- 1College of Agriculture and Biology, Innovative Institute for Plant Health, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Microbial Safety and Health, State Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology, Guangdong Academy of Sciences, Guangzhou, China
The Asian citrus psyllid Diaphorina citri is the transmission vector of Huanglongbing (HLB), a devastating disease of citrus plants. The bacterium “Candidatus Liberibacter asiaticus” (CLas) associated with HLB is transmitted between host plants by D. citri in a circulative manner. Understanding the interaction between CLas and its insect vector is key for protecting citrus cultivation from HLB damage. Here, we used RNA sequencing and liquid chromatography-mass spectrometry (LC-MS) to analyze the transcriptome and metabolome of D. citri interacting with CLas. We identified 662 upregulated and 532 downregulated genes in CLas-infected insects. These genes were enriched in pathways involving carbohydrate metabolism, the insects’ immune system, and metabolism of cofactors and vitamins. We also detected 105 differential metabolites between CLas-infected and non-infected insects, including multiple nucleosides and lipid-related molecules. The integrated analysis revealed nine pathways—including those of the glycine, serine, threonine, and purine metabolism—affected by the differentially expressed genes from both groups. The network for these pathways was subsequently constructed. Our results thus provide insights regarding the cross-talk between the transcriptomic and metabolomic changes in D. citri in response to CLas infection, as well as information on the pathways and genes/metabolites related to the CLas–D. citri interaction.
Introduction
The Asian citrus psyllid Diaphorina citri Kuwayama (Hemiptera: Psyllidae) is the main citrus pest, and it is the most widely distributed insect vector of huanglongbing (HLB, or citrus greening disease). HLB is a serious threat for the citrus industry worldwide (Bové, 2006; Gottwald, 2010; Wang and Trivedi, 2013) as it shortens plant life, sharply reduces fruit yield, and may result in plant death. HLB is present in most citrus-producing countries of Asia, Africa, and America (Graca, 1991, 2008). More than 20% of citrus trees in Brazil and 90% in Florida are currently destroyed by HLB, and HLB is currently the most serious infectious disease affecting global citrus yield (Martínez et al., 2009; Ferrarezi et al., 2020).
HLB is associated with the gram-negative bacterium, Candidatus Liberibacter asiaticus (CLas). Diaphorina citri typically feeds on plants infected with CLas for some time and then migrates to healthy plants; this accelerates the spread of CLas (Mann et al., 2012). Feeding on diseased plants has been shown to increase the fecundity and shorten nymph development duration in D. citri (Pelz-Stelinski et al., 2010; Pelz-Stelinski and Killiny, 2016). A study using an electrical penetration graph showed that the salivary time of D. citri adults feeding on diseased plants was significantly prolonged compared to that of D. citri not feeding on diseased plants (Cen et al., 2012). Moreover, infected D. citri have been shown to migrate to healthy plant earlier and have the ability to fly father than uninfected ones, and as bacterial contents increased, infected females became more attractive to males (Martini et al., 2015; Ren et al., 2016). CLas infection downregulated the gene expression level of a protease and its transporter complex in D. citri nymphs, and the immune system’s ability to synthesize and release immune-related proteases was decreased, allowing the reproduction of CLas (Tiwari et al., 2011). In addition, CLas can affect the behavior, gene expression, and substance metabolism of D. citri, thus promoting its transmission and spread. However, the genetic mechanisms regulating these phenomena are largely unknown.
The draft genome sequence of D. citri is available in the National Center for Biotechnology Information (NCBI). Accordingly, recent studies have continuously provided more information on the transcriptomic and proteomic changes regarding the species’ hemolymph and midgut (Vyas et al., 2015; Gill et al., 2017; Kruse et al., 2017, 2018; Yu et al., 2020), revealing the effects of CLas infection on D. citri’s nutrition metabolism, nymphal development, and immunity. Most aforementioned previous studies have focused on specific tissues of D. citri due to their implication in the insect’s immune system. In the present study, we compared the whole-body transcriptomes and metabolomes between CLas-infected and non-infected D. citri, and we analyzed the DEGs and differentially expressed metabolites to better understand the interaction between CLas and D. citri.
Materials and Methods
Insect Material
The D. citri specimens used in this study were collected from Sun Yat-sen University, Guangzhou, China, in 2014. The strain was reared on Citrus reticulata Blanco cv. Shatangju in a greenhouse under a 14 h: 10 h light: dark cycle at 26 ± 1°C and 65–70% relative humidity for more than 25 generations. The newly emerged female adults were fed for 14 days on the citrus plants infected with HLB, and they were confirmed to carry CLas through sampling detection. Meanwhile, the female adults that was continuously fed on healthy citrus plants for 14 days as control.
Quantitative PCR Detection of CLas
Total RNA (1 μg) was reverse-transcribed into first-strand cDNA using a PrimeScriptTM RT reagent kit (Takara Bio, Inc., Otsu, Shiga, Japan). Quantitative PCR (qPCR) was performed using a 10 μL reaction containing 1 μL cDNA, 0.3 μL each of 10 μmol⋅L–1 forward and reverse primers (Supplementary Table S1), and 5 μL SYBR FAST Universal qPCR mix (KAPA Biosystems, Woburn, MA, United States) in a LightCycler 480 system (Roche Diagnostics GmbH, Mannheim, Germany), with the following amplification conditions: 5 min at 95°C, followed by 45 cycles at 95°C for 10 s, 60°C for 20 s, and 72°C for 20 s. The qPCR experiments were performed for each sample using three biological and three technical replicates. The expression levels of selected genes were normalized to the expression levels of D. citri actin-1. The differential gene expression was calculated using the 2–△ △ Ct method (Livak and Schmittgen, 2001), and the results were expressed as mean ± SE. The differences in gene expression levels between infected and uninfected D. citri were analyzed using t-tests in the SPSS 18.0 statistical software (P < 0.01).
RNA-Seq and Differential Expression Analysis
All 20 insect individuals were pooled and ground in liquid nitrogen. Total RNA was extracted using a total RNA extract kit (Omega, Norcross, GA, United States) according to the manufacturer’s instructions. The cDNA libraries from triplicates of each insect line were generated using the Illumina TruSeq Stranded mRNA kit (Illumina, San Diego, CA, Untied States) and were size-selected to an average fragment size of 450 bp. The libraries were sequenced on the Illumina HiSeq Xten platform producing paired-end (PE) 150 bp reads.
Low quality reads from Illumina sequencing were filtered out using Cutadapt software v1.16 with parameters of—discard-trimmed-n3-e0.1 (Martin, 2011). The obtained clean reads were mapped to the D. citri genome (NCBI accession No. GCA_000475195.1) with the Hisat2 software (Kim et al., 2015). Read counts for all genes were extracted with HTSeq-count (Anders et al., 2015) and normalized using the R package DESeq2 (Love et al., 2014). The differentially expressed genes (DEGs) between the CLas-infected and non-infected lines were estimated by DESeq2 according to the threshold of | log2 ratio| > 1 and adjusted P < 0.05 (BH adjustment). The Blast2GO software was used to generate GO annotation of the DEGs. The DEGs were further annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database by KEGG Automatic Annotation Server. The GO functional enrichment and KEGG pathway enrichment of the DEGs were analyzed using the Fisher’s exact test with Benjamini and Hochberg (BH) adjustment.
