- 1Cotton Research Center, Shandong Academy of Agricultural Sciences, Jinan, China
- 2Institute for the Control of Agrochemicals, Ministry of Agriculture and Rural Affairs, Beijing, China
- 3Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing, China
Aphis craccivora (Koch) is an economically important pest that affects legumes in worldwide. Chemical control is still the primary efficient method for A. craccivora management. However, the mechanism underlying insecticide resistance in A. craccivora has not been elucidated. A previous study observed that piperonyl butoxide (PBO) and diethyl maleate (DEM) significantly synergized imidacloprid in A. craccivora field populations, indicating that cytochrome P450 (P450) and glutathione S-transferase (GST) genes may play pivotal roles in imidacloprid resistance. In this study, 38 P450 genes and 10 GST genes were identified in A. craccivora through transcriptomic analysis. The expression levels of these P450 and GST genes were measured in susceptible (SUS) strains of A. craccivora under imidacloprid treatment with LC15, LC50, and LC85 doses. The expression levels of CYP18A1, CYP6CY21, CYP6DA1, CYP6DA2, CYP4CJ1, CYP4CJ2, and CYP380C6 were up-regulated in the three treatments. Most of these genes belong to CYP3 and CYP4 Clans. In addition, the expression levels of all P450 and GST genes in A. craccivora were also measured in the Juye (JY) and Linqing (LQ) field populations. The expression levels of CYP6DA2, CYP4CJ1, and CYP380C6 were up-regulated in the SUS strain after imidacloprid treatment at three doses, and these genes were overexpressed in the JY population. Furthermore, the sensitivity of A. craccivora to imidacloprid was significantly increased after knockdown of CYP380C6 and CYP6DA2 through RNA interference. These results may help to elucidate the mechanisms underlying of imidacloprid resistance in A. craccivora.
Introduction
The Aphis craccivora (Koch), is one of the most important worldwide, affecting multiple legumes, such as pea, cowpea, and peanut. A. craccivora causes serious yield losses in legumes by sucking the leaf, bud, and flower sap. In addition, A. craccivora can transmit two major plant viruses, bean leaf roll virus and faba bean necrotic yellows virus, which seriously affects the yield and quality of legumes (Li et al., 2013a). Insecticides, including pyrethroids, organophosphates, and neonicotinoids, are currently the primary management tool or controlling A. craccivora (Zhang et al., 2015; Fouad et al., 2016). However, due to the extensive and recurrent use of these insecticides, field populations of A. craccivora have developed insecticide resistance (Li et al., 2013a; Abd-Ella, 2014).
The molecular mechanism underlying insecticide resistance primarily includes the reduction of target insensitivity and the enhancement of metabolic detoxification. Metabolic resistance to insecticides primarily due to the overexpression of three enzymes, including cytochrome P450 monooxygenases (P450s), glutathione S-transferases (GSTs), and carboxylesterase (CarEs; Li et al., 2007). P450s are a multi-gene superfamily which can participate in the synthesis and metabolism of endogenous compounds, and able to metabolize many types of xenobiotics, such as insecticides and plant toxins (Scott and Wen, 2001; Feyereisen, 2005, 2011). The P450 genes are primarily divided into four Clans: CYP2, CYP3, CYP4, and Mitochondrial (Mito) Clan (Feyereisen, 1999). P450s are involved in imidacloprid resistance in several pests; these genes include CYP6AY1, CYP6ER1, CYP6CW1, and CYP4CE1 in Nilaparvata lugens, CYP6CM1 in Bemisia tabaci, CYP9M10 in Culex quinquefasciatus, and CYP6D1 in Musca domestica (Liu and Scott, 1998; Karunker et al., 2008; Bass et al., 2011; Nauen et al., 2013; Zimmer et al., 2014). Also, GSTs, which are involved in the detoxification of exogenous toxins and intracellular transport, are important conjugation enzymes (De and Hemingway, 2010). The GST genes can be divided into seven classes, including Delta, Epsilon, Omega, Sigma, Theta, Zeta, and Microsomal (Enayati et al., 2005). In N. lugens, GSTs could be utilized as antioxidant defense agents to confer resistance to pyrethroid (Vontas et al., 2001). The Delta and Epsilon classes are insect-specific, playing important roles in xenobiotic detoxification (Che-Mendoza et al., 2009). The expression of AgGSTe2, which can metabolize DDT, was more than 5.0-fold in the DDT-resistant strain of Anopheles gambiae compared with the susceptible (SUS) strain (Ranson et al., 2000). Finally, CarEs play important roles in the degradation of insecticides via hydrolysis and catalysis (Li et al., 2007). In Aphis gossypii, the increased expression of CarEs was related to resistance to omethoate (Gong et al., 2014).
Imidacloprid is a representative neonicotinoid insecticide and has been extensively utilized in the management of piercing-sucking pests, such as aphids, planthoppers, leafhoppers, and thrips (Choi et al., 2001; Nauen and Denholm, 2010; Wang et al., 2018). However, different levels of imidacloprid resistance have been developed in different piercing-sucking pests due to the wide application of this insecticide (Karunker et al., 2008; Wen et al., 2009; Andrei et al., 2010; Cui et al., 2016). The populations of A. gossypii, such as Langfang (Hebei Province, China), Akesu (Xinjiang Province, China), Dezhou (Shandong Province, China), and Kuitun (Xinjiang Province, China), exhibited moderate levels of resistance to imidacloprid (Cui et al., 2016). Six populations of Myzus persicae showed high-levels of resistance to imidacloprid in South China (Zhang and Zhou, 2014). In previous studies, the constitutive overexpression of detoxification genes has been demonstrated to be related to insecticide resistance (Li et al., 2007). In addition, the induction of detoxification gene expression by insecticides is a rapid and effective method to identify key genes related to metabolic resistance, especially in the early stages of resistance development (Terriere, 1984; Zhang et al., 2016a).
In previous studies, we found that the resistance ratio of imidacloprid in Juye (JY, Shandong Province, China) and Linqing (LQ, Shandong Province, China) populations was 8.32- and 3.68-fold, exhibiting low resistance and sensitivity levels, respectively (Yang et al., 2020). Furthermore, piperonyl butoxide (PBO) and diethyl maleate (DEM) significantly synergized with imidacloprid in the field populations of A. craccivora. These results indicated that the P450s and GSTs may play important roles in imidacloprid tolerance in field populations of A. craccivora. In this study, the P450 and GST genes were identified via transcriptome analysis, and the changes in the expression levels of these genes were detected in the A. craccivora SUS strain under imidacloprid treatment. The expression levels of the P450 and GST genes were also detected in the field populations of A. craccivora. Furthermore, the functions of the up-regulated genes in imidacloprid treatment were verified through RNA interference (RNAi). These results may establish a foundation for further research exploring the mechanism underlying imidacloprid resistance in A. craccivora.
Materials and Methods
Insects
The susceptible (SUS) strain of A. craccivora was obtained from the China Southern Pesticide Creation Center (Shanghai, China) and reared in the laboratory without any contact with insecticides. The SUS strain of A. craccivora was collected from Shanghai (China) in 1980. The field populations of A. craccivora were collected from Linqing (LQ, Shandong Province, China, 115.702478E, 36.81416N) and Juye (JY, Shandong Province, China, 116.146121E, 35.225549N) in 2019. Insects were reared on Vicia faba plants at 25 ± 1°C with 70–80% humidity and a 16 h light: 8 h dark photoperiod.
Insecticides and Chemicals
Imidacloprid technical (96.7%) was provided by Shandong Lukang Biological Pesticide CO., Ltd. Dimethl sulfoxide (DMSO) and Triton X-100 were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Expression Induction
For expression induction, brachypterous A. craccivora adults were treated with imidacloprid at doses of LC15 (0.02 mg/L), LC50 (0.36 mg/L), and LC85 (4.2 mg/L). The experiment was performed according to the leaf-dipping method (Fouad et al., 2016). Imidacloprid was dissolved in DMSO and subsequently diluted in distilled water containing 0.05% Triton X-100 to generate a given concentration. Vicia faba seeding with brachypterous A. craccivora adults were immersed in imidacloprid for 5 s. Also, seedlings with A. craccivora brachypterous adult dipped in distilled water containing 0.05% Triton X-100 and 1% DMSO as a control. Filter paper was used to absorb excess liquid and transfer the Vicia faba seeding to the feeding box. All insects were reared at 25 ± 1°C, with 70–80% humidity and 16 h light: 8 h dark photoperiod. An aphid was considered dead if it was incapable of coordinated forward movement. Survival insects were collected at 48 h after imidacloprid application. The experiment included three replicates, and every replicate contained 30 adults. The insects were frozen in liquid nitrogen and stored at −80°C.
RNA Isolation, Purification, and Quantification
Total RNA was extracted using TRIzol reagent (Life Technologies, USA) following the manufacturer’s instructions. Spectrophotometry (NanoDrop 2000, Thermo Scientific) and agarose gel electrophoresis were employed to detect the quantity and quality of the RNAs, respectively. The RNA integrity was assessed by Agilent 2100 with RIN number >7.0.
cDNA Library Construction
Poly (A) RNA was purified from total RNA (10 μg) using poly T oligo attached magnetic beads using two rounds of purification. Then the poly (A) RNA was fragmented into small pieces using divalent cations under high temperature. Then the cleaved RNA fragments were reverse transcribed to create the cDNA, which were next used to synthesize U labeled second stranded DNAs with Escherichia coli DNA polymerase I, RNase H and dUTP. An A base was then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contains a T base overhang for ligating the adapter to the A tailed fragmented DNA. Single or dual index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. After the heat labile UDG enzyme treatment of the U labeled second stranded DNAs, the ligated products were amplified with PCR by the following conditions: initial denaturation at 95°C for 3 min; 8 cycles of denaturation at 98°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 30 s; and then final extension at 72°C for 5 min. The average insert size for the final cDNA library was 300 bp (±50 bp). At last, we performed the 150 bp paired end sequencing on an Illumina Hiseq 6000 (LC Bio, China) following the vendor’s recommended protocol.
