- 1Endocrine Division, Hospital de Clínicas de Porto Alegre, Porto Alegre, Brazil
- 2Post-Graduate Program in Medical Sciences: Endocrinology, Faculdade de Medicina, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil
- 3Department of Food Science and Physiology, School of Pharmacy and Nutrition, University of Navarra, Pamplona, Spain
- 4Nephrology Division, Hospital de Clínicas de Porto Alegre, Porto Alegre, Brazil
Introduction: Diabetic kidney disease (DKD) is a common microvascular complication that affects 40% of patients with diabetes mellitus (DM). Emerging evidence suggests a role for several microRNAs (miRNAs) in the development of DKD. In this context, miR-15a-5p and miR-30e-5p have been shown to regulate the expression of the uncoupling protein 2 (UCP2), a mitochondrial protein that decreases reactive oxygen species (ROS) formation by the mitochondria. Since ROS overproduction is a key contributor to the pathogenesis of DKD, dysregulation of these two miRNAs could be involved in DKD pathogenesis. Thus, the aim of this study was to compare the expressions of miR-15a-5p and miR-30e-5p in type 1 DM (T1DM) patients with DKD (cases) and without this complication (controls), and to perform bioinformatics analyses to investigate their putative targets and biological pathways under their regulation.
Methods: MiR-15a-5p and miR-30e-5p expressions were analyzed in plasma and urine of 17 T1DM controls and 23 DKD cases (12 with moderate DKD and 11 with severe DKD) using qPCR. Bioinformatics analyses were performed in Cytoscape software.
Results: MiR-30e-5p expression was downregulated in plasma of patients with moderate and severe DKD compared to T1DM controls. Moreover, this miRNA was also downregulated in urine of patients with severe DKD compared to the other groups. No difference was found in miR-15a-5p expression between groups. Bioinformatics analyses indicated that miR-30e-5p and miR-15a-5p regulate various genes that participate in pathways related to angiogenesis, apoptosis, cell differentiation, oxidative stress, and hypoxia.
Conclusion: MiR-30e-5p seems to be downregulated in plasma and urine of patients with DKD.
Introduction
Diabetic kidney disease (DKD) is a common microvascular complication that occurs in approximately 40% of patients with diabetes mellitus (DM), and that can lead to end-stage renal disease (ESRD) (Gross et al., 2005). DKD is clinically characterized by albuminuria and/or a gradual reduction in the glomerular filtration rate (GFR) (Ritz et al., 2011). Pathological changes in renal cells of DKD patients include glomerular hypertrophy, mesangial expansion, and tubulointerstitial fibrosis due to the accumulation of extracellular matrix (ECM) proteins, thickening of basement membrane, and podocyte dysfunction (Assmann et al., 2018b). At the cellular level, chronic hyperglycemia causes reactive oxygen species (ROS) overproduction by the mitochondria, which then triggers key pathways related to DKD: increased formation of advanced glycation end-products (AGEs) and overexpression of their receptors (RAGEs); activation of protein kinase C isoforms; increased flux of glucose by the polyol pathway; and upregulation of the hexosamine pathway (Brownlee, 2005; Giacco and Brownlee, 2010; Thomas et al., 2015).
The main risk factors for DKD are duration of chronic hyperglycemia, arterial hypertension, dyslipidemia, and genetic and epigenetic components (Brennan et al., 2013; Assmann et al., 2018b). Regarding epigenetic factors, emerging evidence has suggested an important role of microRNAs (miRNAs) in the pathogenesis of DKD (Kato and Natarajan, 2015; Wanner and Bechtel-Walz, 2017; Assmann et al., 2018a,b). MiRNAs are small (≅21–23 nucleotides) non-coding RNAs that regulate gene expression of 60% of protein coding genes (Assmann et al., 2017); thus, regulating many cellular functions and influencing the development and progression of a number of diseases (Kato and Natarajan, 2015; Assmann et al., 2017, 2018b).
In this context, miR-15a-5p and miR-30e-5p target the uncoupling protein 2 (UCP2) gene (Sun et al., 2011; Jiang et al., 2013). UCP2 is a mitochondrial protein that seems to mildly uncouples the oxidative phosphorylation from ATP synthesis by dissipating the proton gradient generated across the mitochondrial inner membrane, consequently decreasing ATP production and reducing ROS formation by the mitochondria (Brand and Esteves, 2005; Fisler and Warden, 2006; Souza et al., 2011). Taking into account the role of UCP2 in decreasing oxidative stress, and that UCP2 gene polymorphisms have been associated with DKD and other diabetic complications (Rudofsky et al., 2006; Tiwari et al., 2009; Crispim et al., 2010; Souza et al., 2011; de Souza et al., 2015), dysregulation of miR-15a-5p and miR-30e-5p might also be involved in DKD pathogenesis.
Accordingly, experimental studies have linked both miRNAs to podocyte injury, epithelial-mesenchymal transition (EMT) in tubular epithelial cells, and kidney fibrosis, which are features related to chronic kidney disease (CKD) and DKD (Jiang et al., 2013; Sun et al., 2014; Wu et al., 2014, 2015; Guo et al., 2017; Zhao D. et al., 2017). In humans, miR-15a-5p was reported as being downregulated in urine of patients with CKD or DKD compared to healthy controls (Khurana et al., 2017; Xie et al., 2017). Expression of miR-30e-5p was also downregulated in urinary exosomes of DKD patients compared to healthy subjects or type 2 DM patients without this complication (Delic et al., 2016). Moreover, expression of this miRNA in urine was correlated with proteinuria levels in DKD patients (Cardenas-Gonzalez et al., 2017). Even though these studies have associated dysregulation of miR15a-5p and miR-30e-5p with DKD, their exact roles and clinical relevance remain unknown. Therefore, in the present study, we analyzed miR-15a-5p and miR-30e-5p expressions in plasma and urine of type 1 DM (T1DM) patients with and without DKD. Moreover, we carried out bioinformatics analyses to investigate the putative targets and biological pathways under regulation of these two miRNAs.
Materials and Methods
Sample and Phenotype Measurements
This case-control study was designed following STROBE guidelines for reporting of association studies (von Elm et al., 2014). The sample comprised 40 T1DM patients, who were divided in 17 patients without DKD (control group) and 23 DKD cases (12 with moderate DKD and 11 with severe DKD). All T1DM patients included in the study were from outpatient clinics at Hospital de Clínicas de Porto Alegre or Instituto da Criança com Diabetes at Grupo Hospitalar Conceição (Rio Grande do Sul, Brazil), and were recruited between August 2014 and July 2018, accordingly to the flowchart showed in the Supplementary Figure 1. T1DM diagnosis was based on the American Diabetes Association criteria (American Diabetes Association, 2018).
Diabetic kidney disease was classified based on the Kidney Disease Improving Global Outcomes (KDIGO) guidelines (Andrassy, 2013). T1DM patients were divided into 3 groups according to their renal function: (1) patients with ≥10 years of T1DM and without DKD [urinary albumin excretion (UAE) <30 mg/g and estimated GFR (eGFR) ≥60 ml/min/1.73 m2; T1DM control]; (2) patients with moderate DKD (UAE <30 and eGFR 30–59 or UAE 30–300 and eGFR ≥45 or UAE >300 and eGFR >60); and (3) patients with severe DKD (UAE <30 and eGFR <29 or UAE 30–300 and eGFR <44 or UAE >300 and eGFR <59). Exclusion criteria were any febrile illness during the last 3 months, chronic inflammatory or rheumatic diseases, hepatitis, HIV-positivity, glucocorticoid treatment, liver or cardiac failure, kidney transplantation, hereditary dyslipidemia, and inborn or acquired errors of metabolism excepting DM. In order to avoid bias, we selected this extensive list of exclusion criteria since they might interfere with miRNA expression. Moreover, all samples were collected during the morning, since the period of the day might also influence miRNA expression.
A standard questionnaire was used to collect information on age, age at diagnosis, T1DM duration, drug treatment, and ethnicity. The ethnic group was defined based on self-classification. All patients underwent physical and laboratory evaluations, as previous described (Assmann et al., 2014). Serum creatinine was measured by the Jaffé reaction and UAE by immunoturbidimetry (Sera- Pak immuno microalbuminuria, Bayer, Tarrytown, NY, United States) (Zelmanovitz et al., 1997). Estimated GFR was calculated using the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation: eGFR = 141 × min (SCR/κ, 1)α × max (SCR/κ, 1)-1,209 × 0,993age × 1,018 (if female) × 1,159 (if black) (Levey et al., 2009). All subjects gave written informed consent in accordance with the Declaration of Helsinki. The study protocol was approved by the Ethic Committees in Research from Hospital de Clínicas de Porto Alegre and Grupo Hospitalar Conceição/Instituto da Criança com Diabetes.
