- 1Division of Molecular Systems for Brain Function, Medical Institute of Bioregulation, Kyushu University Institute for Advanced Study, Fukuoka, Japan
- 2PRESTO, Japan Science and Technology Agency, Saitama, Japan
Synapses are fundamental units of neurotransmission and play a central role in the formation and function of neural circuits. These dynamic structures exhibit morphological and functional plasticity in response to experience and activity, supporting higher brain functions such as learning, memory, and emotion. Their molecular composition includes diverse membrane-associated and cytoskeletal proteins that mediate intercellular signaling, regulate synaptic plasticity, and maintain structural stability. Disruptions in these protein networks, often referred to as synaptopathies, are closely linked to psychiatric and neurological disorders. Such disruptions commonly manifest as region-specific changes in synapse number, morphology, or signaling efficacy. Although a large number of synaptic proteins have been identified through conventional proteomic approaches, our understanding of synaptic specificity and plasticity remains limited. This is primarily due to insufficient spatial resolution, lack of cell-type specificity, and challenges in applying these methods to intact neural circuits in vivo. Recent advances in proximity labeling techniques such as BioID and APEX can spatial proteomics limiting cell compartments and cell-type. BioID also enables proteomic analysis within synaptic compartments under both physiological and pathological conditions in vivo. These technologies allow unbiased, high-resolution profiling of protein networks in specific synapse types, synaptic clefts, and glial-neuronal interfaces, thereby providing new insights into the molecular basis of synaptic diversity and function. In this short review, we summarize recent developments in synaptic proteomics enabled by proximity labeling. We also discuss how these approaches have advanced our understanding of synapse-specific molecular architecture and their potential to inform the mechanisms of synapse-related brain disorders, as well as the development of targeted diagnostic and therapeutic strategies.
Introduction
Synapses are highly specialized subcellular compartments of neurons and represent the fundamental computational units for neurotransmission. Each neuron connects to thousands of others via asymmetric intercellular junctions composed of presynapses and postsynapses, facilitating continuous signal transmission. In the human brain, approximately 150 trillion synapses form intricate neural circuits across various brain regions. These synapses undergo dynamic morphological and functional plasticity throughout life, influenced by environmental stimuli such as sensory experience and behavioral activity besides to the genetic programs. This remarkable plasticity underpins fundamental brain functions such as learning, memory, and emotion by supporting adaptive modifications in synaptic connectivity and strength. Structurally, synapses are composed by synaptic vesicles, the presynaptic active zone, the synaptic cleft, and the postsynaptic density (PSD), each compartment drives the above brain functions in a coordinated manner. Notably, thousands of distinct synaptic proteins orchestrate the brain functions and the structural of synapses, and define the discrete synaptic properties in the different brain regions (O’Rourke et al., 2012; Koopmans et al., 2019; Van Oostrum et al., 2023). Also, synaptic proteins include cytoskeletal proteins, receptors, neurotransmitters, adhesion molecules and scaffold proteins (O’Rourke et al., 2012; Koopmans et al., 2019), and this molecular diversity exemplifies how synaptic components are not merely structural but actively shape signaling integration, synapse specification, and plasticity. This functional complexity is made possible by the spatially confined and molecularly compartmentalized organization of synapses, which enables precise and localized biochemical processing. Thus, each synapse operates as a self-regulating biochemical microdomain capable of adaptive computation within neural circuits. Conversely, the dysfunction of synaptic protein networks leads to impairments in synapse number, morphology, and signal transmission. Accumulating evidence suggests that such synaptic dysfunctions, which are often referred to as synaptopathies, are closely associated with neurodevelopmental and psychiatric disorders as well as with the progression of neurodegenerative diseases (Grant, 2012; Lepeta et al., 2016; Hindley et al., 2023). Notably, these abnormalities often manifest in specific brain regions. Although large-scale efforts have led to the identification of over 2,000 distinct synaptic proteins through conventional proteomic approaches (Bayés et al., 2011; Loh et al., 2016; Koopmans et al., 2019), the molecular mechanisms governing synaptic specificity, diversity, and plasticity remain incompletely understood. This is due, in part, to limitations in spatial resolution, cell-type specificity, and the ability to analyze intact neural circuits in vivo.
In recent years, emerging proximity labeling (PL)-based proteomic techniques such as BioID (biotin ligase-based), APEX (ascorbate peroxidase) and HRP (horseradish peroxidase) have made it possible to profile the local proteome of synaptic compartments with high spatial resolution in living tissue (Han et al., 2018; Takano and Soderling, 2021; Ito et al., 2024). These techniques have facilitated the discovery of proteomes associated with specific neuronal populations (Uezu et al., 2016), synaptic clefts (Loh et al., 2016; Takano et al., 2020), and tripartite synapses formed by astrocyte-neuron connections (Takano et al., 2020; Takano and Soderling, 2021). In this short review, we highlight recent advances in spatial synaptic proteomics enabled by proximity labeling (PL) technologies and discuss how these approaches have advanced our understanding of the molecular mechanisms underlying synapse formation, diversity, and function. We further introduce current insights into the pathophysiology of synapse-related neurological disorders uncovered through PL-based studies and outline future directions for the therapeutic application of these technologies. By enabling precise profiling of synapse-specific molecular networks, PL-based proteomic approaches offer novel insights into brain function and hold considerable promise for the development of targeted diagnostic and therapeutic strategies for synapse-associated disorders.
Proximity labeling approaches for synaptic protein profiling
Traditionally, synaptic proteins have been identified using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of synaptic vesicles and synaptosomes, purified by differential centrifugation, density-gradient centrifugation, immune-purification and affinity-purification (Fernández et al., 2009; Morciano et al., 2009; Grønborg et al., 2010; Boyken et al., 2013; Wilhelm et al., 2014; Dieterich and Kreutz, 2016; Xu et al., 2021; Kaizuka et al., 2024). While these proteomic approaches have proven valuable for detecting synaptic proteins enriched in cultured neurons and brain tissues, they are limited by low spatial resolution and contamination from heterogeneous mixtures derived from multiple synapse types. These limitations hinder the ability to resolve the molecular characteristics of specific cell types, synapse subtypes, synaptic clefts, and tripartite synapses.
