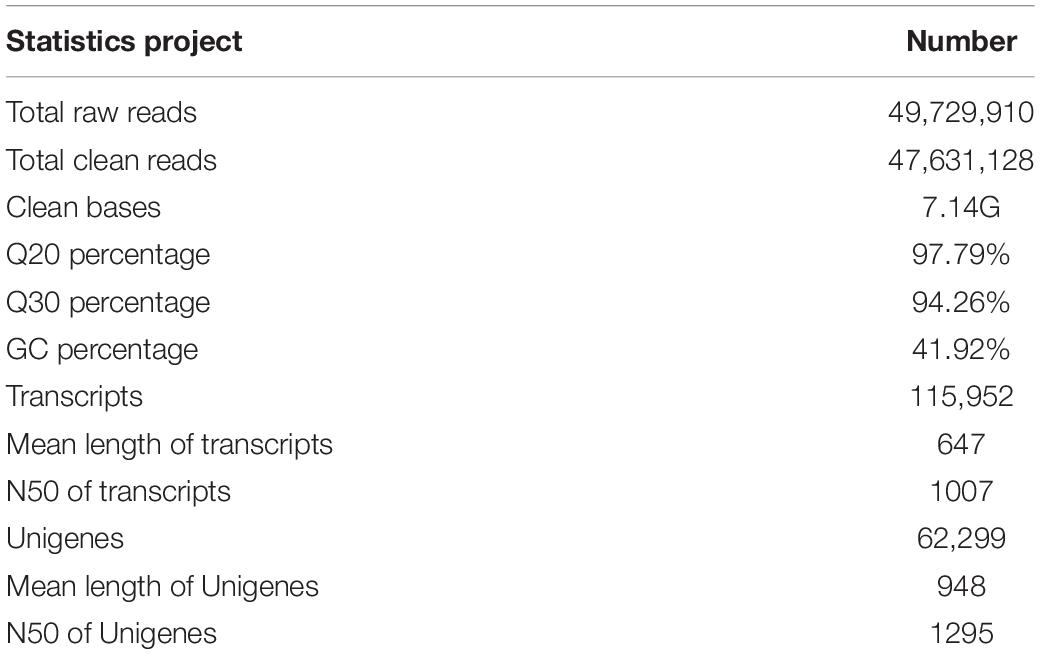
The transcriptome information of the longicorn beetle, A. germari, was characterized by constructing a cDNA library prepared from purified mRNA isolated from the adults’ antennae. HiSeq 2500 system generated a total of 49,729,910 raw reads and 47,631,128 clean reads. The Q20 and Q30 base call accuracies were 97.79 and 94.26%, respectively (Table 1). From these, 62,299 unigenes were screened from 115,952 transcripts. The mean length of the transcript and the unigene was 647 and 948 bp, respectively, with an N50 of 1,007 and 1,295 bp, respectively (Table 1). The length frequency distribution for unigenes and transcripts (Supplementary Figure S1) showed declines in the number of transcripts with increasing length. However, the number of unigenes was increased at first and then declined in the range between 1,001 and 2,000 bp. Less than one-fifth of short reads (<301 bp) were assembled into unigenes, while most reads over 500 bp were assembled into unigenes, indicating that longer reads (>500 bp) are more likely to be assembled into unigenes.
A total of 36,834 unigenes (59.12%) were compared to proteins in the NCBI Nr protein database using the BLASTX algorithm (E-value < 0.00001). As shown in Figure 1, among the annotated unigenes, approximately 44.8% unigenes showed a high homology (E-value < 1e-60) (Figure 1A). The identity comparison showed that 83.9% unigenes have more than 60% identity with other insects (Figure 1B). The top five species distributions were shown in Figure 1C. Approximately 76.1% unigenes were annotated to five top-hit insect species. T. castaneum was the first top-hit species with 53.4% annotated genes. The other top-hit species were Dendroctonus ponderosae (17.1%), Bombyx mori (3.0%), Lasius niger (1.4%), and Danaus plexippus (1.2%) (Figure 1C).
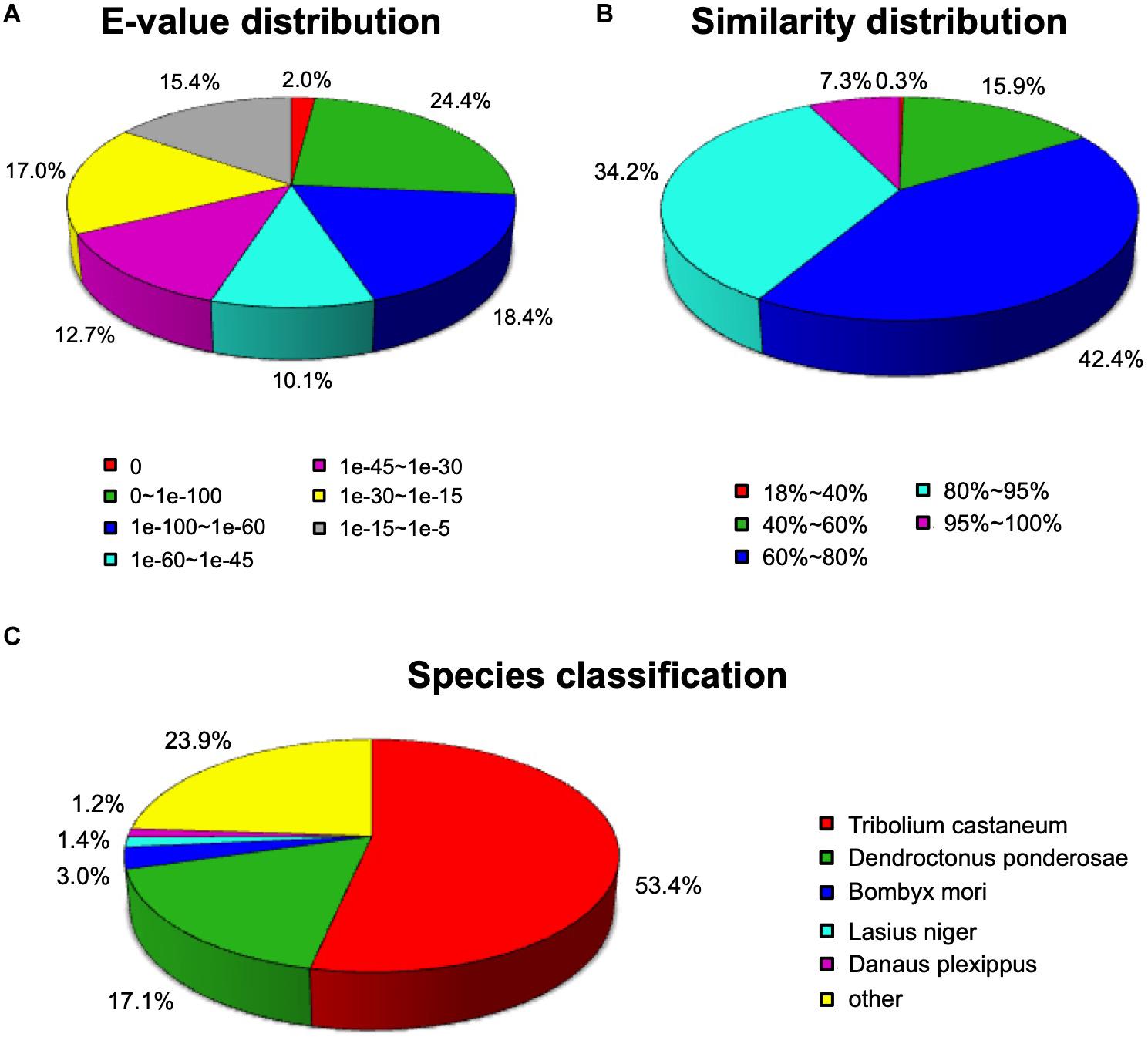
Figure 1. Homology analysis of Apriona germari unigenes. (A) E-value distribution. (B) Similarity distribution. (C) Species distribution. All unigenes that had BLASTX annotations within the NCBI nr database with a cutoff E-value of 10–5 were analyzed. The first hit of each sequence was used for analysis.
