- 1Geomatics Program, Department of Built Environment, North Carolina A&T State University, Greensboro, NC, United States
- 2Department of Electrical and Computer Engineering, North Carolina A&T State University, Greensboro, NC, United States
- 3Department of Chemical, Biological, and Bioengineering, North Carolina A&T State University, Greensboro, NC, United States
- 4Department of Computational Science and Engineering, North Carolina A&T State University, Greensboro, NC, United States
Weeds are among the significant factors that could harm crop yield by invading crops and smother pastures, and significantly decrease the quality of the harvested crops. Herbicides are widely used in agriculture to control weeds; however, excessive use of herbicides in agriculture can lead to environmental pollution as well as yield reduction. Accurate mapping of crops/weeds is essential to determine weeds’ location and locally treat those areas. Increasing demand for flexible, accurate and lower cost precision agriculture technology has resulted in advancements in UAS-based remote sensing data collection and methods. Deep learning methods have been successfully employed for UAS data processing and mapping tasks in different domains. This research investigate, compares and evaluates the performance of deep learning methods for crop/weed discrimination on two open-source and published benchmark datasets captured by different UASs (field robot and UAV) and labeled by experts. We specifically investigate the following architectures: 1) U-Net Model 2) SegNet 3) FCN (FCN-32s, FCN-16s, FCN-8s) 4) DepLabV3+. The deep learning models were fine-tuned to classify the UAS datasets into three classes (background, crops, and weeds). The classification accuracy achieved by U-Net is 77.9% higher than 62.6% of SegNet, 68.4% of FCN-32s, 77.2% of FCN-16s, and slightly lower than 81.1% of FCN-8s, and 84.3% of DepLab v3+. Experimental results showed that the ResNet-18 based segmentation model such as DepLab v3+ could precisely extract weeds compared to other classifiers.
Introduction
The world’s population will increase by 30% by 2050, with a need for requiring a 60% increase in food production to meet the increasing crop demand arising from food and biofuel consumption (Pingali, 2007). Commitment to supply the amount of food demanded using sustainable modules could also facilitate the achievement of the FAO goal of providing food security to undernourished people (FAO and IFAD, 2012). However, as the agricultural sector focuses on increasing productivity, there is the need to solve issues arising concurrently such as weed management, climate change (Radoglou-Grammatikis et al., 2020), and reduction in arable lands, irrigation, and fertilizer application. Among the challenges in crop production, weed control is ranked as one of the influencers of crop yield (Raja et al., 2020). Weeds are any undesirable plants that grow amongst agricultural crops or available surface interfering with crop growth and human activities (Alsherif, 2020). They compete with crops for valuable nutrients, water, sunlight, and carbon dioxide profoundly affecting farm productivity by invading crops and smother pastures and leading to significantly decrease in the quality of harvested crops (Milberg and Hallgren, 2004). Herbicides are widely used in agriculture to control weeds. However, uncontrolled herbicides application in agriculture can lead to environmental pollution as well as yield reduction (Horrigan et al., 2002). Therefore, minimizing the amounts of herbicides and creating a rotational routine calendar are essential steps towards sustainable agriculture. In the conventional weeds control methods, the herbicide is applied over the whole field, even for the area without weeds where no treatment is required. To improve weed and crop control in modern agriculture, it is essential to extract and map weeds locations and locally treat those areas. Precision agriculture techniques should regularly monitor crop growth to maximize yield while minimizing the use of resources such as chemicals and fertilizer and reducing the side effects of herbicides on the environment (Duckett et al., 2018).
Remote sensing technology has been widely used for the classification and mapping purposes in agriculture including soil properties (Coopersmith et al., 2014), classification of crop species (Grinblat et al., 2016), detection of crop water stress (Mehdizadeh et al., 2017; Dorbu et al., 2021), monitoring of weeds and crop diseases (Milioto et al., 2017), and mapping of crop yield (Ramos et al., 2017). Recently, unmanned aerial systems (UASs) have become effective platforms for crop and weeds monitoring due to their abilities to hover close to crops and weeds to acquire high-resolution imagery at a low cost. Compared to other remote sensing platforms such as satellites and aircraft, UASs also offer a higher spatial resolution, are less dependence on weather conditions, and have flexible revisit time (Gebrehiwot et al., 2019; Vinh et al., 2019; Hashemi-Beni et al., 2018). Several studies used weed maps obtained from UAS for variable application with ground sprayers, finding no difference in crop biomass between blanket and precision spraying, while decreasing herbicide use (Castaldi et al., 2017; Pelosi et al., 2015). Despite the increased use of UASs in precision agriculture applications, efficiently processing of the high-resolution imagery data remains a challenge.
