- Center for Human Genetics, University Hospitals Leuven, University of Leuven, Leuven, Belgium
The fragile X syndrome arises from the FMR1 CGG expansion of a premutation (55–200 repeats) to a full mutation allele (>200 repeats) and is the most frequent cause of inherited X-linked intellectual disability. The risk for a premutation to expand to a full mutation allele depends on the repeat length and AGG triplets interrupting this repeat. In genetic counseling it is important to have information on both these parameters to provide an accurate risk estimate to women carrying a premutation allele and weighing up having children. For example, in case of a small risk a woman might opt for a natural pregnancy followed up by prenatal diagnosis while she might choose for preimplantation genetic diagnosis (PGD) if the risk is high. Unfortunately, the detection of AGG interruptions was previously hampered by technical difficulties complicating their use in diagnostics. Therefore we recently developed, validated and implemented a new methodology which uses long-read single-molecule sequencing to identify AGG interruptions in females with a FMR1 premutation. Here we report on the assets of AGG interruption detection by sequencing and the impact of implementing the assay on genetic counseling.
Introduction
Variability of the CGG tandem repeat in the 5′ untranslated region (UTR) of the fragile X mental retardation gene (FMR1; MIM# 309550) is associated with various disorders. Whereas most individuals in the general population have around 30 CGG repeats (<45 repeats), patients with fragile X syndrome (FXS; MIM# 300624) carry large, full expansions sized above 200 repeats (Oberlé et al., 1991; Verkerk et al., 1991). These large expansions are usually epigenetically silenced thereby inhibiting the production of fragile X mental retardation protein (FMRP) (Pieretti et al., 1991). The absence of protein evokes FXS, a neurodevelopmental disorder characterized by intellectual disability, emotional problems, autism, hyperactivity, hypersensitivity and mild dysmorphic features (Penagarikano et al., 2007). Premutation carriers represent yet another group with repeat sizes varying between 55 and 200 repeats, and might be affected by fragile X-associated tremor/ataxia syndrome (FXTAS; MIM# 300623) or fragile X-associated primary ovarian insufficiency (FXPOI; MIM# 311360) amongst other medical problems (Sullivan et al., 2005; Van Esch, 2006; Hagerman and Hagerman, 2015). In between the premutation alleles (55–200 repeats) and normal alleles (<45 repeats) an intermediate zone (45–54 repeats) exists. Although carriers of intermediate alleles are generally believed to be healthy, some reports have shown that these alleles might be associated with parkinsonism (Loesch et al., 2009) and FXTAS, although with a milder phenotype, less frequently and at later age-of-onset (Hall et al., 2012; Liu et al., 2013). Hence, depending on the size of the FMR1 CGG repeat, an individual will be affected by different disorders varying both mechanistically and phenotypically.
The FMR1 CGG repeat is susceptible to meiotic instability which is reflected by repeat size differences between parents and offspring. Normal and intermediate alleles (<55 repeats) are usually inherited stably or differ at most a few repeats (Nolin et al., 2013). In both male and female premutation carriers repeat expansions and contractions are common (Nolin et al., 2003). Furthermore, women carrying a premutation allele are at risk of transmitting a full mutation to their offspring. With a reported frequency of around 1 in 200 females carrying a premutation, a significant fraction of the female population is at risk of having children with FXS (Cronister et al., 2008; Tassone et al., 2012). This risk depends on the repeat size and the number of AGG triplets interrupting the repeat whereby larger repeats with fewer AGGs have the highest expansion risks (Yrigollen et al., 2014a; Nolin et al., 2015). These AGGs intersperse the CGG repeat every 9 or 10 CGG repeats at the 5' end where their presence reduces repeat instability (Eichler et al., 1994; Yrigollen et al., 2014a; Nolin et al., 2015). The influence of AGG interruptions is the most profound for alleles ranging from 60 to 85 repeats. For instance, the risk of transmitting a full mutation for a woman with 75 repeats and two AGG triplets is 12%, but this increases to 77% if no AGG interruptions are present (Yrigollen et al., 2012). Some studies have reported that maternal age can also influence the expansion risk (Yrigollen et al., 2014a), but this could not be confirmed by others (Nolin et al., 2015). Hence, further large scale studies are needed to solve this issue. For small (<60 repeats) or large (>85 repeats) premutation alleles the influence of AGG interruptions on the expansion risk is only minor. Alleles smaller than 60 repeats have only an expansion risk of 2.6%, even in the absence of AGG repeats while large alleles on the contrary have an expansion risk higher than 60%, even when two stabilizing AGGs are present (Yrigollen et al., 2012).
