- 1Departamento de Biología, Facultad de Química y Biología, Universidad de Santiago de Chile, Santiago, Chile
- 2ANID – Millennium Science Initiative Program – Millennium Institute for Integrative Biology (iBIO), Santiago, Chile
The continuous usage of single Saccharomyces cerevisiae strains as starter cultures in fermentation led to the domestication and propagation of highly specialized strains in fermentation, resulting in the standardization of wines and beers. In this way, hundreds of commercial strains have been developed to satisfy producers’ and consumers’ demands, including beverages with high/low ethanol content, nutrient deprivation tolerance, diverse aromatic profiles, and fast fermentations. However, studies in the last 20 years have demonstrated that the genetic and phenotypic diversity in commercial S. cerevisiae strains is low. This lack of diversity limits alternative wines and beers, stressing the need to explore new genetic resources to differentiate each fermentation product. In this sense, wild strains harbor a higher than thought genetic and phenotypic diversity, representing a feasible option to generate new fermentative beverages. Numerous recent studies have identified alleles in wild strains that could favor phenotypes of interest, such as nitrogen consumption, tolerance to cold or high temperatures, and the production of metabolites, such as glycerol and aroma compounds. Here, we review the recent literature on the use of commercial and wild S. cerevisiae strains in wine and beer fermentation, providing molecular evidence of the advantages of using wild strains for the generation of improved genetic stocks for the industry according to the product style.
Introduction
For centuries, humans have mass-produced food and alcoholic beverages using fermentative yeasts, of which wine and beer are the best-known products derived from this process (Steensels and Verstrepen, 2014; Legras et al., 2018; Parapouli et al., 2020). Yeasts are naturally present in fermentation raw materials, such as grape musts (wine fermentation) and cereals (beer fermentation), representing a fraction of the natural microbiota of these carbon-rich environments. However, in most cases, the main microorganisms that could outcompete the others are species belonging to the Saccharomyces genus, mainly Saccharomyces cerevisiae (Sternes et al., 2017; Stefanini and Cavalieri, 2018; Mandakovic et al., 2020). S. cerevisiae is able to dominate the fermentation process, due to its high fermentative capacity and ability to tolerate different stresses, such as high osmotic stress, low pH, high ethanol levels, anerobiosis, and nutrient starvation (Warringer et al., 2011; Brice et al., 2014; García-Ríos et al., 2014; Kitichantaropas et al., 2016; Taymaz-Nikerel et al., 2016; Legras et al., 2018; García-Ríos and Guillamón, 2019). In this sense, most producers use starter cultures in the fermentation process. By the intense re-utilization of yeast, specific S. cerevisiae strains were involuntarily selected as starter cultures, guaranteeing a controlled and precise fermentation process and avoiding stuck or sluggish fermentations (Albergaria and Arneborg, 2016). However, the current set of commercial S. cerevisiae strains and its derived hybrids is insufficient to provide novel properties to beer and wine, stressing the need for new and improved strains for the industry (Aquilani et al., 2015; Alperstein et al., 2020; Gibson et al., 2020). In recent years, bioprospecting effort has increased the isolation of wild strains from different niches, opening the opportunity to use them (or their genetic variability) in the fermentation process (Duan et al., 2018; Peter et al., 2018; Kang et al., 2019; Pontes et al., 2020).
Here, we review the contemporary literature concerning the use of commercial and wild S. cerevisiae strains in the wine and beer industries, highlighting their advantages and disadvantages. Under this scenario, wild strains represent alternative genetic stocks for the industry to overcome current challenges. We review recent progress in the characterization of these strains, new allelic determinants and their performance under wine and beer fermentation conditions, together with their genetic and phenotypic features. In this sense, we defined as “wild strains” those isolated from non-fermentative niches that show no signs of domestication. However, the importance of native strains belonging to domesticated lineages cannot be ignored. In this sense, we highlight the genetic and phenotypic diversity in native and wild strains, providing strategies to improve these genetic stocks for beer and wine fermentations.
All That Glitters is not Gold: Advantages and Disadvantages of Using Commercial S. Cerevisiae Strains
In industrial-scale fermentations, the utilization of starter cultures is preferred over spontaneous inoculations to avoid technical hitches related to slow fermentation rates, end product variability, and yeast contaminants that can spoil the final product (Eliodório et al., 2019; Parapouli et al., 2020). Commercial yeast strains are isolates from fermentation-related environments or are derived from breeding programs, in which they were selected for certain phenotypic traits, such as efficient nitrogen consumption (García-Ríos et al., 2014; Tesnière et al., 2015), fast fermentation rates (Preiss et al., 2018), and pleasant aroma profiles (Eder et al., 2018; Schwarz et al., 2020). These commercial strains usually underwent distinct domestication trajectories, with the exact conditions depending on the traditional practices in the brewery or winery. In this way, traits of industrial interest can differ between distinct lineages, reflecting their specific domestication environment. For example, wine strains have a superior performance in general stress conditions (high sugar and alcohol content) compared to beer strains (Gallone et al., 2016). In contrast, beer strains cover desirable and specific traits for brewing, such as maltotriose utilization, and a lack of production of undesirable off-flavors, such as 4-vinyl guaiacol (Gallone et al., 2016; Krogerus et al., 2017a; Nikulin et al., 2018; Vakirlis et al., 2019). Consequently, industrial beer strains show more notable domestication signatures than wine strains, probably because of the continuous recycling of yeast after each fermentation batch throughout the year (Steensels et al., 2019). In contrast, wine strains only grow and ferment in grape musts during a short period of the year, i.e., 2–3 weeks, and are then forced “back-to-nature.” These differences result in specific genomic features and life cycles in beer and wine strains, as a consequence of selection, domestication, and early hybridization events in ancient lineages.
The genomic characterization of wine and brewing strains demonstrates that their geographical origin and industrial applications have shaped the evolutionary divergence of industrial yeasts (Liti et al., 2009; Borneman et al., 2011; Gallone et al., 2016; Gonçalves et al., 2016; Legras et al., 2018; Peter et al., 2018). In particular, commercial wine strains form a defined phylogenetic cluster, distributed around the world across the Mediterranean, and Mediterranean-like regions (Legras et al., 2007; Liti et al., 2009; Almeida et al., 2015; Peter et al., 2018), and are separated from the S. cerevisiae ale strains (Gallone et al., 2016; Gonçalves et al., 2016). Ale strain divergence is complex and is shaped by their clonal life cycle in an industrial niche and by the geographical location of the brewery. They separate into two clades or lineages, Beer 1 and Beer 2. The Beer 1 clade contains Belgium/Germany, Unites States, Britain and kveik strains (Gallone et al., 2016; Gonçalves et al., 2016; Preiss et al., 2018; Pontes et al., 2020). In contrast, the Beer 2 clade lacks a geographical structure, and strains are closely-related to wine strains (Gallone et al., 2016). Interestingly, kveik yeasts form a distinct group related to the Beer 1 clade. However, these strains have a possible mixed ancestry, suggesting a hybrid origin for kveik strains between Beer 1 and an unknown lineage (Preiss et al., 2018). Indeed, further evidence suggests that polyploid brewing strains originated from ancient admixture events between European wine strains and Asian rice wine strains (Fay et al., 2019; Pontes et al., 2020). Altogether, these findings demonstrate that modern commercial strains are the product of unintended historical hybridization events as a result of human selection for specific traits (Fay et al., 2019; Nespolo et al., 2020; Pontes et al., 2020).
