- 1Department of Veterinary Clinical Medicine, College of Veterinary Medicine, University of Illinois Urbana-Champaign, Urbana, IL, United States
- 2Wildlife Epidemiology Laboratory, College of Veterinary Medicine, University of Illinois Urbana-Champaign, Urbana, IL, United States
- 3Comparative Pathology Laboratory, University of California, Davis, Davis, CA, United States
- 4Zoological Pathology Program, University of Illinois, Brookfield, IL, United States
- 5Brookfield Zoo Chicago, Brookfield, IL, United States
- 6Department of Medicine and Epidemiology, School of Veterinary Medicine, University of California, Davis, Davis, CA, United States
Introduction: Nannizziopsis guarroi causes mycotic dermatitis, colloquially known as “yellow fungus disease”, in lizards. Several fungal diagnostic assays may detect this microbe in clinical cases, but the clinical performance of these assays has yet to be explored.
Methods: Six adult bearded dragons were topically exposed to N. guarroi along the dorsal midline and serially sampled over five months as clinical cutaneous lesions developed.
Results: The median dates of first test positivity for fungal culture, quantitative polymerase chain reaction (qPCR), and commercially available next-generation sequencing (NGS) were 28, 6, and 17.5 days prior to lesion development, respectively. Lesions did not fluoresce under long-wave ultraviolet illumination. Femoral pores were a site of infection in all four male lizards, representing a novel presentation. Additionally, one lizard developed subclinical granulomatous N. guarroi pneumonia. Postmortem assessment of diagnostics identified a high level of agreement between histopathology, fungal culture, and qPCR; however, cytologic agreement with histopathology was poor.
Discussion: Fungal culture, qPCR, and NGS are appropriate screening tools for the detection of N. guarroi prior to the onset of cutaneous lesions and may be used as diagnostic tools to confirm N. guarroi infection in clinically affected bearded dragons.
Introduction
Nannizziopsis guarroi is an onygenalean fungus that causes dermatomycosis, or nannizziomycosis, in reptiles (Gentry et al., 2021). Cutaneous disease in bearded dragons (Pogona vitticeps) has colloquially been called “yellow fungus disease” (Paré et al., 2020). However, clinical disease does not necessarily result in yellow skin discoloration, and N. guarroi colonies are white in culture (Paré et al., 2020; Foltin and Keller, 2022). Cutaneous nannizziomycosis can vary from yellow-to-brown skin discoloration to severe erosive, necrotizing dermatitis (Paré and Sigler, 2016; Paré et al., 2020; Gentry et al., 2021). Deeper lesions may affect underlying muscle and bone, and systemic nannizziomycosis has been reported as intracoelomic granulomas in the lung, liver, and body wall (Paré and Sigler, 2016; Paré et al., 2020; Tournade et al., 2021; Ayers et al., 2022). Systemic N. guarroi has been reported in the absence of cutaneous lesions and generally results in a poor prognosis (Ayers et al., 2022; Schilliger et al., 2023).
There are many diagnostic assays available that may detect fungus as a component of disease manifestations and others that can specifically identify N. guarroi. In culture, N. guarroi cannot be diagnosed based on morphology alone due to microscopic and gross colony morphology similarities to other species of Nannizziopsis and Parannizziopsis (Sigler et al., 2013; Stchigel et al., 2013). Diagnostic tests including cytology, mycological culture, histopathology, long-wave ultraviolet (UV) fluorescence (Wood’s lamp), and panfungal polymerase chain reaction (PCR) assays can screen for the presence of fungus but do not provide definitive genus or species information (Czurda and Lion, 2017; Kain, 2017; White and Barnes, 2017; Willinger, 2017; Wellehan and Divers, 2019; Dyer and Foy, 2022). Fungal speciation requires molecular sequencing or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS); the latter has not been validated for the detection of N. guarroi, but its use has been reported (Bader, 2017; Lackner and Lass-Flörl, 2017; Schneider et al., 2018; Wickes and Wiederhold, 2018; Jiang et al., 2022). An N. guarroi quantitative PCR (qPCR) assay is offered at the authors’ institution and has been utilized in published cases of N. guarroi in lizards (Paré et al., 2020; Tournade et al., 2021; Foltin and Keller, 2022). Next-generation sequencing (NGS) is becoming more frequent in clinical practice as a speciation tool and has the potential to identify genomic antifungal resistance markers (Jiang et al., 2022; Damerum et al., 2023). This study sought to assess the diagnostic performance of several fungal detection assays for the detection of N. guarroi, which has not yet been investigated.
This prospective study experimentally infected bearded dragons with N. guarroi to evaluate the diagnostic performance of fungal culture, qPCR, a commercial NGS assay, and UV fluorescence throughout the clinical course of infection. It was hypothesized that molecular diagnostics (qPCR, NGS) would detect N. guarroi earlier than fungal culture or UV fluorescence.
Materials and methods
Animals
This study was performed with the approval of the University of Illinois Institutional Animal Care and Use Committee (No. 22209). Six adult bearded dragons (Pogona vitticeps; four males and two females) maintained as a research colony were enrolled. The bearded dragons, maintained at the same institution for the prior 4 years, were 5 years of age and weighed 0.29–0.40 kg (0.63–0.88 lbs). They were deemed healthy based on known history and evaluation (physical examination, complete blood count, and fecal analysis) performed 14 days prior to initial N. guarroi exposure. Each lizard had a negative N. guarroi qPCR from a cutaneous swab 14 days prior to the study start.
Lizards were individually housed on newspaper in 91.45 × 71.12 × 45.72-cm enclosures located in the same room for the entirety of the study. Enclosures were spot cleaned daily and deep cleaned weekly with warm soapy water. Room ambient temperature was maintained between 23.3°C and 24.4°C (74.0°F and 76.0°F). Each enclosure contained a heat lamp and UVA/UVB-emitting bulb on for 12 h/day, allowing a basking temperature of 35.0°C–37.8°C (95.0°F–100.0°F). Light from the heat lamp and UVA/UVB-emitting bulbs (ZooMed ReptiSun 10.0 High Output UVB Fluorescent bulb) overlapped at the basking site.
Fungal isolate
An isolate of N. guarroi, obtained from a bearded dragon with naturally occurring dermatomycosis, was utilized in this study (Lab ID: CC1; GenBank No. MT506461). The bearded dragon was euthanized after the development of deep cutaneous lesions histopathologically consistent with cutaneous mycosis. Postmortem fungal culture of cutaneous swabs produced a pure isolate of white fluffy colonies. The initial isolate identity was confirmed molecularly based on prior published methods for other N. guarroi isolates (McEntire et al., 2022). PCR targets for identity confirmation included the D1/D2 domains of the 28S ribosomal DNA (D1/D2 rDNA) (Cano et al., 2004), the 18S–28S ribosomal internal transcribed spacer region including the 5.8S rDNA (ITS) (primers ITS4 and ITS5) (White et al., 1990), the actin gene (Voigt and Wöstemeyer, 2000), and the β-tubulin gene (primers BT2-F and BT2-R) (Gilgado et al., 2005). PCR products were electrophoresed on a 1% agarose gel, then treated with ExoSAP-IT (USB Corporation, Cleveland, OH, USA) and bidirectionally sequenced (ACGT, Wheeling, IL, USA). Chromatograms were visually inspected and trimmed of primer sequences and low-quality base pair calls. The resulting sequences were compared to those in the NCBI GenBank database using BLASTN. The 601-bp ITS product was 100% identical to N. guarroi (OW988362.1, MW617063.1). Similarly, a 573-bp D1/D2 product was 100% identical to N. guarroi (MH874904.1, HF547864.1). The 872-bp actin product was 100% identical to one N. guarroi sequence (HF547897.1). The 429-bp β-tubulin product was 99.75% identical to N. guarroi (HF547900.1, HF547896.1). Multilocus sequencing confirmed the fungal isolate as N. guarroi.
The isolate was propagated on potato dextrose agar (Remel, Thermo Scientific, Lenexa, KS, USA) and incubated under ambient lighting at 23°C (73.4°F) for at least 10 days. A swab of the isolate was suspended in sterilized distilled water. The suspension was vortexed for 2 min and then filtered through glass wool under vacuum. The number of conidia in suspension was estimated by hemocytometer count.
Fungal exposure
For the initial exposure event, a conidial concentration of 7.0919 × 104 conidia/μL was used, aligning with another N. guarroi experimental infection study (Gentry et al., 2021). Conidial suspension (10 μL) was applied to five unabraded sites along the dorsal midline and allowed to dry completely. After failure of clinical lesion formation within 31 days postinitial exposure (DPIE), as reported by Gentry et al. (2021), repeat exposures were performed on days 36, 64, and 99 DPIE if each lizard had not developed clinical cutaneous lesions. During additional exposure events, the highest concentration that could be harvested from the fungal mats grown in culture was utilized (1.12 × 105 conidia/μL), and 20 μL per site of this conidial suspension was applied to the same five dorsal midline sites.
