- 1Department of Orthopedic Surgery, Zhongnan Hospital of Wuhan University, Wuhan, China
- 2Department of Pharmacology, Wuhan University School of Basic Medical Sciences, Wuhan, China
- 3Hubei Provincial Key Laboratory of Developmentally Originated Disease, Wuhan, China
Real-time fluorescence quantitative PCR (RT-qPCR) is widely used to detect gene expression levels, and selection of reference genes is crucial to the accuracy of RT-qPCR results. Minimum Information for Publication of RT-qPCR Experiments (MIQE) proposes that using the panel of reference genes for RT-qPCR is conducive to obtaining accurate experimental results. However, the selection of the panel of reference genes for RT-qPCR in rat developing cartilage has not been well documented. In this study, we selected eight reference genes commonly used in rat cartilage from literature (GAPDH, ACTB, 18S, GUSB, HPRT1, RPL4, RPL5, and SDHA) as candidates. Then, we screened out the optimal panel of reference genes in female and male rat cartilage of fetus (GD20), juvenile (PW6), and puberty (PW12) in physiology with stability analysis software of genes expression. Finally, we verified the reliability of the selected panel of reference genes with the rat model of intrauterine growth retardation (IUGR) induced by prenatal dexamethasone exposure (PDE). The results showed that the optimal panel of reference genes in cartilage at GD20, PW6, and PW12 in physiology was RPL4 + RPL5, which was consistent with the IUGR model, and there was no significant gender difference. Further, the results of standardizing the target genes showed that RPL4 + RPL5 performed smaller intragroup differences than other panels of reference genes or single reference genes. In conclusion, we found that the optimal panel of reference genes in female and male rat developing cartilage was RPL4 + RPL5, and there was no noticeable difference before and after birth.
Introduction
Real-time fluorescence quantitative polymerase chain reaction (RT-qPCR) has the advantages of high sensitivity, strong specificity, and high throughput, so it is widely used for the detection of gene expression levels (Nolan et al., 2006; Zhang et al., 2019). Reference genes are the relatively constant genes for expression in tissues or cells, which can calibrate the experimental errors and ensure the accuracy of the experimental results. In general, researchers use reference genes for RT-qPCR analysis to quantify the relative expression of the target genes. Therefore, the selection of reference genes directly affects the accuracy of the experimental results (Bustin et al., 2009). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and actin beta (ACTB) are frequently used as reference genes in considerable experiments. However, studies found that there was no generality in reference genes (de Oliveira et al., 2012; Pinto et al., 2012), and their stability varied by tissues, treatments, and developmental stages. Therefore, using a fixed reference gene often fails to accurately reflect the gene expression levels of various tissues, periods, and treatments (Sharan et al., 2015; Arya et al., 2017). So, it is necessary to select appropriate reference genes according to the model and actual situation.
Articular cartilage is an essential component of the human and animal skeletal system. In general, rats mainly go through three developmental stages: fetal period (gestational day 20, GD20), juvenile (postnatal week 6, PW6), and puberty (PW12), each of which has different biological characteristics. As the embryo develops into a complete individual, articular cartilage also continues to develop. At various stages of cartilage development, the ratio of chondrocytes and gene expression levels will change (Cleary et al., 2015). Moreover, the expression levels of reference genes differ in treatments (Al-Sabah et al., 2016). This indicates that the stability of reference genes may also change at different developmental stages of cartilage or when the cartilage is treated differently. Besides, Minimum Information for Publication of RT-qPCR (MIQE) proposes that using a single reference gene for RT-qPCR analysis may lead to inaccurate results, and the rigorous PCR analysis needs to select a stable panel of reference genes, a combination of two or more reference genes, as a reference to relatively quantify the expression of the target genes (Bustin et al., 2009; Huggett et al., 2013). Therefore, the screening and identification of the panel of reference genes that stably express at cartilage developmental stages are the basis and premise for carrying out studies on cartilage development. However, there is no related research on which genes can be used as a panel of reference genes of articular cartilage at different developmental stages.
The stability of reference genes possibly differs in various pathological models. Therefore, it is not only necessary to explore the reference genes that are stable during the development in physiology, but also to verify under representative development-related disease models. Due to the simple operation, high incidence of intrauterine growth retardation (IUGR), and high stability of the model, the adverse environmental exposure during pregnancy has become a common pathological model for researching developmental diseases (Rahman et al., 2017; Sabra et al., 2017; Dessì et al., 2018; Zhang et al., 2018). A large number of studies have shown that a variety of prenatal adverse environmental exposures, such as exogenous exposure, malnutrition, infection, hypoxia, and stress, can increase intrauterine maternal glucocorticoid levels, resulting in a series of offspring developmental programming changes (Lesage et al., 2001; Buss et al., 2012; Pawluski et al., 2012; Cuffe et al., 2014; Correia-Branco et al., 2015; Sedaghat et al., 2015). Our previous studies also confirmed that prenatal caffeine, nicotine, and ethanol exposure could cause fetal rat maternal glucocorticoid overexposure, accompanied by IUGR and multiple organ and tissue (including cartilage) developmental disorders (Chen et al., 2007; Liang et al., 2011; He et al., 2017; Qing-Xian et al., 2020). Therefore, fetal-maternal glucocorticoid overexposure under prenatal adverse environmental exposure can be a common phenomenon. Moreover, the IUGR model induced by prenatal dexamethasone exposure (PDE) is a classic method (Tomaszewska et al., 2012; Conti et al., 2017; Chen et al., 2018; Liu et al., 2018; Xiao et al., 2018; Li et al., 2019; Wu et al., 2019). Dexamethasone, as a synthetic glucocorticoid, is easy to enter fetus through the placenta (Morales et al., 1986), so the IUGR model induced by PDE can well simulate the overexposure of glucocorticoid in prenatal adverse environments during the development of offspring rats. By screening the sable reference genes in developmental cartilage in the PDE model and verifying the reliability of reference genes screened in physiology, it can help to further improve the accuracy of research results related to cartilage development.
In this study, we first screened out the optimal panel of reference genes in rat articular cartilage at different developmental stages (GD20, PW6, and PW12) in physiology. Next, to confirm the stability and accuracy of the panel of reference genes screened above, we used PDE to construct a rat IUGR model of glucocorticoid overexposure during pregnancy and further compared the consistency of the panel of reference genes in this model with physiology. Finally, we verify the reliability of the selected panel of reference genes by detecting the expression of cartilage-specific functional genes in the IUGR model induced by PDE. This study explored the optimal panel of reference genes in the cartilage development period for the first time in physiology and PDE-induced IUGR model, providing a methodological basis for studies related to cartilage development.
Materials and Methods
Chemicals and Reagents
Dexamethasone was obtained from the Shuanghe Pharmaceutical Co. (Wuhan, China). A TRIzol kit (CAS No. 5346994) was purchased from Invitrogen Co. (Carlsbad, CA, United States). Isoflurane was obtained from Baxter Healthcare Co. (Deerfield, IL, United States). Reverse transcription and RT-qPCR kits (CAS No. R232) were obtained from Vazyme Biotech Co., Ltd. (Nanjing, China). Power SYBR® Green PCR Master Mix (CAS No. A46109) was obtained from the Thermo Fisher Scientific Co., Ltd. (Shanghai, China). All primers were synthesized by Tianyi Huiyuan Biotech Co., Ltd. (Wuhan, China). The other chemicals and agents were analytical grade.
Animals and Treatments
All experimental Specific pathogen-free Wistar rats [No. 2017–0018, license number: SCXK (Hubei), certification number: 42000600002258] with females weighing 201 ± 20 g and males weighing 270 ± 20 g were obtained from Hubei Provincial Center for Disease Control and Prevention (Hubei, China). All of the experimental animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of the Chinese Animal Welfare Committee. This study was approved by the Animal Experimental Ethics Committee of Wuhan University Medical College (license number: 14016).
The animals were kept in rat cages in an air-conditioned room under standard conditions (room temperature: 18–22°C; humidity: 40–60%; light cycle: 12 h light and dark; and 10–12 air changes per hour) and allowed free access to food and water. All rats were adaptively fed one week before experimentation, and then two female rats were placed together with one male rat overnight in a cage for mating. The vaginal discharge smear of the caged female rat was examined by a microscope the next morning, and if sperm was found, it was confirmed as GD0, as previously described (Dong et al., 2018). Pregnant rats were randomly divided into two groups after confirmation of successful conception: the control and the PDE group. From GD9 to GD20, pregnant rats in the PDE group were injected with dexamethasone (0.2 mg/kg⋅d) subcutaneously at 9 am every day, and the control group was given the same volume of saline.
At GD20, some of the pregnant rats were anesthetized with 2% isoflurane, the fetuses were removed by laparotomy under the ultra-clean platform, and eight pregnant rats with 8 to 14 offspring per litter were included in the experiment. Meanwhile, we recorded the weight and gender of the fetuses. The lower limbs of fetal rats were severed on ice and stored in an ultra-low temperature refrigerator at −80°C for extraction of the total RNA in cartilage. Subsequently, in the postnatal rat experiment, the number of female offspring in each litter was uniformly adjusted to 10 with half males and half females on the first day after birth when the pregnant rats naturally delivered, and each group accommodated eight litters. After weaning at PW4, one female and male rat were selected from each litter and placed into one group and continued to feed to juvenile (PW6), and puberty (PW12). Each group consisted of 8 rats of each sex from 8 litters (n = 8). All animals were anesthetized with 2% isoflurane at each time point, decapitated and sacrificed, and the entire lower limb was retained for extraction of the total RNA in cartilage.
