- 1Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
- 2Institute of Biology of Komi Science Center of Ural Branch of Russian Academy of Sciences, Syktyvkar, Russia
- 3Department of Biological and Medical Physics, Moscow Institute of Physics and Technology, Dolgoprudny, Russia
- 4Insilico Medicine, Baltimore, MD, United States
Some effects of aging in animals are tissue-specific. In D. melanogaster neuronal overexpression of Gclc increases lifespan and improves certain physiological parameters associated with health benefits such as locomotor activity, circadian rhythmicity, and stress resistance. Our previous transcriptomic analyses of Drosophila heads, primarily composed of neuronal tissue, revealed significant changes in expression levels of genes involved in aging-related signaling pathways (Jak-STAT, MAPK, FOXO, Notch, mTOR, TGF-beta), translation, protein processing in endoplasmic reticulum, proteasomal degradation, glycolysis, oxidative phosphorylation, apoptosis, regulation of circadian rhythms, differentiation of neurons, synaptic plasticity, and transmission. Considering that various tissues age differently and age-related gene expression changes are tissue-specific, we investigated the effects of neuronal Gclc overexpression on gene expression levels in the imago thorax, which is primarily composed of muscles. A total of 58 genes were found to be differentially expressed between thoraces of control and Gclc overexpressing flies. The Gclc level demonstrated associations with expression of genes involved in the circadian rhythmicity, the genes in categories related to the muscle system process and the downregulation of genes involved in proteolysis. Most of the functional categories altered by Gclc overexpression related to metabolism including Drug metabolism, Metabolism of xenobiotics by cytochrome P450, Glutathione metabolism, Starch and sucrose metabolism, Citrate cycle (TCA cycle), One carbon pool by folate. Thus, the transcriptomic changes caused by neuron-specific Gclc overexpression in the thorax were less pronounced than in the head and affected pathways also differed from previous results. Although these pathways don't belong to the canonical longevity pathways, we suggest that they could participate in the delay of aging of Gclc overexpressing flies.
Introduction
The aging process is determined by the impairment functioning of tissues and systems of an organism. It was shown that aging is accompanied by significant changes in genome-wide gene expression profiles identified for different tissues of vertebrates and invertebrates (Zhan et al., 2007; Kim et al., 2014; White et al., 2015; Cannon et al., 2017; Chen et al., 2018; Zhang et al., 2018). Furthermore, the age-related changes in gene expression are tissue-specific, suggesting that various tissues age differently (Girardot et al., 2006; Zhan et al., 2007; Yang et al., 2015). Recently, a number of studies showed that muscle tissue undergoes sufficient alterations in gene expression patterns during aging in mammals and flies (Kim et al., 2014; Chen et al., 2018; Zhang et al., 2018; Zhou et al., 2018), and revealed the involvement of several tens of messenger RNAs as well as long non-coding, micro, and circular RNAs in this process (Kim et al., 2014; Chen et al., 2018). In particular, aged rats demonstrated decreased expression of components of AMPK, IGF-1, and CASK pathways that can be a key cause of the loss of muscle mass and function (Zhou et al., 2018). The progressive loss of skeletal muscle function and mass, known as sarcopenia, is one of the useful conditions accompanied the aging process in human and underlied the critical mobility reducing and life quality decline (Herndon et al., 2002; Demontis et al., 2013; Larsson et al., 2019). It must be noted, that the aging-related muscle degeneration are also determined by the progressive loss of motoneurons (Larsson et al., 2019). Therefore, interventions that contribute to the slowing down of aging of nervous tissue (including overexpression of pro-longevity genes) can also have a positive effect on the state of muscle tissue and prevent dangerous structural and cellular changes.
According to the recent experimental data, Drosophila is a convenient and relevant model for the investigation of muscle degeneration disorders (Demontis et al., 2013; Rai et al., 2014; Kreipke et al., 2017). Despite differences in the composition of mammalian and fly muscle systems, the Drosophila model can be used for the identification of key molecular mechanisms of aging-related changes in muscle tissue (Demontis et al., 2013; Rai et al., 2014). Furthermore, the lack of stem cell-based regeneration allows to determine the role of endogenous processes of muscle degeneration (Kreipke et al., 2017). The study of molecular basis of skeletal muscle laminopathies were carried by using of Drosophila melanogaster LamC mutants which demonstrated a numbered of muscular pathologic changes and cellular abnormalities. These negative changes were prevented by regulation of AMP-activated protein kinase (AMPK)/TOR/autophagy signaling pathways that highly conserved between flies and humans (Chandran et al., 2018). Thus, the study of mechanistic basis of muscular disorders using Drosophila allows to reveal potential targets for therapeutic interventions.
The Gclc is a target gene of pro-longevity SKN-1/Nrf2 transcription factor, activated in response to oxidative stress, and implicated in the lifespan extension of flies, worms, and mice (Orr et al., 2005; Luchak et al., 2007; Sykiotis et al., 2011; Mockett and Nobles, 2013; Blackwell et al., 2015). It codes for the catalytic subunit of glutamate-cysteine ligase, that participates in glutathione formation (Orr et al., 2005). Age-dependent downregulation of glutamate-cysteine ligase is associated with a decline in glutathione content, which is involved in multiple cellular functions including antioxidant defense and detoxification of reactive intermediates (Liu et al., 2004; Orr et al., 2005). Previously, it was demonstrated that the neuron- and muscle-specific overexpression of Gclc is associated with increased lifespan in flies, but not the ubiquitous overexpression of Gclc (Orr et al., 2005; Luchak et al., 2007; Mockett and Nobles, 2013). In addition, Gclc overexpression in the nervous system postpones age-associated decline in circadian rhythmicity and a variety of physiological parameters like locomotor activity and resistance to the oxidative, osmotic, and proteotoxic stresses (Moskalev et al., 2016). Gclc overexpression also affects transcription patterns of genes that govern diverse cellular processes such as Jak-STAT, MAPK, FOXO, Notch, mTOR, TGF-beta signaling pathways, translation, protein processing in endoplasmic reticulum, proteasomal degradation, glycolysis, oxidative phosphorylation, apoptosis, regulation of circadian rhythms, differentiation of neurons, synaptic plasticity, and transmission in Drosophila heads (Moskalev et al., 2016).
We supposed, that neuron-specific Gclc overexpression may ameliorate the decline of muscular function and delay the aging of muscle tissue (Moskalev et al., 2016). For the first time, we studied the effect of neuronal overactivation of the pro-longevity gene on age-related transcriptome changes in the muscular system. We used total RNA sequencing and revealed that Gclc overexpression in the head also affected the key signaling pathways implicated in the aging process in the thorax, which is composed primarily of muscle tissue. We identified 58 differentially expressed genes. Among the functional categories [Gene Ontology (GO) terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways] the genes consistently overrepresented were associated with Drug metabolism, Metabolism of xenobiotics by cytochrome P450, Glutathione metabolism, Starch and sucrose metabolism, Neuroactive ligand-receptor interaction, One carbon pool by folate, vesicle, and Cdc73/Paf1 complex. While apparent, the transcriptomic changes in the thorax were less pronounced than in the head.
Materials and methods
Drosophila melanogaster Lines
Flies were separated by sex and maintained in different vials on a standard sugar-yeast medium at constant temperature (25°C) and humidity (60%) in a 12:12 h light-dark cycle in Binder KBF720-ICH (Binder, Germany) climate chamber. The following strains were used: UAS-Gclc (provided by Dr. William C. Orr, Southern Methodist University) and Appl-GAL4 (#32040, Bloomington Drosophila Stock Center). They were backcrossed with w1118 (#3605, Bloomington Drosophila Stock Center, USA) 6–8 times to equilibrate the genetic background. To activate the Gclc constitutive overexpression in the nervous system, the UAS-Gclc was crossed with Appl-GAL4 driver. Newly enclosed progenies were maintained for one, four, and 6 weeks before dissection. The same aged flies from the parental UAS-Gclc line were used as a contro control.
