Abstract
The mature intestinal microbiome is a formidable barrier to pathogen colonization. Day-old chicks seeded with cecal contents of adult hens are resistant to colonization with Salmonella, the basis of competitive exclusion. Competitive exclusion products can include individual microbes but are commonly undefined intestinal communities taken from adult animals and in commercial production is amplified in fermentator and sold commercially in freeze dried lots. While superior to single and multiple species probiotics, reducing Salmonella colonization by multiple logs, undefined products have limited acceptance because of their uncharacterized status. In this study, the bacterial composition of the master stock, preproduction seed stocks and commercial lots of a poultry competitive exclusion product, was defined by 16S rRNA sequence analysis, targeting the 16S rRNA variable region (V1-V3). The samples contained a diversity of genera (22–52 distinct genera) however, the commercial lots displayed less diversity compared to the seeds and the master stock. Community composition varied between seeds and the master stock and was not a good predictor of potency, in terms of log10 reduction in Salmonella abundance. While there was significant correlation in composition between seeds and their commercial lots, this too was a not a good predictor of potency. There was linear correlation between unclassified Actinobacteria, Peptococcus, and unclassified Erysipelotrichaceae, and Salmonella abundance (r2 > .75) for commercial seeds. However, upon review of the literature, these three genera were not consistently observed across studies or between trials that examined the correlation between intestinal community composition and Salmonella prevalence or abundance.
Introduction
Many studies have shown that probiotics or direct-fed microbials can reduce intestinal disease in humans and animals (Gómez-Gallego et al., 2016; Kim et al., 2019; Lai et al., 2019; Renaud et al., 2019; Jakubczyk et al., 2020; Lucey et al., 2021; Xu et al., 2021). This is particularly important in animal production where production costs are directly impacted by intestinal health. The composition of the normal intestinal microbial community plays an important role in animal health and performance through its effect on gut morphology, nutrition, pathogenesis of intestinal disease and the immune response (Hooper et al., 2000). The term competitive exclusion (CE), introduced by Nurmi et al., 1973 is used to describe the process by which beneficial bacteria exclude Salmonella from the intestine (Nurmi et al., 1992).Formulations containing beneficial commensal bacteria have been marketed in some countries as probiotics, competitive exclusion formulations, or direct-fed microbials (Food and Agriculture Organization of the United Nations, 2016; Szott et al., 2022). There has been considerable interest in utilizing competitive exclusion to exclude select pathogens, especially Salmonella, from the gastrointestinal tract of food animals in order to reduce foodborne transmission to humans (Stern et al., 2001; Szott et al., 2022). Competitive exclusion formulations produced from the intestinal content of healthy chickens have been shown to be very effective at eliminating or reducing the prevalence of Salmonella in broiler chicken flocks and on carcasses (Hirn et al., 1992; Corrier et al., 1998; Stern et al., 2001).
The intestinal tract is a novel ecosystem that contains a community of bacteria rivaling the diversity of any other ecosystem on the planet. The density and number of bacteria in the community is higher than the number of host cells. Culture-based studies have suggested that intestinal microbial community is composed primarily of obligate anaerobes (Franks et al., 1998). The predominant cultivatable bacteria present in the chicken ceca are obligate anaerobes at a density of 1011 cells per gram of contents (Barnes et al., 1972) including at least 38 different types of anaerobic bacteria within the chicken cecum (Salanitro et al., 1974a; Barnes, 1979) with more than 200 total bacterial strains identified (Mead, 1989). However only 10%–60% of the bacteria visualized microscopically were cultured indicating a rich community of uncharacterized organisms (Barnes et al., 1972; Salanitro et al., 1974b; Barnes, 1979; Mead, 1989).
Using the DNA sequences of the small subunit ribosome genes present in a bacterial community (16S rRNA clone libraries), the composition of the intestinal community has been evaluated for many animals with very surprising results (Tannock, 1999). Commonly cultured organisms, such as E. coli, have been found to be a minor component of the intestine and novel uncultured organisms have been found to be the most abundant. Applications of molecular ecological profiling on poultry intestinal communities have concurred somewhat with the culture-based studies in that the chicken intestinal communities are primarily composed of Gram-positive bacteria related to mid and low G + C genera such as Clostridia and Lactobacillus (Gong et al., 2002; Lan et al., 2002; Lu et al., 2003; Zhu and Joerger, 2003). While lactobacilli are frequent in the small intestine, Clostridia are abundant throughout the intestinal tract of healthy chickens. The cecal intestinal microbial community is dominated by atypical and novel Clostridia, some of which have high G + C genomes (Apajalahti et al., 2001; Lu et al., 2003; Zhu and Joerger, 2003). The DNA sequences indicate that these normal flora Clostridia are not closely related to pathogenic Clostridium (such as perfringens) and they do not appear to be pathogenic themselves. Only recently have the members of the order Clostridiales been isolated from the chicken cecum and characterized by whole genome sequencing (Medvecky et al., 2018). Approximately half of the chicken gut anaerobes (n = 69) were Clostridiales; consisting of four families (Clostridiaceae, Erysipelotrichaceae, Lachnospiraceae, and Ruminococcaceae) and, at least, 12 distinct genera (Anaerofilum, Anaeromassillibacillus, Anaerotruncus, Blautia, Butyricicoccus, Clostridium, Drancourtella, Eubacterium, Faecalibacterium, Flavonifractor, Gemmiger, and Pseudoflavonifractor). Thirty-six of these isolates show <97% or 95% 16S identity to Clostridia and clostridial species, respectively, in public databases and represent new clostridial genera and species awaiting taxonomic classification or reclassification. Because of their fermentative metabolism and ability to produce short chain fatty acids (SCFAs) (Medvecky et al., 2018), these organisms appear to be important players in developing an exclusive community that reduces the competition or behavior of pathogens.
To explore the genomic features of complex microbial communities, a culture-independent 16S rRNA amplicon sequencing approach has become practical and cost effective due to the advent of high throughput sequencing, a method allowing simultaneous sequencing of hundreds of thousands of individual DNA strands (Shang et al., 2018). The analysis is performed by comparing each 16S rRNA gene sequence to all others that have been detected within the community in order to determine the frequency of occurrence. The abundant organisms are frequently detected while the genes of less abundant organisms are rarely detected. However, if enough sequences are analyzed the method can reveal the presence of rare organisms. The sequence analysis can be performed at the strain level (99% DNA similarity), species level (97% DNA similarity), genus level (95% DNA similarity), and group/family level (90%). While commercial exclusion products are effective at reducing Salmonella colonization in poultry, their market distribution is limited by regulatory restrictions on undefined microbial products. Using a 16S rRNA-based approach, we have characterized the composition of several batches of a competitive exclusion product, marketed for controlling Salmonella in poultry, and used this information to determine if a particular organism or an assemblage of organisms correlated with product efficacy.
Materials and methods
Characterization of master seed and seed batches of a competitive exclusion product
Commercial samples were shipped in sealed plastic bags, in the same packaging sent to customers. Thirteen samples, with potency results supplied by manufacturer were received frozen and were kept frozen until processing. All samples were opened within biosafety cabinet, previously treated with ultraviolet light and 10% bleach, and transferred to microfuge tubes for DNA extraction. Samples consisted of: master seed (sample A), two seed batches with potency <6 log10 reduction of Salmonella abundance (samples E and B), three seed batches with potency >6 log10 reduction (samples C, D, I), three commercial batches with potency <6 log10 reduction (samples H, J, M), and four commercial batches with potency >6 log10 reduction (samples F, G, K, L). Figure 1 shows the derivation of seeds and commercial lots from the master seed (Sample A) and their potency (-log10 reduction in Salmonella abundance). Salmonella reduction data was provided by the manufacturer for seeds and commercial lots; this data is used in their quality control evaluation of product efficacy. Commercial lots were released for sale if they reduced Salmonella colonization by at least 5 log10 compared to the untreated control.
