- 1Computational Biomedicine, Institute of Neuroscience and Medicine INM-9, Forschungszentrum Jülich GmbH, Jülich, Germany
- 2Department of Physics, RWTH Aachen University, Aachen, Germany
- 3Chongqing Key Laboratory of Kinase Modulators as Innovative Medicine, National & Local Joint Engineering Research Center of Targeted and Innovative Therapeutics, College of Pharmacy (International Academy of Targeted Therapeutics and Innovation), Chongqing University of Arts and Sciences, Chongqing, China
- 4Department of Physics, University of Cagliari, Cagliari, Italy
- 5Seaver Autism Center for Research and Treatment, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 6Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 7Icahn School of Medicine at Mount Sinai, The Mindich Child Health and Development Institute, New York, NY, United States
- 8Icahn School of Medicine at Mount Sinai, Friedman Brain Institute, New York, NY, United States
- 9Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 10Alper Center for Neural Development and Regeneration, Icahn School of Medicine at Mount Sinai, Friedman Brain Institute, New York, NY, United States
- 11Dipartimento di Chimica, Università di Pavia, Pavia, Italy
- 12JARA Institute: Molecular Neuroscience and Imaging, Institute of Neuroscience and Medicine INM-11, Forschungszentrum Jülich GmbH, Jülich, Germany
Six variants associated with autism spectrum disorder (ASD) abnormally activate the WASP-family Verprolin-homologous protein (WAVE) regulatory complex (WRC), a critical regulator of actin dynamics. This abnormal activation may contribute to the pathogenesis of this disorder. Using molecular dynamics (MD) simulations, we recently investigated the structural dynamics of wild-type (WT) WRC and R87C, A455P, and Q725R WRC disease-linked variants. Here, by extending MD simulations to I664M, E665K, and D724H WRC, we suggest that all of the mutations weaken the interactions and affect intra-complex allosteric communication between the WAVE1 active C-terminal region (ACR) and the rest of the complex. This might contribute to an abnormal complex activation, a hallmark of WRC-linked ASD. In addition, all mutants but I664M destabilize the ACR V-helix and increase the participation of ACR in large-scale movements. All these features may also abnormally influence the inactive WRC toward a dysfunctional state. We hypothesize that small-molecule ligands counteracting these effects may help restore normal WRC regulation in ASD-related variants.
1 Introduction
Autism spectrum disorder (ASD) is one of the most common neurodevelopmental disorders (NDDs) in childhood, affecting approximately 1% of the population (Zeidan et al., 2022). Individuals with ASD are enriched in de novo missense variants that disrupt protein–protein interactions (PPIs), with estimates that up to 25% of PPIs are disrupted, and genes encoding proteins involved in disrupted PPIs are correspondingly more readily identifiable as ASD risk genes (Chen et al., 2018, 2020). One such gene (Fu et al., 2022; Xie et al., 2025a, 2025b), CYFIP2 (cytoplasmic FMR1-interacting protein 2), encodes a subunit of the WAVE (WASP family verprolin-homologous) regulatory complex (WRC) (Chen et al., 2010) (Figure 1). This large hetero-pentameric complex comprises an elongated, pseudo-symmetrical CYFIP1/2-NCKAP1 (non-catalytic region of tyrosine Kinase Associated Protein 1) dimer and a trimer of ABI1/2/3 (Abelson interactor 1/2/3), HSPC300 (hematopoietic stem/progenitor cell protein 300), and WAVE1/2/3 proteins (Chen et al., 2010) (Figure 1). Activation of WRC has been demonstrated to regulate actin remodeling (Rottner et al., 2021), a process that is critical for brain function and development, such as synapse maturation and formation (De Rubeis et al., 2013; Davenport et al., 2019). Under physiological conditions, WRC remains “inactive” until it binds to cellular partners such as the GTPase Rac1 (Chen et al., 2017). The latter activates the complex by releasing the WAVE1 active C-terminal region (ACR) without apparently affecting the rest of the complex (Figure 1) (Ding et al., 2022).
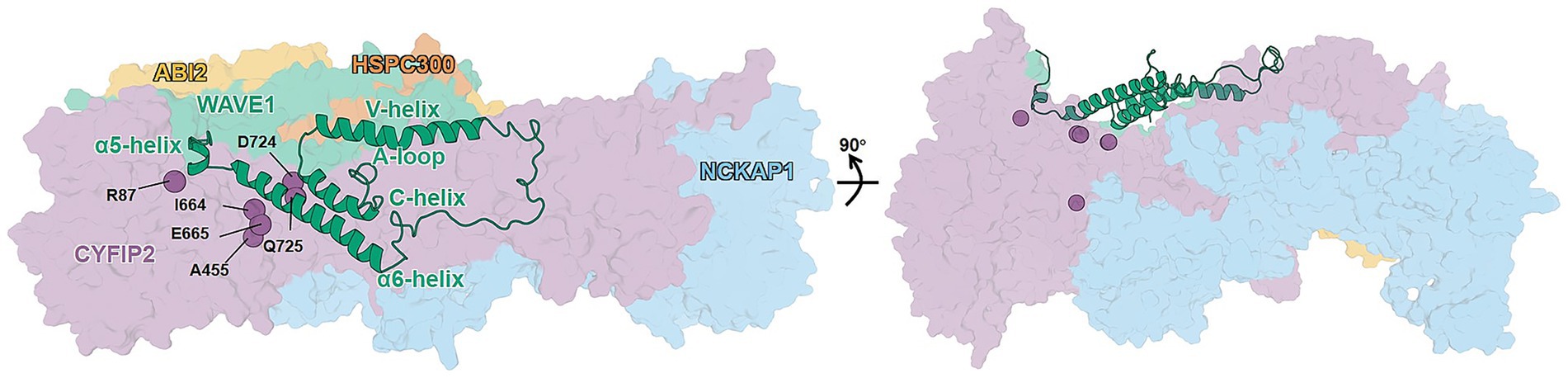
Figure 1. Architecture of WT WRC inactive form. The subunits (CYFIP2, purple; NCKAP1, blue; WAVE1, green; HSPC300, burnt orange; and ABI2, yellow) are shown as surfaces except for ACR (cartoon), which consists of α5, α6, V-, and C-helices, A-loop, and connecting loops. The mutation sites discussed in the text are shown as labeled spheres. The right-hand panel shows the 90° rotated view of the complex, emphasizing the buried location of the A455. The model is taken from our previous work (Xie et al., 2025b).
