Abstract
UpSMART, a research programme involving 24 European cancer centres, aimed to promote digital innovation in early-phase clinical research addressing challenges in recruitment, data collection and analysis. Several open-source digital healthcare products (DHPs) were developed through UpSMART, including eTARGET and trialFinder for trial matching, and PROACT 2.0 for patient-reported data. Lessons learned highlight the importance of multidisciplinary teams, sustainable funding and deployment, and engagement with the research community to maximise impact.
1 Introduction
The global cancer burden is growing—more than 35 million new cases are predicted in 2050, representing a 77% increase compared with 2022 (1). The number of cancer deaths per year is estimated to increase by almost 90% over this period (2). Clinical research in oncology will be crucial to try and mitigate these effects. The number of active cancer trials globally has increased more than 10-fold between 2000 and 2021 (3). However, this increase in trial activity creates a growing need to optimise trial delivery and analysis.
Digital technologies have the potential to improve all stages of the clinical trial process, including recruitment & retention, data collection and analytics (4,5). The benefits could include improved patient experience, cheaper, more efficient trials and more informed decision-making. Many digital tools have become available that could enable these benefits to be realised. However, barriers to the implementation of such technologies include regulatory requirements, data privacy concerns, and resource/infrastructure pressures (6,7).
In order to drive digital innovation in cancer clinical trials, we established UpSMART—a consortium of 24 cancer research centres across the United Kingdom, Italy, Spain and France (8,9). The UpSMART programme aimed to promote adoption of digital healthcare products (DHPs) in order to enhance digital capabilities and enable more informative, more efficient early-phase cancer clinical trials (8). In turn, this would improve the competitiveness of the UK and European clinical trials landscape to commercial trial sponsors and ultimately benefit patients by encouraging more early phase trials (8). We sought to create a repository of DHPs and methods that could be shared with the rest of the clinical trials community. DHPs would be freely distributed through open-source licensing, with the intention that the code could be further modified and improved by a community of users.
UpSMART has been delivered through a ‘hub and spoke’ model with an international coordinating centre based in Manchester and national coordinating centres in Italy and Spain. Together, these centres have managed activities across the other participating sites. The programme was initiated in February 2020 and is due to end in early 2026. Here, we present some of the tools and methods included under UpSMART, describe the challenges we encountered, and outline recommendations for future work to deliver digitised cancer clinical trials.
2 Achievements
Based on surveys conducted during the first year of UpSMART, 29 DHPs had been developed locally by participating centres, of which 8 were prioritised according to clinical need and the resource required for further development in order to release them as open-source products as part of UpSMART. We describe some of the DHPs successfully developed and released, which together address several of the key elements of the clinical trial process (Figure 1).
Figure 1
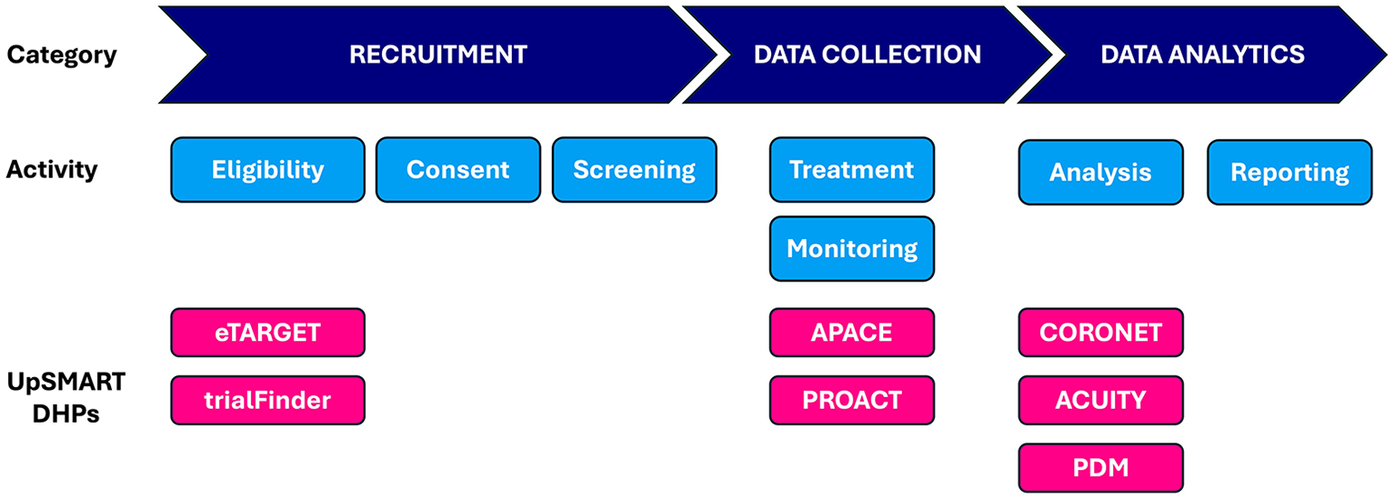
Elements of clinical trial process and corresponding UpSMART DHPs. APACE, Accelerometers to Measure Physical Activity in Cancer Patients on Early Phase Clinical Trials, CORONET, COVID-19 Risk in Oncology Evaluation Tool, DHP, Digital Healthcare Product; PDM: Protocol Deviation Monitoring tool; PROACT, Patient Reported Opinions About Clinical Tolerability.