FIGURE 1
Preparation of bacterial 16S rRNA amplicons for sequencing
DNA was extracted using a MoBio Soil DNA extraction kit (MoBio Laboratories, Carlsbad California, United States) as previously described (Lu et al., 2003). The DNA quality was evaluated by gel electrophoresis with 1 kb ladder molecular weight standards. The DNA was quantified using Nanodrop (Thermo Scientific, Wilmington Delaware, United States). Barcoded PCR primers (Supplementary Tables S1, S2) targeting the 16S rRNA gene variable regions V1-V3 was used to amplify sample DNA (Hamady et al., 2008). Positive and negative, no template controls were included in the 16S PCRs, using Salmonella Typhimurium SR11 genomic DNA or molecular grade dH2O, respectively as template. All PCR reactions were set up in a PCR clean hood, in a confined room, separate from where DNA was extracted in one room and thermocyclers, housed in a physically-separate room. PCR clean hood was previously treated with ultraviolet light and surfaces were wiped down with 10% bleach. Separate set of pipettors are kept in the PCR clean hood, they are only used to set up PCR reactions, and never leave this set up area. Barrier tips are used to dispense PCR reagents and template to prevent contamination of the pipette barrels. No amplicon was observed for the negative, no template control. PCR was performed as described by Garcia et al. (2011). In order to standardize the method and to reduce PCR amplification error, 200 ng of sample DNA was used in each reaction and PCR was run for 20 cycles. Three separate PCR reactions were produced and pooled for sequencing, for each sample. Triplicate trials for each sample were done resulting in 135 independent PCR reactions. Clostridium perfringens ATCC 13124 DNA was used as positive control for amplification and sequencing, and a control lacking template was used to detect reagent contamination.
PCR reactions were run on agarose gels to evaluate quantity and quality of amplicon. Any sample which showed a low amount of DNA was repeated. PCR amplicons were excised from gel, purified using Qiagen Gel extraction kit (Qiagen Inc., Valencia California, United States) and pooled for each sample. Agencourt AMP pure XP kit (Beckman Coulter, Indianapolis, IN, United States) was used to further clean up and concentrate amplicon from gel extractions for 454 pyrosequencing. PCR amplicons were quantified using Nanodrop and their concentration standardized by adjusting to 10 ng/μl. Any sample which contained less than 10 ng/μl was repeated in order to produce the necessary amount of DNA for standardization. Samples were submitted to the Georgia Genomics and Bioinformatics Core (University of Georgia, Athens, GA, United States) for pyrosequencing using 454 protocols established by Roche Inc. (Branford Connecticut, United States) which manufactures the instrument and reagents.
Bioinformatics and statistical analyses
The sequence analysis pipeline was performed using Roche and Mothur software (Schloss et al., 2009). Sequences were sorted by barcodes in order to organize by sample, trimmed based on size (≤500 bp) and quality to remove those sequences with gaps or ambiguous base calls. Sequences were aligned in order to detect and remove chimeras produced from PCR artifact.
The bacterial diversity was calculated comparing the similarity of the sequences and those deposited at Ribosomal Project Database (http://rdp.cme.msu.edu/) using the RDP6 database. The bacterial composition based on genus (95% similarity) and family (90% similarity) was generated in order to determine the phylogenetic composition of each sample. Sørensen’s coefficient of similarity (QS) was used to quantify the similarity of samples (Sorenson, 1948). QS = 2C/A + B where A and B are the number of species in samples A and B, respectively, and C is the number of species shared by the two samples. Sørensen’s coefficient ranges from 0–1. Simple correlation was used to statistically verify the similarity of sample composition. Chi-squared test was used in order to test differences in the proportion of the genus observed among the samples. Linear regression was adopted to determine the correlation between the bacterial genus and the levels of Salmonella reduction reported for commercial lots or seeds. Analysis of covariance was used to identify the effect of the seed on the commercial product. Statistic tests were performed using SAS software.
Screening samples for Salmonella
γ-Proteobacterial 16S sequences do not exhibit enough sequence diversity to reliably identify Salmonella and differentiate it from closely related member species. Therefore, some sequences may be erroneously reported as genus: Salmonella by Mothur, using the RDP6 database. In order to determine if Salmonella was present in commercial samples, a diagnostic PCR, targeting a Salmonella-specific locus, was applied. Fifty ng of sample DNA was used in PCR reactions, as described by Liu et al., 2002 (Liu et al., 2002) using invA primers. The samples were screened by gel electrophoresis using a 1.5% agarose gel containing ethidium bromide in order to visually detect amplicons of the expected size (450 bp). Salmonella Typhimurium genomic DNA served as a positive amplification control. A no template control was included to identify PCR contamination (false positive). The no template control was consistently negative in these Salmonella PCR screens.
Results
Sequencing quality control and culling anomalous sequences, ambiguities and chimeras from final sequence dataset
725,293 total sequences were obtained from the pyrosequencing reactions (Supplementary Table S3). However, after elimination of anomalous long sequences, homopolymers, and ambiguous bases, 703,522 sequences were subjected to chimera analysis for additional quality control. Chimeras are PCR artifacts that erroneously increase sample diversity and alter composition (Ley et al., 2008). After these quality control procedures, 332,559 sequences were of sufficient quality for compositional and statistical analysis. The distribution of sequences among the samples varied from 1,913–64,816 (Supplementary Table S3). The reads were therefore normalized to run data analysis for correlation, chi-square test, linear regression and covariance.
16S-rRNA based compositional analysis of CE seeds and commercial lots
The number of families detected varied from 15 to 28 and the number of genera from 22 to 52 for each sample (Figure 2). The seeds tended to contain the largest number of genera (mean = 45) with commercial lots containing the fewest (mean = 34). There was no correlation between the number of genera or families with Salmonella reduction. The bacterial composition is presented at the phyla (Figure 3), family (Figure 4; Table 1) and genus (Table 2) level. Unclassified bacteria, organisms that have yet been assigned a phylum, made up the smallest proportion, while Firmicutes was the dominant phylum, throughout CE samples, with the order Clostridia representing the most abundant group (58%–95.5%) within the phylum Firmicutes. Twelve different taxonomic families of Clostridia were detected in CE product, indicating that there was high diversity within this order. Firmicutes and the minor proteobacteria were the two phyla present in the master seed. Other phyla were identified in seeds and commercial lots, varying in their proportion, and included phyla Bacteroidetes, Fusobacteria, and Actinobacteria. While CE seed E had the highest proportion of Bacteroidetes (>30%), the proportion of this phyla in commercial lots derived from this seed was low. The same was observed for CE seed I, with regards to the 2nd major phyla, Proteobacteria (∼10%) and abundance of this phyla in resulting commercial lots were sporadic and low. Clostridiaceae, Veillonellaceae, and Lactobacillaceae, were the major families present in the master seed; their proportion in seeds and commercial lots varied. The proportion of these three families in the seeds was not predictive of their abundance in the resulting, commercial lots. Bacteria belonging to the families Clostridiaceae, Bacteroidaceae, Lactobacillaceae, Enterococcaceae, Peptostreptococcaceae, and Veillonellaceae comprised more than 20% of any one sample. The seeds tended to contain higher proportions of Lactobacillaceae and Enterococcaceae while the commercial lots tended to have higher levels of Peptostreptococcaceae. Seed I, which was a progenitor seed stock for commercial lots J–M had a highest proportion of Peptostreptococcaceae, of the seed stocks. This seed stock proved the most efficacious of the seeds at reducing Salmonella. The resulting commercial lots, generated from this seed stock, also had a significantly high proportion of this bacterial family.
