- Research Department, The Self Research Institute, Broken Arrow, OK, United States
Background: Ontologies serve as a foundational bridge between artificial intelligence (AI) and healthcare, enabling structured knowledge frameworks that enhance data interoperability, clinical decision support, and precision medicine.
Objective: This perspective aims to highlight the essential role of ontologies in enabling adaptive, interoperable frameworks that evolve with technological and medical advances to support personalized, accurate, and globally connected healthcare solutions.
Methods: This perspective is based on a targeted literature exploration conducted across PubMed, Scopus, and Google Scholar, prioritizing studies published between 2010 and 2025 and including earlier seminal works where necessary to provide historical context, focusing on ontology-driven AI applications in healthcare. Sources were selected for their relevance to semantic integration, interoperability, and interdisciplinary applicability.
Results: Through the standardization of medical concepts, relationships, and terminologies, ontologies enable semantic integration across diverse healthcare datasets, including clinical, genomic, and phenotypic data. They also address challenges such as fragmented data and inconsistent terminologies. This semantic clarity supports AI applications in clinical decision support, predictive analytics, natural language processing (NLP), and patient-specific disease modeling.
Conclusions: Despite their transformative potential, ontology integration faces significant challenges, including computational complexity, scalability, and semantic mismatches across evolving international standards, such as SNOMED CT and HL7 FHIR. Ethical concerns, particularly around data privacy, informed consent, and algorithmic bias, also require careful consideration. To address these challenges, this perspective outlines strategies including adaptive ontology models, robust governance frameworks, and AI-assisted ontology management techniques. Together, these approaches aim to support personalized, accurate, and globally interoperable healthcare systems.
1 Introduction
Modern healthcare systems continue to face challenges related to fragmented data, lack of standardized vocabularies, and poor semantic alignment. Ontologies, structured frameworks that define standardized concepts and relationships within a domain (1). They enable consistent data interpretation, support automated reasoning, and enhance clinical decision-making. By facilitating semantic interoperability across systems, ontologies allow AI models and healthcare applications to integrate and act upon complex clinical, genomic, and phenotypic data (2–4).
Ontologies, such as SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms) and HL7 FHIR (Fast Healthcare Interoperability Resources), developed by Health Level Seven International (HL7), serve as foundational tools by standardizing medical concepts, terminologies, and data structures (5). These unified frameworks support tailored patient-specific healthcare solutions, improving clinical outcomes and patient experience. They can enhance the efficacy of Clinical Decision Support Systems (CDSS) by providing structured knowledge representation and facilitating rule-based reasoning (6). Ontologies also support personalization of healthcare by adapting intervention plans to individual needs and uncovering hidden comorbidities (7).
By enabling interoperability across Electronic Health Records (EHRs), ontologies facilitate seamless collaboration between healthcare providers and AI systems. They drive advancements in precision medicine and patient-centered care by standardizing medical terminologies and ensuring consistent data exchange. Furthermore, ontologies integrate diverse data sources by aligning terminologies like SNOMED CT, LOINC (Logical Observation Identifiers Names and Codes), and ICD-11 (International Classification of Diseases, 11th Revision) with real-world applications, demonstrating their potential to enhance data-driven decision-making and AI-driven analytics. These attributes make transformative innovations in both AI and healthcare systems (5, 8, 9).
Ontologies offer a solution by bridging artificial intelligence (AI) and healthcare (1). Despite their value, significant challenges remain, particularly in addressing fragmented datasets, lack of standardization, and ethical concerns like data privacy, and adaptive ontology models offer promising solutions by supporting real-time updates, promoting ethical governance, and allowing culturally sensitive customization (10). Through semantic integration and fostering patient-centered design, ontologies are poised to bridge gaps in healthcare systems and advance AI-driven solutions.
The evolution of ontologies, initially rooted in statistical classifications and formal logic, has progressively transformed to address the multifaceted demands of modern healthcare. From supporting clinical operations and reimbursement processes to enabling translational research and reducing errors, improving efficiency, and driving biomedical discovery, ontologies have demonstrated their adaptability and growing importance. Their structural evolution is guided by diverse application areas, allowing them to meet the complex requirements of contemporary healthcare. These applications are particularly significant in supporting international data harmonization and ensuring consistent interpretation across healthcare systems. Currently, ontologies form the foundation for integrating vast biomedical datasets, supporting sophisticated data science inferencing, and enabling advanced analytics and decision-making tools that drive innovation in healthcare systems (5).
By providing a systematic approach to knowledge representation, ontologies enable AI systems to process biomedical data with greater precision and reliability. Their flexibility to adapt to new data and evolving standards reinforces their role in advancing healthcare and AI systems (11, 12).
In this perspective, we highlight the need for adaptive ontology models that evolve with real-time data and emerging standards to support precision medicine, promote ethical AI integration, and address key gaps in semantic interoperability. We outline the historical evolution of ontologies, mechanisms enabling interoperability, real-world applications, and future directions for their development in healthcare innovation. This perspective synthesizes key literature on ontology-driven AI in healthcare, outlines current challenges in semantic interoperability, and proposes a roadmap for advancing adaptive, ethically grounded, and globally interoperable ontology models. It also highlights the reciprocal relationship between ontologies and AI, showing how ontologies enhance AI capabilities while AI methods drive ontology evolution and automation.
2 Methods
We conducted a targeted literature exploration to identify key advancements, current challenges, and gaps in ontology-driven applications of artificial intelligence in healthcare. Searches were performed across PubMed, Scopus, and Google Scholar using combinations of keywords aligned with the article's thematic focus, including “ontology,” “artificial intelligence,” “interoperability,” “semantic integration,” “clinical decision support,” “personalized medicine,” “natural language processing,” and “ethical AI.” These were combined with Boolean operators such as (ontology OR ontologies) AND (“artificial intelligence”) AND (“interoperability” OR “semantic integration” OR “clinical decision support” OR “personalized medicine” OR “natural language processing” OR “ethical AI”).
We prioritized peer reviewed publications published between 2010 and 2025 that addressed the development or application of ontologies in artificial intelligence, healthcare interoperability, and related interdisciplinary domains, while excluding sources lacking direct relevance to biomedical or clinical applications. We also included earlier seminal works published before 2010 when they were considered foundational or essential for understanding the historical evolution of ontology-driven AI applications.
This manuscript was intentionally developed as a perspective rather than a scoping or systematic review. Given the interdisciplinary and conceptual nature of the topic, which spans technical, biomedical, and policy domains, conducting a comprehensive systematic review was beyond the intended scope of this work. Accordingly, this study applies a conceptual grouping of recurring themes to synthesize key insights, emerging trends, and challenges, with the goal of informing future research and development in ontology-driven, AI enabled healthcare systems.
3 Ontologies
3.1 Historical context
Ontologies originated in philosophy, where early thinkers like Aristotle developed taxonomies to classify knowledge. As computer science advanced, ontologies evolved into structured systems for representing domain knowledge. Gruber (1993) defined them as “explicit specifications of a conceptualization” (13), while Guarino (1998) emphasized their value in organizing domain-specific knowledge (14). The introduction of formal ontology languages like RDF (Resource Description Framework) and OWL (Web Ontology Language) (15–17) enabled ontologies to support semantic integration across fragmented datasets in dynamic, multi-user environments (18).
A useful distinction exists between formal ontologies, which provide general upper-level concepts across domains, and application ontologies, which are specifically designed for particular domains such as oncology or cardiology. This distinction is especially relevant in AI and healthcare, where both abstraction and domain specificity are essential.
To remain relevant in fast-evolving fields like AI and healthcare, ontologies must continually adapt to new data, contexts, and standards. During the COVID-19 pandemic, ontologies rapidly incorporated new clinical terminologies and concepts such as “COVID-19-associated pneumonia” or “long COVID” to support data interoperability, surveillance, and research (19). Evolutionary processes, such as alignment, versioning, and meaning negotiation, are critical in distributed healthcare ecosystems where real-time collaboration and interoperability are required (20, 21). Especially in AI-driven healthcare, ontology evolution supports adaptive systems that can integrate diverse knowledge sources, maintain semantic consistency, and ensure accurate decision-making (22, 23).
3.2 Semantic integration
Ontologies play a pivotal role in enabling semantic integration by providing a structured and hierarchical approach to organizing data. They establish a shared vocabulary, enabling diverse systems to communicate seamlessly and interpret information consistently. This hierarchical organization supports advanced reasoning and inference, allowing for the transformation of raw data into actionable insights. To demonstrate their real-world relevance and technical sophistication, the following subsections explore practical applications of semantic integration and the underlying mechanisms that enable its implementation across AI-driven healthcare systems.