Gene Ontology Annotation and KEGG Analysis
According to Gene Ontology (GO) analysis, a total of 27,260 unigenes were assigned to three GO functional categories: biological process, cellular component, and molecular function. Among them, 69,435 times are classified to the category of biological process, 44,677 times to cellular component, and 32,562 times to molecular function (Figure 2). A total of 54 categories were divided into subcategories: biological process (25 subcategories), cellular component (19 subcategories), and molecular function (10 subcategories). Among the 25 subcategories of biological process, the largest proportion of genes is involved in cellular process (14,836; 21.37%), followed by metabolic process (13,995; 20.16%) and single-organism process (11,864; 17.09%). As for the category of cellular component, cell (8,743; 19.57%) and cell part (8,743; 19.57%), followed by organelle (6,240; 13.97%), made up the majority of the proportion. For the category of molecular function, a significant proportion of the genes is assigned to binding (15,075; 46.3%) and catalytic activity (11,594; 35.61%), while no gene was assigned to antioxidant activity and metallochaperone activity (Supplementary Table S2).
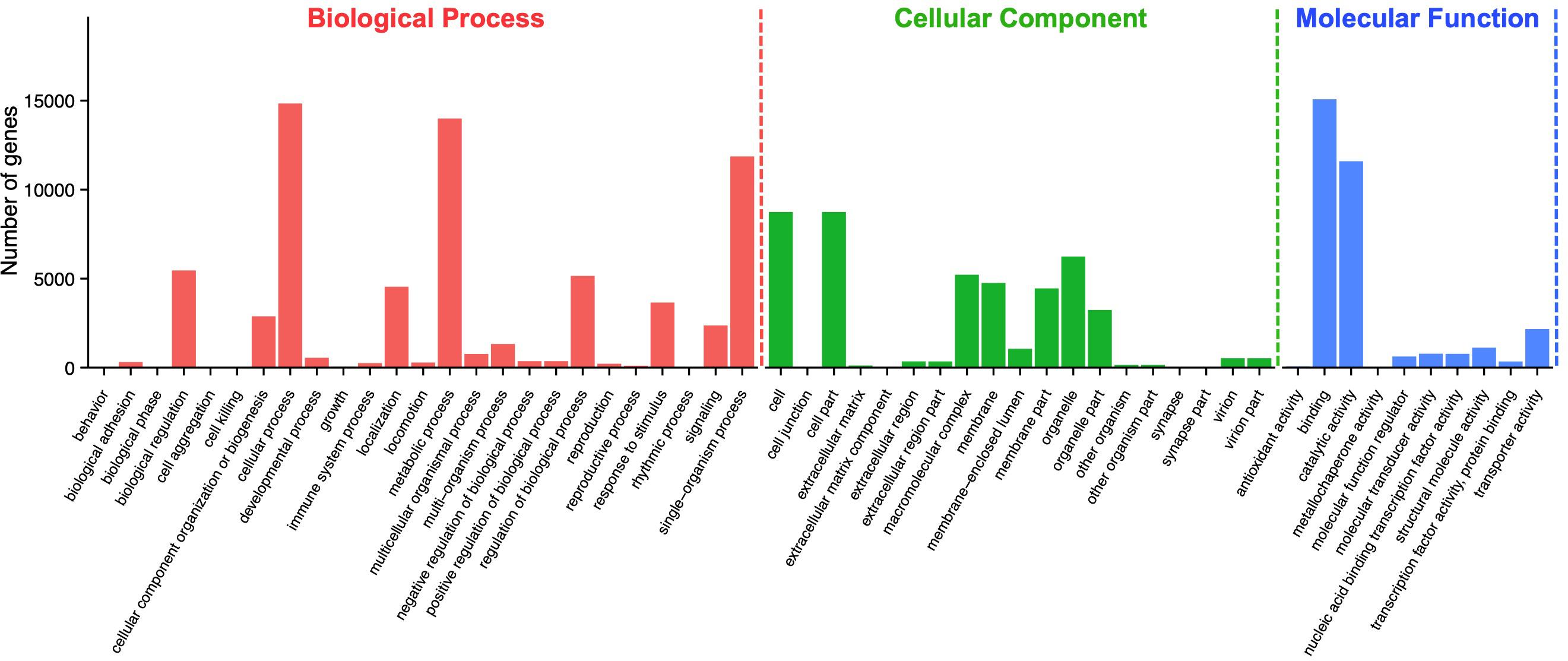
Figure 2. Gene ontology (GO) assignment of Apriona germari unigenes. The GO classification map was done by uploading the GO ID numbers of genes for their involvement in biological processes, cellular components, and molecular functions.
KEGG database was used to analyze the metabolic pathways of unigenes. A total of 16,846 unigenes were assigned to five specific KEGG pathways (Figures 3A–E): cellular processes (Figure 3A), environmental information processing (Figure 3B), genetic information processing (Figure 3C), metabolism (Figure 3D), and organismal systems (Figure 3E). Moreover, 32 KEGG pathways were further assigned to these five pathways, in which the largest number of unigenes (n = 2,100) was assigned to the pathway of signal transduction, followed by transduction (n = 1,510), transport and catabolism (n = 1,310), and endocrine system (n = 1,287).
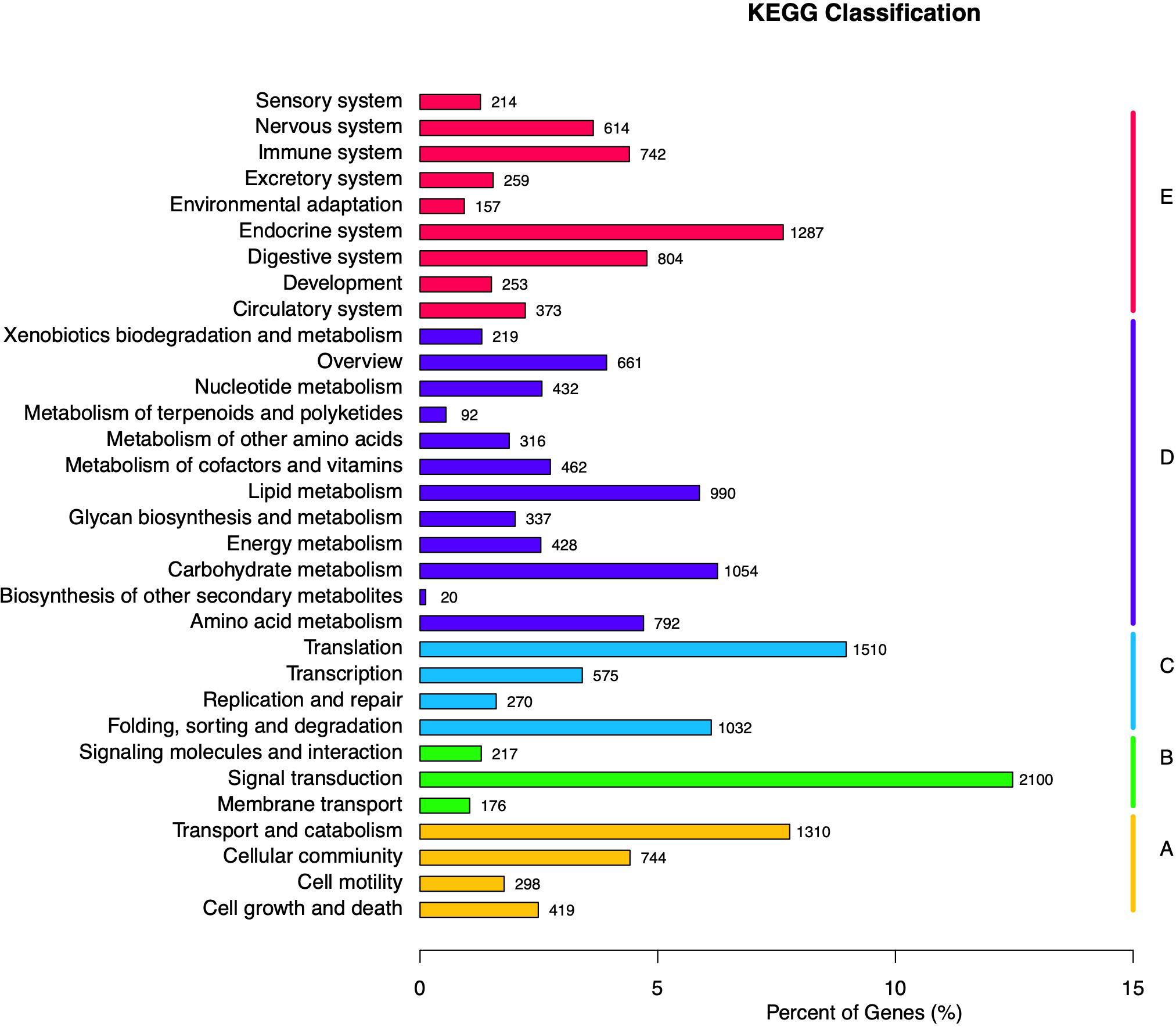
Figure 3. Kyoto Encyclopedia of Genes and Genomes (KEGG) classification of Apriona germari unigene. The x-axis indicates the percentage of annotated genes, and the y-axis indicates the KEGG categories. The capital letters against the colored bars indicate five main categories: (A) cellular processes, (B) environmental information processing, (C) genetic information processing, (D) metabolism, and (E) organism systems.
