- 1Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 2China Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 3Key Laboratory of Agro-Biodiversity and Pest Management of Education Ministry of China, Yunnan Agricultural University, Kunming, China
- 4College of Plant Protection, Yunnan Agricultural University, Kunming, China
- 5College of Biology and Environmental Sciences, Jishou University, Jishou, China
The cation diffusion facilitator (CDF) family is one of the gene families involved in metal ion uptake and transport in plants, but the understanding of the definite roles and mechanisms of most CDF genes remain limited. In the present study, we identified 18 candidate CDF genes from the turnip genome and named them BrrMTP1.1–BrrMTP12. Then, we performed a comparative genomic analysis on the phylogenetic relationships, gene structures and chromosome distributions, conserved domains, and motifs of turnip CDFs. The constructed phylogenetic tree indicated that the BrrMTPs were divided into seven groups (groups 1, 5, 6, 7, 8, 9, and 12) and formed three major clusters (Zn-CDFs, Fe/Zn-CDFs, and Mn-CDFs). Moreover, the structural characteristics of the BrrMTP members in the same group were similar but varied among groups. To investigate the potential roles of BrrMTPs in turnip, we conducted an expression analysis on all BrrMTP genes under Mg, Zn, Cu, Mn, Fe, Co, Na, and Cd stresses. Results showed that the expression levels of all BrrMTP members were induced by at least one metal ion, indicating that these genes may be related to the tolerance or transport of those metal ions. Based on the roles of different metal ions for plants, we hypothesized that BrrMTP genes are possibly involved in heavy metal accumulation and tolerance to salt stress apart from their roles in the maintenance of mineral nutrient homeostasis in turnip. These findings are helpful to understand the roles of MTPs in plants and provide preliminary information for the study of the functions of BrrMTP genes.
Introduction
Normal plant growth requires major and trace elements containing metallic elements, such as K, Ca, Mg, Fe, Na, Cu, Zn, Mn, Ni, and Co, which play various roles in plants. However, these trace elements are highly toxic at excessive amounts. Meanwhile, several non-essential and toxic metal ions, including Cd, Pb, and Hg, have the same chemical structures as some of these trace elements and are thus absorbed by plants (Kim et al., 2004). Salt stress and heavy metal toxicity commonly occur because of the excessive accumulation of various metal ions. Conversely, trace metal element deficiency in poor soils decreases agricultural yield and intake of trace elements in the human body. Therefore, precise metallic element homeostasis is essential in plants (Kim et al., 2004). Several gene families participate in the uptake and transport of metal ions by plants (Lang et al., 2011; Zhang et al., 2013). These metal ion transporters include the cation diffusion facilitator (CDF) family, Zrt/Irt-like protein, P-type ATPase, and ATP-binding cassette transporter, etc., (Zhang et al., 2013).
The CDF family, also called the cation efflux family (Lang et al., 2011), was first described in 1995 (Nies and Silver, 1995). The protein members of the CDF family have been identified in all six kingdoms of living things (Migocka et al., 2015a) and are involved in transport and tolerance to trace elements (Kim et al., 2004). CDF family members have three distinct features, namely, an N-terminal signature sequence, cation efflux domain, and approximately six predicted transmembrane regions (Paulsen and Saier, 1997). Eukaryotic CDFs usually contain a histidine (His)-rich cytoplasmic loop between transmembrane domains 4 and 5 (Paulsen and Saier, 1997). CDF members are classified into three major clusters, namely, Zn-CDFs, Fe/Zn-CDFs, and Mn-CDFs, which differ with respect to selectivity toward the main transported metal ions (Montanini et al., 2007). In plants, CDF members are designated as metal tolerance proteins (MTPs; Migocka et al., 2015b). The MTP family has 12 and 10 members in model plants Arabidopsis and rice, respectively. Unfortunately, their functions remain finitely understood (Migocka et al., 2015b). Recently, MTP proteins from different plant species have been divided into seven groups according to the results of the phylogenetic analysis and annotation performed on Arabidopsis MTPs (Gustin et al., 2011). Among them, groups 1, 5, and 12 belong to the Zn-CDFs, groups 6 and 7 form the Fe/Zn-CDFs, and the Mn-CDF cluster comprises groups 8 and 9 (Gustin et al., 2011). To date, the most studied MTP proteins in plants include the MTP1–MTP4 members of group 1, MTP8 proteins of group 8, and MTP9–MTP11 of group 9 (Migocka et al., 2015b). Most of these MTPs were functionally characterized in diverse plant species. Although group 1 of plant MTPs belongs to Zn-CDFs, its members is capable of transporting different metals, including Zn, Cd, Co, Ni, or Fe, into the vacuole of plant cells (Kim et al., 2004; Xu et al., 2009; Lang et al., 2011; Menguer et al., 2013; Migocka et al., 2015a). Recently, MTP5 has been reported to form a functional complex with MTP12 to transport Zn into the Golgi apparatus in Arabidopsis (Fujiwara et al., 2015). Group 8, comprising only MTP8 proteins, is implicated in Mn homeostasis, which requires a proton gradient when transporting Mn2+ across membranes (Migocka et al., 2015b; Eroglu et al., 2017; Li Q.H. et al., 2017). They are necessary for the active sequestration of excess Mn in intracellular organelles or Mn delivery to Mn-dependent enzymes (Migocka et al., 2015b). Meanwhile, group 9 comprises MTP9–MTP11 proteins (Montanini et al., 2007; Gustin et al., 2011), and MTP 9 and MTP 10 are more closely related and thus form an internal separate clade. This relationship indicates some functional differentiation between MTP9/10 and MTP11 proteins (Gustin et al., 2011). Several MTP11 transporters from different species confer Mn2+ tolerance (Delhaize et al., 2007; Zhang and Liu, 2017), whereas limited evidence of the involvement of MTP9/10 proteins in Mn transport or tolerance to Mn is currently available (Migocka et al., 2015b), though they are also classified as Mn-CDFs. Current studies generally indicated that MTP proteins contribute to the detoxification of heavy metals and promote their accumulation (Kim et al., 2004; Delhaize et al., 2007; Xu et al., 2009; Lang et al., 2011; Menguer et al., 2013; Migocka et al., 2015a,b; Eroglu et al., 2017; Li Q.H. et al., 2017; Zhang and Liu, 2017). The functions of MTPs or other metal transporters in heavy metal hyperaccumultors may improve phytoremediation efficiency (Kim et al., 2004; Gustin et al., 2009). However, knowledge on the functional assignment of plant MTPs remains insufficient. For example, the functional characteristics of groups 6 and 7 have not been elucidated so far.
The Brassicaceae species has shown apparent advantages in the exploration of metal ion transporter functions. These species include the model plant Arabidopsis and many important vegetables and oilseed crops with easily available genomes (Wang et al., 2011; Liu et al., 2014; Kolaj-Robin et al., 2015; Gan et al., 2016; Yang et al., 2016), and have represented a differential range of tolerance to two important abiotic stresses, namely, salinity and heavy metals (Assunção et al., 2003; Prasad and Freitas, 2003; Inan et al., 2004; Megdiche et al., 2007). For example, over half of the identified Ni hyperaccumulators (more than 300 species) were reportedly from Brassicaceae (Assunção et al., 2003). Turnip (Brassica rapa var. rapa), a Brassicaceae biennial plant, has been widely cultivated in Europe, Asia, and America as a vegetable or fodder. In Asia, one of the cultivation centers of turnip is in the Tibetan Plateau and its surrounding countries and regions. Moreover, turnip has been cultivated at the maximum elevation of 4700 m in Nyima County, China. Thus, this species is faced with diverse soil conditions, indicating that turnips from different populations or cultivated landraces may possess various ion absorption or accumulation characteristics to adapt to the corresponding environments. Turnips from Xinjiang, China, reportedly contain abundant mineral elements (Ma et al., 2016), indicating its eminent absorption ability for metal ions. Additionally, turnip has been classified as a high-Cd accumulation plant (Arthur et al., 2000; Li et al., 2016). We found that turnip landraces from China have strong capacities for Cd accumulation, and several landraces were Cd hyperaccumulators (Li et al., 2016, Li X. et al., 2017). To explore the molecular mechanism of metal ion accumulation in turnip, we focused on several important ion transporters based on the sequencing results of turnip (Cheng et al., 2016). In the present study, we systematically analyzed the sequence and structural characteristics of putative MTPs in turnip and preliminarily investigated the potential roles of each BrrMTP member. This study is expected to improve the understanding of the functions of plant MTPs and provide a basis for the analysis of the functions and mechanisms of BrrMTP proteins.
