- State Key Laboratory of Pig Genetic Improvement and Production Technology, Jiangxi Agricultural University, Nanchang, China
The Duroc × (Landrace × Large White) hybrid pig (DLY) is the most popular commercial pig used in the Chinese pig industry. DLY pigs are usually white but sometimes show colored phenotypes. Colored DLY pigs are not favored by slaughterhouses and retailers, thus causing certain economic losses to farmers in China. In this study, we first conducted a genome-wide association study and RNA sequencing to demonstrate that KIT variants are responsible for diversifying coat color phenotypes segregating in a DLY population. We then defined the precise sizes and locations of four duplications (DUP1-4), four candidate causative mutations at the KIT locus, in the pig reference genome using the whole-genome sequence data of representative colored individuals. The sequence data also enabled us to identify a list of new KIT alleles. By investigating the association between these new alleles and coat color phenotypes, we provide further evidence that DUP2 is another causative mutation for the solid white coat color in pigs. DUP1 (the KIT gene duplication), DUP2 and the splice mutation are all required for the manifestation of a solid white coat color. DUP4 had a more significant effect on the formation of the belt phenotype compared with DUP3. Given the necessity of DUP2 for the solid white coat color, we detected IN/IN homozygotes lacking DUP2 in Large White and Landrace pigs and found that French Landrace pigs had the highest frequency (8.98%) of IN/IN individuals. This study not only advances our understanding of the molecular mechanism of the color phenotype in pigs, but also establishes a simple and accurate method for the screening of KIT IN/IN homozygotes in Large White and Landrace that would cause colored DLY pigs.
Introduction
Coat color is one of the most visualized breed features of domestic pigs. After long-term artificial selection, domestic pigs have formed diversifying coat color phenotypes (Porter, 1993; Legault et al., 1998), including black, red, white, spotted, brown, belted, two-end black, etc. The genetic studies of coat color in pigs trace back to the beginning of the last century (Spillman, 1906). To date, several major genes affecting pig coat color phenotypes have been identified. MC1R variants are responsible for the dominant black, black spotted, and red coat colors in both Western and Chinese pigs (Kijas et al., 1998, 2001; Fang et al., 2009). KIT variants determine the dominant white coat color in Western pigs (Johansson et al., 1996; Marklund et al., 1998). MITF mutation causes the recessive white coat color in Chinese Rongchang pigs (Chen et al., 2016). ASIP affects the black-and-tan coat color in Mangalitza pigs (Drögemüller et al., 2006). TYRP1 and EDNRB variants are responsible for the brown (Ren et al., 2010) and white belt coat colors (Ai et al., 2013) in Chinese pigs, respectively.
Multiple alleles for different coat color phenotypes have been identified at the KIT locus, including the recessive wild-type allele i, Patch allele Ip (Johansson et al., 1992, 1996), Belt allele IBe (Giuffra et al., 1999), Roan allele IBe∗ (Pielberg et al., 2002), and the Dominant white alleles I comprising I1, I2, I3 (Pielberg et al., 2002, 2003; Johansson et al., 2005). Recently, whole-genome resequencing has uncovered not only a 450 kb duplication (DUP1) encompassing the entire KIT gene, but also a 4.3-kb duplication (DUP2) ∼100 kb upstream of KIT, a 23-kb duplication (DUP3) ∼100 kb downstream of KIT, and another 4.3-kb duplication (DUP4) within DUP3 (Rubin et al., 2012). The wild-type i allele lacks the four duplications and the splice mutation, causing exon skipping in intron 17 (hereafter refer as to splice mutation). Allele Ip carries DUP1 but not DUP2-4 and the splice mutation. Allele IBe carries DUP2-4 but lacks DUP1 and the splice mutation. The I alleles have variable copy numbers of DUP1-4 and the splice mutation. DUP1, the splice mutation and most likely DUP2 are known to be causative mutations for the dominant white coat color (Giuffra et al., 2002; Rubin et al., 2012). Unequal crossover is prone to occur in duplicated regions during homologous recombination (Zhang, 2003). It is thus likely to create great haplotype diversity due to the variable copy numbers of DUP1-4 at the KIT locus. However, new KIT haplotypes (alleles) in addition to the above-mentioned ones have not been reported yet, and the effect of these new alleles on pig coat color remains elusive.
Here, we first conducted a genome-wide association study (GWAS) and RNA sequencing, illustrating that KIT is a major locus for the coat color phenotypes (Figure 1) segregating in a commercial pig population derived from a three-way cross: Duroc × (Landrace × Large White) (hereafter refer as to DLY). Then, whole-genome sequence data (20× depth) of representative colored individuals from the DLY population identified new alleles (haplotypes) at the KIT locus. Finally, we established the relation of these KIT new alleles to pig coat color phenotypes. These findings advanced our understanding of the molecular mechanism of coat color phenotypes in domestic pigs, and also allowed us to establish a simple but robust PCR-based test for one new KIT allele causing colored individuals in DLY pigs.
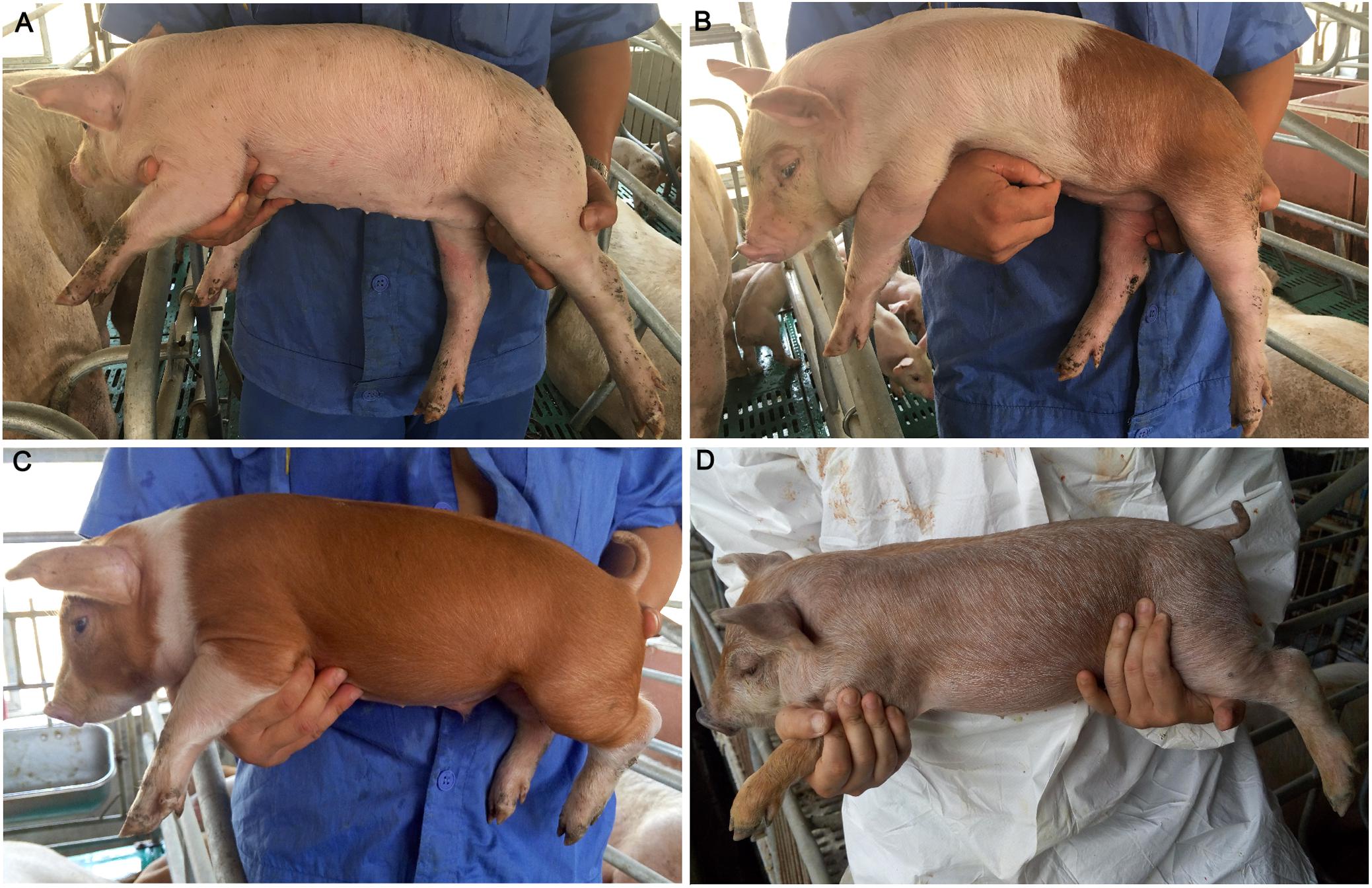
Figure 1. Coat color phenotypes in the tested population. (A) White. (B) White with reddish-brown spots. (C) Reddish-brown with white legs and belts. (D) Roan characterized by brown hairs intermingled with white hairs and most pigs had black spots on the snout.
Materials and Methods
Animals
In this study, ear tissue samples were collected from 190 DLY piglets raised in a commercial pig farm in Xinyu city, Jiangxi province, China. These piglets were derived from a three-way cross between American Duroc boars and French Landrace-Large White hybrid sows. Genomic DNA was extracted from the ear tissue of each pig using a genomic DNA extraction kit (Generay, China) according to the manufacturer’s instructions.
