- 1School of Marine Sciences, Sun Yat-sen University, Zhuhai, China
- 2Southern Marine Science and Engineering Guangdong Laboratory, Zhuhai, China
Yellowfin seabream (Acanthopagrus latus), a protandrous hermaphroditic fish, is a good model for studying the mechanism of sex reversal. However, limited knowledge is known about the genetic information related to reproduction and sex differentiation in this species. Here, we performed de novo transcriptome sequencing analysis of the testis, ovotestis, and ovary to identify sex-related genes in yellowfin seabream. The results assembled 71,765 unigenes in which 16,126 and 17,560 unigenes were differentially expressed in the ovotestis and ovary compared to the testis, respectively. The most differentially expressed gene (DEG)-enriched Kyoto Encyclopedia of Genes and Genomes and GO pathways were closely associated with the synthesis of sex steroid hormones. Functional analyses identified 55 important sex-related DEGs, including 32 testis-biased DEGs (dmrt1, amh, and sox9, etc.), 20 ovary-biased DEGs (cyp19a, foxl2, and wnt4, etc.), and 3 ovotestis-biased DEGs (lhb, dmrt2, and foxh1). Furthermore, the testis-specific expression of dmrt1 and the brain-pituitary-ovary axis expression of foxl2 were characterized, suggesting that they might play important roles in sex differentiation in yellowfin seabream. Our present work provided an important molecular basis for elucidating the mechanisms underlying sexual transition and reproductional regulation in yellowfin seabream.
Introduction
Sex is one of the most intriguing propositions in life science (Mei and Gui, 2015; Schubert, 2015; Li and Gui, 2018). Usually, once the sex is determined, it will be maintained forever in animals. However, sex reversal has been found in a few lower animals, especial in fish species (Todd et al., 2016). Fish are the most species-rich group of vertebrates that have diverse sex strategies. From the discovery of hermaphrodite and sex reversal in rice field eel (Monopterus albus; Liu, 1944), numerous significant achievements have been obtained in basic studies on fish sex (Mei and Gui, 2015). It has been reported that there are about 500 hermaphrodite species (Avise and Mank, 2009). The hermaphrodites could be classified to three types: protogynous in grouper (Epinephelus coioides) and wrasse (Thalassoma bifasciatum), protandrous in Asian seabass (Lates calcarifer), and black seabream (Acanthopagrus schlegelii), and bidirectional hermaphrodites in Gobiodon erythrospilus. Fish have the most diverse sex-determining strategies, including environmental sex determination (ESD), and genotypic sex determination (GSD), such as XX/XY, and ZZ/ZW systems and their variants as well sex-related extra chromosomes (Li et al., 2016, 2017; Li and Gui, 2018).
The SD genes seem to be conserved with Sry for mammals and Dmrt1 for bird (Koopman et al., 1991). However, the SD genes are highly variable in fish species. Recently, advances in fish SD have been fueled by the rapid development of affordable genome and transcriptome technologies (Kamiya et al., 2012; Myosho et al., 2012; Yano et al., 2012; Chen et al., 2014; Dan et al., 2018). Dmy (doublesex- and mab-3-related transcription factor 1-Y) is the first identified SD gene in fish (Matsuda et al., 2002). Subsequently, different SD genes are revealed in other fish species, such as amhy (Y-linked anti-Mullerian hormone) in Odontesthes hatcheri (Hattori et al., 2012), amhr2 (anti-Mullerian hormone receptor type 2) in Takifugu rubripes (Kamiya et al., 2012), sdY (sexually dimorphic on the Y chromosome) in Oncorhynchus mykiss (Yano et al., 2012), and pfpdz1 in Pelteobagrus fulvidraco (Dan et al., 2018). Interestingly, the SD genes might be different even in the same fish subspecies, such as dmy in Oryzias latipes (O. latipes; Matsuda et al., 2002), gsdf (gonadal somatic cell derived factor) in O. luzonensis (Myosho et al., 2012), and sox3-Y (SRY-box transcription factor 3-Y) in O. dancena (Takehana et al., 2014). To date, SD and sexual differentiation are mainly investigated in gonochoristic species; nevertheless, limited knowledge is known in hermaphroditic fish. Recently, several known SD-related genes have been cloned in several hermaphroditic fish, such as foxl2, and gsdf in M. albus (Hu et al., 2014; Zhu et al., 2016), dmrt1 and sox3 in E. coioides (Xia et al., 2007; Yao et al., 2007), and amh and amhr2 in A. schlegelii (Wu et al., 2010). Whereas the sex-related signaling pathways remain unclear, and their conservation should be analyzed in other hermaphroditic animals.
Sparidae is a group of teleost fish with different reproductive strategies, including gonochorism and hermaphroditism. For example, the common dentex (Dentex dentex) is a gonochoristic species (Tsakogiannis et al., 2018a), red porgy (Pagrus pagrus), and common pandora (pagellus erythrinus) are protogynous hermaphrodites (Tsakogiannis et al., 2018b), gilthead seabream (Sparus aurata), and yellowfin seabream (Acanthopagrus latus) are protandrous hermaphrodites (Negintaji et al., 2018; Tsakogiannis et al., 2018a), and sharpsnout seabream (Diplodus puntazzo) is a rudimentary hermaphrodite fish species (Papadaki et al., 2018). Therefore, Sparidae is a good model for studying SD and sex reversal.
Yellowfin seabream (A. latus) belonging to the Sparidae family is a commercially important fish that is popular in Asia because of its beautiful appearance and delicious taste. Yellowfin seabream initially develops a functional testis during the first reproductive cycle; then it undergoes a long term of sexual reversal to develop a functional ovary in the third year. Therefore, yellowfin seabream is considered as a good model for studying sex differentiation and sex reversal. However, there is little information about gonadal development and sex-related genes in this species. In this study, aiming to explore the genetic information associated with gonadal differentiation and sex reversal in yellowfin seabream, we carried out transcriptome sequencing analysis of testis, ovotestis, and ovary. Our results identified a large number of sex-related candidates and the dynamics of signaling pathway during sex reversal, which would provide a molecular basis for studying the mechanism underlying sexual transition and reproduction regulation in this species.
Materials and Methods
Ethical Procedures
All experimental procedures in our study with A. latus were approved by the Ethics Committee of Sun Yat-sen University.
Sample Collection and Histological Examination
Yellowfin seabreams were provided from Hailv aquaculture station (Zhuhai, China). Nine of the yellowfin seabreams were chosen and divided into 3 samples in triplicates for the subsequent experiments, including three 1-year-olds with average body weight (ABW) of 113.3 ± 8.5 g and average body length (ABL) of 15.3 ± 0.3 cm, three 2-year-olds (ABW = 226.7 ± 21.6 g, ABL = 19.5 ± 0.6 cm), and three 3-year-olds (ABW = 519.3 ± 55.3 g, ABL = 26.1 ± 0.8 cm). Each fish was anesthetized by MS-222 (Sigma, United States), and half of the gonads were fixed in 4% PFA/PBS for later histological analysis, and the other was frozen with liquid nitrogen for RNA extraction. Subsequently, the fixed gonad were embedded in paraffin, sectioned, and stained with hematoxylin and eosin (HE). The images were acquired with an inverted microscope (Zeiss, Germany).
RNA Extraction and Illumina Sequencing
Total RNA was isolated with a combined method of trizol (Invitrogen, United States) and SV (Spin or Vacuum) total RNA isolation system (Promega, Unites States), which could yield high-quality RNA. The samples were homogenized with 1 ml trizol reagent for 40 s with BioPrep-24 (Tomos, China), followed by adding 200 μl chloroform and mixed thoroughly. After incubating at room temperature for 5 min, samples were centrifuged at 10,000 × g for 15 min at 4°C; then the upper phase (about 400 μl) was transferred to a new nuclease-free tube and mixed thoroughly with 200 μl ethanol. The mixture was transferred to a new spin column and centrifuged at 14,000 × g for 1 min at room temperature. The following steps were performed according to the manufacturer’s instructions of the SV total RNA isolation system. The amount and quality of isolated RNA were determined by BioSpec-nano Spectrophotometer (Shimadzu, Japan) and Agilent 2100 Bioanalyzer (Agilent Technologies, United States). High-quality RNA (28S/18S ≥ 1.5, OD260/280 ≥ 1.8, and RIN value ≥ 7) was used for cDNA library construction. The library was generated as previously described (Lin et al., 2020). The de novo transcriptome sequencing was performed on the Illumina Hiseq platform (Shanghai Majorbio Bio-pharm Technology Co., Ltd., China). The data have been submitted to the Genbank (SRA accession number: PRJNA622226).
Transcriptome Assembly and Gene Annotation
After sequencing, raw reads were filtered with three steps to get the high-quality clean reads. First, SeqPrep software1 was used to remove adapter sequences. The sickle software2 was used to trim and discard the low-quality reads with a Q-score less than 10. Next, raw reads with poly-N (N means ambiguous base) or length less than 30 bp were discarded. The clean data of the nine samples were pooled together for de novo assembly with Trinity3 with optimization by TransRate4, CD-HIT5, and BUSCO6. The assembled sequences were annotated to public databases of NCBI_Nr, Swiss-Prot, and clusters of orthologous groups of proteins (COG) with a threshold E-value < 10–5. Further functional annotation were performed against the Pfam database with HMMER3, gene ontology (GO) with BLAST2GO (version 2.5.0), and Kyoto Encyclopedia of Genes and Genomes (KEGG) with KOBAS (version 2.1.1), respectively.
Differential Expression Analysis
The gene expression levels were standardized by fragments per kilobases per million (FPKM) read values with RSEM software7. The expressed genes were filtered with a threshold of FPKM value greater than 0.5. The analysis of differentially expressed genes (DEGs) was performed by DESeq2 software with BH method (false discovery rate correction with Benjamini/Hochberg) with “adjusted p-value < 0.05 and the absolute value of log2 fold change ≥ 1” as a threshold to judge the significance of gene expression difference. The DEGs in sex-related GO terms and KEGG pathways were considered as sex-related genes.
Reverse Transcribed and Real-Time Quantitative PCR (RT-qPCR) Verification
To validate the transcriptome data, first-strand cDNAs were synthesized, and Real-Time Quantitative PCR (RT-qPCR) analyses were performed with the RNA template used for transcriptome sequencing. Total RNA from triplicated individuals in each age group were mixed in equal mass for reverse transcription. The first-strand cDNAs were transcribed with the PrimeScriptTM RT reagent Kit (Takara, Japan) according to the manufacturer’s instruction. Primers were designed by Primer38. RT-qPCR was performed as previously described (Lin et al., 2020). Briefly, the thermal cycling conditions were: 94°C for 2 min, 40 cycles of 94°C for 15 s, 57°C for 15 s, 72°C for 15 s, and an additional 72°C for 2 min, followed by a final disassociation curve analysis. All the samples were analyzed in triplicates, and relative expression levels were normalized to β-actin by using the 2–ΔΔCT methods. Primer information is listed in Supplementary Table S1.
Statistics Analysis
All statistics were calculated using SPSS version 20. Differences between control and treatment groups were assessed by one-way ANOVA. p < 0.05 was considered as significant difference.
Results
Morphological and Histological Structure of the Testis, Ovotestis, and Ovary
Although the occurrence of sex reversal in Sparidae is known, the gonadal structure remains unclear in yellowfin seabream. To clarify it, HE staining was performed to characterize the structure of testis, ovotestis, and ovary in 1-, 2-, and 3-year-old yellowfin seabream, respectively. In 1-year-old yellowfin seabream, the milky and swollen gonads were comprised of a huge testicular portion containing all stages of male germ cells and numerous sperm as well as a very small ovarian portion with some primary oocytes (Figures 1A–D). After the first breeding season, the mature testis gradually degenerated; meanwhile, the ovary started to grow and develop. In two-year-old yellowfin seabream, the ovotestis was divided into two distinct portions: a distinct pale orange red and translucent testis comprising a loose structure with many spermatogonias and a faint yellow ovary consisting of many primary oocytes and previtellogenic oocytes (Figures 1E–H). In 3-year-old fish, the mature ovary is filled with lots of mature oocytes, and vitellogenic oocytes as well as a few primary oocytes and previtellogenic oocytes (Figures 1I–K). Next, we performed de novo transcriptome sequencing analysis of the testis, ovotestis, and ovary.