Materials and Methods
Identification and Phylogenetic Analysis of the MTPs in Turnip
The gene sequences of 12 MTPs in Arabidopsis were downloaded from NCBI1 as queries to search against the turnip genome2. The domains and functional sites in each protein were examined with InterProScan3 (Finn et al., 2017). All protein sequences containing any of the typical domains of MTP proteins were extracted as candidates. The candidates were then used to search against the GenBank non-redundant protein database. The ClustalW software was used for the sequence alignment between turnip and Arabidopsis, and phylogenetic trees were constructed using the MEGA 7.0 software. The neighbor-joining method was performed and 1000 bootstrap test replicates were used during the construction (Kumar et al., 2016).
Gene Structure and Location on Chromosomes of Turnip MTP Genes
The diagram of the intron/exon structures of BrrMTP genes was analyzed by using the online Gene Structure Display Server4 (Hu et al., 2015). The chromosomal location of the BrrMTP genes was mapped according to the gene position information using the TBtools5.
Structure Characteristics and Physicochemical Parameters of Turnip MTP Proteins
The Pfam tool6 and the MEME7 program were used to search for conserved domains and motifs in the BrrMTP protein sequences, respectively (Sonnhammer et al., 1998; Bailey et al., 2006); subsequently, the domain and motif diagrams were drawn with the TBtools software. The putative transmembrane regions in proteins were predicted by using the TMHMM Server V. 2.08. The physicochemical parameters of the proteins, including molecular weight (MW), theoretical isoelectric point (pI), and grand average of hydropathicity (GRAVY), were calculated with the ProtParam tool of ExPaSy9 (Artimo et al., 2012).
Plant Growth and Stress Treatments
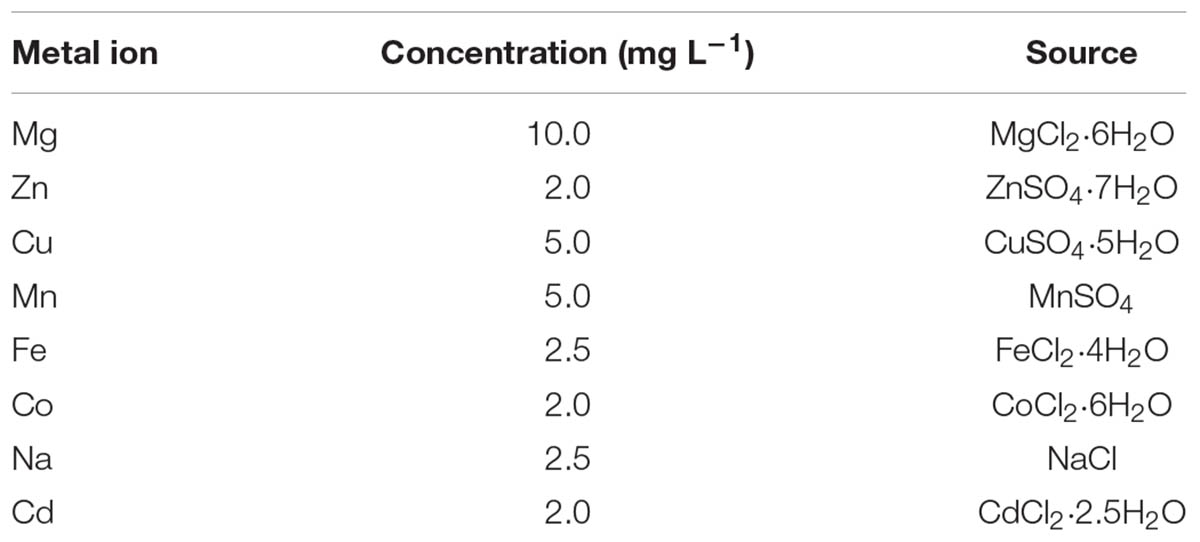
Turnip seeds were sown in soil pots under the natural condition. After 2 weeks of growth, the turnip seedlings were transplanted into uniform pots (d = 9.4 cm, h = 8 cm) with uniform soil (one seedling in a pot) under greenhouse condition (22–25°C, 12-h light/12-h darkness, 50–60% relative humidity). A total of 3 weeks later, the plants were irrigated with eight metal ion solutions. The plant irrigated with water only was used as the control. Each pot was irrigated with a 100 mL solution, and the pots were placed in culture dishes. The ion sources and concentrations are shown in Table 1. The roots and leaves of each treatment were harvested separately, ground to powder and then stored at –80°C for RNA isolation. Three biological replicates were made for each treatment.
RNA Extraction and cDNA Synthesis
The total RNA samples were isolated using the Eastep® Super Total RNA Extraction Kit (Promega, Madison, WI, United States). The RNA concentration was determined by NanoDrop1000 (NanoDrop Technologies, Inc.), with the integrity checked on 0.8% agarose gel. A total of 3 μg of RNA was reverse-transcribed using the GoScript Reverse Transcription System (Promega, Madison, WI, United States) to generate the cDNA.
Quantitative Real-Time PCR (qRT-PCR) Analysis