Phenotypic Measurement
Four coat color phenotypes were observed in the 190 piglets, including (1) white, (2) white with reddish-brown spots, (3) reddish-brown with white legs and belts, (4) roan characterized by brown hairs intermingled with white hairs and most pigs had black spots on the snout (Figure 1). Each individual was recorded for its coat color phenotype using a cell phone camera. Graphic processing was performed to calculate the proportion of reddish-brown hair using PHOTOSHOP CS (Adobe Systems Incorporated, United States) as previously described (Fan et al., 2014). Due to bad photo angles, 87 individuals were discarded for the graphic processing and 103 individuals were used to calculate the proportion of reddish-brown hair. The calculated values were then classified into three scores: 0 for 0% (n = 20), 1 for 1–50% (n = 43), and 2 for 51–100% (n = 40). These scores were used as the coat color phenotypic values of the 103 individuals for subsequent analysis.
Genome Wide Association Study
The 103 DLY piglets were genotyped for 68,528 SNPs using the Porcine 80K Genotyping array (Illumina, United States). SNPs were filtered using PLINK v1.90 (Purcell et al., 2007) by removing those with ambiguous locations, minor allele frequencies of less than 0.01, or genotyping rates of less than 0.95. Individuals with SNP call rates of less than 0.90 were also discarded. After the filtering process, 55,117 SNPs from the 103 individuals were retained for subsequent GWAS.
The GWAS was performed using a mixed linear model implemented in the Genome-wide Efficient Mixed-Model Association (GEMMA) software (Zhou and Stephens, 2012). The phenotypic values were regressed on sex and batch, and the residuals were used as the new phenotypes. Then, a univariate linear mixed model regression was executed with phenotypic trait, genotype, and relatedness matrix files (Wang et al., 2018). The threshold of genomic significance was set by the Bonferroni method: 0.05/the number of tested SNPs (Yang et al., 2005).
RNA Sequencing
Four reddish-brown and four white dermal tissues were collected from DLY piglets raised in the same farm in Xinyu city, Jiangxi province, China. Dermal tissues were preserved in RNA later and stored at -80°C before use. Total RNA was extracted using the Trizol reagent (Invitrogen, United States) following the manufacturer’s protocol. RNA quality was assessed via agarose gel electrophoresis and a NanoDrop-1000 instrument (Thermo Fisher Scientific, United States).
Qualified RNA with an integrity number (RIN) greater than 6.8 were used for cDNA library construction using AmpliSeq for Illumina Library Plus (Illumina, United States). cDNA libraries were sequenced as paired-end 150 bp reads on an HiSeq 4000 instrument (Illumina, United States). RNA-seq reads were aligned against the pig reference genome (Sscrofa11.1) using STAR (Dobin et al., 2013). The counts of all annotated genes were calculated using STRingtie (Pertea et al., 2015) and featureCounts (Liao et al., 2014). Differentially expressed genes (DEGs) between white and reddish-brown skin samples were then determined using the DEseq2 software from Bioconductor (Love et al., 2014), under default parameters.
KIT Genotyping
All DLY piglets were genotyped for the splice mutation via Sanger sequencing after PCR amplification using primers KIT_F (5′-CCCCGACTCTCCTAACAGTGTA-3′) and KIT_R (5′-TGCATGGTATGGCAAAGGTAG-3′). The DUP1 duplication breakpoint (DBP) was amplified using primers DUP1BP_F (5′-ATGTGGAGAAGCAGGAGACC-3′) and DUP1BP_R (5′-TGTTTCACCCGCATCCTACT-3′). PCR reaction was run on a Thermal Cycler (Bio-Rad, United States) at 95°C for 5 min, 30 × (95°C for 30 s, 60°C for 30 s, 72°C 40 s), 72°C 10 min and 12°C forever. PCR products were visualized through 1% agarose gel electrophoresis.
Quantitative PCR and Droplet Digital PCR
Genomic quantitative (qPCR) were performed to detect the copy numbers of DUP1-4 using TaqMan primers and probes as previously descried (Rubin et al., 2012). PCR reactions were conducted using Probe qPCR Mix (TakaRa, Beijing) in a total volume of 10 μl containing 25 ng of genomic DNA, 0.2 μl of target and reference primers (10 μM), 0.1 μl of target and reference probe mix (10 μM), and 0.2 μl ROX Reference Dye. The genomic copy numbers of DUP1-4 were estimated in relation to the single copy locus ESR1 using the ΔΔCt methodology (Livak and Schmittgen, 2001). The Ct value of each sample was obtained from a mean of three runs.
Droplet digital PCR (ddPCR) was explored to verify the genomic qPCR results as previously reported (Hindson et al., 2011; Pinheiro et al., 2012). In brief, genomic DNA was digested with the restriction enzyme SpeI-HF (NEB, United Kingdom) for 1 h at 37°C, then inactivated for 20 min at 80°C. ddPCR reaction mixture was prepared in a volume of 20 μl containing 1 ng digested DNA template, 10 μl 2 × ddPCR Supermix for Probes (Bio-Rad, United States), 1.8 μl each target or reference primer (10 μM, Supplementary Table S1) and 0.5 μl probe mix (10 μM, Supplementary Table S1). The reaction mixture was then emulsified using droplet generator oil (Bio-Rad, United States) and a QX200 Droplet Generator (Bio-Rad, United States). About 20,000 droplets were generated per sample. The droplets were then transferred to a 96-well and were amplified on a Thermal Cycler (Bio-Rad, United States) at 95°C for 10 min, 40 × (94°C for 30 s, 59°C for 1 min), 98°C for 10 min. After amplification, the droplets were read in the QX200 Droplet Reader (Bio-Rad, United States). The copy numbers of DUP1-4 in the tested samples were finally estimated using QuantaSoft version 1.7.4.0917 (Bio-Rad, United States). Each sample was run in duplicate, and unless otherwise specified, the copy numbers were the average of the two measurements.
Whole-Genome Resequencing
Six colored DLY pigs including four individuals with the phenotypic score of 1 and two individuals with the phenotypic score of 2, four Landrace pigs and three Large White pigs were sequenced at 25-fold depth using a whole-genome shotgun strategy. Short-insert (350 bp) DNA libraries were paired-end sequenced on a HiSeq 2500 platform (Illumina, United States). The sequence data for each individual reached more than 25-fold depth. The paired-end short sequence reads were mapped to the Sscrofa11.1 reference genome using the BWA software (v.0.7.10) (Li and Durbin, 2009). Alignment files were then sorted and converted to BAM format via SAMtools software (v.1.6) (Li et al., 2009).
Copy Number Variation Prediction
In addition to whole-genome sequence data of 13 individuals obtained in this study, two publicly available whole-genome sequence data sets were used for the prediction of copy number variations at the KIT locus. One data set included whole-genome sequences (∼10× depth) of 8 Duroc, 7 Pietrain, 4 Hampshire, 9 Landrace, and 17 Large White pigs (Accession numbers: ERP011076 and ERP001813) (Groenen et al., 2012; Frantz et al., 2015). The other one included whole-genome sequence (∼25× depth) data of 6 Chinese wild boars and 62 Chinese domestic pigs (Accession number: SRA096093) (Ai et al., 2015).
Copy number variation regions (CNVRs) on pig chromosome 8 were predicted for 13 re-sequenced individuals using CNVcaller (Wang et al., 2017) under default procedure and parameters in a 800 bp window size. A Gaussian hidden Markov model method was further explored (Miles et al., 2016) to predict the copy number state along the KIT locus for each individual alignment. In brief, we computed the coverage of aligned sequence reads across the DUP1 region encompassing the KIT gene and its 500 kb flanking region on each side (chr8: 40722801-42284000 bp) in 800 bp non-overlapping bins for each sample. Then, we excluded bins where the GC content was lower than 20% to eliminate coverage bias and normalized the coverage values to 2 by median depth for diploid copy number state analysis. At last, copy number state in the KIT region was predicted for all samples by fitting the Gaussian hidden Markov model to the normalized coverage data.
PCR Amplification of Duplication Breakpoints
PCR primers (Supplementary Table S1) were designed to amplify DUP1-4 duplication breakpoint sequences. PCR reaction was performed in a volume of 25 μl containing 1.5 μl genomic DNA (50–100 ng/μl), 12.5 μl 2× Taq Master Mix (Dye) Plus (Vazyme, China), 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM) (Sangon, China), and 10 μl water. PCR reaction was run on a Thermal Cycler (Bio-Rad, United States) at 95°C for 5 min, 30 × (95°C for 30 s, 57°C for 30 s, 72°C 1–2 min), 72°C 10 min and 12°C forever. After amplification, 1% agarose gel electrophoresis was performed to separate the amplified products with a voltage of 160 V and a time of 15 min. PCR products were directly sequenced to verify their identities.
Results and Discussion
GWAS Indicates That KIT Is the Major Gene Responsible for Coat Color Phenotypes in the DLY Population
We genotyped the 103 DLY pigs segregating for four coat color phenotypes using the Illumina 80K chip. GWAS identified a strong association signal for the proportion of reddish-brown hair on chromosome 8 (SSC8). A total of 16 SNPs on this chromosome surpassed the genome-wide significant threshold (P = 9.1E-7), and the strongest associated SNP was rs334600651 (P = 5.51E-10) at 40.78 Mb (Sscrofa11.1, Figure 2A), which was only 618 kb away from the KIT gene. We further determined the DUP1 genotypes (presence or absence) of the 103 DLY pigs by amplification of the DUP1 breakpoints with specific primers (Supplementary Table S1). We then conducted a conditional GWAS analysis in which the DUP1 genotypes were included as a fixed effect and did not observe any significant association signal across the genome afterward (Figure 2B). These results support that KIT variants are responsible for the coat color phenotypes segregating in the DLY population.
