- 1Department of Pharmaceutical Engineering, School of Food and Pharmaceutical Engineering, Zhaoqing University, Zhaoqing, China
- 2Department of Pharmacology of Chinese Medicine, School of Traditional Chinese Medicine, Southern Medical University, Guangzhou, China
- 3Key Laboratory for Research and Utilization of Southern Medicine, Zhaoqing University, Zhaoqing, China
- 4Risk Assessment Laboratory for Agricultural Product Quality and Safety, Ministry of Agriculture and Rural Development, Zhaoqing University, Zhaoqing, China
The emergence of human brain organoids (hBOs) has transformed how we study brain development, disease mechanisms, and therapy discovery. These 3D in vitro neural models closely mimic the cellular diversity, spatial structure, and functional connectivity of the human brain, providing a groundbreaking platform that outperforms traditional 2D cultures and animal models in studying neurodevelopment and neurological disorders. To further explore the potential of hBOs technology, we review current literature focusing particularly on its applications for diagnosing and treating major neurological diseases such as Alzheimer’s disease, Parkinson’s disease, and other related neurological disorders. Using patient-derived induced pluripotent stem cells combined with cutting-edge gene-editing technologies, hBOs enable highly precise mechanistic studies and scalable drug screening. Moreover, we further discuss the advantages and current limitations of hBOs. Despite these challenges, hBOs remain a transformative platform for the development of targeted neurotherapeutics. Collectively, this review offers a solid foundation for advancing neuroscience research and fostering innovative treatment strategies for neurological disorders.
1 Introduction
The human brain is characterized by exceptional cellular diversity and intricate synaptic architecture, presenting considerable challenges for modeling neurological disorders (ND) such as Alzheimer’s disease (AD) and autism spectrum disorders (ASD). Although traditional two-dimensional (2D) cell cultures and animal models have significantly advanced neuroscience research (Wang Z. et al., 2017), they fail to replicate the human brain’s three-dimensional (3D) structure and species-specific features, limiting their translational relevance. Organoid technology, first developed in cancer research in 1946, gained new momentum with the advent of pluripotent stem cell (PSC) technologies in 1998. These breakthroughs enabled the generation of organoids resembling organ-specific structure and function across various systems, including the brain, liver, gut, and kidney (Mishra et al., 2024). In particular, human brain organoids (hBOs), derived from human pluripotent stem cells (hPSCs) such as embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), have the capacity to self-organize into 3D structures that recapitulate key features of the human brain (Lee et al., 2017).
Since 1992, continuous improvements in hBOs have significantly expanded their applications in neuroscience (Figure 1). These models have become essential tools for studying human-specific mechanisms of brain development and neuropathology (Kwak et al., 2024). Compared with 2D cultures and animal models, hBOs demonstrate superior fidelity in replicating human brain architecture, offering broad utility in disease modeling, drug screening, and personalized medicine. However, several limitations persist, including restricted vascularization, inter-organoid heterogeneity, and unresolved ethical concerns (Kanupriya et al., 2025). These factors often lead to hypoxic core regions, increased cellular stress, and limited capacity to model late-stage ND. Moreover, the absence of inter-organ interactions restricts their utility in capturing the systemic complexity of disease pathogenesis. Addressing these challenges is crucial for enhancing the biological fidelity and clinical relevance of hBO-based platforms.
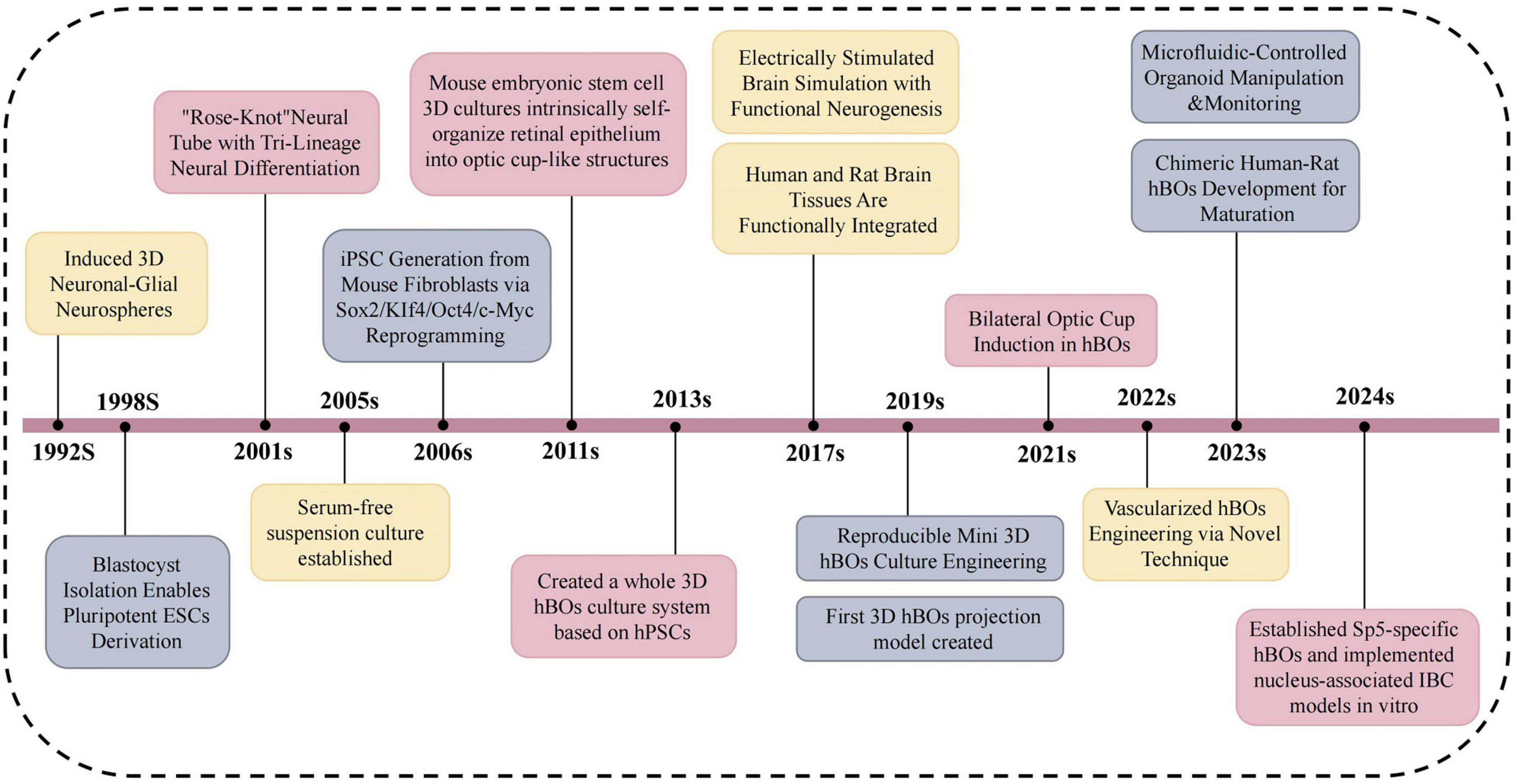
Figure 1. Illustrates major milestones in hBOs research from 1992 to 2024. Yellow denotes progress in culture optimization and functional maturation, enabling the shift from simple neurospheres to functional organoids. Gray indicates innovations in methodology, including reprogramming, engineered platforms, and novel disease models. Pink marks advances in morphogenesis and cell lineage specification, from neural tube-like structures to region- or subtype-specific hBOs. The timeline emphasizes trilineage differentiation, emergence of complex models such as Sp5 and IBC, and enhanced neural circuit formation. This three-decade evolution highlights hBOs as powerful in vitro systems for neuroscience. Sp5, spinal trigeminal nucleus; IBC, inter-brain connection.
2 hBOs modeling the neural microenvironment
Beyond replicating brain structure, human brain organoids (hBOs) closely model the dynamic neural microenvironment. Unlike traditional 2D cultures and animal models, hBOs cultured in 3D systems can produce extracellular matrices (ECMs) that support autocrine and paracrine signaling, thereby enabling more physiologically relevant modeling of cellular proliferation, migration, and differentiation (Mimeault et al., 2007; Acharya et al., 2024). It is important to note that the development and maintenance of hBOs often rely on artificial ECMs such as Matrigel and Geltrex. These materials are commonly used to embed organoids or are included in culture media to provide essential structural support and promote proper tissue organization. Through guided differentiation of iPSCs, hBOs develop structural and functional features resembling early human neural tissue (Qian et al., 2019). Recent advances such as incorporation of vascular-like networks and extended culture stability have improved physiological accuracy (Sun et al., 2022). These enhancements support applications in neurodevelopmental research, disease mechanism elucidation, and therapeutic screening. To better capture complex disease phenotypes, recent bioengineering strategies aim to integrate neuroimmune components, enable in vivo transplantation, and construct multi-regional or multi-organ systems. These innovations improve systemic modeling capabilities and enhance translational relevance. With continued progress in bioengineering and multi-omics integration, hBOs are becoming indispensable tools for neuroscience research, disease modeling, and regenerative medicine (Figure 2).