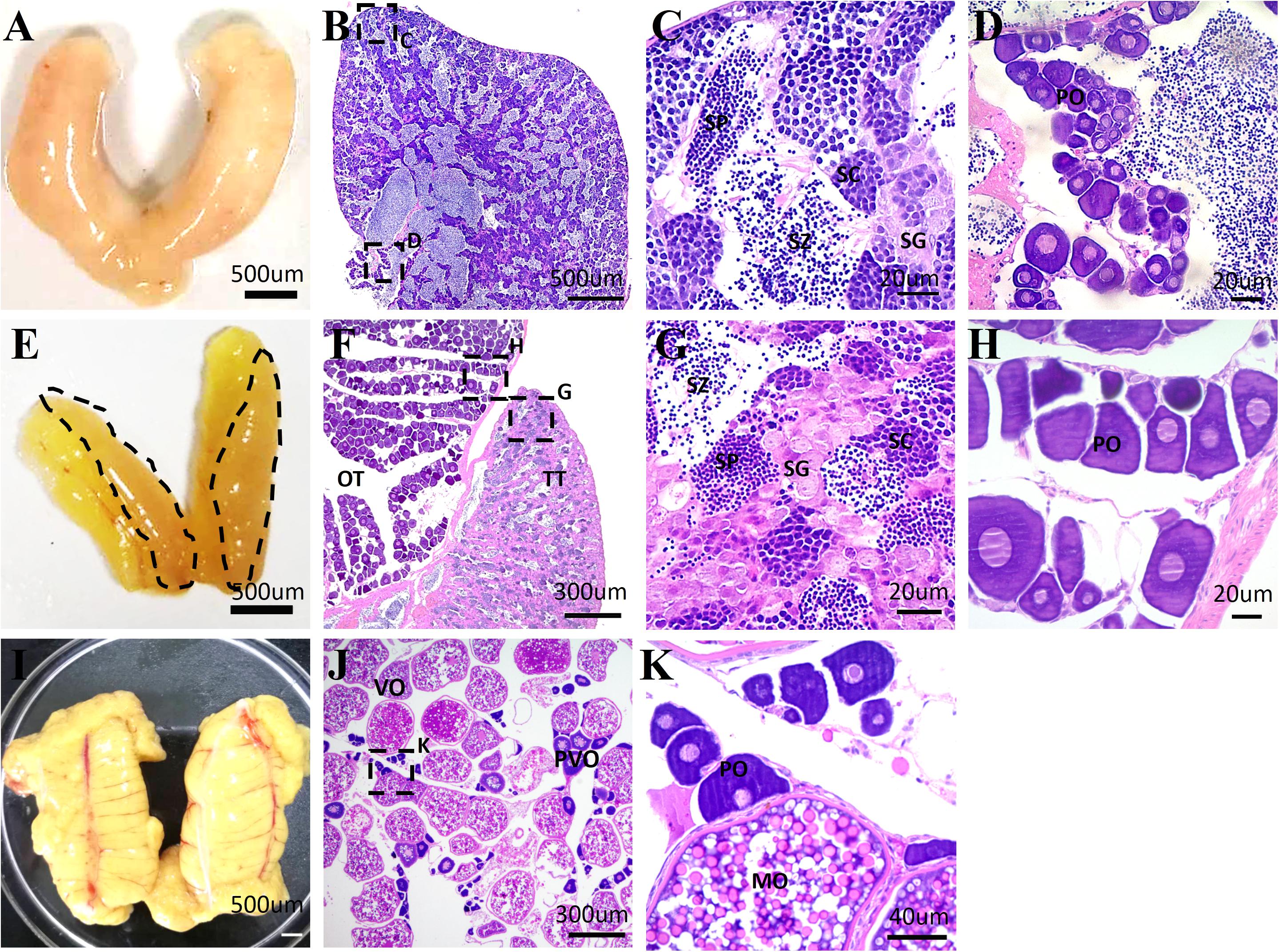
Figure 1. The representative images of the morphology and histological structures of the testis, ovotestis, and ovary of yellowfin seabream. The morphology of testis (A), ovotestis (E), and ovary (I). The dotted line in panel (E) indicates the testicular portion of ovotestis. HE staining of the testis (B–D), ovotestis (F–H), and ovary (J,K). (C,D) The magnified images in panel (B); (G,H) the magnified images in panel (F); and (K) the magnified images in panel (J). MO, mature oocyte; OT, ovarian tissue; PO, primary oocyte; PVO, previtellogenic oocyte; SC, spermatocyte; SG, spermatogonia; SP, spermatid; SZ, spermatozoa; TT, testicular tissue; VO, vitellogenic oocyte.
Transcriptome Sequencing, Assembly, and Annotation
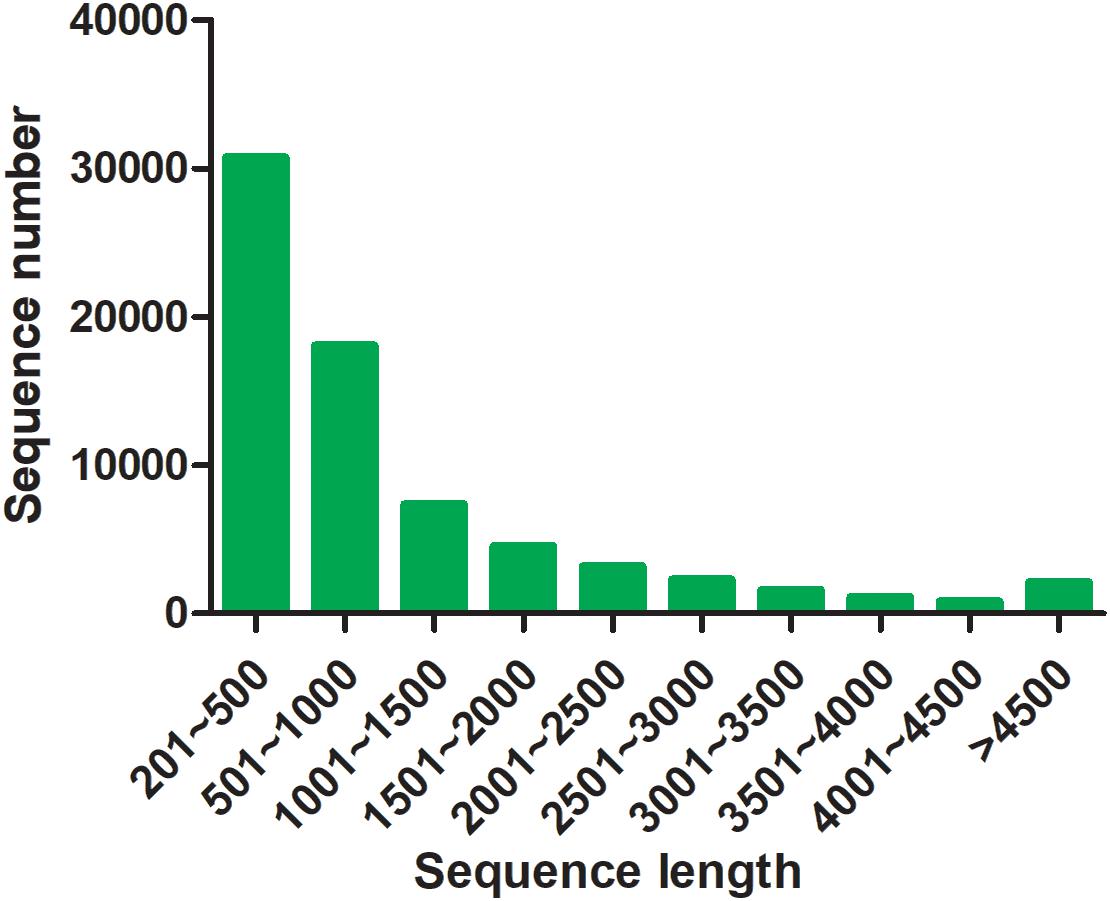
By transcriptome sequencing, 10.1, 7.9, and 8.5 Gb raw data were obtained from the testis, ovotesits, and ovary, respectively. After filtering low-quality reads, 9.8, 7.7, and 8.3 Gb of clean data were obtained in testis, ovotestis, and ovary with Q20 values of 98.68, 98, and 98.25%, respectively (Supplementary Table S2). All of the transcriptomes were grouped into a single file for redundancy elimination. As a result, 71,765 unigenes were assembled, which had an average length of 1201 bp, and an N50 length of 2032 bp (Supplementary Table S3). Length distribution analysis showed that 48,763 unigenes (68%) ranged from 200 to 1000 bp; 17,354 unigenes (24%) ranged from 1000 to 3000 bp, and the remained 5648 unigenes (8%) were longer than 3000 bp in length (Figure 2). The unigenes were functionally annotated to the multiple public databases by BLAST with a threshold e value less than 10–5. A total of 30,584 (42.62%) unigenes were matched with the public databases: in which 29,252 (40.76%), 25,425 (35.43%), 22,732 (31.68%), 13,183 (18.37%), 9522 (13.27%), and 20,057 (27.95%) were matched in NCBI_Nr, Swiss-Prot, Pfam, COG, GO, and KEGG databases, respectively, (Supplementary Table S4).
Quantification and Differentially Expressed Analysis of Unigenes
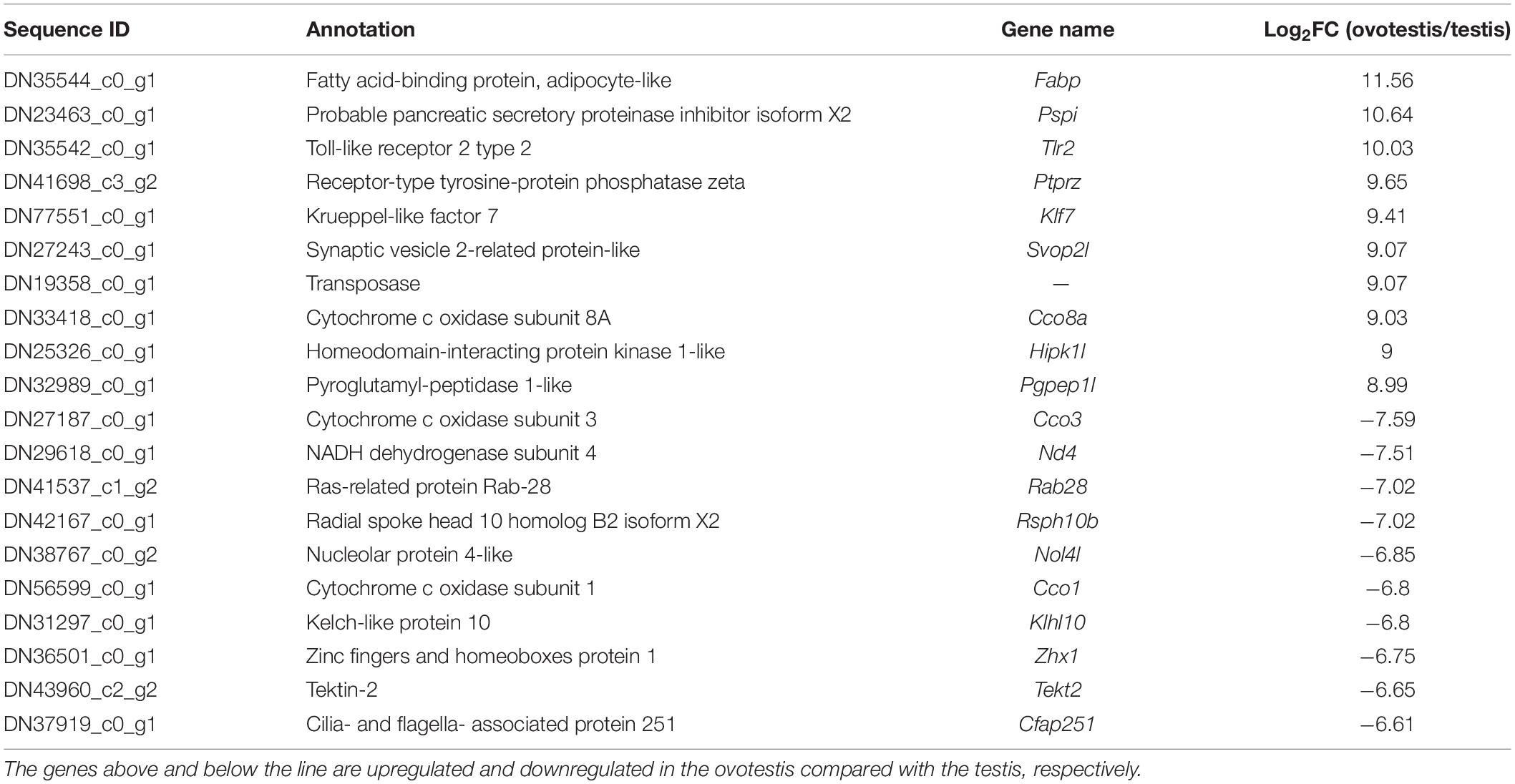
Subsequently, comparative analyses were performed to obtain evidence-based insights into sex reversal. A total of 22,591 DEGs were obtained in the testis, ovotestis, and ovary. Compared to the testis, there were 8453 upregulated DEGs and 7673 downregulated DEGs in the ovotestis and 7808 upregulated DEGs and 9752 downregulated DEGs in the ovary. Compared to the ovotestis, there were 2752 upregulated DEGs and 4131 downregulated DEGs in the ovary (Figures 3A–C). Moreover, there were 313 continuously upregulated DEGs and 822 downregulated DEGs during sex reversal process from testis over ovotestis to ovary (Figures 3D,E). Furthermore, the top 10 up-, or downregulated genes were screened between the ovotestis and testis (Table 1). Several of them were reported to be associated with spermatogenesis. For example, kelch-like 10 (klhl10) is required for the development of elongated spermatids, and epididymal spermatozoa (Yan et al., 2004). Tektin2 is expressed in spermatozoa and might be associated with sperm motility (Xiong et al., 2018). It has been reported that radial spoke head protein (RSPH) family contribute to sperm motility in human (Jeanson et al., 2015). Therefore, rsph10b might also be associated with sperm development in yellowfin seabream. In addition, it has been reported that mutation of cfap251 leads to immotile sperm and infertility in men (Auguste et al., 2018). Our present results reveal that klhl10, rsph10b, tektin2, and cfap251 were highly expressed in testis. In addition, the top 10 up- or downregulated genes between the ovotestis and testis, the ovary and ovotestis are, respectively, listed in Supplementary Tables S5, S6.