Six CYFIP2 variants related to ASD (R87C, A455P, I664M, E665K, D724H, and Q725R) abnormally convert WRC from an “inactive” state to an “active-like” state even when there is no binding to cellular partners (Figure 1) (Schaks et al., 2020)1. This abnormal activation can alter the balance between excitatory and inhibitory, spine morphology, and neuronal excitability, thus increasing the risk of ASD and other NDDs (De Rubeis et al., 2013; Nakashima et al., 2018; Zweier et al., 2019; Zhao and Guan, 2024). Previous all-atom molecular dynamics (MD) studies from us have shown that an internal variant (A455P) and two variants of the ACR/CYFIP2 interface (R87C and Q725R) similarly reduced ACR interactions with the rest of the complex, although located in different regions (Figure 1) (Xie et al., 2025b).
To complete this investigation, here we conducted the same MD protocol on the WRC carrying the remaining CYFIP2 variants (I664M, E665K, and D724H). We then compared the results of all six variants to comprehensively study the impact of ASD-associated variants on PPIs and structural dynamics of the WRC. This might provide mechanistic insights into ASD-associated WRC dysfunctions and a rational basis for therapies that restore normal WRC regulation.
2 Results
For each system, the final 1.5-μs equilibrated trajectories from each of the three independent replicates were pooled for analysis, yielding a total of 4.5-μs trajectories (more details in the Section Materials and Methods and in Supplementary material). In no case was global unfolding observed (Supplementary Table S1 and Supplementary Figures S1–S4)2.
2.1 Positions subject to mutations
In the WT complex, the residues of the mutant site are involved in a series of interactions between subunits: (i) I664 (CYFIP2) forms van der Waals contacts with F157 and W161 (both in ACR), at times of 99 and 94%, respectively (Figure 2A and Supplementary Table S2). (ii) E665 (CYFIP2) forms a salt bridge with K164 (ACR) with an occupancy of 53% (Figure 2B and Supplementary Table S2). (iii) D724 (CYFIP2) forms a hydrogen bond with Q110 (WAVE1 outside the ACR) with 27% occupancy, and van der Waals contacts with V531 (ACR) and L111 (WAVE1 outside the ACR), at times of 100 and 50% (Figure 2C and Supplementary Table S2). The variants (I664M, E665K, and D724H) retain van der Waals contacts but disrupt hydrogen bonds and salt bridges between subunits (Figures 2D–F and Supplementary Table S2).
