- 1Department of Biology, University of Central Florida, Orlando, FL, United States
- 2Southeastern Cooperative Wildlife Disease Study, Department of Population Health, College of Veterinary Medicine, University of Georgia, Athens, GA, United States
- 3Department of Biology, Stetson University, DeLand, FL, United States
- 4Central Florida Zoo & Botanical Gardens’ Orianne Center for Indigo Conservation, Eustis, FL, United States
- 5Deparment of Pathology, College of Veterinary Medicine, University of Georgia, Athens, GA, United States
Raillietiella orientalis, an invasive crustacean pentastome parasite, threatens native snake populations in the southeastern United States, infecting at least 18 species across Florida. Pentastome parasites have complex life cycles, with snakes often serving as definitive hosts for adult parasites that attach to the lungs and shed eggs into the host feces. PCR assays exist that distinguish invertebrate species via amplification and sequencing of mitochondrial DNA fragments. However, no molecular assays specific for R. orientalis or optimized fecal flotation methods for pentastome egg detection are available. We developed a novel PCR assay targeting the R. orientalis cytochrome c oxidase subunit I (CO1) gene and validated it across 611 samples, including cloacal swabs and fecal samples from live and deceased snakes, multiple pentastome species, and confirmed positive and negative control snakes diagnosed from lung dissections, morphology, and sequencing. We also compared the wet mount microscopy and three fecal flotation techniques for egg detection and assessed the impact of aging and drying on the fecal sample effectiveness. Our PCR assay demonstrated 100% specificity for R. orientalis across all sample types with 98% sensitivity for R. orientalis adults, larvae, and eggs (feces). The lowest sensitivity was observed in cloacal swabs (22%). The PCR assay was tested in a separate laboratory with similar results. Wet mount microscopy was more effective than fecal flotation for egg counts, though the false negative rate did not differ significantly between methods. Aging feces reduced egg counts but did not significantly increase the number of false negatives. Based on these results, we recommend using fecal samples from live snakes as the primary detection method, supplemented by cloacal swabs. These optimized methods are critical for improving surveillance of R. orientalis and characterizing the threat of this invasive pentastome to native snake species in the southeastern United States.
Introduction
Emerging infectious diseases are major contributors to wildlife population declines, extirpation events, and extinctions of wildlife (Lorch et al., 2016; Cheng et al., 2021; Akçakaya et al., 2023; Puryear and Runstadler, 2024). Further, the anthropogenic pressures of development, globalization, and climate change may enhance the spread of pathogens, making emerging epidemics a constant challenge for wildlife and informed management action critical (Daszak et al., 2001; Tompkins et al., 2015; Luberto et al., 2024). This creates a consistent need for the development of reliable and cost-effective tools for detection of pathogens that are accessible to the research community as well as conservation practitioners. For example, several detection tools have been developed that are now essential for understanding infection dynamics of fungal pathogens on wildlife species globally. In particular, quantitative PCR assays specific to chytrid fungi (Batrachochytrium dendrobatidis and B. salamandrivorans) (Kerby et al., 2013), the keratinophilic fungus Ophidiomyces ophidiicola (Bohuski et al., 2015), and the psychrophilic fungus Pseudogymnoascus destructans (Muller et al., 2013) have been essential to characterizing fungal diseases in amphibian, snake, and bat populations, respectively. As novel pathogen threats continue to emerge, rapid development of robust detection tools is necessary for surveillance and understanding pathogen spread and epidemiology.
A recently described threat to herpetofauna has emerged with the invasion of the Asia-origin snake lung parasite, Raillietiella orientalis (Hett, 1915), into both Australia and the United States (Kelehear et al., 2011; Miller et al., 2018). Raillietiella orientalis causes a disease known as pentastomiasis (specifically, raillietielliasis). This hematophagous, obligate parasite belongs to the crustacean Subclass Pentastomida, which includes a diverse group of respiratory parasites—125 recognized species—distributed across all continents (Poore, 2012; Curran et al., 2014). These parasites typically have heteroxenous life cycles involving both intermediate and definitive hosts (Paré, 2008). In Florida, R. orientalis is commonly referred to as the invasive snake lungworm due to its vermiform shape and its frequent infection of snakes as the definitive host. In Australia, 38% of 81 wild-caught snakes were infected with R. orientalis, including species in the families Elapidae, Colubridae, and Pythonidae (Kelehear et al., 2014). In both locations of R. orientalis invasion, native snakes experience severe infections with up to 77 individuals in the lung in Australia and up to 107 individuals in the lung in the eastern United States (Kelehear et al., 2014; Palmisano et al., 2025). While the invasion pathway into Australia remains unknown, R. orientalis appears to have been introduced to North America via Burmese pythons (Python bivittatus) in south Florida and was first detected in 2012 (Miller et al., 2018).
Raillietiella orientalis is pervasive in the herpetofauna of much of Florida and is associated with pulmonary lesions (e.g., pneumonia), sepsis, and mortality events in several wild and captive snake species (Farrell et al., 2019; Walden et al., 2020; Bogan et al., 2022; Palmisano et al., 2025). Unlike most pentastome parasites which have two-host life cycles, R. orientalis utilizes a three-host life cycle that includes an initial intermediate host (coprophagous invertebrate), secondary intermediate hosts (lizards and frogs), and a definitive reptile host (Palmisano et al., 2022). The definitive hosts are primarily snakes but adult R. orientalis have also been documented in two invasive lizard species (Fieldsend et al., 2021; Goetz et al., 2021). Lung infections by R. orientalis adults are now widespread in native Florida snake species (18 species documented to date; Metcalf et al., 2019; Miller et al., 2020; Bogan et al., 2024; Horvath et al., 2025). The geographic range of these infections is rapidly expanding throughout Florida, with occurrence data from 2024 showing that the range now extends at least 435 km northwest and 80 km northeast of the previous northernmost documentation from 2020 (Palmisano et al., 2023, 2025). Furthermore, R. orientalis is present in the pet trade and has been documented in reptiles that are commonly sold from the wild including banded water snakes (N. fasciata) and tokay geckos (Gekko gecko) (Fieldsend et al., 2021; Farrell et al., 2023). Ongoing geographic and taxonomic spread of R. orientalis infections combined with a poor understanding of host susceptibility within and among species make reliable detection methods essential for understanding the threat of R. orientalis to native herpetofauna and captive animals.
Fecal samples are often used to diagnose parasitic infections, particularly in species that shed eggs in the host’s feces (Wellehan and Walden, 2019). A variety of techniques are used including fecal smears, sedimentation, and flotation. The best method of fecal analysis depends on the parasite species of concern (reviewed by Ballweber et al., 2014). Unfortunately, the effectiveness of these diagnostic modalities is not well established for detecting pentastomes in reptiles. Recent studies of R. orientalis infection status in snakes (e.g., Kelehear et al., 2014; Miller et al., 2018; Palmisano et al., 2023) have relied heavily on gross observations and sampling road-killed individuals for parasite identification; however, there is a need for methods that allow for non-invasive, antemortem detection. For most host taxa, freshly produced feces are preferred for diagnostic techniques because sample storage duration often corresponds with reduced parasite detection (Nielsen et al., 2010). Documenting the detection efficacy for both fresh and dried fecal samples is important for pentastome detection in snakes because many species, including several that are common in the pet trade, rarely defecate making fresh sample collection challenging (Lillywhite et al., 2002).
In the eastern United States, snakes can become infected with two native pentastome species, Kiricephalus coarctatus and Porocephalus crotali, as well as R. orientalis (Miller et al., 2018; de Luna et al., 2022). Researchers currently use two conventional PCR assays to identify pentastomes from DNA extracted from adult pentastome tissue. One assay targets the 18S ribosomal RNA gene, which identifies pentastomes to genus level with Sanger sequencing of target amplicons (Brookins et al., 2009). The other assay utilizes the cytochrome c oxidase subunit I (CO1) region and can distinguish among species of pentastomes with Sanger sequencing of target amplicons (Folmer et al., 1994). However, neither assay has been tested for effectiveness of amplifying pentastome DNA from non-invasive sample types such as fecal samples and cloacal swab samples from live hosts. Furthermore, pentastomes are metazoans, which share more genetic similarity with their hosts (e.g., snakes) than do viruses or bacteria. This increased genetic similarity heightens the risk of amplifying host DNA and other invertebrate DNA at conserved gene targets, reducing the utility of these targets for accurate parasite taxonomic identification.
In this study, we describe best practices for non-invasive pentastome egg sampling from snakes and present an optimized PCR protocol specific for the R. orientalis CO1 gene. Specifically, we (1) compare the sensitivity of fresh versus aged fecal samples from snakes with confirmed R. orientalis infections and (2) evaluate different fecal flotation techniques to assess the optimal fecal sample collection and flotation techniques to detect the presence of R. orientalis eggs. We also (3) develop a novel, conventional PCR assay targeting the R. orientalis CO1 gene to provide a reliable, cost-effective, and easy-to-implement tool for pathogen detection, and (4) assess the effectiveness of various sample types for PCR. The methods presented offer valuable techniques for non-invasive sampling and detection of R. orientalis infections in definitive hosts, providing efficient tools for pathogen surveillance. These advancements will enhance our ability to monitor for R. orientalis in snake populations of the eastern United States, contributing to a better characterization of the threat it poses to native reptiles and potentially informing management efforts.
Methods
Fecal flotation solution comparison
We collected snakes from a site located in Volusia County, Florida which has a high prevalence of R. orientalis (Farrell et al., 2019). We retained snakes in the Reptile Lab at Stetson University for ~1 week to collect fresh fecal samples to detect patent R. orientalis infections via fecal wet mounts. We returned apparently uninfected specimens to their site of capture, and retained four infected snakes, including a black racer (Coluber constrictor, caught 7 September 2023), two banded water snakes (Nerodia fasciata, caught 26 August and 21 May 2023), and a ribbon snake (Thamnophis saurita, caught 10 April 2023). We confirmed these four snakes with pentastome eggs in their feces were infected by R. orientalis by (1) euthanizing, dissecting, and morphologically verifying the pentastome species after the end of the experiment (three snakes), or (2) examining an adult female pentastome expectorated during the experiment (one snake). Adult R. orientalis are readily distinguished from the native pentastome species infecting snakes in North America by head morphology (de Luna et al., 2022).
