- 1MOE Key Laboratory of Marine Genetics and Breeding, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Laboratory for Marine Fisheries Science and Food Production Processes, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Inhibitors of apoptosis proteins (IAPs) are conserved regulators involved in cell cycle, cell migration, cell death, immunity and inflammation, should be due to the fact that they can assist with the ability to cope with different kinds of extrinsic or intrinsic stresses. Bivalve molluscs are well adapted to highly complex marine environments. As free-living filter feeders that may take toxic dinoflagellates as food, bivalves can accumulate and put up with significant levels of paralytic shellfish toxins (PSTs). PSTs absorption and accumulation could have a deleterious effect on bivalves, causing negative impact on their feeding and digestion capabilities. In the present study, we analyzed IAP genes (PyIAPs) in Yesso scallop (Patinopecten yessoensis), a major fishery and aquaculture species in China. Forty-seven PyIAPs from five sub-families were identified, and almost half of the PyIAP genes were localized in clusters on two chromosomes. Several sites under positive selection was revealed in the significantly expanded sub-families BIRC4 and BIRC5. After exposure to PST-producing dinoflagellates, Alexandrium catenella, fourteen PyIAPs showed significant responses in hepatopancreas and kidney, and more than eighty-five percent of them were from the expanded sub-families BIRC4 and BIRC5. The regulation pattern of PyIAPs was similar between the two tissues, with more than half exhibited expression suppression within three days after exposure. In contrast to hepatopancreas, more acute changes of PyIAPs expression could be detected in kidney, suggesting the possible involvement of these PyIAPs in tissue-specific PST tolerance. These findings also imply the adaptive expansion of bivalve IAP genes in response to algae derived biotoxins.
Introduction
Apoptosis is a form of programmed cell death, which can be triggered by intrinsic cellular genotoxic stress, or by extrinsic signals such as the ligand binding with cell surface death receptors (Pistritto et al., 2016). In cope with different kinds of stresses, organisms could recruit multiple apoptosis regulators in order to balance between the pro- with anti-apoptotic mechanisms, inhibitors of apoptosis proteins (IAPs) for example (Grzybowska-Izydorczyk and Smolewski, 2008). IAPs, also known as baculovirus IAP repeat (BIR)-containing proteins (BIRCs), are functionally conserved in metazoans (Fulda and Vucic, 2012). IAP homologous genes could be widely found in insects, birds, fishes, mammals, and viruses (Luque et al., 2002), however, the IAP gene family vary a lot in gene numbers among species. For instance, in human, a total of eight IAP members known as BIRC1-8 were identified through functional screening and homology searching (Salvesen and Duckett, 2002; Vucic, 2008). In comparison, there are only seven, four and three IAP members were found in sea urchins (Robertson et al., 2006), fruit flies (Jones et al., 2000; Domingues and Ryoo, 2012), sea anemone (Sullivan et al., 2006), respectively, and more than forty IAP members were reported in bivalve mollusc Crassostrea gigas (Zhang et al., 2012), suggesting the adaptive lineage-specific gene gain and loss events.
As their functional diversity and importance are concerned, IAPs have received substantial research interests in a lot of animal species, related to cell fate control as well as body immunity and inflammation. For example, in human, transduction with an adenovirus encoding either HIAP1/c-IAP2 or HIAP2/c-IAP1 was found be able to suppress ER stress-induced apoptosis (Cheung et al., 2006). In the context of a highly inflammatory virus infection environment, IAPs were found could play a crucial role in the expansion of T-cells in mice (Gentle et al., 2014). Similarly, DIAP2 was found functioned in the host immune response to gram-negative bacteria in Drosophila (Leulier et al., 2006). Besides, it was reported that vertebrate IAPs could participate in coping with toxic stresses. In human, IAPs directly ubiquitinate Rac1 at lysine 147 resulting in its proteasomal degradation under CNF1 toxin treatment (Oberoi-Khanuja and Rajalingam, 2012). Meanwhile, in mice, the survivin (BIRC5) was proved to serve as a protector against toxin-induced acute renal failure and IAPs were associated with T-2 toxin-induced mouse chondrocyte damage (Kindt et al., 2008; Wang et al., 2020).
Marine bivalves are recognized for their well adaptation to the highly complex aquatic-environment (Wang et al., 2017). These filter-feeding bivalves can usually accumulate paralytic shellfish toxins (PSTs) by filtering toxic algae or their cysts, especially during the outbreak of red tide (Yang et al., 2018). PSTs, mainly produced by toxic dinoflagellate such as Alexandrium (Anderson et al., 2012), are one kind of potent neurotoxins with inhibitory effects on cell sodium channels (Cusick and Sayler, 2013). PSTs accumulated in bivalves can exert negative impact on their biological function and its sanitary quality, bringing huge economic losses in the aquaculture sector (Landsberg, 2002; Contreras et al., 2011; Vilarino et al., 2013; Borcier et al., 2017). Researchers revealed that the induction of toxic stress by PST accumulation may impact on cellular apoptosis process and immune defense. It has been found that PST could enter the cytoplasm and induce apoptosis of immune cells through a caspase-dependent pathway in oyster (Abi-Khalil et al., 2017). Cellular immunological and inflammatory responses were observed after exposure to Alexandrium catenella in Surf clams Mesodesma donacium (Alvarez et al., 2019) and hemocyte aggregation were found show up after exposed to the PST-producing Gymnodinium catenatum in scallop Nodipecten subnodosus (Estrada et al., 2007). Especially, it has been found that CgIAP1 and CgIAP7B were involved in the regulation of hemocyte immune cell apoptosis with toxic diet A. catenella (Medhioub et al., 2013), implying the possible participation of IAP genes in bivalves facing toxic challenge. However, to our best knowledge, systematic studies on bivalve IAPs in coping with PSTs stress have not been reported yet, and relevant research data may help in-depth understanding of their adaptive features to algae derived toxins.
Yesso scallops (Patinopecten yessoensis) are important fishery and aquaculture species in China, and they are also one type of bivalves that possess outstanding capacity to accumulate and tolerate PSTs (Cheng et al., 2016; Xun et al., 2020). According to our previous work, we found the hepatopancreas and kidney in scallops should be the top-ranked toxin-rich or toxin-tolerant organs, with hepatopancreas mostly accumulating the incoming toxins, and the kidney transforming and/or eliminating them (Tian et al., 2010; Li et al., 2017). In the present study, we provided a systematic characterization of IAP genes in P. yessoensis. In the two most toxic organs, hepatopancreas and kidney, their transcriptional responses to the exposure of PST producing algae A. catenella (strain ACDH) were also analyzed. Our findings will provide helpful resources for further researches to elaborate the functions of the bivalve IAPs and assist in better understanding of the involvement of IAPs in the adaptive response to PST tolerance.
Materials and Methods
Genome-Wide Identification and Characterization of IAP Genes in P. yessoensis
The genome and transcriptome sequences of P. yessoensis (Wang et al., 2017) were searched against the available IAP sequences from representative species, including Homo sapiens, Xenopus laevis, Gallus gallus, Danio rerio, Mus musculus, Lottia gigantean, Octopus bimaculoides. We downloaded the orthologous IAP sequences of these representative species from NCBI1 and Uniprot2 databases (Supplementary Table 1 shown the accession numbers), and used these sequences as queries to perform whole-genome and transcriptome-based blast with the e-value threshold of 1E-05. To ensure the completeness of IAP genes, TBLASTN with the domain sequences of all known IAP proteins were used as queries for additional similarity searches against genomes. Then, BIR HMM searching method was used for sequence analysis to confirm the completeness of IAPs, and the number of conserved BIR Pfam domain (PF00653) (Finn et al., 2016) was summarized for P. yessoensis IAPs in Supplementary Table 2. Next, we used open reading frame (ORF) finder program3 to translate candidate PyIAPs into amino acid (aa) sequences. The translated sequences were submitted to the SMART program4 to verify the presence of the BIR domain and confirm the accuracy of the sequence. The information of isoelectric point (PI) and molecular weight were predicted through Compute PI/Mw tools5. The genetic structure of all the identified PyIAPs and their protein structures were drawn with GSDS2.06 and IBS1.0.3 software (Liu et al., 2015), respectively, and the genomic distribution of PyIAPs were also confirmed.
