- Department of Chemical Engineering, National Institute of Technology Rourkela, Rourkela, Odisha, India
Introduction: Environmental pollution and human exposure to dyes, particularly anthraquinone-based dyes from industries like textiles, paints, plastics, paper, and tanneries (a significant source of toxic waste in India), have increased. These dyes in wastewater pose a serious environmental threat due to their complex structure and resistance to traditional degradation methods. This mini-review explores microbial strategies as a cost-effective and environmentally friendly alternative for decolorizing these dyes, highlighting the limitations of physico-chemical methods.
Methods: This review examines existing literature on microbial decolorization of anthraquinone dyes. It discusses the isolation of microorganisms adapted to dye-contaminated environments as a key strategy. The factors influencing microbial decolorization in batch systems, such as optimal pH, temperature, and inoculum volume, are analyzed. Furthermore, continuous systems like packed bed bioreactors are explored, with a focus on the impact of flow rate and influent dye concentration on treatment efficiency.
Results: The review synthesizes information on the effectiveness of various microbial strategies for anthraquinone dye decolorization in both batch and continuous systems. It highlights the potential of adapted microorganisms for efficient dye removal and discusses the influence of key operational parameters on decolorization performance in different reactor configurations.
Discussion: This mini-review emphasizes the growing importance of biological approaches for anthraquinone dye removal due to their cost-effectiveness and environmental sustainability compared to physico-chemical methods. By providing a comprehensive overview of microbial strategies, this work contributes to the development of sustainable and effective wastewater treatment solutions for dye-containing industrial effluents.
Introduction
The rapid pace of urbanization has led to increased demands for industrialization, which, in turn, poses significant challenges for the environment. The industrial sector releases vast quantities of wastewater, contributing to a decline in freshwater availability and causing global pollution concerns when not adequately treated (Obaideen et al., 2022). Industries such as textiles, printing, tanneries, paint, plastics, and cosmetics rely heavily on synthetic dyes due to their vibrant and long-lasting colors. Sharma et al. noted that significant quantities of these dyes are present in water, often exceeding 1 mg L−1 (Sharma et al., 2021). In the dyeing industry, approximately 100,000 synthetic dyes and dyestuffs are employed, with nearly 10%–15% of unfixed dyes being directly released into water streams without undergoing treatment, as highlighted by Periyasamy (2024). This unregulated discharge of dyes into water bodies poses a serious environmental challenge.
The wastewater discharged from dyeing industries poses a significant environmental concern as it contains toxic contaminants, including dyes and other chemicals. These pollutants can have detrimental effects on human health and aquatic ecosystems (Al-Tohamy et al., 2022; Ding et al., 2022). When untreated effluents enter the water bodies, they can lead to alterations in pH levels, biological oxygen demand (BOD), chemical oxygen demand (COD), and reduced sunlight penetration, ultimately disrupting the biodiversity of the ecosystem (Aragaw, 2024). High COD and total organic carbon (TOC) in dye effluent make their treatment challenging, resulting in ecological damage and alterations in pH levels. Moreover, an increase in COD can lead to the discoloration of water resources into which these effluents are discharged (Al-Tohamy et al., 2023; Periyasamy and Militky, 2020). The complexity of the dye and its breakdown products is a cause of concern, as they are not only toxic but also mutagenic. This toxicity can have adverse effects on living organisms, including carcinogenic and mutagenic effects (Goswami et al., 2024; Patel et al., 2024; Singh et al., 2020). Besides their direct impact, industrial dyes in the environment can accumulate in the food chain, deplete oxygen levels crucial for aquatic organisms, impede the growth of water plants by inhibiting photosynthesis, and even decompose into additional toxic compounds (Sharma et al., 2021; Mehra et al., 2021). Hence, it is important to effectively remove unfixed dyes from industrial effluents before discharging them into the environment.
Dyes are categorized based on their uses and chemical structure, which involves chromophore groups. These chromophores constitute different functional groups like anthraquinone, triphenylmethane, nitro, carbonyl, and various additional groups, which are widely used for the dyeing process (Aboudi et al., 2021; Kumar et al., 2021). The release of dyes into the water stream can vary significantly based on the class of dyes. Their release can range from as low as 2% of the initial concentration for basic dyes to as high as 50% for reactive dyes (Mohanty and Kumar, 2021a; Yaseen and Scholz, 2019). Within the category of reactive dyes, azo and anthraquinone dyes are noteworthy due to their complex structures and high toxicity (Mohanty and Kumar, 2021a). These dyes are resistant to degradation, emphasizing the challenges posed by these dyes in terms of their persistence and toxicity (Dutta et al., 2024; Tripathi et al., 2023). Anthraquinone dyes stand out as the largest class of colored pollutants known for their high resistance to degradation. This resistance is primarily attributed to fused aromatic rings in their structure, which contributes to their long-lasting coloration. The highly recalcitrant anthraquinone dyes, capable of accumulating in an environment, are carcinogenic and mutagenic (Aragaw, 2024; Mohanty and Kumar, 2021b). Anthraquinone dyes account for approximately 15% of the total dye composition. They are characterized by a chromophoric group formed through the conjugation of C=O with C=C, exhibiting water-insoluble properties (Mohanty and Kumar, 2021c; Gupta and Sekar, 2024). Before the dyeing process, it is crucial to reduce the dyes to their water-soluble leuco form. This reduction is achieved by utilizing both an alkali (sodium hydroxide: NaOH) and a reducing agent (Sodium hydrosulfite: Na2S2O4). This chemical reaction results in the formation of a soluble compound known as the leuco dye. These leuco dyes possess a specific affinity for cellulosic fibers, making them suitable for dyeing processes. Reduction agents enable the formation of the desired color-producing form (chromophore) within the dye, leading to richer, brighter colors and improved wash fastness (resistance to fading during washing) (Routoula and Patwardhan, 2020).
Various physicochemical methodologies involving adsorption, oxidation, coagulation, precipitation, and membrane filtration have been utilized to decolorize textile effluents. However, these methods are often cost-intensive, less efficient, and result in the generation of significant quantities of secondary contaminants (Al-Tohamy et al., 2022; Mohanty and Kumar, 2021a). Therefore, there is a burgeoning demand for developing cost-effective approaches to decolorize these environmentally polluting dyes. Biological processes employing bacteria, algae, yeast, and fungi offer excellent alternative approaches to physicochemical techniques. These biological methods enable the decolorization of textile effluents with minimal costs, optimal operating conditions, and in an eco-friendly manner. Moreover, they lead to a reduced volume of sludge formation compared to other procedures (Daph et al., 2022; Moyo et al., 2022). Fungi have extraordinary enzyme systems, notably lignin-degrading enzymes like laccases, manganese peroxidases, and lignin peroxidases, which allow them to efficiently decompose intricate dye molecules. Their mycelium’s structure offers a substantial surface area for biosorption, enabling them to take in and store dyes from wastewater. Furthermore, fungi can endure and adjust to difficult environmental conditions, positioning them as appropriate choices for the biological treatment of dye-polluted wastewater (Dinakarkumar et al., 2024; Kumar and Chandra, 2020). The biodegradation process has indeed emerged as a highly promising approach for the treatment of textile effluent-containing dyes. It not only completely decolorizes the dyes but also transforms them into a non-toxic chemical form, addressing environmental concerns. Moreover, under optimized conditions, whether aerobic or anaerobic, and through the use of microbes, it becomes possible to achieve dye degradation (Aragaw, 2024; Ikram et al., 2021).
Achieving the maximum decolorization efficiency of a bacterium often involves optimizing the process parameters, which can be carried out using various statistical approaches. The traditional method of optimizing operational parameters one factor at a time (OFAT) can be time-consuming and costly (Maniya et al., 2024; Kumar et al., 2019). Response surface methodology (RSM) is an experimental design approach that helps assess the relationships between a defined set of experimental parameters and the observed response (Kumar et al., 2019). RSM employs a Box-Behnken design (BBD) matrix to optimize the decolorization process with a minimal number of experiments (Chauhan et al., 2021). Through the BBD approach, researchers can evaluate the interplay of process parameters and conditions to achieve the maximum desired response. It achieves this by fitting a second-order polynomial equation within specified parameter ranges, allowing for the analysis of responses in a multivariable system (Maniya et al., 2024; Borham et al., 2024). The relationship between the individual process parameters and the response can be quantified using the regression model (Freund et al., 2010; Mohr et al., 2022). RSM is an essential statistical tool to establish novel processes through the optimization and design of new products. In industrial research, RSM presents encouraging results, mainly where a large number of process parameters affect the system (Rani et al., 2024). An efficient decolorization with a bacterium can be recognized by evaluating the degradation kinetics and the operating conditions affecting the decolorization rate. The decolorization can be described by computing the interrelationship of the kinetic properties with the substrate concentration (dye) and other rate-dependent operational parameters like pH and temperature (Kumar et al., 2019; Borham et al., 2024).
The bioremediation of textile wastewater is continuously expanding in the field of environmental biotechnology due to its rapid and efficient nature. However, it’s important to note that pure bacterial strains often fall short of complete dye degradation and have the potential to produce carcinogenic aromatic amines as intermediates. These intermediates must also be effectively decomposed to ensure thorough treatment of wastewater. To address this challenge, the scale-up and maintenance of large-scale pure cultures become crucial in wastewater treatment systems (Jamee et al., 2019; Sridharan and Krishnaswamy, 2023). In recent times, there has been considerable research into the consortia of microorganisms with enhanced degradation capabilities. These consortia show promise in achieving more comprehensive and effective degradation of dye pollutants in textile wastewaters (Das et al., 2023). The use of microbial consortia has proven advantageous over pure cultures for dye degradation due to the enhanced degree of mineralization and biodegradation achieved through synergistic interactions within the bacterial community’s metabolism (Rathour et al., 2024).
In a consortium, different bacterial strains contribute to the degradation process by targeting various positions on the aromatic rings of the dye molecules or by utilizing metabolites produced by other strains to facilitate further degradation (Jamee et al., 2019; Mishra et al., 2021). Microorganisms employ various enzymatic systems for the decolorization of dyes. This process involves the interaction of enzyme molecules with dye molecules. It is facilitated by different enzymes such as lignin peroxidase, laccase, tyrosinase, and azo reductase, which are part of both intracellular and extracellular oxidoreductase enzyme systems (Rathour et al., 2024; Mishra et al., 2021). Utilizing a combination of bacterial and fungal enzymes for the enzymatic transformation of industrial dyes provides a synergistic strategy for effectively breaking down diverse dye structures. Within these combinations, bacterial enzymes like azoreductases, laccases, peroxidases, and dioxygenases start the process of breaking down dye molecules by splitting azo bonds and aromatic rings in both reducing and oxidizing conditions. Concurrently, fungal enzymes, especially those from white-rot fungi such as laccases, manganese peroxidases (MnP), lignin peroxidases (LiP), and versatile peroxidases (VP), further oxidize and break down the intermediate compounds produced by bacterial action into simpler substances. This collaborative effect boosts the efficiency of degradation, particularly for complex dyes, as bacteria often function optimally in the absence or presence of less oxygen, whereas fungi flourish in oxygen-rich settings (Mishra et al., 2021). The metabolic collaboration between bacterial and fungal enzymes not only results in significant decolorization but also the detoxification of dye-containing wastewater, positioning this integrated enzymatic system as highly valuable for environmentally friendly wastewater treatment (Mishra et al., 2021). Nowadays, there is a growing focus on leveraging bacterial-bacterial synergism to develop environmentally friendly technologies for effectively degrading textile dye effluents (Das et al., 2023). These approaches aim to achieve efficient dye removal without the production of toxic metabolites, aligning with the goal of sustainable and eco-friendly wastewater treatment.
Bioreactors serve as the foundation for a wide range of biotransformation processes, including the production of vaccines, enzymes, nutrients, and more, along with biodegradation activities (Mohanty and Kumar, 2021b). While various biodegradation assays for dyes have been conducted using batch reactors, continuous reactors are acknowledged for their enhanced efficiency and suitability for real-time applications (Bharti et al., 2019). In optimal conditions, the efficiency of a continuous reactor can be further improved through the utilization of packing materials (Tripathi et al., 2023; Swain et al., 2021). Developing a continuous method for effluent treatment is essential, especially for addressing the significant removal of anthraquinone dyes, which are commonly found in textile industry wastewater. In this context, continuous-mode bioreactor operations are particularly valuable for efficiently degrading effluents under aerobic conditions (Mohanty and Kumar, 2021a; Tripathi et al., 2023). These bioreactors often involve the use of immobilized biofilms within suspended biological systems. Biofilms can be cultivated by encapsulating microorganisms within the packing material, providing a supportive environment for their growth. This immobilization technique has been demonstrated to be superior to the use of a free-cell system. It offers advantages such as a higher loading rate and prevents the washout of biomass (Mohanty and Kumar, 2021b). Utilizing immobilized cell technology in wastewater treatment processes offers a range of benefits, including increased stability, reduced land usage, lower operational costs, and the potential for waste recycling (Obaideen et al., 2022; Kesari et al., 2021).
Microbial cell immobilization is a key strategy for boosting dye removal in industrial wastewater treatment. Innovative methods like entrapment in polymer matrices (e.g., alginate, PVA), adsorption onto solid supports (e.g., coconut coir), magnetic nanoparticle immobilization, and hybrid matrices enhance microbial stability, reusability, and stress resistance. Electrospun nanofibers and biofilm formation on natural supports further improve efficiency through high surface area and natural microbial activity. These techniques offer operational benefits like easy biomass recovery and reduced sludge, making them promising for treating complex dye effluents (Obaideen et al., 2022; Hassan et al., 2019). Therefore, it has become challenging to find efficient treatment technologies that align with the requirements of dyeing industries. Immobilized cell technology has emerged as a recognized and effective approach for treating dye wastewaters, thereby increasing attention in the field. This review delves into the mechanisms involved in the decolorization of anthraquinone-based dyes by microorganisms. It also explores the isolation and screening of microbes with the capability to effectively decolorize textile effluents, addressing crucial aspects of wastewater treatment in the dyeing industry.
This mini-review highlights the growing importance of microbial decolorization techniques for addressing the environmental challenges posed by anthraquinone dyes. While traditional physicochemical methods are often costly and generate secondary pollutants, biological methods offer a sustainable and cost-effective alternative. The key novelties of this review are:
Focus on anthraquinone dyes: The review specifically addresses the challenge of decolorizing anthraquinone dyes, a class of dyes known for their recalcitrance and toxicity.
Combination of batch and continuous treatment systems: The study explores both batch and continuous treatment systems, emphasizing the advantages and limitations of each approach.
Immobilized cell technology: The review highlights the potential of immobilized cell technology to enhance the efficiency and stability of microbial decolorization processes.
Microbial consortia: The use of microbial consortia is emphasized as a promising strategy to achieve more complete and efficient dye degradation, compared to single-strain cultures.
Optimization techniques: The application of statistical techniques like Response Surface Methodology (RSM) is discussed to optimize decolorization processes and identify optimal operating conditions.
Kinetic studies: The review delves into the kinetic aspects of microbial decolorization, providing valuable insights into the rate and mechanisms of dye degradation.
By combining these novel approaches, researchers can develop more efficient and sustainable bioremediation technologies to mitigate the environmental impact of textile dye pollution.
Dyes and their history
Dyes are organic compounds used to color materials. They have transitioned from natural sources, used since ancient times (first recorded in China, 2600 BC), to predominantly synthetic petrochemical derivatives due to their superior photostability, making them crucial across various industries (Alegbe and Uthman, 2024). While natural dyes faced challenges in processing, color vibrancy, and fastness, W.H. Perkin in the early 20th century, developed the first synthetic dye, i.e., mouve (Aniline purple, Tyrian purple) with excellent dyeing characteristics, leading to their widespread adoption for higher quality and reproducibility. The worldwide dye consumption by textile industries is about 30 million tons, with an annual growth of 3% (Sla et al., 2021). The annual production of synthetic dyes was believed to be more than 7 × 105 tons, and around 10,000 functionally distinct dyes have been utilized in various industrial sectors (Al-Tohamy et al., 2022).
Dye classification
Dyes were classified in several ways, and the main classifications depended on the type of materials, the existence of their respective chromophores, and application methods (Figure 1). They can be categorized according to their structure, properties, and affinity towards the substrate molecules. The affinity of dye molecules towards the cellulosic fiber is through a covalent bond and physical adsorption. All dyes and pigments appear to be colored as they absorb specific wavelengths of light, but differ in their properties and application methods (Aboudi et al., 2021; Benkhaya et al., 2020a). Chromophores are the color-imparting parts of dye molecules, with auxochromes modifying their light absorption. Auxochromes are functional groups linked to a chromophore that alter the chromophore’s capability to absorb light. Most commonly known auxochromic groups are: OH, -NH2, -SO3H, -COOH, -NHR, and–NR2, etc. (Alegbe and Uthman, 2024; Yusuf, 2018). Application-based classes include vat dyes (insoluble, requiring reduction), acid dyes (water-soluble, for protein fibers), reactive dyes (covalent bonds with fibers), direct dyes (water-soluble, weak bonding), basic dyes (cationic, for acrylics), disperse dyes (non-ionic, for polyester), mordant dyes (require fixatives), sulfur dyes (insoluble, for cotton), pigment dyes (insoluble particles), and solvent dyes (non-ionic, for non-polar materials). Each class has distinct chemical structures and application processes suited to different fibers and desired properties (Alegbe and Uthman, 2024; Yusuf, 2018).
