- 1Key Laboratory of Freshwater Aquatic Genetic Resources, Ministry of Agriculture and Rural Affairs, Shanghai Ocean University, Shanghai, China
- 2National Demonstration Center for Experimental Fisheries Science Education, Shanghai, China
- 3Shanghai Engineering Research Center of Aquaculture, Shanghai, China
Transformer-2 (Tra-2) is an upstream regulatory element of the sex regulation mechanism in insects and plays a critical role in sex formation. To understand the role of tra-2 in Hyriopsis cumingii, the full-length Hctra-2 (1867 bp) was obtained from the gonads, and sequence alignment with other species showed that HCTRA-2 protein had a highly conserved RRM domain. Phylogenetic analysis showed that the HCTRA-2 protein was a close relative to of the mollusks TRA-2 protein. The qRT-PCR of tissue-specific expression pattern showed that the Hctra-2 was abundant in gonads, and the expression in testes was higher than that in ovaries (p < 0.01). It suggests that Hctra-2 may play a potential regulatory role in gonadal development of H. cumingii. In the early gonadal development, the Hctra-2 expression was the highest on the third day after fertilization and increased slightly from 4 months to 5 months, which may be related to the embryonic sex determination and early gonadal development. In situ hybridization showed that Hctra-2 mRNA signals were present in both male and female gonads. After silencing Hctra-2 by RNAi, the expression levels of Hcfem-1b and Hcdmrt were changed. It is speculated that there may be a certain relationship between them, which plays an important role in the sex regulation of H. cumingii. Our research will help to deepen our understanding of the shellfish sex determination mechanisms.
Introduction
Hyriopsis cumingii is a pearl-cultured species unique to China. The pearl cultivated by it has smooth and delicate nacre quality and bright color, which is the best one among freshwater mussels (Wang et al., 2007). Interestingly, male and female mussels showed different traits in the culture, and the quality of pearl produced by males was better than that of females (Zhao et al., 2013). Therefore, it is of great significance to raise the pearl yield and quality by using sex control technology. However, the research on the regulation mechanism of H. cumingii sex determination is limited, which hinders the development of sex control breeding technology.
In shellfish, there are androgyny, hermaphroditism, or sex reversal in their sex (Yue et al., 2020). This complex and changeable sex type provides us with a lot of rich research resources and brings some challenges to the study of sex. In recent years, the rapid development of sequencing technology has brought new hope to shellfish sex research. Some genes related to sex determination and differentiation have also been identified in shellfish, such as foxl2, tra, dmrt, fem-1, soxe, soxh, and wnt4 (Naimi et al., 2009; Shi et al., 2015, 2018; Li et al., 2016; Galindo-Torres et al., 2018). Among these genes, the tra was an upstream component of the sex-regulatory mechanism in many species, playing a crucial role in the regulation of sex formation mechanisms. In Caenorhabditis elegans, tra-1 and tra-2 promote female fates (Wang and Kimble, 2001). The regulatory pathway of sex determination is mainly composed of her-1, tra-2, fem-1, fem-2, fem-3, and tra-1. The specific regulatory mechanism is as follows: the ratio of sex chromosome and autosome is the primary signal of sex regulation (Farboud et al., 2013). The signal acts on her-1, which is differentially expressed between two genders. In male XO individuals, her-1 expression is very high, which inhibits the transmembrane receptor protein of TRA-2. Then, the complex formed by the proteins of FEM-1, FEM-2, FEM-3, and CUL-2 binds with the specific site of tra-1, which makes the individual develop into male. In XX hermaphrodite, her-1 expression is very low or even not expressed, so the TRA-2 protein is active. The combination of TRA-2 and the complex containing FEM-1, FEM-2, and FEM-3 proteins makes the expression of TRA-1 protein, and the organism develops toward hermaphroditism (Kuwabara, 2007; Starostina et al., 2007; Mapes et al., 2010). In Drosophila, the regulation of sex is as follows: X: A > sex lethal (sxl) > tra/tra-2 > doublesex (dsx), and fruitless (fru) (Suzuki, 2010). Sex determination is determined by the ratio of X chromosome number to autosomal number. In females, the presence of two X chromosomes activated the expression of sxl. Subsequently, sxl regulates the splicing of tra pre-mRNA transcripts to produce RNA encoding full-length and functional TRA proteins (Sosnowski et al., 1989). Together with tra-2, tra promotes female-specific splicing of dsx and prevents fru synthesis (Camara et al., 2019). In drones, the sxl is inactivated, resulting in the lack of active TRA proteins. Therefore, dsx and fru precursor mRNA default to male-specific splicing, and ontogeny is male (Grmai et al., 2018).
In aquatic species, there are few studies on tra-2, and its action on sex is not clear. In Macrobrachium nipponense, Mntra-2a was highly expressed in both male and female gonads, and its expression was located in oocytes and spermatocytes. It may play an important role in the embryonic development and early gonadal development (Wang et al., 2019). In Fenneropenaeus chinensis, Fctra-2c may participate in female sex determination in a concentration-dependent manner (Li et al., 2012). In medaka (Oryzias latipes), tra-2 transcripts (tra-2a and tra-2b) were mainly expressed in hermaphroditic germ cells before and during their sex differentiation, indicating that both tra2a and tra2b may be involved in the sex differentiation (Shiraishi et al., 2004). Cqtra-2 may play a role in sexual differentiation in the redclaw crayfish (Cherax quadricarinatus), which is mainly expressed in the ovary and gradually increases with embryonic development (Cai et al., 2020).
In this study, we identified Hctra-2 in H. cumingii and analyzed their expression distribution in different tissues and developmental stages of males and females. Besides, the effect of Hctra-2 silencing via RNAi on the expression of Hcfem-1b and Hcdmrt was investigated. These results will be helpful to understand the sex regulation mechanism of H. cumingii, and provide a basis for exploring the sex of shellfish.
Materials and Methods
Animals and Sample Preparation
All the samples used in the study (including juveniles at different developmental stages and mature mussels) were from Zhejiang Weiming aquaculture farm. All experimental processes were approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Ocean University, Shanghai, China.
The samples were taken back from the farm to the laboratory and placed in the water at 26 ± 2°C for 3 days. The gonads, gills, liver, kidney, mantle, foot, and adductor tissues of adult mussels were collected. Embryonic samples were scraped from the gills of H. cumingii at 1 day, 3 days, 5 days, 7 days, 9 days, and 10 days after fertilization. In the juvenile, samples were taken periodically according to different ages. All samples were immediately frozen in liquid nitrogen and stored at - 80°C for RNA extraction.
Full-Length Cloning of Hctra-2
Hctra-2 was derived from the transcriptome library. First, we verified the correctness of partial Hctra-2 sequence. Total RNA was extracted from frozen tissues with RNA prep pure tissue Kit (Tiangen, China). The cDNA was synthesized according to the manufacturer’s protocol of the PrimeScript RT reagent Kit with gDNA Eraser kit (TaKaRa, Japan). Using Tra-F and Tra-R (Table 1) as primers, the sequence of Hctra-2 was amplified by PCR under the following conditions: 94°C for 3 min, followed by 35 cycles of 95°C for 30 s denaturation, 55°C for 30 s of annealing, and 72°C for 2 min extension. After PCR, the amplified targeted-DNA was subcloned into the pMD19 vector (TaKaRa, Japan) for sequence confirmation. The full-length cDNA sequence of Hctra-2 was amplified by the SMARTer RACE 5'/3' Kit (Clontech, United States). The primers used for race amplification were T2-3' and T2-5', and the sequences are shown in Table 1. The reaction products obtained from the RACE kit instructions were purified by 1% agarose gel electrophoresis and connected to the carrier pMD19 vector and then transformed into Escherichia coli DH5α competent cells. Finally, the positive clones were screened for sequencing. Splicing 3' and 5' sequences to obtain the full-length sequence of Hctra-2.