Several studies have attempted to address the problem of vision-based crops and weeds classification (Wu et al., 2021; Osorio et al., 2020). Vision-based weed control systems should detect weeds and map their location to effectively use herbicides only for those areas where weeds are present in the field. Vegetation color index, such as the normalized difference vegetation index (NDVI) is an approach to segment weeds in an agricultural field (Dyrmann and Christiansen, 2014; Osorio et al., 2020). The main challenge of this method is dealing with overlapping plants to separate weeds and crops. Texture-based models have shown promising results in detecting and discriminating plants from images with overlapping leave (Pahikkala et al., 2015). Machine learning (ML) approaches have gained attention for detecting weeds and crops (De Rainville et al., 2014; Haug and Ostermann, 2014; Gašparović et al., 2020; Islam et al., 2020). De Rainville et al. (2014) proposed a Bayesian unsupervised classification method and morphological analysis for separating crops from weeds. Thir method achieved 85% accuracy on segmenting of the weeds without any prior knowledge of the species present in the field. ML algorithms such as Support vector machines (SVMs) and Random Forest (RF) have demonstrated a good performance in remote sensing classification tasks including weed detection for small datasets. Haug and Ostermann (2014) employed a RF classifier method to classify carrot plants and weeds from RGB and near-infrared (NIR) images and achieved an average classification accuracy of 93.8%. Some studies prove that deep convolutional neural networks (CNNs) are efficient methods to deal with the limitations of handcrafted features on classifying weeds, crops and seeds (Lee et al., 2015; Loddo et al., 2021). In recent times, there has been considerable progress in the classification and segmentation of remote sensing data using deep learning for different applications (Chen et al., 2020 and Zhang et al., 2021). Unlike the conventional ML approaches, CNNs have become an increasingly popular approach for remote sensing tasks due to their ability to extracts and learns feature representation directly from big datasets (Ma et al., 2020). This allows extensive learning capabilities and, thus, higher performance and precision (Afza et al., 2021). Several researchers used CNNs in agricultural applications including weed and crop classification (Mortensen et al., 2016; Potena et al., 2016; Di Cicco et al., 2017; Milioto et al., 2017; Grace, 2021; Siddiqui et al., 2021). Mortensen et al. (2016) employed a modified version of the VGG-16 CNN model to classify weeds using mixed crops of an oil radish plot with barley, weed, stump, grass, and background soil images. Milioto et al. (2017) proposed a method that combines vegetation detection and deep learning to classify weeds in an imagery dataset (about 10,000 images) captured by a mobile agricultural robot in a sugar beet field and achieved more than 85% accuracy. Siddiqui et al. (2021) studied data augmentation for separating weeds from crops using a CNN. Potena et al. (2016) presented a perception system for weed-crop classification that uses shallow and deeper CNNs. The shallow network was used to detect vegetation, while the deeper CNN was used to classify weeds and crops. Di Cicco et al. (2017) used an encoder-decoder architecture such as SegNet to procedurally generate large synthetic training datasets randomizing the key features of the target environment (i.e., crop and weed species, type of soil, light conditions). This research aims to employ and compare the performance of state-of-the-art deep learning models such as U-Net Model, SegNet, FCN and DepLabV3+ based on transfer learning for crop and weed separation using a small optical training data and data augmentation methods. The problem of limited labelled images in RS can potentially be overcome by adapting techniques from transfer learning to RS image classification problems for different applications including Agriculture. Transfer learning can reuse the knowledge gained while classifying a dataset and applying it to another dataset having different underlying distributions. In addition, to make the different approaches comparable, which is a major issue in the field of RS data analytics and to identify promising strategies, two different open-source benchmark UAS datasets were used for this research.
Deep Learning Models
Several CNN architectures such as AlexNet (Krizhevsky et al., 2012), VGGNet (Simonyan and Zisserman, 2014), ResNet (He et al., 2016) have been developed and successfully used for image classification tasks. Krizhevsky et al. (2012) developed the first deep CNN in 2012 and showed a large, deep CNN can achieve high-performance results on a big, diverse dataset by using supervised classification. VGG-16 was developed by Simonyan and Zisserman, 2014 to increase the depth to 16 to 19 weight layers while making all filters with 3 × 3 sizes to reduce the number of parameters in the network. However, increasing the number of CNN layers do not simply improve the network’s performance due to the vanishing and exploding gradients issues in deep learning networks. He et al. (2016) introduced skip connection or residual block to overcome this problem and introduced ResNet using residual blocks as basic building blocks allowing training deeper CNN with improved performance. The following sections provide a brief overview about four important deep learning architectures that were trained and employed for crop and weed separation from the UAS datasets: namely SegNet, FCN, U-Net, and DeepLabV3+.
SegNet
SegNet is a CNN architecture proposed by Badrinarayanan et al. (2017) for pixel-wise segmentation applications. It consists of an encoder, decoder, and pixel-wise classification layers (Figure 1).
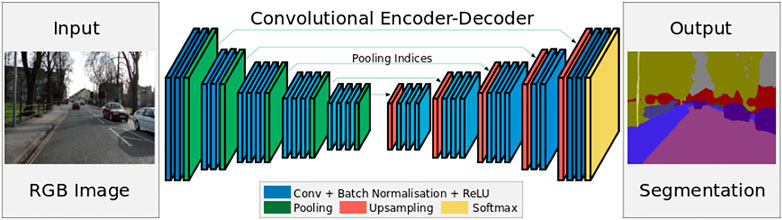
FIGURE 1. SegNet architecture (Badrinarayanan et al., 2017).
The encoder network of SegNet has 13 convolutional layers similar to VGG-16. Convolutions and down-sampling (max pooling) operations are performed at the encoder network. While at the decoder network, convolutions and up-sampling are performed and then, using the probability layer, softmax classifier, each pixel is assigned to a class. This decoder network upsamples the low-resolution feature maps to full resolution using memorized max-pooling indices. The SegNet has a smaller number of trainable parameters and can be trained end-to-end using stochastic gradient descent.
FCNs Models
The FCNs was developed by Long et al. (2015) for semantic segmentation applications by replacing the three fully connected layers of VGG16 with fully convolutional to maintain the 2-D structure of images. Thus, unlike VGG-16, FCNs can take any arbitrary input image size and produce an output with the corresponding size. Furthermore, the FCNs are composed of locally connected layers, such as convolution, pooling, and upsampling, without having any dense layer. This allows reducing the number of parameters and computation time.
Many FCN models were proposed: FCN-8s, FCN-16s, and FCN-32s. The models differ from each other in the last convolution layer and skip connection, as shown in Figure 2.
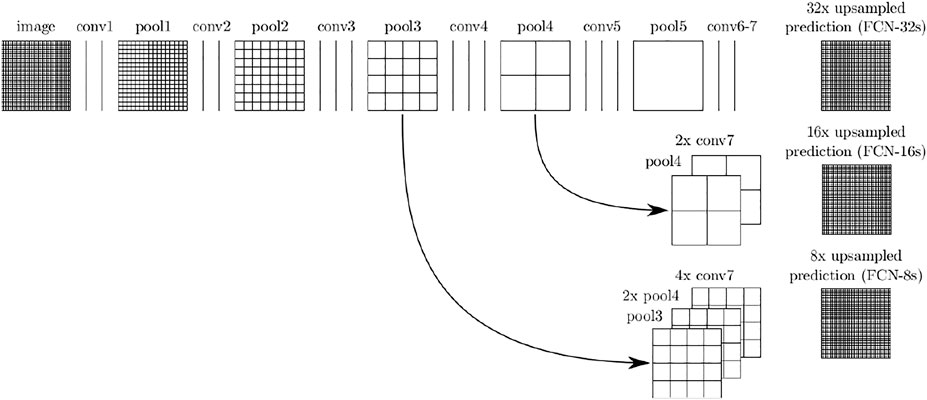
FIGURE 2. FCNs architecture (Long et al., 2015).
U-Net Model
The U-Net is the convolutional neural network proposed by Ronneberger et al. (2015) for biomedical image segmentation. It is based on FCNs, and its architecture was modified to work with fewer training data to give more precise segmentation results. As shown in Figure 3, the U-Net architecture has a “U” shape as implies its name.