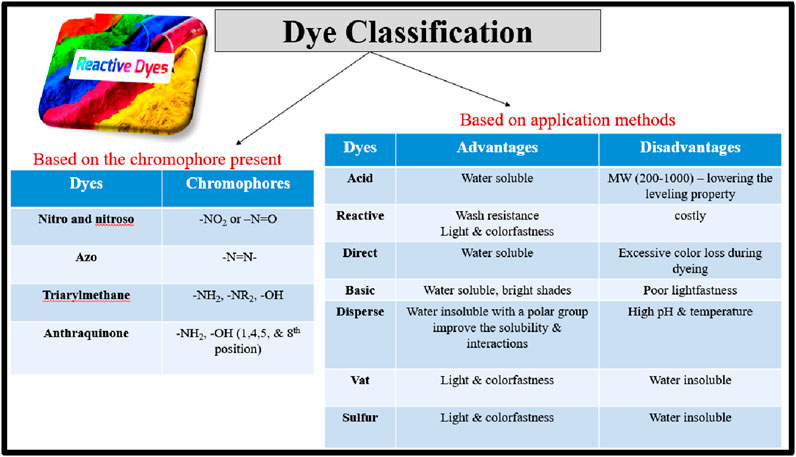
Figure 1. Dyes classification based on the type of materials, the existence of their respective chromophores, and application methods.
Anthraquinone vat dyes
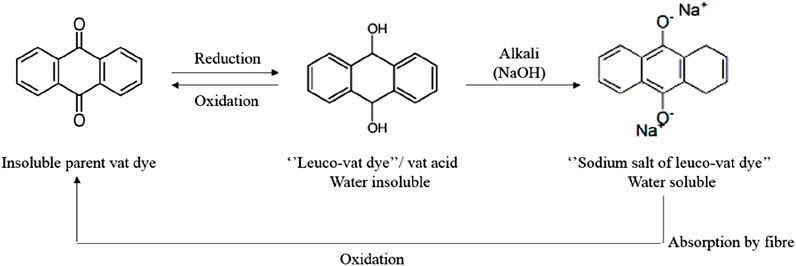
Anthraquinone dyes, the second largest dye group in textiles, are p-benzoquinone derivatives with C=O and C=C chromophores, making them resistant to degradation and known for excellent light-fastness. Vat dyes, including indigo and anthraquinone types, are water-insoluble but become soluble in their leuco form through alkaline reduction with sodium hydrosulfite before application to fabrics (Gupta and Sekar, 2024; Sh et al., 2019). Subsequent oxidation firmly fixes the dye, yielding excellent wash and light fastness. The first anthraquinone vat dye, Indanthrene Blue, was created by Rene Bohn at BASF in 1901 (Sh et al., 2019). The reduction process of vat dyes is represented in Figure 2.
Vat dyeing conditions differ based on the dye, affecting temperature, salt, and alkali needs. IK dyes (cold, 20°C–30°C) have low affinity, needing little alkali and salt. IW dyes (warm, 40°C–45°C) have a higher affinity, requiring more alkali and less salt. IN dyes (normal, 60°C) are highly substantive, needing high alkali but no salt. The IK special class typically requires 45–60 min of dyeing time (Dulo et al., 2021; Yang et al., 2020).
Ecotoxicity of anthraquinone vat dyes
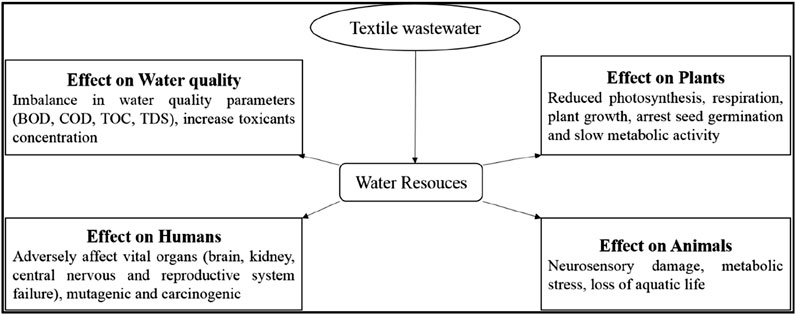
Reactive dyes, including azo, anthraquinone, and triarylmethane, are stable xenobiotic compounds that remain intact throughout conventional wastewater treatment using physicochemical methods (Rane and Joshi, 2021; Shyla et al., 2018). The discharge of dye wastewater into the ecosystem has a significant impact on living organisms. It induces non-aesthetic contamination in the aquatic environment (Sh et al., 2019) as presented in Figure 3. Unusual water coloration can cause neurosensory damage, metabolic stress, and fish death. Stable aromatic compounds in discoloured water can limit plant biomass absorption and respiration. Reduced sunlight penetration due to water discoloration disrupts photosynthesis in aquatic plants, and it may also inhibit germination rates. Reactive dyes can biodegrade aerobically or anaerobically, but some intermediate molecules, especially those lacking hydroxyl and carboxyl groups on aromatic amines, resist degradation (Aragaw, 2024). The persistent, non-degradable toxic amines can be cytotoxic to microorganisms, hindering their activity and potentially accumulating in the ecosystem (Al-Tohamy et al., 2022; Mohanty and Kumar, 2021b). If these amines enter aquatic systems and potable water, their allergic, mutagenic, carcinogenic, and cytotoxic properties can adversely affect vital organs like the brain, liver, reproductive system, kidneys, and central nervous system (Rane and Joshi, 2021).
Treatment technologies for the removal of reactive dyes from wastewater
Textile industries indeed pose a significant environmental threat primarily because of their excessive water usage (Aragaw, 2024). Textile processes are energy-intensive, consuming a large amount of water and producing effluents with hazardous contaminants like surfactants, heavy metals, and chlorinated compounds, as well as increased salt concentrations, TSS, color nutrients (including nitrogen and phosphorus), COD, and TOC (Aragaw, 2024; Al-Tohamy et al., 2023). Over the past decades, various technologies and methods have been developed and employed for textile wastewater treatment. The treatment methods were selected based on the structure, properties, and concentrations of substrate in the wastewater that eliminate the toxic and undesirable contaminants.
Numerous physicochemical and biological methodologies have been employed for the dye’s elimination from textile wastewater. Physical methods include adsorption, membrane filtration, precipitation, coagulation and flocculation, ozonation, UV-irradiation, ion exchange, and advanced oxidation strategies (Benkhaya et al., 2020b). While these techniques were documented to effectively remove dyes from textile wastewater, even though these processes are expensive and inefficient in decolorizing a broad range of colors (Routoula and Patwardhan, 2020).
Chemical techniques play a crucial role in addressing dye-containing wastewater, utilizing various oxidation methods like activated carbon, electrochemical degradation, anion exchange resin, ozonation, cucurbituril, photochemical oxidation (UV with hydrogen peroxide or sodium hypochlorite), and Fenton reagent (Al-Tohamy et al., 2022; Mohanty and Kumar, 2021a). Both treatment methods (physical and chemical) are expensive and involve extensive chemical usage, producing a high amount of sludge and wastewater. It has led to the release of toxic amines with disposal difficulties resulting in secondary contamination, has minimal efficiency, and is less tolerant of varying effluent inputs (Aragaw, 2024; Benkhaya et al., 2020a). Although they have several disadvantages, a few of these methods have proven effective. As a result, various renewable biological sorbents, including rice husk, sugarcane bagasse, brown seaweed biomass, eucalyptus bark, fly ash, coal, peat, coconut husk, chitosan, fungi, yeast, and bacterial biomass, were studied to reduce processing expenses and increase their viability for dye removal efficiency (Al-Tohamy et al., 2022).
Biological approaches utilizing bacteria, algae, yeast, and fungi are exceptional substitutes for physicochemical strategies for textile effluent decolorization. These methods are cost-effective, easily accessible, environment-friendly, require optimal operating conditions, and produce very less volume of sludge compared to other methodologies (Mishra and Maiti, 2018). However, under anaerobic conditions, the fungi and algae do not multiply quickly and have an extended growth period, reducing the treatment efficacy. In the recent past, several bacterial enzymes, including azoreductase, laccase, lignin peroxidase, tyrosinase, and NADH-reductase, were studied to degrade reactive dye in an insoluble or immobilized state. Bilal et al. examined the potential of the agar gel-immobilized manganese peroxidase enzyme to degrade the dye (Bilal et al., 2016). They documented that after immobilization, the enzyme’s thermo-stability was significantly increased, and the decolorization activity was observed up to the 10th operating cycle, where 74.3% decolorization efficiency was achieved after the third cycle. The enzyme demonstrated 78.6%–84.7% decolorization efficiency for all dyes tested within 12 h of incubation. Horseradish peroxidase was immobilized on calcium alginate to examine the degradation of reactive dyes (Farias et al., 2017). Maximum decolorization efficacies of 93% and 75% were documented with the initial dye concentration of 40 mg L−1 of Reactive Blue 221 and Reactive Blue 198 at pH 5.5°C and 30°C.
Mahmood et al. carefully examined the evidence regarding the potential of oxidoreductase enzymes in the detoxification of dye (Mah et al., 2016). They proposed that enzymes are preferred for the treatment of wastewater over other microbes, especially if wastewater includes contaminants that prevent bacterial growth. It has been stated that the dye degradation efficacy using immobilized enzymes is higher than soluble enzymes, and enzymes must be used to treat real textile wastewater needed for a further comprehensive study. The specificity of the enzymes to their active sites and various dyes is still to be examined. The pros and cons of some of the non-biological strategies (dos Santos et al., 2007) in addition to specific biological methods, including fungi, algae, and plants, have been presented in Table 1.
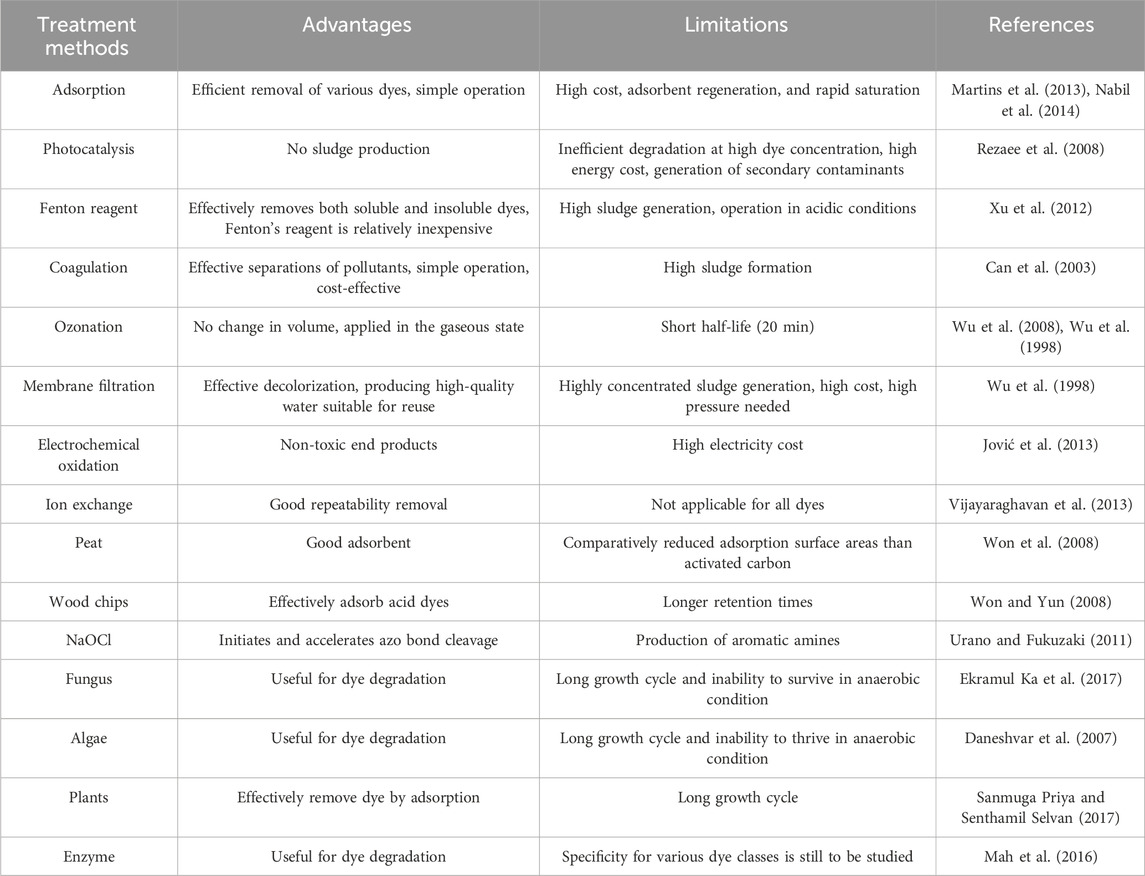
Table 1. Pros and Cons of Existing Strategies used for the decolorization of Textile Wastewater (dos Santos et al., 2007).
Bacterial decolorization of reactive anthraquinone dye
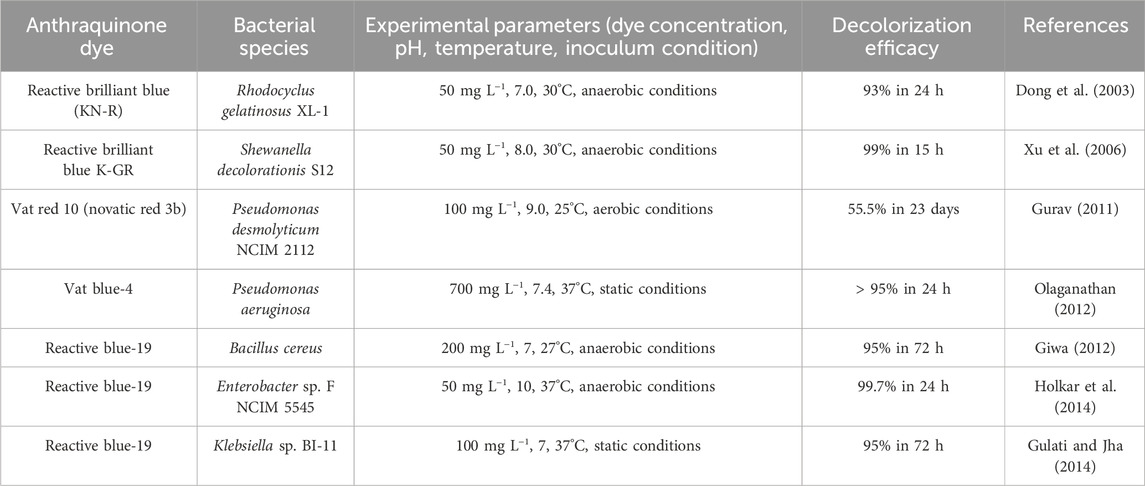
The biodegradation of anthraquinone-based dyes has been an essential subject of research over the past few years (Ekramul Ka et al., 2017). These dyes include the complex aromatic structures that make them recalcitrant to biodegradation (Rane and Joshi, 2021). Nowadays, several researchers have tried to find the microbial strains that could degrade these dyes effectively. Lade et al. isolated Rhodocyclus gelatinosus XL-1 under anaerobic conditions from contaminated wastewater (Lade et al., 2012). They studied the decolorization of reactive brilliant blue (KN-R) using an isolated strain and found that ≥93% decolorization efficacy was achieved. They suggested that the decolorization of the dye resulted from the use of peptone as a substrate. The microbe uses the dye as a co-substrate, and the decolorization efficiency has been improved by adding peptone, where the degradation resulted through mineralization, hydrolysis, and a co-metabolism process. The bacterial decolorization of reactive anthraquinone dyes is presented in Table 2.
Similarly, Xu et al. reported the decolorization of reactive brilliant blue K-GR under anaerobic conditions using Shewanella decolorationis S12 isolated from textile wastewater (Xu et al., 2006). They observed that 99% of decolorization was achieved through flocculation with a dye concentration of 50 mg L−1 within 15 h. The increase in the decolorization was due to the supplementation of various carbon sources such as peptone, lactate, succinate, formate, and yeast extract, whereas the decolorization efficiency decreased with HgCl2.
Gurav et al. investigated the decolorization efficiency of Pseudomonas desmolyticum NCIM 2112 and Galactomyces geotrichum MTCC 1360 for the decolorization of Vat Red 10 dye (0.01%) under aerobic conditions using synthetic wastewater (Gurav, 2011). A maximum decolorization of 55.5% was observed within 23 days of incubation. The formation of 2,6-diisopropyl naphthalene as an end-product is a plant-growth regulator that suggests the conversion of toxic compounds via an oxidative metabolic process to a non-toxic substance.
Bacillus Cereus isolated from contaminated food was used for reactive blue-19 decolorization under anaerobic conditions (Giwa, 2012). They found that with the initial dye concentration of 200 mg L−1, the maximum decolorization efficiency of ≥95% was achieved within 27 h of incubation. An increase in decolorization was induced by adding glucose to the medium, which degrades the dye using the co-metabolic process.