Validation of DEGs by qPCR
Twelve genes were selected for qPCR validation based on the differentially expressed level and specific functions (Supplementary Table S1). Following the user manual, the first cDNA strand fragments were synthesized from total RNA using the PrimeScriptTM RT Master Mix kit (Takara, Japan). Specific primers for selected genes are listed in Supplementary Table S1. qRT-PCR was performed on an ABI7500 real-time fluorescent quantitative PCR instrument (Applied Biosystems, Foster City, United States). The relative mRNA levels of the selected genes were estimated according to the aforementioned method.
Metabolite Extraction and LC-MS/MS Analysis
For metabolite extraction, 20 female adults from each insect line were pooled and surface washes of sterile water for 1 min were performed three times. Then the samples were ground to a powder form with an electronic grinding machine. A total of 20 mg of ground sample was placed in an EP tube, and a 1,000 μL extract solution (acetonitrile: methanol: water = 2: 2: 1, with isotopically labelled internal standard mixture) was added. After 30 s of vortex mixing, the samples were homogenized at 35 Hz for 4 min and sonicated for 5 min in ice-water bath. The homogenization and sonication cycle were repeated three times. Then, the samples were incubated for 1 h at −40°C and centrifuged at 12,000 rpm for 15 min at 4°C. The resulting supernatant was transferred to a fresh glass vial for analysis. The quality control sample was prepared by mixing an equal aliquot of supernatant from all the samples. Six biological replications were performed for each insect line.
LC-MS/MS analyses were performed using an UHPLC system (Vanquish, Thermo Fisher Scientific) with a UPLC BEH Amide column (2.1 mm × 100 mm, 1.7 μm) coupled to a Q Exactive HFX mass spectrometer (Orbitrap MS, Thermo Fisher Scientific). The mobile phase consisted of 25 mmol/L ammonium acetate and 25 ammonia hydroxide in water (pH = 9.75) (A) and acetonitrile (B). The analysis was carried with elution gradient as follows: 0–0.5 min, 95% B; 0.5–7.0 min, 95–65% B; 7.0–8.0 min, 65–40% B; 8.0–9.0 min, 40% B; 9.0–9.1 min, 40–95% B; 9.1–12.0 min, 95% B. Column temperature was 30°C. Auto-sampler temperature was 4°C, and the injection volume was 2 μL. The QE HFX mass spectrometer was used for its ability to acquire MS/MS spectra on information-dependent acquisition mode in the control of the acquisition software (Xcalibur, Thermo Fisher Scientific). In this mode, the acquisition software continuously evaluates the full scan MS spectrum. The electrospray ionization (ESI) source conditions were set as follows: sheath gas flow rate 50 Arb, Aux gas flow rate 10 Arb, capillary temperature 320°C, full MS resolution 60,000, MS/MS resolution 7,500, collision energy 10/30/60 in NCE mode, spray voltage 3.5 kV (positive) or −3.2 kV (negative).
Metabolome Data Processing and Annotation
For peak detection, extraction, alignment, and integration, the raw MS/MS spectra data were converted to the mzXML format using ProteoWizard and processed with an in-house program, developed using R and based on XCMS. Then, an in-house MS2 database (BiotreeDB) was used for metabolite annotation, with a cutoff value of 0.3.
The difference in metabolite contents between the two groups was calculated using analysis of variance (ANOVA) and the Mann-Whitney U-test in SPSS 13.0. Statistical significance was set at P < 0.05. MetaboAnalyst1 was further used for the hierarchical cluster analysis of all samples and overlapped metabolites. PCA and partial least squares-discriminant analysis (PLS-DA) were conducted to investigate the relationships among the test samples. For the pathway enrichment analysis, the differential metabolites between tested groups were assigned to metabolic pathways using the tool MetPA in MetaboAnalyst. The −log(P) and impact values of each metabolic pathway were calculated using the hypergeometric test.
Integrated Analysis
The transcriptome and metabolome pathways that overlapped were considered, and the corresponding KEGG xml files (KGML) were downloaded from the KEGG pathway database. The interactions among these pathways were estimated using a custom Perl script and the KGML files as input files (Chen et al., 2016; Pang et al., 2018). Finally, the pathway–pathway network was visualized using the Cytoscape software (V.3.6.0).
Results
CLas-Infected D. citri
The CLas-specific fragments had 1,167 bp, and the gel imaging results showed that D. citri carried CLas after feeding on CLas-infected plants for 14 days (Figure 1A). The qPCR test also confirmed the CLas infection in the infected insect line, which showed a much higher level of CLas mRNA than the control non-infected line (Figure 1B). Thus, the target insect lines could be used for the subsequent transcriptomic and metabolomic analyses.
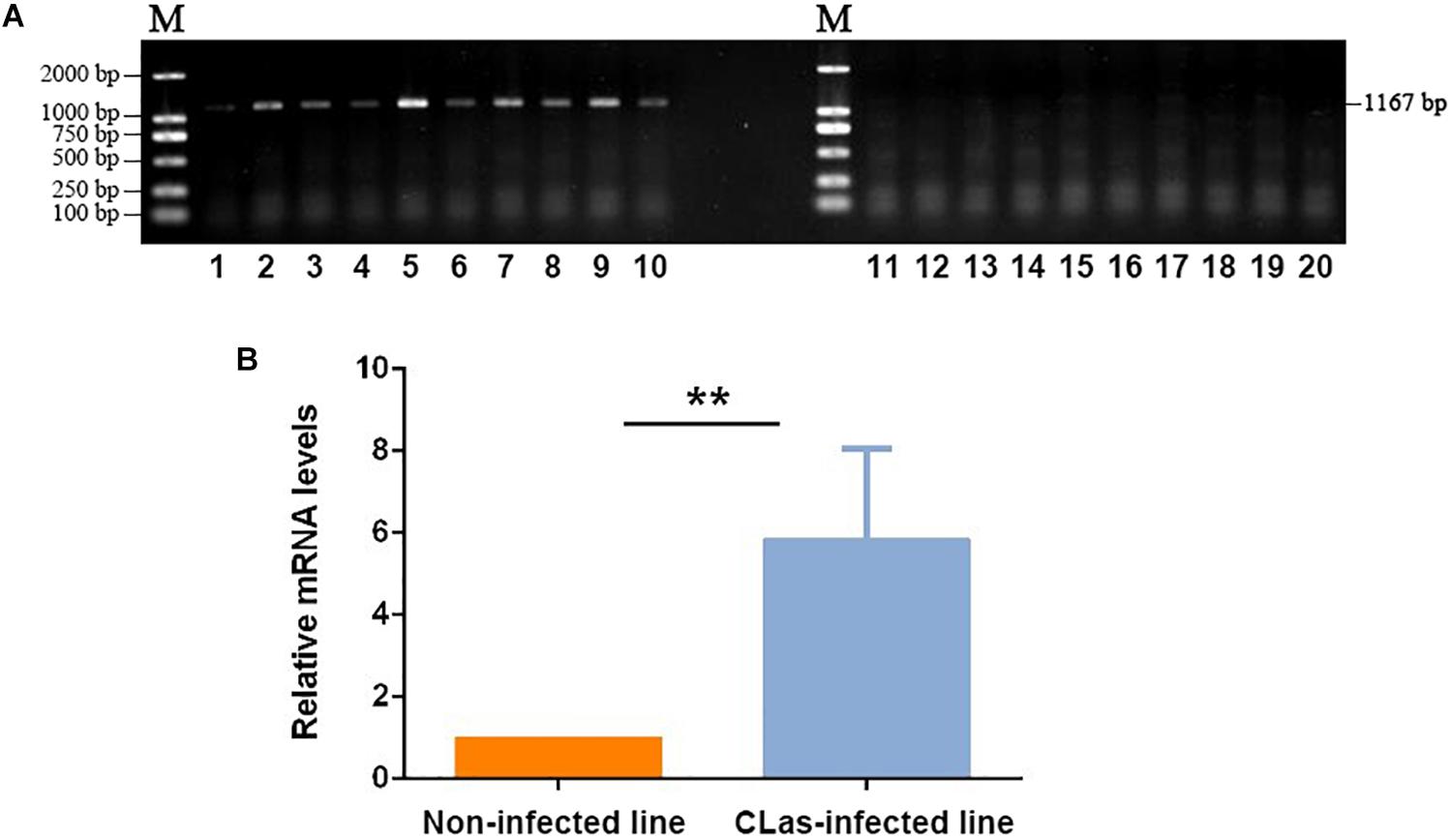
Figure 1. Validation of CLas infection in D. citri individuals. (A) PCR detection of CLas-specific sequence in infected and non-infected insect lines. 1–10: Non-infected line; 11–20: CLas-infected line. (B) qPCR validation of the relative mRNA levels of CLas-specific sequences in infected and non-infected insect lines. The data represent mean + SEM (n = 3), **P < 0.01 (t-test).