FIGURE 2
FIGURE 3
FIGURE 4
TABLE 1
 |
Proportions of bacterial orders or families detected in competitive exclusion seeds and commercial lots.
TABLE 2
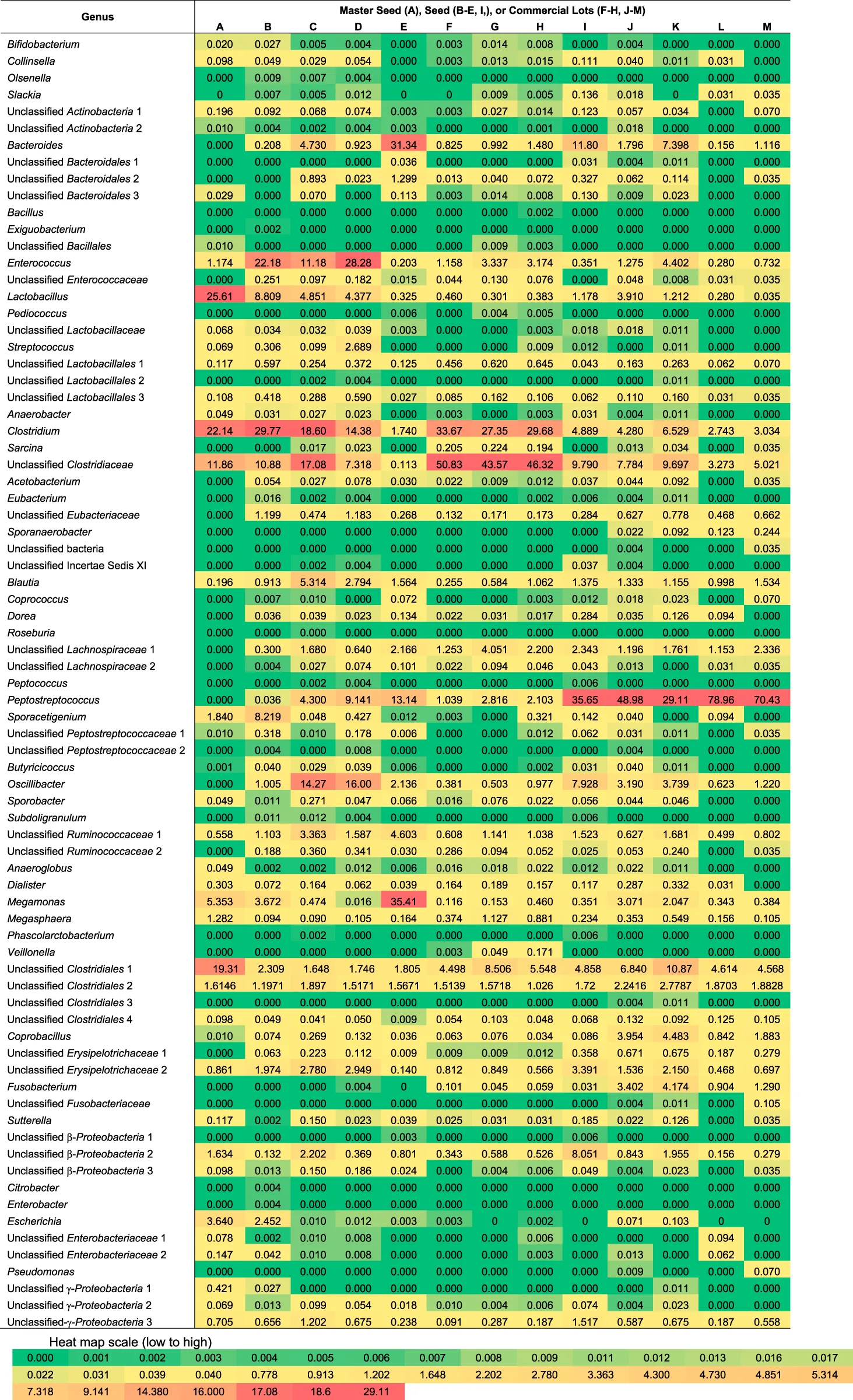 |
Proportion of genera detected in competitive exclusion seeds and commercial lots.
γ-Proteobacteria were detected among all of the samples and in a number of samples the Mothur software reported Salmonella among the genera detected. γ-Proteobacterial 16S sequences, and particularly the Enterobacteriaceae family, do not exhibit enough sequence diversity to be reliably used to report Salmonella because other genera within this group have highly similar 16S sequences. A diagnostic PCR was used to screen commercial samples for Salmonella. This PCR test has been used extensively to screen clinical and environmental samples for the presence of Salmonella (Liu et al., 2002). None of the samples gave a positive reaction therefore the erroneously classified sequences were corrected in the report to reflect their identity as an unknown Enterobacteriaceae species.
Consistency in bacterial composition of commercial lots with their seeds is not a good predictor of product efficacy
Table 3 presents the similarity between the community composition of the master seed, at the family and genus level, and the seeds produced from it. Statistical analysis, using simple correlation, varied across seeds compared to the master (Family: .719–.272; Genus .641–.078). Similarity with the master seed was not a good predictor of efficacy as the seed most similar to the master and with a high correlation value (Seed B) had the lowest Salmonella log10 reduction compared to one of the power seeds, with regards to community similarity (Seed I; Table 3). While individual seeds produced commercial lots with similar community composition at the family and genus level, this was not a good predictor as to which lots would be expected to have the greatest reduction of Salmonella counts (Table 4). Even comparing commercial lots against themselves, varied in their ability to reduce Salmonella abundance, was not a significant predictor of product efficacy. However, community similarity tended to reflect commercial lots origins with seed stocks used to generate CE product (Table 5).
TABLE 3
| Seeds | Log10 reductiona | Family similarity coefficient | Family correlationb,c | Genus similarity coefficient | Genus correlation1,2 |
|---|---|---|---|---|---|
| B | −4.56 | .893 | .719 | .750 | .641 |
| C | −6.89 | .847 | .624 | .727 | .541 |
| D | −6.70 | .678 | .390 | .722 | .320 |
| E | −5.56 | .630 | .360 | .721 | .078 |
| I | −8.44 | .750 | .272 | .696 | .153 |
Similarity in bacterial composition of the master seed A with seeds B, C, D, E, or I.
Reduction in Salmonella abundance.
Pearson’s correlation coefficient.
Not significant.
TABLE 4
| Seeds | Log10 reductiona | Family similarity coefficient | Family correlationb,c | Genus similarity coefficient | Genus correlation1,2 |
|---|---|---|---|---|---|
| I | −8.44 | ||||
| J | −5.45 | .857 | .916 | .796 | .922 |
| K | −7.78 | .885 | .921 | .849 | .933 |
| L | −7.67 | .784 | .873 | .690 | .889 |
| M | −5.07 | .852 | .888 | .731 | .901 |
| E | −5.56 | ||||
| F | −7.83 | .889 | .038 | .835 | −.001 |
| G | −8.22 | .873 | .103 | .826 | .013 |
| H | −5.56 | .881 | .073 | .857 | .017 |
Similarity in bacterial composition between seeds, exhibiting high (Seed I) or low (Seed E) Salmonella reduction, with their commercial lots.