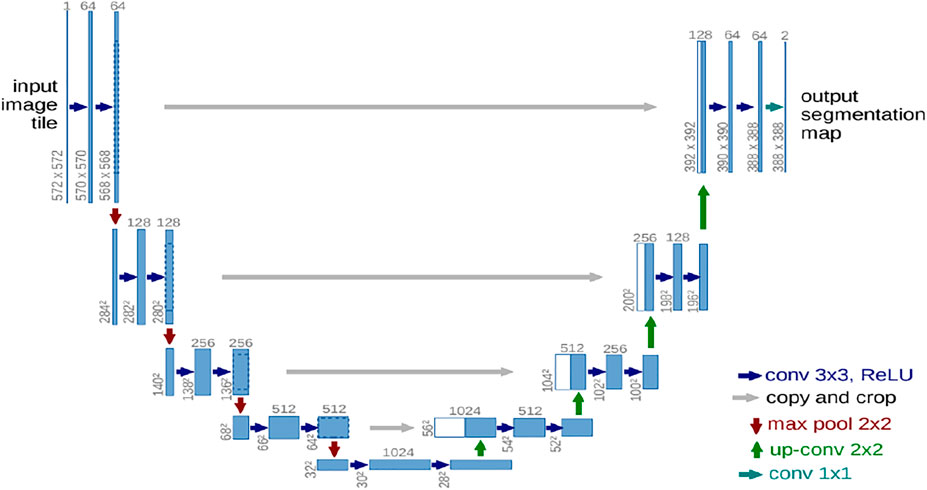
FIGURE 3. U-Net architecture (Ronneberger et al., 2015).
U-Net consists of two structures: a shrinking (contracting) structure and an expanding structure. The shrinking (contracting) structure (also called the encoder) is used to extract more advanced features and reduce the size of feature maps. On the other hand, the expanding or decoder structure is used to map the encoder’s low-resolution features to high resolution.
DeepLabV3+
Deeplab is an encoder-decoder network that was developed by a group of researchers from Google (Chen et al., 2018). It uses Atrous convolutions to overcome the issue related to the excessive downsizing in FCNs due to consecutive pooling operations. DeepLabV3+ has few changes to its predecessors, which are spatial pyramid pooling and encoder-decoder structure (Figure 4). The spatial pyramid pooling module is useful for encoding multiscale object information through multiple atrous convolutions with different rates. With this spatial information, the encoder-decoder can capture the boundary of an object more precisely.
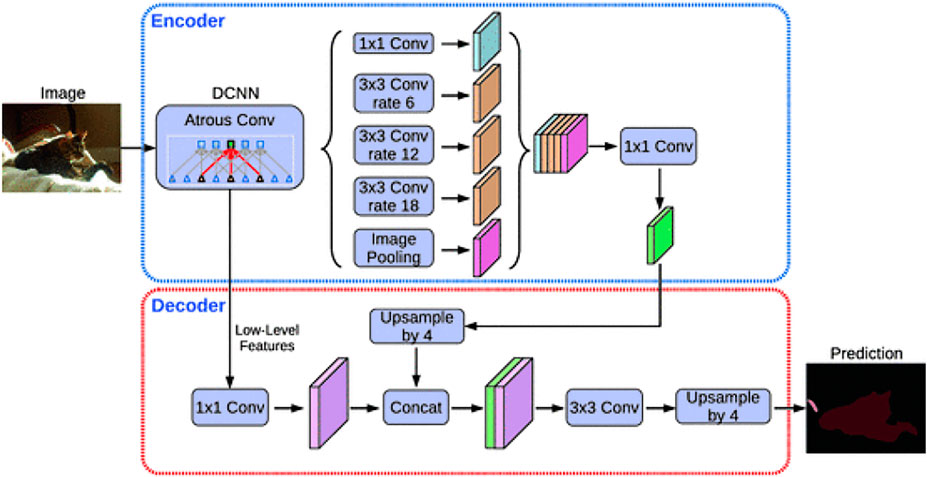
FIGURE 4. DeepLabV3+ architecture (Chen et al., 2018).
DeepLabv3+ uses Xception as the backbone instead of ResNet-101 as the encoder. The input image is down-sampled by a factor of 16. Instead of using bilinear up-sampling by a factor of 16, the decoder up-samples encoded features by a factor of 4 and concatenated with corresponding low-level features. To reduce the number of channels of low-level features, 1 × 1 convolutions are applied before concatenating. After concatenation, the decoder performs few 3 × 3 convolutions, and the features are up-sampled by a factor of 4.
UAS Imagery Datasets
Two published benchmark UAS datasets were used to train and evaluate the deep learning methods: Crop/Weed Field Image Dataset (CWFID) (Haug and Ostermann, 2014) and the Sugar Cane Orthomosaic datasets (Monteiro and Von Wangeheim, 2019).
1. The CWFID dataset was acquired with an autonomous field robot Bonirob mounted with multi-spectral camera in an organic carrot farm in Northern Germany before applying manual weed control in 2013. The CWFID consists of 60 top-down looking images of 1,296 × 966 pixel size during crop growing stages where crop leaves, intra- and inter-row weeds were present. The dataset was fully annotated by experts into three categories: soil or background, crops (162 samples), and weeds (332 samples). An image example is demonstrated in Figure 5. The dataset published for phenotyping and machine vision problems in agriculture and is available through http://github.com/cwfid; more details on the data collection can be found in Haug and Ostermann, 2014.
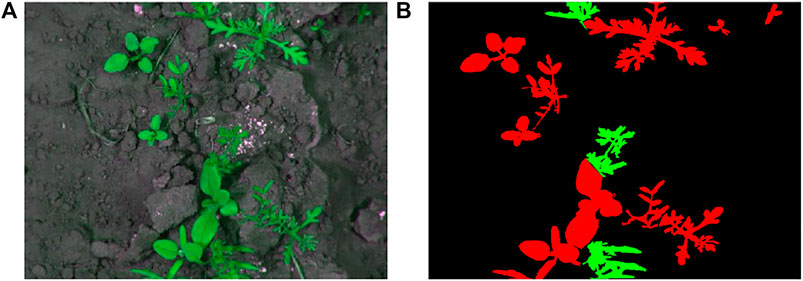
2. The sugarcane orthomosaic data was captured by a UAV from a sugarcane plantation in Northern Brazil. The orthomosaic was acquired and generated from a Horus Aeronaves fixed-wing UAV employing a visible light RGB Canon G9X camera. The UAV captured the data from a flight height of 125–200 m, which resulted in imagery with a spatial resolution of 5 cm/pixel approximately. The dataset was annotated by experts into three categories: soil or background (black), crops (green), and weeds (red). We split this single orthomosic into 60 images for the training purpose. An image example is demonstrated in Figure 6. The Sugar Cane Orthomosaic dataset is available via http://www.lapix.ufsc.br/weed-mapping-sugar-cane.