Recent genetic studies have demonstrated that commercial wine and beer strains lack genetic and phenotypic diversity, where genetically-similar strains are sold with different commercial names (Fernández-Espinar et al., 2001; Schuller et al., 2004; Dunn et al., 2005; Legras et al., 2005; Borneman et al., 2016; Bindon et al., 2019). In particular, wine strains show a lower nucleotide diversity (π = 1 × 10−3) compared to beer strains (π = 2.8 × 10−3) and in the species as a whole (π = 3 × 10−3; Peter et al., 2018). Although beer strains have a greater genetic diversity than wine strains, they originate from only two ancestors, which is estimated to have occurred between AD 1600 and AD 1700 (Gallone et al., 2016, 2018). The low genetic and phenotypic diversity of these strains may negatively-impact the producer’s perception of the substantial role of the chosen yeast over the final product. Therefore, current genetic stocks limit the diversification of novel beverage properties, increasing the need to search for new alternatives to differentiate each fermentation product (Aquilani et al., 2015). This stresses the desire to explore the genetic and phenotypic diversity in other S. cerevisiae lineages to obtain new and novel strains with different and improved fermentative characteristics. In this sense, one source of genetic diversity can be found in non-domesticated S. cerevisiae strains collected from the wild.
Wild S. Cerevisiae Strains: Genetic Stocks for Novel Fermentation Products
For years, scientists believed that S. cerevisiae was primordially found in fruits or fermentation-related environments. However, bioprospecting efforts have established that it is much more widespread; it has been isolated from wasps, flies, oak trees, and their associated substrates, such as bark, soil, and flowers (Figure 1; Zhang et al., 2010; Alsammar and Delneri, 2020; Pontes et al., 2020). In this way, S. cerevisiae follows the nomad model, being able to survive as a generalist at low abundance in a wide range of environments and is not necessarily adapted to a specific niche (Goddard and Greig, 2015). Recent genomic studies, including a large number of wild isolates, demonstrated that the population structure of S. cerevisiae is more complex than previously reported (Liti et al., 2006; Legras et al., 2007), with abundant genomic and phenomic diversity (Liti et al., 2009; Warringer et al., 2011; Wang et al., 2012; Bergström et al., 2014; Legras et al., 2018; Peter et al., 2018). These studies have described more than 20 independent lineages in the species, where 13 correspond to wild or non-domesticated lineages (Duan et al., 2018; Peter et al., 2018; Pontes et al., 2020). Overall, wild strains showed lower heterozygosity levels, higher indels rates and sequence diversity, lower gene duplication levels, lower horizontal gene transfer (HGT) events, and lower genome content variation (median 161 shared ORFs vs. 115 ORFs that are not shared; Peter et al., 2018; Kang et al., 2019). Furthermore, wild strains are generally diploid, and their diversity is dependent on SNPs rather than on genomic rearrangements (Peter et al., 2018).
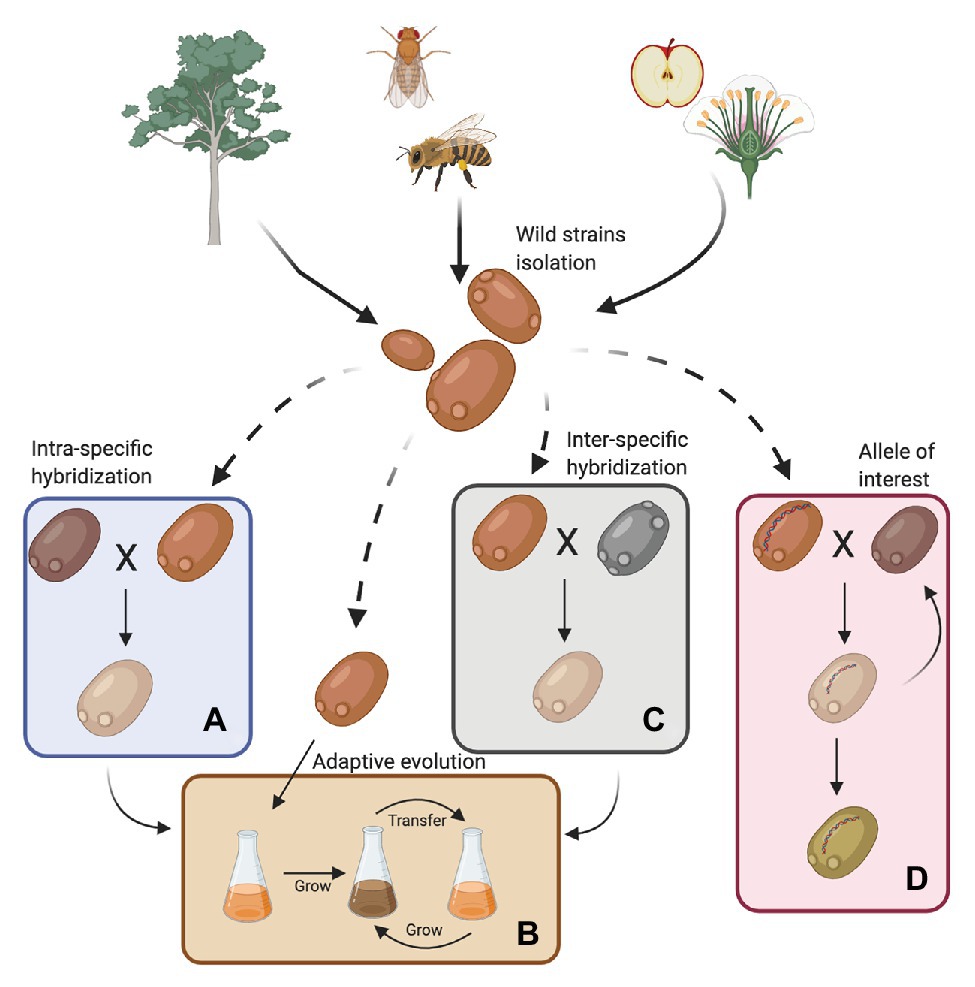
Figure 1. Sources for obtaining wild Saccharomyces cerevisiae strains and their use in the fermentation process. Wild strains may be obtained from various environments such as insects, wasps and flies, flowers, trees, and their surroundings. These strains must be phenotypically characterized under fermentative conditions of interest. In this way, strains can then be selected for genetic improvement programs with commercial strains (A, intra-specific hybridization), subjected to an adaptive evolution process (B) or used in the generation of interspecific hybrids (C). Furthermore, wild alleles may be incorporated into commercial strains by an assisted introgression strategy (D). Finally, strains obtained by hybridization can also be subjected to an adaptive evolution process (Created with BioRender.com).