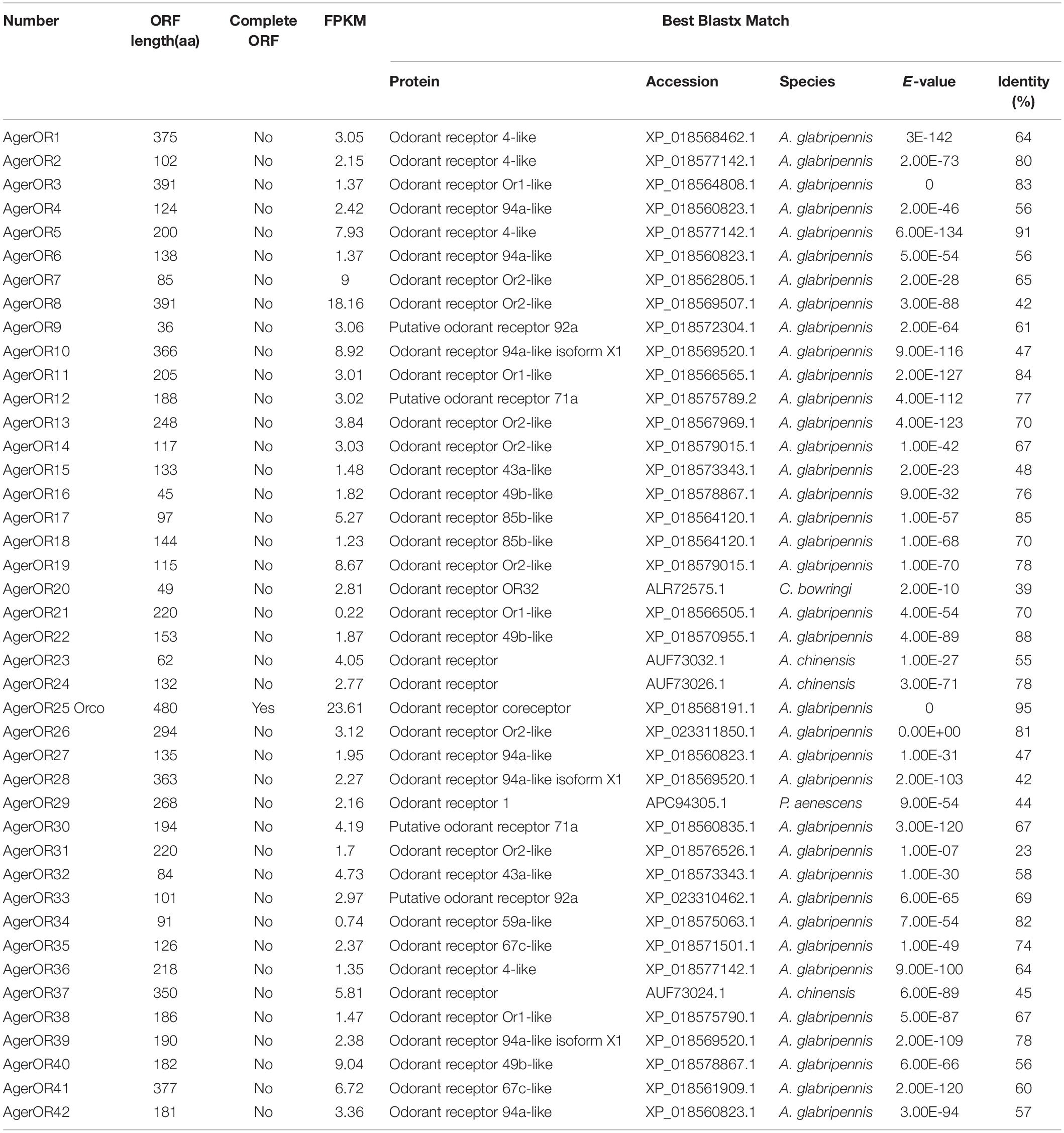
Identification of Putative Odorant Receptors
Based on the comparative analysis of the A. germari antennal transcriptome using Blastx databases, a total of 42 putative AgerORs genes were identified. As shown in Table 2, eight AgerORs (AgerOR1, 3, 8, 10, 25, 28, 37, and 41) were over 1,000 bp in length. However, only the AgerOR25 sequence had complete ORFs. Since AgerOR25 was identified with a high sequence homology with the conserved Orco gene family of other insect species, we therefore designated it as AgerOrco. Next, a phylogenetic tree was constructed to evaluate the relationships of AgerORs with other insects’ ORs (Figure 4). Based on the OR phylogenetic tree analysis, the published OR genes could be divided into multiple subgroups (numbered 1, 2A, 2B, 3, 4, 5A, 5B, 6, and 7). In the current study, except AgerOR25, which was designated as AgerOrco, the other 41 putative AgerORs were classified into five subgroups (group 1, 2A and 2B, 3, 5B, and 7). Seven AgerORs (AgerOR13, 14, 19, 22, 33, 35, and 40) were clustered in group 1, while 14 AgerORs (AgerOR1–3, 5, 10, 11, 17, 26, 28–30, 34, 37, and 39) were assigned to group 2A and 2B. Another 14 AgerORs (AgerOR4, 6, 9, 15, 16, 18, 20, 21, 27, 31, 32, 36, 41, and 42) belonged to group 3. AgerOR12 and AgerOR38 were clustered in group 5B. The other four AgerORs, including AgerOR7, 8, 23, and 24, were classed in group 7. In addition, seven AgerORs (AgerOR2, 3, 5, 13, 33, 34, and 40) were clustered with pheromone receptors from M. caryae (labeled in black square in Figure 4).
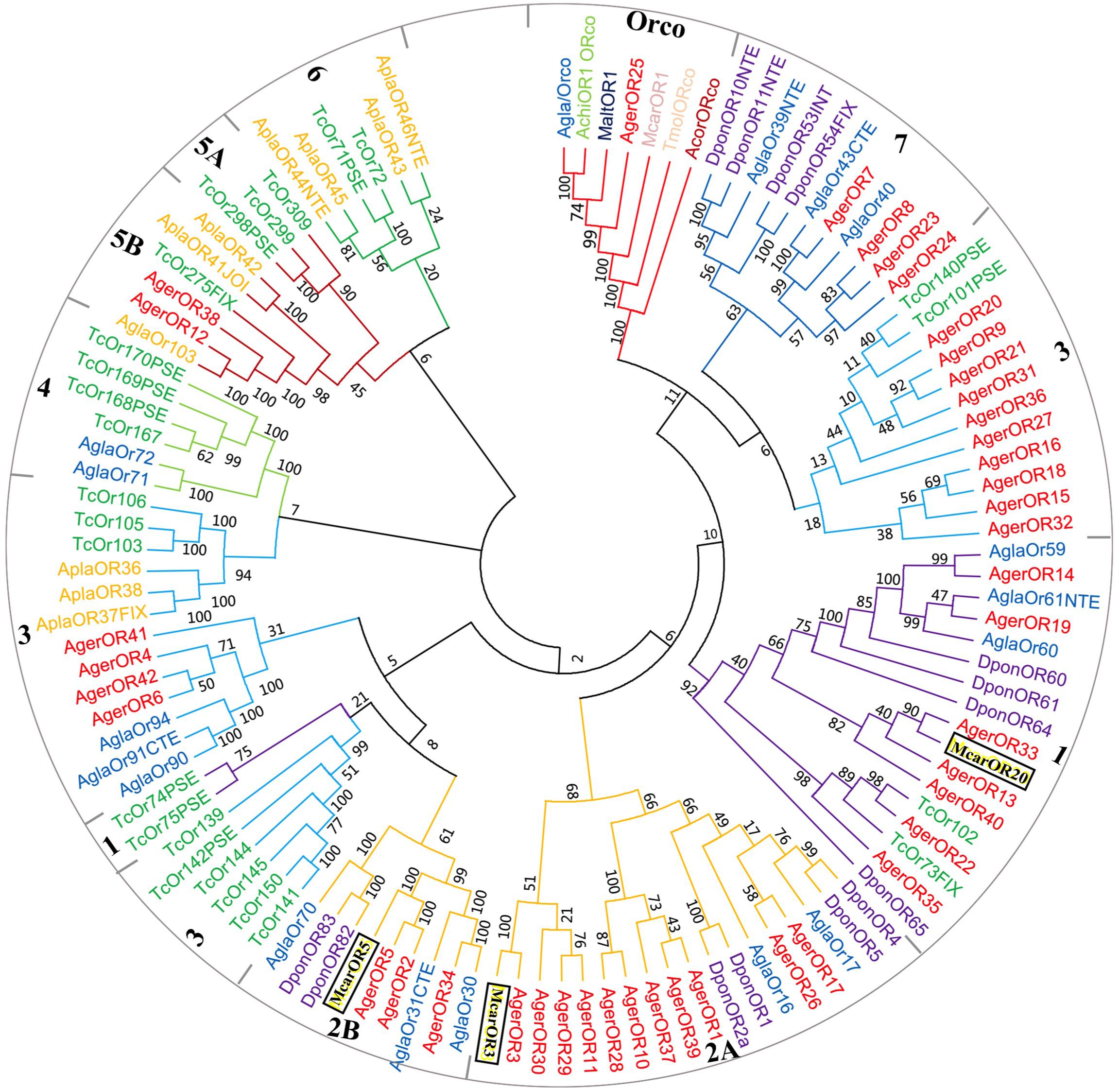
Figure 4. Molecular phylogeny comparing AgerORs with odorant receptors (ORs) from 10 insect species. A total of 42 ORs (AgerOR1–42) from Apriona germari (Ager) and ORs from Anoplophora chinensis (Achi), Anoplophora glabripennis (Agla), Agrilus planipennis (Apla), Dendroctonus ponderosae (Dpon), Tribolium castaneum (Tcas), Anomala corpulenta (Acor), Monochamus alternates (Malt), Megacyllene caryae (Mcar), and Tenebrio molitor (Tmol) were used to construct the phylogenetic tree (see section “Materials and Methods” for details of the phylogenetic analysis).
