Abstract
Craniofacial morphogenesis depends on proper migration of neural crest cells and their interactions with placodes and other cell types. Hox genes provide positional information and are important in patterning the neural crest and pharyngeal arches (PAs) for coordinated formation of craniofacial structures. Hox genes are expressed in the surface ectoderm and epibranchial placodes, their roles in the pharyngeal epithelium and their downstream targets in regulating PA morphogenesis have not been established. We altered the Hox code in the pharyngeal region of the Hoxb3Tg/+ mutant, in which Hoxb3 is driven to ectopically expressed in Hoxb2 domain in the second pharyngeal arch (PA2). In the transgenic mutant, ectopic Hoxb3 expression was restricted to the surface ectoderm, including the proximal epibranchial placodal region and the distal pharyngeal epithelium. The Hoxb3Tg/+ mutants displayed hypoplasia of PA2, multiple neural crest-derived facial skeletal and nerve defects. Interestingly, we found that in the Hoxb3Tg/+ mutant, expression of the Notch ligand Jag1 was specifically up-regulated in the ectodermal pharyngeal epithelial cells of PA2. By molecular experiments, we demonstrated that Hoxb3 could bind to an upstream genomic site S2 and directly regulate Jag1 expression. In the Hoxb3Tg/+ mutant, elevated expression of Jag1 in the pharyngeal epithelium led to abnormal cellular interaction and deficiency of neural crest cells migrating into PA2. In summary, we showed that Hoxb3 regulates Jag1 expression and proposed a model of pharyngeal epithelium and neural crest interaction during pharyngeal arch development.
Introduction
An important phase of mammalian craniofacial development is the formation of the transient pharyngeal arch (PA) structures. These PAs are comprised of an outer surface ectoderm, an inner covering of endoderm, a mesenchymal core, and the cranial neural crest-derived ectomesenchyme. The coordinated development of the different embryonic components give rise to the pharynx, the jaw, the ear and the face. Dysregulation of PA development can lead to many human congenital craniofacial malformations.
The cranial neural crest cells which delaminate from the dorsal region of hindbrain rhombomeres have important structural roles in craniofacial morphogenesis. Neural crest cells originating from different anteroposterior levels of the hindbrain rhombomeres are marked by combinations of Hox genes which specify their identity. Molecular analysis have shown that regulation of Hox expression in the hindbrain rhombomeres and in the neural crest could be independently controlled by separate cis-acting regulatory elements and trans-acting factors (Frasch et al., 1995; Nonchev et al., 1996; Maconochie et al., 1999; Trainor and Krumlauf, 2000). The neural crest cells arising from distinct rhombomere locations are not pre-patterned, but remain plastic, and can respond to signals in the environment they migrate to Trainor and Krumlauf (2000) and Le Douarin et al. (2004). In the PAs, neural crest cells respond to signals from the pharyngeal surface ectoderm and the endoderm in determining their cell fate (Graham et al., 2005). The neural crest cells give rise to cranial ganglia and nerves, muscle and facial bones and cartilages (Frisdal and Trainor, 2014). Defective migration, survival, proliferation, or differentiation of the cranial neural crest cells lead to multiple craniofacial abnormalities (Santagati and Rijli, 2003; Minoux and Rijli, 2010). More importantly, PAs can be formed and their antero-posterior and proximo-distal axes maintained in the absence of neural crest cells, indicating the important roles of pharyngeal epithelium and other cell types during PA development (Veitch et al., 1999; Gavalas et al., 2001; Trainor and Krumlauf, 2001).
The ectodermal pharyngeal epithelium covering the proximal and distal regions of the PAs are developmentally distinct. In the proximal PA region, epibranchial placodal cells not only give rise to neurogenic cranial ganglia, but also non-neuronal epithelial cells which play essential roles in pharyngeal segmentation (Zhang et al., 2017). The invagination of the proximal non-neuronal placode-derived epithelium and the outgrowth of the pharyngeal endoderm form the segmental plates, the contact points of the epithelial layers give rise to the pharyngeal clefts and pouches (Graham and Smith, 2001; Graham, 2003; Kulesa et al., 2010). Defective pharyngeal segmentation as displayed in the Sox3 (Rizzoti and Lovell-Badge, 2007) and Eya1 mutants (Xu et al., 2002; Zhang et al., 2017) would lead to abnormal PA development. Around the pharyngeal clefts, the non-neuronal epibranchial placodal epithelial cells express various signaling factors including Notch and Fgfs that are required for PA morphogenesis (Trokovic et al., 2005; Wang et al., 2020). The roles of the ectodermal pharyngeal epithelium in interacting with neural crest and other cell types in the developing PAs are not well understood.
Notch signaling factors are required for cranial ganglia neurogenesis and craniofacial morphogenesis (Wakeham et al., 1997; Jayasena et al., 2008; Lassiter et al., 2010; Lassiter et al., 2014). Mutations of the Notch ligand JAG1 caused Alagille syndrome with craniofacial defects in human patients (Li et al., 1997; Kamath et al., 2003). Jag1b mutation in zebrafish led to mis-patterning of pharyngeal arch derived skeletons (Barske et al., 2016), while Jag2 mutant mice displayed cleft palate (Vieira et al., 2005; Casey et al., 2006). Neural crest-specific knockout of Jag1 or Notch2 resulted in middle ear bone malformation (Humphreys et al., 2012; Teng et al., 2017). Knockout of Rbpj, a transcriptional cofactor of Notch, in neural crest cells led to shortened mandible formation (Mead and Yutzey, 2012). Deletion of Notch signaling target Hey1 led to deformed pharyngeal arch arteries (Fujita et al., 2016). Interestingly, in Presenilin1/2 null mutant mice which deficient in γ-secretase, the PA2 was absent (Donoviel et al., 1999). The Maml1–/–;Maml3–/– mutant embryos also displayed hypoplastic or even no PA2, indicating that morphogenesis of PA2 is particularly sensitive to Notch signaling activity (Oyama et al., 2011). The underlying cellular and molecular mechanisms for Notch signaling functions in the development of PAs remain elusive. In our previous studies, we showed that Notch1 intracellular domain (N1-ICD) was required in the epibranchial placodal epithelium in the proximal PAs to control cell fate commitment of neuronal and non-neuronal epibranchial placodal cells (Zhang et al., 2017). Moreover, Notch receptor signaling controlled rostral-caudal patterning of the non-neuronal placodal epithelial cells around the pharyngeal clefts, indicating important roles of Notch signaling in epithelial cell fate commitment and differentiation (Wang et al., 2020).
Combinatorial expression of Hox genes in hindbrain rhombomeres, neural crest and pharyngeal surface ectoderm are known to convey positional information in the PAs (Trainor and Krumlauf, 2001; Santagati and Rijli, 2003). Loss-of-function mouse mutant analyses have shown that Hox genes of the first three paralogous groups are required for the development of neural crest derivatives in the pharyngeal region. Mutations of Hoxa genes would lead to neural crest defects and malformation of craniofacial structures. Inactivation of Hoxa2 resulted in homeotic transformation of PA2 components into PA1 (Rijli et al., 1993). Hoxa1 mutation led to deletion of rhombomere 5 and severe reduction of r4, the mutant displayed hypoplasia of PA2 as a result of neural crest deficiency (Chisaka et al., 1992). Hoxa3 mutants showed deformed neural crest derivatives of PA3 including the hyoid and thyroid cartilage (Chojnowski et al., 2016) and endoderm defects (Su et al., 2001; Kameda et al., 2004). Mutations of Hoxb genes led to neurogenic phenotypes. For instance, although double knockout of Hoxa1 and Hoxb1 in mice exacerbated the phenotypes of Hoxa1 mutant and resulted in hypoplasia of PA2 and malformation of middle ear bones (Gavalas et al., 1998), Hoxb1 mutant embryos did not exhibit any defects in neural crest-derived tissues of PA2, but displayed abnormal neuronal identity in hindbrain r4 (Goddard et al., 1996; Studer et al., 1996). Hoxb2 mutant mice displayed retracted lower lip and mild craniofacial features (Barrow and Capecchi, 1996), Hoxb3 null mutant had no craniofacial abnormalities (Manley and Capecchi, 1998). The specific functions of Hox genes in the different cell types of the developing PA are not entirely known.
Insights on the roles of Hox genes in cranial neural crest development have also been obtained by gain-of-function analysis. Cranial neural crest cells rostral to rhombomere 2 do not express Hox genes, and only Hox-free neural crest cells are capable of generating facial skeletal components (Couly et al., 1998; Le Douarin et al., 2004). Gain-of-function experiments by electroporation of Hoxa2 into the Hox-free neural crest domain of chick embryos led to absence of lower jaw and frontonasal structures; defective formation of PA1 derived skeletal structures were also observed in ectopic Hoxa3 and Hoxb4 expression experiments (Creuzet et al., 2002). Heterotopic transplantation experiments with fragments of neural folds have demonstrated that Hox-positive neural crest cells from the posterior region are incapable to replace Hox-free neural crest cells. These studies suggest that cross-talks among Hox genes, neural crest cells and extrinsic signals in the pharyngeal region, including ectodermal and endodermal derived factors, are required for the formation neural crest derived structures (Couly et al., 2002; Creuzet et al., 2002; Le Douarin et al., 2004; Minoux and Rijli, 2010).
We have previously generated a gain-of-function Hoxb3 transgenic mutant Hoxb3Tg and shown that Hoxb3 could transcriptionally suppress Hoxb1 in the hindbrain and maintain anterior-posterior identity of rhombomere 4 and 5 (Wong et al., 2011). Using the Hoxb2-r4 enhancer element (Maconochie et al., 1997; Szeto et al., 2009), we ectopically expressed Hoxb3 to Hoxb2 domains, including hindbrain rhombomere 4 and PA2 in Hoxb3Tg/+, and examined the effect of altering the expression of Hox code in PA development. Although Hoxb3 or Hoxb2 null mutants had no or mild craniofacial defects, the Hoxb3Tg/+ transgenic mutants displayed multiple neural crest-derived abnormalities. We found that ectopic expression of Hoxb3 was restricted to the pharyngeal epithelium in the Hoxb3Tg/+ mutant, allowing the study of interaction between pharyngeal epithelium and migrating neural crest cells. Interestingly, Jag1 was ectopically expressed in the ectodermal epithelium of PA2, in cells which co-expressed Hoxb3. We showed that Hoxb3 could transcriptionally regulate Jag1 expression during PA development. Migration of neural crest cells into the PAs was affected, leading to neural crest deficiency and craniofacial defects at later stages. Our results suggest that Hoxb3 positively regulates the expression of Jag1 in the pharyngeal epithelial cells and affects the colonization and maintenance of neural crest cells in the developing PAs.