3.2.1 Permission to reuse and copyright
In modern healthcare, ontologies are instrumental in bridging diverse datasets such as clinical, genomic, and environmental data into unified and interpretable frameworks. Their structured vocabularies enable systems like the Gene Ontology (GO) and the Foundational Model of Anatomy (FMA) to annotate and cross-reference biological and anatomical information, facilitating precision medicine and real-time clinical decision-making (24).
By aligning genomic, phenotypic, and clinical information, ontologies support diagnostic accuracy, treatment stratification, and predictive analytics in AI applications (25). These capabilities are reflected in practical applications such as the use of geospatial ontologies, which integrate location-based data to enhance public health planning during disease outbreaks or environmental exposures (26). These real-world applications highlight why semantic integration is essential, not only for linking datasets but also for enabling intelligent, adaptive, and context-aware healthcare systems.
3.2.2 Underlying technical mechanisms
This semantic depth is powered by technical mechanisms such as hierarchical taxonomies, logical inference engines, and standardization frameworks. Taxonomies define relationships between broad and specific concepts, enabling structured reasoning and context-aware classification. Ontology-based engines validate data consistency and deduce new knowledge from complex, multimodal datasets (24, 27, 28).
Foundational ontologies such as DOLCE (Descriptive Ontology for Linguistic and Cognitive Engineering) and BFO (Basic Formal Ontology) facilitate alignment across domain-specific ontologies, resolving semantic discrepancies and supporting cross-domain compatibility. BFO, for instance, provides a unifying framework that enables consistent mapping of clinical concepts like “disease onset” in SNOMED CT with biological processes in the Gene Ontology (GO) (25).
Standards such as RDF and OWL support interoperability by enabling shared concept mapping, while SPARQL (SPARQL Protocol and RDF Query Language) improves query performance across large biomedical datasets (29). These technical tools enable inter-ontology reasoning, allowing AI systems to synthesize knowledge across clinical, genomic, and environmental domains, thereby advancing translational research and supporting tailored care strategies (30).
3.3 Ontology development and maintenance
Ontology development is a systematic and iterative process essential for building robust frameworks in complex domains, including healthcare and AI. Effective ontology development includes clearly defined stages: design, validation, alignment, versioning and updates, enrichment, and ontology evolution. These stages collectively address challenges like fragmented datasets, inconsistent terminologies, and evolving domain-specific requirements, ensuring that ontologies remain adaptable and relevant. Such modularity is essential in healthcare AI, where domain knowledge and terminologies rapidly evolve, making this design process vital for AI–healthcare convergence.
3.3.1 Design phase
The design phase begins by identifying domain-specific requirements through consultation with domain experts, existing documentation, and user needs analysis. This phase is crucial to ensure that the ontology accurately represents real-world contexts and aligns with domain-specific standards (31). Techniques like Ontology-Driven Conceptual Modeling (ODCM) provide theoretical guidance, enabling structured representations and conceptual clarity through languages like OntoUML (32).
3.3.2 Validation methodologies
Robust validation ensures that ontologies reliably support AI accuracy in healthcare applications. Competency Questions (CQs) test whether an ontology can answer clinically relevant queries, such as linking symptoms to diagnoses or treatments, which is essential for decision support tools. Metrics like consistency and cohesion assess structural quality, while tools like Protégé automate reasoning and error detection (33). These methods help AI systems make accurate predictions, especially in domains like oncology and cardiology where precise semantic alignment is critical (34).
3.3.3 Alignment
Ontology alignment harmonizes various domain-specific ontologies to facilitate seamless interoperability and consistent data exchange. Semantic mapping techniques link different ontologies, establishing compatibility and coherent data interpretation across healthcare and AI systems. Foundational ontologies including DOLCE and BFO assist this alignment by providing universal frameworks for categorization and ensuring data consistency (35).
3.3.4 Versioning and updates
Ontologies must continually evolve to reflect new data, updated standards, and emerging knowledge. Robust versioning systems are essential for managing changes, maintaining backward compatibility, and ensuring long-term usability. One such method, the Ontology Metadata Vocabulary for Ontology Data Management (ON-ODM), helps track changes across versions by documenting metadata such as authorship, updates, and dependencies. This allows ontology frameworks to adapt to evolving domain needs while preserving accuracy and relevance over time (36).
3.3.5 Enrichment techniques
Enrichment involves refining ontologies by adding new concepts, relationships, and data properties, which enhances their completeness and contextual relevance. Artificial intelligence, particularly NLP and text-mining techniques, accelerates this process by scanning large volumes of biomedical literature, clinical notes, or research databases to identify emerging terms or relationships that may be missing from existing ontologies.
The BioPortal Annotator uses NLP to map free-text inputs to existing biomedical ontologies, enabling automatic detection of relevant concepts and suggesting updates based on current literature. This dramatically improves the speed and accuracy of ontology maintenance by reducing manual curation efforts and ensuring timely integration of new knowledge (37).
Such AI-driven enrichment ensures that ontologies stay current and aligned with evolving healthcare needs, supporting high-quality AI applications and clinical decision support systems (CDSS).
3.3.6 Ontology evolution in collaborative environments
Ontology evolution is essential in collaborative and distributed environments like the Semantic Web, where ontologies dynamically co-evolve alongside user communities. The key processes include meaning negotiation, alignment of multiple organizational ontologies, and management of contextual dependencies. These iterative evolution processes maintain ontology robustness, facilitating real-time knowledge integration essential for modern AI-driven healthcare solutions (18).
3.4 Broader applications of ontologies
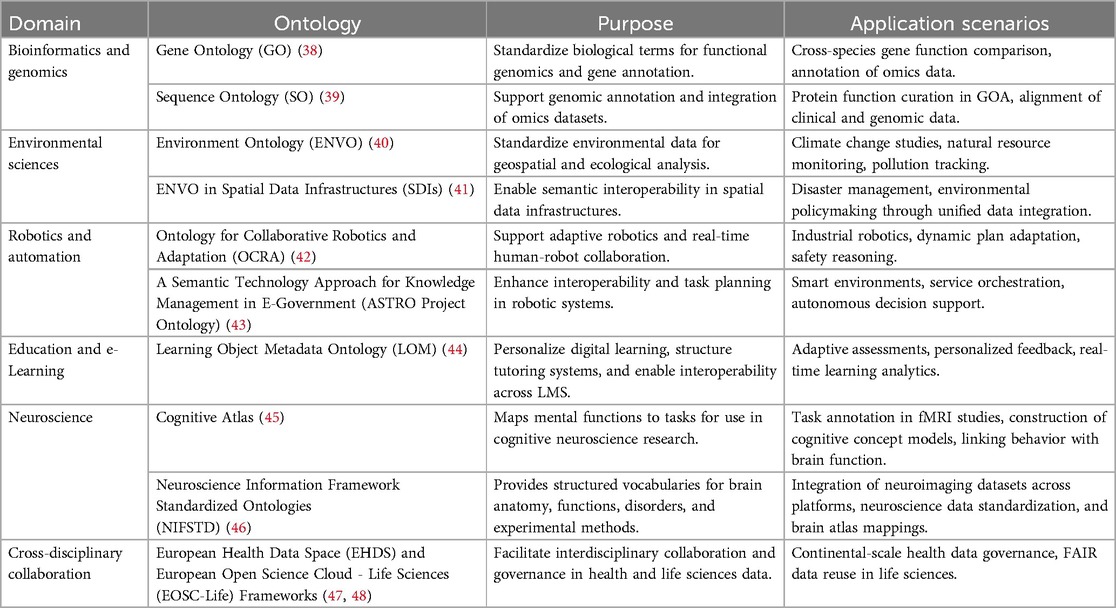
Ontologies are increasingly applied across diverse fields such as bioinformatics, environmental science, robotics, education, and interdisciplinary research. By providing structured vocabularies and semantic frameworks, they enable data integration, automation, and personalized decision-making. This versatility makes ontologies powerful tools for advancing innovation, interoperability, and knowledge-sharing across domains. This cross-domain adaptability underscores the foundational role of ontologies in fostering innovation in complex, data-driven environments like healthcare AI. Table 1 summarizes ontology applications across domains, highlighting their purposes and implementation scenarios.