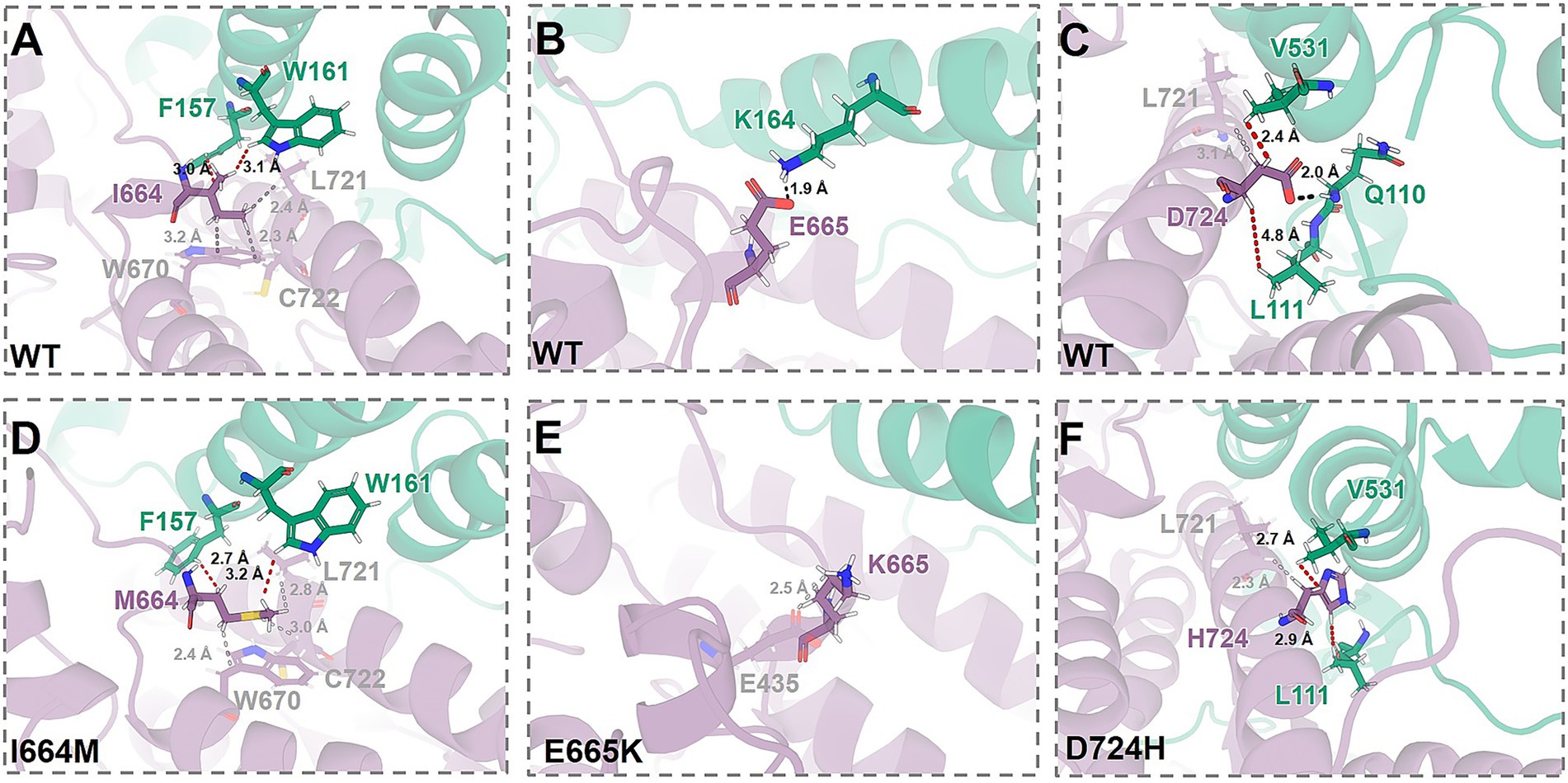
Figure 2. Alterations associated with I664M (A, D), E665K (B, E), and D724H (C, F) in the chemical environments at the mutation sites. The structures of WT WRC, obtained from our previous work (Xie et al., 2025b), and of the variants from MD simulations (see Methods in Supplementary material). CYFIP2 and WAVE1 are depicted as cartoons, colored purple and green, respectively. Mutated residues and contacting groups are represented as sticks. Hydrogen bonds and the shortest van der Waals contact are indicated by black and red dashed lines, respectively.
2.2 ACR/WRC interface contacts
Three variants similarly reduce ACR/WRC interactions [by 10 to 18% for the interface area; from 5 to 11% in the number of contacts (Nc)] compared to the WT complex (Figures 3A,B), while changes to other interfaces are smaller in comparison (interface area between −13 and −2% and Nc between −10 and +6%; Supplementary Figure S5). Thus, all three variants reduce the stability of the ACR/WRC interface (Figures 3A,B). However, they do not affect the overall stability of the complex: Supplementary Figure S4 shows that the changes of radius of gyration in variants range from −0.1 to 0.1% relative to the WT complex.
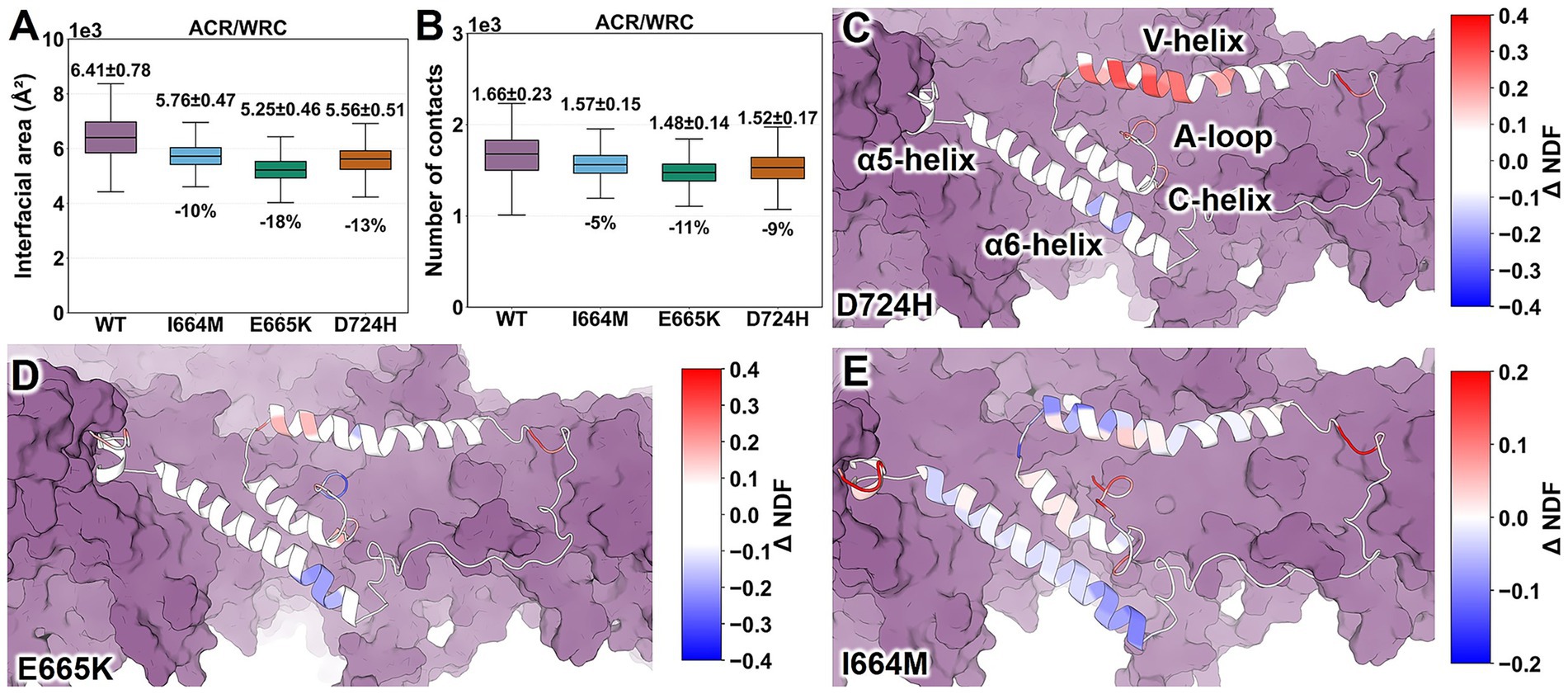
Figure 3. Impact of I664M, E665K, and D724H mutations on interactions between ACR and the rest of the WRC. (A) ACR/WRC interface areas. (B) Number of contacts with heavy atoms within 5 Å. (C–E) ΔNDFi values for ACR residues. They range from −0.4 (blue) to +0.4 (red) for D724H and E665K and from −0.2 (blue) to +0.2 (red) for I664M. ACR is represented as a cartoon, while the rest is depicted as a purple surface. ΔNDFi data for the entire complex are shown in Supplementary Figure S6. Data for WT WRC were obtained from our previous work (Xie et al., 2025b).
2.3 ACR local disorder