In genetic counseling it is important to provide a female premutation carrier with an accurate estimate of her expansion risk because this influences her reproductive planning. When the expansion risk is high, women might opt for preimplantation genetic diagnosis (PGD) where one could select for unaffected males or non-carrier female embryos (Sermon et al., 1999; Burlet et al., 2006). Although the detection of large CGG alleles could be very challenging in a single cell picked from an early embryo, this can now be circumvented by making use of new haplotyping methods (Natesan et al., 2014; Zamani Esteki et al., 2015; Dimitriadou et al., 2017). If the risk of having a child with FXS is low, a women could choose for normal conception, optionally combined with invasive prenatal diagnosis to screen the fragile X status of their fetus (Biancalana et al., 2015). Therefore, it is important that genetic laboratories use both repeat size and the number of AGGs to assess the expansion risk. The repeat size can easily be determined by PCR-based methods and/or Southern blot, but the detection of AGG interruptions is technically challenging. This has hampered the clinical uptake of this information. Interruption analysis can be done by first- and second –generation triplet-primed PCR methods, but unfortunately these only provide an indirect indication of the presence of AGG interruptions (Chen et al., 2010; Hayward and Usdin, 2017). Interpreting the results of these assays is complicated in females who carry two X-chromosomes each containing a different CGG repeat with its own set of AGG triplets. To overcome these problems, a novel methodology was developed based on long-read single-molecule sequencing by Pacific Biosciences. This technology generates long reads (>20 kb), is suited to analyze GC-rich regions (like the FMR1 CGG repeat) and able to detect embedded AGG interruptions (Travers et al., 2010; Loomis et al., 2013; Ardui et al., 2017). The generated single-molecule reads are separated according to the X-chromosome from which they originate, whereafter the exact repeat structure can be determined for each allele individually. Long-read single-molecule sequencing is the only technology so far that can separate the two repeats derived from different X-chromosomes, and hence generates superior results compared to PCR-based assays (Ardui et al., 2017). Furthermore, long-read single-molecule sequencing is diagnostically applicable and different applications are presently being implemented in human genetic diagnostics (Ardui et al., 2018).
The undisputed impact of AGG triplets on the expansion risk estimates of female premutation carriers along with good AGG interruption detection by single-molecule sequencing prompted us to implement this assay for diagnostic use for all female carriers with an intermediate and a premutation allele (45–200 repeats). By doing so we improved the risk assessments for genetic counseling and positively impacted the management of the disorder. We summarize our experience with the use of AGG triplets in the clinic after 1 year during which we analyzed 51 patients.
Materials and Methods
Patients
The 51 female patients were ascertained from January to December 2017 at the Center for Human Genetics, KU Leuven, UZ Leuven, Belgium. The patients were referred for diagnostic testing because of either diagnostic work-up in the fertility clinic, POI, intellectual disability or a family history of FXS. Fifty patients carried a normal allele and an intermediate (26) or premutation allele (24). Also 1 female carrying two premutations was included in this study. The study was approved by the local Ethics Committee and consent was obtained from the patients, both informed and written.
DNA Sampling
For all 51 females DNA was isolated from peripheral white blood cells according to standard procedures. From one pregnant female patient a chorionic villi sample (CVS) sample was received. Villi from CVS were separated from maternal tissue under a microscope to minimize maternal contamination. Two to four villi were provided for DNA extraction.