Implementation and Results
The research implemented and compared the four deep learning models in Matlab for crop and weed separation. The computer was configured with 32 GB memory, an Intel(R) Xeon(R) ES-2620 v3 @ 2.40 GHz × 2 processors memory, and a single NVIDIA Quadro M4000 GPU.
Training the Network Architectures
The provision of training data is a limiting factor for practical applications in remote sensing, however, this problem can be alleviated by exploiting techniques from transfer learning to adapt a classier trained on an image set to another dataset having different underlying distributions. As one approach to transfer learning, we fine-tuned the deep learning models discussed in Deep Learning Models i.e., SegNet, FCN-32s, FCN-16s, FCN-8s, U-Net, and DeepLabV3+ with the both UAS datasets using the same setting parameters. The sugarcane orthomosic was patched into 50 images of 540 × 540 pixels size each for training and testing purposes. We trained the models using Stochastic Gradient Descent (SGD) with a minibatch size of 2, a learning rate of 0.001, and a maximum epoch of 10. A 10-fold cross-validation procedure was used to assess the performance of these models. The significance of this procedure is to assess the predictive performance of the classifiers for classifying a new data set, also known as test data. For this purpose, we partitioned the training images randomly into ten equal parts. At each run, the union of nine parts was put together to form a training set, and the remaining one-part was used as a validation set to measure the classification errors. We repeated the above steps ten times, using a different fold as the testing set each time. Finally, the mean error from all folds was used to estimate the performance of the classifier. We applied the median frequency balancing method to deal with the imbalance problem in the training datasets. A class imbalance in the training images can affect the learning process since the learning is biased in favor of the dominant classes. As a result, the instances that belong to the minority group are misclassified more often than those of the majority group. In the median frequency balancing approach, the weight assigned to each category (ac) in the loss function is the ratio of the class frequencies’ median (median_freq(c)) computed on the entire training set divided by the class frequency (freq(c)). The class frequency is calculated by dividing the number of pixels for each class by the total number of pixels in the image. Therefore, the dominant labels were assigned with the lowest weight, which balances the training process. We also employed data augmentation techniques including random cropping, rotation, and reflection to artificially generate new training data from the existing annotated data and increase the model’s performance during the training stage. We extracted a total of 32 patches (512 ∗512) per image. The patches were inserted into the networks with a batch size of 4. Random translation and rotation data augmentation techniques were implemented by randomly translating the images up to 10 pixels horizontally and vertically and rotating the images with an angle up to 10°.
Prediction Results
This Section Presents the Results of the Weeds/Crops Classification Obtained for all the Networks Trained in This Study
A confusion matrix was used to analyze the performance of the deep learning models for weed/crop classification and the overall accuracy was calculated from the confusion matrix. The accuracy indicates the percentage of correctly identified pixels for each class, while the overall accuracy shows the percentage of correctly identified pixels for all classes. Also, the kappa coefficient (Fleiss et al., 1969) was used in this study to summarize the information provided by the confusion matrix. The Kappa index is a metric that compares an observed accuracy with an expected accuracy or random chance.
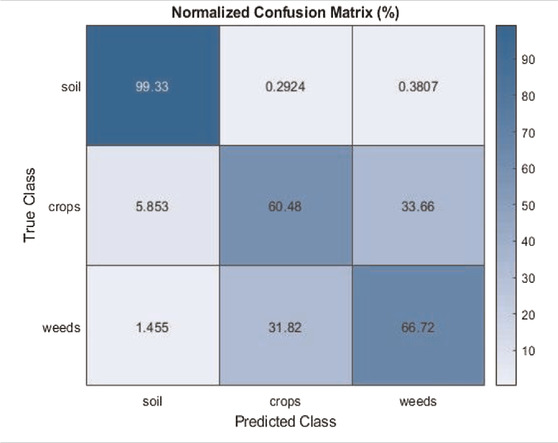
The qualitative classification results of the models are shown in Figures 7, 8. Table 1 is a confusion matrix example evaluating the U-Net prediction for soil, weeds, and crops classification. The U-Net classifier achieved an overall accuracy of 76% on classifying the weed/crop images. As shown in Figure 7, the qualitative evaluation results of DeepLabV3++ obtained a better accuracy in comparison with other models prediction for the CWFID dataset.
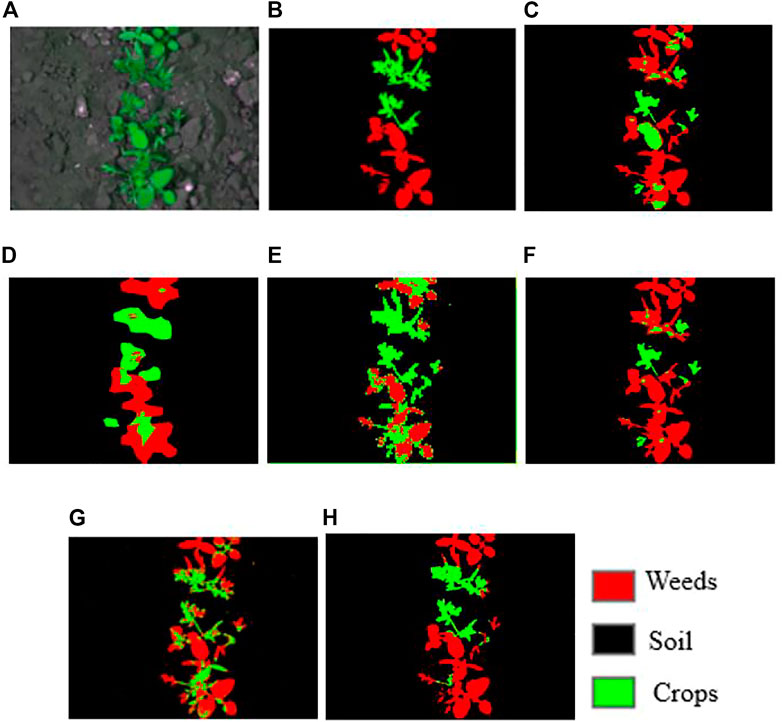
FIGURE 7. Sample qualitative results were achieved for CWFID. (A) input images; (B) groundtruth, (C) SegNet, (D) FCN-32s, (E) FCN-16s, (F) FCN-8s, (G) U-Net, (H) DeepLabV3++.