Tissue- and Sex-Specific Expressions of Odorant Receptors
We next examined the expression of OR genes in adult female and male antennae, maxillary palps, labial palps, and the end part of the abdomen (abdominal end) by qRT-PCR with primers specific for each of the 42 AgerOR genes. AgerOR1, 2, 9, 11, 12, 21, 22, 32, and 36 were found to be ubiquitously expressed in chemosensory organs. In addition, the amplification products of 10, two, two, and two AgerOR genes were identified in the antennae, the maxillary palps, the labial palps, and the abdominal end, respectively (Figure 5 and Supplementary Figure S2).
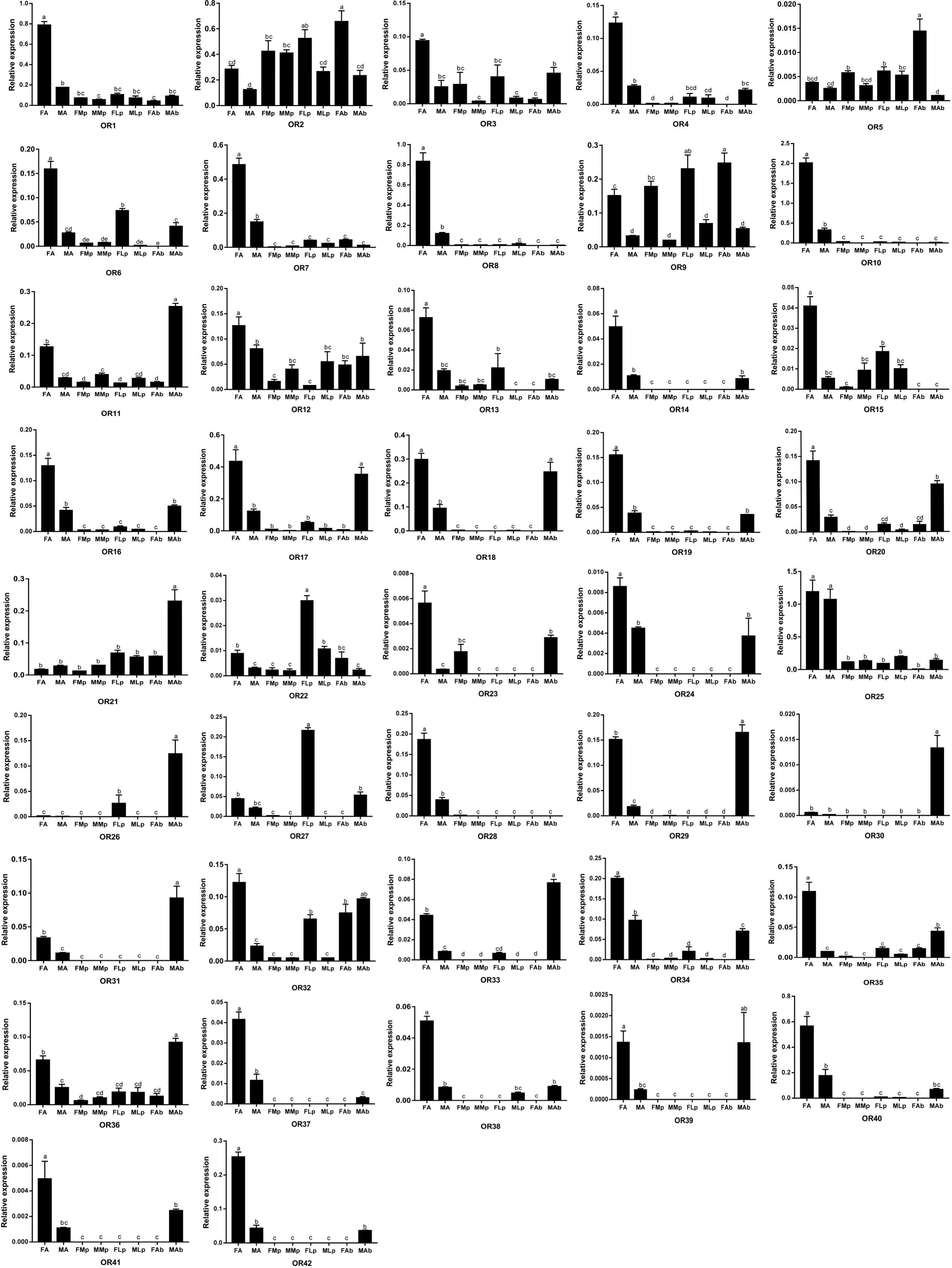
Figure 5. Relative mRNA expression of AgerORs in Apriona germari tissues. The relative mRNA levels were normalized to those of the actin gene and analyzed using the Q-gene method. All values are shown as mean ± SEM normalized. The data were analyzed by least significant difference test after one-way analysis of variance. Different letters (a–e) indicate significant differences between means (P < 0.05). FA, female antennae; MA, male antennae; FMp, female maxillary palps; MMp, male maxillary palps; FLp, female labial palps; MLp, male labial palps; FAb, female abdominal end; MAb, male abdominal end.
Of the 40 AgerORs expressed in the antennae, 27 AgerORs (AgerOR1, 3, 4, 6–8, 10, 12–20, 23, 24, 28, 29, 34, 35, 37, 38, 40, and 42) were female-biased and two AgerORs (AgerOR21 and 25) were, of the same level, expressed in both female and male (Figure 5 and Supplementary Figure S2A). Among the 14 AgerOR genes expressed in the maxillary palps, AgerOR3, AgerOR5, and AgerOR9 were female-biased, while AgerOR11 were expressed at a significantly higher level in the male maxillary palps (Figure 5 and Supplementary Figure S2B). Among the 17 AgerOR genes expressed in the labial palps, AgerOR2, 9, and 27 were highly expressed in the female labial palps (Figure 5 and Supplementary Figure S2C). Finally, among the 38 abdominal end AgerORs analyzed, three AgerORs (AgerOR2, 5, and 9) were highly expressed in the female abdominal end, 13 AgerORs (AgerOR11, 17, 18, 20, 21, 25, 29, 31–34, 36, and 40) showed a high expression in the male end-body (Figure 5 and Supplementary Figure S2D).
Identification of Putative Ionotropic Receptors and Their Expression Pattern
In the current study, three putative AgerIRs were also identified in the combined antennal transcriptome (Table 3). According to the phylogenetic analysis of IRs from one Diptera D. melanogaster and eight species of Coleopterans (Figure 6A), AgerIR3 was clustered with TcasIR25a at high percent identity, suggesting that it belongs to the IR25a coreceptor subfamily.