Wild S. cerevisiae are genetically and phenotypically separated from industrial S. cerevisiae strains, mostly due to the result of human selection (Gallone et al., 2016; Duan et al., 2018; Peter et al., 2018; Pontes et al., 2020). In general, commercial strains produce relatively low levels of extracellular aromatic compounds (such as benzoate, 4-aminobenzoate, and nicotinate) and have low sporulation levels, but exhibit high ethanol and low-pH tolerance (Duan et al., 2018; Kang et al., 2019). In contrast, wild strains have relatively high extracellular levels of different secondary metabolites (such as aspropyl acetate, isoamyl acetate, and ethyl acetate), high fatty acid biosynthesis, and higher energy flux toward the TCA cycle (Kang et al., 2019). These features directly translate into greater production of fatty acids, which are associated with a more robust response to different stresses, such as high salt and ethanol concentrations, high temperatures, and oxidative stress (Qiu et al., 2019). Therefore, wild strains tend to exhibit higher ranges of resistance to multiple conditions, and accordingly, offer significant potential for utilization in a wide range of industrial applications (Kang et al., 2019). Furthermore, industrial strains display selfish behavior with a rapid glucose consumption rate, a large cell size, and nutrient storage inside the cell. In contrast, wild strains display a cooperative behavior and secrete secondary metabolites to poison or cross-feed competitors (Spor et al., 2009). This behavior may be advantageous during fermentation. For example, wild isolates from flowers and sugar-rich sources may produce a different set of volatile compounds, expanding the repertoire of aromas and flavors in alcoholic fermentation (Pontes et al., 2020). In this sense, wine fermentations using indigenous wild strains obtained from oak trees produce earthy and sulfurous organoleptic features, and at the same time, high levels of citrus and floral attributes (Hyma et al., 2011). However, native strains isolated from vineyards, grape, and soil display excellent fermentation properties, but wild isolates from oak barks exhibit stuck fermentation profiles (Camarasa et al., 2011). Therefore, the direct use of wild strains could be limited, and strategies for their improvement are needed.
Although wild isolates may not be useful on their own for fermentative applications, they harbor alleles that could be of interest for breeding programs (Figure 1 and Table 1). For example, low levels of yeast assimilable nitrogen (below 140 mg/L) in grape must are the primary source of sluggish or stuck fermentations (Gobert et al., 2019; Kessi-Pérez et al., 2020a). In this context, recent studies have demonstrated that alleles from a wild S. cerevisiae strain might play a decisive role in the adaptation to low nitrogen concentrations (Molinet et al., 2019; Kessi-Pérez et al., 2020a,b). SAP185, TOR2, SCH9, and NPR1 alleles derived from an oak strain increased amino acid consumption (aspartic acid, histidine, glutamine, and threonine) compared to wine alleles under wine fermentation conditions (Molinet et al., 2019). Another phenotype of interest related to nitrogen consumption is volatile compound production (Swiegers and Pretorius, 2007). In particular, volatile thiols contribute to the typicity of Sauvignon Blanc wines. In this regard, a bulked segregant analysis identified the IRC7 allele from a clinical isolate as responsible for a higher production of 4-mercapto-4-methylpentan-2-one (4MMP), which contributes boxwood and black currant aromas to wines (Swiegers et al., 2009). This allele is an introgression from Saccharomyces paradoxus, while the majority of S. cerevisiae strains (including commercial wine strains) harbor a 38-bp deletion that generates a truncated protein. The overexpression of the beneficial allele in a commercial wine strain (Zymaflore F15) increases 4MMP production during Sauvignon Blanc fermentation from below detectable levels (<10 ng/L) to 1,000 ng/L (Roncoroni et al., 2011). Overall, these studies highlight that there is an emerging opportunity to broaden the genetic and phenotypic variability from natural populations, which are still poorly investigated. Hence, non-domesticated wild isolates harbor a valuable source of genetic diversity, which is useful for domestication and breeding programs to generate novel strains or hybrids for the wine and beer fermentation industries.
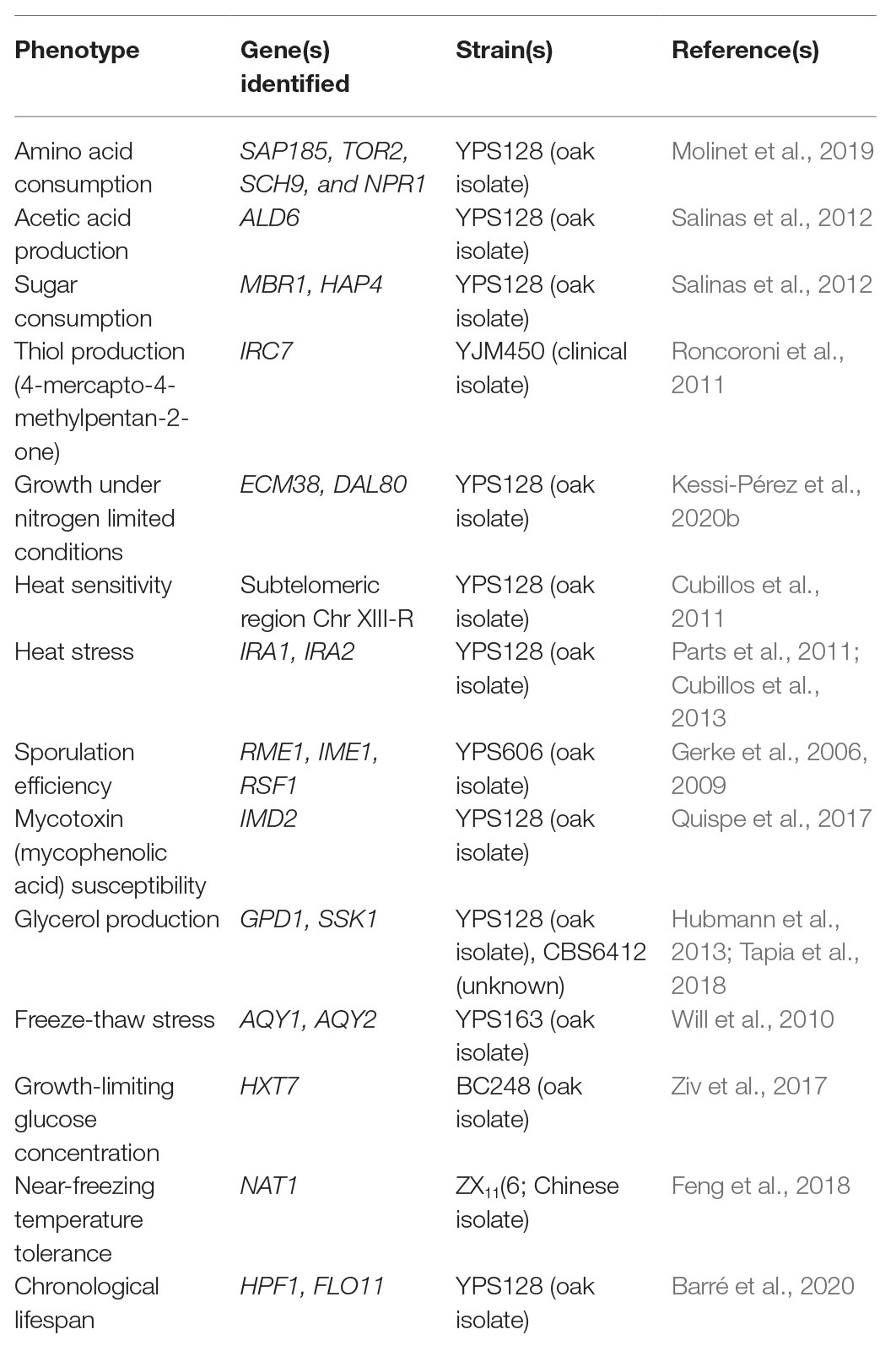
Table 1. Examples of alleles from wild or non-domesticated strains identified in QTL mapping studies with a beneficial impact on different phenotypes of interest.