Bioinformatics Analysis of Hctra-2
The open reading frame (ORF) Finder program performs1 open reading frame determination and amino acid sequence acquisition of Hctra-2. The nucleotide and amino acid sequence similarity between Hctra-2 sequence and homologous species was analyzed by BLAST program2; TMHMM Server v2.0 program3 predicted transmembrane structure of prediction protein; SignalP 4.14 predicted the presence and the location of signal peptides in prediction protein; ProtParam program5 predicted various physical and chemical parameters of protein; and ClustalW 1.8 and BioEdit were used for multiple comparisons of amino acid and coding nucleotide sequences. The phylogenetic tree was constructed by Neighbor-joining (NJ; Zhang and Sun, 2008) method in Mega 6.0, and the confidence values among species were calculated by Bootstrap repeated 1,000 times (Zhang and Sun, 2008).
Expression Studies of Hctra-2
Quantitative real-time reverse transcription PCR was used to analyze the expression level of Hctra-2 in different tissues and years of H. cumingii. The EF-lα was used as the internal reference. qT2-F and qT2-R were used as primers. cDNA synthesis and quantification of each mixed sample were performed using PrimeScript RT reagent Kit with gDNA Eraser kit (TaKaRa, Japan) and TB Green Premix Ex Taq II (Takara, Japan), respectively. The total qRT-PCR volume of 20 ul contained 10 ul TB Green Premix Ex Taq, 0.8 ul forward primer, 0.8 ul reverse primer, 6.8 ul ddH2O, and 1.6 ul cDNA. Amplification was performed using a BIO-RAD CFX96 instrument under the following conditions: pre-denaturation at 95°C for 15 min, degeneration at 95°C for 10 s, annealing at 60°C for 30 s (40 cycles), collecting signals during the extension phase, and each cycle increased 0.5°C for 5 s from 65°C to 95°C, followed collecting the fluorescence signal of the dissolution curve. The target gene and reference gene’s relative expression level was calculated using 2 −△△CT method with three replicates for each group. Use Prism 8.0 software to draw pictures. T-test calculated significant difference; p < 0.05 was considered statistically significant.
In situ Hybridization
In the experimental group, the probe primers of Hctra-2 were qT2-F and IT2-R (Table 1). In the control group, the probe primers were IT2-F and qT2-R (Table 1). In vitro transcription was performed using a T7 High Efficiency Transcription Kit (Transgen, China). DIG RNA Labeling Mix was used for probe labeling. Samples of mature gonads (male and female; 2 years old) were fixed twice in 4% paraformaldehyde, then transferred to 20% sucrose solution, sliced with a cryo slicer, and stored at −20°C. In situ hybridization was performed according to the Enhanced Sensitive ISH Detection Kit II (Boster, United States). Hybridization signals were observed and photographed under the microscope.
Knockdown of Hctra-2 by dsRNA-Mediated RNA Interference
Double-stranded RNA (dsRNA) of Hctra-2 was prepared for the experiment of H. cumingii injection. The primers were dsTra-F1, dsTra-R1, dsTra-F2, and dsTra-R2 (Table 1). Firstly, the target sequence was obtained by PCR amplification, and the single-stranded RNA (ssRNA) was synthesized by T7 transcription kit in vitro. Then, dsRNA was synthesized according to the Ribo RNAMax-T7 in vitro transcription kit. The steps are as follows: mix the same amount of ssRNA into the PCR instrument and heat at 70°C for 10 min resulted in the binding of two reverse complementary ssRNAs to form dsRNA. The excess ssRNA was removed according to the procedure of Ribo RNAMax-T7, and dsRNA was dissolved in 1 × PBS. The control dsRNA was GFP sequence which had no homology with Hctra-2, and the primer sequence is shown in Table 1.
The experiment was divided into two groups: the experimental and the control groups, each with 10 mussels. Farm them in plastic bins. On day 1, day 6, and day 11, 100 μl (400 ng/μl) Hctra-2 dsRNA was injected into the adductor of H. cumingii using a microsyringe of 1 ml, respectively. RNA was extracted from gonads, and the expression of Hctra-2, Hcfem-1b, and Hcdmrt after injection of Hctra-2 dsRNA was detected by qRT-PCR. The primers for Hcfem-1b and Hcdmrt were designed based on the sequencing information of the transcriptome data of H. cumingii. The details are shown in Table 1.
Results
Molecular Identification of Hctra-2 and Sequence Analysis
A partial sequence (about 600 bp) of Hctra-2 cDNA was amplified by PCR using primers Tra-F and Tra-R. Based on the cDNA sequence, 5' and 3' RACE gene-specific primers (T2-3' and T2-5') were designed to amplify the 3' and 5' untranslated region sequence. After sequence splicing, the Hctra-2 full-length cDNA 1867 bp (GenBank no. MH931228) was obtained, in which the ORF was 894 bp, the 5' untranslated region (UTR) was 92 bp, and the 3' UTR was 881 bp (Figure 1). The ORF of Hctra-2 was predicted to encode a protein with 297 amino acids with a predicted molecular weight of 34661.2 and a theoretical isoelectric point of 11.02. The protein has no signal peptide or transmembrane structure. The 65–125 amino acid of Hctra-2 protein is the SR1 domain, and the 128–226 amino acid is a conserved RNA recognition motif (RRM) domain, followed by the linker region (amino acid position 227–243; Figure 1).

Figure 1. Nucleotide and deduced amino acid sequence of Hctra-2 gene from Hyriopsis cumingii. Lowercase indicated 3' and 5' UTRs; uppercases was coding region, where the upper indicated the nucleotides sequence and the lower showed the amino acids. *Represented stop codon; putative polyadenylation signals (AATAAA) were underlined. The gray region was the RRM domain. The green region shows the SR1 domain. The blue region shows the linker.
Multiple Alignment and Phylogenetic Analysis
The amino acid sequence of Hctra-2 was analyzed by blast. The results showed that the similarity of Hctra-2 among different species was high (65–79%), among which Mizuhopecten yessoensis (79.66%) was the highest. Even in humans and mice, the sequence similarity reached 69.6 and 54.7%. TRA-2 sequences of Mytilus coruscus (CAC5410763.1), Crassostrea gigas (XP_034314418.1), M. yessoensis (OWF38207.1), and Pecten maximus (XP_033761757.1) were selected from NCBI database. ClustalW in BioEdit software was used to compare the multi-sequence alignment of TRA-2. The alignment results showed that the TRA-2 of most shellfish was composed of about 300 amino acids (Figure 2), all of which contained the RRM domain, which was highly conserved and located in the same position. Similar to other species, the ribonucleoprotein 1 (RNP-1) has also been found in HCTRA-2, which is very conserved in RRM proteins (Shiraishi et al., 2004; Hu et al., 2020). Phylogenetic tree analysis showed that TRA-2 protein of H. cumingii clustered with C. gigas, M. coruscus, and other bivalves. Mollusks, such as Octopus sinensis and Sepia pharaonis, are classified with fish and mammals (Figure 3).
