Abstract
Introduction:
Parkinson’s disease (PD) is a neurodegenerative disorder characterized by motor symptoms such as bradykinesia, rigidity, and tremor. Despite its prevalence, genetic and epigenetic studies in the Mexican Mestizo population (individuals of mixed Indigenous and European—primarily Spanish—ancestry who represent the majority demographic in Mexico) remain limited. DNA methylation may play a role in PD pathogenesis, with peripheral blood methylation patterns serving as potential biomarkers. This study examines MAPT and SNCA gene methylation in Mexican PD patients to identify epigenetic alterations associated with the disease.
Methods:
In this case-control study, we enrolled 108 PD patients and 108 age- and sex matched controls from Mexico City and Durango. Genomic DNA was extracted from leukocytes, and bisulfite pyrosequencing was performed to assess methylation levels at specific CpG sites within MAPT and SNCA.
Results:
Our analysis revealed a significant reduction in global methylation levels in the MAPT promoter region and SNCA intron 1 in PD patients compared to controls (MAPT, p = 0.0019; SNCA, p = 0.000069). Site-specific analysis showed significant hypomethylation at MAPT CpG sites 1, 4, 10–11 and at SNCA CpG sites 1–3, 5–7 and, 25 in PD cases. Regional analysis (central and northern México) revealed significant differences in MAPT methylation between PD patients and controls exclusively in the northern region (p = 0.0039) and in SNCA methylation only in the central region (p = 0.00001). Gender-based stratification indicated that MAPT methylation was significantly different in men (p = 0.0013), whereas SNCA methylation differences were significant only in women (p = 0.00045). We found an association between global methylation patterns of MAPT (OR = 1.182, 95% CI = 1.029–1.197, p = 0.007) and SNCA (OR = 1.243, 95% CI = 1.067–1.448, p = 0.005) with PD. Gender-stratified regression showed that MAPT methylation was significantly associated with PD exclusively in men (OR = 1.182, 95% CI = 1.041–1.342, p = 0.010), whereas SNCA methylation was significantly associated with PD only in women (OR = 1.337, 95% CI = 1.044–1.713, p = 0.021).
Conclusion:
Our findings reveal significant MAPT and SNCA hypomethylation in PD patients within the Mexican Mestizo population. These epigenetic modifications may contribute to PD pathogenesis and highlight the potential of DNA methylation profiles as biomarkers for PD, particularly in regional- and gender-specific contexts. This study advances the understanding of PD’s molecular mechanisms and underscores the importance of studying diverse populations to identify novel disease biomarkers.
1 Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder affecting approximately 1% of individuals over 60 years of age, characterized by tremor, bradykinesia, and rigidity, all of which significantly impact patients’ quality of life (Balestrino and Schapira, 2020). In Mexico, the prevalence of PD is estimated at 40 to 50 cases per 100,000 individuals annually, contributing to the global burden of an estimated 6.1 million affected individuals (Velázquez-Paniagua et al., 2016; Tysnes and Storstein, 2017; GBD 2016 Parkinson’s Disease Collaborators, 2018). Despite advancements in understanding PD’s motor symptoms, effective curative or neuroprotective treatments remain elusive. Genetic research has identified mutations and polymorphisms in several PD-associated genes, underscoring their role as susceptibility factors. However, the genetic and biochemical mechanisms underlying PD in the Mexican population remain largely unexplored (Salas-Leal et al., 2019; Romero-Gutierrez et al., 2021; Salas-Leal et al., 2021; Salas-Leal et al., 2023). The underrepresentation of Latin American populations in neurogenomic research has hindered the identification of ancestry-specific epigenetic signatures relevant to disease susceptibility. In this context, the Mexican Mestizo population (the predominant demographic group in Mexico, composed of individuals with mixed Indigenous and primarily Spanish European ancestry) provides a genetically distinct landscape shaped by historical admixture, offering an opportunity to uncover novel epigenetic mechanisms underlying PD pathogenesis (Martínez-Cortés et al., 2012; Silva-Zolezzi et al., 2009).
The MAPT and SNCA genes have been implicated in PD pathogenesis. MAPT encodes the microtubule-associated protein tau, which plays a key role in microtubule stabilization and axonal transport. Its misfolding and aggregation have been observed in PD, suggesting a multifaceted role in neuronal dysfunction (Zhang Y. et al., 2018; Zhang X. et al., 2018). In contrast, SNCA encodes α-synuclein, whose abnormal aggregation is a key event for Lewy body formation, a hallmark of PD pathology (Pascale et al., 2016; Goedert et al., 2017; Pedersen et al., 2021).
Epigenetic modifications, particularly DNA methylation changes, have been associated with various neurological disorders, including PD (Urdinguio et al., 2009; Klein and De Jager, 2016). Recent studies on DNA methylation patterns in brain and peripheral blood samples of PD patients have revealed concordant epigenetic landscapes, suggesting that peripheral blood may serve as a viable surrogate for brain tissue in PD epigenetic research (Masliah et al., 2013; Pihlstrom et al., 2015). These insights highlight the potential of epigenetic markers in PD diagnosis and pathogenesis, particularly through non-invasive approaches.
Given the global prevalence of PD and the genetic diversity of the Mexican population, our study aims to characterize the methylation profiles of MAPT and SNCA in leukocytes of Mexican PD patients. This research seeks to bridge the existing knowledge gap in PD’s genetic and epigenetic underpinnings within this demographic, offering new insights into disease mechanisms and potential biomarkers for PD.
2 Materials and methods
2.1 Subjects
This case-control study encompassed 108 Parkinson’s disease (PD) cases and 108 age- and sex-matched controls, devoid of any apparent neurological disorders. Participants were recruited from the Movement Disorders Unit at Dr. Manuel Gea González General Hospital, the Neurology Service of General Hospital 450, and Santiago Ramón y Cajal Hospital at ISSSTE in Durango.
