Department of Pharmacology and Neuroscience, University of Texas Southwestern Medical Center, Dallas, TX, USA
Since the emergence of the first living cells, survival has hinged on the ability to detect and localize chemicals in the environment. Modern animal species ranging from insects to mammals express large odorant receptor repertoires to detect the structurally diverse array of volatile molecules important for survival. Despite the essential nature of chemical detection, there is surprising diversity in the signaling mechanisms that different species use for odorant detection. In vertebrates, odorant receptors are classical G-protein coupled, seven transmembrane receptors that activate downstream effector enzymes that, in turn, produce second messengers that open ion channels. However, recent work reveals that insects have adopted different strategies to detect volatile chemicals. In Drosophila, the odorant receptors, predicted to have seven transmembrane domains, have reversed membrane topology compared to classical G-protein coupled receptors. Furthermore, insect odorant receptors appear to form odorant-gated ion channels. Pheromone detection in insects is even more unusual, utilizing soluble, extracellular receptors that undergo conformational activation. These alternate olfactory signaling strategies are discussed in terms of receptor design principles.
Olfaction, the detection and discrimination of air-borne chemicals, is probably the most important sense for the survival of most animal species. Detection and localization of food, avoidance of toxins and predators, and communication with cohorts and mating partners through volatile pheromones are examples of the range of olfactory-dependent behaviors. In contrast to the visual system, where a handful of receptor genes are sufficient to cover the relevant range of the electromagnetic spectrum, modern animals require large repertoires of receptors to detect the structurally diverse array of odorant molecules important for survival.
All animals detect chemical information with olfactory neurons exposed to the environment. Individual vertebrate olfactory neurons in the olfactory epithelium are tuned to a small fraction of ‘odor space’ (the total range of chemicals that can be detected). The restricted chemical tuning of individual olfactory neurons occurs because these neurons express a single allele of a single odorant receptor gene that is only activated by specific chemical features of odorant molecules (reviewed in Axel, 1995
). In mammals, several hundred receptor genes are present in the genome, and all the olfactory neurons expressing the same receptor gene converge to a single pair of glomeruli in the olfactory bulb (Vassar et al., 1994
; Sullivan et al., 1995
). Thus, activation of a single odorant receptor type corresponds to activation of specific glomeruli. Individual odorants activate subsets of receptors tuned to various facets of chemical structure. Therefore, the unique activity pattern produced among the thousands of glomeruli elicited by a particular odorant is relayed to higher processing centers by the second order mitral cells where a unique odor image is formed.
Odorant signal transduction in vertebrate primary olfactory neurons utilizes a cAMP second messenger mechanism (Figure 1
). Seven-transmembrane odorant receptors activate a Gs heterotrimeric G-protein homolog called Golf that in turn activates adenylyl cyclase III to produce cAMP (Jones and Reed, 1989
; Bakalyar and Reed, 1990
). cAMP, in turn, binds and opens cyclic nucleotide-gated cation channels (Dhallan et al., 1990
). These channels allow sodium and calcium to enter the dendrite, and the calcium influx triggers a second phase of depolarization mediated by calcium activated chloride channels (Lowe and Gold, 1993
). Indeed, these later channels pass the majority of the depolarizing current (Lowe and Gold, 1993
). This multistep signaling cascade is rather slow, requiring hundreds of milliseconds from odor interaction with receptors to full depolarization of the olfactory neurons, but offers the potential advantage that there are multiple steps that can be regulated to control the gain of the neuron.
Figure 1. Model of vertebrate olfactory signal transduction. In the absence of odorant (left), the odorant receptor (OR) is bound to the GDP-bound form of Golf. Activation by odorants (right) causes exchange of GDP for GTP by the alpha subunit of Golf, which activates adenylyl cyclase (AC) to produce cAMP. cAMP binds cyclic nucleotide-gated ion channels that conduct sodium and calcium ions into the neuron. The calcium ions bind calcium-activated chloride channels that allow chloride ions to exit the neurons, further depolarizing the neuron.