Bioinformatic Analysis
Cutadapt and perl scripts in house were used to remove the reads that contained adaptor contamination, low quality bases and undetermined bases (Martin, 2011). The sequence quality was verified using FastQC.1 Trinity 2.4.0 was performed to de novo assembly of the transcripts (Grabherr et al., 2011). The longest transcript in the cluster was chosen as the representative gene sequence. All assembled transcripts were aligned against the non-redundant (Nr) protein database,2 Gene Ontology (GO),3 SwissProt,4 Kyoto Encyclopedia of Genes and Genomes (KEGG),5 and eggnog databases using DIAMOND with a threshold value of E < 10−5 (Buchfink et al., 2015).6
Identification of P450 and GST Genes and Phylogenetic Analysis
The sequences of P450 and GST genes were searched in the transcriptome of A. craccivora according to the annotation information. After removing the short (<300-bp) and repeated sequences, the remaining sequences were aligned in the NCBI database by BLASTx (value of E < 10−5).7 MEGA 5.05 was employed to construct the phylogenetic trees using the neighbor joining method based on the amino acid sequences in A. craccivora and other insects (Tamura et al., 2011). Information on the other insects in the phylogenetic trees was listed in Supplementary Table S1. The P450 genes were named by the P450 nomenclature committee (David R. Nelson, Department of Molecular Sciences, University of Tennessee, Memphis, TN, USA). The GST genes were named by phylogenetic analysis and the BLASTx results. The GenBank accession numbers of these P450 and GST genes were deposited in NCBI and the detailed information is presented in Supplementary Table S2.
Sequence Verification
According to the sequence information obtained from the transcriptome, the specific primer for each gene was designed using Primer5 (Supplementary Table S2). Total RNA was extracted using TRIzol reagent (Life Technologies, USA) following the manufacturer’s instructions. cDNA was synthesized following the manufacturer’s instructions for HiScript III RT SuperMix for qPCR (Vazyme Biotech Co., Ltd.). 2 × Taq Plus Master Mix II (Vazyme Biotech Co., Ltd.) was used for PCR. The amplified products were validated by gel electrophoresis and sequenced at Sangon Biotech (Shanghai, Co., Ltd.).
Detection of P450 and GST Genes by Quantitative Real-Time PCR
RT-qPCR was performed to detect the expression of P450 and GST genes at the mRNA level. A SYBR PrimeScript™ RT-PCR Kit (Takara, Japan) and IQ™ 5 multicolor real-time PCR detection system (BIO-RAD, USA) were utilized. Each experiment included three independent biological replications, and every biological replication included three technical replications. Every biological replication contained 15 insects. The relative expression of each gene was calculated by the 2−ΔΔCt method. RPS8 (Ribosomal protein S8, GenBank No.: GAJW01000269) and RPL14 (Ribosomal protein L14, GenBank No.: GAJW01000046) were used as the internal reference genes (Yang et al., 2015). The Geometric mean of two reference genes was considered as a normalizer (Gharbi et al., 2015). The specific primer of each gene for RT-qPCR were listed in Supplementary Table S2.
RNA Interference
Double-stranded RNA (dsRNA) was synthesized using a T7 high yield transcription kit (Invitrogen, USA) according to the manufacturer’s instructions. The primers for dsRNA synthesis were listed in Supplementary Table S2. The rearing device used for this study were developed based on the methods of Mittler with some modifications (Mittler and Dadd, 1964). The dsRNA of target gene was added to the artificial diet (0.5 M sterile sucrose solution) at a concentration of 150 ng/μl. The artificial diet containing dsRNA-EGFP was employed as the control. The aphids were transferred onto the artificial diet for rearing. The experiments included three biological replications, and each replication included 30 aphids. After 48 h, the efficiency of the dsRNA knockdown of the three P450s expression was analyzed through RT-qPCR. To assess the sensitivity of the A. craccivora to imidacloprid, the brachypterous adults were treated with imidacloprid (LC50 dose) after knockdown of P450s in 48 h. The mortality of A. craccivora was checked after 48 h.
Statistical Analysis
SPSS 20.0 (IBM Corporation, USA) was utilized to perform the statistical analysis. One-way analysis of variance (ANOVA) with the least significant difference (LSD) test was employed in comparing the gene expression levels. The value of p < 0.05 and value of p < 0.01 were regarded as significant and very significant differences, respectively.
Results
Identification of P450 and GST Genes in A. craccivora Through Transcriptome
Approximately 54,778,308 raw reads were generated from Illumina sequencing of the A. craccivora cDNA library (Supplementary Table S3). After removing the low-quality reads, 52,576,182 valid reads were obtained. The clean reads were finally assembled into 39,048 transcripts with a mean length of 689 bp and an N50 length of 1852 bp (Supplementary Table S3). These unigenes were annotated against the NR, COG, KEGG and Swiss-Prot databases with value of E < 10−5. The highest match percentage was to A. gossypii (55.58%), followed by Rhopalosiphum maidis (11.98%), Melanaphis sacchari (5.1%), Acyrthosiphon pisum (4.34%), and M. persicae (3.06%; Supplementary Figure S1). To further elucidate the functions of these unigenes, GO assignments were utilized to classify unigenes into different functional groups according to GO category. Based on sequence homology, 10,383 unigenes were annotated and classified into one or more functional groups corresponding to the three biological processes. Ultimately, 14,860 annotation hits were aligned to biological process, 9,317 to cellular components, and 18,014 to molecular functions (Supplementary Figure S2). The transcriptome data has been deposited in the NCBI database (GEO accession: GSE161346).
P450 and GST sequences were identified by a BLAST search against the transcriptomic database of A. craccivora. The complete coding region was confirmed by ORF finder and protein BLAST results (Supplementary Table S4). These identified genes were classified into different subfamilies via alignment between A. craccivora and other insect species.
In this study, 38 P450 genes were identified in A. craccivora through the transcriptomic database, including five in the CYP2 Clan, 18 in the CYP3 Clan, nine in the CYP4 Clan, and six in the Mito Clan (Figure 1). In addition, 11 P450 genes belonged to the subfamily of CYP6CY in the CYP6 family, and five P450 genes belonged to the subfamily of CYP4CJ in the CYP4 family. Ten GST genes were identified in the transcriptome of A. craccivora, including two Delta class GST, one Omega class GST, three Sigma class GST, two Theta class GST, and two Microsomal class GST (Figure 2).
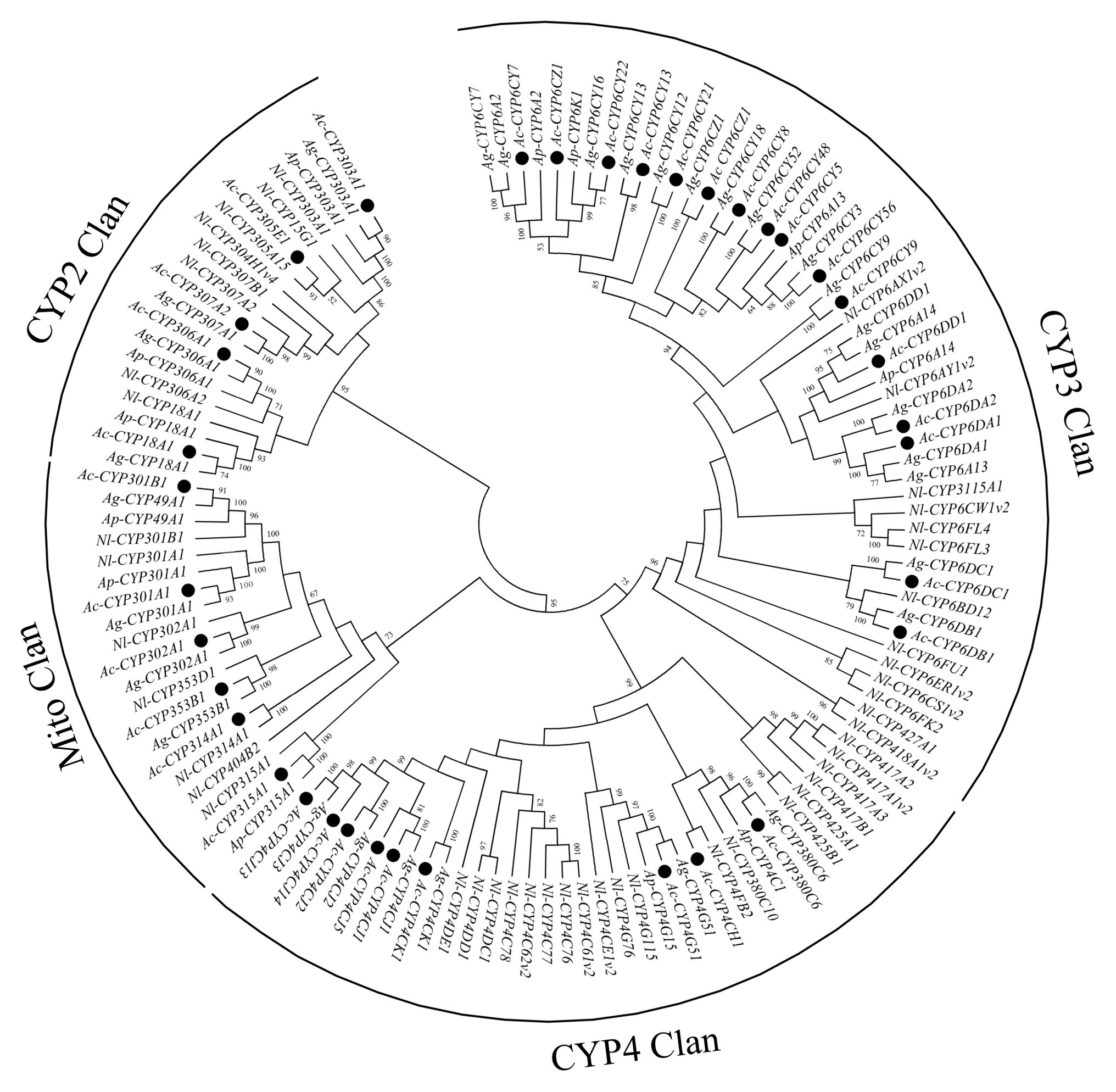
Figure 1. Phylogenetic analysis of P450 genes. The numbers above the branches indicate the support for the phylogenies, and only values above 50% are shown. Blackened circles indicate the Aphis craccivora position. Ac, A. craccivora; Ag, Aphis gossypii; Ap, Acyrthosiphon pisum; Nl, Nilaparvata lugens.