Reduction in Salmonella abundance.
Pearson’s correlation coefficient.
Not significant.
TABLE 5
| Family | F (−7.83)a | G (−8.22)a | H (−5.56)a | J (−5.45)a | K (−7.78)a | L (−7.67)a | M (−5.07)a |
|---|---|---|---|---|---|---|---|
| Genus | |||||||
| F (−7.83)a | .945 (.995) | .915 (.999) | .885 (.215) | .847 (.431) | .857 (.058) | .746 (.094) | |
| G (−8.22)a | 0.899 (.992) | .933 (.998) | .871 (.250) | .833 (.478) | .800 (.085) | .868 (.122) | |
| H (−5.56)a | .843 (.998) | .854 (.997) | .879 (.238) | .844 (.460) | .778 (.077) | .807 (.114) | |
| J (−5.45)a | .808 (.169) | .781 (.213) | .864 (.193) | .879 (.926) | .786 (.970) | .847 (.978) | |
| K (−7.78)a | .825 (.340) | .796 (.400) | .847 (.371) | .885 (.936) | .815 (.824) | .842 (.845) | |
| L (−7.67)a | .769 (.050) | .759 (.090) | .717 (.071) | .702 (.980) | .667 (.870) | .894 (.999) | |
| M (−5.07)a | .660 (.079) | .800 (.119) | .735 (.100) | .800 (.986) | .774 (.886) | .757 (.999) |
Similarity in the bacterial composition of commercial lots produced from seeds E (F–H) and I (J–M) as defined by Sørensen’s coefficient and simple correlation ( ).
−log10_: Reduction in Salmonella abundance. Family comparison is highlighted in light gray. p-values for Pearson’s correlation coefficient were >.05.
Identification of genera within seeds and commercial lots that correlated with reduction of Salmonella colonization in chickens
Linear regression was performed to determine if a particular organism correlated with Salmonella reduction. In the seeds, unclassified Actinobacteria, Peptococcus, and unclassified Erysipelotrichaceae correlated with product efficacy (log10Salmonella reduction) at r2 greater than 75% (Table 6, Supplementary Tables S4, S5). There was a linear correlation among these organisms with Salmonella reduction in the seeds (Figure 5); specifically, an antagonistic relationship for unclassified Erysipelotrichaceae and Peptococcus versus a facultative relationship with Actinobacteria. In contrast, linear regression did not detect an organism correlating significantly with efficacy in the commercial lots themselves (Supplementary Table S5), including the afore mentioned genera. However, because all of the commercial products produced at least a 5 log10 reduction in Salmonella colonization, it may be difficult to detect correlation using this method. Furthermore, Salmonella reduction may not be due to the actions of a particular organism but due to the combined metabolic activity of organisms which produce an exclusive community.
TABLE 6
| Genera | Master seed A (−log10 8.50) vs. | Master seed A (−log10 8.50) vs. | Master seed A (−log10 8.50) vs. | Master seed A (−log10 8.50) vs. | Master seed A (−log10 8.50) vs. |
|---|---|---|---|---|---|
| Seed B (−log10 4.56) | Seed E (−log10 5.56) | Seed D (−log10 6.70) | Seed C (−log10 6.89) | Seed I (−log10 8.44) | |
| Bacteroides | ns | .001 | ns | .03 | .001 |
| Enterococcus | .001 | ns | .001 | .001 | ns |
| Lactobacillus | ns | .001 | .001 | .001 | .001 |
| Anaerobacter | ns | .001 | ns | ns | .001 |
| Sarcina | ns | .001 | ns | ns | .001 |
| unclassified Clostridiaceae | .002 | ns | ns | ns | ns |
| Blautia | ns | ns | ns | .03 | ns |
| Peptostreptococcus | ns | .001 | .001 | ns | .001 |
| Megamonas | ns | .001 | .02 | .04 | .04 |
| Veillonella | .001 | .001 | .001 | .001 | .001 |
Genera shared with the master competitive exclusion stock with seeds as determined by chi-square test.
−log10: Reduction in Salmonella abundance. Ns-not significant. Not statistically significant association of genus (n = 68), in comparison of seed stocks with the master: Bifidobacterium; Collinsella; Olsenella; Slackia; unclassified Actinobacteria 1,2; unclassified Bacteroidales 1–3; Bacillus; Exiguobacterium; unclassified Bacillales; unclassified Enterococcaceae; Pediococcus; unclassified Lactobacillaceae; Streptococcus; unclassified Lactobacillales 1–3; Clostridium; Acetobacterium; Eubacterium; unclassified Eubacteriaceae; Sporanaerobacter; unclassified bacteria; unclassified Incertae Sedis XI; Coprococcus; Dorea; Roseburia; unclassified Lachnospiraceae 1,2; Peptococcus; Sporacetigenium; unclassified Peptostreptococcaceae 1,2; Butyricicoccus; Oscillibacter; Sporobacter; Subdoligranulum; unclassified Ruminococcaceae 1,2; Anaeroglobus; Dialister; Megasphaera; Phascolarctobacterium; unclassified Clostridiales 1–4; Coprobacillus; unclassified Erysipelotrichaceae 1,2; Fusobacterium; unclassified Fusobacteriaceae; Sutterella; unclassified β-Proteobacteria 1–3; Citrobacter; Enterobacter; Escherichia; unclassified Enterobacteriaceae 1,2; Pseudomonas; unclassified γ-Proteobacteria 1–3.
FIGURE 5
Discussion
The host must maintain surveillance over the composition of the microbiota and exhibit control over the abundance and behavior of members that affect the host-microbe homeostasis. Therefore transfer of microbial communities from parent to offspring may have evolved to initially establish ecological health within the lumen of host mucosal systems and reduce susceptibility to mucosal disease (Neish, 2009). A number of studies have demonstrated that administration of complex microbial communities can reduce the ability of Salmonella to colonize young animals; these have been extensively reviewed (Nurmi et al., 1992; Nisbet, 1998; Becker, 2005) including those specifically addressing poultry production (Ferreira et al., 2003). Complex microbial communities, such as those used in competitive exclusion, have been shown in multiple independent studies to be effective in reducing Salmonella in poultry (Cameron et al., 1996; Cameron et al., 1997; Deruyttere et al., 1997; Guillot et al., 1997; Guerra-Garcia Miranda, 2000; Stephan, 2000; Sisak et al., 2001; Nakamura et al., 2002; Ferreira et al., 2003). Products that consist of single bacteria, such as Lactobacillus and Bacillus, are less efficacious (Nurmi et al., 1992). However the complex microbial communities are difficult to characterize using classical bacteriological methods (Hume et al., 1996).