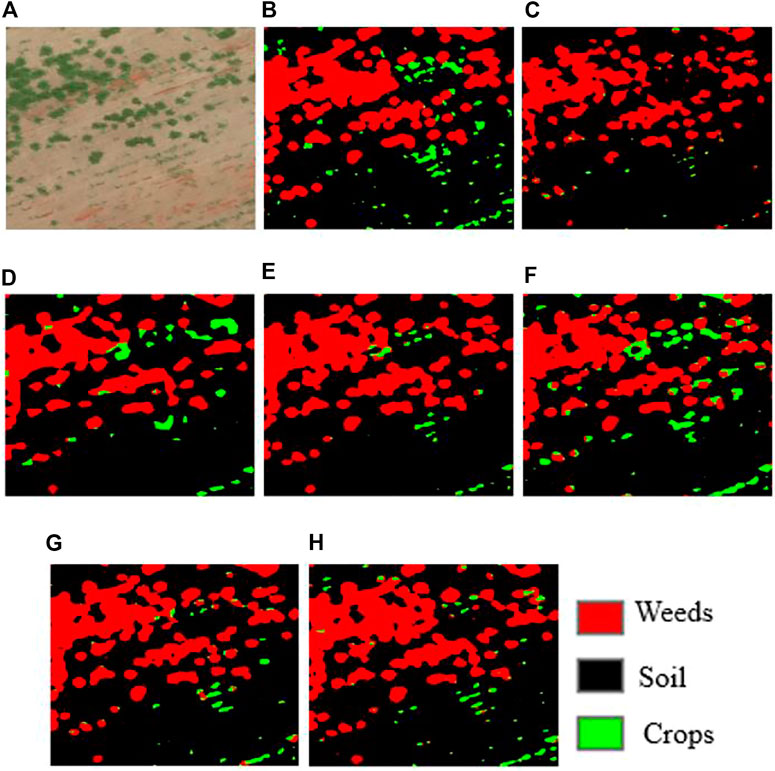
FIGURE 8. Sample qualitative results were achieved for sugarcane dataset. (A) input images; (B) groundtruth, (C) SegNet, (D) FCN-32s, (E) FCN-16s, (F) FCN-8s, (G) U-Net, (H) Deeplabv3++ result.
As shown in Figure 8, the qualitative evaluation results of FCN-8s obtained better accuracy than other models prediction for the sugarcane dataset. Table 1 shows the correspondence between the classification results and the validation images for the CWFID dataset using U-Net. The cells of the confusion matrix represented the percentage of correct and incorrect predictions for all the possible correlations. The cell in the ith row and jth column means the percentage of the ith class samples, which is classified into the jth class. The diagonal cell of the matrix contained the number of correctly identified pixels for each class. As we see from the above table, the U-Net model achieved the classification accuracy of 99.3% for soil, 60.48% for crops, and 66.72% for weeds. The experimental results also show that about 32% of weeds are wrongly classified as crops. This is because weeds and crops have a similar spectral response, which makes it hard to separate them using the U-Net classifier solely from optical imagery.
Table 2 shows the correspondence between the classification results and the validation images for the sugarcane dataset using U-Net. The U-Net model achieved the classification accuracy of 96.63% for soil, 39.19% for crops, and 86.17% for weeds.
Comparison between Classifiers for CWFID Dataset
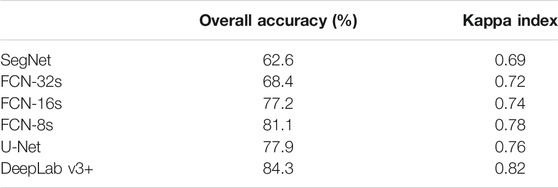
Table 3 illustrates the overall accuracy and Kappa index values for all classifiers on the CWFID dataset. On the other hand, Figure 9 shows the accuracy of the classification approaches for each of the classes for the CWFID dataset.
Based on the results in Table 3, in overall, DeepLabV3+ has a better classification performance than FCN-8s, U-Net, FCN-16s, FCN-32s, and SegNet for weeds and crops discrimination.
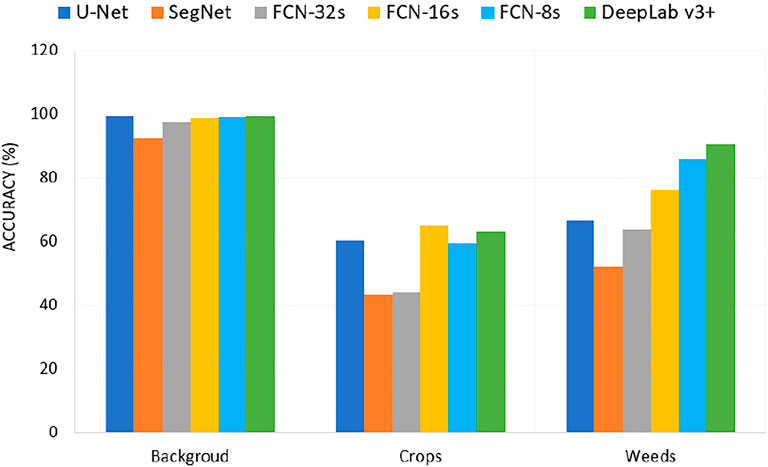
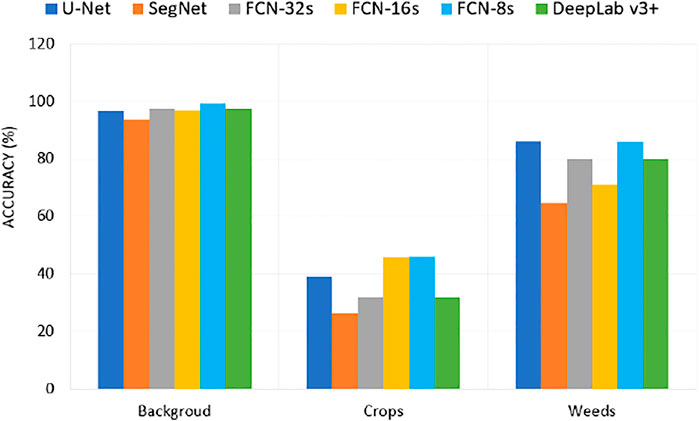
Figure 9 shows the individual classification accuracy of each class (background or soil, crops, and weeds) for the CWFID dataset. The DeepLabV3+ achieved an accuracy of 90.5% on accurately segmenting weeds compared to 85.8% of FCN-8s, 76.1% of FCN-16s, 66.7% of U-Net, 63.6% of FCN-32s, and 52% of SegNet. The results show that DeepLabV3+ has a better performance on detecting weeds from the CWFID dataset. The result shown as FCN-16s and DeepLabV3+ performed better on detecting crops from the input image. However, weeds and crops have a similar spectral response, making it hard to separate them using these classifiers accurately from optical imagery. Using multispectral images might improve segmentation performance using these models.