Antemortem sample collection and processing
Physical examination, cutaneous sample collection, lesion photography, and screening with Wood’s lamp fluorescence were performed weekly. On dates corresponding with exposure events, data collection procedures were performed prior to exposure. Bearded dragons were inspected for development of cutaneous lesions consistent with those described in published cases of N. guarroi infection (Gentry et al., 2021) and photographed if present. Given the absence of a case definition for nannizziomycosis, any dermatologic abnormality was documented and imaged. Suspicious lesions were intermittently sampled by swab or impression smear and routinely stained with Wright–Giemsa (HemaTek 3000, Siemens, Tarrytown, NY, USA) for cytologic evaluation (Sharkey et al., 2014). Microscopic cytologic evaluation of these samples was performed by a board-certified veterinary clinical pathologist to assess for the presence of inflammation and/or fungal elements.
Cutaneous swabs were collected for NGS, fungal culture, and qPCR, in that order. During the preclinical phase, when no lesions were present, whole-body samples were collected by vigorously rubbing swabs 20–30 times each along the dorsal and ventral surfaces of the lizard. After the development of suggestive cutaneous lesions, samples were also obtained from the suspect lesions using the same swabs after collecting the whole-body sample.
Samples collected for NGS used a sterile DNA-free swab (HydraFlock®, Puritan® Cat. No. 25-3406-H, Guilford, ME, USA) and a sterile sample collection tube prefilled with a DNA/RNA preservative (DNA/RNA Shield™ Zymo Research Corp.; Cat. No. R1108, Irvine, CA, USA). Collection was performed with sterile gloves to limit inadvertent sampling of human microbiota. Samples were shipped in batches to a commercial laboratory (MiDog LLC Science, Irvine, CA, USA), which amplifies fungal DNA through ITS-2 targets as previously described in the literature (Tang et al., 2020). NGS results were reported as positive or negative for the detection of Nannizziopsis spp. DNA.
Samples for fungal culture were collected using sterile cotton-tipped applicators (CTAs; Fisherbrand™ Plastic Handled Cotton Swabs and Applicators, Fisher Scientific, Waltham, MA, USA) that were stored in sterile bags without media. Within 2 h of collection, swabs were directly streaked onto a modified agar composed of potato dextrose agar (Remel, Thermo Scientific, Lenexa, KS, USA), penicillin–streptomycin (60,000/60 U/L; Fisher Scientific), gentamicin sulfate (50 mg/L; Sigma-Aldrich, St. Louis, MO, USA), and cycloheximide (400 mg/L; Sigma-Aldrich, St Louis, MO, USA) to reduce bacterial and saprophytic fungal growth, then incubated under ambient lighting at 23°C (73.4°F) for 10–14 days. Fungal culture results were recorded as positive if growth was present that was morphologically consistent with the original pure N. guarroi isolate (white fluffy colonies with microscopic hyphal structures), or negative if no growth consistent with N. guarroi was observed.
Three cutaneous swabs for qPCR testing were collected with sterile CTAs and stored at − 80°C (− 112°F) until collective batched analysis, when an arbitrarily chosen single swab from each time point was analyzed. A commercial nucleic acid purification kit (QIAamp DNA Mini-Kit, Qiagen Inc., Valencia, CA, USA) was used for DNA extraction; the manufacturer’s recommendations were followed, with the addition of a 1-h incubation at 37°C (98.6°F) with 300 U of lyticase (Sigma-Aldrich, St. Louis, MO, USA) prior to the lysis step. Extracted DNA samples were assessed for concentration (measured in nanograms per microliter) and quality (using the ratio of absorbance at 260 to 280 nm) using spectrophotometry (Nanodrop1000, ThermoFisher Scientific, Wilmington, DE, USA), then stored at – 20°C prior to analysis.
A TaqMan qPCR assay (NGEF1_234), targeting a 57-base pair segment of the N. guarroi elongation factor 1 gene (Keller et al., 2022), was performed on extracted DNA. Real-time qPCR was performed using a real-time PCR thermocycler (QuantStudio 3 real-time PCR system; Applied Biosystems, Foster City, CA, USA), and data were analyzed using QuantStudio Design and Analysis Software v.1.5.2 (Applied Biosystems). Each qPCR reaction contained 12.5 μL of 20× TaqMan Platinum PCR Supermix-UDG with ROX (TaqMan Platinum PCR Supermix-UDG with ROX, Invitrogen, Carlsbad, CA, USA), 1.25 μL of 20× TaqMan primer-probe (final concentrations in each PCR reaction: 900 nM forward primer, 900 nM reverse primer, and 250 nM probe), 2.5 μL of plasmid dilution (or nontemplate control: sterile deionized water), and water to a final volume of 25 μL. Cycling parameters were as follows: one cycle at 50°C for 2 min followed by 95°C for 10 min, 40 cycles at 95°C for 15 s and 60°C for 60 s, and a final cycle of 72°C for 10 min. Samples, standard curves, and nontemplate controls were assayed using three technical replicates unless otherwise specified. Positive controls (plasmid-derived standard curves from 1 × 107 to 1 × 100 target copies per reaction) and nontemplate controls were included in all qPCR runs. Mean fungal quantities (measured as copies per ng DNA) were standardized to the total quantity of DNA in each sample by dividing the mean copies/μL of each sample by the DNA concentration. Samples were considered positive if at least two of the three technical replicates amplified with a CT value within 0.3 units of the other positive replicate(s) (Laing et al., 2019). Samples with CT values greater than the CT value for the 1 × 100 standard dilution were considered negative.
Evaluation of long-wave UV fluorescence using a Wood’s lamp (UV502 UV Light With Magnifier, Burton Medical, Addison, IL, USA) was performed weekly. The bearded dragons were manually restrained for dermal illumination in a dark room, and photographs of their dorsal and ventral surfaces were obtained. UV fluorescence of cutaneous lesions was recorded as positive or negative by a single investigator.
Postmortem sample collection and processing
Four days after clinical disease manifested in the final lizard, animals were euthanized on 152 DPIE with pentobarbital (500 mg/kg, transmucosal; EUTHASOL®, Virbac AH Inc., Carros, France) (Wong et al., 2024). The next day, up to three cutaneous lesions were arbitrarily identified and sampled from each lizard, representing the most severe lesions that were previously cytologically confirmed and/or suspected to be dermatomycosis lesions. Cutaneous samples for fungal culture, qPCR, and cytology were collected from each lesion, and UV fluorescence was assessed. The ventral coelom of each lizard was entered, and coelomic organs were evaluated for pathologic changes before the entire carcass was immersed in 10% neutral-buffered formalin for fixation and submitted for evaluation by a board-certified veterinary anatomic pathologist.
Statistical analysis
A time zero was assigned to each bearded dragon on the date the first lesion grossly consistent with nannizziomycosis (Gentry et al., 2021) was noted, and the presence of fungal elements was confirmed cytologically. After cytologic confirmation, each lesion was tracked retroactively using weekly images previously taken to assess for prior presence, and an earlier date of lesion development was assigned if appropriate. The date of the first positive diagnostic result was then assigned for fungal culture, qPCR, NGS, and UV fluorescence in relation to time zero of the date of lesion development.
Although samples were collected weekly, the unexpectedly prolonged time to clinical infection compared to the literature (Gentry et al., 2021) necessitated a reevaluation of data analysis. Thus, samples representing monthly time points throughout the study were submitted for NGS and qPCR to better represent the longitudinal nature of the dataset. Cutaneous samples for qPCR were analyzed from preexposure and at 8, 36, 64, 93, 120, and 148 DPIE, while samples for NGS were run from preexposure and at 8, 36, 64, 93, and 120 DPIE. Fungal cultures and UV fluorescence were performed weekly. Antemortem and postmortem diagnostic testing results are presented descriptively. All descriptive analyses were performed using commercial software (Prism, GraphPad version 10.0.0 [RRID: SCR_002798] or Excel, Microsoft for Mac version 16.86 [RRID: SCR_016137]).