The odorant receptor family in insects proved difficult to identify, because there was virtually no sequence similarity with the vertebrate odorant receptor gene family. Insect odorant receptor genes were finally discovered in Drosophila. A bioinformatic screen of the Drosophila genome sequence identified genes predicted to encode seven transmembrane proteins that were expressed in subsets of antennal neurons (Clyne et al., 1999
; Gao and Chess, 1999
). Large scale sequencing of cDNAs produced from antenna RNA also hit upon this receptor family (Vosshall et al., 1999
). The insect odorant receptors, while predicted to encode seven transmembrane segments, were as similar to ion channels as they were to members of the vertebrate odorant receptor family. Anatomic studies confirmed that Drosophila olfactory neurons expressing the same odorant receptor converge to the same glomerulus in the antennal lobe; the equivalent of the vertebrate olfactory bulb (Vosshall et al., 2000
). Therefore, odorant-specific patterns of glomerular activity probably underlie odorant discrimination in both insect and vertebrates. Despite the conservation in odorant processing implied by the similarity in neuroanatomy and the presence of seven transmembrane receptors, the Drosophila olfactory signal transduction mechanisms turned out to be surprisingly unconventional.
Beginning with the odorant receptors, the first surprise was that these seven-transmembrane receptors are reversed in the membrane compared to all known G-protein-coupled receptors. Benton et al. (2006)
using LacZ fusions and split GFP constructs showed that the topology of the loops between transmembrane domains was reversed relative to the classical G-protein coupled receptor, rhodopsin. The same conclusion was reached by introducing glycosylation sites in different loops and determining which loops were exposed to the glycosylation machinery in the golgi apparatus (Lundin et al., 2007
). While classical G-protein coupled receptors have their C-termini inside the cell, the C-termini of the Drosophila odorant receptors appeared to be outside the cell! If these receptors are reversed, how do they trigger action potentials in the olfactory neurons? Do they activate effector enzymes that produce second messengers?
The second surprise was that the insect receptors are capable of forming odor-activated ion channels capable of depolarizing the olfactory neurons without needing a G-protein-activated second messenger system. Recent work indicates that insect odorant receptors form these odorant-gated ion channels as dimers between a ‘tuning’ receptor that binds odorants, and Or83b, an unusual member of the Or family. (Figure 2
) (Sato et al., 2008
; Smart et al., 2008
; Wicher et al., 2008
).
Figure 2. Two possible models for odorant-gated channels. Left, possible role for G-protein mediated cyclic nucleotides in Or83b activation. Right, direct odorant gating of the Or/Or83b receptor complex. The major monovalent cation in the sensillum lymph is potassium.
Or83b is unusual in several aspects. First, it is the only odorant receptor that is highly conserved among insect species (Jones et al., 2005
). Second, Or83b is expressed in most olfactory neurons. This is in stark contrast to the ‘tuning’ odorant receptors that are each expressed in small subsets of olfactory neurons that innervate a common glomerulus (Vosshall et al., 2000
; Couto et al., 2005
). One function of Or83b is to deliver tuning receptors to the olfactory neuron dendrites. In the absence of Or83b, the tuning receptors are trapped in the cell bodies of the olfactory neurons (Larsson et al., 2004
). In the absence of a tuning receptor, Or83b is still transported to the dendrites of olfactory neurons, but these neurons are unresponsive to odorants, revealing Or83b itself is not an odorant receptor (Dobritsa et al., 2003
; Elmore et al., 2003
; Neuhaus et al., 2005
). Is Or83b a simple chaperone, or does it have a more essential role in olfaction?