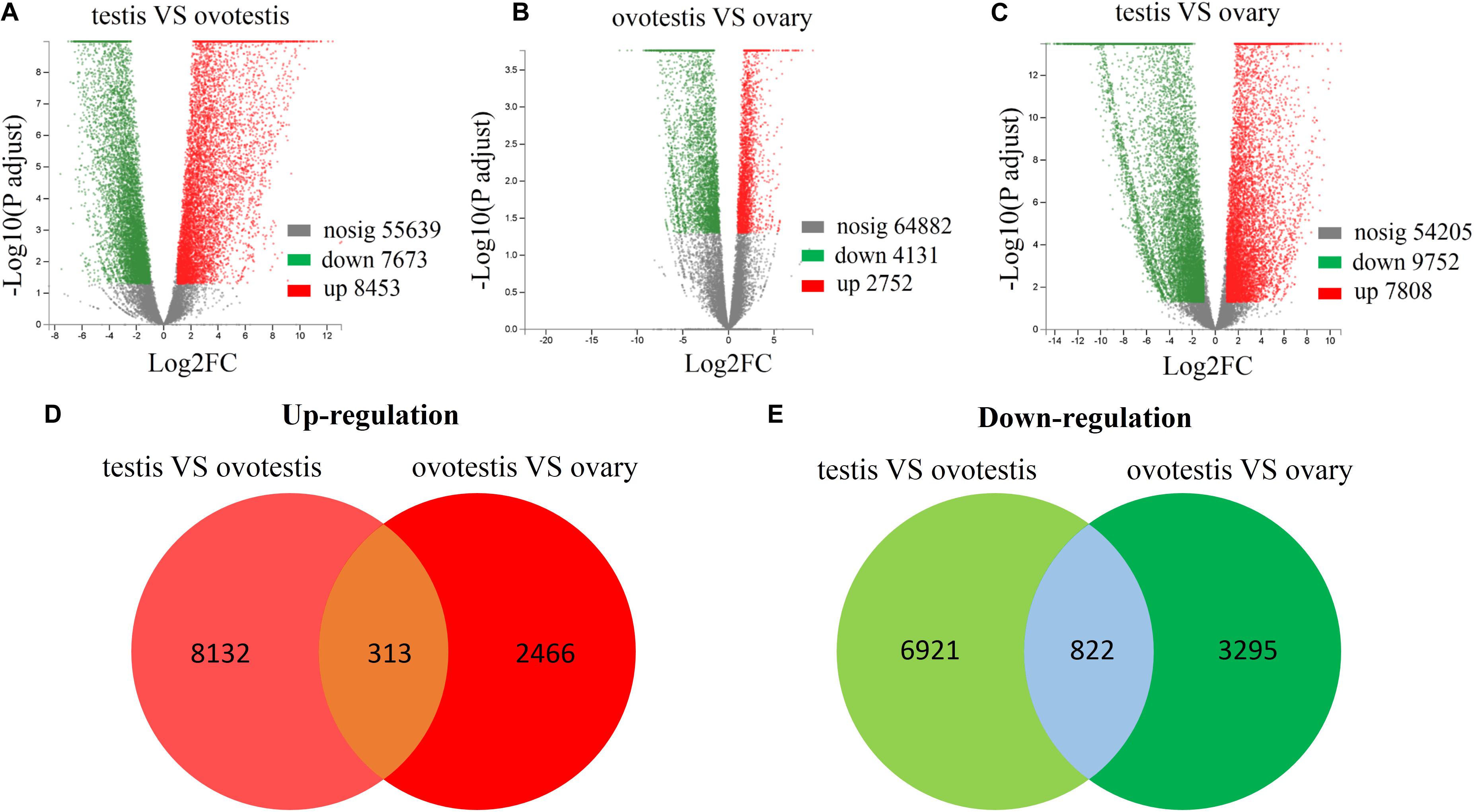
Figure 3. Differential expression analysis of unigenes. Volcano plot of differentially expressed unigenes between testis and ovotestis (A), ovotestis and ovary (B), testis and ovary (C). The up-regulated, down-regulated DEGs were respectively shown in red and green dots, whereas not differentially expressed genes were indicated in gray dots. The venn diagrams of up-regulated (D) or down-regulated (E) DEGs between testis vs ovotestis and ovotestis vs ovary.
Analysis of the DEGs Associated With GO and KEGG Pathways of Sexual Differentiation and Gonadal Development
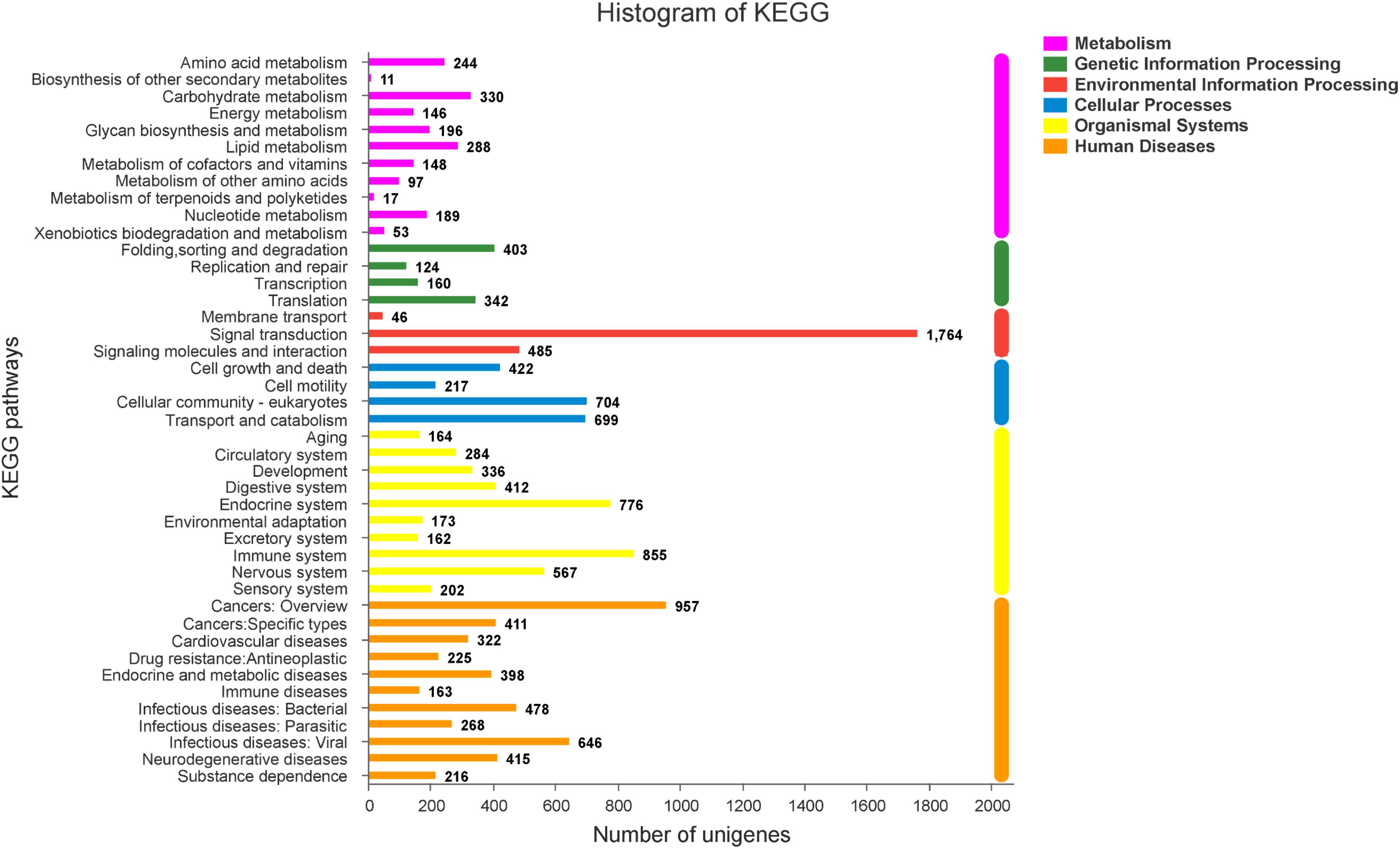
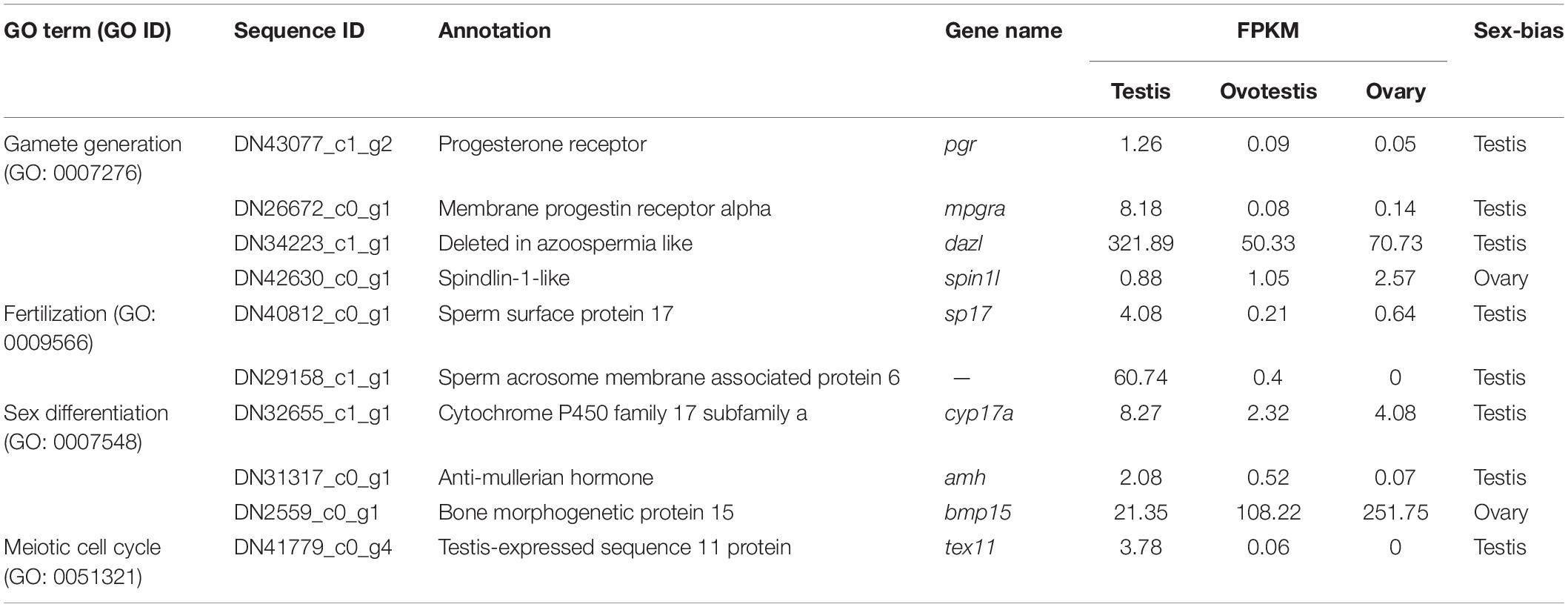
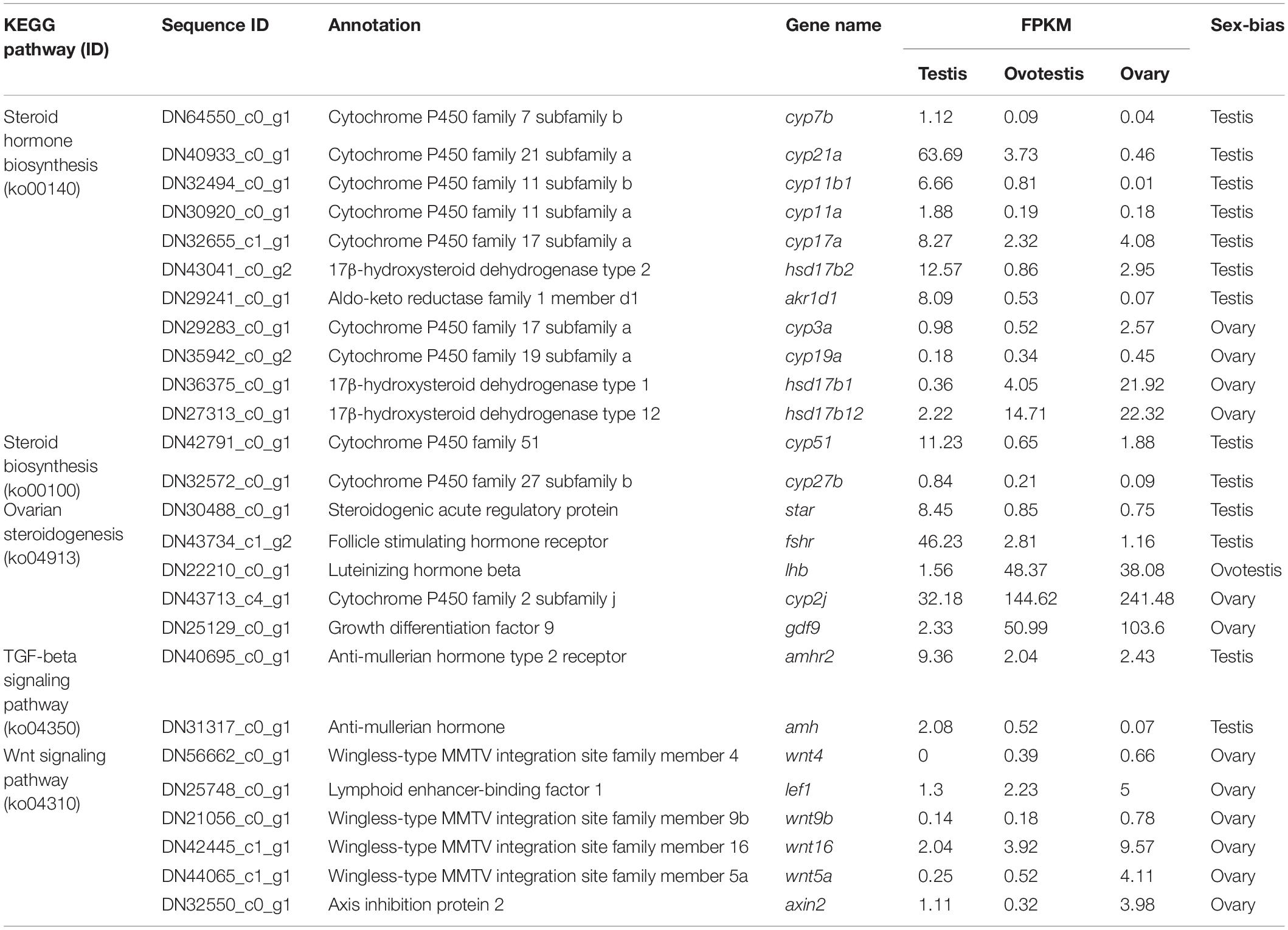
To explore genes associated with sexual differentiation and gonadal development, sex-related GO annotation, and KEGG pathway analysis were searched for all DEGs. The results showed that 4563 and 5797 DEGs were matched to 50 GO and 318 KEGG subcategories, respectively, (Figures 4, 5). Many DEGs were found to be involved with sex-related GO terms, such as gamete generation, fertilization, sex differentiation, and meiotic cell cycle (Table 2). The expression of spermatogenesis-related DEGs were significantly decreased in the ovotestis and ovary, such as deleted in azoospermia-like (dazl), sperm surface protein (sp17), cytochrome P450 family 17 subfamily a (cyp17a), anti-mullerian hormone (amh), and testis-expressed sequence 11 protein (tex11), whereas the expression of oogenesis-related genes, including spindling-1 like (spin1l), and bone morphogenetic protein 15 (bmp15) were significantly upregulated during sex reversal. The most DEG enriched sex-related KEGG pathways were associated with steroid production, including the steroid hormone biosynthesis (ko00140), steroid biosynthesis (ko00100), and ovarian steroidogenesis (ko04913) pathways (Table 3). It is known that sex determination and maintenance are dependent steroid hormones (Senthilkumaran, 2015; Gennotte et al., 2017), the production of which is regulated by cytochrome P450 (cyp), steroid sulfatase, and hydroxysteroid