Results
Clinical signs
All six bearded dragons required re-inoculation of N. guarroi conidial suspension on 36 and 64 DPIE, and four bearded dragons required re-inoculation on 99 DPIE. Cutaneous lesions consistent with N. guarroi infection developed in all lizards, with the first lesion appearing at a median of 99 DPIE (range: 78–120 DPIE). Three of six lizards (Animal IDs: C, E, F) had cytologic confirmation of the presence of fungal elements (hyphae and/or conidia) in a cutaneous lesion at the time of lesion detection, while three (Animal IDs: B, G, J) had cytologic evidence of fungal elements lagging by 1, 2, or 3 weeks after the lesion became clinically apparent. In this latter cohort, the date of disease was assigned as the first date that a lesion was grossly apparent retrospectively on photographs. Cytologically, Wright–Giemsa stain identified fungal elements that appeared as either septate, branching hyphae or solitary conidia, accompanied by variable inflammatory cells and necrotic debris (Figure 1), consistent with nannizziomycosis (Le Donne et al., 2016).
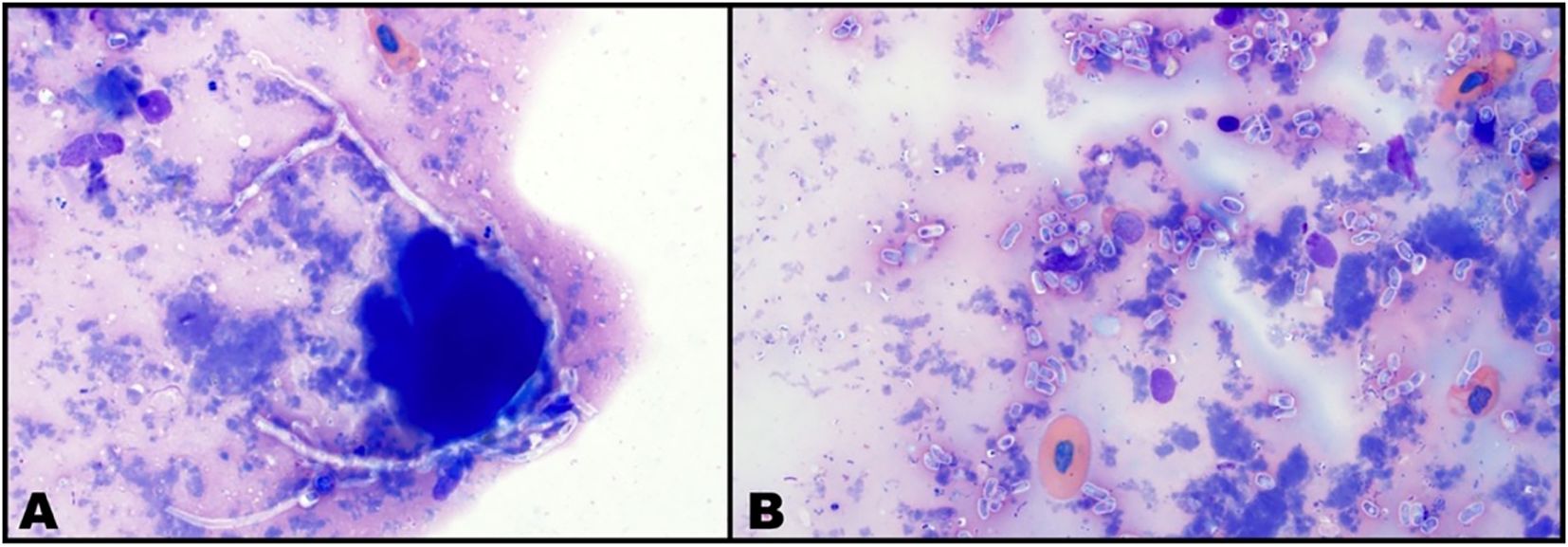
Figure 1. Femoral pore cytology from a male bearded dragon (Pogona vitticeps). Wright–Giemsa stain, × 1,000 magnification. (A) Septate hyphae consistent with Nannizziopsis guarroi and amorphous background material consistent with necrosis. Hyphae are nonstaining to lightly basophilic, occasionally exhibiting undulating contour and right-angle branching (B). Abundant solitary ovoid to cylindrical conidia consistent with N. guarroi and necrotic debris.
No bearded dragon developed lesions at the site of N. guarroi exposure (dorsal midline). The first cutaneous lesions in the four male bearded dragons (Animal IDs: B, C, E, F) were one to three swollen femoral pores per lizard (Figure 2A). Throughout the study, affected femoral pores developed soft tissue swelling, erythema, and eventually scabs. Infected femoral pores readily exfoliated as thick, waxy plugs and frequently resulted in open wounds (Figure 3A). Most male bearded dragons (three of four, 75%; Animal IDs: B, C, F) also developed yellow, diffuse discoloration on the extremities. Initial lesions in the two female lizards (Animal IDs: G, J) were one to two discolored (yellow or black) lesions lateralized on the dorsal or ventral body surfaces (Figures 3B, C). Neither female bearded dragon developed lesions of the femoral pores.

Figure 2. Postmortem images of two male bearded dragons (Pogona vitticeps). (A) Multiple dark, crusted femoral pores (arrowheads) surrounded by yellow discoloration. Three femoral pores contained intracorneal fungal hyphae and conidia. (B) Yellow discoloration (arrow) of the dorsal aspect of the right antebrachium. No fungal elements were identified via histopathology.
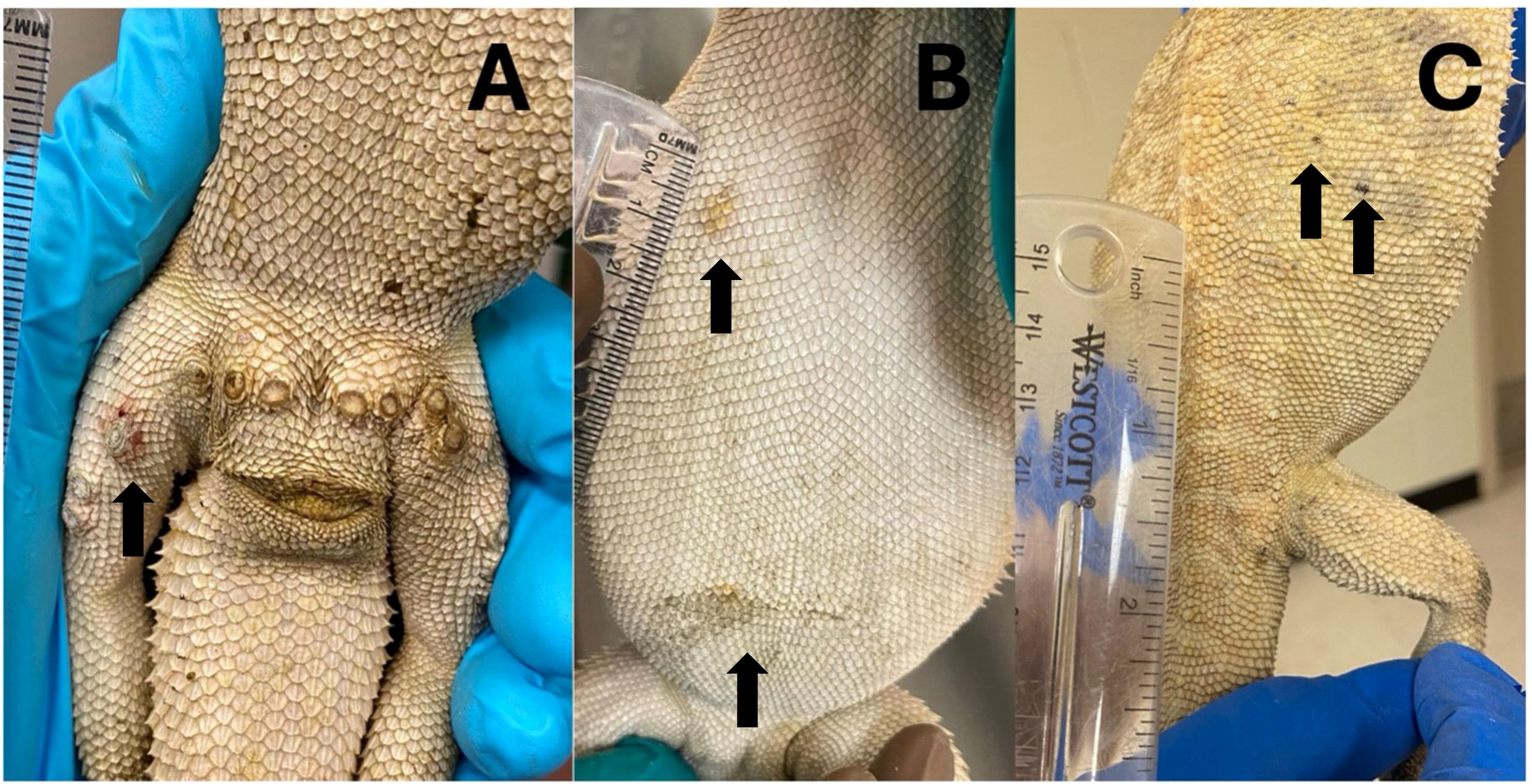
Figure 3. First cutaneous lesions in bearded dragons (Pogona vitticeps) after experimental exposure to Nannizziopsis guarroi. (A) A male bearded dragon (Animal ID: E) on 78 days post initial exposure (DPIE). The fourth lateral right femoral pore is swollen and red. (B) A female bearded dragon (Animal ID: J) with two sites of ventral discoloration on 99 DPIE. (C) A female bearded dragon (Animal ID: G) with multifocal pinpoint grey to black crusts over the right caudal dorsum and caudal to the right pelvic limb on 120 DPIE.