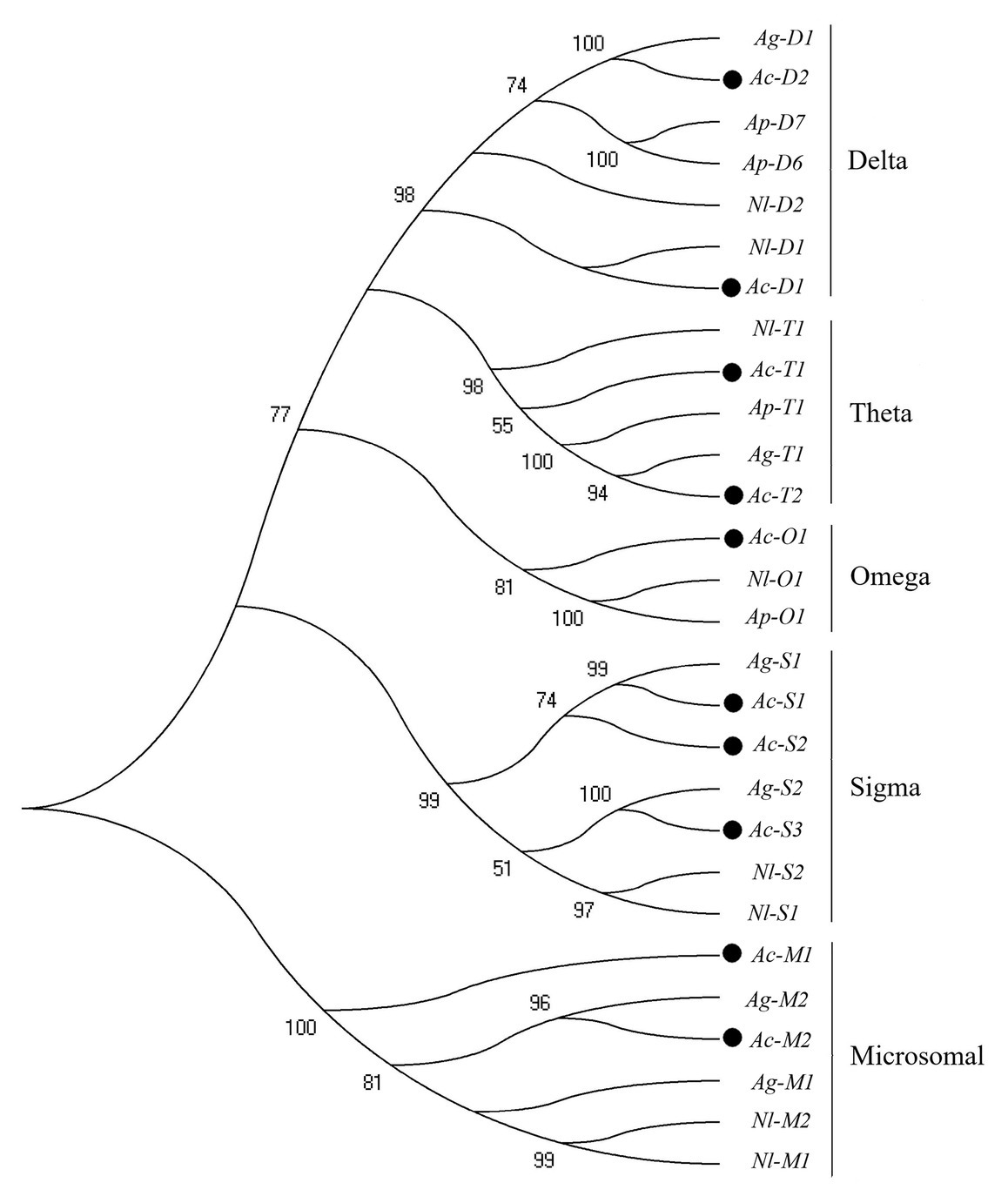
Figure 2. Phylogenetic analysis of GST genes. The numbers above the branches indicate the support for the phylogenies, and only values above 50% are shown. Blackened circles indicate the A. craccivora position. Ac, A. craccivora; Ag, Aphis gossypii; Ap, Acyrthosiphon pisum; Nl, Nilaparvata lugens.
To verify the accuracy of the transcriptome sequence, 50 genes were amplified. The electrophoretic diagram was shown in Supplementary Figure S3, and the sequence information have been submitted to the NCBI database (Supplementary Table S2).
Expression Induction of P450 and GST Genes in the SUS Strain of A. craccivora by Imidacloprid
To obtain potential P450 and GST genes that might be involved in the tolerance of imidacloprid, the expression induction of all P450 and GST genes identified in the transcriptome of A. craccivora, was detected in the SUS strain after imidacloprid treatment with LC15, LC50, and LC85 doses. The amplification efficiency of each pair of primers was presented in Supplementary Table S6.
With the LC15 dose of imidacloprid treatment, 22 genes were significantly up- or down-regulated in the SUS strain compared with the control (Figure 3), including 17 P450 genes (13 up-regulated, and four down-regulated) and five GST genes (three up-regulated, and two down-regulated). Among the P450 genes, one in the CYP2 Clan (Figure 3A), eight in the CYP3 Clan (Figure 3C), and four in the CYP4 Clan (Figure 3B) were up-regulated after imidacloprid treatment. In these P450 genes, the expression level of nine genes changed more than 2.0-fold, among which CYP6CY52, CYP4CJ1, and CYP380C6 were up-regulated more than 4.0-fold. Among the GST genes, two sigma class GST genes and one omega class GST gene were up-regulated after imidacloprid treatment, and sigma two was up-regulated more than two-fold (Figure 3D).
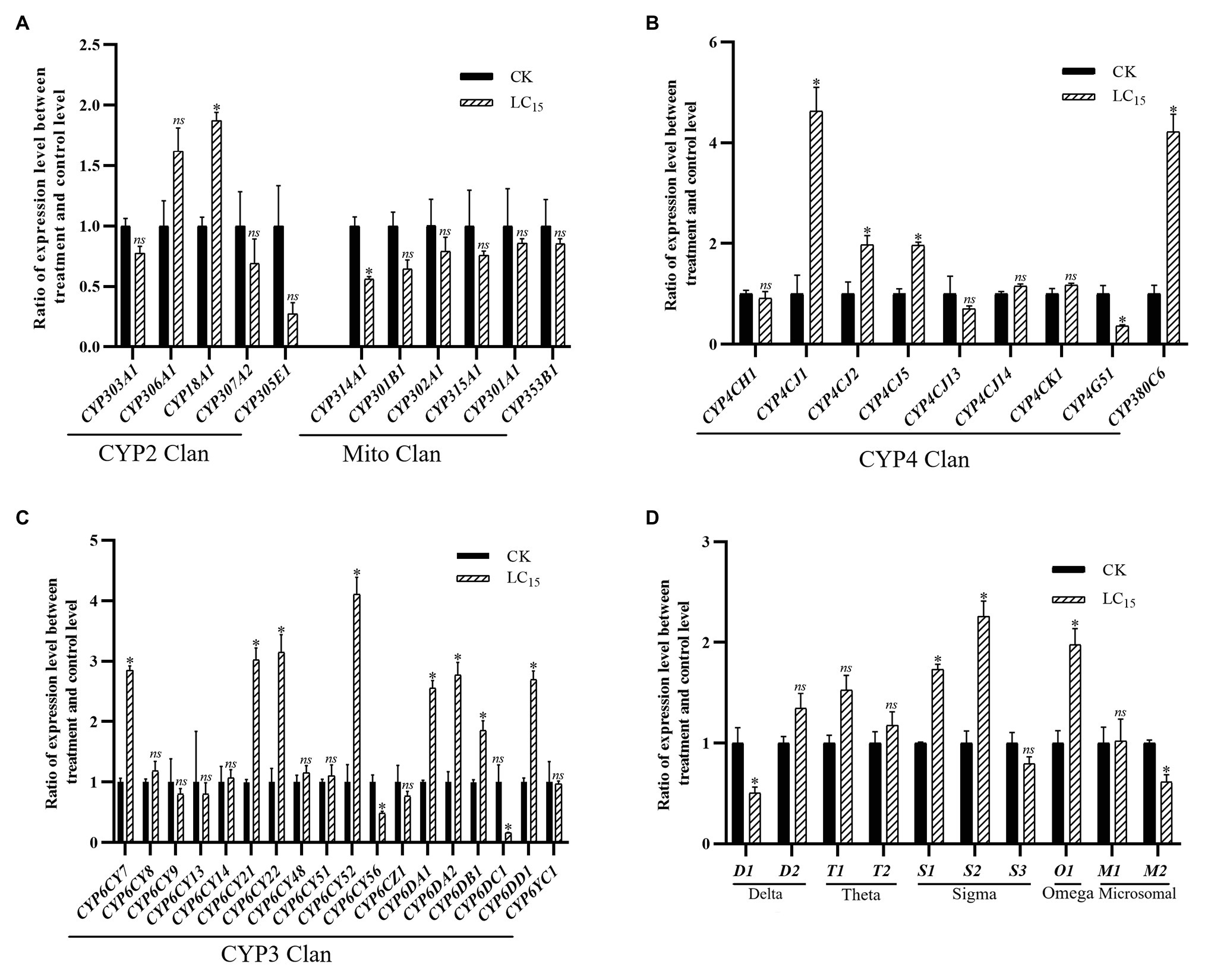
Figure 3. Fold changes in the expression of 38 P450 genes and 10 GST genes in the A. craccivora susceptible strain after the LC15 dose of imidacloprid treatment. The significant differences were marked by asterisks. *Significantly at 0.05 level. The ns represents no significant difference. (A) Fold changes in the expression of P450 genes from CYP2 and Mito Clan. (B) Fold changes in the expression of P450 genes from CYP4 Clan. (C) Fold changes in the expression of P450 genes from CYP3 Clan. (D) Fold changes in the expression of GST genes.
With the LC50 dose of imidacloprid treatment, 15 genes were significant up- or down-regulated in the SUS strain compared with the control (Figure 4), including 11 P450 genes (nine up-regulated and two down-regulated) and four GST genes (two up-regulated and two down-regulated). Among the P450 genes, one in the CYP2 Clan (Figure 4A), five in the CYP3 Clan (Figure 4C), and three in the CYP4 Clan (Figure 4B) were up-regulated after imidacloprid treatment. In these P450 genes, the expression of six genes changed more than 2.0-fold, among which CYP6CY13, CYP6CY21, CYP6DA1, CYP4CJ1, CYP4CJ2, and CYP380C6 were up-regulated more than 4.0-fold. Among the GST genes, one sigma class GST gene and one omega class GST gene were up-regulated after imidacloprid treatment (Figure 4D), and the two were up-regulated more than 2-fold.