Total RNA Extraction and Qualification
The experimental design was the same as in our previous work (Moskalev et al., 2016). Thoraxes in this study and heads in the previous study were collected from the same flies. In brief, we used males and females' thorax of UAS-Gclc flies as control and thoraxes of flies with Gclc overexpression. A comparison of flies with different chronological ages, namely, 1-, 4-, and 6- weeks was conducted. The ages were designated as “young,” “matured,” and “old,” respectively. The flies were collected, frozen and kept at −80°C to prevent RNA degradation. For each experimental variant, 40 flies were selected. Three biological replicates of the experiment were performed. The males and females were analyzed separately to reveal any sex-dependent differences in gene expression caused by neuronal Gclc expression. The RNA used for next-generation sequencing was extracted from 30 flies (10 flies per each replicate) using QIAzol Lysis Reagent (Qiagen, Netherlands) and precipitated with isopropanol. Qubit®2.0 Fluorometer (Invitrogen, USA) and NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies Inc., USA) were used to estimate the RNA concentration and purity. The A260/A280 ratio in the samples was around 1.8–2.0. RNA was additionally purified using DNase I (Promega, USA). The RNA integrity was estimated with Bioanalyzer Agilent 2100 (Agilent Technologies, USA).
Library Preparation and Sequencing Procedure
mRNA was isolated from the total RNA with NEBNext Oligo d(T)25 beads. The fragmentation was performed at 94°C with addition of First Strand Synthesis Reaction Buffer and Random Primer mix (2×). The double-stranded (ds) cDNA was synthetized by using ProtoScript II Reverse Transcriptase and Second Strand Synthesis Enzyme Mix. The blunt ends on the ds cDNA were created with the end-repair reaction. The adapters were ligated using the specific RNA Adapter Indexes supplied in the kit. The PCR reaction was performed to selectively enrich adaptor ligated DNA fragments and create a large amount of DNA, which was then purified using Ampure XP beads. The quantity and quality of libraries were assessed on Rotor-Gene 6000 PCR System (Qiagen, USA) using the qPCR method and on Agilent 2100 Bioanalyzer using a High Sensitivity DNA chip. The final length of the libraries averaged 260 bp. The libraries were normalized to 4 nM, pooled together in equal volumes, and sequenced with 2 × 50 bp paired-end reads on the NextSeq 500 System (Illumina, USA). The obtained data were stored in FASTQ format. The minimum amount of reads for each sample was 20 million.
NGS Data Processing
We used PPLine pipeline to perform quality control, read mapping, and counting (Krasnov et al., 2015). In the details, reads were analyzed using FastQC and trimmed with trimmomatic (Bolger et al., 2014). To evaluate the efficacy of the isolation of polyA fraction and the absence of bacterial contamination, 100.000 randomly selected reads from each sample were mapped to D.Melanogaster rRNA genes and bacterial genomes (all strains that had been submitted to NCBI Genome through 2015), accordingly. Typically, rRNA ratio was 0.3–1.7% (except for 2 samples with 4%). The trimmed reads were mapped to the D.Melanogaster genome (assembly BDGP6, Ensembl release 90) using splice-aware STAR aligner (Dobin et al., 2013). About 90–95% reads were uniquely mapped. Since the samples had demonstrated different RNA integrity numbers (RIN), in order to compare gene expression levels, we take into account a 3′-tail bias as described in Sigurgeirsson et al. (2014) with some modifications. Without a 3′-bias adjustment, one should expect false-positive overexpression of short transcripts (for example, genes of mitochondrial proteins or ribosomal proteins) and under-expression of genes with long transcripts and encoded proteins (for example, multi-domain transmembrane proteins interacting with extracellular matrix).
First, we analyzed read coverage across transcript length using a modified version of geneBody_coverage.py script, part of RSeQC package (Wang et al., 2012). In the samples with lower RIN numbers and more degraded RNA, we observed rapid falling down of read coverage level after 750–1,000 bp from the 3′-tail. The coverage in the region of 0–500 bp from the 3′-tail was almost preserved for all samples.
Then, we quantified transcripts using RSEM (Li and Dewey, 2011) and identified the most abundantly expressed alternative transcript for each gene. The other transcripts were discarded. Then, the transcripts were truncated to the length of 500 bp from the 3′-end and a new GTF file was generated. Finally, using this gene model, we quantified reads using Counts from Subread package (Li and Dewey, 2011). This procedure allowed us to significantly reduce the dependence of the observed expression level fold changes (FC) on the transcript lengths, between the samples with higher and lower RINs.
In order to more effectively eliminate the factor of the 3′-bias, we proceeded the following way. First, we excluded genes with low expression level by introducing a threshold: CPM > 2 for at least 3 samples. About 9,000 genes passed the threshold. Next, we split all the genes into 10 bins, depending on the average gene transcript length, and within each bin we normalized read counts using the TMM (trimmed mean of M-values) method from edgeR (Robinson et al., 2010). Finally, the bins were merged.
A similar procedure was performed to eliminate the dependency of the expression level FC on the “absolute” gene expression level (in terms of read counts per million, CPM). This bias may also occur when comparing samples with different RINs and initial RNA concentration.
Finally, the adjusted read pseudo-counts were analyzed by edgeR using exact test and the quasi-likelihood ratio F-test (Robinson et al., 2010). Gene Ontology, KEGG, and Reactome enrichment was performed using topGO and clusterProfiler Bioconductor packages (Yu et al., 2012). For this purpose, we used Fisher exact test and lists of top 50, 100, 200, 500, and 1,000 up- or down-regulated genes that passed p < 0.05 threshold. Fisher test was applied for each of these top lists, and then the results were merged. As a background, we took only genes that passed the pre-filtering by the expression level. KEGG pathways visualization was performed using modification of pathview package (Luo and Brouwer, 2013) as described earlier (Moskalev et al., 2016).
Reverse Transcription and qPCR Analysis
The total RNA isolated for the RNA-Seq library preparation was used for qPCR analysis. First-strand cDNA was synthesized using 0.5 μg of total RNA, random primers and Mint reverse transcriptase (Evrogen, Russian Federation). Real-time quantitative PCR reactions were performed in triplicate in the presence of the EvaGreenTM dye (Biotium Inc., USA) and gene specific primers (Table 1) on the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, USA). The cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 60 s, and 72°C for 30 s. act5c and tub84B were used as reference genes. The relative expression ratios were calculated using the 2–ΔΔCt method as previously described (Dmitriev et al., 2016). The Wilcoxon and Mann-Whitney tests were applied for assessment of differences in mRNA expression. P ≤ 0.05 were considered statistically significant.
Results
In this study, we analyzed the effects of Gclc overexpression in the neuronal tissues of D. melanogaster on gene expression profiles in the thorax. We derived RNA-Seq expression profiles for 9,000 genes (after eliminating the genes with low expression). Drosophila aging was followed by the differential expression of 2626 genes (p < 0.05). 1488 of 2626 passed FDR < 0.05 threshold. The expression of 667 genes (p < 0.05) was associated with the activation of Gclc in all groups: young/mature/old or males/females (48 of 667 have FDR < 0.05). 58 of 667 genes demonstrated 2-fold or higher expression level changes (Supplementary Table 1). Using the STRING (Search Tool for Genesis/Proteins database) we received some information on the interactions of proteins encoded by these genes and showed that the most enriched pathway is the defense response to Gram-positive bacterium (FDR = 0.0019; Figure 1). As can be seen, the genes included in the proteolysis GO category (highlighted in green) also form a cluster. It is known that there is an increase in proteolysis of myofibril proteins during muscular dystrophy (Costelli et al., 2005; Combaret et al., 2009). According to the results of the DE analysis, overexpression of Gclc in thorax repressed genes involved in the proteolysis process.