The competitive exclusion product contains a diversity of microbial species, many of which are not consistently present in preproduction seeds or commercial lots. In fact, there are some taxa shared between seeds or lots that were not detected in the master by our methods. Peptostreptococcus, abundant in commercial lots, was not detected in the Master Seed, but had to be present to seed these lots and suggests limits of this 16S rRNA gene sequencing approach to detect minor taxa. While certain taxa were identified that seemed to correlate with Salmonella abundance, they are not consistently observed across studies (Azcarate-Peril et al., 2018; Ma et al., 2020; Leyva-Diaz et al., 2021; Pedroso et al., 2021). While others have identified a similar correlation between prevalence or abundance of certain taxa with Salmonella colonization in poultry, these taxa are also not uniformly present in other studies or even between trials, in the same study (Videnska et al., 2013; Azcarate-Peril et al., 2018; Ma et al., 2020; Mon et al., 2020; Pedroso et al., 2021). The bacterial composition of this competitive exclusion product was not reflected in the cecal community composition in animals administered the commercial product (Pedroso et al., 2016). This may be due, in part, to the microbial succession that occurs in the maturation of the intestinal microbiome (Lu et al., 2003). However, the CE product does contain members that appear in an intestinal compartment, ileum or cecum, at some point in the intestine’s development (Lu et al., 2003). Community diversity appears to be key in pathogen exclusion (Pedroso et al., 2021) in that it is not as important who is present as what the community does “collectively” to exclude Salmonella. Others have also noted the importance of community diversity in pathogen exclusion (van Elsas et al., 2012; Antharam et al., 2013; Lone et al., 2013; Stanley et al., 2014; Xu et al., 2014; Zhang et al., 2015; Chopyk et al., 2016). This diversity provides resilience needed to return to homeostasis following some perturbation to the system (Weimer, 2015). As long as there are sufficient members present that collectively perform a specific function, the system is maintained; in this case microbiome’s ability to maintain a barrier to pathogen colonization. Therefore, seeding the chick with the adult intestinal microbiome, provides it with sufficient members, collectively capable of excluding Salmonella while its own microbiome develops as the animal matures.
It is unclear how the products exclude pathogens but it seems unlikely that composition is only one characteristic responsible for their protective effects. The possible mechanisms of action of competitive exclusion formulations have been previously reviewed (Nurmi et al., 1992; Nisbet, 1998; Tuomola et al., 2001; Becker, 2005). Bacteria comprising exclusive communities may: produce bactericidal molecules that damage the cellular integrity of pathogenic bacteria; decrease the growth rate of pathogens by providing competition for nutrients or produce molecules that inhibit processes involved in cell division; produce molecules that reduce expression of, or function of, factors involved in colonization; cause or enhance predation; or physically occupy or modify the ecological niche targeted by the pathogen. Multiple mechanisms are likely responsible for pathogen exclusion. This would explain why diversity is key; why no one specie(s) was consistently associated with Salmonella exclusion in this study. Salmonella is metabolically versatile in that it can utilize some metabolites (ethanolamine, propanediol, etc.) that few other community members can. A metabolic gene(s) involved in competition for substrate A may be distributed across a diversity of bacterial species, where any one, member species could compete for substrate A. This may explain why no one bacterial species has been consistently associated with reduced Salmonella abundance. While an intestinal member species may be able to compete with Salmonella for one substrate or metabolite, Salmonella could turn to another, and another substrate enabling persistent colonization even with low energy substrates. Only organisms with similar metabolic potential could outcompete Salmonella, especially another Salmonella (Cheng et al., 2015). Therefore, if competition were the mechanism of competitive exclusion, it would take a broad array of member species for the community to outcompete Salmonella for all the substrates and metabolites present in the different portions of the intestine.
Similarly, antagonism may be at the heart of competitive exclusion. The antibacterial activity of a competitive bacterial species may be attributed to several factors (Alakomi et al., 2005). For example, the mechanism of antibacterial activity of exclusive lactobacilli might be due to a synergistic action of lactic acid and bacteriocins. Lactate acts as a permeabilizer of the outer membrane of Gram-negative bacteria, increasing their susceptibility to antimicrobial molecules (Fayol-Messaoudi et al., 2005). Lactate also affects the intestinal pH which may affect the surface structures and metabolism of Salmonella (Foster et al., 1994). Other bacterial metabolites, such as acetate, propionate and butyrate, may also contribute to community exclusion of some bacterial species because the undissociated organic acids freely diffuses across the bacterial membrane, lowering the cytoplasmic pH and uncoupling electron transport (Ricke, 2003). The intestinal tract contains high levels of these volatile short chain fatty acids (SCFA) which are produced from the breakdown of complex carbohydrates by anaerobes such as the Clostridia, Bacteroides, and Bifidobacterium. SCFAs have inhibitory effects on Salmonella colonization of the gastrointestinal tract (Bohnhoff et al., 1964); and can modulate expression of Salmonella invasion genes (Durant et al., 1999; Durant et al., 2000a; Durant et al., 2000b). In addition, bile salt deconjugation by Clostridia, to form cholate and deoxycholate, can also synergistically inhibit Salmonella invasion (Ducarmon et al., 2019). The culmination of all of these factors are likely at play in pathogen exclusion requiring collective species metabolic activity.
Community diversity appears to be important in providing an ecosystem with multi-functionality and redundancy of function (Delgado-Baquerizo et al., 2016a; Delgado-Baquerizo et al., 2016b). Diversity maintains function, even when a perturbation is introduced (Isobe et al., 2020). Most juveniles obtain their microbiomes from the parent (Ferretti et al., 2018; Kubasova et al., 2019; Zhu et al., 2021; Chen et al., 2022). The adult microbiome provides the young with pioneer colonizers important in intestinal development, immune function and pathogen exclusion. In poultry production, juveniles are separated from the adult hen, prior to hatch. Seeding chicks with a competitive exclusion product provides an effective barrier to pathogen colonization, that would be otherwise absent in this production environment.
Conclusion
Pathogen exclusion may be a combination of competition and antagonism. In either case, a bacterial census is not likely to identify the magic bullet, the single organism, responsible for excluding Salmonella from a mixed community as exists in the chicken intestine. The key to understanding competitive exclusion will come from comparing communities that permit and exclude Salmonella and associated transcriptome, proteome, or metabolome evidence of competition and antagonism (García et al., 2017). Regulatory agencies are starting to come around to commercial acceptance of these fecal communities for treating or preventing bacterial infections, as evident from US Food and Drug Administration’s approval of fecal microbiota product for the treatment of Clostridium difficile infections (Pharmaceutical Technology Editors, 2022).
Statements
Data availability statement
16S sequence data is available through MG-RAST, metagenomics analysis server (mg-rast.org). The individual data sets can be accessed through https://www.mg-rast.org/linkin.cgi?project = followed by mgp40703 (Commercial Lot M), mgp40704 (Commercial Lot L), mgp4075 (Commercial Lot K), mgp40706 (Commercial Lot J), mgp40707 (Competitive Exclusion Seed I), mgp40708 (Commercial Lot H), mgp40709 (Commercial Lot G), mgp40710 (Commercial Lot F), mgp40711 (Competitive Exclusion Seed E), mgp40712 (Competitive Exclusion Seed D), mgp40713 (Competitive Exclusion Seed C), mgp40714 (Competitive Exclusion B) and mgp40715 (Competitive Exclusion Master Seed A). Therefore, to access Master Seed A, the link is https://www.mg-rast.org/linkin.cgi?project=mgp40715.
Author contributions
ML was responsible for funding acquisition, conceptualization, writing, review, and editing. AP performed the experiments, formal analysis and data curation described in this article. AP and JM were involved in the writing, review and editing.