RNA Extraction and Quantification of miRNA Expressions by qPCR
Peripheral blood samples of all subjects were collected in the morning with at least 8 h of fasting, in EDTA-coated tubes. Midstream 20 ml voided urine samples were also collected from all patients. Immediately after collection, blood and urine samples were centrifuged at 3500 rpm for 15 min at 4°C, and their aliquots were stored at -80°C until quantification of miRNA expressions. Total RNA was isolated from 450 μl of plasma or urine using the MiRVana PARIS miRNA Isolation Kit (Ambion, Thermo Fisher Scientific, DE, United States), according to the manufacturer’s instructions. Purity and concentration of RNA samples were measured using the NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific). Only RNA samples that achieved adequate purity ratios (A260/A280 = 1.9–2.1) were used for subsequent analyses (Bustin et al., 2009).
Real-time quantitative PCR (qPCR) was performed in two separate reactions: first, the total RNA was reverse-transcribed into cDNA and, second, the cDNA was amplified by qPCR. Reverse transcription of 2 ng/μl of RNA into cDNA was carried out using TaqMan miRNA RT assays (Thermo Fisher Scientific) specific for each miRNA of interest (assay reference numbers: 000389 for hsa-miR-15a-5p, and 002223 for hsa-miR-30e-5p). The small nuclear RNA U6 (U6snRNA) gene was used as the reference gene (assay reference number: 001973).
Next, qPCR experiments were carried out in a ViiATM 7 Fast Real-Time PCR System. PCR reactions were performed using 0.5 μl TaqMan miRNA Assays 20× (Thermo Fisher Scientific) for target miRNAs or U6snRNA, 5 μl TaqMan Universal PCR Master Mix II no UNG 2×, and 1 μl of cDNA (10 ng/μl), in a total volume of 10 μl. Each sample was assayed in triplicate and a negative control (without any cDNA) was included in each experiment. Cycling conditions for these genes were an initial cycle of 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 90 s. Quantification of the two target miRNAs was performed using the 2-ΔΔCq method and the U6snRNA gene as the reference and are shown as n-fold changes in relation to the calibrator sample (Bustin et al., 2009). The calibrator sample was a pool of cDNAs from the samples used in the study.
Bioinformatics Analyses
To better understand the functional involvement of miR-15a-5p and miR-30e-5p in DKD, these miRNAs were submitted to bioinformatics analyses to investigate their putative target genes and find possible biological pathways under their regulation. Bioinformatics analyses were performed using the Cytoscape v. 3.2.1 software (Shannon et al., 2003) with two plugin tools: (1) CyTargetLinker (Kutmon et al., 2013), and (2) Biological Networks Gene Ontology (BiNGO) (Maere et al., 2005).
The CyTargetLinker v3.0.1 was used to search for validated and predicted miRNA-target gene interactions (MTI) and visualize them in a graphical way. For this study, we obtained Homo sapiens MTIs from one experimentally validated database (miRTarBase v.4.4) and from two predicted miRNA databases (MicroCosm v.5.0 and TargetScan v.6.2).
Next, functional enrichment analysis of miRNA-target genes was performed to retrieve gene ontology (GO) annotations for miR-15a-5p and miR-30e-5p target genes that were identified with the CyTargetLinker, using the BiNGO plug-in in the Cytoscape environment. This investigation was performed for targets of each individual miRNA as well as for targets of the two miRNAs analyzed together. Significance for GO pathway enrichment was estimated with a hypergeometric test and adjusted for multiple hypotheses using the Benjamini-Hochberg and Hochberg False Discovery Rate (FDR) test. Pathways with a q-value <0.05 were considered strongly enriched for the genes targeted by the two miRNAs analyzed.
Statistical Analyses
Normal distribution of variables was checked using the Kolmogorov Smirnov and Shapiro–Wilk tests. Variables with normal distribution are presented as mean ± SD. Variables with skewed distribution were log-transformed before analyses and are presented as median (25–75th percentiles). Categorical data are shown as percentages. Clinical and laboratory characteristics were compared among groups using One-way ANOVA tests or χ2-tests, as appropriate. MiRNA expressions were compared between groups using Kruskal–Wallis tests. Correlations between quantitative variables were analyzed using Spearman’s correlation tests. All statistical analyses were performed using the SPSS statistical package (v.18.0) for Windows (SPSS Inc., Chicago, IL, United States), and P-values <0.05 were considered statistically significant.
The sample size was calculated in the OpenEpi site1 and based on previous studies (Cardenas-Gonzalez et al., 2017; Xie et al., 2017). Considering a power of 80% (α = 0.05) to detect two fold changes (±1.5 SD) in miRNA expressions between case and control groups, we needed at least 10 patients in each group in order to have an adequate statistical power.
Results
Characteristics of the T1DM Patients
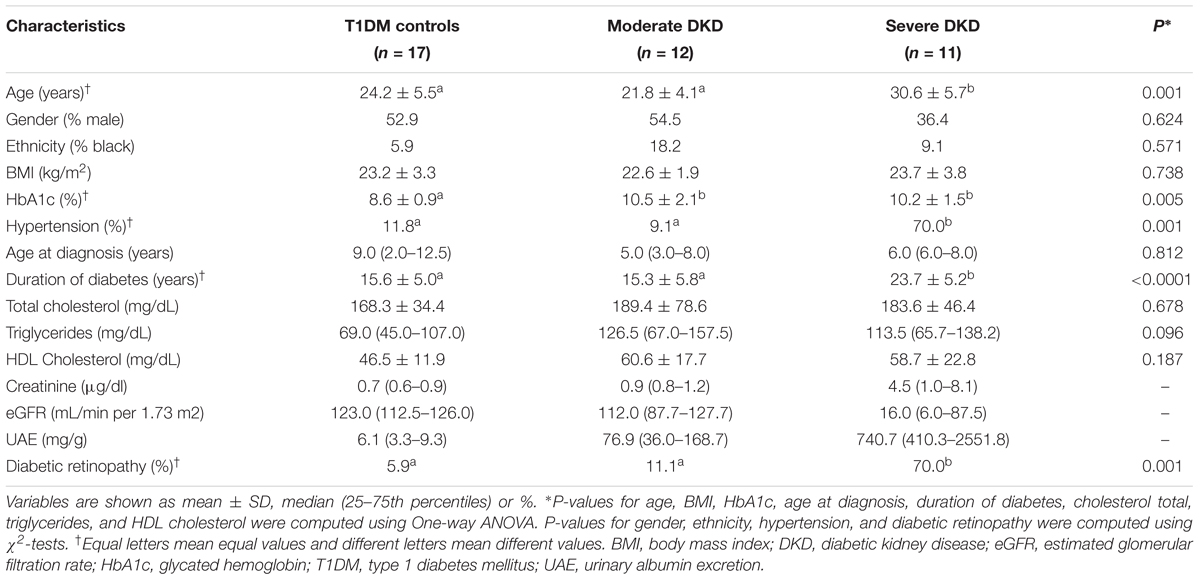
Clinical and laboratorial characteristics of the T1DM control patients and DKD cases included in this study are summarized in Table 1. Gender, ethnicity, BMI, age at T1DM diagnosis, HDL cholesterol, and total cholesterol levels did not differ between cases with moderate or severe DKD and T1DM controls. HbA1c levels were higher in both moderate and severe DKD groups compared to controls (P = 0.005). As expected, age, T1DM duration, prevalence of hypertension and diabetic retinopathy were higher in severe DKD patients compared to patients with moderate DKD and T1DM controls (P < 0.050) (Table 1).