2.1 Digital tools to increase clinical trial recruitment
Although national guidelines strongly encourage participation in cancer clinical trials, enrolment rates remain low (10). One major challenge, particularly for precision oncology trials, is the recruitment and retention of participants (4). With up to 60% of cancer trials now requiring biomarker data for eligibility, next-generation sequencing (NGS) has become essential for identifying potential trials for patients (10). However, the synthesis and interpretation of patients’ molecular and clinical information can present a significant challenge to clinicians due to the volume and complexity of NGS data.
This challenge is exemplified by the TARGET trial, which aimed to match patients to clinical trials based on their tumour genome (measured in either tissue and/or blood) (11). Matching was carried out through a Molecular Tumour Board (MTB)—a multidisciplinary group of clinicians, geneticists and informaticians. However, the TARGET team identified a number of challenges affecting the efficient running of MTB meetings, notably the integration of patients’ clinical and molecular data (11). eTARGET—a web application that integrates patients clinical and genomic information—was developed to address these challenges. eTARGET offers several benefits, including: consolidation of clinical and genomic information into a single, searchable online platform; the ability for members to participate remotely; capture of meeting discussions; and automated generation of template results letters.
Access to matched treatment presents another challenge for MTBs, which are commonly employed in an advanced setting where no further standard of care treatment is available. Patients’ access to treatment may be dependent on a compassionate use or through a clinical trial (12). The lack of matched clinical trials is a common reason for the inability of patients to access treatment (12). The digital ECMT cancer trial matching tool (‘trialFinder’) was developed to support MTB members in the identification of matched trials (13). trialFinder is integrated with eTARGET and uses natural language processing to identify potential genomically matched trials for patients, then ranks the results according to biomarker enrichment and mechanistic reasoning.
Both eTARGET and trialFinder were co-developed with clinicians. They have been deployed to support the TARGET, CUP-COMP and TARGET-National trials, which have together recruited over 3,000 participants to date (11,14,15). The improved efficiency offered by eTARGET and trialFinder have been important factors in enabling these trials to operate at this scale. In addition, a public instance of the trialFinder has been deployed for more widespread use.
2.2 Digital health data collection
Traditional clinical data collection typically occurs during in-person visits, which can create logistical and financial challenges for patients, especially those with high morbidity (16). Furthermore, data collected at study visits represent only a snapshot of the patient’s status. DHPs have the potential to reduce the number of study visits, and can enable the collection of continuous or hard-to-obtain data, such as patient-reported outcomes and biomarkers based on wearables or mobile devices (16).
For example, PROACT is a DHP for communication between participants in early-phase cancer trials and their medical team (17). Participants can communicate via video, audio or text, providing unstructured feedback about their experiences on treatment (17). This type of information is particularly useful for early-phase studies when the potential toxicities of interventions are unknown. Pilot studies showed that PROACT provided a richer set of data that supplemented those data collected through conventional case report forms (17). However, the original version of PROACT included design features that made it difficult and costly to maintain over time, requiring ongoing updates and support that limited its wider usage.