The decolorization of Vat Blue 4 dye under static aerobic conditions by Pseudomonas aeruginosa was estimated by Olaganathan, with the removal of 99% at 700 mg L−1 dye concentration (Olaganathan, 2012). Based on the comparative study with Bacillus sp., Aeromonas sp., Micrococcus sp., and Achromobacter sp., they observed that P. aeruginosa had demonstrated a higher ability for the treatment of textile wastewater using the synergistic effect of bio-bleaching.
Holkar et al. mentioned the degradation kinetics of reactive blue-19 dye under anaerobic conditions by Enterobacter sp. F NCIM 5545 using synthetic wastewater with glucose as a co-substrate (Holkar et al., 2014). They observed that with 400 mg L−1 dye concentration, the strain showed an increased decolorization efficiency of 99.7% relative to any other Enterobacter sp. strain without any toxic effect. In the co-metabolism process, the microbe uses the dye as a co-substrate, in conjunction with a particular substrate, resulting in increased decolorization efficiency under optimal conditions. However, it is necessary to identify potent microbial strains using optimal environmental as well as culture conditions to obtain an enhanced decolorization efficiency in a shorter time.
Bio-decolorization of reactive dyes by bacterial consortium
A microbial consortium was investigated for its ability to degrade synthetic dyes, demonstrating a significant advantage over pure cultures. This consortium achieved substantial mineralization and biodegradation, likely due to synergistic interactions within the microbial community’s metabolic processes (Cao et al., 2022). The microbial strains metabolize the molecular structure of the dye in a consortium by attacking different aromatic ring positions, and for further degradation, it can use metabolites formed by the surviving strains (Aragaw, 2024; Cao et al., 2022). An effective microbial consortium has been established by researchers to improve the decolorization process, which can remove a range of contaminants from textile effluent. Bacillus cereus (BN-7), Pseudomonas fluorescens (BN-5), Pseudomonas putida (BN-4), and Stenotrophomonas acidaminiphila (BN-3) microbial strains were used to develop a consortium by Aragaw et al. (2024). In comparison to the individual microbial strains, the consortium revealed an enhanced (three times higher) decolorization efficacy of 78%–99% with an initial dye concentration of 60 mg L−1 (acid red-88, acid red-97, acid red-119, and acid blue-113) within 24 h of incubation.
Similarly, Dafale et al., reported on the remazol black-B decolorization kinetic in a continuous anoxic-oxic reactor using a developed microbial consortium of Pseudomonas aeruginosa, Proteus mirabilis NAD1 and NAD6, Rhodobacter sphaeroides, and Bacillus circulans. The microbial consortium presented that synthetic wastewater with an initial dye concentration of 100 mg L−1 could decolorize with an efficacy of ≥90% and reduce COD by 80% (Dafale et al., 2008).
The GG-BL consortium was developed combining Galactomyces geotrichum MTCC 1360 and Brevibacillus laterosporus NCIM 2298 to decolorize golden yellow HER under optimal sequential aerobic and microaerophilic conditions was established by (Waghmode et al., 2011). The color, COD, and TOC were effectively reduced by 100%, 84%, and 63%, respectively. The metabolite formed during mineralization indicates the existence of enzymes like laccase, tyrosinase, NADH-DCIP reductase, veratryl alcohol oxidase, azo reductase, and riboflavin reductase that convert dyes into non-toxic compounds. Likewise, specific microbial consortia in aerobic conditions demonstrated different findings for the effective decolorization of reactive red-3BS (90%), acid blue-113 (93.7%), and reactive red-195 (70%).
According to Das et al., a developed consortium with Zobellella taiwanensis AT1–3, and Bacillus pumilus HKG212 was able to decolorize reactive green-19 dye under static conditions (Das et al., 2023). They found that a maximum decolorization of 97% was observed within 24 h of incubation with an initial dye concentration of 100 mg L−1 and yeast extract as co-substrate. The decolorization rate follows first-order kinetics, indicating that a reduction in decolorization was caused by increasing the initial dye concentration.
The available literature demonstrated that the microbial consortia could decolorize the reactive dyes faster and more efficiently than individual microbial strains. It is challenging to assess the influence of experimental parameters on the decolorization mechanism. Therefore, further investigations are required to adequately assess the effect of these conditions through mathematical and statistical modeling by developing a consortium to achieve an efficient dye decolorization. Based on bacterial-bacterial synergism, advanced, eco-friendly treatment strategies for textile wastewater degradation are being developed without generating toxic metabolites. It generates less volume of sludge and non-toxic metabolites than physicochemical approaches (Jadhav et al., 2010).
Mechanism of bacterial decolorization of reactive dyes
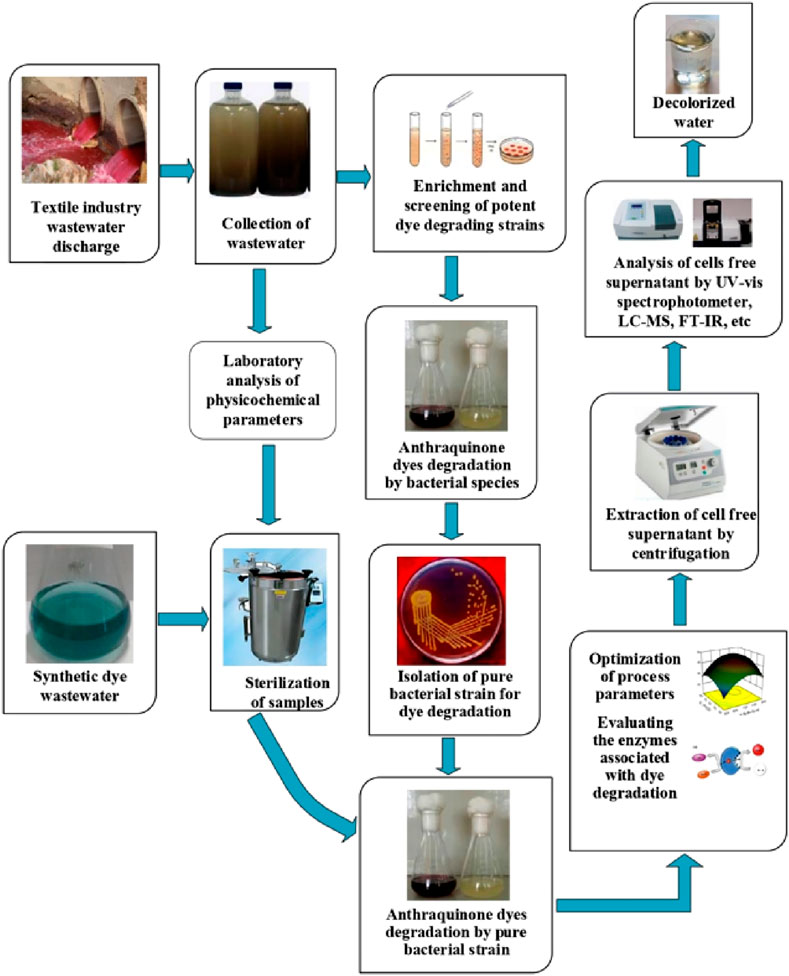
The decolorization of dyes takes place under aerobic, anaerobic, and anaerobic-aerobic conditions, either by adsorption, biochemical reaction, or a combination of the two (Holkar et al., 2014). The treatment method depends on the reactivity and the binding affinity of the dye to the microorganisms (Das et al., 2023). The laboratory-scale treatment of textile wastewater by microorganisms is presented in Figure 4.
The selection of an effective microbial strain is needed for efficient dye degradation, attributed to the variations in the metabolism of various species for the dye and its degradation method (Cao et al., 2022; Ngo and Tischler, 2022). Figure 3 shows the step-by-step process required to decolorize/degrade textile dye wastewater using microorganisms on a laboratory scale. The process of dye decolorization is believed to involve a variety of oxidoreductase enzymes, including laccase, lignin peroxidase, tyrosinase, azoreductase, and NADH-DCIP reductase (Shah et al., 2012).
Microbes operating in aerobic environments need to be acclimated and undergo an extended growth phase. A bacterium in the cytoplasm metabolizes the reductase enzyme, which differs from reactive dyes and weakens the chemical bond under optimum conditions (Thakur et al., 2014). Bacillus halodurans MTCC 865 decolorized acid black-24 within 6 h under aerobic static conditions, with a decolorization efficacy of ≥90% at pH 9°C and 37°C (Prasad and Rao, 2013).
Kumaravel & Shanmugam reported that anaerobic bacteria are nonspecific to reactive dyes and require the reduction process of an extracellular electron transport chain (ETC.) that offers possibilities for the reduction of different dyes. This method involves carrier molecules with a low molecular weight that act as a color shuttle. These carrier molecules are either produced or included separately during the substrate’s metabolism (Kumaravel and Shanmugam, 2024).
Fernandes et al. documented that the metabolites formed during the anaerobic conditions are cytotoxic and mutagenic to the microbial cell, whereas aerobic conditions prevent the process by reoxidizing the metabolites (Fernandes et al., 2015). Therefore, the application of aerobic or anaerobic systems alone will be inefficient for the degradation of reactive dyes. The use of a two-stage procedure that combines the anaerobic and aerobic systems, maybe the best method for reactive dye degradation (Karatas, 2009).
Evidence of anaerobic azo dye reduction by the redox mediator formed through aerobic degradation (Bonakdarpour et al., 2011; Jonstrup et al., 2011). They observed that in anaerobic conditions, the redox intermediate produced by aerobic degradation of aromatic compounds increases the dye decolorization. In two stages of anaerobic and aerobic systems, the anthraquinone dye degradation is quite efficient. When treating wastewater including anthraquinone dyes, microbial decolorization is most effective under anaerobic conditions as compared with aerobic conditions (Holkar et al., 2014). The decolorization mechanism with enzymes and intermediates during anthraquinone dye mineralization is illustrated in Figure 5.
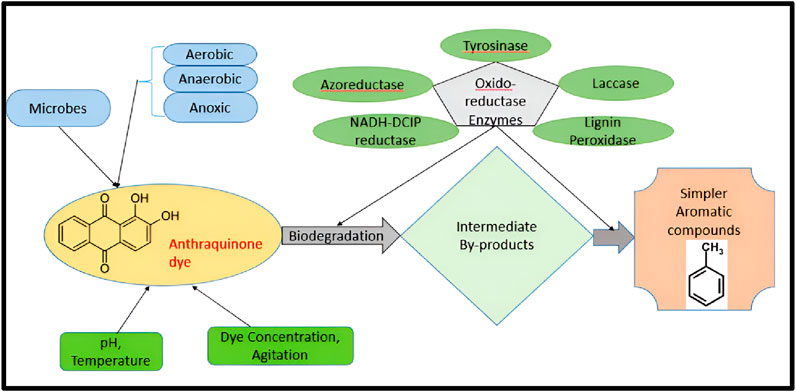
Figure 5. The decolorization mechanism with enzymes and intermediates during anthraquinone dye mineralization.
Oxidoreductase enzymes for dye degradation
Dye decolorization is related to the interaction of the enzyme molecule with the dye molecule through various oxidative enzymes, including lignin peroxidase, laccase, and tyrosinase, as well as reductive enzymes, including azoreductase and NADH-DCIP reductase of the intracellular and extracellular oxidoreductase enzyme system. Enzymatic methods have reduced environmental impacts since they do not include the risk of contamination. Enzymatic processes have been established in textile industries and have proved to be very important as biocatalysts (Goswami et al., 2024; Mishra et al., 2021).
• Lignin peroxidase (LiP): LiP belongs to the oxidoreductases family, mainly those that function as an acceptor of peroxide (peroxidases), and is categorized under the ligninases family. LiP catalyzes the cleavage of an aromatic ring to form reactive radicals by reducing one electron from the side chains of lignin and associated compounds (Kumar and Chandra, 2020; Janusz et al., 2017). The LiP extracted from Brevibacillus laterosporous MTCC 2298 and Acinetobacter calcoaceticus NCIM 2890 effectively decolorized various sulfonated azo dyes (Ghodake et al., 2009; Gomare and Govindwar, 2009). Ligninolytic enzymes are commonly used in the textile industry to remove dye, bio-bleaching of the effluents, and detoxification of toxic compounds. LiP can catalyze both phenolic and non-phenolic lignin compounds by oxidation, leading to the breakdown of the Cα–Cβ bond and the aryl Cα bond, thus opening the aromatic ring, phenolic oxidation, and demethoxylation (Aragaw et al., 2024; Alam et al., 2009). The LiP enzyme has a substantial potential for usage in industrial operations due to its significant redox potential and extended substrate variety in the presence of various mediators. Many studies suggest a substantial induction of lignin peroxidase activity during the decolorization of reactive dyes, indicating their potential for decolorization of dye (Aragaw et al., 2024).
• Laccase: Laccases are versatile enzymes with applications across various industries. They are used in the production of ethanol, food processing, paper and pulp manufacturing, dye bleaching, and even the creation of valuable lignin-based chemicals (Giardina et al., 2010; Shrestha et al., 2016). It is one of the various members of the protein family of multicopper oxidase. They are involved in monomer linkage, fragmentation of polymers, and degradation of aromatic compounds. The enzyme catalyzes the oxidation of modified phenolic and nonphenolic compounds when oxygen is present as an electron donor (Sharma and Kuhad, 2008). Several industrial applications of laccase enzymes, such as dye bleaching, biodegradation, detoxification of the effluent, and alteration of biopolymers, have been reported (Xu et al., 2006; Aravind, 2016; Lu et al., 2010). The first prokaryotic laccase was identified from Azospirillum lipoferum, a rhizospheric bacterium. Another laccase was identified from a melanogenic marine bacterium, Marinomonas mediterranea, that produces two distinct polyphenol oxidases (PPO), a multi-potent PPO that oxidizes substrates characteristic of both tyrosinase and laccase (Claus, 2004). Ridge et al. identified Pseudomonas syringae and Pedomicrobium sp. showing the laccase activity for Cop A protein (Ridge et al., 2007). The laccase enzyme decolorizes the reactive dyes using a nonspecific free radical, thus preventing the generation of toxic amines (Janusz et al., 2020). The laccase extracted from Pseudomonas desmolyticum NCIM 2112 exhibited complete decolorization for various reactive dyes, such as Direct Blue 6, Green HE4B, and Red HE7B (Kalme et al., 2009).
• Tyrosinase: Tyrosinase is a tetramer enzyme composed of four copper atoms per molecule and binding sites for aromatic compounds and oxygen (Singh et al., 2015). The enzyme catalyzes the monophenols hydroxylation to o-diphenols and the subsequent oxidation of o-diphenols to o-quinones (Bibhuti Bhusan Mishra, 2016). The enzyme is, therefore, capable of removing organic contaminants from polluted areas (Singh et al., 2015; Donato et al., 2014). Several researchers have documented the evidence of biodegradation of reactive dyes like Direct Red 5B, Direct Blue GLL, Yellow 5G, Brown R, Reactive Red 141, and Reactive Black5 with tyrosinase (Singh et al., 2015; Pajot et al., 2011). Tyrosinase activity has been reported in microbial strains such as Streptomyces glaucescens, Streptomyces antibioticus, Bacillus licheniformis, Bacillus natto, and Bacillus sphaericus (Zaidi et al., 2014). The extracted tyrosinase enzymes were employed for the oxidation of colored and phenolic compounds.
• Azoreductase: Azoreductases are flavoproteins (NAD (P) H: flavin oxidoreductase) found at cytoplasm (intracellular or extracellular), used as an electron donor for cofactors such as NADH, NADPH, FADH2, FMNH2, and FADH to reduce the azo bonds (Suzuki, 2019). The existence of extracellular oxygen-sensitive azo reductases in anaerobic bacteria, such as Clostridium and Eubacterium, can decolorize the sulfonated azo dyes during their growth (Zahran et al., 2019). The azoreductase substrate specificity is dependent on the functional group in the azo bond.
Patel et al. reported the induction of azoreductase with Pseudomonas sp. KF46, during the decolorization of Orange II, with maximum specificity in the carboxy group eliminating sulfophenyl azo dyes (Patel and Gupte, 2023). The azoreductase induction was previously reported during decolorization of azo dyes under static conditions, and after reducing an azo bond, toxic amines were produced by azoreductase (Dhanve et al., 2008; Kalyani et al., 2008). Several studies have indicated that azoreductase could reduce azo dyes and also have the potential to reduce various compounds of naphtho, benzo, and anthraquinone (Suzuki, 2019). In some instances, these compounds work as a better substrate than methyl red because of increased Km and Vmax values than MR (Leelakriangsak, 2012). These enzymes involve enzymatic detoxification processes, which include reducing quinones, quinone imines, azo dyes, and nitro groups, and protecting cells from the toxic effects of free radicals and reactive oxygen species resulting from reduced electrons (Ito et al., 2008).
• NADH-DCIP reductase and MG reductase: NADH-DCIP reductase are marker enzymes that belong to the oxidase cycle of bacterial and fungal mixed activity and are involved in the detoxification of xenobiotic compounds (Bhosale et al., 2006; Salokhe, 2003). The enzyme uses NADH as an electron donor to reduce the DCIP. Earlier findings suggested that this function is a marker enzyme for azo bond reduction because the 2,6-dichlorophenolindophenol (DCIP) uses this enzyme to accept an NADH electron to form its leuco group. Once reduced, the blue color of DCIP in its oxidized form becomes colorless. Several studies have been reported for the significant induction of NADH-DCIP reductase in the decolorization of Reactive Orange 16, Methyl Red, Red BL1, and Direct Brown MR by Bacillus sp., Brevibacillus laterospores, Pseudomonas sp., and A. calcoaceticus, respectively (Ghodake et al., 2009; Gomare and Govindwar, 2009; Amar et al., 2008).
