- Comprehensive Health Research Centre (CHRC), NOVA Medical School, Faculdade de Ciências Médicas, Universidade NOVA de Lisboa, Lisbon, Portugal
The increasing availability of medical imaging data offers unprecedented opportunities for advancing artificial intelligence (AI)-driven healthcare. However, strict data protection regulations in the European Union (EU), especially the General Data Protection Regulation (GDPR), present significant challenges to data sharing and reuse. Synthetic data—artificially generated data that mimic the statistical properties of real data without revealing sensitive information—have emerged as a promising solution to bridge this gap. This perspective-style review examines the role of synthetic medical imaging data within the European Health Data Space (EHDS), a policy initiative aimed at enabling secure access to health data across the EU. While we briefly reference cross-cutting privacy-enhancing technologies and one non-imaging comparator to illuminate shared governance issues, our analysis and conclusions are scoped to imaging applications. We discuss the technical foundations and types of synthetic data, their potential to enhance reproducibility and innovation, and the complex ethical and legal concerns surrounding their use. Emphasising the need for a risk-based regulatory framework, we advocate for synthetic data governance that ensures utility, transparency, and accountability, especially when such data are generated using generative AI models. This work contributes to ongoing debates on how synthetic imaging data can support a privacy-preserving, data-driven healthcare ecosystem in Europe.
Introduction
The European Health Data Space (EHDS) is a significant initiative that facilitates cross-border data sharing within the European Union (EU). EHDS aims to advance healthcare research, artificial intelligence (AI) development, and patient care by enabling access to healthcare data. Medical imaging data, which are crucial for diagnostics and treatment planning, encounter substantial obstacles to sharing owing to stringent privacy laws, ethical concerns, and security challenges. These barriers pose significant challenges to the development and deployment of innovative AI solutions in healthcare. Synthetic data that replicate realistic imaging datasets without compromising patient privacy have emerged as a promising solution to these challenges, particularly in the early stages of clinical validation. This review examined the potential of synthetic data in medical imaging, focusing on its alignment with EHDS objectives, ethical considerations, regulatory compliance, and the establishment of industry standards. Specifically, it aims to (1) define synthetic data and its applications in medical imaging within the EHDS framework; (2) demonstrate how synthetic data can address barriers to clinical validation, particularly in the early stages of innovative solution development; (3) explore the ethical and regulatory considerations related to the use of synthetic imaging in the development and clinical validation; and (4) propose best practices to ensure synthetic data quality, interoperability, and applicability to support real-world clinical use cases. Our focus is synthetic medical imaging in the EHDS context. We occasionally draw on cross-cutting PETs and one contextual (non-imaging) comparator solely to clarify the same regulatory and ethical questions (e.g., Recital 26 anonymity, controller/processor roles, AI Act risk). Unless explicitly noted, all claims, recommendations, and the governance toolkit are intended for imaging.
Methodology
This literature review employed an established and transparent framework to identify and synthesise relevant literature on the ethical and regulatory considerations of using synthetic medical image data within the EHDS context. Although the structure of this review draws inspiration from established frameworks, it does not fully conform to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines.
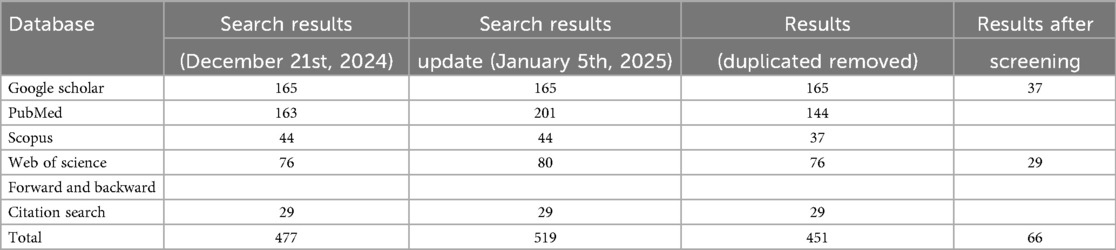
To ensure broad coverage and relevance, a structured search was conducted across four major databases: PubMed, Scopus, Web of Science, and Google Scholar. The initial search was carried out on December 21st, 2024, with an update on January 5th, 2025 to capture any newly added studies. Google Scholar was searched last to capture potential grey literature. In addition to database searches, forward and backward citation tracking was performed by screening the reference lists of included studies to identify further relevant works (leading to the addition of 29 studies).
Search terms were developed to reflect the core themes of the review in consultation with subject-matter experts. Keywords included combinations of terms related to synthetic data, medical images, EHDS, ethics, applications, and clinical relevance. Consistent terms were used across all databases except PubMed, where Medical Subject Headings (MeSH) were applied as appropriate.
The review focused on studies addressing ethical and regulatory issues related to synthetic medical image data within the EHDS framework. The inclusion criteria were:
• English-language publications.
• Published between January 1st, 2020 and January 5th 2025.
• Studies discussing synthetic data, especially in medical imaging, with a link to legal, ethical, or clinical applications.
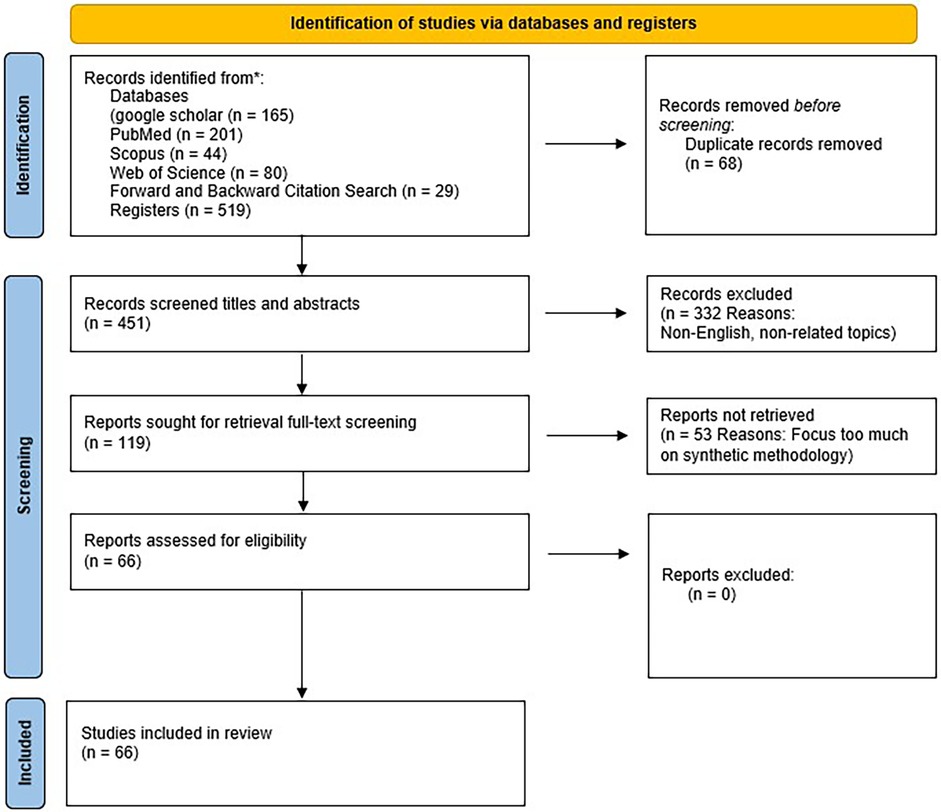
Given the broad scope of the topic, the search also employed multiple subsets of keywords to maximise sensitivity and relevance. From an initial pool of 516 studies, 68 duplicates were removed. Titles and abstracts of the remaining studies were screened against pre-defined inclusion criteria, excluding 332 papers. The full texts of 119 remaining articles were then assessed, of which 53 were excluded for not meeting the eligibility criteria. A total of 66 studies were ultimately included in the final analysis. These were included because relatively few publications addressed the four core objectives of this review in an integrated manner. The complete screening results are summarised in Table 1, and the study selection process is illustrated in Figure 1.
This literature review adopted a structured and rigorous process to identify and analyse studies relevant to using synthetic medical image data ethically within the EHDS framework. Although the review was not formally registered or reported according to PRISMA standards, key elements—such as transparent inclusion criteria, structured searching, and screening by multiple reviewers—were incorporated to enhance robustness.
Given the limited literature directly addressing synthetic image data under EHDS, the included studies were organised into thematic subtopics:
• Introduction to synthetic data;
• Synthetic data within EHDS or related legal/ethical frameworks;
• Applications in medical imaging;
• Implementation and technical considerations.
Each full-text article was assessed independently by the authors. Any disagreements during screening or categorisation were resolved through discussion. Relevant content was highlighted and categorised for use in the manuscript. This structured methodology enabled the development of a comprehensive and credible synthesis, supporting a nuanced discussion of generalisability, ethical compliance, and practical implementation of synthetic image data in the European health research and innovation landscape.
We scoped this review to ethical and regulatory aspects of synthetic medical imaging under the EHDS. A small number of contextual (non-imaging) sources are cited where they inform the same governance questions (e.g., GDPR anonymisation tests, secondary-use access via data access bodies, and high-risk AI obligations).
Synthetic data and its role in EHDS
With an imaging focus, we discuss how the EHDS is designed to enhance the sharing and reuse of health data across the EU. Where we reference LLMs/EHR or non-imaging PETs, these are used as cross-cutting comparators to the same governance issues faced by imaging. The conclusions remain imaging-scoped unless explicitly stated. EHDS establishes a health-specific ecosystem for primary and secondary health data use (1–6). By clarifying the scope of secondary data use, the EHDS aims to facilitate the secure exchange and interoperability of health data throughout Europe (1).
EHDS addresses the challenges associated with increasing the volume and complexity of health data. These challenges include data bottlenecks resulting from the sensitivity of health information, interoperability issues due to a lack of standardisation, and fragmentation, which complicate the navigation of health services and data protection frameworks (6). To mitigate these issues, EHDS proposes a unified market for electronic health record systems, relevant medical devices, and high-risk AI-driven healthcare applications. It establishes governance frameworks, common standards, and infrastructure to ensure seamless data exchange from decentralised sources (1, 6).
A fundamental aspect of EHDS is the assurance that secondary use of data complies with ethical and legal standards, particularly in relation to the General Data Protection Regulation (GDPR). The EHDS enhances data access rights, establishes governance structures at the Member State level, and requires standardised formats for health data systems (1). Nonetheless, a significant challenge persists: facilitating extensive data sharing, while safeguarding patient privacy and mitigating the risks associated with data breaches.
Synthetic data represent a promising solution within the EHDS framework, enabling stakeholders to utilise health data for research, AI model training, and policymaking without compromising patient confidentiality (4). Synthetic data consist of artificially generated datasets that maintain the statistical properties of real-world health data while removing personally identifiable information. This approach aligns with the objective of maximising the utilisation of health information, while ensuring legal compliance (4). Medical images, for instance, fall under the broader category of health data that the EHDS aims to make interoperable and accessible through the digitisation of health records across the EU (7). Synthetic medical imaging data enables researchers and clinicians to develop and test AI modelsat the early stages without accessing sensitive patient information.
Although a legal definition of synthetic data is absent, it is commonly recognised through its generation methodologies. Ianese (4) characterises synthetic data as artificial information derived from real datasets using AI techniques. Despite its fictitious nature, synthetic data retains the statistical properties of the original dataset, rendering it valuable for the research and training of machine learning models. Similarly, Greser (8) contends that synthetic data are not collected, but are instead generated algorithmically to mirror the statistical distributions of real-world data, with its utility contingent upon the accuracy and representativeness of the original dataset.