Materials and Methods
Experimental Animals
The mouse lines used in this study include Hoxb3Tg/+ [Hoxb3Tg2 in Wong et al. (2011)], B2-Cre [Hoxb2-r4-Cre in Szeto et al. (2009)], Wnt1-Cre (Chai et al., 2000), and Z/EG (Novak et al., 2000). The Hoxb3Tg/+ transgenic mice were maintained in FVB genetic background, other mouse lines were maintained in C57BL/6N background. All animals were housed at the Center for Comparative Medicine Research at the University of Hong Kong. The animal experiments were approved by the University of Hong Kong Committee on the Use of Live Animals for Teaching and Research (CULATR No. 4357–17 and 4588–18).
Riboprobe Labeling
Plasmids with target cDNA sequence including Hoxa2 (Nonchev et al., 1996), Hoxb2 (nucleotide 41–1114 of mRNA), Hoxb3 (Wilkinson et al., 1989), Jag1 (Jones et al., 2000) were cloned, linearized (restriction enzymes used were summarized in Table 1) and purified. RNA probes were prepared using 1 μg of purified DNA for reverse transcription and digoxigenin (DIG) labeling (Roche, 11277073910), synthesized RNA probe was precipitated in ethanol, dissolved in DEPC-treated water, and stored at -80°C.
TABLE 1
| Antisense Probe | Restriction enzyme for anti-sense probe | RNA polymerase for RNA transcription | References |
| Hoxa2 | EcoRI | T7 | Nonchev et al., 1996 |
| Hoxb2 | BamHI | T3 | This study |
| Hoxb3 | BamHI | T3 | Wilkinson et al., 1989 |
| Jag1 | EcoRI | T3 | Jones et al., 2000 |
Enzymes used for riboprobe labeling.
Whole-Mount RNA in situ Hybridization
Embryos were harvested, fixed in 4% paraformaldehyde (PFA) at 4°C overnight, dehydrated in a series of methanol/PBST (Phosphate-buffered saline with 0.1% Tween-20) solutions and stored in absolute methanol at -20°C. Before in situ hybridization, samples were rehydrated with gradients of PBST series, treated with 1 μg/ml proteinase K, then post-fixed in 4% PFA, 0.2% glutaraldehyde in PBS. Embryos were then incubated with hybridization mix (50% formamide, 1.3x SSC, 50 μm EDTA, 50 μg/ml, 0.2% Tween20, 0.5% CHAPS, and 100 μg/ml heparin) at 65°C for 2 h, then incubated with the DIG-labeled RNA probe in hybridization mix at 65°C overnight. After hybridization, samples were washed with hybridization mix in maleic acid buffer with Tween-20 (MABT), blocked with 10% blocking reagent (Roche, 11096176001) and 20% heat-inactivated horse serum, and then incubated in 1:2000 alkaline phosphatase conjugated anti-DIG antibody (Roche, 11093274910) in the same blocking buffer at 4°C. After washing with MABT for 24 h, embryos were immersed in BM purple (Roche, 11442074001) for color development. Reaction was stopped by washing with PBST and post-fixed in 4% PFA.
Immunostaining and Antibodies
For whole mount immunostaining, E10.5 mouse embryos were harvested and fixed in 4% PFA for 1.5 h, followed by inactivation of endogenous peroxidase in 0.05% peroxide in PBS at 4°C overnight. Embryos were then blocked in PBS-TS (10% heat inactivated serum, 1% Triton X-100 in PBS) for 3 h at 4°C, incubated with 2H3 anti-neurofilament primary antibody (1:100, Developmental Studies Hybridoma Bank) at 4°C for 3 days. Embryos were then incubated with peroxidase-conjugated rabbit anti-mouse secondary antibody (1:200, Dako) and subjected to 4-chloro-1-napthol (Sigma) color development. The reaction was stopped by washes of 30% ethanol.
For section immunostaining, 10 μm thick cryo-sectioned samples were blocked with 10% heat-inactivated horse serum in PBS for 1 h. The sections were then incubated with primary antibodies including GFP [1:500, Rockland Immunochemicals, 600-101-215), Pax8 (1:500, Proteintech, 10336-1-AP), and Jag1 (1:300, Developmental Studies Hybridoma Bank, TS1.15H)] and E-cadherin (1:500, Cell signaling, 24E10) at 4°C overnight. After three washes of PBS, Alexa Fluor 488- or 594-conjugated secondary antibodies (1:500, Life technologies) were applied. DAPI (Sigma-Aldrich, D9542) was used as a counterstain for the nucleus. Sections were washed with PBS then mounted with mounting medium (Vectashield, Vector Laboratories).
Scanning Electron Microscopy
Embryos were fixed in 2.5% glutaraldehyde for 4 h and then in 1% osmium tetroxide for 1 h, dehydrated in graded ethanol and critical-point dried with transition fluid liquid carbon dioxide in a Ladd critical point dryer. Gold-palladium-coated specimens were examined with a JEOL JSM-6301FE scanning electron microscope operated at 5 KV.
Skeletal Preparations
Newborn mice were fixed in 95% ethanol overnight. After skin removal, the whole-mount skeletons were incubated in 0.015% Alcian Blue and 0.005% Alizarin Red stain diluted in 5% acetic acid in 70% ethanol for 1 week (Mcleod, 1980). The preparations were cleared in 20% glycerol, 1% potassium hydroxide, and then transferred to 50% glycerol/ethanol for photography.
Bioinformatics Analysis of Hoxb3 Binding Sites
The matrix of Hoxb3 binding sequence (PH0058.1) was obtained from the JASPAR database. Hoxb3 binding motif was scanned along 100 kb upstream of Jag1 locus using Find Individual Motif Occurrences v5.0.5 (FIMO) with p < 0.0005 as cutoff. The genomic loci were further analyzed for sequence conservation among human, chicken, lizard and Xenopus with PhyloP score higher than 1. Genomic regions were further aligned with open chromatin regions in developing mouse embryo (E11.5) ATAC-seq data with following accession code: ENCSR150RMQ (facial prominence), ENCSR273UFV (forebrain), ENCSR382RUC (midbrain), ENCSR012YAB (hindbrain), and ENCSR282YTE (neural tube).
P19 Cell Culture
P19 mouse embryonal carcinoma cells were cultured on gelatin-coated (0.1%) tissue culture dishes in Dulbecco’s Modified Eagle Medium (DMEM) medium (Gibco) supplemented with 10% FBS. Retinoic acid (RA) treatment (10–8 M) of P19 cells for activation of hindbrain Hox genes expression were performed as previously described (Okada et al., 2004; Wong et al., 2011). Using 10–8 M RA, expression of anterior Hox genes including Hoxb3 would be activated endogenously.
Quantitative Real-Time PCR
Total RNA samples were extracted according to TRIzol manufacturer’s protocol from RA treated and control P19 cells. cDNA samples were generated from total extracted RNA using Superscript III (ThermoFisher) and polymerase chain reaction (PCR) performed with SYBR Green Premix Ex Taq II (TaKaRa) using the StepOnePlus Real-Time PCR system. Primers for Hoxb3: forward: 5′-GCAGA AAGCC ACCTA CTACG-3′, reverse: 5′-CCATT GAGCT CCTTG CTCTT-3′; for Jag1: forward: 5′-AACAC AGGGA TTGCC CACTT-3′, reverse: 5′-TGTTG CAATC AGGAC CCATC-3′; for Gapdh: forward: 5′-TTCAC CACCA TGGAG AAGGC-3′; reverse: 5′-GGCAT GGACT GTGGT CATGA-3′. Relative expression of Hoxb3 and Jag1 were normalized with Gapdh expression. All the experiments were performed in triplicates.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assay (Hu and Rosenblum, 2005) was performed with the following modifications. For RA-treated P19 cells, 1 × 106 cells were harvested for each assay. For in vivo ChIP, 4 litters of E9.5 wildtype whole embryos were harvested in PBS, fixed in 1% formaldehyde for 20 min at 25°C, and then disintegrated with RIPA buffer. The cross-linked material was sonicated to 200–1,000 bp fragments (Vibracell sonicator; seven times for 10 s at 40% output), anti-Hoxb3 antibody (Santa Cruz, C-20, and sc-17169) or normal rabbit IgG were then used to pull down the chromatin. PCR amplifications were performed using the following primers: for S1: forward: 5′-AGTCA TCGTC TGCTG CCTTT-3′, reverse: 5′-GCAAC GAATT CATTC AGCAA-3′; for S2: forward: 5′-TTTTA GCCCC TGCTT GCTTA-3′, reverse: 5′-TTGGA GAACA GCCCT CATTT-3′; for S3: forward: 5′-CCTAA CCCCT TTCCC ATCAT-3′, reverse: 5′-TTCTT GTTTG GGCTT GCTCT-3′; for B3ARE: forward: 5′-GTAGG TGTGT GGGCA GAGGT-3′, and reverse: 5′-CTGAG TGGAG GATGG GTTGT-3′. The experiments were performed with 3 biological replicates and 3 technical replicates.
In vitro and ex vivo Luciferase Activity Assays
For the in vitro transactivation experiments, pCIG-Hoxb3 cDNA (Yau et al., 2002) or pCIG control expression vector was co-electroporated with Firefly and Renilla luciferase reporters into human embryonic kidney cells 293T. The luciferase reporter constructs were pGL3-Jag1-S2 which contained a 400 bp fragment (Jag1 genomic DNA, mm10 chr2: 136979005-136979405) on the Jag1 locus with the Hoxb3 binding site S2 CTGTAATTAACT, or pGL3-Jag1-mS2 which contained a mutated Hoxb3 binding site mS2 CTGGCCGGCACT, or pGL3 control luciferase vector. After 24 h, the cells were processed for luciferase assay as previously described (Dessaud et al., 2007; Wong et al., 2011). Briefly, cells were homogenized in lysis buffer on ice, Firefly and Renilla luciferase activities were measured with the Dual Luciferase Reporter Assay System (Promega).