Inclusion criteria for all participants included being between 50 and 85 years of age, self-reported Mexican Mestizo ancestry (defined as having at least three generations of ancestors born in Mexico), and willingness to provide written informed consent. In addition, PD patients were required to have a clinical diagnosis of idiopathic Parkinson’s disease according to the United Kingdom Parkinson’s Disease Society Brain Bank criteria (Gibb and Lees, 1988) and a minimum disease duration of 1 year, while healthy controls had to be free of neurological or psychiatric disorders and have no family history of PD. Exclusion criteria for both groups comprised the presence of atypical or secondary parkinsonism (e.g., drug-induced or vascular), other neurodegenerative or psychiatric disorders, active inflammatory or autoimmune diseases, recent infections, or use of immunosuppressive or anti-inflammatory medications within 3 months prior to sample collection.
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Research Ethics Committees of all participating institutions (ISSSTE Hospital General Dr. Santiago Ramon y Cajal: EeI/056/13; Hospital General Dr. Manuel Gea González: CI y CEI/111/15; Hospital General 450 Durango: 23/08/12). Written informed consent was obtained from all participants prior to enrollment. A visual summary of the study design and methodological workflow is provided in Figure 1.
Figure 1

Study design and methodological workflow.
2.2 Clinical assessment
PD cases underwent comprehensive evaluations using the Movement Disorder Society-modified Unified Parkinson’s Disease Rating Scale (UPDRS), Mini-Mental State Examination (MMSE), and Hamilton Rating Scale for Depression (HDRS) to assess motor function, cognitive status, and depression severity, respectively. Controls were assessed for cognitive function and depression using the MMSE and HDRS.
2.3 DNA isolation
To perform DNA extraction from blood samples, the protocol described by Iranpur and Esmailizadeh (2010) was employed. The qualitative assessment of DNA was performed on a 1% agarose gel stained with ethidium bromide. The quantitative assessment was performed using 1 μL of the sample through spectrophotometry at a wavelength of 260 and 280 nm on a Nanodrop 2000 spectrophotometer.
2.4 Methylation profile assessment
The analysis of methylation patterns was performed using the pyrosequencing method following the protocol described by Hogg et al. (2013). Specific probes from the EpiTect Methyl II PCR primer catalog were used for each region under evaluation. For MAPT CpGs 1–11: AGTAAGGAGAAAGGAAGTAGTT (forward), ACTAAATCCCCTACCCTTTACTTTCAAT (reverse), and CTACCCTTTACTTTCAATC (sequencing). For SNCA CpGs 1–7: AGAGAGGGTAGATAGATTTTATGTGAGA (forward), AATAAATACTTATCCCTTTAAAAAACCT (reverse), and AGAGAAAAGTTGGATGT (sequencing). For SNCA CpG25: GATGGTGGAAAGGAG TAGGTAATAGAA (forward), CCAACTTTTCTCTCACATAAAATCTATCTA (reverse), and GTGGAAAGGAGT AGGTAATAGAAT (sequencing). Bisulfite conversion was performed using the EZ DNA Methylation Gold Kit (Zymo Research) following the manufacturer’s protocol. Pyrosequencing was carried out using the Pyromark Q96 MD P instrument (Qiagen Ltd.).
2.5 Statistical analysis
Descriptive statistics were utilized to summarize the data. The student’s t-test (or Mann–Whitney U test for non-normally distributed data) was applied to compare quantitative variables between groups. The chi-square test (or Fisher’s exact test for small sample sizes) assessed differences in categorical variables. Normality of data distribution was verified using Kolmogorov–Smirnov and Shapiro–Wilk tests. Linear regression analysis explored the relationships between variables. All statistical analyses were performed using SPSS v.21.0 (IBM Corp.), and a two-tailed p-value <0.05 was considered statistically significant.
2.6 Ethical considerations
The study received approval from the Ethics Committees of all participating institutions. Informed consent was obtained from all participants, ensuring voluntary participation and confidentiality in line with ethical guidelines for human research.
3 Results
3.1 Study population characteristics
Our study cohort consisted of 216 participants, including 108 PD patients and 108 age- and sex-matched controls, recruited from both central (Mexico City) and northern (Durango) regions of Mexico. Detailed clinical and sociodemographic characteristics are summarized in Table 1. Comparative analysis revealed significant differences between PD patients and controls in body mass index (BMI; p = 0.009), serum uric acid levels (p = 0.010), depression severity assessed by the Hamilton scale (p = 0.034), leukocyte counts (p = 0.009), mean corpuscular hemoglobin (MCH; p = 0.002), and mean corpuscular hemoglobin concentration (MCHC; p < 0.001).