Exploiting Genetic Variants From Wild S. Cerevisiae Isolates to Generate Novel Hybrid Strains for Beer and Wine
Wild strains with genetic variants of interest could be used in breeding-related programs to generate genetically-improved strains (Figure 1; Steensels et al., 2014b). Intra-specific hybridization (two strains from the same species) has been successfully applied in S. cerevisiae, improving different traits for wine fermentation, such as low sulfur compound levels (Agarbati et al., 2020), novel aroma profiles (Marullo et al., 2006; Steensels et al., 2014a), improved stress tolerance (Bonciani et al., 2016), wider temperature tolerance (Marullo et al., 2009), fermentation of nitrogen-deficient musts (Kessi-Pérez et al., 2020c), and lower ethanol production (García et al., 2012). Similarly, breeding strategies have generated hybrids exhibiting high beer fermentation performance and producing different aroma profiles, likely due to transcriptional cross-talk (Gallone et al., 2016).
Interspecific hybrids between S. cerevisiae and S. non-cerevisiae strains are easily isolated from fermentative environments (González et al., 2006, 2008; Querol and Bond, 2009; Peris et al., 2012). Genetic studies highlight that the S. non-cerevisiae parental portion has a wild origin (Gallone et al., 2019; Langdon et al., 2019). In this way, it is possible to use wild Saccharomyces strains for the generation of new commercial hybrid stocks, and thus expand the genetic and phenotypic diversity. Interspecific Saccharomyces hybrids are possible to find due to a weak pre-zygotic barrier between species (Alsammar and Delneri, 2020). In contrast, post-zygotic barriers impede successful meiosis, and spore viabilities in hybrids are typically below 10% (Liti et al., 2006; Hittinger, 2013), limiting the generation of recombinant hybrids between species. Interspecific Saccharomyces hybrids inhabit different fermentation environments, exhibiting interesting and complex genomic compositions (Alsammar and Delneri, 2020). In this sense, recent reports show the existence of four types of hybrids determined by their geographical origin and industrial practices: S. pastorianus associated with beer; S. cerevisiae × Saccharomyces kudriavzevii associated with beer and wine; S. eubayanus × Saccharomyces uvarum associated with different environments; and complex hybrids with three or four parental species: S. cerevisiae × S. kudriavzevii × S. eubayanus × S. uvarum, S. cerevisiae × S. eubayanus × S. uvarum and S. cerevisiae × S. kudriavzevii × S. eubayanus (Gallone et al., 2019; Langdon et al., 2019). In all cases, the S. cerevisiae parental strains belong to three domesticated lineages (Wine, Beer 1, and Beer 2), providing traits and fermentative advantages when used in industrial settings. These reports highlight the importance and relevance of interspecific hybridization in the diversification and adaptation of yeast to industrial niches.
Interspecific hybridization is an evolutionary strategy that allows swift adaptation to new niches (Gallone et al., 2019). In this context, the construction in the laboratory of novel hybrids using wild strains showing improved fermentation performance compared to their parental species, is an attractive strategy for future breeding projects (Figure 1). For example, de novo hybridization of S. cerevisiae × S. eubayanus strains combined the sugar utilization properties of S. cerevisiae and the cryotolerance of S. eubayanus (Hebly et al., 2015; Krogerus et al., 2017b). In this sense, the utilization of all Saccharomyces species for de novo hybridization with S. cerevisiae has the potential to increase the genetic diversity of yeasts for the wine and beer industries (Brickwedde et al., 2018; Nikulin et al., 2018). However, one of the limitations observed in studies that generate de novo hybrids is the use of a restricted number of parental strains. The utilization of a handful of isolates per species is not necessarily representative of the phenotypic spectrum of the progeny generated by hybridizing individuals from two (or more) different species. In this way, taking advantage of the full genetic diversity already described in the Saccharomyces genus would be a promising step toward developing new yeast hybrids. Therefore, wild isolates are strong candidates for innovations in the industry (Cubillos, 2016; Cubillos et al., 2019). However, it is important to note that such polyploid hybrids tend to be genetically unstable, and may undergo extensive changes after hybridization (Gallone et al., 2018), which opens the opportunity to improve and adapt them to conditions found in fermentative processes. Consequently, hybrids have to reach genomic stability under conditions generally encountered during alcoholic fermentation (Krogerus et al., 2018). Experimental evolution approaches are the right strategies to follow in order to reconstruct evolutionary trajectories of novel hybrid strains subjected to fermentation environments (Krogerus et al., 2018). Different studies utilizing experimental evolution provide evidence of the genetic changes taking place during adaptation to fermentation, including partial loss of one of the parental subgenomes, loss of heterozygosity, selection of superior alleles, and the formation of fusion genes following translocations (Piotrowski et al., 2012; Dunn et al., 2013; Hope et al., 2017; Peris et al., 2017; Smukowski Heil et al., 2017).