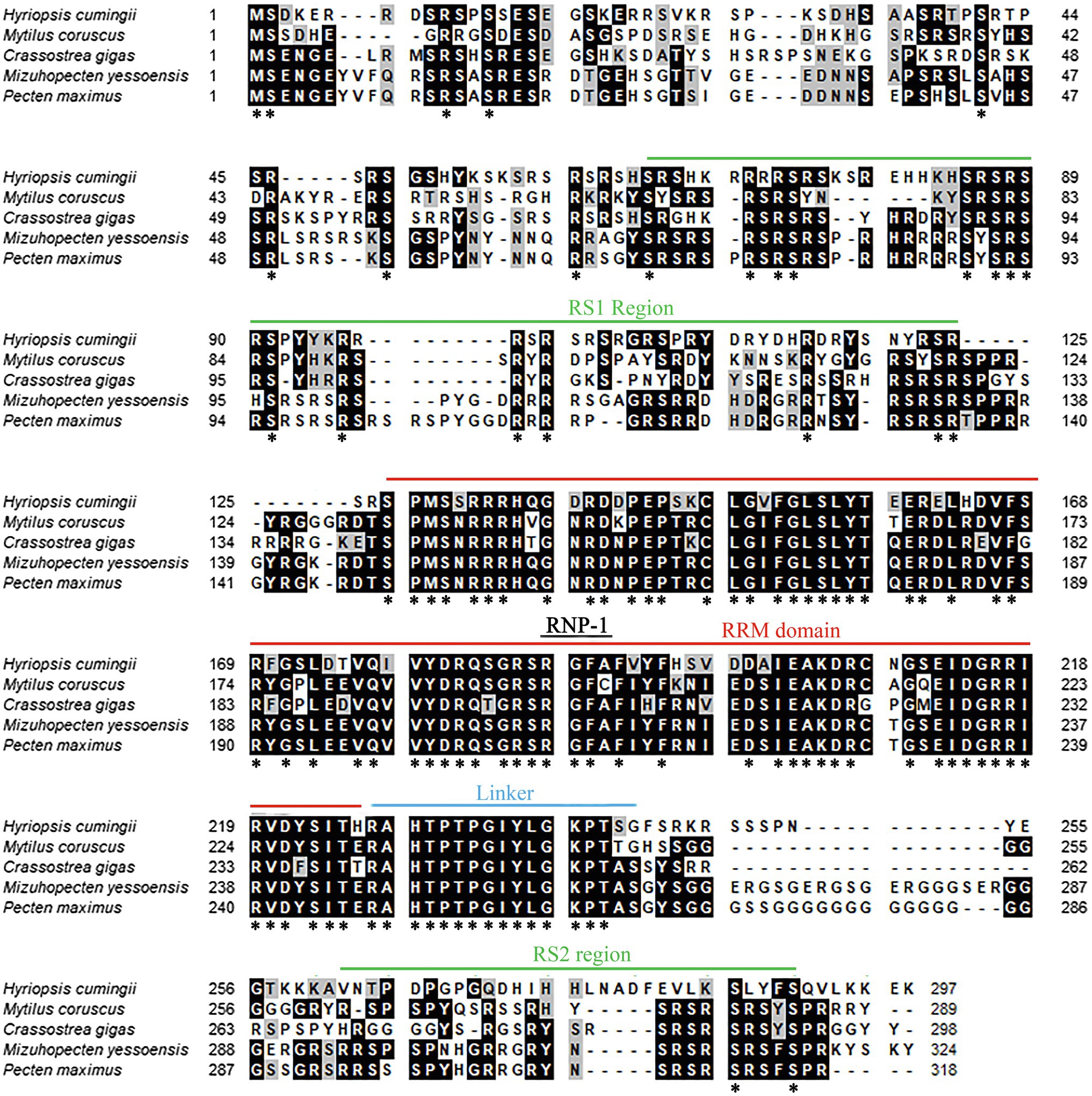
Figure 2. Multiple alignment of Hctra-2 amino acid sequence from H. cumingii with other species. Identical amino acid residues are highlighted in black, and similar amino acids are highlighted in gray. *Means that the amino acids at this site are the same in all the species compared.
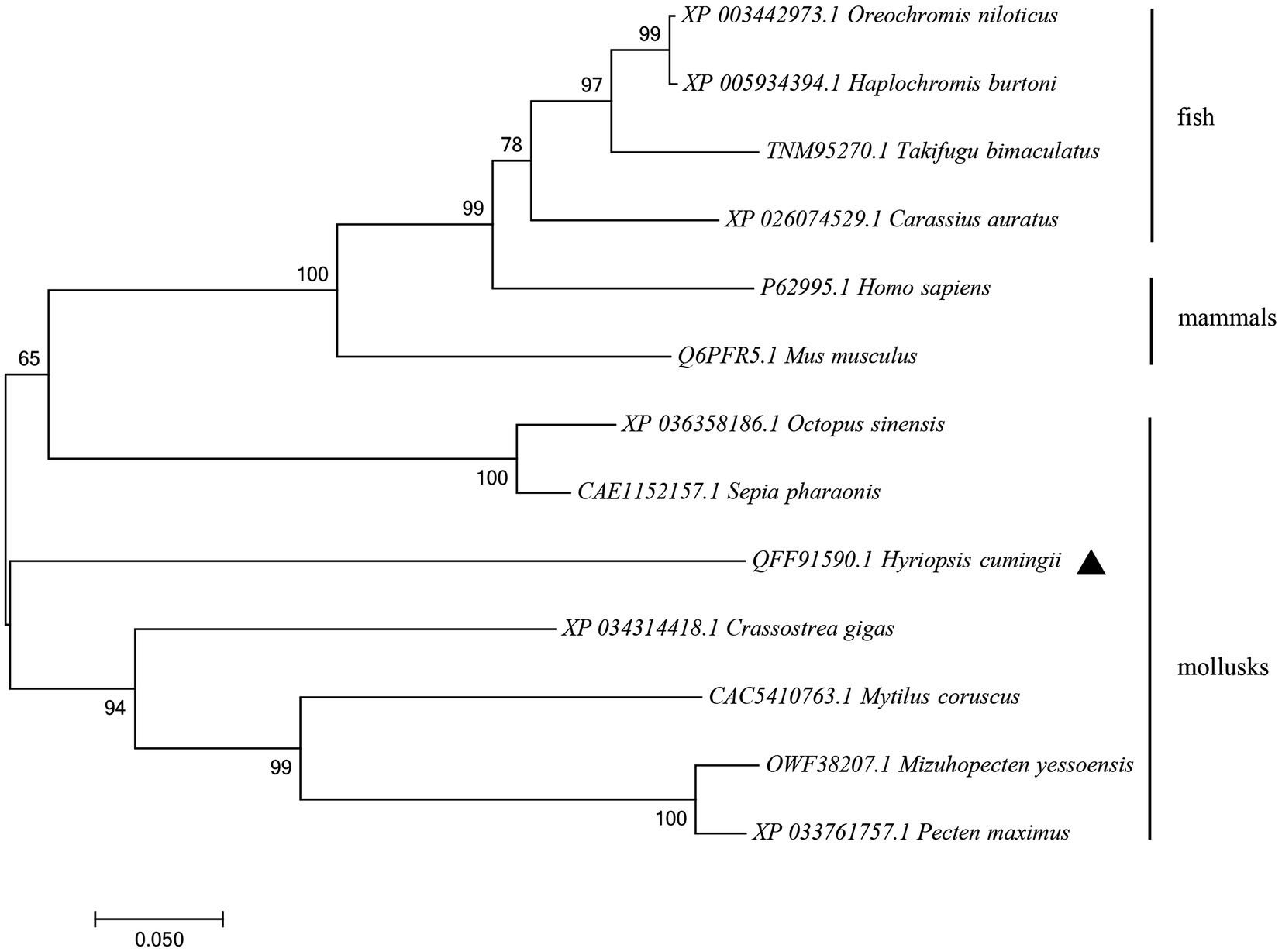
Figure 3. NJ phylogenetic tree of amino acid sequences of Hctra-2 gene in different species. NJ law contribution was made by MEGA6.0; the number on the node was bootstrap test confidence value repeated 1,000 times.
Tissue-Specific Distribution of Hctra-2 in Adult
Quantitative real-time reverse transcription PCR was performed using primers spanning the ORF of Hctra-2. In all tissues of female mussels, the expression of Hctra-2 was high in gonad and liver, followed by gill and kidney, and lower in adductor, mantle, and foot. In male mussels, the expression of Hctra-2 was higher in gonad and liver, then in adductor and gill, and lower in kidney, mantle, and foot. Compared with male and female mussels, except for kidney and foot, the expression of Hctra-2 was significantly different (p < 0.05), especially in gonad, liver, adductor muscle, and mantle (p < 0.01; Figure 4).