For the ex vivo luciferase activity assay, pCIG-Hoxb3 cDNA or pCIG control vector, together with pGL3-Jag1-S2, pGL3-Jag1-mS2, or control luciferase reporter construct, and Renilla luciferase reporter, were electroporated into the hindbrain region of E9.0 mouse embryos. After cultured for 24 h, the pharyngeal portion (Figure 4D) of the embryos were homogenized in lysis buffer on ice, Firefly and Renilla luciferase activities were measured as above.
All the experiments were performed with 3 biological replicates and 3 technical replicates.
Statistical Test
Statistical comparison of two groups was performed by Student’s t-test, where n number represented biological replicates using the same experimental condition. Error bars indicated standard error of the mean (SEM).
Results
Expression of Hox Genes in Pharyngeal Ectoderm and Epibranchial Placodal Cells
The expression of Hox genes in the developing PAs have been well characterized. However, the expression and function of Hox genes in the pharyngeal surface ectoderm and the epibranchial placodal epithelium of the pharyngeal arches are not well understood. We showed that Hoxa2 and Hoxb2 were both expressed in PA2, PA3 and posterior PAs, and Hoxb3 was expressed in PA3 and posterior PAs at E9.5 (Figures 1A,C,E). Interestingly, coronal sections at the level of proximal pharyngeal arches (refer to Figure 1N) of embryos hybridized with Hoxa2 and Hoxb2 probes revealed differential expression of these two genes in PA2. Hoxa2 was expressed strongly in the neural crest derived ectomesenchyme but hardly detectable in the epibranchial placodal epithelium covering PA2 (Figures 1B,B’), although it was expressed in placodal epithelium and neural crest cells of posterior PAs. On the contrary, Hoxb2 was distinctly expressed in the epibranchial placodal epithelium of PA2 and posterior PAs (Figure 1D), while expressed at lower levels in the neural crest cells of these PAs (Figure 1D). Hoxb3 was strongly expressed in the pharyngeal surface ectoderm (Figure 1E), epibranchial placodal epithelium and neural crest cells of PA3 and posterior PAs (Figure 1F).
FIGURE 1
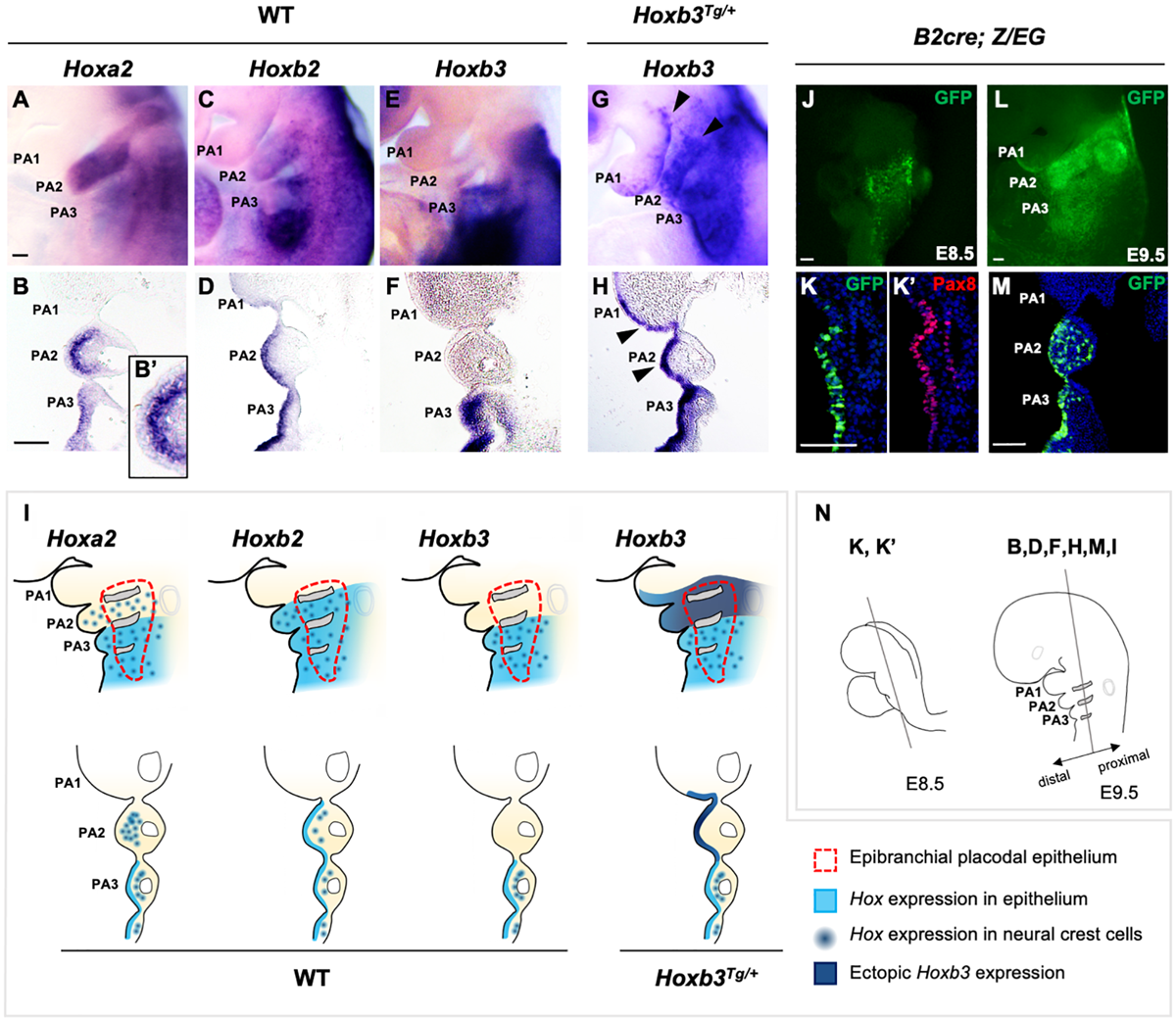
Expression of Hox genes in pharyngeal arches of wildtype and Hoxb3Tg/+ embryos. (A–H) Wholemount in situ hybridization and coronal sections showing expression of Hoxa2, Hoxb2, and Hoxb3 in wildtype and mutant E9.5 embryos (n = 3). An enlarged image in (B’) showing the absence of Hoxa2 expression in the epithelium covering PA2. Arrowheads in (G,H) indicate ectopic expression of Hoxb3 in the pharyngeal epithelium of PA1 and PA2. (I) Schematic summary of Hox gene expression in the pharyngeal ectoderm, epibranchial placodal region, and neural crest cells of E9.5 wildtype and Hoxb3Tg/+ embryos. (J,K) Wholemount GFP autofluorescence (J) and co-immunostaining of GFP (K) and Pax8 (K’) in coronal section of E8.5 B2-Cre;Z/EG embryos (n = 3). (L,M) Wholemount GFP autofluorescence (L) and immunostaining of GFP in coronal section (M) of E9.5 B2-Cre; Z/EG embryos (n = 3). (N) Schematic diagram illustrating the plane of embryo sections for the indicated panels in this figure. PA1, PA2, and PA3 indicate the 1st, 2nd, and 3rd pharyngeal arches. Scale bars: 100 μm.
We have previously demonstrated that in the developing PAs, the proximal pharyngeal epithelium was derived from the posterior placodal area and overlap with the epibranchial placodal cells (Figure 1I, red dotted line; Zhang et al., 2017; Wang et al., 2020), while the distal PAs were enveloped by pharyngeal ectodermal epithelium. Therefore, the proximal and distal pharyngeal epithelium were composed of developmentally distinct epithelial cells. The Hox genes delineate the antero-posterior identity of the pharyngeal arches, including the neural crest cells delaminated from specific hindbrain rhombomeres. Neural crest cells in PA2 expressed Hoxa2 and Hoxb2, while those in PA3 and posterior PAs expressed Hoxb3 in addition to Hoxa2 and Hoxb2. However, for the pharyngeal epithelium, we showed that Hoxb2 marked PA2 (Figures 1D,I) and Hoxb3 marked PA3 (Figures 1F,I). The distinct expression patterns of Hox genes in both the proximal and distal pharyngeal epithelium of specific PAs suggest a role of these genes in maintaining the functions of epibranchial placodal cells in the proximal regions.
Ectopic Expression of Hoxb3 in PA2 of Hoxb3Tg/+ Transgenic Mutants
To investigate the role of the combinatorial Hox code in PA development as well as to understand the potential interactions between placodal cells and underlying neural crest cells, we investigated the Hoxb3Tg/+ gain-of-function transgenic mutant generated with the Hoxb2-r4 enhancer (Wong et al., 2011). The Hoxb2-r4 enhancer could direct reporter gene expression in hindbrain rhombomere 4 and PA2 (Maconochie et al., 1997; Ferretti et al., 2000). We analyzed the enhancer activity in the pharyngeal epithelium using the B2-Cre [Hoxb2-r4-Cre; (Szeto et al., 2009)] mouse line. Wholemount B2-Cre;Z/EG embryos showed expression of the GFP reporter in the pharyngeal region at E8.5 (Figure 1J), and in PA2 at E9.5 (Figure 1L). By immunostaining with Pax8, one of the earliest markers of the posterior placodal area (Schlosser and Ahrens, 2004; Washausen and Knabe, 2017), on coronal sections of E8.5 embryo, we showed that GFP + cells also expressed Pax8 (Figures 1K,K’), confirming the enhancer activity in the placodal epithelial cells. Coronal sections of E9.5 embryos showed B2-Cre activity in the proximal placodal epithelium and pharyngeal ectodermal region from second to posterior arches, as well as the neural crest cells in PA2 and PA3 (Figures 1L,M).