4 AI and ontology applications in healthcare
By addressing challenges linked to fragmented datasets, disconnected systems, and inconsistent medical data representation, ontologies provide a structured framework that enables improved data integration, semantic clarity, and scalable AI-driven solutions in healthcare. They provide a structured framework for organizing diverse healthcare information, ensuring semantic interoperability across clinical applications. By standardizing how data is defined and interpreted, ontologies improve clinical decision-making and support AI models that rely on consistent, high-quality input. Ontologies bridge technology and human-centered care, becoming foundational tools in advancing precision medicine, healthcare automation, and intelligent diagnostics. Their ability to standardize medical data into machine-readable formats empowers healthcare providers and AI-driven systems to derive actionable insights, enhancing patient outcomes and fostering healthcare innovation. Across domains such as EHRs, genomics, pharmacology, and geospatial analytics, ontologies ensure semantic consistency and enable scalable, intelligent applications in real-world care. The following subsections highlight key ontology applications in healthcare AI, from EHR interoperability to population-level predictions. Thus, ontologies are crucial in healthcare and AI.
4.1 Enhancing EHR functionality through ontologies
Ontologies integrated into EHR systems improve data clarity, reduce fragmentation, and enhance care coordination. They go beyond standard interoperability by embedding meaning and relationships directly into data, enabling smarter clinical workflows. Ontology-driven systems and EHR-integrated analytics have demonstrated tangible impacts on clinical efficiency. For instance, Rizzoli Orthopaedic Institute reported a 30% reduction in hospitalizations and over 60% fewer imaging tests by using advanced analytics to manage hereditary bone disorders. Similarly, Kaiser Permanente used EHR-linked analytics to minimize unnecessary antibiotic use in newborns, significantly lowering both side effects and resource utilization. These examples reflect how integrated data systems can optimize patient care and reduce redundant interventions without compromising quality (49). They also enable context-aware decision support by aligning terminologies like SNOMED CT and LOINC with patient records, flagging interactions or missing clinical documentation (50).
Frameworks like openEHR and ISO 13606 use a dual model approach that includes reference models for structure and archetype models for clinical semantics. These models, supported by HL7 FHIR, form the foundation of ontology-aligned systems that can interpret and exchange structured clinical data consistently (51). The canonical ontology model adds a shared conceptual layer that enables different EHR architectures to speak a common language, allowing automated reasoning and intelligent data exchange (52). NHS Digital in the UK utilizes such models to ensure that care records follow patients across systems, reducing duplication and improving continuity (53).
4.2 Clinical decision support and AI-driven diagnostics
Ontologies enhance CDSS by structuring medical knowledge into logically connected frameworks that AI systems can reason over. They enable context-aware AI to connect symptoms, diagnoses, and treatments through explicit semantic links, thereby improving diagnostic accuracy and personalized care.
Ontology-driven systems support intelligent automation and reasoning by aligning terminologies across diverse data sources, such as SNOMED CT, LOINC, and HL7 FHIR. These integrations reduce errors, identify care gaps, and guide clinicians in selecting optimal interventions. By improving semantic clarity and ensuring real-time data alignment, ontologies foster clinical workflows that are both evidence-based and adaptive to patient-specific contexts (54).
Explainable AI (XAI) further builds trust in these systems by showing how decisions are made. This transparency is crucial in clinical settings, where trust and accountability are essential. Addressing data quality and model bias through semantic structuring also increases clinician confidence in AI-driven diagnostics (55).
4.3 Genomic and phenotypic data integration
The Human Phenotype Ontology (HPO) enables AI to connect clinical traits to genetic causes, supporting early diagnosis and precision treatment. A widely adopted example is its use in rare disease diagnostics, where HPO annotations help clinicians prioritize likely genetic causes based on patient-reported symptoms (56).
Whole-genome sequencing (WGS) platforms combined with ontology-based annotation frameworks improve variant interpretation and treatment decisions (57). By standardizing phenotypic descriptors, HPO supports data harmonization across studies, reducing annotation variability and enabling AI systems to draw consistent insights from clinical-genomic data (34).
4.4 Pharmacogenomics and AI-driven treatment recommendations
Ontology frameworks such as PharmGKB (Pharmacogenomics Knowledgebase) and Drug Ontology (DrON) encode drug-gene relationships in structured vocabularies, enabling AI systems to process pharmacogenomic knowledge. These ontologies help AI systems predict optimal drug dosing and identify potential adverse reactions (58).
A real-world application is CPIC (Clinical Pharmacogenetics Implementation Consortium), which uses ontology-based rules to generate treatment recommendations based on genetic test results. By integrating ontologies with EHR and genomic data, CPIC delivers precise, evidence-based recommendations tailored to a patient's genetic profile (34, 59).
4.5 Patient-specific disease modeling
Ontologies support personalized disease modeling by capturing complex relationships among clinical symptoms, genetic variants, and disease progression patterns. Specialized disease-specific ontologies such as the Disease Ontology (DO) and Orphanet Rare Disease Ontology (ORDO) offer structured vocabularies that allow AI systems to assess patient risk profiles more precisely. These ontologies define standardized terms for disease subtypes, stages, and comorbidities, which enhances individualized prognosis modeling.
In clinical practice, ontology-enriched platforms for neurodegenerative diseases integrate imaging, genomic, and behavioral data to detect early signs of disease progression. For example, Parkinson's disease modeling systems use ontology-guided inputs to identify subtle symptom changes over time, improving staging accuracy and treatment response predictions (34, 58).
4.6 Predictive analytics and population health management
Ontology-based predictive analytics enable healthcare systems to anticipate risk and guide preventive interventions. By aligning structured genomic, lifestyle, and environmental data, these models support early detection of chronic diseases such as diabetes or heart failure. For instance, Mount Sinai Health System in New York implemented an ontology-driven predictive model to identify patients at high risk of 30-day readmission, enabling early follow-up and significantly reducing avoidable hospitalizations (55).
In public health, ontologies drive disease surveillance by standardizing how conditions, locations, and risk factors are coded. This allows for faster outbreak detection and supports targeted health campaigns. Clinicians benefit from real-time alerts and visual summaries that support early interventions and risk mitigation strategies (58).
4.7 Geospatial databases in healthcare
Geospatial ontologies bring structure to location-linked health data, allowing integration of patient demographics, environmental exposures, and disease patterns. These models support GeoAI, which applies AI to spatial health analytics for decision-making in resource planning, disaster response, and disease tracking.
A notable example is the use of Virtual Knowledge Graphs (VKGs) in regional COVID-19 tracking platforms, which integrated mobility data, infection rates, and healthcare capacity to guide containment strategies (26). Ontology-driven GIS platforms enhance map-based decision tools, offering public health teams spatial insights that are both contextualized and updated in real time (60).
5 Ontology-enabled applications in healthcare AI
Ontologies empower healthcare AI by offering explicit, logic-based knowledge structures that improve data interoperability, model accuracy, and informed decision-making. They standardize medical concepts, align terminologies, and create meaningful relationships among data points, enabling AI systems to integrate and reason over diverse datasets such as EHRs, genomic profiles, and public health records. Beyond clinical information, ontologies enhance geospatial reasoning and population-level health analysis through spatial semantics. Additionally, ontologies are pivotal in enabling secure data governance, supporting privacy preservation, secure data sharing and patient privacy, thus substantially advancing precision medicine and healthcare innovation.
5.1 Knowledge representation and AI reasoning
In oncology, AI systems supported by ontologies can analyze pathology reports, match patient profiles to treatment guidelines, and flag inconsistencies for clinician review. These real-world applications rely on structured frameworks for representing medical knowledge. Ontologies allow AI to interpret clinical data accurately by embedding formal logic into how conditions, symptoms, and procedures are related.
According to Hoehndorf et al. (2017), knowledge graphs and semantic models built on ontologies significantly improve consistency checks, classification, and deductive inference, allowing AI systems to uncover hidden patterns within medical datasets (61). RDF-based semantic knowledge graphs coupled with inference engines can help AI enforce data consistency and support reasoning across clinical records.
As emphasized by Chudasama et al. (2023), these knowledge structures improve AI interpretability, enabling context-aware recommendations and bridging gaps between clinician knowledge and AI-generated insights (62). By grounding AI decisions in formal ontologies, models become more transparent, traceable, and aligned with medical logic. This structure facilitates rule-based diagnostics and generates clinically relevant explanations, ensuring trust in AI-driven decision-making. Ontologies enhance AI reasoning by structuring clinical knowledge into consistent, computable logic.
5.2 Natural language processing (NLP) in healthcare
Ontology-driven NLP transforms unstructured clinical text into standardized, machine-readable formats that enhance AI performance and consistency. Unlike traditional AI systems that typically work with structured datasets, natural language processing (NLP) tools, particularly large language models (LLMs), are uniquely capable of extracting meaning from unstructured clinical text, such as discharge summaries, referral letters, and observational notes, and converting it into computable, interoperable formats.