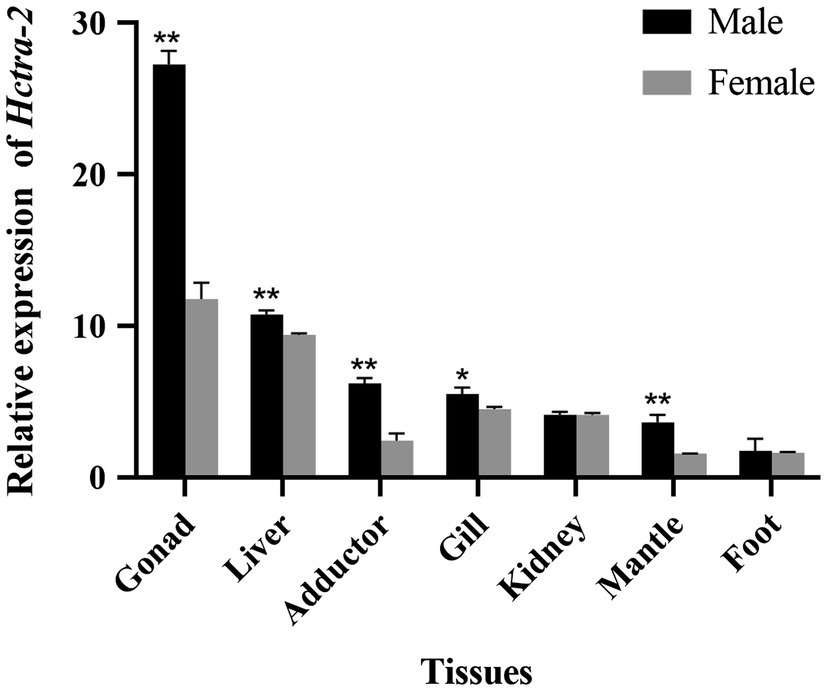
Figure 4. Tissues expression of Hctra-2 gene in adult male and female H. cumingii. Results are expressed as mean ± SD, and significance of comparison is defined as p < 0.05 (*) or p < 0.01 (**) by Student’s t-tests.
Expression Pattern in Different Developmental Stages of Embryos and Juveniles
Hctra-2 expression at different developmental stages in embryos and juveniles (from the fertilized egg up to 8 months old) was detected. The results showed that the expression of Hctra-2 was high in the fertilized eggs, then reached the highest level on the third day, decreased rapidly on the sixth day, and remained at a low steady level until 3 months old. The Hctra-2 expression level increased slightly at the age of 4 to 5 months old and tended to be stable at 6 months old (Figure 5).
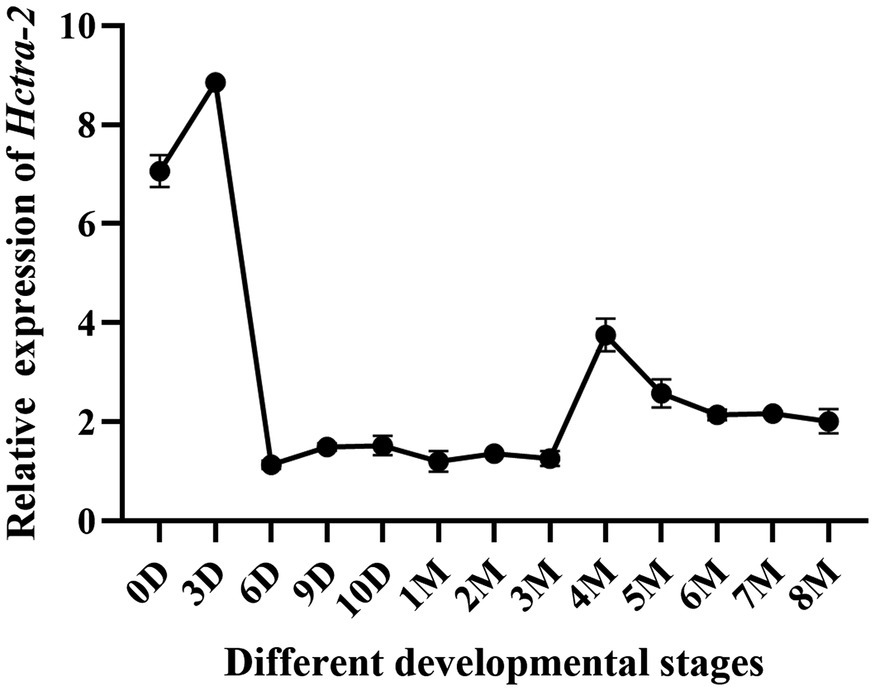
Figure 5. Relative expression of Hctra-2 gene in different developmental periods. (D: days; 0–10 days is embryonic period; M: months; 1–8 mouths is Juveniles).
Localization of Hctra-2 in Mature Gonads
Cell localization of Hctra-2 in testis and ovary was detected by in situ hybridization. Figures 6A,D show the testes and ovaries morphology of H. cumingii stained with HE (hematoxylin-eosin). The male germ cells (from spermatogonia to mature sperm) and female germ cells (from oocyte to mature egg) at different developmental stages can be seen in the figure. Figures 6B,C are the control group and the experimental group of male mussels, respectively. Compared with the control group, the Hctra-2 mRNA purple signals in the experimental group were obvious, and the signals were located on spermatogonia and spermatocytes, but no signal was found on spermatids and sperms. Figures 6E,F are the control group and the experimental group in the female gonad, respectively. Compared with the control group, the hybridized signal of Hctra-2 mRNA appeared in the nucleus and membrane of oocytes and mature ova in the experimental group.
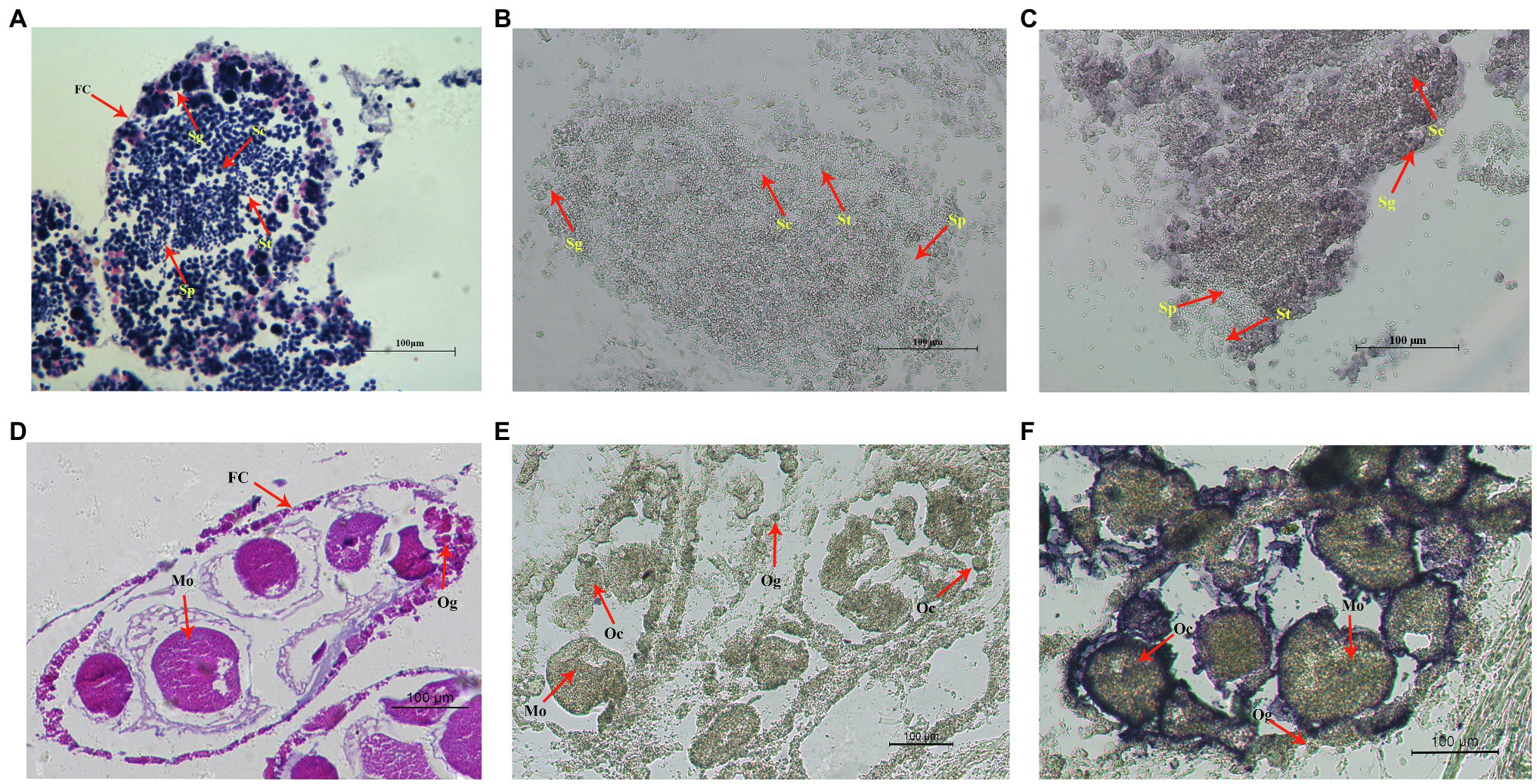
Figure 6. In situ hybridization of Hctra2 gene in testes and ovaries in H. cumingii. (A) testes stained with HE. (B) Male control group. The sense probes acting on testes. (C) Male experience group. The antisense probes acting on testes. (D) Ovaries stained with HE. (E) Female control group. The sense probes acting on ovaries. (F) Female experience group. The antisense probes acting on ovaries. Fc, follicles; Sg, spermatogonia; Sc, spermatocyte; St, spermatid; Sp, sperm; Og, oogonium; Oc, oocyte; and Mo, mature ovum.