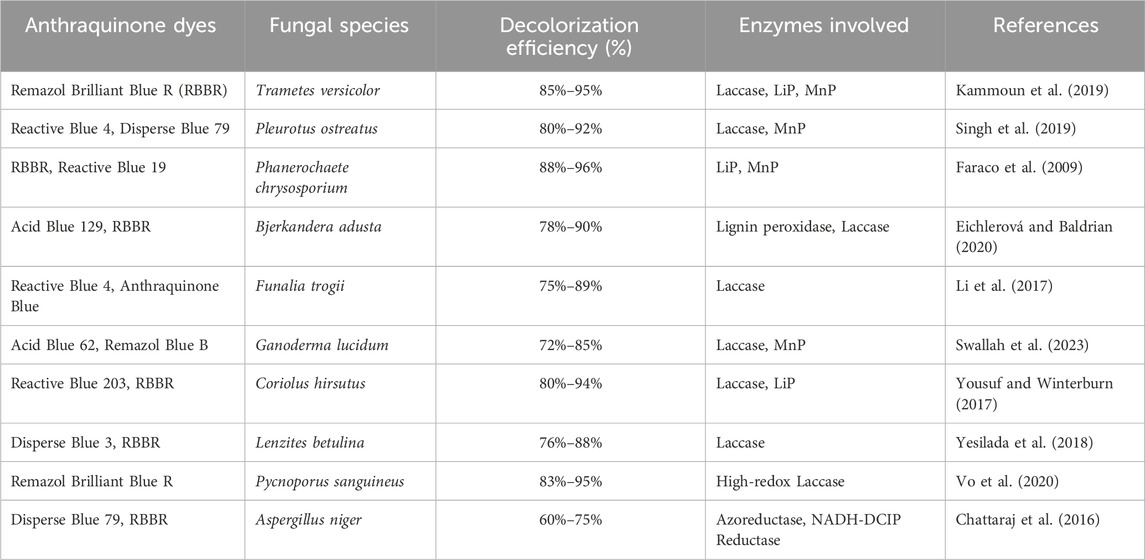
Emerging research highlights further enzymatic systems and strategies beyond common oxidoreductases for enhanced dye degradation. DyP-type peroxidases show promise in degrading recalcitrant dyes at a wider pH range and without mediators. Unspecific peroxygenases (UPOs), with both peroxidative and monooxygenase activity, offer energy-efficient bio-oxidation. Multi-enzyme cascade systems can achieve more complete degradation by sequentially employing oxidative and reductive enzymes. Enzyme immobilization enhances stability and reusability, while directed evolution and protein engineering allow for the creation of enzymes with improved properties. Enzyme-mediated photocatalytic hybrid systems offer synergistic degradation. Finally, omics and metagenomics are facilitating the discovery of novel, robust dye-degrading enzymes from diverse environments, paving the way for advanced bioremediation technologies. These advancements, focusing on enzyme function, system integration, and synthetic biology, are crucial for developing next-generation, eco-compatible solutions for persistent dye pollutants (Colpa et al., 2014; Sugano and Yoshida, 2021). The fungal species and the enzymes involved in anthraquinone dye decolorization is illustrated in Table 3.
Parameters influencing the degradation mechanism
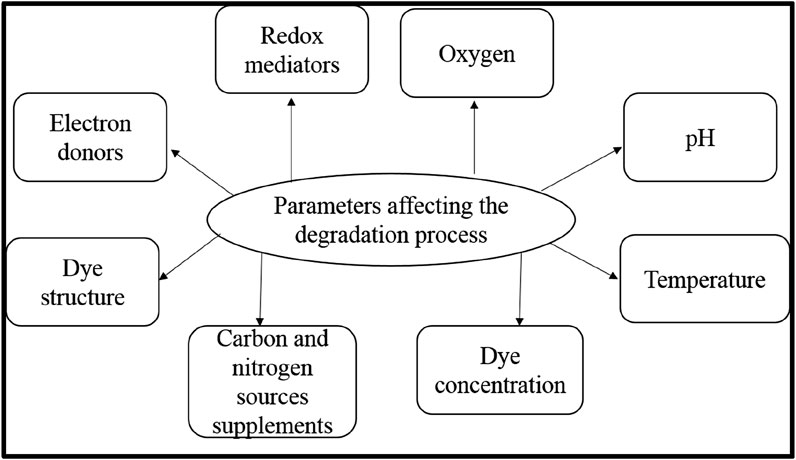
The microbial decolorization of reactive dye is affected by the operating conditions, i.e., both intrinsic and extrinsic parameters. The wastewater generated by the textile industries has different chemical compositions, including salts, nutrients, organics, sulfur compounds, and various toxic substances. Several factors influence the decolorization of dye in the biological treatment method, including different physicochemical parameters like dissolved oxygen, pH, temperature, dye concentration, dye structure, electron donors, and redox mediators, affecting the decolorization efficiency and microbial metabolic activity (Krishnan et al., 2017).
Numerous studies have been performed to investigate the effects of each parameter to increase the effectiveness of the biological treatment method (Figure 6). The impact of the operational conditions is described above.
• Oxygen: It is the most significant parameter that affects cell growth and dye removal in the dye degradation process. Oxygen influences the physiological properties of the cells during the cell cycle and is considered an excellent redox potential electron acceptor. During microbial degradation, once the cells in the medium oxidize the electron donors, resulting in the loss of an electron, that decreases the oxygen concentration relative to the dye molecules. In the reduction product, the water molecules are non-reductive. The oxygen molecules also oxidize the intermediate aromatic compounds formed during dye decolorization (Goswami et al., 2024; Zafar et al., 2022). The intermediates are broken down by hydroxylation and ring-opening when oxygen is present (Pande et al., 2019; Tembhekar et al., 2016). The aerobic environment is necessary for the effective degradation of the dye molecules. An anaerobic-aerobic process can, therefore, treat the dye-containing wastewaters effectively, thus enhancing its biological degradation (Goswami et al., 2024; Tembhekar et al., 2016). Monitoring the oxygen availability during the treatment process is crucial since oxygen is required for the growth and survival of microorganisms.
• pH: Medium pH is also a significant decolorization parameter influencing the dye decolorization efficiency, and the optimum pH range for decolorization is around 6.0 and 10.0 (Chen et al., 2003; Guo et al., 2007). The medium with a highly acidic or basic pH value resulted in reduced decolorization efficiency. The pH effect is assumed to be associated with the transportation of the dye molecules through the cell membrane, which is known to be the rate-limiting phase in the decolorization process (Chang et al., 2001; Kodam et al., 2005). The formation of intermediate aromatic amines as a result of the biodegradation of dye molecules increases the pH to more alkaline than the untreated dye solution (Dönmez et al., 2020). Changing the pH between 7.0 and 9.5 has minimal impact on the decolorization method. Thus, for effective decolorization, the optimum pH of the treatment process is needed (Aragaw, 2024). Wang et al. evaluated the effectiveness of isolated Citrobacter sp. CK3 in the decolorization of Reactive Red 190 under highly acidic (pH 4) and highly alkaline (pH 12) environments (Wang et al., 2009). This pH tolerance of decolorizing microbes is essential to (Chinwendu et al., 2024) make them appropriate for the decolorization of real textile effluents (Wang et al., 2009; Aksu et al., 2003).
• Temperature: The microbes respond by adapting to temperature change through biochemical or enzymatic mechanisms. Therefore, the temperature is a significant factor involving microbial existence-related activities, including water and soil remediation. Some experiments have been carried out on the activation energy of azo dye decolorization, where varying temperatures are established as required for the decolorization using microbial consortium present in active sludge (dos Santos et al., 2007). In 2001, Yu investigated that temperature changes in microbial physiology resulted in abrupt variations in the activation energy (Yu, 2001). In contrast, Angelova et al. reported the impacts of temperature on growth rate, biomass production, and reaction strategy (A et al., 2008). The azo dye decolorization efficacy decreases after reaching the optimum temperature. The reduction of viable cells or denaturation of an azo reductase enzyme may be the cause of the decline in decolorization efficacy at higher or lower temperatures (Steinweg et al., 2013; Rafeeq et al., 2023). Moreover, it was observed that the azoreductase enzyme is quite thermostable at 60°C and can stay effective for a short duration (Rathour et al., 2024; Sanjana et al., 2024). According to Misal et al., the dye would decolorize effectively at a temperature range between 25°C and 35°C which is ideal for microbial growth (Misal et al., 2011).
• Dye concentration: Dye concentration is another significant parameter affecting decolorization efficiency. The increase in the dye concentration reduces the decolorization efficiency due to the toxic effects of dyes on the microbes resulting in an inappropriate cell to dye ratio, and the blocking of azoreductase active sites by dye molecules with different systems (Dhanve et al., 2008; Saratale GD. et al., 2010; Tony et al., 2009a; Tony et al., 2009b). The decolorization of various reactive dyes by microbial strains has been described in similar results (Dhanve et al., 2008; Amar et al., 2008; Saratale et al., 2009a). Reactive dyes containing sulfonic acid (SO3H) groups within their aromatic rings have also been shown to significantly inhibit the microbial growth at increased dye concentrations (Amar et al., 2008; Saratale et al., 2009a). Saratale et al., reported that using microbial consortium, the impact of higher dye concentration gets reduced could be due to the synergistic influence within the microbial community (Saratale et al., 2009b). However, the lack of the impact of dye concentration on decolorization efficiency was observed by Pearce et al., and the result is consistent with a non-enzymatic reduction approach regulated by methods irrespective of the dye concentration (Pearce, 2003). Various studies have revealed a comprehensive kinetic analysis, and the relationship involving substrate-dependent decolorization efficiency and equilibrium transformation (Amar et al., 2008; Pearce, 2003; Chang et al., 2000).
• Carbon and nitrogen sources supplements: The Biodecolorization of reactive dyes without the addition of carbon or nitrogen sources is quite complicated since reactive dyes lack carbon sources (Kebede et al., 2021; Meckenstock et al., 2015). According to Chen et al., and Khehra et al., nitrogen (yeast extract) and carbon (glucose) sources can be added as supplements to aid in the decolorization of dyes by both individual strains and the microbial consortia (Chen et al., 2005; Khehra et al., 2005). In some findings, the supplementation of carbon sources causes a reduction in decolorization efficiency attributed to the cell’s preference to assimilate additional carbon sources over using the dye molecule as the carbon source (Saratale et al., 2009a). Similarly, Chang et al., stated that the supplementation of nitrogen sources might restore NADH, which serves as an electron donor for microbial dye degradation, resulting in effective decolorization (Varjani and Upasani, 2017). Some studies have used lignocellulosic agricultural residue as a substitute for organic sources supplements to make the dye decolorization process economically efficient and effective. The addition of lignocellulosic waste as a substrate has been shown to improve the decolorization efficiency by producing ligninolytic enzymes. Similar findings have also been made for the efficient decolorization of Direct Blue GLL by Comamonas sp. UVS, with the generation of ligninolytic enzymes (Dhanve et al., 2008). Scarlet R dye was decolored by consortium GR and Reactive Green 19A by Micrococcus glutamicus NCIM-2168 using a variety of agricultural wastes as a substrate (wood straw, rice husk, sugarcane bagasse powder, and rice straw). It was discovered that adding rice husk and rice straw extract as a nitrogen source in the synthetic medium increased the decolorization efficiency (Saratale et al., 2009a; Telke et al., 2009).
• Dye structure: Reactive dyes consist of a variety of diverse structures with different functional groups, and any variations in the chemical structures have a significant effect on decolorization efficiency. Simple dye compounds with low molecular mass show increased decolorization efficiency. Reduced decolorization efficiency was observed with dye molecules with higher molecular weights and with electron-withdrawing groups (–SO3H, –SO2NH2) present in the para position of the benzene ring (Pearce, 2003; Hsueh et al., 2009). In 2001, Hu observed an enhanced decolorization efficiency with monoazo dyes related to diazo and triazo dyes (Hu, 2001). The production of azoreductase enzyme associated with specific dye structures was stated by Patel et al. (2023). Hsueh et al. reported that the presence of substituent in the para position affected the decolorization efficiency relative to the ortho and meta positions of the benzene ring (Hsueh et al., 2009). The existence of electron-withdrawing groups on the benzene ring increases the decolorization efficacy (Patel et al., 2023). Suzuki et al. evaluated the aerobic decolorization of 25 sulfonated azo dyes and found that the steric effect of the chemical structure had a significant impact on decolorization efficiency (Suzuki et al., 2001). However, in a non-enzymatic reduction process, decolorization is affected by alteration in the electron density group, resulting in increased decolorization efficiency (Kebede et al., 2021; Meckenstock et al., 2015; Rau and Stolz, 2003).
• Electron donors: Supplementations of electron donors such as yeast extract, peptone, glucose, and lactate cause reductive cleavage of the bond, thereby increasing the dye decolorization efficiency. The addition of a specific electron donor is, therefore, crucial for treating textile wastewater (Al-Amrani et al., 2014). There are different electron donors, and electron acceptors are available in nature; however, it is essential to understand the impact of specific donors and acceptors on microbial decolorization (He and Sanford, 2003). The electron donors such as sodium succinate, sodium citrate, sodium formate, sodium pyruvate, and sodium acetate, were found to increase C.I. Reactive Orange 16 decolorization by isolated Bacillus sp. ADR (Telke et al., 2009) and different azo dyes by isolated Shewanella decolorationis S12 (Hong et al., 2007). It has been reported that microbial decolorization under anaerobic conditions is a biochemical reaction that oxidizes the electron donors, thereby transferring it to the acceptors via a multi-component electron transport chain system. Georgiou et al. reported that the electron transport system was disrupted by the existence of specific electron donors, which could be attributed to the search for donor electrons (Georgiou et al., 2004).
Methanol deserves special consideration among electron donors, as it is commonly utilized as an effective electron donor for treating wastewater. Telke et al. reported that Bacillus sp., ADR needed NADH as an electron donor for NADH-DCIP reductase during the decolorization of C.I. Reactive Orange 16 (Telke et al., 2009). Similarly, the addition of electron donors, like sodium acetate, sodium formate, sodium citrate, and sodium pyruvate induces decolorization activity. Therefore, it is essential to choose biological electron donors for decolorization experiments as they cause degradation and activation of the enzymatic system required for the decolorization activity (Pearce, 2003; van der Zee et al., 2001).
• Redox mediators: The anaerobic azo dye reduction process typically has a rate-limiting step that involves the transfer of reducing equivalents from an electron donor to an electron acceptor (van der Zee et al., 2001). Reducing the electron density in the azo bond can boost decolorization efficiency because the rate of azo dye decolorization is associated with that of the azo bond (Meckenstock et al., 2015). Redox mediators are added, which allows for the transfer of reducing equivalents to the dye’s electron acceptor and reduces the dye’s steric interference (Rau and Stolz, 2003; Moir et al., 2001). Flavin-based substances, such as flavin adenine dinucleotide (FAD) and flavin adenine mononucleotide (FMN)) and quinone-based substances, such as anthraquinone-2,6-disulfonate (AQDS), anthraquinone-2-sulfonate (AQS), cyanocobalamin (vitamin B12), riboflavin (vitamin B2), and lawsone (2-hydroxy-1,4-naphthoquinone)) were widely identified as redox mediators (dos Santos et al., 2007). Redox mediators have a redox potential of 200–350 mV, and a low concentration of the redox mediator is needed for the transfer of electrons (Meckenstock et al., 2015). It was discovered that the decolorization efficacy increases when the redox potential of the system decreases, and as the system’s redox potential increases, the decolorization efficiency decreases (Liu et al., 2009; Rau et al., 2002). Lourenço et al., reported some decolorization in autoclaved cells, indicating the presence of an active, reducing agent that can decolorize dye in the absence of microbial activity (Lourenço et al., 2001). To date, the impact of redox mediators on textile wastewater decolorization remains unclear due to the broad variety of redox potentials amongst reactive dyes (180–430 mV) found in effluents, with increased COD concentrations of around 1.4 g L−1 and various dye characteristics (dos Santos et al., 2007).
• Type of bacterial species: The selection of powerful microbial strains for the treatment procedure should be based on the reactivity or binding affinity of the dye. The physiological behavior of microbial strains differs from the dye concentration and structure, media composition, as well as culture conditions. Hence, the selection of an efficient microbial strain is necessary for the effective decolorization of textile wastewater (Senan and Abraham, 2004).