Expressions of MiR-15a-5p and MiR-30e-5p in Plasma and Urine of T1DM Patients With or Without DKD
Expressions of miR-15a-5p and miR-30e-5p were investigated in plasma and urine of T1DM controls and DKD cases grouped according to the severity of this complication. In plasma, miR-30e-5p expression was downregulated in both severe and moderate DKD patients compared to T1DM controls [severe DKD: 0.53 (0.25–0.84), moderate DKD: 0.25 (0.08–0.82), and T1DM controls: 2.42 (0.51–4.33), P = 0.003; Figure 1B]. In urine samples, miR-30e-5p expression was only downregulated in the severe DKD group compared to moderate DKD and T1DM control groups [severe DKD: 0.34 (0.05–0.85), moderate DKD: 3.92 (0.23–9.66), and T1DM controls: 2.96 (0.99–5.97), P = 0.017; Figure 1D]. MiR-15a-5p expression in plasma and urine did not differ between groups (P > 0.050; Figures 1A,C).
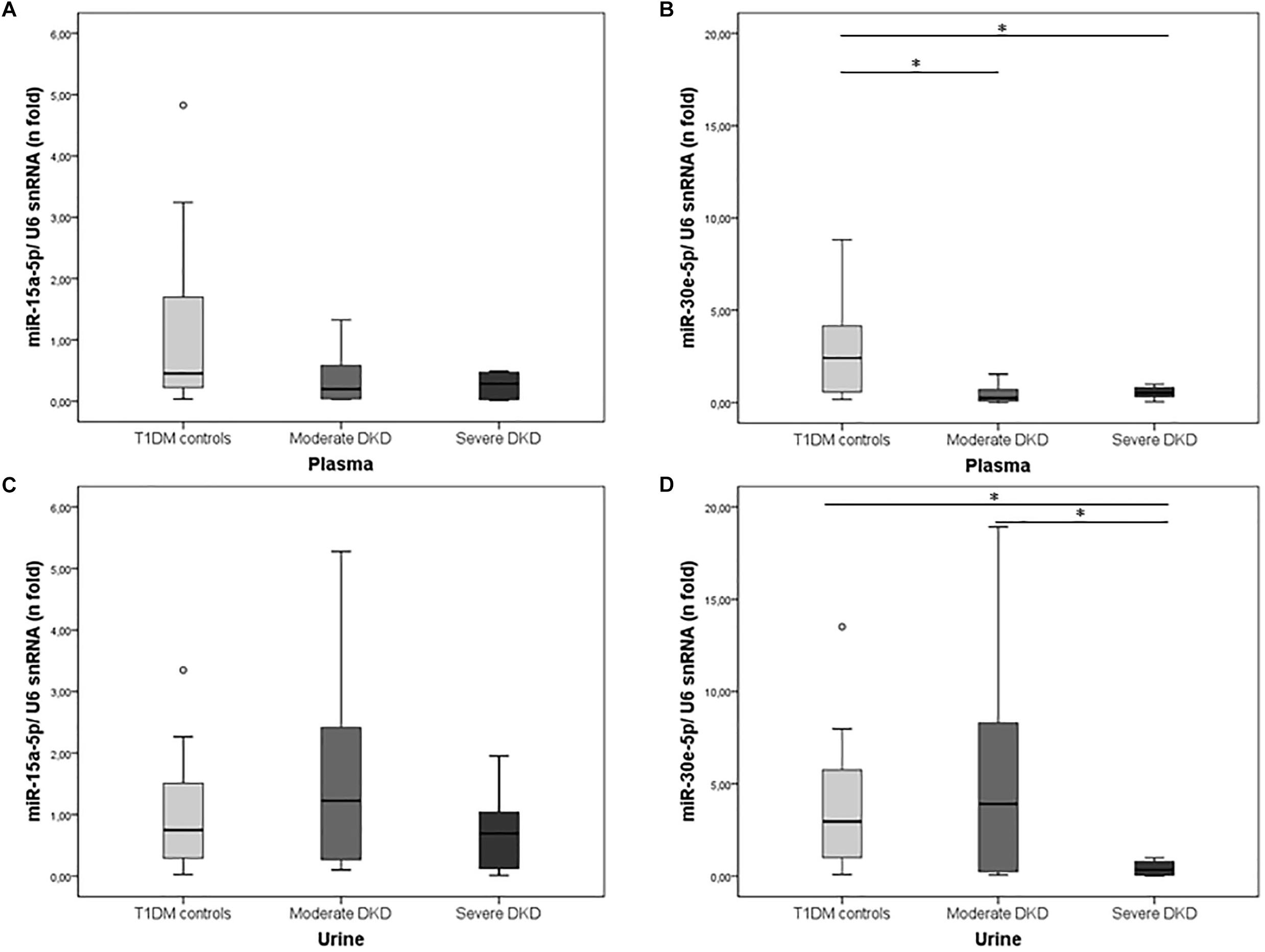
Figure 1. Expression of miR-15a-5p and miR-30e-5p in plasma and urine of T1DM controls and cases with moderate or severe DKD. Relative expressions of (A) miR-15-5p and (B) miR-30e-5p in plasma, and (C) miR-15-5p and (D) miR-30e-5p in urine samples of T1DM controls and patients with moderate or severe DKD. Expressions were evaluated using qPCR. Results are expressed as n-fold changes in relation to the calibrator sample (ΔΔCq method), using the U6 snRNA as the reference gene, and are shown as median (25–75th percentiles). P-values were obtained using Kruskal–Wallis tests. ∗P < 0.050.
We next evaluated possible correlations between miR-15a-5p and miR-30e-5p expressions in plasma and DKD-related measurements (eGFR, creatinine, and UAE levels) and HbA1c. MiR-15a-5p and miR-30e-5p expressions showed significant negative correlations with UAE levels (r = -0.459, P = 0.016 and r = -0.617, P = 0.0001, respectively) and HbA1c levels (r = -0.432, P = 0.009 and r = -0.435, P = 0.004, respectively). No significant correlation was found between the two analyzed miRNAs and eGFR values or creatinine levels.
Target Prediction and Functional Enrichment Analysis for MiR-15a-5p and MiR-30e-5p
Target prediction of the miR-15a-5p and miR-30e-5p was performed using distinct bioinformatics resources in the Cytoscape environment. Using the strategy described in the Material and Methods Section, 2197 genes were identified as putative targets of the miR-15a-5p, while 2208 genes were identified as putative targets of the miR-30e-5p. Of note, 314 targets were modulated by both miRNAs (Figure 2A and Supplementary Figure 2). After that, we analyzed only the experimentally validated target genes of these two miRNAs (Figure 3). As shown in Figure 3A, 23 validated target genes were found for miR-15a-5p and only two validated targets for miR-30e-5p. Among the validated target genes found for miR-15a-5p, some of them have been reported as being associated with kidney dysfunction or DKD pathogenesis, including BCL2, VEGFA, UCP2, BMI1, and NFKB1 and its inhibitor CHUK (IKKA) (Figure 3A). Figure 3B shows those targets of miR-15a-5p and miR-30e-5p that were found in all 3 databases analyzed of MITs (one database of experimentally validated targets and two of computationally predicted targets). As can be observed in this figure, UCP2 is a predicted and validated target of miR-15a-5p, being present in all the 3 analyzed databases (Figure 3B). Of note, most validated MTIs shown in Figure 3A were not computationally predicted (Figure 3B), demonstrating the importance of using distinct databases for target gene analysis.