Table 1
| Variable | Cases PD (n = 108) | Controls (n = 108) | p |
|---|---|---|---|
| Age, years | 70.23 ± 9.16 | 69.97 ± 9.20 | 0.836a |
| Males n (%)/Females n (%) | 56 (51.8)/52 (48.2) | 54 (50)/54 (50) | 0.785c |
| North n (%)/Center n (%) | 69 (63.9)/39 (36.1) | 69 (63.9)/39 (36.1) | 1.000c |
| BMI, kg/m2 | 26.86 ± 6.07 | 23.33 ± 10.92 | 0.009a |
| Glucose, mg/dL | 97.1/ (80.0–117.9) | 93.2 (86.8–106.9) | 0.323b |
| Total cholesterol, mg/dL | 183.58 ± 51.75 | 193.88 ± 36.01 | 0.095a |
| Uric acid, mg/dL | 5.35 ± 2.30 | 6.03 ± 1.31 | 0.010a |
| Creatinine, mg/dL | 0.94 ± 0.55 | 1.04 ± 0.30 | 0.209a |
| MMSE, points | 25.12 ± 4.61 | 25.56 ± 3.21 | 0.475a |
| HDRS, points | 11.40 ± 7.42 | 9.26 ± 5.15 | 0.034a |
| Leukocytes, (×103/μL) | 5.9 (4.35–6.90) | 6.5 (5.4–8.2) | 0.009b |
| Erythrocytes, (×106/μL) | 4.76 (4.45–5.05) | 0.287b | |
| Hemoglobin, g/dL | 14.4 (13.35–15.65) | 13.9 (13.00–15.50) | 0.216b |
| HCT, % | 42.3 (39.20–47.20) | 43.3 (39.70–46.50) | 0.706b |
| MCV, fL | 89.9 (84.70–96.20) | 91.70 (88.00–94.10) | 0.305b |
| MCH, pg | 31.1 (29.55–32.65) | 29.6 (28.50–30.9) | 0.002b |
| MCHC, g/dL | 34.6 (33.10–35.80) | 31.9 (31.00–33.50) | <0.001b |
| Platelets, (×103/μL) | 203.0 (141.0–276.0) | 222.0 (181.0–260.0) | 0.315b |
Clinical and sociodemographic characteristics of the study groups.
HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration.
Student’s t.
Mann–Whitney U.
χ 2.
3.2 Methylation levels in MAPT and SNCA genes
Analysis of global methylation patterns within the MAPT gene promoter region (CpGs 1–11) and the SNCA gene intron 1 (CpGs 1–7 and CpG 25) revealed significant hypomethylation in the PD group compared to controls (MAPT, p = 0.0019; SNCA, p = 0.000069), as illustrated in Figure 2.
Figure 2

Differential global methylation levels in MAPT and SNCA genes between PD patients and controls. Box plots display the median (horizontal line inside the box) and the 25th and 75th percentiles (box borders). Outliers are denoted by closed circles. **p < 0.01, and ***p < 0.001.
Decreased methylation levels were observed across all examined CpG sites in the MAPT gene among PD patients, with significant differences at CpGs 1, 4, 10, and 11 (p = 0.04, p = 0.014, p = 0.018, and p = 0.004, respectively) (Figure 3A). Similarly, in the SNCA gene, all the specific CpG sites exhibited reduced methylation in PD patients, with significant differences at CpGs 1, 2, 3, 5, 6, 7, and 25 (p = 0.007, p = 0.04, p = 0.018, p = 0.016, p = 0.012, p = 0.003, and p = 0.0002 respectively) (Figure 3B).
Figure 3

Comparison of methylation patterns at each CpG site in the MAPT(A) and SNCA(B) genes between PD patients and controls. Box plots display the median (horizontal line inside the box) and the 25th and 75th percentiles (box borders). Outliers are denoted by closed circles. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.3 Regional stratification of methylation patterns
Regional analysis of participants from Mexico City (central) and Durango (northern) showed no significant differences in global methylation levels of the MAPT and SNCA genes between the two regions (Figure 4A). However, stratified comparisons revealed significant differences in MAPT methylation in the northern region (p = 0.0039) and SNCA methylation in the central region (p = 0.00001) between PD patients and controls (Figure 4B). In the central region, significant differences in SNCA methylation between PD patients and controls were observed at CpG sites 1, 2, 3, 5, and 7 (p = 0.0002, p = 0.0075, p = 0.0042, p = 0.0052, and p = 0.00013, respectively), in contrast, MAPT showed differential methylation only at CpG site 1 (p = 0.0306) (Figure 5A). In the northern region, MAPT methylation was significantly different at CpG sites 7, 9, 10, and 11 (p = 0.0184, p = 0.0168, p = 0.0423, and p = 0.00034, respectively), whereas SNCA exhibited differential methylation only at CpG site 25 (p = 0.000283) (Figure 5B).
Figure 4

Regional comparison of MAPT and SNCA gene methylation patterns. Overview of methylation differences in the SNCA and MAPT genes across central and north regions within the study population (A). Comparison of MAPT and SNCA gene methylation levels between PD patients and controls, stratified by region (B). Box plots display the median (horizontal line inside the box) and the 25th and 75th percentiles (box borders). Outliers are denoted by closed circles. *p < 0.05 and **p < 0.01.