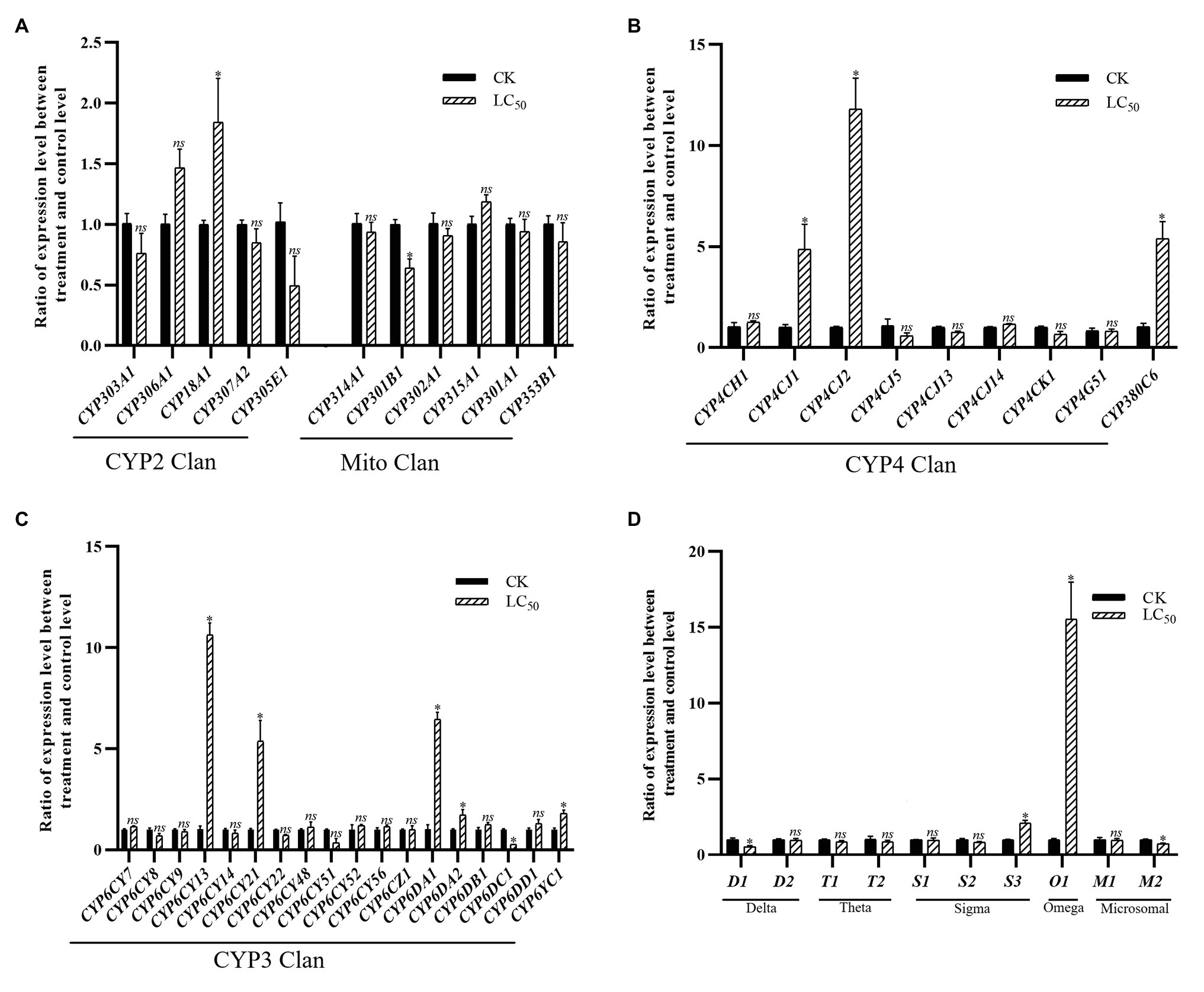
Figure 4. Fold changes in the expression of 38 P450 genes and 10 GST genes in the A. craccivora susceptible strain after the LC50 dose of imidacloprid treatment. The significant differences were marked by asterisks. *Significantly at 0.05 level. The ns represents no significant difference. (A) Fold changes in the expression of P450 genes from CYP2 and Mito Clan. (B) Fold changes in the expression of P450 genes from CYP4 Clan. (C) Fold changes in the expression of P450 genes from CYP3 Clan. (D) Fold changes in the expression of GST genes.
With the LC85 dose of imidacloprid treatment, 15 genes were significant up-or down-regulated in the SUS strain compared with the control (Figure 5), including 12 P450 genes (10 up-regulated, and two down-regulated) and three GST genes (one up-regulated, and two down-regulated). Among the P450 genes, one in the CYP2 Clan (Figure 5A), five in the CYP3 Clan (Figure 5C), and four in the CYP4 Clan (Figure 5B) were up-regulated after imidacloprid treatment. In these P450 genes, the expression of nine genes changed more than 2.0-fold, among which CYP6CY21, CYP4CJ1, CYP4CJ5, and CYP380C6 were up-regulated more than 4.0-fold. Among the GST genes, sigma3 was up-regulated after imidacloprid treatment (Figure 5D), and the fold change more than 2-fold.
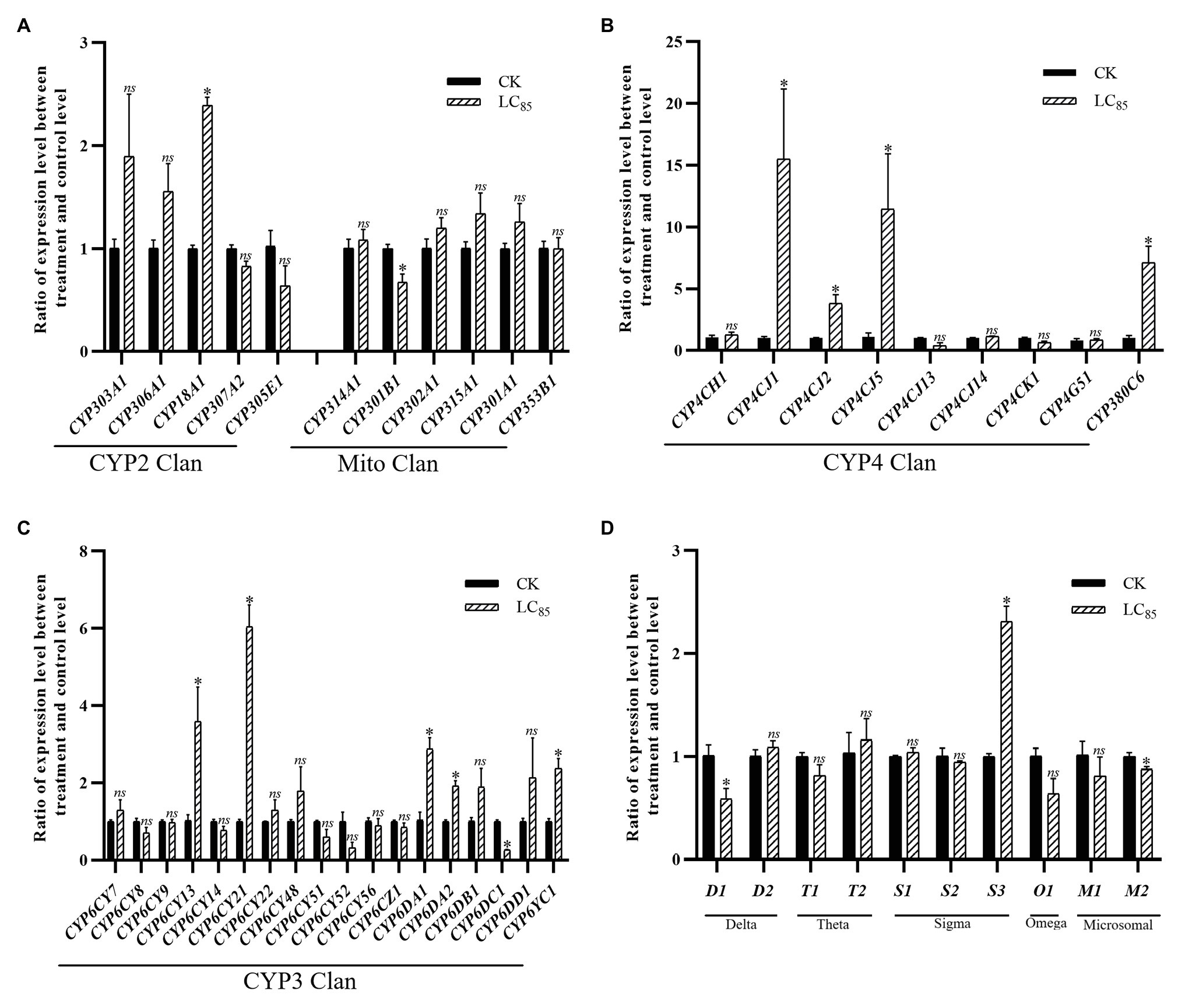
Figure 5. Fold changes in the expression of 38 P450 genes and 10 GST genes in the A. craccivora susceptible strain after the LC85 dose of imidacloprid treatment. The significant differences were marked by asterisks. *Significantly at 0.05 level. The ns represents no significant difference. (A) Fold changes in the expression of P450 genes from CYP2 and Mito Clan. (B) Fold changes in the expression of P450 genes from CYP4 Clan. (C) Fold changes in the expression of P450 genes from CYP3 Clan. (D) Fold changes in the expression of GST genes.
In these three treatments, CYP3 and CYP4 Clans had a higher proportion of genes induced by imidacloprid (Figures 3–5). Among these genes, seven P450 genes (CYP18A1, CYP6CY21, CYP6DA1, CYP6DA2, CYP4CJ1, CYP4CJ2, and CYP380C6) were up-regulated in three doses of imidacloprid treatment.
Expressive Abundance of P450 and GST Genes in the LQ and JY Populations of A. craccivora
To explore more potential stably over-expressing P450 and GST genes involved in the imidacloprid tolerance, the expression levels of all P450 and GST genes were detected in the A. craccivora populations of LQ and JY.
Compared to the SUS strain, 10 genes were significantly up- or down regulated in the JY population (Figure 6), including seven P450 genes (six up-regulated and one down-regulated) and three GST genes (one up-regulated and two down-regulated). Among the P450 genes, one (CYP306A1) in the CYP2 Clan (Figure 6A), three (CYP6DA2, CYP6CY52, and CYP6CY22) in the CYP3 Clan (Figure 6C), and two (CYP4CJ1, CYP380C6) in the CYP4 Clan (Figure 6B) were up-regulated in the JY population. Except for CYP380C6, the fold change of the other five P450 genes were exceeded 2.0-fold. Among the GST genes, sigma3 was up-regulated in the JY population (Figure 6D), and the fold change was more than 2-fold.