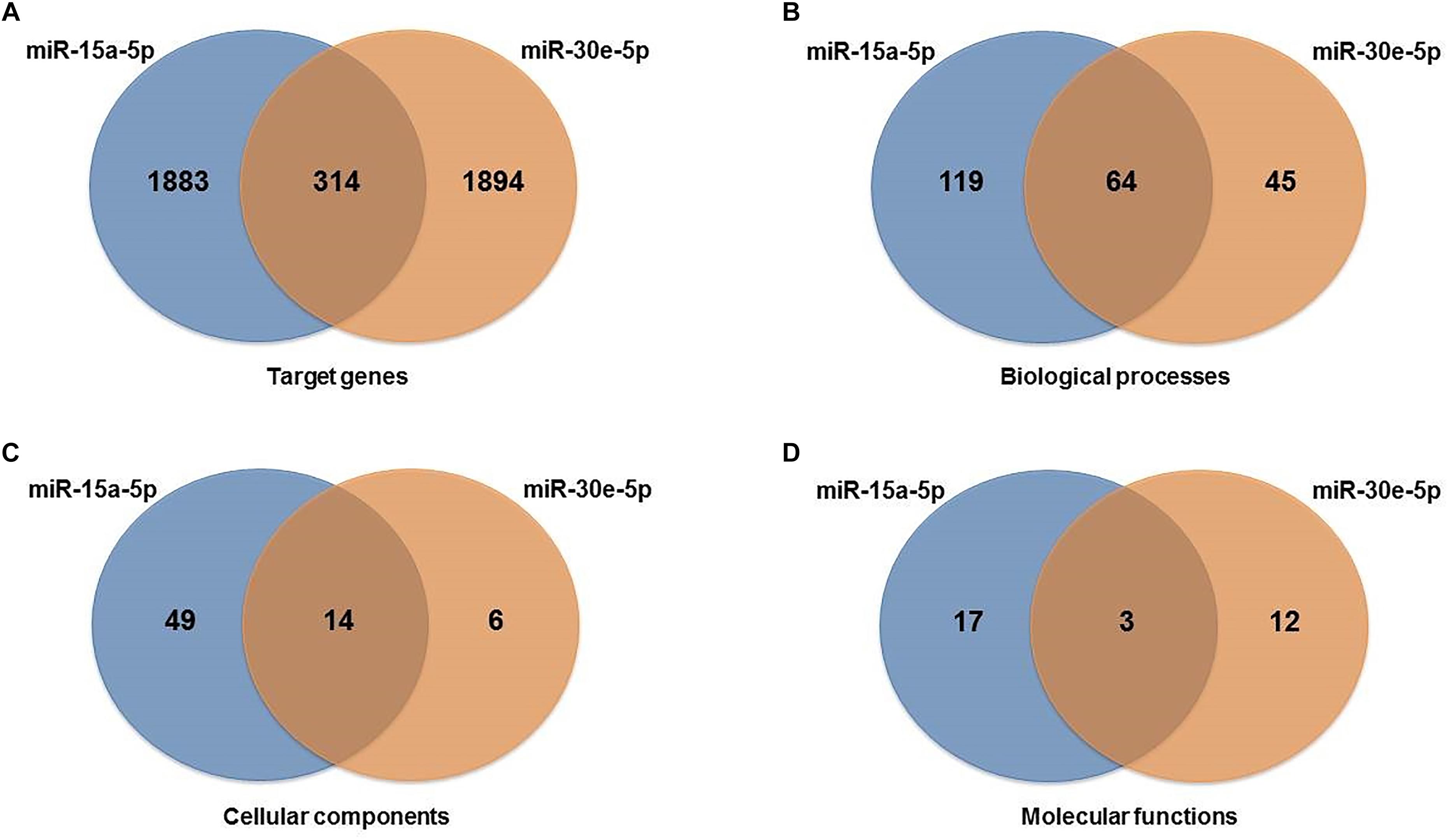
Figure 2. Venn diagrams showing interactions between miR-15a-5p and miR-30e-5p and their target genes and gene ontology pathways: (A) target genes, (B) biological process pathways, (C) cellular component pathways, and (D) molecular function pathways shared between the two miRNAs analyzed.
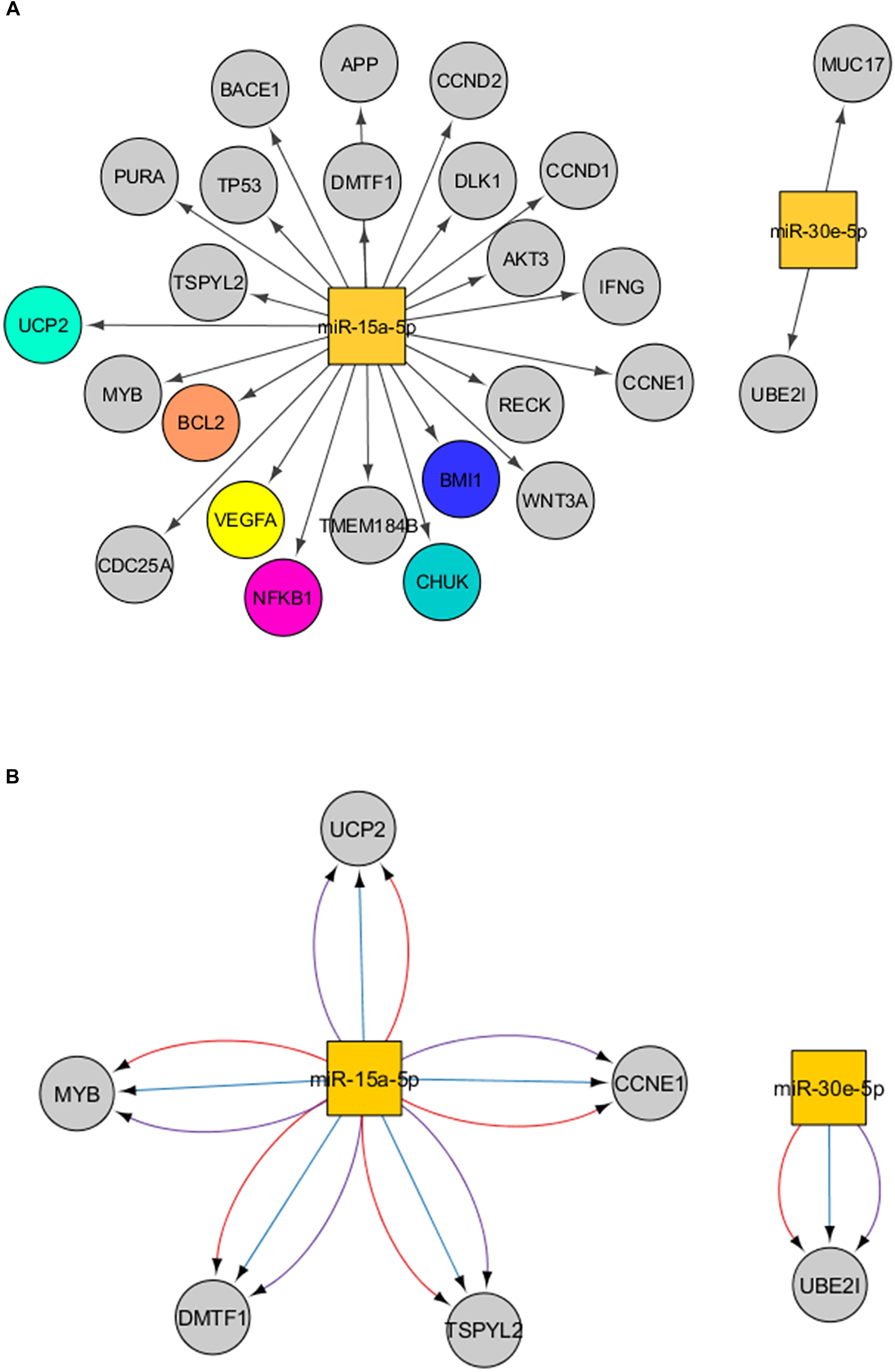
Figure 3. Interactions between miR-15a-5p and miR-30e-5p and their target genes. (A) Experimentally validated target genes of the miRNAs analyzed. Colored genes are those previously associated with DKD. (B) Significant target genes of the miRNAs analyzed found in the three databases analyzed (validated + computationally predicted). Lines in red mean interactions retrieved from miRTarBase, in blue from microcosm, and in purple from TargetScan. Squares represent miRNAs and the circles represent their target genes.
To explore the biological pathways possibly affected by the two miRNAs analyzed, we carried out functional enrichment analysis of their target genes using pathways maps from the BiNGO Database. GO pathways were investigated for biological, cellular, and molecular processes associated with the set of predicted and validated target genes found for miR-15a-5p and miR-30e-5p in the previous analysis. A total of 250 unique significant pathways were enriched for miR-15a-5p, being 183 pathways involved in biological processes, 63 in cellular components, and 20 in molecular functions (Supplementary Table 1). Of note, some of these pathways participate in more than one biological category of BiNGO. For miR-30e-5p, a total of 142 unique significant pathways were enriched, being 109 pathways involved in biological processes, 20 in cellular components, and 15 in molecular functions (Supplementary Table 2).
Moreover, a total of 81 unique pathways were enriched for both miRNAs, being 64 pathways involved in biological process (Figure 2B), 14 in cellular components (Figure 2C), and 3 in molecular functions (Figure 2D). Many of these pathways are well known to be related to DKD pathogenesis, such as transforming growth factor beta receptor, oxidative stress, apoptosis, VEGF and angiogenesis, endoplasmic reticulum stress, hypoxia, and mitochondrial transport pathways (Figure 4A for pathways derived from predicted + validated targets, and Figure 4B for pathways derived only from validated targets).