We housed the snakes individually in glass terraria with heating pads, newspaper substrate, water bowls, and two hiding locations. We fed the snakes once each week using frozen-thawed fish and rodents for N. fasciata, rodents and lizards for C. constrictor, and lizards for T. saurita. We checked terraria daily and collected fresh feces while avoiding urates and portions of feces comprised of mostly fur remnants. We collected 20 fecal samples from 26 September 2023 to 1 February 2024 (eight from C. constrictor, one from T. saurita, seven from N. fasciata #1, and four from N. fasciata #2). Immediately after collection, we mixed the feces with a sterilized (cleaned with 70% ethanol solution) metal spatula and subdivided each fecal sample into four similar-sized subsamples that we then weighed on an analytical balance. The fecal subsamples were small (mean mass=68.6 mg) to facilitate complete pentastome egg counts.
For each fecal sample, we randomly assigned one of four diagnostic procedures to the four subsamples. One subsample was used in its entirety to make a series of fecal smear wet mounts to allow us to count the total number of pentastome eggs per gram of fecal material. The wet mounts were prepared by mixing small amount of fecal material with approximately 0.1 mL of tap water on a glass slide. A cover slip was then placed over the mixture. Forceps were used to break apart any large, hard pieces of fecal matter, allowing them to disperse more evenly in the water for improved visualization on the slide. The other three subsamples were used with three flotation solutions, zinc sulfate (ZnSO4, LabChem, specific gravity=1.18), sodium nitrate (NaNO3, “FECA-Med”, Vedco Inc., specific gravity=1.26), and Sheather’s sugar solution (JorVet, Jorgensen Laboratories, specific gravity=1.27). In all flotation tests we placed a subsample in a fecalizer (BioRx Laboratories, Commerce, CA), which is a device consisting of a well and an insert with screening that grinds up the fecal sample and filters the debris from the eggs. One of the three test solutions was added to the fecalizer to the first fill line and the insert was rotated by hand to break up the feces. More solution was added to the fecalizer until the formation of a positive meniscus, at which point a glass cover slip was placed on top. The cover slide remained in position for ten minutes to allow the eggs to rise from the debris and accumulate on the cover slip, which was then carefully removed and placed on a slide. The slide was viewed under 50x brightfield microscopy and the number of pentastome eggs was recorded.
As these snakes were infected with pentastomes, we classified each test as a true positive result if any pentastome eggs were observed in a fecal subsample or as a false negative if no eggs were observed. To calculate the observed pentastome egg density in a fecal sample, we divided total number of eggs counted in the wet mount subsample by the wet mass of that subsample.
Effectiveness of fecal flotation with aged and dried feces
We checked snake cages daily from 30 December 2023 to 7 February 2024 and collected 18 fresh fecal samples (two from C. constrictor, six from T. saurita, six from N. fasciata #1, and four from N. fasciata #2). After we retrieved the feces, they were thoroughly homogenized with a spatula. Each fecal sample was separated into two subsamples of approximately equal size and then weighed (mean=89.3 mg, SD=4.9 mg). The subsamples were randomly assigned to either the fresh sample group (analyzed within three days of retrieval) or the aged sample group. The aged samples were kept in a desiccator with relative humidity of zero held at approximately 20°C and were held for an average of 16.5 days (range=14–21 days) before flotation. We used sodium nitrate flotation only (methods as described above) and counted the number of pentastome eggs observed. We divided the egg count by the subsample mass to get an observed egg density for each sample. Samples were classified as either true positive (at least one pentastome egg observed after flotation) or a false negative (no eggs observed).
Raillietiella orientalis and reptile sampling
The R. orientalis samples used for genetic analysis were either opportunistically collected from live reptiles and carcasses and shared via the Snake Lungworm Alliance and Monitoring (SLAM) program (https://invasionscience.ufl.edu/slam/) or collected through other studies. Sample sources included frozen-thawed carcasses (i.e., roadkill), live captive-held lizards and frogs (for other studies), and live animals handled for a multitude of other research projects. In total, we tested 611 samples from 27 snake species, four lizard species, and one toad species (Supplementary File 1). These samples were comprised of 412 cloacal swabs and 98 fecal samples from snakes, nine R. orientalis larvae, and 43 R. orientalis adults. Additionally, other pentastome species included 49 adults of two native Florida species, Porocephalus crotali and Kiricephalus coarctatus, collected from snakes, as well as adult Raillietiella females from Hemidactylus spp. These Raillietiella specimens were morphologically consistent with blunt-tipped Raillietiella species (R. orientalis is sharp-tipped), specifically R. indica and R. taegueselfi (de Luna et al., 2022). Their host preference was also consistent with these species and for brevity, we will refer to these specimens as blunt-tipped Raillietiella throughout the manuscript.
Our dataset included 170 R. orientalis-negative controls, 155 R. orientalis-positive controls, and 286 snakes of unknown infection status. The negative controls included 90 cloacal swabs from carcasses that were confirmed negative, 24 fecal samples with no eggs detected, and samples from adults of three other pentastome species: K. coarctatus (n=18), P. crotali (n=20), and blunt-tipped Raillietiella (n=11), which were identified via morphology (all individuals, n=49) and further validated by DNA sequencing (n=18). Species identification of adults relied primarily on body shape, head, and hook morphology and established life cycles. We used CO1 primers (Folmer et al., 1994) to confirm our morphological identification of three P. crotali individuals and 18S rRNA primers (Brookins et al., 2009) to confirm our morphological identification of four K. coarctatus, the three P. crotali, and 11 blunt-tipped Raillietiella (Supplementary File 2).
Negative controls also included an additional 24 fecal samples that we determined were negative through fecal wet mount microscopy, and 97 cloacal swab samples from snakes and lizards examined post-mortem that had no adult R. orientalis in their lungs. Additionally, we extracted DNA from six fecal samples that underwent fecal flotation and kept them separate from our other samples for analysis, due to the potential presence of inhibitors in the flotation solutions (three with sodium nitrate and three with zinc sulfate).
For fecal samples, positive controls included 42 pygmy rattlesnake (Sistrurus miliarius) fecal samples in which we observed pentastome eggs and one carcass that had R. orientalis in the lung. We used Sistrurus miliarius (S. miliarius) fecal samples as positive controls because S. miliarius almost exclusively hosts R. orientalis adult (Miller et al., 2020; Farrell et al., 2019). We confirmed these S. miliarius positive samples via Sanger sequencing (see below). There are currently no known diagnostic features of pentastome eggs that enable species differentiation via our standard wet mount techniques; and therefore, 31 additional fecal samples from other snake host species with pentastome eggs microscopically detected were considered unknowns. For cloacal swabs, the group of positive controls included (1) swabs from snakes that were dissected and found with R. orientalis adults in the lungs, and (2) snakes used in experimental infection studies with confirmed R. orientalis infections via conventional PCR and/or dissections. The positive control pentastome individuals included 9 larval R. orientalis isolated from the viscera of lizards and frogs used in experimental infections, and 43 adult R. orientalis from dissected snakes that were identified morphologically and confirmed via Sanger sequencing.
DNA extractions
For DNA extractions, we used Qiagen DNeasy Tissue kits and adapted the manufacturer’s instructions for each sample type (Qiagen, Hilden, Germany). Adult pentastomes were cut with bead-sterilized forceps and scissors, then the 70% ethanol preservative was evaporated off the cut sections. Larval pentastomes preserved in 70% ethanol were placed in 1.5 mL microcentrifuge tubes and ethanol was evaporated off. DNA was extracted from adult (partial) and larval tissue (whole) using the Qiagen DNeasy Tissue kit, following the manufacturer’s instructions (Qiagen, Hilden, Germany). For the cloacal swab extractions, we followed the manufacturer’s guidelines of placing samples in 20 µL proteinase K and 180 µL of buffer ATL for incubation at 56°C. We incubated the swabs for three to six hours and then proceeded to add 200 µL of 100% ethanol and 200 μL of buffer AL. We used bead-sterilized forceps and scissors to transfer the swabs tipped-end facing up, into the spin columns and then pipetted 600 µL of the liquid into the spin column. Following the initial spin at 8,000 RPM for one minute, we removed the swabs and followed the manufacturer’s guidelines for the remainder of the extractions. The fecal extractions followed the protocol from Ayana et al. (2019), which was developed for the extraction of DNA from helminth eggs in fecal samples. Specifically, we used the modified DNeasy Blood and Tissue Kit protocol without bead beating (Qiagen, Hilden, Germany). We measured the concentrations of all samples using a Nanodrop spectrophotometer, except for three positive fecal samples, one cloacal swab, and one P. crotali sample.
PCR assay development
We initially tested cloacal swabs, fecal samples, and R. orientalis adults with 18S primers (Brookins et al., 2009) and CO1 primers (Folmer et al., 1994) as these were the most recent PCR assays available in the literature. Given the lack of morphological methods for distinguishing pentastome eggs, reliable molecular detection techniques for R. orientalis infections in fecal samples were needed. Because no previous studies had evaluated the effectiveness of cloacal swabs or fecal samples with these assays, we utilized positive control samples confirmed to be infected with R. orientalis via dissection to test these methodologies and determine whether a new assay was needed. Specifically, we ran the 18S assay with 30 adult R. orientalis identified morphologically, 14 fecal samples with pentastomid eggs identified through wet mounts (three were from S. miliarius and considered positive controls, the other 11 were considered unknown), and 16 cloacal swabs, including the four positive control snakes used for the fecal flotation methods. We ran the Folmer CO1 assay with six adult R. orientalis and 16 cloacal swab samples described above. Due to the limited utility of these primer sets (see Results) and because no primers existed that were specific to R. orientalis, we developed an R. orientalis-specific PCR assay.