Various techniques have been used to generate synthetic health data. Generative Adversarial Networks (GAN) consist of a generator and discriminator, which collaboratively generate synthetic samples that closely mimic real data. Conditional GAN (CGAN) enhance this framework by integrating additional information, whereas Conditional Tabular GAN (CTGAN) further refine CGAN for structured health data by addressing rare occurrences through mode-specific normalisation (9). Autoencoders are neural networks that compress the input data into a latent representation before reconstructing them. Variational Autoencoders (VAE) advance this approach by ensuring that the latent space adheres to a Gaussian distribution (9). Alternative methodologies include SynthPop, which generates synthetic data sequentially based on conditional distributions; Maximum Spanning Tree (MST) methods, which preserve noisy marginals of low-dimensional data distributions, and PrivBayes, which constructs Bayesian networks and introduces noise into marginal statistics (9).
Synthetic data can be generated from actual electronic medical records, preserving their statistical properties, while ensuring the exclusion of any real patient data. In contrast to anonymisation techniques, synthetic data retains the essential characteristics of the original dataset while adhering to privacy standards (4).
The increasing deployment of Large Language Models (LLM) in the healthcare sector presents significant challenges related to data availability and quality. Access to high-quality medical data is frequently restricted by paywalls and regulatory constraints, thereby limiting the training of domain-specific AI models. Synthetic data have emerged as a viable solution, as they allow for the expansion of dataset size while ensuring privacy (10). A recent study introduced a medical LLM, GatorTronGPT, which was developed using synthetic data, resulting in enhanced clinical text generation performance compared with models trained solely on real data (11). Consequently, synthetic data generation effectively addresses data scarcity, enhances model performance, and improves generalisation, while safeguarding privacy.
The SECURED (Scaling up Secure Processing, Anonymisation and Generation of Health Data for EU Cross-Border Collaborative Research and Innovation) project, launched in January 2023, aims to enhance multiparty computation, data anonymisation, and synthetic data generation within the health sector. A significant application of this project is the creation of synthetic data for educational purposes, enabling medical students to engage with realistic case studies without compromising actual patient records (12) while ensuring privacy in cross-border data sharing (13).
Research in medical imaging has benefited significantly from the utilisation of synthetic data. Traditional datasets in medical imaging frequently encounter limitations such as insufficient sample sizes, data heterogeneity, and challenges in integration owing to variations in imaging protocols across different institutions. Synthetic medical imaging data can effectively address these issues by generating diverse and high-quality training sets, thereby enhancing deep learning models for diagnostic and treatment planning purposes (10).
Although the integration of synthetic data into the EHDS framework offers numerous benefits, it also presents significant ethical and regulatory challenges. The absence of standardised quality metrics and regulatory classification under the GDPR raises concerns regarding data bias and validity (14, 15). Nevertheless, advancements in deep generative models and privacy-preserving infrastructure, such as EHDS health data nodes, provide potential solutions to these issues. Ensuring transparency in the data generation and validation processes is crucial for maximising the potential of synthetic data while adhering to ethical and legal standards.
Ethical challenges of synthetic data
While synthetic data is increasingly promoted as a privacy-preserving alternative to real-world health data, its use in research and development of innovative health solutions introduces complex ethical considerations beyond data protection compliance. Central among these are concerns related to consent, re-identification risks, bias, scientific integrity, data sovereignty, and cybersecurity (8, 9, 16–19). These issues challenge the assumption that synthetic data is inherently free from ethical scrutiny and underscore the importance of governance frameworks that align with legal requirements and ethical research principles.
Informed consent and data subject autonomy
Although synthetic data does not directly contain identifiable personal information, it is typically generated from real datasets originally contributed by individuals. In many cases, these individuals may not have explicitly consented to the use of their data for synthetic data generation or secondary purposes such as model training, commercial use, or cross-border data sharing (19, 20). This raises important ethical concerns regarding autonomy, transparency, and respect for data subjects’ rights—particularly when synthetic data is used beyond the original purpose for which the real data were collected (21). Ethical research demands that participants be properly informed about how their data may be used, even in derived or non-identifiable forms. This is in line with the 2024 revision of the Declaration of Helsinki, which emphasizes that researchers must obtain free and informed consent for the collection, use, storage, and possible future use of biological materials and identifiable or re-identifiable data. The Declaration also states that if future uses cannot be fully foreseen at the time of consent, they must still be approved by an ethics committee, especially when obtaining new consent is not practical. These updates reflect the growing importance of transparency and ethical oversight in data reuse and secondary research. The European Data Protection Supervisor (EDPS) has emphasised that scientific research cannot serve as a “carte blanche to take irresponsible risks” and must be conducted within an established ethical framework (22).
In response to the tension between strict consent requirements and the need for flexible data use, concepts such as “broad consent” have emerged. Broad consent allows future research uses while upholding ethical standards through ongoing information, oversight, and participant engagement (23). However, this model remains controversial, particularly when applied to data used to generate synthetic datasets that could be reused in ways not foreseen during initial data collection.
Privacy risks and re-identification
Although synthetic data is designed to eliminate personally identifiable information, it does not entirely remove the risk of privacy breaches. According to the G29 Working Party and CNIL, there are three key risks associated with synthetic data derived from anonymised sources: (1) Singling Out, where unique data points lead to the identification of individuals; (2) Linkability, where records across datasets can be connected; and (3) Attribute Inference, where sensitive characteristics of individuals may be deduced (9). These risks are especially pronounced in contexts involving small populations or rare diseases, where the uniqueness of cases increases re-identification potential (19).
Generative Adversarial Networks (GANs), commonly used to generate synthetic medical images, can inadvertently reproduce features of the original dataset if not properly regularised. It may result in membership inference attacks, whereby an adversary can determine whether a particular individual’s data was used in the training set (24). For example, in a study involving adolescents with de novo mutations, a GAN trained on facial phenotype data was shown to be vulnerable to such attacks if model weights were publicly shared (24). Differential privacy techniques have shown promise in mitigating these risks (25). Still, they are not foolproof and can introduce trade-offs with data utility.
The legal ambiguity surrounding the classification of synthetic data adds another layer of complexity. Under the GDPR, data is only considered anonymous if re-identification is not reasonably possible, considering time, cost, and technology (23). However, the threshold for what constitutes “reasonable” varies across jurisdictions. Institutions such as biobanks may not hold re-identification keys. However, they could still be subject to data protection regulations if synthetic data is classified as pseudonymised (1).
Bias and fairness
Synthetic data generation is highly dependent on the quality and diversity of the source data. If the original dataset is unbalanced or reflects systemic inequalities—such as underrepresenting minority populations or biased clinical practices—these issues are replicated or amplified in the synthetic data (6, 18). For instance, chest X-ray datasets have been shown to underrepresent certain demographic groups, leading to reduced diagnostic performance for these populations (26).
While differential privacy techniques are designed to protect sensitive attributes, they may disproportionately affect minority subgroups by introducing statistical noise that distorts already limited representation (19). Several strategies have been proposed to address these challenges, including dataset rebalancing, fairness-aware GANs (such as HealthGAN), adversarial training, and participatory design approaches that involve community stakeholders in model development (14, 27).
Scientific integrity and trust
A critical concern in the use of synthetic data is the potential erosion of scientific integrity and public trust. Unlike real-world datasets, synthetic data lack an intrinsic link to empirical reality, raising questions about their validity for hypothesis testing, model training, and clinical decision-making. The absence of standardised benchmarks for evaluating synthetic data quality further complicates this issue (10, 19).
Studies have shown that different types of synthetic distribution shifts may not improve model robustness and may even obscure performance limitations under real-world conditions (18). Moreover, the use of synthetic data without full disclosure of generation methods, limitations, and validation procedures can mislead users about the reliability of research findings. Transparent reporting and the development of synthetic data quality standards are essential to ensure credibility in research outcomes (17).
Data sovereignty and indigenous data ethics
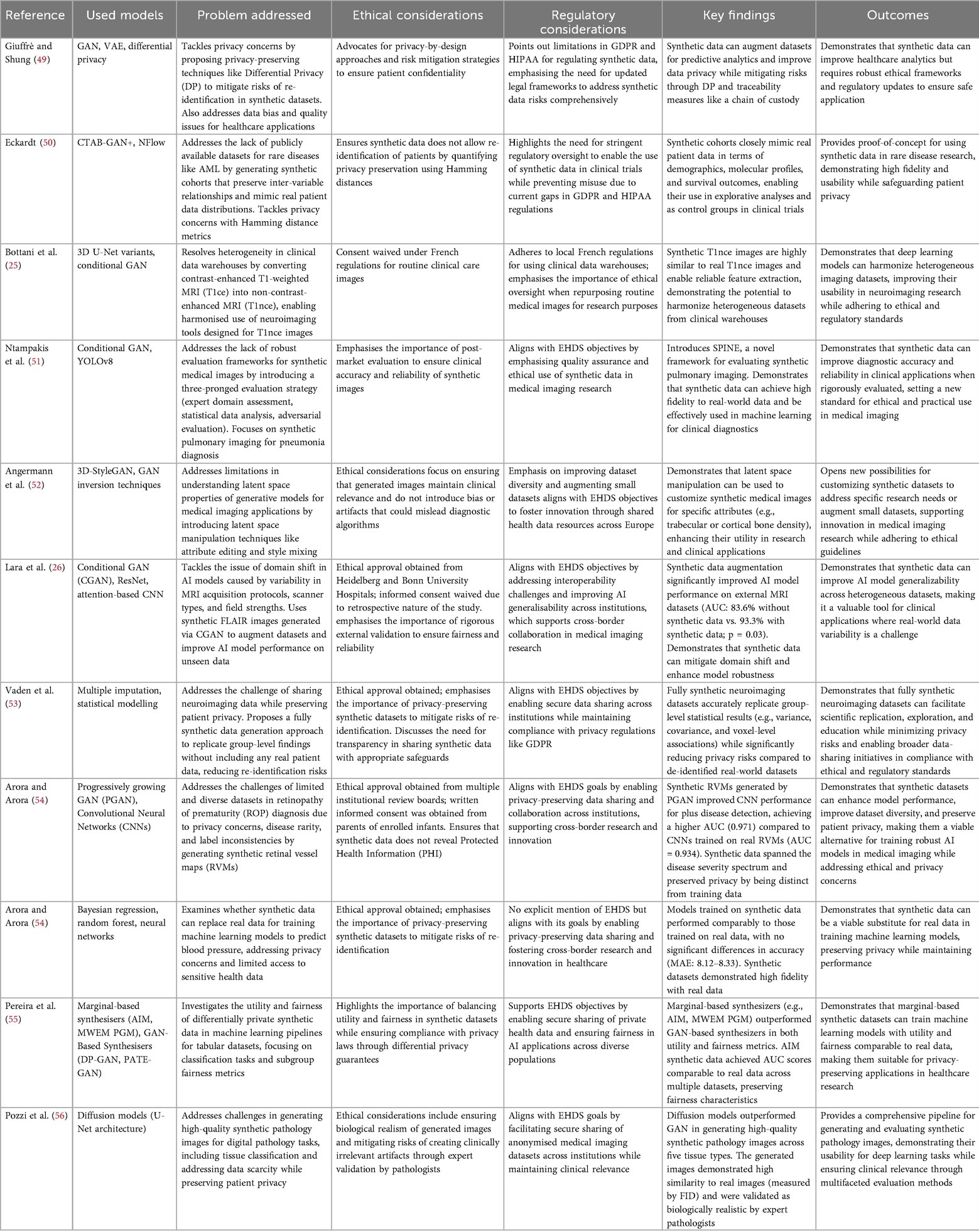
Ethical considerations are particularly salient when synthetic data involves populations with specific historical and cultural vulnerabilities, such as Indigenous communities. Historically, these populations have faced misuse and exploitation of their health data. Using synthetic data does not eliminate the need for appropriate governance; instead, it introduces new complexities related to data sovereignty and community engagement (17).