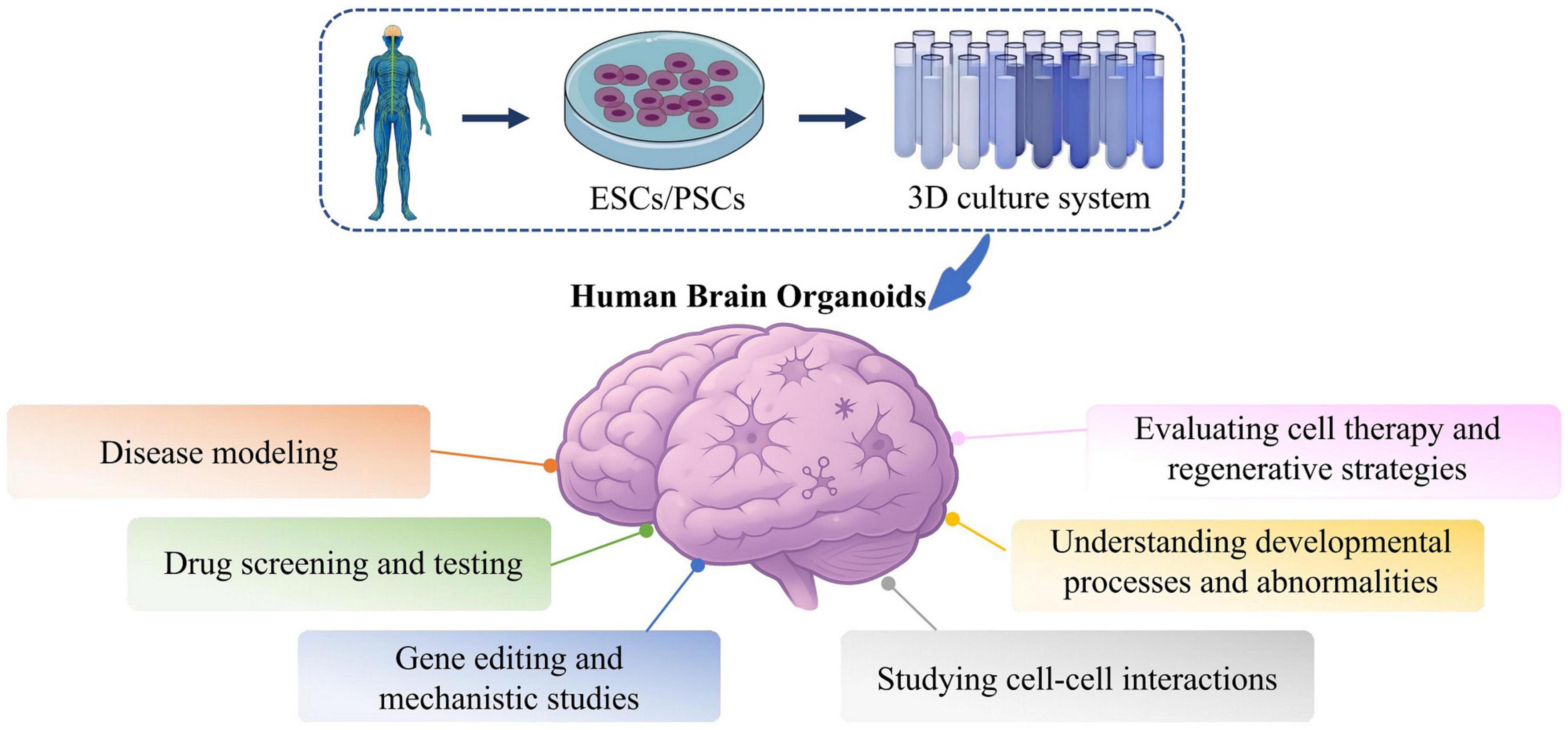
Figure 2. Applications of hBOs in investigating disease mechanisms and conducting drug screening. hBOs are a novel in vitro modeling platform for neurological disorder research and drug development. Using these models, researchers establish disease-specific hBOs, conduct high-throughput drug screening, and employ molecular biology and imaging for phenotype analysis. hBOs effectively replicate disease pathology, demonstrate strong predictability and reproducibility in drug testing, and reveal new molecular insights into disease mechanisms. Embryonic stem cells, ESCs; Pluripotent stem cells, PSCs.
3 Unguided versus guided hBOs in ND modeling
Human brain organoids have become indispensable tools for modeling ND, with a key methodological distinction being the use of unguided or guided differentiation strategies. Unguided hBOs rely on spontaneous self-organization of pluripotent stem cells (PSCs) without exogenous patterning signals, resulting in the generation of heterogeneous brain regions such as the forebrain, midbrain, and hindbrain within a single organoid (Lancaster et al., 2013). This approach recapitulates early brain development and is suitable for modeling disorders such as microcephaly, Zika virus infection, and cortical malformations (Qian et al., 2019). However, unguided organoids suffer from batch variability, inconsistent regional identity, and stochastic architecture, limiting their reproducibility and suitability for region-specific disease modeling and high-throughput drug screening (Velasco et al., 2019).
In contrast, guided hBOs are derived by applying defined patterning cues to direct differentiation toward specific brain regions, such as the cortex, midbrain, or hypothalamus (Qian et al., 2016; Jo et al., 2016). This strategy enhances regional fidelity, reproducibility, and experimental control. For example, midbrain hBOs enriched with dopaminergic neurons are used to model Parkinson’s disease (PD) (Smits and Schwamborn, 2020), while cortical organoids facilitate the study of amyloid and Tau pathologies in AD (Raja et al., 2016). Nevertheless, guided organoids may oversimplify the native brain environment and often lack inter-regional connectivity, limiting their utility in modeling network-level dysfunctions. To address these limitations, hybrid models such as “assembloids” have been developed, fusing region-specific organoids to recreate inter-regional interactions (Bagley et al., 2017). Future advances in single-cell multi-omics, spatial transcriptomics, and bioengineering are expected to integrate the strengths of both approaches, improving the physiological relevance, reproducibility, and translational value of hBO-based models in ND research.
4 hBOs × multi-omics represent a novel strategy
Human brain organoids replicate key structural and functional features of the brain and, when combined with multi-omics technologies including transcriptomics, proteomics, and epigenomics, offer a powerful strategy to decode mechanisms of ND (Taglieri et al., 2025). Single-cell RNA sequencing (scRNA-seq) reveals cell-type heterogeneity and disease-relevant gene expression patterns, such as Wnt signaling disruptions in ASD (Kiaee et al., 2021) and neuroinflammatory markers in AD (Abdelbasset et al., 2024). Because transcript levels do not always reflect protein abundance or activity, proteomics provides essential complementary insights. Mass spectrometry-based profiling can identify post-translational modifications such as phosphorylation, as demonstrated by tau hyperphosphorylation in AD organoids, which is a hallmark of altered signaling pathways (Bracha et al., 2024; Marinho et al., 2023). Epigenomic approaches, including ATAC-seq and DNA methylation analysis, provide information on chromatin accessibility and transcriptional regulation. In AD hBOs, ATAC-seq has revealed reduced enhancer activity in genes associated with neuronal apoptosis, corroborating transcriptomic findings (Marinho et al., 2023). Integrating scRNA-seq with proteomic and phosphoproteomic data improves understanding of transcriptional and post-transcriptional regulation (Pieters et al., 2021), and enables tracking of critical pathways such as NF-κB and PI3K-Akt in disease models (Tanaka, 2024). These multi-omics strategies expand hBOs from static structural models into dynamic systems for mechanistic insight, biomarker discovery, and drug development.
5 Applications and innovations of hBOs in ND research and therapy
Human brain organoids offer enhanced physiological relevance by mimicking the 3D architecture and cellular microenvironment of the human brain. When derived from patient-specific iPSCs, hBOs can reproduce disease-specific phenotypes and provide insights into cell-cell interactions, circuit-level dynamics, and developmental processes underlying ND (Hartlaub et al., 2019). Compared to conventional 2D cultures, which lack spatial complexity and cellular diversity (Lancaster and Knoblich, 2014), hBOs enable more accurate modeling of complex neural processes and offer a robust platform for translational research. As a result, hBOs are increasingly employed in ND research due to their adaptability and biological fidelity (Castiglione et al., 2022; Zhou et al., 2023). Recent advances, including CRISPR/Cas9-based gene editing (Driehuis and Clevers, 2017) and astrocyte-enriched protocols (Bagley et al., 2017), have expanded the scope of mechanistic studies, particularly in modeling neuron-glia interactions and evaluating targeted therapies.
To contextualize the advantages of hBOs, we compared five commonly used neural modeling systems (Table 1), highlighting the superior structural fidelity and translational potential of hBOs. Their ability to reproduce the 3D cytoarchitecture of native brain tissue and capture human-specific features makes them invaluable for high-throughput screening and disease modeling. Nevertheless, batch-to-batch variability caused by differences in stem cell source, reagent quality, and manual handling remains a critical challenge (Humpel, 2015; Quadrato et al., 2017). Other models such as animal chimeras (Bourret et al., 2016), brain-on-a-chip platforms (Amirifar et al., 2022; Kogler et al., 2023), and 2D cultures (Takahashi and Yamanaka, 2006; Zhang et al., 2013) offer unique advantages in terms of scalability or in vivo relevance, but fail to replicate the full complexity of human neurobiology. Standardization in quality control, biomarker-based assessment, and scalable production pipelines is essential to overcome these limitations and improve reproducibility.