Analytical methods for the decolorization studies
The metabolites produced by the mineralization of reactive dyes by microbes were identified using various analytical methods. The predominant method for determining the decolorization of the dye is UV–vis spectroscopy (Prabhakar et al., 2021; Saratale RG. et al., 2010). The dye decolorization is shown by the major peak disappearing at a maximum wavelength and the emergence of new peaks in the UV region (Saratale et al., 2009a). American Dye Manufacturers Institute tristimulus filter method (ADMI 3WL) and ADMI 31WL were used to calculate the actual color rates, independent of hue for dye mixtures and textile effluents (Chen et al., 2003), thus trying to open the way for a more appropriate description of water and wastewater color (dos Santos et al., 2007). The thin-layer chromatography (TLC) method has proven to be the best approach with the separation and spot ability required to separate the dyes and their metabolites with a similar structure. It is feasible by utilizing various stationary phases from indefinite changes in mobile phase mixtures. High-performance thin-layer chromatography (HPTLC) was also used to validate the dye degradation investigation (Bhatt et al., 2005; Mohana et al., 2008). Although the principle of this method is the same as that of TLC, it has certain benefits like automatic application systems, thinner plates, and increased sensitivity. In general, liquid chromatography requires relatively small packing particles with extremely high pressure, which is known as high-performance liquid chromatography (HPLC). The degradation of the dye is indicated by the appearance of additional peaks with varying retention times relative to the original dye, suggesting the formation of metabolites (Chang et al., 2001; Saratale GD. et al., 2010; López-Grimau and Gutiérrez, 2006).
Infrared spectroscopy typically employs Fourier-transform infrared spectroscopy (FTIR), which reveals the degradation of the dye using the FTIR spectrum and establishes the type and extent of interactions in dyes with various functional groups after biodegradation. The mass spectrum of the mixture constituents is acquired in mass spectrometry, which provides a useful tool for qualitative study. The technique is referred to as liquid chromatography-mass spectrometry (LC-MS) when the mobile phase is a liquid, and as gas chromatography-mass spectrometry (GC-MS) when it is a gas. Mass spectroscopy using both methods is essential for determining the metabolites produced following biodegradation (Park and Kim, 2021). It also determines the molecular mass, and structural details of the dye intermediates and can recommend the dye degradation pathways (Amar et al., 2008; Saratale et al., 2009a). Similarly, nuclear magnetic resonance (NMR) is a standard analytical method that enables either liquid or solid-state quantitative study of compounds and is useful in collecting structural details about molecular compounds. Some researchers reported that the decolorization efficiency could be estimated by analyzing the decolorization percentage before and after the treatment through biochemical oxygen demand (BOD), chemical oxygen demand (COD), and total organic carbon (TOC) removal ratio (Saratale et al., 2009a; Telke et al., 2009; Saratale RG. et al., 2010).
Toxicity assessment of the bio-decolorization products
A wide range of synthetic dyes produced by textile manufacturers, particularly anthraquinone dyes, pose a significant ecological threat due to their complex aromatic structures, resistance to degradation, and toxicological impacts. During dyeing processes, an estimated 10%–15% of dyes are released into effluents, leading to the unnatural coloration of surface waters, deterioration of water quality, and disruption of aquatic ecosystems (Pearce, 2003; Robinson et al., 2001). The toxicity of anthraquinone dyes affects various biological systems. In microorganisms, these dyes can inhibit enzymatic activities and disrupt cellular respiration. In plants, they impair growth and physiological processes, as shown in studies where reactive blue-4 (RB4) significantly reduced plumule and radicle lengths by 30%–40% and chlorophyll-a content by 76% in Triticum aestivum (Chaudhari et al., 2017). In animals and humans, anthraquinone dyes and their intermediates have been associated with cytotoxic, genotoxic, and mutagenic effects, possibly through oxidative stress and DNA damage pathways. However, studies like Chaudhari et al. revealed that the biodegradation of RB4 resulted in non-phytotoxic metabolites, indicating the potential for safe bioremediation. Biodegradation of such reactive dyes often requires extended residence times, influencing intracellular retention and processing (Chen, 2002; Moutaouakkil et al., 2004). To evaluate toxicity, various assays are employed: biotoxicity is commonly assessed using microbial respiration inhibition or bioluminescent bacterial assays (e.g., Vibrio fischeri); phytotoxicity is evaluated via seed germination and root/shoot elongation tests in plants; cytotoxicity assays include MTT and neutral red uptake tests on cell cultures; and genotoxicity is measured using assays such as the comet assay, micronucleus test, and Ames test. Despite the promising detoxification potential of fungal and bacterial biodegradation systems, Rawat et al. (2016) highlighted the limited availability of comprehensive toxicity studies and recommended multilevel strategies including molecular, cellular, and ecosystem-level evaluations to ensure the environmental safety of dye degradation products (Rawat et al., 2016).
Advances in bio-decolorization technique
The current research focuses on designing a system that must be environmentally friendly and cost-effective, showing high decolorization efficiency with no or reduced production of sludge, and that it can mineralize the toxic dye to non-toxic compounds. Garg et al. evaluated the decolorization kinetics of reactive red-120 dye by immobilizing Bacillus cohnii RAPT1 on polyurethane in a packed bed reactor (Garg et al., 2016). They observed complete decolorization at 200 mg/L dye concentration within 4 h. Similarly, under immobilized conditions in batch and continuous mode, Chen et al., analyzed the malachite green decolorization by Pandoraea pulmonicola YC32 (Chen et al., 2009). The strain showed 85.2% decolorization efficiency at 50 mg L−1 dye concentration and relatively better recycling in continuous mode than batch mode. Decolorization was attributed to the degradation of the dye by cellular metabolic activity, either through a reducing mechanism/N-demethylation reaction. Tuttolomondo et al., reported the immobilization of Pseudomonas sp. by sol-gel encapsulation (Tuttolomondo et al., 2014). The decolorization efficiency of methyl orange, remazol black, and benzyl orange was reported to be 75, 79, and 83%, respectively.
Nowadays, research is concentrated on using bacterial enzymes to degrade dyes during wastewater treatment in bioreactors. Oxidoreductive enzymes such as laccase, peroxidases, NADH-DCIP reductase, and azoreductase exhibit good potential in dye decolorization (Patel et al., 2023). They reported that bacterial enzymes might be utilized as the primary strategy for decolorization of textile effluents, thus reducing operational costs and sludge generation. Moreover, optimizing the experimental conditions, lack of an engineered approach, and recombinant DNA engineering techniques could be used to achieve the required enzymes with higher efficiency and stability, and the enzymatic treatment makes the process simple to handle (Uday et al., 2016). The decolorization of reactive remazol violet-9 (RRV9), lefavix blue-16 (LB16), and reactive remazol navy-4 (RRN4) dyes was performed using fungal enzymes immobilized in a vertical bioreactor, and an enhanced decolorization efficiency of LB16 > RRV9 > RRN4, respectively (Peinado et al., 2006; Yanto et al., 2014). Similarly, Chhabra et al., observed acid red-27 dye decolorization with immobilized fungal laccase enzymes and exhibited a decolorization efficiency of about 90%–95% (Chhabra et al., 2015).
It is necessary to set up a system to operate using optimal experimental parameters to enhance microbial decolorization efficiency. Response surface methodology (RSM) was used to optimize the experimental parameters for Pseudomonas putida SKG-1 to achieve the highest decolorization as documented by Garg et al. (Garg et al., 2016). With RSM, they obtained an increased dye discoloration efficiency of around 97.8% but only 95.2% without RSM. Therefore, the RSM optimization approach could be an excellent statistical tool for evaluating the optimal experimental conditions relative to the OFAT method. Later, using bio-electrochemical processes, Gao et al., established pure strain cathodes to obtain increased decolorization efficiency of acid orange-7 and observed 95.8% decolorization within 46 h of incubation using the lactate-supplemented medium (Gao et al., 2016).
Recent research is focused on isolating and identifying the microbial strains that could decolorize the azo, anthraquinone, and triphenylmethane dyes rapidly and efficiently. Mishra et al., isolated Bacillus cereus DC11, which exhibits a decolorization efficiency of 95%–98% under anaerobic conditions, and decolorized 100 mM acid blue-25 dye (anthraquinone), 55 mM malachite green dye (triphenylmethane), and 750 mM basic blue X-GRRL dye (azo) within 6 h, 4 h, and 2 h at 20°C–45°C, pH 7 (Mishra and Maiti, 2018). Similarly, Bouraie et al. used Aeromonas hydrophila to decolorize all three dye classes under optimum experimental conditions and observed 90% decolorization efficiency at a dye concentration of 50 mg L−1 (El and El Din, 2016). More effective microbial strains must be identified to remove these three dyes from textile wastewater under optimal experimental conditions. Also, more research should be performed to enhance the potential of microbial strains in removing contaminants from effluent through genetic engineering, hybridization, and other biotechnological methods.
Bioreactors such as fixed/packed bed reactors (FBR/PBR), fluidized beds, sequential batch reactors (SBR), rotating biological contactors (RBC), up-flow anaerobic sludge blanket reactors (UASB), and immobilized cell bioreactor are also being designed for decolorization of dye in large-scale applications. Saratale et al. examined reactive orange-4 decolorization by immobilizing Lysinibacillus sp. RGS in a fixed-bed bioreactor operated in a continuous mode (Saratale et al., 2015). With an initial biomass loading of 4.23 g and flow rate of 25, 30, and 35 mL h−1, they observed a decolorization efficiency of 75%–98% at pH 6.6°C and 30°C within 5 h. Several studies have explained the effective practice of bioreactor systems for textile wastewater treatment (Sharma and Kuhad, 2008; Garg et al., 2016; Chen et al., 2016; Qian et al., 2015). More studies are required to increase the efficacy of these bioreactors to degrade the toxic reactive dyes and their degraded metabolites (Al-Amrani et al., 2014).
Conclusion
Dyes are colored organic substances that add or alter the color of the substrate where they are applied. Most dyes are found naturally, but due to their photo-stability, dyes are synthetically created by humans from petrochemicals. Reactive dyes generated by various industrial wastes are the primary pollutants that are not eliminated using conventional strategies. Biodegradation is the process of removal of any pollutant in the ecosystem through biological approaches using microorganisms to degrade a variety of organic compounds. Applying microbes to biodegrade synthetic dyes is an effective and alternative strategy for treating dye wastewater and thus offers significant advantages. The biological treatment method is inexpensive, requires minimal operating cost, and produces non-toxic end-products on mineralization. This method can also be used with a lower concentration of pollutants that would not be possible through physical and chemical processes. Consideration of the primary and secondary metabolism, a varied range of microbes such as bacteria, fungus, algae, and yeast may decolorize various dye contaminants under varying anaerobic and aerobic environments. Various environmental factors affect the process of biodegradation, such as pH, temperature, inoculum volume, oxygen availability, and nutrients. It is, therefore, necessary to optimize process parameters by observing their synergistic effect on the biodegradation process to obtain enhanced decolorization at the industrial scale.
Pure microbial strains are usually inadequate to completely degrade the dyes, leading to the production of carcinogenic aromatic amines as intermediates that need to be subsequently decomposed. It’s crucial to scale up and maintain large-scale pure cultures for wastewater treatment systems. The microbial consortium has been used to degrade synthetic dyes over pure cultures as it has a higher level of biodegradation and mineralization attributed to the synergetic metabolism within the microbial community. Microbial dye decolorization often involves enzymes, but not exclusively. In some cases, microorganisms physically absorb the dye or break it down through non-enzymatic mechanisms. The presence of various oxidative enzymes (laccase, lignin peroxidase, and other oxidases) and reductive enzymes (azo-reductase, nonspecific reductases) revealed the participation of these enzymes in the decolorization process through oxidative coupling reactions caused by biocatalysts such as lignin peroxidases, laccases, azo-reductases, and tyrosinases. Hence, it is important to understand the enzymatic mechanisms associated with dye degradation that may be effective in solving the important challenges at the industrial level. The effective use of continuous aerobic technology for textile wastewater treatment is based on the bioreactor’s construction. Such reactors can maintain a high loading rate per unit reactor capacity by maintaining the biomass for a longer duration. Therefore, it is important to find a better continuous reactor system based on immobilized bacterial cells for the effective mineralization of textile wastewater. For effective decolorization, it is necessary to identify the optimized process parameters, an appropriate bioreactor system, and a better choice of immobilization support material. Therefore, further studies focusing on the use of genetic engineering to develop recombinant microorganisms capable of degrading mixed anthraquinone-based dyes and simultaneously reducing environmental toxicity may play a crucial role.
Author contributions
SM: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review and editing. AK: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aboudi, K., Ahmed, B., Tyagi, V. K., and Van Lier, J. B. (2021). Occurrence and fate of aromaticity driven recalcitrance in anaerobic treatment of wastewater and organic solid wastes. Clean Energy Resour. Recovery 1 (2021 Jan 1), 203–226. doi:10.1016/b978-0-323-85223-4.00025-7
Aksu, Z., and Dönmez, G. (2003). A comparative study on the biosorption characteristics of some yeasts for Remazol Blue reactive dye. Chemosphere 50 (8), 1075–1083. doi:10.1016/s0045-6535(02)00623-9
Alam, M. Z., Mansor, M. F., and Jalal, K. C. A. (2009). Optimization of decolorization of methylene blue by lignin peroxidase enzyme produced from sewage sludge with Phanerocheate chrysosporium. J. Hazard. Mater. 162 (2–3), 708–715. doi:10.1016/j.jhazmat.2008.05.085
Al-Amrani, W. A., Lim, P.-E., Seng, C.-E., and Wan Ngah, W. S. (2014). Factors affecting bio-decolorization of azo dyes and COD removal in anoxic–aerobic REACT operated sequencing batch reactor. J. Taiwan Inst. Chem. Eng. 45 (2), 609–616. doi:10.1016/j.jtice.2013.06.032
Alegbe, E. O., and Uthman, T. O. (2024). A review of history, properties, classification, applications and challenges of natural and synthetic dyes. Heliyon 10 (13), e33646. doi:10.1016/j.heliyon.2024.e33646
Al-Tohamy, R., Ali, S. S., Li, F., Okasha, K. M., Mahmoud, Y. A. G., Elsamahy, T., et al. (2022). A critical review on the treatment of dye-containing wastewater: ecotoxicological and health concerns of textile dyes and possible remediation approaches for environmental safety. Ecotoxicol. Environ. Saf. 231, 113160. doi:10.1016/j.ecoenv.2021.113160
Al-Tohamy, R., Ali, S. S., Xie, R., Schagerl, M., Khalil, M. A., and Sun, J. (2023). Decolorization of reactive azo dye using novel halotolerant yeast consortium HYC and proposed degradation pathway. Ecotoxicol. Environ. Saf. 263, 115258. doi:10.1016/j.ecoenv.2023.115258
Amar, T., Dayanand, K., Jyoti, P. J., and Sanjay, P. G. (2008). Kinetics and mechanism of reactive red 141 degradation by a bacterial isolate rhizobium radiobacter MTCC 8161. Acta Chem. 55, 320–329.