Phylogenetic Analysis
The whole amino acid sequences of IAP genes from zebrafish (Danio rerio), frog (Xenopus laevis), original chicken (Gallus gallus), human (Homo sapiens), mouse (Mus musculus), lizard (Lottia gigantean), octopus (Octopus bimaculoides) were downloaded from the NCBI (Supplementary Table 1). Multi-sequence alignment for the BIR domains was performed using ClustalW (Larkin et al., 2007) as well as Genedoc software (Nicholas et al., 1997). The phylogenetic analysis was generated through MEGA6 (Tamura et al., 2013), and a neighbor-joining (NJ) tree was constructed using the p-distance method and maximum likelihood (ML) method. The gaps and missing data in positions were eliminated, and we tested the robustness of phylogeny with a bootstrap of 1000 replicates. The resulting phylogenetic tree was output in Newick format, and the online software iTOL7 converted the percentage value to 0-1, and scores below 0.60 were not displayed. Totally, this phylogenetic analysis involved 94 amino acids across 8 animals.
Test of Variable dN/dS Ratios
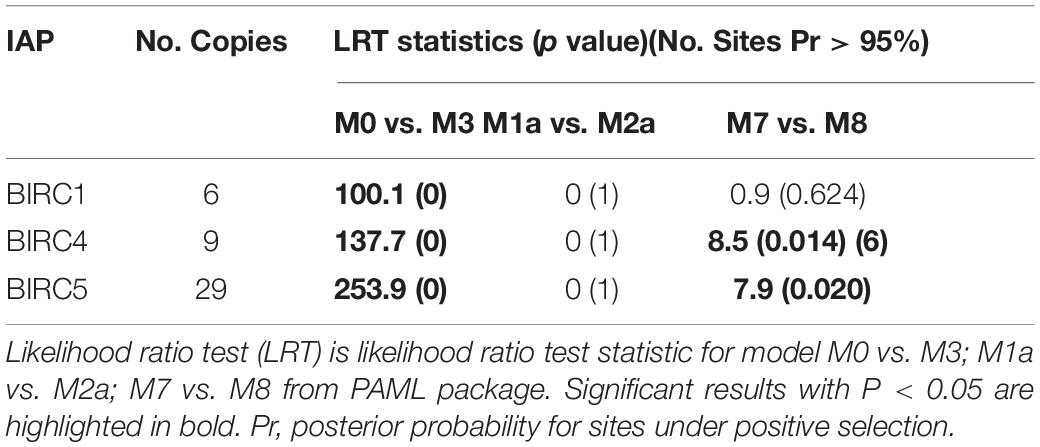
To detect selective constraints and positive selection for the three expanded PyIAP sub-families (BIRC1, BIRC4, and BIRC5), we used a codon-based models of the CodeML application from the PAML 4.9 package (Yang, 2007) with a maximum likelihood ratio test (LRT). First, The PAML format alignment sequence and Newick format evolution tree file of the three sub-families were loaded, with Num of Threads set to 2, and the consistency of the category name and the alignment sequence name in the evolution tree file were checked through the “Check” button. Then, we used various site models that allow the ω ratio to vary among sites of the expanded IAP subfamilies to estimate Non-synonymous (dN) and Synonymous (dS) substitution ratios (dN/dS or ω) and to measure the selective pressure at the protein level, in which a ω of 1, < 1, or > 1 represents neutral evolution, purifying selection/negative selection, or positive selection, respectively. In the site-specific selection analyses, three pairs of codon substitution models (M0 vs. M3, M1a vs. M2a, M7 vs. M8) were conducted (Cheng et al., 2016; Ahmad et al., 2019, 2020; Xun et al., 2020). The model M0 (one ratio) assumes that all sites have the same value, whereas model M3 (discrete) assumes that the ω values of all sites show a simple discrete distribution trend. The model M1a (neutral), allows two classes of codon sites for ω (0 < ω < 1 and ω = 1) and estimates the ratio and ω value of these two types of sites, whereas the model M2a (positive selection), allows one additional site class with ω > 1 and estimates the ratio and ω-value of these three types of sites. The M7 (beta) model assumes that ω (0 < ω < 1) fits a beta distribution, whereas the model M8 (beta and ω) has an extra category with ω > 1. Comparisons between the paired nested site models were performed to evaluate the variation in ω (M0 vs. M3) and to determine the presence of a positively selected class of sites (M1a vs. M2a and M7 vs. M8). After the sites under positive selection were estimated, we applied a Bayes Empirical Bayes (BEB) approach as implemented in PAML 4.9 package to identify the class of sites.
Expression Profiling of PyIAPs After A. catenella Exposure
We choose 2-year-old adult Yesso scallops to perform the toxic diet exposure experiment. These scallops were reared and aerated in filtered seawater at 12–13°C for one week to adapt to the laboratory environment. The A. catenella (ACDH strain) cells were cultivated (Navarro et al., 2006) and collected at the late exponential growth phase (Garcia-Lagunas et al., 2013) by centrifugation (2500 g/10 min). A total of 3 L of A. catenella cells were fed to each scallop once a day, with a final density of 2500 cells/ml. This preliminary experiment of the feeding doses was conducted by setting different cell densities to feed Yesso scallops in order to determine the reasonable food intake. When all 3 L of dinoflagellate cells were eaten up by the exposed scallop in one day, without leftovers in the sea water, the most reasonable cell density was 2500 cells/ml (Hu et al., 2019). In the present study, Yesso scallops were fed continuously, with 3 L dinoflagellate cells each day till their tissues were taken at 0 (control), 1, 3, 5, 10, and 15 days from three scallops for each time point. The hepatopancreases and kidney tissues were collected, rinsed with PBS (1x), immediately frozen with liquid nitrogen, and stored in the refrigerator at −80°C till RNA extraction.
Total RNA was isolated from the sampled hepatopancreas and kidney tissues using phenol- chloroform method. After exposure to toxic ACDH strain, we used RNA-seq data obtained from these tissues to analyze the expression profiles of PyIAP genes (Hu et al., 2019). Briefly, we followed the manufacturer’s instructions of NEB Next mRNA Library Prep Kit to construct the RNA-seq libraries, and these libraries were arranged to PE125 sequencing on the Illumina HiSeq 2000 platform. Then, we mapped RNA-seq reads to the P. yessoensis genome with the help of Tophat 2.0.9, and the expression of all PyIAP genes was normalized and represented in the form of reads per kilobase of exon model per million mapped reads (RPKM). We calculated Fold Change (FC) for each test time point as log2FC among the experimental group (toxin-exposed) with control groups (Supplementary Table 3). The edgeR software (Robinson et al., 2010) was used to analyze the difference of transcriptome data in expression profiles between the experimental group and the control group. For genes screening with significant expression differences, p < 0.05 and—log2FC— ≥ 1 was used as the criterion (Wang et al., 2019). Finally, MeV 4.90 software8 was used to draw a heat map with the log2FC values.