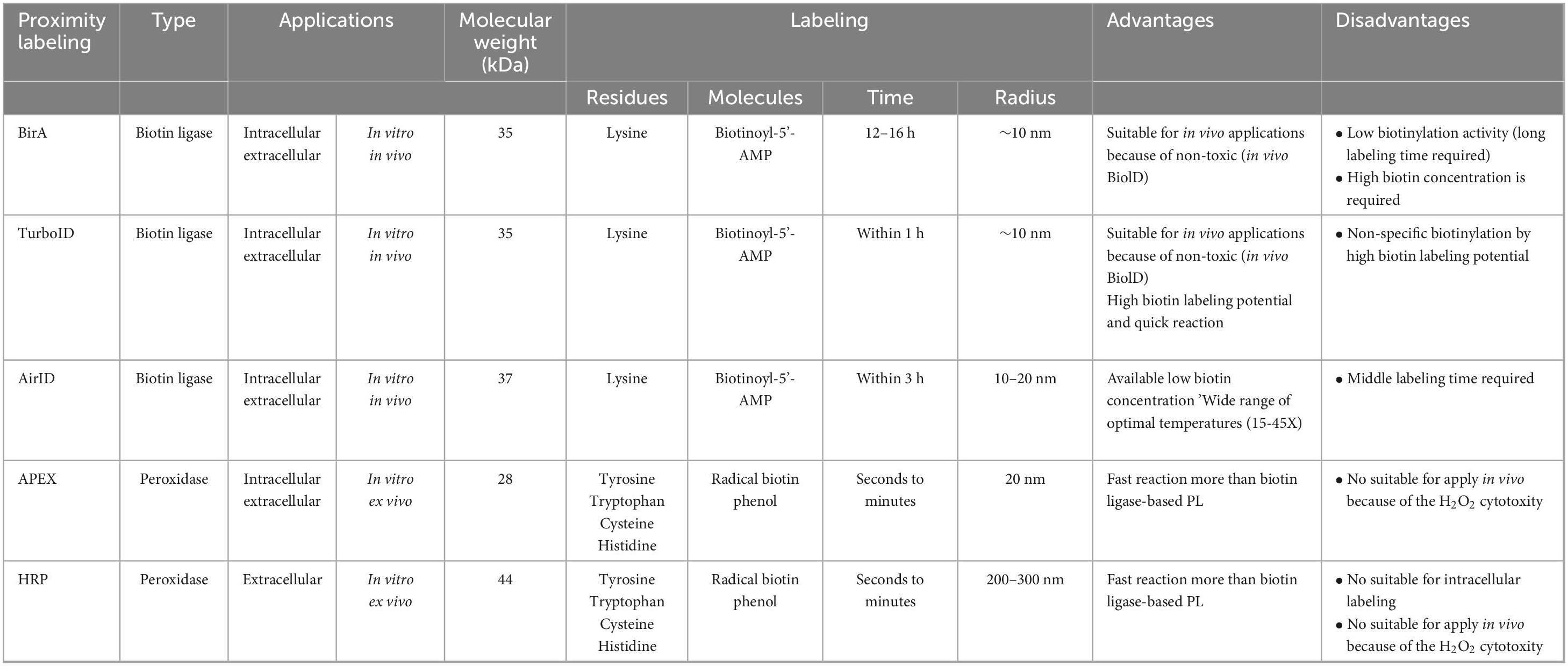
In recent years, PL technologies such as BioID, APEX, and HRP have emerged as powerful biochemical tools for spatially resolved synaptic proteomics (Ito et al., 2024; Table 1). These approaches rely on enzyme-mediated biotinylation of proteins located in the immediate vicinity of a target protein fused to a biotin ligase or peroxidase. Biotinylated proteins are subsequently purified using streptavidin, NeutrAvidin, anti-biotin antibody and Tamavidin 2-REV-coated beads, followed by identification using LC-MS/MS (Figure 1A). BioID, the first biotin ligase-based PL method, uses a mutant Escherichia coli biotin ligase (BirA*-R118G) that generates reactive biotin (biotinoyl-5’-AMP) and biotinylates lysine residues of nearby proteins in the presence of biotin (typically within ∼10–20 nm) (Han et al., 2018; Ito et al., 2024; Table 1). Since the original development of BioID, a broad spectrum of proximity-labeling ligases has been engineered to enhance properties such as molecular size, catalytic efficiency, labeling kinetics, and specificity under various physiological and pathological condition. BioID2 is a truncated variant of BioID that retains proximality labeling capability while offering improved efficiency due to its smaller size (Kim et al., 2016). BASU enables more than 1,000 times faster kinetics and more than 30 times increased signal-to-noise ratio over the prior BioID (Ramanathan et al., 2018). TurboID and miniTurbo show much greater efficiency than BioID and BioID2, and biotinylate proteins for 10 min (Branon et al., 2018). Labeling speed of TurboID (∼1 h) is much faster than BioID (∼12–16 h), but TurboID has strong biotinylation activity, which may cause non-specific labeling (Table 1). Split-BioID is splitting BirA into two parts, fusing each fragment with a different protein, and reactivating the BirA enzyme when the complex is formed (De Munter et al., 2017; Schopp et al., 2017; Cho et al., 2020; Takano et al., 2020). The microID, a truncation variant of BioID, is a small-sized biotin ligase and shows efficient Biotinylation at short labeling times (Kubitz et al., 2022). The ultraID is also the directed evolution of microID (Kubitz et al., 2022). MicroID2 is a modified BioID2 and enables lower background labeling than TurboID (Johnson et al., 2022). AirID was engineered by in silico design and shows low ability to biotinylate proteins non-specifically (Kido et al., 2020; Table 1). In this way, researchers can select appropriate tools based on specific experimental needs. In contrast, APEX and HRP are peroxidase-based PL methods that catalyze biotinylation of tyrosine residues using reactive radicals generated from biotin-phenol and hydrogen peroxide (H2O2) (Table 1). HRP is mainly used for biotinylation of extracellular proteins, because HRP requires intramolecular disulfide bonds, but disulfide bond formation is basically difficult inside cells. On the other hand, APEX can use intracellular labeling because it does not require disulfide bonds. These two peroxidase-based PL methods enable rapid (seconds to minutes) and extensive labeling (APEX: 20 nm, HRP: 200–300 nm) than BioID (∼10 nm). However, the important point to note is that these approaches are mainly restricted to in vitro or ex vivo applications due to the cytotoxicity of H2O2 (Figure 1A and Table 1).