Differences in the Transcriptomic Profile Between CLas-Infected and Non-infected Insects
The RNA-seq experiments yielded an average 42,029,244 raw paired-end reads (range from 39,283,042 to 44,690,980) for each sample with an average Q30 value of 93.94% (Supplementary Table S2). After quality filtering, we obtained an average 39,538,101 high-quality clean reads for each sample. The average mapping rate of the clean reads to the D. citri genome was 81.91% (range from 81.10 to 82.66%), and the mapped reads were used for subsequent analysis.
The correlation test and principal component analysis (PCA) results were consistent regarding the triplicate RNA sequencing (RNA-seq) datasets of CLas-infected and non-infected insect lines (Supplementary Figure S1). The comparative analysis revealed that CLas infection resulted in 1,194 DEGs (4.97% of the total detected genes in the D. citri genome), including 662 up- and 532 downregulated genes (Figure 2A and Supplementary Table S3). Twelve randomly selected DEGs were validated by qPCR (Figure 2B), suggesting the reliability of the RNA-seq results. Among the upregulated genes, we found eight vitellogenin genes and two hemocyanin genes (Supplementary Table S2); the upregulation of these proteins has been observed in previous proteomic studies (Ramsey et al., 2017; Kruse et al., 2018), and our findings further confirmed this at a transcriptomic level.
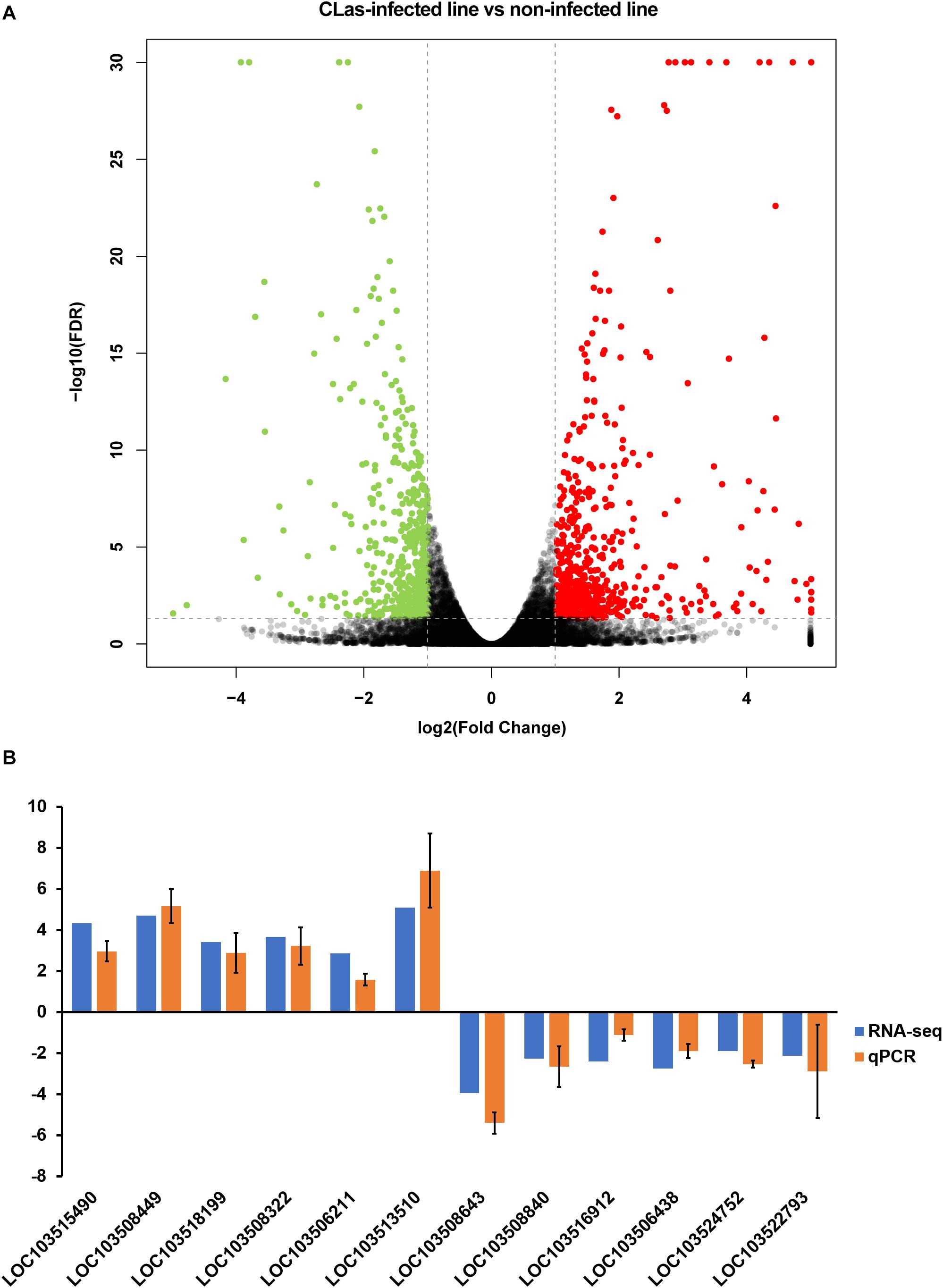
Figure 2. Identification of differentially expressed genes (DEGs) between CLas-infected and non-infected D. citri. (A) Volcano plot for the expression pattern of each gene. Black, red, and green points represent no difference in expression, upregulated genes, and downregulated genes, respectively. (B) Comparison of the transcriptome analysis and qRT-qPCR validation. The data represent mean + SEM (n = 3).
DEGs were mostly enriched in gene ontology (GO) functional categories of carboxylic acid metabolic process, organic acid metabolic process, and cofactor binding (Figure 3A and Supplementary Table S3). However, the functional enrichment of upregulated and downregulated genes was largely different. For example, the downregulated genes were mostly related to functions of photoreceptor activity, protein-chromophore linkage, and hydroxymethyl-, formyl-, and related transferase activity, whereas the upregulated genes were preferentially enriched in the functions of lipid transporter activity and lipid transporter activity (Supplementary Table S4).