PROACT 2.0, developed under the UpSMART programme, has been extensively refactored to address these limitations (
18). PROACT 2.0 uses a simplified architecture to support easier deployment, and can collect both unsolicited (via text, audio or video) and solicited feedback (through customisable questionnaires). After a successful initial pilot study, PROACT 2.0 is currently being used in two further studies:
The PROMOTE study, which monitors adverse events and quality of life in patients with metastatic colorectal cancer treated with anti-VEGF drugs (19)
The Cancer Core Europe DART (CCE-DART) work package 12 sub-study, which aims to evaluate the feasibility of using digital tools to report effects of drugs in patients on phase 1 or 2 anticancer drug trials (20)
PROACT 2.0 has been released under open-source licence, and further development is planned to incorporate new functionality (
21).
Another area of growing interest is the integration of continuous, real-world data streams, which could complement tools like PROACT and further enrich trial datasets. APACE is a multinational study delivered through UpSMART to evaluate the feasibility of collecting continuous physical activity and sleep data from patients with advanced cancer on early phase clinical trials (22). These data could lead to a better understanding of activity levels, sleep and fatigue experienced by participants on such trials, which could in turn help to improve interventions, management, and access to appropriate treatments. The trial aims to recruit 40 participants from 8 centres across three countries (UK, Spain and Italy), demonstrating the UpSMART programme's capability to conduct complex, multinational studies.
2.3 Digital analytics
Health data can be digitally collected on a large scale and in near real-time, offering significant opportunities to enhance healthcare through advanced analytics and clinical decision support. However, the scale of data can also present a challenge. For example, early-phase clinical trials require investigators to analyse and interpret large volumes of emerging data in order to inform decisions regarding dose escalation. Furthermore, investigators may want to explore whether patients’ biomarker status modifies the treatment effect (23).
One way in which DHPs can help address these challenges is by supporting investigators in the exploration of emerging trial data, in order to identify trends and develop hypotheses. ACUITY is a clinical dashboard that presents a set of data-driven interactive visualisations, at either the individual subject or population level. An early version of ACUITY was developed by AstraZeneca and after substantial refactoring and redevelopment through UpSMART, ACUITY has been released under open-source licence for use by the clinical research community (24, 25).
Digital technologies can further support clinical decision-making by identifying patterns within complex, high-dimensional datasets. Machine learning and artificial intelligence can be used to identify latent patterns across datasets such as these (26). The potential benefits of DHPs that use machine learning are illustrated by CORONET, a DHP developed under UpSMART during the COVID pandemic. It was known that cancer patients were at increased risk from COVID-19, but they presented with heterogeneous symptoms that were difficult to distinguish from the complications of cancer and its therapy. CORONET is designed to aid clinicians in deciding whether to admit cancer patients with symptoms of COVID-19 to hospital (27). CORONET uses a Random Forest model trained on real-world data to stratify patients according to their risk of severe complications. The clinical and laboratory tests used by CORONET are routinely available at hospital presentation, supporting its widespread use. The potential benefits are: optimisation of resources by targeting patients most likely to benefit from intensive monitoring, reduction in unnecessary hospitalisations leading to lower healthcare costs and reduced risk of infecting staff or other patients.
Finally, advances in language modelling can enable efficient processing of unstructured data at scale, which could generate novel insights. For example, information about protocol deviations is typically collected as free text, which makes aggregation and analysis within or across trials difficult. The Protocol Deviation Monitoring (PDM) tool, developed under UpSMART, uses advanced language modelling techniques to extract and structure data from protocol deviation reports, and provides an interface for researchers to visualise the results and look for patterns (28).
3 Challenges
UpSMART has delivered on its ambition to design, develop and release DHPs for cancer clinical trials. However, the team encountered a number of challenges during delivery of the programme. Here we describe some of these challenges, as they may be instructive for other teams aiming to introduce digital technologies into clinical trials.
First, availability of staff and infrastructure at research sites to deploy digital solutions. Information technology (IT) resources within hospital research facilities are often limited, and are understandably focussed on clinical care over research activity. We found that participating centres rarely had people with the time and skills needed to implement DHPs, which limited uptake. Infrastructure constraints can also present a barrier to the development and/or deployment of DHPs. For example, we found that limitations on compute resource available at collaborating hospitals hindered our capacity to train AI models locally.
Second, consideration of ethical, privacy, and regulatory factors is critical for the development of DHPs. Access to existing patient data on hospital infrastructure is rightly subject to rigorous data governance process, but this has been reported as a factor limiting the widespread use of real-world patient data (29). Collecting new types of data, or collecting data in new ways, presents additional challenges as there may be a lack of precedent, and governance boards may be reluctant to approve such approaches. For example, before the APACE study could start, hospitals required their own validation of the wearable devices and associated software. Obtaining the necessary approvals introduced delays in study startup, made more challenging due to the multinational nature of the study.