All bearded dragons maintained regular urofecal output and consistency throughout the study. Four lizards (66.67%; Animal IDs: B, F, G, J) had variable appetites that resulted in more than 5% weight loss over two consecutive weeks. Reduced appetite and weight loss were transient and responded to tong feeding and increased diet rations. Body weights at the end of the study ranged from 0.31 to 0.39 kg (0.67–0.85 lbs.). Based on coelomic palpation, one female (Animal ID: G) underwent folliculogenesis after the third exposure event (64 DPIE), then resorbed the follicles after the fourth exposure event (99 DPIE) but before developing cutaneous lesions. During the last month of the study, daily analgesia with meloxicam (Metacam®, Boehringer Ingelheim, Ingelheim am Rhein, Germany; dose: 0.2–0.4 mg/kg PO q24h) was initiated in one male lizard (Animal ID: B) due to perceived discomfort associated with the infected femoral pores. Perceived pain behaviors (abnormal body posture at rest, lethargy) in this lizard ceased once meloxicam was regularly instituted.
Antemortem diagnostic testing
Prior to inoculation, the fungal culture, NGS, and qPCR for each of the bearded dragons was negative. Antemortem, all lizards tested positive for N. guarroi via fungal culture, NGS, and qPCR (Supplementary Table S1). Considering the imbalanced design of antemortem diagnostic testing, with some diagnostics being evaluated weekly (fungal culture and UV fluorescence) and others monthly (qPCR and NGS), paired with intermittent contamination of agar plates that rendered some findings inconclusive, statistical comparisons were not performed. No distinct UV fluorescence was observed at cutaneous lesions or at sites that eventually developed cutaneous lesions via Wood’s lamp examination at any time point. Fluorescence was noted in uninfected male femoral pores, urates, and body sites undergoing ecdysis (Supplementary Figure S1).
Fungal culture was positive for N. guarroi growth before the development of cutaneous lesions in all bearded dragons (Supplementary Table S1). The first positive culture occurred a median of 28 days prior to lesion development, ranging from 50 to 7 days prior. Of the 126 fungal cultures, representing weekly cultures from six lizards, 11 (8.7%) exhibited plate contamination that prevented interpretation. When these contaminated plates are excluded from the dataset, all subsequent weekly cultures were positive for three lizards following their initial positive culture results. One lizard (Animal ID: G) had two intermittent negative cultures following its initial positive, and another (Animal ID: E) had a single negative culture following its initial positive. The remaining individual (Animal ID: C), which had the earliest initial positive culture result (50 days prior to lesion development), exhibited intermittently positive fungal cultures, and for the last five weekly cultures exhibited negative fungal culture growth.
The first positive NGS result occurred before lesion development in five lizards, while it occurred after lesion development in one lizard (Animal ID: E). The first NGS-positive result occurred at a median of 17.5 days prior to lesion development, with a range of 112 days prior to 15 days after lesion development. Five lizards remained consistently NGS positive after their first positive result (100% positive after initial positive). One lizard (Animal ID: G) had a single positive result at 8 DPIE, 112 days prior to developing lesions, then had two negative NGS before testing positive once more, 27 days prior to lesion development. Subsequent NGS testing in this lizard was positive.
The first qPCR-positive result occurred before lesion development in four lizards, on the same date as lesion development in one lizard (Animal ID: C), and after lesion development in another lizard (Animal ID: E). The first qPCR-positive result occurred at a median of 6 days prior to lesion development (range: 27 days preceding to 15 days after lesions developed). Four lizards remained consistently qPCR positive after their first positive. One lizard (Animal ID: F) had a single intermittent negative result following the initial positive, and the final lizard (Animal ID: C) tested qPCR positive only once, then was negative after that time. Of the 15 positive antemortem qPCR samples, copies per reaction ranged from 5.4 to 355.2 with a median of 71.3 and copies per nanogram of DNA ranged from 0.04 to 15 with a median of 0.8 (Supplementary Figure S2).
Postmortem diagnostic testing
Five of six bearded dragons (Animal IDs: B, C, E, F, J) had lesions on their ventral body surfaces, ranging from one to four cutaneous lesions per lizard. Three of the six bearded dragons (Animal IDs: C, F, G) had lesions on their dorsal body surfaces, ranging from one to three lesions per lizard. All male bearded dragons (Animal IDs: B, C, E, F) had lesions associated with their femoral pores, while no female lizard exhibited a femoral pore lesion. While many of the cutaneous lesions exhibited yellow discoloration of the lesion and the surrounding skin, several lesions were darkly colored and showed no yellow discoloration. Histopathology and additional postmortem diagnostic tests (cytology, culture, qPCR, Wood’s lamp) were performed on three arbitrarily chosen cutaneous lesions in four individuals, and on two cutaneous lesions from two individuals with only two cutaneous lesions, for a total of 16 cutaneous lesions (Table 1).
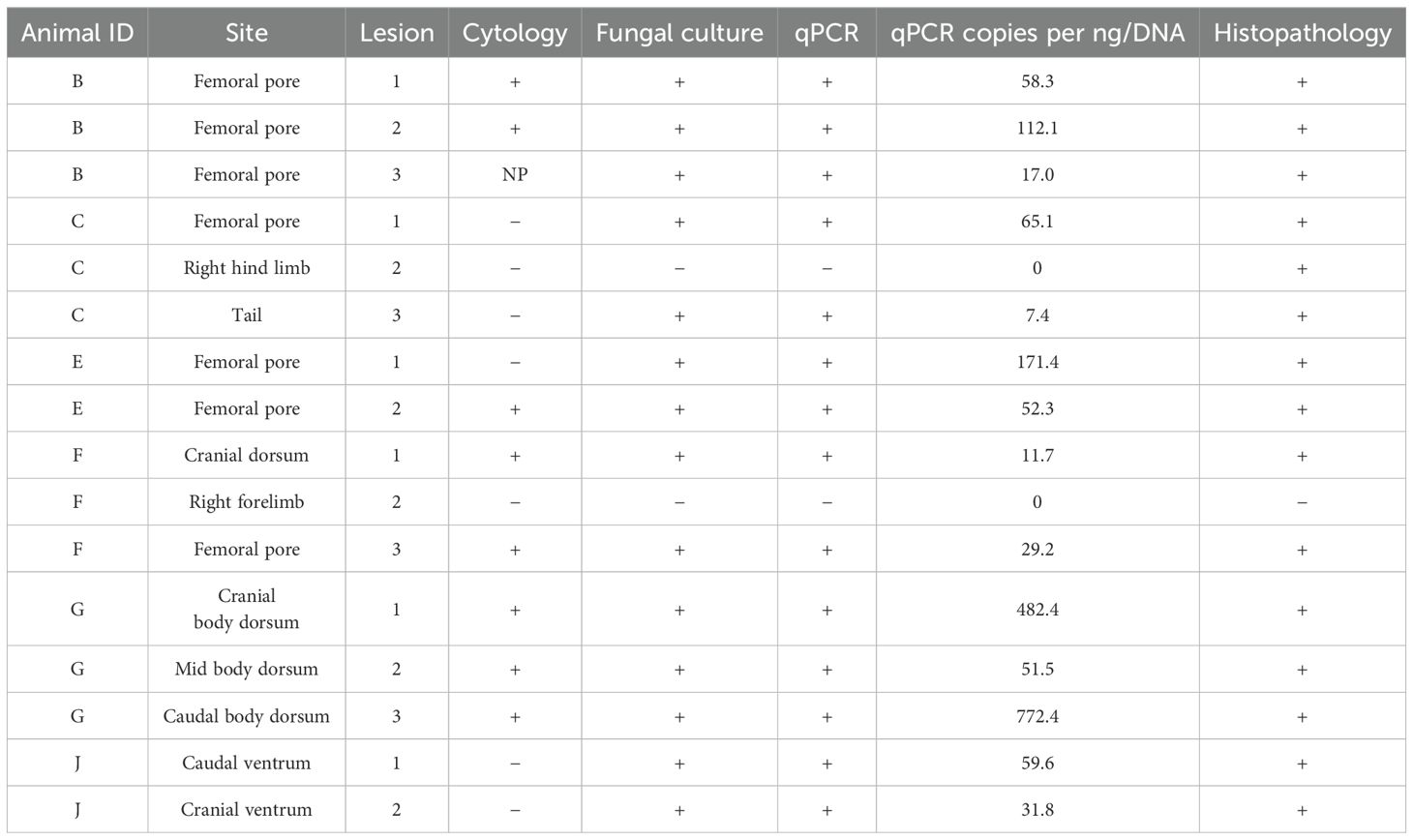
Table 1. Postmortem diagnostic results for lesions from six bearded dragons (Pogona vitticeps) experimentally exposed to Nannizziopsis guarroi and euthanized.