Total RNA Extraction and cDNA Synthesis
The total RNA of cartilage tissue was extracted by TRIzol method following the manufacturer’s instructions. Briefly, after the cartilage (one sample came from one rat, and each group contained 8 samples, i.e., n = 8) was obtained under the ultra-clean platform and ground cartilage with a grinder (No. KZ-II, Servicebio), they were transferred into the 1.5 mL EP tube, and added 1 ml TRIzol and 200 μL chloroform. Then the sample was placed for 10 min on ice after fully mixed. After low temperature high speed centrifugation (Centrifuge 5430R, Eppendorf, Germany) at 12000 × g and 4°C for 15 min, the clarified liquid (400 μL) of the upper layer was transferred into a new EP tube, and placed for 10 min at room temperature after the same amount of isopropanol was added and mixed. The white precipitates containing total RNA were obtained after centrifugation at 12000 × g and 4°C for 10 min. Then washed it with 1 mL of precooled 75% ethanol twice and dissolved it with free RNA enzyme water. The obtained total RNA was treated with DNase to remove DNA pollution after an RNA integrity test with agarose gel electrophoresis showed that it was eligible (Supplementary Figure 1). We used NanoDrop 2000 micronuclei acid analyzer (NanoDrop 2000, Thermo Scientific, United States) to detect the concentration and purity of total RNA to ensure that the A260/A280 ratio of all samples was between 1.8 and 2.0. And we synthesized cDNA according to the instructions of HiScript III RT SuperMix for qPCR (+gDNA wiper) reagent kit. Briefly, after adding 4 μL 4 × gDNA wiper Mix to 1 μg RNA, added RNAase-free ddH2O to 16 μL. After mixing, put it into thermal cycler (Veriti Thermal Cycler): 42°C for 2 min; then added 4 μL 5 × HiScript III RT SuperMix at 37°C for 15 min and 85°C for 5 s. Finally, stored it at −20°C after dilution 1: 4.
Reference Genes Selection, Primers Design, and RT-qPCR
As shown in Supplementary Table 1, we selected eight commonly used reference genes (GAPDH, ACTB, 18S, HPRT1, RPL4, SDHA, GUSB, and RPL5) in cartilage as candidates by consulting the literature (Pombo-Suarez et al., 2008; Zhai et al., 2013; Zhou et al., 2014; Ma et al., 2015; Al-Sabah et al., 2016; He et al., 2018). We searched the complete cDNA sequence of the reference genes from NCBI’s Entrez Nucleotides database. Primer Premier 5.0 (PREMIER Biosoft International) was used for primer design, and then the designed primers were input into the BLAST database for homology comparison. Finally, the specific primer sequences were obtained (Supplementary Table 2). Next, RT-qPCR was used to detect the expression levels of the selected reference genes of rats cartilage tissues at GD20, PW6, and PW12 in each group. The RT-qPCR reaction was performed using the 2–ΔΔCt method according to the AceQ Universal SYBR qPCR Master Mix reagent instructions. The 10 μL reaction system contained 0.2 μL forward and reversed primer, 1 μL cDNA, 3.6 μL ddH2O and 5 μL SYBR. The PCR amplifications were performed using a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). The amplification curve reaction program: 50°C for 2 min, 95°C for 10 min, 95°C for 15 s, 60°C for 1 min, 40 cycles; Melting curve at 95°C for 15 min, 60°C for 30 s, and 95°C for 15 min. Finally, we collected the fluorescence signal and drew the standard curve and melting curve. We repeated the tests for three times each sample.
Standard Curve and Amplification Efficiency
The cDNA product of each sample was divided into eight gradients by four times, and each gradient was set to three replicates. Using the diluted product as the standard, we determined the standard curve of each candidate reference gene. The primer amplification efficiency (E) is related to the slope of the standard curve. According to the slope of the standard curve, the primers amplification efficiency was calculated from the equation .
Software and Statistical Analysis
geNorm, NormFinder, and BestKeeper are three commonly used reference gene stability screening software developed by different teams, and they’re based on different algorithms. GeNorm and NormFinder were, respectively, developed by Vandesompele et al. (2002) and Andersen et al. (2004), and both of them can judge the stability by calculating the stability value (M) of the reference gene. The higher the M value, the more unstable the reference gene, and vice versa. If M ≤ 1.5, it can be considered that the gene is stably expressed in this tissue and can be used as a reference. GeNorm can calculate the paired variation value V of the standardization factor after the introduction of a new reference gene to decide the number of the optimal panel of reference genes, so it can screen the most stable combination of two or more reference genes to correct the data, while NormFinder can only screen the most stable one. When Vn/Vn + 1 < K (K is generally 0.15, but it may be adjusted according to the actual situation), the number of the best panel of reference genes is n. BestKeeper is a gene expression stability analysis software developed by Michael in 2004 for reference genes and target genes by calculating the pairing coefficient (r), standard deviation (SD) and coefficient of variation (CV) produced by the pairing between each gene (Pfaffl et al., 2004). The larger the r is, the smaller the SD and CV are, and the more stable the internal reference genes are (Al-Sabah et al., 2016; Lü et al., 2018; Sarker et al., 2018). The expression level of target genes can be analyzed at the same time in BestKeeper software.
Before the experiment, the minimum animal sample size was calculated as 8 based on a power efficiency β = 0.8 and α = 0.05 for detecting a statistical difference between groups under the maximum failure load. And we used SPSS 20 (SPSS Science Inc., Chicago, IL, United States) and Prism 7 (Graph Pad Software, La Jolla, CA, United States) for data statistics and graphing. Quantitative data were expressed as the mean ± S.E.M. An unpaired two-tailed Student’s t-test was used for comparisons between control and the PDE group. A one-way analysis of variance (ANOVA) was used in BestKeeper to compare the stability of reference genes. P < 0.05 was considered statistically significant for all of the tests.
Results
The Specificity and Amplification Efficiency of the Candidate Genes’ Primers
Before the analysis of reference genes’ stability, all the candidate genes’ primers were detected for melting and standard curves. The results showed that the melting curves of all candidate reference genes’ primers displayed a single peak, and the Tm values ranged from 80 to 90°C (Supplementary Figures 2A–H), indicating that the specificity of the selected primers performed well. The R2 ≥ 0.995 of the standard curves of all candidate reference genes indicated a clear linear relationship between the template cDNA content and the Ct value. According to the slope and R2 of the standard curves, the candidate reference gene primers’ amplification efficiencies were calculated as 97% ∼ 105% (Supplementary Table 3), which were between 90 ∼ 110% recommended by MIQE (Taylor et al., 2010). Therefore, all primers meet the requirements for further experiments.
The Expression Levels and Stability Analysis of the Candidate Reference Genes in Rat Cartilage at Various Developmental Stages in Physiology
To screen the stable reference genes in cartilage at developmental stages in physiology, we used RT-qPCR to detect the expression of the above eight reference genes in cartilage of female rats at the three time-points (GD20, PW6, and PW12), and analyzed their stability. The results showed that the Ct values of candidates in female rat articular cartilage were between 8 and 23 at GD20, PW6 and PW12 (Figure 1A). At GD20 and PW12, geNorm, NormFinder, and BestKeeper stability analysis showed that RPL4, RPL5, and SDHA were the most stable reference genes, while GUSB and 18S were the most unstable reference genes (Figures 1B–D, H–J). At PW6, the three software unanimously showed that RPL4 and RPL5 had the highest stability, but GAPDH ranked third, while GUSB was also the most unstable (Figures 1E–G).
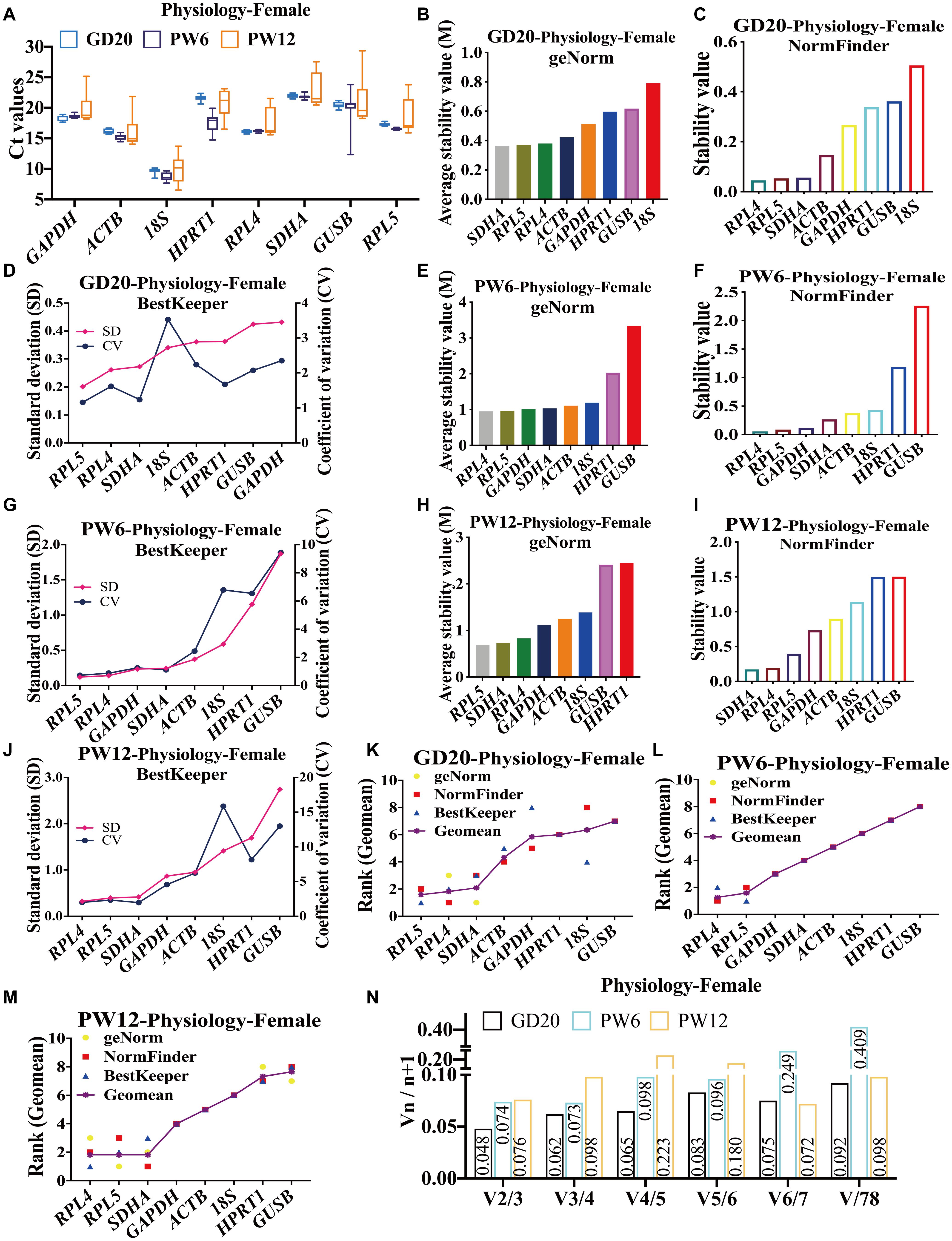
Figure 1. The candidate reference genes’ expression and stability of female rat cartilage in different physiological developmental stages. (A) Distribution of Ct values of candidate reference genes in physiological cartilage of female rat at GD20, PW6 and PW12. GeNorm, NormFinder and BestKeeper analyzed the stability of the candidate reference genes in rat cartilage at GD20 (B–D), PW6 (E–G) and PW12 (H–J) (from most stable to least stable). The comprehensive stability ranking of the candidate reference genes in rat cartilage at GD20 (K), PW6 (L), and PW12 (M) decreased from left to right. (N) Paired coefficient of variation (CV) of standardization factor’s variation after introducing a new reference gene in rat cartilage under physiological conditions at GD20, PW6, and PW12. n = 8.