Optimal forward and reverse primers were designed (Supplementary Table S1) through the online tool Primer-BLAST10 for qRT-PCR analysis. qRT-PCR was conducted in triplicate with different cDNAs from different tissues and treatments. FastStart Universal SYBR Green Master (Rox, Roche, Indianapolis, IN, United States) and a 7500 Sequence Detection System (Applied Biosystems, United States) were used. The reaction parameters for thermal cycling were as follows: 95°C for 10 min, followed by 40 cycles of 94°C for 5 s, 60°C for 15 s and 72°C for 34 s. The turnip beta-tubulin gene was amplified as an internal control. The relative gene expression levels were obtained by dividing the extrapolated transcript levels of the target genes by the levels of the internal control from the same sample.
Statistical Analysis
Statistical analyses were performed using SPSS version 18.0. One-way ANOVA or independent-samples t-test was conducted to analyze significant differences among multiple samples or between each pair of samples at a 0.05 level, respectively.
Results
Identification and Phylogeny of the MTPs in Turnip
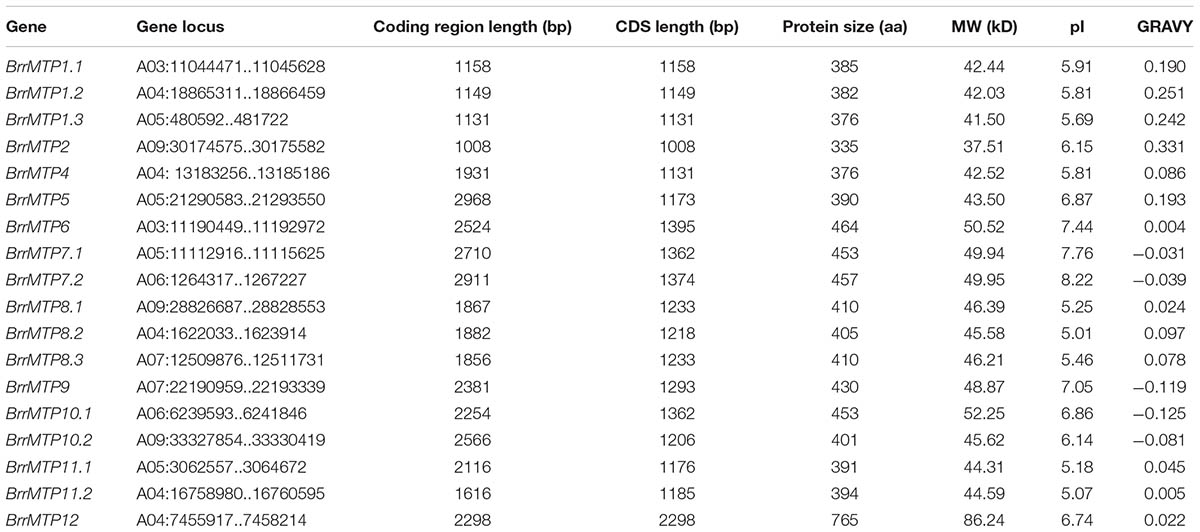
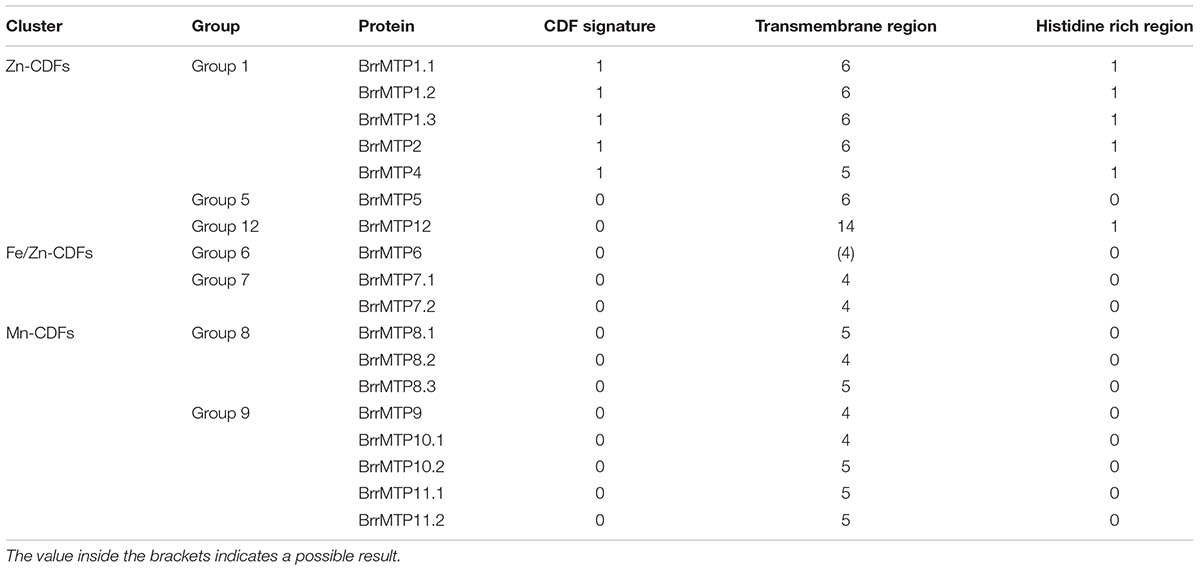
We identified 18 MTPs in turnip on the basis of the published genome result (Table 2). The MTP family in turnip was more expanded than that in Arabidopsis. The complete coding region, CDS, and protein sequences of all turnip MTPs were identified for subsequent information analysis. To gain insights into the phylogenetic relationship of the MTPs between turnip and Arabidopsis, we used the MTP family protein sequences to build a phylogenetic tree. According to the orthologous relationships, the turnip MTPs were designated as BrrMTP1.1 to BrrMTP12 (Figure 1). Interestingly, we found multiple homologous AtMTP1, AtMTP7, AtMTP8, AtMTP10, and AtMTP11 genes in the turnips, whereas no AtMTP3 orthologous gene was found (Figure 1). Accordingly, the 18 BrrMTPs were divided into seven groups (i.e., groups 1, 5, 6, 7, 8, 9, and 12), which formed the three clusters of the phylogenetic tree, namely, Zn-CDFs, Fe/Zn-CDFs, and Mn-CDFs.
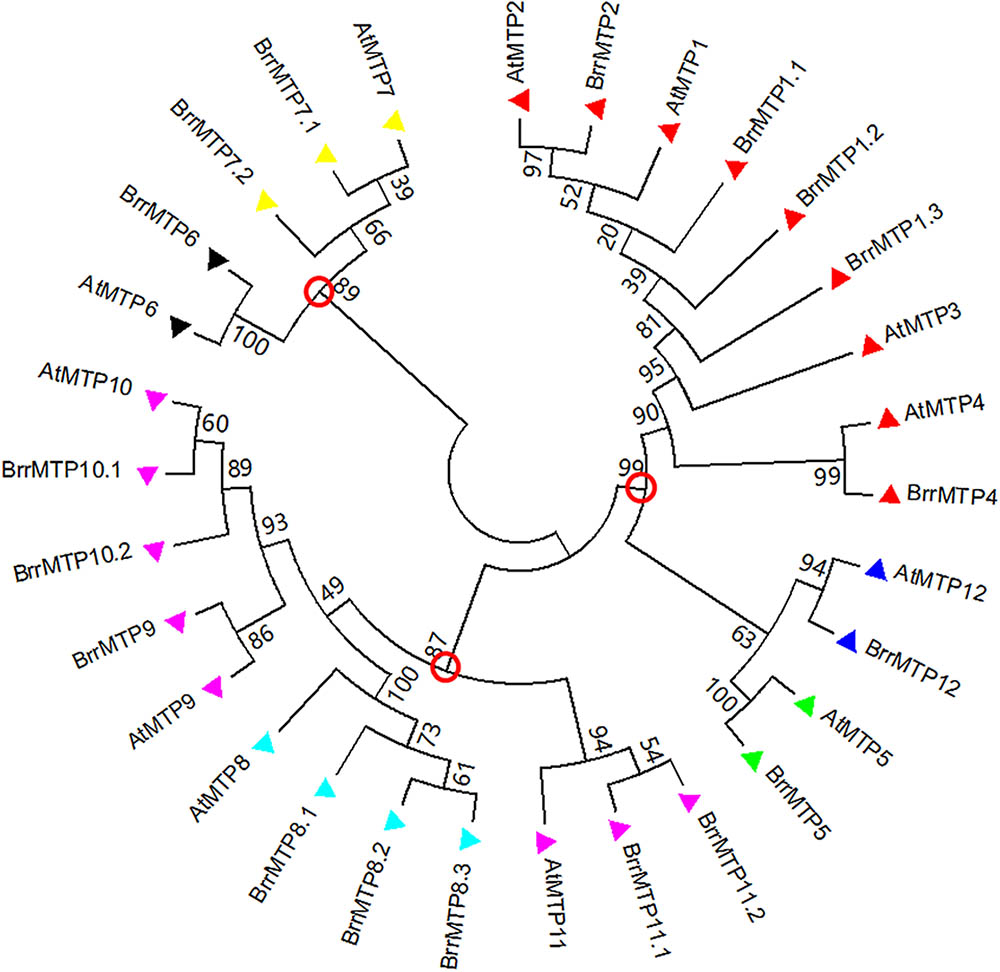
FIGURE 1. Phylogenetic relationship of MTP proteins in turnip and Arabidopsis. The protein sequences were aligned by ClustalW, and phylogenetic trees were constructed using the MEGA 7.0 software with the neighbor-joining method and 1000 bootstrap test. The red circles labeled on the branch roots indicate the divided three clusters of MTP proteins. Triangles with different colors represent the seven groups of MTP proteins (red: group 1; green: group 5; black: group 6; yellow: group 7; light blue: group 8; pink: group 9; blue: group 12).
Gene Structure and Location of Turnip MTP Genes
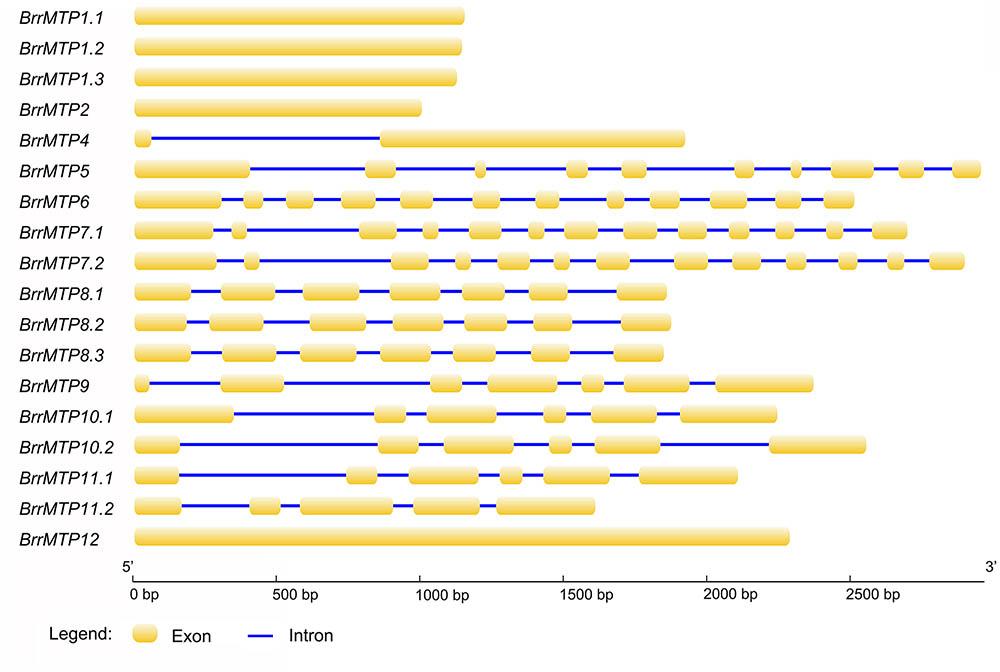
The lengths of the coding region sequences (including exons and introns) of the turnip MTP genes ranged from 1008 bps (BrrMTP2) to 2968 bps (BrrMTP5) (Table 2), and their CDS sequences included 1008–2298 bps (Table 2), which encoded 335–765 amino acids (Table 2). The comparative analysis between the CDS and the gene sequences indicated that the BrrMTPs of the three clusters contained differential introns (Figure 2). The Zn-CDFs contained 0 or 1 intron, except for BrrMTP5 (11), and the Fe/Zn-CDFs included 11 or 12 introns, whereas the members of Mn-CDFs had 4–6 introns (Figure 2).
