- 1National Institute of Plant Genome Research, New Delhi, India
- 2Department of Botany, Gargi College, University of Delhi, New Delhi, India
- 3Department of Plant Molecular Biology, University of Delhi, New Delhi, India
Many quantitative trait loci (QTLs) have been identified by molecular genetic studies which control grain size by regulating grain width, length, and/or thickness. Grain width 2 (GW2) is one such QTL that codes for a RING-type E3 ubiquitin ligase and increases grain size by regulating grain width through ubiquitin-mediated degradation of unknown substrates. A natural variation (single-nucleotide polymorphism at the 346th position) in the functional domain-coding region of OsGW2 in japonica rice genotypes has been shown to cause an increase in grain width/weight in rice. However, this variation is absent in indica rice genotypes. In this study, we report that reduced expression of OsGW2 can alter grain size, even though natural sequence variation is not responsible for increased grain size in indica rice genotypes. OsGW2 shows high expression in seed development stages and the protein localizes to the nucleus and cytoplasm. Downregulation of OsGW2 by RNAi technology results in wider and heavier grains. Microscopic observation of grain morphology suggests that OsGW2 determines grain size by influencing both cell expansion and cell proliferation in spikelet hull. Using transcriptome analysis, upregulated genes related to grain size regulation have been identified among 1,426 differentially expressed genes in an OsGW2_RNAi transgenic line. These results reveal that OsGW2 is a negative regulator of grain size in indica rice and affects both cell number and cell size in spikelet hull.
Introduction
Globally, rice (Oryza sativa L.) is a staple cereal food crop since it is the primary source of caloric intake and is consumed by more than half of the world's population (Sasaki, 2008; Zuo and Li, 2014; Azizi et al., 2019). In upcoming decades, the overall demand of rice will increase, particularly in Asia and Africa (Kubo and Purevdorj, 2004). Hence, there is an urgent need to increase rice grain yield. Rice grain size/weight is the key agronomic trait for the improvement of yield. Rice grain yield is a complex trait, which is determined by four typical quantitative component traits, i.e., number of panicles per plant, number of filled grains per panicle, grain size, and grain weight (Xing and Zhang, 2010; Zuo and Li, 2014). Grain size (grain length and width)/weight is the most vital yield-contributing complex quantitative trait in rice (Fan et al., 2006; Mao et al., 2010; Ying et al., 2012). Therefore, the best approach for enhancing rice yield is to find/develop new high yielding varieties with increased grain size/weight and superior grain nutrient quality (Rosegrant and Cline, 2003). Grain weight and grain size are positively associated with each other. Grain size is defined in terms of its length, width, and thickness. Rice grain size is mainly controlled by genes that determine cell expansion and/or proliferation in spikelet hull and contribute in endosperm development. All these yield component traits are controlled by naturally occurring quantitative trait loci, QTLs (Zuo and Li, 2014). Several QTLs and a number of genes regulating grain size have been identified and functionally characterized by different approaches such as genomics, proteomics, metabolomics, genome editing, and genome-wide association to find out molecular components and genetic regulatory mechanisms controlling grain size trait in rice (Li and Li, 2014; Li et al., 2018; Azizi et al., 2019). Hitherto, much needs to be explored regarding the molecular mechanisms as well as the underlying genes controlling rice grain size/weight to understand the mechanism of grain development, which will provide a way to improve yield and quality.
Ubiquitination has been shown to play a significant role in determining seed size in crop plants (Song et al., 2007; Li et al., 2008; Choi et al., 2018). It is an enzymatic post-translational modification in which ubiquitin protein (76 residues long) covalently attaches with a target protein (Li and Li, 2014), with the consecutive participation of three unique enzymes, E1, E2, and E3 (Moon et al., 2004). E1 is a ubiquitin-activating enzyme. E2 conjugates with activated ubiquitin. Ubiquitin ligase E3 determines substrate specificity (Smalle and Vierstra, 2004). This process regulates the stability, activity, and localization of modified target proteins. Mono-ubiquitination of a target protein affects its function and localization, whereas conjugation of multi-ubiquitin can degrade modified proteins through the ubiquitin−26S proteasome complex (Vierstra, 2009).