Excluding Indigenous populations from training datasets may lead to reduced model accuracy for these groups, while including them—without community oversight—may infringe upon their rights to control their data. Synthetic Indigenous datasets must be developed in consultation with the communities they aim to represent, ensuring alignment with ethical principles such as ownership, control, access, and possession (OCAP).
Cybersecurity and misuse risks
Synthetic data also introduces cybersecurity vulnerabilities that anonymisation may not fully mitigate. Techniques such as data poisoning and adversarial attacks can compromise model integrity at various stages of the AI lifecycle (8). In medical imaging, even minor alterations to input data can cause misclassification, potentially resulting in harmful clinical outcomes.
Partially synthetic datasets combining real and artificial data are particularly susceptible to data leakage and exploitation. Malicious actors may attempt to reverse-engineer synthetic datasets to infer information about real individuals, especially when model architectures and training parameters are publicly available (19).
Synthetic data holds significant promise for enabling innovation while preserving individual privacy. Yet, it is not inherently exempt from ethical scrutiny. Concerns surrounding informed consent, re-identification, bias, scientific reliability, and community engagement highlight the need for robust ethical oversight. Addressing these challenges requires interdisciplinary collaboration, standardised evaluation frameworks, and continuous engagement with stakeholders (28). Only through a deliberate and ethically grounded approach can the benefits of synthetic data be fully realised without compromising individual rights or public trust.
Regulatory frameworks and standards development
Currently, the European Union lacks dedicated regulations that specifically govern the use of synthetic data. While this absence of direct legislative instruments might suggest legal permissibility, it should not be interpreted as an absence of oversight. Synthetic data—particularly in healthcare and AI applications—may still fall under existing frameworks such as the General Data Protection Regulation (GDPR), the Medical Device Regulation (MDR), and the forthcoming AI Act. These instruments impose obligations concerning data quality, privacy, transparency, and accountability, which are highly relevant when synthetic data is used in developing, validating, or deploying medical AI systems (8). This section examines the regulatory, standard, and framework considerations related to the generation and use of synthetic imaging data, emphasising its transformative potential to surmount traditional barriers in healthcare data sharing. The objective is to present a balanced yet supportive perspective on synthetic data, underscoring its alignment with regulatory frameworks and its capacity to drive equitable healthcare innovation across Europe.
European regulatory instruments
1. General Data Protection Regulation (GDPR)Although synthetic data aims to eliminate identifiable personal information, it may still fall under the scope of the GDPR if the risk of re-identification persists. Article 4(1) defines personal data broadly, and Recital 26 clarifies that data is only anonymous if re-identification is not reasonably possible. Consequently, synthetic data must be evaluated case-by-case to determine whether it meets this threshold. Where it does not, obligations related to lawful processing, purpose limitation, and data minimisation remain applicable (29, 30).
2. European Health Data Space (EHDS)The proposed EHDS regulation seeks to establish a harmonised framework for using and reusing health data across the EU. While the regulation prioritises pseudonymised data, it acknowledges anonymised and synthetic data as potential tools for secure secondary use. However, detailed provisions for synthetic data are not yet fully articulated, raising questions about its governance under the EHDS framework (14, 15).
3. AI actThe proposed AI act introduces a risk-based approach to regulating AI systems, including those used in medical and healthcare contexts. While the Act does not refer explicitly to synthetic data, it encompasses training data quality, robustness, and transparency—directly relevant to synthetic dataset generation. Developers using synthetic data to train or validate AI systems may be required to demonstrate the quality and representativeness of their data, especially in high-risk applications (31).
4. Medical device regulation (MDR)Under the MDR, AI systems used for medical purposes may qualify as medical devices, particularly if they support diagnostic or therapeutic decisions. Synthetic data for designing, testing, or validating such systems must be robust, clinically relevant, and traceable. Although the MDR does not yet contain specific provisions for synthetic data, its emphasis on clinical evidence and performance evaluation implies that the origin and quality of training data—including synthetic sources—are subject to scrutiny.
5. Global and comparative contextsGlobally, the practices surrounding synthetic data intersect with regulatory frameworks such as the U.S. Health Insurance Portability and Accountability Act (HIPAA) and China’s Personal Information Protection Law (PIPL). While HIPAA takes a rule-based approach, the GDPR is principle-based, leading to different interpretations and applications of synthetic data protections. In contrast, the PIPL’s limited data set model permits de-identified data to be used for research without explicit consent, demonstrating a more flexible legal framework (32).
6. Standardisation initiativesStandardisation bodies such as ISO, IEC, and CEN are working toward frameworks incorporating synthetic data within broader AI and data governance standards. Emerging standards—such as ISO/IEC 38507 and ISO/IEC TR 24028—provide guidance on the ethical use of AI and risk management, which may be extended to include the generation and validation of synthetic data. The European Commission has played a central role in supporting these efforts by collaborating with CE and CENELEC to develop harmonised standards for AI data quality under the ISO 5259 series (Burden and Stenberg, 2024). The FUTURE-AI initiative also outlines criteria for trustworthy AI and ethical synthetic data use in medical imaging, focusing on fairness, robustness, and explainability (33). Meanwhile, the FAIR (Findable, Accessible, Interoperable, Reusable) and CARE (Collective Benefit, Authority to Control, Responsibility, Ethics) principles offer valuable guidance on ethical and responsible synthetic data practices, particularly in open science and Indigenous contexts (17, 34). Standardisation is also central to tackling data fragmentation, especially in highly sensitive areas such as paediatric oncology. Tozzi et al. (35), in a systematic review of AI in paediatric brain tumour research, underscores the urgent need for harmonised and interoperable data sources across European institutions to improve reproducibility and model performance.Complementary to these efforts, the Fast Healthcare Interoperability Resources (FHIR) standard is key in integrating synthetic datasets into real-world health systems. As discussed by Pereira et al. (10), FHIR defines the structure and semantics of electronic health data, enabling consistent exchange and supporting synthetic data applications through better interoperability, standardisation, and integration across diverse platforms.
Mapping use cases to regulatory and standards obligations
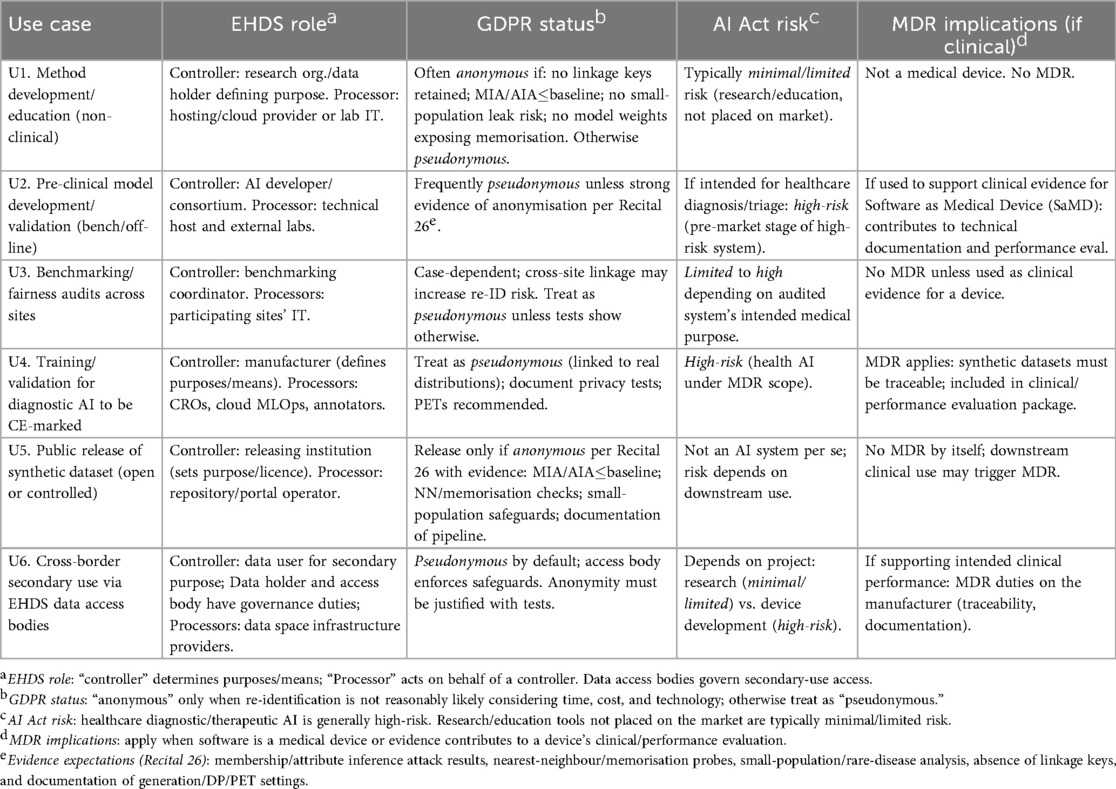
To translate the preceding legal analysis into practice, Table 2 maps common use cases of synthetic imaging data to obligations under the EHDS, GDPR, AI Act, and MDR. Figure 2 provides a Recital 26 decision tree to assess whether a dataset is anonymous or pseudonymous, together with evidence expectations. Unless explicitly noted as cross-cutting, the following mapping is scoped to synthetic medical imaging.
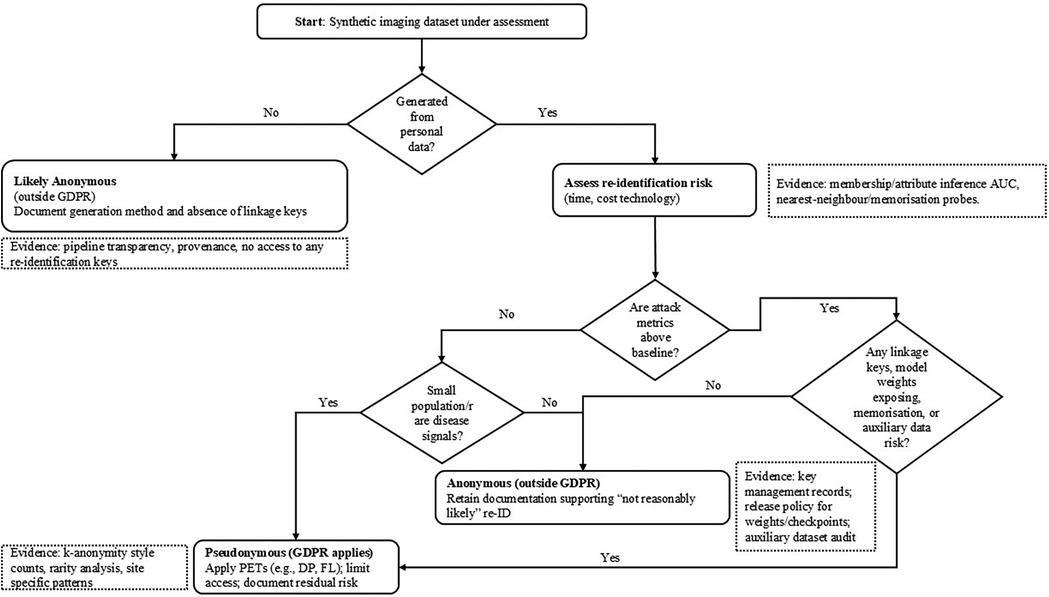
Figure 2. Recital 26 decision tree: is a synthetic imaging dataset anonymous or pseudonymous? Evidence expectations shown in dashed notes.