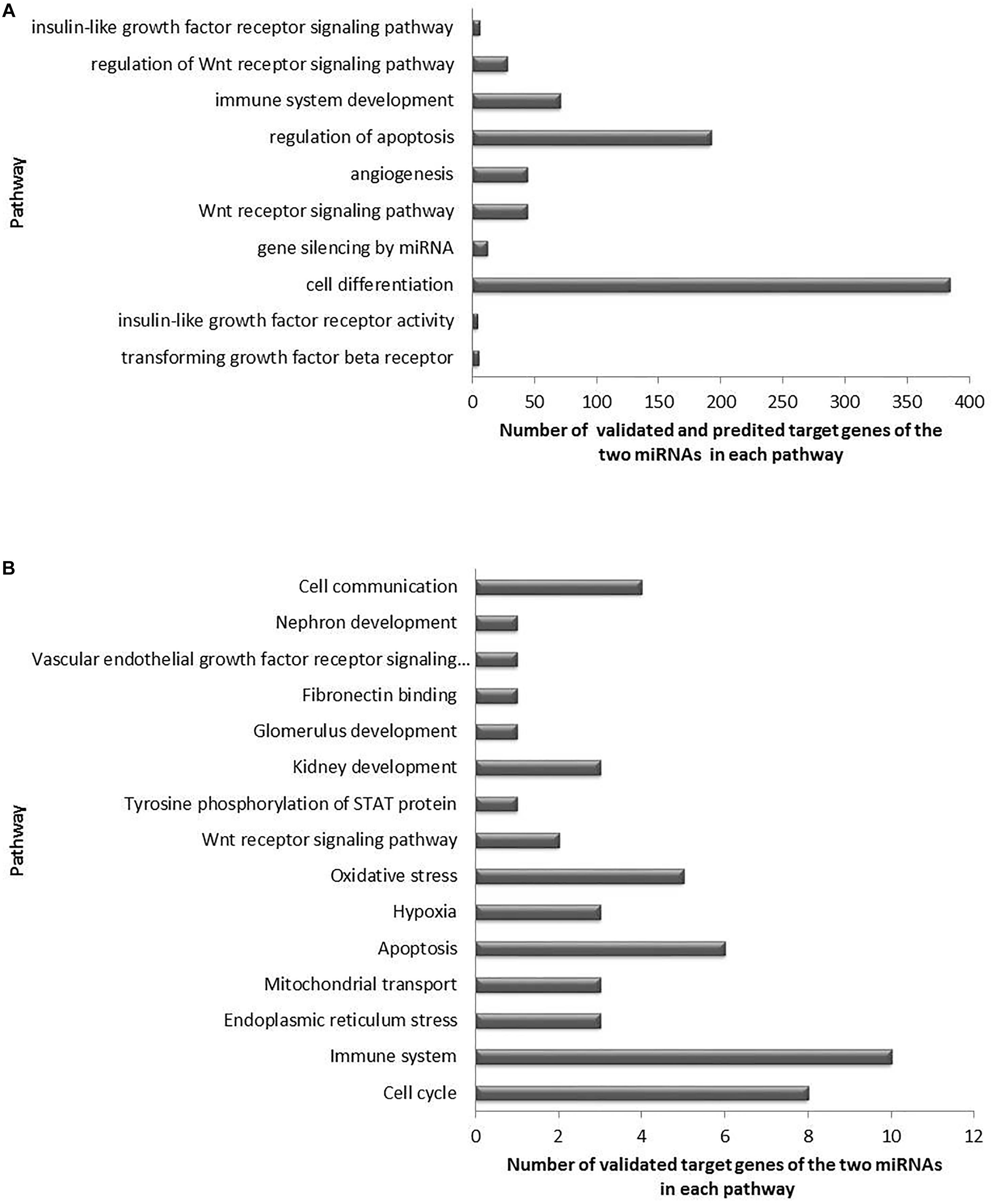
Figure 4. Significant enriched pathways related to DKD pathogenesis and regulated by the target genes of miR-15a-5p and miR-30e-5p. (A) Top 10 selected pathways of predicted and validated target genes of both miRNAs, and (B) selected pathways of validated target genes of both miRNAs.
Discussion
Chronic hyperglycemia may induce cellular damage through increased production of ROS, which then seems to trigger the main pathways related to microvascular diabetic complications, including DKD (Giacco and Brownlee, 2010). Recent studies have shown that miR-15a-5p and miR-30e-5p are associated with podocyte injury, EMT in tubular epithelial cells, and kidney fibrosis (Jiang et al., 2013; Sun et al., 2014; Wu et al., 2014, 2015; Guo et al., 2017; Zhao D. et al., 2017), and also seem to be dysregulated in urine of patients with DKD or CKD (Delic et al., 2016; Cardenas-Gonzalez et al., 2017; Khurana et al., 2017; Xie et al., 2017). Interestingly, both miRNAs target UCP2 (Sun et al., 2011; Jiang et al., 2013), a mitochondrial protein that seems to have a key role in decreasing oxidative stress (Souza et al., 2011). Based on these findings, we therefore analyzed miR-15a-5p and miR-30e-5p expressions in T1DM patients according to DKD stages. Our results indicate that miR-30e-5p is downregulated in plasma and urine of T1DM patients with DKD compared to patients without this complication. No difference was found in the miR-15a-5p expressions between groups.
In agreement with our results, miR-30e-5p expression was also downregulated in urinary exosomes of type 2 DM patients with DKD compared to healthy controls or diabetic patients without this complication (Delic et al., 2016). This association is biologically plausible since all miR-30 family members seem to be essential for structural and functional homeostasis of podocytes, where they are abundantly expressed (Shi et al., 2013; Wu et al., 2014, 2015; Guo et al., 2017). TGF-β1 treatment downregulated miR-30 expression in glomerular podocytes in vivo and in vitro, and the sustained expression of miR-30 inhibited TGF-β1-induced apoptosis of podocytes, while its knockdown aggravated podocyte injury (Shi et al., 2013; Wu et al., 2014). Wu et al. (2014) showed that miR-30 exerts their protective roles by direct inhibition of Notch1 and p53, which mediate podocyte injury. Moreover, miR-30 inhibits the excessive activation of calcium/calcineurin signaling, preventing cytoskeletal damage and apoptosis of podocytes (Wu et al., 2015; Zhao Y. et al., 2017). Zhao D. et al. (2017) reported that in renal tissue of over-8-week-old db/db mice and in human renal tubular epithelial cells (RTECs) cultured for 6 h in high glucose (HG), miR-30e-5p was downregulated while its target GLIPR-2, involved in EMT, was upregulated. Besides, miR-30e-5p overexpression in RTECs promoted proliferation of these cells and inhibited EMT, which could avoid renal fibrosis in DKD (Zhao D. et al., 2017). Accordingly, Jiang et al. (2013) showed that miR-30e-5p was also downregulated in RTECs from mice with ureteral occlusion-induced kidney fibrosis and in TGF-β1-treated NRK-52E renal cells. Transfection of a miR-30e-5p mimic in NRK-52E cells reduced TGF-β1-induced UCP2 expression, inhibiting EMT, whereas a miR-30e inhibitor promoted epithelial cell phenotype changes by loss of E-cadherin, induction of α-SMA, and fibrinogen expression (Jiang et al., 2013). Thus, the downregulation of miR-30e-5p has consistently been shown to be involved in renal fibrosis pathogenesis. Although here we observed that miR-30e-5p was downregulated in plasma of DKD patients independently of the disease severity; in urine, this miRNA was only downregulated in patients with severe DKD, suggesting that it may be a marker of disease progression.
Our bioinformatics analyses indicated that UBE2I (ubiquitin-conjugating enzyme E2 I) and MUC17 (mucin 17) genes are validated target genes of miR-30e-5p (Figure 3A). UBE2I, also known as UCB9, constitutes a core machinery in the sumoylation pathway. Sumoylation is a process in which a small ubiquitin-like modifier (SUMO) is covalently attached to other proteins, modifying their functions (Neyret-Kahn et al., 2013). Important roles for sumoylation were shown in heterochromatin configuration, and sumoylation of core histones negatively regulates transcription (Neyret-Kahn et al., 2013). MiR-30e-5p-induced downregulation of UBE2I inhibited the proliferation and migration of vascular smooth muscle cells (Zong et al., 2017). MUC17 is a glycoprotein characterized as a membrane-bound mucin that provides protection to gut epithelial cells (Gum et al., 2002; Luu et al., 2010). Although UBE2I and MUC17 have not been studied in the context of kidney dysfunction or DKD, a polymorphism in the MUC17 gene was previously associated with protection for CKD in an exome-wide association study (Yamada et al., 2018).