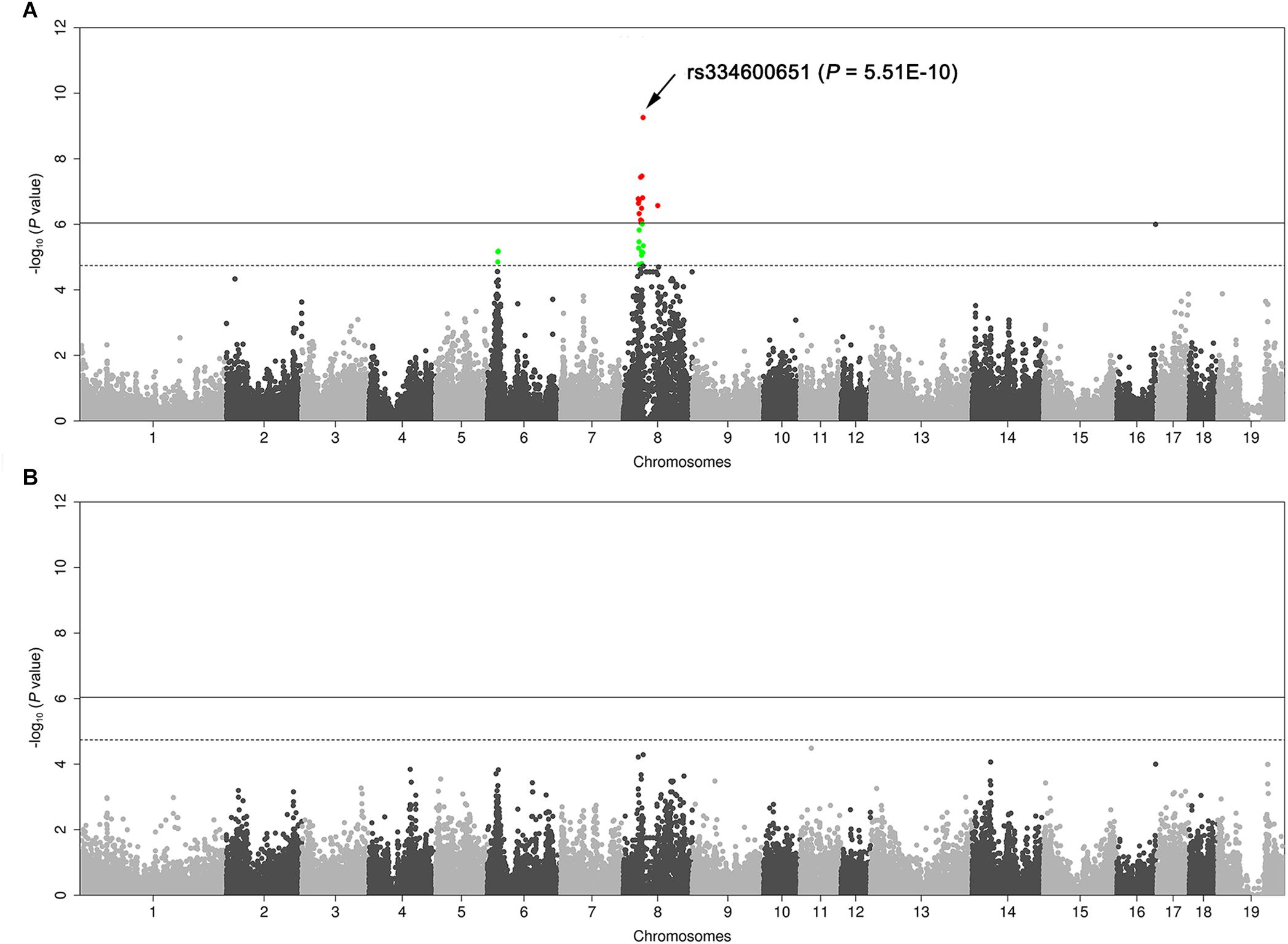
Figure 2. Manhattan plots of genome-wide association study (GWAS) for the proportion of reddish-brown hairs in 103 Duroc × (Landrace × Large White) piglets. (A) Routine GWAS. (B) Conditional GWAS. In the Manhattan plots, genomic positions of qualified SNPs are given in the X-axis, and the –log10 P-values for SNP associations with the phenotype are shown in the Y-axis. The red dots represent SNPs that exceed the 5% genome-wide significance threshold, and green dots represent SNPs surpassing the suggestive significance threshold. Solid and dashed lines indicate the 5% genome-wide and chromosome-wide (suggestive) Bonferroni-corrected thresholds, respectively.
RNA Sequencing Supports That KIT Is the Causative Gene for Coat Color Phenotypes in the DLY Population
We used RNA sequencing to explore the genome-wide mRNA expression profile of the reddish-brown and white skins from DLY pigs. By comparing the transcriptome data of reddish-brown and white hair skin tissues, we identified 71 DEGs (P < 0.05, Figure 3A and Supplementary Table S2) that were over-represented in the KIT-mediated melanin biosynthetic process (Figure 3B). The expression of nine well-known pigmentation genes (TYRP1, TYR, MC1R, PMEL, DCT, TRPM1, SLC24A5, MLANA, and SLC45A2) were up-regulated in reddish brown skin [P < 0.05, Log2 (fold change)]. Given that DEGs between white and reddish-brown skin tissues were significantly enriched in the pigmentation biosynthetic pathway involving KIT, we assume that KIT is the causative gene for coat color phenotypes in the DLY population.
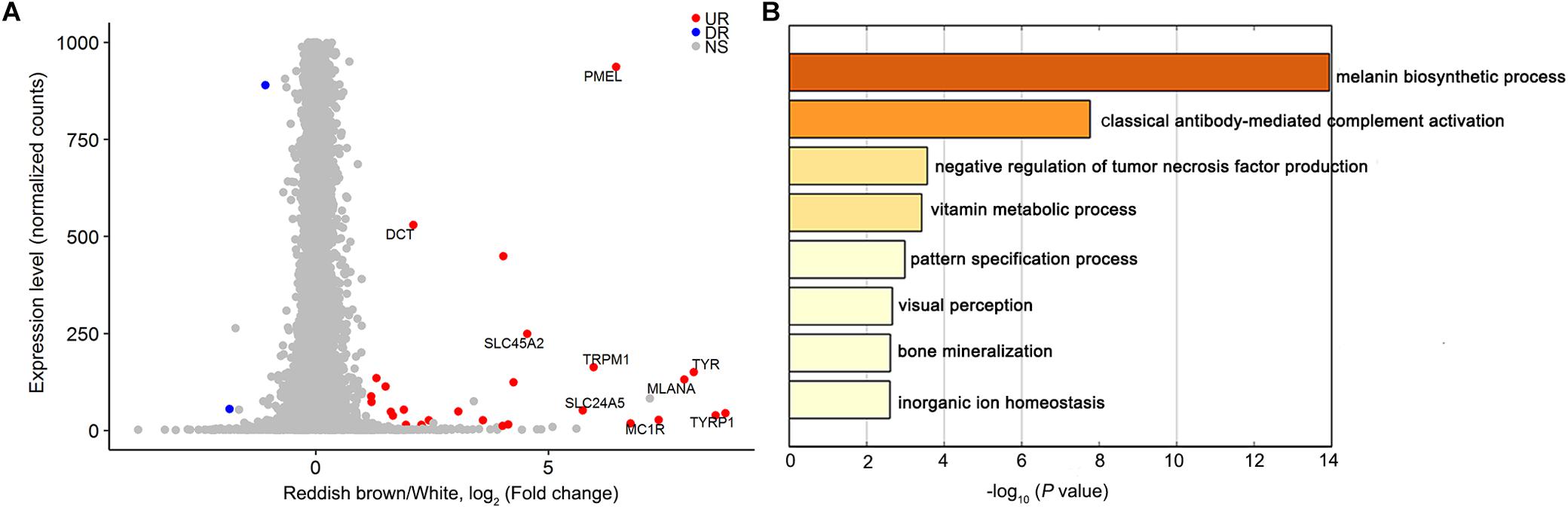
Figure 3. Differentially expressed genes between white and reddish-brown skin in DLY piglets. (A) Transcript levels (normalized gene counts) are plotted as a function of differential expression (log2-transformed fold change) between reddish-brown and white skin. Up-regulated expression genes in reddish-brown skin are shown in red. (B) Heatmap of enriched terms colored by P-value for differentially expressed genes between reddish-brown and white skin.
KIT Genotypes Are Discordant With the Expected Coat Color Phenotypes in the DLY Population and Its Parental Lines
We genotyped the DUP1 and splice mutation of 190 DLY piglets using PCR and Sanger sequencing (see section “Materials and Methods”). At the splicing mutation site, 143 individuals had AG genotypes and 47 were GG individuals (Supplementary Table S3). Of the 143 individuals, 140 were identified as I/i as they carried DUP1 and the AG genotype at the splice mutation site. Unexpectedly, only 64 out of the 143 individuals showed solid white coat color, and the others were colored individuals, which was inconsistent with the expectation that all individuals should be white. Moreover, three DLY piglets lacked DUP1 and showed the AG genotype at the splice mutation site, indicating that these individuals carried the homozygous lethal Dominant white allele IL (Pielberg et al., 2003). The 47 individuals should be i/i at the KIT locus as they lacked DUP1 and had the GG genotype at the splice mutation site. Of the 47 individuals, 18 had a reddish-brown coat color with white belts and 29 showed roan hairs (Supplementary Table S3), suggesting that two different new i alleles are presented in these individuals.