Conclusion
The standardization of wines and beers as a result of the utilization of a genetically reduced set of commercial strains has brought with it the need for new and novel products that can be highlighted and differentiated. In this sense, wild strains and genetically diverse interspecific hybrids are an attractive alternative for the industry. However, challenges persist in adapting and improving wild strains to fermentative environments. Such challenges could be overcome through genetic improvement programs together with adaptive evolution strategies. The generation of new strains and intra‐ and inter-species hybrids could open up new avenues in order to obtain unique strains for the wine and beer industries.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported to FC by the Comisión Nacional de Investigación Científica y Tecnológica CONICYT FONDECYT (Grant N 1180161). This work was funded by ANID – Millennium Science Initiative Program ‐ ICN17_022. JM was supported by Universidad de Santiago de Chile (Grant VRIDEI 022043CR_POSTDOC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Michael Handford (Universidad de Chile) for English proofreading.
References
Agarbati, A., Canonico, L., Comitini, F., and Ciani, M. (2020). Reduction of sulfur compounds through genetic improvement of native Saccharomyces cerevisiae useful for organic and sulfite-free wine. Foods 9:658. doi: 10.3390/foods9050658
Albergaria, H., and Arneborg, N. (2016). Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 100, 2035–2046. doi: 10.1007/s00253-015-7255-0
Almeida, P., Barbosa, R., Zalar, P., Imanishi, Y., Shimizu, K., Turchetti, B., et al. (2015). A population genomics insight into the Mediterranean origins of wine yeast domestication. Mol. Ecol. 24, 5412–5427. doi: 10.1111/mec.13341
Alperstein, L., Gardner, J. M., Sundstrom, J. F., Sumby, K. M., and Jiranek, V. (2020). Yeast bioprospecting versus synthetic biology—which is better for innovative beverage fermentation? Appl. Microbiol. Biotechnol. 104, 1939–1953. doi: 10.1007/s00253-020-10364-x
Alsammar, H., and Delneri, D. (2020). An update on the diversity, ecology and biogeography of the Saccharomyces genus. FEMS Yeast Res. 20:foaa013. doi: 10.1093/femsyr/foaa013
Aquilani, B., Laureti, T., Poponi, S., and Secondi, L. (2015). Beer choice and consumption determinants when craft beers are tasted: an exploratory study of consumer preferences. Food Qual. Prefer. 41, 214–224. doi: 10.1016/j.foodqual.2014.12.005
Barré, B. P., Hallin, J., Yue, J. -X., Persson, K., Mikhalev, E., Irizar, A., et al. (2020). Intragenic repeat expansion in the cell wall protein gene HPF1 controls yeast chronological aging. Genome Res. 30, 697–710. doi: 10.1101/gr.253351.119
Bergström, A., Simpson, J. T., Salinas, F., Barré, B., Parts, L., Zia, A., et al. (2014). A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 31, 872–888. doi: 10.1093/molbev/msu037
Bindon, K. A., Kassara, S., Solomon, M., Bartel, C., Smith, P. A., Barker, A., et al. (2019). Commercial Saccharomyces cerevisiae yeast strains significantly impact shiraz tannin and polysaccharide composition with implications for wine colour and astringency. Biomol. Ther. 9:466. doi: 10.3390/biom9090466
Bonciani, T., Solieri, L., De Vero, L., and Giudici, P. (2016). Improved wine yeasts by direct mating and selection under stressful fermentative conditions. Eur. Food Res. Technol. 242, 899–910. doi: 10.1007/s00217-015-2596-6
Borneman, A. R., Desany, B. A., Riches, D., Affourtit, J. P., Forgan, A. H., Pretorius, I. S., et al. (2011). Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 7:e1001287. doi: 10.1371/journal.pgen.1001287
Borneman, A. R., Forgan, A. H., Kolouchova, R., Fraser, J. A., and Schmidt, S. A. (2016). Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3 (Bethesda) 6, 957–971. doi: 10.1534/g3.115.025692
Brice, C., Sanchez, I., Tesnière, C., and Blondin, B. (2014). Assessing the mechanisms responsible for differences between nitrogen requirements of Saccharomyces cerevisiae wine yeasts in alcoholic fermentation. Appl. Environ. Microbiol. 80, 1330–1339. doi: 10.1128/AEM.03856-13
Brickwedde, A., Brouwers, N., Broek, M.van den, Gallego Murillo, J. S., Fraiture, J. L., Pronk, J. T., et al. (2018). Structural, physiological and regulatory analysis of maltose transporter genes in Saccharomyces eubayanus CBS 12357T. Front. Microbiol. 9:1786. doi: 10.3389/fmicb.2018.01786
Camarasa, C., Sanchez, I., Brial, P., Bigey, F., and Dequin, S. (2011). Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One 6:e25147. doi: 10.1371/journal.pone.0025147
Cubillos, F. A. (2016). Exploiting budding yeast natural variation for industrial processes. Curr. Genet. 62, 745–751. doi: 10.1007/s00294-016-0602-6
Cubillos, F. A., Billi, E., Zörö, E., Parts, L., Fargier, P., Omholt, S., et al. (2011). Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol. Ecol. 20, 1401–1413. doi: 10.1111/j.1365-294X.2011.05005.x
Cubillos, F. A., Gibson, B., Grijalva-Vallejos, N., Krogerus, K., and Nikulin, J. (2019). Bioprospecting for brewers: exploiting natural diversity for naturally diverse beers. Yeast 36, 383–398. doi: 10.1002/yea.3380
Cubillos, F. A., Parts, L., Salinas, F., Bergström, A., Scovacricchi, E., Zia, A., et al. (2013). High-resolution mapping of complex traits with a four-parent advanced intercross yeast population. Genetics 195, 1141–1155. doi: 10.1534/genetics.113.155515
Duan, S. F., Han, P. J., Wang, Q. M., Liu, W. Q., Shi, J. Y., Li, K., et al. (2018). The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat. Commun. 9:2690. doi: 10.1038/s41467-018-05106-7
Dunn, B., Levine, R. P., and Sherlock, G. (2005). Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics 6:53. doi: 10.1186/1471-2164-6-53
Dunn, B., Paulish, T., Stanbery, A., Piotrowski, J., Koniges, G., Kroll, E., et al. (2013). Recurrent rearrangement during adaptive evolution in an interspecific yeast hybrid suggests a model for rapid introgression. PLoS Genet. 9:e1003366. doi: 10.1371/journal.pgen.1003366