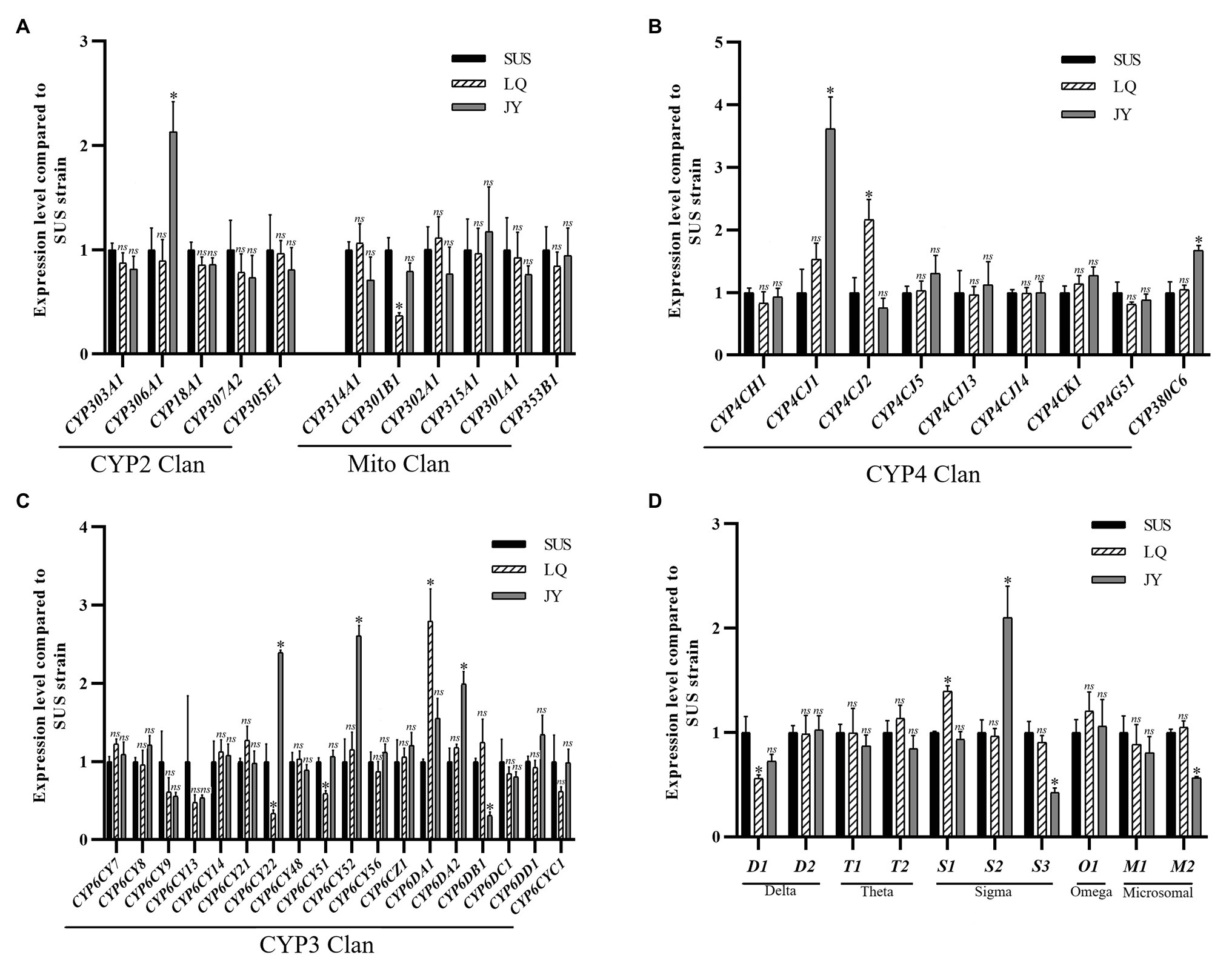
Figure 6. Fold changes of LQ and JY vs. SUS in the expression levels of 10 GST genes the in A. craccivora. The significant differences were marked by asterisks. *Significantly at 0.05 level. The ns represents no significant difference. (A) Fold changes in the expression of P450 genes from CYP2 and Mito Clan. (B) Fold changes in the expression of P450 genes from CYP4 Clan. (C) Fold changes in the expression of P450 genes from CYP3 Clan. (D) Fold changes in the expression of GST genes.
Compared to the SUS strain, seven genes were significantly up- or down-regulated in the LQ population (Figure 6), including five P450 genes (two up-regulated and three down-regulated) and two GST genes (one up-regulated and one down-regulated). Among the P450 genes, one (CYP6DA1) in the CYP3 Clan (Figure 6C), and one (CYP4CJ2) in the CYP4 Clan (Figure 6B) were up-regulated in the LQ population. And the fold change of the two P450 genes were exceeded 2.0-fold. Among the GST genes, sigma1 was up-regulated in the JY population (Figure 6D).
The expression levels of CYP6DA2, CYP4CJ1, and CYP380C6 were up-regulated after imidacloprid treatment at three doses in the SUS strain and up-regulated in the field population of JY. The results indicated that CYP6DA2, CYP4CJ1, and CYP380C6 might play important roles in the imidacloprid tolerance in A. craccivora.
RNA Interference and Effects on Insecticide Sensitivity
The functions of CYP6DA2, CYP4CJ1, and CYP380C6 in imidacloprid tolerance were analyzed by RNAi. The relative expression levels of CYP6DA2, CYP4CJ1, and CYP380C6 were significantly decreased after dsRNA feeding in 48 h (Figure 7A). Furthermore, the mortality was increased, from 62.0% in the control to 79.3, 70.0, and 81.3% in the dsRNA-CYP6DA2, dsRNA-CYP4CJ1, and dsRNA-CYP380C6, with LC50 dose of imidacloprid treatment (Figure 7B). In addition, the mortality was significantly increased in dsRNA-CYP6DA2 and dsRNA-CYP380C6, with LC50 dose of imidacloprid treatment (Figure 7B).
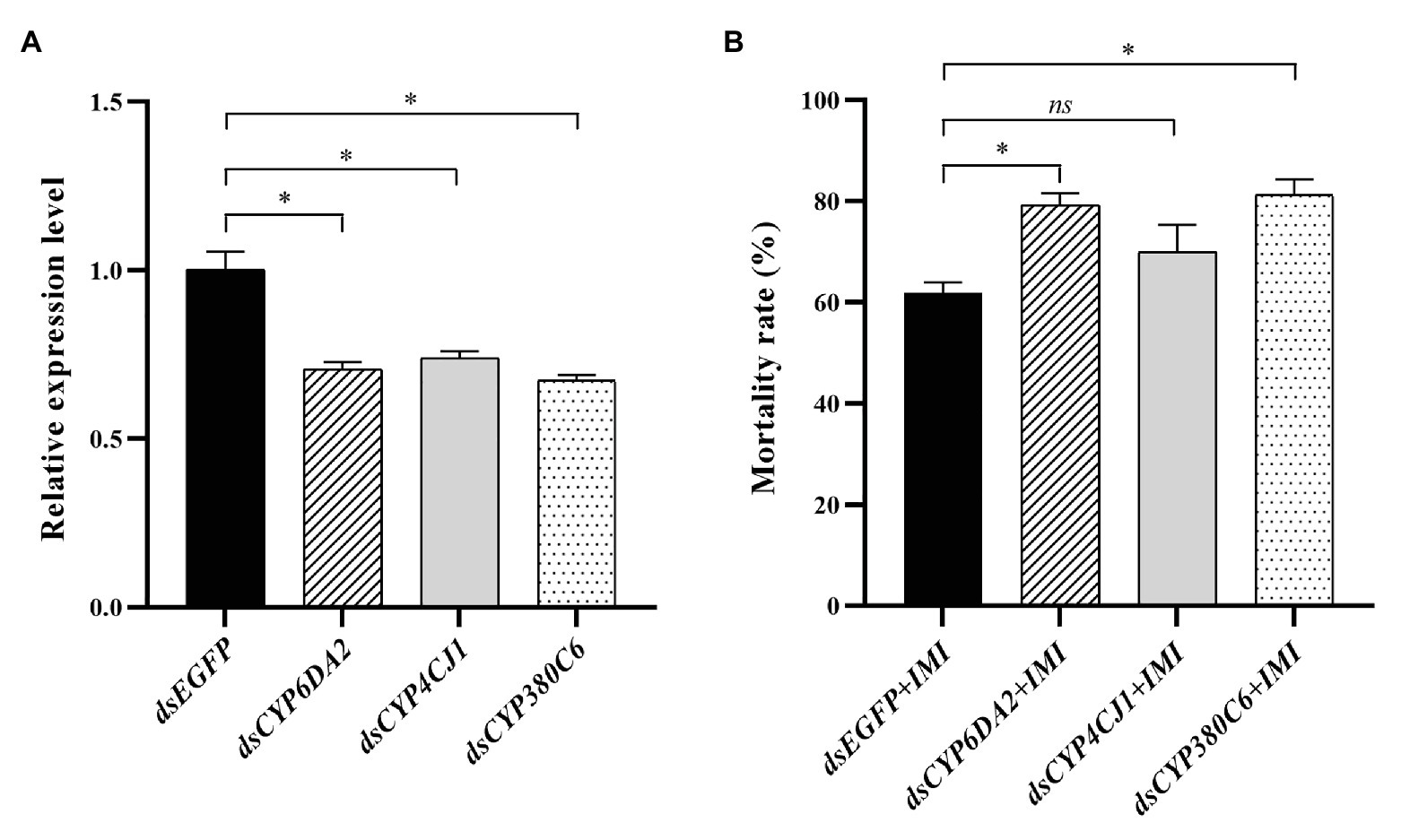
Figure 7. The dsRNA-mediated suppression of P450 genes and effects on imidacloprid toxicity in A. craccivora SUS strain. (A) The dsRNA-mediated suppression the expression of three P450 genes in A. craccivora SUS strain fed on the artificial diet with dsRNA. (B) The mortality of A. craccivora strain with LC50 dose of imidacloprid treatment after knockdown of CYP6DA2, CYP4CJ1, and CYP380C6. The significant differences were marked by asterisks. *Significantly at 0.05 level. The ns represents no significant difference. IMI, imidacloprid.
Discussion
Imidacloprid has been extensively employed in the management of piercing-sucking pests, especially in aphids. Different levels of imidacloprid resistance have been developed in various aphids, such as A. gossypii, M. persicae, and Aphis glycines (Philippou et al., 2010; Yang, 2012; Cui et al., 2016). At present, imidacloprid is still used at lower frequencies in China, which may explain why the resistance of pests to imidacloprid has still increased in recent years.
