- Institute of Nanfan and Seed Industry, Guangdong Academy of Sciences, Guangzhou, China
Spodoptera frugiperda is one of the most harmful pests that attack maize and other major food crops and causes huge economic loss every year in China and other countries and regions. Beauveria bassiana, a kind of entomological fungus that is highly pathogenic to pests, is harmless to the environment and human beings. However, at present, S. frugiperda has gradually developed resistance to many pesticides and microbial insecticides. In this study, transcriptome sequencing was conducted to analyze the differences in gene expression between B. bassiana-infected and -uninfected S. frugiperda. More than 160 Gb of clean data were obtained as 150-bp paired-end reads using the Illumina HiSeq™ 4000 platform, and 2,767 and 2,892 DEGs were identified in LH36vsCK36 and LH144vsCK144, respectively. In order to explore the roles of JAK/STAT, Toll, and Imd signaling pathways in antifungal immune response in S. frugiperda against B. bassiana infection, the expression patterns of those signaling pathway-related genes in B. bassiana-infected S. frugiperda were analyzed by quantitative real-time PCR. In addition, antifungal activity experiments revealed that the suppression of JAK/STAT, Toll, and Imd signaling pathways by inhibitors could inhibit the antifungal activity to a large extent and lead to increased sensitivity of S. frugiperda to B. bassiana infection, indicating that JAK/STAT, Toll, and Imd signaling pathways and their associated genes might be involved in the synthesis and secretion of antifungal substances. This study implied that JAK/STAT, Toll, and Imd signaling pathways played crucial roles in the antifungal immune response of the S. frugiperda larvae, in which the related genes of these signaling pathways could play special regulatory roles in signal transduction. This study would improve our understanding of the molecular mechanisms underlying innate immunity and provide the basis for a wide spectrum of strategies against antifungal resistance of S. frugiperda.
Introduction
The immune mechanism of insects belonging to the invertebrate phylum Arthropoda (Insecta) mainly relies on humoral and cellular immunity. The humoral immunity of insects is mainly composed of antimicrobial peptides and lysozymes against pathogenic fungi and antiviral factors, lectins, and other immune factors against viruses and other pathogens (Lemaitre and Hoffmann, 2007). The cellular immunity of insects is mainly based on the ability of blood cells (hemocytes) of insects, such as granule cells, plasma cells, bead cells, and granulocytes, to recognize and phagocytose pathogenic microorganisms and heal the wound of insects (Kanost et al., 2004; Rao and Yu, 2010).
There are four signaling pathways {JAK/STAT (Janus kinase/signal transducer and activator of transcription) signaling pathway, Toll signaling pathway, Imd (immunodeficiency) signaling pathway, and JNK (c-Jun NH (2)-terminal protein kinase) signaling pathway} in insects that mediate the immune response to different pathogens. The Toll signaling pathway and Imd signaling pathway are classical immune signaling pathways that mediate the transcription and synthesis of most antimicrobial peptides (AMPs) and other active substances (Tanji et al., 2007).
The JAK/STAT signaling pathway is guided by a variety of cytokines and involved in the regulation of many important physiological processes, including growth regulation (Hirano et al., 2000), cell proliferation and differentiation, cell apoptosis, embryonic development (Pires-daSilva and Sommer, 2003), and innate regulation of immunity (Brivanlou et al, 2002a; Karst et al., 2003; Souza-Neto et al., 2009). The extracellular binding of peptide ligands to specific transmembrane receptors initiates the activation of the JAK/STAT signaling pathway. The receptor undergoes conformational transformation and self-phosphorylation by receptor-related Janus kinases, and activated JAKs, in turn, phosphorylate the receptor, which favors the formation of docking sites for cytoplasmic signal transducer and activator of transcription (STAT). Then, the phosphorylated STATs, under the action of JAK, eventually translocate to the nucleus and activate the transcription of target genes (Levy and Darnell, 2002; Li, 2008; Zhang et al., 2023).
In general, the Toll signaling pathway can rapidly mediate the synthesis and secretion of antifungal and antibacterial peptides (Tauszig et al., 2000). When pathogens are recognized, the activated serine protease cascade also activates the Toll signaling pathway. After entering cells, immune signals complete the cytosolic signal transduction process through the core signal factors of the Toll signaling pathway and finally transduce into the nucleus and initiate the expression of target genes (Imler and Hoffmann, 2001; Tauszig-Delamasure et al., 2002; Naitza and Ligoxygakis, 2004; Uvell and Engström, 2007; Ao et al., 2008). The Imd signaling pathway is another important signaling pathway involved in the regulation of the innate immune system, which was first identified and characterized in Drosophila (Corbo and Levine, 1996; Rämet et al., 2002). The Imd signaling pathway mainly regulates antimicrobial peptides, such as aggresin, cecropin, drosocin, and diptericin, that exert effects on Gram-negative bacteria (Levashina et al., 1998; Kaneko et al., 2004a). After Gram-negative bacteria are recognized, the activated Imd proteins act on transcription growth factor kinase 1 (TAK1), which transmits immune signals to IKKs (including IKK-β and IKK-γ) (Rutschmann et al., 2000; Silverman et al., 2003). Then, IKKs activate Relish in two ways. One is that IKKs directly phosphorylate the NF-κB-like signaling factor Relish and activate it (Silverman et al., 2000). Another way is that IKKs function on the DREDD–Fadd–Relish complex, which hydrolyzes and releases the activated Relish protein (Leulier et al., 2002). Relish has two homologs, Relish1 and Relish2, which are activated and then enter the nucleus to regulate the transcription of antimicrobial peptide-related target genes downstream. The cellular immunity of insects mainly involves recognition and phagocytosis of pathogenic microorganisms by insect blood cells, such as granulosa cells, plasma cells, bead cells, and granulocytes, as well as the wound healing of insect bodies.
Spodoptera frugiperda, a lepidopteran of the family Noctuidae, originates from tropical and subtropical areas of the Americas (Todd and Poole, 1980). It is one of the most dangerous agricultural pests in the world and causes serious damage to maize and other major food and economic crops every year (Martinelli et al., 2006). S. frugiperda not only has an adverse impact on social economy and food security but also leads to a significant increase in the use of highly toxic pesticides. The feeding characteristics and development of resistance of S. frugiperda have led to the reuse and overuse of pesticides; these two interrelated problems potentially give rise to food crises and human and environmental health problems (Prasanna et al., 2018). In recent years, the abuse of chemical pesticides has led to a series of problems, such as drug resistance, excessive pesticide residues, and environmental pollution. Compared with conventional chemical control, microbial control has the advantages of specificity and ecological safety. Microbial pesticides, such as bacteria, fungi, and viruses, play an important role in the control of S. frugiperda. At present, entomopathogenic microorganisms such as Beauveria bassiana, Bacillus thuringiensis, and nuclear polyhedrosis viruses (SfMNPVs) have been registered and utilized for the control of S. frugiperda both at home and abroad (Poisot et al., 2018). B. bassiana is a kind of entomological fungus, which can infect more than 700 types of insects belonging to 149 families and 15 orders. At the same time, B. bassiana is harmless to the environment and warm-blooded animals but has a strong virulence for pests; its culture conditions are relatively simple; and the strain is cheap and easy to obtain. Currently, this pathogenic fungus is widely used as a biopesticide to control pests around the world (Ramos et al., 2017).
However, there are few reports on the immune response genes related to signaling pathways of S. frugiperda, an invasive pest in China, after its infection with B. bassiana. Therefore, in this study, high-throughput transcriptome sequencing was performed on S. frugiperda larvae collected at 36 h and 144 h after B. bassiana infection to screen out the genes related to the immune response of S. frugiperda to B. bassiana infection. This study would provide target genes for improving the control effect of B. bassiana against S. frugiperda and lay a foundation for understanding the mechanism of resistance of S. frugiperda to B. bassiana.
Materials and methods
Insects and tested strains
The larvae of S. frugiperda, obtained from a wild field, were reared at 27°C ± 1°C with a relative humidity of 65%∼85% and a photoperiod of 14 L:10 D at the Institute of Nanfan and Seed Industry, Guangdong Academy of Sciences, Guangzhou, China. After at least three generations, the third-instar larvae with the same individual length and instar and moderate epidermal state were selected as the experimental subjects.