In the Hoxb3Tg/+ mutant, driven by the Hoxb2-r4 enhancer element, ectopic Hoxb3 was expressed not only in the entire proximal and distal PA2, but also in posterior region of PA1 (Figures 1G,H). Coronal sections of Hoxb3Tg/+ embryos showed that ectopic expression of Hoxb3 was restricted to the pharyngeal epithelium of posterior PA1 and PA2 (Figure 1H), no expression could be observed in the neural crest cells of PA2. Therefore, in the Hoxb3Tg/+ mutant, we have ectopically expressed Hoxb3 specifically in the pharyngeal epithelium of PA2 and posterior PA1, providing a gain-of-function genetic condition for analysis of epithelial cell function during PA development.
Ectopic Expression of Jag1 in PA2 Epithelium of Hoxb3Tg/+ Mutant
To investigate the consequences of ectopic expression of Hoxb3 in pharyngeal arch patterning, we examined the expression of genes in the distal PAs including dHAND which marked the medial regions (Figure 2A), Gsc which marked the ventral aspects (Figure 2C), and Dlx5 which marked the lateral regions (Figure 2E) of PA1 and PA2 in E10.5 embryos. However, the regional specific expression patterns of dHAND, Gsc, and Dlx3 were not changed in the Hoxb3Tg/+ mutants (Figures 2B,D,F), indicating the patterning of distal PAs remained unaffected with the ectopic expression of Hoxb3.
FIGURE 2
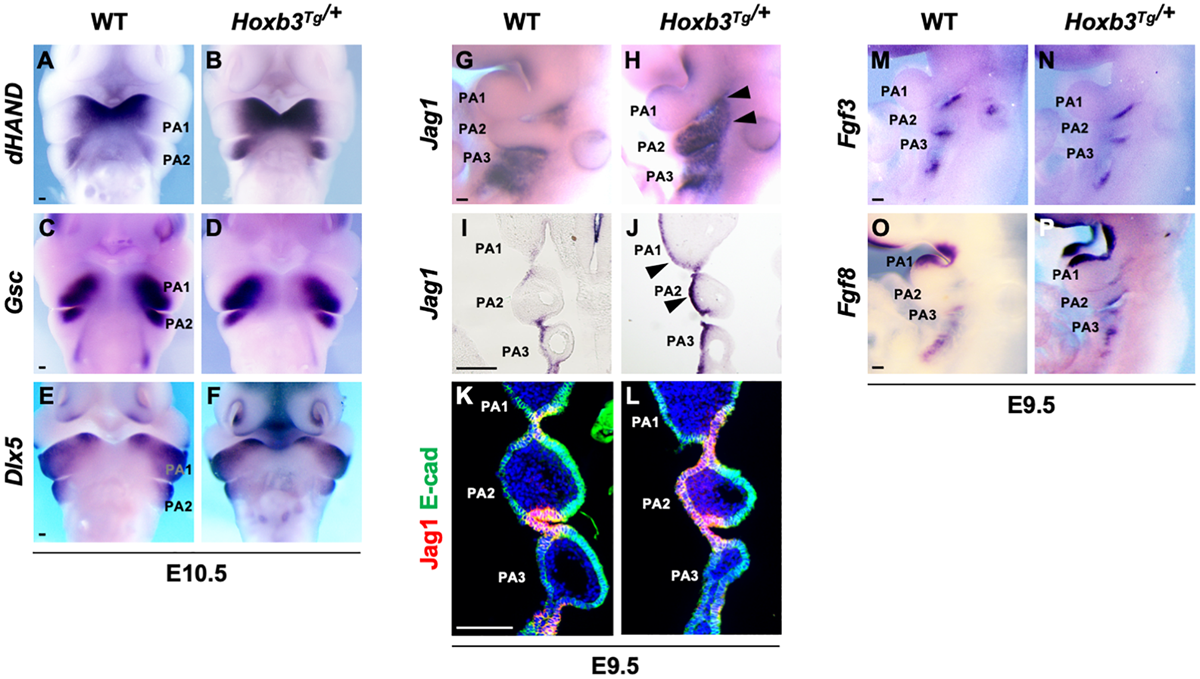
Pharyngeal arch patterning and ectopic expression of Jag1 in the Hoxb3Tg/+ mutants. (A–F) Wholemount in situ hybridization showing expression of dHAND, Gsc, and Dlx5 in wildtype and Hoxb3Tg/+ embryos at E10.5 (n > 3). (G,H) Wholemount in situ hybridization of Jag1 showing ectopic expression in PA1 and PA2 (arrowheads) of E9.5 Hoxb3Tg/+ embryos (n = 4). (I,J) Coronal sections of embryos subjected to wholemount in situ hybridization of Jag1 showing ectopic expression in the ectodermal epithelium of PA1 and PA2 (arrowheads) in Hoxb3Tg/+ embryos (n = 3). (K,L) Immunostaining for Jag1 (red) and E-cadherin (green) on coronal sections of E9.5 wildtype and Hoxb3Tg/+ embryos (n = 5). (M–P) Wholemount in situ hybridization of Fgf3 and Fgf8 on E9.5 wildtype and Hoxb3Tg/+ embryos (n > 3). PA1, PA2, and PA3 indicate the 1st, 2nd, and 3rd pharyngeal arches. Scale bars: 100 μm.
As Notch signaling has been shown to be critical for the development of epibranchial placodal epithelium and proximal PAs (Zhang et al., 2017; Wang et al., 2020), we examined Jag1 which was expressed in the proximal pharyngeal epithelium of developing PAs and restricted to the pharyngeal clefts and pouches as the segmental plates were formed at E9.5 (Figures 2G,I,K). Interestingly, we found that the expression of Jag1 was ectopically activated in the entire pharyngeal ectodermal region of PA2 and posterior PA1 in the Hoxb3Tg/+ mutants at E9.5 (Figures 2H,J). The expression of Jag1 in the epibranchial placodal epithelium of PA1 and PA2 as shown in coronal section of Hoxb3Tg/+ mutant (Figure 2J) was remarkably similar to the ectopic Hoxb3 expression domains (Figure 1H). By immunostaining we could detect Jag1 protein in the pharyngeal epithelium of PA2 of Hoxb3Tg/+ mutant (Figure 2L), suggesting that ectopic expression of Hoxb3 in the pharyngeal epithelium in PA2 could activate Jag1 protein expression in PA2.
We have previously shown that over-expression of the Notch receptor protein N1-ICD in the epibranchial placodal epithelium would lead to misexpression of Fgf ligands and cell fate changes (Wang et al., 2020). Therefore, we examined the expression of Fgf3 and Fgf8, which marked placodal epithelial cells flanking the pharyngeal clefts (Figures 2M,O). However, we found that ectopic expression of the Notch ligand Jag1 did not cause changes of Fgf3 or Fgf8 expression in Hoxb3Tg/+ mutants (Figures 2N,P), suggesting that the pharyngeal epithelial cell fates were not affected.
Malformations of Neural Crest Derived Structures in the Hoxb3Tg Mutants
We first examined the phenotypes of the Hoxb3Tg mutant PAs by scanning electron microscopy. Although the pharyngeal arch phenotypes of the mutant were mild, it was evident that the PA2 was smaller in Hoxb3Tg/+ and significantly reduced in size in the Hoxb3Tg/Tg at E10.5 (Figures 3A–C). To investigate the neural components in the pharyngeal region, wholemount immunohistochemistry was performed using 2H3 anti-neurofilament antibody. In the Hoxb3Tg/+ mutants, the vestibulocochlear nerve (VIIIn), the superficial petrosal nerve (spn), and the chorda tympanic nerve (ct) were absent (Figures 3D,E,G,H). In the Hoxb3Tg/Tg mutants, there was abnormal fusion between the trigeminal ganglion (V) and the facial acoustic ganglia (VII; Figures 3F,I).
FIGURE 3
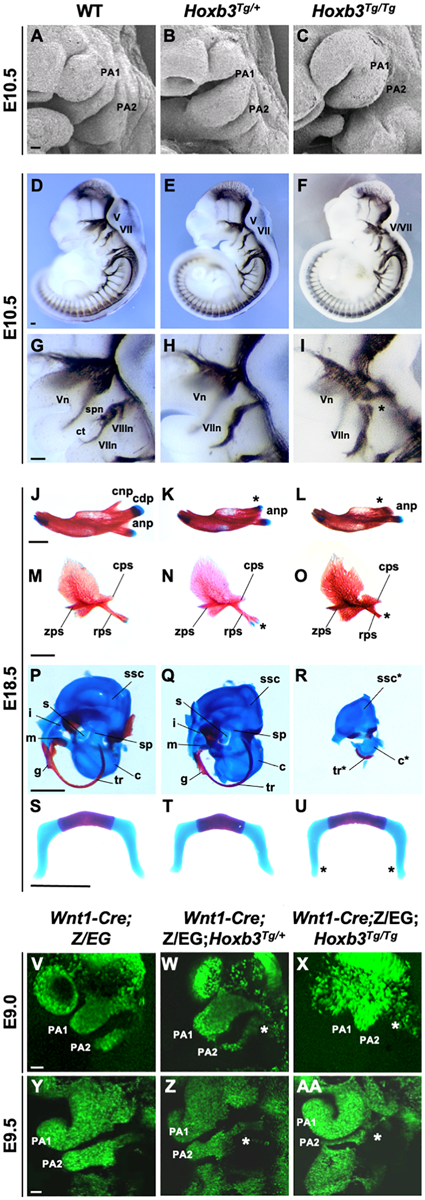
Hypoplastic PA2 and defective neural crest development in Hoxb3Tg mutants. (A–C) Scanning electron microscopy of E10.5 wildtype, Hoxb3Tg/+ and Hoxb3Tg/Tg embryos (n = 3). (D–I) Neurofilament antibody (2H3) staining showing cranial nerve defects in wildtype and Hoxb3Tg mutants at E10.5 (n > 3). (G–I) are enlargements of pharyngeal regions showing abnormal facial nerve branching in Hoxb3Tg/+ and nerve fusion in Hoxb3Tg/Tg (indicated with asterisk). V/Vn, trigeminal nerve; VII/VIIn, facial nerve; VIIIn, vestibuloaoustic nerve; spn, superficial petrosal nerve; and ct, chorda tympani nerve. (J–R) Skeletal preparations showing defective PA1 and PA2 neural crest cell derived structures in the Hoxb3Tg mutants (n > 5). (J–L) Skeletal preparations showing defective mandibles of Hoxb3Tg mutants. Absence of condylar process indicated with asterisks. (M–O) Defective squamous bones in Hoxb3Tg mutants. Abnormal retrotympanic process indicated with asterisks. anp, angular process, cdp and cnp, condylar and coronoid processes of mandible; cps, caudal process of squamous; rps, retrotympanic process of squamous; zps, zygomatic process of squamous; and g, gonial. (P–R) Defective middle ear bones, inner ear otic capsules and associated elements in Hoxb3Tg mutants. c, cochlea; i, incus; m, malleus; s, stapes; sp, styloid process; ssc, semi-circular canals; and tr, tympanic ring. (S–U) Skeletal preparations showing mild defects in the hyoid cartilage of Hoxb3Tg/Tg mutants. *indicates the greater horn. (V–AA) Wholemount GFP autofluorescence of Wnt1-Cre;Z/EG;Hoxb3Tg compound mutant embryos showing abnormal distribution of GFP-positive neural crest cells (asterisks) to PA2 in E9.0 and E9.5 Hoxb3Tg/+ and Hoxb3Tg/Tg embryos. PA1 and PA2 indicate the 1st and 2nd pharyngeal arches (n > 3). Scale bars: A-I and V-AA, 100 μm; J-U, 1,000 μm.