Table 2 translates typical synthetic-imaging use cases into concrete compliance expectations by aligning, for each scenario, (i) the likely EHDS roles (who is the controller defining purposes/means and who acts as processor), (ii) the GDPR posture (whether the dataset should be treated as anonymous, hence outside GDPR, or as pseudonymous, hence regulated), (iii) the anticipated AI Act risk categorisation given the intended use (e.g., research/education vs. diagnostic application), and (iv) any MDR implications where outputs support clinical performance or a medical device dossier. The entries are indicative and context-dependent, but they make explicit the evidence expected to justify an “anonymous” classification (e.g., attack metrics at or below baseline, absence of linkage keys, small-population safeguards) and when to default to a “pseudonymous” treatment with additional safeguards. Complementing the table, Figure 2 operationalises GDPR Recital 26 for synthetic imaging. It begins by asking whether the dataset was generated from personal data. If not, it is likely anonymous (outside GDPR) provided you document the generation process and confirm that no linkage keys exist. If it was generated from personal data, you then assess re-identification risk. First, run membership/attribute-inference attacks and compare their AUC to a random-guess baseline; results meaningfully above baseline indicate pseudonymous data (GDPR applies). Next, examine small-population/rare-disease signals (e.g., k-anonymity counts, site-specific rarity); any such risk also leads to a pseudonymous classification. Finally, check for linkage or memorisation risks—presence of keys, released weights/checkpoints that leak training samples, nearest-neighbour/inversion probes suggesting copying, or auxiliary datasets that enable linkage; any positive finding again implies pseudonymous treatment with safeguards. Only if all three checks (attacks, rarity, linkage/memorisation) are negative or at baseline may the dataset be deemed anonymous, in which case you retain the evidence demonstrating that re-identification is “not reasonably likely.” Dashed callouts in the figure list the concrete evidence expected at each step.
Scholarly perspectives on synthetic data governance
A growing body of scholarship addresses the challenges and opportunities of synthetic data in the EU regulatory context. Scholars agree that while current frameworks allow for synthetic data use, legal ambiguity persists, especially regarding GDPR compliance (8). Synthetic data are increasingly recognised as pivotal for privacy-preserving data sharing under the EHDS, with recent advancements in deep generative models enhancing their fidelity and utility (14, 15). Nevertheless, concerns remain around re-identification risks. As Biasin et al. (29) and van der Wel (30) argue, synthetic data must be rigorously assessed based on how they are generated. Improperly designed datasets could still fall under GDPR provisions, particularly where re-identification is plausible. The GDPR’s flexibility allows Member States to implement additional rules for health data, resulting in regulatory fragmentation and potential obstacles to cross-border collaboration (23). Borissova (36) and Ciminá (37) highlight the tension between Open Science and data protection. While synthetic data offer a privacy-friendly alternative, their classification as anonymised or pseudonymised remains unclear, complicating their use in collaborative research. Cross-border data sharing further intensifies these challenges, particularly around consent, data sovereignty, and integrating privacy-preserving techniques such as differential privacy (15).
Debates also focus on the EHDS framework’s definition of health data. As noted by Foà (1), its scope includes both personal and non-personal data. Yet, it lacks clear guidance on how synthetic data should be classified. For Borissova (36), the legal uncertainty undermines the potential of synthetic data unless clarified within the EHDS.
International comparisons highlight regulatory divergence. Casarosa and Greser (32) contrasts the GDPR’s strict data protection with more permissive frameworks like HIPAA and China’s PIPL. These differences create barriers to interoperability and international collaboration, reinforcing calls by Borissova (36) for alignment through bilateral agreements or global governance initiatives.
In the technical domain, scholars advocate for using Privacy Enhancing Technologies (PETs) such as Federated Learning and Homomorphic Encryption to support secure data sharing (38, 39). These methods allow decentralised collaboration without raw data exchange, aligning with EHDS priorities and addressing data sovereignty concerns. The COVID-19 pandemic provided a testbed for such approaches, demonstrating the utility of synthetic data and the need for interoperable data models (14, 40).
Ethical and procedural transparency remains a central concern. Authors stress the importance of documenting synthetic data generation methods, publishing evaluation metrics, and developing meaningful consent frameworks—particularly when individuals are unaware that their data were used to generate synthetic datasets (20, 29, 36). Aspell et al. (41) argue that evaluation frameworks for synthetic data remain underdeveloped and call for robust metrics to assess fidelity, representativeness, and utility in applied healthcare contexts.
Regulatory misalignment further complicates implementation. The AI Act’s risk-based classification system does not neatly map onto GDPR principles, contributing to uncertainty around the legal status of synthetic data (31). Cybersecurity risks and unclear data ownership also pose challenges, with calls for international coordination to ensure ethical and lawful data use (32, 42). In particular, Davidson and Winter (43) underscore the importance of anticipating emerging governance gaps when multiple legal instruments—such as the GDPR, AI Act, and EHDS—intersect without clearly delineating responsibilities.
The lack of standardised interoperability protocols across Member States impedes the integration of synthetic data into cross-border health systems. Harmonisation of definitions and frameworks is critical to ensuring synthetic data fulfil their potential as safe, equitable tools for healthcare innovation (14, 36).
Synthetic data has an important role in the European digital health ecosystem. While current regulations do not explicitly address their use, synthetic datasets intersect with multiple legal frameworks, including the GDPR, AI Act, MDR, and EHDS. Scholarly contributions reveal a strong consensus on their potential to advance privacy-preserving innovation while highlighting unresolved issues surrounding legal classification, consent, interoperability, and ethical governance. Institutional efforts from the European Commission and scholarly calls for standardisation—such as those by Tozzi et al. (35), Pereira et al. (10), Davidson and Winter (43), and Aspell et al. (41)—underscore the importance of a harmonised and transparent framework. Addressing these challenges through coordinated regulation, technical standards, and ethical practices is essential to unlocking synthetic data’s full value in European healthcare and beyond.
Synthetic image dataset methodology and case studies
Integrating synthetic data into medical imaging research within the European Health Data Space offers transformative potential but raises critical ethical concerns. This section discusses how synthetic data can address privacy preservation, data scarcity, and bias mitigation challenges. Three use cases are presented to illustrate cross-border collaboration in data transfer, the use of synthetic medical image datasets in validation, and the generation of data from Electronic Health Record (EHR) cases aligned with the objectives of the EHDS.
Ethical considerations include ensuring that synthetic datasets do not perpetuate or exacerbate biases present in the original data—particularly concerning under-represented populations—and maintaining representativeness across diverse demographic groups. Moreover, robust validation frameworks are essential to assess synthetic data’s reliability and clinical relevance, especially in sensitive domains such as healthcare diagnostics. Transparency in data generation processes and adherence to privacy regulations, such as the GDPR, foster trust among researchers, clinicians, and patients. By prioritising inclusivity, fairness, and accountability in synthetic data practices, the European Health Data Space can harness these innovative tools to advance medical imaging while upholding ethical standards essential for equitable healthcare innovation.
Synthetic image data
Computer-Aided diagnosis (CAD), driven by AI and deep learning methods, has recently supported the process of medical image analysis—especially MRI—and diagnosis. However, training deep learning models requires large sets of medical imaging data. While Generative Adversarial Networks (GAN) were initially popular for generating synthetic data in the medical imaging domain, they continue to address this challenge effectively (5, 44). GAN are a class of deep learning models composed of two neural networks: a generator and a discriminator. The generator produces synthetic data by creating random noise and generating increasingly realistic outputs (44). At the same time, the discriminator evaluates whether the data are real or synthetic using binary classification (24, 44). The two networks improve iteratively through adversarial training, resulting in highly realistic synthetic outputs (45).
GAN have transformative applications in medical imaging datasets, especially for brain MRI. Clinically, they support advanced analysis by synthesising missing MRI sequences or generating 3D brain MRIs (45). GAN offer significant benefits across clinical research, medical education, and patient privacy protection, among other areas.
Synthetic data generation and diversity
The production of large volumes of synthetic medical images—such as realistic brain scans—can significantly enhance training datasets. This is especially valuable for rare conditions like brain tumours or neurodegenerative diseases, where real data is scarce (19, 44, 45). Synthetic data also benefits medical education by providing abundant training materials and interactive simulations, allowing students to learn more effectively and encounter a broader range of clinical scenarios. GAN, in particular, enable the creation of customised datasets that include both common and rare edge cases, helping students and professionals become familiar with diverse conditions (45). Moreover, synthetic medical image data is increasingly used to augment datasets for AI/ML-based diagnostic tools, predictive screening systems, and other health technologies. This helps mitigate challenges related to limited or imbalanced real-world datasets. Using synthetic data for pre-testing and validation can also reduce costs and accelerate research timelines before transitioning to real-world deployment (19). In summary, synthetic image data can support various aspects of medical research, education, and technology development. Synthetic image data can help with the following aspects:
1. Tumour segmentationGenerative Adversarial Networks (GANs) have shown significant promise in enhancing tumour segmentation tasks by generating highly accurate segmentation masks that delineate tumour boundaries. This is particularly beneficial in brain MRI, where tumours often present with complex shapes and indistinct edges. By synthesizing annotated examples, GANs can augment training data and improve the robustness and sensitivity of segmentation algorithms used in clinical diagnostics (45).
2. Super-resolution and image quality enhancementLow-resolution or poor-quality medical scans can hinder diagnostic accuracy, particularly in resource-limited settings. GANs contribute to super-resolution by learning to enhance these images, producing higher-resolution outputs that retain anatomical fidelity. This refinement aids radiologists and AI systems alike in detecting subtle pathological changes that might otherwise be missed in noisy or compressed images (45).
3. Modality translationGANs can simulate disease progression by translating healthy brain MRIs into their diseased counterparts. This kind of image-to-image translation supports research and clinical training by creating synthetic but realistic progressions of neurodegenerative diseases, tumours, or stroke lesions. Such applications allow for controlled experimentation and help fill gaps where real-world longitudinal imaging data is limited or unavailable (45).
4. Prognosis and image registrationThough less widely explored, GANs are emerging tools in prognosis modeling—predicting how a disease is likely to evolve over time based on imaging patterns. In addition, GANs assist in image registration by aligning images taken at different times or using different modalities, improving longitudinal analysis and multi-modal integration in treatment planning (45).
5. Protection of patient privacyImportantly, because synthetic images do not correspond to actual individuals, they eliminate the risk of patient re-identification. GAN preserve the statistical properties of real datasets without reproducing identifiable features, making them a powerful approach to data sharing that complies with ethical and legal standards such as GDPR and HIPAA (44).
6. Improved model performanceSynthetic image data can substantially enhance the performance of machine learning models, especially in tasks where real data is limited or imbalanced. A notable study by Brugnara et al. (46) demonstrated this in the context of detecting new multiple sclerosis (MS) lesions on brain MRI. By integrating synthetic data into the training pipeline, the ResNet model’s AUC increased from 56% to 77.5%, while an attention-based model achieved a leap from 83.6% to 93.3%. These improvements underscore the potential of synthetic data in boosting generalisability and performance across different AI architectures and clinical settings.
7. Stress-testing AI modelsSynthetic data empowers researchers to replicate rare diseases and simulate challenging imaging conditions that are often underrepresented in real-world datasets. For instance, GANs can generate synthetic histological images of rare cancer subtypes or introduce artefacts such as noise and distortion, enabling robust testing of diagnostic models. Additionally, synthetic data plays a crucial role in mitigating domain shift. By emulating variations in imaging protocols, equipment types, and patient populations across different institutions, it helps uncover and correct biases in AI models trained on data from a single source. This significantly enhances the models’ ability to generalise across diverse clinical environments (24).