Lesion cytology was performed on 15/16 (93.75%) lesions. Cytology was not performed on one lesion due to its extremely small size, lack of easily collected exudate, and a desire to preserve tissue architecture for histologic analysis. Cytology agreed with histopathology regarding the presence or absence of fungal elements in eight of 15 (53.3%) of the lesions. Of the seven lesions with cytologically detectable fungi, five had concurrent nonseptic inflammation and one had septic bacterial inflammation. Of the eight cytologically negative lesions, two exhibited nonseptic inflammation, and one showed septic bacterial inflammation.
Fungal culture and qPCR were assessed in all 16 lesions and agreed with histopathology in 15 (93.8%) lesions. Fungal culture and qPCR agreed with each other 100% of the time. qPCR values, expressed as copies per nanogram of DNA, ranged from 0 to 772.4, with a median of 51.9. No lesions exhibited UV fluorescence under Wood’s lamp illumination.
Six of 15 (37.50%) for which all diagnostics were performed showed complete positive agreement among assays in the detection of N. guarroi (Table 1). In the one lesion for which cytology was not performed, fungal culture, qPCR, and histopathology were in positive agreement. Among the 15 histopathologically positive lesions, cytology was the diagnostic test most commonly in disagreement, with eight of 14 (57.14%) cytology results testing negative. This was followed by fungal culture and qPCR, which were both negative for a single lesion (Animal ID: C, lesion 2). This cutaneous lesion, negative on all diagnostic assays outside of histopathology, corresponded to a flat, firm area just caudal to a femoral pore that oozed orange, malodorous, thick, opaque material when sampled. The cutaneous lesion that was histopathologically negative originated from a yellow-discolored region of the dorsal right antebrachium and demonstrated mild regional hyperkeratosis without infectious agents (Figure 2B; Animal ID: F, lesion 2). Clinically, this lesion lacked scale and was historically the site of a healed bite wound. The site was sampled due to the yellow discoloration that had developed during the infection trial.
Histopathology of cutaneous lesions, stained with hematoxylin and eosin, identified chronic proliferative or erosive dermatitis. All lesions but one (15/16, 93.75%; Figure 2B) were confirmed to have intracorneal fungal hyphae and/or conidia, as confirmed by Grocott–Gomori Methenamine Silver (GMS) stain (Figures 4A, B) (Grocott, 1955). Seven of the 15 histopathologically positive lesions were of femoral pores, five of which had deeply associated subcutaneous or intramuscular granulomas in addition to fungal structures and dermatitis (Figures 4C–F). Subcutaneous or intramuscular granulomas were present in three additional cutaneous lesions from other body sites (ventral hind limb, dorsal interscapular region, and ventral caudal coelom). Staining with GMS identified rare hyphal structures within all deep granulomas (Figures 4C–F), except for one granuloma associated with a femoral pore (seven of eight, 87.5%).
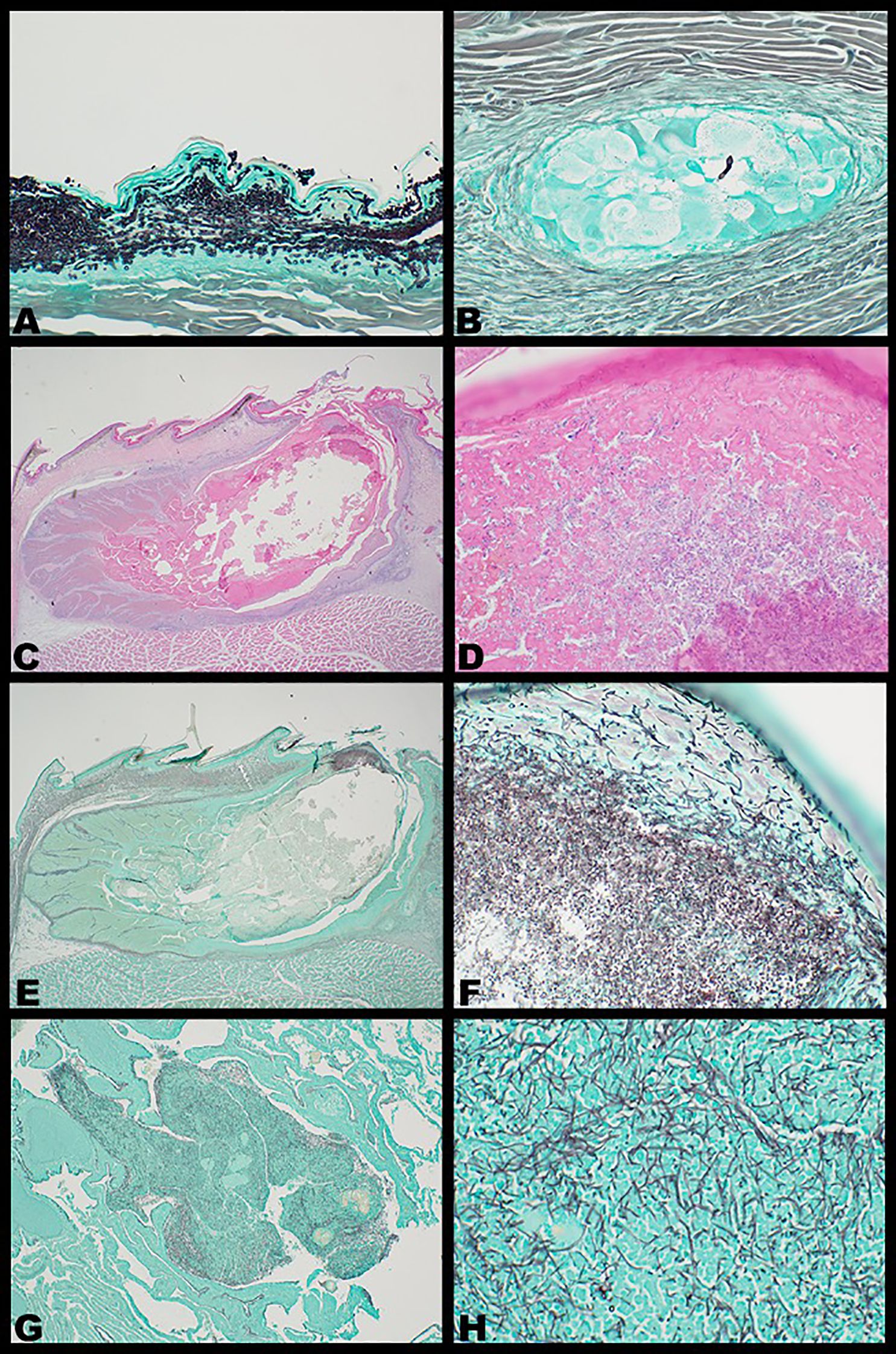
Figure 4. Representative photomicrographs of intralesional Nannizziopsis guarroi in experimentally infected bearded dragons (Pogona vitticeps) stained with hematoxylin and eosin (C, D) or Grocott methenamine silver (GMS) stain (A, B, E–H). Fungi appear black in color under GMS staining. (A) Chronic erosive dermatitis with hyperkeratosis and intracorneal fungal hyphae and conidia; × 40 magnification. (B) Fungal hyphae and conidia within a subcutaneous granuloma; × 40 magnification. (C–F) Inspissated femoral pore with intracorneal fungal hyphae and conidia; × 2 (C) and × 40 magnification (D); × 2 (E) and × 40 magnification (F). (G, H) Pulmonary granuloma with intralesional fungal hyphae; × 4 (G) and × 40 magnification (H).
Systemic N. guarroi infection was identified in one lizard (one of six, 16.67%; Animal ID: B), which had a pinpoint white lesion grossly apparent in the left cranial lung that corresponded to focal chronic granulomatous pneumonia with intralesional fungal hyphae (Figures 4G, H). GMS staining identified fungal structures, and N. guarroi qPCR from this site was positive. Grossly, two bearded dragons (Animal IDs: G, J) had mild to moderate, clear-to-yellow-colored coelomic fluid that was cytologically devoid of infectious agents or neoplastic cells and was of low cellularity. These were both females with resorbing and atretic ovarian follicles.
