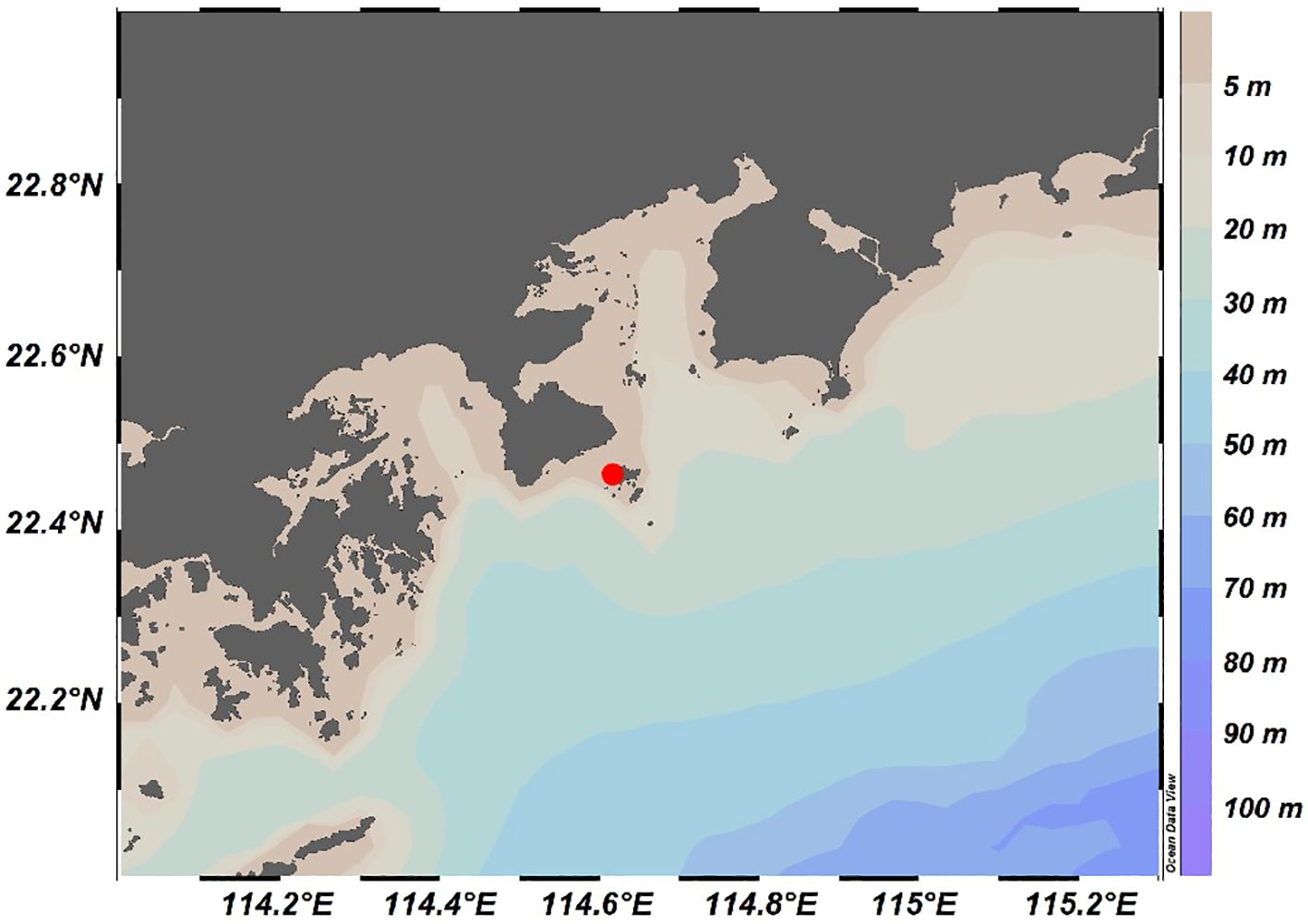
- Department of Ocean Science and Engineering, Southern University of Science and Technology, Shenzhen, China
Nitrogen (N) and phosphorus (P) are essential nutrients for marine phytoplankton, playing a crucial role in shaping the structure of microbial communities. Nutrients in coastal seawater are influenced by multiple factors, including ocean currents, terrestrial runoff, and anthropogenic activities, leading to region-specific patterns of nutrient limitation. This study investigates nutrient limitation in the transitional waters near Sanmen Island, located at the confluence of the Pearl River Estuary (PRE) and the northern South China Sea. Using 4-hourly in situ time-series observations and nutrient addition experiments, we found that nitrogen limitation persists in this region despite its proximity to the nutrient-rich Pearl River. Urea addition significantly enhanced primary productivity, as evidenced by the increased chlorophyll a concentration and the increased relative abundance of cyanobacteria, whereas phosphate addition alone favored the growth of heterotrophic bacteria, yet limited the growth of cyanobacteria and other primary producers. Combined nitrogen-phosphorus treatments revealed serial co-limitation, where nitrogen relief shifted limitation to phosphorus. In conclusion, these findings highlight the complex nutrient dynamics in transitional coastal waters and underscore the impact of anthropogenic nutrient discharge on ecosystem productivity.
1 Introduction
Marine phytoplankton are fundamental to oceanic food webs and global biogeochemical cycles, contributing approximately 50% of global primary productivity (Falkowski et al., 1998; Field et al., 1998). The availability of key inorganic nutrients primarily governs the growth and community structure of marine phytoplankton, and their cellular stoichiometry roughly follows the Redfield ratio, approximating the average ratio of inorganic C, N, and P found in the ocean (Redfield, 1958). Coastal marine environments exhibit complex and spatially variable patterns of nutrient limitation that differ significantly from those in open ocean systems (Elser et al., 2007). While oligotrophic open ocean regions typically experience N limitation or N-P co-limitation, coastal waters present a more heterogeneous picture due to the confluence of terrestrial inputs, anthropogenic activities, and oceanographic processes, leading to a notable deviation in inorganic N:P ratios from the Redfield ratio (Peng et al., 2002; Elser et al., 2007; Lan et al., 2024). According to Liebig’s Law of the Minimum, the primary productivity of marine phytoplankton is limited by the most scarce essential nutrient in the environment (de Baar, 1994). Nitrogen is often the primary limiting nutrient in marine environments, a pattern consistently observed across diverse coastal systems (Vitousek and Howarth, 1991). However, modern understanding recognizes that multiple essential nutrients can be deficient in the system, resulting in simultaneous co-limitation (where multiple nutrients are equally limiting at the same time) or serial co-limitation (where the relief of one limiting nutrient shifts the limitation to another) (de Baar, 1994; Harpole et al., 2011; Moore et al., 2013). A global analysis of ocean phytoplankton nutrient limitation has revealed a high prevalence of co-limitation in coastal regions, where N, P and other nutrients can simultaneously or sequentially limit primary productivity (Elser et al., 2007; Moore et al., 2013; Cao et al., 2024). This complexity is particularly pronounced in transitional coastal waters where multiple water masses converge, creating dynamic conditions that can rapidly alter nutrient availability and limitation patterns (Nixon, 1995), particularly in the face of unprecedented pressures from climate change, eutrophication, and human activities.
The northern South China Sea (SCS) is one of the most dynamic subtropical coastal marine systems globally, characterized by complex interactions between monsoon-driven circulation, river inputs, and anthropogenic activities (Liu et al., 2002). Daya Bay is located in the northern SCS off the coast of Shenzhen and Huizhou. Within the bay, submarine groundwater discharge contributes more nutrients than riverine inputs (Wang et al., 2018). Nitrate and nitrite primarily originate from atmospheric deposition and submarine groundwater discharge, while dissolved inorganic phosphorus (DIP) is mainly derived from riverine sources, and silicate is predominantly supplied by groundwater discharge from the seabed and continental slope (Wang et al., 2018). The rapid economic development along the coast of Shenzhen and Huizhou, such as tourism, fishery, aquaculture, has led to an increasingly anthropogenic impact on Daya Bay and nearby coastal waters, including eutrophication, an increase in the N:P ratio, a shift in the limiting nutrient factor from N to P (Wang et al., 2004, 2008; Wu et al., 2009; Shi and Huang, 2013; Guo et al., 2023). Eutrophication leads to an increase in primary productivity, which has had a significantly negative impact on the aquaculture and tourism industries of Daya Bay (Wu et al., 2009).
Sanmen Island is situated at the mouth of Daya Bay in northern SCS, near the Pearl River Estuary (PRE), making it a unique transitional zone where multiple water masses converge. Seawater environmental parameters near Sanmen Island are influenced by multiple factors, including tidal dynamics from SCS, groundwater discharge from the continent and island, riverine input and human activities (Wang et al., 2018; Li et al., 2021). During the wet season, typically from April to September, the increased flow of the Pearl River can introduce substantial nutrient loads into the nearby estuarine and coastal waters (Yin and Harrison, 2008). The summer riverine input typically enhances nitrogen concentration in the estuary, though phosphorus concentration remains low, and the interaction with seawater from the northern South China Sea further complicates the nutrient limitation in this highly dynamic system (Li et al., 2017). Due to their different geographical locations, the PRE and Daya Bay exhibit distinct patterns of nutrient limitation in summer. The N:P ratio of surface water in the PRE is significantly influenced by Pearl River input and coastal anthropogenic activities (Niu et al., 2020; Tao et al., 2021), while Daya Bay is primarily influenced by Guangdong coastal upwelling brought by the southwest monsoon, with relatively weaker impact from terrestrial runoff and human influence (Han and Ma, 1988; Li et al., 1990; Zhang, 1992; Yang and Tan, 2019). While in the transitional zone offshore Sanmen Island, in the vicinity of the Pearl River Estuary and Daya Bay, the bioavailability of nutrients depends on the mixing of different water bodies, creating a natural laboratory for studying nutrient limitation patterns. The frequent occurrence of phytoplankton blooms in this region during spring and summer is consistent with seasonal variations in nutrient availability and productivity (Lin et al., 2024). However, the effect of nutrient limitation on phytoplankton growth, particularly regarding the role of organic nitrogen sources and the competitive dynamics between autotrophs and heterotrophs, remains poorly understood. Altogether, Sanmen Island’s unique geographical location and complex hydrological conditions make it a special site for studying the ecology of transitional waters.
This study aims to elucidate the primary nutrient limitation in these transitional waters, focusing on the role of organic nitrogen sources like urea on phytoplankton growth, as well as the interactive relationship between nutrient limitation and microbial community structure. Specifically, in July 2022, an in situ observation and an incubation experiment with urea and potassium dihydrogen phosphate (KH2PO4) additions at Sanmen Island were conducted. Through 16S rRNA gene and metagenomic sequencing, the following questions will be addressed: (1) What is the nutrient limitation pattern in the Sanmen Island region? (2) How do microbial communities respond to the addition of urea and KH2PO4, respectively?