dehydrogenases (hsd; Uno et al., 2012; Hilborn et al., 2017). The expression of seven cyp members (cyp7b, cyp21a, cyp11b1, cyp11a, cyp17a, cyp51, and cyp27b) and hsd17b2 were significantly downregulated during sex reversal, whereas the expression of cyp3a, cyp2j, cyp19a, hsd17b1, and hsd17b12 were significantly upregulated. The wnt signaling pathway is required for early ovary development and folliculogenesis in mammal (Hernandez Gifford, 2015). The results indicate that the expression of wnt signaling components, such as wnt4, lef1, wnt9b, wnt16, wnt5a, and axin2, were significantly increased during testis-ovotestis-ovary sex reversal. In addition, two members of TGF-β signaling, amh, and its receptor amhr, were preferentially expressed in the testis.
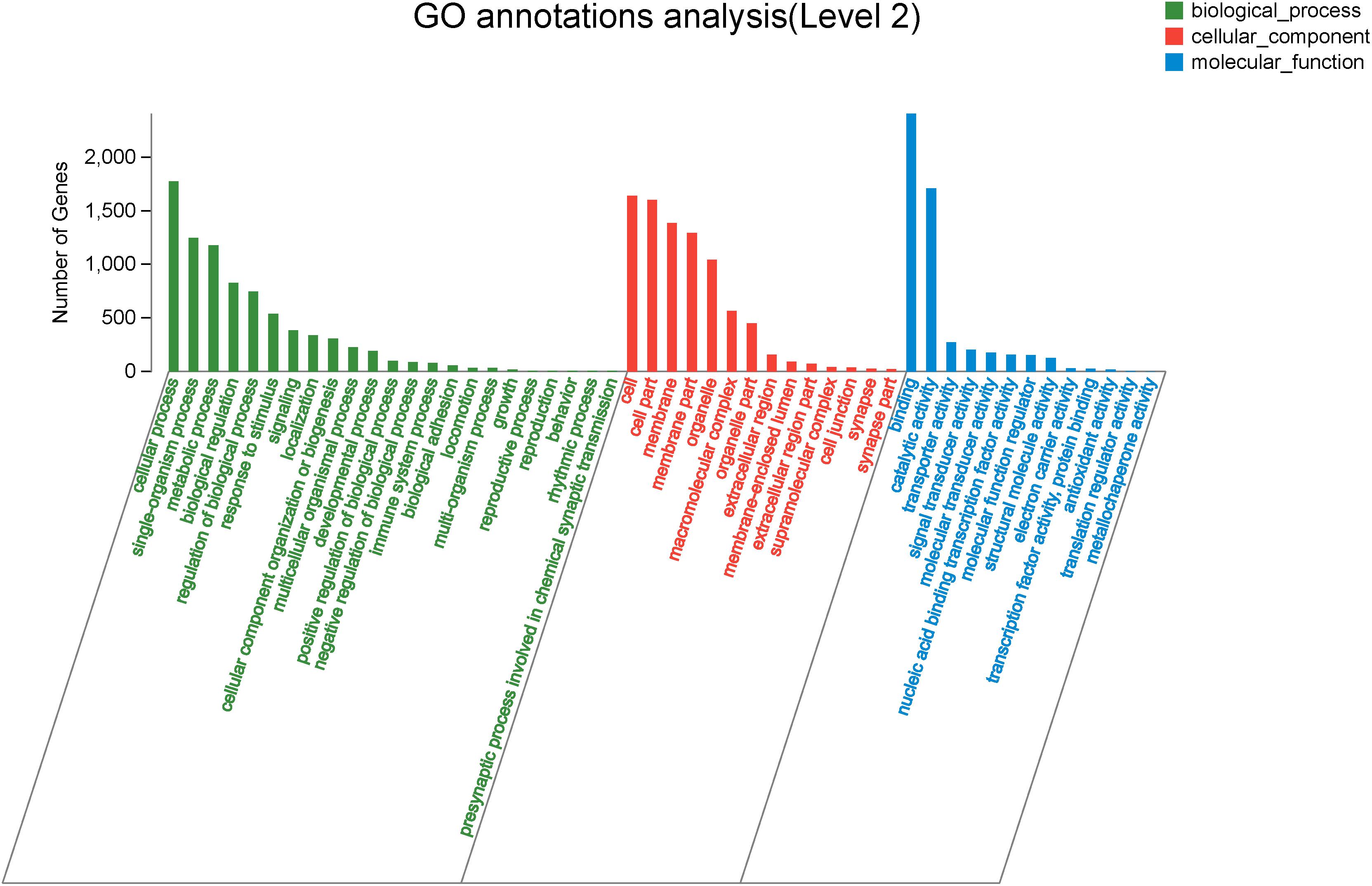
Figure 4. The Gene Ontology annotation of all DEGs. The X-axis represents the 50 subcategories. The Y-axis indicates the number of DEGs in a specific function cluster.
Analysis of DEGs Associated With Sex Determination and Sex Differentiation
Furthermore, genes associated with SD and gametogenesis were comparatively analyzed (Table 4). The expression of testis-determining genes were significantly decreased during sex reversal, such as SRY-box containing protein 9a (sox9a), sox9b, sox10, doublesex, and mab-3 related transcription factor 1 (dmrt1), dmrt3, and dmrt4. Meanwhile, the spermatogenesis-associated markers, piwi-like RNA-mediated gene silencing 1, 2 (piwil1, 2); synaptonemal complex protein 1, 2, 3, (sycp1-3); and outer dense fiber protein 2, 3 (odf2, 3), were also downregulated during the sex reversal. Whereas the expression of known ovarian differentiation-related genes were increased during sex reversal, for example, forkhead transcription factor L2 (foxl2), sox3, and dax1. Two oocyte markers, factor in the germline alpha (figla) and zona pellucida glycoprotein 3 (zp3), were dramatically upregulated in ovotestis and ovary. Overall, we identified 55 sex-related candidate genes, including 32 testis-biased genes, 20 ovary-biased genes, and 3 ovotestis-biased genes through these three strategies.