Results and Discussion
PyIAP Genes Identification and Their Genomic Distribution
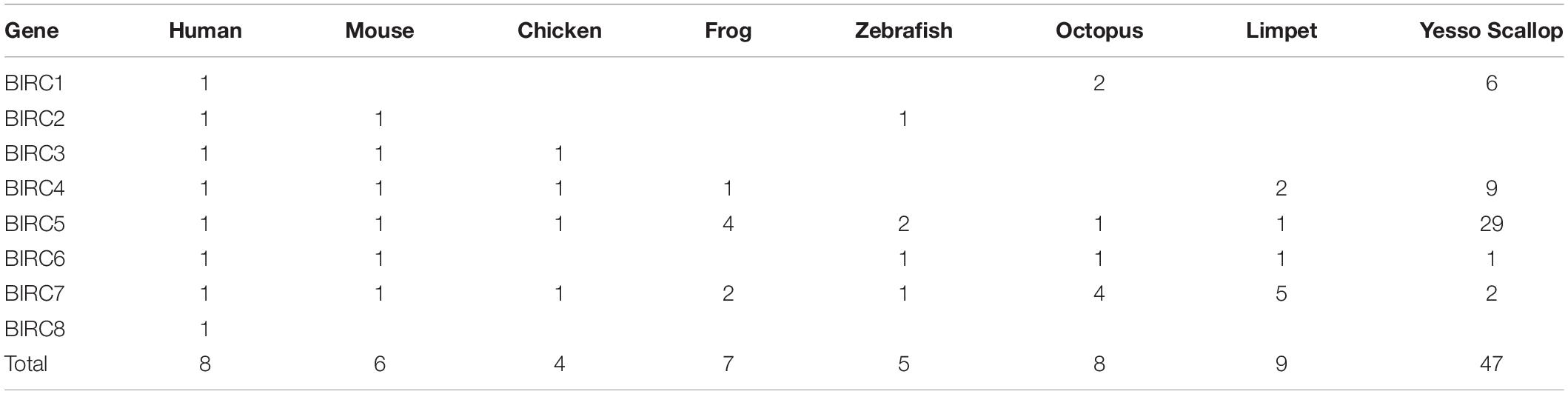
A total of 47 IAP genes were identified from the whole genome of P. yessoensis and they were classified into the five sub-families, including BIRC1, BIRC4, BIRC5, BIRC6 and BIRC7 (Table 1). Specifically, more than half of the identified PyIAP genes (29 members) belong to the BIRC5 sub-family. Six BIRC1 members and nine BIRC4 members were identified for PyIAPs, with the rest three classified into BIRC6 and BIRC7 sub-families, respectively. In comparison with human, from which eight single-copy IAP genes known as BIRC1-8 were identified (Salvesen and Duckett, 2002; Vucic, 2008), the PyIAPs from BIRC1, BIRC4, and BIRC5 sub-families showed clearly gene expansion, whereas the members from sub-family BIRC2, BIRC3 and BIRC8 are found to be absent in the P. yessoensis genome.
The basic information of PyIAPs including gene ID, GeneBank accession number, gene subfamily, genome position, exon number, CDS length, protein length, BIR domain number, PI value as well as molecular weight were summarized in Supplementary Table 2. The ORF of PyIAPs varied from 411 to 14808 bp in length and the protein length ranged from 136 to 4935 amino acids. The exon number of most PyIAPs varied from 2 to 25, except for the shortest PyBIRC5 member (PY.8149.5) with a single exon and for the longest PyBIRC6 member (PY.8833.5) that are comprised with 59 exons.
The 47 PyIAP genes were found to distribute across 27 scaffolds, and a total of 20 PyIAP genes (43%) were found located as gene clusters in form of two or more neighboring members. For example, seven and six PyIAPs were found to locate on scaffold9385 and scaffold383, respectively, showing high distribution density. Besides, among eight chromosomes which PyIAPs located, chromosome NO.3 and NO.7 were found to be the main chromosomes they distributed, with 28 PyIAP genes and seven PyIAP genes, respectively. Of note, the PyIAPs from high density region were usually from the expanded BIRC4 and BIRC5 subfamily. Taken together, we inferred that the expansion of PyIAP genes could be attributed to lineage-specific multiple tandem duplication events. Similar phenomenon has been revealed for PyHsp70s (Cheng et al., 2016; Hu et al., 2019), suggesting the involvement of tandem gene duplication in Yesso scallop’s adaptation to the fluctuating and stressful marine environments.
Conserved Protein Signatures and Phylogenetic Analysis
The IAP proteins are characterized by the presence of Baculovirus IAP Repeat (BIR) at the N-terminal end (Mohamed et al., 2017). According to the analysis of protein structure characteristics (Figure 1), all 47 PyIAP proteins possess one or more repeats of BIR domain. According to the multiple sequence alignment, it showed that the PyBIR domains were composed of about 68–79 amino acid residues, containing conservative cysteine/histidine sequences (Figure 2). This was consistent with the previous findings that BIRs possessed a great quantity of constant amino acid residues, especially including three conserved cysteines and one histidine that coordinated a zinc ion, which was required to stabilize the BIR fold (Silke and Meier, 2013). Besides BIR domains, 27 PyIAPs were found to contain a ring domain at the C-terminus, whereas the longest PyBIRC6 member (PY.8833.5) that mentioned above contain a PFAM as well as a UBCc domain at the C-terminus. Compared with the protein structure of IAPs from various species, including viral, yeast, nematode, fly, and several mammalians (Deveraux and Reed, 1999), previous studies found that the number of BIR domains are usually less than three. Three subtypes of BIR domain (BIR1, BIR2, and BIR3) have been identified so far (Mohamed et al., 2017). In P. yessoensis we found 2 members of PyBIRC4 sub-family containing quadruple BIR domains, and previous study had also shown that the BIR domain expanded in the Crassostrea gigas (Zhang et al., 2011). It has been reported that although the family defining BIR domain is highly conserved, distinct BIR domains may have different functions (Wang and Lin, 2013). Structure of IAPs is closely related to their antiapoptotic ability. The antiapoptotic function of IAPs was affected by interplay among the BIR domains with caspases (Wang and Lin, 2013). For instance, the BIR3 domain of XIAP in human directly binds to the small subunit of caspase-9, but the BIR2 domain interacts with the active-site substrate binding pocket of caspases-3 and -7 (Takahashi et al., 1998). Thus, the various BIR domain changes in PyIAPs, such as the specific quadruple-BIR structure, may contribute to their adaptive functional differentiation in the life activities of scallops which needs to be further studied.
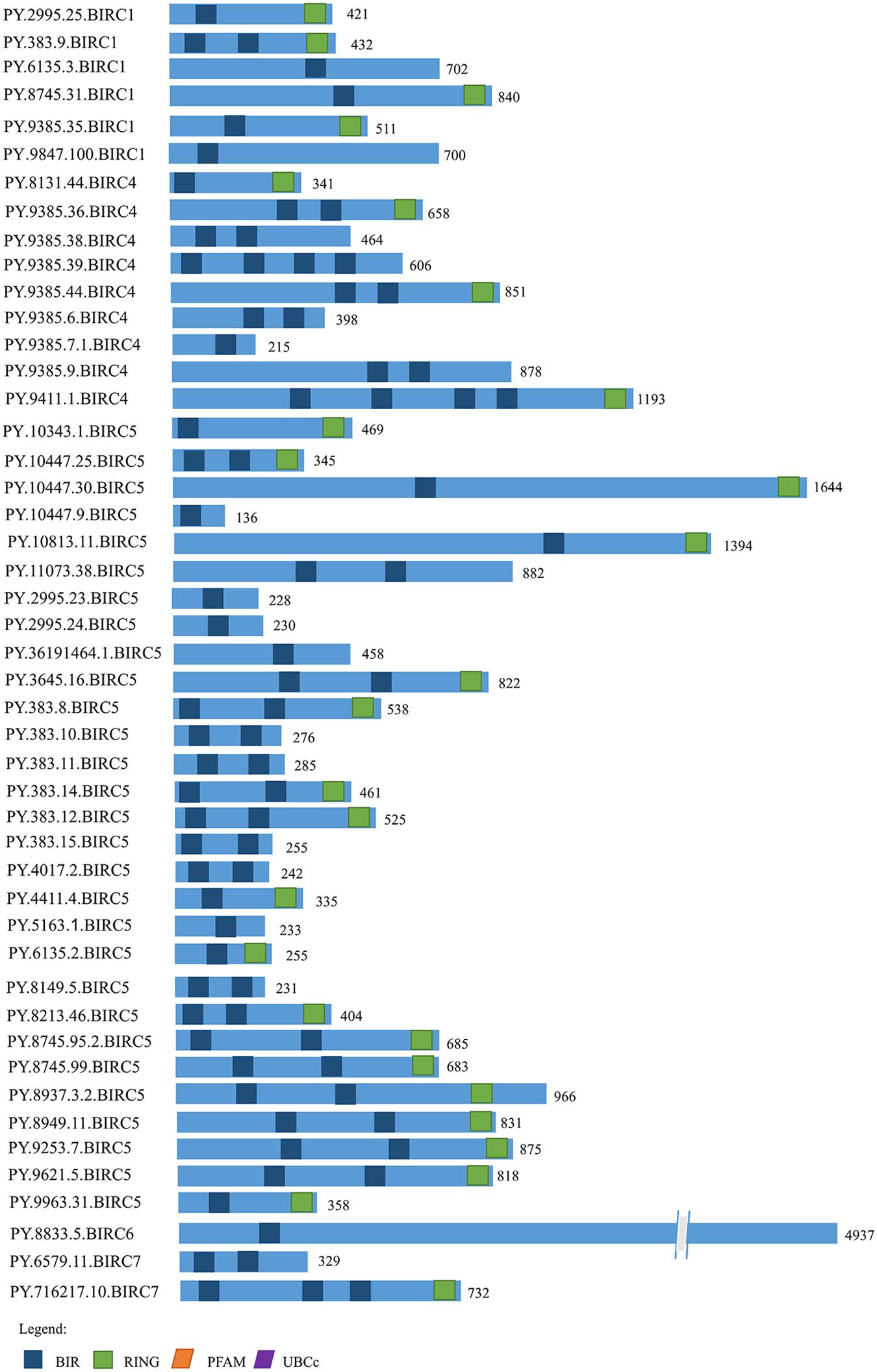
Figure 1. The structure of IAP proteins in Patinopecten yessoensis. The blue boxes indicate the conserved BIR domains. The green boxes indicate RING domains. The orange box indicates PFAM domain and the purple box indicates UBCc domain. Amino acid length is indicated to the right of each protein (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
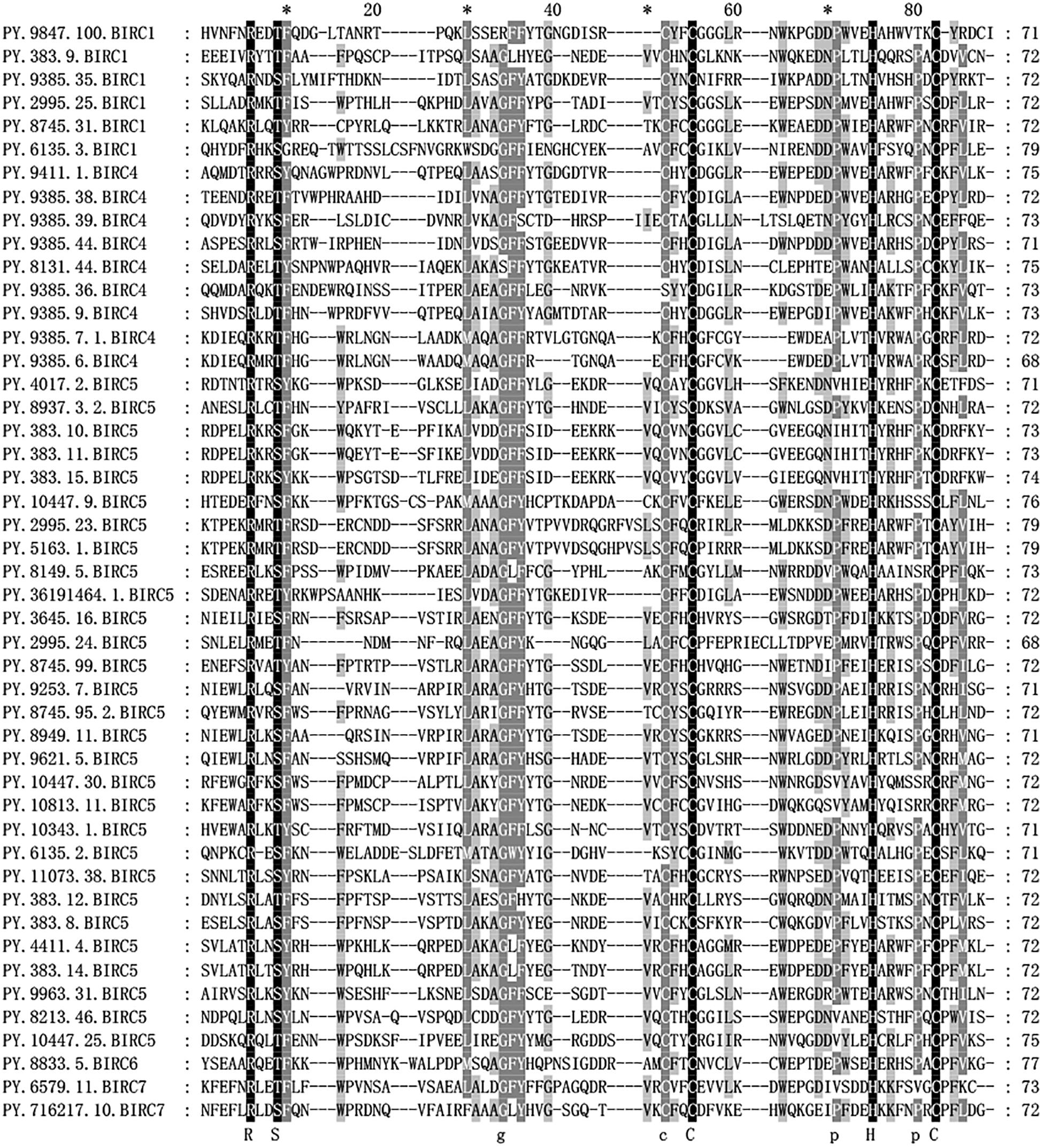
Figure 2. Alignment of Patinopecten yessoensis BIR domain sequences. Black indicates that all sequences have the same amino acid at the same site, gray indicates that most sequences have the same amino acid at the same site (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Inhibitors of apoptosis proteins from eight selected animal species were used to conduct phylogenetic analysis based on NJ method with 1000 bootstrap pseudo-replicates. As shown in Figure 3, all IAP proteins could be subdivided into eight proposed IAP sub-families, including BIRC 1–8 sub-families. The NJ tree clearly showed three distinct cluster of BIRC1, BIRC4, and BIRC5 sub-families (labeled as purple, navy blue, and red, respectively), which were composed of more than half of IAP members from P. yessoensis. The orthologous IAPs of BIRC6 sub-family were clustered together firstly near the phylogenetic root (black labeled clusters), indicating their ancestor state. Two P. yessoensis BIRC7 members together with homologues from other species were first clustered together (light blue), then grouped with the vertebrate BIRC2 (green) and BIRC3 (orange) clusters, implying their closer relationship that may share a same ancestor. Besides, we noticed that, across the species we investigated, there is only one BIRC8 member from human (yellow branch), and it is clustered into BIRC4 sub-family. Previously, it was reported that, within the specific taxonomic lineages, the gene families involved in lineage specific responses to environmental change or microbial attack are usually expanded (Kulmuni et al., 2013; Ellegren, 2014; Zhang et al., 2015). It reminds us that, the obvious gene expansion phenomenon in bivalve IAP gene family, especially in the BIRC1, BIRC4, and BIRC5 sub-families, may indicate the existence of an IAPs-relevant anti-apoptotic system and making IAPs to be good candidates for gene replication and functional diversity research.