While a diverse array of methodologies exists for generating synthetic image data, our focus is on Generative Adversarial Networks (GAN) due to their initial popularity in the medical imaging domain (5, 44). Variations of GAN are particularly advantageous for preserving privacy while effectively generating synthetic tabular data. They are renowned for their ability to handle high-dimensional data; however, hyperparameter tuning is necessary to prevent model collapse (47).
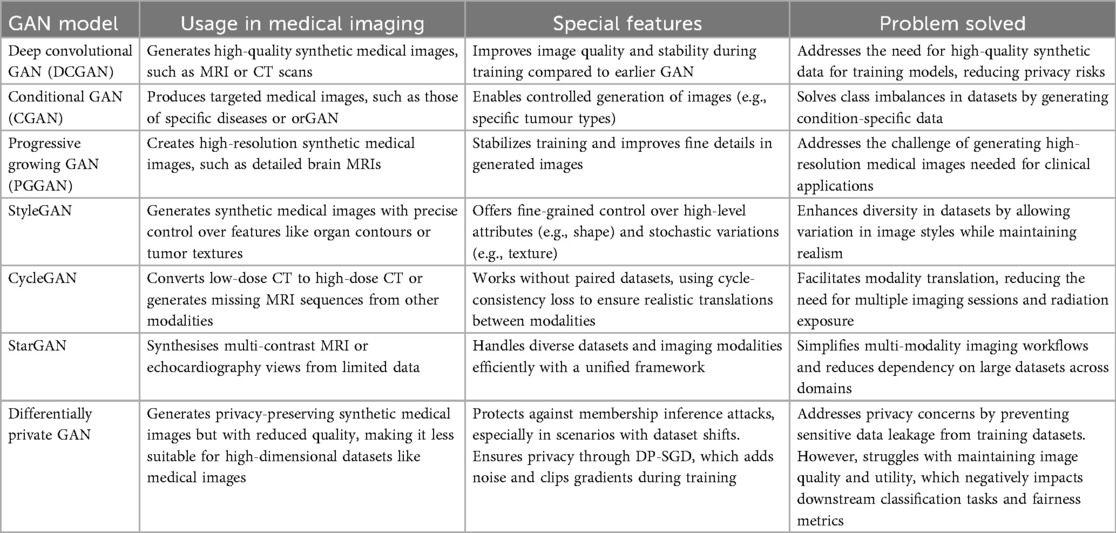
The transformative role of GAN in medical imaging continues to expand, offering benefits that range from data augmentation to enhanced diagnostic precision and privacy preservation. Their versatility extends to generating synthetic data for rare conditions, improving image quality, simulating disease progression, and stress-testing AI models across varied imaging conditions. These capabilities not only enhance model robustness but also contribute to more equitable and generalisable clinical tools. To better understand the landscape of GAN applications in medical imaging, Table 3 summarises key GAN architectures and their corresponding use cases, drawing on recent findings by Kim et al. (5) and Cheng et al. (48).
Case studies in synthetic medical imaging
Several studies demonstrate the application of synthetic image datasets in overcoming barriers in medical research, particularly regarding privacy, data availability, and cross-border collaboration. Table 4 summarises selected case studies, their methodologies, and their impact on medical imaging.
Case studies
Two use cases from Aunón et al. (39) and Yan et al. (57) have been selected as case studies due to their strong alignment with the objectives of the EHDS, particularly in enabling secure cross-border data sharing and reuse, while addressing challenges related to privacy and interoperability.
Aunón (39) comprehensively evaluate Privacy-Enhancing Technologies (PETs), including Federated Learning, Differential Privacy, and Homomorphic Encryption. It demonstrates their applicability within data spaces to support secure collaboration across organisations. The study emphasises the importance of privacy preservation in facilitating data sharing, directly resonating with the EHDS’s goal of establishing interoperable and secure frameworks for health data exchange across the EU and beyond. Furthermore, it highlights practical examples—such as the application of federated learning in healthcare use cases—showing how PETs can help overcome critical barriers like data heterogeneity, quality issues, and privacy concerns, which are key challenges to fulfilling the EHDS’s vision.
In contrast, Yan et al. (57) focus on generating synthetic Electronic Health Record (EHR) data using advanced GAN-based models, such as EMR-WGAN. This paper addresses the EHDS’s objective of standardising and making health records interoperable by offering a transparent tutorial on generating high-quality synthetic EHRs that preserve statistical fidelity while protecting patient privacy. The practical application of synthetic data generation supports secondary uses, including machine learning model development, hypothesis generation, medical education, and AI-driven healthcare innovation. Unlike other studies that concentrate solely on privacy or technical implementation, this paper illustrates how synthetic EHR data can be tailored to specific use cases while remaining accessible and usable across various healthcare systems.
These two papers stand out by offering actionable frameworks that align with EHDS regulatory aims while providing practical solutions to technical barriers in health data sharing. Aunón (39) and Yan et al. (57) uniquely address both technological maturity—such as federated learning protocols—and real-world implementation—such as GAN-based synthetic EHR generation—making them exemplary for illustrating how EHDS objectives can be effectively operationalised across Europe and on a global scale.
Case study 1: cross-border synthetic data transfer
The primary objective of this experiment was to demonstrate how Federated Learning (FL) facilitates collaborative machine learning across multiple healthcare institutions while preserving patient privacy. It aligns with the broader aims of health data spaces, which seek to enable secure and privacy-preserving data sharing to advance medical research and enhance patient outcomes. By leveraging FL, the study tackles critical issues such as data accessibility, heterogeneity, and privacy concerns, showcasing FL’s potential to overcome major barriers in the healthcare domain.
The study employed publicly available datasets of skin lesion images sourced from four institutions in Australia, Austria, Brazil, and the Netherlands. The datasets included HAM10000, PAD-UFES-20, and MED-NODE. Initially, the combined dataset contained eight types of skin lesions, but this was reduced to five (nevus, melanoma, actinic keratosis, basal cell carcinoma, and benign keratosis) due to class imbalance and insufficient sample sizes for certain lesion types. This selection helped mitigate data imbalance by focusing on lesion categories that were adequately represented across the participating institutions.
Several preprocessing steps were carried out to prepare the data for training. First, the HAM10000 dataset was split into subsets corresponding to Australia and Austria, creating four distinct datasets. The images were then normalised to a resolution of 224 224 pixels with RGB channels. Each pixel value was standardised by subtracting the mean and dividing by the standard deviation of each channel. Finally, each dataset was divided into training (75%) and testing (25%) sets, ensuring no overlap. A global test set was constructed from the local test sets to assess the generalisation performance of the FL model.
A pre-trained MobileNet v2 Convolutional Neural Network (CNN) was utilised for transfer learning due to its proven effectiveness in skin lesion classification tasks. Transfer learning reduced the need for extensive labelled data and minimised computational requirements. A weighted cross-entropy loss function was applied to address class imbalances in this multiclass classification task. Class weights were set inversely proportional to the sample sizes of each class, ensuring that under-represented classes had more significant influence during model training. For the FL setup, local models were trained independently on each dataset for 50 epochs with a batch size of 32, using stochastic gradient descent (SGD) and a learning rate 0.001. Models were then aggregated using the FedAvg strategy over 20 communication rounds, each involving 10 epochs of local training per institution.
The performance of both local and federated models was assessed using two key metrics: the F1 score and confusion matrix analysis. F1 scores were calculated per class and as a weighted average across all classes to account for class imbalance. The confusion matrix was normalised by class support to provide a clear visualisation of prediction accuracy for each class.
The findings revealed significant differences between models trained locally and those trained using the federated approach. Locally trained models performed well on lesion types sufficiently represented in their respective datasets but showed poor performance on under-represented classes or those absent from the local data. For instance, the Netherlands dataset lacked certain lesion types, resulting in zero predictive capability for those classes.
In contrast, the federated model demonstrated improved generalisation across all lesion types. Notably, institutions such as the one in the Netherlands could predict lesion types absent from their local training data due to knowledge transfer through model aggregation. However, slight reductions in performance were observed in specific cases—for example, predictions of actinic keratosis in Australia—likely due to averaging effects inherent in the aggregation process. By sharing model parameters instead of raw data, FL preserved patient privacy while enabling collaborative learning across diverse and geographically distributed datasets.
This experiment highlights the transformative potential of FL for healthcare data sharing within health data spaces. It shows that FL can address challenges such as data heterogeneity, class imbalance, and privacy in multiclass classification problems. It also underscores the importance of preprocessing and strategically handling imbalanced datasets for successful model training. From an industry perspective, FL facilitates collaborative research without compromising data ownership or patient confidentiality. It enables wider access to diverse datasets, thereby improving the robustness and generalisability of AI models. Future directions include integrating FL with other PET—such as Differential Privacy or Secure Multi-Party Computation—to bolster security and reduce risks like potential information leakage from model updates. Furthermore, developing advanced aggregation techniques tailored to heterogeneous datasets could further enhance performance in federated environments.
In conclusion, this case study demonstrates how FL aligns with the core objectives of health data spaces by enabling secure, privacy-preserving collaboration among healthcare institutions while addressing essential challenges in AI-driven medical research.
Case study 2: synthetic electronic health record
The primary objective of this experiment (included as non-imaging comparator to clarify governance issues that equally affect imaging) was to create high-quality synthetic EHR data that mimics the statistical properties of real-world patient data while maintaining privacy. It aligns with the objectives of the EHDS, which seeks to promote secure and privacy-preserving data sharing to advance healthcare research and innovation. This approach addresses key challenges in health data accessibility by enabling the generation of realistic synthetic datasets, including privacy concerns, data scarcity, and imbalances in the subpopulation representation.
This study utilised the publicly available MIMIC-IV dataset from the Beth Israel Deaconess Medical Centre, which contains structured EHR data from patients admitted to intensive care units between 2008 and 2019 (Figure 3). The dataset includes demographic information, diagnoses, procedures, and clinical measurements such as BMI and blood pressure. A subset of patients with at least one hospital admission was extracted, resulting in a cohort of approximately 181,000 patients. The diagnoses were converted from ICD-9/10 codes to phecodes for clinical relevance and dimensionality reduction.
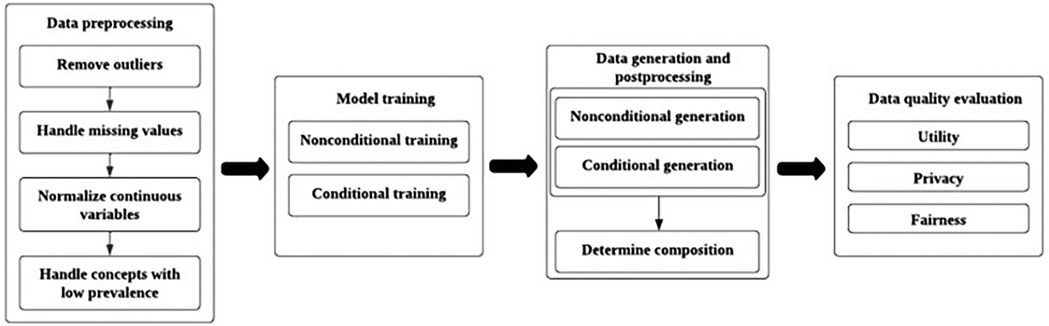
Figure 3. An overview of synthetic electronic health record data generation process through training generative models (Yan et al., 2024).
Preprocessing involves several steps to prepare the data for GAN training. Outliers were identified and removed based on clinically implausible values (e.g. BMIs > 60 or < 10). Missing values were addressed using random sampling based on marginal distributions for variables, such as BMI and blood pressure, which had around 38%–43% missing rates. Continuous variables were normalised to a range of (0,1) to ensure consistent scaling during model training. Low-prevalence concepts were removed or aggregated into higher-level categories to improve the model performance while maintaining clinical relevance.