Preparation of B. bassiana conidial suspension
B. bassiana was cultured on a potato dextrose agar (PDA) medium at 27°C ± 1°C for 1 week. Then, 10 ml 0.15 mol/L NaCl solution was added into the tubes which were then oscillated gently. The conidia and hyphae were transferred into a centrifuge tube with 20 μL Tween-80, vortex-shaken for 10 min, and then filtered. The spores were counted using a blood count plate and diluted with 0.15 mol/L NaCl solution to a concentration of 108 spores/ml. Then, 1 mL of the prepared conidia suspension was added into 10 mL potato dextrose (PD) medium and then cultured at 30°C ± 1°C, 180 rpm for 3 days. The mixture of hyphae and blastospores were autoclaved at 121°C for 20 min to generate the heat-inactivated suspension of B. bassiana. Finally, the B. bassiana conidial suspension and inactivated B. bassiana conidial suspension were used for immune induction in S. frugiperda.
cDNA library construction, transcriptome sequencing, assembly, and functional annotation
The larvae of S. frugiperda were treated with 1 × 108/mL spore suspension of B. bassiana Bb378, while those treated with 0.05% Tween-80 solution were set as controls. The infected larvae were collected at 36 and 144 h after treatment, then flash-frozen with liquid nitrogen, and stored at −80°C. Each treatment was performed using three biological replicates. For every sample, the total RNA was extracted using TRIzol reagent (Invitrogen, United States) according to the manufacturer’s instructions. Contaminating genomic DNA was removed using RNase-free DNase I (TaKaRa Biotechnology Co., Ltd., Dalian, China), and then, the quantity and quality of RNA were assessed. RNA with high concentration, integrity, and purity was chosen for cDNA library construction and final Illumina sequencing at Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The obtained cDNA was then tested and sequenced on the Illumina HiSeq™ 4000 platform as 150-bp paired-end reads.
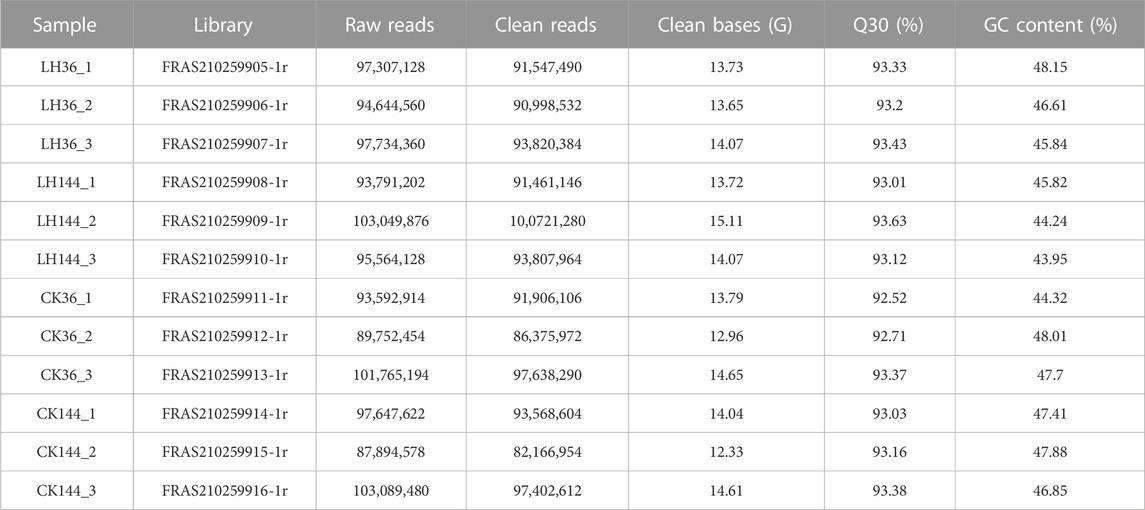
The adapters, primers, ambiguous “N” nucleotides, and low-quality (50% of the bases had a quality value ≤5) sequences were removed from the raw data to obtain clean reads. Then, the quality of the clean reads was assessed by Q30 (percentage of bases with a Phred value of at least 30), the GC content, and sequence duplication level. The clean data were assembled into de novo contigs using Trinity software (Grabherr et al., 2011). Subsequently, transcripts were assembled and obtained by using the de Bruijn graph method. Finally, unigenes were formed from the assembled transcripts using the TGI Clustering tool (Quackenbush et al., 2001; Pertea et al., 2003).
Annotations of all unigenes were performed using BLASTx against a pooled database of the National Center for Biotechnology Information non-redundant (NCBI-NR) protein, UniProt, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) with an E-value <10−5. The annotation of unigenes was obtained using HMMER software (Eddy, 1998), and then, Gene Ontology (GO) annotations were performed on all unigenes using Blast2GO. Subsequently, WEGO was used to determine GO functional classification and evaluate the distribution of gene functions at the macro-level (Ye et al., 2006). Metabolic pathway annotations for the unigenes were predicted based on the KEGG annotations (Kanehisa et al., 2008).
Expression analysis by real-time quantitative PCR
The expression of 28 tested genes related to the Toll signaling pathway, Imd (immunodeficiency) signaling pathway, and JAK/STAT (Janus kinase/signal transducer and activator of transcription) signaling pathway was verified by real-time quantitative PCR (qRT-PCR) with specific primers (Supplementary Table S1). Tissue samples were collected from the infected larvae of S. frugiperda at 12, 24, 36, 48, 60, 72, and 144 h after treatment with three biological replicates. A total of 10 annotated unigenes were selected randomly and quantified by real-time quantitative PCR (qRT-PCR) with specific primers (Supplementary Table S2) to verify the quality of the mRNA-seq data and expression level. The total RNA of S. frugiperda was extracted from samples from LH36, CK36, LH144, and CK144 using the TRIzol method (TaKaRA, Japan). To obtain the first-strand cDNAs, 1 μg of total RNA from the transcriptome samples was reverse-transcribed in a 20-μL reaction system, according to the manufacturer’s instruction (PrimeScript™ RT Reagent Kit, TaKaRa, Japan). qRT-PCR was performed using LightCycler®480 SYBR Green I Master (Roche Diagnostics, Basel, Switzerland) and run on the LightCycler®480 Real-time PCR system (Roche Diagnostics Ltd.) according to the manufacturer’s instructions. Each reaction was conducted in a 10-μL reaction system with 1 μL cDNA (2 ng/μL), 5 μL SYBR Green I Master (LightCycler®480 SYBR Green I Master, Roche Diagnostics Ltd., Lewes, United Kingdom), 0.5 μL/primer, and 3 μL ddH2O. The amplification conditions for qRT-PCR were as follows: denaturation at 95°C for 5 min, followed by 40 cycles for 5 s at 95°C, 20 s at 60°C, and 20 s at 72°C. gapdh was used as the internal reference gene, and each gene was tested in triplicate. The relative expression levels of the candidate chemosensory genes normalized to the internal control gene were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Analysis of the relative gene expression data was carried out using real-time quantitative PCR and the 2−ΔΔCT method.
The effect of signaling pathway inhibitors on antifungal activity in S. frugiperda
The third-instar larvae were injected with the Toll inhibitor (BAY 11-7082), Imd inhibitor (parthenolide), JAK/STAT inhibitor mix (20 μM tyrphostin AG 490, Selleck Chemicals, S1143; 5 μM nifuroxazide, Selleck Chemicals, S4182), heat-inactivated B. bassiana conidial suspension, and 0.15 mol/L NaCl solution (as the control), with a volume of 1 μL. A total of 60 larvae were injected in each treatment and performed in triplicate. After 24 h, the injected S. frugiperda larvae were submerged in B. bassiana conidial suspension (108 spores/mL, with 1% penicillin–streptomycin) for 5 s and then reared with fresh corn at 70% humidity and 26°C ± 2°C. The number of dead larvae was recorded every 12 h. A lethal time of 50% (LT50) was used to show the resistance of S. frugiperda larvae against B. bassiana. LT50 and confidence intervals of 95% of each treatment were analyzed and calculated using SPSS21.0.