It turns out that Or83b is actually an ion channel that dimerizies with tuning receptors to form odorant-gated ion channels! Two groups independently showed that Or83b confers a novel cation conductance when expressed in heterologous tissue culture cells, and when co-expressed with a tuning odorant receptor, made this conductance odorant dependent (Sato et al., 2008
; Wicher et al., 2008
). Mutations in the pore-forming regions of Or83b modulated this conductance, directly implicating this protein in ion flux (Wicher et al., 2008
). These findings suggest insect odorant receptors form odorant-gated ion channels with Or83b and that odorants trigger the opening of the ion channels without requiring a second messenger system. Why do mammals use a G-protein mechanism and insects use a direct ion channel gating mechanism? One possibility is response time. Signaling through a second messenger requires activation of the G protein, activation of the effector enzyme and production and diffusion of a second messenger before the ion channels are opened. A direct gating mechanism bypasses these steps and theoretically should respond faster. This might be relevant to insects that are flying through odorant plumes in the air trying to localize odorant sources.
Is there no role for second messengers in insect olfaction? Controversy lingers. There are a number of reports in the literature suggesting second messenger pathways underlie olfactory transduction in Drosophila. Indeed, olfactory neurons may share components with the phototransduction cascade, a Gq-coupled signaling pathway, as several phototransduction mutants have olfactory defects (Hotta and Benzer, 1969
; Riesgo-Escovar et al., 1995
; Kain et al., 2008
). Furthermore, rapid production of cyclic nucleotides and phosphoinositide (PI) metabolites have been observed in response to odorants in insect olfactory neurons (Zufall and Hatt, 1991
). Together, these studies highlight the importance of PI and possibly cyclic nucleotide signaling for olfactory neuron function, but they do not implicate these second messengers as direct mediators of olfactory signal transduction. For example, these second messengers may underlie long-term homeostatic responses to neuronal activity. Perhaps there is a role for second messengers in insect olfaction by modulating the odorant-gated ion channels.
Work with the insect receptors expressed in heterologous cells showed there is a cytoplasmic rise in cyclic nucleotides that was dependent on expression of a tuning odorant receptor but not Or83b, while there was a cyclic nucleotide-gated conductance that was dependent on expression of Or83b. This suggests the possibility that tuning receptors can activate a cyclase to produce cyclic nucleotides, and that Or83b can be gated by the cyclic nucleotides (Wicher et al., 2008
). GDP-β-S, an inhibitor of G-protein activation, dramatically decreased the odor-activated current. This led to a transduction model in which low odorant concentrations trigger cyclic nucleotide production through the tuning receptor that subsequently gates the Or83b ion channel, while at higher odorant concentrations, the direct gating mechanism operates (Figure 2
). However, work from others showed insect Or/Or83b receptors expressed in heterologous cells loaded with calcium indicators were unaffected by application of inhibitors of G proteins (GDP-β–S), adenylyl cylcase (SQ22536), guanylyl cyclase (ODQ), phosphodiesterases (IBMX) or phospholipase Cβ (U73122) (Smart et al., 2008
). However, it should be noted that none of these studies examined the role of second messengers in insect primary olfactory neurons, and future studies will be required to confirm or exclude a direct role for second messengers in insect odorant detection and to elucidate how their formation is triggered if they are important. What is clear is that Or83b is required for dendritic localization of tuning receptors, and when dimerized with a tuning receptor, forms odorant-gated ion channels.
Recent findings hinted at other types of chemosensory receptors in olfactory organs in Drosophila. CO2 detection by a class of olfactory neuron occurs via two gustatory receptors that function without Or83b (Jones et al., 2007
; Kwon et al., 2007
). Expression mapping of the odorant receptor genes assigned receptors to specific olfactory neurons, allowing a detailed map of the chemosensory system to be established (Couto et al., 2005
; Yao et al., 2005
). However, with the exception of Or35a, none of the neurons located in the coeloconic sensilla expressed a member of the Or gene family. Indeed, Or83b, which is required as an obligate co-receptor for members of the Or family, is not expressed in 20% of the olfactory neurons. Most of the olfactory neurons that lack Or83b expression are located in the coeloconic sensilla, a class of small sensilla located on the antenna that normally respond to general odorants like alcohols, acids, but also to humidity (Yao et al., 2005
). What is the Or83b-independent signaling mechanism in these olfactory neurons?