The Expression Profile of Hctra-2, Hcfem-1b, and Hcdmrt After RNAi
We studied the role of Hctra-2 in the sex regulation of H. cumingii by RNAi. The dsRNA transcribed and synthesized in vitro was injected into the gonads of H. cumingii. The results showed that the silencing efficiency of the dsHctra-2 was 70.6% in males and 55.8% in females (Figures 7A,B), which indicated that the dsRNA-mediated gene silencing was effective. We also observed the changes in expression of Hcfem-1b and Hcdmrt after Hctra-2 knockout. Compared with the negative control group, the expression level of the Hcfem-1b in testes and ovaries decreased significantly, and the silencing efficiency reached 79.9% in males and 79.0% in females (Figure 7C,D). The expression level of the Hcdmrt in the male gonads of the experimental group was 44.7% lower than that of the control group (Figure 7E), and the expression level in female gonads was 16.7% higher than that in the control group (Figure 7F).
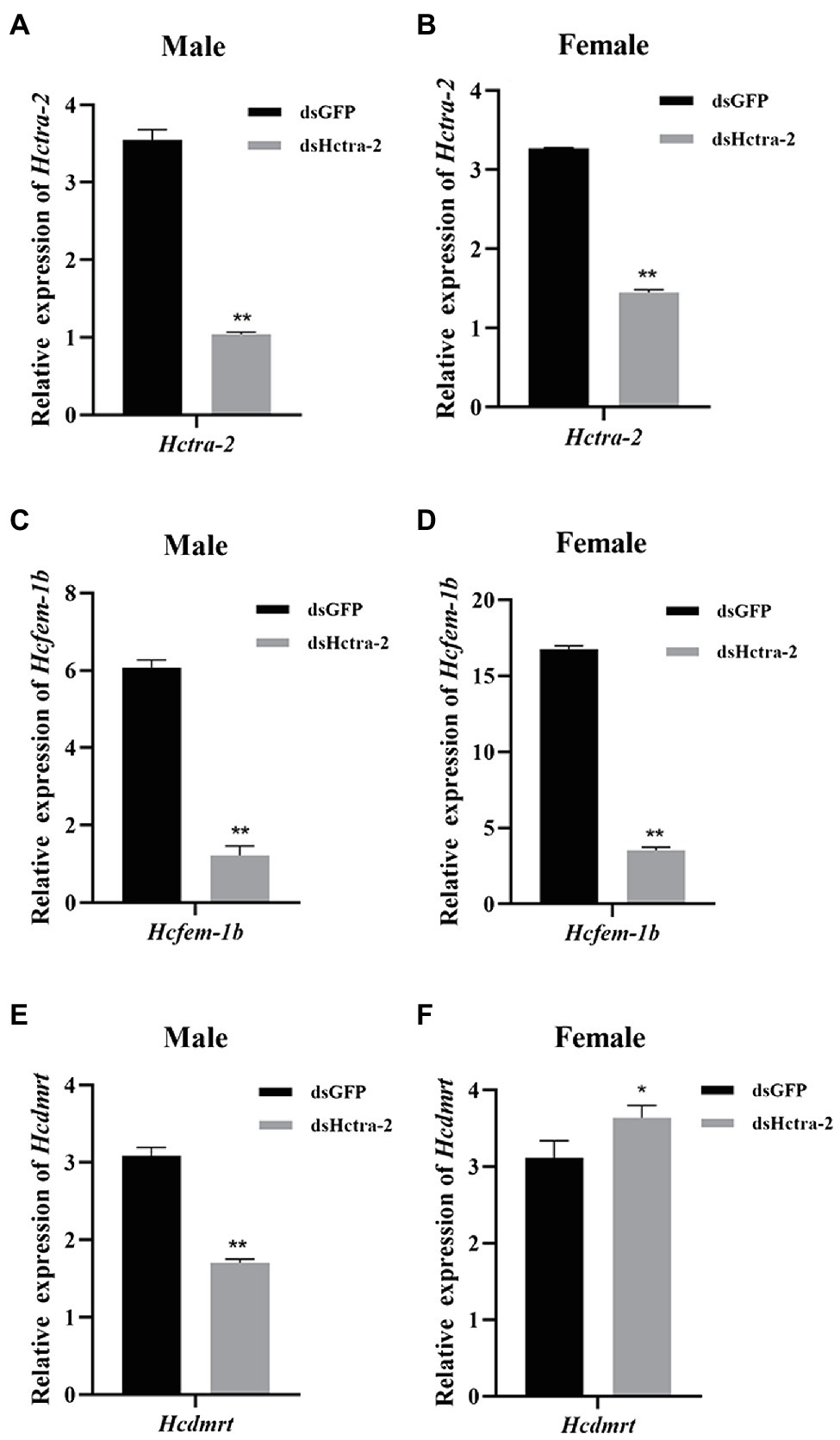
Figure 7. Effects of dsHctra-2 injection on the expression of Hctra-2, Hcfem-1b, and Hcdmrt genes in testes and ovaries of H. cumingii. dsGFP, treated with dsGFP and used as RNAi control group; dsHctra-2, treated with dsRNA of Hctra-2 as RNAi experiment group. (A) The expression of Hctra-2 gene in testes. (B) The expression of Hctra-2 gene in ovaries. (C) The expression of Hcfem-1b gene in testes. (D) The expression of Hcfem-1b gene in ovaries. (E) The expression of Hcdmrt gene in testes. (F) The expression of Hcdmrt gene in ovaries. Results are expressed as mean ± SD, and significance of comparison is defined as p < 0.05 (*) or p < 0.01 (**) by Student’s t-tests.