Data accessibility
The Illumina reads of S. frugiperda were submitted to the NCBI Short Archive (SRA) with BioProject PRJNA987447. Their accession numbers are SAMN35982286, SAMN35982287, SAMN35982288, and SAMN35982289.
Results
Overview of the transcriptome in S. frugiperda
After removing the low-quality reads, trimming off the adapter sequences, sequencing, and a subsequent quality control process, a total of 1,155,833,496 raw reads and 1,111,415,334 clean reads were obtained from those 12 libraries. Furthermore, more than 160 Gb of clean data were obtained. The Q20, Q30, and GC content of each library were over 97.30%, 92.52%, and 43.95%, respectively (Table 1). Unigenes obtained from these transcriptomes were annotated in public databases, including the National Center for Biotechnology Information non-redundant (NCBI-NR) protein, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Based on the annotated results of those transcriptomes, the unigene annotation information from LH36vsCK36 and LH144vsCK144 was selected and analyzed.
Differential gene expression analysis and functional annotation
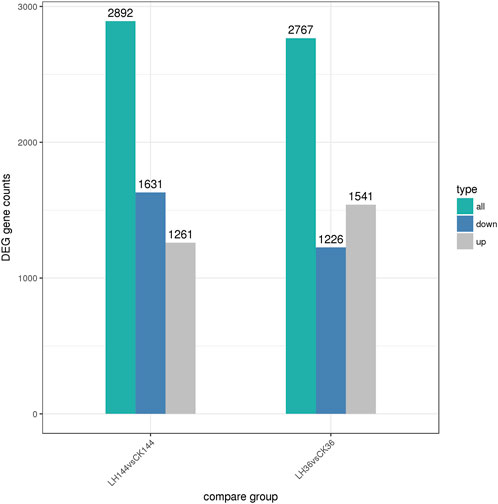
According to the statistical analysis, 2,767 and 2,892 DEGs were identified in LH36vsCK36 and LH144vsCK144, respectively. In LH36vsCK36, 1,541 DEGs were upregulated and 1,226 DEGs were downregulated, while in LH144vsCK144, 1,261 DEGs were upregulated and 1,631 DEGs were downregulated (Figure 1).
GO enrichment analyses for the DEGs were processed in LH36vsCK36 and LH144vsCK144. A total of 623 and 670 DEGs of LH36vsCK36 and LH144vsCK144 were enriched in GO annotation under three main terms, respectively. In LH36vsCK36, 338, 63, and 222 DEGs were enriched in “biological process,” “cellular component,” and “molecular function,” while in LH144vsCK144, 365, 63, and 241 DEGs were enriched, respectively (Figure 2). A corrected p-value <1 was used to screen the significantly enriched GO terms, and the up- and downregulated DEGs in significantly enriched GO terms in LH36vsCK36 and LH144vsCK144 were statistically analyzed.
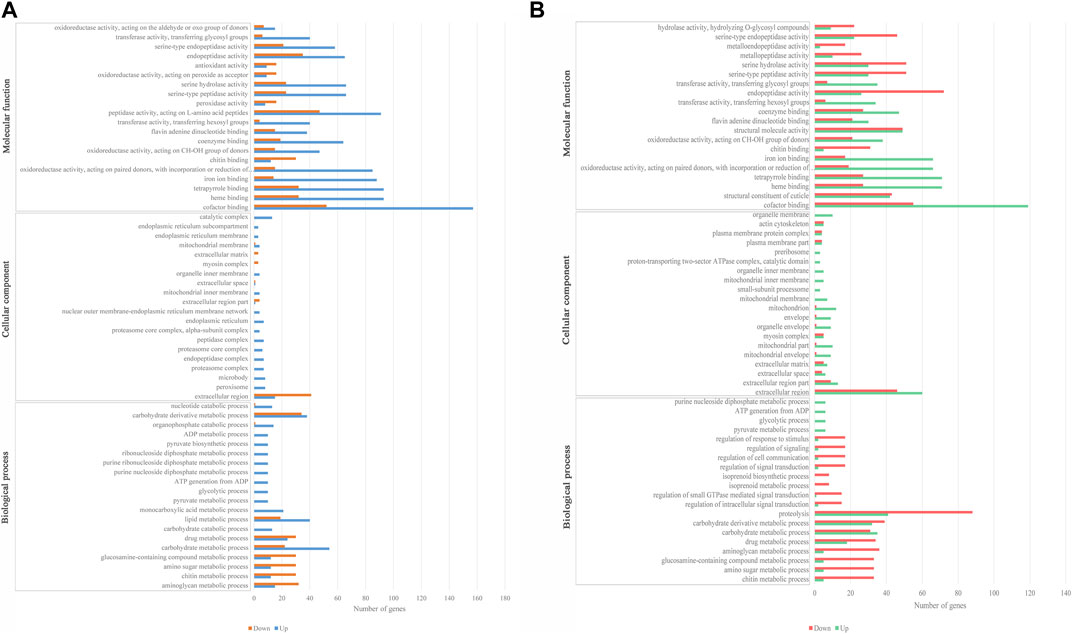
FIGURE 2. Secondary GO annotation of DEGs. (A) Top 20 functional enrichment DEGs in biological process, cellular component, and molecular function in LH36vsCK36; (B) top 20 functional enrichment DEGs in biological process, cellular component, and molecular function in LH144vsCK144.
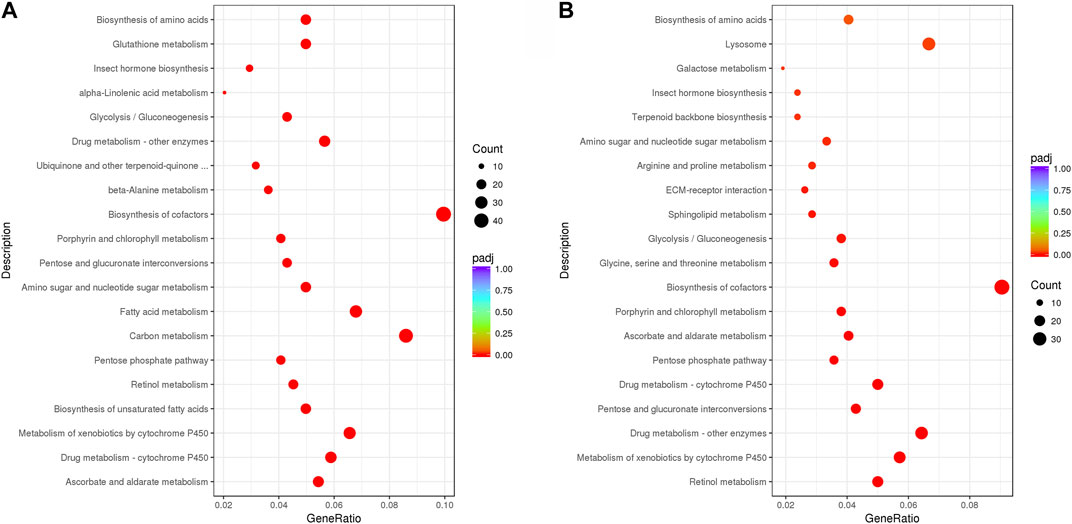
From the KEGG enrichment results, the most significant 20 KEGG pathways were selected to draw a scatter plot. In LH36vsCK36, the DEGs were mainly enriched in the “biosynthesis of cofactors” and “carbon metabolism,” followed by “fatty acid metabolism,” “metabolism of xenobiotics by cytochrome P450,” and “drug metabolism—cytochrome P450,” while in LH144vsCK144, the DEGs were mainly enriched in the “biosynthesis of cofactors,” “lysosome,” and “drug metabolism—other enzymes,” followed by “drug metabolism—cytochrome P450,” “metabolism of xenobiotics by cytochrome P450” and “retinol metabolism” (Figure 3).
Validation of DEG data by qRT-PCR
A total of 10 unigenes were randomly selected for qRT-PCR to confirm the result of the DEG expression using Illumina sequencing in LH36vsCK36 and LH144vsCK144, respectively. Data were presented as log2 values of fold changes in gene expression, normalized to gapdh relative to each sample. In LH36vsCK36, the qRT-PCR results supported the data obtained by DEG analysis (Figure 4). In LH144vsCK144, the results of the changing trend of qRT-PCR underpinned the reliability of the DEG results.