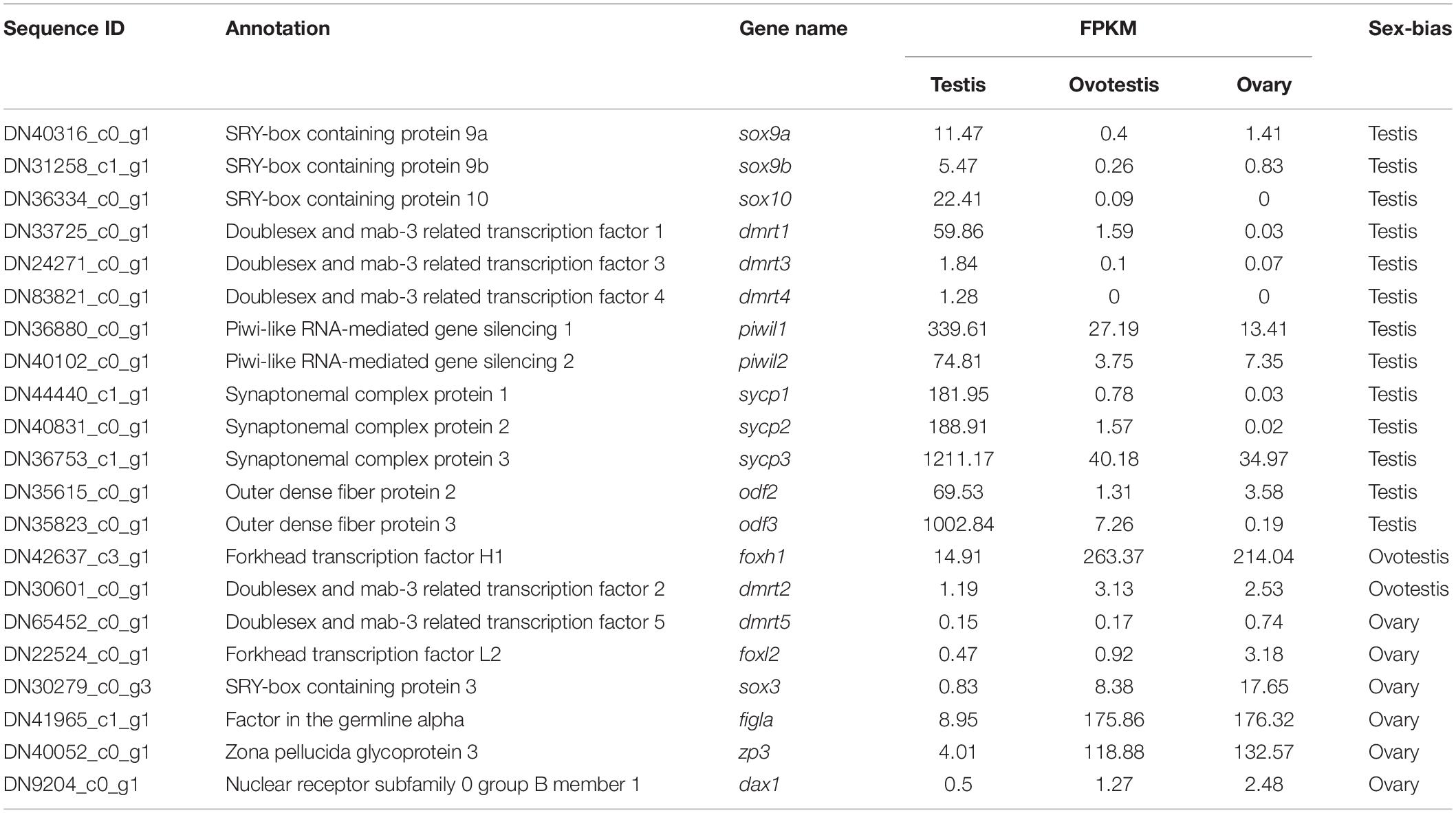
Table 4. Screening of sex determination and sex differentiation associated genes in yellowfin seabream.
Validation of Gene Expression by RT-qPCR
The reliability of the transcriptome data was validated by RT-qPCR analysis of 12 represented sex-related candidate genes. Consistent with our transcriptome data, qPCR analyses indicated that the expression of testicular development–related genes cyp11a, cyp17a, dmrt1, amh, sox9a, and piwil1 were high in testis, gradually decreased in ovotestis, but almost undetectable in mature ovary. Whereas, the expression of ovarian development–related genes cyp19a, hsd17b1, figla, zp3, foxl2, and sox3 were significantly upregulated in ovotestis and mature ovary in comparison with that in mature testis (Figure 6).
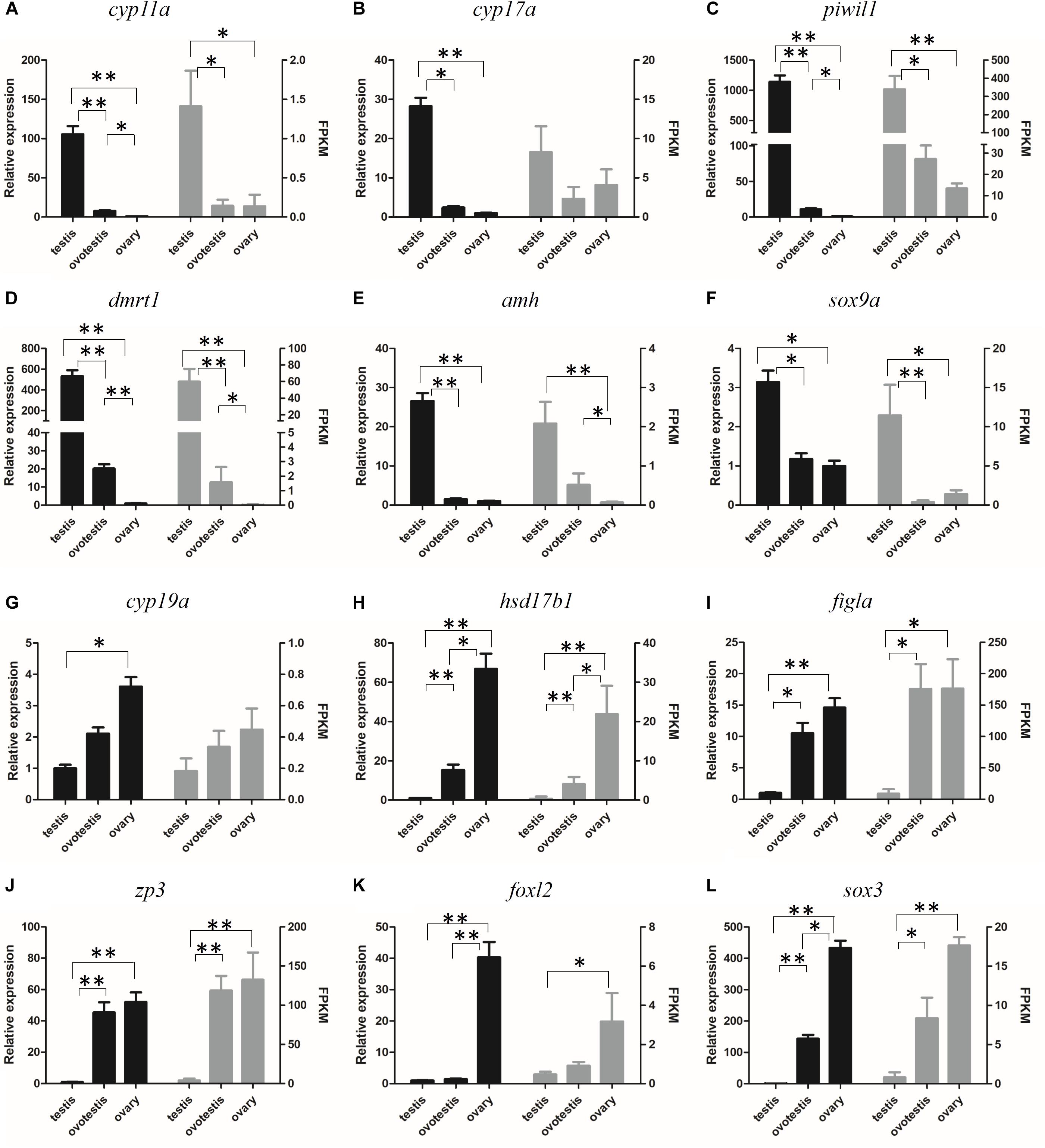
Figure 6. Verification of the expression patterns both in qRT-PCR (left) and RNA-seq (right). (A) cyp11a, (B) cyp17a, (C) piwil1, (D) dmrt1, (E) amh, (F) sox9a, (G) cyp19a, (H) hsd17b1, (I) figla, (J) zp3, (K) foxl2, (L) sox3. Student t-test, ∗p < 0.05 and ∗∗P < 0.01.
Dmrt1 and Foxl2 Are Two Potential Sex Determining Genes in Yellowfin Seabream
It has been demonstrated that dmrt1 and foxl2 are two important SD genes in vertebrates (Huang et al., 2017). Our transcriptome and RT-qPCR validation demonstrated the sexually dimorphic expression of dmrt1 and foxl2, suggesting that they might be associated with sex differentiation in yellowfin seabream. Subsequently, the tissue distributions of dmrt1 and foxl2 transcripts were analyzed in various tissues, including heart, liver, spleen, kidney, pituitary, brain, testis, and ovary. As shown in Figure 7, dmrt1 was exclusively expressed in the testis, indicating that it might play an important role in testis development in yellowfin seabream. The expression of foxl2 was high in pituitary and ovary, moderate in the brain, but low in the testis, implying that foxl2 might regulate ovary development through the brain-pituitary-ovary axis in yellowfin seabream.
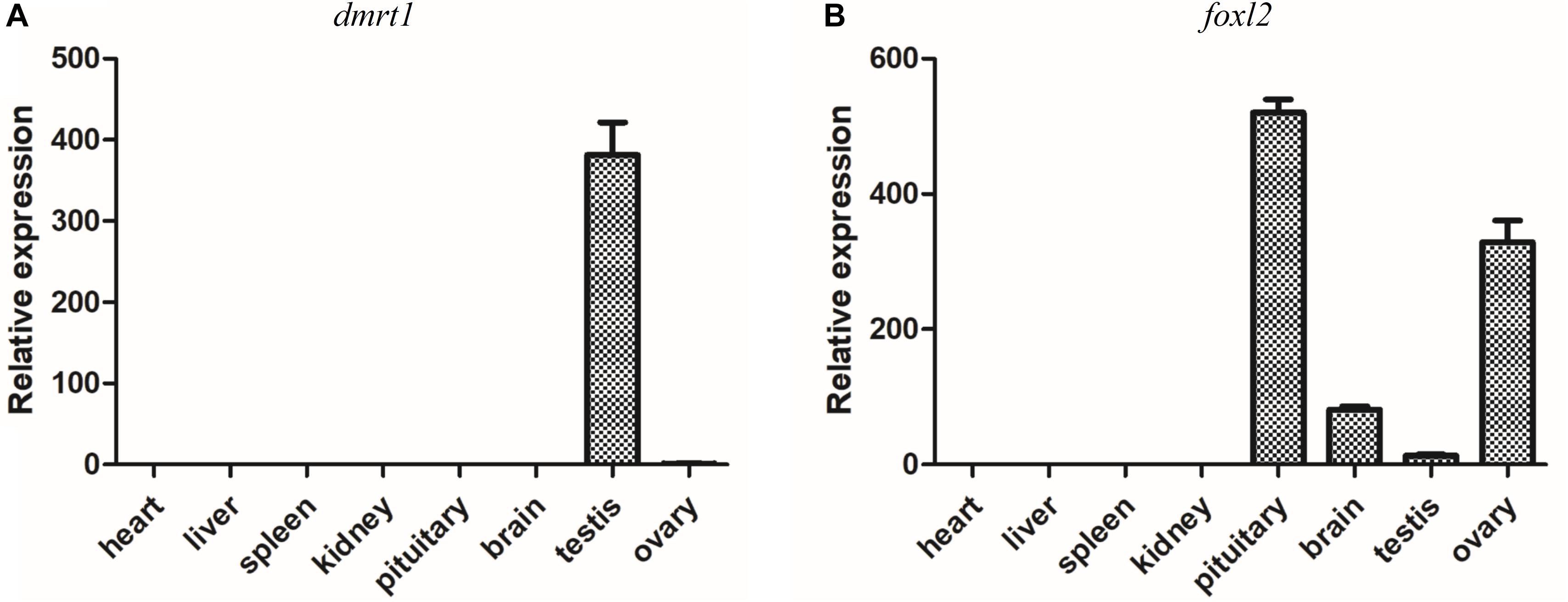
Figure 7. RT-qPCR detection of dmrt1 (A) and foxl2 (B) in different tissues. The relative expression is normalized by β-actin.