Comparison Between Classifiers for Sugarcane Dataset
Table 4 presents the overall accuracy and Kappa index for all classifiers for the sugarcane dataset. On the other hand, Figure 10 shows the classifier’s accuracy for each class for the same dataset.
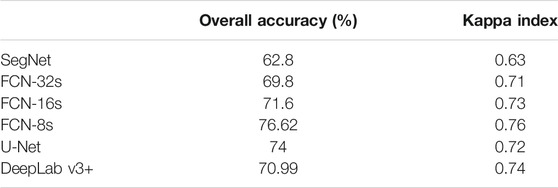
Based on the results in Table 4, FCN-8s has a better overall classification performance (74%) than SegNet (62.8%), FCN-32s (69.8%), FCN-16s (71.6%), U-Net (74%), and DepLabV+ (70.99%) for weeds and crops discrimination.
Figure 10 shows the individual classification accuracy of each class (background or soil, crops, and weeds) for the sugarcane dataset. The U-Net achieved an accuracy of 86.17% on accurately segmenting weeds compared to 85.89% of FCN-8s, 71.04% of FCN-16s, 80.16% of deeplab v3+, 80.16% of FCN-32s, and 64.5% of SegNet. The results showed that U-Net has a better performance on detecting weeds from the sugarcane dataset than other models.
Discussion
The accurate segmentation of crops and weeds has always been the center of attention in precision agriculture. Many methods have been proposed, but it is difficult to properly and sharply segment crops and weeds in images with a high presence of weeds. In this research work, we explored different segmentation models. The SegNet architecture has offered the least promising performance compared to other approaches. This architecture is slower than others because it contains an encoder-decoder structure. On the other hand, FCN-32s and FCN-16s achieved better performance than SegNet but slightly less than U-Net and DeepLab. One of the main issues here is the excessive downsizing operation due to many consecutive pooling operations. Due to this, the input image is downsampled by 32x and then upsampled again to get the classification; this results in loss of information, which is vital for getting a great result in a classification task. Also, the deconvolution to up the sample by 32x is a memory expensive operation. U-Net performed better than SegNet and FCNs- 16s and FCN-32s, but slightly less than DeepLabV3+. Because, unlike the FCNs, U-Net architecture has skip connections from the output to the corresponding input convolution block at the same level to avoid the information loss problem. Also, a key component of this architecture is the operation of connecting the first and second paths. This association allows the network to get very accurate information from the contraction path, producing a segmentation mask that is as close as possible to the desired output. This allows gradients to flow better and give information from multiple scales of the image size, which can help the model classify better. In addition, this architecture uses atrous/dilated convolutions, Atrous Spatial Pyramid Pooling (ASPP), and fully connected CRF, which allows the model to achieve better segmentation results. In general, FCN-8s and DeepLabV3+ showed a great performance in classifying weeds and crops compared to other approaches. DeepLabV3+ uses both the encoder-decoder and the spatial pyramid pooling modules thus arriving at better results.
In terms of the dataset, the study results confirmed that networks trained on the CWFID dataset produced better results than those trained on the sugarcane dataset. This is due to CWFID’s highest image resolution compared to the sugarcane image quality. Although the overall average performance improvement for all models using CWFID was about 6% compared to the sugarcane dataset, it is essential to note that when these models are used to detect weeds for large farms, where thousands of pictures can be taken of the field, this can have a significant impact on handling weeds. Even though we achieved promising results on fine-tuning these pre-trained models using small sample data, they should be trained and tested using more sample data to evaluate their performance further. In addition, more studies should be done to evaluate the effects of image spatial and spectral resolutions on training machine learning models for crop and weed detection.
Conclusion
To improve weed control and precision agriculture, it is essential to extract and map the location of the weeds and locally treat those areas. The similarities between the types of data addressed with classical deep learning applications and RS data, as well as the need for fast processing of huge datasets captured by UAS for smart and precision agriculture, make a compelling argument for integrating deep learning methods into UAS remote sensing. This research explored the application of a deep learning method to quickly process and transform UAS imagery into accurate maps for precision field treatment. The U-Net model, which was initially proposed to segment medical images, was fine-tuned to classify the CWFID and sugarcane orthomosiac UAS datasets into three classes (background, crops, and weeds) and compared its performance with SegNet, FCN-32s, FCN-16s, FCN-8s and DeepLabv3+ deep learning models to discriminate crop from weed. The fine-tuning approach allowed us to overcome the problem of small dataset sizes. The DeepLab v3+ model achieved the classification accuracy of 99.34% for background, 63.24% for crops, and 90.55% for weeds classes for the CWFID dataset. On the other hand, U-Net achieved an accuracy of 86.17% on accurately segmenting weeds compared to 85.8% of FCN-8s, 71.04% of FCN-16s, 80.16% of Deeplab v3+, 80.16% of FCN-32s, and 64.5% of SegNet. In future research, we will incorporate crop geometry constraints to the model to improve classification accuracy.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
LH, AG: developed the methodology, implementation, wrote the majority of the manuscript. AK, AS, FD: Provided input for scientific ideas and contributed to editing and writing draft manuscripts.