The chromosomal location showed that the 18 BrrMTP genes were located in 6 out of the 10 turnip chromosomes (Figure 3). Among them, chromosome A04 contained the maximum number of 5 BrrMTP genes (Figure 3). Chromosomes A03, A05, A06, A07, and A09 contained 2–4 BrrMTPs, whereas chromosomes A01, A02, A08, and A10 carried no BrrMTP genes (Figure 3).
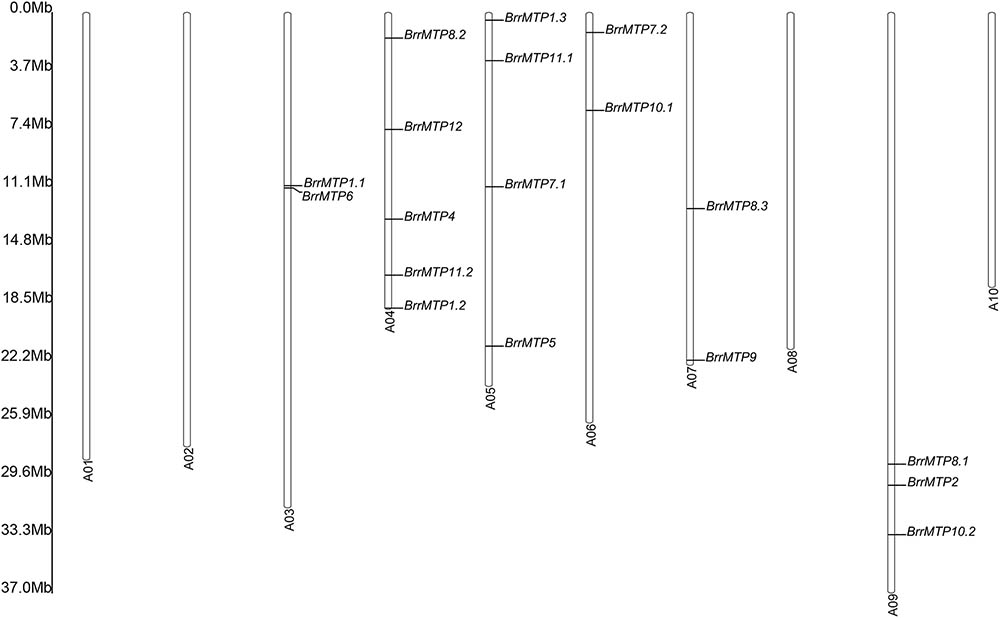
FIGURE 3. Distribution of the MTP genes on turnip chromosomes. A01∼A10 represent the chromosome No, and the rule on the left indicates the physical map distance among genes (Mb).
Structure Characteristics and Physicochemical Parameters of Turnip MTP Proteins
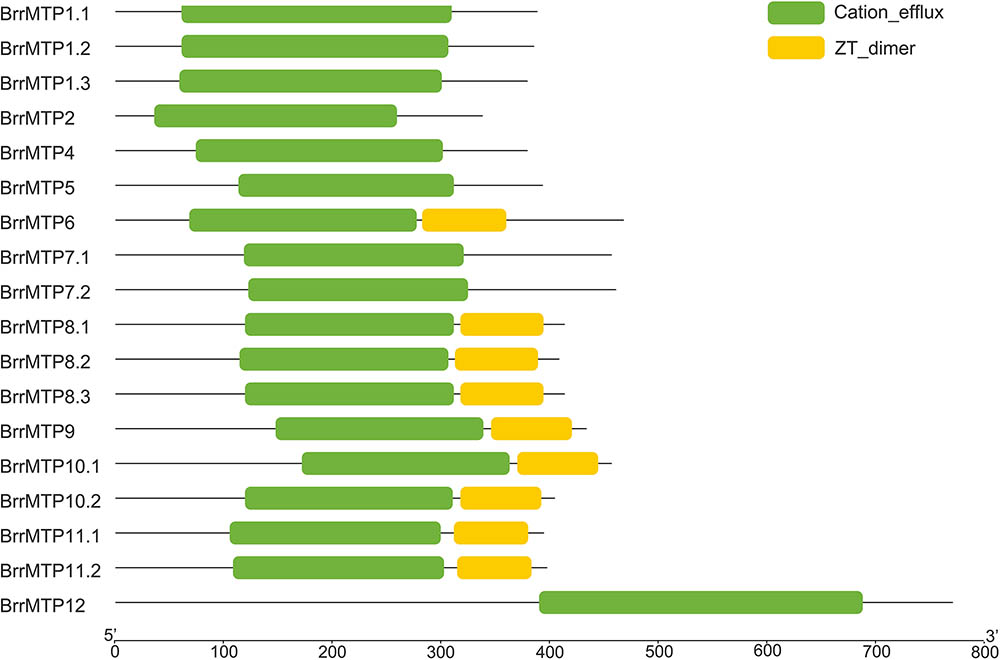
Protein structure analysis showed that all the BrrMTPs contained the cation efflux domain, while the members of groups 6, 8, and 9 had a common ZT dimer structure (Figure 4). Other features of MTP proteins were also observed in different members of BrrMTPs. The CDF signature sequence was observed in the N-terminals of the group 1 BrrMTPs (Table 3 and Supplementary Figure S1). Most BrrMTP proteins contained 4–6 typical transmembrane regions, except for BrrMTP12, which contained 14, and BrrMTP6, which possibly contained 4 (Table 3). Additionally, the BrrMTP members of groups 1 and 12 contained a His-rich region (Table 3 and Supplementary Figure S1). We further analyzed the diversity of the conserved motif compositions in BrrMTPs using the MEME program. A total of 20 conserved motifs, designated as motifs 1–20, were identified within the proteins (Supplementary Figure S2). The members of the same cluster (or the same group) generally contained similar motifs (Supplementary Figure S2). Each cluster included several relatively specific motifs, while motif 4 was shared by all BrrMTPs, except for BrrMTP 5, which only contained two instances of motif 8 (Supplementary Figure S2). Most BrrMTPs contained one or two duplicates of the same motif, whereas BrrMTP12 had five duplicates of motif 15 (Supplementary Figure S2).
The physicochemical parameters, including the MW, pI, and GRAVY values, of the BrrMTP proteins were predicted as shown in Table 2. The MW values ranged from 37.51 kD (BrrMTP2) to 86.24 kD (BrrMTP12), most of which were within 40–50 kD (Table 2). Nearly all the pI values of all BrrMTPs of Zn-CDFs and Mn-CDFs were lower than 7.0, whereas those of the members of Fe/Zn-CDFs were higher than 7.0 (Table 2). The GRAVY results, ranging from -0.119 (BrrMTP9) to 0.331 (BrrMTP2), indicated that the BrrMTPs generally had weak hydrophilicity (Table 2).
Expression Profiles of Turnip MTP Genes in Different Tissues
The qRT-PCR results indicated that the BrrMTP genes had differential tissue expression patterns at the seedling stage in soil. We found that the expressions of BrrMTP1.1, BrrMTP1.2, BrrMTP1.3, BrrMTP2, BrrMTP5, BrrMTP6, BrrMTP7.1, BrrMTP9, BrrMTP11.1, and BrrMTP11.2 in the turnip leaves were much higher than those in the roots (P < 0.05), whereas the expressions of BrrMTP8.1, BrrMTP8.2, BrrMTP8.3, and BrrMTP10.2 showed the opposite results (Figure 5). BrrMTP4, BrrMTP10.1, and BrrMTP12 showed similar expression levels between roots and leaves (Figure 5). Interestingly, the homologous genes broadly showed similar tissue expression characteristics (Figure 5).