Ontology-based NLP enhances semantic interoperability and reduces ambiguity in medical terminology by aligning extracted concepts with structured vocabularies like SNOMED CT and UMLS. A typical case is the term “cold,” which may refer to either a viral illness or a sensation of low temperature. Ontology-driven systems resolve this by evaluating the clinical context and linking the term to the appropriate concept in SNOMED CT (63–65). This is essential for processing physician notes, discharge documents, and patient communications (65, 66). Real-world implementations include AI-powered chatbots and voice-enabled assistants that use ontology-driven NLP to interpret symptoms, retrieve relevant medical facts, and document clinical encounters in real time, which reduces the manual entry burden for providers (62, 67). Fareedi et al. (2025) emphasized the role of ontologies in managing dialogue context and intent recognition in medical chatbot systems. By focusing on language-specific ambiguities and terminology alignment, ontology-driven NLP allows downstream AI tools to function more effectively, supporting real-time documentation, entity recognition, and decision-making across varied clinical environments (67). Ontology-driven NLP transforms clinical text into structured data, enabling real-time AI support.
5.3 Predictive analytics and machine learning
Ontologies improve predictive analytics and machine learning by structuring biomedical data, enhancing diagnostic precision, optimizing early detection, and personalizing treatments. By standardizing inputs across clinical, demographic, and behavioral datasets, ontologies reduce data noise and improve the reliability of predictive models (68, 69). Structured ontology-driven analytics identify high-risk patients, enable early interventions, and optimize population health management (70). Their role extends to model explainability and bias reduction by embedding clinically validated relationships into machine learning pipelines (71). Standardized ontologies improve predictive accuracy and support early, personalized interventions.
5.4 Explainable AI and trustworthy systems
Explainability is crucial for fostering trust, transparency, and regulatory compliance in AI-driven healthcare. Ontologies enhance explainability by structuring knowledge representations that increase model interpretability, aligning AI decisions closely with human reasoning. Neuro-symbolic AI approaches integrate ontologies with deep learning, enhancing both accuracy and interpretability by embedding formal domain knowledge (72). Semantic reasoning, supported by OWL2 description logics and SWRL (Semantic Web Rule Language), allows AI systems to perform automated inference consistent with domain expertise, further improving explainability (73, 74). Additionally, structured ontology-driven methodologies reduce biases and enhance fairness across diverse patient populations. Provenance tracking and semantic knowledge graphs support auditability and accountability, thus reinforcing trust in AI-generated clinical recommendations and regulatory compliance (75, 76).
Ethical and regulatory standards, such as the EU Artificial Intelligence Act, highlight the necessity for interpretable, accountable AI models in healthcare. Ontologies support this need by encoding explicit rules and traceable data relationships, which facilitate transparency in automated predictions. This capability aligns with legal frameworks such as the General Data Protection Regulation (GDPR) and the Health Insurance Portability and Accountability Act (HIPAA) (77, 78). Ontologies strengthen explainability, fairness, and accountability in AI-driven healthcare decisions by providing traceable, interpretable logic that supports compliance with evolving regulatory standards.
5.5 Multi-modal data integration
Ontology-driven AI significantly enhances multi-modal data integration by systematically organizing heterogeneous healthcare datasets, including clinical notes, genomic sequences, imaging scans, and phenotypic observations, into unified, interoperable formats. Ontologies act as the semantic glue that aligns these data types by mapping them to shared vocabularies and hierarchical relationships. For example, ontological standards such as SNOMED CT and HPO enable consistent tagging of diagnoses and phenotypes, while structured imaging metadata is integrated using standard terminologies. This allows AI models to analyze these modalities together for richer clinical insights. A real-world application can be seen in oncology platforms, where SNOMED CT-coded diagnoses, radiology findings, and HPO-based phenotype annotations are combined to build comprehensive patient profiles that support AI-driven treatment recommendations (73, 79). FAIR principles further improve the accessibility and reusability of multi-modal datasets, supporting robust decision-making that aligns with data governance standards (77). Semantic knowledge graphs derived from these ontologies strengthen integration by bridging clinical, imaging, and molecular data. This approach drives innovation in precision medicine and personalized healthcare. As multi-modal AI continues to evolve, ontology-driven frameworks are likely to remain essential for enabling real-time, adaptive, and precision-guided healthcare interventions. Semantic ontologies unify diverse datasets, powering adaptive, precision-guided clinical insights.
6 Challenges and limitations
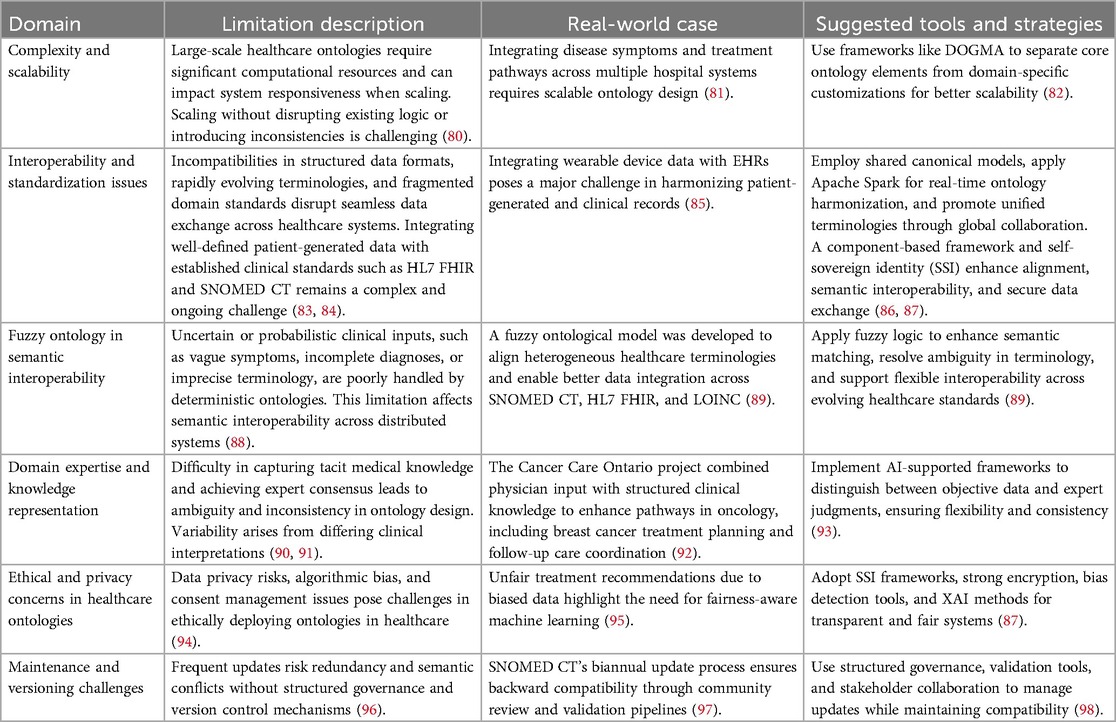
The development and implementation of ontologies in healthcare face a range of practical and conceptual challenges. These include issues related to scalability, interoperability, domain knowledge representation, ethics, and version control. Table 2 summarizes these key challenges, illustrating real-world examples and outlining evidence-based strategies to address them.
7 Enhancing healthcare ontologies for global integration
The integration of healthcare ontologies is essential for achieving semantic interoperability, data standardization, and cross-border collaboration. Leveraging AI-driven automation, modular architectures, and governance frameworks supports scalable, adaptive, and internationally aligned healthcare systems. Ensuring multi-domain compatibility and collaborative knowledge representation will drive innovation in precision medicine, clinical decision support, and biomedical research.
7.1 Collaborative ontology development
Developing robust healthcare ontologies requires collaboration among clinicians, researchers, informaticians, and policymakers, ensuring accurate and globally meaningful knowledge representation. International initiatives, including SNOMED CT adoption, HL7 FHIR integration, and multinational ontology projects, play a crucial role in harmonizing healthcare data globally (99). Open-source platforms like BioPortal, OpenEHR, and Linked Open Data (LOD) further facilitate data accessibility, transparency, and innovation, fostering collaborative knowledge sharing (100).
Cross-institutional alignment, enabled by advanced methods like Multiview Incomplete Knowledge Graph Integration (MIKGI), significantly enhances interoperability across diverse healthcare environments (101). However, persistent barriers such as regional healthcare policies, inconsistent terminology mappings, and large-scale accuracy management continue to pose significant challenges. Overcoming these requires internationally coordinated governance frameworks, automated ontology translation tools, and continuous policy alignment.