We focused on CO1 because 18S data in GenBank revealed no SNPs segregating between R. orientalis and other pentastomes. We designed R. orientalis-specific CO1 primers based on available sequences in GenBank. Specifically, we retrieved all available CO1 sequences across pentastomid species in GenBank, including R. orientalis, R. indica (and junior synonym R. hebitihamata), R. mottae, Porocephalus crotali, Armillifer spp., Kiricephalus coarctatus, Linguatula spp., and Waddycephalus spp. We performed multiple sequence alignment on these sequences to identify regions that were identical among all R. orientalis samples but contained single nucleotide polymorphisms (SNPs) that were fixed between R. orientalis and all other pentastome species. We designed primers within regions that had the highest density of these SNPs, corresponding to a target region spanning 194 base pairs of the CO1 gene: forward primer ROCO1F=5′-GCCTTCTCCATATTACTCCTC-3; reverse primer ROCO1R=5′- CGTATGTTGATGATTGTGGTAGTG-3′. The primers were designed for specificity and efficiency, taking into consideration factors such as melting temperature and potential secondary structures.
We optimized the PCR protocol using temperature gradient cycling protocols with R. orientalis positive controls and negative controls (including the other pentastome species). For our 20 µL reactions, we used 1 µL of template DNA, 0.5 µl of each primer (ROCO1F, ROCO1R) at 10 µM concentration, 13.95 µL of molecular-grade water, 2 µL of NEB OneTaq Buffer (5x), 0.8 µL of dNTPs (10mM/dNTP), 0.8 µL of BSA (10%), 0.2 µL of DMSO (10mM), and 0.25 µL of NEB One Taq enzyme. We amplified our 20 µL mixture in a thermal cycler (Bio-Rad T100) with a 3-minute denaturation period at 94°C, 35 cycles of: denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 60 seconds, and a final extension at 72°C for 7 minutes. To visualize the amplified product, we performed gel electrophoresis using a 2% agarose gel, run at 80 volts for 45 minutes.
We Sanger sequenced 212 PCR products from our R. orientalis protocol that produced bands of the expected size (~200 bp), as well as 22 additional PCR products (7 fecal samples with unknown eggs, 13 unknown cloacal swabs, and three positive control cloacal swabs) that had nontarget bands (larger than expected or smeared). All sequencing was performed by Eurofins Genomics (Louisville, KY, USA).
Statistical analyses
For fecal flotation analysis, we used a 4×2 Fisher’s exact test to determine if there was a statistically significant association between diagnostic test outcome (true positive or false negative) and testing method (wet mount, zinc sulfate, sodium nitrate, or Sheather’s solution). The egg density data exhibited right-skewed distributions, so we log transformed the data to meet assumptions of parametric statistics before data analysis. We used a general linear model with diagnostic method (wet mount, sodium nitrate float, zinc sulfate float, Sheather’s float) as a fixed factor and fecal sample as a random effect to determine if there were significant differences between the mean egg densities observed among the four treatments using JMP v18 (SAS Institute, Cary, NC). A Tukey test was used to determine which methods significantly differed in mean observed egg density. For aged versus fresh fecal analysis, we used a Fisher’s exact test to determine if there was a difference in the number of false positive results between fresh and aged samples. We conducted a Passing-Bablok agreement analysis on the egg counts for fresh and dried subsamples followed a Bland Altman analysis in JMP v18. We also determined if the mean egg density differed between the fresh subsamples and the dried, aged subsample groups using a paired t-test.
To analyze the efficacy of our PCR assay, we separated our samples into groups by sample type (i.e., cloacal swabs, eggs in S. miliarius fecal samples, and pentastome adults and larvae) and by R. orientalis infection status (positive or negative). In this approach, each test result was treated independently based on the known infection status of the animal (confirmed by dissection or microscopy of fecal samples from S. miliarius), which allowed us to evaluate assay performance and sensitivity according to sample type. We calculated the sensitivity, specificity, positive predictive value, and negative predictive value for our R. orientalis assay across the three sample types (pentastomes, cloacal swabs, and fecal samples) using the confirmed positive and negative controls. Sensitivity was calculated as the proportion of animals that were correctly identified as infected by our assay (true positives) out of all animals known to be infected (the sum of true positives and false negatives). Specificity was calculated as the proportion of animals that were correctly identified by our assay as uninfected (true negatives) out of all animals that were uninfected (the sum of true negatives and false positives). The positive predictive value was calculated as the proportion of animals that were correctly identified by our assay as positive (true positives) out of all animals that tested positive, regardless of their true infection status. Finally, the negative predictive value was calculated as the proportion of animals that were correctly identified by our assay as uninfected (true negatives) out of all animals that tested negative, regardless of their true infection status. We quantified differences in detection rates between sample types by calculating Relative Risk for independent groups (i.e., cloacal swabs with paired fecal samples vs. non-paired cloacal swabs) and Odds Ratio for paired samples (i.e., fecal vs. cloacal swabs from the same individual). Confidence intervals were derived using the epitools package in R (Aragon, 2020; Posit, 2024; R Core Team, 2024).
For snakes of unknown infection status (i.e., cloacal swabs of live snakes and fecal samples with pentastome eggs that were not S. miliarius), we assessed the relationship of DNA concentration and amplification with a density plot. For positive control cloacal swabs, we assessed whether sample DNA concentration was associated with amplification probability using logistic regression. We only performed this analysis on positive control cloacal swabs given that almost all other positive control sample types amplified regardless of DNA concentration. In our analysis, we included a density plot to visualize the distribution of amplified samples relative to their DNA concentrations. The density plot was generated using ggplot2 in R (Wickham, 2016; R Core Team, 2024) and displays the likelihood of amplification across varying concentrations, helping to identify potential thresholds that may enhance amplification success.
Independent PCR assay testing
To further validate our PCR assay, we tested the extraction and amplification protocols in a separate research laboratory at the University of Georgia (Athens, GA) using samples from nine snakes collected from Florida which were determined to have R. orientalis infections through postmortem evaluation and morphological pentastome identification. The snake species sampled included the black racer (n=1), common garter snake (n=1, T. sirtalis), S. miliarius (n=5), banded watersnake (n=1), and corn snake (n=1, Pantherophis guttatus). Adult pentastomes were collected and stored in 70% ethanol. Additionally, we collected fecal samples and cloaca swabs from each snake and stored samples at -80°C.
We cut adult pentastomes using sterile razor blades and forceps targeting a midbody section approximately 5 mm in length. Ethanol was evaporated off the cut section and extracted using the Qiagen DNeasy Tissue kit, following the manufacturer’s instructions (Qiagen, Hilden, Germany). We prepared fecal smear wet mounts (n=9) using the exact methods as above. We initially extracted fecal samples (n=6, samples with sufficient fecal content remaining following fecal smear wet mounts) using the Zymo Quick-DNA™ Fecal/Soil Microbe MiniPrep Kit following manufacturer’s instructions (Zymo Research, Irvine, California, USA). We repeated extractions when sufficient fecal material (n=6) remained, using the adapted fecal extraction protocol with the Qiagen DNeasy Tissue kit as detailed above. Cloacal swabs were extracted similarly to the above; however, were vortexed for one minute following the addition of 180 µL of Buffer ATL and 20 µl of Proteinase K and incubated at 56°C for one hour. Additionally, we performed an extra centrifugation step after the 3-minute centrifugation recommended by the manufacturer and used 100 μL of elution buffer. We used the same PCR and thermal cycler protocols however, we used two different thermal cyclers (Bio-rad DNAEngine; MJ Research PTC-200), and a different master mix. The master mix consisted of a 25 µL reaction volume with 5 µL of template DNA, 0.5 µL of 50 µM forward and reverse primers, 11 µL of molecular-grade water, 5 µL of Green GoTaq® Flexi Buffer (5x), 0.25 µL of dNTPS (80mM/dNTP), 2.5 µL of MgCl2 (25mM), and 0.25 µL of GoTaq® Flexi DNA Polymerase (Promega Corporation, Madison, WI, USA). We Sanger sequenced all PCR products at the appropriate base pair length through Genewiz (Genewiz from Azenta Life Sciences, South Plainfield, New Jersey, USA).
Results
Fecal flotation solution comparison
For fecal sample analysis, all four diagnostic methods resulted in a high frequency of pentastome egg detection. The frequency of false negatives ranged from 0% for the wet mounts to 25% for the Sheather’s solution subsamples (Table 1), and these differences were not statistically significant (Fisher’s exact test, P=0.071). The mean pentastome egg density observed was significantly affected by diagnostic method (F3,57 = 32.53, P < 0.0001; Table 1). The mean number of eggs observed was significantly higher in the wet mount subsamples than in the three flotation solutions and the sodium nitrate and zinc sulfate floated subsamples had significantly higher means than Sheather’s solution subsamples (Tukey test, Table 1).
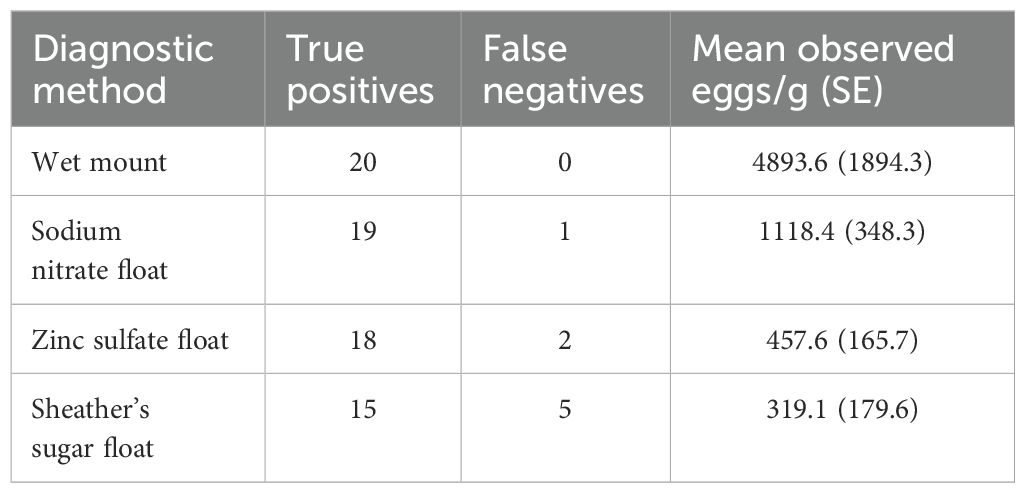
Table 1. Comparison of fecal analyses using wet mounts and three flotation solutions in fecal samples (n=20) for detection of R. orientalis egg shedding in snakes.