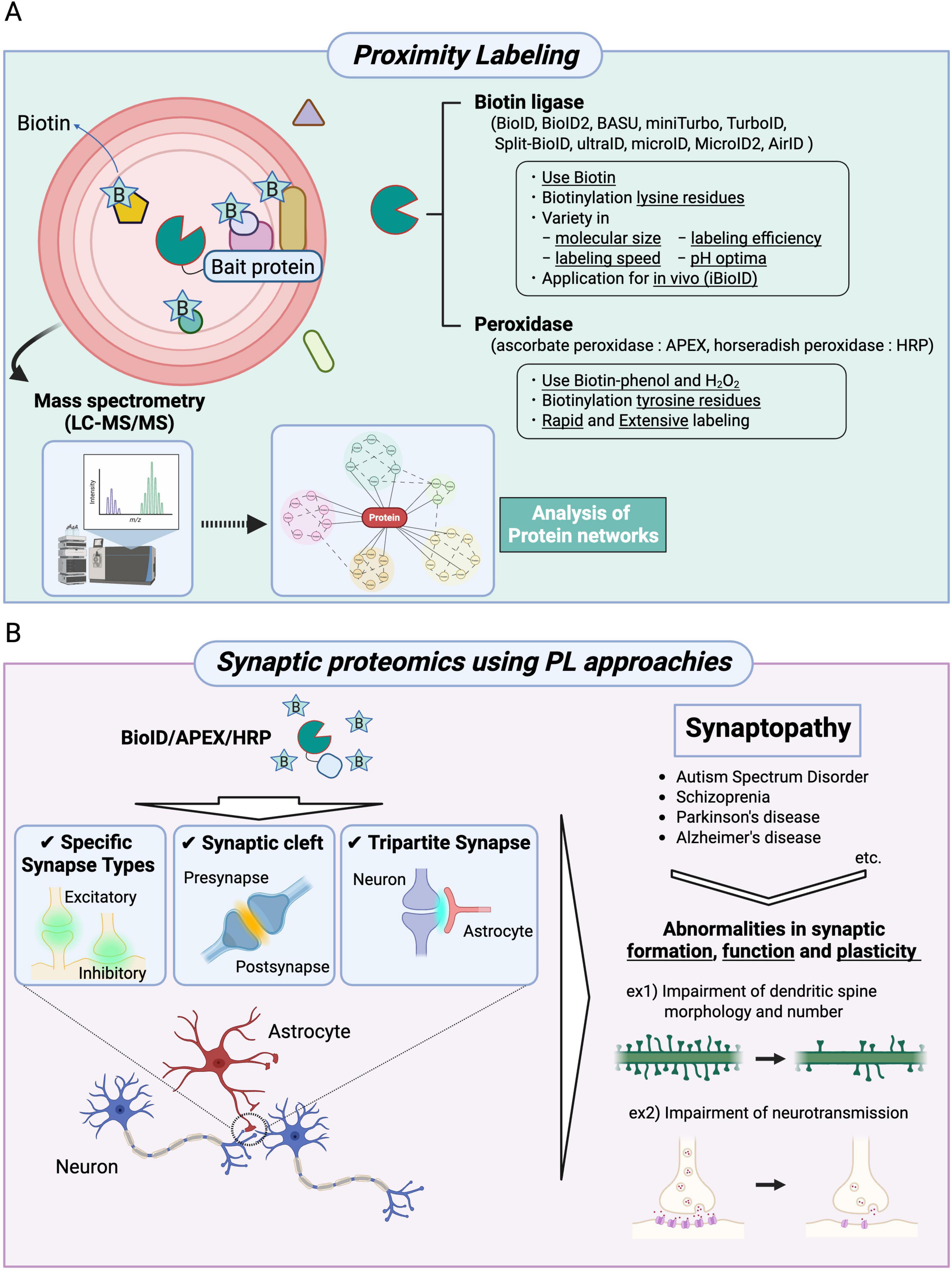
Figure 1. Synaptic proteomics approaches using proximity labeling to uncover physiological and pathological conditions. (A) A schematic diagram and applications of BioID, APEX, and HRP are shown. The proteins of interest (bait proteins) are fused with BioID (a biotin ligase), APEX (ascorbate peroxidase), or HRP (horseradish peroxidase) and expressed in cells. BioID biotinylates lysine residues of proteins in proximity to the bait protein, whereas APEX and HRP biotinylate tyrosine residues of nearby proteins. In the BioID approach, various types of biotin ligases can be selected. Moreover, BioID technologies can be applied to in vivo studies (iBioID). The biotinylated proteins are identified using mass spectrometry, followed by analyses of molecular localization and function based on the constructed protein networks. (B) Proximity labeling methods (BioID, APEX, and HRP) enable high spatial resolution mapping of proteins localized to specific synapse types, the synaptic cleft, and tripartite synapses. These synaptic proteomics approaches have also been applied to the study of synaptopathies, including autism spectrum disorder, schizophrenia, Parkinson’s disease, and Alzheimer’s disease. These neuropsychiatric disorders are characterized by abnormalities in synapse formation, function, and plasticity.
Notably, BioID approaches have enabled the mapping of synaptic proteins in the brain (Uezu et al., 2016). This in vivo BioID (iBioID) technique, in combination with genetic tools such as adeno-associated virus (AAV) vectors and transgenic mice, permits targeted profiling of synaptic proteomes in specific circuits and cell types (Uezu et al., 2016; Ito et al., 2024). However, unlike in vitro, it is necessary to administer biotin continuously for several days. More recently, Cho et al. (2025) introduced a membrane-tethered version of HRP (HRP-TM) that utilizes endogenously generated H2O2 for cell surface biotinylation in vitro, offering potential for application without the need for exogenous H2O2. In another advancement, Zhang et al. (2025) reported TyroID, a novel tyrosinase-based PL technique, that enables non-toxic labeling of various nucleophilic residues both in vitro and in vivo, using reactive o-quinone intermediates derived from phenol-based probes such as alkyne-phenol or biotin-phenol (Zhang et al., 2025). These tools have not yet been applied to spatial synaptic proteomics, but they are expected to be novel in vivo PL tools that compensate for the poor temporal resolution of iBioID. Collectively, PL technologies are rapidly evolving to support in vivo applications and, when combined with virus-based chemogenetic tools such as AAV and transgenic mouse systems, offer a powerful platform for dissecting the molecular mechanisms of synapse formation and function at high spatial resolution.
Synapse-type-specific proteomics using PL approaches
Proximity labeling-based profiling of specific synapse types
Chemical synapses, which serve as the primary sites of neurotransmission, are broadly classified into excitatory or inhibitory synapses in the brain. These synapses exhibit distinct morphological features including synaptic vesicle shape, presynaptic density, and active zone size that vary depending on cell type, brain region, and molecular composition (Van Oostrum et al., 2023; Van Oostrum and Schuman, 2025). Conventional methods for synapse-targeted proteomics lack the spatial resolution necessary to distinguish between specific synapse types. In contrast, PL approaches allow for the precise analysis of defined synapse types both in vitro and in vivo. Using a iBioID strategy, Uezu et al. (2016) identified 121 unique proteins at excitatory synapses and 181 proteins at inhibitory synapses (Uezu et al., 2016; Table 2). Among these synaptic proteins, a previously uncharacterized protein, InSyn1, was found to localize to inhibitory postsynaptic sites. Additionally, they found that InSyn1 regulates miniature excitatory postsynaptic current (mIPSC) by interacting with the dystrophin complex in the hippocampus. Spence et al. (2019) examined the proteome of the developing dendritic filopodia during excitatory synaptogenesis using Wrp (Rac-GAP)-BirA for iBioID labeling. This approach identified 60 synaptic candidate proteins and revealed that CARMIL3, a previously uncharacterized protein, contributes to spine maturation and synapse unsilencing by interacting with WRP and actin capping protein within nascent dendritic spines (Spence et al., 2019; Table 2). Moreover, Falahati et al. (2022) investigated the molecular components of the spine apparatus using synaptopodin-fused BioID2 in the mouse brain (Falahati et al., 2022; Table 2). This approach identified 140 proteins and found that Pdlim7, an actin-binding protein, coassembles with synaptopodin and actin to regulate dendritic spine structure (Falahati et al., 2022). Recently, Rosenthal et al. (2025) explored the development and plasticity of central cholinergic synapses by performing in vivo spatial synaptic proteomics (Table 2). Using CRISPR/Cas9, they inserted miniTurboID into Dα1 and Dα6 subunits of nicotinic acetylcholine receptors (nAchRs) in developing and mature Drosophila brains. Proteomic analysis identified 81 core proteins associated with nAchR function and revealed that the Rho-GTPase regulator Still life (Sif) acts as a key structural organizer of cholinergic synapses through interactions with postsynaptic density components (Rosenthal et al., 2025). In addition to chemical synapses, a recent work has extended PL technologies to electrical synapses in retinal neurons. Tetenborg et al. (2025) employed TurboID-fused Connexin 36 (Cx36), a major neuronal gap junction protein, to profile electrical synapses in zebrafish and mouse retinas. Using two different TurboID strategies in zebrafish and mice, they identified more than 50 novel synaptic proteins and demonstrated that signal-induced proliferation-associated 1-like 3 (SIPA1L3) regulates synaptic density by interacting with Cx36, thereby contributing to electrical synapse formation (Table 2). Together, these studies demonstrate that PL-based approaches enable high-resolution and synapse-type-specific proteomic profiling, including chemical synapses, such as excitatory and inhibitory synapses and electrical synapses, thus providing a powerful platform for dissecting the molecular architecture of diverse synapse types in the brain (Figure 1B).