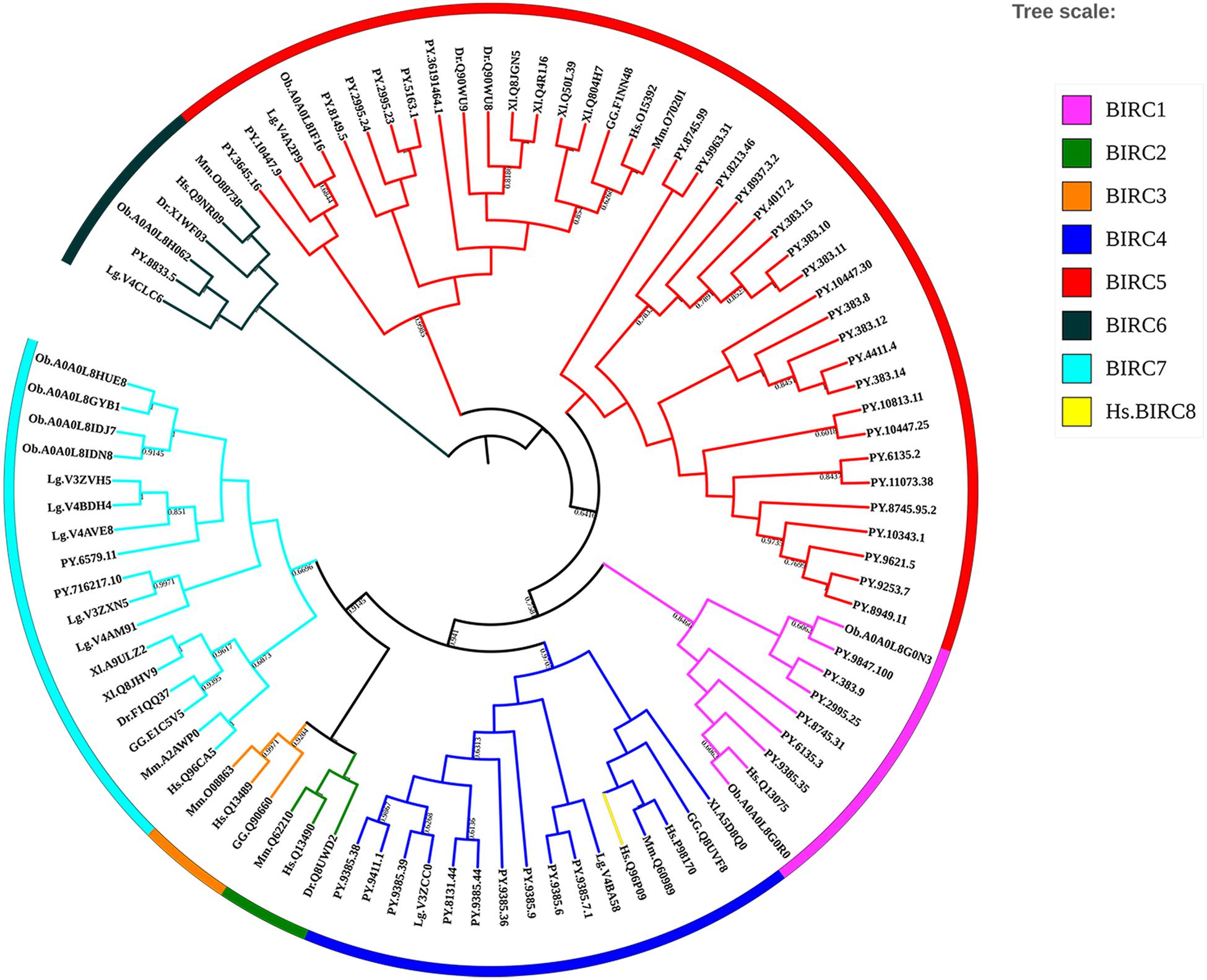
Figure 3. Phylogenetic tree of selected organisms’ IAP amino acid sequences. The trees were constructed using NJ method with bootstrapping of 1000 pseudo replicates, the percentage values were converted to 0-1, and scores below 0.60 were not shown (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article). Species abbreviations: Hs, Homo sapiens; Mm, Mus musculus; GG, Gallus gallus; Xl, Xenopus laevis; Dr, Danio rerio; Lg, Lottia gigantean; Ob, Octopus bimaculoides; PY, Patinopecten yessoensis.
Selective Pressure Analysis of the Expanded PyIAPs Sub-Families
Previous study pointed that, through combination of acquisition of new genes and modification of existing genes driven by positive selection, gene replication and expansion could provide support for the emergence of new functional genes (Kulmuni et al., 2013). These new emerged genes could possibly help organisms adapt to complex living conditions (Zhang et al., 2012; Ellegren, 2014). Here, the three expanded sub-families of PyIAPs (BIRC1, BIRC4 and BIRC5) in Yesso scallops were analyzed for the selective pressure at protein level, using the Non-synonymous/Synonymous ratio (dN/dS or ω) as an important indicator. As the results of Table 2 showed, variable selection pressure was detected to distribute among codons between PyBIRC4 and PyBIRC5 sub-family (p < 0.05). In comparison between the M0 and M3 models, the p-values of the BIRC1, BIRC4 and BIRC5 sub-families are all 0 (less than 0.05), rejecting the M0 model (single ratio) and supporting the M3 model (discrete), demonstrating that it allows for the three sub-families with different ω ratios. Comparisons of M7 (beta) vs. M8 (beta and ω) revealed statistically significance of permitting positive selection for M8 in PyBIRC4 and PyBIRC5 sub-families (p < 0.05), suggesting that it provided consistent evidence for the presence of a small proportion of positively selected sites in the PyBIRC4s and PyBIRC5s. Then, we used the Bayes Empirical Bayes (BEB) approach in PAML to calculate posterior probabilities in order to confirm sites under positive selection (ω > 1) in the M8 model. As shown in Table 2, We have identified six positively selected sites in PyBIRC4 sub-family (p < 0.05) with posterior probability > 95%, suggesting the existence of purifying selection. Scallops are living in highly dynamic marine environment, in order to cope with diverse biotic and abiotic stressors, the scallop’s defense system is expected to change quickly and requires constant adaptive innovations. Gene expansion is a potential source of such innovation (Ellegren, 2014), and the significant expansion of the IAP genes in P. yessoensis may reflect that the requirement of a certain selection pressure for such genes as a strategy of molecular sequence evolution.
Expression Profiles of PyIAP Genes After A. catenella Exposure
As filter feeding species, scallops can accumulate and transform paralytic shellfish toxins (PSTs) from microalgae. Previous reports have shown that, among all organs, hepatopancreas and kidney are the top-ranked tissues with highest accumulation of PST (Tian et al., 2010; Li et al., 2017). In P. yessoensis, compared with hepatopancreas and kidney (∼204 nmol/g), the PSTs accumulation was found far more less in other soft tissues (∼2.8 nmol/g), such as in mantle, gill and muscle, and our preliminary investigation results showed no obvious express change in other soft tissues. Herein, we decided to focus on the responses of PyIAPs in hepatopancreas and kidney. It is reported that hepatopancreas in scallop is the organ mainly to accumulate the incoming toxins directly, while in kidney PSTs can change into higher toxic analogs, such as saxitoxin (STX) and neosaxitoxin (NeoSTX), probably as deterrence against predation (Hu et al., 2019). In order to systematically assess the transcriptional responses of PyIAPs exposed to toxic dinoflagellates, PST-producing A. catenella (strain ACDH) were used as feeding diet and the expression profiling of PyIAPs in both hepatopancreas and kidney was analyzed (Figure 4). After exposure, fourteen PyIAPs showed significant responses in hepatopancreas and kidney, and all of which were from the expanded subfamilies BIRC1, BIRC4 and BIRC5 (two, three and nine members, respectively). The significant regulation pattern of PyIAPs was similar between two tissues, with more than half exhibit expression suppression within three days. Besides, three PyIAPs of BIRC5, one member of BIRC1 and one member of BIRC4 showed significant up-regulation with toxic diet. In contrast to hepatopancreas, more acute responses of PyIAPs to the toxic stress could be detected in kidney, with seventy percent showed acute down-regulation within one day exposure, implying the possible involvement of these PyIAPs in tissue-specific PST tolerance.