Aged versus fresh sample comparison
We observed only a single false negative flotation in the aged fecal subsamples and no false negative results among the fresh subsamples (Table 2) resulting in no significant differences in the frequency of false negative results in the two subsample treatments (Fisher exact test, P=1.00). Agreement analysis for egg counts in the fresh and aged subsamples (Supplementary File 3) indicated a significant bias between the two treatments, with the fresh subsample having higher egg counts than the dried subsample in 14 of 18 comparisons. The mean egg density in the fresh samples (1241.0 eggs/g) was significantly higher than the mean of the aged subsamples (mean = 366.6 eggs/g, t=3.30, df = 17, P=0.004).
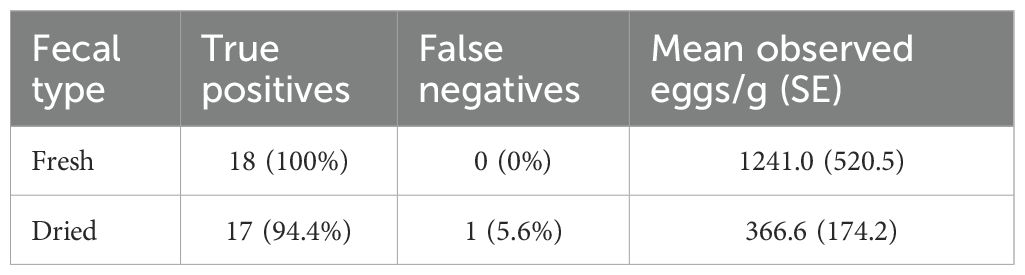
Table 2. The effect of aging and drying on fecal flotation results and observed R. orientalis egg density (all samples done with NaNO3 flotation).
General invertebrate PCR assays
We ran the Folmer et al. (1994) CO1 assay with six R. orientalis adults, which produced amplicons of 710 bp. DNA sequencing confirmed that all six morphologically identified samples were indeed R. orientalis with sequences being 100% identical to other R. orientalis CO1 sequences in GenBank (accession number MG559641). Additionally, the three individuals identified as P. crotali (n=3) were found to be 99–100% identical to P. crotali sequences in GenBank (accession numbers MG559654 and MG559653). We were unable to amplify R. orientalis DNA from the 16 cloacal swabs as well as the 11 blunt-tipped Raillietiella using this protocol, though we ran the PCR three times with positive controls (pentastome adults of other species) that always produced PCR bands of the expected size (710 bp).
For the 18S assay (Brookins et al., 2009), we successfully produced the target PCR amplicon size of ~425 bp in all cloacal swabs (n=16) and 14 fecal samples. Sequencing of these amplicons from cloacal swabs resulted in host DNA for all cases. For the fecal samples, sequencing results were mixed: three samples revealed host DNA (one of the positive control S. miliarius samples), while five samples yielded DNA from helminths (two of the positive control S. miliarius samples), despite confirming the presence of pentastomid eggs with wet mounts. Three fecal samples produced no amplicons, and three others returned sequences that were 100% identical to multiple pentastomid genera.
Notably, all 41 Raillietiellid individuals (R. orientalis and blunt tipped Raillietiella) amplified with the 18S assay produced amplicons of the target size (~425 bp). However, the returned sequences had 99–100% matches to multiple Raillietiella spp. in GenBank. Additionally, 19 of the R. orientalis individuals returned sequences identified as Reighardia spp. which infect seabirds as definitive hosts. For the other pentastome species, four K. coarctatus and three P. crotali samples produced amplicons of the target size (~425 bp). Of the K. coarctatus samples (n=4), two returned sequences with 99–100% identity to K. coarctatus (accession number MG559618), while the other two yielded sequences with 99–100% identity to multiple pentastome genera. Of the P. crotali samples, two yielded sequences with 99–100% identity to multiple pentastome genera, and one returned sequence with 99–100% identity only to P. crotali (accession number MG559598; Supplementary File 2).
Novel R. orientalis specific PCR assay
Using our novel R. orientalis specific PCR assay, we recovered 100% positive predictive value across all positive controls, including R. orientalis adults, R. orientalis larvae, fecal samples with pentastomid eggs from S. miliarius, and cloacal swabs from snakes confirmed to harbor R. orientalis through dissection or inclusion in experimental infections (Table 3). Additionally, all positive control samples had 100% specificity with zero false positives observed. We did not get amplification of adult samples from pentastome species other than R. orientalis, regardless of their DNA concentration (Figure 1; Supplementary File 4, Figure 1). Negative predictive values were 98% for adult and larval R. orientalis, with all samples amplifying except for one larva. Similarly, the negative predictive value for positive control fecal samples was 96%, with only one fecal sample from a confirmed R. orientalis-positive S. miliarius that did not amplify. With this one false negative, the sensitivity for detection in fecal samples was 97.6%, and with one false negative for individuals with known R. orientalis the sensitivity was 98.1%. In contrast, we recovered 47 false negatives from cloacal swab samples, translating to 67.4% negative predictive value and 21.7% sensitivity (Table 3). We failed to amplify R. orientalis DNA in the six fecal samples that underwent fecal flotation.
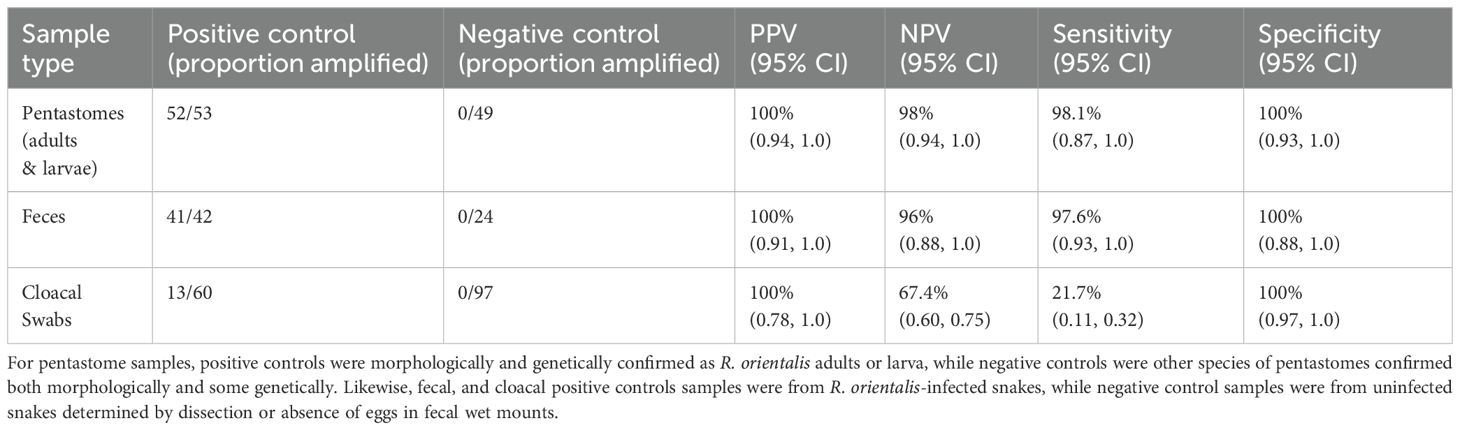
Table 3. The positive predictive values (PPV), negative predictive values (NPV), sensitivity, and specificity for all positive and negative controls used to validate a novel PCR assay for Raillietiella orientalis identification.

Figure 1. The density of DNA concentration across positive controls (Raillietiella orientalis adults and larvae) and three other species of pentastomes with amplification status. One larval R. orientalis that failed to amplify is not shown because of low sample size. This larva had a concentration of 3.3 ng/µL.
Effectiveness of sample type
Raillietiella orientalis individuals (adults and larvae) and fecal samples were more likely to amplify with our assay compared to cloacal swab samples (Table 3). For positive control cloacal swab samples, we did not find a significant effect of DNA concentration on the probability of amplification (Estimate=0.00042, SE=0.0022, z=0.19, P=0.85). The odds ratio for amplification based on DNA concentration was 1.00 (95% CI: 0.99, 1.005) (Figure 2). However, the density plot of amplification from cloacal swabs shows a left skew for samples that did not amplify (Figure 3), suggesting that lower DNA concentrations may reduce the likelihood of amplification (Supplementary File 4, Figure 3). Similarly, fecal samples and cloacal swab samples from hosts with unknown status of R. orientalis infection also show that samples that did not amplify were skewed toward lower DNA concentrations (Figure 4; Supplementary File 4, Figure 2).