Molecular Analysis
The repeat size was determined by (TP-)PCR for all samples. The structure of the CGG repeat in the FMR1 gene was then determined for all intermediate and premutation carriers by sequencing according to the previously published method (Ardui et al., 2017). In brief, the FMR1 CGG repeat was amplified by PCR during which a barcode was incorporated. After pooling different amplicons together, long-read single-molecule sequencing was performed on a PacBio RSII system. This generates long reads which span each PCR molecule multiple times thereby generating a highly accurate consensus sequence. This technology is “single-molecule” which allows the unambiguous separation of the CGG repeats derived from the two different X-chromosomes. Hence, after sequencing, the complete repeat structure could be reconstructed for both X-chromosomes revealing the repeat size and the AGG interruption pattern. The sizes determined by PCR control runs and single-molecule sequencing matched perfectly.
Genetic Counseling
Females with a premutation were offered genetic counseling. If the patient was considering having children, an accurate assessment of the risk that their premutation would expand to a full mutation was provided based on both the FMR1 CGG repeat size and the number of AGG interruptions.
Results
AGG analysis was implemented diagnostically in order to more accurately assess the risk that offspring of premutation carriers will be affected by FXS and thus improve genetic counseling. We report the results of AGG interruption detection in 51 females with intermediate or premutation alleles. Long-read single-molecule sequencing was used to determine the AGG triplets as it allows the determination of the exact FMR1 repeat structure for each individual allele.
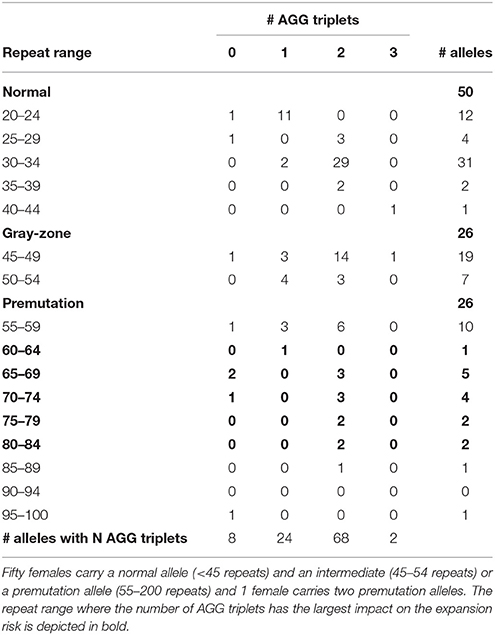
The results of the FMR1 CGG repeat analysis are summarized in Table 1. Fifty females carried a normal allele and an intermediate (26) or a premutation allele (24), while 1 female carried two premutation alleles. Therefore, the total number of premutation alleles is also 26 (Table 1). The normal alleles of all 50 females ranged between 20 and 40 repeats and are interspersed with 0, 1, 2 or 3 AGGs. Two different clusters were identified within the structures of the normal alleles: a smaller group (20–24 repeats) interrupted by 1 AGG (11 patients) and a larger group (30–34 repeats) interrupted by 2 AGGs (29 patients), in line with previously published results (Eichler et al., 1996; Chen et al., 2003). The remaining ten normal alleles are more distributed in size and number of AGGs (Table 1; Eichler et al., 1996). From the 26 intermediate alleles and 26 premutation alleles (from 25 females), the majority (45) were interrupted by 1 or 2 AGG interruptions.