7.2 AI-driven ontology automation
Advancements in AI and NLP enhance ontology development by significantly reducing manual efforts in ontology curation, enabling automated updates, error detection, and efficient semantic alignment. Traditional manual ontology curation is resource-intensive and prone to errors. NLP-driven approaches, including ontology learning from clinical texts and named entity recognition (NER), automate concept extraction, synonym identification, and hierarchical classification (102). AI-assisted tools employing deep learning, neural embeddings, and ontology alignment frameworks efficiently detect inconsistencies and improve semantic alignment (103). However, despite their promise, these AI-driven methods come with limitations. Models may generate hallucinated concepts, introduce misclassifications, or perpetuate bias from training datasets, especially in sensitive clinical contexts. Balancing automation with expert validation is therefore critical. AI-driven semantic mapping capabilities, when combined with human oversight, can enhance data harmonization while preserving accuracy and improving ontology adaptability to evolving medical knowledge (10). Future research should focus on developing adaptive ontology frameworks capable of autonomous evolution in response to emerging healthcare data demands. AI-assisted ontology learning approaches and automated semantic alignment frameworks are crucial for maintaining domain-specific adaptability and real-time update management.
7.3 Standardization for global interoperability
Standardized ontologies establish a structured foundation for data exchange, ensuring AI-driven healthcare solutions remain interoperable and efficient. Aligning healthcare ontologies with international standards such as HL7 FHIR, SNOMED CT, LOINC, and ICD supports consistent medical data representation and enables seamless data exchange across providers and research networks (104). However, key interoperability challenges persist, including semantic mismatches, hierarchical misalignments, and terminology inconsistencies between standards such as ICD-10, ICD-11, and SNOMED CT. These discrepancies complicate integration across domains and increase the risk of misinterpretation.
Efforts from global governance bodies such as HL7 International, ISO TC 215 Committee, WHO Digital Health Technical Advisory Group, and the Joint Initiative Council for Global Health Informatics Standardization are working to align ontologies through harmonized terminologies, formal data models, and coordinated updates. Advances in multi-view alignment, such as BERT-based models and graph neural networks, also improve semantic reconciliation and terminology mapping (105). Ontology standardization is of utmost importance as it ensures consistent data interpretation and exchange across various healthcare systems, regardless of the source or format. This harmonization promotes interoperability and facilitates compliance with dynamic regulatory frameworks, such as GDPR and HIPAA (106). AI-driven compliance monitoring and semantic reconciliation frameworks are essential to manage these complexities effectively. AI-enabled compliance tools and ontology-based reconciliation systems are expected to play an increasingly vital role in managing these complexities and maintaining global interoperability.
7.4 Scalability and adaptability of ontologies
As healthcare data grows in volume and complexity, ontology frameworks must scale efficiently and adapt to evolving medical knowledge. Static ontologies often fall short when faced with rapid developments in fields like genomics, pharmacology, and wearable health data. Modular ontology architectures, which use core structures extended through independent but compatible modules, support flexibility, reuse, and more efficient updates. For instance, Interface-Based Modular Ontology Formalism (IBF) helps tailor ontologies to specific clinical domains while maintaining overall coherence (107). In real-world settings, multi-domain integration that combines clinical records with omics and sensor data is essential for precision medicine. Projects that use FHIR-based ontologies and AI-driven reconciliation frameworks demonstrate how adaptable ontologies improve semantic interoperability and facilitate consistent data exchange across systems (108). However, maintaining continuous updates while ensuring backward compatibility remains a key challenge, especially when integrating legacy systems. Structured versioning protocols, AI-assisted change management, and automated ontology migration tools help address these issues (109). As a result, scalable and adaptable ontologies are critical not just for technical integration but also for enabling responsive, data-driven care models that improve outcomes across healthcare settings.
7.5 Limitations
This perspective reflects a targeted exploration of the literature rather than a comprehensive systematic review. The selection of sources was based on relevance to ontology-driven applications in AI-enabled healthcare, which may not fully represent all relevant literature, as studies outside the chosen databases (PubMed, Scopus, and Google Scholar) were not included. Although the primary focus was on studies published between 2010 and 2025, some earlier seminal works were included to provide historical context, which may slightly broaden the timeframe beyond the initial prioritization. Additionally, this work uses a conceptual grouping of recurring themes to synthesize insights, rather than applying quantitative synthesis or formal quality assessment, which may limit the reproducibility and generalizability of findings. These limitations are inherent to the perspective format, which aims to present key insights and guide future research directions rather than provide an exhaustive or statistically validated review.
8 Conclusion
Ontologies provide the structural backbone for integrating AI into healthcare by supporting semantic interoperability, enhancing decision-making, and enabling scalable, data-driven systems. As healthcare data grows in complexity, the importance of standardized, adaptable frameworks becomes increasingly clear. This perspective underscores the significance of ontologies in enabling AI-driven analytics, personalized care, and seamless data exchange while addressing key challenges such as standardization, governance, and scalability.
Looking ahead, research should prioritize the development of adaptive ontologies capable of evolving with changing healthcare demands and minimizing the burden of manual updates. Future research should also explore approaches that enhance privacy and prioritize patient-centered care in AI-driven systems. The synergy between ontologies and AI is vital for building transparent, explainable, and trustworthy healthcare systems. By fostering interdisciplinary collaboration and applying intelligent automation, ontologies can enable ethically grounded, scalable, and personalized healthcare innovations. Importantly, this perspective underscores the reciprocal relationship between ontologies and AI, highlighting how ontologies empower AI-driven applications while AI advances ontology evolution and maintenance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
RA: Conceptualization, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. RS: Conceptualization, Validation, Visualization, Writing – review & editing. JM: Project administration, Supervision, Validation, Visualization, Writing – review & editing. GT: Formal analysis, Resources, Writing – review & editing. AM: Investigation, Resources, Writing – review & editing. MM-K: Conceptualization, Resources, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to thank the editorial team at Frontiers in Medicine for their valuable guidance and support throughout the publication process.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Saberi MA, Mcheick H, Adda M. From data silos to health records without borders: a systematic survey on patient-centered data interoperability. Information. (2025) 16(2):106. doi: 10.3390/info16020106
2. Guarino N, Oberle D, Staab S. What is an ontology? In: Staab S, Studer R, editors. Handbook on Ontologies. International Handbooks on Information Systems. Berlin, Heidelberg: Springer Nature (2009). p. 1–17. doi: 10.1007/978-3-540-92673-3_0
3. Miller T, Durlik I, Łobodzińska A, Dorobczyński L, Jasionowski R. AI In context: harnessing domain knowledge for smarter machine learning. Appl Sci. (2024) 14(24):11612. doi: 10.3390/app142411612
4. Msheik B, Adda M, Mcheick H, Dbouk M. Survey on knowledge representation models in healthcare. Information. (2024) 15(8):435. doi: 10.3390/info15080435
5. Hardiker NR, Almborg AH, Bracewell L, Chute C, van Gool C, Linton C, et al. Terminologies in the World Health Organization Family of International Classifications (WHO-FIC) (2023). Available online at: https://www.who.int/publications/m/item/terminologies-in-the-world-health-organization-family-of-international-classifications-(who-fic) (Accessed January 05, 2025).
6. Jing X, Min H, Gong Y, Biondich P, Robinson D, Law T, et al. Ontologies applied in clinical decision support system rules: systematic review. JMIR Med Inform. (2023) 11:e43053. doi: 10.2196/43053
7. Riaño D, Real F, López-Vallverdú JA, Campana F, Ercolani S, Mecocci P, et al. An ontology-based personalization of health-care knowledge to support clinical decisions for chronically ill patients. J Biomed Inform. (2012) 45(3):429–46. doi: 10.1016/j.jbi.2011.12.008
8. Varnosfaderani SM, Forouzanfar M. The role of AI in hospitals and clinics: transforming healthcare in the 21st century. Bioengineering. (2024) 11(4):337. doi: 10.3390/bioengineering11040337
9. Stoumpos AI, Talias MA, Ntais C, Kitsios F, Jakovljevic M. Knowledge management and digital innovation in healthcare: a bibliometric analysis. Healthcare. (2024) 12(24):2525. doi: 10.3390/healthcare12242525
10. Dankwa-Mullan I. Health equity and ethical considerations in using artificial intelligence in public health and medicine. Prev Chronic Dis. (2024) 21:E64. doi: 10.5888/pcd21.240245
11. Soori M, Jough FKG, Dastres R, Arezoo B. AI-based decision support systems in industry 4.0: a review. J Econ Technol. (2024) 1:100037. doi: 10.1016/j.ject.2024.08.005
12. Sarker IH. AI-based modeling: techniques, applications and research issues towards automation, intelligent and smart systems. SN Comput Sci. (2022) 3(2):158. doi: 10.1007/s42979-022-01043-x
13. Gruber TR. A translation approach to portable ontology specifications. Knowl Acquis. (1993) 5(2):199–220. doi: 10.1006/knac.1993.1008
14. Guarino N. Formal ontology, conceptual analysis and knowledge representation. Int J Hum Comput Stud. (1995) 43(5–6):625–40. doi: 10.1006/ijhc.1995.1066
15. Bates MJ. Chapter 45Data transmission protocols. In: Bates MJ, editor. Understanding Information Retrieval Systems. Boca Raton, FL: Auerbach Publications, CRC Press, Taylor & Francis (2011). p. 14. Available online at: https://www.taylorfrancis.com/chapters/mono/10.1201/b11499-53/chapter-45data-transmission-protocols-marcia-bates (Accessed January 09, 2025).