The normalized distance fluctuation for residue i (NDF𝑖) quantifies how the i-th residue moves in coordination with the rest of the WRC (Morra et al., 2012). Positive values of the differences in NDF𝑖 values (ΔNDFi; variant minus WT) indicate an increase in local disorder, while negative values suggest the opposite (Morra et al., 2014; Triveri et al., 2023; Castelli et al., 2024; Frasnetti et al., 2024; Torielli et al., 2025). Minor changes are observed in most ACR residues in the loops, α5, and C-helices (|ΔNDF𝑖 | ≤ 0.1; Figures 3C–E). D724H and E665K (Figures 3C,D) increase the propensity for uncoiling of the V-helix segments (ΔNDF𝑖 ranges from 0.12 to 0.30) compared to the WT complex, with only one exception (E515; ΔNDF𝑖 = − 0.12) in E665K. In these two variants, some regions of the α6 helix become more rigid. I664M exerts a weaker effect than other variants (Figure 3E) (Xie et al., 2025b): it destabilizes the α5 helix, stabilizes the α6 helix, and has a mixed impact (both stabilizing and destabilizing) on the V-helix (Figure 3E). Complex and variant-dependent changes are observed in loop regions (Figures 3C–E). The rest of the WRC is not significantly affected (Supplementary Figure S6). Our results suggest that E665K and D724H commonly destabilize the V-helix (Figures 3C,D), while I664M exerts a mild and mixed (both stabilizing and destabilizing) effect (Figure 3E).
2.4 Large-scale movements
Dynamic cross-correlation analysis shows that three variants alter the correlations between residue motions within the WRC, with moderate consistency (Supplementary Figure S7). Pairwise comparisons of variant-induced changes in motion correlation matrices yield cosine similarities ranging from 0.43 to 0.63 (Supplementary Table S3) and Spearman correlations ranging from 0.38 to 0.60 (Supplementary Table S4). Principal component analysis (PCA) reveals that the three largest eigenvectors (PC1–PC3) collectively account for approximately 50% of the total variance (Supplementary Figure S8). PC1–PC3 of E665K and D724H variants feature an increase in ACR contributions relative to the WT complex (from 20 to 42 and 25%, respectively), while I664M shows a smaller decrease (15%)3. These results suggest that E665K and D724H variants may promote ACR detachment by increasing its participation in large-scale movements, whereas I664M does not.