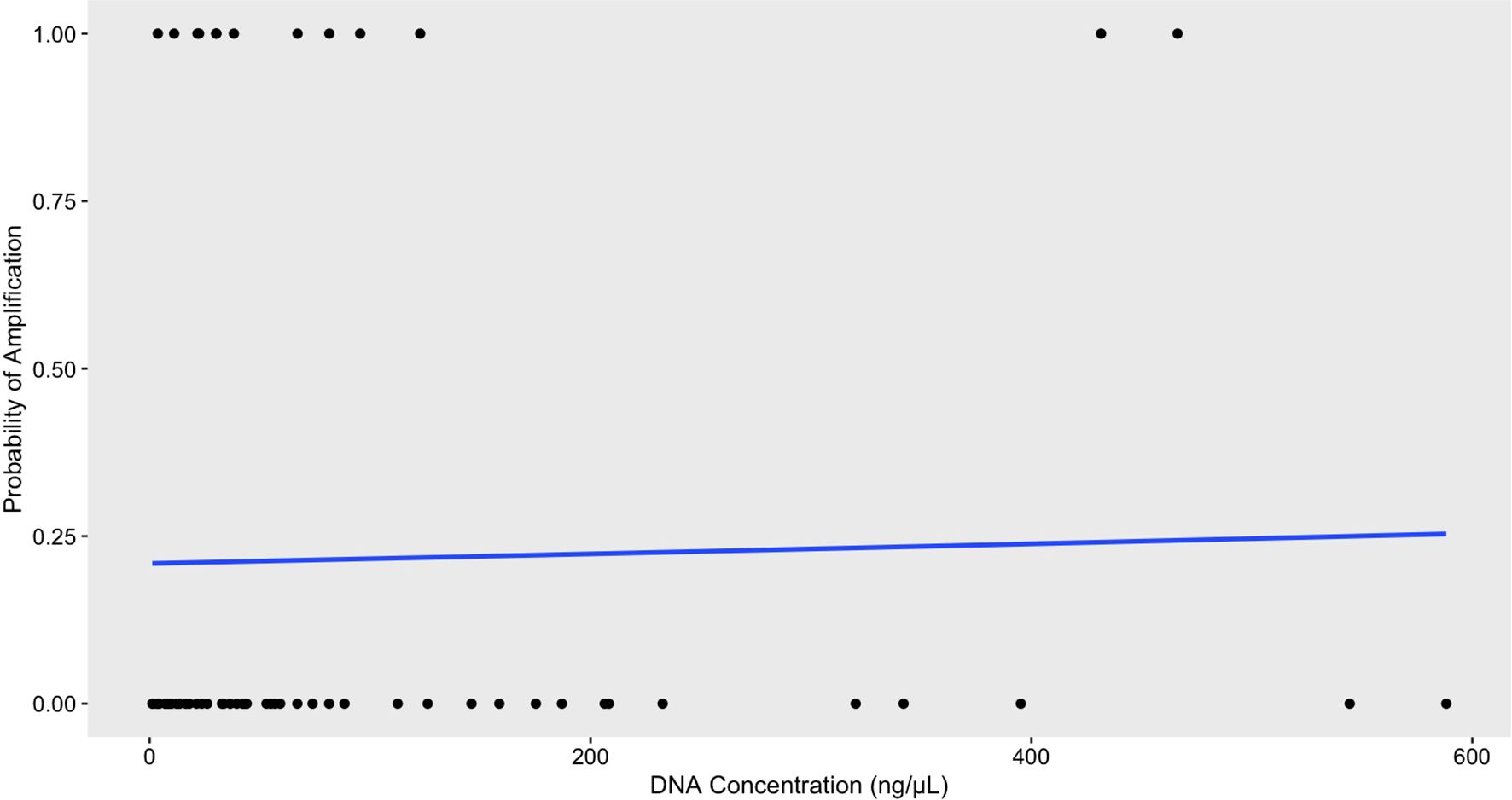
Figure 2. Logistic regression analysis of DNA concentration and its effect on the probability of amplification via conventional PCR for positive control cloacal swab samples using the R. orientalis-specific assay (n=60) revealed no statistically significant effect of concentration (Estimate=0.0004225, SE=0.0022, z=0.193, p=0.847). These results suggest that variations in DNA concentration have little impact on the odds of amplification, as indicated by an odds ratio of 1.000422.
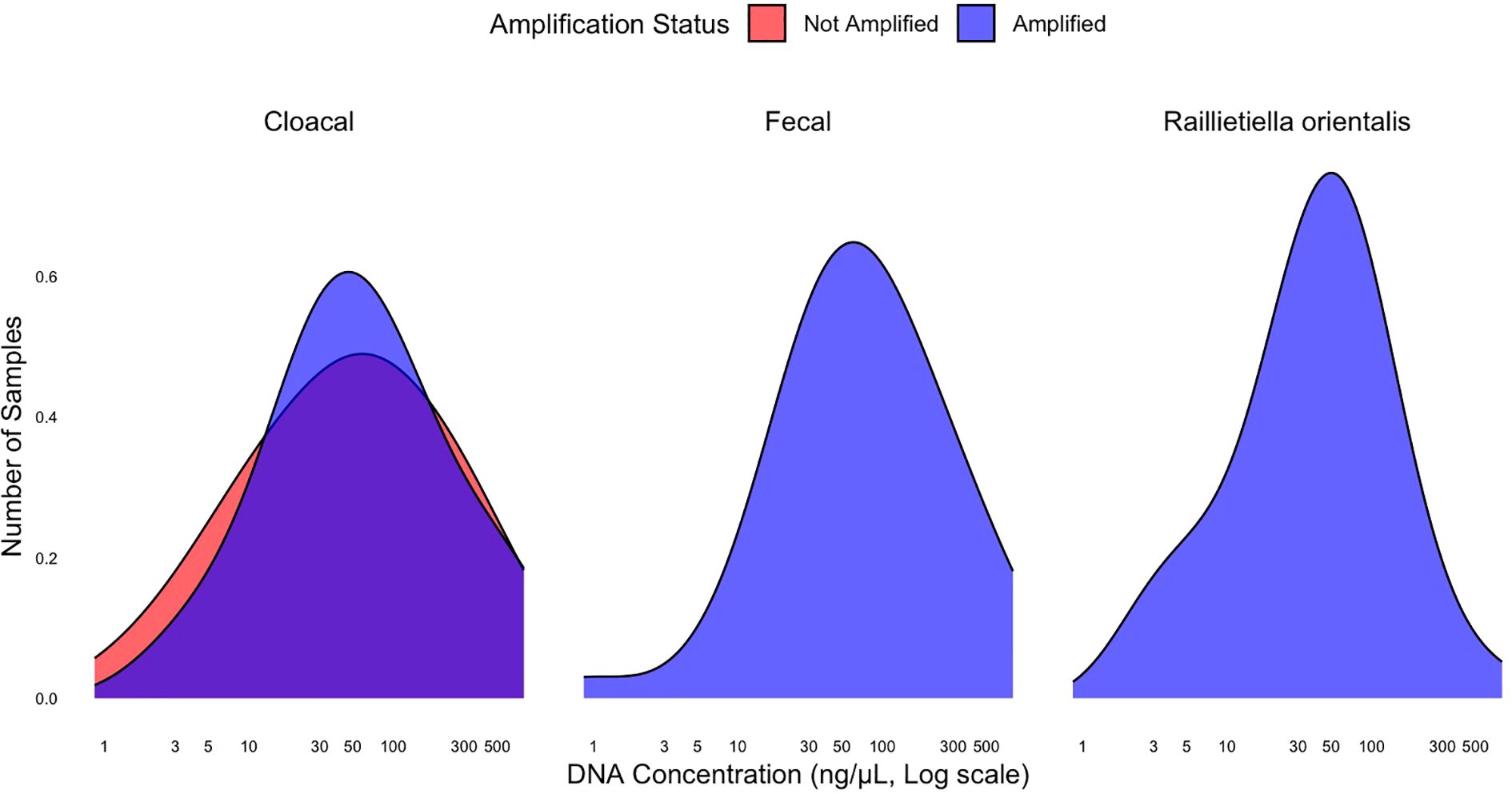
Figure 3. The density of Raillietiella orientalis DNA concentrations among positive controls of three sample types with amplification status. One larval R. orientalis and one fecal sample did not amplify and are not included due to low sample size (n=1). The larva had a concentration of 3.3 ng/µL and the fecal sample had a concentration of 4.9 ng/µL.
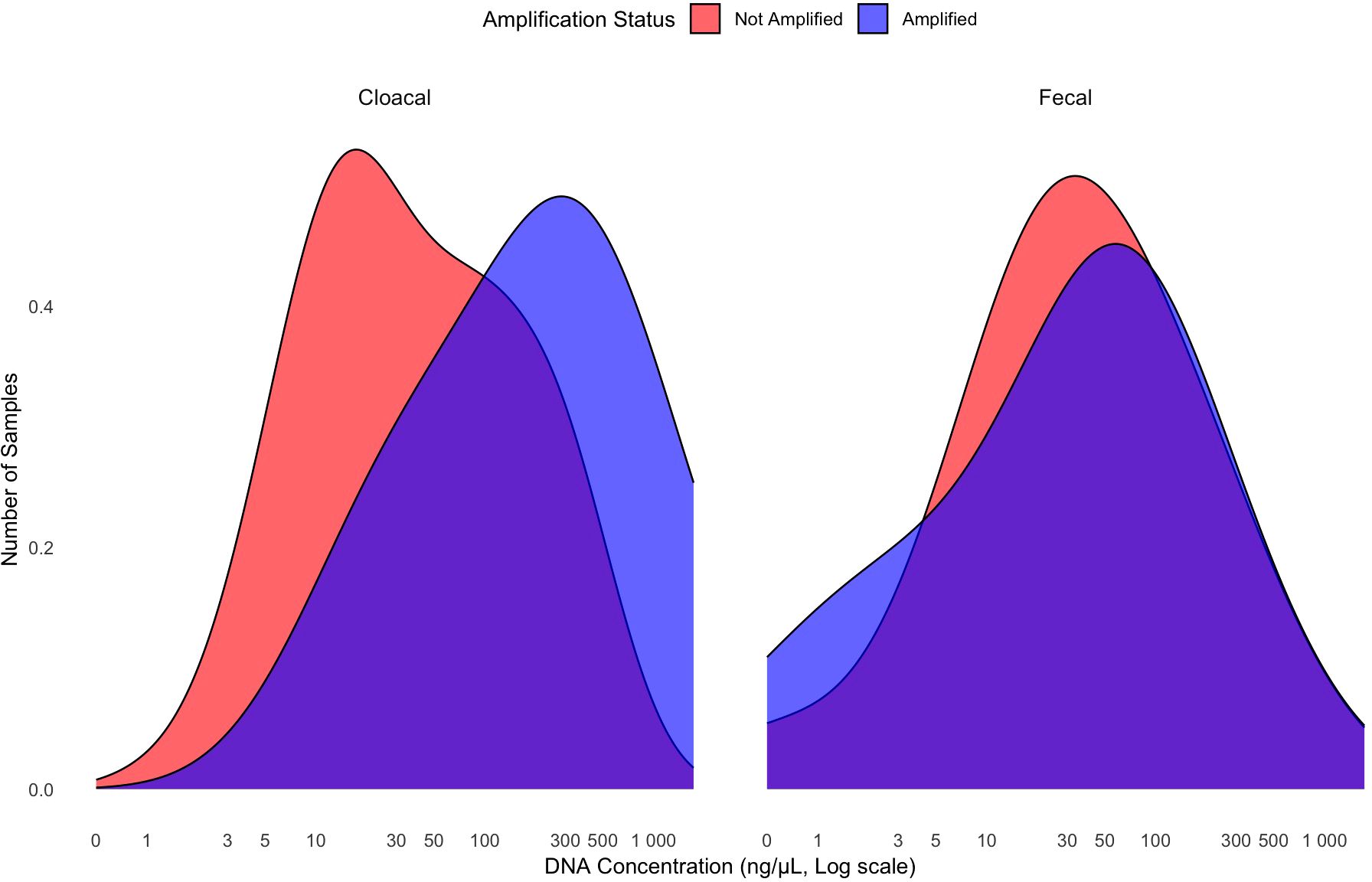
Figure 4. The density of R. orientalis DNA concentration in cloacal swab samples (n=254) and fecal samples (n=31) from hosts of unknown R. orientalis infection status. Nineteen of the fecal samples had recognizable pentastome eggs present (species unknown) and 12 of these 19 amplified. Twelve of the 254 cloacal swabs amplified.