Angelova, B., Avramova, T., Stefanova, L., and Mutafov, S. (2008). Temperature effect on bacterial azo bond reduction kinetics: an Arrhenius plot analysis. Biodegradation 19 (3), 387–393. doi:10.1007/s10532-007-9144-4
Aragaw, T. A. (2024). A review of dye biodegradation in textile wastewater, challenges due to wastewater characteristics, and the potential of alkaliphiles. J. Hazard. Mater. Adv. 16, 100493. doi:10.1016/j.hazadv.2024.100493
Aragaw, T. A., Bogale, F. M., and Tesfaye, E. L. (2024). Ligninolytic oxidative enzymes and their role in textile dye biodegradation: a comprehensive review. Water Pract. Technol. doi:10.2166/wpt.2024.229
Aravind, J. K. P. S. G. B. R. (2016). Optimization of chromium(VI) biosorption using gooseberry seeds by response surface methodology. Glob. J. Environ. Sci. Manag. 2 (1), 61–68. doi:10.7508/gjesm.2016.01.007
Benkhaya, S., M’rabet, S., and El Harfi, A. (2020a). Classifications, properties, recent synthesis and applications of azo dyes. Heliyon 6 (1), e03271. doi:10.1016/j.heliyon.2020.e03271
Benkhaya, S., M’ rabet, S., and El Harfi, A. (2020b). A review on classifications, recent synthesis and applications of textile dyes. Inorg. Chem. Commun. 115, 107891. doi:10.1016/j.inoche.2020.107891
Bharti, V., Vikrant, K., Goswami, M., Tiwari, H., Sonwani, R. K., Lee, J., et al. (2019). Biodegradation of methylene blue dye in a batch and continuous mode using biochar as packing media. Environ. Res. 171, 356–364. doi:10.1016/j.envres.2019.01.051
Bhatt, N., Patel, K. C., Keharia, H., and Madamwar, D. (2005). Decolorization of diazo-dye reactive blue 172 byPseudomonas aeruginosa NBAR12. J. Basic Microbiol. 45 (6), 407–418. doi:10.1002/jobm.200410504
Bhosale, S., Saratale, G., and Govindwar, S. (2006). Biotransformation enzymes in Cunninghamella blakesleeana (NCIM-687). J. Basic Microbiol. 46 (6), 444–448. doi:10.1002/jobm.200510117
Bibhuti Bhusan Mishra, B. X. (2016). Polyphonel oxidases: biochemical and molecular characterization, distribution, role and its control. Enzyme Eng. 05 (01). doi:10.4172/2329-6674.1000141
Bilal, M., Asgher, M., Shahid, M., and Bhatti, H. N. (2016). Characteristic features and dye degrading capability of agar ;agar gel immobilized manganese peroxidase. Int. J. Biol. Macromol. 86, 728–740. doi:10.1016/j.ijbiomac.2016.02.014
Bonakdarpour, B., Vyrides, I., and Stuckey, D. C. (2011). Comparison of the performance of one stage and two stage sequential anaerobic-aerobic biological processes for the treatment of reactive-azo-dye-containing synthetic wastewaters. Int. Biodeterior. and Biodegrad. 65 (4), 591–599. doi:10.1016/j.ibiod.2011.03.002
Borham, A., Haroun, M., Saleh, I. A., Zomot, N., Okla, M. K., Askar, M., et al. (2024). A statistical optimization for almost-complete methylene blue biosorption by Gracilaria bursa-pastoris. Heliyon 10 (15), e34972. doi:10.1016/j.heliyon.2024.e34972
Can, O. T., Bayramoglu, M., and Kobya, M. (2003). Decolorization of reactive dye solutions by electrocoagulation using aluminum electrodes. Ind. Eng. Chem. Res. 42 (14), 3391–3396. doi:10.1021/ie020951g
Cao, Z., Yan, W., Ding, M., and Yuan, Y. (2022). Construction of microbial consortia for microbial degradation of complex compounds. Front. Bioeng. Biotechnol. 10, 1051233. doi:10.3389/fbioe.2022.1051233
Chang, J.-S., Chou, C., and Chen, S.-Y. (2001). Decolorization of azo dyes with immobilized Pseudomonas luteola. Process Biochem. 36 (8–9), 757–763. doi:10.1016/s0032-9592(00)00274-0
Chang, J.-S., Kuo, T.-S., Chao, Y.-P., Ho, J.-Y., and Lin, P.-J. (2000). Azo dye decolorization with a mutant Escherichia coli Strain. Biotechnol. Lett. 22 (9), 807–812. doi:10.1023/a:1005624707777
Chattaraj, S., Johnson, J., and Madamwar, D. (2016). Biotransformation of mixture of dyes by enriched bacterial consortium ASD. Desalination Water Treat. 57 (45), 21585–21597. doi:10.1080/19443994.2015.1124345
Chaudhari, A. U., Paul, D., Dhotre, D., and Kodam, K. M. (2017). Effective biotransformation and detoxification of anthraquinone dye reactive blue 4 by using aerobic bacterial granules. Water Res. 122, 603–613. doi:10.1016/j.watres.2017.06.005
Chauhan, V., Dhiman, V., and Kanwar, S. S. (2021). Combination of classical and statistical approaches to enhance the fermentation conditions and increase the yield of Lipopeptide(s) by Pseudomonas sp. OXDC12: its partial purification and determining antifungal property. Turk. J. Biol. 45 (6), 695–710. doi:10.3906/biy-2106-59
Chen, B.-Y. (2002). Understanding decolorization characteristics of reactive azo dyes by Pseudomonas luteola: toxicity and kinetics. Process Biochem. 38 (3), 437–446. doi:10.1016/s0032-9592(02)00151-6
Chen, B.-Y., Chen, S.-Y., and Chang, J.-S. (2005). Immobilized cell fixed-bed bioreactor for wastewater decolorization. Process Biochem. 40 (11), 3434–3440. doi:10.1016/j.procbio.2005.04.002
Chen, C.-Y., Kuo, J.-T., Cheng, C.-Y., Huang, Y.-T., Ho, I.-H., and Chung, Y.-C. (2009). Biological decolorization of dye solution containing malachite green by Pandoraea pulmonicola YC32 using a batch and continuous system. J. Hazard. Mater. 172 (2–3), 1439–1445. doi:10.1016/j.jhazmat.2009.08.009
Chen, C.-Y., Wang, G.-H., Tseng, I.-H., and Chung, Y.-C. (2016). Analysis of bacterial diversity and efficiency of continuous removal of Victoria Blue R from wastewater by using packed-bed bioreactor. Chemosphere 145, 17–24. doi:10.1016/j.chemosphere.2015.11.061
Chen, K.-C., Wu, J.-Y., Liou, D.-J., and Hwang, S.-C. J. (2003). Decolorization of the textile dyes by newly isolated bacterial strains. J. Biotechnol. 101 (1), 57–68. doi:10.1016/s0168-1656(02)00303-6
Chhabra, M., Mishra, S., and Sreekrishnan, T. R. (2015). Immobilized laccase mediated dye decolorization and transformation pathway of azo dye acid red 27. J. Environ. Health Sci. Eng. 13 (1), 38. doi:10.1186/s40201-015-0192-0
Chinwendu, E. J., Jason, O. C., and Sarah, O. I. (2024). Decolorization and detoxification of synthetic azo dye wastewater by Enterobacter hormaechei MW584986 isolated from textile wastewater. Macedonian J. Ecol. Environ. 26 (1), 39–51. doi:10.59194/mjee24261039c
Claus, H. (2004). Laccases: structure, reactions, distribution. Micron 35 (1–2), 93–96. doi:10.1016/j.micron.2003.10.029
Colpa, D. I., Fraaije, M. W., and Van Bloois, E. (2014). DyP-type peroxidases: a promising and versatile class of enzymes. J. Industrial Microbiol. Biotechnol. 41 (1), 1–7. doi:10.1007/s10295-013-1371-6
Dafale, N., Wate, S., Meshram, S., and Nandy, T. (2008). Kinetic study approach of remazol black-B use for the development of two-stage anoxic–oxic reactor for decolorization/biodegradation of azo dyes by activated bacterial consortium. J. Hazard. Mater. 159 (2–3), 319–328. doi:10.1016/j.jhazmat.2008.02.058
Daneshvar, N., Ayazloo, M., Khataee, A. R., and Pourhassan, M. (2007). Biological decolorization of dye solution containing Malachite Green by microalgae Cosmarium sp. Bioresour. Technol. 98 (6), 1176–1182. doi:10.1016/j.biortech.2006.05.025
Daphedar, A. B., Kakkalameli, S., Faniband, B., Bilal, M., Bhargava, R. N., Ferreira, L. F. R., et al. (2022). Decolorization of various dyes by microorganisms and green-synthesized nanoparticles: current and future perspective. Environ. Sci. Pollut. Res. 30 (60), 124638–124653. doi:10.1007/s11356-022-21196-9
Das, S., Cherwoo, L., and Singh, R. (2023). Decoding dye degradation: microbial remediation of textile industry effluents. Biotechnol. Notes 4, 64–76. doi:10.1016/j.biotno.2023.10.001
Dhanve, R. S., Shedbalkar, U. U., and Jadhav, J. P. (2008). Biodegradation of diazo reactive dye Navy blue HE2R (Reactive blue 172) by an isolated Exiguobacterium sp. RD3. Biotechnol. Bioprocess Eng. 13 (1), 53–60. doi:10.1007/s12257-007-0165-y
Dinakarkumar, Y., Ramakrishnan, G., Gujjula, K. R., Vasu, V., Balamurugan, P., and Murali, G. (2024). Fungal bioremediation: an overview of the mechanisms, applications and future perspectives. Environ. Chem. Ecotoxicol. 6, 293–302. doi:10.1016/j.enceco.2024.07.002
Ding, F., Zhao, F., Jin, R., Rao, Y., Yang, G., Lu, L., et al. (2022). Management of complications in 257 cases of breast augmentation with polyacrylamide hydrogel, using two different strategies: a retrospective study. Aesthetic Plast. Surg. 46, 2107–2121. doi:10.1007/s00266-022-02876-w
Donato, L., Algieri, C., Rizzi, A., and Giorno, L. (2014). Kinetic study of tyrosinase immobilized on polymeric membrane. J. Membr. Sci. 454, 346–350. doi:10.1016/j.memsci.2013.12.029
Dong, X., Zhou, J., and Liu, Y. (2003). Peptone-induced biodecolorization of reactive brilliant blue (KN-R) by Rhodocyclus gelatinosus XL-1. Process Biochem. 39 (1), 89–94. doi:10.1016/s0032-9592(02)00319-9
Dönmez, Ö., Dükkancı, M., and Gündüz, G. (2020). Effects of catalyst preparation method and reaction parameters on the ultrasound assisted Photocatalytic oxidation of reactive yellow 84 dye. J. Environ. Health Sci. Eng. 18 (2), 835–851. doi:10.1007/s40201-020-00507-7
dos Santos, A. B., Cervantes, F. J., and van Lier, J. B. (2007). Review paper on current technologies for decolourisation of textile wastewaters: perspectives for anaerobic biotechnology. Bioresour. Technol. 98 (12), 2369–2385. doi:10.1016/j.biortech.2006.11.013
Dulo, B., Phan, K., Githaiga, J., Raes, K., and De Meester, S. (2021). Natural quinone dyes: a review on structure, extraction techniques, analysis and application potential. Waste Biomass Valorization 12 (12), 6339–6374. doi:10.1007/s12649-021-01443-9
Dutta, S., Adhikary, S., Bhattacharya, S., Roy, D., Chatterjee, S., Chakraborty, A., et al. (2024). Contamination of textile dyes in aquatic environment: adverse impacts on aquatic ecosystem and human health, and its management using bioremediation. J. Environ. Manag. 353, 120103. doi:10.1016/j.jenvman.2024.120103
Eichlerová, I., and Baldrian, P. (2020). Ligninolytic enzyme production and decolorization capacity of synthetic dyes by saprotrophic white rot, Brown rot, and litter decomposing basidiomycetes. J. Fungi 6 (4), 301. doi:10.3390/jof6040301
Ekramul Karim, M., Dhar, K., and Towhid Hossain, M. (2017). Co-metabolic decolorization of a textile reactive dye by Aspergillus fumigatus. Int. J. Environ. Sci. Technol. 14 (1), 177–186. doi:10.1007/s13762-016-1136-7
El, B. M., and El Din, W. S. (2016). Biodegradation of Reactive Black 5 by Aeromonas hydrophila strain isolated from dye-contaminated textile wastewater. Sustain Environ. Res. 26 (5), 209–216. 10.1016/j.serj.2016.04.014
Faraco, V., Pezzella, C., Miele, A., Giardina, P., and Sannia, G. (2009). Bio-remediation of colored industrial wastewaters by the white-rot fungi Phanerochaete chrysosporium and Pleurotus ostreatus and their enzymes. Biodegradation 20 (2), 209–220. doi:10.1007/s10532-008-9214-2
Farias, S., Mayer, D. A., de Oliveira, D., de Souza, SMAGU, and de Souza, A. A. U. (2017). Free and Ca-alginate beads immobilized horseradish peroxidase for the removal of reactive dyes: an experimental and modeling study. Appl. Biochem. Biotechnol. 182 (4), 1290–1306. doi:10.1007/s12010-017-2399-2
Fernandes, F. H., Bustos-Obregon, E., and Salvadori, D. M. F. (2015). Disperse Red 1 (textile dye) induces cytotoxic and genotoxic effects in mouse germ cells. Reprod. Toxicol. 53, 75–81. doi:10.1016/j.reprotox.2015.04.002
Freund, R. J., Wilson, W. J., and Mohr, D. L. (2010). Linear regression. Stat. Methods, 321–374. doi:10.1016/b978-0-12-374970-3.00007-x
Gao, S.-H., Peng, L., Liu, Y., Zhou, X., Ni, B.-J., Bond, P. L., et al. (2016). Bioelectrochemical reduction of an azo dye by a Shewanella oneidensis MR-1 formed biocathode. Int. Biodeterior. and Biodegrad. 115, 250–256. doi:10.1016/j.ibiod.2016.09.005
Garg, A., Sangal, V. K., and Bajpai, P. K. (2016). Decolorization and degradation of Reactive Black 5 dye by photocatalysis: modeling, optimization and kinetic study. Desalination Water Treat. 57 (38), 18003–18015. doi:10.1080/19443994.2015.1086697
Georgiou, D., Metallinou, C., Aivasidis, A., Voudrias, E., and Gimouhopoulos, K. (2004). Decolorization of azo-reactive dyes and cotton-textile wastewater using anaerobic digestion and acetate-consuming bacteria. Biochem. Eng. J. 19 (1), 75–79. doi:10.1016/j.bej.2003.11.003
Ghodake, G., Jadhav, S., Dawkar, V., and Govindwar, S. (2009). Biodegradation of diazo dye Direct brown MR by Acinetobacter calcoaceticus NCIM 2890. Int. Biodeterior. and Biodegrad. 63 (4), 433–439. doi:10.1016/j.ibiod.2008.12.002
Giardina, P., Faraco, V., Pezzella, C., Piscitelli, A., Vanhulle, S., and Sannia, G. (2010). Laccases: a never-ending story. Cell Mol. Life Sci. 67 (3), 369–385. doi:10.1007/s00018-009-0169-1
Giwa, A. (2012). Microbial decolourization of an anthraquinone dye C.I. Reactive blue 19 using Bacillus cereus. Am. Chem. Sci. J. 2 (2), 60–68. doi:10.9734/acsj/2012/1519
Gomare, S. S., and Govindwar, S. P. (2009). Brevibacillus laterosporus MTCC 2298: a potential azo dye degrader. J. Appl. Microbiol. 106 (3), 993–1004. doi:10.1111/j.1365-2672.2008.04066.x
Goswami, D., Mukherjee, J., Mondal, C., and Bhunia, B. (2024). Bioremediation of azo dye: a review on strategies, toxicity assessment, mechanisms, bottlenecks and prospects. Sci. Total Environ. 954, 176426. doi:10.1016/j.scitotenv.2024.176426
Gulati, D., and Jha, I. (2014). Microbial decolourization of dye reactive blue 19 by bacteria isolated from dye effluent contaminated soil. Int. J. Curr. Microbiol. Appl. Sci. 3 (9), 913–922.
Guo, J., Zhou, J., Wang, D., Tian, C., Wang, P., Salah Uddin, M., et al. (2007). Biocalalyst effects of immobilized anthraquinone on the anaerobic reduction of azo dyes by the salt-tolerant bacteria. Water Res. 41 (2), 426–432. doi:10.1016/j.watres.2006.10.022
Gupta, P. O., and Sekar, N. (2024). NLOphoric anthraquinone dyes – a review. ChemistrySelect 9 (20). doi:10.1002/slct.202400736
Gurav, A. A. G. J. S. (2011). KGS. Decolorization of anthroquinone based dye Vat Red 10 by Pseudomonas desmolyticum NCIM 2112 and Galactomyces geotrichum MTCC 1360. Int. J. Biotechnol. Mol. Biol. Res. 2 (6), 93–97.