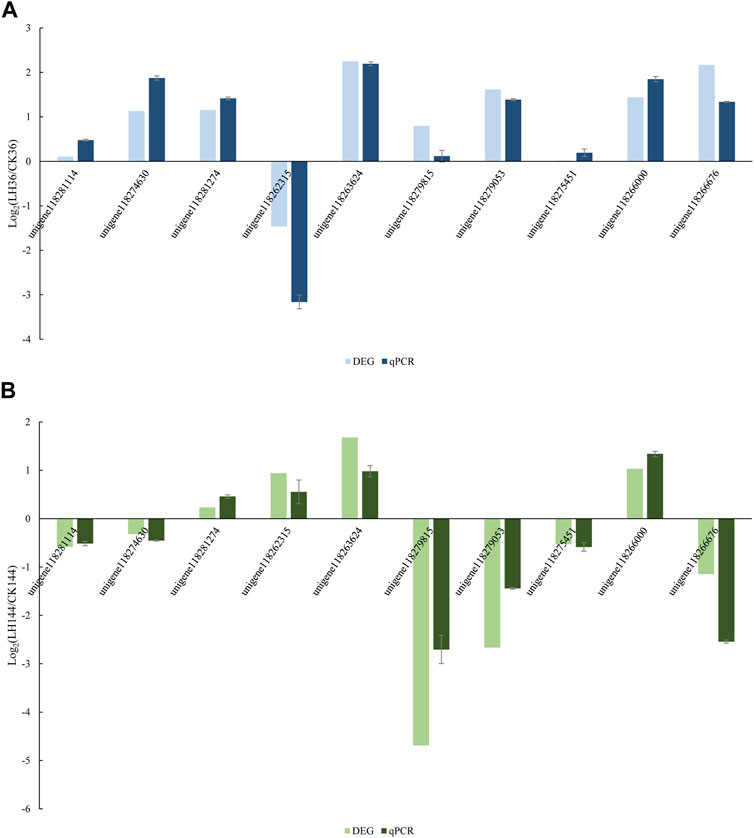
FIGURE 4. Expression ratios of 10 genes in (A) LH36vsCK36; (B) LH144vsCK144. Note: The fold changes in the genes were calculated as the log2 value of each comparison and are shown on the x-axis. Each error bar indicates the standard error with SEMs from the analysis of three replicates (p < 0.05).
Expression patterns of factors related to the Toll signaling pathway in S. frugiperda infected with B. bassiana
Sfruspatzle is an extracellular ligand protein of Toll-like receptors (TLRs). The qPCR result revealed that after being infected with B. bassiana, the relative expression of Sfruspatzle was slightly upregulated in the early stage of infection, while it was sharply upregulated 48 h after infection (Figure 5). The expression patterns of different Toll receptor genes showed significant differences. After being infected with B. bassiana, in S. frugiperda, the peaks of relative expression of Sfrutoll3 occurred at 24, 48, and 144 h; those of Sfrutoll6 occurred at 12 and 36 h; those of Sfrutoll7 appeared at 12 and 60 h; those of Sfrutoll18w occurred at 48 and 72 h; and Sfrutollo showed a high relative expression from 12 to 60 h (Figure 5).
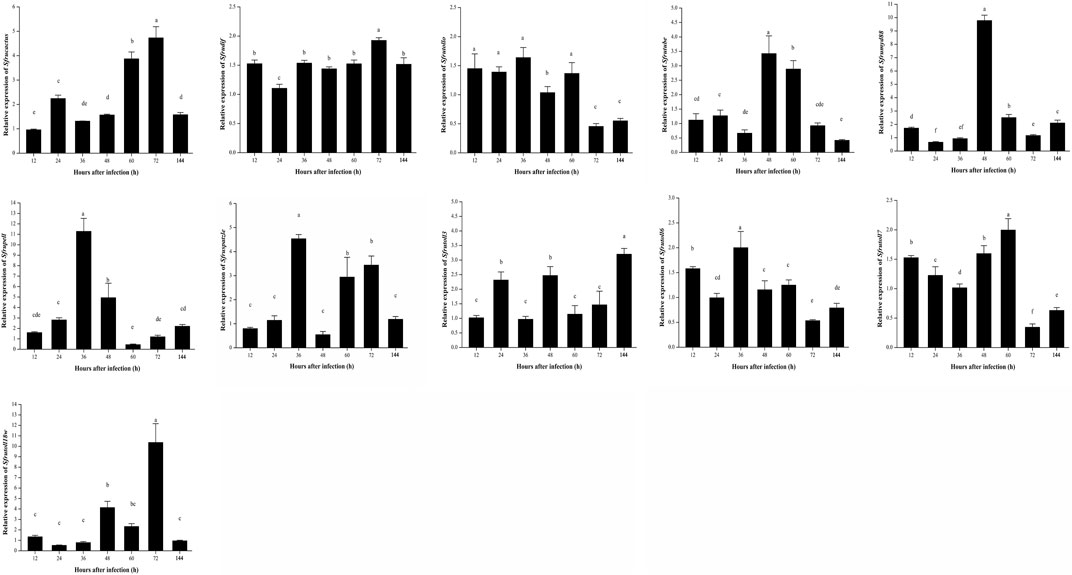
FIGURE 5. Expression patterns of Toll signaling pathway-related genes in S. frugiperda infected by B. bassiana.
The cytoplasmic signal transduction factors, including Sfrutube, Sfrupell, Sfrumyd88, Sfrucactus, and Sfrudif, have become the core signal transfer factors in the Toll signaling pathway. The relative expression of Sfrutube was significantly upregulated within 48–60 h after infection; the relative expression of Sfrucactus was upregulated and peaked within 60–72 h after infection; the relative expression of Sfrupell and Sfrumyd88 peaked at 36 and 48 h, respectively (Figure 5); and there was no significant change in the relative expression of Sfrudif throughout the infection stage. These results suggest that there may be different regulatory modes of intracellular signal transduction factors in the Toll signaling pathway of S. frugiperda to conduct an immune response against B. bassiana infection.
Expression patterns of factors related to the Imd signaling pathway in S. frugiperda infected with B. bassiana
In this study, the expression patterns of three signal factors associated with Imd signaling pathways were detected after infection by B. bassiana. After infection with B. bassiana, the expression of related factors of the Imd signaling pathway was changed, and there were significant differences in expression patterns. The relative expression of Sfrufadd and Sfrupgrp-lb all showed a trend of upregulating and then downregulating, and they reached the peak at 60 and 72 h after infection with B. bassiana, respectively, and then downregulated. On the other hand, the relative expression of Sfrurelish2 was significantly upregulated from 48 to 60 h after being infected with B. bassiana, and in other infection stages, it showed no significant differences (Figure 6).
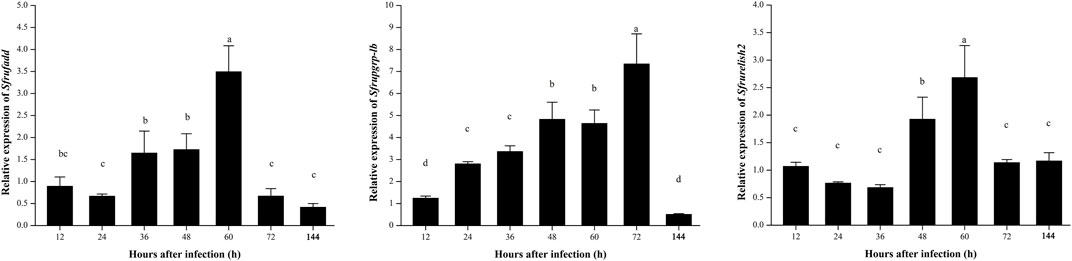
FIGURE 6. Expression patterns of Imd signaling pathway-related genes in S. frugiperda infected by B. bassiana.