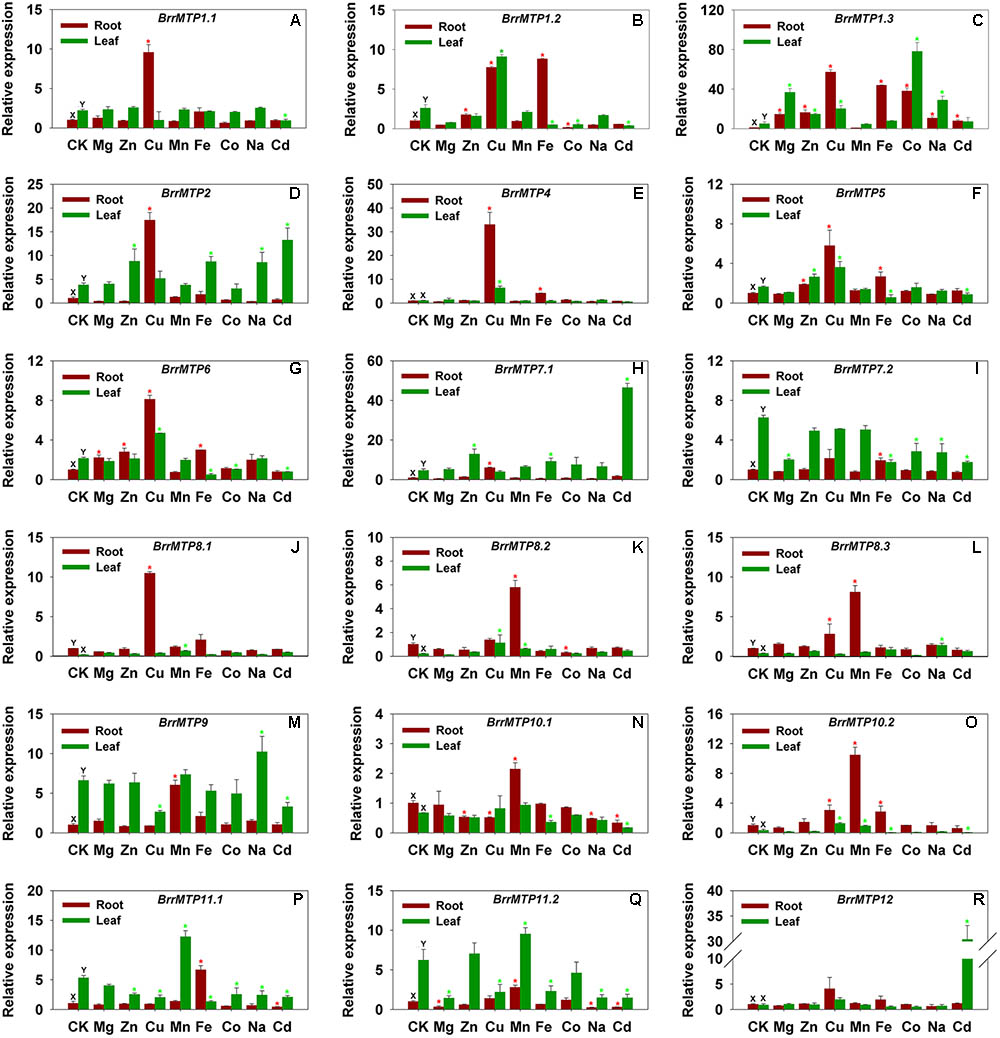
FIGURE 5. Relative expression levels of turnip MTP genes under various metal ion stresses in root or leaf. Data represent means ± SD (A–R). CK represents control samples (A–R). Different letters (X and Y) indicate significant differences between root and leaf under normal condition (n = 3, P < 0.05) (A–R). Red and green asterisks indicate significant differences between the treatment samples and the control samples in root and leaf, respectively (n = 3, P < 0.05) (A–R).
Expression Differences of Turnip MTP Genes Under Diverse Metal Ion Stresses
To explore the potential roles of the BrrMTP genes, we analyzed expression changes in the entire MTP family of turnips in the presence of different metal ions, including macroelements (e.g., Mg), microelements (e.g., Zn, Cu, Mn, Fe, Co, and Na) and non-essential elements (e.g., Cd). The BrrMTP genes showed various responses to the same metal ion either in the root or leaf (Figure 5), and the expression of a certain gene differentially changed under different metal treatments (Figure 5). We summarized the metal ions that significantly induce or inhibit the expression of each BrrMTP gene in either root or leaf (P < 0.05). Each metal ion used in the experiment induced the expression levels of one or several BrrMTP genes in the root or leaf (Figure 5 and Table 4). In detail, Mg induced the expression levels of BrrMTP1.3 and BrrMTP6 (Figure 5 and Table 4); Zn induced the expression levels of BrrMTP1.2, BrrMTP1.3, BrrMTP2, BrrMTP5, BrrMTP6, and BrrMTP7.1 (Figure 5 and Table 4); Cu induced the expression levels of BrrMTP1.1, BrrMTP1.2, BrrMTP1.3, BrrMTP2, BrrMTP4, BrrMTP5, BrrMTP6, BrrMTP7.1, BrrMTP8.1, BrrMTP8.3, and BrrMTP10.2 (Figure 5 and Table 4); Mn induced the expression levels of all Mn-CDFs (BrrMTP8.1–BrrMTP11.2; Figure 5 and Table 4); Fe induced the expression levels of BrrMTP1.2, BrrMTP1.3, BrrMTP2, BrrMTP4, BrrMTP5, BrrMTP6, BrrMTP7.1, BrrMTP7.2, BrrMTP10.2, and BrrMTP11.1 (Figure 5 and Table 4); Co only induced the expression of BrrMTP1.3 (Figure 5 and Table 4); Na induced the expression levels of BrrMTP1.3, BrrMTP2, BrrMTP8.3, and BrrMTP9 (Figure 5 and Table 4); and Cd induced the expression levels BrrMTP1.3, BrrMTP2, BrrMTP7.1, and BrrMTP12 (Figure 5 and Table 4). Meanwhile, the BrrMTP genes showed upregulated expressions under at least one metal ion treatment (Figure 5 and Table 4). In particular, the expression of BrrMTP 1.3 was upregulated under seven metal ion treatments (Figure 5 and Table 4). On the contrary, the expression levels of BrrMTP1.1, BrrMTP1.2, BrrMTP5, BrrMTP6, BrrMTP7.2, BrrMTP8.2, BrrMTP9, BrrMTP10.1, BrrMTP10.2, BrrMTP11.1, and BrrMTP11.2 were downregulated under a single or several metal ion treatments in the roots or leaves and were especially inhibited by Cd (Figure 5 and Table 4). Generally, although the genes of the same group, even the homologous genes, did not show obviously similar expression changes to the same metal ion, the same cluster BrrMTP genes showed some common characteristics (Figure 5 and Table 4). For example, the members of Zn-CDFs showed a common positive response to several metals, including Zn, Cu, Fe, and Cd, whereas the Mn-CDF genes were mainly induced by the Mn ion but widely inhibited by other metal ions (Figure 5 and Table 4).
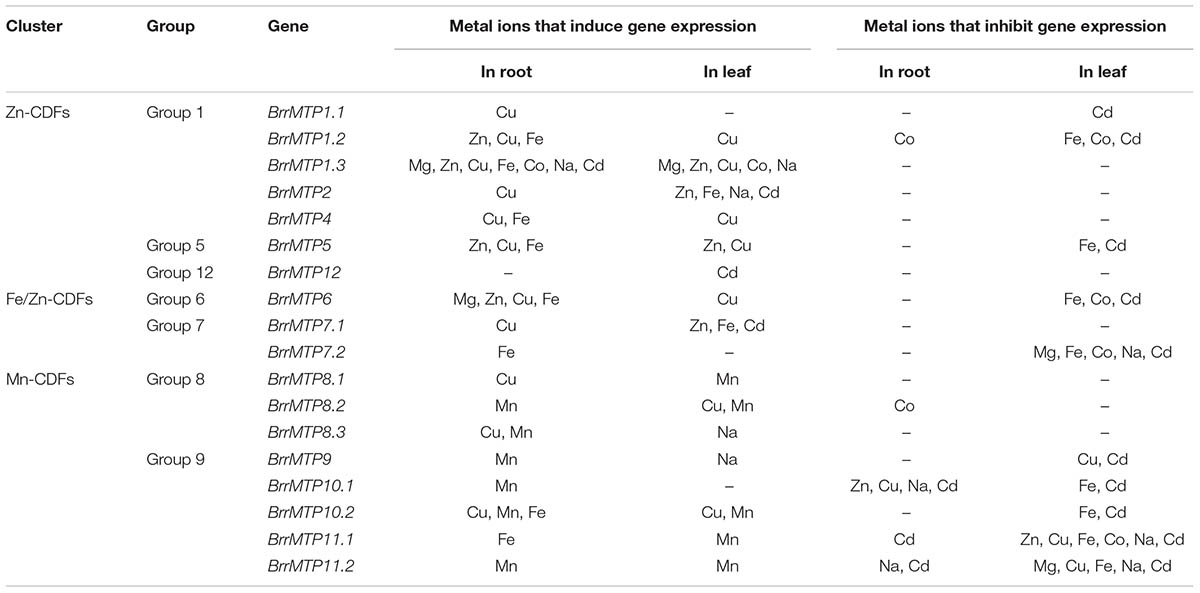
TABLE 4. Summary of the metal ions that either induce or inhibit the expressions of the turnip MTP genes.