Hassan, M. E., Yang, Q., Xiao, Z., Liu, L., Wang, N., Cui, X., et al. (2019). Impact of immobilization technology in industrial and pharmaceutical applications. 3 Biotech. 9 (12), 440. doi:10.1007/s13205-019-1969-0
He, Q., and Sanford, R. A. (2003). Characterization of Fe(III) reduction by chlororespiring Anaeromxyobacter dehalogenans. Appl. Environ. Microbiol. 69 (5), 2712–2718. doi:10.1128/aem.69.5.2712-2718.2003
Holkar, C. R., Pandit, A. B., and Pinjari, D. V. (2014). Kinetics of biological decolorisation of anthraquinone based Reactive Blue 19 using an isolated strain of Enterobacter sp.F NCIM 5545. Bioresour. Technol. 173, 342–351. doi:10.1016/j.biortech.2014.09.108
Hong, Y., Chen, X., Guo, J., Xu, Z., Xu, M., and Sun, G. (2007). Effects of electron donors and acceptors on anaerobic reduction of azo dyes by Shewanella decolorationis S12. Appl. Microbiol. Biotechnol. 74 (1), 230–238. doi:10.1007/s00253-006-0657-2
Hsueh, C.-C., Chen, B.-Y., and Yen, C.-Y. (2009). Understanding effects of chemical structure on azo dye decolorization characteristics by Aeromonas hydrophila. J. Hazard. Mater. 167 (1–3), 995–1001. doi:10.1016/j.jhazmat.2009.01.077
Hu, T.-L. (2001). Kinetics of azoreductase and assessment of toxicity of metabolic products from azo dyes by Pseudomonas luteola. Water Sci. Technol. 43 (2), 261–269. doi:10.2166/wst.2001.0098
Ikram, M., Zahoor, M., and El-Saber, B. G. (2021). Biodegradation and decolorization of textile dyes by bacterial strains: a biological approach for wastewater treatment. Z. fur Phys. Chem. 235 (10), 1381–1393. doi:10.1515/zpch-2020-1708
Ito, K., Nakanishi, M., Lee, W.-C., Zhi, Y., Sasaki, H., Zenno, S., et al. (2008). Expansion of substrate specificity and catalytic mechanism of azoreductase by X-ray crystallography and site-directed mutagenesis. J. Biol. Chem. 283 (20), 13889–13896. doi:10.1074/jbc.m710070200
Jadhav, J. P., Kalyani, D. C., Telke, A. A., Phugare, S. S., and Govindwar, S. P. (2010). Evaluation of the efficacy of a bacterial consortium for the removal of color, reduction of heavy metals, and toxicity from textile dye effluent. Bioresour. Technol. 101 (1), 165–173. doi:10.1016/j.biortech.2009.08.027
Jamee, R., and Siddique, R. (2019). Biodegradation of synthetic dyes of textile effluent by microorganisms: an environmentally and economically sustainable approach. Eur. J. Microbiol. Immunol. 9 (4), 114–118. doi:10.1556/1886.2019.00018
Janusz, G., Pawlik, A., Sulej, J., Świderska-Burek, U., Jarosz-Wilkołazka, A., and Paszczyński, A. (2017). Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 41 (6), 941–962. doi:10.1093/femsre/fux049
Janusz, G., Pawlik, A., Świderska-Burek, U., Polak, J., Sulej, J., Jarosz-Wilkołazka, A., et al. (2020). Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 21 (3), 966. doi:10.3390/ijms21030966
Jonstrup, M., Punzi, M., and Mattiasson, B. (2011). Comparison of anaerobic pre-treatment and aerobic post-treatment coupled to photo-Fenton oxidation for degradation of azo dyes. J. Photochem. Photobiol. A Chem. 224 (1), 55–61. doi:10.1016/j.jphotochem.2011.09.006
Jović, M., Stanković, D., Manojlović, D., Anđelković, I., Milić, A., Dojćinović, B., et al. (2013). Study of the electrochemical oxidation of reactive textile dyes using platinum electrode. Int. J. Electrochem. Sci. 8 (1), 168–183. doi:10.1016/s1452-3981(23)14011-9
Kalme, S., Jadhav, S., Jadhav, M., and Govindwar, S. (2009). Textile dye degrading laccase from Pseudomonas desmolyticum NCIM 2112. Enzyme Microb. Technol. 44 (2), 65–71. doi:10.1016/j.enzmictec.2008.10.005
Kalyani, D. C., Patil, P. S., Jadhav, J. P., and Govindwar, S. P. (2008). Biodegradation of reactive textile dye Red BLI by an isolated bacterium Pseudomonas sp. SUK1. Bioresour. Technol. 99 (11), 4635–4641. doi:10.1016/j.biortech.2007.06.058
Kammoun, S., Loukil, S., and Loukil, Y. B. R. (2019). The impact of fintech on economic performance and financial stability in mena zone. Impact Financ. Technol. Islam Financ. Financ. Stab. doi:10.4018/978-1-7998-0039-2.ch013
Karatas, M. D. S. A. M. E. (2009). Decolorization of reactive dyes under batch anaerobic condition by mixed microbial culture. Afr. J. Biotechnol. 8 (24), 6856–6862. doi:10.4314/ajb.v8i24.68764
Kebede, G., Tafese, T., Abda, E. M., Kamaraj, M., and Assefa, F. (2021). Factors influencing the bacterial bioremediation of hydrocarbon contaminants in the soil: mechanisms and impacts. J. Chem. (1), 9823362. doi:10.1155/2021/9823362
Kesari, K. K., Soni, R., Jamal, Q. M. S., Tripathi, P., Lal, J. A., Jha, N. K., et al. (2021). Wastewater treatment and reuse: a review of its applications and health implications. Water Air Soil Pollut. 232 (5), 208–228. doi:10.1007/s11270-021-05154-8
Khehra, M. S., Saini, H. S., Sharma, D. K., Chadha, B. S., and Chimni, S. S. (2005). Comparative studies on potential of consortium and constituent pure bacterial isolates to decolorize azo dyes. Water Res. 39 (20), 5135–5141. doi:10.1016/j.watres.2005.09.033
Kodam, K. M., Soojhawon, I., Lokhande, P. D., and Gawai, K. R. (2005). Microbial decolorization of reactive azo dyes under aerobic conditions. World J. Microbiol. Biotechnol. 21, 367–370. doi:10.1007/s11274-004-5957-z
Krishnan, J., Arvind Kishore, A., Suresh, A., Madhumeetha, B., and Gnana Prakash, D. (2017). Effect of pH, inoculum dose and initial dye concentration on the removal of azo dye mixture under aerobic conditions. Int. Biodeterior. and Biodegrad. 119, 16–27. doi:10.1016/j.ibiod.2016.11.024
Kumar, A., and Chandra, R. (2020). Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon 6 (2), e03170. doi:10.1016/j.heliyon.2020.e03170
Kumar, A., Dixit, U., Singh, K., Gupta, S. P., Beg, M. S. J., Kumar, A., et al. (2021). Structure and properties of dyes and pigments. Dye Pigment. - Nov. Appl. Waste Treat. doi:10.5772/intechopen.97104
Kumar, N., Sinha, S., Mehrotra, T., Singh, R., Tandon, S., and Thakur, I. S. (2019). Biodecolorization of azo dye Acid Black 24 by Bacillus pseudomycoides: process optimization using Box Behnken design model and toxicity assessment. Bioresour. Technol. Rep. 8, 100311. doi:10.1016/j.biteb.2019.100311
Kumaravel, R., and Shanmugam, V. K. (2024). Biomimetic and ecological perspective towards decolorization of industrial important Azo dyes using bacterial cultures – a review. Sustain. Chem. Environ. 7, 100130. doi:10.1016/j.scenv.2024.100130
Lade, H. S., Waghmode, T. R., Kadam, A. A., and Govindwar, S. P. (2012). Enhanced biodegradation and detoxification of disperse azo dye Rubine GFL and textile industry effluent by defined fungal-bacterial consortium. Int. Biodeterior. and Biodegrad. 72, 94–107. doi:10.1016/j.ibiod.2012.06.001
Leelakriangsak, M. B. S. (2012). Characterization of the decolorizing activity of azo dyes by Bacillus subtilis azoreductase AzoR1. Songklanakarin J. Sci. Technol. 34 (5), 509–516.
Li, G., Liu, X., and Yuan, L. (2017). Improved laccase production by Funalia trogii in absorbent fermentation with nutrient carrier. J. Biosci. Bioeng. 124 (4), 381–385. doi:10.1016/j.jbiosc.2017.05.002
Liu, G., Zhou, J., Wang, J., Zhou, M., Lu, H., and Jin, R. (2009). Acceleration of azo dye decolorization by using quinone reductase activity of azoreductase and quinone redox mediator. Bioresour. Technol. 100 (11), 2791–2795. doi:10.1016/j.biortech.2008.12.040
López-Grimau, V., and Gutiérrez, M. c. (2006). Decolourisation of simulated reactive dyebath effluents by electrochemical oxidation assisted by UV light. Chemosphere 62 (1), 106–112. doi:10.1016/j.chemosphere.2005.03.076
Lourenço, N. D., Novais, J., and Pinheiro, H. (2001). Effect of some operational parameters on textile dye biodegradation in a sequential batch reactor. J. Biotechnol. 89 (2–3), 163–174. doi:10.1016/s0168-1656(01)00313-3
Lu, C., Wang, H., Luo, Y., and Guo, L. (2010). An efficient system for pre-delignification of gramineous biofuel feedstock in vitro: application of a laccase from Pycnoporus sanguineus H275. Process Biochem. 45 (7), 1141–1147. doi:10.1016/j.procbio.2010.04.010
Mahmood, S., Khalid, A., Arshad, M., Mahmood, T., and Crowley, D. E. (2016). Detoxification of azo dyes by bacterial oxidoreductase enzymes. Crit. Rev. Biotechnol. 36 (4), 639–651. doi:10.3109/07388551.2015.1004518
Maniyam, M. N., Gunalan, P., Azman, H. H., Abdullah, H., and Yaacob, N. S. (2024). Enhancing biodecolorization of malachite green by flavobacterium sp. through monothetic analysis. J. Indian Chem. Soc. 101 (12), 101452. doi:10.1016/j.jics.2024.101452
Martins, T. D., Schimmel, D., Dos Santos, J. B. O., and Da Silva, E. A. (2013). Reactive blue 5G adsorption onto activated carbon: kinetics and equilibrium. J. Chem. Eng. Data 58 (1), 106–114. doi:10.1021/je300946j
Meckenstock, R. U., Elsner, M., Griebler, C., Lueders, T., Stumpp, C., Aamand, J., et al. (2015). Biodegradation: updating the concepts of control for microbial cleanup in contaminated aquifers. Environ. Sci. Technol. 49 (12), 7073–7081. doi:10.1021/acs.est.5b00715
Mehra, S., Singh, M., and Chadha, P. (2021). Adverse impact of textile dyes on the aquatic environment as well as on human beings. Toxicol. Int. 28 (2), 165–176. doi:10.18311/ti/2021/v28i2/26798
Misal, S. A., Lingojwar, D. P., Shinde, R. M., and Gawai, K. R. (2011). Purification and characterization of azoreductase from alkaliphilic strain Bacillus badius. Process Biochem. 46 (6), 1264–1269. doi:10.1016/j.procbio.2011.02.013
Mishra, A., Takkar, S., Joshi, N. C., Shukla, S., Shukla, K., Singh, A., et al. (2021). An integrative approach to study bacterial enzymatic degradation of toxic dyes. Front. Microbiol. 12, 802544. doi:10.3389/fmicb.2021.802544
Mishra, S., and Maiti, A. (2018). The efficacy of bacterial species to decolourise reactive azo, anthroquinone and triphenylmethane dyes from wastewater: a review. Environ. Sci. Pollut. Res. 25 (9), 8286–8314. doi:10.1007/s11356-018-1273-2
Mohana, S., Shrivastava, S., Divecha, J., and Madamwar, D. (2008). Response surface methodology for optimization of medium for decolorization of textile dye Direct Black 22 by a novel bacterial consortium. Bioresour. Technol. 99 (3), 562–569. doi:10.1016/j.biortech.2006.12.033
Mohanty, S. S., and Kumar, A. (2021a). Enhanced degradation of anthraquinone dyes by microbial monoculture and developed consortium through the production of specific enzymes. Sci. Rep. 11 (1), 7678–15. doi:10.1038/s41598-021-87227-6
Mohanty, S. S., and Kumar, A. (2021b). Biodegradation of Indanthrene Blue RS dye in immobilized continuous upflow packed bed bioreactor using corncob biochar. Sci. Rep. 11 (1), 13390–13. doi:10.1038/s41598-021-92889-3
Mohanty, S. S., and Kumar, A. (2021c). Biosorption and biodegradation of anthraquinone based dye (Indanthrene blue RS) by the immobilized microbial consortium in a continuous packed bed bioreactor using corn-cob biochar. Available online at: https://www.researchsquare.com/article/rs-447968/v1.
Mohr, D. L., Wilson, W. J., and Freund, R. J. (2022). Linear regression. Stat. Methods, 301–349. doi:10.1016/b978-0-12-823043-5.00007-2
Moir, D., Masson, S., and Chu, I. (2001). Structure–activity relationship study on the bioreduction of AZO dyes by Clostridium paraputrificum. Environ. Toxicol. Chem. 20 (3), 479–484. doi:10.1897/1551-5028(2001)020<0479:sarsot>2.0.co;2
Moutaouakkil, A., Zeroual, Y., Dzayri, F. Z., Talbi, M., Lee, K., and Blaghen, M. (2004). Decolorization of azo dyes with Enterobacter agglomerans immobilized in different supports by using fluidized bed bioreactor. Curr. Microbiol. 48 (2), 124–129. doi:10.1007/s00284-003-4143-0
Moyo, S., Makhanya, B. P., and Zwane, P. E. (2022). Use of bacterial isolates in the treatment of textile dye wastewater: a review. Heliyon 8 (6), e09632. doi:10.1016/j.heliyon.2022.e09632
Nabil, G. M., El-Mallah, N. M., and Mahmoud, M. E. (2014). Enhanced decolorization of reactive black 5 dye by active carbon sorbent-immobilized-cationic surfactant (AC-CS). J. Industrial Eng. Chem. 20 (3), 994–1002. doi:10.1016/j.jiec.2013.06.034
Ngo, A. C. R., and Tischler, D. (2022). Microbial degradation of azo dyes: approaches and prospects for a hazard-free conversion by microorganisms. Int. J. Environ. Res. Public Health 19 (8), 4740. doi:10.3390/ijerph19084740
Obaideen, K., Shehata, N., Sayed, E. T., Abdelkareem, M. A., Mahmoud, M. S., and Olabi, A. G. (2022). The role of wastewater treatment in achieving sustainable development goals (SDGs) and sustainability guideline. Energy Nexus 7, 100112. doi:10.1016/j.nexus.2022.100112
Olaganathan, R. and Patterson. J. (2012). Biodegradation of textile dye anthraquinone vat blue 4 by Pseudomonas aeruginosa. Int. J. Adv. Biotechnol. Res. 3 (4), 743–751.
Pajot, H. F., Fariña, J. I., and de Figueroa, L. I. C. (2011). Evidence on manganese peroxidase and tyrosinase expression during decolorization of textile industry dyes by Trichosporon akiyoshidainum. Int. Biodeterior. Biodegrad. 65 (8), 1199–1207. doi:10.1016/j.ibiod.2011.05.010
Pande, V., Pandey, S. C., Joshi, T., Sati, D., Gangola, S., Kumar, S., et al. (2019). Biodegradation of toxic dyes: a comparative study of enzyme action in a microbial system. Smart Bioremediation Technol., 255–287. doi:10.1016/b978-0-12-818307-6.00014-7
Park, M. K., and Kim, Y.-S. (2021). Mass spectrometry based metabolomics approach on the elucidation of volatile metabolites formation in fermented foods: a mini review. Food Sci. Biotechnol. 30 (7), 881–890. doi:10.1007/s10068-021-00917-9
Patel, D., Singh, A., Ambati, S. R., Singh, R. S., and Sonwani, R. K. (2024). An overview of recent advances in treatment of complex dye-containing wastewater and its techno-economic assessment. J. Environ. Manag. 370, 122804. doi:10.1016/j.jenvman.2024.122804
Patel, J. N., Vaishnav, D. K., Joshi, P. R., and Tipre, D. R. (2023). Bacterial enzymes for azo dye degradation: an insight. Res. J. Chem. Environ. 27 (4), 135–148. doi:10.25303/2704rjce1350148
Patel, Y., and Gupte, A. (2023). Production, partial purification and characterization of intracellular Azoreductase from bacterial isolates during biodecolorization of textile dye Acid Maroon V. Emergent Life Sci. Res. 09 (01), 61–71. doi:10.31783/elsr.2023.916171
Pearce, C. (2003). The removal of colour from textile wastewater using whole bacterial cells: a review. Dyes Pigments 58 (3), 179–196. doi:10.1016/s0143-7208(03)00064-0
Peinado, R. A., Moreno, J. J., Villalba, J. M., González-Reyes, J. A., Ortega, J. M., and Mauricio, J. C. (2006). Yeast biocapsules: a new immobilization method and their applications. Enzyme Microb. Technol. 40 (1), 79–84. doi:10.1016/j.enzmictec.2005.10.040
Periyasamy, A. P. (2024). Recent advances in the remediation of textile-dye-containing wastewater: prioritizing human health and sustainable wastewater treatment. Sustainability 16 (2), 495. doi:10.3390/su16020495
Periyasamy, A. P., and Militky, J. (2020). Sustainability in textile dyeing: recent developments. Sustain. Text. Prod. Process. Manuf. and Chem., 37–79. doi:10.1007/978-3-030-38545-3_2
Prabhakar, Y., Gupta, A., and Kaushik, A. (2021). Microbial degradation of reactive red-35 dye: upgraded progression through Box–Behnken design modeling and cyclic acclimatization. J. Water Process Eng. 40, 101782. doi:10.1016/j.jwpe.2020.101782
Prasad, A. S. A., and Rao, K. V. B. (2013). Aerobic biodegradation of Azo dye by Bacillus cohnii MTCC 3616; an obligately alkaliphilic bacterium and toxicity evaluation of metabolites by different bioassay systems. Appl. Microbiol. Biotechnol. 97 (16), 7469–7481. doi:10.1007/s00253-012-4492-3
Qian, Y., Yang, B., Li, Z., Lei, L., and Zhang, X. (2015)., 23. Declarations, 1194–1199. doi:10.1016/j.cjche.2015.04.007Improving the biodecolorization of reactive blue 13 by sodium anthraquinone-2-sulfonate immobilized on modified polyvinyl alcohol beadsChin. J. Chem. Eng.7
Rafeeq, H., Afsheen, N., Rafique, S., Arshad, A., Intisar, M., Hussain, A., et al. (2023). Genetically engineered microorganisms for environmental remediation. Chemosphere 310, 136751. doi:10.1016/j.chemosphere.2022.136751
Rane, A., and Joshi, S. J. (2021). Biodecolorization and biodegradation of dyes: a review. Open Biotechnol. J. 15 (1), 97–108. doi:10.2174/1874070702115010097
Rani, J., Bera, S., Gaur, V., Singh, J., and Goutam, U. (2024). Response surface methodology for rapid removal of an azo dye methyl orange by indigenous bacterial strain (Bacillus cereus J4). J. Appl. Biol. Biotechnol. 13 (1), 259–266. doi:10.7324/JABB.2024.205683
Rathour, R. K., Sharma, D., Ullah, S., Mahmoud, E.-H. M., Sharma, N., Kumar, P., et al. (2024). Bacterial–microalgal consortia for bioremediation of textile industry wastewater and resource recovery for circular economy. Biotechnol. Environ. 1 (1), 6–21. doi:10.1186/s44314-024-00005-2
Rau, J., and Stolz, A. (2003). Oxygen-insensitive nitroreductases NfsA and NfsB of Escherichia coli function under anaerobic conditions as lawsone-dependent azo reductases. Appl. Environ. Microbiol. 69 (6), 3448–3455. doi:10.1128/aem.69.6.3448-3455.2003
Rau, J. K. H. J. S. A., Knackmuss, H. J., and Stolz, A. (2002). Effects of different quinoid redox mediators on the anaerobic reduction of azo dyes by bacteria. Environ. Sci. Technol. 36 (7), 1497–1504. doi:10.1021/es010227+
Rawat, D., Mishra, V., and Sharma, R. S. (2016). Detoxification of azo dyes in the context of environmental processes. Chemosphere 155, 591–605. doi:10.1016/j.chemosphere.2016.04.068
Rezaee, A., Ghaneian, M. T., Khavanin, A., Hashemian, S. J., and Moussavi, G. (2008). Photochemical oxidation of reactive blue 19 dye (rb19) in textile wastewater by uv/k2s2o8 process. J. Environ. Heal Sci. Eng. 5 (2), 95–100. Available online at: https://ijehse.tums.ac.ir/index.php/jehse/article/view/155.