Funding
This work was supported by Intervet International licenser of the competitive exclusion product Aviguard used in this study. As per contractual agreement between Intervet International and the University of Georgia, the company was provided formal results of this study, prior to submission, but without their input into the study’s findings or writing of said manuscript. JM was supported by United States Department of Agriculture HATCH funds: VA-160130.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CC declared a shared parent affiliation with the author AP to the handling editor at the time of the review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1043383/full#supplementary-material
References
1
AlakomiH. L.MattoJ.VirkajarviI.SaarelaM. (2005). Application of a microplate scale fluorochrome staining assay for the assessment of viability of probiotic preparations. J. Microbiol. Methods62 (1), 25–35. 10.1016/j.mimet.2005.01.005
2
AntharamV. C.LiE. C.IshmaelA.SharmaA.MaiV.RandK. H.et al (2013). Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol.51 (9), 2884–2892. 10.1128/jcm.00845-13
3
ApajalahtiJ. H.KettunenA.BedfordM. R.HolbenW. E. (2001). Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl. Environ. Microbiol.67 (12), 5656–5667. 10.1128/AEM.67.12.5656-5667.2001
4
Azcarate-PerilM. A.ButzN.CadenasM. B.KociM.BallouA.MendozaM.et al (2018). An attenuated Salmonella enterica serovar typhimurium strain and galacto-oligosaccharides accelerate clearance of Salmonella infections in poultry through modifications to the gut microbiome. Appl. Environ. Microbiol.84 (5), e02526. 10.1128/AEM.02526-17
5
BarnesE. M.MeadG. C.BarnumD. A.HarryE. G. (1972). The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br. Poult. Sci.13 (3), 311–326. 10.1080/00071667208415953
6
BarnesE. M. (1979). The intestinal microflora of poultry and game birds during life and after storage. Address of the president of the Society for Applied Bacteriology delivered at a meeting of the society on 10 January 1979. J. Appl. Bacteriol.46 (3), 407–419. 10.1111/j.1365-2672.1979.tb00838.x
7
BeckerP. M. (2005). Physiological Achilles' heels of enteropathogenic bacteria in livestock. Curr. Issues Intest. Microbiol.6 (2), 31–54.
8
BohnhoffM.MillerC. P.MartinW. R. (1964). Resistance of the mouse's intestinal tract to experimental Salmonella infection. I. Factors which interfere with the initiation of infection by oral inoculation. J. Exp. Med.120, 805–816. 10.1084/jem.120.5.805
9
CameronD. M.CarterJ. N.MansellP. (1996). “Evaluation of Aviguard against a Salmonella enteritidis infective model in broiler chickens,” in Western Poultry Disease Conference.
10
CameronD. M.CarterJ. N.MansellP.RedgraveV. A. (1997). “Floor-pen efficacy study with Aviguard against Salmonella typhimurium DT104 colonization in turkeys,” in Proc. International Symposium, 481–485.
11
ChenW.MaJ.JiangY.DengL.LvN.GaoJ.et al (2022). Selective maternal seeding and rearing environment from birth to weaning shape the developing piglet gut microbiome. Front. Microbiol.13, 795101. 10.3389/fmicb.2022.795101
12
ChengY.PedrosoA. A.PorwollikS.McClellandM.LeeM. D.KwanT.et al (2015). rpoS-Regulated core genes involved in the competitive fitness of Salmonella enterica Serovar Kentucky in the intestines of chickens. Appl. Environ. Microbiol.81 (2), 502–514. 10.1128/AEM.03219-14
13
ChopykJ.MooreR. M.DiSpiritoZ.StrombergZ. R.LewisG. L.RenterD. G.et al (2016). Presence of pathogenic Escherichia coli is correlated with bacterial community diversity and composition on pre-harvest cattle hides. Microbiome4 (1), 9–11. 10.1186/s40168-016-0155-4
14
CorrierD. E.NisbetD. J.ByrdJ. A.2ndHargisB. M.KeithN. K.PetersonM.et al (1998). Dosage titration of a characterized competitive exclusion culture to inhibit Salmonella colonization in broiler chickens during growout. J. Food Prot.61 (7), 796–801. 10.4315/0362-028x-61.7.796
15
Delgado-BaquerizoM.GiaramidaL.ReichP. B.KhachaneA. N.HamontsK.EdwardsC.et al (2016b). Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J. Ecol.104 (4), 936–946. 10.1111/1365-2745.12585
16
Delgado-BaquerizoM.MaestreF. T.ReichP. B.JeffriesT. C.GaitanJ. J.EncinarD.et al (2016a). Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun.7 (1), 10541–10548. 10.1038/ncomms10541
17
DeruyttereL.KlaasenJ.FroymanR.DayC. A. (1997). “Field study to demonstrate the efficacy of Aviguard against intestinal Salmonella colonization in broilers,” in Proc. Salmonella and Salmonellosis, Ploufragen, France (Zoopole Development, Institut Superieur des Productions Animales et des Industries Agro-alimentaires), 523–525.
18
DucarmonQ.ZwittinkR.HornungB.Van SchaikW.YoungV.KuijperE. (2019). Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol. Mol. Biol. Rev.83 (3), e00007–e00019. 10.1128/MMBR.00007-19
19
DurantJ. A.CorrierD. E.ByrdJ. A.StankerL. H.RickeS. C. (1999). Feed deprivation affects crop environment and modulates Salmonella enteritidis colonization and invasion of leghorn hens. Appl. Environ. Microbiol.65 (5), 1919–1923. 10.1128/AEM.65.5.1919-1923.1999
20
DurantJ. A.CorrierD. E.RickeS. C. (2000a). Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella typhimurium. J. Food Prot.63 (5), 573–578. 10.4315/0362-028x-63.5.573
21
DurantJ. A.CorrierD. E.StankerL. H.RickeS. C. (2000b). Expression of the hilA Salmonella typhimurium gene in a poultry Salm. enteritidis isolate in response to lactate and nutrients. J. Appl. Microbiol.89 (1), 63–69. 10.1046/j.1365-2672.2000.01089.x
22
Fayol-MessaoudiD.BergerC. N.Coconnier-PolterM. H.Lievin-Le MoalV.ServinA. L. (2005). pH-Lactic acid-and non-lactic acid-dependent activities of probiotic Lactobacilli against Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol.71 (10), 6008–6013. 10.1128/AEM.71.10.6008-6013.2005
23
FerreiraA. J.FerreiraC. S.KnoblT.MorenoA. M.BacarroM. R.ChenM.et al (2003). Comparison of three commercial competitive-exclusion products for controlling Salmonella colonization of broilers in Brazil. J. Food Prot.66 (3), 490–492. 10.4315/0362-028x-66.3.490
24
FerrettiP.PasolliE.TettA.AsnicarF.GorferV.FediS.et al (2018). Mother-to-Infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe24 (1), 133–145. e135. 10.1016/j.chom.2018.06.005
25
Food and Agriculture Organization of the United Nations (2016). in Probiotics in animal nutrition: Production, impact and regulation, F.A.P.a. Health.