In order to further determine the comprehensive stability of the candidate reference genes at various developmental stages (GD20, PW6, and PW12) in female rat cartilage, we calculated the geometric mean of the above three software for ranking of each reference gene. The results showed that under physiological conditions at GD20, the comprehensive gene ranking of the most stable to least stable genes in male rat cartilage was RPL5, RPL4, SDHA, ACTB, GAPDH, HPRT1, 18S, GUSB (Figure 1K). At PW6, it was RPL4, RPL5, GAPDH, SDHA, ACTB, 18S, HPRT1, GUSB (Figure 1L). And at PW12, the ranking was RPL4, RPL5, SDHA, ACTB, 18S, HPRT1, GUSB (Figure 1M). GeNorm further calculated the paired coefficients of standardization factor’s variation after introducing a new reference gene for the three periods and showed that all V2/3 were less than 0.15 (Figure 1N), indicating that the number of the optimal panel of reference genes in female rat cartilage under physiological conditions was two. Therefore, RPL4 and RPL5 could be used as the optimal panel of reference genes in female rats’ cartilage at the development stages under physiological conditions.
In male rats, the Ct values of all the candidate reference genes ranged from 8 to 23 at GD20, PW6 and PW12 (Figure 2A). At GD20, geNorm and NormFinder showed that RPL4 and RPL5 were the most stable reference genes, and GUSB and 18S were the most unstable (Figures 2B,C). BestKeeper showed that RPL5 and ACTB were the top two, and RPL4 was the fifth (Figure 2D). At PW6, geNorm and BestKeeper showed that the stability of RPL4, RPL5, and SDHA ranked in the top three (Figures 2E,G). However, the NormFinder showed that ACTB had the highest stability, and RPL5, SDHA, and RPL4 were ranked 2 to 4 (Figure 2F). Moreover, all believed that 18S and HPRT1 had the worst stability (Figures 2E–G). At PW12, geNorm, NormFinder and BestKeeper all showed that RPL4 and RPL5 were also the most stable reference genes (Figures 2H–J); geNorm and NormFinder showed that GUSB and HPRT1 had the worst stability (Figures 2H,I), while BestKeeper thought it was ACTB (Figure 2J).
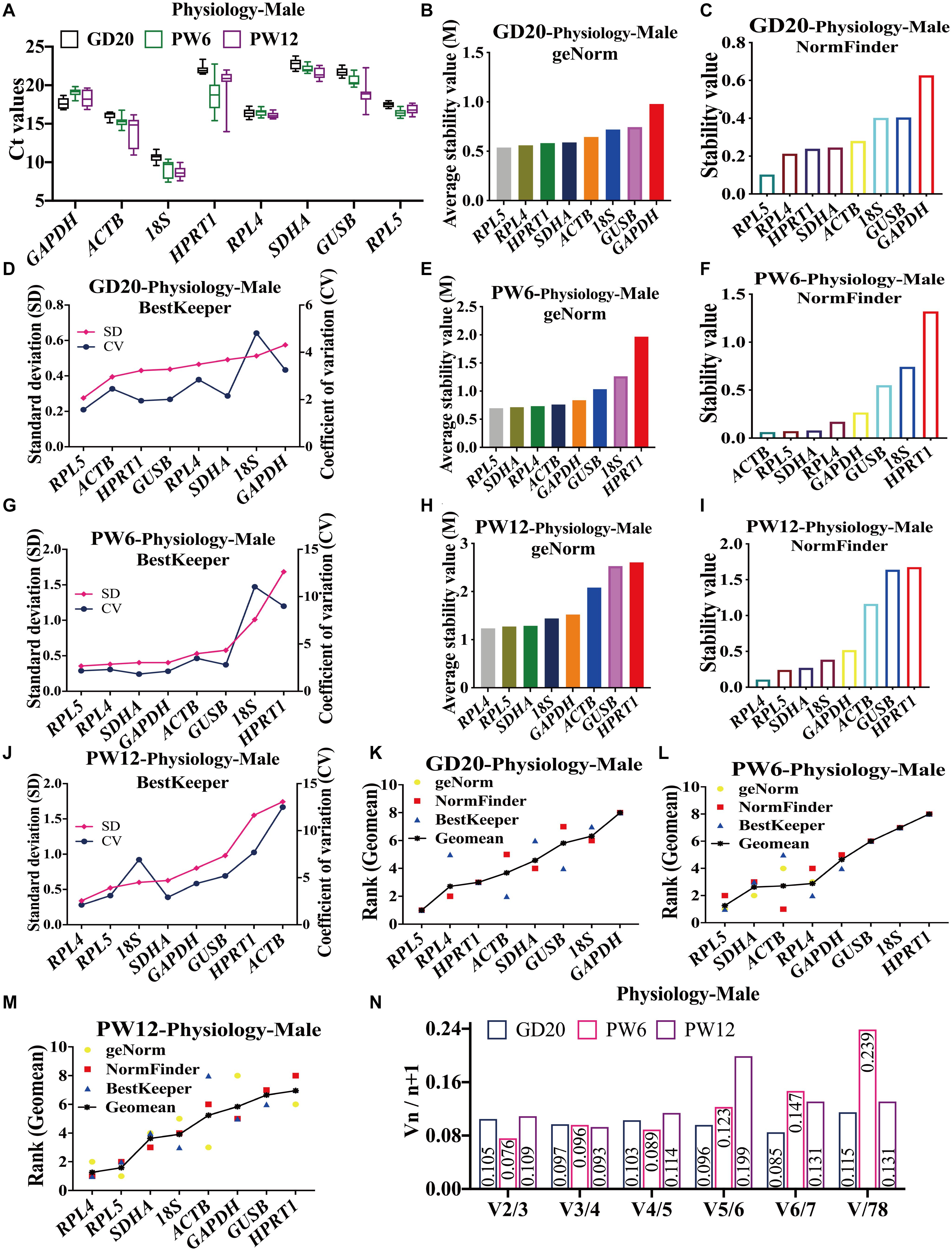
Figure 2. The candidate reference genes’ expression and stability of male rat cartilage in different physiological developmental stages. (A) Distribution of Ct values of the candidate reference genes in physiological cartilage of male rat at GD20, PW6, and PW12. GeNorm, NormFinder and BestKeeper analyzed the stability of the candidate reference genes in male rat cartilage at GD20 (B–D), PW6 (E–G), and PW12 (H–J) (from most stable to least stable). The comprehensive stability ranking of the candidate reference genes in male rat cartilage at GD20 (K), PW6 (L), and PW12 (M) decreased from left to right. (N) Paired coefficient of variation (CV) of standardization factor’s variation after introducing a new reference gene in female rat cartilage under physiological conditions at GD20, PW6, and PW12. n = 8.
Furthermore, under the physiological conditions at GD20, the comprehensive gene ranking in male rat cartilage was RPL5, RPL4, HPRT1, ACTB, SDHA, GUSB, 18S, GAPDH (Figure 2K). At PW6, the ranking was RPL5, SDHA, ACTB, RPL4, GAPDH, GUSB, 18S, HPRT1 (Figure 2L). And at PW12, it was RPL4, RPL5, SDHA, 18S, ACTB, GAPDH, GUSB, HPRT1 (Figure 2M). The same as females, RPL4 and RPL5 still had the highest stability at GD20 and PW12. However, at PW6, RPL5 had the highest stability, while RPL4 ranked fourth. But RPL4 was also suitable for being a reference gene for its stability value M < 1.5. V2/3 was also less than 0.15 (Figure 2N), which indicated that the number of the optimal panel of reference genes in male rat cartilage under physiological conditions was also two. In summary, RPL4 and RPL5 were still the optimal panel of reference genes in male rat cartilage in physiology.