2 Materials and methods
2.1 Experimental design
The sampling site was located at Mawan Wharf, Sanmen Island, with coordinates of 22°27′49.65″N, 114°37′0.73″E (Figure 1). This location was specifically chosen as a representation of transitional coastal waters, as it experiences the confluence of multiple water masses with more pronounced nutrient dynamics compared to inner bay or riverine locations. The experimental design consisted of two complementary approaches, a high-frequent in situ observation and a controlled nutrient incubation experiment. For the in situ observation, samples were collected from surface seawater (~20 cm depth) between 22:00 on July 18, 2022, and 14:00 on July 21, 2022, with the sampling intervals of 4 hours, yielding a total of 17 sample sets. Detailed sampling time for each sample is provided in Supplementary Table 2. Meanwhile, an incubation experiment was initiated at 18:00 on July 18, 2022. Considering that Sanmen Island is directly influenced by anthropogenic activities, and referring to previous incubation experiments, we selected organic nitrogen compound urea and inorganic phosphate KH2PO4 as the added nitrogen and phosphorus sources, respectively (Rondell et al., 2000; Bonnet et al., 2016). Initial surface seawater from the observation site was transferred into four sets of three replicate 20 L transparent low-density polyethylene buckets. The four groups were: (1) Blank group, the control group without nutrient addition; (2) N group, with urea added to reach a final concentration of 0.16 mmol urea-N/L; (3) P group, with KH2PO4 added to a final concentration of 0.01 mmol KH2PO4-P/L; (4) NP group, with both urea and KH2PO4 added at the same concentrations as the N and P groups. Here, the final concentration of phosphate was moderately higher than the highest observed value in surface seawater of Daya Bay during summer (Li et al., 2019). The urea concentration was determined based on the final concentration of phosphate and the Redfield ratio (Redfield, 1958). Due to tidal influence, one replicate each from the Blank and N groups was damaged during the in situ incubation. The remaining samples were collected after 68 hours for productivity and microbial composition analysis. Before sample collection, in situ measurements of temperature, pH, dissolved oxygen (DO), salinity, and chlorophyll a concentration were taken. For microbial sample collection, large particles and zooplankton were first removed by pre-filtering through a 200-mesh (74 μm) nylon sieve. Then, approximately 20 liters of seawater were filtered through 0.22 μm pore size polycarbonate (PC) membranes (Millipore, USA) to capture prokaryotic cells. The filters were used to extract DNA. The remaining filtrate was analyzed to measure the concentrations of nutrients (including nitrate, nitrite, ammonia, phosphate, and silicate) and dissolved organic carbon (DOC).
2.2 Measurement and analysis of environmental parameters
Temperature, pH, DO, salinity, and chlorophyll a concentration were measured in situ using a MultiAnna MTA-6A multi-parameter water quality sonde (Lightsun, China). During measurement, the sonde was immersed in seawater and continuously monitored for at least 3 minutes. After removing outliers, the arithmetic mean of the remaining data was recorded as the result of the environmental parameter.
DOC concentration was measured with a TOC-L total organic carbon analyzer (Shimadzu, Japan) according to the manufacturer’s protocol. Nutrient concentrations, including nitrate, nitrite, ammonia, silicate, and phosphate, were measured using the CleverChem Anna automatic discrete analyzer (DeChem-Tech, Germany) according to the “GB17378.4, 2007 Specifications for oceanographic survey-part 4: Survey of chemical parameters in sea water” and instrument operation manual (GB17378.4, 2007). Data analysis and visualization were conducted using R packages with R version 4.2.3 (R Core Team, 2023). Specifically, in situ samples grouping was determined using the mclust package based on the temporal variation of nutrient concentrations (Scrucca et al., 2016). The Wilcoxon test was used to verify the validity of samples grouping (Wilcoxon, 1945). The ggplot2 package (version 3.5.0) was used for data visualization (Wickham, 2016).
2.3 DNA extraction and sequencing
FastDNA™ SPIN Kit for Soil (MP Biomedical, USA) was used to extract microbial DNA from filter membranes. DNA samples were sent to Novogene Co., Ltd. (Tianjin, China) for amplicon and metagenomic sequencing. The V4-V5 region of prokaryotic 16S rRNA gene was amplified using primers 515Y (5’-GTGYCAGCMGCCGCGGTAA-3’) and 926R (5’-CCGYCAATTYMTTTRAGTTT-3’) (Yeh et al., 2021). The PCR amplification steps include: (1) pre-denaturation at 95°C for 120 seconds; (2) 30 cycles of denaturation at 95°C for 45 seconds, annealing at 50°C for 45 seconds and extension at 68°C for 90 seconds; (3) final extension at 68°C for 300 seconds (Yeh et al., 2021). Amplicon sequencing was carried out on the Illumina NovaSeq PE250 platform (Illumina, USA). Metagenomic sequencing was performed on the Illumina NovaSeq PE150 platform (Illumina, USA).
2.4 16S rRNA gene sequencing analysis
The amplicon sequencing data were processed on the QIIME2 platform (Bolyen et al., 2019). Adapter and primer sequences in the raw reads were trimmed using Cutadapt (version 4.6) (Martin, 2011). The trimmed sequences were classified into prokaryotic and eukaryotic sequences using BBSplit (BBTools version 39.19, https://sourceforge.net/projects/bbmap/) with reference to the SILVA 132 and PR2 4.14.0 databases (Quast et al., 2012; Guillou et al., 2012). For prokaryotic sequences, denoising, paired-end reads merging, and clustering were performed using DADA2 (version 1.22.0) to obtain amplicon sequence variants (ASVs) (Katoh et al., 2002; Callahan et al., 2016). Taxonomic annotation of ASVs was conducted according to the SILVA 138.1 database (Quast et al., 2012). ASVs annotated as chloroplasts or mitochondria were discarded. Default parameters were used for all the software unless otherwise specified. The quality control results are detailed in Supplementary Table 1. Statistical analysis was performed in QIIME2 and R software (R version 4.2.3) (Bolyen et al., 2019; R Core Team, 2023). All samples were rarefied to 140,168 sequences per sample to reduce the impact of varying sequencing depths (Supplementary Figure 1). Alpha diversity of prokaryotes was assessed using the Simpson index. A significant test of alpha diversity was conducted via the Wilcoxon test (Wilcoxon, 1945). Beta diversity was evaluated using non-metric multidimensional scaling (NMDS) analysis via the vegan package (version 2.6-4), and the significance test between different groups was conducted using analysis of similarity (ANOSIM) via the phyloseq package (version 1.42.0) (Clarke and Green, 1988; McMurdie and Holmes, 2013; Oksanen et al., 2022). Differential abundance analysis (DAA) was performed using ANCOM-BC (version 1.0) (Lin and Peddada, 2020). Centered log-ratio (CLR) transformation and Pearson correlation were used to analyze the correlation between cyanobacteria and chlorophyll a concentration (Aitchison, 1982; Rodgers and Nicewander, 1988). Data visualization was conducted using ggplot2 (version 3.5.0) (Wickham, 2016). When plotting the composition of the prokaryotic community, taxa with an average relative abundance <1% across groups were grouped into “Others”. This approach was also used in DAA and correlation analysis between cyanobacteria and chlorophyll a concentration to minimize interference from low-abundance taxa. Given the marine environment, all taxa annotated as freshwater species were labeled with the “-like” suffix to suggest close relatives of marine environments.
2.5 Metagenomic analysis
For raw sequencing data, Readfq V8 (https://github.com/lh3/readfq) and Bowtie2 (version 2.2.4) were used for quality control and host contamination removal, respectively (Langmead and Salzberg, 2012). Clean reads were assembled into contigs using MEGAHIT (version 1.2.9; parameters: –min-count 2 –k-list 21,33,55,77,99,127 –min-contig-len 1000) (Li et al., 2015, 2016). To obtain more functional genes and more precise taxonomy annotation of functional genes, single-sample binning and co-binning were performed with BASALT (version 1.0.0; parameters: –max-ctn 10 –min-cpn 50 –mode continue) to obtain metagenome-assembled genomes (MAGs) (Qiu et al., 2024). Specifically, co-binning was conducted based on the clustering result of sourmash (version 4.8.8) (Brown and Irber, 2016). Samples clustered into the same group were co-binned together to recover more MAGs. MAGs were dereplicated at an average nucleotide identity (ANI) of 95% using dRep (version 3.4.5) (Olm et al., 2017). The completeness and contamination of MAGs were assessed again using CheckM (version 1.2.2) (Parks et al., 2015). 202 MAGs with completeness > 50% and contamination < 10% were retained for downstream analysis. The data preprocessing results are detailed in Supplementary Table 1. Taxonomic annotation of MAGs was conducted using GTDB-Tk (version 2.3.2), and the result was translated into NCBI taxonomy classification via gtdb_to_ncbi_majority_vote.py (v0.2.1, https://github.com/Ecogenomics/GTDBTk) (Chaumeil et al., 2019, 2022). Taxonomic annotation of contigs was conducted via MEGAN6 (version 6.24.20) against the NCBI nt database (downloaded on June 10, 2022) and megan-nucl-Feb2022.db (https://software-ab.cs.uni-tuebingen.de/download/megan6/) (Huson et al., 2016; Sayers et al., 2022). Coding DNA Sequences (CDS) of contigs and MAGs were predicted using Prokka (version 1.14.6), followed by functional annotation against the KEGG database (version 106.0) using KofamScan (version 1.3.0) (Seemann, 2014; Aramaki et al., 2019). CDS and predicted protein sequences were clustered at 95% sequence identity using CD-HIT (version 4.8.1) (Li and Godzik, 2006; Fu et al., 2012). To evaluate the microbial demand for nitrogen and phosphorus in different experimental groups, relative gene abundance related to the uptake and utilization of environmental nitrogen and phosphorus was analyzed. Transcript per million (TPM) was calculated using CoverM (version 0.7.0) with parameters: contig –mapper bwa-mem –min-read-percent-identity 0.95 –min-read-aligned-percent 0.75 -m tpm (Li and Dewey, 2011; Wagner et al., 2012; Li, 2013; Aroney et al., 2025). For phylogenetic tree construction, multiple sequence alignment (MSA) was first constructed using MUSCLE (version 5.1), followed by trimming of MSA using trimAl (version 1.4) with “-automated1” parameter (Capella-Gutiérrez et al., 2009; Edgar, 2022). Finally, the phylogenetic tree was constructed using IQ-TREE (version 2.2.0.3) with the parameter: -m MF (Nguyen et al., 2015). The base R (R version 4.2.3) and ggplot2 (version 3.5.0) packages were used for data visualization (Wickham, 2016; R Core Team, 2023).