2.5 Allosteric analysis
Here, we calculate the allosteric score to quantify the contribution of each residue i to the long-range communication within the complex (ASi; see details in Supplementary material) (Schneider and Antes, 2022). The difference between the scores of the variant and those of the WT, ΔASi (variant minus WT), reflects the impact of the variant on the allosteric pathways within the complex. A positive ΔASi indicates greater allosteric importance. Negative values indicate the opposite effect (Schneider and Antes, 2022). In the WT complex, we identify 50 allosteric hubs (ASi equal to or greater than half of the maximum, 0.3): 28 in CYFIP2, 21 in NCKAP1, and 1 in HSPC300 (Figure 4A). Notably, all variants disrupt allosteric communication within the complex (Figures 4B–G): 10, 11, 22, 25, 15, and 10 allosteric hubs show reduced allosteric importance (ΔASi less than −0.1) in variants R87C, A455P, I664M, E665K, D724H, and Q725R, respectively. In contrast, few hubs show an increased role (ΔASi greater than 0.1): 2, 0, 2, 2, 1, and 0 for the R87C, A455P, I664M, E665K, D724H, and Q725R variants, respectively. The remaining hubs showed only minor variations (|ΔASi| equal to or less than 0.1). The common erosion of the allosteric role in hubs suggests that all variants disrupt the long-range communication between the ACR and the rest of the WRC (Figure 4). This implies that not only A455P (Xie et al., 2025b), but all six variants could decrease ACR/WRC interactions via allosteric changes.
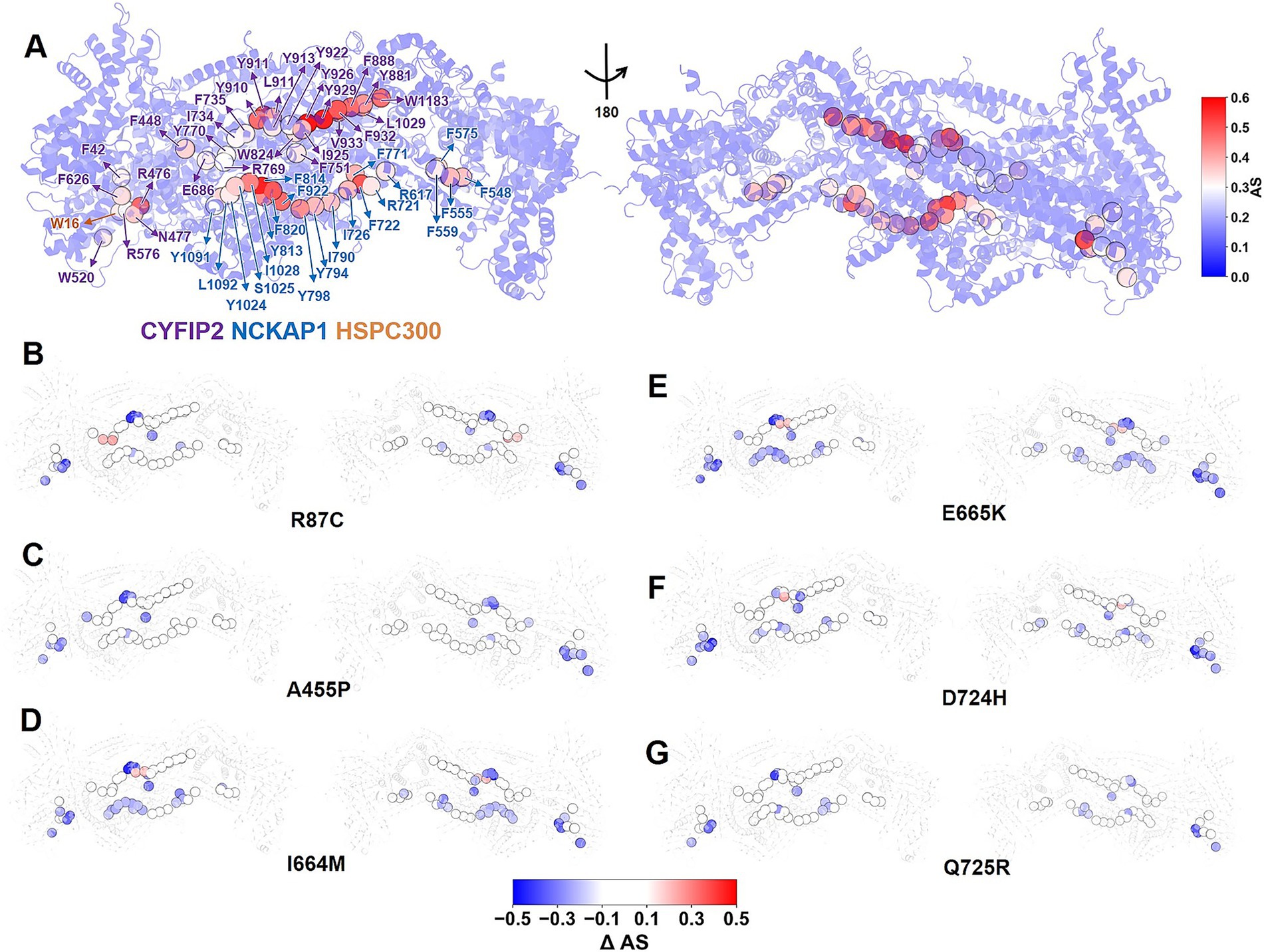
Figure 4. Effect of the six ASD-linked variants on allosteric communication within the complex. (A) Allosteric scores (ASi) for WT WRC residues, ranging from 0 (blue) to 0.6 (red). Residues with ASi greater than 0.3 are designated as allosteric hubs, illustrated by spheres. The residue index of allosteric hubs is labeled as follows: purple for CYFIP2, blue for NCKAP1, and orange for HSPC300. (B–G) ΔASi for allosteric hubs in variants, ranging from −0.5 (blue) to 0.5 (red). The WRC is shown as a cartoon. The suppression of allosteric hubs in all variants indicates that all variants, not only A455P, weaken ACR/WRC interactions via allosteric changes.