The Expression Levels and Stability Analysis of the Candidate Reference Genes in Rat Cartilage at Various Developmental Stages in the PDE-Induced IUGR Model
To further verify the stability of reference genes screened under pathological conditions, we selected the PDE-induced IUGR rat model for similar analysis. The results showed that in female rats in the PDE-induced IUGR model, the average Ct values of the eight candidate genes were between 8 and 23 at GD20, PW6 and PW12 (Figure 3A). At GD20, both geNorm and NormFinder indicated that RPL4, RPL5, and SDHA were the most stable reference genes (Figures 3B,C), and BestKeeper showed that the stability of RPL4, GAPDH, and RPL5 ranked in the top three (Figure 3D), while all the three software showed that GUSB and HPRT1 had the worst stability (Figures 3B–D). At PW6, geNorm and NormFinder also believed that SDHA, RPL4, and RPL5 were ranked top three for stability (Figures 3E,F), while BestKeeper thought that SDHA, RPL5, and GAPDH were ranked top three, and RPL4 was ranked fourth (Figure 3G). GUSB and HPRT1 were also the most unstable at PW6 (Figures 3E–G). At PW12, geNorm showed that the stability of RPL5 and RPL4 were also the top two (Figure 3H). But NormFinder and BestKeeper believed that the stability of ACTB and RPL5 were the top two, while RPL4 ranked 8th and 7th (Figures 3I,J). The different results from the three software may attribute to the algorithm.
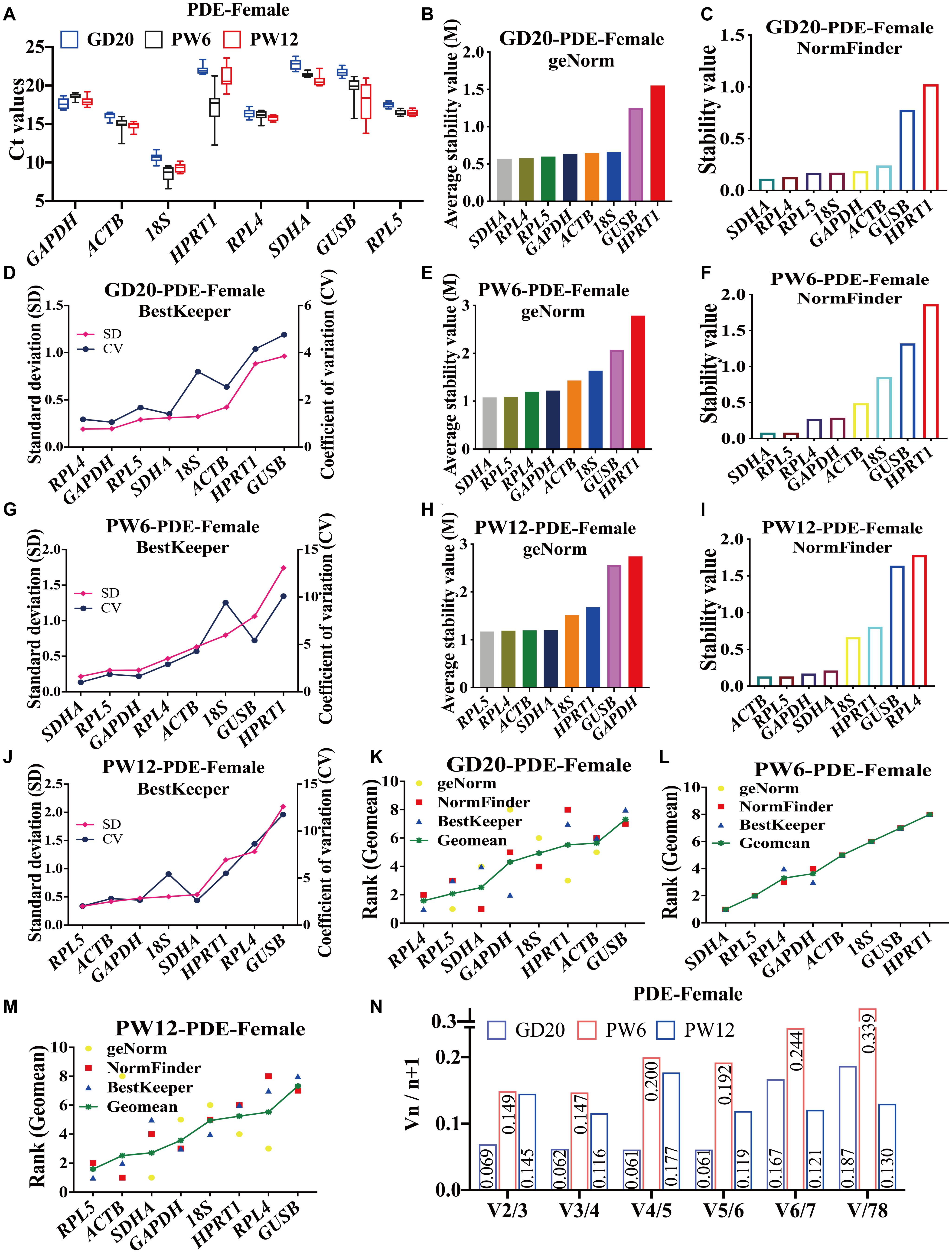
Figure 3. The candidate reference genes’ expression and stability of female rat cartilage in different developmental stages in the PDE-induced IUGR model. (A) Distribution of Ct values of the candidate reference genes in female rat cartilage of PDE model at GD20, PW6, and PW12. GeNorm, NormFinder and BestKeeper analyzed the stability of the candidate reference genes in female rat cartilage of PDE model at GD20 (B–D), PW6 (E–G), and PW12 (H–J) (from most stable to least stable). The comprehensive stability ranking of the candidate reference genes in female rat cartilage at GD20 (K), PW6 (L), and PW12 (M) decreased from left to right. (N) Paired coefficient of variation (CV) of standardization factor’s variation after introducing a new reference gene in female rat cartilage under PDE model at GD20, PW6, and PW12. n = 8.
At GD20, the comprehensive gene ranking in male rat cartilage in the PDE-induced IUGR model was RPL4, RPL5, SDHA, GAPDH, 18S, HPRT1, ACTB, GUSB (Figure 3K); at PW6, the ranking was SDHA, RPL5, RPL4, GAPDH, ACTB, 18S, GUSB, HPRT1 (Figure 3L); and at PW12, it was RPL5, ACTB, SDHA, GAPDH, 18S, HPRT1, RPL4, GUSB (Figure 3M). GeNorm calculated that V2/3 was less than 0.15 (Figure 3N). In summary, at GD20 and PW6, RPL4 and RPL5 were also the best panel of reference genes. However, at PW12, due to the differences in the algorithms of the three software, the results of the comprehensive ranking were different. If only the geNorm ranking was considered at PW6, RPL4 and RPL5 could still be used as the best panel of reference genes in male rat cartilage in the PDE-induced IUGR model.
In male rat cartilage of the PDE-induced IUGR model, all the candidate reference genes showed Ct values also ranged from 8 to 23 at GD20, PW6, and PW12 (Figure 4A). At GD20, the stability analysis of geNorm and NormFinder showed that RPL4 and RPL5 were first and second, respectively, and 18S and GUSB had the worst stability (Figures 4B,C). BestKeeper believed that the stability of RPL4 and GAPDH was in the top two, and RPL5 was ranked third (Figure 4D). At PW6, although the ranking was different, geNorm, NormFinder, and BestKeeper all showed that the stability of RPL4, RPL5, and SDHA was still in the top three. At the same time, GUSB and HPRT1 were the most unstable (Figures 4E–G). At PW12, both geNorm and NormFinder believed that the stability of SDHA and RPL5 was the top two (Figures 4H,I), but BestKeeper considered 18S and RPL4 as the top two, while RPL5 and SDHA, respectively ranked third and fourth (Figure 4J). And all three software considered ACTB to be the most unstable (Figures 4H–J).
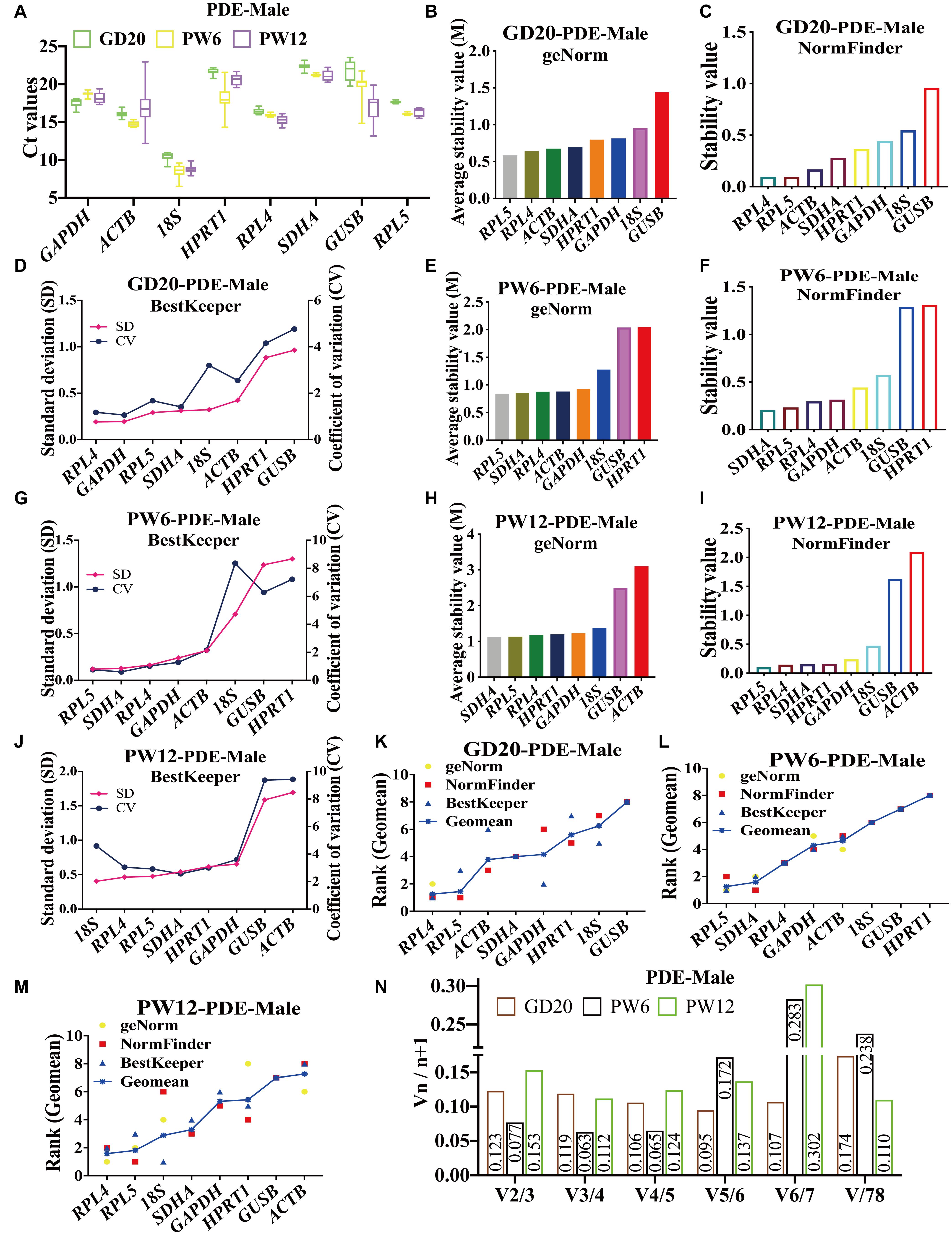
Figure 4. The candidate reference genes’ expression and stability of male rat cartilage in different developmental stages in the PDE model. (A): Distribution of Ct values of the candidate reference genes in cartilage at GD20, PW6 and PW12. GeNorm, NormFinder and BestKeeper analyzed the stability of the candidate reference genes in male rat cartilage at GD20 (B–D), PW6 (E–G) and PW12 (H–J) (from most stable to least stable). The comprehensive stability ranking of the candidate reference genes in the male rat cartilage in PDE model at GD20 (K), PW6 (L), and PW12 (M) decreased from left to right. (N) Paired coefficient of variation (CV) of standardization factor’s variation after introducing a new reference gene in the male rat cartilage in PDE model at GD20, PW6, and PW12. n = 8.