16. Yu L. OWL: web ontology language. In: A Developer’s Guide to the Semantic Web. Berlin, Heidelberg: Springer Nature (2011). p. 155–239. doi: 10.1007/978-3-662-43796-4_5
17. Gibbins N, Shadbolt N. Resource Description Framework (RDF). Southampton, UK: University of Southampton (2009). Available online at: https://eprints.soton.ac.uk/268264/1/gibbins-shadbolt-elis-rdf-v3.pdf (Accessed January 21, 2024).
18. Gazzawe F. Integrating ontology in information science and AI: evolution, applications, and future directions. In: Kontos J, Yannakoudakis EJ, editors. Ontology in Computer Science – State of the Art and Its Future in AI. London: IntechOpen (2025). doi: 10.5772/intechopen.1007650
19. Tayarani MH. Applications of artificial intelligence in battling against COVID-19: a literature review. Chaos Solitons Fractals. (2021) 142:110338. doi: 10.1016/j.chaos.2020.110338
20. Zablith F, Antoniou G, d’Aquin M, Flouris G, Kondylakis H, Motta E, et al. Ontology evolution: a process-centric survey. Knowl Eng Rev. (2015) 30(1):45–75. doi: 10.1017/S0269888913000349
21. Pernisch R, Dell’Aglio D, Bernstein A. Beware of the hierarchy — an analysis of ontology evolution and the materialisation impact for biomedical ontologies. J Web Semant. (2021) 70:100658. doi: 10.1016/j.websem.2021.100658
22. Corcho O, Fernández-López M, Gómez-Pérez A. Ontological engineering: what are ontologies and how can we build them? In: Cardoso J, editor. Semantic Web Services: Theory, Tools and Applications. New York, NY: IGI Global (2007). p. 44–70. doi: 10.4018/978-1-59904-045-5.ch003
23. De Leenheer P, Mens T. Ontology evolution. In: Hepp M, De Leenheer P, De Moor A, Sure Y, editors. Ontology Management of Computing for Human Experience. vol. 7. Boston, MA: Springer Nature (2008). p. 131–76. doi: 10.1007/978-0-387-69900-4_5
24. Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. (2007) 25(11):1251–5. doi: 10.1038/nbt1346
25. Pfaff M, Neubig S, Krcmar H. Ontology for semantic data integration in the domain of IT benchmarking. J Data Semant. (2018) 7:29–46. doi: 10.1007/s13740-017-0084-9
26. Hamdani Y, Xiao G, Ding L, Calvanese D. An ontology-based framework for geospatial integration and querying of raster data cube using virtual knowledge graphs. ISPRS Int J Geo-Inf. (2023) 12(9):375. doi: 10.3390/ijgi12090375
27. Branzi FM, Lambon Ralph MA. Semantic-specific and domain-general mechanisms for integration and update of contextual information. Hum Brain Mapp. (2023) 44(17):5547–66. doi: 10.1002/hbm.26454
28. Thomas PD. The gene ontology and the meaning of biological function. In: Dessimoz C, Škunca N, editors. The Gene Ontology Handbook. Methods Mol Biol. vol. 1446. New York, NY: Humana Press (2017). p. 15–24. doi: 10.1007/978-1-4939-3743-1_2
29. HCL Technologies. The Semantic Web: A new paradigm for integrating healthcare information (2012). Available online at: https://www.hcltech.com/sites/default/files/documents/resources/whitepaper/files/hcl_semantic_web.pdf (Accessed January 28, 2025).
30. Denaxas SC. Integrating bio-ontologies and controlled clinical terminologies: from base pairs to bedside phenotypes. In: Dessimoz C, Škunca N, editors. The Gene Ontology Handbook. Methods in Molecular Biology, vol. 1446. New York, NY: Humana Press (2017). p. 275–87. doi: 10.1007/978-1-4939-3743-1_20
31. Gábor N. Ontology development. In: Studer R, Grimm S, Abecker A, editors. Semantic Web Services: Concepts, Technologies, and Applications. Berlin, Heidelberg: Springer Nature (2007). p. 107–34. doi: 10.1007/3-540-70894-4
32. Masseroli M. Biological and medical ontologies: introduction. In: Ranganathan S, Nakai K, Gribskov M, editors. Reference Module in Life Sciences. vol. 1. London: Academic Press (2019). p. 813–22. doi: 10.1016/B978-0-12-809633-8.20395-6
33. Noy NF, McGuinness DL. Ontology Development 101: A Guide to Creating Your First Ontology. Stanford, CA: Stanford Medical Informatics, Stanford University (2001). Available online at: https://protege.stanford.edu/publications/ontology_development/ontology101.pdf (Accessed February 5, 2025).
34. Shen F, Peng S, Fan Y, Wen A, Liu S, Wang Y, et al. HPO2Vec+: leveraging heterogeneous knowledge resources to enrich node embeddings for the human phenotype ontology. J Biomed Inform. (2019) 96:103246. doi: 10.1016/j.jbi.2019.103246
35. Ameri F, Urbanovsky C, McArthur C. A systematic approach to developing ontologies for manufacturing service modeling. CEUR Workshop Proc. (2012) 886:1–14. Available online at: https://ceur-ws.org/Vol-886/paper_1.pdf
36. Suárez-Figueroa MC, Gómez-Pérez A, Fernández-López M. The NeOn methodology framework: a scenario-based methodology for ontology development. Appl Ontol. (2015) 10(2):107–45. doi: 10.3233/AO-150145
37. Haridy S, Ismail RM, Badr N, Hashem M. An ontology development methodology based on ontology-driven conceptual modeling and natural language processing: tourism case study. Big Data Cogn Comput. (2023) 7(2):101. doi: 10.3390/bdcc7020101
38. Schuurman N, Leszczynski A. Ontologies for bioinformatics. Bioinform Biol Insights. (2008) 2:187–200. doi: 10.4137/BBI.S451
39. Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, et al. The gene ontology annotation (GOA) database: sharing knowledge in uniprot with gene ontology. Nucleic Acids Res. (2004) 32(Database issue):D262–66. doi: 10.1093/nar/gkh021
40. Buttigieg PL, Morrison N, Smith B, Mungall CJ, Lewis SE, the ENVO Consortium. The environment ontology: contextualising biological and biomedical entities. J Biomed Semantics. (2013) 4:43. doi: 10.1186/2041-1480-4-43
41. Lacasta J, Nogueras-Iso J, Béjar R, Muro-Medrano PR, Zarazaga-Soria FJ. A web ontology service to facilitate interoperability within a spatial data infrastructure: applicability to discovery. Data Knowl Eng. (2007) 63(3):947–71. doi: 10.1016/j.datak.2007.06.002
42. Olivares-Alarcos A, Foix S, Borgo S, Alenyà G. OCRA – an ontology for collaborative robotics and adaptation. Comput Ind. (2022) 138:103627. doi: 10.1016/j.compind.2022.103627
43. Saraydaryan J, Jumel F, Guenard A. ASTRO: architecture of services toward robotic objects. Int J Comput Sci Issues. (2014) 11(4.1):1–6. Available online at: https://www.ijcsi.org/papers/IJCSI-11-4-1-1-9.pdf
44. Gligorea I, Cioca M, Oancea R, Gorski AT, Gorski H, Tudorache P. Adaptive learning using artificial intelligence in e-learning: a literature review. Educ Sci. (2023) 13(12):1216. doi: 10.3390/educsci13121216
45. Poldrack RA, Kittur A, Kalar D, Miller E, Seppa C, Gil Y, et al. The cognitive atlas: toward a knowledge foundation for cognitive neuroscience. Front Neuroinform. (2011) 5:17. doi: 10.3389/fninf.2011.00017
46. Bug WJ, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird AR, et al. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics. (2008) 6(3):175–94. doi: 10.1007/s12021-008-9032-z
47. EIT Health. Implementing the European Health Data Space across Europe (2024). Available online at: https://research-and-innovation.ec.europa.eu/strategy/strategy-research-and-innovation/our-digital-future/open-science/european-open-science-cloud-eosc_en (Accessed February 2, 2025).