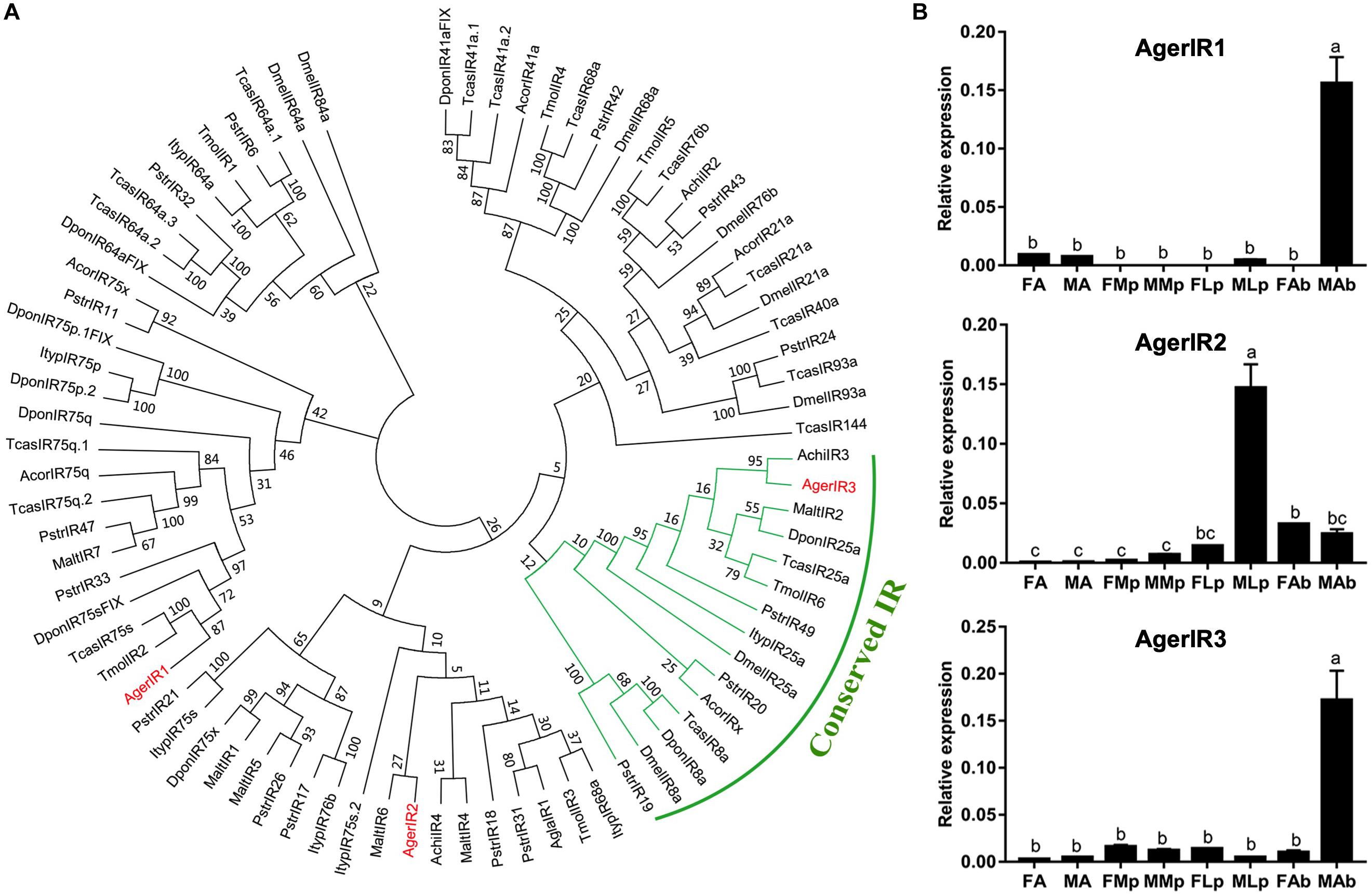
Figure 6. Phylogenetic tree and relative mRNA expression of AgerIRs. (A) A total of 42 ORs (AgerOR1–42) from Apriona germari (Ager) and ORs from Anoplophora chinensis (Achi), Anoplophora glabripennis (Agla), Dendroctonus ponderosae (Dpon), Drosophila melanogaster (Dmel), Phyllotreta striolata (Pstr), Anomala corpulenta (Acor), Monochamus alternates (Malt), Megacyllene caryae (Mcar), Tenebrio molitor (Tmol), and Tribolium castaneum (Tcas) were used to construct the phylogenetic tree. (B) Relative mRNA expression of AgerIRs. The relative mRNA levels were normalized to those of the actin gene and analyzed using the Q-gene method. All values are shown as mean ± SEM normalized. The data were analyzed by least significant difference test after one-way analysis of variance. Different letters (a–c) indicate significant differences between means (P < 0.05). FA, female antennae; MA, male antennae; FMp, female maxillary palps; MMp, male maxillary palps; FLp, female labial palps; MLp, male labial palps; FAb, female abdominal end; MAb, male abdominal end.
To further investigate the expression of these three IR genes, qRT-PCR experiments were also performed using total RNA prepared from male antennae, maxillary palps, labial palps, and the end part of the abdomen (abdominal end) taken from both female and male A. germari adults. As shown in Figure 6B, AgerIR1 and AgerIR3 were found in the male end-body, with a significantly higher expression than in any other tissues. Another AgerIR gene, AgerIR2, showed high expression levels in the male labial palps (Figure 6B and Supplementary Figure S3).
Discussion
Coleoptera is the first order of Insecta and one of the most important orders in forestry, and the longhorn beetle is one of the larger groups of Coleoptera, with 45,000 species in the world and more than 3,100 species known in China (Zhang, 2011). To date, however, olfactory genes have been studied in only 20 species and limited in three species of Cerambycidae (Monochamus alternatus, Batocera horsfieldi, and A. glabripennis) (Li et al., 2014; Gao and Wang, 2015; Hu et al., 2016). A. germari (Hope) (Coleoptera: Cerambycidae) is one of the most serious insect pests that cause damage to economic trees and landscaping trees, resulting in massive environmental damages and economic losses. To reveal the olfactory receptor genes involved in this beetle, we conducted an RNA sequencing analysis of the antennae of adult A. germari.
In the transcriptome sets, we obtained 62,299 unigenes from 115,952 transcripts, with a mean length of 948 bp, and 67.07% of these unigenes were longer than 500 bp. These results indicate the high quality and depth of the transcriptome sequences. The BLASTX homology analysis showed that A. germari antennal transcriptome sequences best match with T. castaneum (53.4%) and D. ponderosae (17.1%), presumably because many homologous genes, including olfactory genes, were present in T. castaneum, D. ponderosae, and A. germari. The above unigenes may play a significant part in antennal chemosensory processes, such as transporter activity and odorant binding.
The 42 AgerORs identified in A. germari are similar to the 43 OR genes in A. corpulenta (Li et al., 2015) and I. typographus (Andersson et al., 2013), fewer than the 57 ORs reported in M. caryae (Mitchell et al., 2012) or the 73 transcripts encoding ORs in the antennal transcriptomes of P. striolata (Wu et al., 2016) and the 53 ORs in A. chinensis but more than the nine ORs identified in M. alternatus (Wang et al., 2014), six ORs in Rhyzopertha dominica (Diakite et al., 2016), or 37 ORs in A. glabripennis (Hu et al., 2016) belonging to seven known coleopteran specific subgroups. There were seven coleopteran specific subgroups reported in previous studies (Engsontia et al., 2008; Andersson et al., 2013, 2019). In the phylogenetic tree of ORs, 42 putative AgerORs sequences were spread into five subgroups (Figure 4). Interestingly, five AgerORs (AgerOR3, 5, 13, 33, and 40) were clustered with three pheromone receptors (PRs: McarOR3, McarOR5, and McarOR20), which are functionally characterized receptors from the cerambycid beetle M. caryae (Mitchell et al., 2012). Among the five AgerORs, AgerOR3 was orthologous to McarOR3, a receptor sensitive to the cerambycid pheromone (S)-2-methyl-1-butanol. Moreover, AgerOR5 was gathered with McarOR5, which is sensitive to 2-phenylethanol. In addition, AgerOR13, 33, and 40 were clustered with McarOR20, which was identified as a receptor of (2S, 3R)-2,3-hexanediol and 3-hydroxyhexan-2-one (Mitchell et al., 2012). Since they have high sequence similarities with the three PRs (McarOR3, McarOR5, and McarOR20) (Supplementary Figures S4–S6), these AgerORs may function to detect the abovementioned pheromones or other behaviorally active compounds.