Using a bioinformatics approach, a set of antenna-specific genes were found, including a family of genes encoding proteins that resembled ionotropic glutamate receptors (iGluR). A total of 61 genes and 2 pseudogenes were discovered. While rather distantly related to classical ionotropic glutamate receptors, there is strong conservation in the pore forming loops and M2 transmembrane domains when compared to the vertebrate iGluR members (Benton et al., 2009
). Fifteen of 60 iGluR mRNAs are expressed in the adult Drosophila antenna and are localized to the dendrites of olfactory neurons located in coeloconic sensilla. Or83b is not expressed in most of the iGluR-expressing neurons, with the exception of IR76b, which is co-expressed with Or35a and Or83b in one coeloconic ORN class. It is not clear if Or35a and the glutamate receptor IR76b operate independently to detect distinct ligands, or if they act in concert to sensitize the neurons to specific odors. However, for the other coeloconic neurons lacking Or83b, the expression of specific glutamate receptors correlated perfectly with the chemical sensitivity of the neurons. Importantly, mis-expression of individual glutamate receptors conferred the odorant sensitivity of the mis-expressed glutamate receptor to other neurons (Benton et al., 2009
). Finally, for at least one iGluR, neurons expressing that receptor project axons to the same glomerulus in the antennal lobe, confirming these neurons are functionally related. Together, these data provide strong evidence that some of these glutamate receptors have evolved to perform as odorant receptors. It will be interesting to determine where the other 45 members of the iGluR family are expressed, and if they also function as chemical detectors, and if any correspond to the humidity detector.
Pheromones are chemicals produced by one individual to influence the behavior of another individual of the same species and are common in animals ranging from C. elegans to mammals. Pheromones are odorants with extraordinary biological significance. In insects, pheromones trigger a number of hardwired behaviors, including mating. Pheromone detection is highly sensitive and exquisitely specific so that low levels of pheromone are detected, and random environmental odorants are not mistaken for pheromone cues. Not surprisingly, specialized machinery has evolved for pheromone detection in insects that is not shared with olfactory neurons that detect food odorants. Recent work indicates that pheromone detection can occur through a unique pathway utilizing secreted, extracellular receptors. Once completely unraveled, knowledge of pheromone signal transduction may lead to new ‘greener’ approaches to control insect pest populations in a species-specific manner.
There is extensive literature describing elegant work with moth sex pheromone detection, a system where single pheromone molecule sensitivity has been reported (Kaissling and Priesner, 1970
). Extracellular pheromone-binding proteins were first identified in male moth antenna as 14–16 kD extracellular proteins that bind directly to pheromones (Vogt and Riddiford, 1981
). However, it was not clear if pheromone-binding proteins were important for detection of pheromone or for removal of pheromone from the extracellular lymph bathing the dendrites of the pheromone-sensitive neurons.
Insight into pheromone signal transduction mechanisms came from a genetic dissection of volatile pheromone detection in Drosophila. The Drosophila pheromone, 11-cis vaccenyl acetate (cVA) is a male-specific pheromone that mediates aggregation and recognition of sex among fruit flies (reviewed in Dickson, 2008
; Vosshall, 2008
). A pheromone-binding protein, LUSH is secreted by non-neuronal support cells into the fluid bathing the pheromone sensitive neuron dendrites (Kim et al., 1998
). The importance of pheromone binding proteins was highlighted when it was shown that cVA detection is abolished in mutants lacking LUSH over all physiological levels of cVA (Xu et al., 2005
; Laughlin et al., 2008
). However, weak responses can still be elicited in lush mutant pheromone-sensitive neurons by intense, supra-physiological cVA doses (Laughlin et al., 2008
). These findings are consistent with models suggesting LUSH acts as a carrier or transporter that shuttles the hydrophobic pheromone through the aqueous sensillum lymph to the olfactory neuron dendrites (Wojtasek and Leal, 1999
; Horst et al., 2001
). However, LUSH has a more interesting role than a simple carrier. In mutants lacking LUSH there is a striking loss of spontaneous activity (i.e. the basal neuronal firing rate in the absence of pheromone) specifically in the cVA sensing neurons (Xu et al., 2005
). Wild type pheromone sensitive neurons have spontaneous firing rates of approximately 1 spike per second in the absence of pheromone (Clyne et al., 1997
; Xu et al., 2005
). However, lush mutants have spontaneous firing rates of only 1 spike every 400 s – a dramatic reduction in the normal spontaneous activity (Xu et al., 2005
). Why would a pheromone carrier alter the firing rate of a neuron in the absence of pheromone? The surprising answer is that an activated conformation of LUSH is the real ligand for pheromone receptors present on pheromone-sensitive neurons.