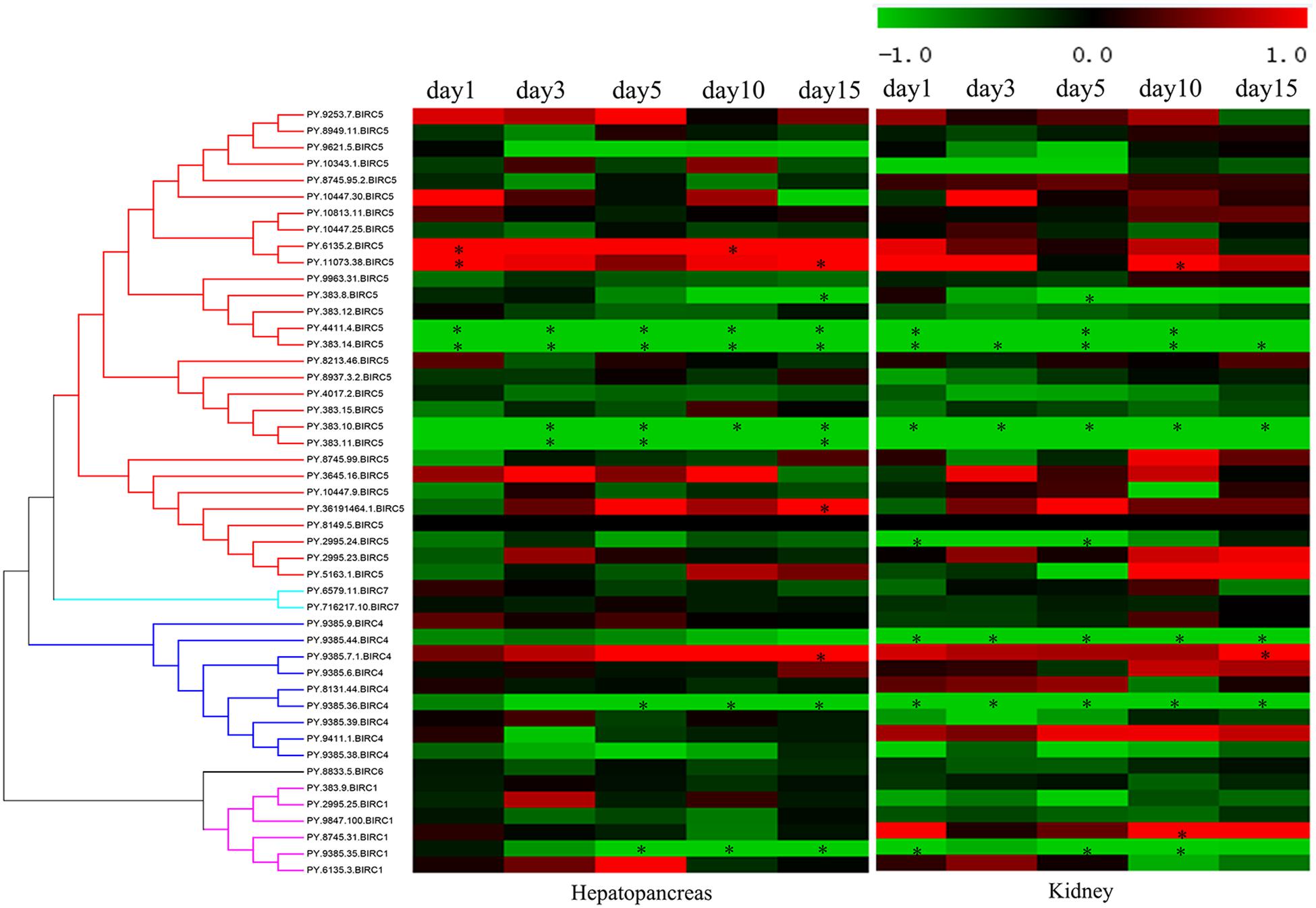
Figure 4. Heatmap of IAPs expression in hepatopancreas and kidney of Patinopecten yessoensis after ACDH exposure at different time points. * indicated significantly up- and down-regulated IAPs from different subfamilies with —log2FC— ≥ 1 and p-value < 0.05 at each test time point (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
In hepatopancreas, the main organ for PST absorption, 46 PyIAPs exhibited detectable expression after ACDH exposure, and seven and four PyIAPs were significantly down- and up-regulated at least at one test time point, respectively (Figure 4, Table 3 and Supplementary Table 3). Among these 11 PyIAPs with significant regulation (| log2FC| ≥ 1, p-value < 0.05), there are eight members from the most expanded BIRC5 subfamily, with six of them showed acute responses within three days. Of note, two members of them (PY.4411.4 and PY.383.14) were rapidly down regulated on day1 and showed continuous suppression till day15, and specially PY.383.14 also showed the highest log2FC changes (| log2FC| reach to 5.91), suggesting their important roles facing toxin accumulation. Besides, three PyBIRC5s were significantly induced, with two members (PY.6135.2, PY.11073.38) being up-regulated on day1 and one member (PY.36191464.1) chronically up-regulated on day15. Other than the BIRC5 subfamily, there were one member of BIRC1 and two members of BIRC4 presented chronic responses, being down-regulated after 5 days exposure or up-regulated on day15.
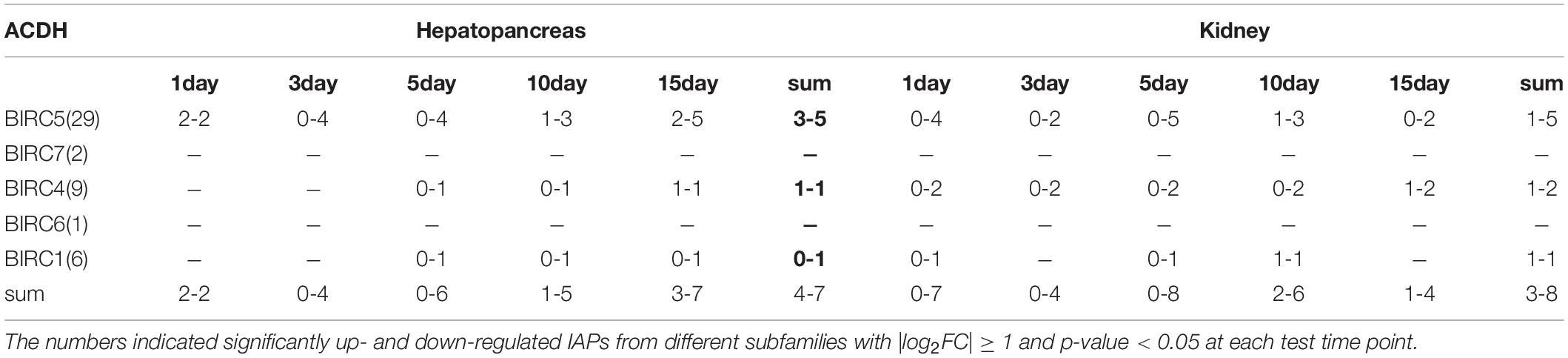
Table 3. Summary of PyIAPs expression in hepatopancreas and kidney after exposure to toxic dinoflagellate ACDH.
In kidney, where PSTs could be transformed to derivatives with higher toxicity, also there were 46 PyIAPs expressed after ACDH exposure, and eight and three PyIAPs were significantly down-regulated and up-regulated at least at one test time point, respectively (Figure 4, Table 3 and Supplementary Table 3). Notably, in comparison with hepatopancreas, more acute down regulations of PyIAPs in kidney could be detected. Seven PyIAPs, including four PyBIRC5s, two PyBIRC4s and one PyBIRC1, reduced their expression significantly within one day exposure and four of them showed continuous suppression till day15, reflecting the sensitive and intensive responses to toxic effects. Especially, PY.383.14 again showed quite high log2FC changes (| log2FC| reach to 5.19), just like it’s performance in hepatopancreas, reinforcing evidence linking its participation during toxic adaption. Besides, three members including PyBIRC1 (PY.8745.31), PyBIRC4 (PY.9385.7.1) and PyBIRC5 (PY.11073.38) were found significantly induced in kidney after five days exposure.
After ACDH exposure, in the two major toxic organs of P. yessoensis, most of the IAP genes with significant altered expression levels were from the expanded BIRC5 and BIRC4 subfamily. Out of the nine significantly down-regulated PyIAPs with toxic diet, four genes are located on scaffold383 and three genes located on scaffold9385. Interestingly, on both of these two scaffolds, a pair of PyIAPs (PY.383.10/383.11 and PY.9385.35/9385.36) that possess the adjacent position could be found, suggesting the IAP genes duplication and expansion in P. yessoensis could have important effects on scallop’s toxin resistance.