Discussion
In this study, we cloned and identified 1867 bp of the Hctra-2, including 881 bp 3'-UTR, 92 bp 5'-UTR and 894 bp ORF corresponding to 297 amino acids. Hctra-2 has a conserved RRM domain (98 amino acids) and linker region (17 amino acids), but only one RS domain (RS1) contains 61 amino acids (Figure 1). The RNP-1 and RRM domains are generally considered to be involved in the recognition of single-stranded RNA and can affect many pre-mRNA selective splicing (Kim et al., 2000; Lee et al., 2017). Like in the Anastrepha fruit flies, the RRM domain of TRA-2 protein confers the tra-tra2 complex to interact with tra and dsx pre-mRNAs to regulate sex-specific splicing (Martín et al., 2011). RS domain is rich in serine and arginine dipeptides (Zhang and Sun, 2008), its main function is to mediate splice site recognition and splicing regulation, and its potency is proportional to the number of repeats of RS dipeptides (Philipps et al., 2003; Long and Caceres, 2009). This indicates that the RS and RRM domains in Hctra-2 endow it with a certain splicing function. The linker region is a characteristic domain of TRA-2 proteins (Dauwalder et al., 1996). We compared the putative HCTRA-2 protein of H. cumingii with M. coruscus, C. gigas, M. yessoensis, and P. maximus. Figure 2 shows the alignment of these TRA2 proteins with different numbers of amino acids: H. cumingii 297, M. coruscus 289, C. gigas 298, M. yessoensis 324, and P. maximus 232. This difference in the number of amino acids was caused by changes in the sequence of TRA-2 amino acids except for the RRM domain and the linker (78 and 17 amino acids, respectively). The homology and conservation of the RRM domain and linker domain in these species suggest the functional similarity of the tra-2 in bivalves. Compared with other species, the linker sequence of HCTRA-2 amino acid has no RS2 domain behind it, which is similar to the Estra-2c of Eriocheir sinensis (Luo et al., 2017). The absence of the RS2 domain in Hctra-2 indicates a weak ability to recognize in the precursor mRNAs.
The expression pattern of Hctra-2 in different tissues and development stages was analyzed by qRT-PCR. Firstly, the tissue distribution showed that the Hctra-2 expression in gonads was higher than in somatic tissues. And Hctra-2 showed sexual dimorphism in gonads (Figure 4). The Hctra-2 expression level in the testis was significantly higher than that in the ovary. This expression pattern is consistent with that of M. nipponense in Arthropoda (Wang et al., 2019). It suggests that Hctra-2 may play a potential regulatory role in gonadal development of H. cumingii. But in many other species, tra-2 has a high expression pattern of ovary, such as P. chinensis (Li et al., 2012), Aedes albopictus (Li et al., 2019), E. chinensis (Luo et al., 2017), and aquatic firefly (Sclerotia aquatilis; Nguantad et al., 2020). The different expression patterns of tra-2 indicate that its regulation of gonadal development is different among species.
The transcripts changes of the Hctra-2 were further observed during embryonic development and early gonadal development (Figure 5). During embryonic development, the expression of Hctra-2 was high at fertilized eggs, peaked at day 3, and then decreased. Studies based on the embryonic development of H. cumingii showed that the embryo on the third day after fertilization is in cleavage stage (Hong et al., 2007). Interestingly, Mntra-2a was also highly expressed in the cleavage stage of M. nipponense embryos (Wang et al., 2019). This indicates that Hctra-2 is very active in embryo division. In a study of Bactrocera dorsalis, the expression level of tra was increased at the fifteenth hour of embryonic development, suggesting that sexual identity may be established at this stage of embryogenesis (Hong et al., 2007). We speculate that Hctra-2 may be involved in sex determination at early embryonic stage. In the early stage of gonadal development, Hctra-2 showed a slight increase at 4 months of age (Figure 5). Ting (2016) found that the gonad tissue of H. cumingii began to appear around 5 months of age, suggesting that Hctra-2 may be involved in the regulation of early gonad development.
The histological location of Hctra-2 in gonad was analyzed by ISH. Hctra-2 mRNA was detected in spermatogonia and spermatocytes in the testes (Figure 6C) and in the membrane and nucleus of ovary oocytes and mature ova (Figure 6F). The expression position of Hctra-2 signals is similar to that of tra-2 in M. nipponense (Wang et al., 2019). These results showed that Hctra-2 was closely related to spermatogonia proliferation, meiosis of spermatocytes and oocytes, and ova maturation, further confirming that Hctra-2 was involved in the gonadal development of H. cumingii. In addition, no Hctra-2 mRNA signal was observed in sperm, while in egg, the signals were concentrated in the nucleus and cell membrane. This suggests that the high expression of Hctra-2 in the fertilized egg is caused by the ova. In Nasonia vitripennis, Nvtra2 was also found in early embryos (<3 h old), suggesting that the mother provides Nvtra2 to the egg and is critical for embryo viability (Geuverink et al., 2017). Studies of Bdtra-2 of Bactrocera dorsalis have shown that Bdtra-2 transcripts are contributed maternally and activated by the zygote in the developing embryos (Laohakieat et al., 2020). Therefore, we hypothesized that in H. cumingii, Hctra-2 might be provided by the mother and involved in embryo sex determination during cleavage. However, whether Hctra-2 is a key gene involved in sex determination needs to be further verified.
In this study, Hctra-2 was silenced by RNAi to explore further the role of Hctra-2 in the sex regulation of H. cumingii. After injection of Hctra-2 dsRNA, the expression of Hcfem-1b and Hcdmrt was observed (Figure 7). These two genes were selected because we know that tra is upstream of fem in C. elegans sex determination process. The activity of TRA-2 protein can directly determine whether it interacts with the FEM protein complex, thus influencing the sex of C. elegans (Zanetti and Puoti, 2013). Secondly, in most insects, tra forms a conserved central axis of sex determination (Verhulst et al., 2010) and plays a key role in sex determination and maintenance by regulating dsx mRNAs (Shukla and Palli, 2013). Both doublesex and mab-3-related transcription factor (dmrt) and dsx belong to the dmrt family, and their roles in sex determination and differentiation have been widely studied (Mawaribuchi et al., 2019; Panara et al., 2019). In addition, our previous study identified the homologous genes of Hcfem-1b (Wang et al., 2021) and Hcdmrt (Ge et al., 2020) of fem and dmrt. Therefore, we wonder whether there is such upstream or downstream regulatory relationship in H. cumingii? With this conjecture in mind, we used RNAi to investigate the interaction of tra-2, fem and dmrt in the regulation mechanism in H. cumingii.
RNA interference of Hctra-2 in both female and male mussels caused the down-regulation of Hcfem-1b expression (Figures 7C,D). This indicates that Hcfem-1b was downstream of Hctra-2 and maintained the consistency of synergistic changes with it. However, when the expression of Hcdmrt was detected, it was found that the Hcdmrt was significantly decreased in males (Figure 7E) and increased in females (Figure 7F). In Bemisia tabaci, RNAi with Bttra2 also affects the expression of Btdsx. And, the silencing of Bttra2 or Btdsx resulted in male genital deformities (Guo et al., 2018). We were also found that Cqdsx was significantly reduced in the RNAi of CqTra2 in redclaw crayfish (Cai et al., 2020). These results indicate that Hcdmrt is also regulated by Hctra-2, but the regulation is different between the sexes. So, is there a regulatory relationship between Hcfem-1b and Hcdmrt? Here, we propose a hypothesis, and its correctness needs further explored. We found that dsx is regulated by fem in honeybee sex determination process. Honeybee sex is determined by the heterozygosity of complementary sex determiner (csd; Beye et al., 2003; Gempe and Beye, 2011). The csd of honeybee is homologous with the drosophila tra (Liu et al., 2012). In females, csd can produce active CSD protein, which can cleave fem to produce functional FEM protein and then promote female splicing of dsx transcript and induce female development. On the contrary, the males lack active CSD protein, the fem transcripts are spliced into male form, dsx pre-mRNA is spliced into the male expressing DSX protein, and the individual develops toward male (Beye et al., 2003; Beye, 2004). Moreover, combined with the silencing efficiency of the genes, the silencing efficiency of Hcfem-1b in testes and ovaries was 79.9 and 79.0%, respectively (Figures 7C,D). The silencing efficiency of Hcdmrt in testes was 44.7% (Figure 7E) and increased by 16.7% in ovaries (Figure 7F). Thus, Hcfem-1b has a higher silencing efficiency, and it is most likely to be directly downstream of Hctra-2, while Hcdmrt is indirectly downstream of it. Therefore, we boldly speculate that there is a cascading regulatory relationship among these three genes, namely, Hctra-2 > Hcfem-1b > Hcdmrt. The difference in this regulatory relationship between males and females is the effect of Hcfem-1b on Hcdmrt. Hcfem-1b may positively regulate Hcdmrt in males and negatively regulate Hcdmrt in females. But at present, this is our preliminary guess, and the specific way of regulation and its role in gender regulation is our goal in the next stage.