According to Rubin et al. (2012), the ratios of DUP2/DUP1 and DUP3/DUP1 in white (I/-) individuals are 1.5–3. We selected 12 DLY individuals representing three coat color phenotypes, i.e., reddish-brown with white belts, reddish-brown with white legs and roan (Figures 1B–D) and determined the splice mutation genotype via Sanger sequencing and the genomic copy numbers of DUP1-4 for each individual using qPCR. We found that both the ratios of DUP2/DUP1 and DUP3/DUP1 were less than 1.5 in the I/i individuals with a reddish-brown coat color with white legs and belts. However, both the ratios of DUP2/DUP1 and DUP3/DUP1 were greater than 1.5 in the i/i individuals with a roan coat color (Supplementary Table S4).
We further selected 10 Landrace and five Large White pigs from the parental farms of these DLY individuals and also detected their copy numbers of DUP1-4 using qPCR. The ratios of DUP2/DUP1 and DUP3/DUP1 were less than 1.5 in four Landrace pigs, and the ratios of DUP3/DUP1 were less than 1.5 in three Landrace and two Large White (Supplementary Table S4). Based on the copy numbers of DUP1-4, we deduced the KIT haplotypes of these tested Large White, Landrace and DLY individuals. The I alleles most likely lacked DUP2-3 in the I/i DLY individuals with the ratios of DUP2/DUP1 and DUP3/DUP1 less than 1.5. These previously unreported I alleles (denoted as IN) could be formed by an unequal crossover at the KIT locus during homologous recombination. For the i/i DLY individuals with the ratios of DUP2/DUP1 and DUP3/DUP1 more than 1.5, these individuals probably carried new i allele (iN) that have multiple copies of DUP2/3 but lack DUP1. Additional KIT new alleles likely exist in Landrace and Large White individuals with the ratios of DUP2/DUP1 and DUP3/DUP1 less than 1.5 (Supplementary Table S4).
Whole-Genome Resequencing Identifies Previously Unreported Alleles of KIT
To verify our speculation of the KIT new alleles, we conducted whole-genome resequencing at 20× depth on 13 pigs including six colored DLY, four Landrace and two Large White pigs that were predicted to carry KIT new alleles as mentioned above and one normal Large White pig. First, to determine the precise position of DUP1 in the pig reference genome (Sscrofa11.1), we used CNVcaller (Wang et al., 2017) to predict the copy number variation regions (CNVRs) of the 13 re-sequenced individuals on chromosome 8. After merging the CNVRs of all individuals, we observed a 561-kb shared CNVR (chr8: 41222801-41784000 bp) encompassing the entire KIT gene (Supplementary Table S5). The CNVR perfectly correspond to the duplicated KIT (DUP1) region (hereafter refer as to the 561-kb region), and its size is larger than the previously reported one (450 kb) (Rubin et al., 2012).
Next, we predicted the copy number variation (CNV) within the 561-kb region using the hidden Markov model (HMM) method described by Miles et al. (2016). To test the reliability of the HMM method, we downloaded the whole-genome sequence data of 113 individuals representing Chinese wild boars, Chinese domestic pigs and Western modern breeds (Duroc, Hampshire, Pietrain, Landrace, and Large White) from the NCBI public database. The diploid copy numbers were predicted in an 800 bp window for the 561-kb region and its 500-kb flanking region (chr8: 40722801-42284000 bp). We found that all Chinese wild boars and domestic pigs and Western Duroc pigs did not have CNV within this region. Pietrain had 3–4 copies of DUP1, Landrace and Large White had 3–6 copies of DUP1, and Hampshire carried DUP2-4 but not DUP1 (Figure 4). This observation was consistent with the previous report by Rubin et al. (2012) and thus supports the reliability of HMM method for CNV prediction.
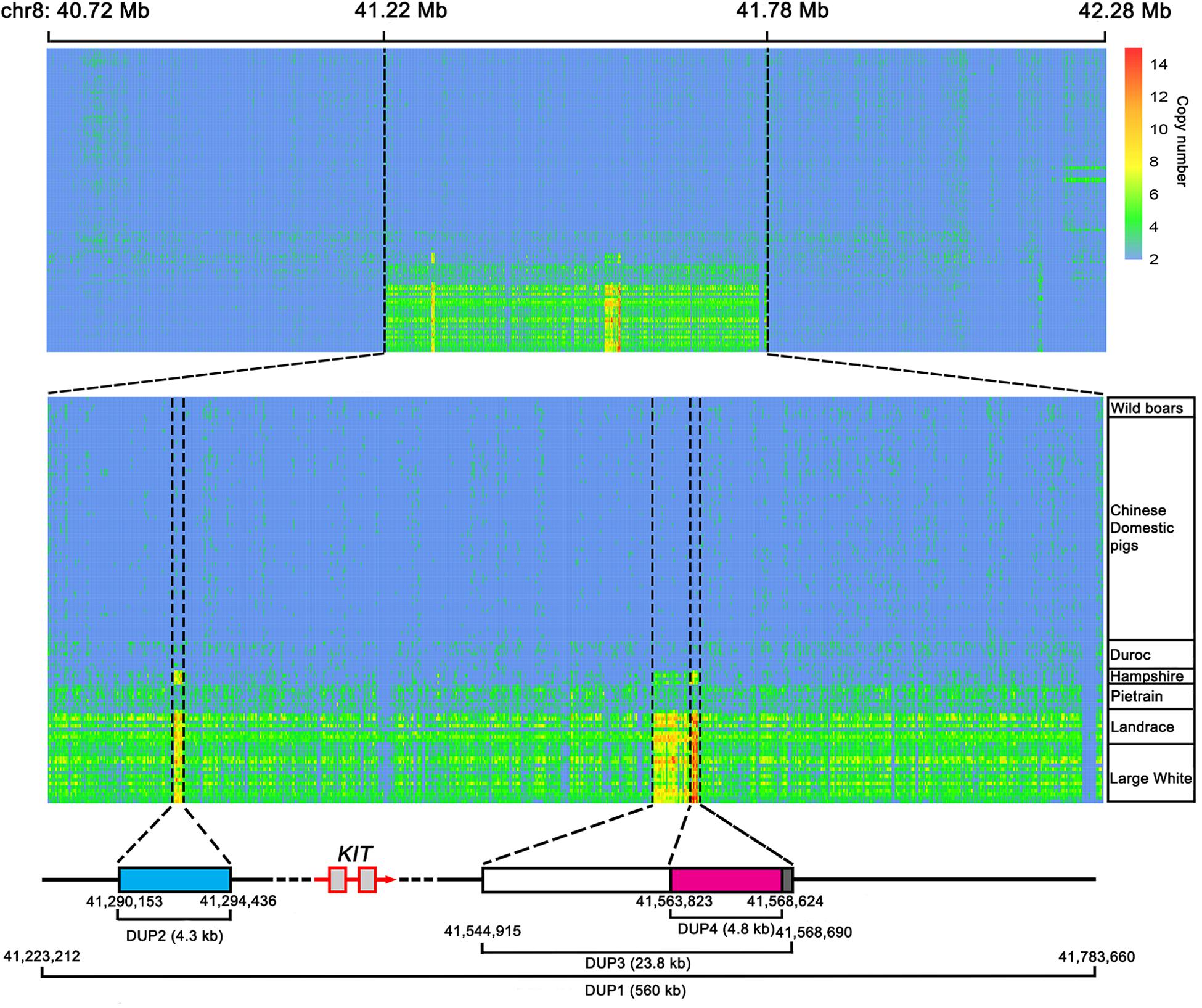
Figure 4. Heatmap of copy number prediction along the KIT locus for 113 individually sequenced pigs retrieved from the NCBI database. For each individual, diploid copy numbers were predicted in 800 bp non-overlapping bins by normalized coverage of DUP1 and its 500 kb flanking region on each side. The copy numbers of DUP1 are shown in magnifications to the middle. The accurate starting and ending positions of DUP1-4 were confirmed by Sanger sequencing.
Further, we designed specific primers to amplify duplication breakpoints of DUP1-4 based on the CNV prediction results in the 561-kb region. PCR amplification was performed using the genomic DNA of Large White pigs having DUP1-4. Sanger sequencing defined the accurate start and stop positions of DUP1-4 in the Sscrofa11.1 genome assembly into chr8: 41223212-41783660 bp (560 kb), chr8: 41290153-41294436 bp (4.3 kb), chr8: 41544915-41568690 bp (23.8 kb), and chr8: 41563823-41568624 bp (4.8 kb), respectively (Figure 4).