3 Results
3.1 Nitrogen limitation controls the primary productivity in seawater near Sanmen Island
Phased variations were observed in nitrate, nitrite, and silicate concentrations in the in situ environment. Nitrate and nitrite concentrations remained relatively low before 06:00 on July 20 but increased significantly after 10:00 (p-value < 0.001), while the silicate concentration exhibited the opposite trend (Figure 2A). After sampling clustering using nitrate, nitrite, and silicate concentrations (see Methods), nine samples collected from 22:00 July 18 to 06:00 July 20 (L1822, L1902, L1906, L1910, L1914, L1918, L1922, L2002, L2006) were grouped as Phase 1, while eight samples from 10:00 July 20 to 14:00 July 21 (L2010, L2014, L2018, L2022, 2102, L2106, L2110, L2114) were grouped as Phase 2 (Figure 2B). Significant differences (p-value < 0.01) were observed between Phase 1 and Phase 2 for nitrite, nitrate, and silicate concentrations, strengthening the rationality of grouping (Figures 2C–E). The concurrent increases in pH, dissolved oxygen, and chlorophyll a are consistent with enhanced photosynthetic activity and primary productivity during Phase 2 (Hamdhani, 2024). The silicate depletion in Phase 2 may indicate that diatoms made a significant contribution to primary productivity. Phosphate and ammonia concentrations were below the detection limit of the instrument (Figure 2A). In the incubation experiment, urea addition alone or combined with KH2PO4 enhanced primary productivity. Chlorophyll a concentration in the Blank group ranged from 2.1997 to 3.1007 μg/L, while in the N group, they rose to between 5.3352 and 12.1482 μg/L, demonstrating that urea addition enhanced primary productivity. In the P group, chlorophyll a concentration remained almost unchanged compared to the Blank group, with a slight decrease in average value (Supplementary Table 2). The NP group yielded substantially higher chlorophyll a concentrations (16.0793-21.2664 μg/L), suggesting NP co-addition further improved the primary productivity than N addition alone. These results indicate that coastal surface seawater near Sanmen Island exhibited serial nitrogen-phosphate co-limitation, dominated by nitrogen limitation.
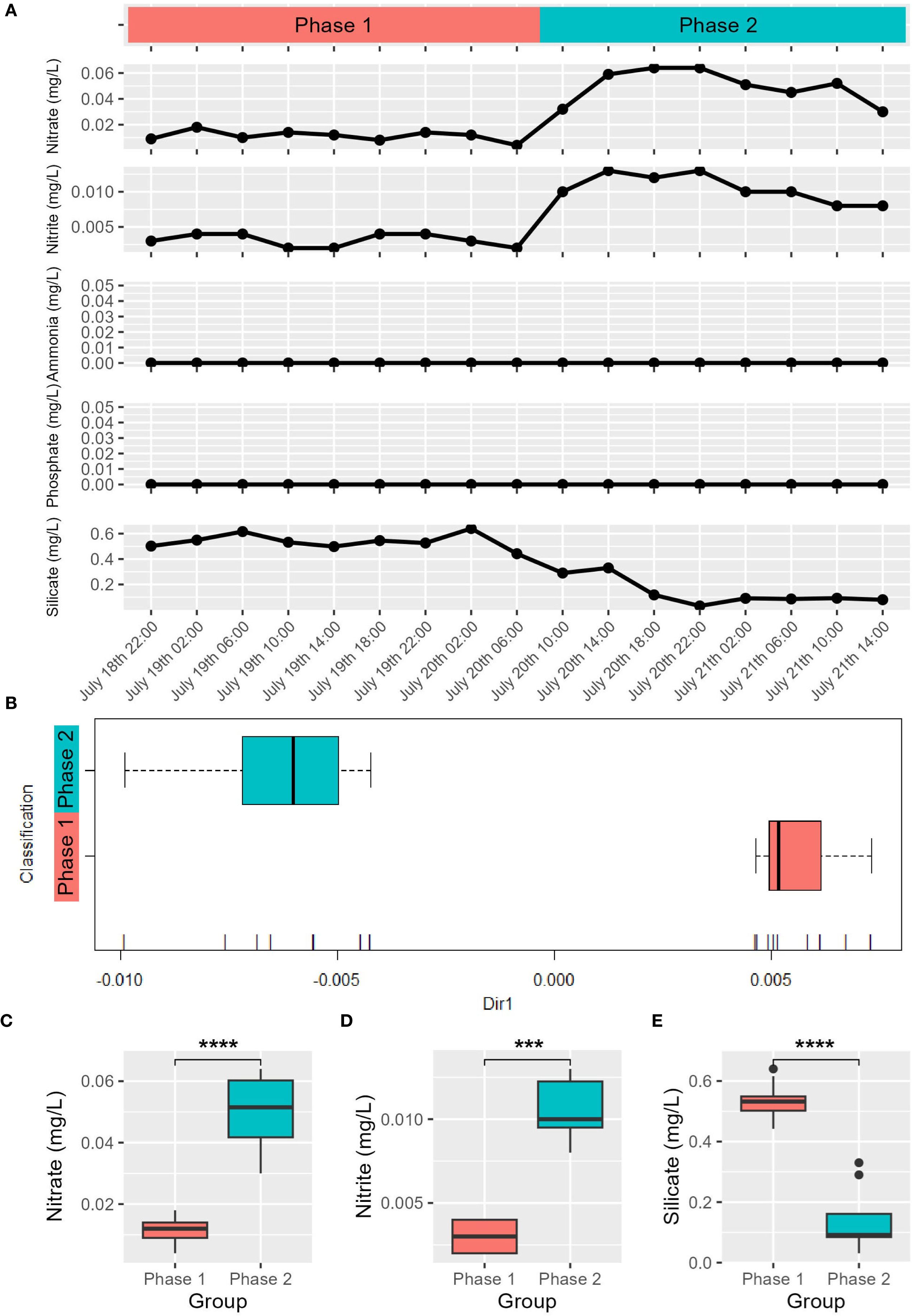
Figure 2. Variations in nutrient concentrations in the in situ environment (A) Grouping result of the in situ samples based on nitrate, nitrite and silicate concentrations (B) The horizontal axis represents the coordinate projection of data on a specific dimension (Dir1). The 17 vertical lines at the bottom of the B panel represent the projection of 17 in situ samples on Dir1, ordered left to right as: L2022, L2018, L2106, L2102, L2114, L2014, L2110, L2010, L1922, L1918, L2006, L1822, L1902, L1906, L1914, L1910, L2002; Wilcoxon test results of nitrate (C), nitrite (D) and silicate (E) concentrations between Phase 1 and Phase 2. “***”p-value < 0.001, “****”p-value < 0.0001.
3.2 Distinct microbial diversity and composition patterns between groups
At the phylum level, Bacteroidota, Cyanobacteria, and Proteobacteria were the most abundant prokaryotes (Figures 3A, B). In situ observations showed that Cyanobacteria consistently dominated the prokaryotic community, while Actinobacteriota showed a significant increase in relative abundance in Phase 2 (q-value < 0.001) (Figure 3A; Supplementary Table 3). At the genus level, Synechococcus_CC9902 had the highest relative abundance, whereas Cyanobium_PCC-6307-like significantly increased in Phase 2 (q-value < 0.001), suggesting an ecotype transition between Phase 1 and Phase 2. Low-abundance taxa (Others) initially increased in Phase 2 but rapidly declined thereafter (Figure 3C). In the incubation experiment, Cyanobacteria did not always dominate the prokaryotic community. In the P group, the mean relative abundance of Bacteroidota and Proteobacteria both exceeded that of Cyanobacteria (Figure 3B). Additionally, Unclassified Saprospiraceae displayed higher mean abundance in the P group compared to the other three groups (Figure 3D). Unlike in the in situ environment, Cyanobium_PCC-6307-like remained at low relative abundance (<1.5%) across all groups (Figure 3D).
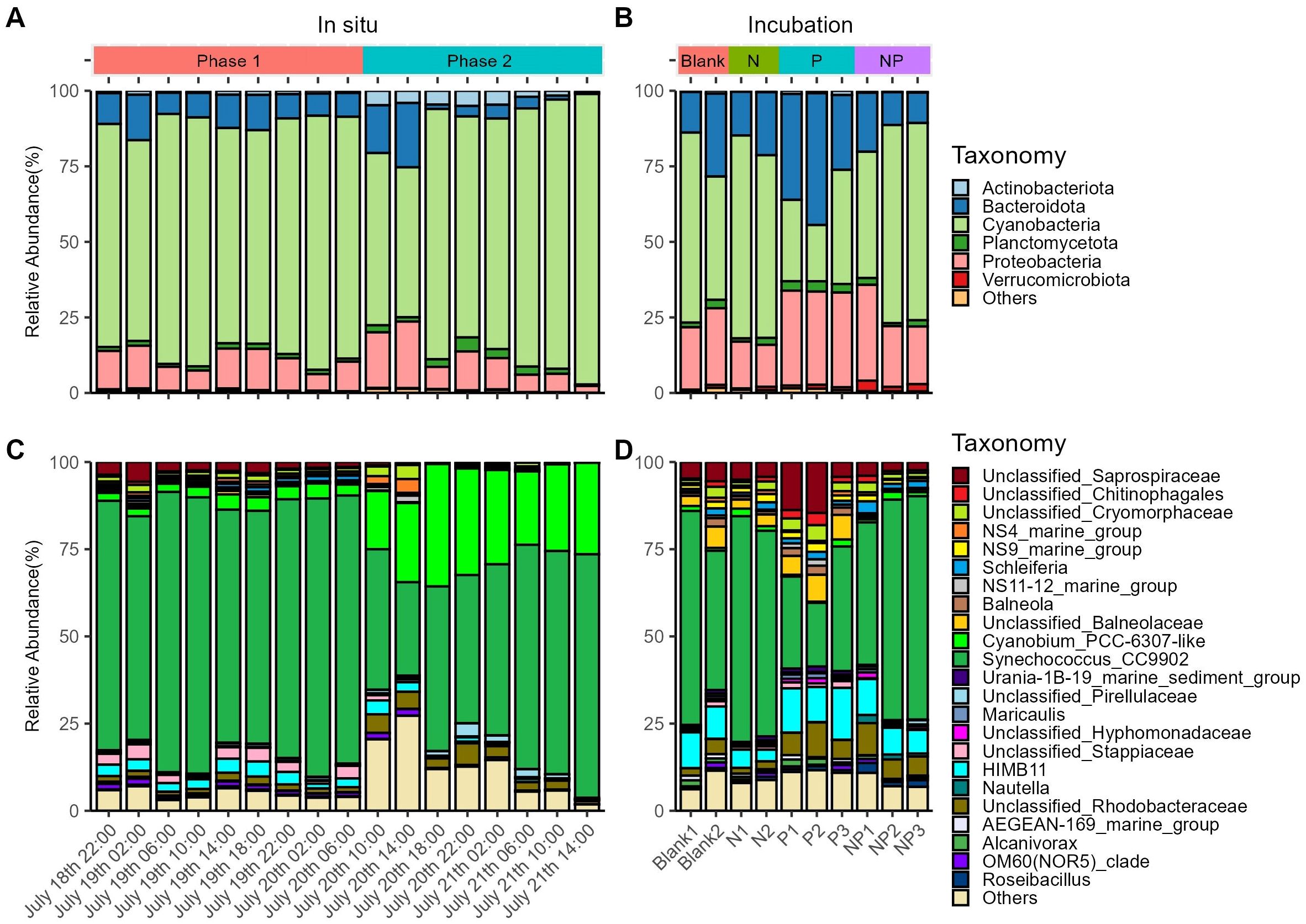
Figure 3. Prokaryotic community structure of the in situ environment (A, C) and the incubation experiment (B, D) at the phylum level (A, B) and other taxonomic levels (C, D).