MiR-15a-5p regulates several genes involved in cell division, metabolism, stress response, apoptosis, and angiogenesis (Finnerty et al., 2010). It is abundantly expressed in human and mouse renal tissue (Finnerty et al., 2010), although only few studies have evaluated its function regarding CKD and DKD. In contrast with our results, miR-15a-5p expression was decreased in urinary exosomes of patients with DKD or CKD compared to controls (Khurana et al., 2017; Xie et al., 2017). In addition, this miRNA was downregulated by HG in RTECs (Sun et al., 2014). Treatment of RTECs with a miR-15a-5p mimic was able to reverse HG-induced EMT in these cells, since it inhibited α-SMA and collagen I expressions, and restored E-cadherin expression (Sun et al., 2014). Although our study did not demonstrate an association between miR-15a-5p expression in plasma and urine of patients with DKD, we observed a negative correlation with UAE levels, suggesting an undescribed role of miR-15a-5p in the glomerular basement membrane integrity. MiR-15a-5p was also negatively correlated with HbA1c levels, which is in accordance with the study by Flowers et al. (2015) that reported a negative correlation between circulating miR-15a-5p expression and blood glucose levels in Asian Indians who had glycemic increment after 2.5 years of follow up compared to those who remained stable.
Our bioinformatics analysis using only an experimentally validated database showed that miR-15a-5p regulates genes from several pathways involved in kidney dysfunction mechanisms and DKD development (Kanwar et al., 2011): VEGFA, BCL2, NFKB1 and its inhibitor CHUK (IKKA), UCP2, and BMI1 (Figure 3A). When we used both experimentally validated and computationally predicted tools, UCP2, CCNE1, TSPYL2, DMTF1, and MYB remained as significant targets of miR-15a-5p (Figure 3B). To date, only UCP2 and TSPYL2 genes have been previously investigated regarding kidney dysfunction.
As already mentioned, UCP2 seems to decrease oxidative stress, being a candidate gene for DKD. Sun et al. (2011) confirmed experimentally in MIN-6 cells that miR-15a-5p directly targets UCP2, decreasing mitochondrial uncoupling and, thus, increasing insulin biosynthesis in this beta-cell line since UCP2 is a negative regulator of insulin secretion. Regarding kidney dysfunction, Qiu et al. (2012) reported that oral administration of genipin (a UCP2 inhibitor) partially prevented the progression of DKD in C57BL/6J mice by improving podocyte function. Accordingly, UCP2 was induced in RTECs after unilateral ureteral obstruction in mice, while those mice with ablated UCP2 resisted obstruction-induced EMT and kidney fibrosis (Jiang et al., 2013). Additionally, UCP2 knockdown in NRK-52E tubular cells abolished the effect of TGF-β1 treatment, decreasing ECM production (Jiang et al., 2013). In contrast, Chen et al. (2014) showed that inhibition of UCP2 by genipin increased oxidative stress in rat RTECs treated with HG medium, leading to increased apoptosis. UCP2 knockdown in renal mesangial cells of rats also increased oxidative stress, inflammation and apoptosis in vitro (Di Castro et al., 2013). Therefore, it is still not clear if UCP2 has a protective or deleterious effect on renal function.
Regarding TSPYL2 (testis-specific protein Y-encoded like 2), also known as CDA1, it acts in chromatin remodeling and as inhibitor of cell proliferation in response to DNA damage (Tao et al., 2011). Interestingly TSPYL2 is a regulator of cell-cycle arrest induced by TGF-β1 (Epping et al., 2015), which is a major player in DKD pathogenesis mainly because of its potent pro-fibrotic actions (Assmann et al., 2018b). Accordingly, TSPYL2 expression was upregulated in the aorta of a murine diabetic model of atherosclerosis (Pham et al., 2010). In vitro studies in vascular cells showed that TGF-β1 treatment increased TSPYL2 protein, which then amplified TGF-β1 signaling leading to upregulation of ECM genes (Pham et al., 2010). Chai et al. (2013) reported that TSPYL2 knockout in diabetic mice reduced expression of TGF-β1 receptors in the kidney as well as reduced renal matrix accumulation and attenuated glomerular and tubulointerstitial injury. Therefore, this gene might be a new candidate gene for DKD.
This study has a few limitations. First, we cannot exclude the possibility of type II error when comparing expressions of the two analyzed miRNAs between groups. However, this bias was minimized since our sample size has a power of 80% (α = 0.05) to detect two fold changes (±1.5 SD) in miRNA expressions between case and control groups. Second, duration of T1DM was significantly higher in patients with severe DKD compared to the other two groups, which was expected since diabetes duration is associated with increased prevalence of diabetic chronic complications (Gross et al., 2005). Although this bias could have influenced our results, it is a conservative bias since if some control patients have already some predisposing factor for later development of DKD, this would only decrease the observed association. Third, many factors, such as presence of inflammatory diseases as well as use of medications, can interfere on miRNA expression. In order to reduce this possibility, we used an extensive list of exclusion criteria, as described in the Material and Methods Section. Lastly, as already mentioned many studies have suggested that hyperglycemia induces oxidative stress, which then triggers the main pathways related to diabetic chronic complications; however, few recent studies have challenged the increased mitochondrial ROS production in diabetic kidney disease (Dugan et al., 2013; Sharma, 2015; Coughlan and Sharma, 2016). The reduction of ROS production by UCP2 is also a matter of discussion that needs further investigation (Couplan et al., 2002; Nedergaard and Cannon, 2003). Even though these controversial premises, our main result showing that miR-30e-5p expression is downregulated in patients with DKD is not necessarily influenced by this limitation since this miRNA has many other targets, including TGF-β1. Thus, despite these limitations, our present data is important to be reported since this is the first study that evaluated miR-30e-5p and miR-15a-5p in both plasma and urine samples and in a Brazilian population.
Conclusion
We demonstrated that miR-30e-5p is downregulated in plasma and urine of DKD patients. MiR-15e-5p was only negatively correlated with UAE levels in T1DM patients. Bioinformatics analyses suggest that both analyzed miRNAs regulate genes involved in key mechanisms related to DKD pathogenesis, such as TGF-β receptor, angiogenesis, apoptosis, and hypoxia. Moreover, they are involved in oxidative stress pathway (probably by targeting UCP2), which is an important mechanism linking hyperglycemia to diabetic chronic complications. Our study also suggests that CCNE1, DMTF1, TSPYL2, MYB and UBE2I might constitute new potential candidate genes for DKD.
Ethics Statement
This study was carried out in accordance with the recommendations of the Ethic Committees in Research from Hospital de Clínicas de Porto Alegre and Grupo Hospitalar Conceição/Instituto da Criança com Diabetes with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Ethic Committees in Research from Hospital de Clínicas de Porto Alegre and Grupo Hospitalar Conceição/Instituto da Criança com Diabetes.
Author Contributions
CD designed the study, researched the data, collected the samples, performed the experiments, and wrote the manuscript. AC collected the samples, researched the data, and reviewed the manuscript. TA researched the data, collected the samples, performed the bioinformatics analyses, contributed to discussion, and reviewed the manuscript. AB, BdS, and LC contributed to discussion and reviewed the manuscript. DC designed the study, contributed to the discussion, and wrote the manuscript.