Expression patterns of factors related to the JAK/STAT signaling pathway in S. frugiperda infected with B. bassiana
In the JAK/STAT signaling pathway, STAT is the core signal transduction factor that is the pattern recognition receptor which can recognize the β-l,3-glucan of fungi (the main component of the fungal cell wall) and lead to the subsequent immune response. When S. frugiperda was infected with B. bassiana, both Sfructl4 and Sfrustat5B showed a significantly upregulated pattern at 72 and 144 h (Figure 7); Sfrusocs2, Sfrusocs4, Sfrusocs5, Sfrutab1, and Sfruken are negative regulatory factors of the JAK/STAT signaling pathway, but the mode of regulation remains unclear. After infection with B. bassiana, the expression patterns of these five regulatory factors were significantly different. The relative expression levels of Sfrusocs2 and Sfrusocs4 presented a similar trend during the whole infection stages; Sfrusocs5 was significantly upregulated 48 h after infection with B. bassiana and peaked at 60 h; and the relative expression levels of Sfrutab1 were significantly upregulated at 24 and 48 h (Figure 7).
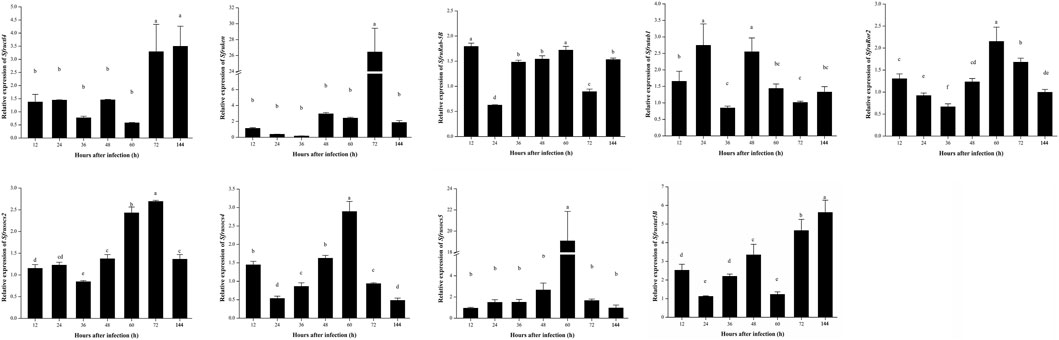
FIGURE 7. Expression patterns of JAK/STAT signaling pathway-related genes in S. frugiperda infected by B. bassiana.
The effect of signaling pathway inhibitors on the survival rate of S. frugiperda infected with B. bassiana
In order to confirm the functions of Toll, Imd, and JAK/STAT signaling pathways in S. frugiperda against B. bassiana infection, the effect of signaling pathway inhibitors on the survival rate of S. frugiperda infected by B. bassiana was tested. The results showed that dead larvae could be found earliest at 48 h and overgrown by the fungus approximately 144 h after injection. In Figure 8, compared to the control group, the survival rate curve was significantly shifted leftward in the group injected with inhibitors (Figure 8). Moreover, the LT50 analysis confirmed this observation. As shown in the tables, LT50 of the group injected with the Toll inhibitor was 20 h less than that of the control group (Supplementary Table S3); LT50 of the group injected with the Imd inhibitor was 21 h less than that of the control group (Supplementary Table S4); and LT50 of the group injected with the JAK/STAT inhibitor mix was 24 h less than that of the control group (Supplementary Table S5), indicating that the inhibition of Toll, Imd, and JAK/STAT signaling pathways could accelerate the pathogenicity of B. bassiana against S. frugiperda.
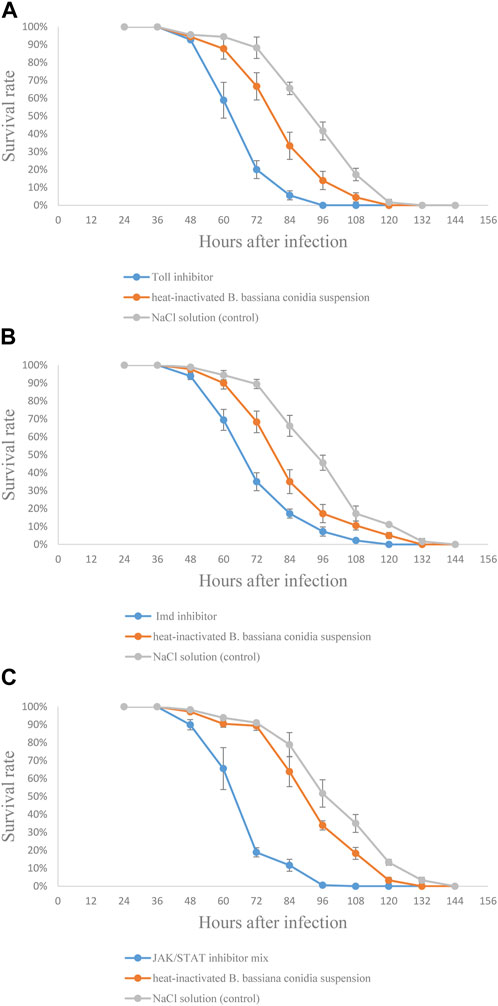
FIGURE 8. Effects of signaling pathway inhibitors on resistance of S. frugiperda larvae against B. bassiana infection. (A) Test with the Toll signaling pathway inhibitor; (B) test with the Imd signaling pathway inhibitor; and (C) test with JAK/STAT signaling pathway inhibitor mix.
Expression patterns of some genes of antibacterial peptides in S. frugiperda infected with B. bassiana
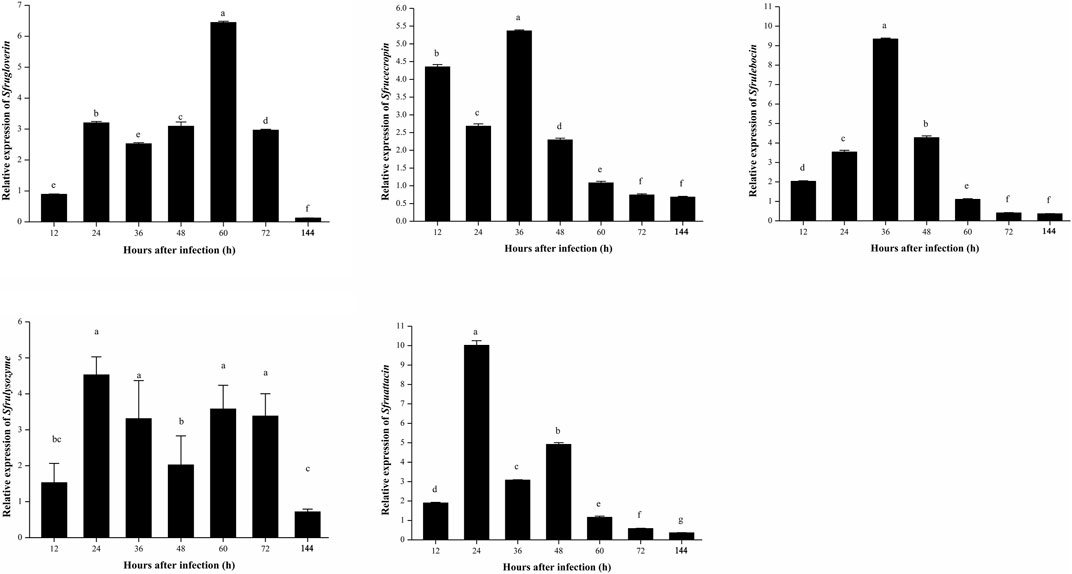
The relative expression levels of five genes of antibacterial peptides, Sfrugloverin, Sfrucecropin, Sfrulebocin, Sfrulysozyme, and Sfruattacin, were analyzed. Sfrugloverin and Sfrulysozyme showed a similar relative expression trend; they all showed relative high expression from 24 to 72 h after being infected with B. bassiana; the relative expression of Sfrucecropin and Sfruattacin was first upregulated, then downregulated and again upregulated, and again downregulated; and the relative expression of Sfrulebocin showed the highest relative expression at 36 h after being infected with B. bassiana (Figure 9).