Figure 5
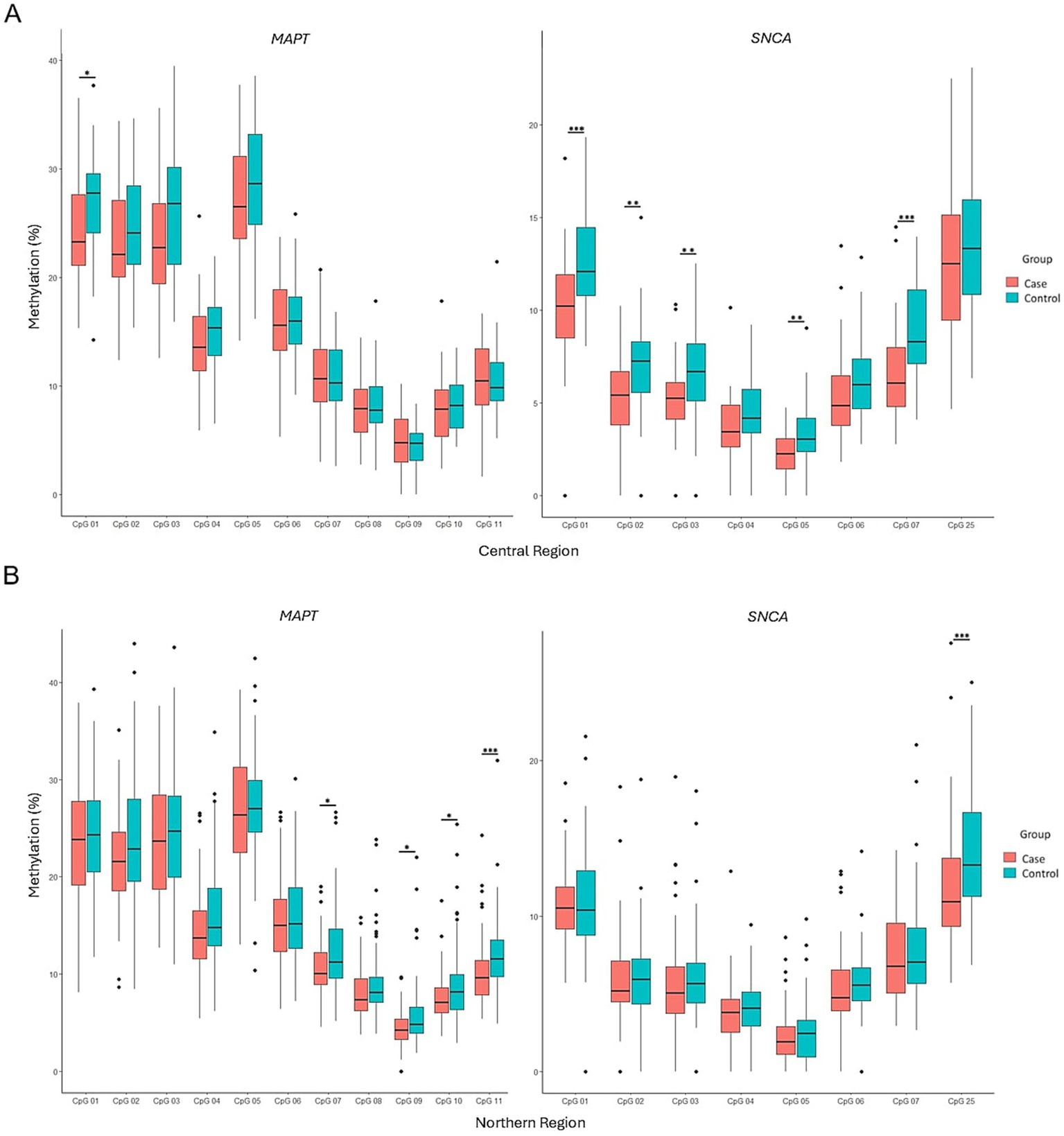
Comparison of methylation patterns at each CpG site in the MAPT and SNCA genes between PD patients and controls in the central (A) and northern (B) regions. Box plots display the median (horizontal line inside the box) and the 25th and 75th percentiles (box borders). Outliers are denoted by closed circles. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.4 Gender-based stratification of methylation patterns
The influence of gender on MAPT and SNCA gene methylation was analyzed. The results indicate that significant differences in MAPT methylation were observed only in men (p = 0.0013), whereas significant differences in SNCA methylation were exclusive to women (p = 0.00045). Hypomethylation of both MAPT and SNCA remained a prominent feature in PD patients (Figure 6). CpG site-specific comparisons showed that, within the male subgroup, significant differences between PD patients and controls were observed at CpG2, CpG4, CpG8, and CpG11 of the MAPT gene (p = 0.0223, p = 0.0185, p = 0.0346, and p = 0.00882, respectively) (Figure 7A). Conversely, in the female subgroup, significant differences were detected at CpG1, CpG2, CpG3, CpG6, CpG7, and CpG25 of the SNCA gene (Figure 7B).
Figure 6

Comparison of MAPT and SNCA genes methylation patterns stratified by gender. The box plots show the median (horizontal line inside the box) and the 25th and 75th percentiles (horizontal borders of the box). Outliers are denoted by closed circles. **p < 0.01 and ***p < 0.001.
Figure 7

Comparison of methylation patterns at each CpG site in the MAPT and SNCA genes between PD patients and controls in females (A) and males (B). Box plots display the median (horizontal line inside the box) and the 25th and 75th percentiles (box borders). Outliers are denoted by closed circles. *p < 0.05, **p < 0.01, and ***p < 0.001.
3.5 Association of methylation profile with Parkinson’s disease
Binary logistic regression analyses demonstrate an association between global methylation patterns of the MAPT (OR = 1.182, 95% CI = 1.029–1.197, p = 0.007) and SNCA (OR = 1.243, 95% CI = 1.067–1.448, p = 0.005) genes and PD. Gender-stratified regression further revealed that MAPT methylation was significantly associated with PD exclusively in men (OR = 1.182, 95% CI = 1.041–1.342, p = 0.010). In contrast, SNCA methylation was significantly associated with PD only in women (OR = 1.337, 95% CI = 1.044–1.713, p = 0.021).
4 Discussion
Our study contributes to the growing body of evidence supporting the role of epigenetic mechanisms, particularly DNA methylation, in the pathogenesis of PD. By focusing on the Mexican Mestizo population, which remains largely underrepresented in epigenetic and genomic studies, we address a significant gap in current research that has been predominantly centered on European and Asian cohorts. Recent multi-ancestry genome-wide association studies have emphasized the need for ethnic diversity in PD epigenetic research, as the genetic architecture and epigenomic regulation of risk loci like MAPT and SNCA differ markedly across populations (Kim et al., 2024; Park et al., 2023; Zhang Y. et al., 2018; Zhang X. et al., 2018).