We collected fecal samples from 139 of the 296 live snakes with cloacal swabs collected. Therefore, these 139 snakes were known to have active fecal content in their cloaca at the time of swabbing. Of these paired samples (fecal and cloacal), 25.2% fecal samples (35/139, 95% CI: 0.18, 0.33) and 7.9% cloacal swabs (11/139, 95% CI: 0.04, 0.14) produced PCR bands of the expected size using our novel assay (Supplementary File 4, Table 1). Fecal samples were ~6 times more likely to test positive than their paired cloacal swabs (OR = 6.27; 95% CI: 2.79–14.06). Among the live snakes with non-paired cloacal swabs, 5.1% (8/157, 95% CI: 0.02, 0.1) yielded PCR bands of the expected size. Among cloacal swabs paired with positive fecal samples, 22.9% were also PCR positive (8/35; 95% CI: 0.10–0.40), compared to only 5.1% in the non-paired group. This represents a 4.94 times higher likelihood of detection via swabs from snakes with paired, positive fecal samples (95% CI: 2.20–11.08).
Sanger sequencing
DNA sequencing with the R. orientalis-specific assay confirmed that all samples that produced PCR amplicons of the expected size (Supplementary File 5, Figure 4) produced CO1 sequence data specific to R. orientalis (all were 100% identical to other R. orientalis CO1 sequences in GenBank). Further, all sequences were 100% identical to each other, suggesting little sequence variation in this gene region, at least within the Florida population of R. orientalis (GenBank accession number PQ621873). All samples that amplified nontarget bands returned host (snake) DNA sequences. Further, we duplicated the reaction, gel, and sequencing for 95 positive control samples and received the sample results for all but one (98.9%). When compared with the results from the University of Georgia laboratory, results from our R. orientalis-specific assay were consistent with our initial findings, with 100% of R. orientalis adults (n=9), 44.4% of cloacal swabs (n=9), and 85.7% of fecal samples (n=7) producing the target amplicon, and all other samples producing no amplicons. Sequences from these samples were 100% identical to R. orientalis sequences in GenBank (Table 4).
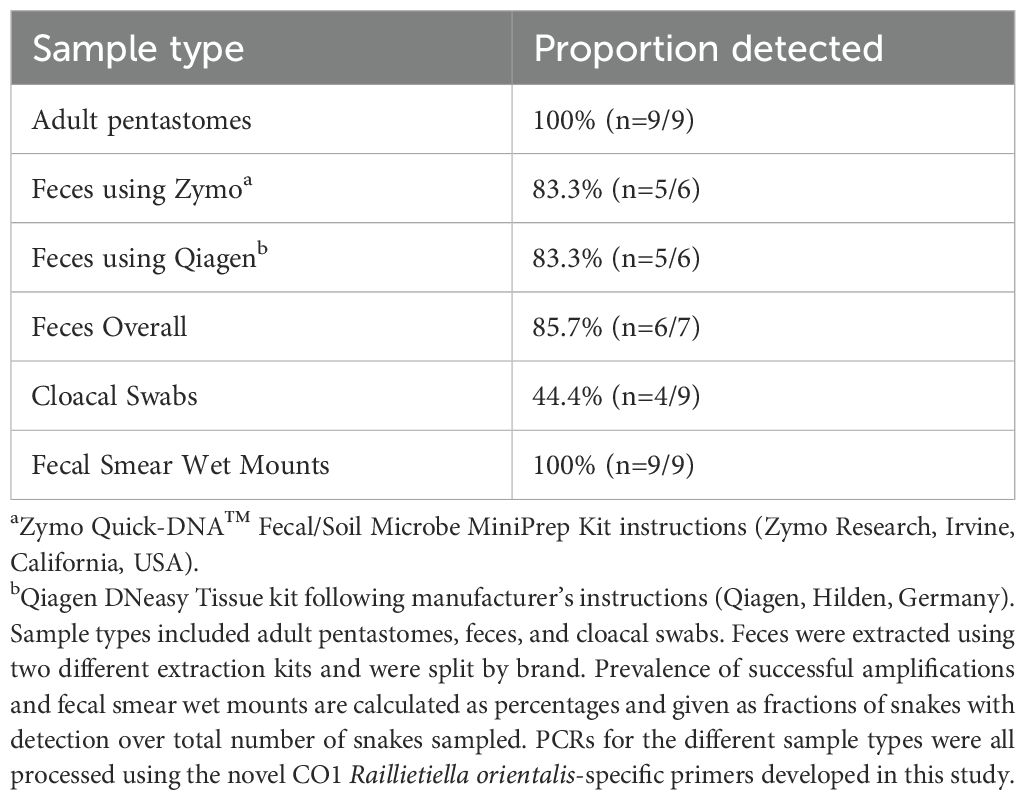
Table 4. PCR and fecal smear wet mount results from the nine independent snakes used for external validation at the University of Georgia.
Discussion
Our study describes optimized pentastome sampling methodology for snake feces and a novel R. orientalis-specific PCR assay that produces ~200 bp PCR amplicons only if R. orientalis DNA is present (Supplementary File 4, Figure 4). Our PCR protocol is a robust and cost-effective detection tool that has 100% specificity across all sample types. We validated this protocol for use in detecting R. orientalis by testing 611 samples from 27 snake species, four lizard species, and one anuran species, including 155 confirmed positive control samples and 170 confirmed negative control samples. Consistent results obtained from a second laboratory, where different reagents, equipment, and researchers were involved, further validate the protocol’s reproducibility, and highlight its potential for widespread adoption across diverse research settings.
This assay is inexpensive and straightforward to use for anyone with access to DNA extraction and PCR equipment. Quantitative (q)PCR assays are incredibly useful for microparasitic pathogens as they are sensitive and provide information on pathogen burdens that cannot otherwise be observed. However, qPCR reagents are significantly more expensive than conventional PCR, qPCR requires more expensive equipment, and qPCR is less relevant for macroparasitic pathogens where quantification is not plausible from indirect samples (and can be measured visually from direct samples). Thus, we focused on a simple diagnostic that reliably confirms the presence or absence of R. orientalis.
If the goal is to identify R. orientalis presence in fecal samples, adult or larval pentastomes, or cloacal swabs in the eastern United States, we recommend using our protocol. If amplification does not occur via our protocol with adult or larval pentastomes, we recommend using the Folmer CO1 assay for parasite species identification outside of R. orientalis. However, nontarget amplification is possible based on our CO1 endosymbiont sequences produced from unknown Raillietiellid samples. More data are needed on use of the Folmer CO1 assay with cloacal swabs and fecal samples to know if it is reliable in amplifying only pentastome DNA. We recommend using the 18S protocol for exploratory measures of unknown pentastomes but acknowledge its high rate of amplifying non-target DNA due to the lack of genetic differentiation among species at this target region. While our assay is reliable for diagnostic purposes, it is not useful for analysis of R. orientalis genetic variation as it contains no polymorphisms. For examining different genetic lineages, the Folmer CO1 primers are a better tool.
Our study had several limitations. First, we had a low rate of amplification from positive control cloacal swabs. However, considering the relative ease and non-invasive nature of cloacal swab collection, and the 100% specificity from cloacal swabs, we recommend further investigation into the potential use of pooled cloacal swab samples as a tool for R. orientalis surveillance. While we did not assess the impact of pooling on sensitivity or specificity, we believe this approach may be worth considering in future studies, especially for large-scale surveillance efforts. However, because cloacal swabs have a high false negative rate, many swabs should be analyzed before determining a location is free of the parasite. We recommend the use of fecal samples when available, as our data showed that fecal samples from infected snakes were more likely to detect R. orientalis than their paired cloacal swabs. The higher prevalence of R. orientalis detected from cloacal swabs in snakes with paired fecal samples indicates that the presence of fecal content in the cloaca may increase sensitivity. Further research should investigate the effectiveness of cloacal swabs with varied DNA extraction methods and their temporal relation to the last defecation, as fecal samples are often not available from wild, field-sampled snakes.
Another limitation of our study is that one of nine R. orientalis larvae and one of 44 fecal samples with R. orientalis eggs did not amplify using our assay. Since we tested a small sample of larvae (all from the secondary intermediate host), our results raise questions about the reliability of larval amplification. Future research should investigate how sensitive the assay is with larvae and adults that have lower than five µL of DNA, as we only had six out of 52 samples that had lower than five µL, and across fecal samples with various egg densities upon wet mount. Additionally, further studies are needed to determine whether fecal flotation techniques inhibit PCR amplification, or if our small sample size led to a possible type II statistical error. For now, we recommend saving a portion of the fecal samples before flotation for DNA extraction and prioritizing fecal samples over cloacal swabs.