Discussion
Experimental infection of adult bearded dragons with N. guarroi was used to assess the performance of several diagnostic methods for fungal detection. Our results demonstrate that fungal culture, qPCR, and NGS can detect N. guarroi prior to the onset of cutaneous lesions, but lesions did not fluoresce under long-wave UV illumination.
Our infection model was based on Gentry et al. (2021), which fulfilled Koch’s postulates criteria for nannizziomycosis. In that study, juvenile bearded dragons topically inoculated with a conidial suspension at a dose of approximately two million conidia per lizard developed cutaneous lesions within 31 days; lesions developed on both abraded and unabraded sites (Gentry et al., 2021). In our study, unabraded adult bearded dragons were exposed to a greater initial dose of approximately 3.5 million conidia per lizard but failed to develop cutaneous lesions within the first 5 weeks. This prompted repeated, increased conidia exposure events (each additional exposure approximating 11.2 million conidia per lizard) to encourage disease manifestation. It was not until 11 weeks into the study (78 DPIE) and three exposure events that the first clinical lesion developed. This extended timeline of lesion development was unexpected, and the prolonged study length required alterations in the frequency of cutaneous swab analyses: although weekly analysis of all diagnostics was initially planned, submission of qPCR and NGS at monthly time points was elected to best represent the longitudinal dataset.
Possible explanations for the differences between our study and that of Gentry et al. (2021) may lie in the study design. Our study utilized a different isolate of N. guarroi, and variable pathogenicity may exist between strains, as has been identified in other fungal organisms such as Aspergillus fumigatus (Rokas, 2022). Additionally, our study animals were apparently healthy adult bearded dragons, while the prior study utilized juvenile lizards, which may have resulted in differences in intrinsic immunocompetence. The immunocompetence of our study animals may have also differed due to the provision of artificial UVB lighting, since vitamin D is a selective regulator of the immune system (Vergneau-Grosset and Péron, 2020). The effects of UV radiation on reptile immunity have been investigated in broad-snouted caimans (Caiman latirostris) and green anoles (Anolis carolinensis) with mixed results: while caimans exposed to artificial UV demonstrated reduced innate immune activity, anoles were resistant to UV radiation-induced immunosuppression (Cope et al., 2001; Siroski et al., 2012). Finally, our study utilized different environmental temperatures. While Gentry et al. (2021) provided environmental temperatures without reporting a heat gradient to optimize fungal growth (30°C to 35°C), our study provided the bearded dragons with basking temperatures that exceeded the high end of the optimal fungal growth range (> 35°C) to meet the bearded dragon’s preferred optimum temperature zone (Raiti, 2012). Regardless of the cause, the unexpected challenges encountered during our infection trial indicate that future studies striving to replicate experimental N. guarroi infection of bearded dragons should endeavor to minimize differences in methodology where possible.
Another unexpected finding was the location of N. guarroi lesion development. Gentry et al. (2021) reported yellow-to-dark lesions at most exposed sites, in addition to occasional nonexposed sites. In our study, no bearded dragon developed cutaneous lesions along the dorsal midline where conidial suspension was directly applied. Five of six lizards developed lesions on their ventral surfaces, and only three developed lesions on their dorsal surfaces. This may indicate environmental presence of N. guarroi being seeded onto the ventral surfaces that are in near-constant contact with the substrate. Seeding of N. guarroi from lizards to the substrate likely occurred secondary to ecdysis, which occurred several times in every lizard throughout our infection trial. In our study, weekly environmental cleaning comprised soapy water in enclosures and changing of the newspaper substrate when soiled. This was performed despite recent studies demonstrating that in vitro inactivation of N. guarroi requires diluted commercial bleach or off-label use of a commercial disinfectant containing 5.4% benzalkonium chloride and 0.4% polyhexanide (Jourdan et al., 2023; Seth et al., 2024). Given that the goal of this study was to induce disease to allow for evaluation of diagnostic assays, environmental decontamination with these disinfectants was not performed. It should also be considered that the weekly sampling method—swabbing the cutaneous surfaces of all aspects of the body—likely contributed to the introduction of N. guarroi organisms to distant sites on the body.
The initial cutaneous lesions in all four males presented as infection of the femoral pores, dermatologic structures on the ventral aspect of the pelvic limbs of bearded dragons and other lizards. Initially, the lesions were clinically assessed as being “impacted” and unlikely to be associated with dermatomycosis. However, after lesion progression, exudate from the pores was assessed cytologically and found to contain fungal elements consistent with N. guarroi. Femoral pore involvement has not been reported in association with N. guarroi infection, and impacted femoral pores are considered a common dermatologic finding in male bearded dragons (Sollom and Baron, 2023). The high incidence of femoral pore infection in our study raises concern that impacted femoral pores could be a clinical sign of nannizziomycosis that may be easily overlooked. Neither female lizard developed lesions at this site. Case criteria and detailed lesion descriptions for N. guarroi infection are not currently defined, and the variety of clinical manifestations in our study underscores the importance of further characterizing lesion distribution to aid clinical detection.
Antemortem, limited comparisons can be made between the diagnostic performance of fungal culture, qPCR, and NGS given the unbalanced nature of the ultimate design. While fungal culture had the earliest median time to first positive result before lesion development, this may be skewed by the weekly data available. If NGS and qPCR analyses had occurred weekly, this might have resulted in differences in diagnostic performance and more rapid detection of N. guarroi via these assays. The earliest positive test result was from NGS at 8 DPIE (112 days preceding lesion development); however, this lizard had several subsequent negative NGS results after this first positive result. Assessment of the clinical sensitivity and specificity of these assays would require assay evaluation in both infected and noninfected bearded dragons at the same points, which was not possible with our dataset. Translation of our findings to clinical recommendations should be made in the context of the advantages and disadvantages of each of the diagnostics.
Fungal culture requires the isolation of a viable organism that can propagate on culture media. It has previously been used to diagnose infection with Nannizziopsis spp. in reptiles, with some negative culture results reported in the literature (Paré and Sigler, 2016; Paré et al., 2020; Tournade et al., 2021). This aligns with our findings that, even after developing clinical lesions, some lizards had negative culture results. Another drawback of culture is that when used in clinical cases, it must be followed by molecular and/or proteomic techniques to identify genus and species, given that colony morphology similarities are shared with other fungi (Paré and Sigler, 2016; Paré et al., 2020). As fungal culture results were positive in all lizards prior to the onset of lesion development, this assay may be considered for screening lizards for N. guarroi cutaneous carriage, as well as for use as a diagnostic tool in the detection of N. guarroi in clinically affected bearded dragons.
Both qPCR and NGS detect DNA, meaning they can detect both viable and nonviable organisms. The qPCR assay used in this study is not validated at this time and has limited availability; however, it has been used to diagnose nannizziomycosis in lizards (Tournade et al., 2021; Foltin and Keller, 2022). The use of NGS has been increasing in veterinary medicine with the recent commercialization of this product, but it has not yet been shown in the literature to detect N. guarroi. In this study, both qPCR and NGS performed well and were able to detect N. guarroi DNA prior to the development of cutaneous lesions in most lizards. Both technologies provide quantification of the fungal organisms, which may be beneficial in monitoring treatment progression. Furthermore, NGS may be able to detect additional organism DNA that could be involved in secondary infection of the wound bed. Both assays were able to detect N. guarroi and may be considered for clinical screening and diagnosis of N. guarroi infections.
Long-wave UV illumination with a Wood’s lamp is commonly utilized as a rapid, painless, patient-side screening tool for dermatologic fungal infections (Klatte et al., 2015; Moskaluk and VandeWoude, 2022). Lesions infected with N. guarroi did not fluoresce at any point, making it a poor diagnostic tool for nannizziomycosis. This contrasts with the yellow-green fluorescence emitted by another onygenalean fungus, Ophidiomyces ophidiicola, a cause of cutaneous mycosis in snakes (Vivirito et al., 2021; Lizarraga et al., 2023). This methodology should not be used in the detection or screening of N. guarroi lesions in bearded dragons.
One of the six lizards had evidence of systemic mycosis in the form of fungal pneumonia. While systemic nannizziomycosis has been reported (Paré and Sigler, 2016; Paré et al., 2020; Tournade et al., 2021; Ayers et al., 2022), the pathogenesis behind the spread of the fungus is unknown. This finding highlights the importance of considering systemic implications of N. guarroi in addition to cutaneous lesions. Given these findings, clinicians should be aware that lizards infected with N. guarroi may develop systemic lesions that could alter their prognosis for recovery. In addition to cutaneous diagnostics, systemic diagnostics, such as advanced imaging, should be considered.