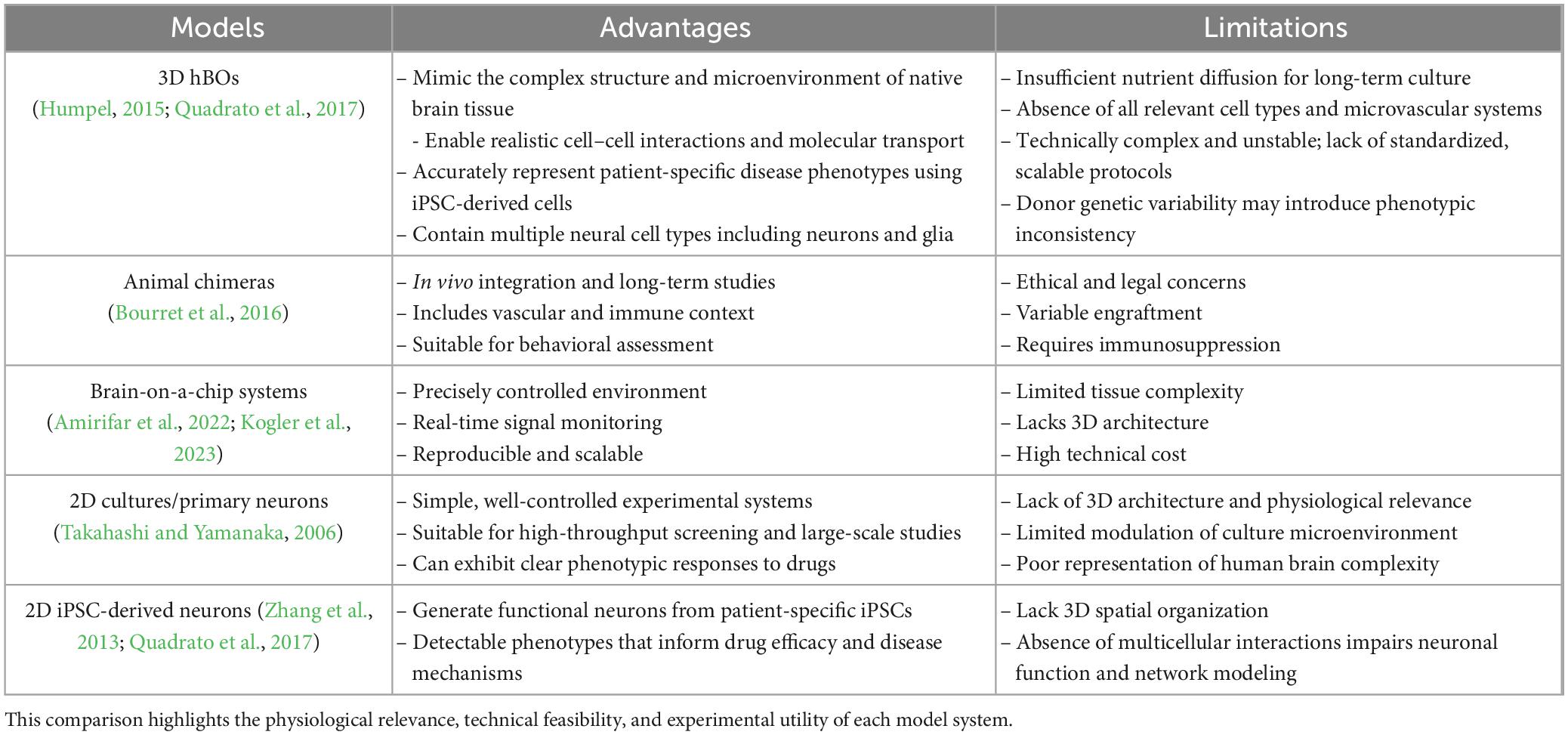
Table 1. Summary of key advantages and limitations of three commonly used neural modeling platforms: 3D human brain organoids (hBOs), animal chimera, brain-on-a-chip systems, 2D cultures/primary neurons, and 2D iPSC-derived neurons.
Despite their promise, hBOs still face major limitations, including the lack of functional vasculature, mature microglia, and complete neural circuitry (Bao et al., 2021; Cao et al., 2023). Furthermore, conventional monoculture hBOs do not replicate systemic inter-organ communication, an increasingly recognized contributor to ND pathogenesis. To overcome these issues, several innovative strategies have been developed (Table 2). Incorporation of microglia enables the establishment of neuroimmune models simulating brain-specific immune responses (Ao et al., 2021; Sabate-Soler et al., 2022; Samudyata et al., 2022). Transplantation of hBOs into rodent brains supports in vivo maturation and therapeutic validation (Dong et al., 2021; Revah et al., 2022), while multi-region organoid fusion approaches (assembloids) allow for the study of interregional connectivity and systemic interactions (Kasai et al., 2020; Zhu et al., 2023). These advances have significantly improved the biological and translational relevance of hBO-based platforms. hBOs can now model a broad range of disorders, including AD, PD, amyotrophic lateral sclerosis (ALS), autism spectrum disorder (ASD) and achalasia-microcephaly syndrome (AMS), faithfully reproducing hallmark pathologies in vitro (Figure 3). As evidence mounts that many ND originate from early developmental disruptions (Barnat et al., 2020; Yeh et al., 2018), and with a rising incidence of early-onset neurological symptoms in younger populations (Jia et al., 2023), the utility of hBOs in modeling disease mechanisms and personalizing pharmacological interventions becomes increasingly evident. With continued refinement, hBOs are poised to become cornerstone tools in precision neurology.
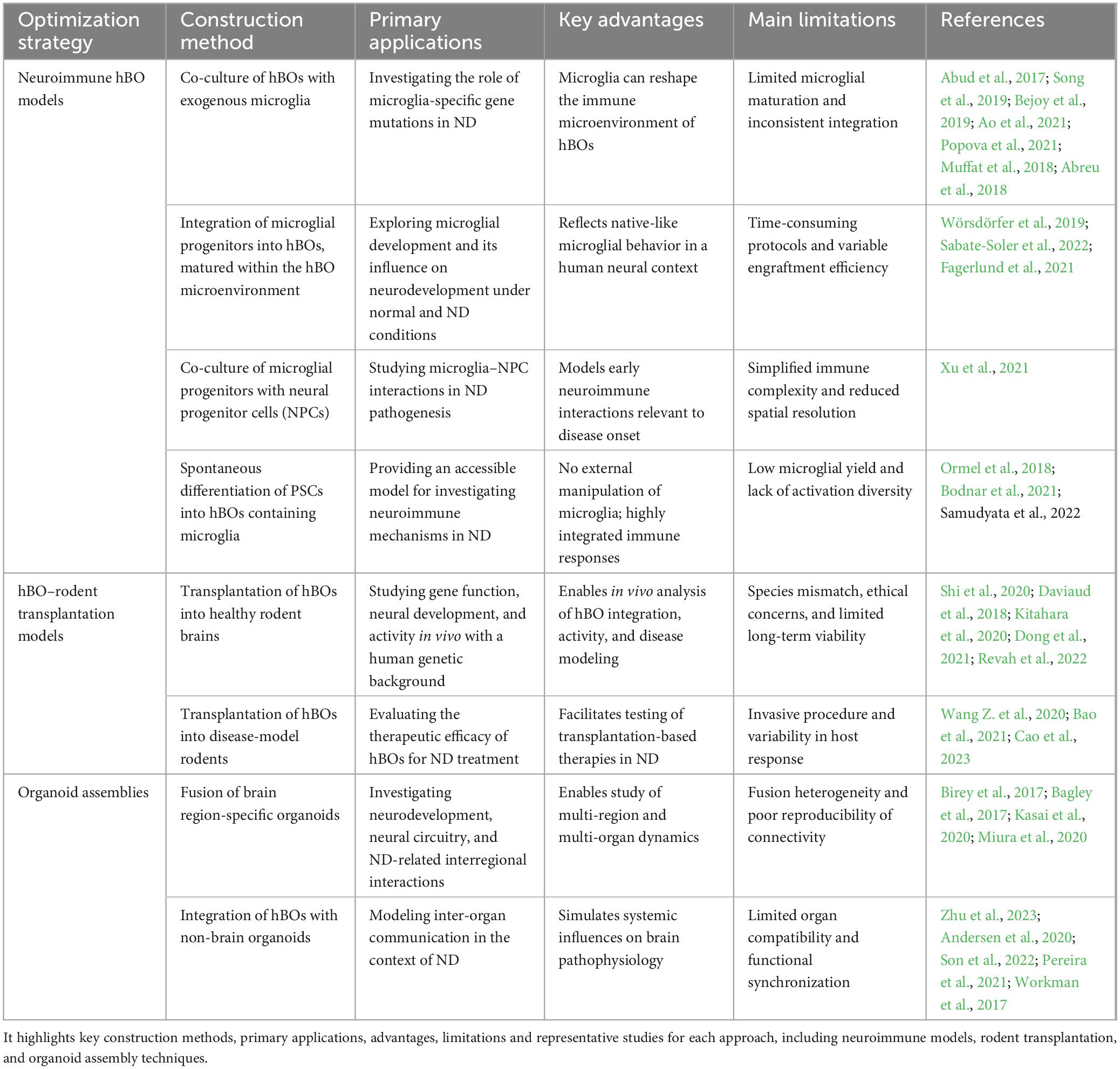
Table 2. Summarizes current bioengineering strategies used to enhance the physiological relevance of hBOs in neurological disease (ND) research.