A third limitation to our study and for all pentastome parasite research is the lack of genetic and taxonomic information for this group. Of the 42 recognized Raillietiella species (Poore, 2012), most lack comprehensive morphological descriptions and only four species have publicly available molecular data: R. orientalis (with two potential but not confirmed junior synonyms, R. piscator and R. gowrii), R. indica (junior synonyms, R. hebitihamata and R. frenata), R. hemidactyli, and R. mottae. While we assessed the use of our assay with the congener blunt-tipped Raillietiella, further studies are needed using type specimens from additional Raillietiella species. This will be essential to confirm the specificity of the assay for R. orientalis and to ensure its broad applicability outside of the eastern United States. We need further molecular characterization of pentastomes and the continued development of specific assays to be used for reliable speciation.
Our fecal smear analyses yielded no false negative results, indicating that the infected snakes used in our egg detection studies consistently had eggs detected in their fecal samples. All three of the flotation solutions we used also typically detected eggs in known infected snakes (i.e., true positive test results). We had false negative results in only 9.4% (9/96) of the flotation tests run in the fecal float comparison experiment and the aged versus fresh sample experiment with five of these false negative tests occurring when we used Sheather’s sugar solution. Our estimates of the percent of eggs in a sample that were observed after flotation indicated that Sheather’s sugar was significantly less effective in harvesting eggs than sodium nitrate and zinc sulfate. Flotation in sodium nitrate (specific gravity 1.25-1.27) was effective in detecting pentastome infection given both the low number of false negative results and the high effectiveness in harvesting eggs. For sodium nitrate flotation, the false negative rate was only 3.6% (2 of 56 tests) for the two experiments. Sheather’s solution might be more effective if combined with centrifugation, but this technique requires more equipment and is time intensive compared to the rapid and simple flotation methods we used. We also used small amounts of fecal material, (~<0.1 g), in our flotation methods as we subdivided the feces and intentionally used small samples to facilitate complete egg counts on fecal smear slides. The probability of false negative results may have been lower had we used greater mass of fecal samples.
An additional limitation to this study was the lack of fecal examination through sedimentation techniques. While fecal sedimentation is the preferred technique for detecting trematode larvae and ova (Hayider et al., 2018) and acanthocephalan ova (Nabi et al., 2015), it has not been shown to be better than flotation for nematode ova or protozoan oocysts (Ines et al., 2016). In reptiles, a comparison of fecal examination techniques has been performed for multiple parasites, including pentastomes (Wolf et al., 2014). While Wolf et al. (2014) detected pentastomid ova in both the flotation and the sedimentation techniques, the number of animals with pentastomiasis was too small to allow for adequate statistical comparison. However, we feel that the likelihood of Raillietiella ova detection through a sedimentation technique may be lower than that of the flotation technique since the ova of R. orientalis demonstrated positive buoyancy with a high rate of detection. Further investigation is warranted to evaluate the utility of fecal sedimentation as it relates to fecal flotation for the detection of R. orientalis.
Aging and drying fecal samples for several weeks significantly reduced pentastome egg counts but did not significantly increase the frequency of false negative results. However, this method provides an alternative for use in captive snakes, in which collection of wet fecal samples can be challenging. This is because the period between successive defecation events typically exceeds one month, especially in large constrictors commonly used in the pet trade (Lillywhite et al., 2002). Furthermore, our results indicate that fecal samples from snakes used in field studies can be stored at ambient temperature until processing later. A limitation of the fecal analysis we used is that it will only work for patent (egg bearing) infections and, therefore, cannot detect juvenile-only or male-only infections.
The R. orientalis-specific assay and optimized fecal sample processing will facilitate new research on detecting R. orientalis in wild herpetofauna across the eastern United States. The PCR assay has already expanded our understanding of R. orientalis epizoology, identifying infections in 18 additional Florida counties (Palmisano et al., 2022, 2025) and several new host taxa (Horvath et al., 2025; Palmisano et al., 2025). We recommend fecal samples as the primary detection method in live snakes, and if fecal flotation is used, splitting samples for both flotation and molecular analysis. While wet mounts are effective, they can be difficult with clumped feces containing mammalian hair; in such cases, flotation may be a simpler alternative. If the sample consists mainly of urates, the absence of feces makes it difficult to draw conclusions about infection, as eggs are typically found in fecal material. Fecal samples can be preserved either by freezing (following our University of Georgia protocol) or by storing in 70-90% ethanol at room temperature, protected from direct sunlight. If fecal samples are unavailable, cloacal swabs can be used, but negative results should not be interpreted as indicating the absence of infection. Our study presents robust fecal sampling and PCR protocols for detecting and monitoring R. orientalis infections, contributing valuable data for pathogen surveillance in the eastern United States. These tools can enhance reptile disease research and inform proactive biosecurity measures.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by University of Central Florida, Stetson, and University of Georgia Institutional Animal Care and Use Committees. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JP: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. CH: Data curation, Investigation, Methodology, Validation, Visualization, Writing – review & editing. JG: Data curation, Writing – review & editing. JH: Data curation, Writing – review & editing. TF: Data curation, Methodology, Resources, Writing – review & editing, Formal analysis, Investigation, Writing – original draft. JB: Resources, Validation, Writing – review & editing, Data curation, Methodology. NN: Resources, Supervision, Validation, Writing – review & editing. AS: Data curation, Formal analysis, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing, Conceptualization, Supervision, Validation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was partially funded by the Orianne Society Grant Program, the American Society of Ichthyologists and Herpetologists Gaige Fund Award, American Museum of Natural History Theodore Roosevelt Memorial Grant, UCF Department of Biology Graduate Student Research Award, and The Rattlesnake Conservancy Venomous Reptile Conservation Grant awarded to Jenna N. Palmisano. Additionally, the North Carolina Herpetological Society David L. Stephan Grants in Herpetology program awarded to Snake Lungworm Alliance and Monitoring.
Acknowledgments
We would like to express our sincere gratitude to our colleagues within the SLAM network for their valuable contributions of samples, which aided in the validation of our PCR assay. Special thanks go to Benjamin S. Stegenga and Kim Titterington for their continued support and collaboration.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/famrs.2025.1531792/full#supplementary-material
References
Akçakaya H. R., Neam K., Hobin L., Lötters S., Martel A., and Pasmans F. (2023). Assessing the extinction risks of amphibians impacted by infectious diseases. Biol. Conserv. 284, 110205. doi: 10.1016/j.biocon.2023.110205
Aragon T. J. (2020). Epitools: Epidemiology tools (R package version 0.5-10.1) [Computer software] (Comprehensive R Archive Network (CRAN). Available online at: https://CRAN.R-project.org/package=epitools (Accessed May 2025).
Ayana M., Cools P., Mekonnen Z., Biruksew A., Dana D., Rashwan N., et al. (2019). Comparison of four DNA extraction and three preservation protocols for the molecular detection and quantification of soil-transmitted helminths in stool. PloS Neglected Trop. Dis. 13, e0007778. doi: 10.1371/journal.pntd.0007778
Ballweber L. R., Beugnet F., Marchiondo A. A., and Payne P. A. (2014). American Association of Veterinary Parasitologists’ review of veterinary fecal flotation methods and factors influencing their accuracy and use—Is there really one best technique? Veterinary Parasitol. 204, 73–80. doi: 10.1016/j.vetpar.2014.05.009
Bogan J. E. Jr., O’Hanlon B. M., Steen D. A., Horan T., Taylor R., Mason A. K., et al. (2024). Health assessment of free-ranging eastern indigo snakes (Drymarchon couperi) from hydrologic restoration construction sites in south Florida, USA. J. Wildlife Dis. 60, 39–51. doi: 10.7589/JWD-D-22-00184
Bogan J. E., Steen D. A., O’Hanlon B., Garner M. M., Walden H. D. S., and Wellehan J. F. X. (2022). Drymarchon couperi (eastern indigo snake). Death associated with Raillietiella orientalis. Herpetological Rev. 53, 147.
Bohuski E., Lorch J. M., Griffin K. M., and Blehert D. S. (2015). TaqMan real-time polymerase chain reaction for detection of Ophidiomyces ophiodiicola, the fungus associated with snake fungal disease. BMC Veterinary Res. 11, 95. doi: 10.1186/s12917-015-0407-8
Brookins M. D., Wellehan J. F. X. Jr., Roberts J. F., Allison K., Curran S. S., Childress A. L., et al. (2009). Massive visceral pentastomiasis caused by Porocephalus crotali in a dog. Veterinary Pathol. 46, 460–463. doi: 10.1354/vp.46-3-460
Cheng T. L., Reichard J. D., Coleman J. T., Weller T. J., Thogmartin W. E., Reichert B. E., et al. (2021). The scope and severity of white-nose syndrome on hibernating bats in North America. Conserv. Biol. 35, 1586–1597. doi: 10.1111/cobi.13739
Curran S. S., Overstreet R. M., Collins D. E., and Benz G. W. (2014). Levisunguis subaequalis n. g., n. sp., a tongue worm (Pentastomida: Porocephalida: Sebekidae) infecting softshell turtles, Apalone spp. (Testudines: Trionychidae) in the southeastern United States. Systematic Parasitol. 87, 33–45. doi: 10.1007/s11230-013-9453-1
Daszak P., Cunningham A. A., and Hyatt A. D. (2001). Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Tropica 78, 103–116. doi: 10.1016/S0001-706X(00)00179-0
de Luna M., García-Barrios R., Barton D. P., and Guajardo-Martínez G. (2022). Tongue worm (Pentastomida) parasites of North American herpetofauna: Checklist of species, identification key, and new state and host records from Mexico. J. Parasitol. 108, 582–594. doi: 10.1645/21-137
Farrell T. M., Agugliaro J., Walden H. D. S., Wellehan J. F. X., Childress A. L., and Lind C. M. (2019). Spillover of pentastome parasites from invasive Burmese pythons (Python bivittatus) to pygmy rattlesnakes (Sistrurus miliarius), extending parasite range in Florida, USA. Herpetological Rev. 50, 73–76.