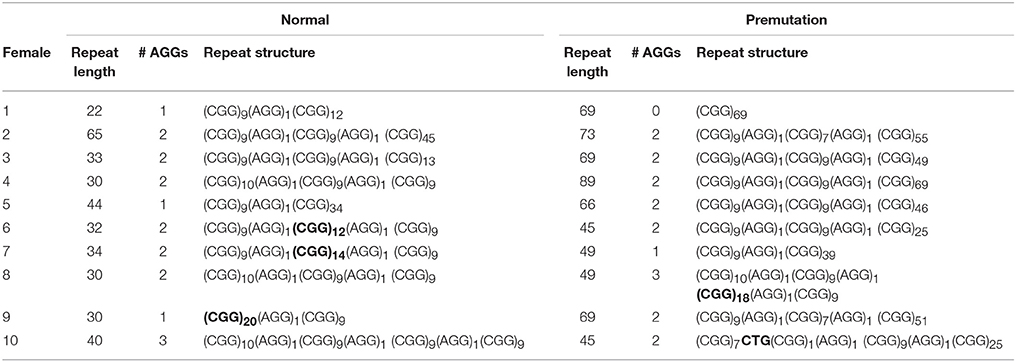
Information on AGG interruptions is vital for females carrying a premutation allele because of the risk of transmitting a full mutation to their children. For the 13 females with a premutation allele ranging between 60 and 84 repeats, knowing the AGG status was reassuring for these carrying 2 AGGs, while it alerted females without any AGG triplets. For example, female 1 (Table 2) presented with an allele of 69 CGG repeats without interruptions. This female opted for PGD because she has a relatively high expansion risk (22,9%) in combination with a reduced fertility. Female 2 (Table 2) carried two premutation alleles (65 and 73 repeats) and chose for PGD because she also carried a translocation. For this female, AGG interruption analysis was performed to determine which allele had the lowest expansion risk. As sequencing revealed that two AGGs were present in each allele, embryos carrying the smallest allele of 65 repeats with 2 AGG's could be prioritized for transfer during PGD. Female 3 (Table 2) carried a premutation allele of 69 repeats interrupted with 2 AGG triplets. This female only has a low expansion risk of 0.5% and hence decided to choose for a natural pregnancy. Invasive prenatal follow-up of the ensuing pregnancy showed that the fetus inherited the normal copy. Two females carried a repeat above 85 repeats and hence invoked a high expansion risks: 100% for the female with 95 repeats without AGGs and 60% for the female with 89 repeats and 2 AGGs (female 4, Table 2). Although the high expansion risk of female 4, she became pregnant naturally. Fortunately, invasive prenatal follow-up showed the premutation allele even contracted in the female fetus (female 5, Table 2), most likely to the allele containing 66 repeats and 2 AGG's. A contraction to the allele with 44 repeats and 1 AGG is less likely, but cannot be excluded because the DNA of the father was not available.
Sequencing of FMR1 CGG repeats not only identified AGG triplets, but any sequence variation at this locus. The FMR1 structure is most commonly built up from (CGG)9AGG and (CGG)10AGG building blocks. This was confirmed in most of the females of our cohort. However, 4 females (female 6–9, Table 2) carried a less common AGG interruption pattern. Female 10 (Table 2) carries an intermediate allele with 45 repeats and two AGG triplets, but also harbored a CTG interruption (Table 2).
Discussion
Technical limitations have hampered the diagnostic uptake of AGG analysis so far, but this is now overcome by a novel single-molecule sequencing approach (Ardui et al., 2017). Long-read single-molecule sequencing generates high quality results and permits to construct unambiguously the repeat structure for both X-chromosomes in females. Incorporating AGG analysis into FMR1 diagnostic work-up allows accurate risk estimates for having a child with FXS which greatly improved genetic counseling for woman carrying a premutation (Yrigollen et al., 2014a; Biancalana et al., 2015; Nolin et al., 2015). Here, we report the results of AGG interruption analysis of the first 51 females with an intermediate or premutation allele which have been collected at the Center of Human Genetics, KU Leuven, UZ Leuven (Belgium) during 1 year.
The impact of AGG interruptions is the most profound for females carrying a premutation sized between 60 and 84 repeats within which 13 females of our cohort fitted. From these 13 females, 3 females carried pure CGG repeats and hence have relatively high expansion risks ranging from 23 to 50% (Yrigollen et al., 2012, 2014a). These females with a high risk of having a child with FXS might opt for PGD where one can select for non-affected male- or carrier female embryos. The other 10 females had either 1 but most often 2 AGG interruptions and hence have more moderate expansion risks, except the two females with 80–84 repeats and 2 AGGs who also have around 30% chance their allele will expand into a full mutation (Yrigollen et al., 2012, 2014a). We conclude that the more accurate risk estimates provided to the females with a premutation simplified choosing the most appropriate reproductive strategy.