The significant differences observed in BMI, uric acid levels, Hamilton scale scores, leukocyte counts, MCH, and MCHC between PD patients and controls underscore the multifactorial nature of the disease. Our findings on uric acid levels align with global research, including the meta-analysis by Wen et al. (2017), which reinforces the antioxidant and neuroprotective properties of uric acid, as well as its role in modulating iron accumulation—a well-established contributor to dopaminergic neuron degeneration (Waring, 2002; Berg and Hochstrasser, 2006; Schlesinger and Schlesinger, 2008; Sian-Hulsmann et al., 2011; Dusek et al., 2016; Wang et al., 2017; Liu et al., 2019).
The association between PD and alterations in hematological parameters, including leukocyte counts and hemoglobin content, may reflect underlying inflammatory processes and oxidative stress that contribute to neuronal loss (Lin et al., 2014; Freed and Chakrabarti, 2016). These observations suggest that systemic changes in PD extend beyond the central nervous system, offering potential biomarkers for disease detection and monitoring.
Our analysis of MAPT and SNCA methylation revealed a distinct pattern of hypomethylation in PD patients, consistent with previous studies linking epigenetic modifications to disease pathogenesis (Pihlstrom et al., 2015; Schmitt et al., 2015). Masliah et al. (2013) provided direct evidence that concordant DNA methylation changes can be observed in both brain and blood samples from PD patients, reinforcing the utility of peripheral tissues for biomarker discovery and mechanistic inference.
The observed hypomethylation across MAPT CpGs sites, particularly CpGs 1, 4, 10, and 11, adds new insight to previously mixed findings, underscoring the complex interplay between tau protein dysregulation and PD. While Coupland et al. (2014) reported differential methylation of MAPT across brain regions and peripheral blood, they did not assess the specific CpG sites examined in our study. Furthermore, Masliah et al. (2013) demonstrated concordant methylation changes between brain and blood in PD, but did not investigate the MAPT locus. Thus, our findings provide, to our knowledge, the first site-specific evidence of MAPT hypomethylation in peripheral blood of PD patients. These discrepancies support the notion of tissue- and ancestry-specific dynamics in MAPT epigenetic regulation. Notably, these epigenetic changes were identified in the Mexican Mestizo population, highlighting the possibility that epigenetic marks may vary in frequency or regulatory function across diverse genetic backgrounds (Romero-Gutierrez et al., 2021).
Similarly, the hypomethylation at SNCA CpGs sites 1, 2, 3, 5, 6, and 7, including CpG25, a previously unexplored site, suggests a nuanced regulatory mechanism affecting α-synuclein levels. These CpG sites are located within intron 1 and, according to the GRCh38/hg38 annotation, are predicted to reside within a transcriptional control region. Notably, CpG25 exhibited the most pronounced difference between PD patients and controls, indicating its potential role in SNCA gene regulation. Our findings are consistent with previous reports of intron 1 hypomethylation in peripheral blood mononuclear cells from PD patients (Guhathakurta et al., 2017), as well as in cerebellum, putamen, and frontal cortex from post-mortem brain tissue (Jowaed et al., 2010). More recently, Gu et al. (2021) demonstrated that this hypomethylation is specific to neuronal cells. These epigenetic alterations align with functional studies indicating that reduced methylation at intron 1 increases SNCA expression, and have been associated with earlier onset and more rapid progression of PD (Fedotova et al., 2023; Jowaed et al., 2010).
Geographic stratification revealed that MAPT hypomethylation was more pronounced among patients from northern Mexico, whereas SNCA hypomethylation was more significant in those from central Mexico, indicating potential regional influences on epigenetic regulation. These differences may be driven by environmental or lifestyle factors (such as exposure to pesticides or air pollutants, both recognized contributors to PD) that could affect DNA methylation patterns in a region-specific manner (Dorsey and Bloem, 2024; Nandipati and Litvan, 2016). Supporting this hypothesis, a study using a mouse model chronically exposed to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine), a prototypical neurotoxin used to model PD, reported altered methylation at intron 1 of SNCA in brain tissue, providing mechanistic evidence for toxin-induced epigenetic modulation of this gene (Schaffner and Kobor, 2022).
Additionally, our findings indicate a gender-specific pattern of hypomethylation: MAPT was significantly hypomethylated in men, while SNCA hypomethylation was predominant in women. This may contribute to sex-based differences in PD susceptibility and progression. In men, MAPT hypomethylation likely enhances its expression, which may disrupt cytoskeletal integrity and accelerate neurodegeneration. Overexpression of MAPT and other PD-related genes has been associated with increased cellular energy demands, particularly in mitochondrial function and calcium homeostasis (Simunovic et al., 2010). This supports the hypothesis that elevated metabolic activity in dopaminergic neurons may accelerate neuronal aging and increase PD risk (Flores-Ponce and Velasco, 2024). Hypomethylation of intron 1 in SNCA has been linked to increased gene expression in post-mortem brain tissue from patients with sporadic PD (Ai et al., 2014), potentially leading to alpha-synuclein accumulation and heightened neuronal vulnerability in women. In SNCA-knockout mice, reduced bone loss following ovariectomy suggests that this protein may interact with hormonal processes (Lisofsky et al., 2015). Additionally, women with PD are reported to experience tremors more frequently and exhibit more rapid nigrostriatal degeneration than men (Haaxma et al., 2007), which may be linked to differential SNCA expression in dopaminergic neurons of the substantia nigra compacta and potentially modulated by estrogen activity.
Several limitations should be considered when interpreting our findings. This study focused exclusively on two candidate genes (MAPT and SNCA), limiting the ability to capture broader epigenomic alterations associated with PD. Although significant methylation differences were observed, the lack of functional validation (such as gene expression analysis or mechanistic assays) precludes conclusions about their biological relevance. The cross-sectional design further limits causal inference, as methylation changes may reflect disease progression or treatment effects rather than etiological mechanisms. Moreover, analyses were conducted on peripheral blood leukocytes without adjustment for potential cellular heterogeneity, and other confounding factors such as age, medication use, or environmental exposures were not accounted for. The absence of correction for multiple testing across CpG sites increases the risk of false positives. Lastly, findings were derived from a single, region-specific cohort without replication in an independent sample, which may limit the generalizability of the results to other populations.