26
FosterJ. W.ParkY. K.BangI. S.KaremK.BettsH.HallH. K.et al (1994). Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology140 (2), 341–352. 10.1099/13500872-140-2-341
27
FranksA. H.HarmsenH. J.RaangsG. C.JansenG. J.SchutF.WellingG. W. (1998). Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol.64 (9), 3336–3345. 10.1128/AEM.64.9.3336-3345.1998
28
GarcíaC.TebbjiF.DaigneaultM.LiuN. N.KöhlerJ. R.Allen-VercoeE.et al (2017). The human gut microbial metabolome modulates fungal growth via the TOR signaling pathway. mSphere2 (6), e00555. 10.1128/mSphere.00555-17
29
GarciaS. L.JangidK.WhitmanW. B.DasK. C. (2011). Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresour. Technol.102 (15), 7249–7256. 10.1016/j.biortech.2011.04.098
30
Gómez-GallegoC.JunnilaJ.MännikköS.HämeenojaP.ValtonenE.SalminenS.et al (2016). A canine-specific probiotic product in treating acute or intermittent diarrhea in dogs: A double-blind placebo-controlled efficacy study. Vet. Microbiol.197, 122–128. 10.1016/j.vetmic.2016.11.015
31
GongJ.ForsterR. J.YuH.ChambersJ. R.SabourP. M.WheatcroftR.et al (2002). Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett.208 (1), 1–7. S0378109701005213 [pii]. 10.1111/j.1574-6968.2002.tb11051.x
32
Guerra-Garcia MirandaM. H. (2000). Aviguard: The competitive exclusion as control method of salmonella in poultry. Trabajo Monografico (Ing Zootecnista). 46p.
33
GuillotJ. F.SalmonA.MoulineC.DelaporteJ.MagninM. (1997). “Effect of a gut microflora (Aviguard) against controlled Salmonella enteritidis contamination in chickens,” in Proceedings of the Salmonella and Salmonellosis Symposium, 521.
34
HamadyM.WalkerJ. J.HarrisJ. K.GoldN. J.KnightR. (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods5 (3), 235–237. 10.1038/nmeth.1184
35
HirnJ.NurmiE.JohanssonT.NuotioL. (1992). Long-term experience with competitive exclusion and salmonellas in Finland. Int. J. Food Microbiol.15 (3-4), 281–285. 10.1016/0168-1605(92)90059-c
36
HooperL. V.FalkP. G.GordonJ. I. (2000). Analyzing the molecular foundations of commensalism in the mouse intestine. Curr. Opin. Microbiol.3 (1), 79–85. 10.1016/s1369-5274(99)00055-7
37
HumeM. E.HollisterA. G.NisbetD. J.CorrierD. E.DeLoachJ. R. (1996). Effect of a characterized continuous-flow culture of cecal bacteria on Salmonella typhimurium crop colonization in broiler chicks. Avian Dis.40 (2), 391–397. 10.2307/1592237
38
IsobeK.IseY.KatoH.OdaT.VincenotC. E.KobaK.et al (2020). Consequences of microbial diversity in forest nitrogen cycling: Diverse ammonifiers and specialized ammonia oxidizers. ISME J.14 (1), 12–25. 10.1038/s41396-019-0500-2
39
JakubczykD.LeszczyńskaK.GórskaS. (2020). The effectiveness of probiotics in the treatment of inflammatory bowel disease (IBD)-A critical review. Nutrients12 (7), 1973. 10.3390/nu12071973
40
KimK.HeY.XiongX.EhrlichA.LiX.RaybouldH.et al (2019). Dietary supplementation of Bacillus subtilis influenced intestinal health of weaned pigs experimentally infected with a pathogenic E. coli. J. Animal Sci. Biotechnol.10 (1), 52. 10.1186/s40104-019-0364-3
41
KubasovaT.KollarcikovaM.CrhanovaM.KarasovaD.CejkovaD.SebkovaA.et al (2019). Contact with adult hen affects development of caecal microbiota in newly hatched chicks. PLoS One14 (3), e0212446. 10.1371/journal.pone.0212446
42
LaiH. H.ChiuC. H.KongM. S.ChangC. J.ChenC. C. (2019). Probiotic Lactobacillus casei: Effective for managing childhood diarrhea by altering gut microbiota and attenuating fecal inflammatory markers. Nutrients11 (5), 1150. 10.3390/nu11051150
43
LanP. T.HayashiH.SakamotoM.BennoY. (2002). Phylogenetic analysis of cecal microbiota in chicken by the use of 16S rDNA clone libraries. Microbiol. Immunol.46 (6), 371–382. 10.1111/j.1348-0421.2002.tb02709.x
44
LeyR. E.HamadyM.LozuponeC.TurnbaughP. J.RameyR. R.BircherJ. S.et al (2008). Evolution of mammals and their gut microbes. Science320 (5883), 1647–1651. 10.1126/science.1155725
45
Leyva-DiazA. A.Hernandez-PatlanD.Solis-CruzB.AdhikariB.KwonY. M.LatorreJ. D.et al (2021). Evaluation of curcumin and copper acetate against Salmonella Typhimurium infection, intestinal permeability, and cecal microbiota composition in broiler chickens. J. Anim. Sci. Biotechnol.12 (1), 23. 10.1186/s40104-021-00545-7
46
LiuT.LiljebjelkeK.BartlettE.HofacreC.SanchezS.MaurerJ. J. (2002). Application of nested polymerase chain reaction to detection of Salmonella in poultry environment. J. Food Prot.65 (8), 1227–1232. 10.4315/0362-028x-65.8.1227
47
LoneA. G.SelingerL. B.UwieraR. R.XuY.InglisG. D. (2013). Campylobacter jejuni colonization is associated with a dysbiosis in the cecal microbiota of mice in the absence of prominent inflammation. PLoS One8 (9), e75325. 10.1371/journal.pone.0075325
48
LuJ.IdrisU.HarmonB.HofacreC.MaurerJ. J.LeeM. D. (2003). Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol.69 (11), 6816–6824. 10.1128/aem.69.11.6816-6824.2003
49
LuceyP. M.LeanI. J.AlyS. S.GolderH. M.BlockE.ThompsonJ. S.et al (2021). Effects of mannan-oligosaccharide and Bacillus subtilis supplementation to preweaning Holstein dairy heifers on body weight gain, diarrhea, and shedding of fecal pathogens. J. Dairy Sci.104 (4), 4290–4302. 10.3168/jds.2020-19425
50
MaB.MeiX.LeiC.LiC.GaoY.KongL.et al (2020). Enrofloxacin shifts intestinal microbiota and metabolic profiling and hinders recovery from Salmonella enterica subsp. enterica serovar typhimurium infection in neonatal chickens. mSphere5 (5), e00725. 10.1128/mSphere.00725-20
51
MeadG. C. (1989). Microbes of the avian cecum: Types present and substrates utilized. J. Exp. Zool. Suppl.3, 48–54. 10.1002/jez.1402520508
52
MedveckyM.CejkovaD.PolanskyO.KarasovaD.KubasovaT.CizekA.et al (2018). Whole genome sequencing and function prediction of 133 gut anaerobes isolated from chicken caecum in pure cultures. BMC Genomics19 (1), 561. 10.1186/s12864-018-4959-4
53
MonK. K. Z.ZhuY.ChanthavixayG.KernC.ZhouH. (2020). Integrative analysis of gut microbiome and metabolites revealed novel mechanisms of intestinal Salmonella carriage in chicken. Sci. Rep.10 (1), 4809. 10.1038/s41598-020-60892-9
54
NakamuraA.OtaY.MizukamiA.ItoT.NgwaiY. B.AdachiY. (2002). Evaluation of aviguard, a commercial competitive exclusion product for efficacy and after-effect on the antibody response of chicks to Salmonella. Poult. Sci.81 (11), 1653–1660. 10.1093/ps/81.11.1653
55
NeishA. S. (2009). Microbes in gastrointestinal health and disease. Gastroenterology136 (1), 65–80. [pii]. 10.1053/j.gastro.2008.10.080
56
NisbetD. J. (1998). Use of competitive exclusion in food animals. J. Am. Vet. Med. Assoc.213 (12), 1744–1746.