Discussion
In this study, we performed comprehensive transcriptomic analyses of the testis, ovotestis, and ovary in yellowfin seabream. By function analysis, we identified 55 key sex-related genes associated with steroidogenic hormones, wnt signaling pathway, germ cell development, and sex differentiation. Moreover, the testis-specific expression of dmrt1 and brain-pituitary-ovary axis expression of foxl2 indicates that they might play important roles in sexual differentiation in yellowfin seabream. Our results provide primary genetic information for gonadal development during sex reversal in yellowfin seabream. The identification of sex-related genes was the foundation for studying the sex differentiation and sex reversal in this species.
Transcriptomic Overview of Different Gonad Developmental Stages in Yellowfin Seabream
Zhang et al. (2019) report de novo transcriptome data of the gonads in black seabream, a fish species that has a close phylogenetic relationship with yellowfin seabream. The N50 length (2194 bp) and average length (1062 bp) are similar in these two species, whereas the number of assembled unigenes (71,765) in yellowfin seabream is less than that (109,504) in black seabream, suggesting that their genetic information is different. Meanwhile, many unigenes were not annotated in the public databases, suggesting that there might be more novel transcripts in yellowfin seabream. Interestingly, two of the top 10 DEGs (rsph and klhl10) are found in both black seabream, and yellowfin seabream, suggesting they might play important roles in seabream sex reversal.
The Expression Change of Sex-Related Signaling Pathway During Sex Reversal
Sex reversal is tightly regulated by steroid hormones, such as estrogen and testosterone (Gennotte et al., 2017; Otani et al., 2017; Han et al., 2018). Steroid hormones are synthesized through a series of isomerization and hydroxylation reaction catalyzed by many enzymes, such as cyp, and hydroxysteroid dehydrogenases families (Hilborn et al., 2017). It has been demonstrated that some CYP components play important roles in sexual differentiation (Uno et al., 2012). Cyp11a plays an important role in converting cholesterol to pregnenolone, which is the first step for the synthesis of steroid hormones. The sexual dimorphic expression of cyp11a has been characterized in several fish species (Nishiyama et al., 2015; Liang et al., 2018). Cyp17 has both 17a-hydroxylase and 17, 20-lyase activities, which is required for producing androgens and estrogens. The sexual dimorphism of cyp17a has been observed in several fish species (Chen et al., 2010; Xiong et al., 2015). In zebrafish, cyp17a1 mutants develop to all fertile males due to the decreased level of estrogen, demonstrating that cyp17a1 is essential for estrogen production and ovarian maintenance (Zhai et al., 2018). Cyp19a1a plays a pivotal role in ovary development by converting the testosterone into estradiol (Guiguen et al., 2010; Fowler et al., 2011; Lambeth et al., 2013). Here, the downregulated expression of cyp11a and cyp17a and upregulated expression of cyp19a1a was found during testis-ovotestis-ovary sex reversal, indicating that they might be involved in sex differentiation in yellowfin seabream. hsd17b1 and hsd17b12 were essential for ovarian development by converting inactive estrone to active estradiol, whereas hsd17b2 regulates testicular development by modification of testosterone (Hilborn et al., 2017, Hiltunen et al., 2019). Consistently, the testis-biased expression of hsd17b2, and ovary-biased expression of hsd17b1 and hsd17b12 were revealed in yellowfin seabream, indicating that they also might be involved in sexual differentiation as in the above studies.
Wnt4, one of the best characterized components in wnt/β-catenin signaling pathway, is required for initiating ovarian differentiation in mammal (Chassot et al., 2014; Roly et al., 2018). Recently, the critical role of wnt4 in folliculogenesis has been characterized in teleost, such as medaka (Li et al., 2012), spotted scat (Chen et al., 2016), orange-spotted grouper (Chen et al., 2015), and black seabream (Wu and Chang, 2009). During yellowfin seabream testis-to-ovary sex reversal, the expression of the wnt signaling pathway, including wnt4, wnt5a, wnt9b, wnt16, axin2, and lef1, were significantly upregulated, indicating that the wnt signaling played important roles in ovarian development in yellowfin seabream.
Sex-Biased Expression of Germ Cell Markers in Yellowfin Seabream
The germ cell–specific piwi plays a crucial role in mammal spermatogenesis, male germ cell proliferation, differentiation, and meiosis control (Deng and Lin, 2002; Carmell et al., 2007; Houwing et al., 2007, 2008). Odfs are the components of sperm cytoskeletal structure, which are required for male fertility in mice and zebrafish (Hsu et al., 2010; Yang et al., 2014; Lehti and Sironen, 2017). Zp3 encodes the zona pellucida glycoprotein, an important component that is required for oogenesis and fertilization in vertebrates (Mold et al., 2009; Sun et al., 2010; Chuang-Ju et al., 2011). It has been reported that figla plays an important role in folliculogenesis and ovarian differentiation (Nishimura et al., 2018; Qin et al., 2018). During yellowfin seabream sex reversal, the expression of piwil1, piwil2, odf1, and odf2 were significantly decreased; meanwhile, the expression of figla, and zp3 were dramatically increased, suggesting that they should contribute in sex differentiation in yellowfin seabream.
Candidates of Sex Determination-Related and Sex Differentiation-Related Genes in Yellowfin Seabream
The DM-domain-containing genes might be the most important SD genes in animals (Matson and Zarkower, 2012). dmrt1 plays a robust role in SD, male germ cell differentiation, and maintenance in mammal, bird, and fish species (Smith et al., 2009; Matson et al., 2011; Li et al., 2013; Lin et al., 2017; Zhang et al., 2016; Webster et al., 2017). dmrt2 was highly expressed in male germ cells in Chinese tongue sole (Zhu et al., 2019); dmrt2b and dmrt3 were dominantly expressed in the ovotestis of orange-spotted grouper (Lyu et al., 2019); dmrt2, dmrt3, dmrt4, and dmrt5 were significantly upregulated during ovary-to-testis sex reversal in the swamp eel (Sheng et al., 2014). In yellowfin seabream, the testis-specific expression of dmrt1, dmrt3, and dmrt4; ovotestis-biased expression of dmrt2; and ovary-biased expression of dmrt5 suggest that the dmrt family might play important roles in sex reversal (Table 4 and Figure 6D). Furthermore, the testis-specific tissue distribution of dmrt1 implies its potential role in testis differentiation (Figure 7A).
The sox super family play multiple roles in many biological processes, including gonad development (Kamachi and Kondoh, 2013). The germ cell–specific sox3 plays a significant role in gametogenesis, sex determination, and gonad differentiation in vertebrates (Takehana et al., 2014); sox9 cooperates with sox8 to protect the adult testis from male-to-female genetic reprogramming and complete degeneration (Vidal et al., 2001; Barrionuevo et al., 2016); sox10 is an important factor in human disorders of sex development (Polanco et al., 2010); and sox17 is critical for PGC formation (Irie et al., 2015). It has been implied that the expression of sox3 was dynamic along with the process of sex reversal in protogynous hermaphroditic grouper (Yao et al., 2007) and protandrous hermaphroditic black seabream (Shin et al., 2009). sox9a, sox9b, and sox10 were highly expressed in the testis, whereas sox3 was mainly expressed in the ovary, suggesting that sox genes play complicated roles in sex differentiation in yellowfin seabream.
foxl2, the hallmark of granulosa cell, is one of earliest sexually dimorphic genes during ovarian development (Bertho et al., 2016; Okada et al., 2017), which plays a pivotal role in estradiol production through the endocrine system along brain-pituitary-gonad axis (Sridevi et al., 2012; Georges et al., 2014; Liu et al., 2019). Loss of foxl2 results in dysregulation of estradiol and somatic cell reprogramming (Uhlenhaut et al., 2009) and even ovary-to-testis sex reversal (Li et al., 2013; Yang et al., 2017). Therefore, the brain-pituitary-ovary axis expression of foxl2 indicated that foxl2 might regulate ovarian differentiation in yellowfin seabream.
The Mechanisms of Natural Sex Reversal in Fish Species
Natural sexual reversal is found in many fish species (Avise and Mank, 2009). This interesting phenomena and its potential underlying molecular mechanism have been characterized in some fish species. In orange-spotted grouper, the female-to-male sex reversal could be induced by overexpression of amh, which results in upregulation of testis development–associated genes, especially for those genes closely related with the synthesis of 11-ketotestosterone. Meanwhile, the decreased level of serum 17β-estradiol and the repression of ovarian development and maintenance-related genes were also found in amh overexpressed orange-spotted grouper (Han et al., 2018). In rice-field eel, there is a high expression level of foxl2 and cyp19a1a in the ovary prior to sex reversal. However, at the beginning of the ovary-to-testis sex reversal, a sharp decrease of foxl2 and cyp19a1a in the ovotestis (Liu et al., 2009; Hu et al., 2014). In black seabream, the sex switch is controlled by a series of male-related genes (dmrt1, piwi1, piwi2, sox9, sox30, and amh) and female-related genes (jnk1, vasa, wnt4, figla, and foxl2), most of which were involved in the signaling pathway of sex steroid hormones. The sex reversal is initiated by the decrease of male-related genes and the increase of female-related genes (Zhang et al., 2019). In yellowfin seabream, our results indicate that the sex reversal might be closely associated with 55 sex-related genes, most of which were involved in the signaling network of sex steroid hormones, such as estradiol and testosterone. Therefore, the sex reversal of yellowfin seabream should be controlled by the change of sex hormone and the decrease of androgens but elevated levels of estrogens. On the basis of the present work and previous studies in other fish species, we could conclude that the sex switch is determined and maintained by the balance of sex hormones. Once the original balance (estrogens in protogynous fish and androgens in protandrous fish) is broken out, the sex reversal might be induced by the occurrence of the upregulation of opposite hormones (androgens in protogynous fish and estrogens in protandrous fish).