Eder, M., Sanchez, I., Brice, C., Camarasa, C., Legras, J. L., and Dequin, S. (2018). QTL mapping of volatile compound production in Saccharomyces cerevisiae during alcoholic fermentation. BMC Genomics 19:166. doi: 10.1186/s12864-018-4562-8
Eliodório, K. P., Cunha, G. C. D. G. E., Müller, C., Lucaroni, A. C., Giudici, R., Walker, G. M., et al. (2019). Advances in yeast alcoholic fermentations for the production of bioethanol, beer and wine. Adv. Appl. Microbiol. 109, 61–119. doi: 10.1016/bs.aambs.2019.10.002
Fay, J. C., Liu, P., Ong, G. T., Dunham, M. J., Cromie, G. A., Jeffery, E. W., et al. (2019). A polyploid admixed origin of beer yeasts derived from European and Asian wine populations. PLoS Biol. 17:e3000147. doi: 10.1371/journal.pbio.3000147
Feng, L., Jia, H., Qin, Y., Song, Y., Tao, S., and Liu, Y. (2018). Rapid identification of major QTLS associated with near-freezing temperature tolerance in Saccharomyces cerevisiae. Front. Microbiol. 9:2110. doi: 10.3389/fmicb.2018.02110
Fernández-Espinar, M. T., López, V., Ramón, D., Bartra, E., and Querol, A. (2001). Study of the authenticity of commercial wine yeast strains by molecular techniques. Int. J. Food Microbiol. 70, 1–10. doi: 10.1016/S0168-1605(01)00502-5
Gallone, B., Mertens, S., Gordon, J. L., Maere, S., Verstrepen, K. J., and Steensels, J. (2018). Origins, evolution, domestication and diversity of Saccharomyces beer yeasts. Curr. Opin. Biotechnol. 49, 148–155. doi: 10.1016/j.copbio.2017.08.005
Gallone, B., Steensels, J., Mertens, S., Dzialo, M. C., Gordon, J. L., Wauters, R., et al. (2019). Interspecific hybridization facilitates niche adaptation in beer yeast. Nat. Ecol. Evol. 3, 1562–1575. doi: 10.1038/s41559-019-0997-9
Gallone, B., Steensels, J., Prahl, T., Soriaga, L., Saels, V., Herrera-Malaver, B., et al. (2016). Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166, 1397.e16–1410.e16. doi: 10.1016/j.cell.2016.08.020
García, V., Rivera, J., Contreras, A., Ganga, M. A., and Martínez, C. (2012). Development and characterization of hybrids from native wine yeasts. Braz. J. Microbiol. 43, 482–489. doi: 10.1590/S1517-83822012000200008
García-Ríos, E., and Guillamón, J. M. (2019). Sulfur dioxide resistance in Saccharomyces cerevisiae: beyond SSU1. Microb. Cell 6, 509–523. doi: 10.15698/mic2019.12.699
García-Ríos, E., Gutiérrez, A., Salvadó, Z. Z., Arroyo-López, F. N., and Guillamon, J. M. (2014). The fitness advantage of commercial wine yeasts in relation to the nitrogen concentration, temperature, and ethanol content under microvinification conditions. Appl. Environ. Microbiol. 80, 704–713. doi: 10.1128/AEM.03405-13
Gerke, J. P., Chen, C. T. L., and Cohen, B. A. (2006). Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics 174, 985–997. doi: 10.1534/genetics.106.058453
Gerke, J., Lorenz, K., and Cohen, B. (2009). Genetic interactions between transcription factors cause natural variation in yeast. Science 323, 498–501. doi: 10.1126/science.1166426
Gibson, B., Dahabieh, M., Krogerus, K., Jouhten, P., Magalhães, F., Pereira, R., et al. (2020). Adaptive laboratory evolution of ale and lager yeasts for improved brewing efficiency and beer quality. Annu. Rev. Food Sci. Technol. 11, 23–44. doi: 10.1146/annurev-food-032519-051715
Gobert, A., Tourdot-Maréchal, R., Sparrow, C., Morge, C., and Alexandre, H. (2019). Influence of nitrogen status in wine alcoholic fermentation. Food Microbiol. 83, 71–85. doi: 10.1016/j.fm.2019.04.008
Goddard, M. R., and Greig, D. (2015). Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 15:fov009. doi: 10.1093/femsyr/fov009
Gonçalves, M., Pontes, A., Almeida, P., Barbosa, R., Serra, M., Libkind, D., et al. (2016). Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 26, 2750–2761. doi: 10.1016/j.cub.2016.08.040
González, S. S., Barrio, E., Gafner, J., and Querol, A. (2006). Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 6, 1221–1234. doi: 10.1111/j.1567-1364.2006.00126.x
González, S. S., Barrio, E., and Querol, A. (2008). Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl. Environ. Microbiol. 74, 2314–2320. doi: 10.1128/AEM.01867-07
Hebly, M., Brickwedde, A., Bolat, I., Driessen, M. R. M., de Hulster, E. A. F., van den Broek, M., et al. (2015). S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 15:fov005. doi: 10.1093/femsyr/fov005
Hittinger, C. T. (2013). Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 29, 309–317. doi: 10.1016/j.tig.2013.01.002
Hope, E. A., Amorosi, C. J., Miller, A. W., Dang, K., Heil, C. S., and Dunham, M. J. (2017). Experimental evolution reveals favored adaptive routes to cell aggregation in yeast. Genetics 206, 1153–1167. doi: 10.1534/genetics.116.198895
Hubmann, G., Foulquié-Moreno, M. R., Nevoigt, E., Duitama, J., Meurens, N., Pais, T. M., et al. (2013). Quantitative trait analysis of yeast biodiversity yields novel gene tools for metabolic engineering. Metab. Eng. 17, 68–81. doi: 10.1016/j.ymben.2013.02.006
Hyma, K. E., Saerens, S. M., Verstrepen, K. J., and Fay, J. C. (2011). Divergence in wine characteristics produced by wild and domesticated strains of Saccharomyces cerevisiae. FEMS Yeast Res. 11, 540–551. doi: 10.1111/j.1567-1364.2011.00746.x
Kang, K., Bergdahl, B., Machado, D., Dato, L., Han, T. -L., Li, J., et al. (2019). Linking genetic, metabolic, and phenotypic diversity among Saccharomyces cerevisiae strains using multi-omics associations. Gigascience 8:giz015. doi: 10.1093/gigascience/giz015
Kessi-Pérez, E., Molinet, J., García, V., Aguilera, O., Cepeda, F., López, M., et al. (2020c). Generation of a non-transgenic genetically improved yeast strain for wine production from nitrogen-deficient musts. Microorganisms 8:1194. doi: 10.3390/microorganisms8081194
Kessi-Pérez, E. I., Molinet, J., and Martínez, C. (2020a). Disentangling the genetic bases of Saccharomyces cerevisiae nitrogen consumption and adaptation to low nitrogen environments in wine fermentation. Biol. Res. 53:2. doi: 10.1186/s40659-019-0270-3