To examine the phenotypes of neural crest derived skeletal components, neonates were examined by skeletal staining. Among the PA1 derived structures, the mandible was reduced in length and poorly ossified and the coronoid process was absent in both Hoxb3Tg/+ and Hoxb3Tg/Tg mutants. The condylar process was present in the Hoxb3Tg/+ embryos, but missing in the Hoxb3Tg/Tg embryos (Figures 3J–L). The maxillary arch derived squamous bone was slightly affected with a duplication found in the retrotympanic process in the Hoxb3Tg/+ embryos (Figures 3M,N). The squamous bone was more severely affected in the Hoxb3Tg/Tg mutants (Figure 3O). Among the PA1 derived middle ear structures, the malleus and incus were hypoplastic in the Hoxb3Tg/+ embryos (Figures 3P,Q), and absent in the Hoxb3Tg/Tg mutants (Figure 3R). The otic capsule and tympanic ring were hypoplastic in the Hoxb3Tg/Tg mutants (Figure 3R). For the PA2 derived skeletal elements, the styloid process was slightly truncated in Hoxb3Tg/+, while both the stapes and styloid process were severely reduced in the Hoxb3Tg/Tg mutants (Figures 3Q,R). The hyoid cartilage appeared normal in Hoxb3Tg/+, the morphology of the greater horn of the hyoid cartilage was mildly affected in the Hoxb3Tg/Tg mutants (Figures 3S–U). Taken together, we showed that the development of both PA1 and PA2 neural crest derived skeletal structures were affected in the Hoxb3Tg transgenic mutants.
Deficient Neural Crest Cell Migration Into PA2 of Hoxb3Tg Mutant
We hypothesized that the hypoplastic PA2 and abnormal neural crest derived structures in the Hoxb3Tg mutants could be due to neural crest cell deficiencies. By using the Wnt1-cre;Z/EG mice as neural crest cell lineage marker, we generated compound mutants and traced the migrating neural crest cells at E9.0 and E9.5. We observed consistently fewer neural crest cells migrating to PA2 of the Hoxb3Tg/+, and significantly few cells entering PA2 of the Hoxb3Tg/Tg mutants at E9.0 and E9.5 (Figures 3V–AA). Therefore, the multiple neural and skeletal phenotypes observed in the mutant pharyngeal regions were likely due to insufficient neural crest cells populating PA2.
Hoxb3 Activates the Expression of Jag1 by Direct Binding to Specific Genomic Sites
To investigate the possibility that Hoxb3 may directly activate Jag1 expression by binding to genomic regulatory elements, we performed bioinformatics analyses to identify possible Hoxb3 binding sites around the Jag1 gene. Using the consensus Hoxb3 binding matrix from JASPAR database, we searched the conserved regions within 100 kb upstream of Jag1 among six vertebrate species including mouse, human, chicken, lizard, Xenopus, and zebrafish. Furthermore, by comparing the open chromatin regions upstream of Jag1 from several E11.5 mouse tissues, we identified three potential regulatory sites, named S1, S2, and S3. The S1 and S2 sites were located upstream of Jag1 gene (mm10 chr2:137190579-137190732; chr2:137153318-137153475), the S3 site was in the intron between Exon2 and Exon3 of Jag1 (mm10 chr2:137110522-137110680; Figure 4A).
FIGURE 4
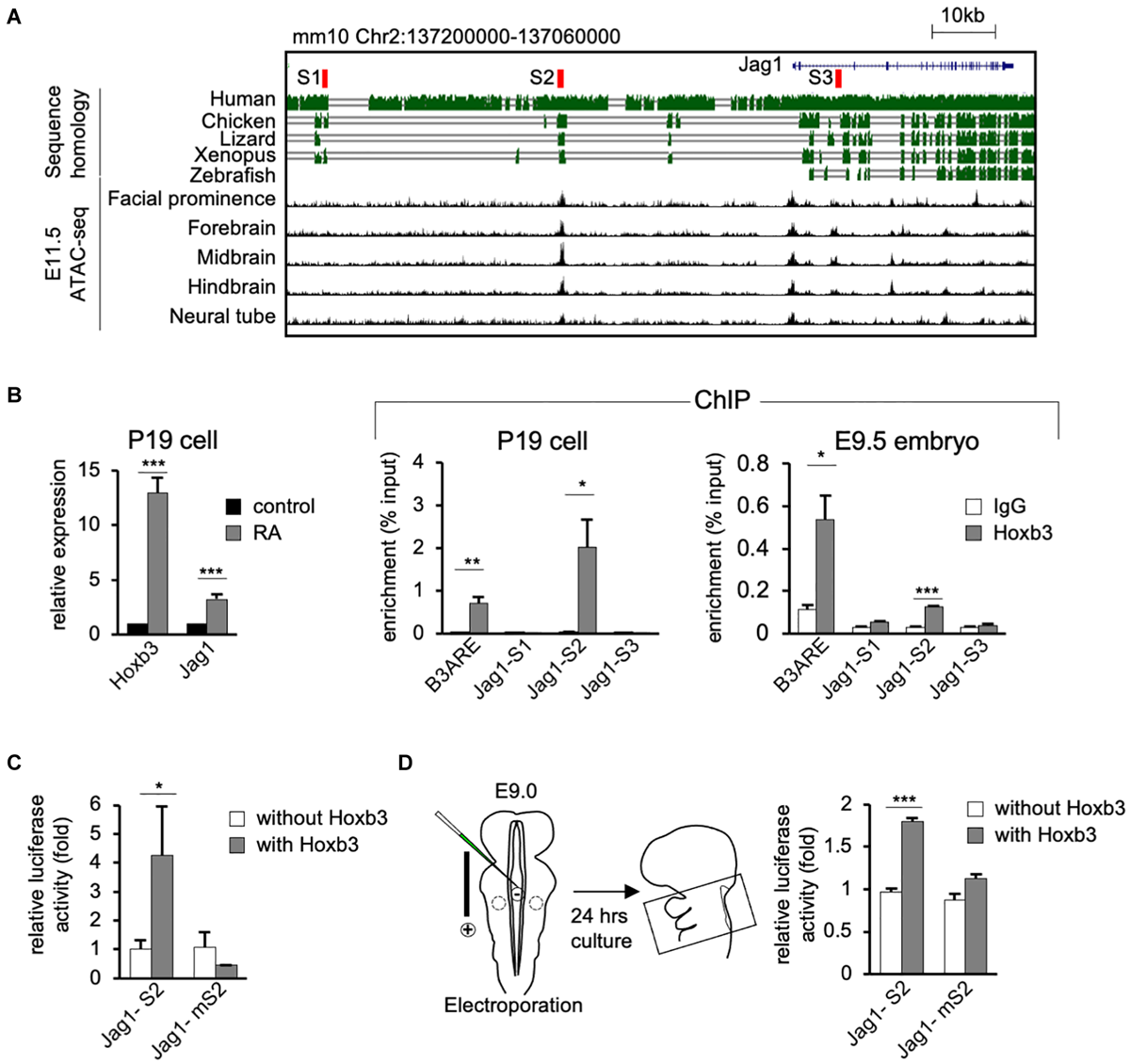
Hoxb3 regulates Jag1 expression by binding to cis-regulatory region S2. (A) Genome browser tracks showing mouse Jag1 locus including 100 Kb upstream genomic region. Alignment of genomic regions of human, chicken, lizard and xenopus revealed conserved regions and identification of potential Hoxb3 binding regions S1, S2, and S3 (highlighted in red). ATAC-seq tracks from ENCODE database of multiple E11.5 mouse embryo tissues showed an open chromatin region (S2) 40 kb upstream of Jag1 transcription start site. (B)In vitro and in vivo chromatin immunoprecipitation (ChIP) analyses using RA treated P19 cells and mouse embryos. qRT-PCR analysis of P19 cells showed elevated expression of Hoxb3 and Jag1 after RA treatment. ChIP qRT-PCR analysis showed that Hoxb3 could bind to S2 site of Jag1, but not S1 or S3 sites. Hoxb3 could also bind to the B3ARE site, an autoregulatory element which served as a positive control. ChIP qRT-PCR analysis using E9.5 embryos also showed that Hoxb3 could bind to the S2 site and the positive control B3ARE site. (C)In vitro luciferase reporter assay using 293T cells. Expression of Hoxb3 could activate luciferase activity when co-transfected with Jag1-S2 reporter, the activity was much lower when co-transfected with Jag1-mS2 reporter containing mutated S2 site. (D)Ex vivo luciferase reporter assay using E9.0 mouse embryos. Co-electroporation of Hoxb3 expression vector with Jag1-S2 or Jag1-mS2 luciferase reporter showed that Hoxb3 could elevate luciferase activity via the normal S2 site. Data in (B–D) are shown as mean ± SE (n = 3 biological replicates). *P < 0.05, **P < 0.01, and ***P < 0.001.