57
NurmiE.NuotioL.SchneitzC. (1992). The competitive exclusion concept: Development and future. Int. J. Food Microbiol.15 (3-4), 237–240. 10.1016/0168-1605(92)90054-7
58
PedrosoA. A.BatalA. B.LeeM. D. (2016). Effect of in ovo administration of an adult-derived microbiota on establishment of the intestinal microbiome in chickens. Am. J. Vet. Res.77 (5), 514–526. 10.2460/ajvr.77.5.514
59
PedrosoA. A.LeeM. D.MaurerJ. J. (2021). Strength lies in diversity: How community diversity limits Salmonella abundance in the chicken intestine. Front. Microbiol.12, 694215. 10.3389/fmicb.2021.694215
60
Pharmaceutical Technology Editors (2022). FDA approves first fecal microbiota product. United States Food and Drug Administration. Available at: https://www.fda.gov/news-events/press-announcements/fda-approves-first-fecal-microbiota-product (Accessed December 19, 2022).
61
RenaudD. L.KeltonD. F.WeeseJ. S.NobleC.DuffieldT. F. (2019). Evaluation of a multispecies probiotic as a supportive treatment for diarrhea in dairy calves: A randomized clinical trial. J. Dairy Sci.102 (5), 4498–4505. 10.3168/jds.2018-15793
62
RickeS. C. (2003). Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult. Sci.82 (4), 632–639. 10.1093/ps/82.4.632
63
SalanitroJ. P.BlakeI. G.MuirheadP. A. (1974a). Studies on the cecal microflora of commercial broiler chickens. Appl. Microbiol.28 (3), 439–447. 10.1128/am.28.3.439-447.1974
64
SalanitroJ. P.FairchildsI. G.ZgornickiY. D. (1974b). Isolation, culture characteristics, and identification of anaerobic bacteria from the chicken cecum. Appl. Microbiol.27 (4), 678–687. 10.1128/am.27.4.678-687.1974
65
SchlossP. D.WestcottS. L.RyabinT.HallJ. R.HartmannM.HollisterE. B.et al (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol.75 (23), 7537–7541. 10.1128/AEM.01541-09
66
ShangY.KumarS.OakleyB.KimW. K. (2018). Chicken gut microbiota: Importance and detection Technology. Front. Vet. Sci.5, 254. 10.3389/fvets.2018.00254
67
SisakF.HavlickovaH.KolacekJ. (2001). Prophylactic efficacy of Aviguard against salmonellosis in laying hens, 51.Veterinarstvi
68
SorensonT. (1948). A method of establishing groups of equal amplitude in plant sociology based on similarity of species content. K. Dan. Vidensk. Selsk.5 (1-34), 4–7.
69
StanleyD.WuS. B.RodgersN.SwickR. A.MooreR. J. (2014). Differential responses of cecal microbiota to fishmeal, Eimeria and Clostridium perfringens in a necrotic enteritis challenge model in chickens. PLoS One9 (8), e104739. 10.1371/journal.pone.0104739
70
StephanB. (2000). The efficacy of a competitive exclusion flora (Aviguard) against experimental Salmonella challenge in Turkey poults, 98–100.
71
SternN. J.CoxN. A.BaileyJ. S.BerrangM. E.MusgroveM. T. (2001). Comparison of mucosal competitive exclusion and competitive exclusion treatment to reduce Salmonella and Campylobacter spp. colonization in broiler chickens. Poult. Sci.80 (2), 156–160. 10.1093/ps/80.2.156
72
SzottV.ReicheltB.FrieseA.RoeslerU. (2022). A complex competitive exclusion culture reduces Campylobacter jejuni colonization in broiler chickens at slaughter age in vivo. Vet. Sci.9 (4), 181. 10.3390/vetsci9040181
73
TannockG. W. (1999). Analysis of the intestinal microflora: A renaissance. Ant. Van Leeuwenhoek76 (1-4), 265–278. 10.1023/a:1002038308506
74
TuomolaE.CrittendenR.PlayneM.IsolauriE.SalminenS. (2001). Quality assurance criteria for probiotic bacteria. Am. J. Clin. Nutr.73 (2), 393S–398S. 10.1093/ajcn/73.2.393s
75
van ElsasJ. D.ChiurazziM.MallonC. A.ElhottovaD.KristufekV.SallesJ. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U. S. A.109 (4), 1159–1164. 10.1073/pnas.1109326109
76
VidenskaP.SisakF.HavlickovaH.FaldynovaM.RychlikI. (2013). Influence of Salmonella enterica serovar Enteritidis infection on the composition of chicken cecal microbiota. BMC Vet. Res.9, 140. 10.1186/1746-6148-9-140
77
WeimerP. J. (2015). Redundancy, resilience, and host specificity of the ruminal microbiota: Implications for engineering improved ruminal fermentations. Front. Microbiol.6, 296. 10.3389/fmicb.2015.00296
78
XuX.YangS.OlajideJ. S.QuZ.GongZ.WangJ.et al (2021). Clostridium butyricum supplement can ameliorate the intestinal barrier roles in broiler chickens experimentally infected with Clostridium perfringens. Front. Physiol.12, 737481. 10.3389/fphys.2021.737481
79
XuY.Dugat-BonyE.ZaheerR.SelingerL.BarbieriR.MunnsK.et al (2014). Escherichia coli O157:H7 super-shedder and non-shedder feedlot steers harbour distinct fecal bacterial communities. PLoS One9 (5), e98115. 10.1371/journal.pone.0098115
80
ZhangL.DongD.JiangC.LiZ.WangX.PengY. (2015). Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe34, 1–7. 10.1016/j.anaerobe.2015.03.008
81
ZhuH.YangM.LoorJ. J.ElolimyA.LiL.XuC.et al (2021). Analysis of cow-calf microbiome transfer routes and microbiome diversity in the newborn holstein dairy calf hindgut. Front. Nutr.8, 736270. 10.3389/fnut.2021.736270
82
ZhuX. Y.JoergerR. D. (2003). Composition of microbiota in content and mucus from cecae of broiler chickens as measured by fluorescent in situ hybridization with group-specific, 16S rRNA-targeted oligonucleotide probes. Poult. Sci.82 (8), 1242–1249. 10.1093/ps/82.8.1242
Summary
Keywords
avian, microbiome, Salmonella, exclusion, competition
Citation
Lee MD, Pedroso AA and Maurer JJ (2023) Bacterial composition of a competitive exclusion product and its correlation with product efficacy at reducing Salmonella in poultry. Front. Physiol. 13:1043383. doi: 10.3389/fphys.2022.1043383
Received
13 September 2022
Accepted
20 December 2022
Published
09 January 2023
Volume
13 - 2022
Edited by
Brian B. Oakley, Western University of Health Sciences, United States
Reviewed by
Chongxiao (Sean) Chen, University of Georgia, United States
Sundus Javed, COMSATS University, Islamabad Campus, Pakistan
Updates
Copyright
© 2023 Lee, Pedroso and Maurer.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margie D. Lee, mlee2@vt.edu
† Present address: Adriana A. Pedroso, Ventura Coastal, LLC, Tipton, CA, United States
This article was submitted to Avian Physiology, a section of the journal Frontiers in Physiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.