Kessi-Pérez, E. I., Ponce, B., Li, J., Molinet, J., Baeza, C., Figueroa, D., et al. (2020b). Differential gene expression and allele frequency changes favour adaptation of a heterogeneous yeast population to nitrogen-limited fermentations. Front. Microbiol. 11:1204. doi: 10.3389/fmicb.2020.01204
Kitichantaropas, Y., Boonchird, C., Sugiyama, M., Kaneko, Y., Harashima, S., and Auesukaree, C. (2016). Cellular mechanisms contributing to multiple stress tolerance in Saccharomyces cerevisiae strains with potential use in high-temperature ethanol fermentation. AMB Express 6:107. doi: 10.1186/s13568-016-0285-x
Krogerus, K., Holmström, S., and Gibson, B. (2018). Enhanced wort fermentation with de novo lager hybrids. Appl. Environ. Microbiol. 84:e02302–17. doi: 10.1128/AEM.02302-17
Krogerus, K., Magalhães, F., Vidgren, V., and Gibson, B. (2017a). Novel brewing yeast hybrids: creation and application. Appl. Microbiol. Biotechnol. 101, 65–78. doi: 10.1007/s00253-016-8007-5
Krogerus, K., Seppänen-Laakso, T., Castillo, S., and Gibson, B. (2017b). Inheritance of brewing-relevant phenotypes in constructed Saccharomyces cerevisiae×Saccharomyces eubayanus hybrids. Microb. Cell Factories 16:66. doi: 10.1186/s12934-017-0679-8
Langdon, Q. K., Peris, D., Baker, E. C. P., Opulente, D. A., Nguyen, H. V., Bond, U., et al. (2019). Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 3, 1576–1586. doi: 10.1038/s41559-019-0998-8
Legras, J. -L., Galeote, V., Bigey, F., Camarasa, C., Marsit, S., Nidelet, T., et al. (2018). Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 35, 1712–1727. doi: 10.1093/molbev/msy066
Legras, J. -L., Merdinoglu, D., Cornuet, J. -M., and Karst, F. (2007). Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 16, 2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x
Legras, J. -L., Ruh, O., Merdinoglu, D., and Karst, F. (2005). Selection of hypervariable microsatellite loci for the characterization of Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 102, 73–83. doi: 10.1016/j.ijfoodmicro.2004.12.007
Liti, G., Barton, D. B. H., and Louis, E. J. (2006). Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174, 839–850. doi: 10.1534/genetics.106.062166
Liti, G., Carter, D. M., Moses, A. M., Warringer, J., Parts, L., James, S. A., et al. (2009). Population genomics of domestic and wild yeasts. Nature 458, 337–341. doi: 10.1038/nature07743
Mandakovic, D., Pulgar, R., Maldonado, J., Mardones, W., González, M., Cubillos, F. A., et al. (2020). Fungal diversity analysis of grape musts from central valley-Chile and characterization of potential new starter cultures. Microorganisms 8:956. doi: 10.3390/microorganisms8060956
Marullo, P., Bely, M., Masneuf-Pomarède, I., Pons, M., Aigle, M., and Dubourdieu, D. (2006). Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 6, 268–279. doi: 10.1111/j.1567-1364.2006.00034.x
Marullo, P., Mansour, C., Dufour, M., Albertin, W., Sicard, D., Bely, M., et al. (2009). Genetic improvement of thermo-tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res. 9, 1148–1160. doi: 10.1111/j.1567-1364.2009.00550.x
Molinet, J., Cubillos, F. A., Salinas, F., Liti, G., and Martínez, C. (2019). Genetic variants of TORC1 signaling pathway affect nitrogen consumption in Saccharomyces cerevisiae during alcoholic fermentation. PLoS One 14:e0220515. doi: 10.1371/journal.pone.0220515
Nespolo, R. F., Solano-Iguaran, J. J., Paleo-López, R., Quintero-Galvis, J. F., Cubillos, F. A., and Bozinovic, F. (2020). Performance, genomic rearrangements, and signatures of adaptive evolution: lessons from fermentative yeasts. Ecol. Evol. 10, 5240–5250. doi: 10.1002/ece3.6208
Nikulin, J., Krogerus, K., and Gibson, B. (2018). Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 35, 113–127. doi: 10.1002/yea.3246
Parapouli, M., Vasileiadis, A., Afendra, A. S., and Hatziloukas, E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31. doi: 10.3934/microbiol.2020001
Parts, L., Cubillos, F. a., Warringer, J., Jain, K., Salinas, F., Bumpstead, S. J., et al. (2011). Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 21, 1131–1138. doi: 10.1101/gr.116731.110
Peris, D., Lopes, C. A., Belloch, C., Querol, A., and Barrio, E. (2012). Comparative genomics among Saccharomyces cerevisiae × Saccharomyces kudriavzevii natural hybrid strains isolated from wine and beer reveals different origins. BMC Genomics 13:407. doi: 10.1186/1471-2164-13-407
Peris, D., Moriarty, R. V., Alexander, W. G., Baker, E. C., Sylvester, K., Sardi, M., et al. (2017). Hybridization and adaptive evolution of diverse Saccharomyces species for cellulosic biofuel production. Biotechnol. Biofuels 10:78. doi: 10.1186/s13068-017-0763-7
Peter, J., De Chiara, M., Friedrich, A., Yue, J. -X., Pflieger, D., Bergström, A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. doi: 10.1038/s41586-018-0030-5
Piotrowski, J. S., Nagarajan, S., Kroll, E., Stanbery, A., Chiotti, K. E., Kruckeberg, A. L., et al. (2012). Different selective pressures lead to different genomic outcomes as newly-formed hybrid yeasts evolve. BMC Evol. Biol. 12:46. doi: 10.1186/1471-2148-12-46
Pontes, A., Hutzler, M., Brito, P. H., and Sampaio, J. P. (2020). Revisiting the taxonomic synonyms and populations of Saccharomyces cerevisiae—phylogeny, phenotypes, ecology and domestication. Microorganisms 8:903. doi: 10.3390/microorganisms8060903
Preiss, R., Tyrawa, C., Krogerus, K., Garshol, L. M., and van Der Merwe, G. (2018). Traditional Norwegian Kveik are a genetically distinct group of domesticated Saccharomyces cerevisiae brewing yeasts. Front. Microbiol. 9:2137. doi: 10.3389/fmicb.2018.02137
Qiu, X., Zhang, J., Zhou, J., Fang, Z., Zhu, Z., Li, J., et al. (2019). Stress tolerance phenotype of industrial yeast: industrial cases, cellular changes, and improvement strategies. Appl. Microbiol. Biotechnol. 103, 6449–6462. doi: 10.1007/s00253-019-09993-8