Discussion
Although increasing functional studies on plant MTP members have been reported in diverse species, such as Arabidopsis thaliana (Desbrosses-Fonrouge et al., 2005; Delhaize et al., 2007; Eroglu et al., 2016, 2017), Oryza sativa (Menguer et al., 2013; Zhang and Liu, 2017), Brassica juncea (Xu et al., 2009; Lang et al., 2011), Cucumis sativus (Migocka et al., 2015a,b), Thlaspi goesingense (Kim et al., 2004), and Camellia sinensis (Li Q.H. et al., 2017), the understanding of the roles of the MTP family is still significantly limited. On the basis of the increasing genome information, analyzing gene families via comparative genomics is an efficient method for modern functional genomics research (Xu et al., 2016). However, unlike many other gene families, including some transcription factors and kinases (Iftikhar et al., 2017; Ma et al., 2017; Xi et al., 2017), studies on the characteristics and functional analysis of the plant MTP gene family are just reported in few species recently (Fu et al., 2017; Vatansever et al., 2017). As MTP genes have been demonstrated to participate in tolerating and transporting various heavy metals, including plant trace elements and non-essential elements (Kim et al., 2004; Delhaize et al., 2007; Xu et al., 2009; Lang et al., 2011), they might thus play significant roles in plant mineral nutrition maintenance and resistance to stresses caused by metals. Turnip has shown relatively high capacities toward absorbing trace elements and heavy metal Cd (Li et al., 2016; Ma et al., 2016). Thus, exploring the functional characteristics of the metal ion transporters in turnip is of great interest. In the present study, 18 MTP family members in turnip were successfully identified by bioinformatics analysis. These 18 BrrMTPs were located at different positions on 6 chromosomes and were unevenly distributed within the genome. Compared with Arabidopsis, gene family expansion occurred in the BrrMTP gene family. This might be because the polyploidization events occurred in the evolutionary history of Brassica rapa (Yin et al., 2017). These events were followed by chromosomal reduction and rearrangement and numerous gene losses (Yin et al., 2017). In the present results, MTP3 was not observed in turnip; this was likely because of gene loss. However, the incomplete genomes factor was not excluded. The increase in the number of MTP genes during plant evolution was likely related to the functional evolution of metal tolerance and accumulation.
The phylogenetic relationship indicated that BrrMTPs could be divided into three clusters and seven groups as in Arabidopsis (Montanini et al., 2007; Kolaj-Robin et al., 2015). Clusters Zn-CDF, Fe/Zn-CDF, and Mn-CDF contained 7, 3, and 8 members, respectively. This classification was supported by the subsequent analysis of gene structure and protein features. Although the coding regions of most BrrMTPs (except BrrMTP12) encoded polypeptides of 335–464 amino acids in length, the intron numbers of BrrMTPs had significant differences among different groups, ranging from 0 to 12. Variations in intron lengths also existed in the BrrMTPs. However, their functions and evolutionary process require further verification. Like the protein size, the MW, pI, and GRAVY of most BrrMTPs were relatively conserved or similar but closer within groups. Particularly, BrrMTP12 possessed significantly larger protein size (765 amino acids) and MW (86.24 kD) than the other BrrMTP members. This result was consistent with the difference between AtMTP12 and other AtMTPs, indicating the distinctive function and evolutionary process of MTP12. Undoubtedly, all the BrrMTPs contained the conserved cation efflux domain in their sequences. Moreover, a zinc transporter dimerization (ZT dimer) domain was detected in the members of groups 6, 8, and 9, but whether this domain is correlated with the functions of these BrrMTP is unknown. The CDF signature sequence, transmembrane region and His-rich loop are three significant structural features of MTP proteins (Montanini et al., 2007; Kolaj-Robin et al., 2015), which are closely related to their functional characteristics. However, these features showed obvious differences among different groups (or clusters). The complete CDF signature sequences were only observed in the BrrMTPs of group 1. The His-rich loop in the MTP protein sequences has been considered responsible for metal selectivity (Kang and Carey, 1999; Podar et al., 2012; Kolaj-Robin et al., 2015). In the present study, groups 1 and 12 from the Zn-CDF cluster possessed individual His-rich sequences. However, BrrMTP1.1–BrrMTP4 included 17–40 amino acid residues in length, whereas the sequence length of BrrMTP12 was 115 amino acid residues. The results might indicate the differential abilities of different BrrMTPs to transport metal ions. Unlike the above two features, most BrrMTPs contained 4–6 conserved transmembrane regions, except for BrrMTP12, which possessed 14 ones, similar to 15 ones in AtMTP12 (Zhang et al., 2013). Generally, these results were consistent with the structure characteristics of AtMTPs. In Arabidopsis, AtMTP5–AtMTP12 proteins only contained one of the features of the MTP family, such as the transmembrane domains, whereas AtMTP1–AtMTP4 included all the MTP features (Paulsen and Saier, 1997; Zhang et al., 2013). However, three different clusters were divided by the researchers, which included AtMTP6–AtMTP11 (cluster I), AtMTP5 and AtMTP12 (cluster II), and AtMTP1–AtMTP4 (cluster III) (Paulsen and Saier, 1997; Zhang et al., 2013). The results of motif compositions also showed the structural similarity within the BrrMTP groups and the particularity among different groups, indicating the possible functional diversity of the entire family of BrrMTPs.
To date, members of plant MTPs have been reported to be involved in tolerating and transporting different metals, including Zn, Cd, Co, Ni, Fe, and Mn (Kim et al., 2004; Xu et al., 2009; Lang et al., 2011; Menguer et al., 2013; Migocka et al., 2015a,b; Eroglu et al., 2017; Li Q.H. et al., 2017; Zhang and Liu, 2017). Thus, these transporters have shown potential applications in the phytoremediation of heavy metal-polluted soils, especially the members from heavy metal hyperaccumulators (Kim et al., 2004; Gustin et al., 2009). Eroglu et al. (2016) recently has reported that MTP8 in Arabidopsis determines tolerance to iron deficiency-induced chlorosis; this study provides a new insight into roles of MTP proteins in plants. However, whether the MTP family contributes to the plant response toward salt stress is unclear. Moreover, whether the different members have function division or cooperation when transporting various metal ions remains undefined. To expand the information for these issues, we predicted the potential substrate metal ions of each member of the BrrMTPs by performing gene-induced expression. We primarily examined the spatial expression patterns of the BrrMTP genes under normal nutrient conditions. The results showed that all members of groups 1, 5, 6, and 7 and several members of group 9 (BrrMTP9, BrrMTP11.1 and BrrMTP11.2) were mainly expressed in the leaves, whereas most of the others were expressed in the roots, indicating that different BrrMTP members play specific roles or run different mechanisms in turnips. The results are partly consistent with the reports regarding their individual homologous genes (Lang et al., 2011; Chen et al., 2013; Vatansever et al., 2017). This might be related to the special nutrient transport during growth and development of turnips, which form fleshy roots. To investigate the potential roles of BrrMTP members, we detected the expression dynamics of the BrrMTP genes under eight metal ion treatments. These metal ions include a macroelement (Mg), microelement (Zn, Cu, Mn, Fe, Co, or Na) or non-essential element (Cd) for plant growth and also represent heavy metals (e.g., Zn, Cu, Mn, and Cd) or salt ions (e.g., Mg and Na). The results showed that each metal ion induced the expression levels of at least one BrrMTP gene in the roots and leaves, indicating that the BrrMTPs may be involved in the tolerance or transport of corresponding macroelements and microelements. In that case, the BrrMTPs are of significance for turnip plants to maintain mineral element balance under normal condition or to improve tolerance to heavy metals and salts when faced with stresses. Cd is a non-essential element to plants, and plants do not possess specific mechanisms for the uptake of Cd2+, but Cd can be absorbed and transported through some of the carriers used for the uptake of essential metals for plant development (Li L.Z. et al., 2017). We found that BrrMTP1.3, BrrMTP2, BrrMTP7.1, and BrrMTP12 were increasingly expressed when treated by Cd, indicating that these genes may be related to the high-Cd accumulation characteristics of turnip (Li et al., 2016). Interestingly, our results are consistent with those of previous studies wherein the MTPs of group 1 were reported to transport multiple metal ions, whereas the members of group 8 or 9 showed a relative specificity to Mn (Kim et al., 2004). However, our results possibly indicated that the MTP genes of turnip have extended functions during the evolution or domestication process. For instance, we found that BrrMTP1.3, an expansion member of group 1 in turnip, showed a positive response (upregulated expression) to seven metal ions in contrast to Arabidopsis. We also found that several metal ions induced the expression levels of groups 6 and 7 in the roots and leaves. Given that the functions of these MTP genes are rarely reported, this study provided a basis for their functional investigation. Additionally, although Cu has not been identified as substrates for MTP transporters even in model plants, we found that Cu induced the expression levels of most BrrMTP genes. The results are supported by a recent study in sweet orange (Fu et al., 2017); thus these findings provide valuable information for future studies on the role of MTPs in Cu detoxification. Nevertheless, we also found that some BrrMTP genes responded to Mg and Na, which have not been used as substrates for MTP transporters in plants either (Kolaj-Robin et al., 2015). Thus, whether our results are related to the special evolution, domestication, or adaptation process of turnips remains unknown. The reasons and mechanisms for these questions need further studies.