Our cohort included both male and female bearded dragons; however, our sample size was not large enough to draw conclusions about the effect of sex and reproductive status on N. guarroi infection. Both female lizards (Animal IDs: G, J) required four exposure events, and the individual (Animal ID: G) whose physical examination findings were consistent with folliculogenesis was the last one to develop clinical infection. The effect of reproductive status on the immune status of bearded dragons is not fully elucidated; however, a study performed in ornate tree lizards (Urosaurus ornatus) found that vitellogenic females demonstrated similar wound healing rates compared to nonreproductive females and suggested that these animals had comparable immune function despite increased physiologic demands (French and Moore, 2008). Further investigations into the effects of vitellogenesis and sex-dependent host immune dynamics are warranted.
Our study was limited by a small sample size and the inability to pursue the same timepoints for every diagnostic test assessed, which precluded statistical comparisons and necessitated a solely descriptive presentation of results. Furthermore, due to the repeated fungal exposures required, infection development could not be clearly linked to a specific exposure event, and cutaneous lesions may have resulted from an accumulation of fungal organisms over time. Given these repeated exposure events, it is possible that the bearded dragons could have survived a single N. guarroi exposure without developing nannizziomycosis, and this represents an interesting question for future research. Lastly, the location and frequency of shedding in the bearded dragons were not recorded during this project, and this information may have been valuable for understanding how recent sheds affect cutaneous positivity status on diagnostic tests.
In conclusion, our findings support that fungal culture, NGS, and qPCR can often successfully detect N. guarroi prior to lesion development. The ability to confirm N. guarroi carriage in bearded dragons through multiple methods before cutaneous lesion manifestation may permit early disease intervention and reduce infection spread. Future studies evaluating natural N. guarroi exposure in bearded dragons are needed, along with published case criteria for nannizziomycosis. Given the experimental design, our study exposed bearded dragons to higher doses of N. guarroi and potentially created higher environmental burdens of the fungus than would be encountered under natural conditions. Additional studies investigating the lowest number of viable fungi required to induce disease are warranted.
Samples were collected within 24 h of euthanasia, at 153 days postinitial exposure. “+” positive; “−” negative; NP, not performed.
Data availability statement
The data presented in the study are deposited in the Dryad repository (https://doi.org/10.5061/dryad.n02v6wx9c).
Ethics statement
The animal study was approved by University of Illinois Champaign Urbana. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AW: Conceptualization, Data curation, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. LA: Visualization, Investigation, Methodology, Writing – review & editing. JD: Writing – review & editing, Investigation. AB: Writing – review & editing, Investigation. MR: Writing – review & editing, Visualization, Investigation. DI: Writing – review & editing, Visualization, Investigation. KT: Conceptualization, Methodology, Writing – review & editing. JR: Writing – review & editing, Methodology, Conceptualization. MA: Methodology, Resources, Writing – review & editing, Conceptualization, Formal analysis. KK: Project administration, Visualization, Funding acquisition, Data curation, Resources, Writing – review & editing, Formal analysis, Supervision, Conceptualization, Methodology, Investigation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Funding for this project included start-up funds for Dr. Keller from both the University of Illinois College of Veterinary Medicine and the University of California Davis School of Veterinary Medicine.
Acknowledgments
The content of this manuscript has previously appeared as a Master’s thesis (Wong, 2024). The authors thank the “Dragon Team” of the Wildlife Epidemiology Laboratory for the daily care and devotion to these bearded dragons: Dr. Salma Abdel Mageed, Lexi Franz, QianQian Huang, Nainoa Kapele, Dr. Nicholas Liszka, Dr. Kelly Loeb, Dr. Paul Orr, Dr. Varun Seth, and Dr. Maural Sowlat. We are also thankful to Amber Simmons and Maris Daleo for their technical assistance and support.
Conflict of interest
KK currently serves on the scientific advisory board for MiDog, LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/famrs.2025.1607686/full#supplementary-material
References
Ayers K. A., Keller K. A., Webb J. K., and Samuelson J. P. (2022). Nannizziopsis guarroi as a cause of multisystemic granulomatous disease with clinical cutaneous lesions in a bearded dragon (Pogona vitticeps). J. Exot Pet Med. 40, 8–9. doi: 10.1053/j.jepm.2021.10.001
Bader O. (2017). “Fungal species identification by MALDI-ToF mass spectrometry,” in Human Fungal Pathogen Identification Methods and Protocols. Ed. Lion T. (Springer, New York, NY), 323–338.
Cano J., Guarro J., and Gené J. (2004). Molecular and morphological identification of Colletotrichum species of clinical interest. J. Clin. Microbiol. 42, 2450–2454. doi: 10.1128/JCM.42.6.2450–2454.2004
Cope R. B., Fabacher D. L., Lieske C., and Miller C. A. (2001). Resistance of a lizard (the green anole, Anolis carolinensis; Polychridae) to ultraviolet radiation–induced immunosuppression. Photochem. Photobiol. 74, 46–54. doi: 10.1562/0031-8655(2001)074<0046:roaltg>2.0.co;2
Czurda S. and Lion T. (2017). “Broad-spectrum molecular detection of fungal nucleic acids by PCR-based amplification techniques,” in Human Fungal Pathogen Identification Methods and Protocols. Ed. Lion T. (Springer, New York, NY), 257–266.
Damerum A., Malka S., Lofgrern N., Vecere G., and Krumbeck J. A. (2023). Next-generation DNA sequencing offers diagnostic advantages over traditional culture testing. Am. J. Vet. Res. 84, 1–9. doi: 10.2460/ajvr.23.03.0054
Dyer J. M. and Foy V. M. (2022). Revealing the unseen: a review of Wood’s lamp in dermatology. J. Clin. Aesthet Dermatol. 15, 25–30. Available at: https://pmc.ncbi.nlm.nih.gov/articles/PMC9239119/
Foltin E. T. and Keller K. A. (2022). Successful treatment of Nannizziopsis guarroi infection using systemic terbinafine in a central bearded dragon (Pogona vitticeps). J. Herpetol Med. Surg. 32, 20–25. doi: 10.5818/JHMS-D-21-00026.2
French S. S. and Moore M. C. (2008). Immune function varies with reproductive stage and context in female and male tree lizards, Urosaurus ornatus. Gen. Comp. Endocrinol. 155, 148–156. doi: 10.1016/j.ygcen.2007.04.007
Gentry S., Lorch J. M., Lankton J. S., and Pringle A. (2021). Koch’s postulates: confirming Nannizziopsis guarroi as the cause of yellow fungus disease in Pogona vitticeps. Mycologia. 113, 1253–1263. doi: 10.1080/00275514.2021.195444
Gilgado F., Cano J., Gené J., and Guarro J. (2005). Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J. Clin. Microbiol. 43, 4930–4942. doi: 10.1128/JCM.43.10.4930-4942.2005
Grocott R. G. (1955). A stain for fungi in tissue sections and smears using Gomori’s methenamine-silver nitrate technic. Am. J. Clin. Pathol. 25, 975–979. doi: 10.1093/ajcp/25.8_ts.0975
Jiang S., Chen Y., Han S., Lv L., and Li L. (2022). Next-generation sequencing applications for the study of fungal pathogens. Microorganisms. 10, 1882. doi: 10.3390/microorganisms10101882
Jourdan B., Hemby C., Allender M. C., Levy I., Foltin E., and Keller K. A. (2023). Effectiveness of common disinfecting agents against isolates of Nannizziopsis guarroi. J. Herpetol Med. Surg. 33, 40–44. doi: 10.5818/JHMS-D-22-00004
Kain R. (2017). “Histopathology,” in Human Fungal Pathogen Identification Methods and Protocols. Ed. Lion T. (Springer, New York, NY), 185–194.
Keller K. A., Allender M. C., Guzman D. S. M., Paul-Murphy J., Byrne B., Raudabaugh D. B., et al. (2022). “Application of a newly developed real-time qPCR assay for the detection of N. guarroi in companion lizards,” in Proceedings of the International Conference on Avian Herpetological and Exotic Mammal Medicine.
Klate K. L., van der Beek N., and Kemperman P. M. J. H. (2015). 100 years of Wood's lamp revised. J Eur Acad Dermatol Venereol. 29(5), 842–7. doi: 10.1111/jdv.12860
Lackner M. and Lass-Flörl C. (2017). “Commercial molecular tests for fungal diagnosis from a practical point of view,” in Human Fungal Pathogen Identification Methods and Protocols. Ed. Lion T. (Springer, New York, NY), 85–106.