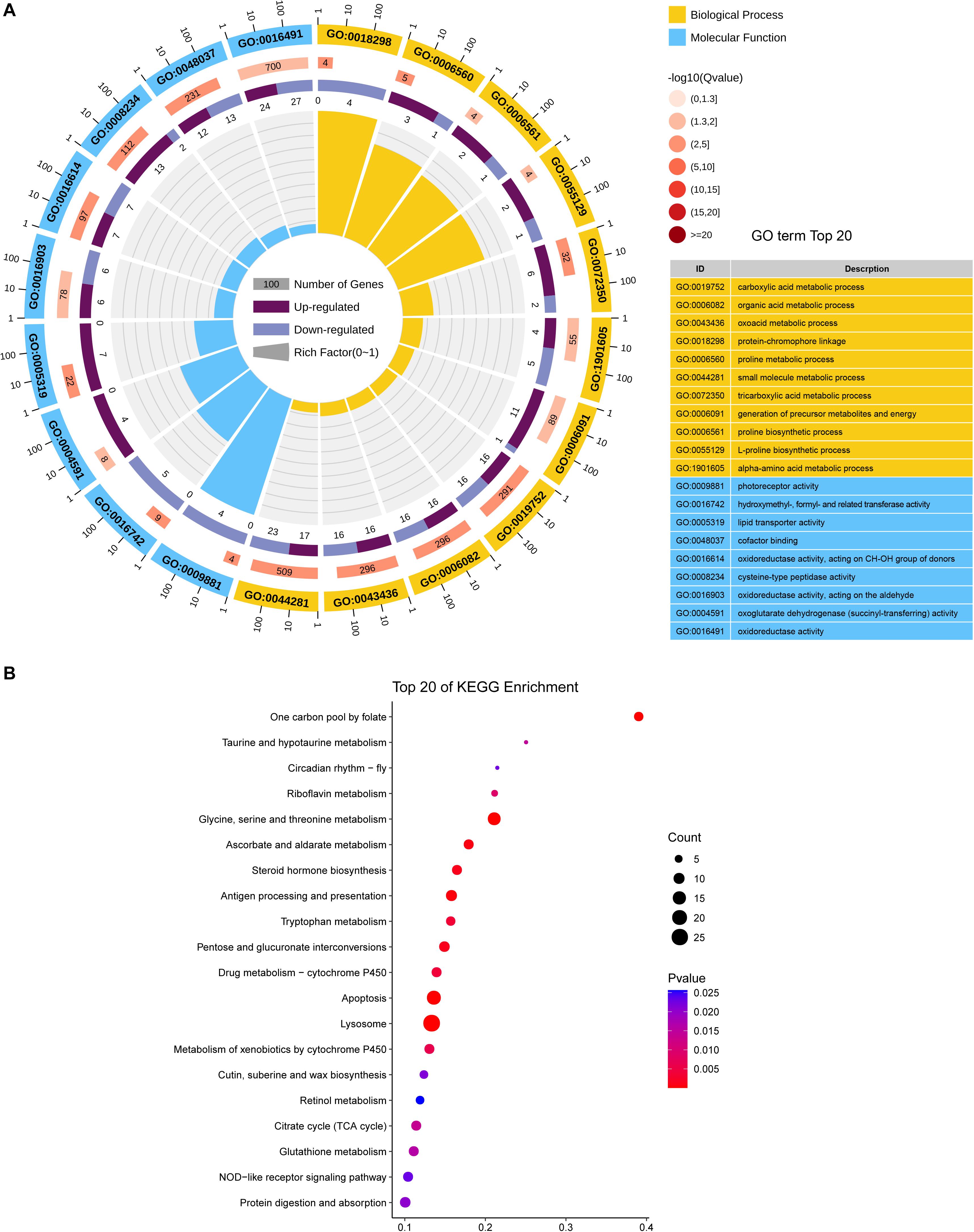
Figure 3. Functional CLassification of DEGs between CLas-infected and non-infected D. citri. (A) GO enrichment analysis of the DEGs. The outer ring represents the number of the top 20 enriched GO terms, and different colors indicate different ontologies. Ring 2 represents the gene counts in the whole genome background, and the color changes according to the Q-value of the enrichment analysis. Ring 3 represents the counts for upregulated and downregulated genes in corresponding GO terms. Ring 4 indicates the rich factor (the number of DEGs divided by the number of background genes in the corresponding term). (B) Enriched pathways annotated in the KEGG database of DEGs.
We further analyzed the pathways affected by the DEGs. The analysis showed the enrichment of DEGs in multiple carbohydrate metabolism pathways, including the citrate cycle (TCA cycle), ascorbate and aldarate metabolism, pentose, and glucuronate interconversions, and pyruvate metabolism (Figure 3B and Supplementary Table S5). This indicated that CLas infection may affect the carbon utilization of D. citri. Moreover, two immune system-related pathways, antigen processing, and presentation and NOD-like receptor signaling pathway, were found to be significantly enriched with DEGs, suggesting that CLas inevitably modulated D. citri immunity. Additionally, we noticed that several cofactor and vitamin metabolism pathways were significantly affected by the DEGs. This is in accordance with the GO enrichment in the functional category of cofactor binding. The affected cofactors and vitamins included folate, riboflavin, and retinol.
Metabolic Differences Conferred by CLas Infection
The metabolome of insects from CLas-infected and non-infected lines was measured using LC-MS/MS, which identified a total of 315 metabolites across all samples (Supplementary Table S6). The PCA showed that almost all samples were located within the 95% confidence interval, and these samples were distinctly separated into two groups according to their grouping situation (Figure 4A). The substantial difference between CLas-infected and non-infected insect lines was further confirmed by OPLS-DA score plots (Supplementary Figure S2). Of all detected metabolites, 105 showed significant difference in abundance between CLas-infected and non-infected lines (Mann-Whitney U-test, P < 0.05, Figure 4B). This result revealed that infection with CLas led to higher level of steroids (ergosterol), flavonoids (castavinol), and nucleosides such as cytosine, inosine, 5′-methylthioadenosine and 2-methylguanosine. In addition, we observed the upregulation of fatty acid molecules (linoleic acid, polyoxyethylene dioleate, 2-methylbutyroylcarnitine, L-acetylcarnitine, butyrylcarnitine, and polyoxyethylene dioleate) but the downregulation of fatty amides (linoleamide, palmitic amide, and oleamide) in CLas-infected insects (Figure 4B).