Farrell T. M., Walden H. D. S., and Ossiboff R. J. (2023). The invasive pentastome Raillietiella orientalis in a banded water snake from the pet trade. J. Veterinary Diagn. Invest. 35, 201–203. doi: 10.1177/10406387231153924
Fieldsend T., Harman M., and Miller M. (2021). First record of an Asian tongueworm, Raillietiella orientalis (Pentastomida: Raillietiellidae), parasitizing a Tokay gecko (Gekko gecko, Squamata: Gekkonidae): A novel interaction between two non-native species in Florida. Reptiles Amphibians 28, 255–256. doi: 10.17161/randa.v28i2.15238
Folmer O., Black M., Hoeh W., Lutz R., and Vrijenhoek R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol. 3, 294–299.
Goetz S. M., Steen D. A., Miller M. A., Guyer C., Kottwitz J., Roberts J. F., et al. (2021). Argentine black and white tegu (Salvator merianae) can survive the winter under semi-natural conditions well beyond their current invasive range. PloS One 16, e0245877. doi: 10.1371/journal.pone.0245877
Hayider N., Mekuria S., and Mekibib B. (2018). Major trematodes of cattle slaughtered at Hirna municipal abattoir: Prevalence, associated risk factors and test agreement of sedimentation technique in Ethiopia. J. Parasitol. Vector Biol. 10, 51–57. doi: 10.5897/JPVB2017.0318
Horvath S., Palmisano J. N., Savage A. E., Martin E., Donini J., Metcalf M. F., et al. (2025). Farancia abacura (red-bellied mudsnake). Parasites. Herpetological Rev. 54, 449.
Hett M. L. (1915). On some new pentastomids from the Zoological Society’s Garden, London. Proceedings of the Zoological Society of London. 1915, 115–121.
Inês E. D., Figueiredo Pacheco F. T., Carneiro Pinto M., Silva de Almeida Mendes P., da Costa-Ribeiro H., Matos Soares N., et al. (2016). Concordance between the zinc sulphate flotation and centrifugal sedimentation methods for the diagnosis of intestinal parasites. Biomédica 36, 519–524. doi: 10.7705/biomedica.v36i4.3052
Kelehear C., Spratt D. M., Dubey S., Brown G. P., and Shine R. (2011). Using combined morphological, allometric, and molecular approaches to identify species of the genus Raillietiella (Pentastomida). PloS One 6, e24936. doi: 10.1371/journal.pone.0024936
Kelehear C., Spratt D. M., O’Meally D., and Shine R. (2014). Pentastomids of wild snakes in the Australian tropics. Int. J. Parasitol.: Parasites Wildlife 3, 20–31. doi: 10.1016/j.ijppaw.2014.03.003
Kerby J. L., Schieffer A., Brown J. R., and Whitfield S. (2013). Utilization of fast qPCR techniques to detect the amphibian chytrid fungus: A cheaper and more efficient alternative method. Methods Ecol. Evol. 4, 162–166. doi: 10.1111/2041-210X.12008
Lillywhite H. B., de Delva P. I., and Noonan B. P. (2002). “Patterns of gut passage time and the chronic retention of fecal mass in viperid snakes,” in Biology of the Vipers. Eds. Schuett G. W., Höggren M., Douglas M. E., and Greene H. W. (Eagle Mountain, UT: Eagle Mountain Publishing), 497–506.
Lorch J. M., Knowles S., Lankton J. S., Michell K., Edwards J. L., Kapfer J. M., et al. (2016). Snake fungal disease: An emerging threat to wild snakes. Philos. Trans. R. Soc. B: Biol. Sci. 371, 20150457. doi: 10.1098/rstb.2015.0457
Luberto E. T., Ramsey M. L., and Kollath D. R. (2024). The ecology of pathogenic Onygenales fungi and the impacts of climate change. Curr. Clin. Microbiol. Rep. 11, 62–69. doi: 10.1007/s40588-024-00234-6
Metcalf M. F., Marsh A., Brosse W., and Herman J. E. (2019). Crotalus adamanteus: endoparasite. Herpetological Rev. 50, 389.
Miller M. A., Kinsella J. M., Snow R. W., Falk B. G., Reed R. N., Goetz S. M., et al. (2020). Highly competent native snake hosts extend the range of an introduced parasite beyond its invasive Burmese python host. Ecosphere 11, e03153. doi: 10.1002/ecs2.3153
Miller M. A., Kinsella J. M., Snow R. W., Hayes M. M., Falk B. G., Reed R. N., et al. (2018). Parasite spillover: Indirect effects of invasive Burmese pythons. Ecol. Evol. 8, 830–840. doi: 10.1002/ece3.3746
Muller L. K., Lorch J. M., Lindner D. L., O’Connor M., Gargas A., and Blehert D. S. (2013). Bat white-nose syndrome: A real-time TaqMan polymerase chain reaction test targeting the intergenic spacer region of Geomyces destructans. Mycologia 105, 253–259. doi: 10.3852/12-242
Nabi S., Tanveer S., Ganaie S. A., Niyaz U., and Abdullah I. (2015). Acanthocephalan infestation in fishes – A review. J. Zool. Stud. 2, 33–38.
Nielsen M. K., Vidyashankar A. N., Andersen U. V., DeLisi K., Pilegaard K., and Kaplan R. M. (2010). Effects of fecal collection and storage factors on strongylid egg counts in horses. Veterinary Parasitol. 167, 55–61. doi: 10.1016/j.vetpar.2009.09.017
Palmisano J. N., Bockoven C., McPherson S. M., Ossiboff R. J., Walden H. D., and Farrell T. M. (2022). Infection experiments indicate that common Florida anurans and lizards may serve as intermediate hosts for the invasive pentastome parasite, Raillietiella orientalis. J. Herpetol. 56, 355–361. doi: 10.1670/21-067
Palmisano J. N., Brennan M., Durso A. M., Kesselring J. H., Morgan T., Lepera Z., et al. (2025). Rapid spread of the invasive parasite Raillietiella orientalis in 14 new Florida counties and in pet trade snakes. BioInvasion Records 14, 261–269. doi: 10.3391/bir.2025.14.1.20
Palmisano J. N., Farrell T. M., Hazelrig C. M., and Brennan M. N. (2023). Documenting range expansion of the invasive pentastome parasite, Raillietiella orientalis, using southern black racer and eastern coachwhip road mortality. Southeastern Nat. 22, N17–N22. doi: 10.1656/058.022.0110
Paré J. A. (2008). An overview of pentastomiasis in reptiles and other vertebrates. J. Exotic Pet Med. 17, 285–294. doi: 10.1053/j.jepm.2008.09.003
Poore G. C. (2012). The nomenclature of the recent Pentastomida (Crustacea), with a list of species and available names. Systematic Parasitol. 82, 211–240. doi: 10.1007/s11230-012-9362-0
Posit. (2024). RStudio: Integrated development environment for R (Version 2024.12.0 + 467) (Posit Software, PBC). Available online at: https://posit.co (Accessed May 2025).
Puryear W. B. and Runstadler J. A. (2024). High-pathogenicity avian influenza in wildlife: A changing disease dynamic that is expanding in wild birds and having an increasing impact on a growing number of mammals. J. Am. Veterinary Med. Assoc. 264, 1–9. doi: 10.2460/javma.24.0007
R Core Team (2024). R: A language and environment for statistical computing (Version 4.4.2) (R Foundation for Statistical Computing). Available online at: https://www.R-project.org/ (Accessed May 2025).
Tompkins D. M., Carver S., Jones M. E., Krkošek M., and Skerratt L. F. (2015). Emerging infectious diseases of wildlife: A critical perspective. Trends Parasitol. 31, 149–159. doi: 10.1016/j.pt.2015.01.003
Walden H. D. S., Iredale M. E., Childress A., Wellehan J. F. X. Jr., and Ossiboff R. J. (2020). Case report: Invasive pentastomes, Raillietiella orientalis (Sambon 1922), in a free-ranging banded water snake (Nerodia fasciata) in north central Florida, USA. Front. Veterinary Sci. 7. doi: 10.3389/fvets.2020.00467
Wellehan J. F. X. and Walden H. D. S. (2019). “Parasitology (including hemoparasites),” in Mader’s Reptile and Amphibian Medicine and Surgery. Eds. Divers S. J. and Stahl S. J. (New York: Springer), 281–300.
Wickham H. (2016). ggplot2: Elegant graphics for data analysis (Springer-Verlag). doi: 10.1007/978-3-319-24277-4
Keywords: pentastome, reptiles, snake parasites, parasite detection, fecal sampling, emerging disease
Citation: Palmisano JN, Hazelrig CM, Gazil JA, Hanco JK, Farrell TM, Bogan JE Jr., Nemeth NM and Savage AE (2025) A novel PCR assay and sampling techniques for the detection of Raillietiella orientalis. Front. Amphib. Reptile Sci. 3:1531792. doi: 10.3389/famrs.2025.1531792
Received: 20 November 2024; Accepted: 12 May 2025;
Published: 02 June 2025.
Edited by:
Kurt k Sladky, University of Wisconsin-Madison, United StatesReviewed by:
John Abramyan, University of Michigan–Dearborn, United StatesThomas Fieldsend, King’s College London, United Kingdom
Copyright © 2025 Palmisano, Hazelrig, Gazil, Hanco, Farrell, Bogan, Nemeth and Savage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jenna N. Palmisano, amVubmEucGFsbWlzYW5vQHVjZi5lZHU=
 Jenna N. Palmisano
Jenna N. Palmisano Corinna M. Hazelrig
Corinna M. Hazelrig Jack A. Gazil3
Jack A. Gazil3 Jennifer K. Hanco
Jennifer K. Hanco Terence M. Farrell
Terence M. Farrell Anna E. Savage
Anna E. Savage