In the PDE-induced IUGR model, at GD20, the comprehensive ranking in male rat cartilage was RPL4, RPL5, ACTB, SDHA, GAPDH, HPRT1, 18S, GUSB (Figure 4K); at PW6, it was RPL5, SDHA, RPL4, GAPDH, ACTB, 18S, GUSB, HPRT1 (Figure 4L); at PW12, it was RPL4, RPL5, 18S, SDHA, GAPDH, HPRT1, GUSB, ACTB (Figure 4M). The same as female rats, geNorm calculated that V2/3 in all three periods was also less than 0.15 (Figure 4N). In summary, at GD20 and PW12, RPL4 and RPL5 had the highest stability. While at PW6, RPL5, and SDHA had the highest stability, and RPL4 ranked third. If the stability of the reference gene and the simplicity and economy of the experiment were considered comprehensively, RPL4 and RPL5 could still be selected as the panel of reference genes at PW6. As a result, RPL4 and RPL5 were always the optimal panel of reference genes in male rats under the PDE-induced IUGR model.
Verification of Stability and Accuracy of Cartilage Reference Genes in the PDE-Induced IUGR Model
To further verify the stability and reliability of the screened panel of reference genes, we used the PDE-induced IUGR rat model to confirm the influence of using a single-reference gene or panel of reference genes on the mRNA expression by standardizing cartilage functional gene ACAN at GD20, PW6, and PW12. A large number of early animal and cell experiments had shown that the matrix content of cartilage in each of the above periods decreased significantly in the PDE-induced IUGR model, and the expression levels of ACAN mRNA and protein were also considerably reduced (Iwaniak et al., 2016; Chen et al., 2018; Xiao et al., 2020). The verification results showed that when normalized with the two single reference genes screened above (RPL4 and RPL5), the mRNA expression level of ACAN was significantly reduced in cartilage of male and female rats in the above three periods (P = 0.084, P < 0.05, P < 0.01; Figures 5A–C). It should be noted that the mRNA expression of ACAN in rat cartilage standardized by SDHA was also significantly reduced (P = 0.069, P < 0.05, P < 0.01; Supplementary Figure 3A). However, except for the decrease of ACAN mRNA expression in female rats cartilage standardized by GAPDH and ACTB (P < 0.05, P < 0.01; Figures 5D,E), there was no significant change in the expression of ACAN mRNA when normalized with other single reference genes (ACTB, 18S, HPRT1, GAPDH, and GUSB) (Figures 5D–F and Supplementary Figure 3B). It showed that the target gene expression could also show statistical differences when GAPDH and ACTB were used for standardization, but RPL4 and RPL5 had higher stability and reliability than GAPDH and ACTB.
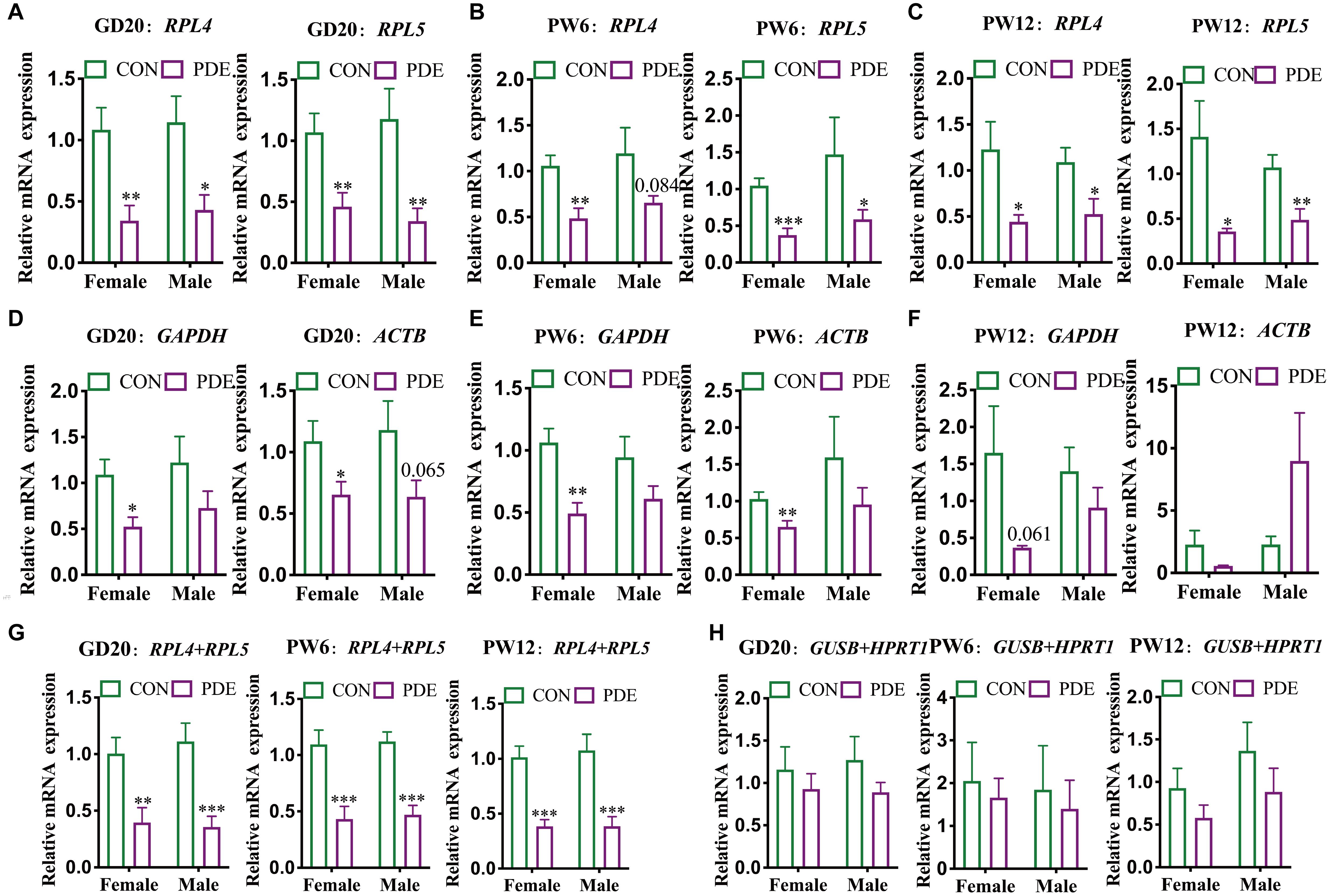
Figure 5. Different reference genes normalized ACAN mRNA expression in cartilage in different developmental periods. RPL4 and RPL5 normalized ACAN mRNA expression in cartilage at GD20 (A), PW6 (B), and PW12 (C). GAPDH and ACTB normalized ACAN mRNA expression in cartilage at GD20 (D), PW6 (E), and PW12 (F). RPL4 + RPL5 (G) and GUSB + HPRT1 (H), respectively, normalized ACAN mRNA expression in cartilage at GD20, PW6, and PW12. The values were the means ± S.E.M, n = 8. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. control.
Besides, we further verified the mRNA expression levels of ACAN with the panel of reference genes. When the two reference genes (RLP4 + RPL5) screened above were used as the panel of reference genes, the mRNA expression levels of ACAN in the cartilage of male and female rats in all the three stages were significantly reduced (P < 0.01, P < 0.001; Figure 5G). Because of the relative stability of SDHA indicated from the above results, we also verified that when RPL4 + SDHA and RPL5 + SDHA were used as the panel of reference genes, the mRNA expression levels of ACAN in the cartilage of male and female rats in the three periods were also significantly reduced (P < 0.01, P < 0.001; Supplementary Figures 4A–C). When other panels of reference genes (GUSB + HPRT1; RPL4 + HPRT1) were selected, there were no significant changes except for the reduction of ACAN mRNA expression in the cartilage of female rats normalized by RPL4 + HPRT1 at PW12 (P < 0.05; Figure 5H and Supplementary Figure 4D).