Conclusion
In this study, the full length of Hctra-2 of H. cumingii was cloned. The results of gene sequence analysis and multiple sequence alignment showed that Hctra-2 was highly conserved in RRM domain and linker region, and the amino acid sequence similarity among different species was high. The phylogenetic tree showed that tra-2 was closely related to mollusks. The qRT-PCR results indicated that Hctra-2 may play an important role in gonadal development and early embryonic development. The probe signal of Hctra-2 mRNA was found in both male and female gonads, which further indicated that Hctra-2 was involved in gonadal development. RNAi experiments explored the relationship between two sex-related genes (Hcfem-1b and Hcdmrt) and Hctra-2. We speculated that there was a cascade regulatory relationship between them, and the way of action was different between males and females. However, the specific way of this regulatory relationship and its impact on gender regulation still to be further studied.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MH931228.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Ocean University, Shanghai, China.
Author Contributions
XW completed the sample collection. YW and JG conceived and designed the experiments, analyzed the data, interpreted the results, and wrote the manuscript. GW and JL provided feedback on discussion and results. All authors have given approval to the final version of the manuscript.
Funding
This study was supported by the National Key R&D Program of China (grant number 2018YFD0901406), the National Natural Science Foundation of China (grant number 31772835), and the China Agriculture Research System of MOF and MARA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful for the samples provided by the Weiming aquaculture farm.
Abbreviations
tra-1 and tra-2, transformer-1 and transformer-2; qRT-PCR, quantitative real-time reverse transcription PCR; RNAi, RNA interference; fem-1, fem-2, and fem-3, feminization-1, feminization-2, and feminization-3; dmrt, doublesex and mab-3-related transcription factor; foxl2, forkhead box l2; her-1, hermaphroditization-1; cul-2, cullin 2; dsx, doublesex; fru, fruitless; sxl, sex lethal; dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; ORF, open reading frame; UTR, untranslated region; HE, hematoxylin-eosin; RRM, RNA recognition motif; csd, complementary sex determiner; Fc, follicles; Sg, spermatogonia; Sc, spermatocyte; St, spermatid; Sp, sperm; Og, oogonium; Oc, oocyte; Mo, mature ovum; ISH, in situ hybridization.
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/orffinder/
2. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
3. ^http://www.cbs.dtu.dk/services/TMHMM/
References
Beye, M. (2004). The dice of fate: the csd gene and how its allelic composition regulates sexual development in the honey bee, Apis mellifera. BioEssays 26, 1131–1139. doi: 10.1002/bies.20098
Beye, M., Hasselmann, M., Fondrk, M. K., Page, R. E., and Omholt, S. W. (2003). The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114, 419–429. doi: 10.1016/S0092-8674(03)00606-8
Cai, L., Zheng, J., Jia, Y., Gu, Z., Liu, S., Chi, M., et al. (2020). Molecular characterization and expression profiling of three transformer-2 splice isoforms in the Redclaw crayfish, Cherax quadricarinatus. Front. Physiol. 11:00631. doi: 10.3389/fphys.2020.00631
Camara, N., Whitworth, C., Dove, A., and Van Doren, M. (2019). Doublesex controls specification and maintenance of the gonad stem cell niches in drosophila. Development 146:170001. doi: 10.1242/dev.170001
Dauwalder, B., Amaya-Manzanares, F., and Mattox, W. (1996). A human homologue of the drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc. Natl. Acad. Sci. U. S. A. 93, 9004–9009. doi: 10.1073/pnas.93.17.9004
Farboud, B., Nix, P., Jow, M. M., Gladden, J. M., and Meyer, B. J. (2013). Molecular antagonism between X-chromosome and autosome signals determines nematode sex. Genes Dev. 27, 1159–1178. doi: 10.1101/gad.217026.113
Galindo-Torres, P., García-Gasca, A., Llera-Herrera, R., Escobedo-Fregoso, C., Abreu-Goodger, C., and Ibarra, A. M. (2018). Sex determination and differentiation genes in a functional hermaphrodite scallop. Mar. Genomics 37, 161–175. doi: 10.1016/j.margen.2017.11.004
Ge, J., Wu, C., Xia, S., Guo, P., Wang, G., and Li, J. (2020). Identification and expression analysis of Dmrta2-2 Gene in Hyriopsis cumingii. Genomics Appl. Biol. 39, 33–41. doi: 10.13417/j.gab.039.003941
Gempe, T., and Beye, M. (2011). Function and evolution of sex determination mechanisms, genes and pathways in insects. BioEssays 33, 52–60. doi: 10.1002/bies.201000043
Geuverink, E., Rensink, A. H., Rondeel, I., Beukeboom, L. W., Louis, V., and Verhulst, E. C. (2017). Maternal provision of transformer-2 is required for female development and embryo viability in the wasp Nasonia vitripennis. Insect Biochem. Mol. Biol. 90, 23–33. doi: 10.1016/j.ibmb.2017.09.007
Grmai, L., Hudry, B., Miguel-Aliaga, I., and Bach, E. A. (2018). Chinmo prevents transformer alternative splicing to maintain male sex identity. PLoS Genet. 14:e1007203. doi: 10.1371/journal.pgen.1007203
Guo, L., Xie, W., Liu, Y., Yang, Z., Yang, X., Xia, J., et al. (2018). Identification and characterization of doublesex in Bemisia tabaci. Insect Mol. Biol. 27, 620–632. doi: 10.1111/imb.12494
Hong, W., Yi, B., Jiale, L., and Jianju, W. (2007). A primary study on the morphological changes of embryo of Hyriopsis cumingii in nurturing pouch of outer gill. J. Shanghai Fish. Univ. 16, 219–223.