X-ray crystal structures of LUSH with and without cVA bound were solved by John Laughlin and David Jones at the University of Colorado Heath Sciences Center (Laughlin et al., 2008
). These structures revealed that LUSH undergoes a conformational shift upon binding cVA. Mutations in LUSH that enhanced or inhibited that conformational shift without altering cVA binding had large effects on the activity of LUSH, suggesting the conformational shift in LUSH is the true signal activating receptors on pheromone sensitive neurons (Laughlin et al., 2008
). This was confirmed when a particular LUSH mutant, LUSHD118A, was found to adopt the activated conformation in the absence of cVA and constitutively activate pheromone-sensitive neurons in the absence of cVA (Laughlin et al., 2008
). Thus, the actual cVA pheromone receptor appears to be an extracellular binding protein.
How is the conformational shift in LUSH transduced into activation of the pheromone-sensitive olfactory neurons? There must be a specific receptor complex expressed exclusively by the pheromone-sensitive neurons, because dominant LUSHD118A only activates pheromone-sensitive neurons, and not any other class of olfactory neuron (Laughlin et al., 2008
). Like detection of general odorants, cVA signaling requires Or83b (Jin et al., 2008
) and a specific odorant receptor, Or67d (Ha and Smith, 2006
; Kurtovic et al., 2007
). Loss of either of these factors results in low spontaneous activity in the pheromone sensitive neurons and loss of cVA sensitivity, as observed in lush mutants. Further, dominant LUSHD118A fails to activate pheromone sensitive neurons missing either of these components (Jin et al., 2008
; Laughlin et al., 2008
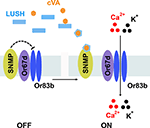
). However, there is at least one additional factor required for activation of pheromone sensitive neurons, SNMP.
SNMP was identified in moths as a dendritic protein expressed in a subset of pheromone-sensitive neurons (Rogers et al., 2001a
,b
). SNMP is a homolog of CD36, a protein family important for many biological processes, including cholesterol uptake by macrophages (reviewed in Vogt et al., 2009
). CD36 has also been implicated in the signal to convert macrophages into foam cells (Guest et al., 2007
; Thorne et al., 2007
), possibly through tyrosine kinase signaling (Rahaman et al., 2006
). Mice lacking CD36 are defective for uptake of free fatty acids by adipose tissue and muscle (Coburn et al., 2000
). Drosophila SNMP has the domain structure common to this family-a large extracellular domain flanked by two transmembrane domains with two short intracellular domains. SNMP is essential for pheromone signal transduction. When mutants lacking this gene product were analyzed they were insensitive to cVA at all concentrations, yet had normal responses to food odorants (Benton et al., 2007
; Jin et al., 2008
). Interestingly, unlike mutants lacking Or67d, Or83b or LUSH that have reduced spontaneous activity when absent, SNMP mutants have increased spontaneous activity. This suggests that SNMP may be an inhibitory subunit in the receptor complex (Benton et al., 2007
; Jin et al., 2008
). A working model is cVA-activated LUSH binds to SNMP, releasing Or67d/Or83b from SNMP inhibition, resulting in activation of the neurons (Figure 3
). However, there are likely to be addition factors required for pheromone signaling that remain unidentified that are not required for general odorants. Expression of Or67d, Or83b, SNMP together with LUSH in food-sensing olfactory sensilla fails to confer cVA sensitivity to these neurons (Laughlin et al., 2008
). Thus, there are likely additional components yet to be discovered in this pheromone signaling mechanism.