Discussion
In this study, we used the three common gene expression stability analysis software to screen the optimal panel of reference genes (Al-Sabah et al., 2016; Lü et al., 2018; Sarker et al., 2018). The results showed that under physiological conditions, at the three stages of rat articular cartilage development (GD20, PW6, and PW12), the expression of 18S was the highest, and the expression of SDHA was the lowest. All the three software showed that RPL4, RPL5, and SDHA had high stability, but there were specific differences, which may be related to the different algorithms used by each software. After comprehensively sorting the results of the three software, we found that RPL4 and RPL5 were the most stable genes at GD20, PW6, and PW12 in female rat cartilage and at GD20 and PW12 in male under physiological conditions. However, at PW6 in male rats cartilage, RPL5 and SDHA had the highest stability, and RPL4 ranked fourth, which was different from females. Although the reason for the gender difference in RPL4 stability at PW6 is unknown, RPL4 can still be used as a reference gene due to its M < 0.15. In addition, although the stability of SDHA and ACTB in male rat cartilage at PW6 ranked in the top three, the stability in other groups was worse, and the expression levels of SDHA was lower. Considering the stability of the reference genes and the simplicity and economy of the experiments, SDHA and ACTB were not the best reference genes.
It was worth noting that under physiological conditions, SDHA was quite stable in male and female rat cartilage at all development stages. The reason for that, maybe, the cartilage belongs to a hypoxic tissue whose metabolism process is mainly anaerobic respiration, but the succinate ubiquinone oxidoreductase encoded by the SDHA is the main component in the mitochondrial respiratory chain (Lane et al., 1977; Holzer et al., 2019; Zhao et al., 2019). However, in this study, the comprehensive stability of RPL4 and RPL5 is still higher than that of SDHA, which may be related to RPL4 and RPL5 as ribosomal protein-coding genes. A large number of studies have shown that the expression of various ribosomal protein genes in tissues is very stable, and it has been widely used as a reference gene for RT-qPCR in human, animal and plant (Al-Sabah et al., 2016; Mitra et al., 2016; Sadangi et al., 2017; Xiang et al., 2018). Ribosomal protein is a crucial component of the basic physiological process in the cell, which is the key to maintaining cell development and tissue homeostasis (Robledo et al., 2008; Perucho et al., 2014; Cedraz de Oliveira et al., 2017). Therefore, to maintain the basic physiological function of cells, the expression of ribosomal protein is often quite stable in various tissues and developmental stages. Wang et al. considered that ribosomal protein coding gene could replace the traditional reference gene as a commonly used candidate in RT-qPCR assay (Wan et al., 2011). RPL4 and RPL5 encode the component proteins of ribosomal 60S large subunit (Pillet et al., 2015; Hofman et al., 2017). Studies from Kolkova et al. (2013) and Brattelid et al. (2010) have shown that RPL4 is the most stable reference gene in ovarian tumors and rat myocardium. Al-Sabah also pointed out that RPL4 is also one of the most stable reference genes in articular cartilage under mechanical stress (Al-Sabah et al., 2016). At the same time, there are still a large number of studies showing that RPL5 is the most stable reference gene in various tissues, such as murine cornea (Ren et al., 2010), right heart failure tissue from human (Li et al., 2017), breast muscle of chicken (Marciano et al., 2020) and broilers cartilage (Hul et al., 2020). The above results are consistent with our results, indicating that RPL4 and RPL5 have high stability for RT-qPCR assay in a variety of tissues, including cartilage in the development. Besides, in this study, geNorm calculated that V2/3 in all three periods in female and male rat cartilage was less than 0.15, which indicated that two reference genes could be selected as the optimal panel of reference genes. In summary, under physiological conditions, there was no significant difference in the optimal panel of reference genes in male and female rat cartilage between in utero (GD20) and postnatal (PW6, PW12). The expression levels of RPL4 and RPL5 were quite stable in the articular cartilage of male and female rats at all stages of development. They could be used as the optimal panel of reference genes.
To further verify the reliability of the selected reference genes in physiology, we detect the expression of the candidate reference genes in the cartilage development of male and female rats in the PDE-induced IUGR model which is a common experimental model to study cartilage development-derived diseases (Rowas et al., 2012; Tie et al., 2016; Xie et al., 2018; Cai et al., 2019). The results showed that RPL4 and RPL5 were also the optimal panel of reference genes in female rat cartilage at GD20 and PW6, which was consistent with physiological conditions; at PW12, geNorm also believed that RPL4 and RPL5 were the most stable reference genes in female rat cartilage, but NormFinder and Bestkeeper believed that RPL5 and ACTB were the most stable reference genes, while RPL4 was relatively unstable. The difference may be due to the different algorithms of the three software. Therefore, if only geNorm ranking was considered, RPL4 and RPL5 were still the optimal panel of reference genes in female rat cartilage at PW12. In the male rats of PDE model, RPL4 and RPL5 still had the highest stability at GD20 and PW12 and were the optimal panel of reference genes. However, at PW6, RPL5 and SDHA had the highest stability, but SDHA expression was low, and RPL4 ranked third. Also, considering the stability of the reference gene and the simplicity and economy of the experiments, RPL4 and RPL5 were still selected as the most proper panel of reference genes in the cartilage of male rats at PW6. In summary, consistent with physiological conditions, RPL4 and RPL5 were still the best panel of reference genes in the articular cartilage of male and female rats at developmental stages in the PDE model. It suggested that the reference gene selected in the cartilage at the developmental stages in physiology was still suitable for the PDE-induced IUGR rat model. Because fetal-maternal glucocorticoid overexposure may be a common mechanism of IUGR and multiple organ developmental disorders caused by the poor environment during pregnancy (Lesage et al., 2001; Chen et al., 2007; Liang et al., 2011; Buss et al., 2012; Pawluski et al., 2012; Xu et al., 2012; Cuffe et al., 2014; Correia-Branco et al., 2015; Sedaghat et al., 2015; He et al., 2017), the panel of reference genes selected under the PDE-induced rats IUGR model may also be applicable to rats IUGR models induced by other factors, but further verification is required.
Minimum Information for Publication of RT-qPCR Experiments proposes that a single reference gene to standardize the target genes will affect the accuracy of the results (Huggett et al., 2013). Therefore, to ensure the reliability and rigor of RT-qPCR analysis, it is of considerable significance to screen the stable panel of reference genes in rat developmental periods and verify their reliability. Previous studies showed that the matrix content of cartilage in each of the above periods decreased significantly in the PDE model, and the mRNA and protein expression levels of ACAN were also significantly reduced (Iwaniak et al., 2016; Chen et al., 2018; Xiao et al., 2020). To further verify the reliability of reference genes screened above, we used different reference genes to standardize the mRNA expression of ACAN in offspring rat cartilage in the PDE model. The results showed that the mRNA expression levels of ACAN were significantly reduced using RPL4 and RPL5 to normalize the mRNA expression. When using GAPDH and ACTB commonly used previously, the ACAN expression only decreased in some groups, indicating that their stability was not equal to RPL4 and RPL5. By other single reference genes (GAPDH, ACTB, 18S, GUSB, and HPRT1), there were no significant differences. Further, using a panel of reference genes for normalization, the results showed that the expression levels of ACAN in male and female rat cartilage at various developmental stages were more considerably reduced when we used RLP4 + RPL5 than a single reference gene. And there was no obvious difference by other panel of reference genes (GUSB + HPRT1; RPL4 + HPRT1), but they were better than their respective single reference genes. These results suggested that selecting the optimal panel of reference genes as a reference could reduce the differences within the group and was more conducive to obtaining a stable and accurate experimental conclusion.
Conclusion
In summary, the present study screened and determined that the optimal panel of reference genes was RPL4 + RPL5 in the developmental period of male and female rats by detecting the expression of the candidate reference genes in the cartilage at different developmental stages (GD20, PW6, and PW12) in physiology and PDE-induced IUGR model. Furthermore, it was confirmed that there was no noticeable difference in gender and selection of the optimal panel of reference genes in the cartilage before and after birth. The present study provided a theoretical and experimental basis for further research on developmental cartilage diseases.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Animal Experimental Ethics Committee of Wuhan University Medical College.
Author Contributions
LL, QL, HH, MC, and SZ designed and performed the experiments. LC and HW analyzed the data. LL, HH, and HW were involved in writing the manuscript and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81972036, 81673490, 81673524, and 81703241), the National Key Research and Development Program of China (Grant Nos. 2020YFA0803900 and 2017YFC1001300), the Major Technological Innovation Projects of Hubei Province (Grant No. 2019ACA140), and Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (Grant No. TFJC2018001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members from Hubei Provincial Key Laboratory of Developmentally Originated Disease for their help in animal experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.590124/full#supplementary-material
Abbreviations
18S: 18s ribosomal RNA; ACAN: aggrecan; ACTB: actin beta; Ct: circulation threshold; CV: coefficient of variation; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; GD: gestational day; GUSB: beta-glucuronidase; HPRT1: phosphoribosyltransferase 1; IUGR: intrauterine growth retardation; MIQE: Minimum Information for Publication of RT-qPCR; mRNA: messenger RNA; PDE: prenatal dexamethasone exposure; PW: postnatal week; RPL4: ribosomal protein L4; RPL5: ribosomal protein L5; RT-qPCR: real-time fluorescence quantitative polymerase chain reaction; SD: standard deviation; SDHA: complex flavoprotein subunit A.