Funding
This material is based upon work supported by the National Science Foundation under Grant No. 1832110: Developing a Robust, Distributed, and Automated Sensing and Control System for Smart Agriculture and Grant No. 1800768: Remote Sensing for Flood Modeling and Management.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afza, F., Sharif, M., Mittal, M., Khan, M. A., and Hemanth, D. J. (2021). A Hierarchical Three-step Superpixels and Deep Learning Framework for Skin Lesion Classification. Methods. doi:10.1016/j.ymeth.2021.02.013
Alsherif, E. A. (2020). Cereal Weeds Variation in Middle Egypt: Role of Crop Family in weed Composition. Saudi J. Biol. Sci. 27 (9), 2245–2250. doi:10.1016/j.sjbs.2020.07.001
Badrinarayanan, V., Kendall, A., and Cipolla, R. (2017). Segnet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39 (12), 2481–2495. doi:10.1109/tpami.2016.2644615
Castaldi, F., Pelosi, F., Pascucci, S., and Casa, R. (2017). Assessing the Potential of Images from Unmanned Aerial Vehicles (UAV) to Support Herbicide Patch Spraying in maize. Precision Agric. 18 (1), 76–94. doi:10.1007/s11119-016-9468-3
Chen, H., Miao, F., and Shen, X. (2020). Hyperspectral Remote Sensing Image Classification with CNN Based on Quantum Genetic-Optimized Sparse Representation. IEEE Access 8, 99900–99909. doi:10.1109/access.2020.2997912
Chen, L. C., Zhu, Y., Papandreou, G., Schroff, F., and Adam, H. (2018). “Encoder-decoder with Atrous Separable Convolution for Semantic Image Segmentation,” in Proceedings of the European conference on computer vision (Munich, Germany: Springer), 801–818. doi:10.1007/978-3-030-01234-2_49
Coopersmith, E. J., Minsker, B. S., Wenzel, C. E., and Gilmore, B. J. (2014). Machine Learning Assessments of Soil Drying for Agricultural Planning. Comput. Electron. Agric. 104, 93–104. doi:10.1016/j.compag.2014.04.004
De Rainville, F.-M., Durand, A., Fortin, F.-A., Tanguy, K., Maldague, X., Panneton, B., et al. (2014). Bayesian Classification and Unsupervised Learning for Isolating Weeds in Row Crops. Pattern Anal. Applic 17 (2), 401–414. doi:10.1007/s10044-012-0307-5
Di Cicco, M., Potena, C., Grisetti, G., and Pretto, A. (2017). “Automatic Model Based Dataset Generation for Fast and Accurate Crop and Weeds Detection,” in 2017 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS) (IEEE), 5188–5195. doi:10.1109/iros.2017.8206408
Dorbu, F., Hashemi-Beni, L., Karimoddini, A., and Shahbazi, A. (2021). UAV Remote Sensing Assessment of Crop Growth. Photogrammetric Eng. Remote Sensing 87 (129), 891–899. doi:10.14358/pers.21-00060r2
Duckett, T., Pearson, S., Blackmore, S., Grieve, B., Chen, W. H., Cielniak, G., et al. (2018). Agricultural Robotics: the Future of Robotic Agriculture. arXiv preprint arXiv:1806.06762.
Dyrmann, M., and Christiansen, P. (2014). Automated Classification of Seedlings Using Computer Vision: Pattern Recognition of Seedlings Combining Features of Plants and Leaves for Improved Discrimination. Aarhus University.
Fao, W. F. P., and Ifad, (2012). The State of Food Insecurity in the World 2012. Economic Growth Is Necessary but Not Sufficient to Accelerate Reduction of Hunger and Malnutrition. Rome: FAO.
Fleiss, J. L., Cohen, J., and Everitt, B. S. (1969). Large Sample Standard Errors of Kappa and Weighted Kappa. Psychol. Bull. 72 (5), 323–327. doi:10.1037/h0028106
Gašparović, M., Zrinjski, M., Barković, Đ., and Radočaj, D. (2020). An Automatic Method for weed Mapping in Oat fields Based on UAV Imagery. Comput. Electronics Agric. 173, 105385.
Gebrehiwot, A., Hashemi-Beni, L., Thompson, G., Kordjamshidi, P., and Langan, T. (2019). Deep Convolutional Neural Network for Flood Extent Mapping Using Unmanned Aerial Vehicles Data. Sensors 19 (7), 1486. doi:10.3390/s19071486
Grace, R. K. (2021). Crop and Weed Classification Using Deep Learning. Turkish J. Computer Mathematics Education (Turcomat) 12 (7), 935–938.
Grinblat, G. L., Uzal, L. C., Larese, M. G., and Granitto, P. M. (2016). Deep Learning for Plant Identification Using Vein Morphological Patterns. Comput. Electronics Agric. 127, 418–424. doi:10.1016/j.compag.2016.07.003
Hashemi-Beni, L., Jones, J., Thompson, G., Johnson, C., and Gebrehiwot, A. (2018). Challenges and Opportunities for UAV-Based Digital Elevation Model Generation for Flood-Risk Management: A Case of princeville, north carolina. Sensors 18 (11), 3843. doi:10.3390/s18113843
Haug, S., and Ostermann, J. (2014). “September)A Crop/weed Field Image Dataset for the Evaluation of Computer Vision Based Precision Agriculture Tasks,” in European conference on computer vision (Cham: Springer), 105–116.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep Residual Learning for Image Recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 770–778. doi:10.1109/cvpr.2016.90
Horrigan, L., Lawrence, R. S., and Walker, P. (2002). How Sustainable Agriculture Can Address the Environmental and Human Health Harms of Industrial Agriculture. Environ. Health Perspect. 110 (5), 445–456. doi:10.1289/ehp.02110445
Islam, N., Rashid, M. M., Wibowo, S., Wasimi, S., Morshed, A., Xu, C., et al. (2020). “Machine Learning Based Approach for Weed Detection in Chilli Field Using RGB Images,” in The International Conference on Natural Computation, Fuzzy Systems and Knowledge Discovery (Cham: Springer), 1097–1105.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Imagenet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 25, 1097–1105.
Lee, S. H., Chan, C. S., Wilkin, P., and Remagnino, P. (2015). “September)Deep-plant: Plant Identification with Convolutional Neural Networks,” in 2015 IEEE international conference on image processing (ICIP) (IEEE), 452–456.