5 Conclusion
This study reveals significant hypomethylation in the MAPT and SNCA genes in peripheral blood leukocytes of Mexican Mestizo patients with Parkinson’s disease. These epigenetic alterations are associated with the disease in a region- and sex-specific manner, highlighting the potential utility of DNA methylation profiles as accessible, non-invasive biomarkers for PD.
Our findings support the growing recognition that epigenetic mechanisms contribute to neurodegeneration and underscore the relevance of population-specific studies. The observed hypomethylation patterns, particularly in MAPT among men and SNCA among women, offer novel insights into the sex-dependent epigenetic regulation of PD risk.
Future investigations are warranted to validate the functional consequences of the observed methylation changes through gene expression and protein assays, thereby confirming their biological relevance. Longitudinal studies are essential to elucidate how these epigenetic patterns evolve throughout disease progression. Moreover, expanding analyses to brain tissue or neural cell models could clarify their relevance to central neuropathology. Finally, genome-wide approaches and replication in diverse populations will be critical to identify broader and more generalizable epigenetic signatures.
Statements
Data availability statement
The data supporting the findings of this study have been deposited in Figshare and are publicly available at the following DOI: https://doi.org/10.6084/m9.figshare.29441972.
Ethics statement
The studies involving humans were approved by Ethics Committees of the General Hospital Dr. Manuel Gea González, General Hospital 450, and Santiago Ramón y Cajal Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
EM-M: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. DR-R: Writing – original draft, Software, Validation, Formal analysis. AS-L: Investigation, Writing – review & editing, Methodology. SS-P: Validation, Supervision, Writing – review & editing, Software. FC-J: Methodology, Investigation, Supervision, Writing – review & editing. EM-H: Writing – review & editing, Investigation, Conceptualization, Supervision. OL-L: Supervision, Formal analysis, Writing – review & editing, Methodology. MP: Data curation, Methodology, Writing – review & editing, Supervision. GQ-C: Validation, Writing – review & editing, Supervision, Methodology, Resources. OA-C: Resources, Funding acquisition, Conceptualization, Supervision, Writing – review & editing. AS-C: Project administration, Funding acquisition, Investigation, Writing – review & editing, Resources, Supervision. JS-P: Conceptualization, Supervision, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Council for Science and Technology (CONACYT) under the grants FOSISS 2014:233092 and CB 2015:253857 and by the Science and Technology Council of the State of Durango (COCYTED) under the grant F-101.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. During the preparation of this work the authors used ChatGPT in order to improve language and readability. After using this tool, the authors reviewed and edited the content as needed and takes full responsibility for the content of the publication.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Ai S. X. Xu Q. Hu Y. C. Song C. Y. Guo J. F. Shen L. et al . (2014). Hypomethylation of SNCA in blood of patients with sporadic Parkinson’s disease. J. Neurol. Sci.337, 123–128. doi: 10.1016/j.jns.2013.11.033
2
Balestrino R. Schapira A. H. V. (2020). Parkinson disease. Eur. J. Neurol.27, 27–42. doi: 10.1111/ene.14108
3
Berg D. Hochstrasser H. (2006). Iron metabolism in parkinsonian syndromes. Mov. Disord.21, 1299–1310. doi: 10.1002/mds.21020
4
Coupland K. G. Mellick G. D. Silburn P. A. Mather K. Armstrong N. J. Sachdev P. S. et al . (2014). DNA methylation of the MAPT gene in Parkinson’s disease cohorts and modulation by vitamin E in vitro. Mov. Disord.29, 1606–1614. doi: 10.1002/mds.25784
5
Dorsey E. R. Bloem B. R. (2024). Parkinson’s disease is predominantly an environmental disease. J. Parkinsons Dis.14, 451–465. doi: 10.3233/JPD-230357
6
Dusek P. Schneider S. A. Aaseth J. (2016). Iron chelation in the treatment of neurodegenerative diseases. J. Trace Elem. Med. Biol.38, 81–92. doi: 10.1016/j.jtemb.2016.03.010
7
Fedotova E. Y. Iakovenko E. V. Abramycheva N. Y. Illarioshkin S. N. (2023). SNCA gene methylation in Parkinson’s disease and multiple system atrophy. Epigenomes7:5. doi: 10.3390/epigenomes7010005
8
Flores-Ponce X. Velasco I. (2024). Dopaminergic neuron metabolism: relevance for understanding Parkinson’s disease. Metabolomics20:116. doi: 10.1007/s11306-024-02181-4
9
Freed J. Chakrabarti L. (2016). Defining a role for hemoglobin in Parkinson’s disease. npj Parkinsons Dis.2:16021. doi: 10.1038/npjparkd.2016.21
10
GBD 2016 Parkinson’s Disease Collaborators (2018). Global, regional, and national burden of Parkinson’s disease, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol.17, 939–953. doi: 10.1016/S1474-4422(18)30295-3
11
Gibb W. R. Lees A. J. (1988). The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry51, 745–752. doi: 10.1136/jnnp.51.6.745
12
Goedert M. Jakes R. Spillantini M. G. (2017). The synucleinopathies: twenty years on. J. Parkinsons Dis.7, S51–S69. doi: 10.3233/JPD-179005
13
Gu J. Barrera J. Yun Y. Murphy S. K. Beach T. G. Woltjer R. L. et al . (2021). Cell-type specific changes in DNA methylation of SNCA intron 1 in synucleinopathy brains. Front. Neurosci.15:652226. doi: 10.3389/fnins.2021.652226
14
Guhathakurta S. Evangelista B. A. Ghosh S. Basu S. Kim Y. S. (2017). Hypomethylation of intron1 of α-synuclein gene does not correlate with Parkinson’s disease. Mol. Brain10:6. doi: 10.1186/s13041-017-0285-z
15
Haaxma C. A. Bloem B. R. Borm G. F. Oyen W. J. Leenders K. L. Eshuis S. et al . (2007). Gender differences in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry78, 819–824. doi: 10.1136/jnnp.2006.103788
16
Hogg K. Blair J. D. McFadden D. E. von Dadelszen P. Robinson W. P. (2013). Early onset pre-eclampsia is associated with altered DNA methylation of cortisol-signalling and steroidogenic genes in the placenta. PLoS One8:e62969. doi: 10.1371/journal.pone.0062969
17
Iranpur V. M. Esmailizadeh A. K. . (2010). Rapid extraction of high quality DNA from whole blood stored at 4°C for long period. Available online at: https://www.protocol-online.org/prot/Protocols/Rapid-Extraction-of-High-Quality-DNA-from-Whole-Blood-Stored-at-4-C-for-Long-Period-4175.html (Accessed June 11, 2019).