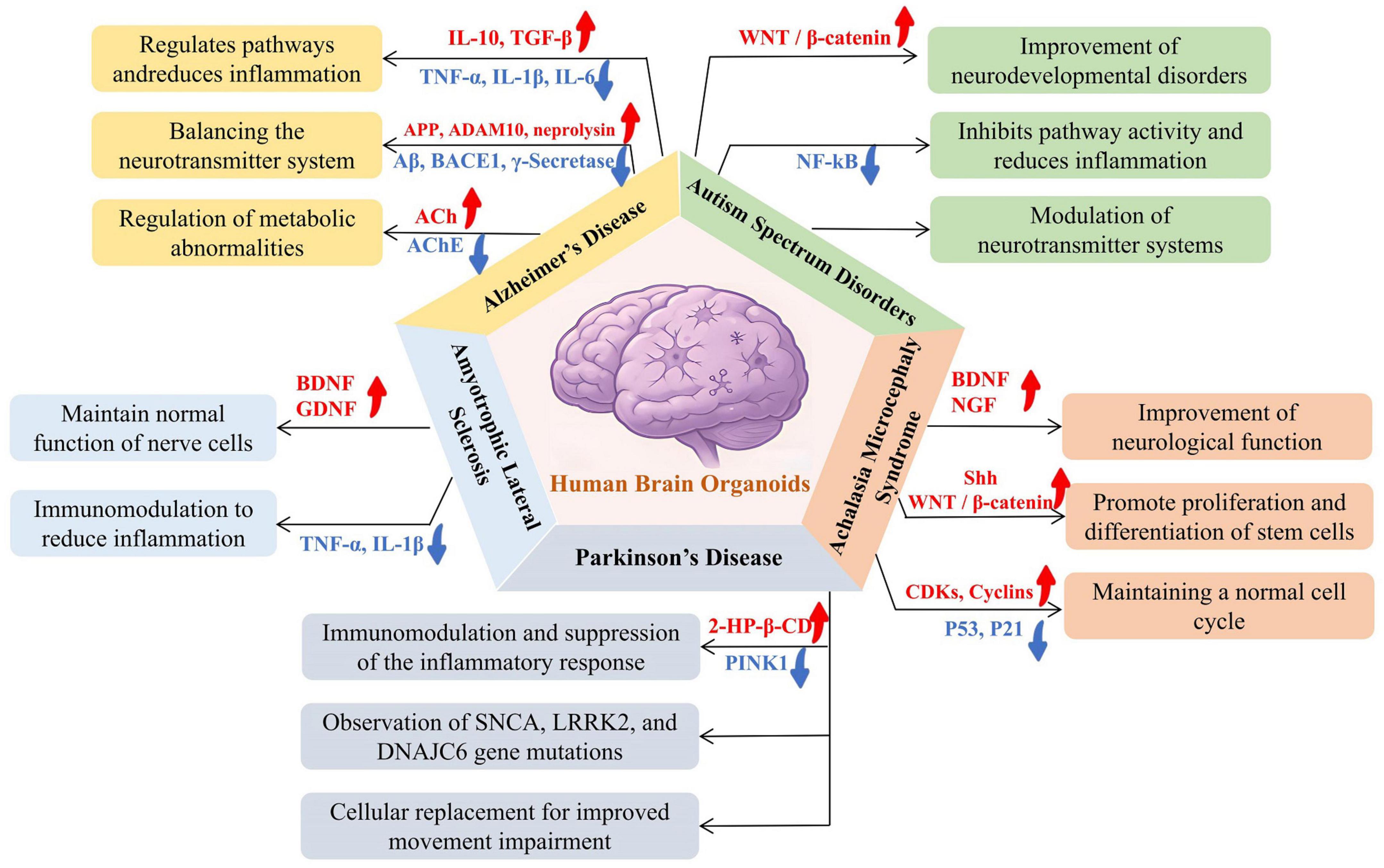
Figure 3. Applications of hBOs in modeling disease mechanisms across five common neurological disorders: Alzheimer’s disease (AD), autism spectrum disorder (ASD), achalasia microcephaly syndrome (AMS), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS). The research demonstrated specific applications in disease modeling, including simulating beta-amyloid plaque and tau tangles in AD, studying neurodevelopmental abnormalities and synaptic dysfunction in ASD, observing brain developmental disorders and microcephaly in AMS, recapitulating dopaminergic neuron degeneration in PD, and modeling motor neuron degeneration and neuroinflammation in ALS. The hBOs provide a valuable in vitro model system for investigating the pathogenesis of neurological diseases, capable of recapitulating key pathological features and aiding in disease mechanism research and drug development.
5.1 Alzheimer’s disease (AD)
Alzheimer’s disease is a progressive neurodegenerative diseases (NDGD) primarily characterized by cognitive decline and behavioral disturbances. Its key pathological hallmarks include extracellular amyloid-beta (Aβ) plaque deposition and intracellular accumulation of hyperphosphorylated tau (P-Tau), ultimately leading to neurofibrillary tangles (Arvanitakis et al., 2019). Chen et al. (2021) demonstrated that hBOs exposed to serum from AD patients reproduced core AD pathologies, including elevated P-Tau expression, Aβ aggregation, disrupted neural networks, and synaptic degeneration. Similarly, Pavoni et al. (2018) showed that exogenous administration of Aβ42 to hBOs led to time-dependent Aβ accumulation and plaque formation. Mechanistic studies by Lee et al. (2022) revealed that Zika virus–induced activation of the PERK/eIF2α signaling pathway in hBOs could trigger AD-like pathological features. Notably, pharmacological inhibition of PERK significantly alleviated these abnormalities, offering valuable therapeutic insights (Lee et al., 2022). Zhao et al. (2020) demonstrated that hBOs derived from AD patients carrying the APOEε4/ε4 genotype exhibited increased apoptosis, reduced synaptic integrity, and exacerbated tau phosphorylation compared to those from APOEε3/ε3 individuals. Notably, isogenic conversion of APOE4 to APOE3 significantly attenuated these pathological phenotypes, supporting APOE4’s role in driving neurodegenerative processes in AD (Zhao et al., 2020). Perez-Corredor et al. (2024) applied CRISPR-Cas9 to convert the APOE3Ch allele to wild-type APOE3 in hBOs, finding that APOE3Ch substantially reduced tau pathology in AD organoid models. Additionally, astrocytes and microglia derived from APOE4-genotyped hBOs demonstrated impaired Aβ42 uptake, while APOE4-to-APOE3 conversion markedly improved these pathological features (Kim et al., 2015). Penney et al. (2020) highlighted that hBOs derived from iPSCs can successfully recapitulate key cellular dysfunctions observed in AD, including impaired neuron–glia interactions and AD-associated molecular phenotypes, underscoring the utility of hBO-based platforms in modeling complex neurodegenerative processes. Choi et al. (2014) established a 3D hBO model harboring familial AD mutations that faithfully recapitulated both extracellular amyloid-β plaque deposition and intracellular tau aggregation. Treatment with β- and γ-secretase inhibitors significantly reduced amyloid and tau pathologies, demonstrating the model’s potential for therapeutic screening (Choi et al., 2014).
5.2 Parkinson’s disease (PD)
Parkinson’s disease, the second most prevalent NDGD, is clinically characterized by bradykinesia, resting tremors, and muscular rigidity. Its pathological hallmarks include progressive dopaminergic neuron loss in the substantia nigra and the formation of Lewy bodies. Mohamed et al. detected α-synuclein aggregates in hBOs derived from SNCA-mutant cells, leading to dopaminergic neuronal degeneration (Mohamed et al., 2021). Kano et al. (2020) reported a significant reduction in astrocyte populations in PD-hBOs harboring PRKN mutations, mirroring neuropathological changes observed in PD patients with these variants. Furthermore, LRRK2 mutations introduced into healthy PSCs successfully recapitulated hallmark PD features, including dopaminergic neuron loss and Lewy body formation (Zagare et al., 2022). Wulansari et al. (2021) demonstrated that DNAJC6 mutations impaired WNT–LMX1A signaling, increased α-synuclein accumulation, and disrupted autophagy–lysosomal function in hBOs. Significantly, treatment of PINK1-mutant hBOs with 2-hydroxypropyl-β-cyclodextrin (2-HP-β-CD) improved mitochondrial function and neuronal autophagy, reducing dopaminergic neuron degeneration and necrosis. These findings suggest that 2-HP-β-CD may serve as a promising disease-modifying therapy for PD (Jarazo et al., 2022). Zheng et al. (2023) transplanted hBOs derived from healthy human cells into the striatum of immunodeficient PD model mice. The organoids successfully engrafted, matured, and significantly improved motor function, underscoring their potential for cell-replacement therapy in PD (Zheng et al., 2023). Additionally, CRISPR-generated LRRK2-knockout hBOs reproduced PD-related pathology, further confirming LRRK2’s pivotal role in PD pathogenesis (Kim et al., 2019).