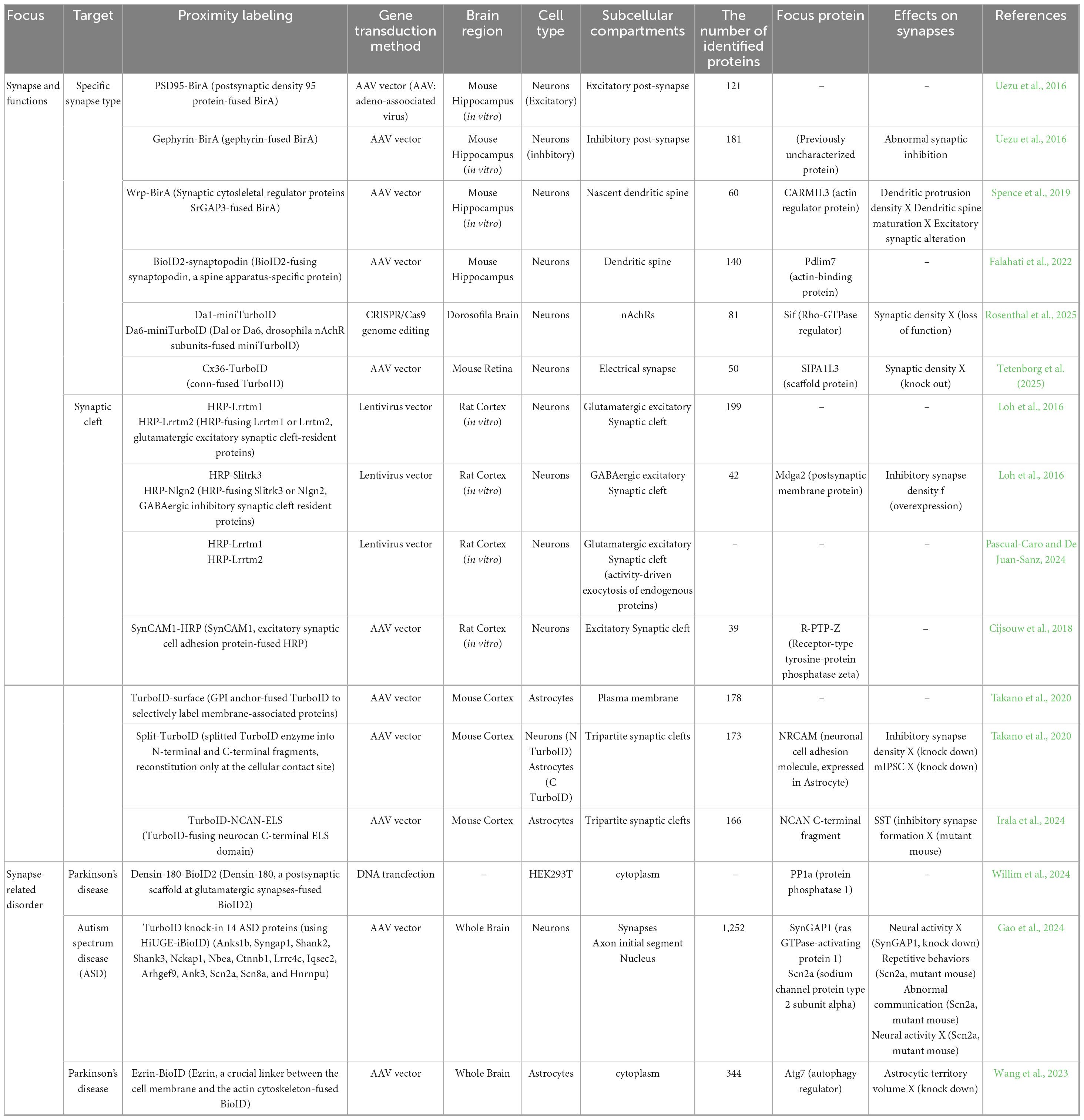
Table 2. Summary of synaptic proteomics using proximity labeling to uncover molecular orchestrations in the brain.
Proximity labeling-based profiling of synaptic cleft and tripartite synapse
The synaptic cleft is a highly specialized extracellular compartment, approximately 20 nm in width, formed between the presynaptic and postsynaptic membranes. It plays a critical role in neurotransmission by mediating cell-cell communication through a dense array of adhesion molecules, receptors, and neurotransmitters. Accurate characterization of the protein components within synaptic clefts is therefore essential for understanding the molecular basis of synaptic function and plasticity. Loh et al. (2016) developed a peroxidase-based synaptic cleft proteomes in cultured neurons by expressing HRP-tagged versions of cleft-resident adhesion molecules, including Lrrtm1, Lrrtm2, Slitrk3 and Nlgn2, which are selectively enriched in excitatory or inhibitory synapse (Table 2). This proteome analysis identified 199 glutamatergic and 42 GABAergic proteins, including Mdga2, a previously uncharacterized protein localized at inhibitory synaptic clefts. Further, functional analysis revealed that Mdga2 regulates recruitment of presynaptic terminals to inhibitory postsynapses through interaction with Neureglin-2 (Loh et al., 2016). Also, Pascual-Caro and De Juan-Sanz (2024) introduced an HRP-based approach to label neural activity-driven trafficking of endogenous synaptic proteins by fusing HRP to Lrrtm1 and Lrrtm2 (Pascual-Caro and De Juan-Sanz, 2024; Table 2). Similarly, Cijsouw et al. (2018) used HRP-tagged SynCAM1, an excitatory synaptic adhesion molecule, to identify receptor-type tyrosine-protein phosphatase zeta (R-PTP-ζ) as a novel candidate synaptic cleft protein in cortical neurons (Table 2). These studies demonstrate that PL-based strategies can selectively label proteins localized within the synaptic cleft, thereby minimizing contamination from intracellular components and enabling precise mapping of extracellular synaptic interfaces (Figure 1B).