Orco is the conserved subunit of ORs in insects and has been suggested as an attractive target for the manipulation of insect pest control programs (Kepchia et al., 2019). AgerOR25 could be the Orco in A. germari due to its specific Orco subgroup. Consistent with our previous report (Sun et al., 2018), the potential Orco (AgerOR25) showed a high sequence identity with MaltOR1 (Wang et al., 2014), McarOR1 (Mitchell et al., 2012), AglaOR1 (Mitchell et al., 2017), and AchiOR1 (Sun et al., 2018) (Supplementary Figure S7), indicating the conserved attribute of the Orco gene. Since the Orco plays a critical role in odorant detection, to disrupt this gene expression could be a potential utilization for control strategies. Orco silencing through RNA interference may hinder their ability to use olfactory cues to locate host plants and/or mates and to reduce pest populations. Overall, the highly conserved Orco genes of several Cerambycidae species could be anticipated as a potential interfering target for manipulation of the control programs.
Odor receptors are generally expressed in the antennae, but the specific function of odor receptors is different. In a recent study, 45 ORs were identified from the transcriptome of A. chinensis antennae. The qRT-PCR analysis showed that 41 ORs were highly expressed in the antennae; however, one OR was found to be highly expressed in the maxillary palp and the other three ORs were found to be highly expressed in the male body (Sun et al., 2018). Among the transcriptome of Adelphocoris lineolatus antennae with 57 randomly selected ORs, 26 ORs were found to be specifically expressed in male antennae, 16 ORs were found to be specifically expressed in female antennae, and the other ORs showed complex expression patterns, such as high expression in the body except the head or equal expression in male and female antennae (Xiao et al., 2017). In the current study, 27 AgerORs showed an antenna-specific expression profile. Those with female-biased expression may play a vital role in some female-specific behaviors such as oviposition site seeking. Notably, AgerOR3, 5, 13, 33, and 40 showed a clear female-biased expression profile (Figure 5). These five AgerORs were clustered with the pheromone receptors of M. caryae on the phylogenetic tree and have relatively similar amino acid sequences (Figure 4 and Supplementary Figures S4, S6). These findings allowed us to speculate that these receptors could be a potential pheromone receptor for sensing a male-produced sex pheromone in A. germari. In addition, we found that some AgerORs were highly expressed in the maxillary or the labial palp, suggesting that they may be involved in host selection for both sexes and oviposition site selection for females. We also found some AgerORs to be highly expressed in end-body tissues, which is consistent with what has been reported in other insects (Li et al., 2015; Zhang et al., 2016). In particular, 13 AgerORs showed a high expression in male end-body. Previous studies showed that some species of coleopteran, including Callosobruchus chinensis (Bruchidae), Aleochara curtula (Staphylinidae), and Adalia bipunctata (Coccinellidae), utilize cuticular hydrocarbons as contact sex pheromones (Tanaka et al., 1981; Peschke and Metzler, 1987; Hemptinne et al., 1998). We speculated that these AgerORs may play an important role related to function as the contact sex pheromone of A. germari.
IRs is a conserved family of synaptic ligand-gated ion channels, and its function has been reported to be involved not only in olfaction and gustation but also in thermosensation and hygrosensation (Missbach et al., 2014; Rimal and Lee, 2018). Three AgerIRs (AgerIR1–3) were identified in this study. The number of IRs of A. germari was less than that of D. ponderosae (n = 15) or P. striata (n = 49) (Wu et al., 2016) but relatively similar to A. chinensis (n = 4) (Sun et al., 2018), A. glabripennis (n = 4) (Hu et al., 2016), A. corpulenta (n = 5), and M. alternates (n = 7) (Wang et al., 2014). In the IR phylogenetic tree, IR25a and IR8a formed a conserved IR ortholog; AgerIR3 was in the conserved IR orthologs, which indicates that AgerIR3 is the conserved IR of A. germari, and AgerIR2 clustered with IR76b clade. In D. melanogaster, IRs were found to be expressed in the olfactory and the gustatory organs with the detection of acids, amines, and aldehydes (Benton et al., 2009; Abuin et al., 2011; Depetris-Chauvin et al., 2015; Ganguly et al., 2017). In A. glabripennis, a previous study suggested that the maxillary and the labial palps of the male beetle play an important role in detecting this beetles’ female-deposited trail-sex pheromone blend (Graves et al., 2016). Based on the results that AgerIR2 was highly expressed in the male labial palps (Figure 6B and Supplementary Figure S3), we speculate that this receptor may function to detect the female-deposited trail-sex pheromone blend of A. germari. In addition, AgerIR1 and AgerIR3 were found to be expressed in the end-body, indicating that IRs are expressed mainly, but not limited, in the olfactory and the gustatory organs. In the current study, however, the IR8a clade in A. germari was missing, which may be attributed to the sequencing depth and coverage. Further studies by adopting next-generation sequencing technology would be needed to clarify the question.
In summary, we firstly conducted an RNA sequencing analysis of the antennae of adult A. germari. Forty-five putative receptor genes were identified from the olfactory receptor gene families, including 42 AgerORs and three AgerIRs, through bioinformatic analysis. The qRT-PCR demonstrated that most olfactory receptors were prominently expressed in the antennae, especially in female antennae, suggesting that they were involved in female-specific behaviors. The functions of these receptors are unknown but can be inferred from the receptors of other longhorns. AgerOR3, 5, 13, 33, and 40 were clustered with the pheromone receptors of M. caryae, implying that they may be sensitive to structurally related chemicals that are pheromones of M. caryae. AgerIR2 was highly expressed in the male labial palps, suggesting that it may function to detect the female-deposited trail-sex pheromone blend of A. germari. In addition, four known Orcos of currently reported cerambycidae species and a new Orco (AgerOR25) of A. germari identified in this study were highly conservative, which could be anticipated as a potential interfering target for the manipulation of control programs. Overall, our findings provide a theoretical basis for subsequent studies on the olfactory mechanisms of A. germari and offer some new insights into the functions and the evolutionary characteristics of ORs and IRs in Coleoptera insects.
Data Availability Statement
The datasets generated for this study can be found in NCBI Genbank. The Genbank accession numbers were assigned as MT598219 to MT598257 for ORs and MT591677 to MT591679 for IRs.
Author Contributions
L-WZ, J-LQ, and Z-QL conceived and designed the experiments. J-LQ, G-CL, JY, and BC performed the experiments. J-LQ, L-WZ, D-ZM, BC, and LS analyzed the data. J-LQ, L-WZ, D-ZM, and Z-QL wrote the manuscript. All the authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key Research and Development Program (2017YFD0600101), the Forest-Scientific Research in the Public Interest (No. 201204506), the National Natural Science Foundation of China (31700569 and 31170616), and the Research Innovation Program for College Graduates of Anhui Agricultural University (Grant No. 2019YSJ-16).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Hong Huat Hoh (OIST Graduate University, Japan) for editing the manuscript. We also thank Dr. Le-tian Xu (Hubei University, China) for his assistance in data processing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00807/full#supplementary-material
FIGURE S1 | Sequence length distribution of transcripts and unigenes assembled from Illumina reads for the mantle transcriptome of Apriona germari. The x-axis indicates the length interval of transcripts and unigenes, and the y-axis indicates the number of transcripts and unigenes for each size.
FIGURE S2 | Relative mRNA expression of AgerORs in Apriona germari tissues. The relative mRNA levels were normalized to those of the actin gene and analyzed using the Q-gene method. All values are shown as mean ± SEM normalized. The data were analyzed by least significant difference test after one-way analysis of variance.
FIGURE S3 | Relative mRNA expression of AgerIRs in Apriona germari tissues. The relative mRNA levels were normalized to those of the actin gene and analyzed using the Q-gene method. All values are shown as mean ± SEM normalized. The data were analyzed by least significant difference test after one-way analysis of variance.
FIGURE S4 | Multiple sequence alignment of Megacyllene caryae and Apriona germari ORs. The multiple alignment and the homology of each OR were calculated using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
FIGURE S5 | Sequence alignment of McarOR3 and AgerOR3. The multiple alignment and the homology of each OR were calculated using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
FIGURE S6 | Sequence alignment of McarOR5 and AgerOR5. The multiple alignment and the homology of each OR were calculated using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
FIGURE S7 | Alignment of four known Orcos and a new Orco (AgerOR25) of Apriona germari identified in this study. Malter, M. alternates; Mcar, M. caryae; Agla, A. glabripennis; Achi, A. chinensis; Ager, A. germari. The multiple alignment and the homology of each Orco were calculated using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
TABLE S1 | Primers of Apriona germari olfactory receptor genes used for quantitative real-time polymerase chain reaction.