5.3 Amyotrophic lateral sclerosis (ALS)
Amyotrophic lateral sclerosis is a progressive NDGD marked by the degeneration of both upper and lower motor neurons. Genetic studies have identified 42 ALS-associated genes, including C9ORF72, ATXN2, and TAR DNA-binding protein 43 (TDP-43). Pathogenic mechanisms include excitotoxicity, oxidative stress imbalance, and mitochondrial dysfunction (Brown and Al-Chalabi, 2017). Szebényi et al. (2021) developed an hBO slice culture model from iPSCs of C9ORF72 ALS patients and identified early pathological features, including P62 accumulation in astroglia, poly(GA) dipeptide aggregates, DNA damage, and nuclear pyknosis in deep-layer neurons, which were partially rescued by treatment with the PERK inhibitor GSK2606414. Tamaki et al. (2023) reported that TDP-43 aggregates spread intercellularly within hBOs. This spread triggers astrocyte proliferation, DNA double-strand breaks, and cell death, which are hallmarks of ALS pathology. de Majo et al. (2023) demonstrated that GRN-deficient astrocytes induced TDP-43 hyperphosphorylation and misfolding in hBOs, a molecular signature of TDP-43 proteinopathy. Furthermore, co-culture models of hBOs and microglia have offered deeper insights into how glial cells, especially microglia and astrocytes, interact in ALS pathogenesis (Hong et al., 2023).
5.4 Autism spectrum disorders (ASD)
Autism spectrum disorders is a complex neurodevelopmental condition characterized by deficits in social communication, language impairments, and repetitive behaviors, with highly heterogeneous genetic and environmental etiologies. Zhang et al. (2020) reported that mutations in RAB39B led to increased hBO volume and excessive neural progenitor cell (NPC) proliferation, resulting in thickened SOX2+ ventricular zones and impaired neuronal differentiation. These abnormalities were attributed to hyperactivation of the PI3K-AKT-mTOR signaling pathway following RAB39B deletion (Zhang et al., 2020). Wang et al. utilized CRISPR-Cas9 to generate CHD8-deficient iPSCs, which were subsequently differentiated into hBOs. Their study revealed that CHD8 regulates ASD-associated genes, such as TCF4 and AUTS2, affecting Wnt/β-catenin signaling and GABAergic neuron differentiation–key processes implicated in ASD pathogenesis (Wang P. et al., 2017). Mariani et al. (2015) demonstrated that hBOs derived from ASD patients exhibited accelerated NPC cell cycle progression during early neurodevelopment, leading to overproduction of GABAergic neurons and resulting in an excitatory/inhibitory (E/I) imbalance. This phenotype was potentially driven by dysregulation of the FOXG1 gene (Mariani et al., 2015). Schafer et al. (2019) analyzed patient-derived hBOs and identified asynchronous disruptions in gene regulatory networks during early NPC development, which prematurely promoted neuronal differentiation. Additionally, Marchetto et al. (2017) showed that aberrant regulation of the β-catenin/BRN2 transcriptional axis resulted in synaptic transmission deficits and functional impairments in neuronal networks derived from ASD hBOs.
5.5 Achalasia-microcephaly syndrome (AMS)
Achalasia-microcephaly syndrome is a neurodevelopmental disorder (NDVD) characterized by imbalanced NPC proliferation and apoptosis, leading to reduced neuronal and glial populations and resulting in structural abnormalities of the brain. Key genes implicated in AMS pathogenesis include NARS1, WDR62, CDK5RAP2, and CPAP. Lancaster et al. (2013) were among the first to model AMS using hBOs derived from patient-specific iPSCs carrying CDK5RAP2 mutations. These organoids exhibited key pathological features, including impaired NPC proliferation and premature neuronal differentiation. Moreover, by employing RNA interference and patient-derived iPSCs, hBOs were generated that recapitulated the core characteristics of microcephaly, such as disrupted progenitor zone organization and early neurogenesis, providing critical insights into the cellular mechanisms contributing to the reduced brain size observed in AMS patients (Lancaster et al., 2013). Wang L. et al. (2020) developed cortical hBOs from AMS patients with NARS1 mutations and observed diminished proliferative capacity of radial glial cells (RGCs) and disrupted lineage specification of both RGCs and astrocytes. These findings underscore the critical role of NARS1 in RGC regulation during brain development (Wang L. et al., 2020). Gabriel et al. (2016) used iPSC-derived hBOs to show that loss of CPAP, another AMS-associated gene, induced a premature shift from symmetric to asymmetric NPC division, ultimately impairing neurogenesis. Similarly, Zhang et al. (2019) modeled primary AMS using hBOs bearing WDR62 mutations and reported defects in NPC cell cycle progression and reduced outer radial glia proliferation.
5.6 Epilepsy (EP) and brain tumors (BT)
Human brain organoids have emerged as powerful platforms for investigating the pathogenesis of EP and BT. Adeyeye et al. (2024) demonstrated the utility of integrating hBOs with microelectrode array (MEA) technology to study impaired plasticity and aberrant information processing in epileptic neural circuits. Brown et al. (2024) demonstrated that hBOs replicate key developmental and electrophysiological features of genetic epilepsies, including hyperexcitability dynamics and responsiveness to antiepileptic drugs, thus providing a physiologically relevant 3D model to investigate EP and screen therapeutic compounds. By engineering diverse hBO-based epilepsy models, researchers have explored mechanisms underlying EP and the interplay between neuronal firing patterns, cellular maturation, and subtype-specific vulnerability (Gross, 2022). Moreover, hBOs deficient in CDKL5 successfully recapitulated epilepsy-related phenotypes. These models revealed that CDKL5 mutations cause early-stage cortical neuron hyperexcitability, which is followed by late-stage hypoexcitability. Importantly, both hyperexcitability and hypoexcitability were reversed by pharmacological or gene therapy interventions (Negraes et al., 2021). In BT research, Linkous et al. (2019) developed a glioblastoma (GBM) model by co-culturing patient-derived glioblastoma stem cells (GSCs) with embryonic stem cells (ESCs) to generate GSC-hBOs. These GSCs exhibited deep tissue infiltration, proliferated within host tissue, and formed tumor-like masses that closely mimicked primary GBM pathology (Linkous et al., 2019). In studies of medulloblastoma (MB), Ballabio et al. (2020) demonstrated that SMARCA4 suppresses the oncogenic activity of the OTX2/MYC axis in both patient-derived tissues and hBO models. Treatment with an EZH2-specific inhibitor significantly reduced OTX2/MYC-driven tumorigenesis in hBOs, underscoring the potential of hBOs for modeling genetic drivers and therapeutic responses in MB.
6 Conclusion and future perspectives
Human brain organoids represent powerful tools for modeling ND including AD, PD, ASD, ALS, AMS and others. By providing representative examples, this review highlights how hBOs are being applied to investigate disease-specific mechanisms and therapy. Compared to 2D cultures and animal models, hBOs better replicate the structural complexity and cellular diversity of the human brain, enhancing their translational relevance. However, several limitations persist, including limited vascularization, incomplete neuronal maturation, batch variability, lack of microglia and mature oligodendrocytes, and ethical concerns. While emerging technologies such as multi-omics integration, gene editing, and biomaterial engineering hold great promise, this mini-review does not provide comprehensive coverage of those aspects. Continued progress in standardization and bioengineering will be essential to overcome current challenges and unlock the full diagnostic and therapeutic potential of hBOs in neuroscience research.