Third, the pace of development for artificial intelligence (AI) and other digital technologies is typically faster that than the pace of clinical evaluation in healthcare. This disparity could lead to challenges in integrating rapidly evolving AI tools into clinical practice, where thorough evaluation is essential to ensure safety and efficacy (30). Recent progress in the field of large language models (LLMs) illustrates the rapid pace of AI development—at the start of UpSMART, OpenAI's GPT-1 model had 117 m parameters and was limited to relatively simple tasks (31). In contrast, GPT-4 (published in 2023) had over 1 trillion parameters and was capable of complex tasks, including coding assistance, medical reasoning and multimodal AI (32). Whilst LLMs offer great potential to improve DHP performance, careful consideration of how to safely deploy them in the healthcare setting is required. For example, LLMs remain prone to ‘hallucinations’ –plausible-sounding content that is factually incorrect, unsupported by source data, or entirely fabricated (33). Furthermore, protection of patient data presents another challenge when using LLMs: either substantial local computational resources are needed to run models on-site, or patient data must be transmitted to cloud-based models, raising concerns about data privacy, security, and regulatory compliance (34).
4 Discussion
4.1 Recommendations
Based on our experience with UpSMART, we outline the following recommendations that could benefit future development and implementation of DHPs.
4.1.1 Start with the clinical use case
Whilst digital skills are essential for the technical development and deployment of DHPs, input from clinicians and patients—who are typically the end-users—is vital to ensure that these tools address genuine clinical use cases and are compatible with existing healthcare workflows. (35) We recommend involving clinicians and/or patients from the earliest stages of development and throughout the implementation process. Their engagement ensures that DHPs are not only methodologically sound but also practically usable and relevant. Moreover, they can serve as effective champions among their peers, promoting uptake and use of DHPs. For example, clinical input and advocacy have been crucial in developing and promoting usage of eTARGET and trialFinder, and patients were consulted from the outset of PROACT 2.0 development to ensure the useability of the application.
4.1.2 Invest in the digital workforce
Successful implementation of DHPs requires the right IT infrastructure, integration with existing IT systems, training for end users and continued support following deployment (36, 37). The UpSMART programme brought together a multidisciplinary central team that included oncologists, study managers, AI researchers and software engineers. The collaborative efforts of this team played a significant role in developing the chosen DHPs and making them available for use. However, recent studies report that although digital and analytics transformation are a high priority for healthcare organisations, most lack the necessary resources to implement these changes (38).
One way to address this could be to promote the hiring of dedicated digital teams at research sites. Funders could play a pivotal role in this regard, similar to their support for research nurses. There is some evidence from UpSMART that access to digital skills is improving—participating centres have reported that access to clinical informatician resource has become more common over the course of the programme. Nevertheless, assembling research teams that include people with digital skills is likely to be an important enabling factor for the wider implementation of DHPs.
4.1.3 Provide software as a service (SaaS) to increase adoption
The goal of UpSMART was to release source code for DHPs, with the expectation that healthcare providers deploy and maintain the DHPs, ideally supported by a community of developers. However, the lack of dedicated digital teams available to perform these tasks limited the adoption and use of DHPs by participating centres, even where clinicians recognised the potential benefits.
Adoption of a SaaS model could mitigate this issue. SaaS delivered by a dedicated digital team from a central site could represent a more efficient allocation of resources compared with individual deployments at each site, by reducing duplication of roles and infrastructure. We recognise that SaaS puts the onus for support onto the hosting organisation and that funding such support is difficult within academia. Nevertheless, we expect that SaaS would substantially reduce barriers against adoption and increase clinical uptake of DHPs. To support SaaS provision, academic teams may need to explore alternative funding mechanisms, such as licensing agreements (e.g., royalties from commercial users to subsidise access for non-profit stakeholders) or partnerships with digital health companies that offer app-related services including technical support and maintenance.
4.1.4 Engage with the research community
Having developed a DHP, it is important to engage with the research community in order to increase awareness and drive uptake. Without such efforts, the research community may remain unaware of available DHPs and their potential benefits, limiting their clinical impact. This is particularly important for products developed under the open-source model, which relies on collaboration between research groups to adapt and improve existing tools for the benefit of all.