In insects, metabolic resistance to insecticides is mainly due to the long-term selective pressure of exogenous compounds along with the gene amplification or overexpression of detoxification enzymes (Li et al., 2007). The detoxification enzymes, primarily P450s, GSTs, and CarEs, can metabolize insecticides to low-toxic or non-toxic substances (Enayati et al., 2005; Yan et al., 2009; Feyereisen, 2011). Transcriptome sequencing has been shown to be an efficient means of obtaining a large number of gene sequences in insect species, such as A. gossypii (Li et al., 2013b), and Locusta migratoria (Zhang et al., 2018). In the A. craccivora transcriptome, 38 P450 genes were identified, and most of the genes belonged to the CYP3 and CYP4 Clans, which is in keeping line with results the finding observed in other insect species (Zhang et al., 2010, 2016b; Yang and Liu, 2011). In addition, although a large number of P450 genes have been obtained in different insect species, the number of P450s in different insects varies considerably, for instance, there are 100 and 80 P450 genes in C. quinquefasciatus (Yang and Liu, 2011), 64 P450 genes in Acyrthosiphon pisum (Zhang et al., 2010), and only 36 P450 genes in Pediculus humans (Lee et al., 2010). In this study, only 38 P450 genes were obtained from A. craccivora transcriptome. Similarly, the number of P450s in A. gossypii obtaining from transcriptome sequencing, was only 35 (Pan et al., 2018). However, in another A. gossypii study, 54 P450 genes, including the sequences (<300 bp), were sequenced in transcriptome, and only 34 P450 genes in the phylogenetic analysis (Li et al., 2017). In addition, an ortholog of the juvenile hormone epoxidase, CYP15A1, was not identified in A. craccivora. Therefore, the differences of P450 gene numbers in insects may be attributable to the depth of sequencing or the insect adaptation to the environment. In insects, the CYP3 and CYP4 Clans are highly related to the development of insecticide resistance. For instance, in Helicoverpa armigera and Cydia pomonella, CYP3 and CYP4 Clans played important roles in pyrethroid insecticides, especially CYP6 and CYP9 families belonged to CYP3 Clan (Brun-Barale et al., 2010; Yang et al., 2013; Wang et al., 2019). However, in A. craccivora, relatively fewer P450s in CYP3 Clan lesser than in other species, were induced by insecticide. This might be related with the lack of P450 genes, or the difference of A. craccivora’s feeding and adaptation to the environment. However, the number of detoxification genes was few, the A. mellifera and P. humans can resist much insecticides (Claudianos et al., 2006; Lee et al., 2010).
Besides, 10 GST genes were identified in the A. craccivora transcriptome, including two Delta genes which play important roles in xenobiotic detoxification. In addition, the number of GST genes was relatively low in A. craccivora, compared with Acyrthosiphon pisum (Ramsey et al., 2010), Drosophila melanogaster (Adams et al., 2000), and Tribolium castaneum (Kim et al., 2010), which was 32, 40, and 41, respectively. However, only 13 protein sequences were obtained in NCBI through BLAST A. gossypii GST. The Delta and Epsilon classes of GSTs are insects specific and are thought to contribute to the as adaptation of insects to environmental variation (Ranson et al., 2000). In D. melanogaster, C. pomonella, and A. pisum, the number of The Delta and Epsilon classes were 11 and 14 (Adams et al., 2000), 4 and 5 (Ramsey et al., 2010), 10 and 0 (Hu et al., 2020). However, the number of the Delta and Epsilon classes were one and zero in A. craccivora. And the number of these two classes GST number were one and zero, four and zero, respectively in A. mellifera and P. humans (Claudianos et al., 2006; Lee et al., 2010). On the contrary, the number of sigma class in both fly and mosquito is only one. However, the number in A. craccivora is three. The Papilio multicaudatus has multiple sigma GSTs and are postulated to play a catalytic role, in the metabolism of plant allelochemicals (Claudianos et al., 2006). This indicated that the functional differentiation of GSTs in insect evolution and the adaptation to the environment (Hu et al., 2020).
At present, the study of insecticide resistance has changed from single gene analysis to the whole genome analysis, from detecting the expression change of a single gene to detecting the expression of multiple genes, and revealing that insecticide resistance is mediated by multiple genes (Zhu et al., 2008a,b; Feyereisen, 2011; Zhang et al., 2016a). To obtain potential P450 and GST genes that might be involved in imidacloprid resistance, all genes were detected in the SUS strain after imidacloprid treatment at three doses and detected in the field populations of JY and LQ. With imidacloprid treatment at doses of LC15, LC50, and LC85, 22 genes, 15 genes, and 15 genes were significantly regulated in the SUS strain compared with control, respectively. The expression levels of eight P450 genes were significantly changed under imidacloprid treatment at three doses; one (CYP6DC1) of them was down-regulated, while seven of them were up-regulated (CYP18A1, CYP6CY21, CYP6DA1, CYP6DA2, CYP4CJ1, CYP4CJ2, and CYP380C6). Six-sevenths of up-regulated P450 genes belonged to the CYP3 and CYP4 Clans, which play important roles in insecticide resistance. The over-expression of two P450 genes, CYP6G1 and CYP12D1, was found to be associated with DDT resistance in D. melanogaster (Festucci-Buselli et al., 2005). In M. domestica, CYP6A1 and CYP6D1 have been related to pyrethroid resistance (Carino et al., 1994; Liu and Scott, 1996). The JY populations showed low level resistance to imidacloprid, and five P450 genes (CYP6CY22, CYP6CY52, CYP6DA2, CYP4CJ1, and CYP380C6) which belong to CYP3 and CYP4 Clans were overexpressed in the JY population compared with the SUS strain. Meanwhile, three (CYP6DA2, CYP4CJ1, and CYP380C6) of these genes were also up-regulated after imidacloprid treatment with three doses in the SUS strain. Genes in CYP6CY subfamily participate in neonicotinoid insecticides. In this study, CYP6CY22 and CYP6CY52 were up-regulated after imidacloprid treatment at three doses. In the A. gossypii field population, CYP6CY22 and CYP6CY13 were significantly up-regulated compared with the insecticide-susceptible strain. CYP6CY22 and CYP6CY13 can metabolize seven tested neonicotinoid insecticides (Hirata et al., 2017). Therefore, it is deduced that the two P450 genes in the CYP6CY subfamily may play important role in imidacloprid resistance. In A. gossypii, the expression level of CYP4CJ1 was significantly induced by gossypol and tannic acid, and knockdown of CYP4CJ1 could increase the sensitivity of A. gossypii to these two plant allelochemicals (Ma et al., 2019). Meanwhile, the expression levels of CYP6DA2 in A. gossypii was significantly induced by gossypol (Peng et al., 2016). And in resistant adult A. gossypii, CYP380C6 was up-regulated under spirotetramat stress (Pan et al., 2018). Besides, the expression levels of several GST genes were significantly up-regulated in the SUS strain after imidacloprid treatment at three doses. Sigma1 and Sigma2 were up-regulated after LC15 imidacloprid treatment. Meanwhile, Sigma3 was up-regulated after LC50 and LC85 imidacloprid treatment. Sigma class GSTs show low activity with typical GST substrates, but have high affinity for the lipid peroxidation product, 4-hydroxynonenal, and are localized in metabolically active tissues in flies (Claudianos et al., 2010). However, it is observed that these GSTs also play a role in xenobiotic detoxification and insecticide resistance (Gawande et al., 2014). Omega1 was up-regulated after LC15 and LC50 imidacloprid treatment, and the fold change in LC50 was 15.5-fold. In Leptinotarsa decemlineata, the Omega5 was significantly overexpressed after exposure to cyhalothrin, fipronil and endosulfan (Han et al., 2016).
The in vivo suppression of P450 gene expression by RNAi, resulting in increased sensitivity to insecticide, has been widely used to research the contribution of P450s to insecticide resistance. In Spodoptera frugiperda, the second-instar larvae became more sensitive to chlorantraniliprole with a higher mortality rate than the control, after silencing CYP321A8, CYP321A9, and CYP321B1 (Zhang et al., 2020). In Blattella germanica, RNAi-mediated knockdown of CYP4G19 significantly decreased its expression, which resulted in a non-uniform array of the lipid layer, enhanced cuticle permeability, and compromised insecticide tolerance (Chen et al., 2020). In Rhynchophorus ferrugineus, silencing of CYP345J1 and CYP6NR1 significantly decreased the survival rate treated with imidacloprid, indicating that overexpression of these two P450s may play an important role in developing tolerance to imidacloprid in a date palm field (Antony et al., 2019). In this study, the suppression of CYP6DA2 and CYP380C6 could increase the sensitivity to imidacloprid. These results indicated that, CYP6DA2 and CYP380C6 may contribute to resisting imidacloprid in A. craccivora. Meanwhile, the expression levels of CYP6DA2 in A. gossypii was significantly induced by gossypol, and knockdown of CYP6DA2 could significantly increase the toxicity of gossypol (Peng et al., 2016). Knockdown of CYP380C6 significantly increased the sensitivity to spirotetramat in A. gossypii resistance strain (Pan et al., 2018). In addition, CYP380C6 and CYP380C9 in M. persicae play a crucial role in mitigating indole glucosinolate-mediated plant defense, and such effect is transgenerational (Jib et al., 2020). Therefore, it is concluded that the CYP6DA2 and CYP380C6 in A. craccivora may play important roles in insecticide tolerance and defense of plant secondary substances.
In conclusion, these results may establish a foundation for further research exploring the metabolic mechanism underlying imidacloprid resistance in A. craccivora. Further research may attempt to select a resistant strain in the laboratory, and perform functional expression of important genes to study the insecticide resistance in A. craccivora.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Y-XY, J-HZ, and MZ conceived the study. Y-XY, R-HL, ZL, A-YW, CX, and A-LD conducted the experiments. Y-XY and R-HL analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by Natural Science Foundation of Shandong Province, China (ZR2019BC055); Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2018E06); and National Key Research and Development Program of China (2017YFD0201906).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank David Nelson (Department of Molecular Science, University of Tennessee, Memphis, TN, United States) and the P450 nomenclature committee for naming P450s identified in this study. We also thank Zhibing Xu (East China University of Science and Technology, Shanghai, China) for providing the SUS strain of A. craccivora.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.624287/full#supplementary-material
Supplementary Figure S1 | Nr homologous species distribution in A. craccivora.
Supplementary Figure S2 | Gene Ontology (GO) annotation and classification of the A. craccivora.