Funding
This study was partially supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundo de Incentivo à Pesquisa e Eventos (FIPE) at Hospital de Clínicas de Porto Alegre (Grant No. 2017-0095), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) – Finance Code 001, and Post-Graduated Program in Medical Sciences: Endocrinology – Federal University of Rio Grande do Sul. DC and LC are recipients of scholarships from CNPq, while CD, TA, and BdS are recipients of scholarships from the CAPES.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00563/full#supplementary-material
Footnotes
References
American Diabetes Association (2018). 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2018. Diabetes Care 41(Suppl. 1), S13–S27. doi: 10.2337/dc18-S002
Andrassy, K. M. (2013). Comments on KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease’. Kidney Int. 84, 622–623. doi: 10.1038/ki.2013.243
Assmann, T. S., Brondani Lde, A., Bauer, A. C., Canani, L. H., and Crispim, D. (2014). Polymorphisms in the TLR3 gene are associated with risk for type 1 diabetes mellitus. Eur. J. Endocrinol. 170, 519–527. doi: 10.1530/EJE-13-0963
Assmann, T. S., Recamonde-Mendoza, M., Costa, A. R., Punales, M., Tschiedel, B., Canani, L. H., et al. (2018a). Circulating miRNAs in diabetic kidney disease: case-control study and in silico analyses. Acta Diabetol. 56, 55–65. doi: 10.1007/s00592-018-1216-x
Assmann, T. S., Recamonde-Mendoza, M., de Souza, B. M., Bauer, A. C., and Crispim, D. (2018b). MicroRNAs and diabetic kidney disease: systematic review and bioinformatic analysis. Mol. Cell. Endocrinol. 477, 90–102. doi: 10.1016/j.mce.2018.06.005
Assmann, T. S., Recamonde-Mendoza, M., De Souza, B. M., and Crispim, D. (2017). MicroRNA expression profiles and type 1 diabetes mellitus: systematic review and bioinformatic analysis. Endocr. Connect. 6, 773–790. doi: 10.1530/EC-17-0248
Brand, M. D., and Esteves, T. C. (2005). Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2, 85–93. doi: 10.1016/j.cmet.2005.06.002
Brennan, E., McEvoy, C., Sadlier, D., Godson, C., and Martin, F. (2013). The genetics of diabetic nephropathy. Genes 4, 596–619. doi: 10.3390/genes4040596
Brownlee, M. (2005). The pathobiology of diabetic complications: a unifying mechanism. Diabetes 54, 1615–1625. doi: 10.2337/diabetes.54.6.1615
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cardenas-Gonzalez, M., Srivastava, A., Pavkovic, M., Bijol, V., Rennke, H. G., Stillman, I. E., et al. (2017). Identification, confirmation, and replication of novel urinary microrna biomarkers in lupus nephritis and diabetic nephropathy. Clin. Chem. 63, 1515–1526. doi: 10.1373/clinchem.2017.274175
Chai, Z., Dai, A., Tu, Y., Li, J., Wu, T., Wang, Y., et al. (2013). Genetic deletion of cell division autoantigen 1 retards diabetes-associated renal injury. J. Am. Soc. Nephrol. 24, 1782–1792. doi: 10.1681/ASN.2013010060
Chen, X. L., Tang, W. X., Tang, X. H., Qin, W., and Gong, M. (2014). Downregulation of uncoupling protein-2 by genipin exacerbates diabetes-induced kidney proximal tubular cells apoptosis. Ren. Fail. 36, 1298–1303. doi: 10.3109/0886022X.2014.930650
Coughlan, M. T., and Sharma, K. (2016). Challenging the dogma of mitochondrial reactive oxygen species overproduction in diabetic kidney disease. Kidney Int. 90, 272–279. doi: 10.1016/j.kint.2016.02.043
Couplan, E., del Mar Gonzalez-Barroso, M., Alves-Guerra, M. C., Ricquier, D., Goubern, M., and Bouillaud, F. (2002). No evidence for a basal, retinoic, or superoxide-induced uncoupling activity of the uncoupling protein 2 present in spleen or lung mitochondria. J. Biol. Chem. 277, 26268–26275. doi: 10.1074/jbc.M202535200
Crispim, D., Fagundes, N. J., dos Santos, K. G., Rheinheimer, J., Boucas, A. P., de Souza, B. M., et al. (2010). Polymorphisms of the UCP2 gene are associated with proliferative diabetic retinopathy in patients with diabetes mellitus. Clin. Endocrinol. 72, 612–619. doi: 10.1111/j.1365-2265.2009.03684.x
de Souza, B. M., Michels, M., Sortica, D. A., Boucas, A. P., Rheinheimer, J., Buffon, M. P., et al. (2015). Polymorphisms of the UCP2 gene are associated with glomerular filtration rate in type 2 diabetic patients and with decreased UCP2 gene expression in human kidney. PLoS One 10:e0132938. doi: 10.1371/journal.pone.0132938
Delic, D., Eisele, C., Schmid, R., Baum, P., Wiech, F., Gerl, M., et al. (2016). Urinary exosomal miRNA signature in Type II diabetic nephropathy patients. PLoS One 11:e0150154. doi: 10.1371/journal.pone.0150154
Di Castro, S., Scarpino, S., Marchitti, S., Bianchi, F., Stanzione, R., Cotugno, M., et al. (2013). Differential modulation of uncoupling protein 2 in kidneys of stroke-prone spontaneously hypertensive rats under high-salt/low-potassium diet. Hypertension 61, 534–541. doi: 10.1161/HYPERTENSIONAHA.111.00101
Dugan, L. L., You, Y. H., Ali, S. S., Diamond-Stanic, M., Miyamoto, S., DeCleves, A. E., et al. (2013). AMPK dysregulation promotes diabetes-related reduction of superoxide and mitochondrial function. J. Clin. Invest. 123, 4888–4899. doi: 10.1172/JCI66218
Epping, M. T., Lunardi, A., Nachmani, D., Castillo-Martin, M., Thin, T. H., Cordon-Cardo, C., et al. (2015). TSPYL2 is an essential component of the REST/NRSF transcriptional complex for TGFbeta signaling activation. Cell Death Differ. 22, 1353–1362. doi: 10.1038/cdd.2014.226
Finnerty, J. R., Wang, W. X., Hebert, S. S., Wilfred, B. R., Mao, G., and Nelson, P. T. (2010). The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J. Mol. Biol. 402, 491–509. doi: 10.1016/j.jmb.2010.07.051
Fisler, J. S., and Warden, C. H. (2006). Uncoupling proteins, dietary fat and the metabolic syndrome. Nutr. Metab. 3:38.
Flowers, E., Gadgil, M., Aouizerat, B. E., and Kanaya, A. M. (2015). Circulating micrornas associated with glycemic impairment and progression in Asian Indians. Biomark Res. 3:22. doi: 10.1186/s40364-015-0047-y
Giacco, F., and Brownlee, M. (2010). Oxidative stress and diabetic complications. Circ. Res. 107, 1058–1070. doi: 10.1161/CIRCRESAHA.110.223545
Gross, J. L., de Azevedo, M. J., Silveiro, S. P., Canani, L. H., Caramori, M. L., and Zelmanovitz, T. (2005). Diabetic nephropathy: diagnosis, prevention, and treatment. Diabetes Care 28, 164–176.
Gum, J. R. Jr., Crawley, S. C., Hicks, J. W., Szymkowski, D. E., and Kim, Y. S. (2002). MUC17, a novel membrane-tethered mucin. Biochem. Biophys. Res. Commun. 291, 466–475. doi: 10.1006/bbrc.2002.6475
Guo, Y., Deng, X., Chen, S., Yang, L., Ni, J., Wang, R., et al. (2017). MicroRNA-30e targets BNIP3L to protect against aldosterone-induced podocyte apoptosis and mitochondrial dysfunction. Am. J. Physiol. Renal. Physiol. 312, F589–F598. doi: 10.1152/ajprenal.00486.2016
Jiang, L., Qiu, W., Zhou, Y., Wen, P., Fang, L., Cao, H., et al. (2013). A microRNA-30e/mitochondrial uncoupling protein 2 axis mediates TGF-beta1-induced tubular epithelial cell extracellular matrix production and kidney fibrosis. Kidney Int. 84, 285–296. doi: 10.1038/ki.2013.80
Kanwar, Y. S., Sun, L., Xie, P., Liu, F. Y., and Chen, S. (2011). A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu. Rev. Pathol. 6, 395–423. doi: 10.1146/annurev.pathol.4.110807.092150
Kato, M., and Natarajan, R. (2015). MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann. N. Y. Acad. Sci. 1353, 72–88. doi: 10.1111/nyas.12758
Khurana, R., Ranches, G., Schafferer, S., Lukasser, M., Rudnicki, M., Mayer, G., et al. (2017). Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA 23, 142–152. doi: 10.1261/rna.058834.116
Kutmon, M., Kelder, T., Mandaviya, P., Evelo, C. T., and Coort, S. L. (2013). CyTargetLinker: a cytoscape app to integrate regulatory interactions in network analysis. PLoS One 8:e82160. doi: 10.1371/journal.pone.0082160
Levey, A. S., Stevens, L. A., Schmid, C. H., Zhang, Y. L., Castro, A. F. III, Feldman, H. I., et al. (2009). A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 150, 604–612.