In the brain, the astrocyte, which is the most abundant glial cell in the brain, interact with neurons at specialized contact sites to modulate synaptic function and circuit remodeling. These astrocyte-synapse junctions, referred to as tripartite synapses, play crucial roles in the regulation of neurotransmission, synaptic plasticity, and brain homeostasis (Takano and Soderling, 2021; Farizatto and Baldwin, 2023; Raghunathan and Eroglu, 2025). However, conventional proteomic approaches often struggle to resolve such contact-dependent molecular interactions due to the high degree of cellular heterogeneity and the complex intermingling of neural structures within the brain. To overcome these challenges, Takano et al. (2020) developed two innovative proximity labeling (PL)-based techniques: TurboID-surface and Split-TurboID (Takano et al., 2020; Takano and Soderling, 2021). TurboID-surface utilizes a glycosylphosphatidylinositol (GPI) anchor-fused TurboID to selectively label membrane-associated proteins. Split-TurboID separates the TurboID enzyme into N-terminal and C-terminal fragments, which are individually expressed in distinct cell types and become functionally reconstituted only at the cellular contact site (Takano et al., 2020). By integrating these tools with cell type-specific AAV, they performed spatial proteomic profiling of tripartite synapses in the mouse brain, identifying 118 proteins enriched at astrocyte-neuron junctions. Interestingly, neuronal cell adhesion molecule (NRCAM) was found to be strongly localized at perisynaptic astrocytic processes and was shown to facilitate the formation and function of inhibitory postsynapses. This effect is mediated through the recruitment of gephyrin via homophilic interactions between neuronal and astrocytic NRCAM (Takano et al., 2020; Table 2). In addition to this approach, Irala et al. (2024) investigated astrocyte-derived secreted factors that influence the development of inhibitory synapses (Irala et al., 2024). They engineered a secreted form of TurboID fused to the neurocan (NCAN)-ELS domain, which contains synaptogenic protein interaction motifs, and introduced it into astrocytes using an AAV vector. This approach revealed that the C-terminal fragment of astrocyte-secreted NCAN plays a key role in regulating the formation and functional maturation of somatostatin-positive inhibitory synapses in the developing mouse cortex (Irala et al., 2024; Table 2). Together, these studies demonstrate the high versatility and spatial precision of PL-based approaches such as TurboID-surface and Split-TurboID for analyzing protein networks at specialized subcellular and intercellular sites. When coupled with viral gene delivery and cell type-specific expression systems, these methods provide a powerful experimental platform for elucidating the molecular architecture of complex cellular interactions, including those at synaptic clefts and tripartite synapses, under both physiological and pathological conditions (Figure 1B).
Synaptopathy-focused proteomics using PL approaches
A previous great number of studies demonstrate that synaptopathies, defined as abnormalities in synaptic formation, functions, and plasticity, are common pathological features of neurodevelopmental disorders such as autism spectrum disorder (ASD), psychiatric disorders like schizophrenia and neurodegenerative diseases such as Parkinson’s disease (PD) and Alzheimer’s disease (Grant, 2012; Lepeta et al., 2016; Hindley et al., 2023; Figure 1B). Therefore, elucidating the molecular basis of synaptopathies represents a crucial step toward a comprehensive understanding of their pathophysiology and the development of targeted therapeutic strategies. In recent years, spatial synaptic proteomics using PL technologies has emerged as a powerful and versatile approach to uncover the molecular architecture and dynamic regulation of synapses under both physiological and pathological conditions. These technologies enable high-resolution mapping of protein interactions and local proteomes in defined synaptic compartments and cell types, thereby offering novel insights into the mechanisms underlying synapse-related neurological disorders (Figure 1B).
Previous studies have shown that Densin-180, a PSD protein encoded by LRRC7, is highly expressed at excitatory synapses. Densin-180 deficient mice show impaired long-term depression and memory formation and aggressive behavior (Strack et al., 2000; Carlisle et al., 2011; Chong et al., 2019). Recently, Willim et al. (2024) reported that human variants in LRRC7 are associated with neurodevelopmental disorders including intellectual disability, autism, aggression and abnormal eating behaviors (Willim et al., 2024). In this study, PL screening using BioID2-fused Densin-180 identified protein phosphatase (PP1α), another PSD component, as a strong interactor with the leucine rich repeat (LRR) domain of Densin-180 in HEK293T cells. Functional analysis revealed that disease-associated LRR domain variants disrupt binding to PP1α. These findings suggest that Densin-180 scaffolds PP1α to its postsynaptic substrates, and that disruption of this interaction impairs synaptic signaling, contributing to the observed behavioral and cognitive abnormalities (Willim et al., 2024; Table 2). Gao et al. (2024) developed a high-throughput PL screening platform that combines iBioID using TurboID with Homology independent Universal Genome Engineering (HiUGE) approach, which is a CRISPR/Cas9-based genome editing system to investigate 14 risk genes of ASD. Using these approaches, they identified 1,252 interacting proteins. Among these, they focused on Syngap1, a synaptopathy-related protein, and Scn2a, a channelopathy-related protein. Notably, PL and immunoblot analysis showed that an autism-associated mutation of Syngap1 disrupts its interaction with Anks1b, and this interaction is essential for the formation of neural activity in the crucial period of synaptogenesis (Gao et al., 2024; Table 2). Additionally, a patient-derived mutation of Scn2a exhibited repetitive behaviors and deficits in social communication. Proteomic analysis showed that these mutants displayed downregulation of Scn1b and Fgf12, both key modulators of Scn2a function. Importantly, restoring the expression of these proteins rescued the abnormal electrophysiological phenotypes, highlighting their therapeutic potential (Gao et al., 2024; Table 2). Wang et al. (2023) investigated the effect of PD-associated G2019S mutation in the leucine-rich repeat kinase 2 (LRRK2) on the synaptic functions (Wang et al., 2023). Using BioID-fused Ezrin, a protein highly expressed in astrocytes, they identified autophagy-related 7 (Atg7) as an binding partner. Further analysis using LRRK2 G2019Ski/ki mice revealed that phosphorylation of Ezrin disrupts its interaction with Atg7, leading to dysregulated astrocyte morphology and impaired synaptic connectivity. These findings suggest that astrocyte dysfunction caused by LRRK2 mutation contributes to synaptic pathophysiology in PD (Wang et al., 2023; Table 2). Collectively, these studies demonstrate that PL-based proteomics can illuminate the molecular pathways underlying synaptopathies (Figure 1B). By mapping protein interactions in disease-relevant synaptic contexts, PL approaches offer new avenues for therapeutic development.
Concluding remarks and outlook
Proximity labeling approaches have significantly advanced the field of synaptic proteomics by enabling molecular profiling of subcellular compartments with high spatial precision. Applications of BioID, APEX, and HRP have facilitated the identification of proteomes in various synaptic environments, including excitatory and inhibitory synapses (Uezu et al., 2016), cholinergic (Rosenthal et al., 2025) and electrical synapses (Tetenborg et al., 2025), as well as glial interfaces such as tripartite synapses (Takano et al., 2020). These studies have expanded the catalog of synaptic proteins and provided valuable insights into how synapses are assembled, maintained, and modified in both healthy and diseased brains (Figure 1 and Table 2).