For in situ samples, a significant difference in prokaryotic diversity was observed between Phase 1 and Phase 2. The diversity (Simpson index) of prokaryotes was significantly higher in Phase 2 compared to Phase 1 (p-value < 0.05) (Figure 4A). NMDS analysis also confirmed distinct clustering between Phase 1 and Phase 2 (Significance < 0.05) (Figure 4B). In the incubation experiment, the Blank, N, and NP groups exhibited larger fluctuations in diversity index, whereas the P group maintained relatively stable values and had the highest mean Simpson index (Figure 4C).
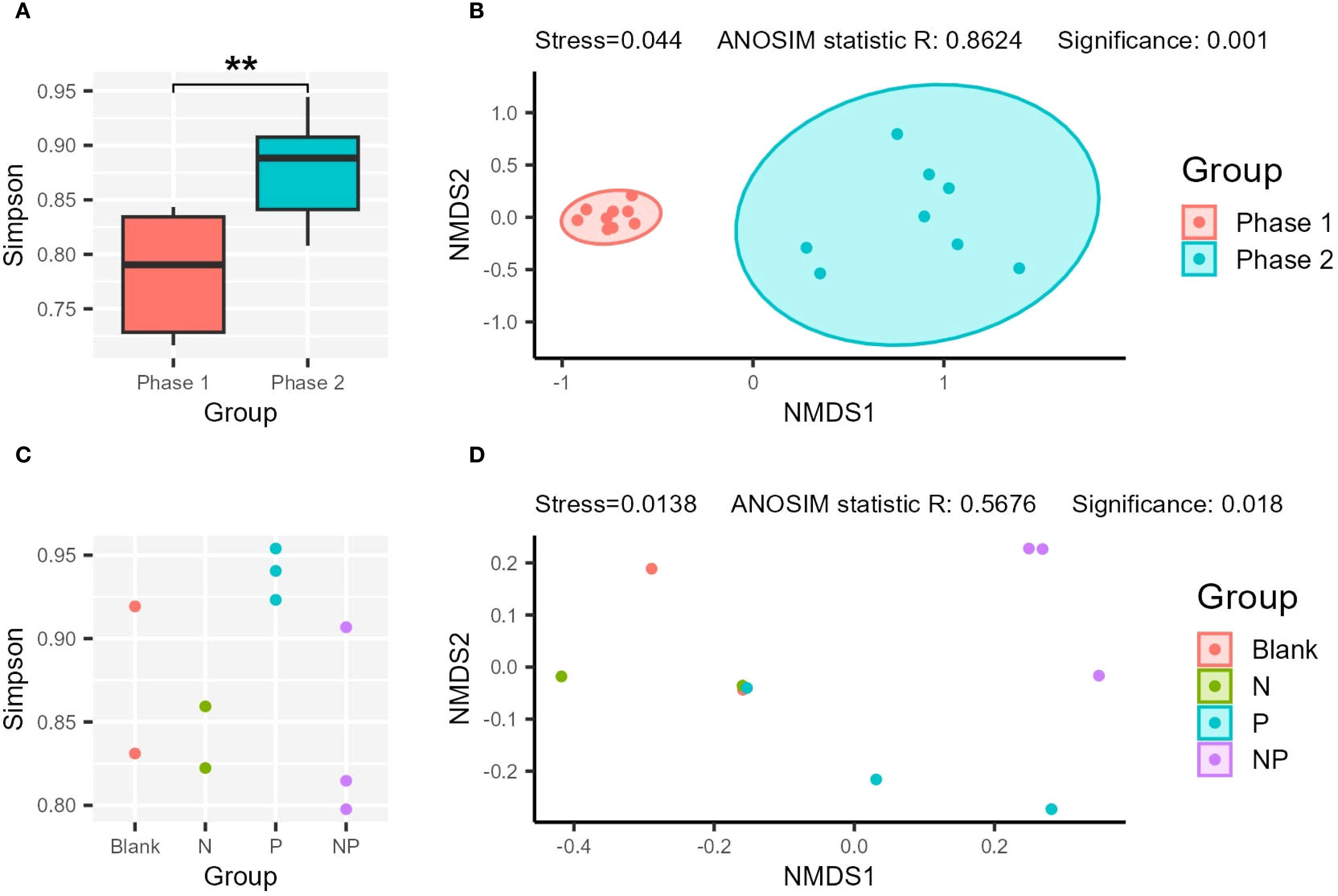
Figure 4. Comparison of the alpha and beta diversity. The comparison of Simpson index (A, C), and NMDS analysis (B, D) of the in situ (A, B) and incubation samples (C, D). “**”: p-value < 0.01.
3.3 Contrasting microbial responses to urea and phosphate addition
The addition of urea promoted the utilization potential of nutrients by autotrophic microorganisms. In both the N and NP groups, the relative gene abundances of ferredoxin-nitrite reductase (nirA), ferredoxin-nitrate reductase (narB), alkaline phosphatase D (phoD), urease (ureABCDEFG), phosphate transport system (pstSCAB), phosphonate transport system (phnC, phnD, phnE), nitrate/nitrite transporters (NRT2, narK, nrtP, nasA), phosphate starvation-inducible proteins (phoH, phoL), urea transport system (urtABCDE), phosphonate dehydrogenase (ptxD), Trk/Ktr system potassium uptake proteins (trkA, trkG, trkH, ktrA, ktrB, ktrC, ktrD), and potassium/hydrogen antiporter (cvrA, nhaP2) derived from cyanobacteria increased. In contrast, this phenomenon was not observed in the P group. Instead, the abundances of the above-mentioned genes derived from cyanobacteria decreased slightly in the P group, indicating that the competitiveness of cyanobacteria in the P group decreased (Figure 5 and Supplementary Table 4). Urea addition stimulated cyanobacteria to enhance their transport and utilization potential of dissolved inorganic nitrogen (DIN), urea, DIP, and phosphonate, thereby increasing primary productivity. The phosphate transport system substrate-binding protein (pstS), a key component of the phosphate transport system, was found to be upregulated under phosphorus starvation (Willsky and Malamy, 1980; Ames, 1986; Hussein et al., 2020). In the N group, the relative gene abundance of cyanobacterial pstS increased, indicating a higher potential demand for phosphorus by cyanobacteria. This result also supports the notion that the nutrient limitation pattern in the seawater environment of Sanmen Island is dominated by the serial nitrogen limitation.
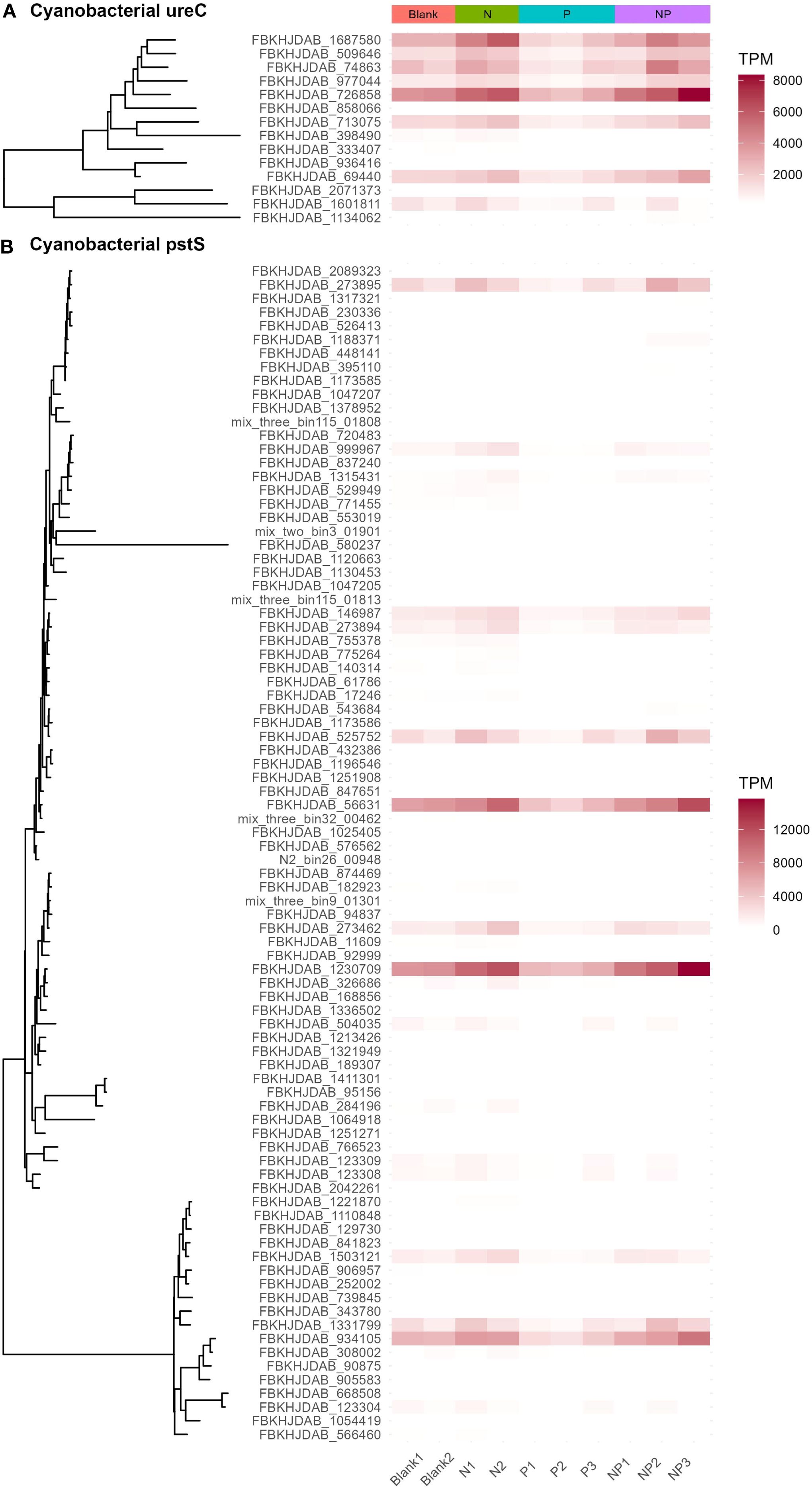
Figure 5. Taxonomic annotations and relative gene abundances of cyanobacterial ureC (A) and pstS (B). Urease subunit alpha (ureC) and phosphate transport system substrate-binding protein (pstS) are marker genes for urea hydrolysis and phosphate transmembrane transport, respectively (Hung et al., 2013; Abdo et al., 2022).