Then, we applied the HMM method to predict the diploid copy number variation in the 560-kb region for the 13 re-sequenced individuals. By comparing the CNV prediction results of these individuals with those of the 113 individuals from NCBI public database, we found abnormal copy numbers of DUP2 and DUP3 in one Large White pig from public database and 12 re-sequenced individuals, which could carry unreported KIT new alleles (Supplementary Figure S1). To clarify the KIT haplotypes of these individuals, we calculated the copy number ratios of DUP2-4 to DUP1 for each individual, confirming that one Large white pig from the public database and 12 re-sequenced individuals carried unreported KIT haplotypes. The ratios of DUP2/DUP1 and DUP3/DUP1 were 1.0 and the ratios of DUP4/DUP1 were 2.0–3.0 in four DLY individuals, two Landrace pigs and one Large White pig from public database. Two Large White and two Landrace pigs had 2.0–3.0 ratios of DUP2/DUP1 and DUP4/DUP1 and 1.0 ratio of DUP3/DUP1. The ratios of DUP2/DUP1 were 3.0 and the ratios of DUP3/DUP1 and DUP4/DUP1 were 2.0 in two DLY individuals with the i/i genotype (DLY-5 and DLY-6) (Figure 5).
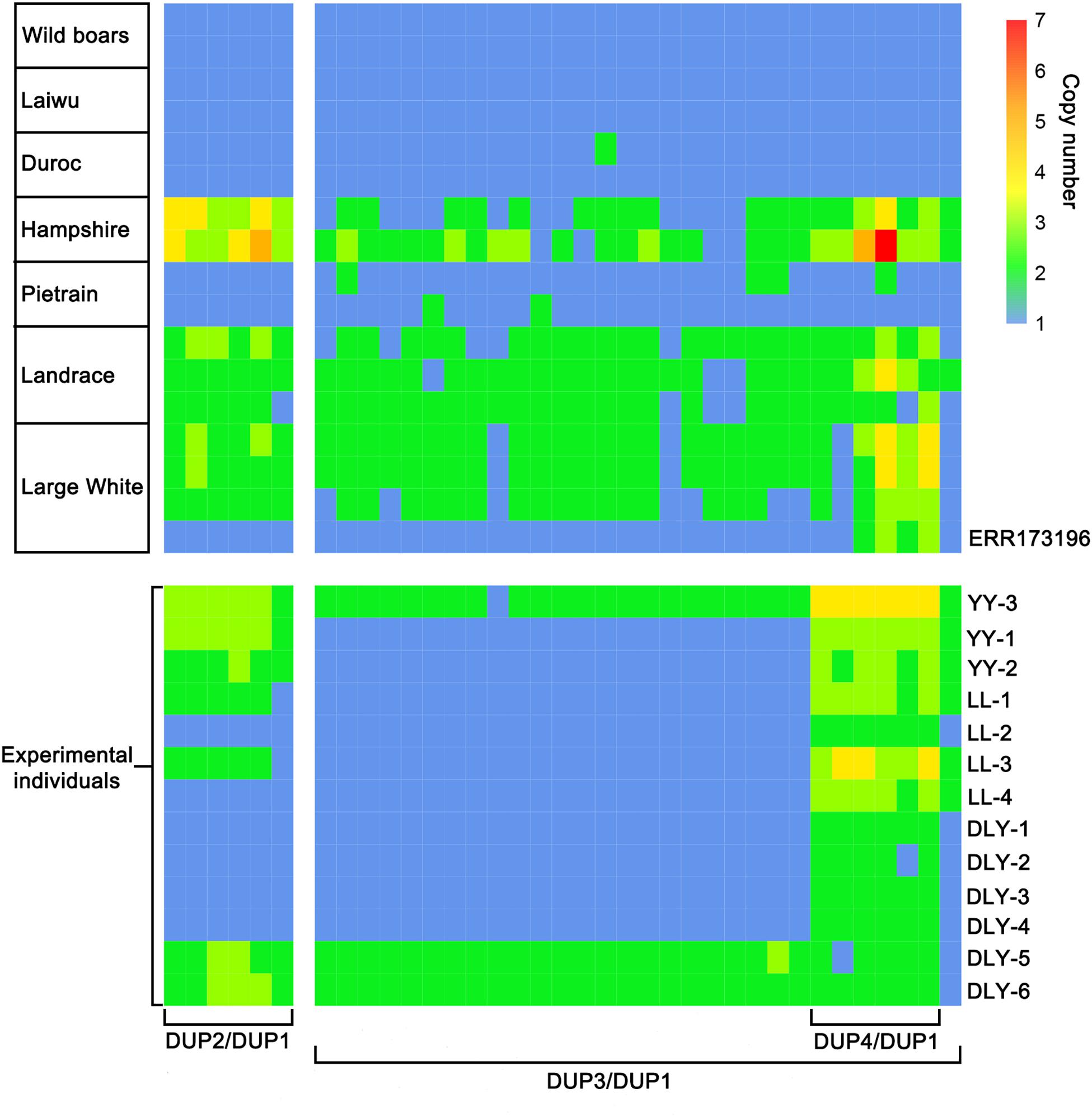
Figure 5. Heatmap of the ratios of DUP2-4 to DUP1 in 17 representative individuals retrieved from the NCBI database and 13 individuals re-sequenced in this study. ERR173196, Accession number in the NCBI database; LL, Landrace; YY, Large White; DLY, Duroc × (Landrace × Large White).
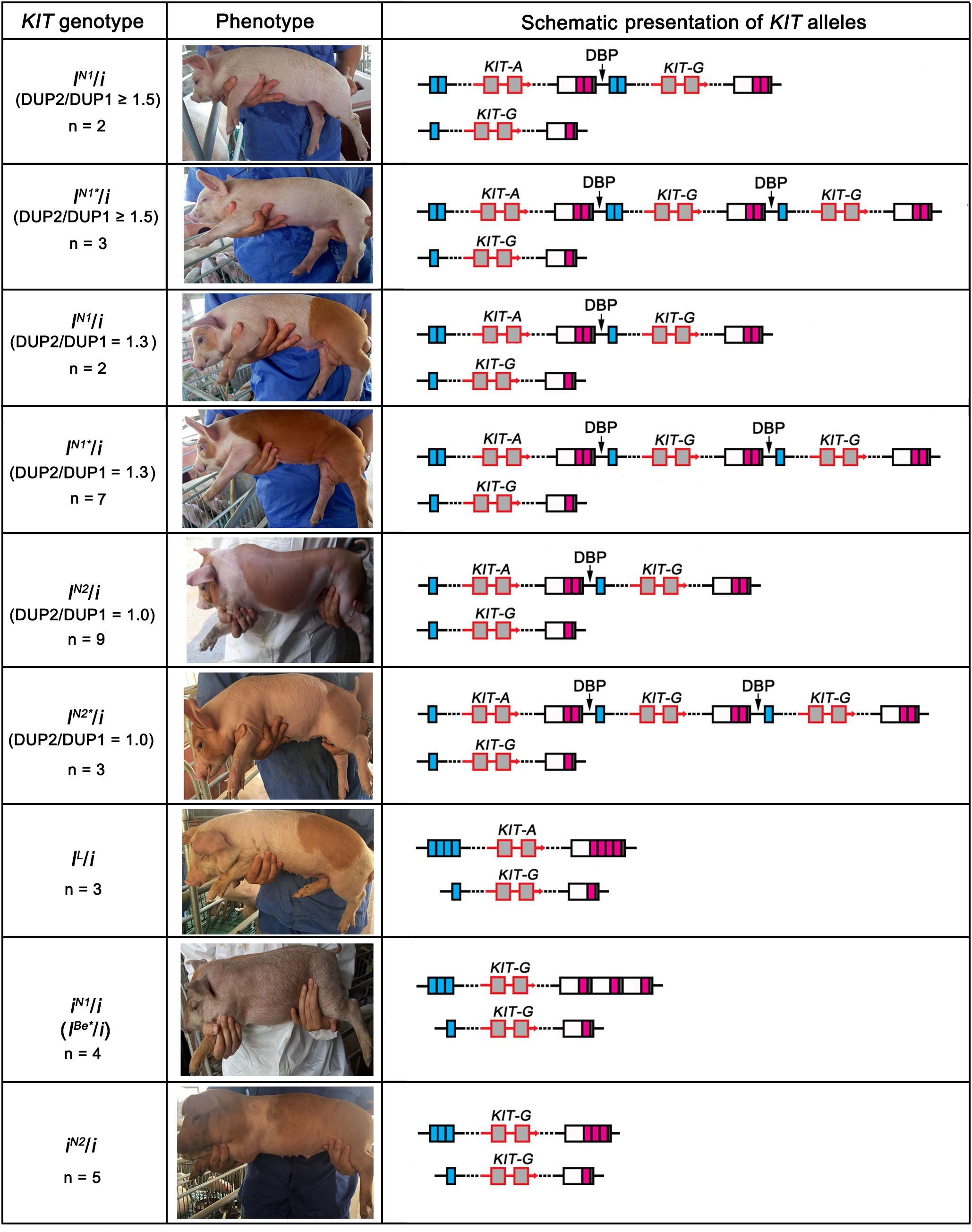
To accurately decipher the allelic structure of the re-sequenced individuals at the KIT locus, we determined the copy numbers of DUP1-4 (Figure 6A) and the ratios of G to A at the splice mutation (data not shown) in these individuals using Droplet digital PCR (ddPCR) (see section “Materials and Methods”) and amplified DUP1-4 breakpoint sequences (Figure 6B). In comparison with the KIT allelic structure reported by Rubin et al. (2012; Figure 6C), we identified five KIT new alleles: iN1 with multiple copies of DUP2/3 but lacking DUP1/4 and the splice mutation, iN1 and IN1∗ carrying multiple copies of DUP1, DUP2, DUP4 and the splice mutation but not DUP3, IN2 and IN2∗ with multiple copies of DUP1, DUP4 and the splice mutation but without DUP2 and DUP3 (Figure 6D).