Author contributions
HL: Writing – review & editing, Investigation, Conceptualization, Writing – original draft. JZ: Investigation, Writing – original draft, Conceptualization, Writing – review & editing. JL: Software, Investigation, Writing – review & editing, Writing – original draft. YW: Data curation, Conceptualization, Writing – original draft, Writing – review & editing. CL: Writing – original draft, Data curation, Investigation, Writing – review & editing. YH: Data curation, Writing – original draft, Investigation, Writing – review & editing. JW: Data curation, Investigation, Writing – review & editing, Writing – original draft. WL: Writing – review & editing, Funding acquisition, Conceptualization, Writing – original draft. HW: Investigation, Writing – review & editing, Writing – original draft, Funding acquisition, Conceptualization. ZM: Conceptualization, Investigation, Funding acquisition, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by National Natural Science Foundation of China (81873030), Science Project of Education Department of Guangdong Province (2024KSYS007, and 2020ZDZX2045), Innovative Research Team Funding Project of Zhaoqing University (TD202414), Zhaoqing Science and Technology Innovation Guidance Project (ZhaoKe[2025]4-No.272), Innovation and Entrepreneurship Training Program for College Students of Zhaoqing University (202410580022, X202510580154, and X202510580156), Zhaoqing University Research Startup Project for Recruited Talents (611/240024), and Zhaoqing University Research Fund for Young Faculty (504/2025013337).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelbasset, M., Saron, W. A. A., Ma, D., Rathore, A. P. S., Kozaki, T., Zhong, C., et al. (2024). Differential contributions of fetal mononuclear phagocytes to Zika virus neuroinvasion versus neuroprotection during congenital infection. Cell 187, 7511–7532. doi: 10.1016/j.cell.2024.10.028
Abreu, C. M., Gama, L., Krasemann, S., Chesnut, M., Odwin-Dacosta, S., Hogberg, H. T., et al. (2018). Microglia increase inflammatory responses in iPSC-Derived Human BrainSpheres. Front. Microbiol. 9:2766. doi: 10.3389/fmicb.2018.02766
Abud, E. M., Ramirez, R. N., Martinez, E. S., Healy, L. M., Nguyen, C. H. H., Newman, S. A., et al. (2017). iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293. doi: 10.1016/j.neuron.2017.03.042
Acharya, P., Choi, N. Y., Shrestha, S., Jeong, S., and Lee, M. Y. (2024). Brain organoids: A revolutionary tool for modeling neurological disorders and development of therapeutics. Biotechnol. Bioeng. 121, 489–506. doi: 10.1002/bit.28606
Adeyeye, A., Mirsadeghi, S., Gutierrez, M., and Hsieh, J. (2024). Integrating adult neurogenesis and human brain organoid models to advance epilepsy and associated behavioral research. Epilepsy Behav. 159:109982. doi: 10.1016/j.yebeh.2024.109982
Amirifar, L., Shamloo, A., Nasiri, R., de Barros, N. R., Wang, Z. Z., Unluturk, B. D., et al. (2022). Brain-on-a-chip: Recent advances in design and techniques for microfluidic models of the brain in health and disease. Biomaterials 285:121531. doi: 10.1016/j.biomaterials.2022.121531
Andersen, J., Revah, O., Miura, Y., Thom, N., Amin, N. D., Kelley, K. W., et al. (2020). Generation of functional human 3D cortico-motor assembloids. Cell 183, 1913–1929. doi: 10.1016/j.cell.2020.11.017
Ao, Z., Cai, H., Wu, Z., Song, S., Karahan, H., Kim, B., et al. (2021). Tubular human brain organoids to model microglia-mediated neuroinflammation. Lab. Chip. 21, 2751–2762. doi: 10.1039/d1lc00030f
Arvanitakis, Z., Shah, R. C., and Bennett, D. A. (2019). Diagnosis and management of dementia: Review. JAMA 322, 1589–1599. doi: 10.1001/jama.2019.4782
Bagley, J. A., Reumann, D., Bian, S., Lévi-Strauss, J., and Knoblich, J. A. (2017). Fused cerebral organoids model interactions between brain regions. Nat. Methods 14, 743–751. doi: 10.1038/nmeth.4304
Ballabio, C., Anderle, M., Gianesello, M., Lago, C., Miele, E., Cardano, M., et al. (2020). Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat. Commun. 11:583. doi: 10.1038/s41467-019-13989-3
Bao, Z., Fang, K., Miao, Z., Li, C., Yang, C., Yu, Q., et al. (2021). Human cerebral organoid implantation alleviated the neurological deficits of traumatic brain injury in mice. Oxid. Med. Cell. Longev. 2021:6338722. doi: 10.1155/2021/6338722
Barnat, M., Capizzi, M., Aparicio, E., Boluda, S., Wennagel, D., Kacher, R., et al. (2020). Huntington’s disease alters human neurodevelopment. Science 369, 787–793. doi: 10.1126/science.aax3338
Bejoy, J., Yuan, X., Song, L., Hua, T., Jeske, R., Sart, S., et al. (2019). Genomics analysis of metabolic pathways of human stem cell-derived microglia-like cells and the integrated cortical spheroids. Stem Cells Int. 2019:2382534. doi: 10.1155/2019/2382534
Birey, F., Andersen, J., Makinson, C. D., Islam, S., Wei, W., Huber, N., et al. (2017). Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59. doi: 10.1038/nature22330
Bodnar, B., Zhang, Y., Liu, J., Lin, Y., Wang, P., Wei, Z., et al. (2021). Novel scalable and simplified system to generate microglia-containing cerebral organoids from human induced pluripotent stem cells. Front. Cell. Neurosci. 15:682272. doi: 10.3389/fncel.2021.682272
Bourret, R., Martinez, E., Vialla, F., Giquel, C., Thonnat-Marin, A., and De Vos, J. (2016). Human-animal chimeras: Ethical issues about farming chimeric animals bearing human organs. Stem. Cell. Res. Ther. 7:87. doi: 10.1186/s13287-016-0345-9
Bracha, S., Johnson, H. J., Pranckevicius, N. A., Catto, F., Economides, A. E., Litvinov, S., et al. (2024). Engineering Toxoplasma gondii secretion systems for intracellular delivery of multiple large therapeutic proteins to neurons. Nat. Microbiol. 9, 2051–2072. doi: 10.1038/s41564-024-01750-6
Brown, R., Rabeling, A., and Goolam, M. (2024). Progress and potential of brain organoids in epilepsy research. Stem Cell Res. Ther. 15:361. doi: 10.1186/s13287-024-03944-5
Brown, R. H., and Al-Chalabi, A. (2017). Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172. doi: 10.1056/NEJMra1603471
Cao, S. Y., Yang, D., Huang, Z. Q., Lin, Y. H., Wu, H. Y., Chang, L., et al. (2023). Cerebral organoids transplantation repairs infarcted cortex and restores impaired function after stroke. NPJ Regen. Med. 8:27. doi: 10.1038/s41536-023-00301-7
Castiglione, H., Vigneron, P. A., Baquerre, C., Yates, F., Rontard, J., and Honegger, T. (2022). Human brain organoids-on-chip: Advances, challenges, and perspectives for preclinical applications. Pharmaceutics 14:2301. doi: 10.3390/pharmaceutics14112301
Chen, X., Sun, G., Tian, E., Zhang, M., Davtyan, H., Beach, T. G., et al. (2021). Modeling sporadic Alzheimer’s Disease in human brain organoids under serum exposure. Adv. Sci. 8:e2101462. doi: 10.1002/advs.202101462
Choi, S. H., Kim, Y. H., Hebisch, M., Sliwinski, C., Lee, S., D’Avanzo, C., et al. (2014). A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278. doi: 10.1038/nature13800
Daviaud, N., Friedel, R. H., and Zou, H. (2018). Vascularization and engraftment of transplanted human cerebral organoids in mouse cortex. eNeuro 5, 1–18. doi: 10.1523/ENEURO.0219-18.2018
de Majo, M., Koontz, M., Marsan, E., Salinas, N., Ramsey, A., Kuo, Y. M., et al. (2023). Granulin loss of function in human mature brain organoids implicates astrocytes in TDP-43 pathology. Stem Cell Rep. 18, 706–719. doi: 10.1016/j.stemcr.2023.01.012
Dong, X., Xu, S. B., Chen, X., Tao, M., Tang, X. Y., and Fang, K. H. (2021). Human cerebral organoids establish subcortical projections in the mouse brain after transplantation. Mol. Psychiatry 26, 2964–2976. doi: 10.1038/s41380-020-00910-4
Driehuis, E., and Clevers, H. (2017). CRISPR/Cas 9 genome editing and its applications in organoids. Am. J. Physiol. Gastrointest. Liver. Physiol. 312, G257–G265. doi: 10.1152/ajpgi.00410.2016
Fagerlund, I., Dougalis, A., Shakirzyanova, A., Gómez-Budia, M., Pelkonen, A., Konttinen, H., et al. (2021). Microglia-like cells promote neuronal functions in cerebral organoids. Cells 11:124. doi: 10.3390/cells11010124
Gabriel, E., Wason, A., Ramani, A., Gooi, L. M., Keller, P., Pozniakovsky, A., et al. (2016). CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J. 35, 803–819. doi: 10.15252/embj.201593679