The addition of phosphate enhanced the potential of heterotrophic bacteria to compete for nitrogen and phosphorus. Chlorophyll a concentration did not increase in the P group, nor did the abundances of cyanobacterial genes relate to nitrogen and phosphorus transport and utilization. However, in the P group, heterotrophic bacteria exhibited an enhanced potential for nutrient competition under phosphate stimulation. Compared with the Blank group, the relative gene abundances of alkaline phosphatases (phoA, phoB), alkaline phosphatase D (phoD), urease (ureABCDEFG), phosphate transport system (pstSCAB), phosphate transport system protein (phoU), phosphonate transport system (phnC, phnD, phnE, phnF, phnK), phosphate-Na+ symporter (yjbB), phosphate starvation-inducible proteins (phoH, phoL), phosphate regulon sensor histidine kinase (phoR), phosphate regulon response regulator (phoB), nitrate/nitrite transport system (nrtA, nasF, cynA, nrtB, nasE, cynB), phosphonate dehydrogenase (ptxD), Trk/Ktr system potassium uptake proteins (trkA, trkG, trkH, ktrA, ktrB, ktrC, ktrD), KUP system potassium uptake protein (kup), potassium-dependent mechanosensitive channel (mscK, kefA, aefA), inward rectifier potassium channel (IRPC), voltage-gated potassium channel (kch, trkA, mthK, pch), and glutathione-regulated potassium-efflux system ancillary protein KefG (kefG) derived from heterotrophic bacteria, mainly Rhodobacterales and Maricaulales, increased in the P group. Among these genes, nitrate/nitrite transport system related genes and some phoD genes also increased in the NP group, while other genes remained at the same level or decreased (Supplementary Table 4). In contrast, the abundances of the above genes derived from heterotrophic bacteria decreased in the N group, indicating that the heterotrophic competition for nutrients was weaker in the N group.
4 Discussion
Studies on marine eutrophication confirm that increases in nitrogen levels can significantly boost algal growth, potentially triggering harmful algal blooms (Lan et al., 2024). Urea has gained recognition as a significant nitrogen source for marine phytoplankton, including the picocyanobacteria Synechococcus (Solomon et al., 2010). The ability of marine microorganisms to utilize urea through urease enzymes provides them with a competitive advantage in nitrogen-limited environments, contributing to their ecological success in coastal waters (Dyhrman and Anderson, 2003; Mulholland and Lee, 2009). While nitrogen is often the primary limiting nutrient in marine systems, phosphorus plays a critical role in freshwater and can significantly influence marine ecosystems, particularly in coastal lagoons and estuaries with elevated phosphorus inputs from agriculture and sewage (Sylvan et al., 2006; Elser et al., 2007; Conley et al., 2009). Phosphorus limitation is less common in marine systems but becomes more prominent when nitrogen levels are already elevated, making freshwater-influenced marine systems vulnerable to phosphorus-driven changes in community structure (Sylvan et al., 2006; Elser et al., 2007). Heterotrophic bacteria exhibit higher phosphorus demand compared to autotrophic organisms, with their phosphate uptake accounting for a substantial portion of total phosphate uptake in marine environments (Cotner and Wetzel, 1992). This competitive advantage allows heterotrophic bacteria to thrive when phosphate is added, potentially limiting the growth of primary producers like cyanobacteria. These dynamics highlight the significance of understanding phosphorus cycling and competition in influencing microbial community structure and nutrient availability in coastal ecosystems.
Daya Bay exhibits distinct nutrient limitation factors between the inner and outer bay. Prior to China’s Reform and Opening-Up, the inner bay was less impacted by human activities and showed nitrogen limitation (Wang et al., 2004, 2008; Shi and Huang, 2013). With subsequent coastal economic development and increased anthropogenic influences, the limiting nutrient factor in the inner bay shifted from nitrogen to phosphorus (Wang et al., 2004, 2008; Shi and Huang, 2013; Guo et al., 2023). However, in areas less affected by human activities, such as the bay mouth, nitrogen remains the limiting element factor (Ma et al., 2023; Zhao et al., 2025). The Pearl River Estuary and Daya Bay, located west and east of Shenzhen, respectively, demonstrate different patterns of nutrient limitation. In summer, the N/P ratio of surface water in the PRE exhibits a “high at north and south, low in the middle” pattern, which is significantly influenced by the Pearl River input and coastal anthropogenic activities (Li et al., 2017; Ke et al., 2022). Although Daya Bay also receives terrestrial runoff and human influence, their impacts are relatively weaker and diluted by Guangdong coastal upwelling (Han and Ma, 1988; Li et al., 1990; Zhang, 1992; Yang and Tan, 2019). During summer, influenced by the southwest monsoon, Guangdong coastal upwelling invades Daya Bay (Han and Ma, 1988; Li et al., 1990; Han and Ma, 1991; Zhang, 1992; Xu et al., 2014; Yang and Tan, 2019). Given the relatively small discharge from Daya Bay’s coastal rivers, the intensity of Guangdong coastal upwelling primarily determines the spatial distribution pattern of nutrients in Daya Bay during summer (Han and Ma, 1988; Li et al., 1990; Zhang, 1992; Yang and Tan, 2019). Therefore, the distribution pattern of nutrients in the surface seawater of Daya Bay during summer may be different annually (Shi and Huang, 2013; Wu et al., 2019; Yang and Tan, 2019; Yang et al., 2020; Zhou et al., 2024; Shi et al., 2025; Zhao et al., 2025). Sanmen Island is located in the mouth of Daya Bay and experiences additional influence from the Pearl River plume in summer (Ou et al., 2009; Yang and Tan, 2019; Ma et al., 2025). Compared with historical data, in situ environmental parameters showed characteristics of “high temperature, low salinity, low DIN, extremely low ammonia, and extremely low phosphate”, which were distinct from previous data in Daya Bay but frequently in the open sea side of the PRE (Shi and Huang, 2013; Li et al., 2017; Wu et al., 2019; Yang and Tan, 2019; Yang et al., 2020; Tao et al., 2021; Ke et al., 2022; Zhou et al., 2024; Shi et al., 2025; Zhao et al., 2025). Therefore, during the observation, the marine environment around Sanmen Island was likely predominantly influenced by the Pearl River plume, with a relatively weak intrusion of Guangdong coastal upwelling. Furthermore, the unique geographical position of Sanmen Island makes it a potentially critical site for assessing the relative strengths of the Pearl River plume and Guangdong coastal upwelling.
The different response patterns of microorganisms to urea and phosphate addition confirm the serial N limitation pattern in surface seawater near Sanmen Island during summer. Due to the extremely low phosphate concentration during in situ observation, the nutrient limitation factor could not be directly calculated from the DIN: DIP ratio. However, chlorophyll a concentrations increased in the N group, with a higher increase in the NP group, indicating the nutrient limitation pattern in Sanmen Island was serial N limitation. A similar phenomenon was also observed in the in situ observation, where the concentration of chlorophyll a increased in Phase 2, coinciding with higher nitrate and nitrite concentrations. In contrast, phosphate addition alone (P group) not only failed to observe increased primary productivity but showed slightly reduced chlorophyll a concentration. Since the relative abundance of cyanobacteria did not show a significant correlation with chlorophyll a concentrations (p-value > 0.05), suggesting the chlorophyll a concentration was not only determined by cyanobacteria (Supplementary Figure 2). Other phytoplankton also contribute significantly to primary production, such as diatoms. The significant decrease in silicate after the bloom indicates diatoms play an important role in the increase in chlorophyll a concentration. Therefore, the rise in chlorophyll a concentration may reflect the combined contributions of cyanobacteria, diatoms, and possibly other eukaryotic phytoplankton. However, the increased relative abundance of functional genes derived from cyanobacteria in both N and NP groups confirms that urea addition promoted the growth of primary producers (Supplementary Table 4). This provides genetic evidence supporting the serial N limitation pattern observed in surface waters near Sanmen Island during summer.
The bottle effect can alter microbial interactions. The “you produce while I clean up” theory describes an interaction where heterotrophic bacteria (e.g., Roseobacter) mineralize organic matter and provide inorganic nutrients, which are then utilized by autotrophs, such as cyanobacteria, for organic matter production (Christie-Oleza et al., 2015). However, in all experimental groups, the addition of exogenous nutrients only enhanced the nutrient uptake potential of microorganisms belonging to one nutritional type of microorganism (autotroph or heterotroph). Even in N and NP groups with higher primary productivity, the nutrient uptake and utilization potential of heterotrophic bacteria were suppressed, likely due to the bottle effect. Since the closed system had almost no material exchange with the outside environment, nutrient supply remained finite over time (Ionescu et al., 2015). As a result, competition for limited resources between autotrophs and heterotrophs inevitably reduced each other’s ecological niches (Calvo-Díaz et al., 2011; Ionescu et al., 2015). Notably, the competitive potential of heterotrophic bacteria was inactivated in the Blank group. Previous studies suggest heterotrophic bacteria exhibit higher phosphorus demand (Biddanda et al., 2001; Makino et al., 2003; Godwin and Cotner, 2015). Phosphate requirements among marine heterotrophic bacteria vary greatly, with their phosphate uptake accounting for 5% to over 90% of total phosphate uptake in the marine environments, depending on the study region (Faust and Correll, 1976; Harrison, 1983; Kirchman, 1994). Overall, heterotrophs dominate phosphate assimilation in aquatic systems (Kirchman, 1994). Therefore, in the P group, the addition of phosphate likely triggered the nutrient competition of heterotrophic bacteria, while cyanobacteria were at a competitive disadvantage due to the lack of available nitrogen sources. In fact, the bottle effect also contributed to differences between the in situ environment and the incubation experiment after the increase in primary productivity. During the transition from Phase 1 to Phase 2 in the in situ environment, two phenomena occurred that were not observed in the incubation experiment: an increase in DIN concentration and a bloom of Cyanobium_PCC-6307-like. These two phenomena were likely caused by the intrusion of an exogenous water mass with higher DIN concentration and another strain of cyanobacteria. This water mass passed through Sanmen Island and mixed with the local water mass. The incubation buckets’ water mass was isolated and therefore unmixed and unaffected.