48. EOSC-Life. European Open Science Cloud for Life Sciences (2024). https://www.eosc-life.eu/ (Accessed February 7, 2025).
49. Razzak MI, Imran M, Xu G. Big data analytics for preventive medicine. Neural Comput Appl. (2020) 32(9):4417–51. doi: 10.1007/s00521-019-04095-y
50. European Commission. Connected Health: Quality and Safety for European Citizens. European Commission, Directorate-General for Information Society and Media. Luxembourg: Office for Official Publications of the European Communities (2013). Available online at: https://ec.europa.eu/information_society/activities/ict_psp/documents/connected-health.pdf (Accessed February 7, 2025).
51. Pedrera-Jiménez M, García-Barrio N, Frid S, Moner D, Boscá-Tomás D, Lozano-Rubí R, et al. Can OpenEHR, ISO 13606, and HL7 FHIR work together? An agnostic approach for the selection and application of electronic health record standards to the next-generation health data spaces. J Med Internet Res. (2023) 25(1):e48702. doi: 10.2196/48702
52. Garde S, Knaup P, Hovenga EJS, Heard S. Towards semantic interoperability for electronic health records. Methods Inf Med. (2007) 46(3):332–43. doi: 10.1160/ME5001
53. Benson T, Grieve G. Principles of Health Interoperability: FHIR, HL7 and SNOMED CT. 4th ed. Cham, Switzerland: Springer (2021). p. 475. Available from: https://catalog.nlm.nih.gov/permalink/01NLM_INST/1o1phhn/alma9918382982706676 (Accessed February 10, 2025).
54. Xu Q, Zhai JC, Huo CQ, Li Y, Dong XJ, Li DF, et al. OncoPDSS: an evidence-based clinical decision support system for oncology pharmacotherapy at the individual level. BMC Cancer. (2020) 20(1):740. doi: 10.1186/s12885-020-07221-5
55. Mukamurera PN. The role of artificial intelligence in clinical decision support systems. Res Inven J Public Health Pharm. (2024) 3(2):14–7. doi: 10.59298/RIJPP/2024/321417
56. Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, et al. The human phenotype ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. (2014) 42(D1):D966–74. doi: 10.1093/nar/gkt1026
57. Austin-Tse CA, Jobanputra V, Perry DL, Bick D, Taft RJ, Venner E, et al. Best practices for the interpretation and reporting of clinical whole genome sequencing. npj Genom Med. (2022) 7(1):27. doi: 10.1038/s41525-022-00295-z
58. Taherdoost H, Ghofrani A. AI’s role in revolutionizing personalized medicine by reshaping pharmacogenomics and drug therapy. Intell Pharm. (2024) 2(5):643–50. doi: 10.1016/j.ipha.2024.08.005
59. Zhu Y, Elemento O, Pathak J, Wang F. Drug knowledge bases and their applications in biomedical informatics research. Brief Bioinform. (2019) 20(4):1308–21. doi: 10.1093/bib/bbx169
60. Boulos MNK, Peng G, VoPham T. An overview of GeoAI applications in health and healthcare. Int J Health Geogr. (2019) 18(1):7. doi: 10.1186/s12942-019-0171-2
61. Hoehndorf R, Queralt-Rosinach N. Data science and symbolic AI: synergies, challenges and opportunities. Data Sci. (2017) 1(1):1–12. doi: 10.3233/DS-170004
62. Chudasama Y, Huang H, Purohit D, Vidal ME. Toward interpretable hybrid AI: integrating knowledge graphs and symbolic reasoning in medicine. IEEE Access. (2025) 13:39489–509. doi: 10.1109/ACCESS.2025.3529133
63. Kersloot MG, van Putten FJP, Abu-Hanna A, Cornet R, Arts DL. Natural language processing algorithms for mapping clinical text fragments onto ontology concepts: a systematic review and recommendations for future studies. J Biomed Semantics. (2020) 11(1):14. doi: 10.1186/s13326-020-00231-z
64. Senderov V, Simov K, Franz N, Stoev P, Catapano T, Agosti D, et al. OpenBiodiv-O: ontology of the OpenBiodiv knowledge management system. J Biomed Semantics. (2018) 9(1):5. doi: 10.1186/s13326-017-0174-5
65. Li J. Ontology-based clinical information extraction using SNOMED CT (dissertation). University of Texas Health Science Center at Houston, School of Biomedical Informatics, Houston, TX (2018). Available online at: https://digitalcommons.library.tmc.edu/uthshis_dissertations/43/ (Accessed February 17, 2025).
66. Lopez C, Tucker S, Salameh T, Tucker C. An unsupervised machine learning method for discovering patient clusters based on genetic signatures. J Biomed Inform. (2018) 85:30–9. doi: 10.1016/j.jbi.2018.07.004
67. Fareedi AA, Ismail M, Gagnon S, Ghazanweh A, Arooj Z. Digital health transformation: leveraging a knowledge graph reasoning framework and conversational agents for enhanced knowledge management. Systems. (2025) 13(2):72. doi: 10.3390/systems13020072
68. Chakraborty C, Bhattacharya M, Pal S, Lee SS. From machine learning to deep learning: advances of the recent data-driven paradigm shift in medicine and healthcare. Curr Res Biotechnol. (2024) 7:100164. doi: 10.1016/j.crbiot.2023.100164
69. Singh AV, Chandrasekar V, Paudel N, Laux P, Luch A, Gemmati D, et al. Integrative toxicogenomics: advancing precision medicine and toxicology through artificial intelligence and OMICs technology. Biomed Pharmacother. (2023) 163:114784. doi: 10.1016/j.biopha.2023.114784
70. Ahuja SK, Shrimankar DD, Durge AR. A study and analysis of disease identification using genomic sequence processing models: an empirical review. Curr Genomics. (2023) 24(4):207–35. doi: 10.2174/0113892029269523231101051455
71. Mishra R, Li B. The application of artificial intelligence in the genetic study of Alzheimer’s disease. Aging Dis. (2020) 11(6):1567–84. doi: 10.14336/AD.2020.0312
72. Smirnov A, Ponomarev A, Agafonov A. Ontology-based neuro-symbolic AI: effects on prediction quality and explainability. IEEE Access. (2024) 12:156609–26. doi: 10.1109/ACCESS.2024.3485185
73. Ghidalia S, Narsis OL, Bertaux A, Nicolle C. Combining machine learning and ontology: a systematic literature review. Preprint (2024). Available online at: https://hal.science/hal-04373122v2 (Accessed February 26, 2025).
74. Saeed W, Omlin C. Explainable AI (XAI): a systematic meta-survey of current challenges and future opportunities. Knowl Based Syst. (2023) 263:110273. doi: 10.1016/j.knosys.2023.110273
75. Chen H, He F, Lei S, Tao D. Spectral complexity-scaled generalisation bound of complex-valued neural networks. Artif Intell. (2023) 322:103951. doi: 10.1016/j.artint.2023.103951
76. Díaz-Rodríguez N, Del Ser J, Coeckelbergh M, López de Prado M, Herrera-Viedma E, Herrera F. Connecting the dots in trustworthy artificial intelligence: from AI principles, ethics, and key requirements to responsible AI systems and regulation. Inf Fusion. (2023) 99:101896. doi: 10.1016/j.inffus.2023.101896
77. Huerta EA, Blaiszik B, Brinson LC, Bouchard KE, Diaz D, Doglioni C, et al. FAIR for AI: an interdisciplinary and international community building perspective. Sci Data. (2023) 10(1):487. doi: 10.1038/s41597-023-02298-6
78. Amann J, Blasimme A, Vayena E, Frey D, Madai VI. Explainability for artificial intelligence in healthcare: a multidisciplinary perspective. BMC Med Inform Decis Mak. (2020) 20(1):310. doi: 10.1186/s12911-020-01332-6
79. Martínez-García A, Alvarez-Romero C, Román-Villarán E, Bernabeu-Wittel M, Parra-Calderón CL. FAIR principles to improve the impact on health research management outcomes. Heliyon. (2023) 9(5):e15733. doi: 10.1016/j.heliyon.2023.e15733
80. Chandrasegaran SK, Ramani K, Sriram RD, Horváth I, Bernard A, Harik RF, et al. The evolution, challenges, and future of knowledge representation in product design systems. Comput Aided Des. (2013) 45(2):204–28. doi: 10.1016/j.cad.2012.08.006
81. Stoutenburg SK, Kalita J, Ewing K, Hines LM. Scaling alignment of large ontologies. Int J Bioinform Res Appl. (2010) 6(4):384–401. doi: 10.1504/IJBRA.2010.036001
82. Jarrar M, Meersman R. Scalability and knowledge reusability in ontology modeling. In: International Conference on Infrastructure for e-Business, e-Education, e-Science, and e-Medicine (SSGRR 2002s); 2002 Aug; Rome, Italy. Brussels: VUB STARlab (2002). p. 1–8. Available online at: https://www.jarrar.info/publications/Scalability_and_Reusable_in_Ontology_Modeling_SSGRR2002s_Published.pdf (Accessed March 04, 2025).