Luu, Y., Junker, W., Rachagani, S., Das, S., Batra, S. K., Heinrikson, R. L., et al. (2010). Human intestinal MUC17 mucin augments intestinal cell restitution and enhances healing of experimental colitis. Int. J. Biochem. Cell Biol. 42, 996–1006. doi: 10.1016/j.biocel.2010.03.001
Maere, S., Heymans, K., and Kuiper, M. (2005). BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. doi: 10.1093/bioinformatics/bti551
Nedergaard, J., and Cannon, B. (2003). The ‘novel’ ‘uncoupling’ proteins UCP2 and UCP3: what do they really do? Pros and cons for suggested functions. Exp. Physiol. 88, 65–84. doi: 10.1113/eph8802502
Neyret-Kahn, H., Benhamed, M., Ye, T., Le Gras, S., Cossec, J. C., Lapaquette, P., et al. (2013). Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 23, 1563–1579. doi: 10.1101/gr.154872.113
Pham, Y., Tu, Y., Wu, T., Allen, T. J., Calkin, A. C., Watson, A. M., et al. (2010). Cell division autoantigen 1 plays a profibrotic role by modulating downstream signalling of TGF-beta in a murine diabetic model of atherosclerosis. Diabetologia 53, 170–179. doi: 10.1007/s00125-009-1555-9
Qiu, W., Zhou, Y., Jiang, L., Fang, L., Chen, L., Su, W., et al. (2012). Genipin inhibits mitochondrial uncoupling protein 2 expression and ameliorates podocyte injury in diabetic mice. PLoS One 7:e41391. doi: 10.1371/journal.pone.0041391
Ritz, E., Zeng, X. X., and Rychlik, I. (2011). Clinical manifestation and natural history of diabetic nephropathy. Contrib. Nephrol. 170, 19–27. doi: 10.1159/000324939
Rudofsky, G. Jr., Schroedter, A., Schlotterer, A., Voron’ko, O. E., Schlimme, M., Tafel, J., et al. (2006). Functional polymorphisms of UCP2 and UCP3 are associated with a reduced prevalence of diabetic neuropathy in patients with type 1 diabetes. Diabetes Care 29, 89–94. doi: 10.2337/diacare.29.1.89
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, K. (2015). Mitochondrial hormesis and diabetic complications. Diabetes 64, 663–672. doi: 10.2337/db14-0874
Shi, S., Yu, L., Zhang, T., Qi, H., Xavier, S., Ju, W., et al. (2013). Smad2-dependent downregulation of miR-30 is required for TGF-beta-induced apoptosis in podocytes. PLoS One 8:e75572. doi: 10.1371/journal.pone.0075572
Souza, B. M., Assmann, T. S., Kliemann, L. M., Gross, J. L., Canani, L. H., and Crispim, D. (2011). The role of uncoupling protein 2 (UCP2) on the development of type 2 diabetes mellitus and its chronic complications. Arq. Bras. Endocrinol. Metabol. 55, 239–248. doi: 10.1590/s0004-27302011000400001
Sun, L. L., Jiang, B. G., Li, W. T., Zou, J. J., Shi, Y. Q., and Liu, Z. M. (2011). MicroRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res. Clin. Pract. 91, 94–100. doi: 10.1016/j.diabres.2010.11.006
Sun, T., Yang, J., Dong, W., Wang, R., Ma, P., Kang, P., et al. (2014). Down-regulated miR-15a mediates the epithelial-mesenchymal transition in renal tubular epithelial cells promoted by high glucose. Biosci. Biotechnol. Biochem. 78, 1363–1370. doi: 10.1080/09168451.2014.936345
Tao, K. P., Fong, S. W., Lu, Z., Ching, Y. P., Chan, K. W., and Chan, S. Y. (2011). TSPYL2 is important for G1 checkpoint maintenance upon DNA damage. PLoS One 6:e21602. doi: 10.1371/journal.pone.0021602
Thomas, M. C., Brownlee, M., Susztak, K., Sharma, K., Jandeleit-Dahm, K. A., Zoungas, S., et al. (2015). Diabetic kidney disease. Nat. Rev. Dis. Primers 1:15018. doi: 10.1038/nrdp.2015.18
Tiwari, A. K., Prasad, P., Thelma, B. K., Kumar, K. M., Ammini, A. C., Gupta, A., et al. (2009). Oxidative stress pathway genes and chronic renal insufficiency in asian indians with type 2 diabetes. J. Diabetes Complications 23, 102–111. doi: 10.1016/j.jdiacomp.2007.10.003
von Elm, E., Altman, D. G., Egger, M., Pocock, S. J., Gotzsche, P. C., Vandenbroucke, J. P., et al. (2014). The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Int. J. Surg. 12, 1495–1499. doi: 10.1016/j.ijsu.2014.07.013
Wanner, N., and Bechtel-Walz, W. (2017). Epigenetics of kidney disease. Cell Tissue Res. 369, 75–92. doi: 10.1007/s00441-017-2588-x
Wu, J., Zheng, C., Fan, Y., Zeng, C., Chen, Z., Qin, W., et al. (2014). Downregulation of microRNA-30 facilitates podocyte injury and is prevented by glucocorticoids. J. Am. Soc. Nephrol. 25, 92–104. doi: 10.1681/ASN.2012111101
Wu, J., Zheng, C., Wang, X., Yun, S., Zhao, Y., Liu, L., et al. (2015). MicroRNA-30 family members regulate calcium/calcineurin signaling in podocytes. J. Clin. Invest. 125, 4091–4106. doi: 10.1172/JCI81061
Xie, Y., Jia, Y., Cuihua, X., Hu, F., Xue, M., and Xue, Y. (2017). Urinary exosomal MicroRNA profiling in incipient type 2 diabetic kidney disease. J. Diabetes Res. 2017:6978984. doi: 10.1155/2017/6978984
Yamada, Y., Kato, K., Oguri, M., Horibe, H., Fujimaki, T., Yasukochi, Y., et al. (2018). Identification of 13 novel susceptibility loci for early-onset myocardial infarction, hypertension, or chronic kidney disease. Int. J. Mol. Med. 42, 2415–2436. doi: 10.3892/ijmm.2018.3852
Zelmanovitz, T., Gross, J. L., Oliveira, J. R., Paggi, A., Tatsch, M., and Azevedo, M. J. (1997). The receiver operating characteristics curve in the evaluation of a random urine specimen as a screening test for diabetic nephropathy. Diabetes Care 20, 516–519. doi: 10.2337/diacare.20.4.516
Zhao, D., Jia, J., and Shao, H. (2017). miR-30e targets GLIPR-2 to modulate diabetic nephropathy: in vitro and in vivo experiments. J. Mol. Endocrinol. 59, 181–190. doi: 10.1530/jme-17-0083
Zhao, Y., Wu, J., Zhang, M., Zhou, M., Xu, F., Zhu, X., et al. (2017). Angiotensin II induces calcium/calcineurin signaling and podocyte injury by downregulating microRNA-30 family members. J. Mol. Med. 95, 887–898. doi: 10.1007/s00109-017-1547-z
Keywords: microRNA expression, miR-15a-5p, miR-30e-5p, diabetic kidney disease, bioinformatics analysis, type 1 diabetes mellitus
Citation: Dieter C, Assmann TS, Costa AR, Canani LH, de Souza BM, Bauer AC and Crispim D (2019) MiR-30e-5p and MiR-15a-5p Expressions in Plasma and Urine of Type 1 Diabetic Patients With Diabetic Kidney Disease. Front. Genet. 10:563. doi: 10.3389/fgene.2019.00563
Received: 31 January 2019; Accepted: 29 May 2019;
Published: 12 June 2019.
Edited by:
Rui Henrique, Portuguese Oncology Institute, PortugalReviewed by:
Ling-Qing Yuan, Central South University, ChinaCharles Affourtit, University of Plymouth, United Kingdom
Copyright © 2019 Dieter, Assmann, Costa, Canani, de Souza, Bauer and Crispim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daisy Crispim, ZGNtb3JlaXJhQGhjcGEuZWR1LmJy
 Cristine Dieter
Cristine Dieter Taís Silveira Assmann
Taís Silveira Assmann Aline Rodrigues Costa1
Aline Rodrigues Costa1 Daisy Crispim
Daisy Crispim