Nevertheless, several technical and conceptual challenges must be addressed to fully harness the potential of PL-based approaches. One major limitation is the low temporal resolution of current labeling systems. BioID-based methods typically require several days of biotin supplementation to achieve effective labeling in vivo, which makes it difficult to capture rapid or transient protein interactions that occur in response to neuronal activity or environmental changes. In contrast, APEX and HRP allow for much faster labeling (≤1 min) but depend on hydrogen peroxide, which is cytotoxic and unsuitable for applications in intact brain tissue. Although new methods such as HRP-TM, which utilizes endogenous hydrogen peroxide (Cho et al., 2025), and TyroID, which employs non-toxic o-quinone chemistry (Zhang et al., 2025), offer promising alternatives, their specificity and applicability in complex brain tissue remain to be fully validated. Further in vivo studies are expected to assess their performance under physiological and pathological conditions, particularly in identifying activity-dependent or circuit-specific proteomic changes within intact neural networks. Another important issue is the limited spatial resolution of current labeling methods. In the brain, synapses are highly compact structures where proteins from presynaptic neurons, postsynaptic neurons, and surrounding glial cells are densely intermingled. Although, TurboID can label proteins within a radius of approximately 10 nm, its biotynilation activity is so potent compared to BioID (Branon et al., 2018). Therefore, it may biotinylate not only the intended molecular targets but also nearby proteins from adjacent compartments. This overlap makes it difficult to determine exactly where the labeled proteins are localized within the synapse. To overcome this limitation, recent methods such as Split-HRP (Martell et al., 2016) and Split-TurboID (Cho et al., 2020; Takano et al., 2020) have been developed. This technique divides the labeling enzyme into two inactive fragments that only reconstitute and become active when two different cell types are in direct contact. Using this strategy, we successfully identified molecules such as NRCAM that localize specifically to astrocyte-neuron interfaces and play a critical role in organizing inhibitory synapses (Takano et al., 2020). These findings highlight how cell-contact-dependent labeling can improve spatial precision and uncover new mechanisms of synaptic regulation.
Quantitative interpretation of PL data also remains a challenge. Currently, there is no widely accepted standard for normalization, statistical comparison, or integration of PL proteomes across different developmental stages or disease models. Combining PL-based proteomic data with complementary approaches such as single-cell transcriptomics (Yao et al., 2023; Zhang et al., 2023), spatial transcriptomics (Lein et al., 2017; Yuan et al., 2025), and high-resolution imaging (Newman et al., 2022; Unterauer et al., 2024) will likely be necessary to interpret the data in a biologically meaningful context. For instance, a recently published single-cell mass cytometry-based atlas of the developing mouse brain provides a valuable resource for anchoring synaptic proteomic data within a broader cellular and developmental framework (Van Deusen et al., 2025). The combined analysis of this advanced technology and PL approaches could also offer a key resource for future novel “single-synapse proteome” research field. Additionally, PL methods have provided important insights into the molecular mechanisms of neurological and psychiatric disorders (Table 2). Recent studies using disease models or patient-derived mutations have shown how alterations in protein–protein interactions can impair synaptic signaling and lead to behavioral and cognitive deficits. For example, disrupted interactions between Densin-180 and PP1α (Willim et al., 2024) as well as changes in the Scn2a-associated proteome (Gao et al., 2024) have been linked to neurodevelopmental disorders. These findings highlight the utility of PL techniques for mechanistic investigations and therapeutic target identification besides for mapping molecular discovery (Figure 1B). In summary, proximity labeling-based synaptic proteomics represents a powerful platform for investigating the molecular logic of synapse formation, function, and dysfunction. Also, synaptic proteomics has greatly advanced our understanding of molecular diversity within synapses, and revealing a number of unknown molecular mechanisms involved in this diversity may provide cues to decoding the intricate brain functions induced by diverse neural circuits. Future improvements in the temporal control, spatial accuracy, and quantitative robustness of these technologies will be crucial for advancing both basic neuroscience and clinical applications. By integrating molecular, cellular, and circuit-level information, PL approaches have the potential to reshape our understanding of the brain and inform the development of targeted therapies for complex brain disorders.
Author contributions
JM: Conceptualization, Visualization, Writing – original draft, Writing – review and editing. TT: Funding acquisition, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by a Grant-Aid for Scientific Research B (21380936, 22512442 and 24935004) and a Grant-in-Aid for Transformative Research Areas A (24984754) from the JSPS (TT), PRESTO (21461219 and 1274608) from JST (TT), a Brain Mind 2.0 from AMED (24019528 and 24019272) (TT). Ono Pharmaceutical Foundation for Oncology, Immunology, and Neurology (TT) and The Takeda Science Foundation (TT).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bayés, À, Van De Lagemaat, L. N., Collins, M. O., Croning, M. D. R., Whittle, I. R., Choudhary, J. S., et al. (2011). Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 14, 19–21. doi: 10.1038/nn.2719
Boyken, J., Grønborg, M., Riedel, D., Urlaub, H., Jahn, R., and Chua, J. J. E. (2013). Molecular profiling of synaptic vesicle docking sites reveals novel proteins but few differences between glutamatergic and GABAergic synapses. Neuron 78, 285–297. doi: 10.1016/j.neuron.2013.02.027
Branon, T. C., Bosch, J. A., Sanchez, A. D., Udeshi, N. D., Svinkina, T., Carr, S. A., et al. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887. doi: 10.1038/nbt.4201