Single-molecule sequencing provides a direct read-out of the FMR1 CGG repeat and hence allows to grasp the complete repeat sequence. Most of our normal, intermediate and premutation alleles are constructed with CGG9AGG or CGG10AGG building blocks, concordant with previously published reports (Eichler et al., 1996; Yrigollen et al., 2014b). However, the repeats from 4 females deviated from these common building blocks and are more rare in the general population (Eichler et al., 1996; Yrigollen et al., 2014b). PCR-based assays might struggle to generate the correct repeat structure for these females as they use common haplotypes to infer the repeat structure of females whose X chromosomes camouflage each other's repeat structure (Chen et al., 2010). In another female a CTG interruption was detected within the CGG repeat which has not been reported so far. Most interruptions are AGG triplets, although also a TGG interruption was discovered by Kunst and Warren (1994) in a male sample. Possibly, also these alternative interruptions might stabilize the CGG repeat. Systematic mapping and collection on the transmission of repeats carrying those rare interruptions would provide insights in the stability of such repeats. It remains unfortunate that the PCR-based AGG assays cannot detect novel interruptions and hence impede further characterization of these unusual interruptions.
To conclude, AGG analysis by single-molecule sequencing generates clear results which contribute to determining an accurate expansion risk for females with a premutation and hence positively impacts genetic counseling. By incorporating AGG analysis into our diagnostic setting we also plan to investigate mother-daughter transmissions which can further help fine-tuning the risk estimates belonging to specific repeat classes.
Author Contributions
SA designed and performed all laboratory work and analyzed the data. HV, TdR, and KD ascertained the patients included in the study, interpreted the data and provided genetic counseling. VR and GM facilitated the diagnostic implementation. SA wrote the article which was critically revised and approved by VR, TdR, HV, KD, GM, and JV. JV coordinated the study and supervised the organization of the whole process.
Funding
SA received a personal grant from the Agency for Innovation by Science and Technology (IWT) (PhD grant: SB/131787) and a grant from KU Leuven, SymBioSys (PFV/10/016) and the Hercules Foundation (ZW11-14).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Wim Meert and Steve Smekens for their help with the laboratory work.
References
Ardui, S., Ameur, A., Vermeesch, J. R., and Hestand, M. S. (2018). Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 46, 2159–2168. doi: 10.1093/nar/gky066
Ardui, S., Race, V., Zablotskaya, A., Hestand, M., Van Esch, H., Devriendt, K., et al. (2017). Detecting AGG Interruptions in Male and Female FMR1 premutation carriers by single-molecule sequencing. Hum. Mutat. 38, 324–331. doi: 10.1002/humu.23150
Biancalana, V., Glaeser, D., McQuaid, S., and Steinbach, P. (2015). EMQN best practice guidelines for the molecular genetic testing and reporting of fragile X syndrome and other fragile X-associated disorders. Eur. J. Hum. Genet. 23, 417–425. doi: 10.1038/ejhg.2014.185
Burlet, P., Frydman, N., Gigarel, N., Kerbrat, V., Tachdjian, G., Feyereisen, E., et al. (2006). Multiple displacement amplification improves PGD for fragile X syndrome. Mol. Hum. Reprod. 12, 647–652. doi: 10.1093/molehr/gal069