Discussion
In this study, the transcriptome of S. frugiperda infected with B. bassiana was analyzed using Illumina HiSeq™ 4000 technology. More than 160 Gb of clean data were obtained. Different DEGs were identified in LH36vsCK36 and LH144vsCK144 libraries, and the upregulated and downregulated genes also showed differences, which might be attributed to the length of time that S. frugiperda was infected with B. bassiana. This transcriptome sequencing dataset provides a repository for future studies on the interaction mechanism between S. frugiperda and B. bassiana.
Insects defend against pathogens, such as bacteria, fungi, and viruses, via some evolutionarily conserved signaling pathways, including Toll, Imd, and JAK/STAT (Janeway and Medzhitov, 2002; Nakamoto et al., 2012). These pathways are based on different pattern recognition receptors (PRRs) which recognize characteristic molecules of pathogens and then induce downstream effectors against viral infection (Kumar et al., 2009; Palmer et al., 2018). The Toll signaling pathway is a classical signaling pathway of insect innate immunity. It has been reported that the Toll signaling pathway is activated by fungi and Lys-type peptidoglycan (PG) of Gram-positive bacteria via peptidoglycan recognition protein (PGRP)-SA, PGRP-SD, and Gram-negative binding protein 1 (GNBP1) (Michel et al., 2001; Gobert et al., 2003; Bischoff et al., 2004; Kiyoshi and Shizuo, 2013). In this study, we investigated the role of five Toll-like receptors (TLRs) (Sfrutollo, Sfrutoll3, Sfrutoll6, Sfrutoll7, and Sfrutoll18w), five cytoplasmic signal transduction factors (Sfrumyd88, Sfrupell, Sfrutube, Sfrucactus, and Sfrudif), and Sfruspatzle in the Toll signaling pathway in S. frugiperda against B. bassiana infection. The Toll pathway is activated by the activation of Spätzle (Spz), the Toll receptor ligand (Morisato and Anderson, 1994; Schneider et al., 1994).
The relative expression levels of Toll-like receptors (Sfrutollo, Sfrutoll3, Sfrutoll6, Sfrutoll7, and Sfrutoll18w) and Sfruspatzle, Sfrumyd88, Sfrutube, and Sfrupell all showed a significant upregulation at 36 or 48 h after infection, indicating that the Toll signaling pathway of S. frugiperda was activated after B. bassiana infection and the relative high expression of their corresponding proteins. We speculated that this might be related to the formation of a receptor-proximal oligomeric complex assembled by proteins Spz, Toll receptors and MyD88, Tube, and Pelle, and this complex will further trigger the phosphorylation and regulation of expression of different antimicrobial peptides (AMPs) (Towb et al., 2001). The relative expression levels of Sfrutollo, Sfrutoll6, and Sfrutoll7 presented a similar trend, which could be supported by the previous research on Bombyx mori (Imamuraand and Yamakawa, 2002; Mita et al., 2004), while the relative expression levels of Sfrutoll3 and Sfrutoll18w showed a difference. The variations in the expression levels of S. frugiperda TLRs after B. bassiana infection suggested that S. frugiperda TLRs might play different roles in antifungal immune responses, and the cytoplasmic signal transduction process of the Toll signaling pathway in S. frugiperda had a complex regulatory mechanism, which needed to be further studied.
In order to further verify the role of the Toll signaling pathway in defense against B. bassiana in S. frugiperda, the antifungal activity in vivo was conducted by inhibition assay. The result showed that BAY 11-7082 increased the sensitivity of S. frugiperda to B. bassiana infection and decreased the antifungal activity of the hemolymph of S. frugiperda, suggesting that the Toll signaling pathway plays a crucial role in the synthesis of antifungal substances against B. bassiana infection.
The Imd signaling pathway is one of the important signaling pathways in the insect innate immune system. The activated Imd protein acts on TAK 1, which then sends immune signals to IKKs that activate Relish. The activated Relish proteins enter the nucleus to regulate the transcription of target genes such as antimicrobial peptides. It is generally believed that in Drosophila, the Imd pathway is activated by meso-diaminopimelic acid (DAP)-type PG of Gram-negative bacteria and some bacilli species (Choe et al., 2002; Gottar et al., 2002; Gobert et al., 2003; Kaneko et al., 2004b), while the results of this experiment show that the Imd signaling pathway of S. frugiperda is different from that of Drosophila (Zhong et al., 2012). In our study, the relative expression levels of Sfrufadd, Sfrupgrp-lb, and Sfrurelish2 all presented a gradual upregulated trend after infection with B. bassiana, suggesting that B. bassiana could activate the Imd signaling pathway in S. frugiperda. In a previous study on B. mori, Staphylococcus aureus, Escherichia coli, and B. bassiana were all able to activate the Imd signaling pathway (Cheng et al., 2006). A relatively high expression level of Sfrulysozyme was observed at 24 h after infection by B. bassiana, which was also found in the previous research on Galleria mellonella larvae (Wojda and Jakubowicz, 2007). The variation in the trend of the relative expression level of the rest of the AMP genes in S. frugiperda was similar to that in B. mori in a previous study (Geng et al., 2016), indicating that they might act as antifungal effectors in S. frugiperda. The upregulated expression of antimicrobial peptide genes may be due to the active regulation of S. frugiperda antimicrobial peptide expression by B. bassiana to inhibit the growth of other bacteria to protect its own nutritional and parasitic requirements (Lee et al., 2005).
The JAK/STAT signaling pathway, which is involved in the regulation of a variety of important physiological processes, is the key signal pathway of immune regulation (Brivanlou et al., 2002b). STAT is the core signal transduction factor of the JAK/STAT signaling pathway, and CTL is a pattern recognition receptor that recognizes fungal β-l,3-glucan (a major component of the fungal cell wall) and guides subsequent immune responses. After infection with B. bassiana, the relative expression pattern of Sfructl4 was synchronized with that of Sfrustat5B, indicating that in S. frugiperda, the JAK/STAT signaling pathway was activated after infection with B. bassiana, and Sfructl4 and Sfrustat5B might play an important role in antifungal immune response (Liu et al., 2015). At the later stage of infection progress, Sfrustat5B was upregulated, which might be due to the production of a large number of spores and vegetative hyphae in the late stage of infection, and β-l,3-glucan, the main component of the fungal cell wall, may induce stronger immune responses in S. frugiperda. SOCS and Ken are negative regulators of the JAK/STAT signaling pathway. SOCS inhibits the activity of JaK kinase or cytokine receptor (such as STAT) (Cooney, 2002), and Ken selectively regulates the expression of some target genes (Arbouzova et al., 2006). In this study, the relative expression of Sfrusocs2, Sfrusocs4, Sfrusocs5, and Sfruken was maintained at a low level in the early stages of infection progress, suggesting that these genes might be involved in the immune response in S. frugiperda infected by B. bassiana, which indicates that S. frugiperda could activate JAK/STAT-mediated immune responses by downregulating the repressors Sfrusocs2, Sfrusocs4, Sfrusocs5, and Sfruken (Kausar et al., 2022). The result of the antifungal activity in vivo experiment showed that, compared with the control group, the test group (injected with inhibitors) had a lower survival rate and a 20 h reduction in LT50. So the JAK/STAT signaling pathway might regulate the synthesis of antifungal substances in S. frugiperda, which further indicates that the JAK/STAT signaling pathway plays an important role in the resistance of S. frugiperda to B. bassiana infection. However, there are also complex regulatory mechanisms in the JAK/STAT signaling pathway, which need to be further studied.
AMPs play an irreplaceable role in the process of innate immunity in insects, and a wide range of antimicrobial peptides display both antibacterial and antifungal functions (Goto et al., 2001; Cerenius et al., 2010; Loof et al., 2011; Browne et al., 2013; Ragland and Criss, 2017; Sheehan et al., 2020). In this study, the relative expression of Sfrulysozyme showed a high level from 24 to 72 h after infection by B. bassiana (Figure 9); a similar finding was also found in a previous study on G. mellonella larvae (Wojda and Jakubowicz, 2007), suggesting that it might be an antifungal function in S. frugiperda. The relative expression of Sfrucecropin, Sfruattacin, Sfrugloverin, and Sfrulebocin all showed upregulated expression patterns to a varying degree after S. frugiperda was infected with B. bassiana, indicating that these antimicrobial peptides were involved in the immune response of S. frugiperda to B. bassiana infection.