In recognition of the importance of engaging with the research community, we convened a dedicated conference to present our findings. This event brought together a diverse audience, including healthcare providers, pharmaceutical companies, regulatory bodies, and patients. The event sparked substantial dialogue, and highlighted an interest in the development of digital healthcare products among the cancer research community.
5 Conclusions
The UpSMART programme has provided a rich set of DHPs that have been co-developed with patients and clinicians. The lessons learned from the programme and the recommendations outlined here provide guidelines for navigating complexity within the rapidly evolving field of digital healthcare, realising the potential offered by advances in technology, and ultimately improving patient outcomes.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
PO’R: Writing – original draft, Software, Resources, Methodology, Writing – review & editing, Supervision, Project administration. FB: Project administration, Writing – original draft, Methodology, Writing – review & editing, Resources. LC: Formal analysis, Writing – review & editing, Conceptualization, Funding acquisition, Writing – original draft, Supervision, Methodology. DG: Supervision, Funding acquisition, Conceptualization, Writing – review & editing, Formal analysis, Methodology, Writing – original draft. AL: Writing – review & editing, Supervision, Writing – original draft, Software. RH: Supervision, Writing – review & editing, Writing – original draft, Software. LS: Methodology, Resources, Writing – original draft, Funding acquisition, Conceptualization, Supervision, Project administration, Writing – review & editing. AP: Resources, Writing – review & editing, Writing – original draft, Project administration. MS: Writing – review & editing, Software, Writing – original draft. DE: Writing – review & editing, Project administration, Software, Writing – original draft. SS: Writing – review & editing, Project administration, Resources, Writing – original draft. AV: Funding acquisition, Resources, Supervision, Conceptualization, Writing – review & editing, Project administration, Writing – original draft, Software, Methodology. LA: Writing – original draft, Funding acquisition, Resources, Software, Project administration, Supervision, Conceptualization, Methodology, Writing – review & editing. SD: Project administration, Funding acquisition, Resources, Conceptualization, Methodology, Writing – review & editing, Supervision, Software, Writing – original draft. CG: Software, Writing – review & editing, Supervision, Writing – original draft. GP: Funding acquisition, Resources, Project administration, Writing – original draft, Supervision, Writing – review & editing, Software, Methodology, Conceptualization. EG: Project administration, Supervision, Methodology, Writing – review & editing, Software, Funding acquisition, Writing – original draft, Resources, Conceptualization. HF: Writing – original draft, Project administration, Methodology, Software, Writing – review & editing, Resources. Fd: Project administration, Methodology, Supervision, Funding acquisition, Writing – review & editing, Conceptualization, Writing – original draft, Resources. AF: Resources, Software, Project administration, Supervision, Writing – review & editing, Methodology, Writing – original draft, Funding acquisition, Conceptualization. CD: Writing – original draft, Conceptualization, Supervision, Funding acquisition, Writing – review & editing. HU: Supervision, Writing – review & editing, Writing – original draft, Resources, Project administration, Methodology.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by Cancer Research UK (CRUK), Associazione Italiana per la Ricerca sul Cancro (AIRC) and Fundacion Científica-Asociacion Espanola Contra el Cancer (FC-AECC), via an Accelerator Award [A29374] through the CRUK National Biomarker Centre [CTRNBC-2022/100001]. This work was supported by Azure sponsorship credits granted by Microsoft’s AI for Good Research Lab. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Acknowledgments
We gratefully acknowledge all the participants in the UpSMART Accelerator Consortium for their participation, sharing insights from their centres and for the use of the digital healthcare products available across the Consortium. We also thank the UpSMART Scientific Advisory Board and the UpSMART Steering Committee for their valuable guidance and support. Finally, we thank all the patients who participated in UpSMART related studies and their families. We are grateful for the support from the Christie Hospital NHS Foundation Trust, and from the NIHR Manchester Biomedical Research Centre (BRC).