Ridge, J. P., Lin, M., Larsen, E. I., Fegan, M., McEwan, A. G., and Sly, L. I. (2007). A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ. Microbiol. 9 (4), 944–953. doi:10.1111/j.1462-2920.2006.01216.x
Robinson, T., McMullan, G., Marchant, R., and Nigam, P. (2001). Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative. Bioresour. Technol. 77 (3), 247–255. doi:10.1016/s0960-8524(00)00080-8
Routoula, E., and Patwardhan, S. V. (2020). Degradation of anthraquinone dyes from effluents: a review focusing on enzymatic dye degradation with industrial potential. Environ. Sci. Technol. 54 (2), 647–664. doi:10.1021/acs.est.9b03737
Salokhe, M. D. (2003). Effect of carbon source on the biotransformation enzymes in Serratia marcescens. World J. Microbiol. Biotechnol. 19 (2), 199–200. doi:10.1023/a:1023263613480
Sanjana, M., Prajna, R., S Katti, U., and Kavitha, R. V. (2024). Bioremediation – the recent drift towards a sustainable environment. Environ. Sci. Adv. 3 (8), 1097–1110. doi:10.1039/d3va00358b
Sanmuga Priya, E., and Senthamil Selvan, P. (2017). Water hyacinth (Eichhornia crassipes) – an efficient and economic adsorbent for textile effluent treatment – a review. Arabian J. Chem. 10, S3548–S3558. doi:10.1016/j.arabjc.2014.03.002
Saratale, G. D., Chien, L.-J., and Chang, J. S. (2010a). “Enzymatic treatment of lignocellulosic wastes for anaerobic digestion and bioenergy production,” in Environmental anaerobic technology (Singapaore: IMPERIAL COLLEGE PRESS), 279–308.
Saratale, R. G., Saratale, G. D., Chang, J. S., and Govindwar, S. P. (2009a). Ecofriendly degradation of sulfonated diazo dye C.I. Reactive Green 19A using Micrococcus glutamicus NCIM-2168. Bioresour. Technol. 100 (17), 3897–3905. doi:10.1016/j.biortech.2009.03.051
Saratale, R. G., Saratale, G. D., Chang, J. S., and Govindwar, S. P. (2010b). Decolorization and biodegradation of reactive dyes and dye wastewater by a developed bacterial consortium. Biodegradation 21 (6), 999–1015. doi:10.1007/s10532-010-9360-1
Saratale, R. G., Saratale, G. D., Govindwar, S. P., and Kim, D. S. (2015). Exploiting the efficacy of Lysinibacillus sp. RGS for decolorization and detoxification of industrial dyes, textile effluent and bioreactor studies. J. Environ. Sci. Health, Part A 50 (2), 176–192. doi:10.1080/10934529.2014.975536
Saratale, R. G., Saratale, G. D., Kalyani, D. C., Chang, J. S., and Govindwar, S. P. (2009b). Enhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium-GR. Bioresour. Technol. 100 (9), 2493–2500. doi:10.1016/j.biortech.2008.12.013
Senan, R. C., and Abraham, T. E. (2004). Bioremediation of textile azo dyes by aerobic bacterial consortium aerobic degradation of selected azo dyes by bacterial consortium. Biodegradation 15 (4), 275–280. doi:10.1023/b:biod.0000043000.18427.0a
Shah, P. D., Dave, S. R., and Rao, M. S. (2012). Enzymatic degradation of textile dye Reactive Orange 13 by newly isolated bacterial strain Alcaligenes faecalis PMS-1. Int. Biodeterior. and Biodegrad. 69, 41–50. doi:10.1016/j.ibiod.2012.01.002
Shahid, M., Wertz, J., Degano, I., Aceto, M., Khan, M. I., and Quye, A. (2019). Analytical methods for determination of anthraquinone dyes in historical textiles: a review. Anal. Chim. Acta 1083, 58–87. doi:10.1016/j.aca.2019.07.009
Sharma, J., Sharma, S., and Soni, V. (2021). Classification and impact of synthetic textile dyes on Aquatic Flora: a review. Regional Stud. Mar. Sci. 45, 101802. doi:10.1016/j.rsma.2021.101802
Sharma, K. K., and Kuhad, R. C. (2008). Laccase: enzyme revisited and function redefined. Indian J. Microbiol. 48 (3), 309–316. doi:10.1007/s12088-008-0028-z
Shrestha, P., Joshi, B., Joshi, J., Malla, R., and Sreerama, L. (2016). Isolation and physicochemical characterization of laccase from Ganoderma lucidum -CDBT1 isolated from its native habitat in Nepal. BioMed Res. Int. 2016, 1–10. doi:10.1155/2016/3238909
Shyla, H., Saha, P., and Rao, K. V. B. (2018). Biodegradation and decolorization of two different azo dyes, reactive blue 221 and direct black 38, and assessment of the degraded dye metabolites. Desalination Water Treat. 123, 338–347. doi:10.5004/dwt.2018.22624
Singh, A., Mittal, A., and Jangid, N. K. (2020). Toxicology of dyes. Adv. Hum. Serv. Public Health, 50–69. doi:10.4018/978-1-7998-0311-9.ch003
Singh, R. L., Singh, P. K., and Singh, R. P. (2015). Enzymatic decolorization and degradation of azo dyes – a review. Int. Biodeterior. and Biodegrad. 104, 21–31. doi:10.1016/j.ibiod.2015.04.027
Singh, V., Lehri, A., and Singh, N. (2019). Assessment and comparison of phytoremediation potential of selected plant species against endosulfan. Int. J. Environ. Sci. Technol. 16 (7), 3231–3248. doi:10.1007/s13762-018-1880-y
Slama, H. B., Chenari Bouket, A., Pourhassan, Z., Alenezi, F. N., Silini, A., Cherif-Silini, H., et al. (2021). Diversity of synthetic dyes from textile industries, discharge impacts and treatment methods. Appl. Sci. 11 (14), 6255. doi:10.3390/app11146255
Sridharan, R., and Krishnaswamy, V. G. (2023). Bioremediation of textile dyes for sustainable environment—a review. Mod. Approaches Waste Bioremediation, 447–460. doi:10.1007/978-3-031-24086-7_21
Steinweg, J. M., Dukes, J. S., Paul, E. A., and Wallenstein, M. D. (2013). Microbial responses to multi-factor climate change: effects on soil enzymes. Front. Microbiol. 4 (JUN), 47630. doi:10.3389/fmicb.2013.00146
Sugano, Y., and Yoshida, T. (2021). DyP-type peroxidases: recent advances and perspectives. Int. J. Mol. Sci. 22 (11), 5556. doi:10.3390/ijms22115556
Suzuki, H. (2019). Remarkable diversification of bacterial azoreductases: primary sequences, structures, substrates, physiological roles, and biotechnological applications. Appl. Microbiol. Biotechnol. 103 (10), 3965–3978. doi:10.1007/s00253-019-09775-2
Suzuki, Y., Yoda, T., Ruhul, A., and Sugiura, W. (2001). Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-2 isolated from soil. J. Biol. Chem. 276 (12), 9059–9065. doi:10.1074/jbc.m008083200
Swain, G., Singh, S., Sonwani, R. K., Singh, R. S., Jaiswal, R. P., and Rai, B. N. (2021). Removal of Acid Orange 7 dye in a packed bed bioreactor: process optimization using response surface methodology and kinetic study. Bioresour. Technol. Rep. 13, 100620. doi:10.1016/j.biteb.2020.100620
Swallah, M. S., Bondzie-Quaye, P., Wu, Y., Acheampong, A., Sossah, F. L., Elsherbiny, S. M., et al. (2023). Therapeutic potential and nutritional significance of Ganoderma lucidum - a comprehensive review from 2010 to 2022. Food Funct. 14 (4), 1812–1838. doi:10.1039/d2fo01683d
Telke, A. A., Kalyani, D. C., Dawkar, V. V., and Govindwar, S. P. (2009). Influence of organic and inorganic compounds on oxidoreductive decolorization of sulfonated azo dye C.I. Reactive Orange 16. J. Hazard. Mater. 172 (1), 298–309. doi:10.1016/j.jhazmat.2009.07.008
Tembhekar, P., Padoley, K., Chandra, T. S., Malik, S., Sharma, A., Gupta, S., et al. (2016). Advanced oxidative pretreatment of complex effluents for biodegradability enhancement and color reduction. Environ. Waste Manag., 299–339.
Thakur, J. K., Paul, S., Dureja, P., Annapurna, K., Padaria, J. C., and Gopal, M. (2014). Degradation of sulphonated azo dye red HE7B by Bacillus sp. and elucidation of degradative pathways. Curr. Microbiol. 69 (2), 183–191. doi:10.1007/s00284-014-0571-2
Tony, B. D., Goyal, D., and Khanna, S. (2009a). Decolorization of Direct Red 28 by mixed bacterial culture in an up-flow immobilized bioreactor. J. Ind. Microbiol. Biotechnol. 36 (7), 955–960. doi:10.1007/s10295-009-0574-3
Tony, B. D., Goyal, D., and Khanna, S. (2009b). Decolorization of textile azo dyes by aerobic bacterial consortium. Int. Biodeterior. and Biodegrad. 63 (4), 462–469. doi:10.1016/j.ibiod.2009.01.003
Tripathi, M., Singh, S., Pathak, S., Kasaudhan, J., Mishra, A., Bala, S., et al. (2023). Recent strategies for the remediation of textile dyes from wastewater: a systematic review. Toxics 11 (11), 940. doi:10.3390/toxics11110940
Tuttolomondo, M. V., Alvarez, G. S., Desimone, M. F., and Diaz, L. E. (2014). Removal of azo dyes from water by sol–gel immobilized Pseudomonas sp. J. Environ. Chem. Eng. 2 (1), 131–136. doi:10.1016/j.jece.2013.12.003
Uday, U. S. P., Bandyopadhyay, T. K., and Bhunia, B. (2016). “Bioremediation and detoxification technology for treatment of dye(s) from textile effluent,” in Textile wastewater treatment (London, United Kingdom: InTech).
Urano, H., and Fukuzaki, S. (2011). The mode of action of sodium hypochlorite in the decolorization of azo dye orange II in aqueous solution. Biocontrol Sci. 16 (3), 123–126. doi:10.4265/bio.16.123
van der Zee, F. P., Bouwman, R. H. M., Strik, DPBTB, Lettinga, G., and Field, J. A. (2001). Application of redox mediators to accelerate the transformation of reactive azo dyes in anaerobic bioreactors. Biotechnol. Bioeng. 75 (6), 691–701. doi:10.1002/bit.10073
Varjani, S. J., and Upasani, V. N. (2017). A new look on factors affecting microbial degradation of petroleum hydrocarbon pollutants. Int. Biodeterior. and Biodegrad. 120, 71–83. doi:10.1016/j.ibiod.2017.02.006
Vijayaraghavan, J., Basha, S. J. S., and Jegan, J. (2013). A Review on efficacious methods to decolorize reactive azo dye. J. Urban Environ. Eng. 7 (1), 30–47. Available online at: https://www.jstor.org/stable/26203388.
Vo, T. S., Kim, Y.-S., Ngo, D.-N., and Ngo, D.-H. (2020). Myricetin from Rhodomyrtus tomentosa (Aiton) Hassk fruits attenuates inflammatory responses in histamine-exposed endothelial cells. Process Biochem. 92, 457–463. doi:10.1016/j.procbio.2020.02.004
Waghmode, T. R., Kurade, M. B., Khandare, R. V., and Govindwar, S. P. (2011). A sequential aerobic/microaerophilic decolorization of sulfonated mono azo dye Golden Yellow HER by microbial consortium GG-BL. Int. Biodeterior. and Biodegrad. 65 (7), 1024–1034. doi:10.1016/j.ibiod.2011.08.002
Wang, H., Zheng, X.-W., Su, J.-Q., Tian, Y., Xiong, X.-J., and Zheng, T.-L. (2009). Biological decolorization of the reactive dyes Reactive Black 5 by a novel isolated bacterial strain Enterobacter sp. EC3. J. Hazard. Mater. 171 (1–3), 654–659. doi:10.1016/j.jhazmat.2009.06.050
Won, S. W., Han, M. H., and Yun, Y. S. (2008). Different binding mechanisms in biosorption of reactive dyes according to their reactivity. Water Res. 42 (19), 4847–4855. doi:10.1016/j.watres.2008.09.003
Won, S. W., and Yun, Y. S. (2008). Biosorptive removal of Reactive Yellow 2 using waste biomass from lysine fermentation process. Dyes Pigments 76 (2), 502–507. doi:10.1016/j.dyepig.2006.10.011
Wu, J., Doan, H., and Upreti, S. (2008). Decolorization of aqueous textile reactive dye by ozone. Chem. Eng. J. 142 (2), 156–160. doi:10.1016/j.cej.2007.11.019
Wu, J., Eiteman, M. A., and Law, S. E. (1998). Evaluation of membrane filtration and ozonation processes for treatment of reactive-dye wastewater. J. Environ. Eng. 124 (3), 272–277. doi:10.1061/(asce)0733-9372(1998)124:3(272)
Xu, M., Guo, J., Zeng, G., Zhong, X., and Sun, G. (2006). Decolorization of anthraquinone dye by Shewanella decolorationis S12. Appl. Microbiol. Biotechnol. 71 (2), 246–251. doi:10.1007/s00253-005-0144-1
Xu, T., Lou, L., Luo, L., Cao, R., Duan, D., and Chen, Y. (2012). Effect of bamboo biochar on pentachlorophenol leachability and bioavailability in agricultural soil. Sci. Total Environ. 414, 727–731. doi:10.1016/j.scitotenv.2011.11.005
Yang, J., Monnot, M., Ercolei, L., and Moulin, P. (2020). Membrane-based processes used in municipal wastewater treatment for water reuse: state-of-the-art and performance analysis. Membranes 10 (6), 131. doi:10.3390/membranes10060131
Yanto, D. H. Y., Tachibana, S., and Itoh, K. (2014). Biodecolorization of textile dyes by immobilized enzymes in a vertical bioreactor system. Procedia Environ. Sci. 20, 235–244. doi:10.1016/j.proenv.2014.03.030
Yaseen, D. A., and Scholz, M. (2019). Textile dye wastewater characteristics and constituents of synthetic effluents: a critical review. Int. J. Environ. Sci. Technol. 16 (2), 1193–1226. doi:10.1007/s13762-018-2130-z
Yesilada, O., Birhanli, E., and Geckil, H. (2018). Bioremediation and decolorization of textile dyes by white rot fungi and laccase enzymes, 121–153.
Yousuf, R. G., and Winterburn, J. B. (2017). Waste date seed oil extract as an alternative feedstock for Poly(3-hydroxybutyrate) synthesis. Biochem. Eng. J. 127, 68–76. doi:10.1016/j.bej.2017.08.007
Yu, J. (2001). Optimal decolorization and kinetic modeling of synthetic dyes by Pseudomonas strains. Water Res. 35 (15), 3579–3586. doi:10.1016/s0043-1354(01)00100-2
Yusuf, M. (2018). Handbook of renewable materials for coloration and finishing. Handb. Renew. Mater Color Finish. Available online at: https://www.researchgate.net/publication/327812624_Handbook_of_Renewable_Materials_for_Coloration_and_Finishing.
Zafar, S., Bukhari, D. A., and Rehman, A. (2022). Azo dyes degradation by microorganisms – an efficient and sustainable approach. Saudi J. Biol. Sci. 29 (12), 103437. doi:10.1016/j.sjbs.2022.103437
Zahran, S. A., Ali-Tammam, M., Hashem, A. M., Aziz, R. K., and Ali, A. E. (2019). Azoreductase activity of dye-decolorizing bacteria isolated from the human gut microbiota. Sci. Rep. 9 (1), 5508. doi:10.1038/s41598-019-41894-8
Keywords: anthraquinone vat dyes, biodecolorization, consortium, optimization, kinetics, bioreactor
Citation: Mohanty SS and Kumar A (2025) Microbial decolorization of anthraquinone dyes: batch and continuous treatment systems - a mini-review. Front. Environ. Eng. 4:1553712. doi: 10.3389/fenve.2025.1553712
Received: 31 December 2024; Accepted: 12 May 2025;
Published: 11 June 2025.
Edited by:
Sandhya Patidar, Heriot-Watt University, United KingdomReviewed by:
Zhijie Chen, University of New South Wales, AustraliaKamila Rybczyńska-Tkaczyk, University of Life Sciences of Lublin, Poland
Copyright © 2025 Mohanty and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Swati Sambita Mohanty, c3dhdGlzYW1iaXRhQGdtYWlsLmNvbQ==
†ORCID: Swati Sambita Mohanty, orcid.org/0000-0002-2928-986X
 Swati Sambita Mohanty
Swati Sambita Mohanty Arvind Kumar
Arvind Kumar