In summary, the different expression patterns of Toll, Imd, and JAK/STAT signaling pathway-related genes revealed their different and specific functions in immune signal transduction in S. frugiperda. Further studies are needed to investigate the resistance mechanism of S. frugiperda to pathogenic microorganisms. This study not only lays a solid foundation for the research and development of new fungal agents to control S. frugiperda and other pests but also provides an important scientific basis for the development and use of new biocontrol agents.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JL, JY, HW, and JL conceived, coordinated, and designed the research. YM, YC, ML, and HX assembled and analyzed the transcriptome datasets. JL and YM performed the experiments. JY, JL, and HW drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the GDAS Project of Science and Technology Development (Grant No. 2020GDASYL-20200103056), the National Natural Science Foundation of China (Grant No. 32172471), and the Guangzhou Science and Technology Program (Grant No. 2023A04J0845). The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
The authors thank Xiaoshan Chen of the Institute of Nanfan and Seed Industry, Guangdong Academy of Sciences, for rearing insects.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1249662/full#supplementary-material
References
Ao, J. Q., Ling, E. J., and Yu, X. Q. (2008). A Toll receptor from Manduca sexta is in response to Escherichia coli infection. Mol. Immunol. 45, 543–552. doi:10.1016/j.molimm.2007.05.019
Arbouzova, N. I., Bach, E. A., and Zeidler, M. P. (2006). Ken & Barbie selectively regulates the expression of a subset of JAK/STAT pathway target genes. Curr. Biol. 16 (1), 80–88. doi:10.1016/j.cub.2005.11.033
Bischoff, V., Vignal, C., Boneca, I. G., Michel, T., Hoffmann, J. A., and Royet, J. (2004). Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 5 (11), 1175–1180. doi:10.1038/ni1123
Brivanlou, A. H., and Darnell, J. E. (2002a). Signal transduction and the control of gene expression. Science 295 (5556), 813–818. doi:10.1126/science.1066355
Brivanlou, A. H., and Darnell, J. E. (2002b). Signal transduction and the control of gene expression. Science 295, 813–818. doi:10.1126/science.1066355
Browne, N., Heelan, M., and Kavanagh, K. (2013). An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence 4 (7), 597–603. doi:10.4161/viru.25906
Cerenius, L., Kawabata, S., Lee, B. L., Nonaka, M., and Söderhäll, K. (2010). Proteolytic cascades and their involvement in invertebrate immunity. Trends biochem. Sci. 35 (10), 575–583. doi:10.1016/j.tibs.2010.04.006
Cheng, T., Zhao, P., Liu, C., Xu, P., Gao, Z., Xia, Q., et al. (2006). Structures, regulatory regions and inductive expression patterns of antimicrobial peptide genes in the silkworm, Bombyx mori. Genomics 87 (3), 356–365. doi:10.1016/j.ygeno.2005.11.018
Choe, K. M., Werner, T., Stoven, S., Hultmark, D., and Anderson, K. V. (2002). Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296 (5566), 359–362. doi:10.1126/science.1070216
Cooney, R. N. (2002). Suppressors of cytokine signaling (SOCS): inhibitors of the JAK/STAT pathway. Shock 17 (2), 83–90. doi:10.1097/00024382-200202000-00001
Corbo, J. C., and Levine, M. (1996). Characterization of all immunodeficiency mutant in Drosophila. Mech. Dev. 55 (2), 211–220. doi:10.1016/0925-4773(96)00506-0
Geng, T., Huang, Y., Hou, C., Qin, G., Lv, D., and Guo, X. (2016). Inductive expression patterns of genes related to toll signaling pathway in silkworm (Bombyx mori) upon Beauveria bassiana infection. J. Asia-Pac. Entomol. 19 (3), 861–868. doi:10.1016/j.aspen.2016.08.001
Gobert, V., Gottar, M., Matskevich, A. A., Rutschmann, S., Royet, J., Belvin, M., et al. (2003). Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 302 (5653), 2126–2130. doi:10.1126/science.1085432
Goto, A., Kumagai, T., Kumagai, C., Hirose, J., Narita, H., Mori, H., et al. (2001). A Drosophila haemocyte-specific protein, hemolectin, similar to human von willebrand factor. Biochem. J. 359 (Pt 1), 99–108. doi:10.1042/0264-6021:3590099
Gottar, M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoffmann, J. A., et al. (2002). The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416 (6881), 640–644. doi:10.1038/nature734
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. doi:10.1038/nbt.1883
Hirano, T., Ishihara, K., and Hibi, M. (2000). Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 19 (21), 2548–2556. doi:10.1038/sj.onc.1203551
Imamura, M., and Yamakawa, M. (2002). Molecular cloning and expression of a Toll receptor gene homologue from the silkworm, Bombyx mori. Biochim. Biophys. Acta. 1576 (3), 246–254. doi:10.1016/s0167-4781(02)00336-6
Imler, J. L., and Hoffmann, J. A. (2001). Toll receptors in innate immunity. Trends Cell. Biol. 11 (7), 304–311. doi:10.1016/s0962-8924(01)02004-9
Janeway, C. A., and Medzhitov, R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20, 197–216. doi:10.1146/annurev.immunol.20.083001.084359
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic acids. Res. 36, 480–484. doi:10.1093/nar/gkm882
Kaneko, T., Goldman, W. E., Mellroth, P., Steiner, H., Fukase, K., Kusumoto, S., et al. (2004a). Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20 (5), 637–649. doi:10.1016/s1074-7613(04)00104-9
Kaneko, T., Goldman, W. E., Mellroth, P., Steiner, H., Fukase, K., Kusumoto, S., et al. (2004b). Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20 (5), 637–649. doi:10.1016/s1074-7613(04)00104-9
Kanost, M. R., Jiang, H., and Yu, X. Q. (2004). Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 198, 97–105. doi:10.1111/j.0105-2896.2004.0121.x
Karst, S. M., Wobus, C. E., Lay, M., Davidson, J., and Virgin, H. Wt. (2003). STAT1-dependent innate immunity to a Norwalk-like virus. Science 299 (5612), 1575–1578. doi:10.1126/science.1077905
Kausar, S., Gul, I., Liu, R., Ke, X. X., Dong, Z., Abbas, M. N., et al. (2022). Antheraea pernyi suppressor of cytokine signaling 2 negatively modulates the JAK/STAT pathway to attenuate microbial infection. Int. J. Mol. Sci. 23(18), 10389. doi:10.3390/ijms231810389
Kiyoshi, T., and Shizuo, A. (2013). Toll-like receptors in innate immunity. Int. Immunol. 17 (1), 1–14. doi:10.1093/intimm/dxh186
Kumar, H., Kawai, T., and Akira, S. (2009). Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 388 (4), 621–625. doi:10.1016/j.bbrc.2009.08.062
Lee, S. Y., Nakajima, I., IharaKinoshita, H., and Nihira, T. (2005). Cultivation of entomopathogenic fungi for the search of antibacterial compounds. Mycopathologia 160 (4), 321–325. doi:10.1007/s11046-005-0179-y
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. doi:10.1146/annurev.immunol.25.022106.141615
Leulier, F., Vidal, S., Saigo, K., Ueda, R., and Lemaitre, B. (2002). Inducible expression of double-stranded RNA reveals a role for dFADD in the regulation of the antibacterial response in Drosophila adults. Curr. Biol. 12 (12), 996–1000. doi:10.1016/s0960-9822(02)00873-4
Levashina, E. A., Ohresser, S., Lemaitre, B., and Imler, J. L. (1998). Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. J. Mol. Biol. 278 (3), 515–527. doi:10.1006/jmbi.1998.1705
Levy, D. E., and Darnell, J. E. (2002). Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 3 (9), 651–662. doi:10.1038/nrm909
Li, W. X. (2008). Canonical and non-canonical JAK-STAT signaling. Trends Cell. Biol. 18 (11), 545–551. doi:10.1016/j.tcb.2008.08.008