Conflict of interest
LC consultancy role/attended advisory boards for Bicycle Therapeutics, Athenex and Boehringer Ingelheim. She has received institutional funding from Sierra Oncology, Athenex, Takeda, CellCentric, CytomX Therapeutics, Eli Lilly, Boehringer Ingelheim, Bicycle Therapeutics, Lupin Pharmaceuticals, Repare Therapeutics, ADC Therapeutics, Merck Serono, Moma Therapeutics, Loxo Oncology, Epkin and Nurix Therapeutics. DG Received institutional research funding from AstraZeneca, Merck, Carrick Therapeutics, Corbus Pharmaceuticals, Bayer, Incyte, Exscientia plc, Step Pharma, Starpharma, Debiopharm, Amgen, Faron Pharmaceuticals, Janssen. LS employed by AstraZeneca R&D. GP employed by Vall d'Hebron Institute of Oncology (VHIO), funded by the UpSMART Accelerator award. EG Consultancy/Advisory for Roche, Ellipses Pharma, Boehringer Ingelheim, Janssen Global Services, Seattle Genetics, Skypta, Sotio, Sanofi, Anaveon, Abbvie, Astex Therapeutics, Alentis Therapeutics, Marengo Therapeutics, Hengrui, Incyte, Medpace, Pfizer, Amgen, GenMab, GreyWolf, Gilead and Daiichi Sankyo. Institutional funding from Novartis, Roche, Thermo Fisher, AstraZeneca, Taiho, BeiGene, Janssen and Anaveon. Employed by Vall d'Hebron Institute of Oncology (VHIO), funded by the UpSMART Accelerator award. CD Research funding to CRUK NBC from: Amgen, AstraZeneca, Astex Pharmaceuticals, Angle PLC, Boehringer Ingelheim, Biomodal, Carrick Therapeutics, Celgene, Clearbridge Biomedics, Epigene Therapeutics Inc, Guardant, Menarini, Merck AG, Neomed Therapeutics, Taiho Oncology, Thermo Fisher Scientific, UCB Pharma, RedX Pharma and CV6 Therapeutics (NI) Ltd. Honoraria for consultancy and/or advisory boards received from: Merck, AstraZeneca, GRAIL, Boehringer Ingelheim, VHIO, CIC bioGUNE, The Centre for Discovery in Cancer Research (CDCR).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Parts of this manuscript were prepared with assistance from ChatGPT (GPT-5, August 2025 version), a large language model developed by OpenAI (https://openai.com).
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1.
World Health Organization. Global cancer burden growing, amidst mounting need for services [Internet]. (2024). Available online at: https://www.who.int/news/item/01-02-2024-global-cancer-burden-growing-amidst-mounting-need-for-services(Accessed June 4, 2025).
2.
Bizuayehu HM Ahmed KY Kibret GD Dadi AF Belachew SA Bagade T et al Global disparities of cancer and its projected burden in 2050. JAMA Netw Open. (2024) 7(11):e2443198. 10.1001/jamanetworkopen.2024.43198
3.
Izarn F Henry J Besle S Ray-Coquard I Blay JY Allignet B . Globalization of clinical trials in oncology: a worldwide quantitative analysis. ESMO Open. (2025) 10(1):104086. 10.1016/j.esmoop.2024.104086
4.
Inan OT Tenaerts P Prindiville SA Reynolds HR Dizon DS Cooper-Arnold K et al Digitizing clinical trials. NPJ Digit Med. (2020) 3(1):101. 10.1038/s41746-020-0302-y
5.
Beyor N Close K Hakim N Shapiro M Ringel MS Smith B . A Digital Redesign for Clinical Trials [Internet]. Boston: Boston Consulting Group (2019). Available online at: https://web-assets.bcg.com/img-src/BCG-A-Digital-Redesign-for-Clinical-Trials-June-2019-rev_tcm9-222141.pdf
6.
Rosa C Campbell ANC Miele GM Brunner M Winstanley EL . Using e-technologies in clinical trials. Contemp Clin Trials. (2015) 45:41–54. 10.1016/j.cct.2015.07.007
7.
Rosa C Marsch LA Winstanley EL Brunner M Campbell ANC . Using digital technologies in clinical trials: current and future applications. Contemp Clin Trials. (2021) 100:106219. 10.1016/j.cct.2020.106219
8.
Butt F Laura S Luca A Xenia VA Louise C Rachel C et al The UpSMART accelerator: driving digital innovation to change the conduct of early phase cancer medicine trials. Digital Medicine. (2022) 8(1). 10.4103/digm.digm_3_21
9.
UpSMART | Improving experimental cancer research [Internet]. (2025). Available online at: https://upsmart.digitalecmt.com/(Accessed March 31, 2025).
10.