Chen, L., Hadd, A., Sah, S., Filipovic-Sadic, S., Krosting, J., Sekinger, E., et al. (2010). An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J. Mol. Diagn. 12, 589–600. doi: 10.2353/jmoldx.2010.090227
Chen, L. S., Tassone, F., Sahota, P., and Hagerman, P. J. (2003). The (CGG)n repeat element within the 5' untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum. Mol. Genet. 12, 3067–3074. doi: 10.1093/hmg/ddg331
Cronister, A., Teicher, J., Rohlfs, E. M., Donnenfeld, A., and Hallam, S. (2008). Prevalence and instability of fragile X alleles: implications for offering fragile X prenatal diagnosis. Obstet. Gynecol. 111, 596–601. doi: 10.1097/AOG.0b013e318163be0b
Dimitriadou, E., Melotte, C., Debrock, S., Esteki, M. Z., Dierickx, K., Voet, T., et al. (2017). Principles guiding embryo selection following genome-wide haplotyping of preimplantation embryos. Hum. Reprod. 32, 687–697. doi: 10.1093/humrep/dex011
Eichler, E. E., Holden, J. J., Popovich, B. W., Reiss, A. L., Snow, K., Thibodeau, S. N., et al. (1994). Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat. Genet. 8, 88–94. doi: 10.1038/ng0994-88
Eichler, E. E., Macpherson, J. N., Murray, A., Jacobs, P. A., Chakravarti, A., and Nelson, D. L. (1996). Haplotype and interspersion analysis of the FMR1 CGG repeat identifies two different mutational pathways for the origin of the fragile X syndrome. Hum. Mol. Genet. 5, 319–330. doi: 10.1093/hmg/5.3.319
Hagerman, P. J., and Hagerman, R. J. (2015). Fragile X-associated tremor/ataxia syndrome. Ann. N. Y. Acad. Sci. 1338, 58–70. doi: 10.1111/nyas.12693
Hall, D. A., Tassone, F., Klepitskaya, O., and Leehey, M. (2012). Fragile X-Associated tremor Ataxia Syndrome in FMR1 gray zone allele carriers. Mov. Disord. 27, 296–300. doi: 10.1002/mds.24021
Hayward, B. E., and Usdin, K. (2017). Improved assays for AGG interruptions in Fragile X premutation carriers. J. Mol. Diagnostics 19, 828–835. doi: 10.1016/j.jmoldx.2017.06.008
Kunst, C. B., and Warren, S. T. (1994). Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell 77, 853–861. doi: 10.1016/0092-8674(94)90134-1
Liu, Y., Winarni, T., Zhang, L., Tassone, F., and Hagerman, R. (2013). Fragile X-associated tremor/ataxia syndrome (FXTAS) in grey zone carriers. Clin. Genet. 84, 74–77. doi: 10.1111/cge.12026
Loesch, D. Z., Khaniani, M. S., Slater, H. R., Rubio, J. P., Bui, Q. M., Kotschet, K., et al. (2009). Small CGG repeat expansion alleles of FMR1 gene are associated with parkinsonism. Clin. Genet. 76, 471–476. doi: 10.1111/j.1399-0004.2009.01275.x
Loomis, E. W., Eid, J. S., Peluso, P., Yin, J., Hickey, L., Rank, D., et al. (2013). Sequencing the unsequenceable: expanded CGG-repeat alleles of the fragile X gene. Genome Res. 23, 121–128. doi: 10.1101/gr.141705.112
Natesan, S. A., Bladon, A. J., Coskun, S., Qubbaj, W., Prates, R., Munne, S., et al. (2014). Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet. Med. 16, 838–845. doi: 10.1038/gim.2014.45
Nolin, S. L., Brown, W. T., Glicksman, A., Houck, G. E., Gargano, A. D., Sullivan, A., et al. (2003). Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. Am. J. Hum. Genet. 72, 454–464. doi: 10.1086/367713
Nolin, S. L., Glicksman, A., Ersalesi, N., Dobkin, C., Brown, W. T., Cao, R., et al. (2015). Fragile X full mutation expansions are inhibited by one or more AGG interruptions in premutation carriers. Genet. Med. 17, 358–364. doi: 10.1038/gim.2014.106
Nolin, S. L., Sah, S., Glicksman, A., Sherman, S. L., Allen, E., Berry-Kravis, E., et al. (2013). Fragile X AGG analysis provides new risk predictions for 45-69 repeat alleles. Am. J. Med. Genet. Part A 161, 771–778. doi: 10.1002/ajmg.a.35833