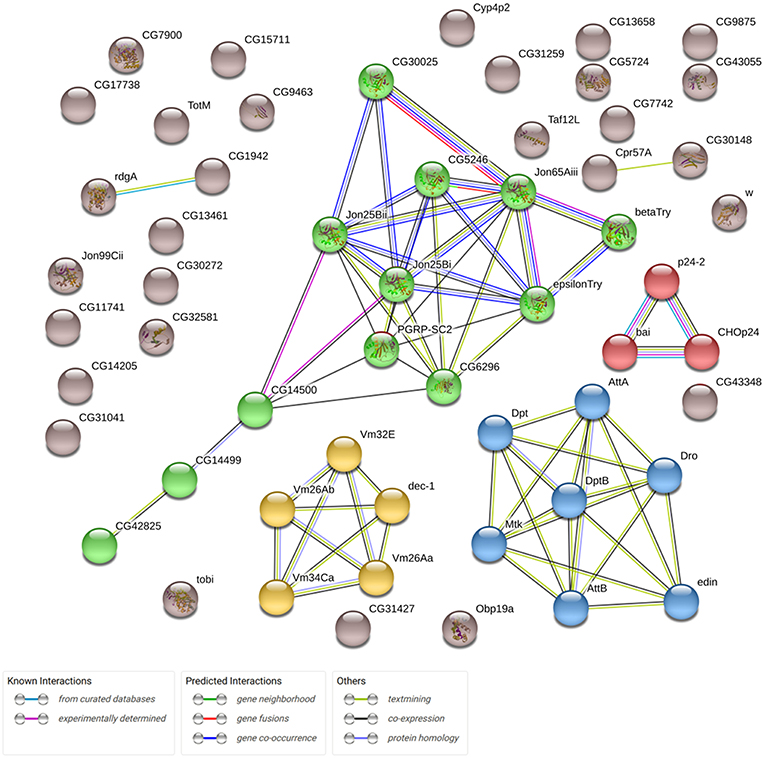
Figure 1. Network demonstrating associations (protein interactions, co-expression, etc.) between 58 genes, expression of which is altered in transgenic flies (overexpressing Gclc; STRING database). Clusters formed by genes are marked different colors.
We also verified the expression levels of some muscle-related genes using the qPCR method (Supplementary Table 2 and Supplementary Figure 1), the results showed a similar change in expression of Mlp60A, Act88F, and Arc1 genes compared with the DE analysis.
The survival analysis of the examined flies demonstrated that the increase of median and maximum lifespan of transgenic Drosophila is more pronounced in females than in males (Moskalev et al., 2016). In this study, we compared the resulting DE genes list (old vs. young comparisons for males and female's groups) as depicted in the Venn diagram of Figure 2, which resulted in 19 common DE genes shared by all 4 lists, 25 DE genes shared by Tg males and Tg females. There are only 6 genes common in NTg males and NTg females lists of DE genes. These genes are presented in Supplementary Table 3.
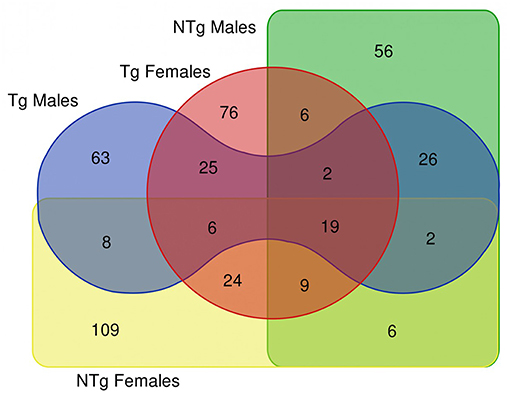
Figure 2. Venn diagram illustrating the results of differential gene expression analysis (old vs. young flies) in various groups of samples. Tg-transgenic, Gclc-overexpressing; NTg-non-transgenic. In Venn diagram, we included differentially expressed genes that passed thresholds: fold change > 4 and p < 0.05.
KEGG pathway enrichment analysis suggested that the activation of transgenic Gclc is followed with the differential expression of genes involved in a variety of cellular processes including Drug metabolism, Metabolism of xenobiotics by cytochrome P450, Glutathione metabolism, Starch and sucrose metabolism, Neuroactive ligand-receptor interaction, One carbon pool by folate, vesicle, and Cdc73/Paf1 complex (Fisher test p < 0.05).
Considering age-associated expression changes, we found predominant up-regulation of genes participating “One carbon pool by folate” pathway and a slight down-regulation of genes involved in Porphyrin metabolism and Neuroactive ligand-receptor interaction in Appl-GAL4 > UAS-Gclc flies (Supplementary Figure 2). For UAS-Gclc flies, we found aging-associated up-regulation of Genetic Information Processing, including the RNA polymerase, Ribosome, Protein export, and Protein processing in endoplasmic reticulum, whereas in the experimental group these changes were not observed (Figure 3 and Supplementary Figure 2).
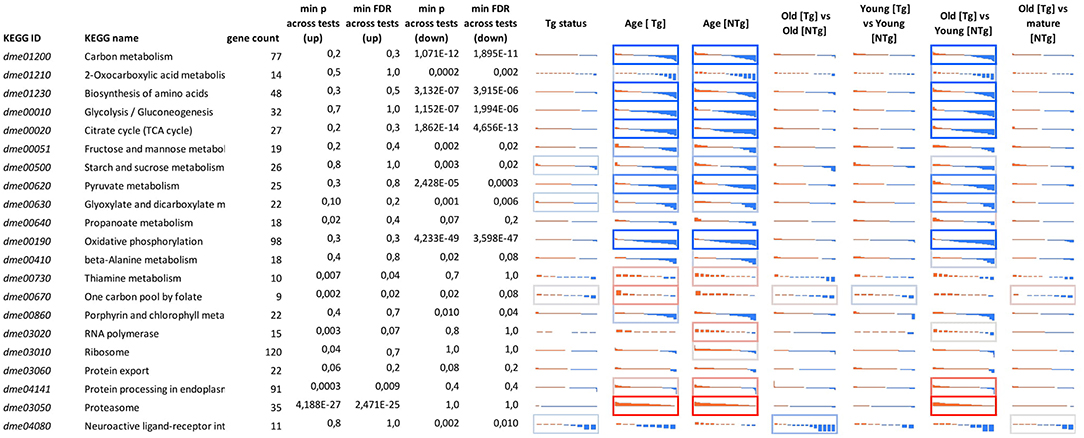
Figure 3. Differential expression profiles for genes participating in cellular pathways for different 641 genotypes (UAS-Gclc flies designated as Tg, Appl-GAL4 > UAS-Gclc flies—as NTg) and ages (old, mature, young). Logarithmic Fold Change (LogFC) (vertical axis) is ranged from −2 to +2, i.e., from four-fold decrease (blue) to 4-fold increase (red). Cell borders demonstrate the statistical significance of gene set enrichment analysis (Fisher test p-value): blue—enriched with down—regulated genes; and red—enriched with overexpressed ones min p-value across tests (up)—minimal enrichment p-value (Fisher exact test) across tests, only for up-regulated genes; min p-value across tests (down)—minimal enrichment p-value (Fisher exact test) across tests, only for down-regulated genes. DE genes that passed threshold LogCPM > 3 were presented.
Using the Reactome pathway database, we also identified several pathways enriched with genes, the expression of which is altered by Gclc overexpression. Among them: response to CaM pathway, DAG and IP3 signaling, Amino acid synthesis and interconversion (transamination), Peroxisomal lipid metabolism, Pyrimidine, and Purine catabolism (Supplementary Table 4).
Discussion
Overall, our data demonstrate that the neuronal Gclc overexpression resulted in less pronounced effects on the gene expression, the key signaling pathways involved in the aging process in the thorax than in the head. For example, the differential expression (DE) analysis revealed that only 58 genes (|LogFC|>1, p < 0.05) were differentially expressed in the thorax, while in the head, the neuronal Gclc overexpression altered the expression level of 188 genes. In heads, the Gclc overexpression influenced the following pathways: oxidative phosphorylation, ribosome biogenesis, Wnt, mTOR/PI3K, FOXO pathways, and enhanced cAMP signaling (Moskalev et al., 2016). In the thorax, no statistically significant effects of the neuronal Gclc overexpression were observed.