The variation in relative gene abundances of potassium transporter and potassium channel reflects the importance of potassium ions (K+) in microbial survival. K+ is involved in many physiological processes, such as microbial osmoregulation, pH maintenance, and enzyme activation (Stautz et al., 2021). Trk/Ktr system is one of the three primary bacterial K+ transporter systems (Tanudjaja et al., 2023). In N and NP groups, the relative abundance of cyanobacterial Trk/Ktr genes increased, indicating that the potential demand of cyanobacteria for K+ was stimulated by urea addition rather than by the exogenous supply of K+. In contrast to Trk/Ktr, the potassium/hydrogen antiporter mediates K+ efflux and helps bacteria adapt to alkaline environments (Plack and Rosen, 1980; Radchenko et al., 2006). Given the pH increase in N and NP groups, the increase in relative abundance of potassium/hydrogen antiporter genes derived from cyanobacteria suggests their enhanced potential to alleviate intracellular alkalization stress induced by seawater alkalinization. In the P group, when heterotrophic bacteria (mainly Alphaproteobacteria) gained competitive advantage, the relative abundance of their genes related to K+ uptake and efflux increased (Supplementary Table 4). Overall, regardless of K+ addition, or whether cyanobacteria or heterotrophic bacteria were stimulated, microorganisms with competitive advantage exhibited enhanced potential for both K+ uptake and efflux (Supplementary Table 4). Considering the important role of K+ in maintaining cellular homeostasis, preserving the capacity for K+ equilibrium is indispensable for microbial survival (Stautz et al., 2021).
Unclassified Cryomorphaceae and NS4_marine_group had the potential to serve as predictors of eutrophication in the in situ environment. Unclassified Cryomorphaceae and NS4_marine_group belong to Flavobacteriales, which have the ability to degrade and utilize algae-derived organic matter, with their abundance showing strong correlations with eutrophication (DeLong et al., 1993; Teeling et al., 2012; Fontanez et al., 2015). In other coastal environments, such as Cam Ranh Bay in Vietnam and the Gulf of Trieste in the northern Mediterranean, NS4_marine_group was identified as a significant predictor of eutrophication or algal blooms, showing strong associations with ammonia and phosphate concentrations (Kolda et al., 2020; Kopprio et al., 2021; Tsang et al., 2021; Celussi et al., 2024). In the first two samples of Phase 2 (L2010 and L2014), the relative abundances of Unclassified Cryomorphaceae and NS4_marine_group increased simultaneously with those of Cyanobium_PCC-6307-like, followed by rapid declines. Although object genes derived from Flavobacteriales didn’t dominate in the incubation experiment, the relative gene abundances of phoD and yjbB increased in the P and NP groups, indicating greater potential for phosphorus acquisition and utilization (Martinez et al., 1996; Clark et al., 1998; Luo et al., 2009; Motomura et al., 2011). Therefore, in the in situ environment, Unclassified Cryomorphaceae and NS4_marine_group responded to the bloom of primary productivity, exhibiting a temporary increase in relative abundance.
Cyanobacteria have the potential to promote seawater alkalization through urea decomposition. As dominant primary producers in the ocean, cyanobacteria can utilize urea as a nitrogen source (Esteves-Ferreira et al., 2018; Veaudor et al., 2019; Díez et al., 2023; Li et al., 2023). Urease catalyzes urea hydrolysis to produce ammonia, which could increase environmental pH (Collier et al., 2009; Carlini and Ligabue-Braun, 2016; Veaudor et al., 2019; Li et al., 2023). Urease subunit alpha (ureC) is widely used as a representative gene due to its crucial role in urease activity (Reed, 2001; Abdo et al., 2022). In the incubation experiment, both the abundance of ureC genes derived from Synechococcus and seawater pH increased in N and NP groups (Supplementary Tables 2, 4). Previous studies have confirmed that under acidification conditions, Synechococcus strains utilizing urea showed significantly enhanced survival when exogenous urea was provided, whereas ureC-mutant strains exhibited reduced tolerance to acidification stress (Li et al., 2023). Although Synechococcus were not directly exposed to acidification conditions in the incubation experiment, the increase in ureC relative gene abundance and seawater pH in N and NP groups suggests their potential to reduce ocean acidification.
It should be noted that due to the geographical location of Sanmen Island and the variable inter-annual and seasonal dynamics of these transitional waters, the conclusion of Sanmen Island’s nutrient limitation pattern derived from this study, which focused on one site at one season, may not be applicable throughout Daya Bay or across different years and seasons. In addition, due to experimental biases in DNA extraction and sequencing, eukaryotic sequences are exceedingly rare in the sequencing dataset. Therefore, to ensure the reliability of downstream analyses, we excluded eukaryotic sequences. In this study, the absence of eukaryotic sequences limited the exploration of drivers contributing to primary productivity. In future research, the inclusion of eukaryotic sequences will provide a more comprehensive understanding of biogeochemical cycles in transitional water mass.
5 Conclusion
In the summer season, as represented by transitional waters off Sanmen Island, the nutrient limitation pattern is characterized by serial nitrogen limitation. The addition of urea enhanced the potential of cyanobacteria to uptake and utilize nutrients, stimulating the bloom in primary productivity. This stimulatory effect was more pronounced under the co-addition of urea and phosphate. In contrast, phosphate addition alone not only failed to increase primary productivity but also increased the competitive potential of heterotrophic bacteria for nutrient acquisition, thereby exacerbating resource competition and constraining the ecological niche of cyanobacteria.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1282918.
Author contributions
YL: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft. XZ: Formal analysis, Investigation, Writing – review & editing. RN: Data curation, Investigation, Writing – original draft. RY: Investigation, Methodology, Writing – review & editing. SX: Methodology, Project administration, Writing – review & editing. KG: Investigation, Project administration, Writing – review & editing. JG: Project administration, Resources, Writing – review & editing. JT: Investigation, Methodology, Writing – review & editing. SW: Project administration, Resources, Writing – review & editing. SH: Conceptualization, Funding acquisition, Methodology, Resources, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by the National Natural Science Foundation of China (grant numbers: 42476109, 42276163) and the Shenzhen Science, Technology and Innovation Commission Program (grant number: JCYJ20220530115401003).
Acknowledgments
We thank Professor Xinxin Li, Xin Zhao for their guidance on DOC measurement. Additionally, we thank Yiman Peng, Zhengting Wu, and Yongjuan Shi for their assistance with sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frmbi.2025.1655960/full#supplementary-material
References
Abdo A. I., Xu Y., Shi D., Li J., Li H., El-Sappah A. H., et al. (2022). Nitrogen transformation genes and ammonia emission from soil under biochar and urease inhibitor application. Soil Tillage Res. 223, 105491. doi: 10.1016/j.still.2022.105491
Aitchison J. (1982). The statistical analysis of compositional data. J. R. Stat. Soc Ser. B (Methodol). 44, 139–160. doi: 10.1111/j.2517-6161.1982.tb01195.x
Ames G. F. L. (1986). Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu. Rev. Biochem. 55, 397–425. doi: 10.1146/annurev.bi.55.070186.002145
Aramaki T., Blanc-Mathieu R., Endo H., Ohkubo K., Kanehisa M., Goto S., et al. (2019). KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 36, 2251–2252. doi: 10.1093/bioinformatics/btz859
Aroney S. T. N., Newell R. J. P., Nissen J. N., Camargo A. P., Tyson G. W., and Woodcroft B. J. (2025). CoverM: read alignment statistics for metagenomics. Bioinformatics. 41, btaf147. doi: 10.1093/bioinformatics/btaf147
Biddanda B., Ogdahl M., and Cotner J. (2001). Dominance of bacterial metabolism in oligotrophic relative to eutrophic waters. Limnol. Oceanogr. 46, 730–739. doi: 10.4319/lo.2001.46.3.0730
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bonnet S., Moutin T., Rodier M., Grisoni J. M., Louis F., Folcher E., et al. (2016). Introduction to the project VAHINE: VAriability of vertical and tropHIc transfer of diazotroph derived N in the south wEst Pacific. Biogeosci. 13, 2803–2814. doi: 10.5194/bg-13-2803-2016
Brown C. T. and Irber L. (2016). sourmash: a library for MinHash sketching of DNA. J. Open Source Software 1, 27. doi: 10.21105/joss.00027
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Calvo-Díaz A., Díaz-Pérez L., Suárez L. Á., Morán X. A. G., Teira E., and Marañón E. (2011). Decrease in the autotrophic-to-heterotrophic biomass ratio of picoplankton in oligotrophic marine waters due to bottle enclosure. Appl. Environ. Microbiol. 77, 5739–5746. doi: 10.1128/AEM.00066-11
Cao W., Yang Q., Ji F., and Liu C. (2024). Influence of N, P, and Fe availability on Braarudosphaera bigelowii, Trichodesmium, Crocosphaera, and noncyanobacterial diazotrophs: a review. Front. Mar. Sci. 11. doi: 10.3389/fmars.2024.1467599
Capella-Gutiérrez S., Silla-Martínez J. M., and Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carlini C. R. and Ligabue-Braun R. (2016). Ureases as multifunctional toxic proteins: A review. Toxicon. 110, 90–109. doi: 10.1016/j.toxicon.2015.11.020
Celussi M., Manna V., Banchi E., Fonti V., Bazzaro M., Flander-Putrle V., et al. (2024). Annual recurrence of prokaryotic climax communities in shallow waters of the North Mediterranean. Environ. Microbiol. 26, e16595. doi: 10.1111/1462-2920.16595
Chaumeil P. A., Mussig A. J., Hugenholtz P., and Parks D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chaumeil P. A., Mussig A. J., Hugenholtz P., and Parks D. H. (2022). GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics. 38, 5315–5316. doi: 10.1093/bioinformatics/btac672
Christie-Oleza J. A., Scanlan D. J., and Armengaud J. (2015). You produce while I clean up”, a strategy revealed by exoproteomics during Synechococcus-Roseobacter interactions. Proteomics. 15, 3454–3462. doi: 10.1002/pmic.201400562
Clark L. L., Ingall E. D., and Benner R. (1998). Marine phosphorus is selectively remineralized. Nature. 393, 426–426. doi: 10.1038/30881
Clarke K. R. and Green R. H. (1988). Statistical design and analysis for a 'biological effects' study. Mar. Ecol. Prog. Ser. 46, 213–226. doi: 10.3354/meps046213
Collier J. L., Baker K. M., and Bell S. L. (2009). Diversity of urea-degrading microorganisms in open-ocean and estuarine planktonic communities. Environ. Microbiol. 11, 3118–3131. doi: 10.1111/j.1462-2920.2009.02016.x
Conley D. J., Paerl H. W., Howarth R. W., Boesch D. F., Seitzinger S. P., Havens K. E., et al. (2009). Controlling eutrophication: nitrogen and phosphorus. Science. 323, 1014–1015. doi: 10.1126/science.1167755
Cotner J. B. and Wetzel R. G. (1992). Uptake of dissolved inorganic and organic bphosphorus compounds by phytoplankton and bacterioplankton. Limnol. Oceanogr. 37, 232–243. doi: 10.4319/lo.1992.37.2.0232
de Baar H. J. W. (1994). von Liebig's law of the minimum and plankton ecology (1899-1991). Prog. Oceanogr. 33, 347–386. doi: 10.1016/0079-6611(94)90022-1
DeLong E. F., Franks D. G., and Alldredge A. L. (1993). Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934. doi: 10.4319/lo.1993.38.5.0924
Díez J., López-Lozano A., Domínguez-Martín M. A., Gómez-Baena G., Muñoz-Marín M. C., Melero-Rubio Y., et al. (2023). Regulatory and metabolic adaptations in the nitrogen assimilation of marine picocyanobacteria. FEMS Microbiol. Rev. 47, fuac043. doi: 10.1093/femsre/fuac043
Dyhrman S. T. and Anderson D. M. (2003). Urease activity in cultures and field populations of the toxic dinoflagellate Alexandrium. Limnol. Oceanogr. 48, 647–655. doi: 10.4319/lo.2003.48.2.0647
Edgar R. C. (2022). Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 13, 6968. doi: 10.1038/s41467-022-34630-w
Elser J. J., Bracken M. E. S., Cleland E. E., Gruner D. S., Harpole W. S., Hillebrand H., et al. (2007). Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 10, 1135–1142. doi: 10.1111/j.1461-0248.2007.01113.x
Esteves-Ferreira A. A., Inaba M., Fort A., Araújo W. L., and Sulpice R. (2018). Nitrogen metabolism in cyanobacteria: metabolic and molecular control, growth consequences and biotechnological applications. Crit. Rev. Microbiol. 44, 541–560. doi: 10.1080/1040841X.2018.1446902
Falkowski P. G., Barber R. T., and Smetacek V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science. 281, 200–206. doi: 10.1126/science.281.5374.200
Faust M. A. and Correll D. L. (1976). Comparison of bacterial and algal utilization of orthophosphate in an estuarine environment. Mar. Biol. 34, 151–162. doi: 10.1007/BF00390757
Field C. B., Behrenfeld M. J., Randerson J. T., and Falkowski P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 281, 237–240. doi: 10.1126/science.281.5374.237
Fontanez K. M., Eppley J. M., Samo T. J., Karl D. M., and DeLong E. F. (2015). Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00469
Fu L., Niu B., Zhu Z., Wu S., and Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
GB17378.4 (2007). “The specification for marine monitoring-part 4: Seawater analysis,” in General Administration of Quality Supervision. Inspection and Quarantine of the People’s Republic of China; Standardization Administration (Inspection and Quarantine of the People’s Republic of China; Standardization Administration, China).