Overall, the expression responses to common metal ions of all identified MTP family genes in turnip were explored for the first time in the present study. Our results were partly consistent with those of previous studies (Zhang et al., 2013; Fu et al., 2017). Thus, we provided some original information for the subsequent research of several MTP members that have been seldom studied. However, it is incredible to predict the substrate ions for transporters only from the gene expression response, as ion interaction (e.g., antagonism and synergism) or some physiological or metabolic processes affected by certain ion may indirectly cause changes in other ions and the expression dynamic of their corresponding transporters. Thus, concrete gene function studies by modern molecular biology technology are urgently required for each BrrMTP gene member.
Conclusion
We identified 18 candidate CDF genes from the turnip genome and presented a comparative genomic analysis of CDFs (or MTPs) in turnip, particularly their phylogenetic relationships, gene structures and chromosome distributions, conserved domains, and motifs. The CDF gene family in turnip generated expansions in MTP1, MTP7, MTP8, MTP10, and MTP11 in contrast to those of A. thaliana, although MTP3 might be lost during the evolutionary process. Based on the constructed phylogenetic tree, we divided the BrrMTPs into seven groups (groups 1, 5, 6, 7, 8, 9, and 12), which formed three major clusters (Zn-CDFs, Fe/Zn-CDFs, and Mn-CDFs). The BrrMTP proteins showed similar structural characteristics within groups but significant differences among different groups. Afterward, we performed an expression analysis on all BrrMTP members under eight metal treatments (Mg, Zn, Cu, Mn, Fe, Co, Na, and Cd) to investigate the potential roles of BrrMTPs in turnip. The results showed that the expression levels of all BrrMTP genes can be induced by at least one metal ion, indicating that these genes may be related to the tolerance or and transport of corresponding metal ions. We found first-hand that two metal ions, namely, Mg and Na, significantly induce the expression levels of several BrrMTP genes. According to the different roles of these metal ions for plants, we hypothesized that the BrrMTP family genes are possibly involved in heavy metal accumulation and tolerance to salt stress apart from their role in the maintenance of mineral nutrient homeostasis in turnip. However, these conclusions need further verification by concrete gene function analysis.
Author Contributions
YpY and YhY conceived and designed the experiments. XL, YW, and WH analyzed the data. YW, XL, and BL performed the experiments. XL wrote the manuscript.
Funding
This work was financially supported by the Western Youth Project B of the “Light of West China” Program of Chinese Academy of Sciences (Grant Number Y7260411W1) and the National Natural Science Foundation of China (Grant Number 31590823).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00103/full#supplementary-material
Abbreviations
CDF, cation diffusion facilitator; GRAVY, grand average of hydropathicity; His, histidine; MTP, metal tolerance protein; MW, molecular weight; pI, isoelectric point.
Footnotes
- ^https://www.ncbi.nlm.nih.gov/
- ^http://www.bioinformatics.nl/brassica/index.html?data=bras_tp%2Fdata&loc=A01%3A11420701..17130662&tracks=DNA&highlight=
- ^http://www.ebi.ac.uk/InterProScan/
- ^http://gsds.cbi.pku.edu.cn/
- ^https://github.com/CJ-Chen/TBtools
- ^http://pfam.xfam.org/search#tabview=tab1
- ^http://meme-suite.org/tools/meme
- ^http://www.cbs.dtu.dk/services/TMHMM/
- ^http://web.expasy.org/protparam/
- ^https://www.ncbi.nlm.nih.gov/tools/primer-blast/
References
Arthur, E., Crews, H., and Morgan, C. (2000). Optimizing plant genetic strategies for minimizing environmental contamination in the food chain. Int. J. Phytoremediation 2, 1–21. doi: 10.1080/15226510008500027
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Assunção, A. G. L., Schat, H., and Aarts, M. G. M. (2003). Thlaspi caerulescens, an attractive model species to study heavy metal hyperaccumulation in plants. New Phytol. 159, 351–360. doi: 10.1046/j.1469-8137.2003.00820.x
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Chen, Z. H., Fujii, Y., Yamaji, N., Masuda, S., Takemoto, Y., Kamiya, T., et al. (2013). Mn tolerance in rice is mediated by MTP8.1, a member of the cation diffusion facilitator family. J. Exp. Bot. 64, 4375–4387. doi: 10.1093/jxb/ert243
Cheng, F., Sun, R. F., Hou, X. L., Zheng, H. K., Zhang, F. L., Zhang, Y. Y., et al. (2016). Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 48, 1218–1224. doi: 10.1038/ng.3634
Delhaize, E., Gruber, B. D., Pittman, J. K., White, R. G., Leung, H., Miao, Y. S., et al. (2007). A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 51, 198–210. doi: 10.1111/j.1365-313X.2007.03138.x
Desbrosses-Fonrouge, A. G., Voigt, K., Schröder, A., Arrivault, S., Thomine, S., and Krämer, U. (2005). Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 579, 4165–4174. doi: 10.1016/j.febslet.2005.06.046
Eroglu, S., Giehl, R. F. H., Meier, B., Takahashi, M., Terada, Y., Ignatyev, K., et al. (2017). Metal tolerance protein 8 mediates manganese homeostasis and iron reallocation during seed development and germination. Plant Physiol. 174, 1633–1647. doi: 10.1104/pp.16.01646
Eroglu, S., Meier, B., von Wirén, N., and Peiter, E. (2016). The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in arabidopsis. Plant Physiol. 170, 1030–1045. doi: 10.1104/pp.15.01194
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Fu, X. Z., Tong, Y. H., Zhou, X., Ling, L. L., Chun, C. P., Cao, L., et al. (2017). Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene 629, 1–8. doi: 10.1016/j.gene.2017.07.072
Fujiwara, T., Kawachi, M., Sato, Y., Mori, H., Kutsuna, N., Hasezawa, S., et al. (2015). A high molecular mass zinc transporter MTP12 forms a functional heteromeric complex with MTP5 in the Golgi in Arabidopsis thaliana. FEBS J. 282, 1965–1979. doi: 10.1111/febs.13252
Gan, X. C., Hay, A., Kwantes, M., Haberer, G., Hallab, A., Dello Ioio, R., et al. (2016). The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat. Plants 2:16167. doi: 10.1038/nplants.2016.167
Gustin, J. L., Loureiro, M. E., Kim, D., Na, G., Tikhonova, M., and Salt, D. E. (2009). MTP1-dependent Zn sequestration into shoot vacuoles suggests dual roles in Zn tolerance and accumulation in Zn-hyperaccumulating plants. Plant J. 57, 1116–1127. doi: 10.1111/j.1365-313X.2008.03754.x
Gustin, J. L., Zanis, M. J., and Salt, D. E. (2011). Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol. Biol. 11:76. doi: 10.1186/1471-2148-11-76
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Iftikhar, H., Naveed, N., Virk, N., Bhatti, M. F., and Song, F. M. (2017). In silico analysis reveals widespread presence of three gene families, MAPK, MAPKK and MAPKKK, of the MAPK cascade from crop plants of Solanaceae in comparison to the distantly-related syntenic species from Rubiaceae, coffee. Peer J. 5:e3255. doi: 10.7717/peerj.3255