The GAN architecture consists of two neural networks: a generator that produces synthetic records and a discriminator that distinguishes between real and synthetic data. The EMR-WGAN model was employed because of its ability to capture complex statistical relationships in EHR data while stabilising the training through Wasserstein divergence. The training used a non-conditional paradigm where all variables were treated equally without explicit labels guiding the generation. Multiple runs were performed with checkpoints evaluated for optimal synthetic data quality to address challenges, such as mode collapse and instability in GAN training. A SoftMax layer was added to ensure that categorical variables adhered to the one-hot encoding constraints. While record-level clinical constraints (e.g. prohibiting male patients from having female-specific diagnoses) were not enforced during training, these violations were analysed during evaluation.
The quality of the synthetic data was assessed across three dimensions: utility, privacy, and fairness. The utility metrics include:
• Dimension-wise Distribution: Evaluated how well synthetic data preserved the variable distributions.
• Column-wise Correlation: Measured correlation consistency between real and synthetic variables.
• Latent Cluster Analysis: Assessed structural similarity in the latent space.
• Prediction performance: Comparison of model performance when trained on real vs. synthetic data.
• Feature Importance: Examined overlap in key predictive features.
Privacy risks were evaluated using membership and attribute inference attacks, whereas fairness focused on equitable representation across patient subpopulations. The results demonstrated that the EMR-WGAN model effectively generated high-quality synthetic EHR data while reducing privacy risks compared with real datasets. Dimension-wise distribution analysis showed that the second run achieved the lowest absolute prevalence difference (APD), indicating better preservation of variable distributions. Column-wise correlation and latent cluster analysis also highlighted strong alignment with real data structures.
However, this study had some limitations. Male-specific diagnoses were occasionally assigned to female records in synthetic datasets because of the insufficient preservation of sex-diagnosis correlations. Privacy evaluations revealed significantly reduced membership inference risks compared to real data but highlighted minor variations across runs. This experiment underscores the potential of GAN-based synthetic EHR generation as a transformative tool for healthcare research within the EHDS framework. This approach addresses critical barriers, such as privacy concerns and limited access to diverse patient populations, by enabling the secure sharing of realistic yet anonymised datasets. Synthetic EHRs offer opportunities for hypothesis testing, model training, and educational purposes without compromising patient confidentiality. In the health care industry, they facilitate software development, medical education, and system testing under realistic conditions.
Future directions include incorporating temporal information into synthetic records for longitudinal analyses, improving fairness across subpopulations, and exploring hybrid models that combine GAN with other privacy-enhancing technologies such as differential privacy. These advancements align with the EHDS goals by promoting equitable access to high-quality health data while safeguarding individual privacy.
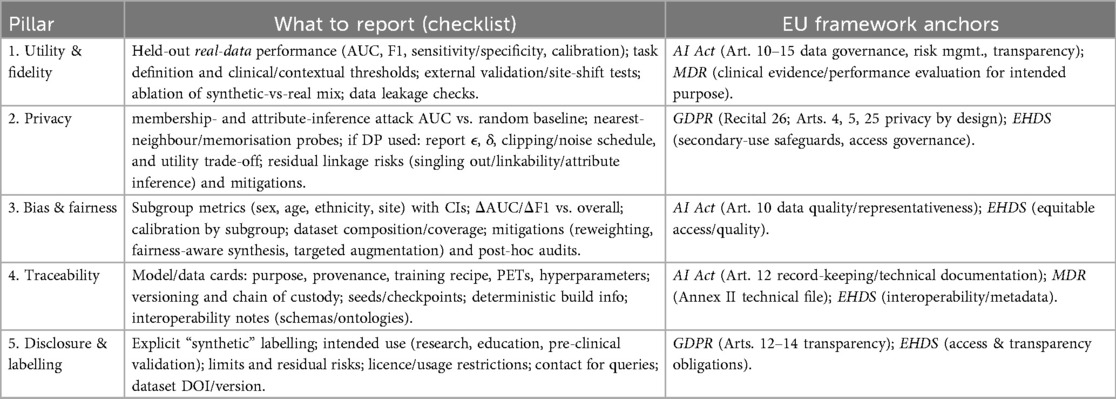
Failure modes and a minimum validation battery for synthetic medical imaging
From methodological overview to quality assurance, we now specify imaging-specific failure modes and a minimum validation battery. Although synthetic data can mitigate access and privacy barriers, generative pipelines are prone to well-documented pitfalls, such as mode collapse and diversity loss, identity leakage/memorisation that elevates re-identification risk, overfitting and site/style shortcuts that undermine external validity, and hallucinated anatomical artefacts that erode clinical credibility. Because these issues directly intersect with the privacy (e.g., membership/attribute inference) and fairness concerns discussed earlier, a clear set of pre-release checks is needed before any secondary use under the EHDS. Below, we enumerate imaging-specific failure modes and propose a minimum validation battery that covers utility/fidelity on held-out real data, privacy attack testing, subgroup fairness, and documentation/traceability, establishing a pragmatic evidentiary floor. This battery is designed to complement the regulatory mapping (Table 2, Figure 2) and to operationalise the governance dimensions formalised in the SID-GT toolkit, enabling data stewards, developers, and access bodies to apply consistent acceptance criteria aligned with EHDS objectives.
Minimum validation battery before secondary use under EHDS
We recommend that the following baseline battery be completed and reported before secondary use or sharing within EHDS. Items map to the governance pillars (utility, privacy, fairness, traceability, and disclosure) and to EU frameworks, as summarised elsewhere in the manuscript.
A. Utility & fidelity (held-out real data)
• Task performance on held-out real data: report AUC/F1, sensitivity/specificity, and calibration (e.g., ECE/Brier), comparing (i) real-only, (ii) real+synthetic, and (iii) synthetic-only training.
• External/site validation: evaluate across at least one external site/scanner/protocol to test for domain shift; include ablations of the synthetic proportion.
• Expert review: double-blinded radiologist rating (e.g., 5-point realism/anatomical plausibility) with inter-rater agreement (e.g., Cohen’s ); flag systematic artefacts.
• Distributional alignment: report simple but informative shifts (intensity histograms, lesion size/location distributions); include nearest-neighbour distance distributions to detect collapse.
B. Privacy (link to “Privacy Risks and Re-identification”)
• Membership/attribute inference: report attack AUC vs. random baseline; describe attacker knowledge. If above-baseline, treat as pseudonymous and apply additional safeguards.
• Memorisation probes: nearest-neighbour search in a perceptual feature space; duplicate detection; generator inversion tests; report minimum distances and exemplar pairs.
• If using differential privacy (DP): report , clipping/noise schedule, and induced utility trade-offs; document residual risks in the release notes.
C. Bias & fairness (link to “Bias and Fairness”)
• Subgroup metrics with uncertainty: report AUC/F1 and calibration with 95% CIs for sex, age bands, ethnicity (where lawful), and site; pre-specify acceptable disparity margins (e.g., AUC within a narrow, justified range).
• Coverage summary: provide subgroup counts/percentages in the synthetic set; describe any targeted augmentation or reweighting applied.
D. Traceability
• Data/model cards: document generator architecture, training data provenance and inclusion/exclusion criteria, PETs/DP settings, seeds/checkpoints, and versioning; maintain chain-of-custody records.
E. Disclosure & labelling
• Synthetic labelling & intended use: mark datasets as synthetic; state intended scope (research, education, pre-clinical validation), limits (e.g., under-represented phenotypes), and residual risks. Provide the licence and contact.
Acceptance guidance
• Release for broader secondary use (e.g., public/consortium sharing): only if attack metrics are at or near baseline, no memorisation evidence is found, expert review shows no systematic anatomical artefacts, and subgroup disparities are within pre-specified margins. Otherwise, treat as regulated pseudonymous data with restricted access and safeguards.
• Clinical evidence contribution: when synthetic data contributes to device performance documentation, ensure full traceability and external validation; prominently disclose synthetic proportions and any fairness mitigations.
This validation battery is designed to be used in conjunction with the governance checklist (SID-GT, cf. Table 5), providing a pragmatic baseline for quality, privacy, and equity before secondary use under EHDS.
Critical discussion and future directions
Current challenges
The rapid advancement of AI in healthcare presents significant ethical, technical, and regulatory challenges. As Federico and Trotsyuk (62) argue, there is a dual obligation to foster innovation while safeguarding individual rights, with particular attention paid to unintended consequences, such as privacy breaches and the amplification of existing biases. Their call for a globally harmonised regulatory framework reflects a growing consensus that balancing innovation with public welfare requires coordinated, anticipatory governance. A similar perspective is echoed by Aucouturier and Grinbaum (63), who advocates a shift from compliance-driven assessments to ethics-by-design methodologies. Such an approach would embed ethical considerations directly into the development pipeline of AI systems, thereby enhancing their long-term accountability and societal alignment. Collaborative governance is critical in this context. Baumgartner et al. (15) emphasise the importance of interdisciplinary teams in managing the complexities of modern health data systems, particularly where data sovereignty, interoperability, and privacy intersect. Colonna and Submitter (64) further note that as private actors increasingly influence public AI research, governance frameworks must evolve to counter regulatory arbitrage and ensure that societal interests are prioritised over commercial gain.
Cross-national data sharing introduces additional complexity owing to the heterogeneous nature of datasets. When data originate from multiple countries, format, quality, and completeness variations can negatively impact the performance of privacy-enhancing technologies (PETs). This highlights the need for rigorous pre-processing and homogenisation techniques. He (2) points out that without a clear EU-level interpretation of relevant laws, data access bodies and holders may apply inconsistent standards, potentially undermining the EHDS goals. Therefore, EHDS implementation must be closely monitored to ensure that data minimisation and ethical use are consistently upheld. The quality and representativeness of datasets remain a persistent concern. Biasin et al. (29) and Burden et al. (38) warn of the dangers of “data contamination” from synthetic content and stress the need for diverse, high-quality datasets to prevent AI systems from reinforcing existing inequities. Without rigorous standards and continuous auditing, AI-driven systems risk perpetuating harmful biases and eroding public trust.
Transparency is a fundamental element in building trust. Baumgartner et al. (15) underscore the value of open-source methodologies and documentation for fostering confidence in synthetic data and AI applications. However, as Federico and Trotsyuk (62) cautioned, existing regulations often fail to enforce transparency, leaving significant gaps in accountability. Another pressing challenge relates to the resource demands of synthetic data generation, particularly when using models, such as GAN, for medical image synthesis. As Arora and Arora (44) noted, the generation of high-quality synthetic images requires substantial computational power. Asadi et al. (65) similarly highlights the significant trade-offs between the costs of training GAN measured in time, energy, and memory, and the practical benefits of using synthetic data. This raises a critical question: are the performance gains from synthetic datasets sufficient to justify their resource intensity?
Future directions for synthetic data
The future of AI in healthcare will depend mainly on addressing the aforementioned challenges, particularly regarding the ethical, technical, and governance aspects of synthetic data. Bertl et al. (66) identified several key barriers, including a lack of standardised interoperability frameworks, ethical uncertainties in data reuse, and insufficient collaboration among stakeholders. These challenges are exacerbated by the speed of technological change, which often outpaces the adaptability of the existing regulatory frameworks. Therefore, robust yet flexible standards are needed to ensure AI systems remain equitable, safe, and effective (15, 62, 64, 66).