Gross, C. (2022). Epilepsy research now in 3D: Harnessing the power of brain organoids in epilepsy. Epilepsy. Curr. 22, 135–136. doi: 10.1177/15357597211070391
Hartlaub, A. M., McElroy, C. A., Maitre, N. L., and Hester, M. E. (2019). Modeling human brain circuitry using pluripotent stem cell platforms. Front. Pediatr. 7:57. doi: 10.3389/fped.2019.00057
Hong, Y., Dong, X., Chang, L., Xie, C., Chang, M., Aguilar, J. S., et al. (2023). Microglia-containing cerebral organoids derived from induced pluripotent stem cells for the study of neurological diseases. iScience 26:106267. doi: 10.1016/j.isci.2023.106267
Humpel, C. (2015). Organotypic brain slice cultures: A review. Neuroscience 305, 86–98. doi: 10.1016/j.neuroscience.2015.07.086
Jarazo, J., Barmpa, K., Modamio, J., Saraiva, C., Sabaté-Soler, S., Rosety, I., et al. (2022). Parkinson’s disease phenotypes in patient neuronal cultures and brain organoids improved by 2-hydroxypropyl-β-cyclodextrin treatment. Mov. Disord. 37, 80–94. doi: 10.1002/mds.28810
Jia, J., Zhang, Y., Shi, Y., Yin, X., Wang, S., Li, Y., et al. (2023). A 19-year-old adolescent with probable Alzheimer’s Disease. J. Alzheimers. Dis. 91, 915–922. doi: 10.3233/JAD-221065
Jo, J., Xiao, Y., Sun, A. X., Cukuroglu, E., Tran, H. D., Göke, J., et al. (2016). Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 19, 248–257. doi: 10.1016/j.stem.2016.07.005
Kano, M., Takanashi, M., Oyama, G., Yoritaka, A., Hatano, T., Shiba-Fukushima, K., et al. (2020). Reduced astrocytic reactivity in human brains and midbrain organoids with PRKN mutations. NPJ Park. Dis. 6:33. doi: 10.1038/s41531-020-00137-8
Kanupriya, K., Pal Verma, S., Sharma, V., Mishra, I., and Mishra, R. (2025). Advances in human brain organoids: Methodological innovations and future directions for drug discovery. Curr. Drug Res. Rev. 17, 360–374. doi: 10.2174/0125899775369286250206050006
Kasai, T., Suga, H., Sakakibara, M., Ozone, C., Matsumoto, R., Kano, M., et al. (2020). Hypothalamic contribution to pituitary functions is recapitulated in vitro using 3D-cultured human iPS cells. Cell. Rep. 30, 18–24. doi: 10.1016/j.celrep.2019.12.009
Kiaee, K., Jodat, Y. A., Bassous, N. J., Matharu, N., and Shin, S. R. (2021). Transcriptomic mapping of neural diversity, differentiation and functional trajectory in iPSC-Derived 3D brain organoid models. Cells 10:3422. doi: 10.3390/cells10123422
Kim, H., Park, H. J., Choi, H., Chang, Y., Park, H., Shin, J., et al. (2019). Modeling G2019S-LRRK2 sporadic Parkinson’s Disease in 3D midbrain organoids. Stem Cell Rep. 12, 518–531. doi: 10.1016/j.stemcr.2019.01.020
Kim, Y. H., Choi, S. H., D’Avanzo, C., Hebisch, M., Sliwinski, C., Bylykbashi, E., et al. (2015). A 3D human neural cell culture system for modeling Alzheimer’s disease. Nat. Protoc. 10, 985–1006. doi: 10.1038/nprot.2015.065
Kitahara, T., Sakaguchi, H., Morizane, A., Kikuchi, T., Miyamoto, S., and Takahashi, J. (2020). Axonal extensions along corticospinal tracts from transplanted human cerebral organoids. Stem Cell Rep. 15, 467–481. doi: 10.1016/j.stemcr.2020.06.016
Kogler, S., Kmurcu, K., Olsen, C., Shoji, J. Y., Skottvoll, F. S., Krauss, S., et al. (2023). Organoids, organ-on-a-chip, separation science and mass spectrometry: An update. TrAC 161:116996. doi: 10.1016/j.trac.2023.116996
Kwak, T., Park, S. H., Lee, S., Shin, Y., Yoon, K. J., Cho, S. W., et al. (2024). Guidelines for manufacturing and application of organoids: Brain. Int. J. Stem Cells. 17, 158–181. doi: 10.15283/ijsc24056
Lancaster, M. A., and Knoblich, J. A. (2014). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340. doi: 10.1038/nprot.2014.158
Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Lee, C. T., Bendriem, R. M., Wu, W. W., and Shen, R. F. (2017). 3D brain Organoids derived from pluripotent stem cells: Promising experimental models for brain development and neurodegenerative disorders. J. Biomed. Sci. 24:59. doi: 10.1186/s12929-017-0362-8
Lee, S. E., Choi, H., Shin, N., Kong, D., Kim, N. G., Kim, H. Y., et al. (2022). Zika virus infection accelerates Alzheimer’s disease phenotypes in brain organoids. Cell Death Discov. 8:153. doi: 10.1038/s41420-022-00958-x
Linkous, A., Balamatsias, D., Snuderl, M., Edwards, L., Miyaguchi, K., Milner, T., et al. (2019). Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 26, 3203–3211. doi: 10.1016/j.celrep.2019.02.063
Marchetto, M. C., Belinson, H., Tian, Y., Freitas, B. C., Fu, C., Vadodaria, K., et al. (2017). Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry 22, 820–835. doi: 10.1038/mp.2016.95
Mariani, J., Coppola, G., Zhang, P., Abyzov, A., Provini, L., Tomasini, L., et al. (2015). FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390. doi: 10.1016/j.cell.2015.06.034
Marinho, L. S. R., Chiarantin, G. M. D., Ikebara, J. M., Cardoso, D. S., de Lima-Vasconcellos, T. H., Higa, G. S. V., et al. (2023). The impact of antidepressants on human neurodevelopment: Brain organoids as experimental tools. Semin. Cell Dev. Biol. 144, 67–76. doi: 10.1016/j.semcdb.2022.09.007
Mimeault, M., Hauke, R., Mehta, P. P., and Batra, S. K. (2007). Recent advances in cancer stem/progenitor cell research: Therapeutic implications for overcoming resistance to the most aggressive cancers. J. Cell. Mol. Med. 11, 981–1011. doi: 10.1111/j.1582-4934.2007.00088.x
Mishra, I., Gupta, K., Mishra, R., Chaudhary, K., and Sharma, V. (2024). An exploration of organoid technology: Present advancements, applications, and obstacles. Curr. Pharm. Biotechnol. 25, 1000–1020. doi: 10.2174/0113892010273024230925075231
Miura, Y., Li, M. Y., Birey, F., Ikeda, K., Revah, O., Thete, M. V., et al. (2020). Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. biotechnol. 38, 1421–1430. doi: 10.1038/s41587-020-00763-w
Mohamed, N. V., Sirois, J., Ramamurthy, J., Mathur, M., Lépine, P., Deneault, E., et al. (2021). Midbrain organoids with an SNCA gene triplication model key features of synucleinopathy. Brain Commun. 3:fcab223. doi: 10.1093/braincomms/fcab223
Muffat, J., Li, Y., Omer, A., Durbin, A., Bosch, I., Bakiasi, G., et al. (2018). Human induced pluripotent stem cell-derived glial cells and neural progenitors display divergent responses to Zika and dengue infections. Proc. Natl. Acad. Sci. U. S. A. 115, 7117–7122. doi: 10.1073/pnas.1719266115
Negraes, P. D., Trujillo, C. A., Yu, N. K., Wu, W., Yao, H., Liang, N., et al. (2021). Altered network and rescue of human neurons derived from individuals with early-onset genetic epilepsy. Mol. Psychiatry 26, 7047–7068. doi: 10.1038/s41380-021-01104-2
Ormel, P. R., Vieira de Sá, R., van Bodegraven, E. J., Karst, H., Harschnitz, O., Sneeboer, M. A. M., et al. (2018). Microglia innately develop within cerebral organoids. Nat. Commun. 9:4167. doi: 10.1038/s41467-018-06684-2
Pavoni, S., Jarray, R., Nassor, F., Guyot, A. C., Cottin, S., Rontard, J., et al. (2018). Small-molecule induction of Aβ-42 peptide production in human cerebral organoids to model Alzheimer’s disease associated phenotypes. PLoS One 13:e0209150. doi: 10.1371/journal.pone.0209150
Penney, J., Ralvenius, W. T., and Tsai, L. (2020). Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol. Psychiatry 25, 148–167. doi: 10.1038/s41380-019-0468-3
Pereira, J. D., DuBreuil, D. M., Devlin, A. C., Held, A., Sapir, Y., Berezovski, E., et al. (2021). Human sensorimotor organoids derived from healthy and amyotrophic lateral sclerosis stem cells form neuromuscular junctions. Nat. Commun. 12:4744. doi: 10.1038/s41467-021-24776-4
Perez-Corredor, P., Vanderleest, T. E., Vacano, G. N., Sanchez, J. S., Villalba-Moreno, N. D., Marino, C., et al. (2024). APOE3 Christchurch modulates β-catenin/Wnt signaling in iPS cell-derived cerebral organoids from Alzheimer’s cases. Front. Mol. Neurosci. 17:1373568. doi: 10.3389/fnmol.2024.1373568
Pieters, V. M., Co, I. L., Wu, N. C., and McGuigan, A. P. (2021). Applications of omics technologies for three-dimensional in vitro disease models. Tissue Eng. Part C Methods 27, 183–199. doi: 10.1089/ten.TEC.2020.0300
Popova, G., Soliman, S. S., Kim, C. N., Keefe, M. G., Hennick, K. M., Jain, S., et al. (2021). Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell 28, 2153–2166. doi: 10.1016/j.stem.2021.08.015