To test whether Hoxb3 can directly bind to the three identified binding sites around Jag1 gene, we performed in vitro and in vivo chromatin immunoprecipitation assays using P19 cells and mouse embryos. We treated P19 cells with RA to activate the endogenous expression of Hoxb3. The Quantitative real-time PCR (qRT-PCR) results showed that the expression of Hoxb3 and Jag1 were significantly increased after RA treatment (Figure 4B). Chromatin fragments from RA treated P19 cells were immunoprecipitated with a Hoxb3 antibody (Supplementary Figure 1). Several sets of primers were used to detect potential binding to the S1, S2, and S3 sites. A known Hoxb3 binding site, B3ARE in the auto-regulatory element (Yau et al., 2002; Wong et al., 2011), was used as a positive control. The qRT-PCR results of ChIP assays showed that Hoxb3 antibody could immunoprecipitate the B3ARE and S2 sites but not S1 or S3 sites. We further performed in vivo ChIP assays using wildtype E9.5 embryos. The entire embryos were lysed and the chromatin fragments were immunoprecipitated with IgG and Hoxb3 antibodies. The PCR results showed that endogenous Hoxb3 protein could form complex with chromatin fragments containing the S2 binding site (Figure 4B). To further investigate whether Hoxb3 could positively regulate the expression of Jag1 through binding to the S2 site, we performed luciferase assay on human embryonic kidney cells 293T. Luciferase activity was significantly increased in the presence of Hoxb3, but no activation could be observed when Jag-S2 site was mutated (Figure 4C). To further investigate whether Hoxb3 could positively regulate the expression of Jag1 through binding to the S2 site, we performed mouse ex vivo electroporation experiment (Figure 4D). Co-expression of the luciferase reporter containing S2 binding site of Jag1 (Jag1-S2) and Hoxb3 expression vector, luciferase activity could be detected in the embryo lysates. Without Hoxb3, luciferase activity was significantly reduced. However, with the S2 site mutated in the Jag1 luciferase reporter (Jag1-mS2), electroporation of Hoxb3 expression vector could not increase the luciferase activity.
In conclusion, by molecular studies we demonstrated that endogenous Hoxb3 protein could directly activate the expression of Jag1 through specific binding to the S2 regulatory site. This study revealed a novel regulatory mechanism for Jag1 gene expression. During normal development, cell type-specific expression of Jag1 could be regulated by Hoxb3.
Discussion
The roles of Hox genes in providing positional information in axial development have long been established. In the pharyngeal region, combinatorial Hox expression defines the positional identify of the hindbrain rhombomeres and associated neural crest cells. It has been suggested that the surface ectoderm also expresses combinatorial Hox genes, presenting segmentally patterned “ectomeres” where the ectodermal epithelium is regionally specified and associated with the Hox-patterned pharyngeal arches (Couly and Le Douarin, 1990; Hunt et al., 1991). However, the antero-posterior segmental specific functions of the ectodermal pharyngeal epithelium in the PAs remain elusive. The pharyngeal epithelium is important for providing environment cues for cranial neural crest cell homing, survival and differentiation. Our gain-of-function Hoxb3Tg mutant analysis here suggests that the cross-talk of Hox genes with molecular signaling in the pharyngeal epithelium is required for interaction and maintenance of neural crest cells during PA development.
Expression of Hox Genes and Neural Crest Defects in Hoxb3Tg Mutant PAs
In the Hoxb3Tg mutant, we have previously shown that hindbrain neurogenesis defect was observed as a result of the loss of Hoxb1 expression in hindbrain r4 (Wong et al., 2011). However, here we found that the generation, delamination and migration of r4-derived neural crest was not affected, except that the population of neural crest cells entering the mutant PA2 were reduced (Figures 3W,X,Z,AA). The expression patterns of Hoxa1, Hoxa2, and Hoxb1 in the PAs were not changed in the Hoxb3Tg/+ mutant (Supplementary Figure 2). Therefore, in the Hoxb3Tg/+ mutant, normal combinatorial Hox genes were expressed in the neural crest cells. The dorso-ventral patterning of PAs also remained unaffected in the mutant (Figures 3A–F). The specific changes in the Hoxb3Tg/+ mutant were with the surface ectoderm, the entire proximal-distal pharyngeal epithelium of PA2 and posterior PA1 were the only cells expressing Hoxb3 ectopically. Although PA1 is normally a Hox-free zone, and the enhancer activity of the Hoxb2-r4 element used to drive ectopic gene expression is expected to cover PA2 only (Figures 1L,M), as a result of transgene insertion activity, ectopic Hoxb3 expression was unexpectedly also found in PA1 epithelium. The ectopic expression patterns of Hoxb3 would explain that neural crest structures derived from both PA1 and PA2 were affected in the Hoxb3Tg mutants. The malformations could be a direct effect of ectopic Hoxb3 expression in the ectodermal pharyngeal epithelium, or through other signaling molecules including Jag1 as discussed below.
In the heterozygous Hoxb3Tg/+ transgenic mutant, the PA phenotypes were mild, but the hypoplasia of PA2 and craniofacial malformations were more evident in the homozygous Hoxb3Tg/Tg with a higher dosage of the transgene. The fusion of the trigeminal and facioacoustic ganglia observed in Hoxb3Tg/Tg suggests that there might be mixing or mis-routing of neural crest cells. As visualized by Wnt1-Cre;Z/EG lineage tracing experiments, in E9.0 Hoxb3Tg/Tg mutant embryo the neural crest cells appeared aggregated around the proximal PA1 and PA2 region (Figure 3X). As many of the neural crest cells did not reach PA2, a deficiency of neural crest cells would explain the pharyngeal skeletal phenotype in the Hoxb3Tg mutants.
Hoxb3 Directly Regulates Jag1 Expression
To address how ectopic Hoxb3 expression in the pharyngeal epithelium could affect interaction with neural crest cells, we examined the Notch signaling ligand Jag1 and found that Hoxb3 could directly regulate Jag1 expression. Jag1 is required for craniofacial morphogenesis and other developmental processes (Xue et al., 1999; Humphreys et al., 2012), but the spatial-temporal regulation of Jag1 expression during development is unclear. By bioinformatics analyses, we identified several Hoxb3 binding sites in the highly conserved genomic regions around Jag1 gene. Using in vivo ChIP assays and in vitro/ex vivo luciferase reporter assays, we demonstrated that Hoxb3 could directly bind to the S2 site and trans-activate gene expression. From ENCODE databases, many other transcription factors binding sites could be identified around S2, indicating that this 40 Kb upstream genomic region could be a cis-regulatory enhancer element for activating Jag1 expression. In the Hoxb3Tg gain-of-function mutant, we showed that Jag1 expression could be specifically activated by ectopic Hoxb3 expression in the pharyngeal epithelium of PA1 and PA2 (Figure 2J). It is possible that during normal craniofacial development, Hoxb3 is also required to maintain Jag1 expression in the posterior pharyngeal arches and clefts to coordinate with the pharyngeal segmentation process.
The expression of Jag1 has been reported to be activated by other Hox3 genes. In hemogenic endothelium, overexpression of Hoxa3 could activate the expression of Jag1 (Sanghez et al., 2017). In HEL and K563 cells, overexpression of Hoxd3 also led to elevated Jag1 expression. Moreover, the expression level of Jag1 was dependent on transfected Hoxd3 expression (Taniguchi et al., 2001). These studies confirmed that the Hox3 genes could regulate Jag1 expression and Notch signaling in broader developmental contexts.
The Role of Jag1 in PA Development
Recent studies of the proximal PA and epibranchial placode suggest that pharyngeal segments are gateways for neural crest cell migration into the PAs. Entering between the pharyngeal clefts, the neural crest cells populate the PAs distally, where subsequent morphogenetic events take place. For example, Sox3 knockout mice with defective epibranchial placodal cells in the proximal pharyngeal epithelium displayed mis-migration of neural crest cells into the PAs and resulted in cranial skeletal defects (Rizzoti and Lovell-Badge, 2007). On the other hand, in the Eya1 mutant which failed to maintain the epibranchial placodal cells, defective neural crest-derived maxilla and mandible were observed (Xu et al., 1999; Zhang et al., 2017). Importantly, both Sox3 and Eya1 are not expressed in neural crest but in the epibranchial placodal cells, indicating a non-cell autonomous role of epibranchial placodal epithelium in controlling the migration of neural crest cells to the PAs. In the Hoxb3Tg/+ mutants described in this study, ectopic expression of Hoxb3 and Jag1 were found only in the pharyngeal epithelium, the neural crest-derived skeletal defects could be the consequence of misexpression of Hoxb3 in the epibranchial placodal cells in the proximal PA.
We have previously shown that Jag1 is initially broadly expressed in the proximal pharyngeal region in the epibranchial placodal cells at early stage (E8.5), but its expression is down-regulated in PA epithelium and became restricted to the pharyngeal clefts at around E9.5 (Zhang et al., 2017). While the functional role of Jag1 in pharyngeal segmentation is not known, it appears that Jag1 expressing regions serve as boundaries for neural crest cell migration. In the Hoxb3Tg/+ mutant, persisted expression of Jag1 in the proximal pharyngeal epithelium of PA2 could inhibit the colonization of neural crest cells into the arch (Figure 5), leading to the craniofacial abnormalities. Our genetic study has revealed the importance of the pharyngeal epithelial cells in interacting with cranial neural crest for proper craniofacial development.