TABLE S2 | Gene Ontology classification count of Apriona germari antennae transcriptome.
Footnotes
References
Abuin, L., Bargeton, B., Ulbrich, M. H., Isacoff, E. Y., Kellenberger, S., and Benton, R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. doi: 10.1016/j.neuron.2010.11.042
Andersson, M. N., Ewald, G. W., Keeling, C. I., Bengtsson, J. M., Yuen, M. M. S., Li, M., et al. (2013). Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetle, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytine). BMC Genom. 14:198. doi: 10.1186/1471-2164-14-198
Andersson, M. N., Keeling, C. I., and Mitchell, R. F. (2019). Genomic content of chemosensory genes correlates with host range in wood-boring beetles (Dendroctonus ponderosae, Agrilus planipennis, and Anoplophora glabripennis). BMC Genom. 20:690. doi: 10.1186/s12864-019-6054-x
Benton, R., Sachese, S., Michnick, S. W., and Vosshall, L. B. (2006). Atypical membrane topology and heteromeric funcation of Drosophila odorant receptors in vivo. PLoS Biol. 4:e20. doi: 10.1371/journal.pbio.0040020
Benton, R., Vannice, K. S., Gomez Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Bi, Y. G., Guo, L., Wang, Z. G., Lü, F., and Yan, A. H. (2017). Electroantennogram and behavioral responses of Apriona germari to five plant volatiles and their mixtures. Sci. Sericult. 43, 32–38. doi: 10.13441/j.cnki.cykx.2017.01.005
Bohbot, J. D., and Dickens, J. C. (2012). Odorant receptor modulation: ternary paradigm for mode of action of insect repellents. Neuropharmacology 62, 2086–2059. doi: 10.1016/j.neuropharm.2012.01.004
Budelli, G., Ni, L., Berciu, C., Van, G. L., Knecht, Z. A., Chang, E. C., et al. (2019). Ionotropic receptors specify the morphogenesis of phasic sensors controlling rapid thermal preference in Drosophila. Neuron 101, 738.e–747.e. doi: 10.1016/j.neuron.2018.12.022
Cattaneo, A. M., Gonzalez, F., Bengtsson, J. M., Corey, E. A., Jacquin-Joly, E., Montagné, N., et al. (2017). Candidate pheromone receptors of codling moth Cydia pomonella respond to pheromone and kairomones. Sci. Rep. 7:41105. doi: 10.1038/srep41105
Chen, Y., and Amrein, H. (2017). Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Curr. Biol. 27, 2741.e–2750.e. doi: 10.1016/j.cub.2017.08.003
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6:e1001064. doi: 10.1371/journal.pgen.1001064
Depetris-Chauvin, A., Galagovsky, D., and Grosjean, Y. (2015). Chemicals and chemoreceptors: ecologically relevant signals driving behavior in Drosophila. Front. Ecol. Evol. 3:41. doi: 10.3389/fevo.2015.00041
Diakite, M. N., Wang, J., Ali, S., and Wang, M. Q. (2016). Identification of chemosensory gene families in Rhyzopertha dominica (Coleoptera: Bostrichidae). Can. Entomol. 148, 8–21. doi: 10.4039/tce.2015.13
Durand, N., Carot-Sans, G., Bozzolan, F., Rosell, G., Siaussat, D., Debernard, S., et al. (2011). Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS One 6:e29147. doi: 10.1371/journal.pone.0029147
Dweck, H. K. M., Ebrahim, S. A. M., Kromann, S., Bown, D., Hillbur, Y., Sachse, S., et al. (2013). Olfactory preference for egg laying on citrus substrates in Drosophila. Curr. Biol. 23, 2472–2480. doi: 10.1016/j.cub.2013.10.047
Engsontia, P., Sanderson, A. P., Cobb, M., Walden, K. K. O., Robertson, H. M., and Brown, S. (2008). The red flour beetle’s large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect. Biochem. Mol. Biol. 38, 387–397. doi: 10.1016/j.ibmb.2007.10.005
Ganguly, A., Pang, L., Duong, V.-K., Lee, A., Schoniger, H., Varady, E., et al. (2017). A molecular and celluar context-dependent role for Ir76b in detection of amino acid taste. Cell Rep. 18, 737–750. doi: 10.1016/j.celrep.2016.12.071
Gao, X., and Wang, M. Q. (2015). A cDNA library from the antennae of Monochamus alternatus Hope and binding properties of odorant-binding proteins. J. Appl. Entomol. 139, 229–236. doi: 10.1111/jen.12136
Getahun, M. N., Wicher, D., Hansson, B. S., and Olsson, S. (2012). Temporal response dynamics of Drosophila olfactory sensory neurons depends on receptor type and response polarity. Front. Cell Neurosci. 6:54. doi: 10.3389/fncel.2012.00054
Gong, Z. Z., Zhou, W. W., Zhu, Z. R., and Cheng, J. A. (2008). Advances in the studies of insect olfactory receptors. Acta Entomol. Sinica 51, 761–768. doi: 10.1016/S1005-9040(08)60003-3
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Graves, F., Baker, T. C., Zhang, A., Keena, M., and Hoover, K. (2016). Sensory aspects of trail-following behaviors in the asian longhorned beetle, Anoplophora glabripennis. J. Insect Behav. 29, 615–628. doi: 10.1007/s10905-016-9587-8
Hemptinne, J.-L., Lognay, G., and Dixon, A. F. G. (1998). Mate recognition in the two-spot ladybird beetle, Adalia bipunctata: role of chemical and behavioural cues. J. Insect Physiol. 44, 1163–1171. doi: 10.1016/s0022-1910(98)00081-x
Hu, P., Wang, J. Z., Cui, M. M., Tao, J., and Luo, Y. Q. (2016). Antennal transcriptome analysis of the Asian longhorned beetle Anoplophora glabripennis. Sci. Rep. 6, 26652. doi: 10.1038/srep26652
Huang, D. Z. (1999). The Regional Dynamic Law and Comprehensive Management of Mulberry Longhorn Cattle. Harbin: Northeast Forestry University Press.