Supplementary Figure S3 | Electrophoretic images of 50 genes. M: marker, the marker from top to bottom are 2,000 bp, 1,500 bp, 1,000 bp, 750 bp, 250 bp, and 100 pp. 1, CYP301B1; 2, CYP315A1; 3, CYP353B1; 4, CYP6CY22; 5, CYP6YC1; 6, CYP18A1; 7, CYP6DB1; 8, CYP306A1; 9, CYP6CY22; 10, CYP6CY7; 11, CYP4CJ2; 12, CYP6CY52; 13, CYP380C6; 14, CYP4CJ1; 15, GSTM1; 16, GSTD1; 17, CYP6CY21; 18, CYP6CY8; 19, CYP6CY5; 20, CYP6CY48; 21, CYP4CJ14; 22, CYP4CJ5; 23, CYP6DA1; 24, CYP6DA2; 25, CYP6CY14; 26, CYP6CZ1; 27, CYP314A1; 28, CYP6DC1; 29, CYP301A1; 30, CYP305E1; 31, CYP4G51; 32, CYP6CY13; 33, CYP307A2; 34, CYP302A1; 35, CYP6DD1; 36, CYP303A1; 37, CYP4CH1; 38, CYP4CK1; 39, CYP4CJ13; 40, CYP6CY56; 41, RPL11; 42, RPS8; 43, GSTM2; 44, CYPGSTD2; 45, GSTS1; 46, GSTS3; 47, GSTS2; 48, GSTT2; 49, GSTO1; 50, GSTT1.
Footnotes
1. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
2. http://www.ncbi.nlm.nih.gov/
3. http://www.geneontology.org
References
Abd-Ella, A. A. (2014). Toxicity and persistence of selected neonicotinoid insecticides on cowpea aphid, Aphis craccivora Koch (Homoptera: Aphididae). Arch. Phytopathol. Plant Protect. 47, 366–376. doi: 10.1080/03235408.2013.809919
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Andrei, A., Galen, D., Megan, P., Christopher, C., David, R., Matthew, M., et al. (2010). Resistance and cross-resistance to imidacloprid and thiamethoxam in the Colorado potato beetle Leptinotarsa decemlineata. Pest Manag. Sci. 63, 32–41. doi: 10.1002/ps.1305
Antony, B., Johny, J., Abdelazim, M. A., Jakse, J., Al-Saleh, M. A., and Pain, A. (2019). Global transcriptome profiling and functional analysis reveal that tissue-specific constitutive overexpression of cytochrome P450s confers tolerance to imidacloprid in palm weevils in date palm fields. Genome Biol. 20, 1–23. doi: 10.1186/s12864-019-5837-4
Bass, C., Carvalho, R. A., Oliphant, L., Puinean, A. M., Field, L. M., Nauen, R., et al. (2011). Overexpression of a cytochrome P450 monooxygenase, CYP6ER1, is associated with resistance to imidacloprid in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 20, 763–773. doi: 10.1111/j.1365-2583.2011.01105.x
Brun-Barale, A., Hema, O., Martin, T., Suraporn, S., Audant, P., Sezutsu, H., et al. (2010). Multiple P450 genes overexpressed in deltamethrin-resistant strains of Helicoverpa armigera. Pest Manag. Sci. 66, 900–909. doi: 10.1002/ps.1960
Buchfink, B., Xie, C., and Huson, H. D. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Carino, F. A., Konener, J. F., Plapp, F. W. Jr., and Feyereisen, R. (1994). Constitutive overexpression of the cytochrome P450 gene CYP6A1 in a house fly strain with metabolic resistance to insecticides. Insect Biochem. Mol. Biol. 24, 411–418. doi: 10.1016/0965-1748(94)90034-5
Che-Mendoza, A., Penilla, R. P., and Rodríguez, D. A. (2009). Insecticide resistance and glutathione S-transferases in mosquitoes: a review. Q. J. Med. 8, 1386–1397.
Chen, N., Pei, X. J., Li, S., Fan, Y. L., and Liu, T. X. (2020). Involvement of integument-rich CYP4G19 in hydrocarbon biosynthesis and cuticular penetration resistance in Blattella germanica (L). Pest Manag. Sci. 76, 215–226. doi: 10.1002/ps.5499
Choi, B. R., Lee, S. W., and Yoo, J. K. (2001). Resistance mechanisms of green peach aphid, Myzus persicae (Homoptera: Aphididae), to imidacloprid. Korean J. Appl. Entomol. 40, 265–271.
Claudianos, C., Ranson, H., Johnson, R. M., Biswas, S., Schuler, M. A., Berenbaum, M. R., et al. (2006). A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 15, 615–636. doi: 10.1111/j.1365-2583.2006.00672.x
Claudianos, C., Ranson, H., Johnson, R. M., Biswas, S., Schuler, M. A., Berenbaum, M. R., et al. (2010). A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 15, 615–636. doi: 10.1111/j.1365-2583.2006.00672.x
Cui, L., Zhang, J., Qi, H. L., Wang, Q. Q., Lu, Y. H., and Rui, C. H. (2016). Monitoring and mechanisms of imidacloprid resistance in Aphis gossypii (Hemiptera: Aphidiidae) in the main cotton production areas of China. Acta Entomol. Sin. 11, 1246–1253.
De, S. D., and Hemingway, J. (2010). Structural organization of the estα3 (1) gene in a Colombian strain of Culex quinquefasciatus differs from that in Cuba. Med. Vet. Entomol. 16, 99–105. doi: 10.1046/j.0269-283x.2002.00347.x
Enayati, A. A., Ranson, H., and Hemingway, J. (2005). Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14, 3–8. doi: 10.1111/j.1365-2583.2004.00529.x
Festucci-Buselli, R. A., Carvalho-Dias, A. S., de Oliveira-Andrade, M., Caixeta-Nunes, C., Li, H. M., Stuart, J. J., et al. (2005). Expression of Cyp6g1 and Cyp12d1 in DDT resistant and susceptible strains of Drosophila melanogaster. Insect Mol. Biol. 14, 69–77. doi: 10.1111/j.1365-2583.2005.00532.x
Feyereisen, R. (1999). Insect P450 enzyme. Annu. Rev. Entomol. 44, 507–533. doi: 10.1146/annurev.ento.44.1.507
Feyereisen, R. (2005). 4.1 - insect cytochrome P450. Compr. Molec. Insect Sci. 4, 1–77. doi: 10.1016/B0-44-451924-6/00049-1
Feyereisen, R. (2011). Insect CYP genes and P450 enzymes. Insect Mol. Biol. Biochem. 8, 236–316. doi: 10.1016/B978-0-12-384747-8.10008-X
Fouad, E. A., Abou-Yousef, H. M., Abdallah, I. S., and Kandil, M. A. (2016). Resistance monitoring and enzyme activity in three field populations of cowpea aphid (Aphis craccivora) from Egypt. Crop Prot. 81, 163–167. doi: 10.1016/j.cropro.2015.12.015
Gawande, N. D., Subashini, S., Murugan, M., and Subbarayalu, M. (2014). Molecular screening of insecticides with sigma glutathione S-transferases (GST) in cotton aphid Aphis gossypii using docking. Bioinformation 11, 679–683. doi: 10.6026/97320630010679
Gharbi, S., Shamsara, M., Khateri, S., Soroush, R. M., Ghorbanmehr, N., Tavallaie, M., et al. (2015). Identification of reliable reference genes for quantification of micrornas in serum samples of sulfur mustard-exposed veterans. Cell J. 17, 494–501.
Gong, Y. H., Yu, X. R., Shang, Q. L., Shi, X. Y., and Gao, X. W. (2014). Oral delivery mediated RNA interference of a carboxylesterase gene results in reduced resistance to organophosphorus insecticides in the cotton aphid, Aphis gossypii Glover. PLoS One 9:e102823. doi: 10.1371/journal.pone.0102823
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Han, J. B., Li, G. Q., Wan, P. J., Zhu, T. T., and Meng, Q. W. (2016). Identification of glutathione S-transferase genes in Leptinotarsa decemlineata and their expression patterns under stress of three insecticides. Pestic. Biochem. Physiol. 133, 26–34. doi: 10.1016/j.pestbp.2016.03.008
Hirata, K., Jouraku, A., Kuwazaki, S., Shimomura, H., and Iwasa, T. (2017). Studies on Aphis gossypii cytochrome P450s CYP6CY22 and CYP6CY13 using an in vitro system. J. Pest. Sci. 42, 97–104. doi: 10.1584/jpestics.D17-006
Hu, C., Wang, W., Ju, D., Chen, G. M., Tan, X. L., Mota-Sanchez, D., et al. (2020). Functional characterization of a novel λ-cyhalothrin metabolising glutathione S-transferase, CpGSTe3, from the codling moth Cydia pomonella. Pest Manag. Sci. 76, 1039–1047. doi: 10.1002/ps.5614
Ji, R., Lei, J. X., Chen, I. W., Sang, W., Yang, S., Fang, J. C., et al. (2020). Cytochrome P450s CYP380C6 and CYP380C9 in green peach aphid facilitate its adaptation to indole glucosinolate-mediated plant defense. Pest Manag. Sci. 77, 148–158. doi: 10.1002/ps.6002
Karunker, I., Benting, J., Lueke, B., Ponge, T., Nauen, R., Roditakis, E., et al. (2008). Over-expression of cytochrome P450 CYP6CM1 is associated with high resistance to imidacloprid in the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Biochem. Mol. Biol. 38, 634–644. doi: 10.1016/j.ibmb.2008.03.008
Kim, H. S., Murphy, T., Xia, J., Caragea, D., Park, Y., Beeman, R. W., et al. (2010). BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 38, D437–D442. doi: 10.1093/nar/gkp807
Lee, S. H., Kang, J. S., Min, J. S., Yoon, K. S., Strycharz, J. P., Johnson, R., et al. (2010). Decreased detoxification genes and genome size make the human body louse an efficient model to study xenobiotic metabolism. Insect Mol. Biol. 19, 599–615. doi: 10.1111/j.1365-2583.2010.01024.x
Li, X., Ju, Q., Zhao, Z. Q., Xie, H. X., Wang, J. N., and Qu, M. J. (2013a). Control effects and safety assessment of four insecticides on Aphis craccivora. Shandong Agr. Sci. 4, 93–95.