FIGURE 5
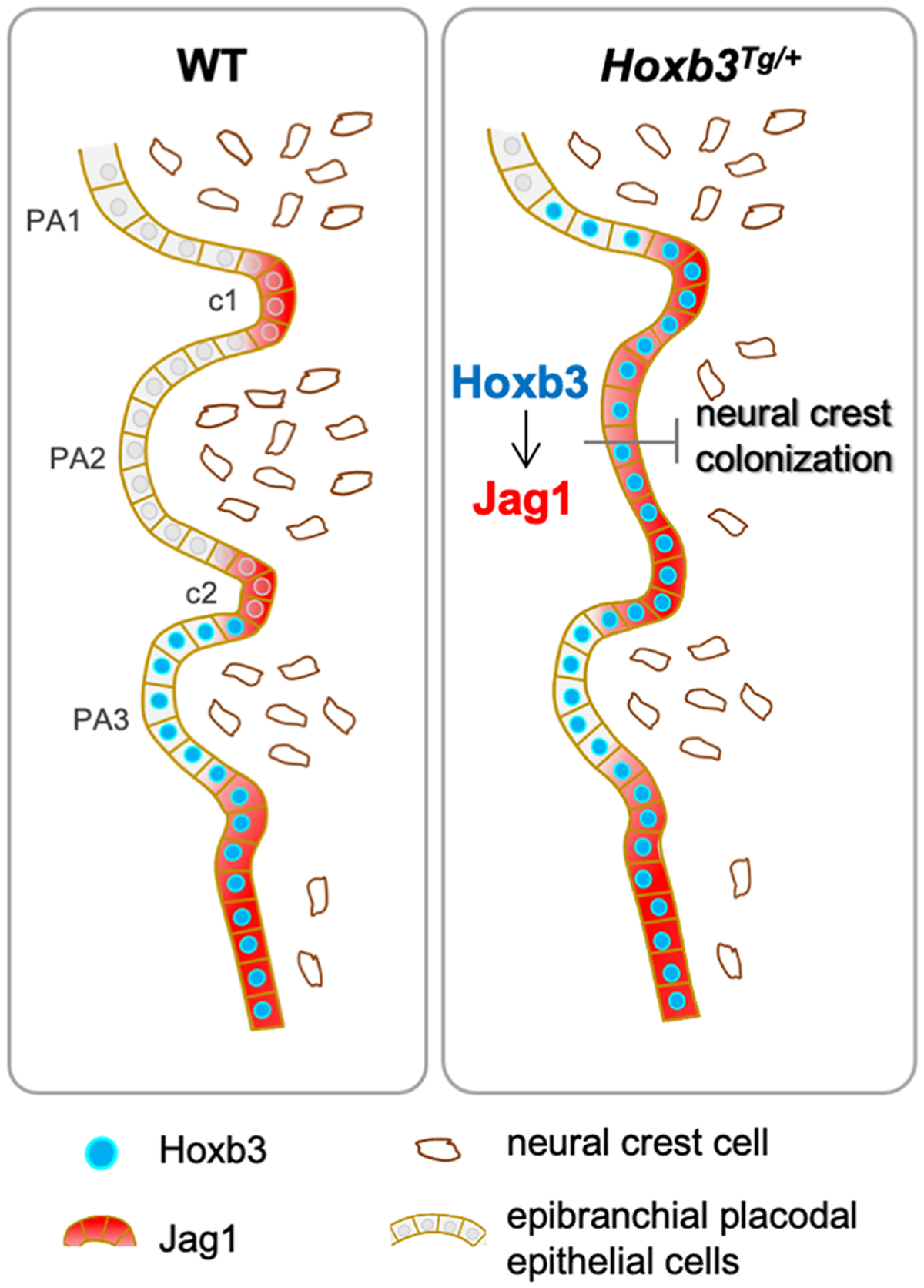
Hoxb3 regulate Jag1 expression and affect pharyngeal epithelium and neural crest interactions. Schematic diagram showing expression of Hoxb3 and Jag1 in epibranchial placodal epithelial cells of wildtype and Hoxb3Tg mutant. In wildtype embryos, Jag1 expression is restricted to the pharyngeal clefts which function as boundaries for neural crest migration into the pharyngeal arches. In the Hoxb3Tg mutant, ectopic Hoxb3 expressed in the epibranchial placodal epithelium of PA2 could activate Jag1 expression directly. Elevated Jag1 expression is incompatible with neural crest migration into the pharyngeal arch. As a result, deficiency of neural crest colonization into PA2 led to abnormal development of multiple neural crest derivatives in the pharyngeal region of the Hoxb3Tg mutant. PA1, PA2, and PA3 indicate the 1st, 2nd, and 3rd pharyngeal arches; c1 and c2 indicate the 1st and 2nd pharyngeal clefts.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by The University of Hong Kong Committee on the Use of Live Animals for Teaching and Research (CULATR Nos. 4357–17 and 4588–18).
Author contributions
HZ, EW, and MS conceived the study, designed the experiments, and wrote the manuscript. HZ, JX, KS, KT, JS-P, LW, EW, and WC conducted the experiments. ST, JS-P, EW, and HZ managed the experimental animals. JX, KS, KT, and MS reviewed and revised the manuscript and edited the figures. All authors participated in data analysis and revisions of the manuscript. MS obtained resources and funding for the study.
Funding
This work was supported by a research grant from the Hong Kong Research Grant Council (RGC GRF 777411) to MS.
Acknowledgments
We thank Kathryn S.E. Cheah and Urban Lendahl for helpful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.612230/full#supplementary-material
References
1
BarrowJ. R.CapecchiM. R. (1996). Targeted disruption of the Hoxb-2 locus in mice interferes with expression of Hoxb-1 and Hoxb-4.Development1223817–3828.
2
BarskeL.AskaryA.ZunigaE.BalczerskiB.BumpP.NicholsJ. T.et al (2016). Competition between Jagged-Notch and Endothelin1 Signaling Selectively Restricts Cartilage Formation in the Zebrafish Upper Face.PLoS Genet12:e1005967. 10.1371/journal.pgen.1005967
3
CaseyL. M.LanY.ChoE. S.MaltbyK. M.GridleyT.JiangR. (2006). Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development.Dev. Dyn.2351830–1844. 10.1002/dvdy.20821
4
ChaiY.JiangX.ItoY.BringasP.Jr.HanJ.RowitchD. H.et al (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis.Development1271671–1679.
5
ChisakaO.MusciT. S.CapecchiM. R. (1992). Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-# 150; 1.6.Nature355516–520.
6
ChojnowskiJ. L.TrauH. A.MasudaK.ManleyN. R. (2016). Temporal and spatial requirements for Hoxa3 in mouse embryonic development.Dev. Biol.41533–45. 10.1016/j.ydbio.2016.05.010
7
CoulyG.Le DouarinN. (1990). Head morphogenesis in embryonic avian chimeras: evidence for a segmental pattern in the ectoderm corresponding to the neuromeres.Development108543–558.
8
CoulyG.CreuzetS.BennaceurS.VincentC.Le DouarinN. M. (2002). Interactions between Hox-negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head.Development1291061–1073.
9
CoulyG.Grapin-BottonA.ColteyP.RuhinB.Le DouarinN. M. (1998). Determination of the identity of the derivatives of the cephalic neural crest: incompatibility between Hox gene expression and lower jaw development.Development1253445–3459.
10
CreuzetS.CoulyG.VincentC.Le DouarinN. M. (2002). Negative effect of Hox gene expression on the development of the neural crest-derived facial skeleton.Development1294301–4313.
11
DessaudE.YangL. L.HillK.CoxB.UlloaF.RibeiroA.et al (2007). Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism.Nature450717–720. 10.1038/nature06347
12
DonovielD. B.HadjantonakisA.-K.IkedaM.ZhengH.HyslopP. S. G.BernsteinA. (1999). Mice lacking both presenilin genes exhibitearlyembryonic patterningdefects.Genes Dev.132801–2810. 10.1101/gad.13.21.2801
13
FerrettiE.MarshallH.PopperlH.MaconochieM.KrumlaufR.BlasiF. (2000). Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1. Pbx and Hox proteins.Development127155–166.
14
FraschM.ChenX.LufkinT. (1995). Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila.Development121957–974.
15
FrisdalA.TrainorP. (2014). Development and evolution of the pharyngeal apparatus.Wiley Interdiscip. Rev. Dev. Biol.3403–418. 10.1002/wdev.147
16
FujitaM.SakabeM.IokaT.WatanabeY.Kinugasa-KatayamaY.TsuchihashiT.et al (2016). Pharyngeal arch artery defects and lethal malformations of the aortic arch and its branches in mice deficient for the Hrt1/Hey1 transcription factor.Mech. Dev.13965–73. 10.1016/j.mod.2015.11.002
17
GavalasA.StuderM.LumsdenA.RijliF. M.KrumlaufR.ChambonP. (1998). Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch.Development1251123–1136.
18
GavalasA.TrainorP.Ariza-McnaughtonL.KrumlaufR. (2001). Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest.Development1283017–3027.
19
GoddardJ. M.RosselM.ManleyN. R.CapecchiM. R. (1996). Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VIIth nerve.Development1223217–3228.
20
GrahamA. (2003). Development of the pharyngeal arches.Am. J. Med. Genet. A119A251–256. 10.1002/ajmg.a.10980
21
GrahamA.SmithA. (2001). Patterning the pharyngeal arches.Bioessays2354–61. 10.1002/1521-1878(200101)23:1<54::AID-BIES1007<3.0.CO;2-5
22
GrahamA.OkabeM.QuinlanR. (2005). The role of the endoderm in the development and evolution of the pharyngeal arches.J. Anat.207479–487. 10.1111/j.1469-7580.2005.00472.x
23
HuM. C.RosenblumN. D. (2005). Smad1, β-catenin and Tcf4 associate in a molecular complex with the Myc promoter in dysplastic renal tissue and cooperate to control Myc transcription.Development132215–225. 10.1242/dev.01573
24
HumphreysR.ZhengW.PrinceL. S.QuX.BrownC.LoomesK.et al (2012). Cranial neural crest ablation of Jagged1 recapitulates the craniofacial phenotype of Alagille syndrome patients.Hum. Mol. Genet.211374–1383. 10.1093/hmg/ddr575
25
HuntP.WhitingJ.MuchamoreI.MarshallH.KrumlaufR. (1991). Homeobox genes and models for patterning the hindbrain and branchial arches.Dev. Suppl.1187–196.