Figure 3. Model for pheromone detection. The extracellular receptor LUSH binds cVA pheromone and undergoes an activating conformational shift. Activated LUSH binds SNMP and relieves SNMP-mediated inhibition of the Or67d/Or83b receptor complex, allowing cations to enter the neurons.
What is the logic for using an extracellular binding protein in pheromone detection? We suggest this strategy has the potential to increase the sensitivity and specificity of the pheromone detection process. For example, if pheromone binding induces a stable, activated conformation in LUSH, this species could diffuse in the sensillum lymph until it interacts with a receptor complex on the dendrites and induces action potentials. This could, in theory, robustly increase pheromone detection to single molecule sensitivity. Utilizing an extracellular binding protein could also increase the specificity of pheromone detection. LUSH is able to bind to a wide variety of chemicals (Zhou et al., 2004
), but only cVA interacts with LUSH in just the right way to induce the activated conformation of the binding protein. Thus a bona fide pheromone must not only bind LUSH, but also induce the relevant conformational shift in the binding protein in order to activate the pheromone-sensitive neurons. This mechanism may prevent pheromone-like odorants from activating pheromone-sensitive neurons.
Olfactory neurons in vertebrates use second messenger signaling to amplify odorant-triggered signals, whereas insects appear to use odorant-gated ion channels for general odorants with a possible role for second messengers as well. Insect pheromone detection utilizes conformational activation of soluble pheromone receptors to confer sensitivity and specificity to pheromone perception. Recent studies indicate vertebrate pheromones may also be detected through binding proteins (Chamero et al., 2007
; Sherborne et al., 2007
). While extracellular binding proteins functioning as odorant receptors were only recently uncovered, we note that bacteria produce periplasmic receptors that work in a similar manner. Thus, bacteria appear to have discovered this elegant solution for detecting rare chemicals in the environment long ago.
In summary, the neuronal strategy for odorant discrimination appears to be conserved between vertebrates and insects, but the underlying signal transduction mechanisms are surprisingly different. From a design standpoint, the biochemistry of how specific odorant cues are transduced by an olfactory neuron is not as important as having specific receptors to detect essential compounds expressed in labeled lines and a neuronal network to integrate this information so the animal can respond appropriately. Olfactory neurons in both insects and vertebrates converge onto glomeruli where multiple primary olfactory neurons synapse onto a relatively small number of second-order neurons. Convergence converts the relatively noisy, stochastic signals from individual primary olfactory neurons into a high fidelity information transfer by summing simultaneous inputs (Bhandawat et al., 2007
). Individual odorants activate reproducible subsets of olfactory neurons expressing single tuning receptors, allowing the nervous system to deconstruct odorants into receptor-activating epitopes in both mammals and insects. How this information is processed into the sensation of ‘odor’ remains a mystery.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors apologize to our colleagues whose contributions could not be cited here due to limited space requirements. We thank Helmut Kramer and Robin Hiesinger for critical review of the manuscript.
Kain, P., Chakraborty, T. S., Sundaram, S., Siddiqi, O., Rodrigues, V., and Hasan, G. (2008). Reduced odor responses from antennal neurons of G(q)alpha, phospholipase Cbeta, and rdgA mutants in Drosophila support a role for a phospholipid intermediate in insect olfactory transduction. J. Neurosci. 28, 4745–4755.
Smart, R., Kiely, A., Beale, M., Vargas, E., Carraher, C., Kralicek, A. V., Christie, D. L., Chen, C., Newcomb, R. D., and Warr. C. G. (2008). Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem. Mol. Biol. 38, 770–780.