Conclusion
To our best knowledge, this is the first report of large-scale RNA sequencing of testis, ovotestis, and ovary in yellowfin seabream. Our present work provides an important genetic resource and a global view of DEGs and signaling pathway change during sex reversal of yellowfin seabream. A total of 55 sex differentiation–associated candidates has been obtained from the transcriptome data, including steroid biosynthesis–related genes (Cyp family and Hsd family), germ cell markers (piwi, odf, figla, and zp3), and the known SD factors, such as the DM domain containing family and Sox family. In addition, the testis-specific expression of dmrt1 and the brain-pituitary-ovary axis expression of foxl2 were characterized, suggesting that they might play important roles in sex differentiation in yellowfin seabream, and the specific function of these two genes deserve to be further investigated.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of Sun Yat-sen University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
JL and SL conceived and designed the experiments. SL and WF performed the experiments. GL, PH, and DG performed bioinformatics analysis. JH and JX contributed to discussion of the final results. SL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Postdoctoral Science Foundation of China (2018M631016), Fundamental Research Funds for the Central Universities (19lgpy94), Guangzhou Science and Technology Project (201803020017), and National Natural Science Foundation of China (31902427).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00709/full#supplementary-material
Footnotes
- ^ http://github.com/jstjohn/SeqPrep
- ^ http://github.com/najoshi/sickle
- ^ http://github.com/trinityrnaseq/trinityrnaseq
- ^ http://hibberdlab.com/transrate
- ^ http://weizhongli-lab.org/cd-hit/
- ^ http://busco.ezlab.org
- ^ http://deweylab.github.io/RSEM/
- ^ http://primer3.ut.ee
References
Auguste, Y., Delague, V., Desvignes, J. P., Longepied, G., Gnisci, A., Besnier, P., et al. (2018). Loss of calmodulin- and radial-spoke-associated complex protein CFAP251 leads to immotile spermatozoa lacking mitochondria and infertility in men. Am. J. Hum. Genet. 103, 413–420. doi: 10.1016/j.ajhg.2018.07.013
Avise, J. C., and Mank, J. E. (2009). Evolutionary perspectives on hermaphroditism in fishes. Sex. Dev. 3, 152–163. doi: 10.1159/000223079
Barrionuevo, F. J., Hurtado, A., Kim, G. J., Real, F. M., and Bakkali, M. (2016). Sox9 and Sox8 protect the adult testis from male-to-female genetic reprogramming and complete degeneration. eLife 5:e15635. doi: 10.7554/eLife.15635
Bertho, S., Pasquier, J., Pan, Q., Le Trionnaire, G., Bobe, J., Postlethwait, J. H., et al. (2016). Foxl2 and its relatives are evolutionary conserved players in gonadal sex differentiation. Sex. Dev. 10, 111–129. doi: 10.1159/000447611
Carmell, M. A., Girard, A., Van De Kant, H. J., Bourc’his, D., Bestor, T. H., De Rooij, D. G., et al. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514. doi: 10.1016/j.devcel.2007.03.001
Chassot, A. A., Gillot, I., and Chaboissier, M. C. (2014). R-spondin1, WNT4, and the CTNNB1 signaling pathway: strict control over ovarian differentiation. Reproduction 148, R97–R110. doi: 10.1530/rep-14-0177
Chen, C. F., Wen, H. S., Wang, Z. P., He, F., Zhang, J. R., Chen, X. Y., et al. (2010). Cloning and expression of P450c17-I (17alpha-hydroxylase/17,20-lyase) in brain and ovary during gonad development in Cynoglossus semilaevis. Fish Physiol. Biochem. 36, 1001–1012. doi: 10.1007/s10695-009-9378-7
Chen, H., Li, S., Xiao, L., Zhang, Y., Li, G., Liu, X., et al. (2015). Wnt4 in protogynous hermaphroditic orange-spotted grouper (Epinephelus coioides): identification and expression. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 183, 67–74. doi: 10.1016/j.cbpb.2015.01.002
Chen, J., Yinglei, L., Zhang, J., Liu, H., and Li, Y. (2016). Identification and expression analysis of two Wnt4 genes in the spotted scat (Scatophagus argus). Electron. J. Biotechnol. 19, 20–27.
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., et al. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46, 253–260. doi: 10.1038/ng.2890
Chuang-Ju, L., Qi-Wei, W., Xi-Hua, C., Li, Z., Hong, C., Fang, G., et al. (2011). Molecular characterization and expression pattern of three zona pellucida 3 genes in the Chinese sturgeon, Acipenser sinensis. Fish Physiol. Biochem. 37, 471–484. doi: 10.1007/s10695-010-9448-x
Dan, C., Lin, Q., Gong, G., Yang, T., Xiong, S., Xiong, Y., et al. (2018). A novel PDZ domain-containing gene is essential for male sex differentiation and maintenance in yellow catfish (Pelteobagrus fulvidraco). Sci. Bull. 63, 1420–1430.
Deng, W., and Lin, H. (2002). miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830.
Fowler, P. A., Anderson, R. A., Saunders, P. T., Kinnell, H., Mason, J. I., Evans, D. B., et al. (2011). Development of steroid signaling pathways during primordial follicle formation in the human fetal ovary. J. Clin. Endocrinol. Metab. 96, 1754–1762. doi: 10.1210/jc.2010-2618
Gennotte, V., Akonkwa, B., Melard, C., Denoel, M., Cornil, C. A., and Rougeot, C. (2017). Do sex reversal procedures differentially affect agonistic behaviors and sex steroid levels depending on the sexual genotype in Nile tilapia? J. Exp. Zool. A Ecol. Integr. Physiol. 327, 153–162. doi: 10.1002/jez.2080
Georges, A., L’hote, D., Todeschini, A. L., Auguste, A., Legois, B., Zider, A., et al. (2014). The transcription factor FOXL2 mobilizes estrogen signaling to maintain the identity of ovarian granulosa cells. eLife 3:e04207.
Guiguen, Y., Fostier, A., Piferrer, F., and Chang, C. F. (2010). Ovarian aromatase and estrogens: a pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 165, 352–366. doi: 10.1016/j.ygcen.2009.03.002
Han, Y., Peng, C., Wang, L., Guo, J., Lu, M., Chen, J., et al. (2018). Female-to-male sex reversal in orange-spotted grouper (Epinephelus coioides) caused by overexpressing of Amh in vivo. Biol. Reprod. 99, 1205–1215. doi: 10.1093/biolre/ioy157
Hattori, R. S., Murai, Y., Oura, M., Masuda, S., Majhi, S. K., Sakamoto, T., et al. (2012). A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. U.S.A. 109, 2955–2959.
Hernandez Gifford, J. A. (2015). The role of WNT signaling in adult ovarian folliculogenesis. Reproduction 150, R137–R148. doi: 10.1530/rep-14-0685
Hilborn, E., Stal, O., and Jansson, A. (2017). Estrogen and androgen-converting enzymes 17beta-hydroxysteroid dehydrogenase and their involvement in cancer: with a special focus on 17beta-hydroxysteroid dehydrogenase type 1, 2, and breast cancer. Oncotarget 8, 30552–30562. doi: 10.18632/oncotarget.15547
Hiltunen, J. K., Kastaniotis, A. J., Autio, K. J., Jiang, G., Chen, Z., and Glumoff, T. (2019). 17B-hydroxysteroid dehydrogenases as acyl thioester metabolizing enzymes. Mol. Cell. Endocrinol. 489, 107–118. doi: 10.1016/j.mce.2018.11.012
Houwing, S., Berezikov, E., and Ketting, R. F. (2008). Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27, 2702–2711. doi: 10.1038/emboj.2008.204
Houwing, S., Kamminga, L. M., Berezikov, E., Cronembold, D., Girard, A., Van Den Elst, H., et al. (2007). A role for piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82.
Hsu, C. C., Hou, M. F., Hong, J. R., Wu, J. L., and Her, G. M. (2010). Inducible male infertility by targeted cell ablation in zebrafish testis. Mar. Biotechnol. 12, 466–478. doi: 10.1007/s10126-009-9248-4
Hu, Q., Guo, W., Gao, Y., Tang, R., and Li, D. (2014). Molecular cloning and analysis of gonadal expression of Foxl2 in the rice-field eel Monopterus albus. Sci. Rep. 4:6884. doi: 10.1038/srep06884
Huang, S., Ye, L., and Chen, H. (2017). Sex determination and maintenance: the role of DMRT1 and FOXL2. Asian J. Androl. 19, 619–624. doi: 10.4103/1008-682x.194420
Irie, N., Weinberger, L., Tang, W. W., Kobayashi, T., Viukov, S., Manor, Y. S., et al. (2015). SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253–268. doi: 10.1016/j.cell.2014.12.013
Jeanson, L., Copin, B., Papon, J. F., Dastot-Le Moal, F., Duquesnoy, P., Montantin, G., et al. (2015). RSPH3 mutations cause primary ciliary dyskinesia with central-complex defects and a near absence of radial spokes. Am. J. Hum. Genet. 97, 153–162. doi: 10.1016/j.ajhg.2015.05.004
Kamachi, Y., and Kondoh, H. (2013). Sox proteins: regulators of cell fate specification and differentiation. Development 140, 4129–4144. doi: 10.1242/dev.091793
Kamiya, T., Kai, W., Tasumi, S., Oka, A., Matsunaga, T., Mizuno, N., et al. (2012). A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8:e1002798. doi: 10.1371/journal.pgen.1002798
Koopman, P., Gubbay, J., Vivian, N., Goodfellow, P., and Lovell-Badge, R. (1991). Male development of chromosomally female mice transgenic for Sry. Nature 351, 117–121. doi: 10.1038/351117a0
Lambeth, L. S., Cummins, D. M., Doran, T. J., Sinclair, A. H., and Smith, C. A. (2013). Overexpression of aromatase alone is sufficient for ovarian development in genetically male chicken embryos. PLoS One 8:e68362. doi: 10.1371/journal.pone.0068362
Lehti, M. S., and Sironen, A. (2017). Formation and function of sperm tail structures in association with sperm motility defects. Biol. Reprod. 97, 522–536. doi: 10.1093/biolre/iox096
Li, J.-Z., Liu, Q., Wang, D.-S., Zhou, L.-Y., Sakai, F., and Nagahama, Y. (2012). Molecular cloning and identification of two Wnt4 genes from the medaka (Oryzias latipes). Acta Hydrobiol. Sin. 36, 983–986.
Li, M. H., Yang, H. H., Li, M. R., Sun, Y. L., Jiang, X. L., Xie, Q. P., et al. (2013). Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology 154, 4814–4825. doi: 10.1210/en.2013-1451
Li, X. Y., and Gui, J. F. (2018). Diverse and variable sex determination mechanisms in vertebrates. Sci. China Life Sci. 61, 1503–1514. doi: 10.1007/s11427-018-9415-7
Li, X.-Y., Liu, X.-L., Ding, M., Li, Z., Zhou, L., Zhang, X.-J., et al. (2017). A novel male-specific SET domain-containing gene setdm identified from extra microchromosomes of gibel carp males. Sci. Bull. 62, 528–536.
Li, X. Y., Zhang, Q. Y., Zhang, J., Zhou, L., Li, Z., Zhang, X. J., et al. (2016). Extra microchromosomes play male determination role in polyploid gibel carp. Genetics 203, 1415–1424. doi: 10.1534/genetics.115.185843
Liang, D., Fan, Z., Zou, Y., Tan, X., Wu, Z., Jiao, S., et al. (2018). Characteristics of Cyp11a during gonad differentiation of the olive flounder Paralichthys olivaceus. Int. J. Mol. Sci. 19:2641. doi: 10.3390/ijms19092641
Lin, G., Zheng, M., Gao, D., Li, S., Fang, W., Huang, J., et al. (2020). Hypoosmotic stress induced tissue-specific immune responses of yellowfin seabream (Acanthopagrus latus) revealed by transcriptomic analysis. Fish Shellfish Immunol. 99, 473–482. doi: 10.1016/j.fsi.2020.02.028
Lin, Q., Mei, J., Li, Z., Zhang, X., Zhou, L., and Gui, J. F. (2017). Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics 207, 1007–1022. doi: 10.1534/genetics.117.300274
Liu, C. (1944). Rudimentary hermaphroditism in the synbranchoid eel, Monopterus javanensis. Sinensia 15, 1–8.
Liu, J. F., Guiguen, Y., and Liu, S. J. (2009). Aromatase (P450arom) and 11beta-hydroxylase (P45011beta) genes are differentially expressed during the sex change process of the protogynous rice field eel, Monopterus albus. Fish Physiol. Biochem. 35, 511–518. doi: 10.1007/s10695-008-9255-9
Liu, W., Zhang, H., Xiang, Y., Jia, K., Luo, M., and Yi, M. (2019). A novel germline and somatic cell expression of two sexual differentiation genes, Dmrt1 and Foxl2 in marbled goby (Oxyeleotris marmorata). Aquaculture 516:734619.