Liu, W., Liu, J., Lu, Y., Gong, Y., Zhu, M., Chen, F., et al. (2015). Immune signaling pathways activated in response to different pathogenic micro-organisms in Bombyx mori. Mol. Immunol. 65 (2), 391–397. doi:10.1016/j.molimm.2015.02.018
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt, method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Loof, T. G., Schmidt, O., Herwald, H., and Theopold, U. (2011). Coagulation systems of invertebrates and vertebrates and their roles in innate immunity: the same side of two coins? J. Innate. Immun. 3 (1), 34–40. doi:10.1159/000321641
Martinelli, S., Barata, R. M., Zucchi, M. I., Silva-Filho Mde, C., and Omoto, C. (2006). Molecular variability of Spodoptera frugiperda (Lepidoptera: noctuidae) populations associated to maize and cotton crops in Brazil. J. Econ. Entomol. 99 (2), 519–526. doi:10.1603/0022-0493-99.2.519
Michel, T., Reichhart, J. M., Hoffmann, J. A., and Royet, J. (2001). Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414 (6865), 756–759. doi:10.1038/414756a
Mita, K., Kasahara, M., Sasaki, S., Nagayasu, Y., Yamada, T., Kanamori, H., et al. (2004). The genome sequence of silkworm, Bombyx mori. DNA Res. 11 (1), 27–35. doi:10.1093/dnares/11.1.27
Morisato, D., and Anderson, K. V. (1994). The spatzle gene encodes a component of the extracellular signaling pathway establishing the dorsal-ventral pattern of the Drosophila embryo. Cell. 76 (4), 677–688. doi:10.1016/0092-8674(94)90507-x
Naitza, S., and Ligoxygakis, P. (2004). Antimicrobial defences in Drosophila: the story so far. Mol. Immunol. 40, 887–896. doi:10.1016/j.molimm.2003.10.008
Nakamoto, M., Moy, R. H., Xu, J., Bambina, S., Yasunaga, A., Shelly, S. S., et al. (2012). Virus recognition by toll-7 activates antiviral autophagy in Drosophila. Immunity 36 (4), 658–667. doi:10.1016/j.immuni.2012.03.003
Palmer, W. H., Varghese, F. S., and van Rij, R. P. (2018). Natural variation in resistance to virus infection in Dipteran insects. Viruses 10 (3), 118. doi:10.3390/v10030118
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi:10.1093/bioinformatics/btg034
Pires-daSilva, A., and Sommer, R. J. (2003). The evolution of signalling pathways in animal development. Nat. Rev. Genet. 4 (1), 39–49. doi:10.1038/nrg977
Poisot, A., Hruska, A., Fredrix, M., et al. (2018). Integrated management of the fall armyworm on maize: A guide for farmer field schools in africa.
Prasanna, B. M., Huesing, J. E., Eddy, R., and Peschke, V. M. (2018). Fall armyworm in africa: A guide for integrated pest management.
Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., et al. (2001). The tigr gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic acids. Res. 29, 159–164. doi:10.1093/nar/29.1.159
Ragland, S. A., and Criss, A. K. (2017). From bacterial killing to immune modulation: recent insights into the functions of lysozyme. PLoS Pathog. 13 (9), e1006512. doi:10.1371/journal.ppat.1006512
Rämet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B., and Ezekowitz, R. A. B. (2002). Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E.coli. Nature 416, 644–648. doi:10.1038/nature735
Ramos, Y., Portal, O., Lysøe, E., and Klingen, I. (2017). Diversity and abundance of Beauveria bassiana in soils, stink bugs and plant tissues of common bean from organic and conventional fields. J. Invertebr. Pathol. 150, 114–120. doi:10.1016/j.jip.2017.10.003
Rao, X. J., and Yu, X. Q. (2010). Lipoteichoic acid and lipopolysaccharide can activate antimicrobial peptide expression in the tobacco hornworm Manduca sexta. Dev. Comp. Immunol. 34 (10), 1119–1128. doi:10.1016/j.dci.2010.06.007
Rutschmann, S., Jung, A. C., Zhou, R., Silverman, N., Hoffmann, J. A., and Ferrandon, D. (2000). Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat. Immunol. l (4), 342–347. doi:10.1038/79801
Schneider, D. S., Jin, Y., Morisato, D., and Anderson, K. V. (1994). A processed form of the Spatzle protein defines dorsal-ventral polarity in the Drosophila embryo. Development 120 (5), 1243–1250. doi:10.1242/dev.120.5.1243
Sheehan, G., Farrell, G., and Kavanagh, K. (2020). Immune priming: the secret weapon of the insect world. Virulence 11 (1), 238–246. doi:10.1080/21505594.2020.1731137
Silverman, N., Zhou, R., Erlich, R. L., Hunter, M., Bernstein, E., Schneider, D., et al. (2003). Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J. Biol. Chem. 278 (49), 48928–48934. doi:10.1074/jbc.M304802200
Silverman, N., Zhou, R., Stöven, S., Pandey, N., Hultmark, D., and Maniatis, T. (2000). A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes. Dev. 14 (19), 2461–2471. doi:10.1101/gad.817800
Souza-Neto, J. A., Sim, S., and Dimopoulos, G. (2009). An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U. S. A. 106 (42), 17841–17846. doi:10.1073/pnas.0905006106
Tanji, T., Hu, X., Weber, A. N., and Ip, Y. T. (2007). Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell. Biol. 27 (12), 4578–4588. doi:10.1128/MCB.01814-06
Tauszig, S., Jouanguy, E., Hoffmann, J. A., and Imler, J. L. (2000). Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 97 (19), 10520–10525. doi:10.1073/pnas.180130797
Tauszig-Delamasure, S., Bilak, H., Capovilla, M., Hoffmann, J. A., and Imler, J. L. (2002). Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 3 (1), 91–97. doi:10.1038/ni747
Todd, E. L., and Poole, R. W. (1980). Keys and illustrations for the armyworm moths of the noctuid genus Spodoptera guenée from the western hemisphere. Ann. Entomol. Soc. Am. 1980 (6), 722–738. doi:10.1093/aesa/73.6.722
Towb, P., Bergmann, A., and Wasserman, S. A. (2001). The protein kinase Pelle mediates feedback regulation in the Drosophila Toll signaling pathway. Development 128 (23), 4729–4736. doi:10.1242/dev.128.23.4729
Uvell, H., and Engström, Y. (2007). A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet. 23 (7), 342–349. doi:10.1016/j.tig.2007.05.003
Wojda, I., and Jakubowicz, T. (2007). Humoral immune response upon mild heat-shock conditions in Galleria mellonella larvae. J. Insect Physiol. 53 (11), 1134–1144. doi:10.1016/j.jinsphys.2007.06.003
Ye, J., Fang, L., Zheng, H., Zhang, Y., Chen, J., Zhang, Z., et al. (2006). Wego: A web tool for plotting GO annotations. Nucleic acids. Res. 34, 293–297. doi:10.1093/nar/gkl031
Zhang, Y., Li, B. X., Mao, Q. Z., Zhuo, J. C., Huang, H. J., Lu, J. B., et al. (2023). The JAK-STAT pathway promotes persistent viral infection by activating apoptosis in insect vectors. PLoS Pathog. 19 (3), e1011266. doi:10.1371/journal.ppat.1011266
Keywords: Spodoptera frugiperda, Beauveria bassiana, transcriptome, signaling pathways, gene expression
Citation: Li J, Mao Y, Yi J, Lin M, Xu H, Cheng Y, Wu H and Liu J (2023) Induced expression modes of genes related to Toll, Imd, and JAK/STAT signaling pathway-mediated immune response in Spodoptera frugiperda infected with Beauveria bassiana. Front. Physiol. 14:1249662. doi: 10.3389/fphys.2023.1249662
Received: 29 June 2023; Accepted: 11 August 2023;
Published: 24 August 2023.
Edited by:
Ya-Nan Zhang, Huaibei Normal University, ChinaCopyright © 2023 Li, Mao, Yi, Lin, Xu, Cheng, Wu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Wu, YW50ZW5uYTEyMTdAMTYzLmNvbQ==; Jianbai Liu, bGl1amI1QG1haWwyLnN5c3UuZWR1LmNu
 Jihu Li
Jihu Li Yongkai Mao
Yongkai Mao Jiequn Yi
Jiequn Yi Han Wu
Han Wu Jianbai Liu
Jianbai Liu