Only two genes: cytochrome P450 4p2 (Cyp4p2) and white (w), were differentially expressed in both the head and in the thorax. The Cyp4p2 gene encodes a member of the cytochrome P450 superfamily of enzymes. The w gene, famous for its role as the importer of eye pigments precursor, also plays a role in memory and cGMP transport (Evans et al., 2008; Sitaraman et al., 2008). The null mutation of this gene in D. melanogaster was associated with decreased lifespan, stress-resistance, and locomotor activity (Ferreiro et al., 2017). Using the GML model “~Gender + Genotype,” we indicated that the effect of Gclc expression on the transcriptional profile of the thorax is increased with age (Figure 4). In contrast, comparing the transcriptional profile of the Drosophila heads with Gclc overexpression demonstrated that the differences in gene expression are more pronounced in young flies than in old flies (Radyuk et al., 2012). There is data that the aging process in muscles is more dramatic than in the nervous system (Herndon et al., 2002). These results demonstrate a different contribution of Gclc overexpression in tissue-specific aging. According to our results, the DE genes in the thoraxes associated with aging are less than the age-related DE genes of the heads. On the other hand, the Zhan et al. in their work showed that the muscle had the largest number of age-related genes than the heads and other tissues of Drosophila (Zhan et al., 2007). The authors suggest that a characteristic of aging in muscle is the modulation of expression of genes involved in proteasomal and mitochondrial functions. Our data showed that the most of age-dependent up-regulated genes in thoraxes were associated with Protein processing in endoplasmic reticulum, Proteasome, RNA polymerase pathways, while down-regulated genes are related to metabolism and biosynthesis, such as Oxidative phosphorylation, Citrate cycle (TCA cycle), Biosynthesis of amino acids, Glycolysis/Gluconeogenesis, Glyoxylate and dicarboxylate metabolism, 2-Oxocarboxylic acid metabolism, Pyruvate metabolism, and carbohydrate metabolism. The age-related disorder of oxidative phosphorylation is well-known for different Drosophila body parts and other model organisms (Kim et al., 2005; Lesnefsky and Hoppel, 2006; Cannon et al., 2017). Cannon et al. also showed that carbohydrate metabolism was down-regulated in aged fly hearts, consisting of cardiac muscle (Cannon et al., 2017). This is consistent with our data. In addition, the age-dependent decrease in the expression level of citric acid cycle genes was observed in the heads of D. melanogaster and C. elegans (Mccarroll et al., 2004).
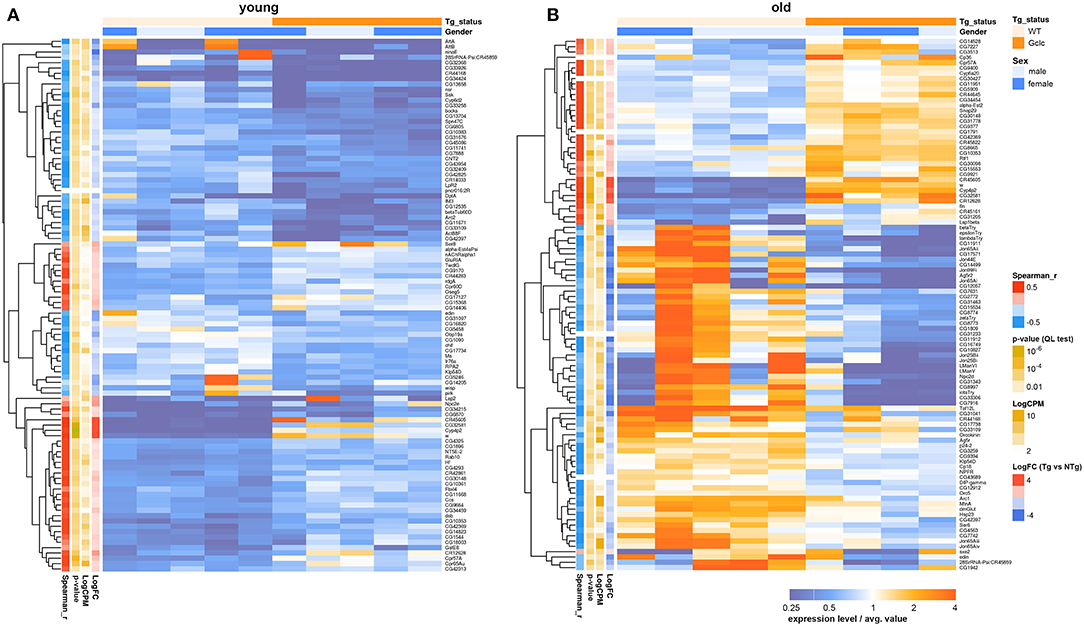
Figure 4. Heatmap illustrating differential gene expression profiles between transgenic (Tg) and non-transgenic (NTg) organisms: young (A) and old (B) flies. Heatmap colors indicate gene expression level in each sample, normalized to the average gene expression level across all the samples. Orange-above the average, blue-below the average value. LogFC-binary logarithm of gene expression level fold change between Tg and NTg organisms; p-value—edgeR's quasi-likelihood F-test p-value; LogCPM—binary logarithm of read counts per million; Spearman_r—Spearman rank correlation coefficient between Tg/NTg status and gene expression is presented.
It is known that aging is accompanied by the decline of muscle mass and strength (Demontis et al., 2013). This phenomenon is called sarcopenia and is associated with impaired physical performance, cognitive function, and mortality (Gonzalez-Freire et al., 2018; Landi and Calvani, 2018; Van Ancum et al., 2018). One of the major factors contributing to the development of sarcopenia may be the decline in antioxidant protection by glutathione (Mosoni et al., 2004; Sullivan-Gunn and Lewandowski, 2013). Previously, we demonstrated that neurospecific overexpression of Gclc delay aging-associated decline of the spontaneous locomotor activity of D. melanogaster males and females, compared to controls (Moskalev et al., 2016). The slowing down of aging-associated decline of muscular function may be linked with 16–29% increase in glutathione content that was detected by Orr et al. in the Drosophila headless bodies (thoraces plus abdomens) due to neurospecific Gclc overexpression (Orr et al., 2005).
While the imago thorax is primarily composed of muscle tissues, several DE genes are muscle-specific. We observed a 1.3–2-fold decrease in the level of muscle-specific protein genes in D. melanogaster thorax with Gclc overexpression (Figure 5). Using microarray analysis, Montana et al. demonstrated that the expression of muscle-specific proteins (Act88F, Act57B, Mlp60A, Msp300) and the homolog of the neuronal activity-regulated protein, ARC is up-regulated in Drosophila hypercontraction muscle mutants (Montana and Littleton, 2006). The authors concluded that the hypercontraction of muscles resulted in activation of actin cytoskeleton re-modulation mechanisms (Montana and Littleton, 2006). Among the differentially expressed genes in categories related to the muscle system process, we also observed that the Sclp gene was slightly decreased. Kuelzer et al. showed that Sclp plays an important role in programmed cell death of muscle (Kuelzer et al., 1999). In comparisons of old transgenic flies with the appropriate control group, we found a 1.8-fold increase in the expression of the genes (hbs and tum) involved in the fusion between myoblasts for the formation of syncytial myofibrils (Guerin and Kramer, 2009; Shelton et al., 2009). Sohn et al. showed that nephrin, the vertebrate homolog of the hbs, affects the development of the muscles and absence of nephrin results incompletely fused myotubes (Sohn et al., 2009). The tum gene also is conserved in C. elegans, Drosophila, humans, and involved in somatic muscle patterning (Guerin and Kramer, 2009). In the same comparison, the fln gene encoding muscle-specific protein, which is necessary for adult flies, was up-regulated more than two times (Henkin et al., 2004; Schnorrer et al., 2010). In old transgenic flies, we found an increase in the expression of the rotated abdomen (rt) gene encoding the POMT1 protein. POMT1 has a highly conserved homolog in nematodes and humans (Lyalin et al., 2006). It has been shown that the mutation of POMT1 results in muscular dystrophy and associated neural defects in the mammals (Akasaka-Manya et al., 2004; Haines et al., 2007). These results shed light on the mechanisms underlying delay of age-related locomotor activity decline in flies with Gclc overexpression (Moskalev et al., 2016).