Querol, A., and Bond, U. (2009). The complex and dynamic genomes of industrial yeasts. FEMS Microbiol. Lett. 293, 1–10. doi: 10.1111/j.1574-6968.2008.01480.x
Quispe, X., Tapia, S. M., Villarroel, C., Oporto, C., Abarca, V., García, V., et al. (2017). Genetic basis of mycotoxin susceptibility differences between budding yeast isolates. Sci. Rep. 7:9173. doi: 10.1038/s41598-017-09471-z
Roncoroni, M., Santiago, M., Hooks, D. O., Moroney, S., Harsch, M. J., Lee, S. A., et al. (2011). The yeast IRC7 gene encodes a β-lyase responsible for production of the varietal thiol 4-mercapto-4-methylpentan-2-one in wine. Food Microbiol. 28, 926–935. doi: 10.1016/j.fm.2011.01.002
Salinas, F., Cubillos, F. a., Soto, D., Garcia, V., Bergström, A., Warringer, J., et al. (2012). The genetic basis of natural variation in oenological traits in Saccharomyces cerevisiae. PLoS One 7:e49640. doi: 10.1371/journal.pone.0049640
Schuller, D., Valero, E., Dequin, S., and Casal, M. (2004). Survey of molecular methods for the typing of wine yeast strains. FEMS Microbiol. Lett. 231, 19–26. doi: 10.1016/S0378-1097(03)00928-5
Schwarz, L. V., Marcon, A. R., Delamare, A. P. L., Agostini, F., Moura, S., and Echeverrigaray, S. (2020). Selection of low nitrogen demand yeast strains and their impact on the physicochemical and volatile composition of mead. J. Food Sci. Technol. 57, 2840–2851. doi: 10.1007/s13197-020-04316-6
Smukowski Heil, C. S., DeSevo, C. G., Pai, D. A., Tucker, C. M., Hoang, M. L., and Dunham, M. J. (2017). Loss of heterozygosity drives adaptation in hybrid yeast. Mol. Biol. Evol. 34, 1596–1612. doi: 10.1093/molbev/msx098
Spor, A., Nidelet, T., Simon, J., Bourgais, A., De Vienne, D., and Sicard, D. (2009). Niche-driven evolution of metabolic and life-history strategies in natural and domesticated populations of Saccharomyces cerevisiae. BMC Evol. Biol. 9:296. doi: 10.1186/1471-2148-9-296
Steensels, J., Gallone, B., Voordeckers, K., and Verstrepen, K. J. (2019). Domestication of industrial microbes. Curr. Biol. 29, R381–R393. doi: 10.1016/j.cub.2019.04.025
Steensels, J., Meersman, E., Snoek, T., Saels, V., and Verstrepen, K. J. (2014a). Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 80, 6965–6975. doi: 10.1128/AEM.02235-14
Steensels, J., Snoek, T., Meersman, E., Nicolino, M. P., Voordeckers, K., and Verstrepen, K. J. (2014b). Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 38, 947–995. doi: 10.1111/1574-6976.12073
Steensels, J., and Verstrepen, K. J. (2014). Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annu. Rev. Microbiol. 68, 61–80. doi: 10.1146/annurev-micro-091213-113025
Stefanini, I., and Cavalieri, D. (2018). Metagenomic approaches to investigate the contribution of the vineyard environment to the quality of wine fermentation: potentials and difficulties. Front. Microbiol. 9:991. doi: 10.3389/fmicb.2018.00991
Sternes, P. R., Lee, D., Kutyna, D. R., and Borneman, A. R. (2017). A combined meta-barcoding and shotgun metagenomic analysis of spontaneous wine fermentation. Gigascience 6, 1–10. doi: 10.1093/gigascience/gix040
Swiegers, J. H., Kievit, R. L., Siebert, T., Lattey, K. A., Bramley, B. R., Francis, I. L., et al. (2009). The influence of yeast on the aroma of Sauvignon Blanc wine. Food Microbiol. 26, 204–211. doi: 10.1016/j.fm.2008.08.004
Swiegers, J. H., and Pretorius, I. S. (2007). Modulation of volatile sulfur compounds by wine yeast. Appl. Microbiol. Biotechnol. 74, 954–960. doi: 10.1007/s00253-006-0828-1
Tapia, S. M., Cuevas, M., Abarca, V., Delgado, V., Rojas, V., García, V., et al. (2018). GPD1 and ADH3 natural variants underlie glycerol yield differences in wine fermentation. Front. Microbiol. 9:1460. doi: 10.3389/fmicb.2018.01460
Taymaz-Nikerel, H., Cankorur-Cetinkaya, A., and Kirdar, B. (2016). Genome-wide transcriptional response of Saccharomyces cerevisiae to stress-induced perturbations. Front. Bioeng. Biotechnol. 4:17. doi: 10.3389/fbioe.2016.00017
Tesnière, C., Brice, C., and Blondin, B. (2015). Responses of Saccharomyces cerevisiae to nitrogen starvation in wine alcoholic fermentation. Appl. Microbiol. Biotechnol. 99, 7025–7034. doi: 10.1007/s00253-015-6810-z
Vakirlis, N., Monerawela, C., McManus, G., Ribeiro, O., McLysaght, A., James, T., et al. (2019). Evolutionary journey and characterisation of a novel pan-gene associated with beer strains of Saccharomyces cerevisiae. Yeast 36, 425–437. doi: 10.1002/yea.3391
Wang, Q. -M., Liu, W. -Q., Liti, G., Wang, S. -A., and Bai, F. -Y. (2012). Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol. Ecol. 21, 5404–5417. doi: 10.1111/j.1365-294X.2012.05732.x
Warringer, J., Zörgö, E., Cubillos, F. a., Zia, A., Gjuvsland, A., Simpson, J. T., et al. (2011). Trait variation in yeast is defined by population history. PLoS Genet. 7:e1002111. doi: 10.1371/journal.pgen.1002111
Will, J. L., Kim, H. S., Clarke, J., Painter, J. C., Fay, J. C., and Gasch, A. P. (2010). Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet. 6:e1000893. doi: 10.1371/journal.pgen.1000893
Zhang, H., Skelton, A., Gardner, R. C., and Goddard, M. R. (2010). Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 10, 941–947. doi: 10.1111/j.1567-1364.2010.00681.x
Keywords: wine, beer, fermentation, Saccharomyces cerevisiae, wild strains, hybrids
Citation: Molinet J and Cubillos FA (2020) Wild Yeast for the Future: Exploring the Use of Wild Strains for Wine and Beer Fermentation. Front. Genet. 11:589350. doi: 10.3389/fgene.2020.589350
Edited by:
Irene Stefanini, University of Turin, ItalyReviewed by:
Jan Steensels, Flanders Institute for Biotechnology, BelgiumVittorio Capozzi, Italian National Research Council, Italy
Copyright © 2020 Molinet and Cubillos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francisco A. Cubillos, ZnJhbmNpc2NvLmN1YmlsbG9zLnJAdXNhY2guY2w=
 Jennifer Molinet
Jennifer Molinet Francisco A. Cubillos
Francisco A. Cubillos