Fox JL Daniel S Abrol M . NGS panels’ capture of biomarkers associated with targeted therapies in clinical trials. J Clin Oncol. (2024) 42(16_suppl):e13508. 10.1200/JCO.2024.42.16_suppl.e13508
11.
Rothwell DG Ayub M Cook N Thistlethwaite F Carter L Dean E et al Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. (2019) 25:21. 10.1038/s41591-019-0380-z
12.
Crimini E Repetto M Tarantino P Ascione L Antonarelli G Rocco EG et al Challenges and obstacles in applying therapeutical indications formulated in molecular tumor boards. Cancers (Basel). (2022) 14(13):3193. 10.3390/cancers14133193
13.
O’Regan P Hoskins R Grave C Stevenson JA Frost H Graham DM et al Digital ECMT cancer trial matching tool: an open source research application to support oncologists in the identification of precision medicine clinical trials. JCO Clin Cancer Inform. (2023) 7:e2200137. 10.1200/CCI.22.00137
14.
Study Details | Carcinoma of Unknown Primary (CUP): a Comparison Across Tissue and Liquid Biomarkers | ClinicalTrials.gov [Internet]. (2025). Available online at: https://clinicaltrials.gov/study/NCT04750109(Accessed July 3, 2025).
15.
The Christie NHS Foundation Trust. Tumour Characterisation to Guide Experimental Targeted Therapy—National [Internet]. clinicaltrials.gov; 2022. Report No.: NCT04723316. Available online at: https://clinicaltrials.gov/ct2/show/NCT04723316(Accessed July 20, 2025).
16.
Mittermaier M Venkatesh KP Kvedar JC . Digital health technology in clinical trials. NPJ Digit Med. (2023) 6(1):88. 10.1038/s41746-023-00841-8
17.
Hughes A Landers D Arkenau HT Shah S Stephens R Mahal A et al Development and evaluation of a new technological way of engaging patients and enhancing understanding of drug tolerability in early clinical development: PROACT. Adv Ther. (2016) 33(6):1012–24. 10.1007/s12325-016-0335-4
18.
Agnelli L Villa A Butt F Duca M Guidi A Carapezza M et al PROACT 2.0: a new open-source tool to improve patient-doctor communication in clinical trials. Tumori. (2024) 110(5):304–8. 10.1177/03008916241248007
19.
Study Details | Appropriateness of Colonoscopy Indication: an Evaluation of the Clinical and Economic Impact. | ClinicalTrials.gov [Internet]. (2025). Available online at: https://clinicaltrials.gov/study/NCT06775951(Accessed June 12, 2025).
20.
Project – CCE_DART [Internet]. (2024). Available online at: https://cce-dart.com/project/(Accessed March 20, 2025).
21.
GitHub [Internet]. (2025). Proact 2 Team. Available online at: https://github.com/Proact2(Accessed June 13, 2025).
22.
Study Details | APACE - Feasibility of Using Accelerometers to Measure Physical Activity in Cancer Patients on Early Phase Clinical Trials | ClinicalTrials.gov [Internet]. (2025). Available online at: https://clinicaltrials.gov/study/NCT06868355(Accessed June 6, 2025).
23.
Janiaud P Serghiou S Ioannidis JPA . New clinical trial designs in the era of precision medicine: an overview of definitions, strengths, weaknesses, and current use in oncology. Cancer Treat Rev. (2019) 73:20–30. 10.1016/j.ctrv.2018.12.003
24.
Tautz S . REACTing to data: the use of data visualisation within early clinical statistical programming at AZ. PHUSE Computational Science Symposium 2017 [Internet]; London (2017). Available online at: https://www.lexjansen.com/phuse/2017/dv/DV03.pdf
25.
Acuity | UpSMART [Internet]. (2025). Available online at: https://upsmart.digitalecmt.com/acuity/(Accessed June 6, 2025).
26.
Kang J Chowdhry A Pugh S Park J . Integrating artificial intelligence and machine learning into cancer clinical trials. Semin Radiat Oncol. (2023) 33(4):386–94. 10.1016/j.semradonc.2023.06.004
27.
Lee RJ Wysocki O Zhou C Shotton R Tivey A Lever L et al Establishment of CORONET, COVID-19 risk in oncology evaluation tool, to identify patients with cancer at low versus high risk of severe complications of COVID-19 disease on presentation to hospital. JCO Clin Cancer Inform. (2022) 6:e2100177. 10.1200/CCI.21.00177
28.