Carlisle, H. J., Luong, T. N., Medina-Marino, A., Schenker, L., Khorosheva, E., Indersmitten, T., et al. (2011). Deletion of Densin-180 results in abnormal behaviors associated with mental illness and reduces mGluR5 and DISC1 in the postsynaptic density fraction. J. Neurosci. 31, 16194–16207. doi: 10.1523/JNEUROSCI.5877-10.2011
Cho, K. F., Branon, T. C., Rajeev, S., Svinkina, T., Udeshi, N. D., Thoudam, T., et al. (2020). Split-TurboID enables contact-dependent proximity labeling in cells. Proc. Natl. Acad. Sci. 117, 12143–12154. doi: 10.1073/pnas.1919528117
Cho, Y., Jeong, I., Kim, K., and Rhee, H.-W. (2025). Painting cell–cell interactions by horseradish peroxidase and endogenously generated hydrogen peroxide. ACS Chem. Biol. 20, 86–93. doi: 10.1021/acschembio.4c00419
Chong, C. H., Li, Q., Mak, P. H. S., Ng, C. C. P., Leung, E. H. W., Tan, V. H., et al. (2019). Lrrc7 mutant mice model developmental emotional dysregulation that can be alleviated by mGluR5 allosteric modulation. Transl. Psychiatry 9:244. doi: 10.1038/s41398-019-0580-9
Cijsouw, T., Ramsey, A. M., Lam, T. T., Carbone, B. E., Blanpied, T. A., and Biederer, T. (2018). Mapping the proteome of the synaptic cleft through proximity labeling reveals new cleft proteins. Proteomes 6:48. doi: 10.3390/proteomes6040048
De Munter, S., Görnemann, J., Derua, R., Lesage, B., Qian, J., Heroes, E., et al. (2017). Split-BioID: A proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett. 591, 415–424. doi: 10.1002/1873-3468.12548
Dieterich, D. C., and Kreutz, M. R. (2016). Proteomics of the synapse – a quantitative approach to neuronal plasticity. Mol. Cell. Proteomics 15, 368–381. doi: 10.1074/mcp.R115.051482
Falahati, H., Wu, Y., Feuerer, V., Simon, H.-G., and De Camilli, P. (2022). Proximity proteomics of synaptopodin provides insight into the molecular composition of the spine apparatus of dendritic spines. Proc. Natl. Acad. Sci. 119:e2203750119. doi: 10.1073/pnas.2203750119
Farizatto, K. L. G., and Baldwin, K. T. (2023). Astrocyte-synapse interactions during brain development. Curr. Opin. Neurobiol. 80:102704. doi: 10.1016/j.conb.2023.102704
Fernández, E., Collins, M. O., Uren, R. T., Kopanitsa, M. V., Komiyama, N. H., Croning, M. D. R., et al. (2009). Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol. Syst. Biol. 5:269. doi: 10.1038/msb.2009.27
Gao, Y., Hisey, E., Bradshaw, T. W. A., Erata, E., Brown, W. E., Courtland, J. L., et al. (2019). Plug-and-play protein modification using homology-independent universal genome engineering. Neuron 103, 583–597.e8. doi: 10.1016/j.neuron.2019.05.047
Gao, Y., Shonai, D., Trn, M., Zhao, J., Soderblom, E. J., Garcia-Moreno, S. A., et al. (2024). Proximity analysis of native proteomes reveals phenotypic modifiers in a mouse model of autism and related neurodevelopmental conditions. Nat. Commun. 15:6801. doi: 10.1038/s41467-024-51037-x
Grant, S. G. (2012). Synaptopathies: Diseases of the synaptome. Curr. Opin. Neurobiol. 22, 522–529. doi: 10.1016/j.conb.2012.02.002
Grønborg, M., Pavlos, N. J., Brunk, I., Chua, J. J. E., Münster-Wandowski, A., Riedel, D., et al. (2010). Quantitative comparison of glutamatergic and GABAergic synaptic vesicles unveils selectivity for few proteins including MAL2, a novel synaptic vesicle protein. J. Neurosci. 30, 2–12. doi: 10.1523/JNEUROSCI.4074-09.2010
Han, S., Li, J., and Ting, A. Y. (2018). Proximity labeling: Spatially resolved proteomic mapping for neurobiology. Curr. Opin. Neurobiol. 50, 17–23. doi: 10.1016/j.conb.2017.10.015
Hindley, N., Sanchez Avila, A., and Henstridge, C. (2023). Bringing synapses into focus: Recent advances in synaptic imaging and mass-spectrometry for studying synaptopathy. Front. Synaptic Neurosci. 15:1130198. doi: 10.3389/fnsyn.2023.1130198
Irala, D., Wang, S., Sakers, K., Nagendren, L., Ulloa Severino, F. P., Bindu, D. S., et al. (2024). Astrocyte-secreted neurocan controls inhibitory synapse formation and function. Neuron 112, 1657–1675.e10. doi: 10.1016/j.neuron.2024.03.007
Ito, Y., Nagamoto, S., and Takano, T. (2024). Synaptic proteomics decode novel molecular landscape in the brain. Front. Mol. Neurosci. 17:1361956. doi: 10.3389/fnmol.2024.1361956
Johnson, B. S., Chafin, L., Farkas, D., Adair, J., Elhance, A., Farkas, L., et al. (2022). MicroID2: A novel biotin ligase enables rapid proximity-dependent proteomics. Mol. Cell. Proteomics 21:100256. doi: 10.1016/j.mcpro.2022.100256
Kaizuka, T., Suzuki, T., Kishi, N., Tamada, K., Kilimann, M. W., Ueyama, T., et al. (2024). Remodeling of the postsynaptic proteome in male mice and marmosets during synapse development. Nat. Commun. 15:2496. doi: 10.1038/s41467-024-46529-9
Kido, K., Yamanaka, S., Nakano, S., Motani, K., Shinohara, S., Nozawa, A., et al. (2020). AirID, a novel proximity biotinylation enzyme, for analysis of protein–protein interactions. eLife 9:e54983. doi: 10.7554/eLife.54983
Kim, D. I., Jensen, S. C., Noble, K. A., Kc, B., Roux, K. H., Motamedchaboki, K., et al. (2016). An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196. doi: 10.1091/mbc.E15-12-0844
Koopmans, F., Van Nierop, P., Andres-Alonso, M., Byrnes, A., Cijsouw, T., Coba, M. P., et al. (2019). SynGO: An evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234.e4. doi: 10.1016/j.neuron.2019.05.002
Kubitz, L., Bitsch, S., Zhao, X., Schmitt, K., Deweid, L., Roehrig, A., et al. (2022). Engineering of ultraID, a compact and hyperactive enzyme for proximity-dependent biotinylation in living cells. Commun. Biol. 5:657. doi: 10.1038/s42003-022-03604-5
Lein, E., Borm, L. E., and Linnarsson, S. (2017). The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69. doi: 10.1126/science.aan6827
Lepeta, K., Lourenco, M. V., Schweitzer, B. C., Martino Adami, P. V., Banerjee, P., Catuara-Solarz, S., et al. (2016). Synaptopathies: Synaptic dysfunction in neurological disorders – A review from students to students. J. Neurochem. 138, 785–805. doi: 10.1111/jnc.13713
Loh, K. H., Stawski, P. S., Draycott, A. S., Udeshi, N. D., Lehrman, E. K., Wilton, D. K., et al. (2016). Proteomic analysis of unbounded cellular compartments: Synaptic clefts. Cell 166, 1295–1307.e21. doi: 10.1016/j.cell.2016.07.041
Martell, J. D., Yamagata, M., Deerinck, T. J., Phan, S., Kwa, C. G., Ellisman, M. H., et al. (2016). A split horseradish peroxidase for the detection of intercellular protein–protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 34, 774–780. doi: 10.1038/nbt.3563