83. Torab-Miandoab A, Samad-Soltani T, Jodati A, Rezaei-Hachesu P. Interoperability of heterogeneous health information systems: a systematic literature review. BMC Med Inform Decis Mak. (2023) 23(1):18. doi: 10.1186/s12911-023-02115-5
84. Bodenreider O, Cornet R, Vreeman DJ. Recent developments in clinical terminologies – SNOMED CT, LOINC, and RxNorm. Yearb Med Inform. (2018) 27:129–39. doi: 10.1055/s-0038-1667077
85. Kilintzis V, Chouvarda I, Beredimas N, Natsiavas P, Maglaveras N. Supporting integrated care with a flexible data management framework built upon linked data, HL7 FHIR and ontologies. J Biomed Inform. (2019) 94:103179. doi: 10.1016/j.jbi.2019.103179
86. Yang X, Huang K, Yang D, Zhao W, Zhou X. Biomedical big data technologies, applications, and challenges for precision medicine: a review. Glob Chall. (2023) 8(1):2300163. doi: 10.1002/gch2.202300163
87. Weigl L, Barbereau T, Fridgen G. The construction of self-sovereign identity: extending the interpretive flexibility of technology towards institutions. Gov Inf Q. (2023) 40(4):101873. doi: 10.1016/j.giq.2023.101873
88. Zhao L, Lee S-W. Integrating ontology-based approaches with deep learning models for fine-grained sentiment analysis. Comput Mater Continua. (2024) 81(1):1855–77. doi: 10.32604/cmc.2024.056215
89. Okemwa J, Owoche PO, Mbuguah S. A fuzzy ontological model for semantic interoperability in distributed healthcare information systems. Int J Res Innov Appl Sci. (2025) 9(12):478–83. doi: 10.51584/IJRIAS.2024.912043
90. Zheng K, Padman R, Johnson MP, Hasan S. Guideline representation ontologies for evidence-based medicine practice. In: Khroumbati K, Dwivedi YK, Srivastava A, editors. Handbook of Research on Advances in Health Informatics and Electronic Healthcare Applications: Global Adoption and Impact of Information Communication Technologies. Hershey, PA: IGI Global (2010). p. 234–54. Available online at: https://www.igi-global.com/gateway/chapter/36385
91. Sicilia JJ, Sicilia MA, Sánchez-Alonso S, García-Barriocanal E, Pontikaki M. Knowledge representation issues in ontology-based clinical knowledge management systems. International Journal of Technology Management. (2009) 47(1–3):191–206. doi: 10.1504/IJTM.2009.024122
92. Nahm M, Nguyen VD, Razzouk E, Zhu M, Zhang J. Distributed cognition artifacts on clinical research data collection forms. Summit Transl Bioinform. (2010) 2010:36–40. Available online at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3041537/21347145
93. Luschi A, Petraccone C, Fico G, Pecchia L, Iadanza E. Semantic ontologies for Complex healthcare structures: a scoping review. IEEE Access. 2023;11:19228–46. doi: 10.1109/ACCESS.2023.3248969
94. Yigzaw KY, Olabarriaga SD, Michalas A, Marco-Ruiz L, Hillen C, Verginadis Y, et al. Chapter 14 – health data security and privacy: challenges and solutions for the future. In: Hovenga E, Grain H, editors. Roadmap to Successful Digital Health Ecosystems: A Global Perspective. London: Academic Press (2022). p. 335–62. doi: 10.1016/B978-0-12-823413-6.00014-8
95. Grande D, Marti XL, Feuerstein-Simon R, Merchant RM, Asch DA, Lewson A, et al. Health policy and privacy challenges associated with digital technology. JAMA Network Open. (2020) 3(7):e208285. doi: 10.1001/jamanetworkopen.2020.8285
96. Putrama IM, Martinek P. Heterogeneous data integration: challenges and opportunities. Data Brief. (2024) 56:110853. doi: 10.1016/j.dib.2024.110853
97. Ochs C, Perl Y, Geller J, Haendel M, Brush M, Arabandi S, et al. Summarizing and visualizing structural changes during the evolution of biomedical ontologies using a diff abstraction network. J Biomed Inform. (2015) 56:127–44. doi: 10.1016/j.jbi.2015.05.018
98. Lin AY, Arabandi S, Beale T, Duncan WD, Hicks A, Hogan WR, et al. Improving the quality and utility of electronic health record data through ontologies. Standards. (2023) 3(3):316–40. doi: 10.3390/standards3030023
99. Lehne M, Sass J, Essenwanger A, Schepers J, Thun S. Why digital medicine depends on interoperability. npj Digit Med. (2019) 2:79. doi: 10.1038/s41746-019-0158-1
100. Del Carmen Legaz-García M, Miñarro-Giménez JA, Menárguez-Tortosa M, Fernández-Breis JT. Generation of open biomedical datasets through ontology-driven transformation and integration processes. J Biomed Semantics. (2016) 7:32. doi: 10.1186/s13326-016-0075-z
101. Zhou D, Gan Z, Shi X, Patwari A, Rush E, Bonzel CL, et al. Multiview incomplete knowledge graph integration with application to cross-institutional EHR data harmonization. J Biomed Inform. (2022) 133:104147. doi: 10.1016/j.jbi.2022.104147
102. Liu K, Hogan WR, Crowley RS. Natural language processing methods and systems for biomedical ontology learning. J Biomed Inform. (2011) 44(1):163–79. doi: 10.1016/j.jbi.2010.07.006
103. Makin A. Ontology-Driven Knowledge Management Systems Enhanced by Large Language Models. ResearchGate [Preprint] (2024). doi: 10.13140/RG.2.2.24648.23043
104. Abraham B. Comparative study of healthcare messaging standards for interoperability in eHealth systems (master’s thesis). Western Sydney University (2017). http://hdl.handle.net/1959.7/uws:47389 (Accessed March 5, 2025).
105. Hu P, Ye Q, Zhang W, Liu J, Ruan T. Integration of multiple terminology bases: a multi-view alignment method using the hierarchical structure. Bioinformatics. (2023) 39(11):btad689. doi: 10.1093/bioinformatics/btad689
106. Yusuff M. Ensuring compliance with GDPR, CCPA, and other data protection regulations: challenges and best practices (2023). p. 1–5. Available online at: https://www.researchgate.net/publication/387224965 (Accessed March 7, 2025).
107. Ensan F, Du W. A Modular Approach to Scalable Ontology Development. Canadian Semantic Web: Technologies and Applications. Boston, MA: Springer (2010). p. 79–103. doi: 10.1007/978-1-4419-7335-1_4
108. Tong L, Shi W, Isgut M, Zhong Y, Lais P, Gloster L, et al. Integrating multi-omics data with EHR for precision medicine using advanced artificial intelligence. IEEE Rev Biomed Eng. (2024) 17:80–97. doi: 10.1109/RBME.2023.3324264
109. Khabouze R. Modernization of Legacy Information Technology Systems. Minneapolis: Walden University (2022). Available online at: https://scholarworks.waldenu.edu/dissertations/12743 (Accessed March 7, 2025).
Keywords: ontologies, semantic interoperability, artificial intelligence, healthcare integration, clinical decision support, natural language processing, ethical AI, multi-modal data integration
Citation: Ambalavanan R, Snead RS, Marczika J, Towett G, Malioukis A and Mbogori-Kairichi M (2025) Ontologies as the semantic bridge between artificial intelligence and healthcare. Front. Digit. Health 7:1668385. doi: 10.3389/fdgth.2025.1668385
Received: 17 July 2025; Accepted: 18 August 2025;
Published: 29 August 2025.
Edited by:
Gunnar Piho, Tallinn University of Technology, EstoniaReviewed by:
Marcin Golec, Heidelberg University Hospital, GermanyCopyright: © 2025 Ambalavanan, Snead, Marczika, Towett, Malioukis and Mbogori-Kairichi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radha Ambalavanan, cmFkaGFAc2VsZnJlc2VhcmNoLm9yZw==
 Radha Ambalavanan
Radha Ambalavanan R Sterling Snead
R Sterling Snead Julia Marczika
Julia Marczika Gideon Towett
Gideon Towett Alex Malioukis
Alex Malioukis Mercy Mbogori-Kairichi
Mercy Mbogori-Kairichi