3 Discussion
Disruption of ACR’s V-helix contacts with the rest of the complex abnormally activates the WRC (Chen et al., 2010). Here, we used molecular dynamics simulations to examine how ASD-associated variants influence the structural dynamics of ACR, particularly those of the V-helix. All mutations are located in CYFIP2: five are at the interface with ACR, and one (A455P) is buried internally (Figure 1). Our main findings are as follows (Supplementary Table S5)4:
1. None of the variants show global unfolding or loss of complex stability (Supplementary Figures S4,S5).
2. All of the mutations weaken ACR/WRC contacts relative to the WT complex, regardless of their location or chemical properties (Figures 1, 3A,B). The mutations disrupt long-range communication between the ACR (including its V-helix) and the rest of the WRC (Figure 4), which may promote aberrant ACR detachment and WRC activation. A455P does so through allosteric effects (Figure 4C), while the others do so by disrupting interfacial hydrogen bonds and/or salt bridges at their respective sites (Figure 2 and Supplementary Table S2). These findings are consistent with the ~50% reduction in CYFIP2 binding to ACR’s V-helix, C-helix, and A-loop for R87C WRC (Nakashima et al., 2018).
3. Most variants increase their propensity for unwinding in the V-helix relative to WT (Figures 3C,D). This may indicate a process leading to ACR detachment (Chen et al., 2010). However, I664M exerts a weaker mixed effect (both stabilizing and destabilizing; see Figure 3E). Additionally, I664M decreases ACR participation in large-scale movements compared to WT, while the others show an opposite trend.
In summary, we propose that ASD-linked mutations facilitate ACR detachment (particularly the V-helix) by weakening its contacts with the rest of the complex and eroding long-range allosteric communication within the complex. Additionally, all but I664M may favor ACR detachment by increasing V-helix disorder in ACR and enlarging ACR participation in large-scale movements. The present findings are consistent with those of previous in vivo experiments, which demonstrated that these six mutations cause aberrant lamellipodia without the binding of their cellular partners (Schaks et al., 2020). The lamellipodia are cellular hallmarks of ACR detachment and WRC activation (Schaks et al., 2018).
Unfortunately, no ligand or therapeutic strategy exists yet for the WRC dysfunction associated with ASD-linked mutations. In an effort at identifying new therapeutic agents counteracting the deranged effect of the disease, we hypothesize here that ligands stabilizing the ACR/WRC interface or reconstructing allosteric communication hold the potential to restore normal WRC regulation in these ASD-linked variants. The ACR/CYFIP2 interface is a promising target region for ligands that stabilize ACR/WRC interactions, since CYFIP2 is the primary interactor within the WRC (Figure 1). In vivo studies could determine whether such ligand candidates restore the function of the WT, namely the formation of lamellipodia only after Rac1 binding (Schaks et al., 2020).
4 Materials and methods
Details of model construction and property calculations are described in the Supplementary material.
4.1 Molecular dynamics simulation
MD simulations were performed using AMBER 22 software (Case et al., 2005). Long-range electrostatic interactions were calculated using the Particle Mesh Ewald (PME) method (Darden et al., 1993). A cutoff distance of 10 Å was applied to short-range non-bonded interactions, which include Lennard-Jones forces and the short-range component of the PME calculation. Periodic boundary conditions were applied. The systems underwent three successive minimization cycles: (i) 10,000 steep descent steps followed by 10,000 conjugate gradient minimization steps with a 100 kcal/(mol·Å2) constraint applied to the entire solute; (ii) the same protocol with the same constraints, but applied only to heavy atoms; and (iii) the same protocol without any constraints. The systems were then heated from 100 K to 310 K in 0.5 ns using Langevin dynamics (Lemons and Gythiel, 1997). A constraint of 100 kcal/(mol·Å2) was applied to the heavy atoms. The systems were then subjected to another 0.5 ns at 310 K without restrictions. An integration time interval of 1 fs was used during heating. Subsequently, each system was subjected to three independent isobaric-isothermal (NPT) simulations lasting 2 μs, each started at different velocities. The temperature (310 K) and pressure (1 atm) were maintained using Langevin dynamics (Lemons and Gythiel, 1997) and a Monte Carlo barostat (Åqvist et al., 2004), respectively. A time step of 2 fs was used during NPT simulations. The trajectories were output at a frequency of 10 ps. The data for the MD simulations, including the input files, parameter files, and analysis scripts, can be found in the Zenodo repository: https://zenodo.org/record/15481836.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
SX: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. KZ: Formal analysis, Investigation, Writing – review & editing. SD: Writing – review & editing. GB: Methodology, Writing – review & editing, Formal analysis. GC: Writing – review & editing, Methodology. PR: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing – review & editing. PC: Conceptualization, Formal analysis, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by: (i) the China Scholarship Council (Project ID 202306650006); (ii) Italy’s National Recovery and Resilience Plan (NRRP), Mission 4-Component 2-Investment 1.1, Call No. 104 (issued Feb 2, 2022 by MUR), financed by the EU-NextGenerationEU, for the project “PpiAsd: Protein–protein interactions in Autism Spectrum Disorder” (CUP F53D23001170006), with the Grant Assignment Decree adopted on June 30, 2023; (iii) the HEAL ITALIA partnership (Health Extended ALliance for Innovative Therapies, Advanced Lab-research, and Integrated Approaches of Precision Medicine), funded by MUR under PNRR Mission 4, Component 2, Investment 1.3, project PE00000019 (University of Cagliari); (iv) the NRRP, Mission 4-Component 2-Investment 1.5, Call No. 3277 (Dec 30, 2021, MUR), EU-NextGenerationEU, Project Code ECS0000038, “eINS-Ecosystem of Innovation for Next Generation Sardinia” (CUP MUR: ECS00000038; CUP: F53C22000430001), Concession Decree No. 1056 adopted on June 23, 2022.