References
Al-Sabah, A., Stadnik, P., Gilbert, S. J., Duance, V. C., and Blain, E. J. (2016). Importance of reference gene selection for articular cartilage mechanobiology studies. Osteoarthritis Cartilage 24, 719–730. doi: 10.1016/j.joca.2015.11.007
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.Can-04-0496
Arya, S. K., Jain, G., Upadhyay, S. K., Sarita, Singh, H., Dixit, S., et al. (2017). Reference genes validation in Phenacoccus solenopsis under various biotic and abiotic stress conditions. Sci. Rep. 7:13520. doi: 10.1038/s41598-017-13925-9
Brattelid, T., Winer, L. H., Levy, F. O., Liestøl, K., Sejersted, O. M., and Andersson, K. B. (2010). Reference gene alternatives to Gapdh in rodent and human heart failure gene expression studies. BMC Mol. Biol. 11:22. doi: 10.1186/1471-2199-11-22
Buss, C., Entringer, S., and Wadhwa, P. D. (2012). Fetal programming of brain development: intrauterine stress and susceptibility to psychopathology. Sci. Signal. 5:pt7. doi: 10.1126/scisignal.2003406
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cai, X., Gao, L., Cucchiarini, M., and Madry, H. (2019). Association of nicotine with osteochondrogenesis and osteoarthritis development: the state of the art of preclinical research. J. Clin. Med. 8:1699. doi: 10.3390/jcm8101699
Cedraz de Oliveira, H., Pinto Garcia, A. A. J., Gonzaga Gromboni, J. G., Vasconcelos Farias, Filho, R., Souza, et al. (2017). Influence of heat stress, sex and genetic groups on reference genes stability in muscle tissue of chicken. PLoS One 12:e0176402. doi: 10.1371/journal.pone.0176402
Chen, M., Wang, T., Liao, Z. X., Pan, X. L., Feng, Y. H., and Wang, H. (2007). Nicotine-induced prenatal overexposure to maternal glucocorticoid and intrauterine growth retardation in rat. Exp. Toxicol. Pathol. 59, 245–251. doi: 10.1016/j.etp.2007.05.007
Chen, Z., Zhao, Z., Li, Y., Zhang, X., Li, B., Chen, L., et al. (2018). Course-, dose-, and stage-dependent toxic effects of prenatal dexamethasone exposure on fetal articular cartilage development. Toxicol. Lett. 286, 1–9. doi: 10.1016/j.toxlet.2018.01.008
Cleary, M. A., van Osch, G. J., Brama, P. A., Hellingman, C. A., and Narcisi, R. (2015). FGF, TGFβ and Wnt crosstalk: embryonic to in vitro cartilage development from mesenchymal stem cells. J. Tissue Eng. Regen. Med. 9, 332–342. doi: 10.1002/term.1744
Conti, M., Spulber, S., Raciti, M., and Ceccatelli, S. (2017). Depressive-like phenotype induced by prenatal dexamethasone in mice is reversed by desipramine. Neuropharmacology 126, 242–249. doi: 10.1016/j.neuropharm.2017.09.015
Correia-Branco, A., Keating, E., and Martel, F. (2015). Maternal undernutrition and fetal developmental programming of obesity: the glucocorticoid connection. Reprod. Sci. 22, 138–145. doi: 10.1177/1933719114542012
Cuffe, J. S., Walton, S. L., Singh, R. R., Spiers, J. G., Bielefeldt-Ohmann, H., Wilkinson, L., et al. (2014). Mid- to late term hypoxia in the mouse alters placental morphology, glucocorticoid regulatory pathways and nutrient transporters in a sex-specific manner. J. Physiol. 592, 3127–3141. doi: 10.1113/jphysiol.2014.272856
de Oliveira, L. A., Breton, M. C., Bastolla, F. M., Camargo Sda, S., Margis, R., Frazzon, J., et al. (2012). Reference genes for the normalization of gene expression in eucalyptus species. Plant Cell Physiol. 53, 405–422. doi: 10.1093/pcp/pcr187
Dessì, A., Corona, L., Pintus, R., and Fanos, V. (2018). Exposure to tobacco smoke and low birth weight: from epidemiology to metabolomics. Expert Rev. Proteomics 15, 647–656. doi: 10.1080/14789450.2018.1505508
Dong, W., Xu, D., Hu, Z., He, X., Guo, Z., Jiao, Z., et al. (2018). Low-functional programming of the CREB/BDNF/TrkB pathway mediates cognitive impairment in male offspring after prenatal dexamethasone exposure. Toxicol. Lett. 283, 1–12. doi: 10.1016/j.toxlet.2017.10.020
He, T., Huang, Y., Chak, J. C., and Klar, R. M. (2018). Recommendations for improving accuracy of gene expression data in bone and cartilage tissue engineering. Sci. Rep. 8:14874. doi: 10.1038/s41598-018-33242-z
He, Z., Lv, F., Ding, Y., Zhu, C., Huang, H., Zhang, L., et al. (2017). Insulin-like growth factor 1 mediates adrenal development dysfunction in offspring rats induced by prenatal food restriction. Arch. Med. Res. 48, 488–497. doi: 10.1016/j.arcmed.2017.11.013
Hofman, I. J. F., van Duin, M., De Bruyne, E., Fancello, L., Mulligan, G., Geerdens, E., et al. (2017). RPL5 on 1p22.1 is recurrently deleted in multiple myeloma and its expression is linked to bortezomib response. Leukemia 31, 1706–1714. doi: 10.1038/leu.2016.370
Holzer, T., Probst, K., Etich, J., Auler, M., Georgieva, V. S., Bluhm, B., et al. (2019). Respiratory chain inactivation links cartilage-mediated growth retardation to mitochondrial diseases. J. Cell Biol. 218, 1853–1870. doi: 10.1083/jcb.201809056
Huggett, J. F., Foy, C. A., Benes, V., Emslie, K., Garson, J. A., Haynes, R., et al. (2013). The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 59, 892–902. doi: 10.1373/clinchem.2013.206375
Hul, L. M., Ibelli, A. M. G., Peixoto, J. O., Souza, M. R., Savoldi, I. R., Marcelino, D. E. P., et al. (2020). Reference genes for proximal femoral epiphysiolysis expression studies in broilers cartilage. PLoS One 15:e0238189. doi: 10.1371/journal.pone.0238189
Iwaniak, P., Dobrowolski, P., Tomaszewska, E., Hułas-Stasiak, M., Tomczyk, A., and Gawron, A. (2016). The influence of dexamethasone administered prenatally on cartilage of newborn spiny mouse (Acomys cahirinus) offspring. J. Dev. Orig. Health Dis. 7, 298–305. doi: 10.1017/s2040174415007874
Kolkova, Z., Arakelyan, A., Casslén, B., Hansson, S., and Kriegova, E. (2013). Normalizing to GADPH jeopardises correct quantification of gene expression in ovarian tumours - IPO8 and RPL4 are reliable reference genes. J. Ovarian Res. 6, 60. doi: 10.1186/1757-2215-6-60
Lane, J. M., Brighton, C. T., and Menkowitz, B. J. (1977). Anaerobic and aerobic metabolism in articular cartilage. J. Rheumatol. 4, 334–342.
Lesage, J., Blondeau, B., Grino, M., Breant, B., and Dupouy, J. P. (2001). Maternal undernutrition during late gestation induces fetal overexposure to glucocorticoids and intrauterine growth retardation, and disturbs the hypothalamo-pituitary adrenal axis in the newborn rat. Endocrinology 142, 1692–1702. doi: 10.1210/endo.142.5.8139
Li, B., Zhu, Y., Chen, H., Gao, H., He, H., Zuo, N., et al. (2019). Decreased H3K9ac level of AT2R mediates the developmental origin of glomerulosclerosis induced by prenatal dexamethasone exposure in male offspring rats. Toxicology 411, 32–42. doi: 10.1016/j.tox.2018.10.013
Li, M., Rao, M., Chen, K., Zhou, J., and Song, J. (2017). Selection of reference genes for gene expression studies in heart failure for left and right ventricles. Gene 620, 30–35. doi: 10.1016/j.gene.2017.04.006
Liang, G., Chen, M., Pan, X. L., Zheng, J., and Wang, H. (2011). Ethanol-induced inhibition of fetal hypothalamic-pituitary-adrenal axis due to prenatal overexposure to maternal glucocorticoid in mice. Exp. Toxicol. Pathol. 63, 607–611. doi: 10.1016/j.etp.2010.04.015
Liu, M., Chen, B., Pei, L., Zhang, Q., Zou, Y., Xiao, H., et al. (2018). Decreased H3K9ac level of StAR mediated testicular dysplasia induced by prenatal dexamethasone exposure in male offspring rats. Toxicology 408, 1–10. doi: 10.1016/j.tox.2018.06.005
Lü, J., Chen, S., Guo, M., Ye, C., Qiu, B., Wu, J., et al. (2018). Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 9:1614. doi: 10.3389/fphys.2018.01614
Ma, H., Yang, Q., Li, D., and Liu, J. (2015). Validation of suitable reference genes for quantitative polymerase chain reaction analysis in rabbit bone marrow mesenchymal stem cell differentiation. Mol. Med. Rep. 12, 2961–2968. doi: 10.3892/mmr.2015.3776
Marciano, C. M. M., Ibelli, A. M. G., Peixoto, J. O., Savoldi, I. R., do Carmo, K. B., Fernandes, L. T., et al. (2020). Stable reference genes for expression studies in breast muscle of normal and white striping-affected chickens. Mol. Biol. Rep. 47, 45–53. doi: 10.1007/s11033-019-05103-z
Mitra, T., Bilic, I., Hess, M., and Liebhart, D. (2016). The 60S ribosomal protein L13 is the most preferable reference gene to investigate gene expression in selected organs from turkeys and chickens, in context of different infection models. Vet. Res. 47:105. doi: 10.1186/s13567-016-0388-z
Morales, W. J., Diebel, N. D., Lazar, A. J., and Zadrozny, D. (1986). The effect of antenatal dexamethasone administration on the prevention of respiratory distress syndrome in preterm gestations with premature rupture of membranes. Am. J. Obstet. Gynecol. 154, 591–595. doi: 10.1016/0002-9378(86)90607-1
Nolan, T., Hands, R. E., and Bustin, S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582. doi: 10.1038/nprot.2006.236
Pawluski, J. L., Brain, U. M., Underhill, C. M., Hammond, G. L., and Oberlander, T. F. (2012). Prenatal SSRI exposure alters neonatal corticosteroid binding globulin, infant cortisol levels, and emerging HPA function. Psychoneuroendocrinology 37, 1019–1028. doi: 10.1016/j.psyneuen.2011.11.011
Perucho, L., Artero-Castro, A., Guerrero, S., Ramón y Cajal, S., Me, L. L., and Wang, Z. Q. (2014). RPLP1, a crucial ribosomal protein for embryonic development of the nervous system. PLoS One 9:e99956. doi: 10.1371/journal.pone.0099956
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/b:bile.0000019559.84305.47
Pillet, B., García-Gómez, J. J., Pausch, P., Falquet, L., Bange, G., de la Cruz, J., et al. (2015). The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to Its nuclear Pre-60S assembly site. PLoS Genet. 11:e1005565. doi: 10.1371/journal.pgen.1005565
Pinto, F., Pacheco, C. C., Ferreira, D., Moradas-Ferreira, P., and Tamagnini, P. (2012). Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS One 7:e34983. doi: 10.1371/journal.pone.0034983
Pombo-Suarez, M., Calaza, M., Gomez-Reino, J. J., and Gonzalez, A. (2008). Reference genes for normalization of gene expression studies in human osteoarthritic articular cartilage. BMC Mol. Biol. 9:17. doi: 10.1186/1471-2199-9-17
Qing-Xian, L., Lin-Long, W., Yi-Zhong, W., Liang, L., Hui, H., Liao-Bin, C., et al. (2020). Programming changes in GLUT1 mediated the accumulation of AGEs and matrix degradation in the articular cartilage of female adult rats after prenatal caffeine exposure. Pharmacol. Res. 151:104555. doi: 10.1016/j.phrs.2019.104555
Rahman, M. L., Valeri, L., Kile, M. L., Mazumdar, M., Mostofa, G., Qamruzzaman, Q., et al. (2017). Investigating causal relation between prenatal arsenic exposure and birthweight: Are smaller infants more susceptible? Environ. Int. 108, 32–40. doi: 10.1016/j.envint.2017.07.026
Ren, S., Zhang, F., Li, C., Jia, C., Li, S., Xi, H., et al. (2010). Selection of housekeeping genes for use in quantitative reverse transcription PCR assays on the murine cornea. Mol. Vis. 16, 1076–1086.