Morciano, M., Beckhaus, T., Karas, M., Zimmermann, H., and Volknandt, W. (2009). The proteome of the presynaptic active zone: From docked synaptic vesicles to adhesion molecules and maxi-channels. J. Neurochem. 108, 662–675. doi: 10.1111/j.1471-4159.2008.05824.x
Newman, Z. L., Bakshinskaya, D., Schultz, R., Kenny, S. J., Moon, S., Aghi, K., et al. (2022). Determinants of synapse diversity revealed by super-resolution quantal transmission and active zone imaging. Nat. Commun. 13:229. doi: 10.1038/s41467-021-27815-2
O’Rourke, N. A., Weiler, N. C., Micheva, K. D., and Smith, S. J. (2012). Deep molecular diversity of mammalian synapses: Why it matters and how to measure it. Nat. Rev. Neurosci. 13, 365–379. doi: 10.1038/nrn3170
Pascual-Caro, C., and De Juan-Sanz, J. (2024). Monitoring of activity-driven trafficking of endogenous synaptic proteins through proximity labeling. PLoS Biol. 22:e3002860. doi: 10.1371/journal.pbio.3002860
Raghunathan, K., and Eroglu, C. (2025). Developmental roles of astrocytes in circuit wiring. Curr. Opin. Neurobiol. 92:103042. doi: 10.1016/j.conb.2025.103042
Ramanathan, M., Majzoub, K., Rao, D. S., Neela, P. H., Zarnegar, B. J., Mondal, S., et al. (2018). RNA–protein interaction detection in living cells. Nat. Methods 15, 207–212. doi: 10.1038/nmeth.4601
Rosenthal, J. S., Zhang, D., Yin, J., Long, C., Yang, G., Li, Y., et al. (2025). Molecular organization of central cholinergic synapses. Proc. Natl. Acad. Sci. 122:e2422173122. doi: 10.1073/pnas.2422173122
Schopp, I. M., Amaya Ramirez, C. C., Debeljak, J., Kreibich, E., Skribbe, M., Wild, K., et al. (2017). Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 8:15690. doi: 10.1038/ncomms15690
Spence, E. F., Dube, S., Uezu, A., Locke, M., Soderblom, E. J., and Soderling, S. H. (2019). In vivo proximity proteomics of nascent synapses reveals a novel regulator of cytoskeleton-mediated synaptic maturation. Nat. Commun. 10:386. doi: 10.1038/s41467-019-08288-w
Strack, S., Robison, A. J., Bass, M. A., and Colbran, R. J. (2000). Association of calcium/calmodulin-dependent kinase II with developmentally regulated splice variants of the postsynaptic density protein densin-180. J. Biol. Chem. 275, 25061–25064. doi: 10.1074/jbc.C000319200
Takano, T., and Soderling, S. H. (2021). Tripartite synaptomics: Cell-surface proximity labeling in vivo. Neurosci. Res. 173, 14–21. doi: 10.1016/j.neures.2021.05.002
Takano, T., Wallace, J. T., Baldwin, K. T., Purkey, A. M., Uezu, A., Courtland, J. L., et al. (2020). Chemico-genetic discovery of astrocytic control of inhibition in vivo. Nature 588, 296–302. doi: 10.1038/s41586-020-2926-0
Tetenborg, S., Shihabeddin, E., Kumar, E. O. A. M., Sigulinsky, C. L., Dedek, K., Lin, Y. P., et al. (2025). Uncovering the electrical synapse proteome in retinal neurons via in vivo proximity labeling. BioRxiv[Preprint] doi: 10.1101/2024.11.26.625481
Uezu, A., Kanak, D. J., Bradshaw, T. W. A., Soderblom, E. J., Catavero, C. M., Burette, A. C., et al. (2016). Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129. doi: 10.1126/science.aag0821
Unterauer, E. M., Shetab Boushehri, S., Jevdokimenko, K., Masullo, L. A., Ganji, M., Sograte-Idrissi, S., et al. (2024). Spatial proteomics in neurons at single-protein resolution. Cell 187, 1785–1800.e16. doi: 10.1016/j.cell.2024.02.045
Van Deusen, A. L., Kumar, S., Calhan, O. Y., Goggin, S. M., Shi, J., Williams, C. M., et al. (2025). A single-cell mass cytometry-based atlas of the developing mouse brain. Nat. Neurosci. 28, 174–188. doi: 10.1038/s41593-024-01786-1
Van Oostrum, M., Blok, T. M., Giandomenico, S. L., Tom Dieck, S., Tushev, G., Fürst, N., et al. (2023). The proteomic landscape of synaptic diversity across brain regions and cell types. Cell 186, 5411–5427.e23. doi: 10.1016/j.cell.2023.09.028
Van Oostrum, M., and Schuman, E. M. (2025). Understanding the molecular diversity of synapses. Nat. Rev. Neurosci. 26, 65–81. doi: 10.1038/s41583-024-00888-w
Wang, S., Baumert, R., Séjourné, G., Sivadasan Bindu, D., Dimond, K., Sakers, K., et al. (2023). PD-linked LRRK2 G2019S mutation impairs astrocyte morphology and synapse maintenance via ERM hyperphosphorylation. BioRxiv [Preprint] doi: 10.1101/2023.04.09.536178
Wilhelm, B. G., Mandad, S., Truckenbrodt, S., Kröhnert, K., Schäfer, C., Rammner, B., et al. (2014). Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028. doi: 10.1126/science.1252884
Willim, J., Woike, D., Greene, D., Das, S., Pfeifer, K., Yuan, W., et al. (2024). Variants in LRRC7 lead to intellectual disability, autism, aggression and abnormal eating behaviors. Nat. Commun. 15:7909. doi: 10.1038/s41467-024-52095-x
Xu, Y., Song, X., Wang, D., Wang, Y., Li, P., and Li, J. (2021). Proteomic insights into synaptic signaling in the brain: The past, present and future. Mol. Brain 14:37. doi: 10.1186/s13041-021-00750-5
Yao, Z., Van Velthoven, C. T. J., Kunst, M., Zhang, M., McMillen, D., Lee, C., et al. (2023). A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332. doi: 10.1038/s41586-023-06812-z
Yuan, C., Patel, K., Shi, H., Wang, H.-L. V., Wang, F., Li, R., et al. (2025). mcDETECT: Decoding 3D spatial synaptic transcriptomes with subcellular-resolution spatial transcriptomics. BioRxiv[Preprint] doi: 10.1101/2025.03.27.645744
Zhang, M., Pan, X., Jung, W., Halpern, A. R., Eichhorn, S. W., Lei, Z., et al. (2023). Molecularly defined and spatially resolved cell atlas of the whole mouse brain. Nature 624, 343–354. doi: 10.1038/s41586-023-06808-9
Keywords: synapse, proteomics, BioID, spine formation, cytoskeleton, synaptopathy
Citation: Matsubayashi J and Takano T (2025) Proximity labeling uncovers the synaptic proteome under physiological and pathological conditions. Front. Cell. Neurosci. 19:1638627. doi: 10.3389/fncel.2025.1638627
Received: 31 May 2025; Accepted: 02 July 2025;
Published: 23 July 2025.
Edited by:
Robert M. Hughes, East Carolina University, United StatesReviewed by:
Pirta Elina Hotulainen, Minerva Foundation Institute for Medical Research, FinlandGeorge Leondaritis, University of Ioannina, Greece
Prateek Kumar, Yale University, United States
Copyright © 2025 Matsubayashi and Takano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tetsuya Takano, dGV0c3V5YS50YWthbm9AYmlvcmVnLmt5dXNodS11LmFjLmpw
 Junpei Matsubayashi
Junpei Matsubayashi Tetsuya Takano
Tetsuya Takano