Li, F., Ma, K. S., Liang, P. Z., Chen, X. W., Liu, Y., and Gao, X. W. (2017). Transcriptional responses of detoxification genes to four plant allelochemicals in Aphis gossypii. J. Econ. Entomol. 110, 624–631. doi: 10.1093/jee/tow322
Li, X., Schuler, M. A., and Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253. doi: 10.1146/annurev.ento.51.110104.151104
Li, Z.-Q., Zhang, S., Luo, J.-Y., Wang, C.-Y., Lv, L.-M., Dong, S.-L., et al. (2013b). Ecological adaption analysis of the cotton aphid (Aphis gossypii) in different phenotypes by transcriptome comparison. PLoS One 8:e83180. doi: 10.1371/journal.pone.0083180
Liu, N. N., and Scott, J. G. (1996). Genetic analysis of factors controlling high-level expression of cytochrome P450, CYP6D1, cytochrome b5, P450 reductase, and monooxygenase activities in LPR house flies, Musca domestica. Biochem. Genet. 34, 133–148. doi: 10.1007/bf00553609
Liu, N. N., and Scott, J. G. (1998). Increased transcription of CYP6D1 causes cytochrome P450-mediated insecticide resistance in house fly. Insect Biochem. Mol. Biol. 28, 531–535. doi: 10.1016/S0965-1748(98)00039-3
Ma, K. S., Li, F., Tang, Q. L., Liang, P. Z., Liu, Y., Zhang, B. Z., et al. (2019). CYP4CJ1-mediated gossypol and tannic acid tolerance in Aphis gossypii Glover. Chemosphere 219, 961–970. doi: 10.1016/j.chemosphere.2018.12.025
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mittler, T. E., and Dadd, R. H. (1964). An improved method for feeding aphids on artificial diets. Soc. Am. 57, 139–140. doi: 10.1093/aesa/57.1.139a
Nauen, R., and Denholm, I. (2010). Resistance of insect pests to neonicotinoid insecticides: current status and future prospects. Arch. Insect Biochem. Physiol. 8, 200–215. doi: 10.1002/arch.20043
Nauen, R., Vontas, J., Kaussmann, M., and Wolfel, K. (2013). Pymetrozine is hydroxylated by CYP6CM1, a cytochrome P450 conferring neonicotinoid resistance in Bemisia tabaci. Pest Manag. Sci. 69, 457–461. doi: 10.1002/ps.3460
Pan, Y. O., Chai, P., Zheng, C., Xu, H. F., Wu, Y. Q., Gao, X. W., et al. (2018). Contribution of cytochrome P450 monooxygenase CYP380C6 to spirotetramat resistance in Aphis gossypii Glover. Pestic. Biochem. Physiol. 148, 182–189. doi: 10.1016/j.pestbp.2018.04.015
Peng, T., Pan, Y., Gao, X., Xi, J., and Shang, Q. (2016). Cytochrome P450 CYP6DA2 regulated by cap “n” collar isoform C (CncC) is associated with gossypol tolerance in Aphis gossypii Glover. Insect Mol. Biol. 25, 450–459. doi: 10.1111/imb.12230
Philippou, D., Field, L., and Moores, G. (2010). Metabolic enzyme(s) confer imidacloprid resistance in a clone of Myzus persicae (Sulzer) (Hemiptera: Aphididae) from Greece. Pest Manag. Sci. 66, 390–395. doi: 10.1002/ps.1888
Ramsey, J. S., Rider, D. S., Walsh, T. K., De Vos, M., Gordon, K. H. J., Ponnala, S. L., et al. (2010). Comparative analysis of detoxification enzymes in Acyrthosiphon pisum and Myzus persicae. Insect Mol. Biol. 19, 155–164. doi: 10.1111/j.1365-2583.2009.00973.x
Ranson, H., Jensen, B., Wang, X., Prapanthadara, L., and Collins, F. H. (2000). Genetic mapping of two loci affecting DDT resistance in the malaria vector Anopheles gambiae. Insect Mol. Biol. 9, 499–507. doi: 10.1046/j.1365-2583.2000.00214.x
Scott, J., and Wen, Z. (2001). Cytochromes P450 of insects: the tip of the iceberg. Pest Manag. Sci. 57, 958–967. doi: 10.1002/ps.354
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Terriere, L. C. (1984). Induction of detoxication enzymes in insects. Annu. Rev. Entomol. 29, 71–88. doi: 10.1146/annurev.en.29.010184.000443
Vontas, J. G., Small, G. J., and Hemingway, J. (2001). Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem. J. 357, 65–72. doi: 10.1042/0264-6021:3570065
Wang, K., Zhang, M., Huang, Y. Y., Yang, Z. L., Su, S., and Chen, M. H. (2018). Characterisation of imidacloprid resistance in the bird cherry-oat aphid, Rhopalosiphum padi, a serious pest on wheat crops. Pest Manag. Sci. 74, 1457–1465. doi: 10.1002/ps.4834
Wang, Z. G., Jiang, S. S., Mota-Sanchez, D., Wang, W., Li, X. R., Gao, Y. L., et al. (2019). Cytochrome P450-mediated λ-cyhalothrin-resistance in a field strain of Helicoverpa armigera from Northeast China. J. Agric. Food Chem. 67, 3546–3553. doi: 10.1021/acs.jafc.8b07308
Wen, Y. C., Liu, Z. W., Bao, H. B., and Han, Z. J. (2009). Imidacloprid resistance and its mechanisms in field populations of brown planthopper, Nilaparvata lugens Stål in China. Pestic. Biochem. Physiol. 94, 36–42. doi: 10.1016/j.pestbp.2009.02.009
Yan, S. G., Cui, F., and Qiao, C. L. (2009). Structure, function and applications of carboxylesterases from insects for insecticide resistance. Protein Pept. Lett. 16, 1181–1188. doi: 10.2174/092986609789071243
Yang, C. X., Pan, H. P., Liu, Y., Zhou, X. G., and Doucet, D. (2015). Temperature and development impacts on housekeeping gene expression in cowpea aphid, Aphis craccivora (Hemiptera: Aphidiae). PLoS One 10:e0130593. doi: 10.1371/journal.pone.0130593
Yang, S. (2012). Study on monitoring and mechanisms of imidacloprid resistance in soybean aphid, Aphis clycines (Matsumura). Northeast Agricultural University.
Yang, T., and Liu, N. (2011). Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS One 6:e29418. doi: 10.1371/journal.pone.0029418
Yang, X. Q., Li, X. C., and Zhang, Y. L. (2013). Molecular cloning and expression of CYP9A61: a chlorpyrifos-ethyl and lambda-cyhalothrin-inducible cytochrome P450 cDNA from Cydia pomonella. Int. J. Mol. Sci. 14, 24211–24229. doi: 10.3390/ijms141224211
Yang, Y. X., Li, Z., Duan, A. L., Xue, C., Xu, Z. P., and Zhao, M. (2020). Preliminary study on metablic resistance mechanism of Aphis craccivora in intercropping field of cotton and peanut to imidacloprid. Cotton Sci. 32, 151–157.
Zhang, B. Z., Su, X., Zhen, C. A., Lu, L. Y., Li, Y. S., Ge, X., et al. (2020). Silencing of cytochrome P450 in Spodoptera frugiperda (Lepidoptera: Noctuidae) by RNA interference enhances susceptibility to chlorantraniliprole. J. Insect Sci. 20, 1–7. doi: 10.1093/jisesa/ieaa047
Zhang, J. H., Zhang, Y. X., Wang, Y. C., Yang, Y. X., Cang, X. Z., and Liu, Z. W. (2016a). Expression induction of P450 genes by imidacloprid in Nilaparvata lugens: a genome-scale analysis. Pestic. Biochem. Physiol. 132, 59–64. doi: 10.1016/j.pestbp.2015.10.016
Zhang, P. Y., and Zhou, X. M. (2014). Resistance of Myzus persicae populations in South China to seven pesticides. Guangdong Agr. Sci. 41, 81–77.
Zhang, W. D., Liu, L., Qu, C., and Xue, M. (2015). Toxicities and control effects of clothianidin and other insecticides to Aphis craccivora by the seed dressing. J. Peanut Sci. 44, 29–33.
Zhang, X., Kang, X., Wu, H., Silver, K., Zhang, J., Ma, E., et al. (2018). Transcriptome-wide survey, gene expression profiling and exogenous chemical-induced transcriptional responses of cytochrome P450 superfamily genes in migratory locust (Locusta migratoria). Insect Biochem. Mol. Biol. 100, 66–77. doi: 10.1016/j.ibmb.2018.06.006
Zhang, Y. H., Wang, Q., Liu, J., Zhang, P. F., and Chen, J. Q. (2010). A genome-wide analysis of P450 gene families in the pea aphid, Acyrthosiphon pisum (Hemiptera: Aphidoidea). Acta Entomol. Sin. 53, 849–856. doi: 10.1016/S1002-0721(10)60377-8
Zhang, Y. X., Yang, Y. X., Sun, H. H., and Liu, Z. W. (2016b). Metabolic imidacloprid resistance in the brown planthopper, Nilaparvata lugens, relies on multiple P450 enzymes. Insect Biochem. Mol. Biol. 79, 50–56. doi: 10.1016/j.ibmb.2016.10.009
Zhu, F., Feng, J. N., Zhang, L., and Liu, N. (2008a). Characterization of two novel cytochrome P450 genes in insecticide-resistant house-flies. Insect Mol. Biol. 17, 27–37. doi: 10.1111/j.1365-2583.2008.00777.x
Zhu, F., Li, T., Zhang, L., and Liu, N. N. (2008b). Co-up-regulation of three P450 genes in response to permethrin exposure in permethrin resistant house flies, Musca domestica. BMC Physiol. 8:18. doi: 10.1186/1472-6793-8-18
Zimmer, C. T., Bass, C., Williamson, M. S., Kaussmann, M., Wölfel, K., Gutbrod, O., et al. (2014). Molecular and functional characterization of CYP6BQ23, a cytochrome P450 conferring resistance to pyrethroids in European populations of pollen beetle, Meligethes aeneus. Insect Biochem. Mol. Biol. 45, 18–29. doi: 10.1016/j.ibmb.2013.11.008
Keywords: Aphis craccivora, P450 genes, GST genes, imidacloprid, expression induction, RNA interference
Citation: Yang Y-X, Lin R-H, Li Z, Wang A-Y, Xue C, Duan A-L, Zhao M and Zhang J-H (2021) Function Analysis of P450 and GST Genes to Imidacloprid in Aphis craccivora (Koch). Front. Physiol. 11:624287. doi: 10.3389/fphys.2020.624287
Edited by:
Ya-Nan Zhang, Huaibei Normal University, ChinaReviewed by:
Ran Wang, Beijing Academy of Agricultural and Forestry Sciences, ChinaXue-Qing Yang, Shenyang Agricultural University, China
Yixi Zhang, Nanjing Agricultural University, China
Su Liu, Anhui Agricultural University, China
Copyright © 2021 Yang, Lin, Li, Wang, Xue, Duan, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Zhao, c2NyY3pobUAxNjMuY29t; Jian-Hua Zhang, emhhbmdqaWFuaHVhMTk4OTA0QDE2My5jb20=
 Yuan-Xue Yang
Yuan-Xue Yang Rong-Hua Lin
Rong-Hua Lin Zhuo Li1
Zhuo Li1 Jian-Hua Zhang
Jian-Hua Zhang