Hu, Y., Jin, S., Fu, H., Qiao, H., Zhang, W., Jiang, S., et al. (2020). Functional analysis of a SoxE gene in the oriental freshwater prawn, Macrobrachium nipponense by molecular cloning, expression pattern analysis, and in situ hybridization (de Haan, 1849). 3 Biotech 10:10. doi: 10.1007/s13205-019-1996-x
Kim, I., Muto, Y., Watanabe, S., Kitamura, A., Futamura, Y., Yokoyama, S., et al. (2000). Interactions of a didomain fragment of the drosophila sex-lethal protein with single-stranded uridine-rich oligoribonucleotides derived from the transformer and sex-lethal messenger RNA precursors: NMR with residue-selective [5-2H] uridine substitutions. J. Biomol. NMR 17, 153–165. doi: 10.1023/A:1008357028116
Kuwabara, P. E. (2007). A complex solution to a sexual dilemma. Dev. Cell 13, 6–8. doi: 10.1016/j.devcel.2007.06.004
Laohakieat, K., Isasawin, S., and Thanaphum, S. (2020). The transformer-2 and fruitless characterisation with developmental expression profiles of sex-determining genes in Bactrocera dorsalis and B. correcta. Sci. Rep. 10:17938. doi: 10.1038/s41598-020-74856-6
Lee, K. C., Jang, Y. H., Kim, S. K., Park, H. Y., Thu, M. P., Lee, J. H., et al. (2017). RRM domain of arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes. Plant Cell Rep. 36, 1083–1095. doi: 10.1007/s00299-017-2140-1
Li, X., Jin, B., Dong, Y., Chen, X., Tu, Z., and Gu, J. (2019). Two of the three transformer-2 genes are required for ovarian development in Aedes albopictus. Insect Biochem. Mol. Biol. 109, 92–105. doi: 10.1016/j.ibmb.2019.03.008
Li, S., Li, F., Wen, R., and Xiang, J. (2012). Identification and characterization of the sex-determiner transformer-2 homologue in Chinese shrimp, Fenneropenaeus chinensis. Sex. Dev. 6, 267–278. doi: 10.1159/000341377
Li, Y., Zhang, L., Sun, Y., Ma, X., Wang, J., Li, R., et al. (2016). Transcriptome sequencing and comparative analysis of ovary and testis identifies potential key sex-related genes and pathways in scallop Patinopecten yessoensis. Mar. Biotechnol. 18, 453–465. doi: 10.1007/s10126-016-9706-8
Liu, Z. Y., Wang, Z. L., Yan, W. Y., Wu, X. B., Zeng, Z. J., and Huang, Z. Y. (2012). The sex determination gene shows no founder effect in the giant honey bee, Apis dorsata. PLoS One 7, 1–5. doi: 10.1371/journal.pone.0034436
Long, J. C., and Caceres, J. F. (2009). The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417, 15–27. doi: 10.1042/BJ20081501
Luo, D., Liu, Y., Hui, M., Song, C., Liu, H., and Cui, Z. (2017). Molecular characterization and expression profiles of four transformer-2 isoforms in the Chinese mitten crab Eriocheir sinensis. Chin. J. Oceanol. Limnol. 35, 782–791. doi: 10.1007/s00343-017-6071-z
Mapes, J., Chen, J. T., Yu, J. S., and Xue, D. (2010). Somatic sex determination in Caenorhabditis elegans is modulated by SUP-26 repression of tra-2 translation. Proc. Natl. Acad. Sci. U. S. A. 107, 18022–18027. doi: 10.1073/pnas.1004513107
Martín, I., Ruiz, M. F., and Sánchez, L. (2011). The gene transformer-2 of sciara (diptera, nematocera) and its effect on drosophila sexual development. BMC Dev. Biol. 11:19. doi: 10.1186/1471-213X-11-19
Mawaribuchi, S., Ito, Y., and Ito, M. (2019). Independent evolution for sex determination and differentiation in the DMRT family in animals. Biol. Open 8:041962. doi: 10.1242/bio.041962
Naimi, A., Martinez, A. S., Specq, M. L., Mrac, A., Diss, B., Mathieu, M., et al. (2009). Identification and expression of a factor of the DM family in the oyster Crassostrea gigas. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 152, 189–196. doi: 10.1016/j.cbpa.2008.09.019
Nguantad, S., Chumnanpuen, P., Thancharoen, A., Vongsangnak, W., and Sriboonlert, A. (2020). Identification of potential candidate genes involved in the sex determination cascade in an aquatic firefly, Sclerotia aquatilis (coleoptera, lampyridae). Genomics 112, 2590–2602. doi: 10.1016/j.ygeno.2020.01.025
Panara, V., Budd, G. E., and Janssen, R. (2019). Phylogenetic analysis and embryonic expression of panarthropod Dmrt genes. Front. Zool. 16:23. doi: 10.1186/s12983-019-0322-0
Philipps, D., Celotto, A. M., Wang, Q. Q., Tarng, R. S., and Graveley, B. R. (2003). Arginine/serine repeats are sufficient to constitute a splicing activation domain. Nucleic Acids Res. 31, 6502–6508. doi: 10.1093/nar/gkg845
Shi, J., Hong, Y., Sheng, J., Peng, K., and Wang, J. (2015). De novo transcriptome sequencing to identify the sex-determination genes in Hyriopsis schlegelii. Biosci. Biotechnol. Biochem. 79, 1257–1265. doi: 10.1080/09168451.2015.1025690
Shi, Y., Liu, W., and He, M. (2018). Proteome and transcriptome analysis of ovary, intersex gonads, and testis reveals potential key sex reversal/differentiation genes and mechanism in scallop Chlamys nobilis. Mar. Biotechnol. 20, 220–245. doi: 10.1007/s10126-018-9800-1
Shiraishi, E., Imazato, H., Yamamoto, T., Yokoi, H., Abe, S. I., and Kitano, T. (2004). Identification of two teleost homologs of the drosophila sex determination factor, transformer-2 in medaka (Oryzias latipes). Mech. Dev. 121, 991–996. doi: 10.1016/j.mod.2004.04.013
Shukla, J. N., and Palli, S. R. (2013). Tribolium castaneum transformer-2 regulates sex determination and development in both males and females. Insect Biochem. Mol. Biol. 43, 1125–1132. doi: 10.1016/j.ibmb.2013.08.010
Sosnowski, B. A., Belote, J. M., and Yckeown, M. (1989). Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 59, 449–459. doi: 10.1016/0092-8674(89)90426-1
Starostina, N. G., Lim, J., Schvarzstein, M., Wells, L., Spence, M. A., and Kipreos, E. T. (2007). A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell 13, 127–139. doi: 10.1016/j.earlhumdev.2006.05.022
Suzuki, M. G. (2010). Sex determination: insights from the silkworm. J. Genet. 89, 357–363. doi: 10.1007/s12041-010-0047-5
Ting, X. (2016). Study on DUI occurring and gonad development of freshwater pearl mussels. master’s thesis. China: Shanghai Ocean University.
Verhulst, E. C., van de Zande, L., and Beukeboom, L. W. (2010). Insect sex determination: it all evolves around transformer. Curr. Opin. Genet. Dev. 20, 376–383. doi: 10.1016/j.gde.2010.05.001
Wang, Y. Y., Duan, S. H., Dong, S. S., Cui, X. Y., Wang, G. L., and Li, J. L. (2021). Comparative proteomic study on fem-1b in female and male gonads in Hyriopsis cumingii. Aquac. Int. 29, 1–18. doi: 10.1007/s10499-020-00605-1
Wang, Y., Jin, S., Fu, H., Qiao, H., Sun, S., Zhang, W., et al. (2019). Molecular cloning, expression pattern analysis, and in situ hybridization of a transformer-2 gene in the oriental freshwater prawn, Macrobrachium nipponense (de Haan, 1849). 3 Biotech 9:205. doi: 10.1007/s13205-019-1737-1
Wang, S., and Kimble, J. (2001). The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO J. 20, 1363–1372. doi: 10.1093/emboj/20.6.1363
Wang, G., Yuan, Y., and LI, J.-L. (2007). SSR analysis of genetic diversity and phylogenetic relationships among different populations of Hyriopsis cumingii from the five lakes of China. J. Fish. China 12, 12–18.
Yue, C., Li, Q., Yu, H., Liu, S., and Kong, L. (2020). Restriction site-associated DNA sequencing (RAD-seq) analysis in pacific oyster Crassostrea gigas based on observation of individual sex changes. Sci. Rep. 10:9873. doi: 10.1038/s41598-020-67007-4
Zanetti, S., and Puoti, A. (2013). Sex determination in the Caenorhabditis elegans germline. Adv. Exp. Med. Biol. 757, 41–69. doi: 10.1007/978-1-4614-4015-4_3
Zhang, W., and Sun, Z. (2008). Random local neighbor joining: a new method for reconstructing phylogenetic trees. Mol. Phylogenet. Evol. 47, 117–128. doi: 10.1016/j.ympev.2008.01.019
Keywords: Hyriopsis cumingii, transformer-2, gonadal development, sex determination, in situ hybridization, RNA interference
Citation: Wang Y, Wang X, Ge J, Wang G and Li J (2021) Identification and Functional Analysis of the Sex-Determiner Transformer-2 Homologue in the Freshwater Pearl Mussel, Hyriopsis cumingii. Front. Physiol. 12:704548. doi: 10.3389/fphys.2021.704548
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Radha Chaube, Banaras Hindu University, IndiaAshis Saha, Central Institute of Freshwater Aquaculture (ICAR), India
Copyright © 2021 Wang, Wang, Ge, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guiling Wang, eWh3dXdhbmcyMDA4QDEyNi5jb20=
 Yayu Wang
Yayu Wang Xiaoqiang Wang
Xiaoqiang Wang Jingyuan Ge
Jingyuan Ge Guiling Wang
Guiling Wang Jiale Li
Jiale Li