Protocol Deviation Monitoring | UpSMART [Internet]. (2025). Available online at: https://upsmart.digitalecmt.com/protocol-deviation-monitoring-dhp/(Accessed June 6, 2025).
29.
Jones MC Stone T Mason SM Eames A Franklin M . Navigating data governance associated with real-world data for public benefit: an overview in the UK and future considerations. BMJ Open. (2023) 13(10):e069925. 10.1136/bmjopen-2022-069925
30.
Kelly CJ Karthikesalingam A Suleyman M Corrado G King D . Key challenges for delivering clinical impact with artificial intelligence. BMC Med. (2019) 17(1):195. 10.1186/s12916-019-1426-2
31.
Radford A Narasimhan K . Improving Language Understanding by Generative Pre-Training. (2018). Available online at: https://www.semanticscholar.org/paper/Improving-Language-Understanding-by-Generative-Radford-Narasimhan/cd18800a0fe0b668a1cc19f2ec95b5003d0a5035(Accessed November 11, 2025).
32.
Achiam J Adler S Agarwal S Ahmad L Akkaya I Aleman FL et al GPT-4 Technical Report [Internet]. arXiv. (2024). Available online at: http://arxiv.org/abs/2303.08774(Accessed March 27, 2025).
33.
Jung KH . Large language models in medicine: clinical applications, technical challenges, and ethical considerations. Healthc Inform Res. (2025) 31(2):114–24. 10.4258/hir.2025.31.2.114
34.
Dennstädt F Hastings J Putora PM Schmerder M Cihoric N . Implementing large language models in healthcare while balancing control, collaboration, costs and security. NPJ Digit Med. (2025) 8(143). 10.1038/s41746-025-01476-7
35.
Doyen S Dadario NB . 12 Plagues of AI in healthcare: a practical guide to current issues with using machine learning in a medical context. Front Digit Health. (2022) 4. Available online at: https://www.frontiersin.org/journals/digital-health/articles/10.3389/fdgth.2022.765406/full(Accessed October 23, 2025). 10.3389/fdgth.2022.765406
36.
Preti LM Ardito V Compagni A Petracca F Cappellaro G . Implementation of machine learning applications in health care organizations: systematic review of empirical studies. J Med Internet Res. (2024) 26:e55897. 10.2196/55897
37.
Borges Do Nascimento IJ Abdulazeem H Vasanthan LT Martinez EZ Zucoloto ML Østengaard L et al Barriers and facilitators to utilizing digital health technologies by healthcare professionals. NPJ Digit Med. (2023) 6(1):161. 10.1038/s41746-023-00899-4
38.
Digital transformation in healthcare: Investment priorities | McKinsey [Internet]. (2024). Available online at: https://www.mckinsey.com/industries/healthcare/our-insights/digital-transformation-health-systems-investment-priorities(Accessed June 11, 2025).
Summary
Keywords
digital healthcare products, cancer clinical trials, clinical trial innovation, digital health technologies, open-source
Citation
O’Regan P, Butt F, Carter L, Graham DM, Le Blanc A, Hoskins R, Stephenson L, Patil A, Shabbir M, Eken D, Singh S, Villa A, Agnelli L, Damian S, Grave C, Pretelli G, Garralda E, Frost H, de Braud F, Freitas A, Dive C and Unsworth H (2025) UpSMART: five years of digital innovation in cancer clinical research—achievements, challenges, and recommendations. Front. Digit. Health 7:1708067. doi: 10.3389/fdgth.2025.1708067
Received
18 September 2025
Revised
03 November 2025
Accepted
04 November 2025
Published
27 November 2025
Volume
7 - 2025
Edited by
Stephane Meystre, University of Applied Sciences and Arts of Southern Switzerland, Switzerland
Reviewed by
Nicole Caston, University of North Carolina at Chapel Hill, United States
Updates
Copyright
© 2025 O’Regan, Butt, Carter, Graham, Le Blanc, Hoskins, Stephenson, Patil, Shabbir, Eken, Singh, Villa, Agnelli, Damian, Grave, Pretelli, Garralda, Frost, de Braud, Freitas, Dive and Unsworth.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
* Correspondence: Paul O’Regan paul.oregan@cruk.manchester.ac.uk
† Present Address: Laura Stephenson, Clinical Pharmacology and Safety Sciences, AstraZeneca R&D, Macclesfield, United Kingdom
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.