Robledo, S., Idol, R. A., Crimmins, D. L., Ladenson, J. H., Mason, P. J., and Bessler, M. (2008). The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14, 1918–1929. doi: 10.1261/rna.1132008
Rowas, S. A., Haddad, R., Gawri, R., Al Ma’awi, A. A., Chalifour, L. E., Antoniou, J., et al. (2012). Effect of in utero exposure to diethylstilbestrol on lumbar and femoral bone, articular cartilage, and the intervertebral disc in male and female adult mice progeny with and without swimming exercise. Arthritis Res. Ther. 14:R17. doi: 10.1186/ar3696
Sabra, S., Malmqvist, E., Saborit, A., Gratacós, E., and Gomez Roig, M. D. (2017). Heavy metals exposure levels and their correlation with different clinical forms of fetal growth restriction. PLoS One 12:e0185645. doi: 10.1371/journal.pone.0185645
Sadangi, C., Rosenow, F., and Norwood, B. A. (2017). Validation of reference genes for quantitative gene expression analysis in experimental epilepsy. J. Neurosci. Res. 95, 2357–2366. doi: 10.1002/jnr.24089
Sarker, N., Fabijan, J., Emes, R. D., Hemmatzadeh, F., Meers, J., Moreton, J., et al. (2018). Identification of stable reference genes for quantitative PCR in koalas. Sci. Rep. 8:3364. doi: 10.1038/s41598-018-21723-0
Sedaghat, K., Zahediasl, S., and Ghasemi, A. (2015). Intrauterine programming. Iran. J. Basic Med. Sci. 18, 212–220.
Sharan, R. N., Vaiphei, S. T., Nongrum, S., Keppen, J., and Ksoo, M. (2015). Consensus reference gene(s) for gene expression studies in human cancers: end of the tunnel visible? Cell Oncol. 38, 419–431. doi: 10.1007/s13402-015-0244-6
Taylor, S., Wakem, M., Dijkman, G., Alsarraj, M., and Nguyen, M. (2010). A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50, S1–S5. doi: 10.1016/j.ymeth.2010.01.005
Tie, K., Zhang, X., Tan, Y., Deng, Y., Li, J., Ni, Q., et al. (2016). Intrauterine low-functional programming of IGF1 by prenatal nicotine exposure mediates the susceptibility to osteoarthritis in female adult rat offspring. FASEB J. 30, 785–797. doi: 10.1096/fj.15-274399
Tomaszewska, E., Dobrowolski, P., and Wydrych, J. (2012). Postnatal administration of 2-oxoglutaric acid improves articular and growth plate cartilages and bone tissue morphology in pigs prenatally treated with dexamethasone. J. Physiol. Pharmacol. 63, 547–554.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Wan, Q., Whang, I., Choi, C. Y., Lee, J. S., and Lee, J. (2011). Validation of housekeeping genes as internal controls for studying biomarkers of endocrine-disrupting chemicals in disk abalone by real-time PCR. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 153, 259–268. doi: 10.1016/j.cbpc.2010.11.009
Wu, T., Huang, Y., Gong, Y., Xu, Y., Lu, J., Sheng, H., et al. (2019). Treadmill exercise ameliorates depression-like behavior in the rats with prenatal dexamethasone exposure: the role of hippocampal mitochondria. Front. Neurosci. 13:264. doi: 10.3389/fnins.2019.00264
Xiang, Q., Li, J., Qin, P., He, M., Yu, X., Zhao, K., et al. (2018). Identification and evaluation of reference genes for qRT-PCR studies in Lentinula edodes. PLoS One 13:e0190226. doi: 10.1371/journal.pone.0190226
Xiao, H., Wen, Y., Pan, Z., Shangguan, Y., Qin, J., Tan, Y., et al. (2018). Increased H3K27ac level of ACE mediates the intergenerational effect of low peak bone mass induced by prenatal dexamethasone exposure in male offspring rats. Cell Death Dis. 9:638. doi: 10.1038/s41419-018-0701-z
Xiao, H., Xie, X., Wen, Y., Tan, Y., Shangguan, Y., Li, B., et al. (2020). Subchondral bone dysplasia partly participates in prenatal dexamethasone induced-osteoarthritis susceptibility in female offspring rats. Bone 133, 115245. doi: 10.1016/j.bone.2020.115245
Xie, Z., Zhao, Z., Yang, X., Pei, L., Luo, H., Ni, Q., et al. (2018). Prenatal nicotine exposure intergenerationally programs imperfect articular cartilage via histone deacetylation through maternal lineage. Toxicol. Appl. Pharmacol. 352, 107–118. doi: 10.1016/j.taap.2018.03.018
Xu, D., Zhang, B., Liang, G., Ping, J., Kou, H., Li, X., et al. (2012). Caffeine-induced activated glucocorticoid metabolism in the hippocampus causes hypothalamic-pituitary-adrenal axis inhibition in fetal rats. PLoS One 7:e44497. doi: 10.1371/journal.pone.0044497
Zhai, Z., Yao, Y., and Wang, Y. (2013). Importance of suitable reference gene selection for quantitative RT-PCR during ATDC5 cells chondrocyte differentiation. PLoS One 8:e64786. doi: 10.1371/journal.pone.0064786
Zhang, G., Zhou, J., Huang, W., Yu, L., Zhang, Y., and Wang, H. (2018). Placental mechanism of prenatal nicotine exposure-reduced blood cholesterol levels in female fetal rats. Toxicol. Lett. 296, 31–38. doi: 10.1016/j.toxlet.2018.07.022
Zhang, L., Jing, X., Chen, W., Bai, J., Vasseur, L., He, W., et al. (2019). Selection of reference genes for expression analysis of plant-derived microRNAs in Plutella xylostella using qRT-PCR and ddPCR. PLoS One 14:e0220475. doi: 10.1371/journal.pone.0220475
Zhao, W. P., Wang, H. W., Liu, J., Zhang, Z. H., Zhu, S. Q., and Zhou, B. H. (2019). Mitochondrial respiratory chain complex abnormal expressions and fusion disorder are involved in fluoride-induced mitochondrial dysfunction in ovarian granulosa cells. Chemosphere 215, 619–625. doi: 10.1016/j.chemosphere.2018.10.043
Keywords: intrauterine growth retardation, real-time quantitative polymerase chain reaction, cartilage development, prenatal dexamethasone exposure, the panel of reference genes
Citation: Liu L, Han H, Li Q, Chen M, Zhou S, Wang H and Chen L (2020) Selection and Validation of the Optimal Panel of Reference Genes for RT-qPCR Analysis in the Developing Rat Cartilage. Front. Genet. 11:590124. doi: 10.3389/fgene.2020.590124
Received: 03 August 2020; Accepted: 24 November 2020;
Published: 16 December 2020.
Edited by:
Y-h. Taguchi, Chuo University, JapanReviewed by:
Khyati Shah, University of California, San Francisco, United StatesMing-Xing Lu, Yangzhou University, China
Copyright © 2020 Liu, Han, Li, Chen, Zhou, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liaobin Chen, bGJjaGVuQHdodS5lZHUuY24=; Hui Wang, d2FuZ2h1aTE5QHdodS5lZHUuY24=
 Liang Liu
Liang Liu Hui Han1
Hui Han1 Hui Wang
Hui Wang