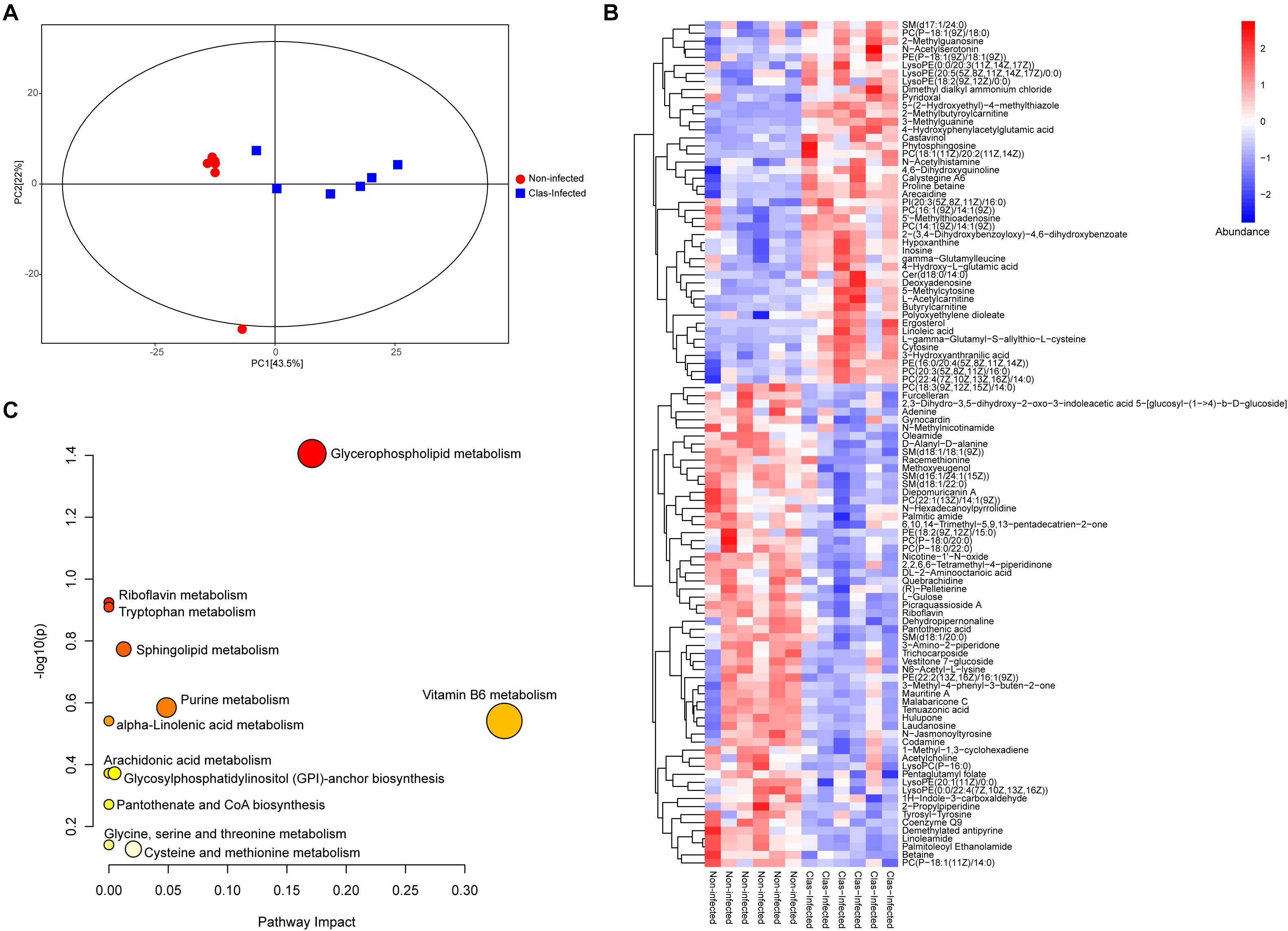
Figure 4. Impact of CLas infection on the metabolic profile of D. citri. (A) Principal component analysis (PCA) plots of the metabolite composition of samples from CLas-infected and non-infected D. citri. Each symbol represents a sample, different symbol shapes denote different groups. The circular line indicates the 95% confidence interval (Hotelling’s T-squared ellipse). (B) Hierarchical cluster and heatmap displaying the abundance levels of the differential metabolites in each sample. (C) KEGG pathway annotation of differential metabolites between CLas-infected and non-infected D. citri. The point size represents the significant compound number in the corresponding pathway.
The metabolic pathway analysis revealed that the pathways of vitamin B6 metabolism, glycerophospholipid metabolism, and purine metabolism were largely affected by the differentially expressed metabolites under CLas infection (Figure 4C and Supplementary Table S7). Notably, most of the detected metabolites in these pathways were significantly upregulated, indicating the potentially positive effect of CLas in the metabolism of these compounds.
Regulatory Network Associated With CLas Infection
We further determined the gene regulatory network associated with CLas infection using an integrated analysis of the transcriptome and metabolome. Nine pathways were found to be affected by both differentially expressed genes and metabolites (Supplementary Table S8), and these pathways could be linked to each other through differential factors except the tryptophan metabolism pathway (Figure 5). The glycine, serine, and threonine metabolism contained the most differential factors, and this pathway interacted with those of the cysteine and methionine metabolism, glycerophospholipid metabolism, sphingolipid metabolism, and purine metabolism. The purine metabolism pathway also harbored multiple differential factors, most of which were significantly upregulated under CLas infection.
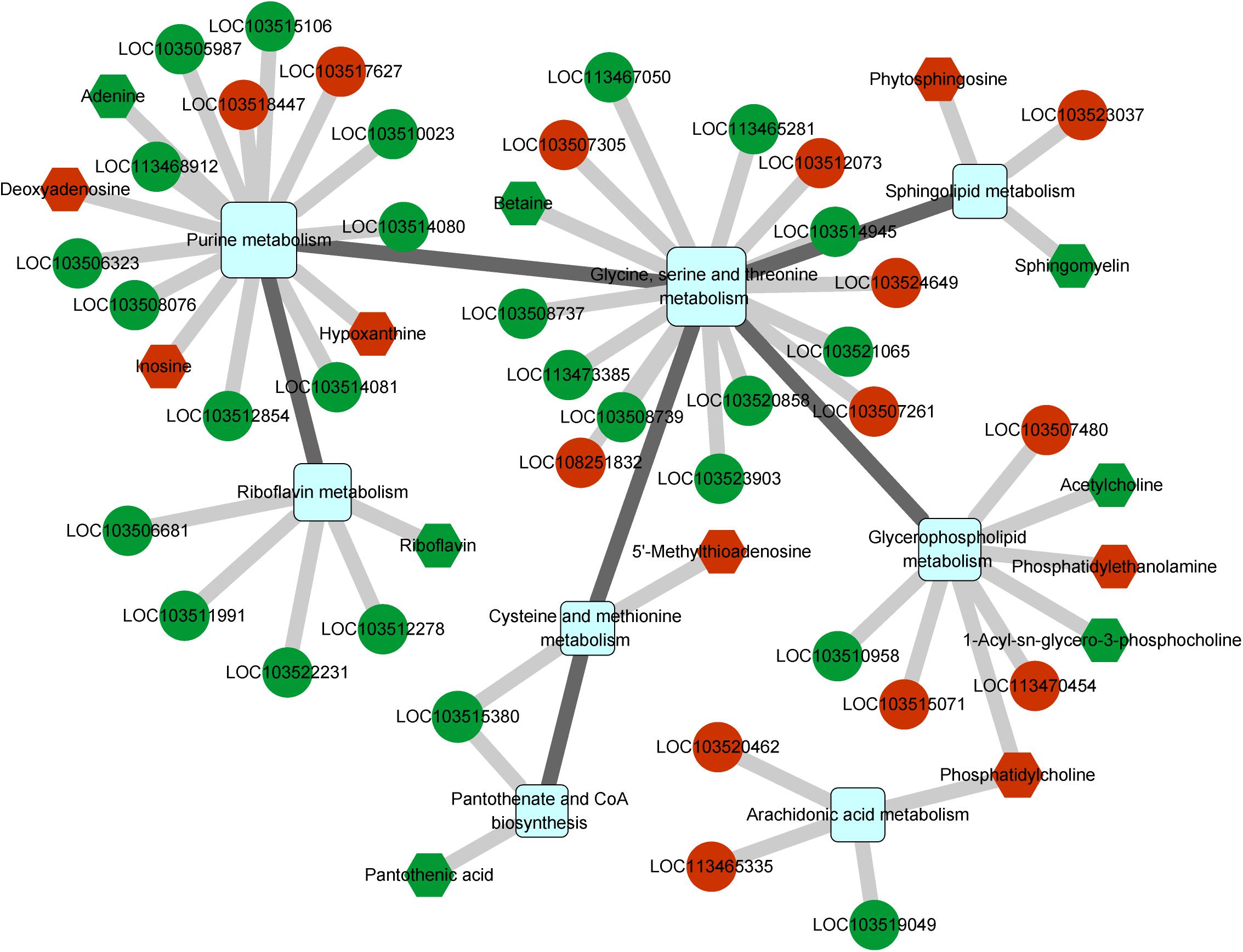
Figure 5. Integrated analysis of regulated pathways identified by transcriptomic and metabolomic analyses. Squares, circles, and hexagons represent pathways, DEGs, and differential metabolites, respectively. For DEGs and differential metabolites, red and green indicate the upregulated and downregulated factors, respectively.