Conclusion
In this study, a comprehensive identification and characterization of IAP genes were performed for the first time in Yesso scallop P. yessoensis. Forty-seven PyIAPs from five subfamilies were identified, and significant expansion of subfamilies BIRC4, BIRC5 that under the pressure of purifying selection were revealed. After exposure to PST-producing dinoflagellates, A. catenella, fourteen PyIAPs showed significant responses in hepatopancreas and kidney, and two, three and nine members were from the expanded subfamilies BIRC1, BIRC4 and BIRC5, respectively. Significant regulation pattern of PyIAPs was similar between two tissues, with more than half exhibit expression suppression within three days, and most of which are usually located as gene clusters that distributed on the same scaffold, suggesting the PyIAPs duplication and expansion could have important effects on scallop’s toxin resistant. Besides, in comparison with hepatopancreas, more acute responses of PyIAPs to the toxic stress could be detected in kidney, implying the possible involvement of these PyIAPs in tissue-specific PST tolerance. These findings could assist in a better understanding of the adaptive expansion of bivalve IAP genes to the marine environments with algae derived biotoxins, and providing good candidate members of IAP genes for their further functional study in marine bivalves.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SL, SW, and ZB conceived and designed the study. XZ, FZ, YW, and NH performed the experiments. XZ, XC, and XD participated in data analysis. XZ, SL, and XH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the grant support from National Natural Science Foundation of China (U1706203), Fundamental Research Funds for the Central Universities (202064008), Sanya Yazhou Bay Science and Technology City (SKJC-KJ-2019KY01), and Taishan Scholar Project Fund of Shandong Province of China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.633301/full#supplementary-material
Supplementary Table 1 | IAP protein sequences accession ID of different species used in this study. Sequences were retrieved from Uniprot and NCBI database.
Supplementary Table 2 | Summarized information of IAP genes identified in P. yessoensis genome.
Supplementary Table 3 | Different expression of PyIAPs in hepatopancreas and kidney after A. catenella exposure analyzed using edgeR package.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://www.uniprot.org/
- ^ http://www.ncbi.nlm.nih.gov/orffinder/
- ^ http://smart.embl-heidelberg.de/
- ^ https://www.expasy.org/
- ^ http://gsds.gao-lab.org/index.php
- ^ http://itol.embl.de/
- ^ http://mev.tm4.org
References
Abi-Khalil, C., Finkelstein, D. S., Conejero, G., Du Bois, J., Destoumieux-Garzon, D., and Rolland, J. L. (2017). The paralytic shellfish toxin, saxitoxin, enters the cytoplasm and induces apoptosis of oyster immune cells through a caspase-dependent pathway. Aquat. Toxicol. 190, 133–141. doi: 10.1016/j.aquatox.2017.07.001
Ahmad, H. I., Asif, A. R., Ahmad, M. J., Jabbir, F., Adnan, M., Ahmed, S., et al. (2020). Adaptive evolution of peptidoglycan recognition protein family regulates the innate signaling against microbial pathogens in vertebrates. Microb. Pathog. 147:104361. doi: 10.1016/j.micpath.2020.104361
Ahmad, M. J., Ahmad, H. I., Adeel, M. M., Liang, A., Hua, G., Murtaza, S., et al. (2019). Evolutionary analysis of makorin ring finger protein 3 reveals positive selection in mammals. Evol. Bioinform. Online 15:1176934319834612. doi: 10.1177/1176934319834612
Alvarez, G., Diaz, P. A., Godoy, M., Araya, M., Ganuza, I., Pino, R., et al. (2019). Paralytic shellfish toxins in surf clams Mesodesma donacium during a large bloom of Alexandrium catenella dinoflagellates associated to an intense shellfish mass mortality. Toxins 11:188. doi: 10.3390/toxins11040188
Anderson, D. M., Alpermann, T. J., Cembella, A. D., Collos, Y., Masseret, E., and Montresor, M. (2012). The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 14, 10–35. doi: 10.1016/j.hal.2011.10.012
Borcier, E., Morvezen, R., Boudry, P., Miner, P., Charrier, G., Laroche, J., et al. (2017). Effects of bioactive extracellular compounds and paralytic shellfish toxins produced by Alexandrium minutum on growth and behaviour of juvenile great scallops Pecten maximus. Aquat. Toxicol. 184, 142–154. doi: 10.1016/j.aquatox.2017.01.009
Cheng, J., Xun, X., Kong, Y., Wang, S., Yang, Z., Li, Y., et al. (2016). Hsp70 gene expansions in the scallop Patinopecten yessoensis and their expression regulation after exposure to the toxic dinoflagellate Alexandrium catenella. Fish Shellfish Immunol. 58, 266–273. doi: 10.1016/j.fsi.2016.09.009
Cheung, H. H., Kelly, N. L., Liston, P., and Korneluk, R. G. (2006). Involvement of caspase-2 and caspase-9 in endoplasmic reticulum stress-induced apoptosis: a role for the IAPs. Exp. Cell Res. 312, 2347–2357. doi: 10.1016/j.yexcr.2006.03.027
Contreras, A. M., Marsden, I. D., and Munro, M. H. G. (2011). Physiological effects and biotransformation of PSP Toxins in the New Zealand Scallop, Pecten novaezelandiae. Mar. Freshwater Res. 31, 1151–1159. doi: 10.2983/035.031.0426
Cusick, K. D., and Sayler, G. S. (2013). An overview on the marine neurotoxin, saxitoxin: genetics, molecular targets, methods of detection and ecological functions. Mar. Drugs 11, 991–1018. doi: 10.3390/md11040991
Deveraux, Q. L., and Reed, J. C. (1999). IAP family proteins—suppressors of apoptosis. Genes Dev. 13, 239–252. doi: 10.1101/gad.13.3.239
Domingues, C., and Ryoo, H. D. (2012). Drosophila BRUCE inhibits apoptosis through non-lysine ubiquitination of the IAP-antagonist REAPER. Cell Death Differ. 19, 470–477. doi: 10.1038/cdd.2011.116
Ellegren, H. (2014). Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 29, 51–63. doi: 10.1016/j.tree.2013.09.008
Estrada, N., de Jesus Romero, M., Campa-Cordova, A., Luna, A., and Ascencio, F. (2007). Effects of the toxic dinoflagellate, Gymnodinium catenatum on hydrolytic and antioxidant enzymes, in tissues of the giant lions-paw scallop Nodipecten subnodosus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 146, 502–510. doi: 10.1016/j.cbpc.2007.06.003
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Fulda, S., and Vucic, D. (2012). Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 11, 109–124. doi: 10.1038/nrd3627
Garcia-Lagunas, N., Romero-Geraldo, R., and Hernandez-Saavedra, N. Y. (2013). Genomics study of the exposure effect of Gymnodinium catenatum, a paralyzing toxin producer, on Crassostrea gigas’ defense system and detoxification genes. PLoS One 8:e72323. doi: 10.1371/journal.pone.0072323
Gentle, I. E., Moelter, I., Lechler, N., Bambach, S., Vucikuja, S., Hacker, G., et al. (2014). Inhibitors of apoptosis proteins (IAPs) are required for effective T-cell expansion/survival during antiviral immunity in mice. Blood 123, 659–668. doi: 10.1182/blood-2013-01-479543
Grzybowska-Izydorczyk, O., and Smolewski, P. (2008). The role of the inhibitor of apoptosis protein (IAP) family in hematological malignancies. Postepy Hig. Med. Dosw. (Online) 62, 55–63. doi: 10.1117/12.764110