Godwin C. M. and Cotner J. B. (2015). Aquatic heterotrophic bacteria have highly flexible phosphorus content and biomass stoichiometry. ISME J. 9, 2324–2327. doi: 10.1038/ismej.2015.34
Guillou L., Bachar D., Audic S., Bass D., Berney C., Bittner L., et al. (2012). The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. doi: 10.1093/nar/gks1160
Guo Z., Xiao Y., Liu Y., Wu P., and Li C. (2023). Long-term variations of biogenic elements and nutritional status in Daya Bay, northern South China Sea. J. Mar. Sci. Eng. 11, 904. doi: 10.3390/jmse11050904
Hamdhani H. (2024). Relationship between chlorophyll-a, pH, and dissolved oxygen in a tropical urban lake waters: a case study from Air Hitam Lake, Samarinda City, Indonesia. Water Conserv. Manage. 8, 185–189. doi: 10.26480/wcm.02.2024.185.189
Han W. and Ma K. (1988). Study on offshore upwelling off eastern guangdong. Acta Oceanolog. Sin. 10, 52–59.
Han W. and Ma K. (1991). Study on the process of sea water exchange in Daya Bay. Mar. Sci. 2), 64–67.
Harpole W. S., Ngai J. T., Cleland E. E., Seabloom E. W., Borer E. T., Bracken M. E. S., et al. (2011). Nutrient co-limitation of primary producer communities. Ecol. Lett. 14, 852–862. doi: 10.1111/j.1461-0248.2011.01651.x
Harrison W. G. (1983). Uptake and recycling of soluble reactive phosphorus by marine microplankton. Mar. Ecol. Prog. Ser. 10, 127–135. doi: 10.3354/meps010127
Hung S. H., Chung C. C., Liao C. W., Gong G. C., and Chang J. (2013). Sequence diversity and expression levels of Synechococcus phosphate transporter gene in the East China Sea. J. Exp. Mar. Biol. Ecol. 440, 90–99. doi: 10.1016/j.jembe.2012.11.018
Huson D. H., Beier S., Flade I., Górska A., El-Hadidi M., Mitra S., et al. (2016). MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PloS Comput. Biol. 12, e1004957. doi: 10.1371/journal.pcbi.1004957
Hussein F. B., Venkiteshwaran K., and Mayer B. K. (2020). Cell surface-expression of the phosphate-binding protein PstS: System development, characterization, and evaluation for phosphorus removal and recovery. J. Environ. Sci. 92, 129–140. doi: 10.1016/j.jes.2020.02.016
Ionescu D., Bizic-Ionescu M., Khalili A., Malekmohammadi R., Morad M. R., de Beer D., et al. (2015). A new tool for long-term studies of POM-bacteria interactions: overcoming the century-old Bottle Effect. Sci. Rep. 5, 14706. doi: 10.1038/srep14706
Katoh K., Misawa K., Kuma K., and Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Ke S., Zhang P., Ou S., Zhang J., Chen J., and Zhang J. (2022). Spatiotemporal nutrient patterns, composition, and implications for eutrophication mitigation in the Pearl River Estuary, China. Estuar. Coast. Shelf Sci. 266, 107749. doi: 10.1016/j.ecss.2022.107749
Kirchman D. L. (1994). The uptake of inorganic nutrients by heterotrophic bacteria. Microb. Ecol. 28, 255–271. doi: 10.1007/BF00166816
Kolda A., Gavrilović A., Jug-Dujaković J., Ljubešić Z., El-Matbouli M., Lillehaug A., et al. (2020). Profiling of bacterial assemblages in the marine cage farm environment, with implications on fish, human and ecosystem health. Ecol. Indic. 118, 106785. doi: 10.1016/j.ecolind.2020.106785
Kopprio G. A., Cuong L. H., Luyen N. D., Duc T. M., Ha T. H., Huong L. M., et al. (2021). Carrageenophyte-attached and planktonic bacterial communities in two distinct bays of Vietnam: Eutrophication indicators and insights on ice-ice disease. Ecol. Indic. 121, 107067. doi: 10.1016/j.ecolind.2020.107067
Lan J., Liu P., Hu X., and Zhu S. (2024). Harmful algal blooms in eutrophic marine environments: causes, monitoring, and treatment. Water. 16, 2525. doi: 10.3390/w16172525
Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 1303.3997. doi: 10.48550/arXiv.1303.3997
Li B. and Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li W. and Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Li S. Q., Huang H. L., Sun T. R., Gao H. Y., Wang X. W., Fu F. X., et al. (2023). Cyanobacteria using urea as a nitrogen source can overcome acid stress. bioRxiv. 2023.03.29.534730. doi: 10.1101/2023.03.29.534730
Li D., Liu C. M., Luo R., Sadakane K., and Lam T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Li D., Liu J., Zhang R., Chen M., Yang W., Li J., et al. (2019). N2 fixation impacted by carbon fixation via dissolved organic carbon in the changing Daya Bay, South China Sea. Sci. Total Environ. 674, 592–602. doi: 10.1016/j.scitotenv.2019.04.176
Li D., Luo R., Liu C. M., Leung C. M., Ting H. F., Sadakane K., et al. (2016). MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 102, 3–11. doi: 10.1016/j.ymeth.2016.02.020
Li Y. H., Qiu Y., Yang L. Q., and Hu J. Y. (2021). Distribution characteristics of temperature and salinity in Daya Bay and its adjacent waters in summer. J. Appl. Oceanogr. 40, 284–292. doi: 10.3969/J.ISSN.2095-4972.2021.02.011
Li R., Xu J., Li X., Shi Z., and Harrison P. J. (2017). Spatiotemporal variability in phosphorus species in the Pearl River Estuary: Influence of the river discharge. Sci. Rep. 7, 13649. doi: 10.1038/s41598-017-13924-w
Li L., Zeng G., Xu J., and State Oceanic Administration The Third Institute of Oceanography (1990). “Phenomena of cold water invasion into Daya Bay during summer 1987,” in Collections of papers on marine ecology in the Daya Bay (Ocean Press, Xiamen), 95–99.
Lin X., Li K., Zhao H., Gao Y., Zhang Z., Wang L., et al. (2024). Seasonal changes of plankton community and its influencing factors in subtropical coastal marine areas revealed by eDNA-based network analysis. Front. Mar. Sci. 11. doi: 10.3389/fmars.2024.1416359
Lin H. and Peddada S. D. (2020). Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514. doi: 10.1038/s41467-020-17041-7
Liu K. K., Chao S. Y., Shaw P. T., Gong G. C., Chen C. C., and Tang T. Y. (2002). Monsoon-forced chlorophyll distribution and primary production in the South China Sea: observations and a numerical study. Deep Sea Res. Part I. 49, 1387–1412. doi: 10.1016/S0967-0637(02)00035-3
Luo H., Benner R., Long R. A., and Hu J. (2009). Subcellular localization of marine bacterial alkaline phosphatases. Proc. Natl. Acad. Sci. 106, 21219–21223. doi: 10.1073/pnas.0907586106
Ma C., He W., Zhang G., Li X., and Zhao J. (2025). Spatiotemporal variations in Pearl River plume dispersion over the last decade based on VIIRS-derived sea surface salinity. Mar. pollut. Bull. 218, 118179. doi: 10.1016/j.marpolbul.2025.118179
Ma Y., Wang X., Xu X., Liu W., Wang Z., Shi H., et al. (2023). Distributions characteristics of nutrients in sea water and its ecological environment effects in Daya Bay. Environ. Monit. China. 39, 110–122. doi: 10.19316/j.issn.1002-6002.2023.06.12
Makino W., Cotner J. B., Sterner R. W., and Elser J. J. (2003). Are bacteria more like plants or animals? Growth rate and resource dependence of bacterial C: N: P stoichiometry. Funct. Ecol. 17, 121–130. doi: 10.1046/j.1365-2435.2003.00712.x
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3. doi: 10.14806/ej.17.1.200
Martinez J., Smith D. C., Steward G. F., and Azam F. (1996). Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat. Microb. Ecol. 10, 223–230. doi: 10.3354/ame010223
McMurdie P. J. and Holmes S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Moore C. M., Mills M. M., Arrigo K. R., Berman-Frank I., Bopp L., Boyd P. W., et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710. doi: 10.1038/ngeo1765
Motomura K., Hirota R., Ohnaka N., Okada M., Ikeda T., Morohoshi T., et al. (2011). Overproduction of YjbB reduces the level of polyphosphate in Escherichia coli: a hypothetical role of YjbB in phosphate export and polyphosphate accumulation. FEMS Microbiol. Lett. 320, 25–32. doi: 10.1111/j.1574-6968.2011.02285.x
Mulholland M. R. and Lee C. (2009). Peptide hydrolysis and the uptake of dipeptides by phytoplankton. Limnol. Oceanogr. 54, 856–868. doi: 10.4319/lo.2009.54.3.0856
Nguyen L. T., Schmidt H. A., von Haeseler A., and Minh B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Niu L., Luo X., Hu S., Liu F., Cai H., Ren L., et al. (2020). Impact of anthropogenic forcing on the environmental controls of phytoplankton dynamics between 1974 and 2017 in the Pearl River estuary, China. Ecol. Indic. 116, 106484. doi: 10.1016/j.ecolind.2020.106484
Nixon S. W. (1995). Coastal marine eutrophication: A definition, social causes, and future concerns. Ophelia. 41, 199–219. doi: 10.1080/00785236.1995.10422044
Oksanen J., Simpson G. L., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., et al. (2022). vegan: Community Ecology Package. R package version 2.6-4. Available online at: https://cran.r-project.org (Accessed June 24, 2025).