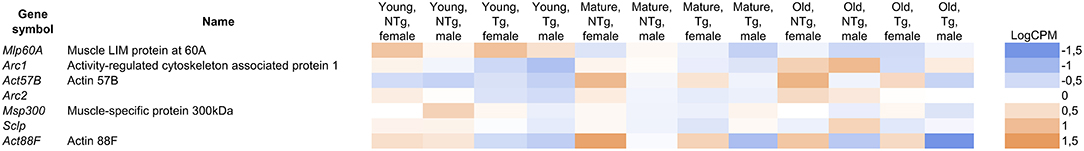
Figure 5. The differentially expressed genes in the categories related to the muscle system process. LogCPM (Counts per Million) is ranged from −1.5 to +1.5. Shades of different colors show the range of expression values (blue-down-regulated; orange-up-regulated). UAS-Gclc flies designated as Tg, Appl-GAL4 > UAS-Gclc flies—as NTg.
The analysis of KEGG pathways demonstrated that the One carbon pool by folate KEGG pathway was up-regulated during aging of D. melanogaster with Gclc overexpression compared with age-related changes in the control group (Supplementary Figure 2). These results correspond with the current paradigms. One-carbon metabolism is a crucial metabolic pathway that is connected with multiple physiological processes like de novo nucleotide synthesis and DNA methylation, and also provides a variety of metabolic intermediates which react either with pro-oxidants or promote antioxidant defense (Cavallaro et al., 2010; Vijaya Lakshmi et al., 2013).
Majumdar et al. demonstrated that folic acid and vitamin B12 can reduce the level of oxidative stress induced by the administration of arsenic trioxide. The authors consider that the prevention of mitochondrial dysfunction occurs due to the activation of antioxidant defense enzymes such as superoxide dismutase and catalase, and level of antioxidant glutathione (Majumdar et al., 2009). Vitamin B12 and folic acid play an important role in metabolism of homocysteine (Sahin et al., 2003). Elderly people with lower levels of folate and vitamin B12 have high homocysteine concentration, which is seen as a predictor of potential health problems such as Alzheimer's disease, cardiovascular disease, and loss of cognitive function (Koehler et al., 1997; Selhub, 2002). The up-regulation of this pathway was also observed by us in Drosophila flies exposed to different stress conditions like starvation, cold shock, or irradiation (Moskalev et al., 2015).
In our prior work, we demonstrated that the neuronal overexpression of Gclc gene resulted in a more pronounced increase of the median and maximum lifespan in Drosophila females than in males (Moskalev et al., 2016). Female longevity is observed in different taxa, but not well understood. In the present study, we found that overexpression of Gclc gene causes slight upregulation of Sod2 gene (in females), which participates Longevity regulating KEGG pathway. SOD acting in conjunction with glutathione provide a protective mechanism against destructive superoxide radicals and were shown to be involved in lifespan regulation of various model organisms including Drosophila species (Sohal et al., 2002; Tower, 2015). The Sod2 gene activity was associated with increased lifespan in Saccharomyces cerevisiae (Fabrizio et al., 2003; Unlu and Koc, 2007; Laschober et al., 2010), Drosophila melanogaster (Kirby et al., 2002; Curtis et al., 2007), and Mus musculus (Treiber et al., 2011). Male groups overexpressing Gclc exhibited no significant increases in Sod2 expression in comparison with their respective controls. We also showed that among the 25 DE genes found only in the Gclc-overexpressing female's comparison group (Supplementary Table 3), most of the genes are participants of the carbohydrate metabolism. Disruption of the carbohydrate metabolism and high-sugar intake are implicated in altering glucose homeostasis, influencing in a negative way the development, fertility, and lifespan in yeasts, worms, fruit flies, mammals (Wagner et al., 2015; Ravichandran et al., 2017; Alcantar-Fernandez et al., 2018). It is noteworthy that there are 3 Maltase genes (Mal-A3 Mal-A4, Mal-A7), the differential expression of which were found only in transgenic females. A recent study conducted by Inomata and co-workers identified that the maltase enzymes are responsible for dietary carbohydrate changes and could increase the capacity of the response associated with environmental changes (Inomata et al., 2019).
The sleep process is a physiological behavior observed in almost all organisms. The sleep process is controlled by two processes: sleep homeostasis and circadian rhythm (Borbely and Achermann, 1999). It is known that these two processes can work independently, but if they directly influence each other is still questionable (Deboer, 2018). In our prior studies, analysis demonstrated that the neuronal Gclc gene overexpression declined age-dependent alterations in circadian rhythmicity (Moskalev et al., 2016). These effects were accompanied by increased mRNA levels of a number of genes, which products are involved in regulation of circadian rhythms. It is known that the expression of these genes is regulated by the mechanism of a negative feedback loop. In brief, this loop can be described as follows: The CLOCK/BMAL1 heterodimers bind to the Per and Cry promoters to initiate their transcription. The protein products of Per and Cry genes form a complex in the cytoplasm with other proteins, such as CK1ε, and translocate back to the nucleus, where they inhibit the CLOCK/BMAL1 complex and repress transcription of their own and other genes (Yu et al., 2006; Lowrey and Takahashi, 2011). In contrast, in this study, the Reactome pathway analysis revealed several down-regulated pathways involved in circadian rhythms, such as Degradation of PER, Degradation of CRY, and Circadian Clock pathway. We found that heimdall (hll) gene encoding the long-chain fatty acid-CoA ligase, was significantly overexpressed (threshold of 4-fold) in Appl-GAL4 > UAS-Gclc flies. Recently, Thimgan et al. demonstrated that the expression of hll alters lipid metabolism (Thimgan et al., 2015). Interestingly, it was also demonstrated that hll can modulate sleep homeostasis (Thimgan et al., 2015). Lipid metabolism and sleep are related to each other according to Taheri et al. (2004) and Nedeltcheva et al. (2009). A number of genes associated with lipid metabolism can modulate lifespan of model organisms (Bolger et al., 2014). Thus, Gclc overexpression had some effects on both processes that control sleep.
Gclc overexpression in the D. melanogaster thorax up-regulated the rdgA gene. Inoue et al. revealed that the rdgA gene codes for an ATP-dependent Diacylglycerol kinase (DAGK) that converts DAG to phosphatidic acid (Inoue et al., 1989). According to the Reactome pathway analysis, the elements of phosphatidic acid biosynthetic process were significantly up-regulated in the thorax of flies with Gclc overexpression. Acting as signaling molecules, members of the DAG kinase family can take part in various biological processes such as synaptic transmission, photoreceptor transduction, and acquired and innate immunity (Merida et al., 2008). However, Lin et al. observed that the knockdown of rdgA gene in D. melanogaster and its ortholog dgk-5 in C. elegans both resulted in increased lifespan of model organisms. They assumed that the effects were associated with reduced TOR signaling (Lin et al., 2014).
In D. melanogaster with Gclc overexpression, we observed 1.5–4-fold increase in expression of some genes involved in eggshell chorion formation (Cp18, Vm26Aa, Vm34Ca). Several genome-wide transcriptome studies showed that the expression levels of genes, which participates in eggshell chorion formation, decline with age (Pletcher et al., 2002; Carlson et al., 2015).
Using the GLM model “~Age + Gender + Genotype,” we compared the expression profiles of old Appl-GAL4 > UAS-Gclc flies with control groups of different ages. The old flies with Gclc overexpression consistently demonstrated expression profiles more similar to control mature flies than control flies of the same age. We found that the Dsh gene involved in canonical and non-canonical Wnt signaling pathways was up-regulated in old Appl-GAL4 > UAS-Gclc flies. Disheveled (Dsh) acts directly downstream of frizzled receptors and serves as an intermediary between frizzled and glycogen synthase kinase-3β. Dsh also participates in the planar cell polarity (PCP) pathway and the Wnt/Ca2+ signaling pathway. The PCP pathway regulates the proper orientation of wing hairs and thoracic bristles in Drosophila (Kramer, 2016).
Thus, neuronal Gclc overexpression in Drosophila induces tissue-specific transcriptomic changes in the thorax that may be associated with anti-aging effects such as delay of age-related alterations in locomotor activity and circadian rhythmicity. However, for more substantiated conclusions about the causative link between overexpression of Gclc in neurons, tissue-specific transcriptomic changes in muscles and anti-aging effects on the organismal level, more experimental evidences may be required.
Data Availability
The sequencing data are available through the NCBI Sequence Read Archive (project ID PRJNA474212).