Among all related differential factors, the downregulated gene LOC103515380 (branched-chain-amino-acid aminotransferase) was involved in both pantothenate and CoA biosynthesis and cysteine and methionine metabolism pathways, indicating its reduced function in the regulatory network under CLas infection. In addition, we observed increased levels of the metabolite phosphatidylcholine, an intermediate product of the glycerophospholipid metabolism and the origin of the arachidonic acid metabolism (Supplementary Figure S3). Overall, the network analysis showed that CLas infection mainly impacted the metabolism of amino acids, lipids, and cofactors of D. citri.
Discussion
As a key vector of CLas transmission, D. citri interacts with CLas in a circulative and propagative manner; moreover, CLas can be distributed in various insect tissues, including the hemolymph and salivary glands (Ammar et al., 2011; Ghanim et al., 2017). Previous studies on the transcriptomic and proteomic changes in CLas-infected and non-infected D. citri have indicated the involvement of CLas infection in the nutrition metabolism, nymphal development, and immunity of D. citri (Vyas et al., 2015; Gill et al., 2017; Kruse et al., 2017, 2018; Yu et al., 2020). Here, we focused on the transcriptomic changes in the metabolism pathways conferred by CLas infection in D. citri by using a combined transcriptomic and metabolic approach. The transcriptomic analysis revealed 662 up- and 532 downregulated genes. The DEG pattern was similar to that previously obtained from the gut transcriptome (Kruse et al., 2017; Yu et al., 2020). The LC-MS/MS analysis identified 315 metabolites in D. citri, a number much larger than those obtained in previous GC-MS analyses (Killiny and Jones, 2018). These differential factors represent the body-wide transcriptomic and metabolomic responses to CLas infection, which should be compared to the values from specific tissues.
Previous transcriptomic and proteomic analyses of D. citri showed an upregulation of transcripts and proteins involved in defense and immunity (including phenoloxidase, vitellogenin, and hemocyanin) in response to CLas infection (Kruse et al., 2017, 2018; Ramsey et al., 2017). Our data further showed that genes in the pathways of antigen processing and presentation and NOD-like receptor signaling were significantly affected (Supplementary Table S5). The heat shock proteins (HSP) coding genes in these two pathways were upregulated, as reported by Kruse et al. (2017) and Yu et al. (2020). HSPs have been recognized as important immune response factors to biological and environmental stresses in multiple insect systems (Pockley, 2003; Aguilar et al., 2005; Gotz et al., 2012). Thus, the upregulation of HSPs indicated the activation of D. citri’s immune system conferred by CLas infection. The draft genome sequence indicated that D. citri presented the Toll immune signaling pathway, as shown by previous studies (Ramsey et al., 2015; Kruse et al., 2017; Yu et al., 2020). In the transcriptome herein obtained, we also observed the upregulation of Ras-related C3 botulinum toxin substrate (LOC103512003), a gene induced by Toll-like receptor 2 in the Toll immune signaling pathway (Arbibe et al., 2000).
Another significant change conferred by CLas infection was with respect to amino acid metabolism. At the transcriptomic level, the metabolism pathways of glycine, serine, threonine, tryptophan, phenylalanine, tyrosine, and glutathione were significantly affected (Supplementary Table S5). Genomic comparisons of CLas with cultivable L. crescens revealed a lack of genes related to the production of fatty acids and aromatic amino acids (phenylalanine, tryptophan, and tyrosine) (Duan et al., 2009; Tyler et al., 2009; Lai et al., 2016). This suggests that CLas must rely on either the host plant or the insect vector for these essential nutrients, and therefore they may use these amino acids by modulating the corresponding metabolism pathways in D. citri. The metabolomic analysis also revealed the significant change in the tryptophan metabolism pathway (Figure 4). Moreover, CLas infection elevated the abundance of several aliphatic amino acids (glycine, L-asparagine, and beta-alanine), the metabolism of which is closely associated with the fatty acid metabolism.
Our results showed that CLas-infected D. citri tended to be enriched in fatty acids (Supplementary Table S6), which is in accordance with the results of previous studies (Killiny et al., 2017; Killiny and Jones, 2018). The increased abundance of linoleic acid was also observed in CLas-infected D. citri nymphs (Killiny and Jones, 2018). In contrast, the abundance of some fatty amides, derivatives of fatty acids, was significantly reduced. This means that CLas infection promotes the accumulation of fatty acids in D. citri through the modulation of fatty acid biosynthesis and the reduced transformation of fatty acids to the respective derivatives. The increasing abundance of fatty acids may also occur because of their important roles in the innate immunity of insects (Stanley et al., 2012). Remarkably, we observed the upregulation of lipid transporter activity and the significant overexpression of almost all vitellogenin genes (Supplementary Tables S3, S4). Vitellogenin is a lipoprotein with functions in insect fecundity and immunity, and it is synthesized in the fat body and secreted into the hemolymph (Arrese and Soulages, 2010). Vitellogenin plays an important nutritional role during vitellogenesis (Tufail and Takeda, 2008) and has been shown to function in insect immunity by binding to pathogen lipopolysaccharides and peptidoglycans (Li et al., 2009). Previous proteomic data have shown that CLas infection causes the upregulation of vitellogenin proteins in the D. citri hemolymph, gut, or whole body (Ramsey et al., 2017; Kruse et al., 2017, 2018). Our finding further supports the upregulation of vitellogenin in the CLas-infected D. citri, and this is in accordance with a previous report that showed the increasing reproductive fitness in CLas-infected D. citri (Pelz-Stelinski and Killiny, 2016).
The transcriptomic results also suggested that CLas modulated the cofactor and vitamin metabolism in D. citri (Figures 3B, 4C). Pantothenate, the starting compound in CoA biosynthesis, was found to be significantly reduced by CLas infection. This may result from the interaction between CLas metabolism and host proteins (Ramsey et al., 2017). Our study also indicated the decrease in riboflavin in CLas-infected insects. Riboflavin provisioning has been previously reported in bacteria–insect endosymbiotic relationships (Moriyama et al., 2015; Ju et al., 2020). Therefore, the observed reduction in riboflavin may result from the impact of CLas on other symbionts that confer fitness in riboflavin provisioning to D. citri, as CLas infection not only modulates host immunity but is also related to other microbes in the host insect (Ramsey et al., 2017; Kruse et al., 2018). In fact, bacteria benefiting from the host insect’s cofactor and vitamin biosynthesis is a common phenomenon (Bauer et al., 2014; Salem et al., 2014; Ju et al., 2020), and this may also explain the potential downregulation of folate biosynthesis and retinol metabolism pathways inferred from the transcriptomic analysis herein shown (Supplementary Table S5).