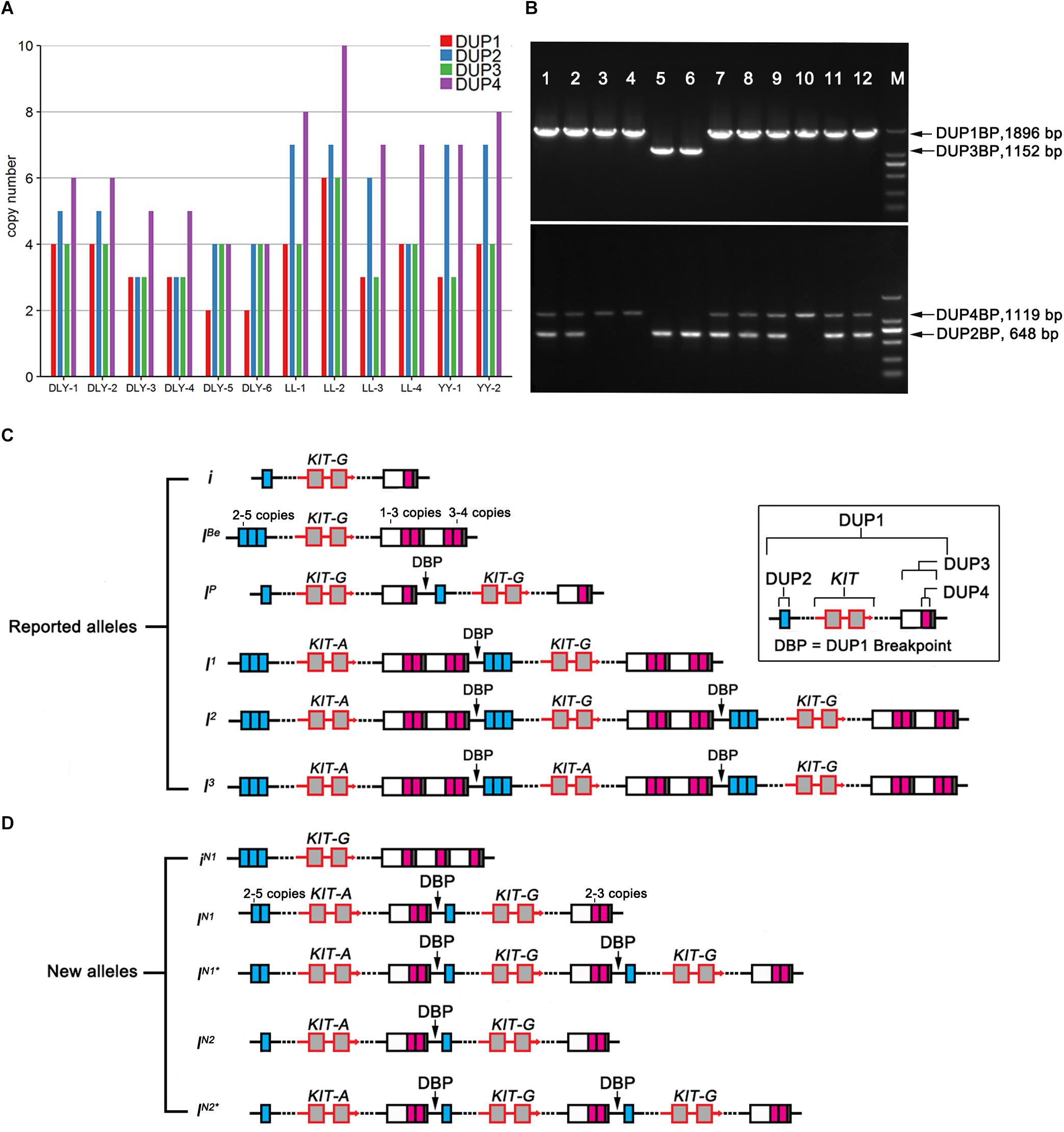
Figure 6. Copy number variations and deduced alleles at the KIT locus. (A) Genomic copy numbers of DUP1-4 in 12 re-sequenced individuals determined by ddPCR. (B) Amplification results of duplication breakpoint sequences DUP1 (DUP1BP, 1896 bp), DUP2 (DUP2BP, 648 bp), DUP3 (DUP3BP, 1152 bp), and DUP4 (DUP4BP, 1119 bp) in 12 pigs, Marker 2000 bp. (C) Schematic presentation of the previously reported KIT alleles adopted from Rubin et al. (2012) with slight modifications. (D) Schematic presentation of new KIT alleles identified in 12 re-sequenced individuals.
The Effect of KIT New Alleles on Pig Coat Color Phenotypes
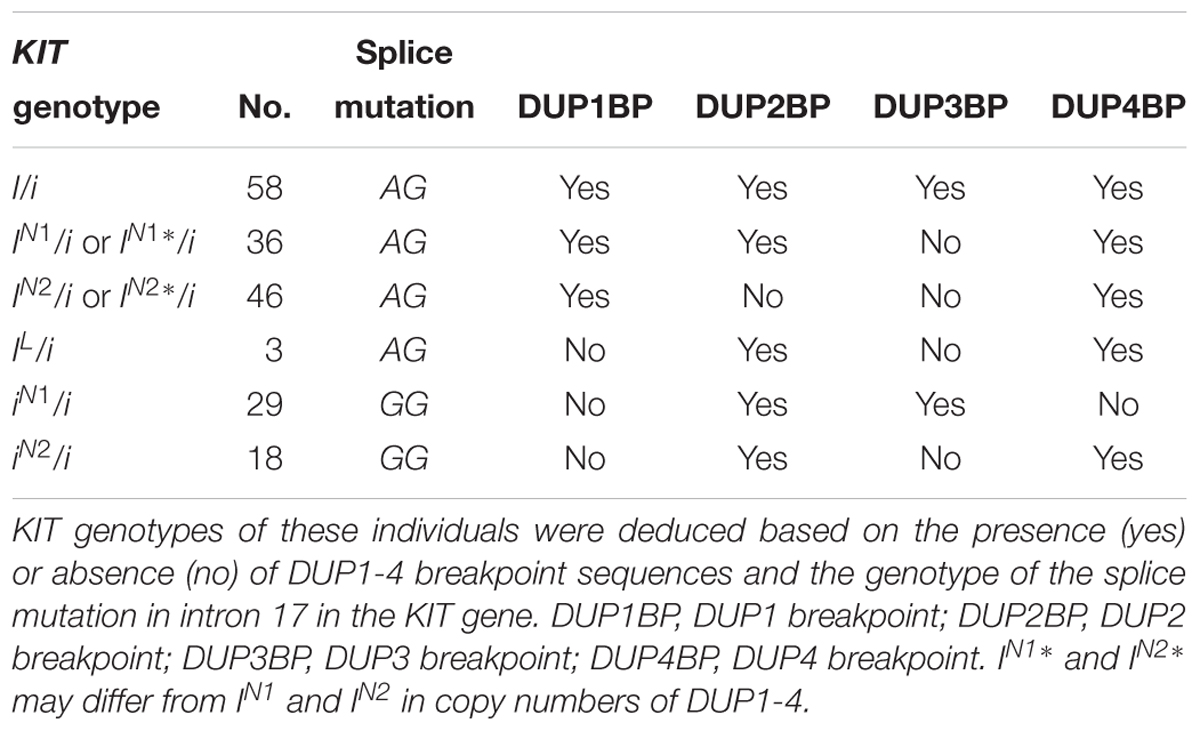
To investigate the effect of KIT new alleles on coat color phenotype, we determined the KIT genotypes of the 190 DLY pigs via amplification of DUP1-4 breakpoint sequences and the splice mutation (Table 1). In addition to 58 I/i individuals carrying the normal KIT Dominant White alleles I (including I1, I2, or I3), we identified 36 iN1/i or IN∗/i individuals lacking DUP3, 46 IN2/i or IN2∗/i animals lacking both DUP2 and DUP3, three IL/i pigs having the splice mutation but not DUP1, 29 iN1/i and 18 IN2/i pigs. The IN2 allele differs from iN1 in that it had multiple copies of DUP2 and DUP4 but not DUP1, DUP3 and the splice mutation (Figure 7).
We then analyzed the association between the KIT genotypes and the coat color phenotypes of these 190 individuals. As expected, the 58 I/i DLY individuals had a solid white coat color. We randomly selected five individuals to perform ddPCR for quantifying the copy numbers of DUP1-4, and found that the ratios of DUP2/DUP1 and DUP3/DUP1 were 1.7–2.0 in all I/i individuals (Figure 7 and Supplementary Table S6), which is consistent with the previous report (Rubin et al., 2012).
We observed three coat color phenotypes in the 36 iN1/i or IN1∗/i individuals: white, white with reddish-brown spots and reddish-brown with white legs and belts. The ratios of DUP3/DUP1 were 1.0 and the ratios of DUP4/DUP1 were greater than 1.5 in all 36 individuals as revealed by ddPCR. However, the ratio of DUP2/DUP1 varied in these individuals. All individuals (n = 5) with a DUP2/DUP1 ratio of no less than 1.5 had the solid white coat color. When individuals had a DUP2/DUP1 ratio of 1.3 (i.e., most likely having one DUP2, n = 9), they displayed colored phenotypes even these animals carried DUP1, DUP4 and the splice mutation (Figure 7 and Supplementary Table S6). This indicates that more than one DUP2 is required for the formation of the solid white coat color in pigs.