Author Contributions
AM, MS, GK, ZG, EL, and AZ wrote the manuscript text. EP, MS, LK, ZG, and EL carried out the experiments. GK and ZG carried out the bioinformatic analysis. AM and AK supervised the bioinformatic research and text of the manuscript. All authors read and approved the final manuscript.
Funding
This work and publication costs were funded by the Russian Science Foundation grant N 17-74-30030.
Conflict of Interest Statement
AZ was employed by company Insilico Medicine.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Institute of Biology of Komi Science Center of Ural Branch of Russian Academy of Sciences and Moscow Institute of Physics and Technology for scientific advice, Drosophila strains, and for assistance in the experiments with Drosophila and Insilico Medicine, Inc. for assistance in data analysis. This work was performed using EIMB RAS Genome center equipment (http://www.eimb.ru/RUSSIAN_NEW/INSTITUTE/ccu_genome_c.php).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00149/full#supplementary-material
Abbreviations
bp, base pairs; DAG, Diacylglycerol; DAGK, Diacylglycerol kinase; ds cDNA, double-stranded cDNA; Gclc, Glutamate-cysteine ligase catalytic subunit; GO, Gene Ontology; DE, differentially expressed; FC, fold changes; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCP, planar cell polarity pathway.
References
Akasaka-Manya, K., Manya, H., and Endo, T. (2004). Mutations of the POMT1 gene found in patients with Walker-Warburg syndrome lead to a defect of protein O-mannosylation. Biochem. Biophys. Res. Commun. 325, 75–79. doi: 10.1016/j.bbrc.2004.10.001
Alcantar-Fernandez, J., Navarro, R. E., Salazar-Martinez, A. M., Perez-Andrade, M. E., and Miranda-Rios, J. (2018). Caenorhabditis elegans respond to high-glucose diets through a network of stress-responsive transcription factors. PLoS ONE 13:e0199888. doi: 10.1371/journal.pone.0199888
Blackwell, T. K., Steinbaugh, M. J., Hourihan, J. M., Ewald, C. Y., and Isik, M. (2015). SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88, 290–301. doi: 10.1016/j.freeradbiomed.2015.06.008
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borbely, A. A., and Achermann, P. (1999). Sleep homeostasis and models of sleep regulation. J. Biol. Rhythms 14, 557–568.
Cannon, L., Zambon, A. C., Cammarato, A., Zhang, Z., Vogler, G., Munoz, M., et al. (2017). Expression patterns of cardiac aging in Drosophila. Aging Cell 16, 82–92. doi: 10.1111/acel.12559
Carlson, K. A., Gardner, K., Pashaj, A., Carlson, D. J., Yu, F., Eudy, J. D., et al. (2015). Genome-wide gene expression in relation to age in large laboratory cohorts of Drosophila melanogaster. Genet. Res. Int. 2015:835624. doi: 10.1155/2015/835624
Cavallaro, R. A., Fuso, A., Nicolia, V., and Scarpa, S. (2010). S-adenosylmethionine prevents oxidative stress and modulates glutathione metabolism in TgCRND8 mice fed a B-vitamin deficient diet. J. Alzheimers. Dis. 20, 997–1002. doi: 10.3233/JAD-2010-091666
Chandran, S., Suggs, J. A., Wang, B. J., Han, A., Bhide, S., Cryderman, D. E., et al. (2018). Suppression of myopathic lamin mutations by muscle-specific activation of AMPK and modulation of downstream signaling. Hum. Mol. Genet. 28, 351–371. doi: 10.1093/hmg/ddy332
Chen, J., Zou, Q., Lv, D., Wei, Y., Raza, M. A., Chen, Y., et al. (2018). Comprehensive transcriptional landscape of porcine cardiac and skeletal muscles reveals differences of aging. Oncotarget 9, 1524–1541. doi: 10.18632/oncotarget.23290
Combaret, L., Dardevet, D., Bechet, D., Taillandier, D., Mosoni, L., and Attaix, D. (2009). Skeletal muscle proteolysis in aging. Curr. Opin. Clin. Nutr. Metab. Care 12, 37–41. doi: 10.1097/MCO.0b013e32831b9c31
Costelli, P., Reffo, P., Penna, F., Autelli, R., Bonelli, G., and Baccino, F. M. (2005). Ca2+-dependent proteolysis in muscle wasting. Int. J. Biochem. Cell Biol. 37, 2134–2146. doi: 10.1016/j.biocel.2005.03.010
Curtis, C., Landis, G. N., Folk, D., Wehr, N. B., Hoe, N., Waskar, M., et al. (2007). Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol. 8:R262. doi: 10.1186/gb-2007-8-12-r262
Deboer, T. (2018). Sleep homeostasis and the circadian clock: do the circadian pacemaker and the sleep homeostat influence each other's functioning? Neurobiol. Sleep Circ. Rhythms 5, 68–77. doi: 10.1016/j.nbscr.2018.02.003
Demontis, F., Piccirillo, R., Goldberg, A. L., and Perrimon, N. (2013). Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis. Model. Mech. 6, 1339–1352. doi: 10.1242/dmm.012559
Dmitriev, A. A., Kudryavtseva, A. V., Krasnov, G. S., Koroban, N. V., Speranskaya, A. S., Krinitsina, A. A., et al. (2016). Gene expression profiling of flax (Linum usitatissimum L.) under edaphic stress. BMC Plant Biol. 16:237. doi: 10.1186/s12870-016-0927-9
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Evans, J. M., Day, J. P., Cabrero, P., Dow, J. A., and Davies, S. A. (2008). A new role for a classical gene: white transports cyclic GMP. J. Exp. Biol. 211, 890–899. doi: 10.1242/jeb.014837
Fabrizio, P., Liou, L. L., Moy, V. N., Diaspro, A., Valentine, J. S., Gralla, E. B., et al. (2003). SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 163, 35–46.
Ferreiro, M. J., Perez, C., Marchesano, M., Ruiz, S., Caputi, A., Aguilera, P., et al. (2017). Drosophila melanogaster White Mutant w1118 Undergo Retinal Degeneration. Front. Neurosci. 11:732. doi: 10.3389/fnins.2017.00732
Girardot, F., Lasbleiz, C., Monnier, V., and Tricoire, H. (2006). Specific age-related signatures in Drosophila body parts transcriptome. BMC Genomics 7:69. doi: 10.1186/1471-2164-7-69
Gonzalez-Freire, M., Adelnia, F., Moaddel, R., and Ferrucci, L. (2018). Searching for a mitochondrial root to the decline in muscle function with ageing. J. Cachexia Sarcopenia Muscle. 9, 435–440. doi: 10.1002/jcsm.12313
Guerin, C. M., and Kramer, S. G. (2009). RacGAP50C directs perinuclear gamma-tubulin localization to organize the uniform microtubule array required for Drosophila myotube extension. Development 136, 1411–1421. doi: 10.1242/dev.031823
Haines, N., Seabrooke, S., and Stewart, B. A. (2007). Dystroglycan and protein O-mannosyltransferases 1 and 2 are required to maintain integrity of Drosophila larval muscles. Mol. Biol. Cell 18, 4721–4730. doi: 10.1091/mbc.e07-01-0047
Henkin, J. A., Maughan, D. W., and Vigoreaux, J. O. (2004). Mutations that affect flightin expression in Drosophila alter the viscoelastic properties of flight muscle fibers. Am. J. Physiol. Cell Physiol. 286, C65–72. doi: 10.1152/ajpcell.00257.2003
Herndon, L. A., Schmeissner, P. J., Dudaronek, J. M., Brown, P. A., Listner, K. M., Sakano, Y., et al. (2002). Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419, 808–814. doi: 10.1038/nature01135
Inomata, N., Takahasi, K. R., and Koga, N. (2019). Association between duplicated maltase genes and the transcriptional regulation for the carbohydrate changes in Drosophila melanogaster. Gene 686, 141–145. doi: 10.1016/j.gene.2018.11.007
Inoue, Y., Bong-Young, C., Murata, K., and Kimura, A. (1989). Sexual response in Saccharomyces cerevisiae: alteration of enzyme activity in the glyoxalase system by mating factor. Biochem. Biophys. Res. Commun. 165, 1091–1095.