Oberlé, I., Rousseau, F., Heitz, D., Kretz, C., Devys, D., Hanauer, A., et al. (1991). Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 252, 1097–1102. doi: 10.1126/science.252.5009.1097
Penagarikano, O., Mulle, J. G., and Warren, S. T. (2007). The pathophysiology of fragile x syndrome. Annu. Rev. Genomics Hum. Genet. 8, 109–129. doi: 10.1146/annurev.genom.8.080706.092249
Pieretti, M., Zhang, F., Fu, Y.-H., Warren, S. T., Oostra, B. A., Caskey, C. T., et al. (1991). Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66, 817–822. doi: 10.1016/0092-8674(91)90125-I
Sermon, K., Seneca, S., Vanderfaeillie, A., Lissens, W., Joris, H., Vandervorst, M., et al. (1999). Preimplantation diagnosis for fragile X syndrome based on the detection of the non-expanded paternal and maternal CGG. Prenat. Diagn. 19, 1223–1230. doi: 10.1002/(SICI)1097-0223(199912)19:13<1223::AID-PD724>3.0.CO;2-0
Sullivan, A. K., Marcus, M., Epstein, M. P., Allen, E. G., Anido, A. E., Paquin, J. J., et al. (2005). Association of FMR1 repeat size with ovarian dysfunction. Hum. Reprod. 20, 402–412. doi: 10.1093/humrep/deh635
Tassone, F., Iong, K. P., Tong, T.-H., Lo, J., Gane, L. W., Berry-Kravis, E., et al. (2012). FMR1 CGG allele size and prevalence ascertained through newborn screening in the United States. Genome Med. 4:100. doi: 10.1186/gm401
Travers, K. J., Chin, C. S., Rank, D. R., Eid, J. S., and Turner, S. W. (2010). A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res. 38:e159. doi: 10.1093/nar/gkq543
Van Esch, H. (2006). The Fragile X premutation: new insights and:clinical consequences. Eur. J. Med. Genet. 49, 1–8. doi: 10.1016/j.ejmg.2005.11.001
Verkerk, A. J., Pieretti, M., Sutcliffe, J. S., Fu, Y. H., Kuhl, D. P., Pizzuti, A., et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914. doi: 10.1016/0092-8674(91)90397-H
Yrigollen, C. M., Durbin-Johnson, B., Gane, L., Nelson, D. L., Hagerman, R., Hagerman, P. J., et al. (2012). AGG interruptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genet. Med. 29, 729–736. doi: 10.1038/gim.2012.34
Yrigollen, C. M., Martorell, L., Durbin-Johnson, B., Naudo, M., Genoves, J., Murgia, A., et al. (2014a). AGG interruptions and maternal age affect FMR1 CGG repeat allele stability during transmission. J. Neurodev. Disord. 6:24. doi: 10.1186/1866-1955-6-24
Yrigollen, C. M., Sweha, S., Durbin-Johnson, B., Zhou, L., Berry-Kravis, E., Fernandez-Carvajal, I., et al. (2014b). Distribution of AGG interruption patterns within nine world populations. Intractable Rare Dis. Res. 3, 153–161. doi: 10.5582/irdr.2014.01028
Keywords: AGG interruptions, fragile X syndrome, single-molecule real-time sequencing, fragile X premutation, genetic counseling, FMR1, CGG repeat
Citation: Ardui S, Race V, de Ravel T, Van Esch H, Devriendt K, Matthijs G and Vermeesch JR (2018) Detecting AGG Interruptions in Females With a FMR1 Premutation by Long-Read Single-Molecule Sequencing: A 1 Year Clinical Experience. Front. Genet. 9:150. doi: 10.3389/fgene.2018.00150
Received: 07 February 2018; Accepted: 10 April 2018;
Published: 16 May 2018.
Edited by:
Shai E. Elizur, Sheba Medical Center, IsraelReviewed by:
Maria Paola Lombardi, University of Amsterdam, NetherlandsSarah L. Nolin, Institute for Basic Research in Developmental Disabilities (IBR), United States
Copyright © 2018 Ardui, Race, de Ravel, Van Esch, Devriendt, Matthijs and Vermeesch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joris R. Vermeesch, am9yaXMudmVybWVlc2NoQHV6bGV1dmVuLmJl
 Simon Ardui
Simon Ardui Valerie Race
Valerie Race Thomy de Ravel
Thomy de Ravel