18
Jowaed A. Schmitt I. Kaut O. Wüllner U. (2010). Methylation regulates alpha-synuclein expression and is decreased in Parkinson's disease patients’ brains. J. Neurosci.30, 6355–6359. doi: 10.1523/JNEUROSCI.6119-09.2010
19
Kim J. J. Vitale D. Otani D. V. Lian M. M. Heilbron K. Iwaki H. et al . (2024). Multi-ancestry genome-wide association meta-analysis of Parkinson’s disease. Nat. Genet.56, 27–36. doi: 10.1038/s41588-023-01584-8
20
Klein H. U. De Jager P. L. (2016). Uncovering the role of the methylome in dementia and neurodegeneration. Trends Mol. Med.22, 687–700. doi: 10.1016/j.molmed.2016.06.008
21
Lin W. C. Tsai N. W. Huang Y. C. Cheng K. Y. Chen H. L. Li S. H. et al . (2014). Peripheral leukocyte apoptosis in patients with parkinsonism: correlation with clinical characteristics and neuroimaging findings. Biomed. Res. Int.2014:635923. doi: 10.1155/2014/635923
22
Lisofsky N. Martensson J. Eckert A. Lindenberger U. Gallinat J. Kuhn S. (2015). Hippocampal volume and functional connectivity changes during the female menstrual cycle. NeuroImage118, 154–162. doi: 10.1016/j.neuroimage.2015.06.012
23
Liu C. Liang M. C. Soong T. W. (2019). Nitric oxide, iron and neurodegeneration. Front. Neurosci.13:114. doi: 10.3389/fnins.2019.00114
24
Martínez-Cortés G. Salazar-Flores J. Fernández-Rodríguez L. G. Rubi-Castellanos R. Rodríguez-Loya C. Velarde-Félix J. S. et al . (2012). Admixture and population structure in Mexican-Mestizos based on paternal lineages. J. Hum. Genet.57, 568–574. doi: 10.1038/jhg.2012.67
25
Masliah E. Dumaop W. Galasko D. Desplats P. (2013). Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics8, 1030–1038. doi: 10.4161/epi.25865
26
Nandipati S. Litvan I. (2016). Environmental exposures and Parkinson’s disease. Int. J. Environ. Res. Public Health13:881. doi: 10.3390/ijerph13090881
27
Park K. W. Ryu H. S. Shin E. Park Y. Jeon S. R. Kim S. Y. et al . (2023). Ethnicity- and sex-specific genome wide association study on Parkinson’s disease. npj Parkinsons Dis9:141. doi: 10.1038/s41531-023-00580-3
28
Pascale E. Di Battista M. E. Rubino A. Purcaro C. Valente M. Fattapposta F. et al . (2016). Genetic architecture of MAPT gene region in Parkinson disease subtypes. Front. Cell. Neurosci.10:96. doi: 10.3389/fncel.2016.00096
29
Pedersen C. C. Lange J. Forland M. G. G. Macleod A. D. Alves G. Maple-Grodem J. (2021). A systematic review of associations between common SNCA variants and clinical heterogeneity in Parkinson’s disease. npj Parkinsons Dis.7:54. doi: 10.1038/s41531-021-00196-5
30
Pihlstrom L. Berge V. Rengmark A. Toft M. (2015). Parkinson’s disease correlates with promoter methylation in the alpha-synuclein gene. Mov. Disord.30, 577–580. doi: 10.1002/mds.26073
31
Romero-Gutierrez E. Vazquez-Cardenas P. Moreno-Macias H. Salas-Pacheco J. Tusie-Luna T. Arias-Carrion O. (2021). Differences in MTHFR and LRRK2 variant’s association with sporadic Parkinson’s disease in Mexican Mestizos correlated to native American ancestry. npj Parkinsons Dis.7:13. doi: 10.1038/s41531-021-00157-y
32
Salas-Leal A. C. Salas-Pacheco S. M. Gavilan-Ceniceros J. A. P. Castellanos-Juarez F. X. Mendez-Hernandez E. M. La Llave-Leon O. et al . (2021). Alpha-syn and SNP rs356219 as a potential biomarker in blood for Parkinson’s disease in Mexican Mestizos. Neurosci. Lett.754:135901. doi: 10.1016/j.neulet.2021.135901
33
Salas-Leal A. C. Salas-Pacheco S. M. Hernandez-Cosain E. I. Velez-Velez L. M. Antuna-Salcido E. I. Castellanos-Juarez F. X. et al . (2023). Differential expression of PSMC4, SKP1, and HSPA8 in Parkinson's disease: insights from a Mexican Mestizo population. Front. Mol. Neurosci.16:1298560. doi: 10.3389/fnmol.2023.1298560
34
Salas-Leal A. C. Sandoval-Carrillo A. Romero-Gutierrez E. Castellanos-Juarez F. X. Mendez-Hernandez E. M. La Llave-Leon O. et al . (2019). rs3764435 associated with Parkinson’s disease in Mexican Mestizos: case-control study reveals protective effects against disease development and cognitive impairment. Front. Neurol.10:1066. doi: 10.3389/fneur.2019.01066
35