Kim, J. Y., Park, Y. K., Lee, K. P., Lee, S. M., Kang, T. W., Kim, H. J., et al. (2014). Genome-wide profiling of the microRNA-mRNA regulatory network in skeletal muscle with aging. Aging 6, 524–544. doi: 10.18632/aging.100677
Kim, S. N., Rhee, J. H., Song, Y. H., Park, D. Y., Hwang, M., Lee, S. L., et al. (2005). Age-dependent changes of gene expression in the Drosophila head. Neurobiol. Aging 26, 1083–1091. doi: 10.1016/j.neurobiolaging.2004.06.017
Kirby, K., Hu, J., Hilliker, A. J., and Phillips, J. P. (2002). RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc. Natl. Acad. Sci. USA. 99, 16162–16167. doi: 10.1073/pnas.252342899
Koehler, K. M., Pareo-Tubbeh, S. L., Romero, L. J., Baumgartner, R. N., and Garry, P. J. (1997). Folate nutrition and older adults: challenges and opportunities. J. Am. Diet. Assoc. 97, 167–173. doi: 10.1016/S0002-8223(97)00044-8
Kramer, I. M. (ed.). (2016). “WNT signaling and the regulation of cell adhesion and differentiation,” in Signal Transduction (Third Edition) (Boston: Academic Press), 703–740.
Krasnov, G. S., Dmitriev, A. A., Kudryavtseva, A. V., Shargunov, A. V., Karpov, D. S., Uroshlev, L. A., et al. (2015). PPLine: an automated pipeline for SNP, SAP, and splice variant detection in the context of proteogenomics. J. Proteome Res. 14, 3729–3737. doi: 10.1021/acs.jproteome.5b00490
Kreipke, R. E., Kwon, Y. V., Shcherbata, H. R., and Ruohola-Baker, H. (2017). Drosophila melanogaster as a Model of Muscle Degeneration Disorders. Curr. Top. Dev. Biol. 121, 83–109. doi: 10.1016/bs.ctdb.2016.07.003
Kuelzer, F., Kuah, P., Bishoff, S. T., Cheng, L., Nambu, J. R., and Schwartz, L. M. (1999). Cloning and analysis of small cytoplasmic leucine-rich repeat protein (SCLP), a novel, phylogenetically-conserved protein that is dramatically up-regulated during the programmed death of moth skeletal muscle. J. Neurobiol. 41, 482–494.
Landi, F., and Calvani, R. (2018). Editorial: protein and sarcopenia: experimental data and clinical evidence. Curr. Protein Pept. Sci. 19:632. doi: 10.2174/138920371907180514125341
Larsson, L., Degens, H., Li, M., Salviati, L., Lee, Y. I., Thompson, W., et al. (2019). Sarcopenia: Aging-Related Loss of Muscle Mass and Function. Physiol. Rev. 99, 427–511. doi: 10.1152/physrev.00061.2017
Laschober, G. T., Ruli, D., Hofer, E., Muck, C., Carmona-Gutierrez, D., Ring, J., et al. (2010). Identification of evolutionarily conserved genetic regulators of cellular aging. Aging Cell 9, 1084–1097. doi: 10.1111/j.1474-9726.2010.00637.x
Lesnefsky, E. J., and Hoppel, C. L. (2006). Oxidative phosphorylation and aging. Ageing Res. Rev. 5, 402–433. doi: 10.1016/j.arr.2006.04.001
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Lin, Y. H., Chen, Y. C., Kao, T. Y., Lin, Y. C., Hsu, T. E., Wu, Y. C., et al. (2014). Diacylglycerol lipase regulates lifespan and oxidative stress response by inversely modulating TOR signaling in Drosophila and C. elegans. Aging Cell 13, 755–764. doi: 10.1111/acel.12232
Liu, H., Wang, H., Shenvi, S., Hagen, T. M., and Liu, R. M. (2004). Glutathione metabolism during aging and in Alzheimer disease. Ann. N. Y. Acad. Sci. 1019, 346–349. doi: 10.1196/annals.1297.059
Lowrey, P. L., and Takahashi, J. S. (2011). Genetics of circadian rhythms in Mammalian model organisms. Adv. Genet. 74, 175–230. doi: 10.1016/B978-0-12-387690-4.00006-4
Luchak, J. M., Prabhudesai, L., Sohal, R. S., Radyuk, S. N., and Orr, W. C. (2007). Modulating longevity in Drosophila by over- and underexpression of glutamate-cysteine ligase. Ann. N. Y. Acad. Sci. 1119, 260–273. doi: 10.1196/annals.1404.000
Luo, W., and Brouwer, C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. doi: 10.1093/bioinformatics/btt285
Lyalin, D., Koles, K., Roosendaal, S. D., Repnikova, E., Van Wechel, L., and Panin, V. M. (2006). The twisted gene encodes Drosophila protein O-mannosyltransferase 2 and genetically interacts with the rotated abdomen gene encoding Drosophila protein O-mannosyltransferase 1. Genetics 172, 343–353. doi: 10.1534/genetics.105.049650
Majumdar, S., Mukherjee, S., Maiti, A., Karmakar, S., Das, A. S., Mukherjee, M., et al. (2009). Folic acid or combination of folic acid and vitamin B(12) prevents short-term arsenic trioxide-induced systemic and mitochondrial dysfunction and DNA damage. Environ. Toxicol. 24, 377–387. doi: 10.1002/tox.20442
Mccarroll, S. A., Murphy, C. T., Zou, S., Pletcher, S. D., Chin, C. S., Jan, Y. N., et al. (2004). Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 36, 197–204. doi: 10.1038/ng1291
Merida, I., Avila-Flores, A., and Merino, E. (2008). Diacylglycerol kinases: at the hub of cell signalling. Biochem. J. 409, 1–18. doi: 10.1042/BJ20071040
Mockett, R. J., and Nobles, A. C. (2013). Lack of robustness of life extension associated with several single-gene P element mutations in Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 68, 1157–1169. doi: 10.1093/gerona/glt031
Montana, E. S., and Littleton, J. T. (2006). Expression profiling of a hypercontraction-induced myopathy in Drosophila suggests a compensatory cytoskeletal remodeling response. J. Biol. Chem. 281, 8100–8109. doi: 10.1074/jbc.M512468200
Moskalev, A., Shaposhnikov, M., Proshkina, E., Belyi, A., Fedintsev, A., Zhikrivetskaya, S., et al. (2016). The influence of pro-longevity gene Gclc overexpression on the age-dependent changes in Drosophila transcriptome and biological functions. BMC Genomics 17:1046. doi: 10.1186/s12864-016-3356-0
Moskalev, A., Zhikrivetskaya, S., Krasnov, G., Shaposhnikov, M., Proshkina, E., Borisoglebsky, D., et al. (2015). A comparison of the transcriptome of Drosophila melanogaster in response to entomopathogenic fungus, ionizing radiation, starvation and cold shock. BMC Genomics 16(Suppl. 13):S8. doi: 10.1186/1471-2164-16-S13-S8
Mosoni, L., Breuille, D., Buffiere, C., Obled, C., and Mirand, P. P. (2004). Age-related changes in glutathione availability and skeletal muscle carbonyl content in healthy rats. Exp. Gerontol. 39, 203–210. doi: 10.1016/j.exger.2003.10.014
Nedeltcheva, A. V., Kilkus, J. M., Imperial, J., Kasza, K., Schoeller, D. A., and Penev, P. D. (2009). Sleep curtailment is accompanied by increased intake of calories from snacks. Am. J. Clin. Nutr. 89, 126–133. doi: 10.3945/ajcn.2008.26574
Orr, W. C., Radyuk, S. N., Prabhudesai, L., Toroser, D., Benes, J. J., Luchak, J. M., et al. (2005). Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J. Biol. Chem. 280, 37331–37338. doi: 10.1074/jbc.M508272200
Pletcher, S. D., Macdonald, S. J., Marguerie, R., Certa, U., Stearns, S. C., Goldstein, D. B., et al. (2002). Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12, 712–723. doi: 10.1016/S0960-9822(02)00808-4
Radyuk, S. N., Gambini, J., Borras, C., Serna, E., Klichko, V. I., Viña, J., et al. (2012). Age-dependent changes in the transcription profile of long-lived Drosophila over-expressing glutamate cysteine ligase. Mech. Ageing Dev. 133, 401–413. doi: 10.1016/j.mad.2012.04.006
Rai, M., Nongthomba, U., and Grounds, M. D. (2014). Skeletal muscle degeneration and regeneration in mice and flies. Curr. Top. Dev. Biol. 108, 247–281. doi: 10.1016/B978-0-12-391498-9.00007-3
Ravichandran, M., Grandl, G., and Ristow, M. (2017). Dietary Carbohydrates Impair Healthspan and Promote Mortality. Cell Metab. 26, 585–587. doi: 10.1016/j.cmet.2017.09.011
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sahin, K., Onderci, M., Sahin, N., Gursu, M. F., and Kucuk, O. (2003). Dietary vitamin C and folic acid supplementation ameliorates the detrimental effects of heat stress in Japanese quail. J. Nutr. 133, 1882–1886. doi: 10.1093/jn/133.6.1882
Schnorrer, F., Schonbauer, C., Langer, C. C., Dietzl, G., Novatchkova, M., Schernhuber, K., et al. (2010). Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464, 287–291. doi: 10.1038/nature08799
Selhub, J. (2002). Folate, vitamin B12 and vitamin B6 and one carbon metabolism. J. Nutr. Health Aging 6, 39–42.