Lyu, Q., Hu, J., Yang, X., Liu, X., Chen, Y., Xiao, L., et al. (2019). Expression profiles of dmrts and foxls during gonadal development and sex reversal induced by 17alpha-methyltestosterone in the orange-spotted grouper. Gen. Comp. Endocrinol. 274, 26–36. doi: 10.1016/j.ygcen.2018.12.014
Matson, C. K., Murphy, M. W., Sarver, A. L., Griswold, M. D., Bardwell, V. J., and Zarkower, D. (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101–104. doi: 10.1038/nature10239
Matson, C. K., and Zarkower, D. (2012). Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163–174. doi: 10.1038/nrg3161
Matsuda, M., Nagahama, Y., Shinomiya, A., Sato, T., Matsuda, C., Kobayashi, T., et al. (2002). DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417, 559–563. doi: 10.1038/nature751
Mei, J., and Gui, J. F. (2015). Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 58, 124–136. doi: 10.1007/s11427-014-4797-9
Mold, D. E., Dinitz, A. E., and Sambandan, D. R. (2009). Regulation of zebrafish zona pellucida gene activity in developing oocytes. Biol. Reprod. 81, 101–110. doi: 10.1095/biolreprod.108.071720
Myosho, T., Otake, H., Masuyama, H., Matsuda, M., Kuroki, Y., Fujiyama, A., et al. (2012). Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191, 163–170. doi: 10.1534/genetics.111.137497
Negintaji, A., Safahieh, A., Zolgharnein, H., and Matroodi, S. (2018). Short-term induction of vitellogenesis in the immature male yellowfin seabream (Acanthopagrus latus) exposed to bisphenol A and 17 beta-estradiol. Toxicol. Ind. Health 34, 119–127. doi: 10.1177/0748233717748099
Nishimura, T., Yamada, K., Fujimori, C., Kikuchi, M., Kawasaki, T., Siegfried, K. R., et al. (2018). Germ cells in the teleost fish medaka have an inherent feminizing effect. PLoS Genet. 14:e1007259. doi: 10.1371/journal.pgen.1007259
Nishiyama, M., Uchida, K., Abe, N., and Nozaki, M. (2015). Molecular cloning of cytochrome P450 side-chain cleavage and changes in its mRNA expression during gonadal development of brown hagfish, Paramyxine atami. Gen. Comp. Endocrinol. 212, 1–9. doi: 10.1016/j.ygcen.2015.01.014
Okada, H., Hagihara, S., Yamashita, K., Ijiri, S., and Adachi, S. (2017). Expression pattern of foxl2 and dmrt1 in gonad of Amur sturgeon Acipenser schrenckii in relation to sex differentiation. Aquaculture 479, 712–720.
Otani, A., Nakajima, T., Okumura, T., Fujii, S., and Tomooka, Y. (2017). Sex reversal and analyses of possible involvement of sex steroids in scallop gonadal development in newly established organ-culture systems. Zool. Sci. 34, 86–92. doi: 10.2108/zs160070
Papadaki, M., Mazzella, D., Santinelli, V., Fakriadis, I., Sigelaki, I., and Mylonas, C. C. (2018). Hermaphroditism and reproductive function of hatchery-produced sharpsnout seabream (Diplodus puntazzo) under attenuated annual thermal cycles. Aquaculture 482, 231–240.
Polanco, J. C., Wilhelm, D., Davidson, T. L., Knight, D., and Koopman, P. (2010). Sox10 gain-of-function causes XX sex reversal in mice: implications for human 22q-linked disorders of sex development. Hum. Mol. Genet. 19, 506–516.
Qin, M., Zhang, Z., Song, W., Wong, Q. W., Chen, W., Shirgaonkar, N., et al. (2018). Roles of Figla/figla in juvenile ovary development and follicle formation during zebrafish gonadogenesis. Endocrinology 159, 3699–3722. doi: 10.1210/en.2018-00648
Roly, Z. Y., Backhouse, B., Cutting, A., Tan, T. Y., and Sinclair, A. H. (2018). The cell biology and molecular genetics of Mullerian duct development. Wiley Interdiscip. Rev. Dev. Biol. 7:e310. doi: 10.1002/wdev.310
Schubert, C. (2015). Making females male with Dmrt1. Biol. Reprod. 92:134. doi: 10.1095/biolreprod.115.129601
Senthilkumaran, B. (2015). Pesticide- and sex steroid analogue-induced endocrine disruption differentially targets hypothalamo-hypophyseal-gonadal system during gametogenesis in teleosts - A review. Gen. Comp. Endocrinol. 219, 136–142. doi: 10.1016/j.ygcen.2015.01.010
Sheng, Y., Chen, B., Zhang, L., Luo, M., Cheng, H., and Zhou, R. (2014). Identification of Dmrt genes and their up-regulation during gonad transformation in the swamp eel (Monopterus albus). Mol. Biol. Rep. 41, 1237–1245. doi: 10.1007/s11033-013-2968-6
Shin, H. S., An, K. W., Park, M. S., Jeong, M. H., and Choi, C. Y. (2009). Quantitative mRNA expression of sox3 and DMRT1 during sex reversal, and expression profiles after GnRHa administration in black porgy, Acanthopagrus schlegeli. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 154, 150–156.
Smith, C. A., Roeszler, K. N., Ohnesorg, T., Cummins, D. M., Farlie, P. G., Doran, T. J., et al. (2009). The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267–271. doi: 10.1038/nature08298
Sridevi, P., Chaitanya, R. K., Dutta-Gupta, A., and Senthilkumaran, B. (2012). FTZ-F1 and FOXL2 up-regulate catfish brain aromatase gene transcription by specific binding to the promoter motifs. Biochim. Biophys. Acta 1819, 57–66. doi: 10.1016/j.bbagrm.2011.10.003
Sun, Y., Yu, H., Zhang, Q., Qi, J., Zhong, Q., Chen, Y., et al. (2010). Molecular characterization and expression pattern of two zona pellucida genes in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155, 316–321. doi: 10.1016/j.cbpb.2009.11.016
Takehana, Y., Matsuda, M., Myosho, T., Suster, M. L., Kawakami, K., Shin, I. T., et al. (2014). Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5:4157. doi: 10.1038/ncomms5157
Todd, E. V., Liu, H., Muncaster, S., and Gemmell, N. J. (2016). Bending genders: the biology of natural sex change in fish. Sex. Dev. 10, 223–241. doi: 10.1159/000449297
Tsakogiannis, A., Manousaki, T., Lagnel, J., Papanikolaou, N., Papandroulakis, N., Mylonas, C. C., et al. (2018a). The gene toolkit implicated in functional sex in sparidae hermaphrodites: inferences from comparative transcriptomics. Front. Genet. 9:749. doi: 10.3389/fgene.2018.00749
Tsakogiannis, A., Manousaki, T., Lagnel, J., Sterioti, A., Pavlidis, M., Papandroulakis, N., et al. (2018b). The transcriptomic signature of different sexes in two protogynous hermaphrodites: insights into the molecular network underlying sex phenotype in fish. Sci. Rep. 8:3564. doi: 10.1038/s41598-018-21992-9
Uhlenhaut, N. H., Jakob, S., Anlag, K., Eisenberger, T., Sekido, R., Kress, J., et al. (2009). Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130–1142. doi: 10.1016/j.cell.2009.11.021
Uno, T., Ishizuka, M., and Itakura, T. (2012). Cytochrome P450 (CYP) in fish. Environ. Toxicol. Pharmacol. 34, 1–13. doi: 10.1016/j.etap.2012.02.004
Vidal, V. P., Chaboissier, M. C., De Rooij, D. G., and Schedl, A. (2001). Sox9 induces testis development in XX transgenic mice. Nat. Genet. 28, 216–217. doi: 10.1038/90046
Webster, K. A., Schach, U., Ordaz, A., Steinfeld, J. S., Draper, B. W., and Siegfried, K. R. (2017). Dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 422, 33–46. doi: 10.1016/j.ydbio.2016.12.008
Wu, G. C., and Chang, C. F. (2009). wnt4 Is associated with the development of ovarian tissue in the protandrous black Porgy, Acanthopagrus schlegeli. Biol. Reprod. 81, 1073–1082. doi: 10.1095/biolreprod.109.077362
Wu, G. C., Chiu, P. C., Lyu, Y. S., and Chang, C. F. (2010). The expression of amh and amhr2 is associated with the development of gonadal tissue and sex change in the protandrous black porgy, Acanthopagrus schlegeli. Biol. Reprod. 83, 443–453. doi: 10.1095/biolreprod.110.084681
Xia, W., Zhou, L., Yao, B., Li, C. J., and Gui, J. F. (2007). Differential and spermatogenic cell-specific expression of DMRT1 during sex reversal in protogynous hermaphroditic groupers. Mol. Cell. Endocrinol. 263, 156–172. doi: 10.1016/j.mce.2006.09.014
Xiong, S., Jing, J., Wu, J., Ma, W., Dawar, F. U., Mei, J., et al. (2015). Characterization and sexual dimorphic expression of Cytochrome P450 genes in the hypothalamic-pituitary-gonad axis of yellow catfish. Gen. Comp. Endocrinol. 216, 90–97. doi: 10.1016/j.ygcen.2015.04.015
Xiong, Z., Zhang, H., Huang, B., Liu, Q., Wang, Y., Shi, D., et al. (2018). Expression pattern of prohibitin, capping actin protein of muscle Z-line beta subunit and tektin-2 gene in Murrah buffalo sperm and its relationship with sperm motility. Asian Australas. J. Anim. Sci. 31, 1729–1737. doi: 10.5713/ajas.18.0025
Yan, W., Ma, L., Burns, K. H., and Matzuk, M. M. (2004). Haploinsufficiency of kelch-like protein homolog 10 causes infertility in male mice. Proc. Natl. Acad. Sci. U.S.A. 101, 7793–7798. doi: 10.1073/pnas.0308025101
Yang, K., Grzmil, P., Meinhardt, A., and Hoyer-Fender, S. (2014). Haplo-deficiency of ODF1/HSPB10 in mouse sperm causes relaxation of head-to-tail linkage. Reproduction 148, 499–506. doi: 10.1530/rep-14-0370
Yang, Y. J., Wang, Y., Li, Z., Zhou, L., and Gui, J. F. (2017). Sequential, divergent, and cooperative requirements of Foxl2a and Foxl2b in ovary development and maintenance of zebrafish. Genetics 205, 1551–1572. doi: 10.1534/genetics.116.199133
Yano, A., Guyomard, R., Nicol, B., Jouanno, E., Quillet, E., Klopp, C., et al. (2012). An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22, 1423–1428. doi: 10.1016/j.cub.2012.05.045
Yao, B., Zhou, L., Wang, Y., Xia, W., and Gui, J. F. (2007). Differential expression and dynamic changes of SOX3 during gametogenesis and sex reversal in protogynous hermaphroditic fish. J. Exp. Zool. A Ecol. Genet. Physiol. 307, 207–219. doi: 10.1002/jez.361
Zhai, G., Shu, T., Xia, Y., Lu, Y., Shang, G., Jin, X., et al. (2018). Characterization of sexual trait development in cyp17a1-deficient zebrafish. Endocrinology 159, 3549–3562. doi: 10.1210/en.2018-00551
Zhang, K., Xu, J., Zhang, Z., Huang, Y., Ruan, Z., Chen, S., et al. (2019). A comparative transcriptomic study on developmental gonads provides novel insights into sex change in the protandrous black porgy (Acanthopagrus schlegelii). Genomics 111, 277–283. doi: 10.1016/j.ygeno.2018.11.006
Zhang, T., Oatley, J., Bardwell, V. J., and Zarkower, D. (2016). DMRT1 is required for mouse spermatogonial stem cell maintenance and replenishment. PLoS Genet. 12:e1006293. doi: 10.1371/journal.pgen.1006293
Zhu, Y., Cui, Z., Yang, Y., Xu, W., Shao, C., Fu, X., et al. (2019). Expression analysis and characterization of dmrt2 in Chinese tongue sole (Cynoglossus semilaevis). Theriogenology 138, 1–8. doi: 10.1016/j.theriogenology.2019.06.035
Keywords: yellowfin seabream, gonad transcriptome, sex reversal, sex-biased genes, protandrous hermaphroditic fish
Citation: Li S, Lin G, Fang W, Huang P, Gao D, Huang J, Xie J and Lu J (2020) Gonadal Transcriptome Analysis of Sex-Related Genes in the Protandrous Yellowfin Seabream (Acanthopagrus latus). Front. Genet. 11:709. doi: 10.3389/fgene.2020.00709
Received: 13 April 2020; Accepted: 11 June 2020;
Published: 16 July 2020.
Edited by:
Peng Xu, Xiamen University, ChinaReviewed by:
Changwei Shao, Yellow Sea Fisheries Research Institute (CAFS), ChinaJie Mei, Huazhong Agricultural University, China
Copyright © 2020 Li, Lin, Fang, Huang, Gao, Huang, Xie and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianguo Lu, bHVqaWFuZ3VvQG1haWwuc3lzdS5lZHUuY24=; amlhbmd1b25rQGdtYWlsLmNvbQ==
 Shizhu Li
Shizhu Li Genmei Lin1
Genmei Lin1 Jianguo Lu
Jianguo Lu