Conclusion
In conclusion, by using transcriptomic and metabolomic analyses, we identified the genes and metabolites that differentially expressed between CLas-infected and non-infected D. citri. The integrated analysis of these two omics data revealed that CLas infection had a remarkable impact on pathways associated with the metabolism of amino acids, lipids, and cofactors. The present study provides a foundation for further investigation on the mechanisms of CLas–D. citri interaction.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
KL, RP, and QH conceived and designed the study and wrote the manuscript. JH, ZG, and MZ contributed to materials. KL and JH performed the experiments. RP performed the data analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Science and Technology Planning Project of Guangdong Province, China (2016A050502063), Characteristic Innovation Project of Ordinary University of Guangdong Province, China (KA1548807), and Ecological Control of Agricultural Pests Construction Fund (KA200540404).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.630037/full#supplementary-material
Supplementary Figure 1 | The correlation of the RNA-seq datasets from CLas-infected and non-infected D. citri. The correlation was calculated according to Spearman’s rank correlation coefficient (A) and principal component analysis (B).
Supplementary Figure 2 | Orthogonal projections tolatent structures- discriminant analysis (OPLS-DA) plots of metabolite composition of samples from CLas-infected and non-infected D. citri. Each symbol represents a sample, different symbol shapes denote different groups. The circle line indicates the 95% confidence interval (Hotelling’s T-squared ellipse).
Supplementary Figure 3 | Pathways of Glycerophospholipid metabolism (A) and Arachidonic acid metabolism (B) affected by CLas infection. Red indicates that the gene/compound is up-regulated, blue indicates that the gene/compound is down-regulated.
Supplementary Table 1 | The sequences of primers used in this study.
Supplementary Table 2 | The summary of RNA-seq datasets.
Supplementary Table 3 | The expression statistics of all genes obtained by RNA-seq.
Supplementary Table 4 | Results of the GO enrichment analysis for the DEGs.
Supplementary Table 5 | Results of the KEGG pathway enrichment analysis for the DEGs.
Supplementary Table 6 | The information of compounds identified by the metabolome analysis.
Supplementary Table 7 | The pathways affected by the differential metabolites.
Supplementary Table 8 | The overlapped pathways identified from transcriptome and metabolome analysis.
Footnotes
References
Aguilar, R., Jedlicka, A. E., Mintz, M., Mahairaki, V., Scott, A. L., and Dimopoulos, G. (2005). Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem. Mol. Biol. 35, 709–719. doi: 10.1016/j.ibmb.2005.02.019
Ammar, E.-D., Shatters, R. G. Jr., and Hall, D. G. (2011). Localization of Candidatus Liberibacter asiaticus, associated with Citrus Huanglongbing Disease, in its Psyllid vector using fluorescence in situ hybridization. J. Phytopathol. 159, 726–734. doi: 10.1111/j.1439-0434.2011.01836.x
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Arbibe, L., Mira, J. P., Teusch, N., Kline, L., Guha, M., Mackman, N., et al. (2000). Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533–540. doi: 10.1038/82797
Arrese, E. L., and Soulages, J. L. (2010). Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 55, 207–225. doi: 10.1007/978-1-4615-9221-1_8
Bauer, E., Salem, H., Marz, M., Vogel, H., and Kaltenpoth, M. (2014). Transcriptomic immune response of the cotton stainer Dysdercus fasciatus to experimental elimination of vitamin-supplementing intestinal symbionts. PLoS One 9:e114865. doi: 10.1371/journal.pone.0114865
Bové, J. M. (2006). Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37.
Cen, Y., Yang, C., Holford, P., Beattie, G.a.C., Spooner-Hart, R. N., Liang, G., et al. (2012). Feeding behaviour of the Asiatic citrus psyllid, Diaphorina citri, on healthy and huanglongbing-infected citrus. Entomol. Exp. Appl. 143, 13–22. doi: 10.1111/j.1570-7458.2012.01222.x
Chen, D., Zhang, H., Lu, P., Liu, X., and Cao, H. (2016). Synergy evaluation by a pathway-pathway interaction network: a new way to predict drug combination. Mol. Biosyst. 12, 614–623. doi: 10.1039/c5mb00599j
Duan, Y., Zhou, L., Hall, D. G., Li, W., Doddapaneni, H., Lin, H., et al. (2009). Complete genome sequence of citrus Huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 22, 1011–1020. doi: 10.1094/mpmi-22-8-1011
Ferrarezi, R. S., Vincent, C. I., Urbaneja, A., and Machado, M. A. (2020). Editorial: unravelling citrus huanglongbing disease. Front. Plant. Sci. 11:609655. doi: 10.3389/fpls.2020.609655
Ghanim, M., Achor, D., Ghosh, S., Kontsedalov, S., Lebedev, G., and Levy, A. (2017). ‘Candidatus Liberibacter asiaticus’ accumulates inside endoplasmic reticulum associated vacuoles in the gut cells of Diaphorina citri. Sci. Rep. 7:16945.
Gill, T. A., Chu, C., and Pelz-Stelinski, K. S. (2017). Comparative proteomic analysis of hemolymph from uninfected and Candidatus Liberibacter asiaticus-infected Diaphorina citri. Amino Acids 49, 389–406. doi: 10.1007/s00726-016-2373-2
Gottwald, T. R. (2010). Current epidemiological understanding of citrus Huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/annurev-phyto-073009-114418
Gotz, M., Popovski, S., Kollenberg, M., Gorovits, R., Brown, J. K., Cicero, J. M., et al. (2012). Implication of Bemisia tabaci heat shock protein 70 in Begomovirus-whitefly interactions. J. Virol. 86, 13241–13252. doi: 10.1128/jvi.00880-12
Graca, J. V. (2008). “Biology, history and world status of huanglongbing,” in Proceedings of the I Taller Internacional Sobre Huanglongbing de los Cítricos (Candidatus Liberibacter spp.) y el Psílido Asiático de los Cítricos (Diaphorina citri), Hermosillo, 1–7. doi: 10.1079/9781786394088.0001
Ju, J. F., Bing, X. L., Zhao, D. S., Guo, Y., Xi, Z., Hoffmann, A. A., et al. (2020). Wolbachia supplement biotin and riboflavin to enhance reproduction in planthoppers. ISME J. 14, 676–687. doi: 10.1038/s41396-019-0559-9
Killiny, N., Hijaz, F., El-Shesheny, I., Alfaress, S., Jones, S. E., and Rogers, M. E. (2017). Metabolomic analyses of the haemolymph of the Asian citrus psyllid Diaphorina citri, the vector of huanglongbing. Physiol. Entomol. 42, 134–145. doi: 10.1111/phen.12183
Killiny, N., and Jones, S. E. (2018). Metabolic alterations in the nymphal instars of Diaphorina citri induced by Candidatus Liberibacter asiaticus, the putative pathogen of huanglongbing. PLoS One 13:e0191871. doi: 10.1371/journal.pone.0191871
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kruse, A., Fattah-Hosseini, S., Saha, S., Johnson, R., Warwick, E., Sturgeon, K., et al. (2017). Combining ‘omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS One 12:e0179531. doi: 10.1371/journal.pone.0179531
Kruse, A., Ramsey, J. S., Johnson, R., Hall, D. G., MacCoss, M. J., and Heck, M. (2018). Candidatus Liberibacter asiaticus minimally alters expression of immunity and metabolism proteins in hemolymph of Diaphorina citri, the insect vector of Huanglongbing. J. Proteome Res. 17, 2995–3011. doi: 10.1021/acs.jproteome.8b00183
Lai, K. K., Davis-Richardson, A. G., Dias, R., and Triplett, E. W. (2016). Identification of the genes required for the culture of Liberibacter crescens, the closest cultured relative of the Liberibacter plant pathogens. Front. Microbiol. 7:547. doi: 10.3389/fmicb.2016.00547
Li, Z., Zhang, S., Zhang, J., Liu, M., and Liu, Z. (2009). Vitellogenin is a cidal factor capable of killing bacteria via interaction with lipopolysaccharide and lipoteichoic acid. Mol. Immunol. 46, 3232–3239. doi: 10.1016/j.molimm.2009.08.006
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550.