An equally pressing concern is the fair distribution of the benefits of AI. Federico and Trotsyuk (62) stressed the importance of ensuring that AI advancements serve both underrepresented and underserved populations. Their view aligns with that of Aucouturier and Grinbaum (63), emphasising ethical governance structures that prioritise inclusivity and societal well-being. The challenges and opportunities surrounding synthetic medical imaging within the EHDS framework require a coordinated set of actions. These must address technical hurdles and the ethical and legal complexities of the implementation. Central to this effort is the need to clarify data ownership, especially when synthetic datasets are derived from personal health data. Intellectual property concerns must be resolved, and individuals should retain agency over how their real or synthetic data are used. It includes the development of harmonised consent frameworks that enable patients to make informed decisions about their data and provide mechanisms to withdraw consent when needed.
Bias mitigation must also be prioritised in synthetic data development. This can be achieved by integrating fair auditing and demographic balancing mechanisms into the data generation pipeline. Ensuring that synthetic datasets reflect a wide range of populations is critical for achieving equitable AI outcomes. Interoperability is another major hurdle to overcome. Unified protocols and standards across EU member states are essential for facilitating seamless data exchange while respecting national data sovereignty. Harmonising these frameworks will be key to enabling collaborative AI innovation throughout Europe. To prevent fragmentation and inconsistency, the EU must take the lead in establishing a unified standard for synthetic-data governance. It could be modelled after initiatives such as FUTURE-AI, which propose benchmarks for technical quality, ethical integrity, and interoperability. Such frameworks should promote transparency, fairness, and robustness, which are essential criteria for ensuring the reliability of synthetic data and their integration into clinical workflow.
Embedding ethics-by-design principles at every stage of synthetic data development is vital. From the outset, privacy, consent, and fairness should be considered and supported by the interdisciplinary ethics committees. Regular ethical audits should be institutionalised to address risks and proactively build public trust. Transparency in the data-generation process is crucial. Developers should provide detailed documentation of their methodologies, algorithms, and validation metrics. Open sourcing of these frameworks, where possible, would facilitate peer review, replication, and broader collaboration, aligning with Open Science principles and enhancing trust in synthetic data tools. In essence, ethical frameworks should not be viewed as regulatory hurdles but as enablers of trust and innovation. Within EHDS, robust governance mechanisms are essential for unlocking the full potential of synthetic imaging data. Addressing consent, ownership, bias, and interoperability issues is fundamental to this vision. Synthetic data offers the potential to transform healthcare by enabling secure, privacy-preserving data sharing and supporting the development of AI applications that are accessible and equitable. With careful alignment of technical innovation and ethical regulations, synthetic data can support advances in diagnostics, personalised treatment, and public health, while maintaining public confidence and compliance with fundamental rights.
One of the most exciting developments in this space is the integration of synthetic medical image generation with natural language processing (NLP), mainly through vision-language models (VLM). Systems such as LLaVA-Med and Med-PaLM merge the interpretive strengths of computer vision and text-based AI to unlock powerful new applications (5):
• Guided image synthesis: VLM can create synthetic medical images based on clinical text inputs. For example, given a description such as “an MRI scan showing glioblastoma with a 5 cm lesion in the frontal lobe,” the system can generate a corresponding synthetic image.
• Multimodal dataset creation: By pairing synthetic images with automated text annotations, VLM can create large-scale multimodal datasets for tasks such as classification, segmentation, or anomaly detection.
• Visual question answering (VQA): These models can generate synthetic visual responses to clinical queries, thereby supporting medical education and training by providing tailored examples.
• Cross-modality synthesis: VLM facilitate conversion across imaging modalities (e.g. MRI to CT), supporting diagnostic flexibility, and reducing redundant scanning.
• Automated annotation and captioning: VLM streamline the process of labelling images by generating consistent, detailed captions, saving time and ensuring annotation quality.
• Interactive data generation: Through conversational interfaces, clinicians can iteratively refine synthetic images by adjusting input specifications, enhancing usability, and tailoring outputs to clinical needs (5).
Towards a governance toolkit for synthetic imaging data
While the ethical and regulatory considerations discussed above provide a conceptual foundation, their translation into actionable practices remains a key challenge. To support alignment with the European Health Data Space (EHDS) objectives, we propose a Synthetic Imaging Data Governance Toolkit (SID-GT). This toolkit serves as a practical checklist for researchers, developers, and data custodians to evaluate synthetic datasets along five essential governance pillars: utility, privacy, fairness, traceability, and disclosure. The toolkit and checklist presented here target synthetic medical imaging; when guidance generalises beyond imaging, this is indicated explicitly. Each pillar is explicitly linked to European regulatory frameworks. including the GDPR, AI Act, MDR, and EHDS regulation, ensuring that governance practices are not only technically robust but also legally grounded. For clarity, Table 5 summarises the proposed governance pillars, associated reporting requirements, and their alignment with key EU regulatory instruments. By embedding these dimensions into evaluation protocols and reporting standards, the SID-GT promotes transparency, accountability, and comparability across projects, facilitating trust and interoperability within EHDS.
In conclusion, when developed and governed responsibly, synthetic data can revolutionise medical research and healthcare delivery across Europe. The EHDS provides a promising foundation, but its success depends on concerted efforts to integrate ethics, transparency, and standardisation into every phase of synthetic data innovation. The introduction of a practical governance toolkit (Table 5) illustrates how these principles can be operationalised, providing a concrete pathway to ensure that synthetic imaging data are used in a manner that is legally robust, ethically sound, and clinically meaningful.
Author contributions
JJ: Writing – review & editing, Data curation, Writing – original draft, Investigation, Visualization, Formal analysis, Resources. LD: Methodology, Validation, Visualization, Formal analysis, Supervision, Conceptualization, Writing – original draft, Investigation, Writing – review & editing, Funding acquisition. JM: Project administration, Supervision, Formal analysis, Visualization, Methodology, Conceptualization, Validation, Funding acquisition, Writing – review & editing, Writing – original draft, Investigation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The present publication was funded by Fundação Ciência e Tecnologia, IP national support through UID/04923 – Comprehensive Health Research Centre.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Foà D. Secondary use of health data and its regulation: impact assessment of the proposed European health data space on scientific research. Pers Ammin. (2024) 14:803–37. https://journals.uniurb.it/index.php/pea/article/view/4861
2. He Z. From privacy-enhancing to health data utilisation: the traces of anonymisation and pseudonymisation in EU data protection law. Digit Soc. (2023) 2:17. doi: 10.1007/s44206-023-00043-5
3. Hernãndez-Peñaloza G, Uribe S, García FM, Graf N, Álvarez F. General context and relevant public datasets available for improving pathways in paediatric cancer applying artificial intelligence. A review. EJC Paediatr Oncol. (2024) 4:100196. doi: 10.1016/j.ejcped.2024.100196
4. Ianese S. Challenging data protection in the age of data spaces. Eur J Priv Law Technol. (2024). https://universitypress.unisob.na.it/ojs/index.php/ejplt/article/viewFile/2002/1525
5. Kim K, Cho K, Jang R, Kyung S, Lee S, Ham S, et al. Updated primer on generative artificial intelligence and large language models in medical imaging for medical professionals. Korean J Radiol. (2024) 25:224–42. doi: 10.3348/kjr.2023.0818
6. Lianos I. Access to health data: competition and regulatory alternatives – three dimensions of fairness. SSRN Electron J. (2024). doi: 10.2139/ssrn.4962599
7. Marques IC. The elephant in the room: rethinking GDPR enforcement in the context of genetic data (master’s thesis). Tilburg: Tilburg University (2022). Available online at: https://arno.uvt.nl/show.cgi?fid=159614
8. Greser J. Cybersecurity framework for synthetic data in training medical AI. Eur J Risk Regul. (2024) 15:903–11. doi: 10.1017/err.2024.74
9. Lebrun T. Health data: exploring and enhancing emerging privacy-protection mechanisms (Ph.D. thesis). Lyon: INSA Lyon (2024). Available online at: https://www.insa-lyon.fr/en/mots-cl%C3%A9s/infomaths
10. Pereira T, Morgado J, Silva F, Pelter MM, Dias VR, Barros R, et al. Sharing biomedical data: strengthening AI development in healthcare. Healthcare. (2021) 9:827. doi: 10.3390/healthcare9070827
11. Filippini F. Large language models in Dutch healthcare (master’s thesis). Utrecht: Utrecht University (2024). Available online at: https://studenttheses.uu.nl/handle/20.500.12932/46149
12. Regazzoni F, Acs G, Aszalos AZ, Avgerinos C, Bakalos N, Berral JL, et al. Secured for health: scaling up privacy to enable the integration of the European health data space. In: 2024 Design, Automation & Test in Europe Conference & Exhibition (25–27 March 2024). (2024). p. 1–4.
13. Arora A, Arora A. Synthetic patient data in health care: a widening legal loophole. Lancet. (2022) 399:1601–2. doi: 10.1016/S0140-6736(22)00232-X
14. Bartell JA, Valentin SB, Krogh A, Langberg H, Bøgsted M. A primer on synthetic health data. arXiv [Preprint]. (2024). doi: 10.48550/arXiv.2401.17653
15. Baumgartner M, Kreiner K, Lauschensky A, Jammerbund B, Donsa K, Hayn D, et al. Health data space nodes for privacy-preserving linkage of medical data to support collaborative secondary analyses. Front Med. (2024) 11:1301660. doi: 10.3389/fmed.2024.1301660
16. Chlap P, Min H, Vandenberg N, Dowling J, Holloway L, Haworth A. A review of medical image data augmentation techniques for deep learning applications. J Med Imaging Radiat Oncol. (2021) 65:545–63. doi: 10.1111/1754-9485.13261
17. Hägglund M, Blusi M, Bonacina S, Nilsson L, Madsen IC, Pelayo S, et al. Studies in health technology and informatics. In: Caring is Sharing - Exploiting the Value in Data for Health and Innovation - Proceedings of MIE 2023. Studies in Health Technology and Informatics. Vol. 302. (2023). Available online at: https://ebooks.iospress.nl/volume/caring-is-sharing-exploiting-the-value-in-data-for-health-and-innovation-proceedings-of-mie-2023 (Accessed January 05, 2025).
18. Petersen E, Potdevin Y, Mohammadi E, Zidowitz S, Breyer S, Nowotka D, et al. Responsible and regulatory conform machine learning for medicine: a survey of challenges and solutions. IEEE Access. (2022) 10:58375–418. doi: 10.1109/ACCESS.2022.3178382
19. Susser D, Schiff DS, Gerke S, Cabrera LY, Cohen IG, Doerr M, et al. Synthetic health data: real ethical promise and peril. Hastings Cent Rep. (2024) 54:8–13. doi: 10.1002/hast.4911
20. Shanley D, Hogenboom J, Lysen F, Wee L, Gomes AL, Dekker A, et al. Getting real about synthetic data ethics. EMBO Rep. (2024) 25:2152–5. doi: 10.1038/s44319-024-00101-0
21. Floridi L, Cowls J, Beltrametti M, Chatila R, Chazerand P, Dignum V, et al. AI4People-an ethical framework for a good AI society: opportunities, risks, principles, and recommendations. Minds Mach. (2018) 28:689–707. doi: 10.1007/s11023-018-9482-5
22. EDPS. Opinion 3/2020 on the European strategy for data: scientific research and the GDPR. Tech. Rep. 3/2020. European Data Protection Supervisor (2020). Available online at: https://www.edps.europa.eu/sites/default/files/publication/20-01-06_opinion_research_en.pdf (Accessed April 18, 2025).
23. EC. Common infrastructure for national cohorts in Europe, Canada, and Africa. Tech. Rep. (2019). Available online at: https://www.cineca-project.eu/ (Accessed March 12, 2025).