Shelton, C., Kocherlakota, K. S., Zhuang, S., and Abmayr, S. M. (2009). The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development 136, 1159–1168. doi: 10.1242/dev.026302
Sigurgeirsson, B., Emanuelsson, O., and Lundeberg, J. (2014). Sequencing degraded RNA addressed by 3' tag counting. PLoS ONE 9:e91851. doi: 10.1371/journal.pone.0091851
Sitaraman, D., Zars, M., Laferriere, H., Chen, Y. C., Sable-Smith, A., Kitamoto, T., et al. (2008). Serotonin is necessary for place memory in Drosophila. Proc. Natl. Acad. Sci. USA. 105, 5579–5584. doi: 10.1073/pnas.0710168105
Sohal, R. S., Mockett, R. J., and Orr, W. C. (2002). Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic. Biol. Med. 33, 575–586. doi: 10.1016/S0891-5849(02)00886-9
Sohn, R. L., Huang, P., Kawahara, G., Mitchell, M., Guyon, J., Kalluri, R., et al. (2009). A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc Natl Acad Sci U.S.A. 106, 9274–9279. doi: 10.1073/pnas.0904398106
Sullivan-Gunn, M. J., and Lewandowski, P. A. (2013). Elevated hydrogen peroxide and decreased catalase and glutathione peroxidase protection are associated with aging sarcopenia. BMC Geriatr. 13:104. doi: 10.1186/1471-2318-13-104
Sykiotis, G. P., Habeos, I. G., Samuelson, A. V., and Bohmann, D. (2011). The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr. Opin. Clin. Nutr. Metab. Care 14, 41–48. doi: 10.1097/MCO.0b013e32834136f2
Taheri, S., Lin, L., Austin, D., Young, T., and Mignot, E. (2004). Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 1:e62. doi: 10.1371/journal.pmed.0010062
Thimgan, M. S., Seugnet, L., Turk, J., and Shaw, P. J. (2015). Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila. Sleep 38, 801–814. doi: 10.5665/sleep.4680
Tower, J. (2015). “Superoxide dismutase (SOD) genes and aging in Drosophila,” in Life Extension. Healthy Ageing and Longevity, eds A. Vaiserman, A. Moskalev, and E. Pasyukova. (New York, NY; Dordrecht; London: Springer), 67–81.
Treiber, N., Maity, P., Singh, K., Kohn, M., Keist, A. F., Ferchiu, F., et al. (2011). Accelerated aging phenotype in mice with conditional deficiency for mitochondrial superoxide dismutase in the connective tissue. Aging Cell 10, 239–254. doi: 10.1111/j.1474-9726.2010.00658.x
Unlu, E. S., and Koc, A. (2007). Effects of deleting mitochondrial antioxidant genes on life span. Ann. N. Y. Acad. Sci. 1100, 505–509. doi: 10.1196/annals.1395.055
Van Ancum, J. M., Pijnappels, M., Jonkman, N. H., Scheerman, K., Verlaan, S., Meskers, C. G. M., et al. (2018). Muscle mass and muscle strength are associated with pre- and post-hospitalization falls in older male inpatients: a longitudinal cohort study. BMC Geriatr. 18:116. doi: 10.1186/s12877-018-0812-5
Vijaya Lakshmi, S. V., Naushad, S. M., Seshagiri Rao, D., and Kutala, V. K. (2013). Oxidative stress is associated with genetic polymorphisms in one-carbon metabolism in coronary artery disease. Cell Biochem. Biophys. 67, 353–361. doi: 10.1007/s12013-011-9322-1
Wagner, A. E., Piegholdt, S., Rabe, D., Baenas, N., Schloesser, A., Eggersdorfer, M., et al. (2015). Epigallocatechin gallate affects glucose metabolism and increases fitness and lifespan in Drosophila melanogaster. Oncotarget 6, 30568–30578. doi: 10.18632/oncotarget.5215
Wang, L., Wang, S., and Li, W. (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. doi: 10.1093/bioinformatics/bts356
White, R. R., Milholland, B., Macrae, S. L., Lin, M., Zheng, D., and Vijg, J. (2015). Comprehensive transcriptional landscape of aging mouse liver. BMC Genomics 16:899. doi: 10.1186/s12864-015-2061-8
Yang, J., Huang, T., Petralia, F., Long, Q., Zhang, B., Argmann, C., et al. (2015). Synchronized age-related gene expression changes across multiple tissues in human and the link to complex diseases. Sci. Rep. 5:15145. doi: 10.1038/srep15145
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Yu, W., Zheng, H., Houl, J. H., Dauwalder, B., and Hardin, P. E. (2006). PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 20, 723–733. doi: 10.1101/gad.1404406
Zhan, M., Yamaza, H., Sun, Y., Sinclair, J., Li, H., and Zou, S. (2007). Temporal and spatial transcriptional profiles of aging in Drosophila melanogaster. Genome Res. 17, 1236–1243. doi: 10.1101/gr.6216607
Zhang, S., Ratliff, E. P., Molina, B., El-Mecharrafie, N., Mastroianni, J., Kotzebue, R. W., et al. (2018). Aging and Intermittent Fasting Impact on Transcriptional Regulation and Physiological Responses of Adult Drosophila Neuronal and Muscle Tissues. Int. J. Mol. Sci. 19:1140. doi: 10.3390/ijms19041140
Keywords: Gclc, glutathione, lifespan, aging, gene expression, thorax, Drosophila melanogaster
Citation: Moskalev A, Guvatova Z, Shaposhnikov M, Lashmanova E, Proshkina E, Koval L, Zhavoronkov A, Krasnov G and Kudryavtseva A (2019) The Neuronal Overexpression of Gclc in Drosophila melanogaster Induces Life Extension With Longevity-Associated Transcriptomic Changes in the Thorax. Front. Genet. 10:149. doi: 10.3389/fgene.2019.00149
Received: 25 June 2018; Accepted: 12 February 2019;
Published: 05 March 2019.
Edited by:
Elena G. Pasyukova, Institute of Molecular Genetics (RAS), RussiaReviewed by:
Kyung-Jin Min, Inha University, South KoreaJohn Tower, University of Southern California, United States
Copyright © 2019 Moskalev, Guvatova, Shaposhnikov, Lashmanova, Proshkina, Koval, Zhavoronkov, Krasnov and Kudryavtseva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexey Moskalev, YW1vc2thbGV2QGxpc3QucnU=
 Alexey Moskalev
Alexey Moskalev Zulfiya Guvatova
Zulfiya Guvatova Mikhail Shaposhnikov
Mikhail Shaposhnikov Ekaterina Lashmanova
Ekaterina Lashmanova Ekaterina Proshkina
Ekaterina Proshkina Liubov Koval
Liubov Koval Alex Zhavoronkov
Alex Zhavoronkov George Krasnov
George Krasnov Anna Kudryavtseva
Anna Kudryavtseva