Inan, G., Zhang, Q., Li, P. H., Wang, Z. L., Cao, Z. Y., Zhang, H., et al. (2004). Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 135, 1718–1737. doi: 10.1104/pp.104.041723
Kang, X. S., and Carey, J. (1999). Role of heme in structural organization of cytochrome c probed by semisynthesis. Biochemistry 38, 15944–15951. doi: 10.1021/bi9919089
Kim, D., Gustin, J. L., Lahner, B., Persans, M. W., Baek, D., Yun, D. J., et al. (2004). The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. Plant J. 39, 237–251. doi: 10.1111/j.1365-313X.2004.02126.x
Kolaj-Robin, O., Russell, D., Hayes, K. A., Pembroke, J. T., and Soulimane, T. (2015). Cation diffusion facilitator family: structure and function. FEBS Lett. 589, 1283–1295. doi: 10.1016/j.febslet.2015.04.007
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lang, M. L., Hao, M. Y., Fan, Q. W., Wang, W., Mo, S. J., Zhao, W. C., et al. (2011). Functional characterization of BjCET3 and BjCET4, two new cation-efflux transporters from Brassica juncea L. J. Exp. Bot. 62, 4467–4480. doi: 10.1093/jxb/err137
Li, L. Z., Tu, C., Wu, L. H., Peijnenburg, W. J. G. M., Ebbs, S., and Luo, Y. M. (2017). Pathways of root uptake and membrane transport of Cd2+ in the zinc/cadmium hyperaccumulating plant Sedum plumbizincicola. Environ. Toxicol. Chem. 36, 1038–1046. doi: 10.1002/etc.3625
Li, Q. H., Li, Y., Wu, X. Y., Zhou, L., Zhu, X. J., and Fang, W. P. (2017). Metal transport protein 8 in Camellia sinensis confers superior manganese tolerance when expressed in yeast and Arabidopsis thaliana. Sci. Rep. 7:39915. doi: 10.1038/srep39915
Li, X., Zhang, X., Li, B., Wu, Y., Sun, H., and Yang, Y. (2017). Cadmium phytoremediation potential of turnip compared with three common high Cd-accumulating plants. Environ. Sci. Pollut. Res. 24, 21660–21670. doi: 10.1007/s11356-017-9781-z
Li, X., Zhang, X. M., Yang, Y., Li, B. Q., Wu, Y. S., Sun, H., et al. (2016). Cadmium accumulation characteristics in turnip landraces from China and assessment of their phytoremediation potential for contaminated soils. Front. Plant Sci. 7:1862. doi: 10.3389/fpls.2016.01862
Liu, S. Y., Liu, Y. M., Yang, X. H., Tong, C. B., Edwards, D., Parkin, I. A. P., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
Ma, G. C., Wang, Y. R., and Xuan, Z. Y. (2016). Analysis and comparison of nutritional compositions in Xinjiang turnip (Brassica rapa L.). Sci. Technol. Food Ind. 37, 360–364.
Ma, J. Q., Jian, H. J., Yang, B., Lu, K., Zhang, A. X., Liu, P., et al. (2017). Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus L.). Gene 620, 36–45. doi: 10.1016/j.gene.2017.03.030
Megdiche, W., Ben Amor, N., Debez, A., Hessini, K., Ksouri, R., Zuily-Fodil, Y., et al. (2007). Salt tolerance of the annual halophyte Cakile maritima as affected by the provenance and the developmental stage. Acta Physiol. Plant. 29, 375–384. doi: 10.1007/s11738-007-0047-0
Menguer, P. K., Farthing, E., Peaston, K. A., Ricachenevsky, F. K., Fett, J. P., and Williams, L. E. (2013). Functional analysis of the rice vacuolar zinc transporter OsMTP1. J. Exp. Bot. 64, 2871–2883. doi: 10.1093/jxb/ert136
Migocka, M., Kosieradzka, A., Papierniak, A., Maciaszczyk-Dziubinska, E., Posyniak, E., Garbiec, A., et al. (2015a). Two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. J. Exp. Bot. 66, 1001–1015. doi: 10.1093/jxb/eru459
Migocka, M., Papierniak, A., Kosieradzka, A., Posyniak, E., Maciaszczyk-Dziubinska, E., Biskup, R., et al. (2015b). Cucumber metal tolerance protein CsMTP9 is a plasma membrane H+-coupled antiporter involved in the Mn2+ and Cd2+ efflux from root cells. Plant J. 84, 1045–1058. doi: 10.1111/tpj.13056
Montanini, B., Blaudez, D., Jeandroz, S., Sanders, D., and Chalot, M. (2007). Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8:107. doi: 10.1186/1471-2164-8-107
Nies, D. H., and Silver, S. (1995). Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. Biotechnol. 14, 186–199. doi: 10.1007/BF01569902
Paulsen, I. T., and Saier, M. H. (1997). A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156, 99–103. doi: 10.1007/s002329900192
Podar, D., Scherer, J., Noordally, Z., Herzyk, P., Nies, D., and Sanders, D. (2012). Metal selectivity determinants in a family of transition metal transporters. J. Biol. Chem. 287, 3185–3196. doi: 10.1074/jbc.M111.305649
Prasad, M. N. V., and Freitas, H. M. D. (2003). Metal hyperaccumulation in plants - biodiversity prospecting for phytoremediation technology. Electron. J. Biotechnol. 6, 285–321. doi: 10.2225/vol6-issue3-fulltext-6
Sonnhammer, E. L., Eddy, S. R., Birney, E., Bateman, A., and Durbin, R. (1998). Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26, 320–322. doi: 10.1093/nar/26.1.320
Vatansever, R., Filiz, E., and Eroglu, S. (2017). Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification. Biometals 30, 217–235. doi: 10.1007/s10534-017-9997-x
Wang, X. W., Wang, H. Z., Wang, J., Sun, R. F., Wu, J., Liu, S. Y., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Xi, Y., Liu, J. Y., Dong, C., and Cheng, Z. M. (2017). The CBL and CIPK gene family in grapevine (Vitis vinifera): genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 8:978. doi: 10.3389/fpls.2017.00978
Xu, J., Chai, T. Y., Zhang, Y. X., Lang, M. L., and Han, L. (2009). The cation-efflux transporter BjCET2 mediates zinc and cadmium accumulation in Brassica juncea L. leaves. Plant Cell Rep. 28, 1235–1242. doi: 10.1007/s00299-009-0723-1
Xu, L., Zhang, D. Y., Xu, Z. L., Huang, Y. H., He, X. L., Wang, J. Y., et al. (2016). Comparative expression analysis of Calcineurin B-like family gene CBL10A between salt-tolerant and salt-sensitive cultivars in B. oleracea. Sci. Total Environ. 571, 1–10. doi: 10.1016/j.scitotenv.2016.07.130
Yang, J. H., Liu, D. Y., Wang, X. W., Ji, C. M., Cheng, F., Liu, B. N., et al. (2016). The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 48, 1225–1232. doi: 10.1038/ng.3657
Yin, X., Wang, Q. L., Chen, Q., Xiang, L., Yang, Y. Q., and Yang, Y. P. (2017). Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in turnip (Brassica rapa var. rapa). Front. Plant Sci. 8:1191. doi: 10.3389/fpls.2017.01191
Zhang, B. J., Zhang, X. X., and Luo, L. G. (2013). The major gene families related to cadmium absorption and transportation in plants. Genomics Appl. Biol. 32, 127–134.
Keywords: phylogenetic analysis, protein domain, metallic element, ion transport, gene expression
Citation: Li X, Wu Y, Li B, He W, Yang Y and Yang Y (2018) Genome-Wide Identification and Expression Analysis of the Cation Diffusion Facilitator Gene Family in Turnip Under Diverse Metal Ion Stresses. Front. Genet. 9:103. doi: 10.3389/fgene.2018.00103
Received: 16 October 2017; Accepted: 15 March 2018;
Published: 04 April 2018.
Edited by:
Yi Zhao, Institute of Computing Technology (CAS), ChinaReviewed by:
Mehar Hasan Asif, National Botanical Research Institute (CSIR), IndiaYan Zhang, Institute of Biophysics (CAS), China
Copyright © 2018 Li, Wu, Li, He, Yang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonghong Yang, eXloODMxOTk0QDE2My5jb20= Yongping Yang, eWFuZ3lwQG1haWwua2liLmFjLmNu
 Xiong Li
Xiong Li Yuansheng Wu3,4
Yuansheng Wu3,4 Boqun Li
Boqun Li Yonghong Yang
Yonghong Yang Yongping Yang
Yongping Yang