Mann, R. S., Ali, J. G., Hermann, S. L., Tiwari, S., Pelz-Stelinski, K. S., Alborn, H. T., et al. (2012). Induced release of a plant-defense volatile ‘deceptively’ attracts insect vectors to plants infected with a bacterial pathogen. PLoS Pathog. 8:e1002610. doi: 10.1371/journal.ppat.1002610
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Martínez, Y., Llauger, R., Batista, L., Luis, M., Iglesia, A., and Collazo, C. (2009). First report of ‘Candidatus Liberibacter asiaticus’ associated with Huanglongbing in Cuba. Plant Pathol. 58, 389–389.
Martini, X., Hoffmann, M., Coy, M. R., Stelinski, L. L., and Pelz-Stelinski, K. S. (2015). Infection of an insect vector with a bacterial plant pathogen increases its propensity for dispersal. PLoS One 10:e0129373. doi: 10.1371/journal.pone.0129373
Moriyama, M., Nikoh, N., Hosokawa, T., and Fukatsu, T. (2015). Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. mBio 6, e01732-15.
Pang, R., Chen, M., Yue, L., Xing, K., Li, T., Kang, K., et al. (2018). A distinct strain of Arsenophonus symbiont decreases insecticide resistance in its insect host. PLoS Genet. 14:e1007725. doi: 10.1371/journal.pgen.1007725
Pelz-Stelinski, K. S., Brlansky, R. H., Ebert, T. A., and Rogers, M. E. (2010). Transmission parameters for Candidatus liberibacter asiaticus by Asian citrus psyllid (Hemiptera: Psyllidae). J. Econ. Entomol. 103, 1531–1541. doi: 10.1603/ec10123
Pelz-Stelinski, K. S., and Killiny, N. (2016). Better together: association with ‘Candidatus Liberibacter Asiaticus’ increases the reproductive fitness of its insect vector, Diaphorina citri (Hemiptera: Liviidae). Ann. Entomol. Soc. Am. 109, 371–376. doi: 10.1093/aesa/saw007
Pockley, A. G. (2003). Heat shock proteins as regulators of the immune response. Lancet 362, 469–476. doi: 10.1016/s0140-6736(03)14075-5
Ramsey, J. S., Chavez, J. D., Johnson, R., Hosseinzadeh, S., Mahoney, J. E., Mohr, J. P., et al. (2017). Protein interaction networks at the host-microbe interface in Diaphorina citri, the insect vector of the citrus greening pathogen. R. Soc. Open Sci. 4:160545. doi: 10.1098/rsos.160545
Ramsey, J. S., Johnson, R. S., Hoki, J. S., Kruse, A., Mahoney, J., Hilf, M. E., et al. (2015). Metabolic interplay between the Asian citrus psyllid and its Profftella symbiont: an Achilles’ heel of the citrus greening insect vector. PLoS One 10:e0140826. doi: 10.1371/journal.pone.0140826
Ren, S. L., Li, Y. H., Zhou, Y. T., Xu, W. M., Cuthbertson, A. G. S., Guo, Y. J., et al. (2016). Effects of Candidatus Liberibacter asiaticus on the fitness of the vector Diaphorina citri. J. Appl. Microbiol. 121, 1718–1726.
Salem, H., Bauer, E., Strauss, A. S., Vogel, H., Marz, M., and Kaltenpoth, M. (2014). Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. Biol. Sci. 281:20141838. doi: 10.1098/rspb.2014.1838
Stanley, D., Haas, E., and Miller, J. (2012). Eicosanoids: exploiting insect immunity to improve biological control programs. Insects 3, 492–510. doi: 10.3390/insects3020492
Tiwari, S., Gondhalekar, A. D., Mann, R. S., Scharf, M. E., and Stelinski, L. L. (2011). Characterization of five CYP4 genes from Asian citrus psyllid and their expression levels in Candidatus Liberibacter asiaticus-infected and uninfected psyllids. Insect Mol. Biol. 20, 733–744. doi: 10.1111/j.1365-2583.2011.01103.x
Tufail, M., and Takeda, M. (2008). Molecular characteristics of insect vitellogenins. J. Insect Physiol. 54, 1447–1458. doi: 10.1016/j.jinsphys.2008.08.007
Tyler, H. L., Roesch, L. F. W., Gowda, S., Dawson, W. O., and Triplett, E. W. (2009). Confirmation of the sequence of ‘Candidatus Liberibacter asiaticus’ and assessment of microbial diversity in Huanglongbing-infected citrus phloem using a metagenomic approach. Mol. Plant Microbe Interact. 22, 1624–1634. doi: 10.1094/mpmi-22-12-1624
Vyas, M., Fisher, T. W., He, R., Nelson, W., Yin, G., Cicero, J. M., et al. (2015). Asian citrus psyllid expression profiles suggest Candidatus Liberibacter Asiaticus-mediated alteration of adult nutrition and metabolism, and of nymphal development and immunity. PLoS One 10:e0130328. doi: 10.1371/journal.pone.0130328
Wang, N., and Trivedi, P. (2013). Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 103, 652–665. doi: 10.1094/phyto-12-12-0331-rvw
Keywords: Diaphorina citri, Candidatus Liberibacter asiaticus, interaction, transcriptome, metabolome
Citation: Liu K, He J, Guan Z, Zhong M, Pang R and Han Q (2021) Transcriptomic and Metabolomic Analyses of Diaphorina citri Kuwayama Infected and Non-infected With Candidatus Liberibacter Asiaticus. Front. Physiol. 11:630037. doi: 10.3389/fphys.2020.630037
Received: 16 November 2020; Accepted: 21 December 2020;
Published: 24 February 2021.
Edited by:
Bin Tang, Hangzhou Normal University, ChinaReviewed by:
Yanjing Wang, Zhejiang Agriculture and Forestry University, ChinaDandan Wei, Southwest University, China
Sheila Ons, National University of La Plata, Argentina
Copyright © 2021 Liu, He, Guan, Zhong, Pang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Pang, cHI4MzlAMTYzLmNvbQ==; cGFuZ3I2QG1haWwuc3lzdS5lZHUuY24=; Qunxin Han, aHF4OTlAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Kai Liu
Kai Liu Jiawei He1†
Jiawei He1† Rui Pang
Rui Pang