24. Chen RJ, Lu MY, Chen TY, Williamson DF, Mahmood F. Synthetic data in machine learning for medicine and healthcare. Nat Biomed Eng. (2021) 5:493–7. doi: 10.1038/s41551-021-00751-8
25. Bottani S, Thibeau-Sutre E, Maire A, Ströer S, Dormont D, Colliot O, et al. Contrast-enhanced to non-contrast-enhanced image translation to exploit a clinical data warehouse of T1-weighted brain MRI. BMC Med Imaging. (2024) 24:67. doi: 10.1186/s12880-024-01242-3
26. Lara MAR, Echeveste R, Ferrante E. Addressing fairness in artificial intelligence for medical imaging. Nat Commun. (2022) 13:4581. doi: 10.1038/s41467-022-32186-3
27. de Vries K. A researcher’s guide for using personal data and non-personal data surrogates: synthetic data and data of deceased people. In: Lege D, editor. Law, AI and Digitalisation. Vol. 2021. Uppsala: Iustus förlag (2022). p. 117–40. Available online at: https://urn.kb.se/resolve?urn=urn:nbn:se:uu:diva-501668
28. Abujaber AA, Nashwan AJ. Ethical framework for artificial intelligence in healthcare research: a path to integrity. World J Methodol. (2024) 14:94071. doi: 10.5662/wjm.v14.i3.94071
29. Biasin E, Harcus AMC, Vedder A. In silico world D9.2 in-depth analysis of legal and ethical requirements. Tech. Rep. KUL Centre for IT and IP Law (KUL-CTP) (2023).
30. van der Wel I. Synthetic data generation for research – enabler for privacy-enhancing health data sharing? (master thesis). Delft: Delft University of Technology, M.Sc. Complex Systems Engineering and Management (2024). Available online at: https://repository.tudelft.nl/
31. Bogucki A, Engler A, Perarnaud C, Renda A. The AI act and emerging EU digital acquis: overlaps, gaps and inconsistencies. Tech. Rep. Funding support received from the Future of Life Institute. Centre for European Policy Studies (CEPS) (2022).
32. Casarosa F, Greser J. The challenges of cybersecurity in the health sector. Eur J Risk Regul. (2024) 15:872–5. doi: 10.1017/err.2024.94
33. Colcelli V. The European union legal framework for using artificial intelligence and imaging databases and imaging biobanks for research purposes: applying the notion of fairness. Cad Ibero-Am Direito Sanit. (2024) 13:124–40. doi: 10.17566/ciads.v13i4.1288
34. Ritoré Á, Jiménez CM, González JL, Rejón-Parrilla JC, Hervás P, Toro E, et al. The role of open access data in democratizing healthcare AI: a pathway to research enhancement, patient well-being and treatment equity in Andalusia, Spain. PLOS Digit Health. (2024) 3:e0000599. doi: 10.1371/journal.pdig.0000599
35. Tozzi AE, Croci I, Voicu P, Dotta F, Colafati GS, Carai A, et al. A systematic review of data sources for artificial intelligence applications in pediatric brain tumors in Europe: implications for bias and generalizability. Front Oncol. (2023) 13:1285775. doi: 10.3389/fonc.2023.1285775
36. Borissova G. Balancing open data and data protection: harnessing synthetic data for responsible scientific research under the European union data protection legal framework (LLM master’s thesis). Tilburg: Tilburg University (2024). Available online at: https://hdl.handle.net/1814/77765
37. Ciminá V. The protection of personal data for the purpose of scientific research and the open science framework: ongoing challenges and future opportunities. J Open Access Law. (2023) 11:1–10. doi: 10.63567/ta33bj95
38. Burden J, Maurice MC, Ruse-Khan HG, Markschies L, Müller D. Legal aspects of access to human-generated data and other essential inputs for AI training. Research paper 35/2024. University of Cambridge Faculty of Law (2024). doi: 10.2139/ssrn.5045155
39. Aunón J.M., Hurtado-Ramírez D., Porras-Díaz L., Irigoyen-Peña B., Rahmian S., Al-Khazraji Y., et al. Evaluation and utilisation of privacy enhancing technologies-a data spaces perspective. Data Brief. (2024) 55:110560. doi: 10.1016/j.dib.2024.110560
40. Barbanotti L, Cicchetti M, Varriale G. Drug discovery and big data: from research to the community. In: Coletti S, Bernardi G, editors. Exscalate4CoV. Cham: Springer (2023). SpringerBriefs in Applied Sciences and Technology.
41. Aspell N, Goldsteen A, Renwick R. Dicing with data: the risks, benefits, tensions and tech of health data in the iToBoS project. Front Digit Health. (2024) 6:1272709. doi: 10.3389/fdgth.2024.1272709
42. de Bienassis K, Fujisawa R, Hashiguchi TCO, Klazinga N, Oderkirk J. Health data and governance developments in relation to COVID-19: how OECD countries are adjusting health data systems for the new normal. OECD Health Working Papers 138. Paris: OECD Publishing (2022).
43. Davidson E, Winter JS. Consumer health data: regulation, governance, and innovation. In: Proceedings of the 58th Hawaii International Conference on System Sciences. (2025).
44. Arora A, Arora A. Generative adversarial networks and synthetic patient data: current challenges and future perspectives. Future Healthc J. (2022) 9:190–3. doi: 10.3389/fdgth.2024.1272709
45. Ali H, Biswas MR, Mohsen F, Shah U, Alamgir A, Mousa O, et al. The role of generative adversarial networks in brain MRI: a scoping review. Insights Imaging. (2022) 13:98. doi: 10.1186/s13244-022-01237-0
46. Brugnara G, Preetha CJ, Deike K, Haase R, Pinetz T, Foltyn-Dumitru M, et al. Addressing the generalizability of AI in radiology using a novel data augmentation framework with synthetic patient image data: proof-of-concept and external validation for classification tasks in multiple sclerosis. Radiol Artif Intell. (2024) 6:e230514. doi: 10.1148/ryai.230514
47. Pezoulas VC, Zaridis DI, Mylona E, Androutsos C, Apostolidis K, Tachos NS, et al. Synthetic data generation methods in healthcare: a review on open-source tools and methods. Comput Struct Biotechnol J. (2024) 23:2892–910. doi: 10.1016/J.CSBJ.2024.07.005
48. Cheng V, Suriyakumar VM, Dullerud N, Joshi S, Ghassemi M. Can you fake it until you make it? Impacts of differentially private synthetic data on downstream classification fairness. In: Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency. New York (NY), USA: Association for Computing Machinery (2021). FAccT ’21. p. 149–60.
49. Giuffrè M, Shung DL. Harnessing the power of synthetic data in healthcare: innovation, application, and privacy. npj Digit Med. (2023) 6:186. doi: 10.1038/s41746-023-00927-3
50. Eckardt JN, Hahn W, Röllig C, Stasik S, Platzbecker U, Müller-Tidow C, et al. Mimicking clinical trials with synthetic acute myeloid leukemia patients using generative artificial intelligence. npj Digit Med. (2024) 7:76. doi: 10.1038/s41746-024-01076-x
51. Ntampakis N, Argyriou V, Diamantaras K, Goulianas K, Sarigiannidis P, Siniosoglou I. Introducing spine: a holistic approach to synthetic pulmonary imaging evaluation through end-to-end data and model management. IEEE Open J Eng Med Biol. (2024) 5:576–88. doi: 10.1109/OJEMB.2024.3426910
52. Angermann C, Baumgartner L, Becker F, Bühler K, Egger J. Three-dimensional bone-image synthesis with generative adversarial networks. J Imaging. (2024) 10:318. doi: 10.3390/jimaging10120318
53. Vaden KI, Norton IH, Callahan TJ, Miller MI, Mori S. The incorporation of synthetic data methods in neuroimaging. NeuroImage. (2020) 219:117171. doi: 10.1016/j.neuroimage.2020.117171
54. Arora A, Arora A. Machine learning models trained on synthetic datasets of multiple sample sizes for the use of predicting blood pressure from clinical data in a national dataset. PLoS One. (2023) 18:e0283094. doi: 10.1371/journal.pone.0283094
55. Pereira M, Kshirsagar M, Mukherjee S, Dodhia R, Ferres JL, de Sousa R. Assessment of differentially private synthetic data for utility and fairness in end-to-end machine learning pipelines for tabular data. PLoS One. (2024) 19:e0297271. doi: 10.1371/journal.pone.0297271
56. Pozzi A, Cerrai F, Fontanella S, Marzullo A, Marchesin S, Caputo B. Generating and evaluating synthetic data in digital pathology through diffusion models. Sci Rep. (2024) 14:1234. doi: 10.1038/s41598-024-79602-w
57. Yan C, Zhang Z, Nyemba S, Li Z. Generating synthetic electronic health record data using generative adversarial networks: tutorial. JMIR AI. (2024) 3:e52615. doi: 10.2196/52615
58. Schütte AD, Hetzel J, Gatidis S, Hepp T, Dietz B, Bauer S, et al. Overcoming barriers to data sharing with medical image generation: a comprehensive evaluation. npj Digit Med. (2021) 4:141. doi: 10.1038/s41746-021-00507-3
59. Coyner AS, Chen JS, Chang K, Singh P, Ostmo S, Chan RV, et al. Synthetic medical images for robust, privacy-preserving training of artificial intelligence: application to retinopathy of prematurity diagnosis. Ophthalmol Sci. (2022) 2:100126. doi: 10.1016/j.xops.2022.100126
60. Zheng C, Bian F, Li L, Xie X, Liu H, Liang J, et al. Assessment of generative adversarial networks for synthetic anterior segment optical coherence tomography images in closed-angle detection. Transl Vis Sci Technol. (2021) 10:34. doi: 10.1167/tvst.10.4.34
61. Krause J, Grabsch HI, Kloor M, Jendrusch M, Echle A, Buelow RD, et al. Deep learning detects genetic alterations in cancer histology generated by adversarial networks. J Pathol. (2021) 254:70–9. doi: 10.1002/path.5638
62. Federico CA, Trotsyuk AA. Biomedical data science, artificial intelligence, and ethics: navigating challenges in the face of explosive growth. Annu Rev Biomed Data Sci. (2024) 7:1–14.38598860
63. Aucouturier E, Grinbaum A. iRECS deliverable 2.2: recommendations to address ethical challenges from research in new technologies. Tech. Rep. CEA – Commissariat à l'énergie atomique et aux énergies alternatives (2023). Available online at: https://cea.hal.science/cea-04293426 (Accessed January 05, 2025).
64. Colonna L, Submitter SL. The AI act’s research exemption: a mechanism for regulatory arbitrage? SSRN Electron J. (2023). doi: 10.2139/ssrn.4575971
65. Asadi F, Angsuwatanakul T, O’Reilly JA. Evaluating synthetic neuroimaging data augmentation for automatic brain tumour segmentation with a deep fully-convolutional network. IBRO Neurosci Rep. (2024) 16:57–66. doi: 10.1016/J.IBNEUR.2023.12.002
Keywords: synthetic data, medical imaging, European Health Data Space (EHDS), data privacy, generative AI, GDPR compliance, AI act, Medical Device Regulation (MDR)
Citation: Jiang J, Domingues L and Mendes JM (2025) Synthetic data in medical imaging within the EHDS: a path forward for ethics, regulation, and standards. Front. Digit. Health 7:1620270. doi: 10.3389/fdgth.2025.1620270
Received: 29 April 2025; Accepted: 8 September 2025;
Published: 8 October 2025.
Edited by:
Lisette van van Gemert-Pijnen, University of Twente, NetherlandsCopyright: © 2025 Jiang, Domingues and Mendes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jorge M. Mendes, am9yZ2UubWVuZGVzQG5tcy51bmwucHQ=
 Junying Jiang
Junying Jiang Lúcia Domingues
Lúcia Domingues Jorge M. Mendes
Jorge M. Mendes