Loddo, A., Loddo, M., and Di Ruberto, C. (2021). A Novel Deep Learning Based Approach for Seed Image Classification and Retrieval. Comput. Electronics Agric. 187, 106269. doi:10.1016/j.compag.2021.106269
Long, J., Shelhamer, E., and Darrell, T. (2015). “Fully Convolutional Networks for Semantic Segmentation,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 3431–3440. doi:10.1109/cvpr.2015.7298965
Ma, H., Liu, Y., Ren, Y., Wang, D., Yu, L., and Yu, J. (2020). Improved CNN Classification Method for Groups of Buildings Damaged by Earthquake, Based on High Resolution Remote Sensing Images. Remote Sensing 12 (2), 260. doi:10.3390/rs12020260
Mehdizadeh, S., Behmanesh, J., and Khalili, K. (2017). Using MARS, SVM, GEP and Empirical Equations for Estimation of Monthly Mean Reference Evapotranspiration. Comput. Electron. Agric. 139, 103–114. doi:10.1016/j.compag.2017.05.002
Milberg, P., and Hallgren, E. (2004). Yield Loss Due to Weeds in Cereals and its Large-Scale Variability in Sweden. Field crops Res. 86 (2-3), 199–209. doi:10.1016/j.fcr.2003.08.006
Milioto, A., Lottes, P., and Stachniss, C. (2017). REAL-TIME BLOB-WISE SUGAR BEETS VS WEEDS CLASSIFICATION FOR MONITORING FIELDS USING CONVOLUTIONAL NEURAL NETWORKS. ISPRS Ann. Photogrammetry, Remote Sensing Spat. Inf. Sci. 4. doi:10.5194/isprs-annals-iv-2-w3-41-2017
Monteiro, A. A. O., and von Wangenheim, A. (2019). Orthomosaic Dataset of RGB Aerial Images for Weed Mapping. INCoD Datasets Repository. Available: http://www.lapix.ufsc.br/weed-mapping-sugar-cane.
Mortensen, A. K., Dyrmann, M., Karstoft, H., Jørgensen, R. N., and Gislum, R. (2016). “Semantic Segmentation of Mixed Crops Using Deep Convolutional Neural Network,” in CIGR-AgEng ConferenceAarhus, Denmark. Abstracts and Full papers, 26-29 June 2016 (Organising Committee, CIGR), 1–6.
Osorio, K., Puerto, A., Pedraza, C., Jamaica, D., and Rodríguez, L. (2020). A Deep Learning Approach for weed Detection in Lettuce Crops Using Multispectral Images. AgriEngineering 2 (3), 471–488. doi:10.3390/agriengineering2030032
Pahikkala, T., Kari, K., Mattila, H., Lepistö, A., Teuhola, J., Nevalainen, O. S., et al. (2015). Classification of Plant Species from Images of Overlapping Leaves. Comput. Electronics Agric. 118, 186–192. doi:10.1016/j.compag.2015.09.003
Pelosi, F., Castaldi, F., and Casa, R. (2015). Operational Unmanned Aerial Vehicle Assisted post-emergence Herbicide Patch Spraying in maize: a Field Study. Precision Agric. 15, 159–166. doi:10.3920/978-90-8686-814-8_19
Pingali, P. (2007). Westernization of Asian Diets and the Transformation of Food Systems: Implications for Research and Policy. Food policy 32 (3), 281–298. doi:10.1016/j.foodpol.2006.08.001
Potena, C., Nardi, D., and Pretto, A. (2016). “July). Fast and Accurate Crop and weed Identification with Summarized Train Sets for Precision Agriculture,” in International Conference on Intelligent Autonomous Systems (Cham: Springer), 105–121.
Radoglou-Grammatikis, P., Sarigiannidis, P., Lagkas, T., and Moscholios, I. (2020). A Compilation of UAV Applications for Precision Agriculture. Computer Networks 172, 107148. doi:10.1016/j.comnet.2020.107148
Raja, R., Nguyen, T. T., Slaughter, D. C., and Fennimore, S. A. (2020). Real-time weed-crop Classification and Localisation Technique for Robotic weed Control in Lettuce. Biosyst. Eng. 192, 257–274. doi:10.1016/j.biosystemseng.2020.02.002
Ramos, P. J., Prieto, F. A., Montoya, E. C., and Oliveros, C. E. (2017). Automatic Fruit Count on Coffee Branches Using Computer Vision. Comput. Electronics Agric. 137, 9–22. doi:10.1016/j.compag.2017.03.010
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-net: Convolutional Networks for Biomedical Image Segmentation,” in International Conference on Medical image computing and computer-assisted intervention (Cham: Springer), 234–241. doi:10.1007/978-3-319-24574-4_28
Siddiqui, S. A., Fatima, N., and Ahmad, A. (2021). “Neural Network based Smart Weed Detection System,” in 2021 International Conference on Communication, Control and Information Sciences (ICCISc), June, 2021 (IEEE) Vol. 1, 1–5.
Simonyan, K., and Zisserman, A. (2014). Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv preprint arXiv:1409.1556.
Vinh, K., Gebreyohannes, S., and Karimoddini, A. (2019). “March). An Area-Decomposition Based Approach for Cooperative Tasking and Coordination of Uavs in a Search and Coverage mission,” in 2019 IEEE Aerospace Conferenc (IEEE), 1–8.
Wu, Z., Chen, Y., Zhao, B., Kang, X., and Ding, Y. (2021). Review of Weed Detection Methods Based on Computer Vision. Sensors 21 (11), 3647. doi:10.3390/s21113647
Keywords: remote sensing, image segmentation, precision agriculture, automation, high resolution
Citation: Hashemi-Beni L, Gebrehiwot A, Karimoddini A, Shahbazi A and Dorbu F (2022) Deep Convolutional Neural Networks for Weeds and Crops Discrimination From UAS Imagery. Front. Remote Sens. 3:755939. doi: 10.3389/frsen.2022.755939
Received: 09 August 2021; Accepted: 17 January 2022;
Published: 11 February 2022.
Edited by:
Costas Armenakis, York University, CanadaReviewed by:
Eleni Mangina, University College Dublin, IrelandMarija Popovic, Imperial College London, United Kingdom
Copyright © 2022 Hashemi-Beni, Gebrehiwot, Karimoddini, Shahbazi and Dorbu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leila Hashemi-Beni, bGhhc2hlbWliZW5pQG5jYXQuZWR1
 Leila Hashemi-Beni
Leila Hashemi-Beni Asmamaw Gebrehiwot1
Asmamaw Gebrehiwot1 Freda Dorbu
Freda Dorbu