Hu, B., Li, M., Yu, X., Xun, X., Lu, W., Li, X., et al. (2019). Diverse expression regulation of Hsp70 genes in scallops after exposure to toxic Alexandrium dinoflagellates. Chemosphere 234, 62–69. doi: 10.1016/j.chemosphere.2019.06.034
Jones, G., Jones, D., Zhou, L., Steller, H., and Chu, Y. (2000). Deterin, a new inhibitor of apoptosis from Drosophila melanogaster. J. Biol. Chem. 275, 22157–22165. doi: 10.1074/jbc.M000369200
Kindt, N., Menzebach, A., Van de Wouwer, M., Betz, I., De Vriese, A., and Conway, E. M. (2008). Protective role of the inhibitor of apoptosis protein, survivin, in toxin-induced acute renal failure. FASEB J. 22, 510–521. doi: 10.1096/fj.07-8882com
Kulmuni, J., Wurm, Y., and Pamilo, P. (2013). Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity (Edinb) 110, 538–547. doi: 10.1038/hdy.2012.122
Landsberg, J. H. (2002). The effects of harmful algal blooms on aquatic organisms. Rev. Fish. Sci. 10, 113–390. doi: 10.1080/20026491051695
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leulier, F., Lhocine, N., Lemaitre, B., and Meier, P. (2006). The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol. Cell Biol. 26, 7821–7831. doi: 10.1128/MCB.00548-06
Li, Y., Sun, X., Hu, X., Xun, X., Zhang, J., Guo, X., et al. (2017). Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 8:1721. doi: 10.1038/s41467-017-01927-0
Liu, W., Xie, Y., Ma, J., Luo, X., Nie, P., Zuo, Z., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Luque, L. E., Grape, K. P., and Junker, M. (2002). A highly conserved arginine is critical for the functional folding of inhibitor of apoptosis (IAP) BIR domains. Biochemistry 41, 13663–13671. doi: 10.1021/bi0263964
Medhioub, W., Ramondenc, S., Vanhove, A. S., Vergnes, A., Masseret, E., Savar, V., et al. (2013). Exposure to the neurotoxic dinoflagellate, Alexandrium catenella, induces apoptosis of the hemocytes of the oyster, Crassostrea gigas. Mar. Drugs 11, 4799–4814. doi: 10.3390/md11124799
Mohamed, M. S., Bishr, M. K., Almutairi, F. M., and Ali, A. G. (2017). Inhibitors of apoptosis: clinical implications in cancer. Apoptosis 22, 1487–1509. doi: 10.1007/s10495-017-1429-4
Navarro, J. M., Muñoz, M. G., and Contreras, A. M. (2006). Temperature as a factor regulating growth and toxin content in the dinoflagellate Alexandrium catenella. Harmful Algae 5, 762–769. doi: 10.1016/j.hal.2006.04.001
Nicholas, K. B., Nicholas, H. B., and Deerfield, D. W. (1997). GeneDoc: analysis and visualization of genetic variation. Embnew News 4, 28–30. doi: 10.11118/actaun201361041061
Oberoi-Khanuja, T. K., and Rajalingam, K. (2012). IAPs as E3 ligases of Rac1: shaping the move. Small GTPases 3, 131–136. doi: 10.4161/sgtp.19988
Pistritto, G., Trisciuoglio, D., Ceci, C., Garufi, A., and D’Orazi, G. (2016). Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 8, 603–619. doi: 10.18632/aging.100934
Robertson, A. J., Croce, J., Carbonneau, S., Voronina, E., Miranda, E., McClay, D. R., et al. (2006). The genomic underpinnings of apoptosis in Strongylocentrotus purpuratus. Dev. Biol. 300, 321–334. doi: 10.1016/j.ydbio.2006.08.053
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Salvesen, G. S., and Duckett, C. S. (2002). IAP proteins: blocking the road to death’s door. Nat. Rev. Mol. Cell Biol. 3, 401–410. doi: 10.1038/nrm830
Silke, J., and Meier, P. (2013). Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation. Cold Spring Harb Perspect. Biol. 5, a008730. doi: 10.1101/cshperspect.a008730
Sullivan, J. C., Ryan, J. F., Watson, J. A., Webb, J., Mullikin, J. C., Rokhsar, D., et al. (2006). StellaBase: the nematostella vectensis genomics database. Nucleic Acids Res. 34, D495–D499. doi: 10.1093/nar/gkj020
Takahashi, R., Deveraux, Q., Tamm, I., Welsh, K., Assa-Munt, N., Salvesen, G. S., et al. (1998). A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem. 273, 7787–7790. doi: 10.1074/jbc.273.14.7787
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tian, H., Gao, C., Wang, Z., Sun, P., Fan, S., and Zhu, M. (2010). Comparative study on in vitro transformation of paralytic shellfish poisoning (PSP) toxins in different shellfish tissues. Acta Oceanol. Sinica 29, 120–126. doi: 10.1007/s13131-010-0015-1
Vilarino, N., Louzao, M. C., Fraga, M., Rodriguez, L. P., and Botana, L. M. (2013). Innovative detection methods for aquatic algal toxins and their presence in the food chain. Anal. Bioanal. Chem. 405, 7719–7732. doi: 10.1007/s00216-013-7108-6
Vucic, D. (2008). Targeting IAP (inhibitor of apoptosis) proteins for therapeutic intervention in tumors. Curr. Cancer Drug Targets 8, 110–117. doi: 10.2174/156800908783769373
Wang, H., Huang, R., Yang, K., Xu, M., Fan, D., Liu, M., et al. (2019). Identification of differentially expressed genes and preliminary validations in cardiac pathological remodeling induced by transverse aortic constriction. Int. J. Mol. Med. 44, 1447–1461. doi: 10.3892/ijmm.2019.4291
Wang, K., and Lin, B. (2013). Inhibitor of apoptosis proteins (IAPs) as regulatory factors of hepatic apoptosis. Cell Signal 25, 1970–1980. doi: 10.1016/j.cellsig.2013.06.003
Wang, S., Zhang, J., Jiao, W., Li, J., Xun, X., Sun, Y., et al. (2017). Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1:120. doi: 10.1038/s41559-017-0120
Wang, W., Ji, Y., Yang, W., Zhang, C., Angwa, L., Jin, B., et al. (2020). Inhibitors of apoptosis proteins (IAPs) are associated with T-2 toxin-induced decreased collagen II in mouse chondrocytes in vitro. Toxicon 176, 34–43. doi: 10.1016/j.toxicon.2020.01.002
Xun, X., Cheng, J., Wang, J., Li, Y., Li, X., Li, M., et al. (2020). Solute carriers in scallop genome: gene expansion and expression regulation after exposure to toxic dinoflagellate. Chemosphere 241:124968. doi: 10.1016/j.chemosphere.2019.124968
Yang, S., Wei, J., and He, F. (2018). Risk assessment indexes for shellfish poisoning outbreak caused by red tide. J. Zhejiang Univ. Med. Sci. 47, 111–117.
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhang, L., Li, L., Guo, X., Litman, G. W., Dishaw, L. J., and Zhang, G. (2015). Massive expansion and functional divergence of innate immune genes in a protostome. Sci. Rep. 5:8693. doi: 10.1038/srep08693
Keywords: Patinopecten yessoensis, IAPs, gene expansion, PST exposure, expression regulation
Citation: Zhu X, Zhang F, Lian S, Wang Y, Hu N, Chen X, Dai X, Hu X, Wang S and Bao Z (2021) IAPs Gene Expansion in the Scallop Patinopecten yessoensis and Their Expression Profiles After Exposure to the Toxic Dinoflagellate. Front. Physiol. 12:633301. doi: 10.3389/fphys.2021.633301
Received: 25 November 2020; Accepted: 15 January 2021;
Published: 05 February 2021.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Chunde Wang, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences (CAS), ChinaGuangxu Liu, Zhejiang University, China
Copyright © 2021 Zhu, Zhang, Lian, Wang, Hu, Chen, Dai, Hu, Wang and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanshan Lian, bGlhbnNoYW5zaGFuQG91Yy5lZHUuY24=
†These authors have contributed equally to this work
 Xiaomei Zhu1†
Xiaomei Zhu1† Shanshan Lian
Shanshan Lian Xiaoting Dai
Xiaoting Dai Xiaoli Hu
Xiaoli Hu Shi Wang
Shi Wang Zhenmin Bao
Zhenmin Bao