Hussain, A., Zhang, M., Üçpunar, H. K., Svensson, T., Quillery, E., Gompel, N., et al. (2016). Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 14:e1002454. doi: 10.1371/journal.pbio.1002454
Jacquin-Joly, E., and Merlin, C. (2005). Insect olfactory receptors: contributions of molecular biology to chemical ecology. J. Chem. Ecol. 30, 2359–2397. doi: 10.1007/s10886-004-7941-3
Kepchia, D., Xu, P. X., Terryn, R., Castro, A., Schürer, S. C., Leal, W. S., et al. (2019). Use of machine learning to identify novel, behaviorally active antagonists of the insect odorant receptor co-receptor (Orco) subunit. Sci. Rep. 9:4055. doi: 10.1038/s41598-019-40640-4
Knecht, Z. A., Silbering, A. F., Cruz, J., Yang, L., Croset, V., Benton, R., et al. (2017). Ionotropic receptor-dependent mosit and dry cells control hygrosensation in Drosophila. eLife 6:e26654. doi: 10.7554/eLife.26654
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Li, H., Zhang, A., Chen, L. Z., Zhang, G., and Wang, M. Q. (2014). Construction and analysis of cDNA libraries from the antennae of Batocera horsfieldi and expression pattern of putative odorant binding proteins. J. Insect Sci. 14:57. doi: 10.1093/jis/14.1.57
Li, X., Ju, Q., Jie, W. C., Li, F., Jiang, X. J., Hu, J. J., et al. (2015). Chemosensory gene families in adult antennal of Anomala corpulenta motschulsky (Coleoptera: Scarabaeidae: Rutelinae). PLoS One 10:e0144214. doi: 10.1371/journal.pone.0121504
Liu, S., Rao, X. J., Li, M. Y., Feng, M. F., He, M. Z., and Li, S. G. (2015). Identification of candidate chemosensory genes in the antennal transcriptome of Tenebrio molitor (Coleoptera: Tenebrionidae). Comp. Biochem. Physiol. Part D Genomics Proteomics 13, 44–51. doi: 10.1016/j.cbd.2015.01.004
Lu, D. G., Kang, W., Li, Y. J., Wang, E. D., Liu, X. H., Liu, Q. R., et al. (2001). The feasibility of control of Anoplophora glabripennis with sterile insect technique. Acta Agric. Nucleatae Sinica 15, 302–307. doi: 10.3969/j.issn.1000-8551.2001.05.010
Lundin, C., Käll, L., Kreher, S. A., Kapp, K., Sonnhammer, E. L., Carlson, J. R., et al. (2007). Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 581, 5601–5604. doi: 10.1016/j.febslet.2007.11.007
Martin, J. P., Beyerlein, A., Dacks, A. M., Reisenman, C. E., Riffell, J. A., Lei, H., et al. (2011). The neurobiology of insect olfaction: sensory processing in a comparative context. Prog. Neurobiol. 95, 427–447. doi: 10.1016/j.pneurobio.2011.09.007
Missbach, C., Dweck, H. K., Vogel, H., Vilcinskas, A., Stensmyr, M. C., Hansson, B. S., et al. (2014). Evolution of insect olfactory receptors. eLife 3:e02115. doi: 10.1146/annurev-genet-110410-132440
Mitchell, R. F., Hall, L. P., Reagel, P. F., Mckenna, D. D., Baker, T. C., and Hildebrand, J. G. (2017). Odorant receptors and antennal lobe morphology offer a new approach to understanding olfaction in the Asian longhorned beetle. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 203, 99–109. doi: 10.1007/s00359-016-1138-4
Mitchell, R. F., Hughes, D. T., Luetje, C. W., Millar, J. G., Soriano-Agatón, F., Hanks, L. M., et al. (2012). Sequencing and characterzing odorant receptors of cerambycid beetle Megacyllene caryae. Insect Biochem. Mol. Biol. 42, 499–505. doi: 10.1016/j.ibmb.2012.03.007
Mitchell, R. F., Schneider, T. M., Schwartz, A. M., Andersson, M. N., and McKenna, D. D. (2020). The diversity and evolution of odorant receptors in beetles (Coleoptera). Insect Mol. Biol. 29, 77–91. doi: 10.1111/imb.12611
Nichols, A. S., Chen, S., and Luetje, C. W. (2011). Subunit contributions to insect olfactory receptor funcation: channel block and odorant recognition. Chem. Senses. 36, 781–790. doi: 10.1093/chemse/bjr053
Nichols, A. S., and Luetje, C. W. (2010). Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interaction. J. Biol. Chem. 285, 11854–11862. doi: 10.1074/jbc.M109.058321
Peschke, K., and Metzler, M. (1987). Cuticular hydrocarbons and female sex pheromones of the rove beetle, Aleochara curtula (Goeze) (Coleoptera: Staphylinidae). Insect Biochem. 17, 167–178. doi: 10.1016/0020-1790(87)90157-0
Prieto-Godino, L. L., Rytz, R., Cruchet, S., Bargeton, B., Abuin, L., Silbering, A. F., et al. (2017). Evolution of acid-sensing olfactory circuits in Drosophilids. Neuron 93, 661.e6–676.e6. doi: 10.1016/j.neuron.2016.12.024
Rimal, S., and Lee, Y. (2018). The multidimensional ionotropic receptors of Drosophila melanogaster. Insect Mol. Biol. 27, 1–7. doi: 10.1111/imb.12347
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sato, K., and Touhara, K. (2009). Insect olfaction: receptors, signal transduction, and behavior. Results Probl. Cell Differ. 47, 121–138. doi: 10.1007/400_2008_10
Simon, P. (2003). Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19, 1439–1440. doi: 10.1093/bioinformatics/btg157
Sun, L., Zhang, Y. N., Qian, J. L., Kang, K., Zhang, X. Q., Deng, J. D., et al. (2018). Identification and expression patterns of Anoplophora chinensis (Forster) chemosensory receptor genes from the antennal transcriptome. Front. Physiol. 9:90. doi: 10.3389/fphys.2018.00090
Tanaka, N., Ohsawa, K., Hiroshi, H., and Yamamoto, I. (1981). Copulation release pheromone, Erectin, from the Azuki bean weevil (Callosobruchus chinensis L.). J. Pesticide Sci. 6, 75–82.
Tang, Y. P., Ding, Y. Z., Wang, T. S., Xie, F., Su, Y. D., Zhang, L. W., et al. (2013). Preferences of Apriona germari for different plants and the effectiveness of two pesticides in controlling this pest. Chin. Bull. Entomol. 50, 1109–1114. doi: 10.7679/j.issn.2095-1353.2013.153
Vogt, R. G., and Riddiford, L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi: 10.1038/293161a0
Wang, D., Pentzold, S., Kunert, M., Groth, M., Brandt, W., Pasteels, J. M., et al. (2018). A subset of chemosensory genes differs between two populations of a specialized leaf beetle after host plant shift. Ecol. Evol. 8, 8055–8075. doi: 10.1002/ece3.4246
Wang, J., Li, D. Z., Min, S. F., Mi, F., Zhou, S. S., and Wang, M. Q. (2014). Analysis of chemosensory gene families in the beetle Monochamus alternates and its parasitoid Dastarcus helophoroides. Comp. Biochem. Physiol. Part D Genomics. Proteomics. 11, 1–8. doi: 10.1016/j.cbd.2014.05.001
Wu, H., Li, R. T., Dong, J. F., Jiang, N. J., Huang, L. Q., and Wang, C. Z. (2018). An odorant receptor and glomerulus responding to farnesene in Helicoverpa assulta (Lepidoptera: Noctuidae). Insect Biochem. Mol. Biol. 115:103106. doi: 10.1016/j.ibmb.2018.11.006
Wu, Z. Z., Bin, S. Y., He, H. L., Wang, Z. B., Li, M., and Lin, J. T. (2016). Differential expression analysis of chemoreception genes in the striped flea beetle Phyllotreta striolata using a transcriptomic approach. PLoS One 11:e0153067. doi: 10.1371/journal.pone.0153067
Xiao, Y., Sun, L., Ma, X. Y., Dong, K., Liu, H. W., Wang, Q., et al. (2017). Identification and characterization of distinct expression profiles of candidate chemosensory membrane proteins in the antennal transcriptome of Adelphocoris lineolatus (Goeze). Insect Mol. Biol. 26, 74–91. doi: 10.1111/imb.12272
Yao, C. A., Ignell, R., and Carlson, J. R. (2005). Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J. Neurosci. 25, 8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005
Zhan, W. H., Zhang, S. S., Geng, H. W., Wang, Y. Y., Guo, K., and Chen, J. (2018). Research progress on olfactory recognition proteins of longhorn beetle. J. Henan Agricul. Sci. 47, 1–6. doi: 10.15933/j.cnki.1004-3268.2018.03.001
Zhang, J. (2011). Taxonomic Study on the Cerambycidae in Jilin Province, dissertation [China (ChangChun)] Northeast Normal University, Changchun.
Zhang, L., Wang, B. D., and Xu, Z. C. (2011). Volatile constituents of four moraceous host plants of Apriona germari. Acta Ecol. Sinica 31, 7479–7485.
Zhang, L. W., Kang, K., Jiang, S. C., Zhang, Y. N., Wang, T. T., Zhang, J., et al. (2016). Analysis of antennal transcriptome and insights into oflactory genes in Hyphantria cunea (Drury). PLoS One 11:e164729. doi: 10.1371/journal.pone.0164729
Keywords: odorant receptor, ionotropic receptor, expression pattern, antennal transcriptome, Apriona germari
Citation: Qian J-L, Mang D-Z, Lv G-C, Ye J, Li Z-Q, Chu B, Sun L, Liu Y-J and Zhang L-W (2020) Identification and Expression Profile of Olfactory Receptor Genes Based on Apriona germari (Hope) Antennal Transcriptome. Front. Physiol. 11:807. doi: 10.3389/fphys.2020.00807
Received: 19 March 2020; Accepted: 17 June 2020;
Published: 22 July 2020.
Edited by:
Petros T. Damos, Aristotle University of Thessaloniki, GreeceReviewed by:
Yang Liu, Institute of Plant Protection (CAAS), ChinaNai-Yong Liu, Southwest Forestry University, China
Copyright © 2020 Qian, Mang, Lv, Ye, Li, Chu, Sun, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long-Wa Zhang, emhhbmdsd0BhaGF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Jia-Li Qian
Jia-Li Qian Ding-Ze Mang
Ding-Ze Mang Guo-Chang Lv
Guo-Chang Lv Jia Ye
Jia Ye Zhao-Qun Li
Zhao-Qun Li Bo Chu
Bo Chu Long Sun
Long Sun Yu-Jun Liu
Yu-Jun Liu Long-Wa Zhang
Long-Wa Zhang