26
JayasenaC. S.OhyamaT.SegilN.GrovesA. K. (2008). Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode.Development1352251–2261. 10.1242/dev.017905
27
JonesE. A.Clement-JonesM.WilsonD. I. (2000). JAGGED1 expression in human embryos: correlation with the Alagille syndrome phenotype.J. Med. Genet.37658–662. 10.1136/jmg.37.9.658
28
KamathB.BasonL.PiccoliD.KrantzI.SpinnerN. (2003). Consequences of JAG1 mutations.J. Med. Genet.40891–895. 10.1136/jmg.40.12.891
29
KamedaY.AraiY.NishimakiT.ChisakaO. (2004). The role of Hoxa3 gene in parathyroid gland organogenesis of the mouse.J. Histochem. Cytochem.52641–651. 10.1177/002215540405200508
30
KulesaP. M.BaileyC. M.Kasemeier-KulesaJ. C.MclennanR. (2010). Cranial neural crest migration: new rules for an old road.Dev. Biol.344543–554. 10.1016/j.ydbio.2010.04.010
31
LassiterR. N.BallM. K.AdamsJ. S.WrightB. T.StarkM. R. (2010). Sensory neuron differentiation is regulated by notch signaling in the trigeminal placode.Dev. Biol.344836–848. 10.1016/j.ydbio.2010.05.514
32
LassiterR. N.StarkM. R.ZhaoT.ZhouC. J. (2014). Signaling mechanisms controlling cranial placode neurogenesis and delamination.Dev. Biol.38939–49. 10.1016/j.ydbio.2013.11.025
33
Le DouarinN. M.CreuzetS.CoulyG.DupinE. (2004). Neural crest cell plasticity and its limits.Development1314637–4650. 10.1242/dev.01350
34
LiL.KrantzI. D.DengY.GeninA.BantaA. B.CollinsC. C.et al (1997). Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1.Nat. Genet.16243–251. 10.1038/ng0797-243
35
MaconochieM.KrishnamurthyR.NonchevS.MeierP.ManzanaresM.MitchellP. J.et al (1999). Regulation of Hoxa2 in cranial neural crest cells involves members of the AP-2 family.Development1261483–1494.
36
MaconochieM.NonchevS.StuderM.ChanS.-K.PöpperlH.ShamM. H.et al (1997). Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1.Genes Dev.111885–1895. 10.1101/gad.11.14.1885
37
ManleyN. R.CapecchiM. R. (1998). Hox group 3 paralogs regulate the development and migration of the thymus, thyroid, and parathyroid glands.Dev. Biol.1951–15. 10.1006/dbio.1997.8827
38
McleodM. J. (1980). Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S.Teratology22299–301. 10.1002/tera.1420220306
39
MeadT. J.YutzeyK. E. (2012). Notch pathway regulation of neural crest cell development in vivo.Dev. Dyn.241376–389. 10.1002/dvdy.23717
40
MinouxM.RijliF. M. (2010). Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development.Development1372605–2621. 10.1242/dev.040048
41
NonchevS.VesqueC.MaconochieM.SeitanidouT.ArizamcnaughtonL.FrainM.et al (1996). Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20.Development122543–554.
42
NovakA.GuoC.YangW.NagyA.LobeC. G. (2000). Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre−mediated excision.Genesis28147–155. 10.1002/1526-968x(200011/12)28:3/4<147::aid-gene90>3.0.co;2-g
43
OkadaY.ShimazakiT.SobueG.OkanoH. (2004). Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells.Dev. Biol.275124–142. 10.1016/j.ydbio.2004.07.038
44
OyamaT.HarigayaK.SasakiN.OkamuraY.KokuboH.SagaY.et al (2011). Mastermind-like 1 (MamL1) and mastermind-like 3 (MamL3) are essential for Notch signaling in vivo.Development1385235–5246. 10.1242/dev.062802
45
RijliF. M.MarkM.LakkarajuS.DierichA.DolleP.ChambonP. (1993). A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene.Cell751333–1349. 10.1016/0092-8674(93)90620-6
46
RizzotiK.Lovell-BadgeR. (2007). SOX3 activity during pharyngeal segmentation is required for craniofacial morphogenesis.Development1343437–3448. 10.1242/dev.007906
47
SanghezV.LuzziA.ClarkeD.KeeD.RuxD.OsawaM.et al (2017). Notch activation is required for downregulation of HoxA3-dependent endothelial cell phenotype during blood formation.PLoS One12:e0186818. 10.1371/journal.pone.0186818
48
SantagatiF.RijliF. M. (2003). Cranial neural crest and the building of the vertebrate head.Nat. Rev. Neurosci.4806–818. 10.1038/nrn1221
49
SchlosserG.AhrensK. (2004). Molecular anatomy of placode development in Xenopus laevis.Dev. Biol.271439–466. 10.1016/j.ydbio.2004.04.013
50
StuderM.LumsdenA.Ariza-McnaughtonL.BradleyA.KrumlaufR. (1996). Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1.Nature384630–634. 10.1038/384630a0
51
SuD.EllisS.NapierA.LeeK.ManleyN. R. (2001). Hoxa3 and pax1 regulate epithelial cell death and proliferation during thymus and parathyroid organogenesis.Dev. Biol.236316–329. 10.1006/dbio.2001.0342
52
SzetoI. Y.LeungK. K.ShamM. H.CheahK. S. (2009). Utility of HoxB2 enhancer-mediated Cre activity for functional studies in the developing inner ear.Genesis47361–365. 10.1002/dvg.20507
53
TaniguchiY.SatoM.TanakaO.SekiguchiM.InokoH.KimuraM. (2001). ”HOXD3 regulates expression of JAGGED1, a ligand for Notch receptors”, in: Nucleic acids symposium series.Oxford: Oxford University Press, 43–44.
54
TengC. S.YenH. Y.BarskeL.SmithB.LlamasJ.SegilN.et al (2017). Requirement for Jagged1-Notch2 signaling in patterning the bones of the mouse and human middle ear.Sci. Rep.7:2497. 10.1038/s41598-017-02574-7
55
TrainorP.KrumlaufR. (2000). Plasticity in mouse neural crest cells reveals a new patterning role for cranial mesoderm.Nat. Cell Biol.296–102. 10.1038/35000051
56
TrainorP.KrumlaufR. (2001). Hox genes, neural crest cells and branchial arch patterning.Curr. Opin. Cell Biol.13698–705. 10.1016/s0955-0674(00)00273-8
57
TrokovicN.TrokovicR.PartanenJ. (2005). Fibroblast growth factor signalling and regional specification of the pharyngeal ectoderm.Int. J. Dev. Biol.49797–805. 10.1387/ijdb.051976nt
58
VeitchE.BegbieJ.SchillingT. F.SmithM. M.GrahamA. (1999). Pharyngeal arch patterning in the absence of neural crest.Curr. Biol.91481–1484. 10.1016/s0960-9822(00)80118-9
59
VieiraA. R.AvilaJ. R.Daack-HirschS.DraganE.FelixT. M.RahimovF.et al (2005). Medical sequencing of candidate genes for nonsyndromic cleft lip and palate.PLoS Genet1:e64. 10.1371/journal.pgen.0010064
60
WakehamA.CorreiaK.SamperE.BrownS.AguileraR.NakanoT.et al (1997). Conservation of the Notch signalling pathway in mammalian neurogenesis.Development1241139–1148.
61
WangL.XieJ.ZhangH.TsangL. H.TsangS. L.BrauneE. B.et al (2020). Notch signalling regulates epibranchial placode patterning and segregation.Development147:dev183665. 10.1242/dev.183665
62
WashausenS.KnabeW. (2017). Pax2/Pax8-defined subdomains and the occurrence of apoptosis in the posterior placodal area of mice.Brain Struct. Funct.2222671–2695. 10.1007/s00429-016-1364-0
63
WilkinsonD. G.BhattS.CookM.BoncinelliE.KrumlaufR. (1989). Segmental expression of Hox-2 homoeobox-containing genes in the developing mouse hindbrain.Nature341405–409. 10.1038/341405a0
64
WongE. Y.WangX. A.MakS. S.Sae-PangJ. J.LingK. W.FritzschB.et al (2011). Hoxb3 negatively regulates Hoxb1 expression in mouse hindbrain patterning.Dev. Biol.352382–392. 10.1016/j.ydbio.2011.02.003
65
XuP.-X.AdamsJ.PetersH.BrownM. C.HeaneyS.MaasR. (1999). Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia.Nat. Genet.23113–117. 10.1038/12722
66
XuP.-X.ZhengW.LaclefC.MaireP.MaasR. L.PetersH.et al (2002). Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid.Development1293033–3044.
67
XueY.GaoX.LindsellC. E.NortonC. R.ChangB.HicksC.et al (1999). Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1.Hum. Mol. Genet.8723–730. 10.1093/hmg/8.5.723
68
YauT. O.KwanC. T.JaktL. M.StallwoodN.CordesS.ShamM. H. (2002). Auto/cross-regulation of Hoxb3 expression in posterior hindbrain and spinal cord.Dev. Biol.252287–300. 10.1006/dbio.2002.0849
69
ZhangH.WangL.WongE. Y. M.TsangS. L.XuP. X.LendahlU.et al (2017). An Eya1-Notch axis specifies bipotential epibranchial differentiation in mammalian craniofacial morphogenesis.Elife6:e30126. 10.7554/eLife.30126
Summary
Keywords
Hoxb3, JAG1, pharyngeal arch, epibranchial placodes, pharyngeal epithelium, cranial neural crest, craniofacial development
Citation
Zhang H, Xie J, So KKH, Tong KK, Sae-Pang JJ, Wang L, Tsang SL, Chan WY, Wong EYM and Sham MH (2021) Hoxb3 Regulates Jag1 Expression in Pharyngeal Epithelium and Affects Interaction With Neural Crest Cells. Front. Physiol. 11:612230. doi: 10.3389/fphys.2020.612230
Received
30 September 2020
Accepted
09 December 2020
Published
11 January 2021
Volume
11 - 2020
Edited by
Jean-Pierre Saint-Jeannet, New York University, United States
Reviewed by
Heather C. Etchevers, INSERM U1251 Centre de Génétique Médicale de Marseille (MMG), France; Juhee Jeong, New York University, United States
Updates
Copyright
© 2021 Zhang, Xie, So, Tong, Sae-Pang, Wang, Tsang, Chan, Wong and Sham.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mai Har Sham, mhsham@cuhk.edu.hk
This article was submitted to Craniofacial Biology and Dental Research, a section of the journal Frontiers in Physiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.