Acknowledgments
We would like to thank Forschungszentrum Jülich for the computing time granted (project ID CYFIP2WRC on JURECA-DC) and the EBRAINS2.0 project.
Conflict of interest
SX, KZ, and PC were employed by Forschungszentrum Jülich GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. ChatGPT-5 and DeepL were used for language polishing to improve the manuscript's readability and clarity.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncom.2025.1704350/full#supplementary-material
Footnotes
1. ^Other ASD-linked variants have been summarized in our previous work (Xie et al., 2025a). Y108H in CYFIP2 increases Rac1 binding abnormally (Schaks et al., 2020), but it is unclear if the remaining variants also promote aberrant WRC activation.
2. ^The WT, R87C, A455P, and Q725R WRC trajectories for analysis were taken from our previous work (Xie et al., 2025b).
3. ^The convergence of these results is affected by the timescale of the simulations (6 μs). Consequently, we restrict our discussion to qualitative trends.
4. ^Except for the allosteric analysis, the findings of WT, R87C, A455P, and Q725R WRC discussed here were obtained from our previous work (Xie et al., 2025b).
References
Åqvist, J., Wennerström, P., Nervall, M., Bjelic, S., and Brandsdal, B. (2004). Molecular dynamics simulations of water and biomolecules with a Monte Carlo constant pressure algorithm. Chem. Phys. Lett. 384, 288–294. doi: 10.1016/j.cplett.2003.12.039
Case, D. A., Cheatham, T. E. III, Darden, T., Gohlke, H., Luo, R., Merz, K. M. Jr., et al. (2005). The amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688. doi: 10.1002/jcc.20290
Castelli, M., Magni, A., Bonollo, G., Pavoni, S., Frigerio, F., Oliveira, A. S. F., et al. (2024). Molecular mechanisms of chaperone-directed protein folding: insights from atomistic simulations. Protein Sci. 33:e4880. doi: 10.1002/pro.4880
Chen, Z., Borek, D., Padrick, S. B., Gomez, T. S., Metlagel, Z., Ismail, A. M., et al. (2010). Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538. doi: 10.1038/nature09623
Chen, B., Chou, H., Brautigam, C. A., Xing, W., Yang, S., Henry, L., et al. (2017). Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. eLife 6:e29795. doi: 10.7554/eLife.29795
Chen, S., Fragoza, R., Klei, L., Liu, Y., Wang, J., Roeder, K., et al. (2018). An interactome perturbation framework prioritizes damaging missense mutations for developmental disorders. Nat. Genet. 50, 1032–1040. doi: 10.1038/s41588-018-0130-z
Chen, S., Wang, J., Cicek, E., Roeder, K., Yu, H., and Devlin, B. (2020). De novo missense variants disrupting protein–protein interactions affect risk for autism through gene co-expression and protein networks in neuronal cell types. Mol. Autism. 11:76. doi: 10.1186/s13229-020-00386-7
Darden, T., York, D., and Pedersen, L. (1993). Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. doi: 10.1063/1.464397
Davenport, E. C., Szulc, B. R., Drew, J., Taylor, J., Morgan, T., Higgs, N. F., et al. (2019). Autism and schizophrenia-associated CYFIP1 regulates the balance of synaptic excitation and inhibition. Cell Rep. 26, 2037–51.e6. doi: 10.1016/j.celrep.2019.01.092
De Rubeis, S., Pasciuto, E., Li, K., Fernández, E., Di Marino, D., Buzzi, A., et al. (2013). CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron 79, 1169–1182. doi: 10.1016/j.neuron.2013.06.039
Ding, B., Yang, S., Schaks, M., Liu, Y., Brown, A. J., Rottner, K., et al. (2022). Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase. Nat. Commun. 13:5444. doi: 10.1038/s41467-022-33174-3
Frasnetti, E., Cucchi, I., Pavoni, S., Frigerio, F., Cinquini, F., Serapian, S. A., et al. (2024). Integrating molecular dynamics and machine learning algorithms to predict the functional profile of kinase ligands. J. Chem. Theory Comput. 20, 9209–9229. doi: 10.1021/acs.jctc.4c01097
Fu, J. M., Satterstrom, F. K., Peng, M., Brand, H., Collins, R. L., Dong, S., et al. (2022). Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 54, 1320–1331. doi: 10.1038/s41588-022-01104-0