For the 46 IN2/i or IN2∗/i individuals, all of them showed colored phenotypes including white with reddish-brown spots and reddish-brown with white legs and belts. The ddPCR analysis showed that the ratios of DUP2/DUP1 and DUP3/DUP1 were 1.0 and the ratios of DUP4/DUP1 were greater than 1.5 in these individuals (Figure 7 and Supplementary Table S6). This again highlights the importance of DUP2 in determining the solid white coat color in pigs and supports the previous assumption that DUP2 is another causative mutation for the Dominant White coat color in addition to DUP1 (the KIT gene duplication) and the splice mutation (Rubin et al., 2012).
We noticed that three DLY individuals carried the homozygous lethal allele IL (Figure 7 and Supplementary Table S6), which lacked DUP1 and DUP3 but had DUP2, DUP4 and the splice mutation. The coat color phenotypes of these three individuals were all white with reddish-brown spots (Figure 7 and Supplementary Table S6), suggesting that the splice mutation, DUP2 and DUP4 can collectively cause white coat color even without the KIT gene duplication (DUP1). When we looked at the 29 iN1/i individuals, all of which had the roan phenotype (Figure 7 and Supplementary Table S6). Allele iN1 carries DUP2 and DUP3 but not DUP1, DUP4 and the splice mutation, which is similar to the IBe allele carrying DUP2-4 but not DUP1 and the splice mutation, and is most likely the previously reported IBe∗ allele causing the roan coat color in IBe∗/i individuals (Pielberg et al., 2002). This finding indicates that DUP4 is required for manifestation of the Belt phenotype of the IBe allele.
We then focused on the 18 IN2/i individuals that carried DUP2 and DUP4 but not DUP1, DUP3 and the splice mutation. These individuals all showed a reddish-brown coat color with white belts (Figure 7 and Supplementary Table S6), suggesting that DUP2 and DUP4 could result in a belt coat color in the absence of DUP1 and the splice mutation. Rubin et al. (2012) measured the copy numbers of DUP1-4 in four belted breeds including Angler Sattelschwein, British Saddleback, Cinta Senese, and Hampshire pigs. They found that all individuals had DUP2 and DUP4 but lacked DUP1, and a few individuals did not have DUP3, which allowed them to assume that both DUP2 and DUP4 contribute to the belt coat color phenotype. Our findings are consistent with their assumption. We noticed that both alleles iN1 and IN2 lack DUP1 and the splice mutation. The difference between the two alleles was that the iN1 allele has DUP2 and DUP3 but not DUP4, and is responsible for the roan phenotype, while the IN2 allele carries DUP2 and DUP4 but not DUP3 and leads to the reddish-brown coat color with white belts. This clearly suggests that DUP4 had a more significant effect on the formation of the belt phenotype compared with DUP3.
To validate the effect of the KIT new alleles on coat color phenotype, we selected two IN2/IL Landrace sows to cross with Duroc boars (i/i), generating a total of 23 Duroc × Landrace (DL) hybrid offspring. We further determined the KIT genotypes of the 23 DL individuals using ddPCR of DUP1-4, amplification of DUP1-4 breakpoints and Sanger sequencing of the splice mutation. Of the 23 individuals, four had the IL/i genotype and white coat color with reddish-brown spots. Nineteen DL individuals were colored pigs with a DUP3/DUP1 ratio of 1.0 and DUP2/DUP1, DUP4/DUP1 ratios of 1.3 (Supplementary Table S7). These observation were consistent with our finding of the coat color phenotypes in IL/i and IN2/i DLY pigs.
Homozygotes of IN/IN Lacking DUP2 and Causing Colored DLY Pigs Are Presented in French Landrace Pigs at a Higher Frequency
Considering that more than one DUP2 is essential for the manifestation of the solid white coat color, and IN (IN2 or IN2∗)/i DLY pigs lacking DUP2 always show colored phenotypes, we detected the proportion of IN/IN homozygotes in 1504 Landrace and 775 Large White pigs from different countries using PCR amplification of DUP2 breakpoint sequences. We found that the frequency of IN/IN individuals was generally higher in Landrace than in Large White pigs, and French Landrace pigs had the highest frequency (8.98%) of IN/IN individuals (Figure 8). In this study, we sampled DLY pigs that were derived from Duroc boars × (French Landrace boars × French Large White sows) in a commercial pig farm. This explains why a large number of colored DLY individuals were observed in the farm where we collected samples. The PCR-based test for DUP2 established in this study can be explored to efficiently detect the IN/IN homozygotes in Landrace and Large White pigs, which would be helpful in reducing the number of colored DLY pigs and consequently prevent the economic loss for commercial farms in China where solid white DLY commercial pigs are favored.
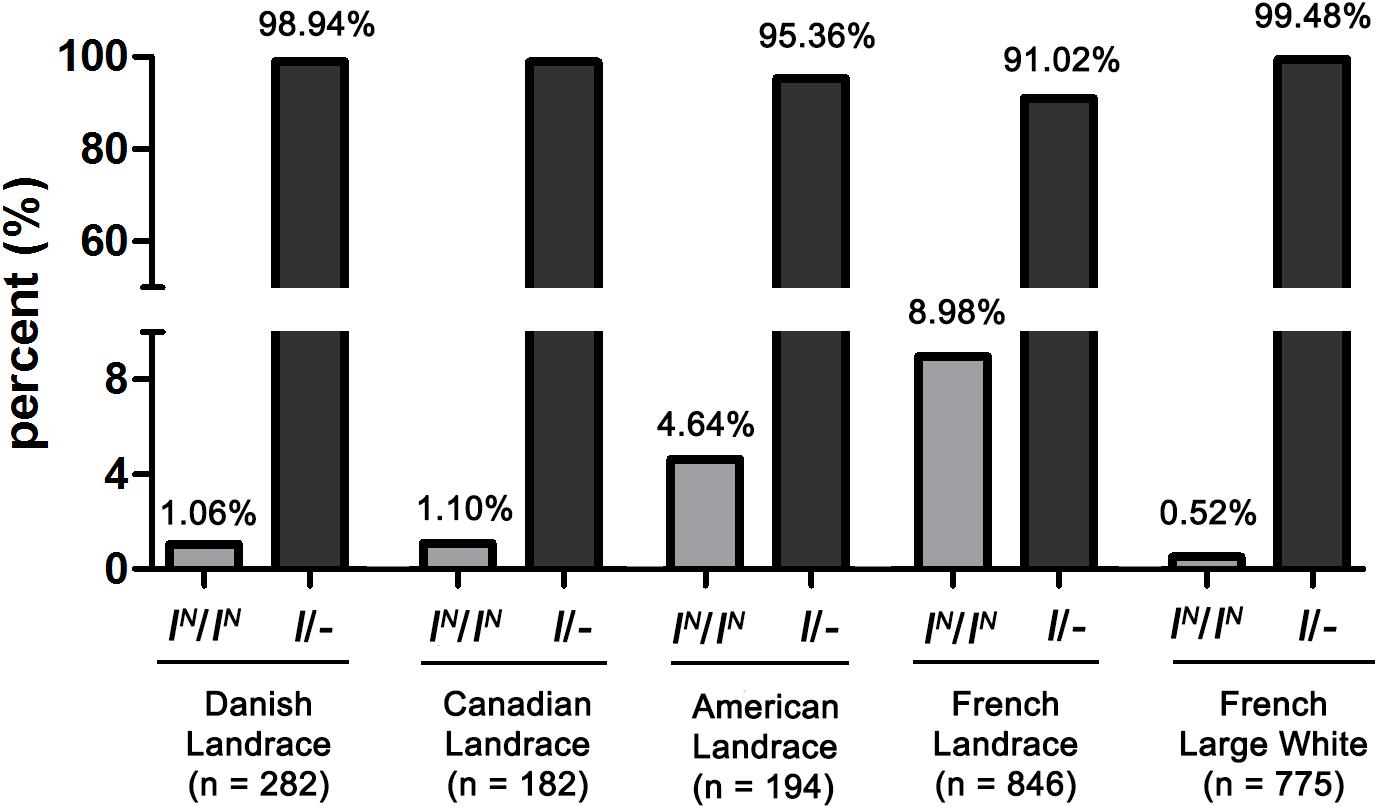
Figure 8. The frequency of IN(IN2 or IN2∗)/IN homozygotes lacking DUP2 in Landrace and Large White pigs.
Conclusion
KIT variants are responsible for the coat color phenotypes segregating in the DLY population. DUP1, DUP2 and the splice mutation are all required for manifestation of a solid white coat color phenotype. In DLY pigs, DUP2 and the splice mutation can cause a white coat color with reddish-brown spots even in the absence of DUP1. DUP2 and DUP4 can result in a belt coat color phenotype in DLY pigs even without DUP1 and the splice mutation. Moreover, the solid white coat color cannot be formed in DLY pigs lacking DUP2 even if they carry DUP1 and the splice mutation. The PCR-based test for DUP2 provides a robust and simple tool for detecting IN(IN2 or IN2∗)/IN homozygotes in Large White and Landrace pigs. These homozygotes cause colored DLY pigs and segregate in French Landrace pigs at the highest frequency in comparison to other Large White and Landrace populations.