Qian, X., Nguyen, H. N., Song, M. M., Hadiono, C., Ogden, S. C., Hammack, C., et al. (2016). Brain-region-specific organoids using mini-bioreactors for modeling ZIKV Exposure. Cell 165, 1238–1254. doi: 10.1016/j.cell.2016.04.032
Qian, X., Song, H., and Ming, G. L. (2019). Brain organoids: Advances, applications and challenges. Development 146:dev166074. doi: 10.1242/dev.166074
Quadrato, G., Nguyen, T., Macosko, E. Z., Sherwood, J. L., Min Yang, S., Berger, D. R., et al. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53. doi: 10.1038/nature22047
Raja, W. K., Mungenast, A. E., Lin, Y. T., Ko, T., Abdurrob, F., Seo, J., et al. (2016). Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS One 11:e0161969. doi: 10.1371/journal.pone.0161969
Revah, O., Gore, F., Kelley, K. W., Andersen, J., Sakai, N., and Chen, X. (2022). Maturation and circuit integration of transplanted human cortical organoids. Nature 610, 319–326. doi: 10.1038/s41586-022-05277-w
Sabate-Soler, S., Nickels, S. L., Saraiva, C., Berger, E., Dubonyte, U., Barmpa, K., et al. (2022). Microglia integration into human midbrain organoids leads to increased neuronal maturation and functionality. Glia 70, 1267–1288. doi: 10.1002/glia.24167
Samudyata, Oliveira, A. O., Malwade, S., Rufino de Sousa, N., Goparaju, S. K., Gracias, J., et al. (2022). SARS-CoV-2 promotes microglial synapse elimination in human brain organoids. Mol. Psychiatry 27, 3939–3950. doi: 10.1038/s41380-022-01786-2
Schafer, S. T., Paquola, A. C. M., Stern, S., Gosselin, D., Ku, M., Pena, M., et al. (2019). Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat. Neurosci. 22, 243–255. doi: 10.1038/s41593-018-0295-x
Shi, Y., Sun, L., Wang, M., Liu, J., Zhong, S., Li, R., et al. (2020). Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 18:e3000705. doi: 10.1371/journal.pbio.3000705
Smits, L. M., and Schwamborn, J. C. (2020). Midbrain organoids: A new tool to investigate Parkinson’s Disease. Front. Cell. Dev. Biol. 8:359. doi: 10.3389/fcell.2020.00359
Son, J., Park, S. J., Ha, T., Lee, S. N., Cho, H. Y., and Choi, J. W. (2022). Electrophysiological monitoring of neurochemical-based neural signal transmission in a human brain-spinal cord assembloid. ACS. Sens. 7, 409–414. doi: 10.1021/acssensors.1c02279
Song, L., Yuan, X., Jones, Z., Vied, C., Miao, Y., Marzano, M., et al. (2019). Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Sci. Rep. 9:11055. doi: 10.1038/s41598-019-47444-6
Sun, X. Y., Ju, X. C., Li, Y., Zeng, P. M., Wu, J., Zhou, Y. Y., et al. (2022). Generation of vascularized brain organoids to study neurovascular interactions. ELife 11:e76707. doi: 10.7554/eLife.76707
Szebényi, K., Wenger, L. M. D., Sun, Y., Dunn, A. W. E., Limegrover, C. A., Gibbons, G. M., et al. (2021). Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nat. Neurosci. 24, 1542–1554. doi: 10.1038/s41593-021-00923-4
Taglieri, M., Di Gregorio, L., Matis, S., Uras, C. R. M., Ardy, M., Casati, S., et al. (2025). Colorectal organoids: Models, imaging, omics, therapy, immunology, and ethics. Cells 14:457. doi: 10.3390/cells14060457
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Tamaki, Y., Ross, J. P., Alipour, P., Castonguay, C. É, Li, B., Catoire, H., et al. (2023). Spinal cord extracts of amyotrophic lateral sclerosis spread TDP-43 pathology in cerebral organoids. PLoS Genet. 19:e1010606. doi: 10.1371/journal.pgen.1010606
Tanaka, Y. (2024). Lessons about physiological relevance learned from large-scale meta-analysis of co-expression networks in brain organoids. PLoS Biol. 22:e3002965. doi: 10.1371/journal.pbio.3002965
Velasco, S., Kedaigle, A. J., Simmons, S. K., Nash, A., Rocha, M., Quadrato, G., et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527. doi: 10.1038/s41586-019-1289-x
Wang, L., Li, Z., Sievert, D., Smith, D. E. C., Mendes, M. I., Chen, D. Y., et al. (2020). Loss of NARS1 impairs progenitor proliferation in cortical brain organoids and leads to microcephaly. Nat. commun. 11:4038. doi: 10.1038/s41467-020-17454-4
Wang, P., Mokhtari, R., Pedrosa, E., Kirschenbaum, M., Bayrak, C., Zheng, D., et al. (2017). CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism. 8:11. doi: 10.1186/s13229-017-0124-1
Wang, Z., Wang, S. N., Xu, T. Y., Hong, C., Cheng, M. H., Zhu, P. X., et al. (2020). Cerebral organoids transplantation improves neurological motor function in rat brain injury. CNS Neurosci. Ther. 26, 682–697. doi: 10.1111/cns.13286
Wang, Z., Wang, S. N., Xu, T. Y., Miao, Z. W., Su, D. F., and Miao, C. Y. (2017). Organoid technology for brain and therapeutics research. CNS Neurosci. Ther. 23, 771–778. doi: 10.1111/cns.12754
Workman, M. J., Mahe, M. M., Trisno, S., Poling, H. M., Watson, C. L., Sundaram, N., et al. (2017). Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23, 49–59. doi: 10.1038/nm.4233
Wörsdörfer, P., Dalda, N., Kern, A., Krüger, S., Wagner, N., Kwok, C. K., et al. (2019). Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci. Rep 9:15663. doi: 10.1038/s41598-019-52204-7
Wulansari, N., Darsono, W. H. W., Woo, H. J., Chang, M. Y., Kim, J., Bae, E. J., et al. (2021). Neurodevelopmental defects and neurodegenerative phenotypes in human brain organoids carrying Parkinson’s disease-linked DNAJC6 mutations. Sci. Adv. 7:eabb1540. doi: 10.1126/sciadv.abb1540
Xu, R., Boreland, A. J., Li, X., Erickson, C., Jin, M., Atkins, C., et al. (2021). Developing human pluripotent stem cell-based cerebral organoids with a controllable microglia ratio for modeling brain development and pathology. Stem Cell Rep. 16, 1923–1937. doi: 10.1016/j.stemcr.2021.06.011
Yeh, T. H., Liu, H. F., Li, Y. W., Lu, C. S., Shih, H. Y., Chiu, C. C., et al. (2018). C9orf72 is essential for neurodevelopment and motility mediated by Cyclin G1. Exp. Neurol. 304, 114–124. doi: 10.1016/j.expneurol.2018.03.002
Zagare, A., Barmpa, K., Smajic, S., Smits, L. M., Grzyb, K., Grünewald, A., et al. (2022). Midbrain organoids mimic early embryonic neurodevelopment and recapitulate LRRK2-p.Gly2019Ser-associated gene expression. Am. J. Hum. Genet. 109, 311–327. doi: 10.1016/j.ajhg.2021.12.009
Zhang, W., Ma, L., Yang, M., Shao, Q., Xu, J., Lu, Z., et al. (2020). Cerebral organoid and mouse models reveal a RAB39b-PI3K-mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev. 34, 580–597. doi: 10.1101/gad.332494.119
Zhang, W., Yang, S. L., Yang, M., Herrlinger, S., Shao, Q., Collar, J. L., et al. (2019). Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 10:2612. doi: 10.1038/s41467-019-10497-2
Zhang, Y., Pak, C., Han, Y., Ahlenius, H., Zhang, Z., Chanda, S., et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798. doi: 10.1016/j.neuron.2013.05.029
Zhao, J., Fu, Y., Yamazaki, Y., Ren, Y., Davis, M. D., Liu, C. C., et al. (2020). APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat. Commun. 11:5540. doi: 10.1038/s41467-020-19264-0
Zheng, X., Han, D., Liu, W., Wang, X., Pan, N., Wang, Y., et al. (2023). Human iPSC-derived midbrain organoids functionally integrate into striatum circuits and restore motor function in a mouse model of Parkinson’s disease. Theranostics 13, 2673–2692. doi: 10.7150/thno.80271
Zhou, J. Q., Zeng, L. H., Li, C. T., He, D. H., Zhao, H. D., Xu, Y. N., et al. (2023). Brain organoids are new tool for drug screening of neurological diseases. Neural. Regen. Res. 18, 1884–1889. doi: 10.4103/1673-5374.367983
Keywords: human brain organoids, neurological disorders, disease modeling, therapeutic innovation, induced pluripotent stem cells
Citation: Li H, Zhu J, Li J, Wu Y, Luo C, Huang Y, Wu J, Liu W, Wang H and Mo Z (2025) Human brain organoids: an innovative model for neurological disorder research and therapy. Front. Cell. Neurosci. 19:1658074. doi: 10.3389/fncel.2025.1658074
Received: 03 July 2025; Accepted: 08 August 2025;
Published: 26 August 2025.
Edited by:
Fabiola M. Ribeiro, Federal University of Minas Gerais, BrazilReviewed by:
Pablo Leal Cardozo, Federal University of Minas Gerais, BrazilRaghav Mishra, Lloyd Institute of Management & Technology, India
Copyright © 2025 Li, Zhu, Li, Wu, Luo, Huang, Wu, Liu, Wang and Mo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongwu Wang, aHd3YW5nQHpxdS5lZHUuY24=; Zhixian Mo, Y2hlcnJ5bW8wMDFAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Hancheng Li
Hancheng Li Junxiao Zhu1,3†
Junxiao Zhu1,3† Jieyu Li
Jieyu Li Hongwu Wang
Hongwu Wang