Olm M. R., Brown C. T., Brooks B., and Banfield J. F. (2017). dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868. doi: 10.1038/ismej.2017.126
Ou S., Zhang H., and Wang D. X. (2009). Dynamics of the buoyant plume off the Pearl River Estuary in summer. Environ. Fluid Mech. 9, 471–492. doi: 10.1007/s10652-009-9146-3
Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., and Tyson G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Peng Y., Sun L., Chen H., and Wang Z. (2002). Study on eutrophication and change of nutrients in the Daya Bay. Mar. Sci. Bull. 21, 44–49.
Plack R. H. and Rosen B. P. (1980). Cation/proton antiport systems in Escherichia coli. Absence of potassium/proton antiporter activity in a pH-sensitive mutant. J. Biol. Chem. 255, 3824–3825. doi: 10.1016/S0021-9258(19)85594-1
Qiu Z., Yuan L., Lian C. A., Lin B., Chen J., Mu R., et al. (2024). BASALT refines binning from metagenomic data and increases resolution of genome-resolved metagenomic analysis. Nat. Commun. 15, 2179. doi: 10.1038/s41467-024-46539-7
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Radchenko M. V., Tanaka K., Waditee R., Oshimi S., Matsuzaki Y., Fukuhara M., et al. (2006). Potassium/proton antiport system of Escherichia coli. J. Biol. Chem. 281, 19822–19829. doi: 10.1074/jbc.M600333200
R Core Team (2023). The R Project for Statistical Computing. Available online at: https://www.r-project.org/ (Accessed June 24, 2025).
Redfield A. C. (1958). The biological control of chemical factors in the environment. Am. Sci. 46, 230A–2221.
Reed K. E. (2001). Restriction enzyme mapping of bacterial urease genes: using degenerate primers to expand experimental outcomes. Biochem. Mol. Biol. Educ. 29, 239–244. doi: 10.1016/S1470-8175(01)00095-9
Rodgers J. L. and Nicewander W. A. (1988). Thirteen ways to look at the correlation coefficient. Am. Stat. 42, 59–66. doi: 10.1080/00031305.1988.10475524
Rondell J. B., Finster K. W., and Lomstein B. A. (2000). Urea and DON uptake by a Lyngbya gracialis dominated microbial mat: a controlled laboratory experiment. Aquat. Microb. Ecol. 21, 169–175. doi: 10.3354/ame021169
Sayers E. W., Bolton E. E., Brister J. R., Canese K., Chan J., Comeau D. C., et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Res. 50, D20–d26. doi: 10.1093/nar/gkab1112
Scrucca L., Fop M., Murphy T. B., and Raftery A. E. (2016). mclust 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 8, 289–317. doi: 10.32614/RJ-2016-021
Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shi Z. and Huang X. (2013). Structure of N, P, Si and their temporal-spatial distribution in Daya Bay, South China. Mar. Environ. Sci. 32, 916–921.
Shi X., Ye F., Wu Y., Li J., and Wei G. (2025). Seasonal and spatial variability of dissolved organic nitrogen concentration and composition in Daya Bay, China. Mar. Environ. Res. 204, 106911. doi: 10.1016/j.marenvres.2024.106911
Solomon C. M., Collier J. L., Berg G. M., and Glibert P. M. (2010). Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquat. Microb. Ecol. 59, 67–88. doi: 10.3354/ame01390
Stautz J., Hellmich Y., Fuss M. F., Silberberg J. M., Devlin J. R., Stockbridge R. B., et al. (2021). Molecular mechanisms for bacterial potassium homeostasis. J. Mol. Biol. 433, 166968. doi: 10.1016/j.jmb.2021.166968
Sylvan J. B., Dortch Q., Nelson D. M., Maier Brown A. F., Morrison W., and Ammerman J. W. (2006). Phosphorus limits phytoplankton growth on the louisiana shelf during the period of hypoxia formation. Environ. Sci. Technol. 40, 7548–7553. doi: 10.1021/es061417t
Tanudjaja E., Hoshi N., Yamamoto K., Ihara K., Furuta T., Tsujii M., et al. (2023). Two Trk/Ktr/HKT-type potassium transporters, TrkG and TrkH, perform distinct functions in Escherichia coli K-12. J. Biol. Chem. 299, 102846. doi: 10.1016/j.jbc.2022.102846
Tao W., Niu L., Dong Y., Fu T., and Lou Q. (2021). Nutrient pollution and its dynamic source-sink pattern in the Pearl River Estuary (South China). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.713907
Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 336, 608–611. doi: 10.1126/science.1218344
Tsang H. H., Domingos J. A., Westaway J. A. F., Kam M. H. Y., Huerlimann R., and Bastos Gomes G. (2021). Digital droplet PCR-based environmental DNA tool for monitoring Cryptocaryon irritans in a marine fish farm from Hong Kong. Divers. 13, 350. doi: 10.3390/d13080350
Veaudor T., Cassier-Chauvat C., and Chauvat F. (2019). Genomics of urea transport and catabolism in cyanobacteria: Biotechnological implications. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02052
Vitousek P. M. and Howarth R. W. (1991). Nitrogen limitation on land and in the sea: How can it occur? Biogeochem. 13, 87–115. doi: 10.1007/BF00002772
Wagner G. P., Kin K., and Lynch V. J. (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285. doi: 10.1007/s12064-012-0162-3
Wang X., Li H., Zheng C., Yang J., Zhang Y., Zhang M., et al. (2018). Submarine groundwater discharge as an important nutrient source influencing nutrient structure in coastal water of Daya Bay, China. Geochim. Cosmochim. Acta 225, 52–65. doi: 10.1016/j.gca.2018.01.029
Wang Y. S., Lou Z. P., Sun C. C., and Sun S. (2008). Ecological environment changes in Daya Bay, China, from 1982 to 2004. Mar. pollut. Bull. 56, 1871–1879. doi: 10.1016/j.marpolbul.2008.07.017
Wang Y., Wang Z., and Huang L. (2004). Environment changes and trend in Daya Bay in recent 20 years. J. Trop. Oceanogr. 5, 85–95.
Wilcoxon F. (1945). Individual comparisons by ranking methods. Biom. Bull. 1, 80–83. doi: 10.2307/3001968
Willsky G. R. and Malamy M. H. (1980). Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J. Bacteriol. 144, 356–365. doi: 10.1128/jb.144.1.356-365.1980
Wu J., Tang J., Li Y., Hong Z., and Zhao Z. (2019). Restudy of constitution and distribution of inorganic nitrogen and the eutrophication in Daya bay, Shenzhen. Environ. Sci. Surv. 38, 79–83. doi: 10.13623/j.cnki.hkdk.2019.03.017
Wu M. L., Wang Y. S., Sun C. C., Wang H., Dong J. D., and Han S. H. (2009). Identification of anthropogenic effects and seasonality on water quality in Daya Bay, South China Sea. J. Environ. Manage. 90, 3082–3090. doi: 10.1016/j.jenvman.2009.04.017
Xu J. D., Cai S. Z., Xuan L. L., Qiu Y., Zhou X. W., and Zhu D. Y. (2014). Observational study on summertime upwelling in coastal seas between eastern Guangdong and southern Fujian. J. Trop. Oceanogr. 33, 1–9. doi: 10.11978/j.issn.1009-5470.2014.02.001
Yang W., Huang D., Chen J., Chen X., and Wang Y. (2020). Spatio-temporal distribution and eutrophication assessment of nutrients in Daya Bay during 2009-2015. South China Fish. Sci. 16, 54–61. doi: 10.12131/20190244
Yang X. and Tan Y. (2019). Effects of shelf seawater intrusion on phytoplankton community structure in Daya Bay in the summer. Mar. Sci. 43, 96–105. doi: 10.11759/hykx20190321003
Yeh Y. C., McNichol J., Needham D. M., Fichot E. B., Berdjeb L., and Fuhrman J. A. (2021). Comprehensive single-PCR 16S and 18S rRNA community analysis validated with mock communities, and estimation of sequencing bias against 18S. Environ. Microbiol. 23, 3240–3250. doi: 10.1111/1462-2920.15553
Yin K. and Harrison P. J. (2008). Nitrogen over enrichment in subtropical Pearl River estuarine coastal waters: Possible causes and consequences. Cont. Shelf Res. 28, 1435–1442. doi: 10.1016/j.csr.2007.07.010
Zhang B. (1992). Difference of seawater characteristics in Daya Bay in 1987 and 1989. J. Oceanogr. Taiwan Str. 11, 35–41.
Zhao J., He J., Li C., Yang Z., Liu C., Zhang X., et al. (2025). Seasonal and spatial variations of nutrients and environmental effects under anthropogenic influence in Daya Bay, China. Front. Mar. Sci. 12. doi: 10.3389/fmars.2025.1560930
Keywords: nitrogen limitation, Pearl River Estuary, South China Sea, primary productivity, cyanobacteria
Citation: Liu Y, Zhou X, Niu R, Yan R, Xu S, Guo K, Guo J, Tao J, Wu S and Hou S (2025) Serial nitrogen-phosphate co-limitation controls the primary productivity in the transitional waters of northern South China Sea and the Pearl River Estuary. Front. Microbiomes 4:1655960. doi: 10.3389/frmbi.2025.1655960
Received: 29 June 2025; Accepted: 22 September 2025;
Published: 08 October 2025.
Edited by:
Chandni Sidhu, Max Planck Society, GermanyReviewed by:
Guangshan Wei, Ministry of Natural Resources of the PR China, ChinaDu Su, Hebei Normal University of Science and Technology, China
Copyright © 2025 Liu, Zhou, Niu, Yan, Xu, Guo, Guo, Tao, Wu and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengwei Hou, aG91c3dAc3VzdGVjaC5lZHUuY24=
 Yuanhao Liu
Yuanhao Liu Xunying Zhou
Xunying Zhou Ruoyu Niu
Ruoyu Niu Shuaishuai Xu
Shuaishuai Xu Sha Wu
Sha Wu Shengwei Hou
Shengwei Hou