Laing O., Halliwell J., and Barbaric I. (2019). Rapid PCR assay for detecting common genetic variants arising in human pluripotent stem cell cultures. Curr. Protoc. Stem Cell Bio. 49, e83. doi: 10.1002/cpsc.83
Le Donne V., Crossland N., Brandão J., Sokolova Y., Fowlkes N., Nevarez J. G., et al. (2016). Nannizziopsis guarroi infection in 2 inland bearded dragons (Pogona vitticeps): clinical, cytologic, histologic, and ultrastructural aspects. Vet. Clin. Pathol. 45, 368–375. doi: 10.1111/vcp.12345
Lizarraga A. J., Hart L., Wright R. M., Williams L. R., and Glavy J. S. (2023). Incidents of snake fungal disease caused by the fungal pathogen Ophiodiomyces ophidiicola in Texas. Front. Fungal Biol. 4. doi: 10.3389/ffunb.2023.1064939
McEntire M. S., Reinhart J. M., Cox S. K., and Keller K. A. (2022). Single-dose pharmacokinetics of orally administered terbinafine in bearded dragons (Pogona vitticeps) and the antifungal susceptibility patterns of Nannizziopsis guarroi. Am. J. Vet. Res. 83, 256–263. doi: 10.2460/ajvr.21.02.0023
Moskaluk A. E. and VandeWoude S. (2022). Current topics in dermatophyte classification and clinical diagnosis. Pathogens. 11, 957. doi: 10.3390/pathogens11090957
Paré J. A. and Sigler L. (2016). An overview of reptile fungal pathogens in the genera Nannizziopsis, Paranannizziopsis, and Ophidiomyces. J. Herpetol Med. Surg. 26, 46–53. doi: 10.5818/1529-9651-26.1-2.46
Paré J. A., Wellehan J., Perry S. M., Scheelings T. F., Keller K., and Boyer T. (2020). Onygenalean dermatomycoses (formerly yellow fungus disease, snake fungal disease) in reptiles. J. Herpetol. 30, 198–209. doi: 10.5818/19-12-221.1
Raiti P. (2012). Husbandry, diseases, and veterinary care of the bearded dragon (Pogona vitticeps). J. Herpetol. 22, 117–131. doi: 10.5818/1529-9651-22.3.117
Rokas A. (2022). Evolution of the human pathogenic lifestyle in fungi. Nat. Microbiol. 7, 607–619. doi: 10.1038/s41564-022-01112-0
Schilliger L., Paillusseau C., François C., and Bonwitt J. (2023). Major emerging fungal diseases of reptiles and amphibians. Pathogens. 12, 429. doi: 10.3390/pathogens12030429
Schneider J., Heydel T., Klasen L., Pees M., Schrödl W., and Schmidt V. (2018). Characterization of Nannizziopsis guarroi with genomic and proteomic analysis in three lizard species. Med. Mycol. 56, 610–620. doi: 10.1093/mmy/myx083
Seth V., Adamovicz L., and Keller K. A. (2024). Benzalkonium chloride and polyhexanide disinfectant (F10 SC) requires off-label use for environmental disinfection to be effective against Nannizziopsis guarroi. J. Exot Pet Med. 49, 1–4. doi: 10.1053/j.jepm.2024.01.005
Sharkey L. C., Seelig D. M., and Overmann J. (2014). All lesions great and small, part 1: Diagnostic cytology in veterinary medicine. Diagn. Cytopathol. 42, 535–543. doi: 10.1002/dc.23097
Sigler L., Hambleton S., and Paré J. A. (2013). Molecular characterization of reptile pathogens currently known as members of the Chrysoporium anamorph of Nannizziopsis vriesii complex and relationship with some human-associated isolates. J. Clin. Microbiol. 51, 3338–3357. doi: 10.1128/JCM.01465-13
Siroski P. A., Poletta G. L., Fernandez L., Ortega H. H., and Merchant M. E. (2012). Ultraviolet radiation on innate immunity and growth of broad-snouted caiman (Caiman latirostris): implications for facilities design. Zoo Biol. 31, 523–533. doi: 10.1002/zoo.20417
Sollom H. J. and Baron H. R. (2023). Clinical presentation and disease prevalence of captive central bearded dragons (Pogona vitticeps) at veterinary clinics in Australia. Aust. Vet. J. 101, 200–207. doi: 10.1111/avj.13234
Stchigel A. M., Sutton D. A., Cano-Lira J. F., Cabañes F. J., Abarca L., Tintelnot K., et al. (2013). Phylogeny of chrysoporia infecting reptiles: proposal of the new family Nannizziopsiaceae and five new species. Persoonia. 31, 86–100. doi: 10.3767/003158513X669698
Tang S., Prem A., Tjokrosurjo J., Sary M., Van Bel M. A., Rodrigues-Hoffman A., et al. (2020). The canine skin and ear microbiome: a comprehensive survey of pathogens implicated in canine skin and ear infections using a novel next-generation-sequencing-based assay. Vet. Microbiol. 247, 108764. doi: 10.1016/j.vetmic.2020.108764
Tournade C. M., Doss G. A., Adamovicz L., Ambar N., Allender M. C., Lennox A. M., et al. (2021). Antemortem diagnosis of Nannizziopsis guarroi fungal pneumonia in a green iguana (Iguana iguana). J. Exot Pet Med. 38, 44–47. doi: 10.1053/j.jepm.2021.04.006
Vergneau-Grosset C. and Péron F. (2020). Effect of ultraviolet radiation on vertebrate animals: update from ethological and medical perspectives. Photochem. Photobiol. 19, 752. doi: 10.1039/c9pp00488b
Vivirito K., Haynes E., Adamovicz L., Wright A., Durante K., Stanford K., et al. (2021). Ultraviolet fluorescence as a field-applicable screening tool for lesions consistent with ophidiomycosis in Lake Erie water snakes (Nerodia spideon insularum). J. Wild Dis. 57, 380–385. doi: 10.7589/JWD-D-20-00013
Voigt K. and Wöstemeyer J. (2000). Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiol. Res. 155, 179–195. doi: 10.1016/S0944-5013(00)80031-2
Wellehan J. F. X. and Divers S. J. (2019). “Mycology,” in Mader’s Reptile and Amphibian Medicine and Surgery. Eds. Divers S. and Stahl S. (Saunders, St. Louis, MO), 270–280.
White P. L. and Barnes R. A. (2017). “Isolation of nucleic acids for fungal diagnosis,” in Human Fungal Pathogen Identification Methods and Protocols. Ed. Lion T. (Springer, New York, NY), 223–248.
White T. J., Bruns T., Lee S., and Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications. Eds. Innis M. A., Gelfand D. H., Sninsky J. J., and White T. L. (Academic Press, San Diego, CA), 315–322.
Wickes B. L. and Wiederhold N. P. (2018). Molecular diagnostics in medical mycology. Nat. Commun. 9, 5135. doi: 10.1038/s41467-018-07556-5
Willinger B. (2017). “Culture-based techniques,” in Human Fungal Pathogen Identification Methods and Protocols. Ed. Lion T. (Springer, New York, NY), 195–208.
Wong A. D. (2024). Evaluation of diagnostic modalities for the detection of Nannizziopsis guarroi in experimentally infected bearded dragons (Pogona vitticeps). University of Illinois Urbana-Champaign, Champaign (IL.
Keywords: bearded dragon, Pogona vitticeps, Nannizziopsis guarroi, dermatomycoses, Onygenales, fungi
Citation: Wong AD, Adamovicz L, Dalen JP, Bender AM, Rosser MF, Imai DM, Terio KA, Reinhart JM, Allender MC and Keller KA (2025) Multiple diagnostic modalities are appropriate for detecting Nannizziopsis guarroi in experimentally infected bearded dragons (Pogona vitticeps). Front. Amphib. Reptile Sci. 3:1607686. doi: 10.3389/famrs.2025.1607686
Received: 07 April 2025; Accepted: 30 July 2025;
Published: 09 September 2025.
Edited by:
Franciscus Scheelings, The University of Melbourne, AustraliaReviewed by:
Ana Cristina Sampaio, Universidade de Trás-os-Montes e Alto Douro, PortugalVolker Schmidt, Leipzig University, Germany
Savannah Gentry, University of Wisconsin-Madison, United States
Copyright © 2025 Wong, Adamovicz, Dalen, Bender, Rosser, Imai, Terio, Reinhart, Allender and Keller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krista A. Keller, a2FrZWxsQHVjZGF2aXMuZWR1
 Amanda D. Wong
Amanda D. Wong Laura Adamovicz
Laura Adamovicz Jacob P. Dalen2
Jacob P. Dalen2 Alexander M. Bender
Alexander M. Bender Michael F. Rosser
Michael F. Rosser Denise M. Imai
Denise M. Imai Karen A. Terio
Karen A. Terio Jennifer M. Reinhart
Jennifer M. Reinhart Matthew C. Allender
Matthew C. Allender Krista A. Keller
Krista A. Keller