Schaffner S. L. Kobor M. S. (2022). DNA methylation as a mediator of genetic and environmental influences on Parkinson’s disease susceptibility: impacts of alpha-synuclein, physical activity, and pesticide exposure on the epigenome. Front. Genet.13:971298. doi: 10.3389/fgene.2022.971298
36
Schlesinger I. Schlesinger N. (2008). Uric acid in Parkinson’s disease. Mov. Disord.23, 1653–1657. doi: 10.1002/mds.22139
37
Schmitt I. Kaut O. Khazneh H. deBoni L. Ahmad A. Berg D. et al . (2015). L-dopa increases alpha-synuclein DNA methylation in Parkinson’s disease patients in vivo and in vitro. Mov. Disord.30, 1794–1801. doi: 10.1002/mds.26319
38
Sian-Hulsmann J. Mandel S. Youdim M. B. Riederer P. (2011). The relevance of iron in the pathogenesis of Parkinson’s disease. J. Neurochem.118, 939–957. doi: 10.1111/j.1471-4159.2010.07132.x
39
Silva-Zolezzi I. Hidalgo-Miranda A. Estrada-Gil J. Fernández-López J. C. Uribe-Figueroa L. Contreras A. et al . (2009). Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc. Natl. Acad. Sci. U.S.A.106, 8611–8616. doi: 10.1073/pnas.0903045106
40
Simunovic F. Yi M. Wang Y. Stephens R. Sonntag K. C. (2010). Evidence for gender-specific transcriptional profiles of nigral dopamine neurons in Parkinson disease. PLoS One5:e8856. doi: 10.1371/journal.pone.0008856
41
Tysnes O. B. Storstein A. (2017). Epidemiology of Parkinson’s disease. J. Neural Transm.124, 901–905. doi: 10.1007/s00702-017-1686-y
42
Urdinguio R. G. Sanchez-Mut J. V. Esteller M. (2009). Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol.8, 1056–1072. doi: 10.1016/S1474-4422(09)70262-5
43
Velázquez-Paniagua M. Vázquez-Álvarez A. M. Valverde-Aguilar G. Vergara-Aragón P. (2016). Current treatments in Parkinson's including the proposal of an innovative dopamine microimplant. Rev. Méd. Hosp. Gen. México79, 79–87. doi: 10.1016/j.hgmx.2015.10.006
44
Wang N. Jin X. Guo D. Tong G. Zhu X. (2017). Iron chelation nanoparticles with delayed saturation as an effective therapy for Parkinson disease. Biomacromolecules18, 461–474. doi: 10.1021/acs.biomac.6b01547
45
Waring W. S. (2002). Uric acid: an important antioxidant in acute ischaemic stroke. QJM95, 691–693. doi: 10.1093/qjmed/95.10.691
46
Wen M. Zhou B. Chen Y. H. Ma Z. L. Gou Y. Zhang C. L. et al . (2017). Serum uric acid levels in patients with Parkinson’s disease: a meta-analysis. PLoS One12:e0173731. doi: 10.1371/journal.pone.0173731
47
Zhang X. Gao F. Wang D. Li C. Fu Y. He W. et al . (2018). Tau pathology in Parkinson’s disease. Front. Neurol.9:809. doi: 10.3389/fneur.2018.00809
48
Zhang Y. Shu L. Sun Q. Pan H. Guo J. Tang B. (2018). A comprehensive analysis of the association between SNCA polymorphisms and the risk of Parkinson’s disease. Front. Mol. Neurosci.11:391. doi: 10.3389/fnmol.2018.00391
Summary
Keywords
Parkinson’s disease, DNA methylation, SNCA , MAPT , Mexican Mestizos
Citation
Miranda-Morales EG, Ramos-Rosales DF, Salas-Leal AC, Salas-Pacheco SM, Castellanos-Juárez FX, Méndez-Hernández EM, La Llave-León O, Peñaherrera MS, Quiñones-Canales G, Arias-Carrión O, Sandoval-Carrillo AA and Salas-Pacheco JM (2025) Differential methylation of SNCA and MAPT genes associated with Parkinson’s disease in Mexican Mestizos. Front. Aging Neurosci. 17:1612544. doi: 10.3389/fnagi.2025.1612544
Received
15 April 2025
Accepted
24 June 2025
Published
09 July 2025
Volume
17 - 2025
Edited by
Andrea Fuso, Sapienza University of Rome, Italy
Reviewed by
Dalapathi Gugulothu, Delhi Pharmaceutical Sciences and Research University, India
Khairiah Razali, International Islamic University Malaysia, Malaysia
Abeer Eissa, Ain Shams University, Egypt
Updates
Copyright
© 2025 Miranda-Morales, Ramos-Rosales, Salas-Leal, Salas-Pacheco, Castellanos-Juárez, Méndez-Hernández, La Llave-León, Peñaherrera, Quiñones-Canales, Arias-Carrión, Sandoval-Carrillo and Salas-Pacheco.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ada A. Sandoval-Carrillo, adda-sandoval@hotmail.com; José M. Salas-Pacheco, jsalas_pacheco@hotmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.