Lemons, D. S., and Gythiel, A. (1997). Paul Langevin’s 1908 paper “on the theory of brownian motion” [“Sur la théorie du mouvement brownien,” C. R. Acad. Sci. (Paris) 146, 530–533 (1908)]. Am. J. Phys. 65, 1079–1081. doi: 10.1119/1.18725
Morra, G., Genoni, A., and Colombo, G. (2014). Mechanisms of differential allosteric modulation in homologous proteins: insights from the analysis of internal dynamics and energetics of PDZ domains. J. Chem. Theory Comput. 10, 5677–5689. doi: 10.1021/ct500326g
Morra, G., Potestio, R., Micheletti, C., and Colombo, G. (2012). Corresponding functional dynamics across the Hsp90 chaperone family: insights from a multiscale analysis of MD simulations. PLoS Comput. Biol. 8:e1002433. doi: 10.1371/journal.pcbi.1002433
Nakashima, M., Kato, M., Aoto, K., Shiina, M., Belal, H., Mukaida, S., et al. (2018). De novo hotspot variants in CYFIP2 cause early-onset epileptic encephalopathy. Ann. Neurol. 83, 794–806. doi: 10.1002/ana.25208
Rottner, K., Stradal, T. E. B., and Chen, B. (2021). WAVE regulatory complex. Curr. Biol. 31, R512–R517. doi: 10.1016/j.cub.2021.01.086
Schaks, M., Reinke, M., Witke, W., and Rottner, K. (2020). Molecular dissection of neurodevelopmental disorder-causing mutations in CYFIP2. Cells 9:1355. doi: 10.3390/cells9061355
Schaks, M., Singh, S. P., Kage, F., Thomason, P., Klünemann, T., Steffen, A., et al. (2018). Distinct interaction sites of Rac GTPase with WAVE regulatory complex have non-redundant functions in vivo. Curr. Biol. 28, 3674–84.e6. doi: 10.1016/j.cub.2018.10.002
Schneider, M., and Antes, I. (2022). SenseNet, a tool for analysis of protein structure networks obtained from molecular dynamics simulations. PLoS One 17:e0265194. doi: 10.1371/journal.pone.0265194
Torielli, L., Guarra, F., Shao, H., Gestwicki, J. E., Serapian, S. A., and Colombo, G. (2025). Pathogenic mutation impairs functional dynamics of Hsp60 in mono- and oligomeric states. Nat. Commun. 16:3158. doi: 10.1038/s41467-025-57958-5
Triveri, A., Casali, E., Frasnetti, E., Doria, F., Frigerio, F., Cinquini, F., et al. (2023). Conformational behavior of SARS-Cov-2 spike protein variants: evolutionary jumps in sequence reverberate in structural dynamic differences. J. Chem. Theory Comput. 19, 2120–2134. doi: 10.1021/acs.jctc.3c00077
Xie, S., Zuo, K., De Rubeis, S., Bonollo, G., Colombo, G., Ruggerone, P., et al. (2025b). Impact of genetic variants associated with neurodevelopmental disorders on the WAVE regulatory complex. J. Chem. Inf. Model. 65, 7399–7405. doi: 10.1021/acs.jcim.5c01162
Xie, S., Zuo, K., De Rubeis, S., Ruggerone, P., and Carloni, P. (2025a). Molecular basis of the CYFIP2 and NCKAP1 autism-linked variants in the WAVE regulatory complex. Protein Sci. 34:e5238. doi: 10.1002/pro.5238
Zeidan, J., Fombonne, E., Scorah, J., Ibrahim, A., Durkin, M. S., Saxena, S., et al. (2022). Global prevalence of autism: a systematic review update. Autism Res. 15, 778–790. doi: 10.1002/aur.2696
Zhao, F., and Guan, W. (2024). Defects of parvalbumin-positive interneurons are implicated in psychiatric disorders. Biochem. Pharmacol. 230:116599. doi: 10.1016/j.bcp.2024.116599
Keywords: WAVE regulatory complex, neurodevelopmental disorder, autism spectrum disorder, missense variants, molecular dynamics, allosteric analysis
Citation: Xie S, Zuo K, De Rubeis S, Bonollo G, Colombo G, Ruggerone P and Carloni P (2025) Common characteristics of variants linked to autism spectrum disorder in the WAVE regulatory complex. Front. Comput. Neurosci. 19:1704350. doi: 10.3389/fncom.2025.1704350
Edited by:
Ruben A. Tikidji-Hamburyan, George Washington University, United StatesReviewed by:
Shula Shazman, Open University of Israel, IsraelSadhna Rao, Genentech Inc., United States
Copyright © 2025 Xie, Zuo, De Rubeis, Bonollo, Colombo, Ruggerone and Carloni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ke Zuo, ay56dW9Ac3RpbXVsYXRlLWVqZC5ldQ==; Paolo Ruggerone, cGFvbG8ucnVnZ2Vyb25lQHVuaWNhLml0; Paolo Carloni, cC5jYXJsb25pQGZ6LWp1ZWxpY2guZGU=
 Song Xie
Song Xie Ke Zuo
Ke Zuo Silvia De Rubeis
Silvia De Rubeis Giorgio Bonollo11
Giorgio Bonollo11 Giorgio Colombo
Giorgio Colombo Paolo Carloni
Paolo Carloni