Ethics Statement
All procedures used for this study and involving animals were in compliance with guidelines for the care and utility of experimental animals established by the Ministry of Agriculture of China.
Author Contributions
JR designed the study and analyzed the data. JR and ZW wrote the manuscript. ZW, MH, and HC performed the bioinformatic analyses. ZW, ZD, YH, and HZ collected the data and performed the sequencing and genotyping experiments.
Funding
This research was supported by the National Natural Science Foundation of China (Grant Number: 31525023).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00218/full#supplementary-material
FIGURE S1 | Heatmap of copy number prediction in the KIT region. For each individual, diploid copy numbers were predicted in 800 bp non-overlapping bins by normalized coverage of DUP1 and its 500 kb flanking region on each side. The heatmap of DUP2-4 and the KIT gene of the representative individuals retrieved from the NCBI database and 13 re-sequenced individuals are shown in the top and bottom, respectively. ERR173196, Accession number in the NCBI database; LL, Landrace; YY, Large White; DLY, Duroc × (Landrace × Large White).
TABLE S1 | Primers and probes for detecting genomic copy numbers and amplifying duplication breakpoint sequences at the KIT locus. ∗5′ labeled with HEX and 3′ labeled with BHQ1. The other probes were 5′ labeled with 6-FAM and 3′ labeled with BHQ1.
TABLE S2 | Differentially expressed genes between reddish brown and white skin of DLY piglets.
TABLE S3 |KIT genotypes and coat color phenotypes in the tested DLY population.
TABLE S4 | Genomic copy numbers of DUP1-4 in 27 pigs estimated by qPCR. DLY, Duroc × (Landrace × Large White); LL, Landrace; YY, Large White.
TABLE S5 | Copy number variations in the 561-kb region containing the KIT gene in 13 re-sequenced individuals. DLY, Duroc × (Landrace × Large White); LL, Landrace; YY, Large White.
TABLE S6 | Association of KIT genotypes and alleles with coat color phenotypes. Genomic copy numbers of DUP1-4 of these individuals were measured by ddPCR.
TABLE S7 |KIT genotypes and coat color phenotypes in 23 Duroc × Landrace hybrid piglets.
References
Ai, H., Fang, X., Yang, B., Huang, Z., Chen, H., Mao, L., et al. (2015). Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet. 47, 217–225. doi: 10.1038/ng.3199
Ai, H., Huang, L., and Ren, J. (2013). Genetic diversity, linkage disequilibrium and selection signatures in chinese and western pigs revealed by genome-wide SNP markers. PLoS One 8:e56001. doi: 10.1371/journal.pone.0056001
Chen, L., Guo, W., Ren, L., Yang, M., Zhao, Y., Guo, Z., et al. (2016). A de novo silencer causes elimination of MITF-M expression and profound hearing loss in pigs. BMC Biol. 14:52. doi: 10.1186/s12915-016-0273-2
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Drögemüller, C., Giese, A., Martins-Wess, F., Wiedemann, S., Andersson, L., Brenig, B., et al. (2006). The mutation causing the black-and-tan pigmentation phenotype of Mangalitza pigs maps to the porcine ASIP locus but does not affect its coding sequence. Mamm. Genome 17, 58–66. doi: 10.1007/s00335-005-0104-1
Fan, Y., Wang, P., Fu, W., Dong, T., Qi, C., Liu, L., et al. (2014). Genome-wide association study for pigmentation traits in Chinese Holstein population. Anim. Genet. 45, 740–744. doi: 10.1111/age.12189
Fang, M., Larson, G., Ribeiro, H. S., Li, N., and Andersson, L. (2009). Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5:e1000341. doi: 10.1371/journal.pgen.1000341
Frantz, L. A. F., Schraiber, J. G., Madsen, O., Megens, H.-J., Cagan, A., Bosse, M., et al. (2015). Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet. 47, 1141–1148. doi: 10.1038/ng.3394
Giuffra, E., Evans, G., Törnsten, A., Wales, R., Day, A., Looft, H., et al. (1999). The belt mutation in pigs is an allele at the dominant white (I/KIT) locus. Mamm. Genome 10, 1132–1136.
Giuffra, E., Törnsten, A., Marklund, S., Bongcam-Rudloff, E., Chardon, P., Kijas, J. M. H., et al. (2002). A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm. Genome 13, 569–577. doi: 10.1007/s00335-002-2184-5
Groenen, M. A. M., Archibald, A. L., Uenishi, H., Tuggle, C. K., Takeuchi, Y., Rothschild, M. F., et al. (2012). Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398. doi: 10.1038/nature11622
Hindson, B. J., Ness, K. D., Masquelier, D. A., Belgrader, P., Heredia, N. J., Makarewicz, A. J., et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610. doi: 10.1021/ac202028g
Johansson, A., Pielberg, G., Andersson, L., and Edfors-Lilja, I. (2005). Polymorphism at the porcine dominant white/KIT locus influence coat colour and peripheral blood cell measures. Anim. Genet. 36, 288–296. doi: 10.1111/j.1365-2052.2005.01320.x
Johansson, M., Chaudhary, R., Hellmén, E., Höyheim, B., Chowdhary, B., and Andersson, L. (1996). Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm. Genome 7, 822–830.
Johansson, M., Ellegren, H., Marklund, L., Gustavsson, U., Ringmar-Cederberg, E., Andersson, K., et al. (1992). The gene for dominant white color in the pig is closely linked to ALB and PDGRFRA on chromosome 8. Genomics 14, 965–969.
Kijas, J. M., Moller, M., Plastow, G., and Andersson, L. (2001). A frameshift mutation in MC1R and a high frequency of somatic reversions cause black spotting in pigs. Genetics 158, 779–785.
Kijas, J. M., Wales, R., Törnsten, A., Chardon, P., Moller, M., and Andersson, L. (1998). Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 150, 1177–1185.
Legault, C., Rothschild, M. F., and Ruvinsky, A. (1998). “Genetics of colour variation,” in THE Genetics of the Pig, eds M. F. Rothschild and A. Ruvinsky (Oxfordshire: CAB International).
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Marklund, S., Kijas, J., Rodriguezmartinez, H., Rönnstrand, L., Funa, K., Moller, M., et al. (1998). Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 8, 826–833.
Miles, A., Iqbal, Z., Vauterin, P., Pearson, R., Campino, S., Theron, M., et al. (2016). Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 26, 1288–1299. doi: 10.1101/gr.203711.115
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Pielberg, G., Day, A. E., Plastow, G. S., and Andersson, L. (2003). A sensitive method for detecting variation in copy numbers of duplicated genes. Genome Res. 13, 2171–2177.
Pielberg, G., Olsson, C., Syvänen, A., and Andersson, L. (2002). Unexpectedly high allelic diversity at the KIT locus causing dominant white color in the domestic pig. Genetics 160, 305–311.
Pinheiro, L. B., Coleman, V. A., Hindson, C. M., Herrmann, J., Hindson, B. J., Bhat, S., et al. (2012). Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 84, 1003–1011. doi: 10.1021/ac202578x
Porter, V. (1993). Pigs. A Handbook to the Breeds of the World. Ithaca, NY: Comstoc Publishing Associates.
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Ren, J., Mao, H., Zhang, Z., Xiao, S., Ding, N., and Huang, L. (2010). A 6-bp deletion in the TYRP1 gene causes the brown colouration phenotype in Chinese indigenous pigs. Heredity 106, 862–868. doi: 10.1038/hdy.2010.129
Rubin, C. J., Megens, H. J., Barrio, A. M., Maqbool, K., Sayyab, S., Schwochow, D., et al. (2012). Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. U.S.A. 109, 19529–19536. doi: 10.1073/pnas.1217149109
Wang, C., Wang, X., Tang, J., Chen, H., Zhang, J., Li, Y., et al. (2018). Genome-wide association studies for two exterior traits in chinese dongxiang spotted pigs. Anim. Sci. J. 89, 868–875. doi: 10.1111/asj.13003
Wang, X., Zheng, Z., Cai, Y., Chen, T., Li, C., Fu, W., et al. (2017). CNVcaller: highly efficient and widely applicable software for detecting copy number variations in large populations. GigaScience 6, 1–12. doi: 10.1093/gigascience/gix115
Yang, Q., Cui, J., Chazaro, I., Cupples, L. A., and Demissie, S. (2005). Power and type I error rate of false discovery rate approaches in genome-wide association studies. BMC Genet. 6(Suppl. 1):S134. doi: 10.1186/1471-2156-6-s1-s134
Zhang, J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/s0169-5347(03)00033-8
Keywords: pig, coat color, KIT, genome resequencing, allele diversity
Citation: Wu Z, Deng Z, Huang M, Hou Y, Zhang H, Chen H and Ren J (2019) Whole-Genome Resequencing Identifies KIT New Alleles That Affect Coat Color Phenotypes in Pigs. Front. Genet. 10:218. doi: 10.3389/fgene.2019.00218
Received: 15 November 2018; Accepted: 27 February 2019;
Published: 18 March 2019.
Edited by:
Meng-Hua Li, Chinese Academy of Sciences, ChinaReviewed by:
Mingzhou Li, Sichuan Agricultural University, ChinaYaofeng Zhao, China Agricultural University, China
Copyright © 2019 Wu, Deng, Huang, Hou, Zhang, Chen and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ren, cmVuanVuanhhdUBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Zhongping Wu
Zhongping Wu Zheng Deng†
Zheng Deng† Min Huang
Min Huang Hao Chen
Hao Chen Jun Ren
Jun Ren