Grain yield and quality are controlled by several QTLs and genes. A number of grain size related QTLs (GW2, GW5, GS3, GS5, qGL3, TGW6, GW8, etc.) and genes have been functionally characterized in the last few decades. They regulate either cell division or cell expansion, or both, during the process of seed development, by being involved in signaling pathways facilitated by ubiquitin-mediated proteasomal degradation, transcription factors, guanine nucleotide-binding proteins (G-proteins), protein kinases and phytohormones (Zuo and Li, 2014; Wang et al., 2019; Zhao et al., 2019). GW2 is one such QTL that regulates grain weight/yield by regulating grain width in japonica rice genotypes. It codes for a RING-type protein having E3 ubiquitin ligase activity that binds with its substrates and targets them for degradation through the ubiquitin−26S proteasomal complex. A natural variation in long grain WY3 cultivar results in truncation of the functional domain of GW2. This results in a wider spikelet hull and an increase in the number of cells in the outer parenchyma cell layer, without affecting cell numbers in the endosperm. Hence, it has been identified that GW2 negatively regulates grain size and weight in japonica rice cultivars (Song et al., 2007). Chitinase 14 (CHT14) and phosphoglycerate kinase (PGK) are involved in carbohydrate metabolism and show strong interaction with GW2, but are not involved in ubiquitin-mediated degradation (Lee et al., 2018). GW2 also targets expansin-like 1 (EXPLA1), which is a cell wall-releasing protein, for degradation through the proteasome pathway and hence participates in determining seed size and weight (Choi et al., 2018). WIDE AND THICK GRAIN 1 (WTG1) also controls rice grain size through the ubiquitin–proteasome pathway. WTG1 is a deubiquitinating enzyme, an otubain-like protease. WTG1/OsOTUB1 regulates seed size/shape by affecting only cell expansion in the hull, suggesting a different pathway than GW2 (Jiao et al., 2010; Huang et al., 2017; Li and Li, 2019). DA2 is the homolog of GW2 in Arabidopsis thaliana that also controls seed and organ size. Rice GW2 homologs have been identified in maize and wheat, where they regulate kernel size (Li et al., 2010; Sestili et al., 2019).
Although the specific natural variation in GW2, causing its inactivation, results in higher grain width/weight in japonica rice genotypes, such a sequence variation is absent in indica genotypes (Dixit et al., 2013). The aim of the current study was to examine if, despite the absence of this variation, downregulation of GW2 in indica rice would alter grain size. Downregulation in transgenic rice plants, by RNAi, revealed that the grain width/weight in indica rice genotypes increases due to higher cell division as well as expansion in spikelet hull which in turn is controlled by differential gene expression of many seed development-related genes.
Materials and Methods
Plant Material and Growth Conditions
Rice indica and japonica genotypes (PB1, IR64, Sonasal, LGR, and Nipponbare) were grown in experimental fields of NIPGR (New Delhi, India) during the months of June to October (Tmax 35–40°C; Tmin 25–28°C). Rice transgenic plants along with the wild-type lines (WT) were grown in a mix of soil and compost (3:2), which was supplemented with NPK, in a greenhouse with a 12-h/12-h light/dark cycle, at 28/23°C temperature, 65% relative humidity, and 250 μmol m−2 s−1 photosynthetic active radiation (PAR) light. The complete development of rice seed occurs in five different stages based on morphological changes occurring in the embryo and endosperm categorized as S1 (0–2 DAP, days after pollination), S2 (3–4 DAP), S3 (5–10 DAP), S4 (11–20 DAP), and S5 (21–29 DAP) (Agarwal et al., 2007, 2011). In accordance, rice panicles were tagged and harvested as mentioned previously (Sharma et al., 2012). Flag leaf and seed tissue from each variety, at each stage, were harvested and frozen in liquid nitrogen and stored at −80°C until further use.
RNA Isolation, cDNA Preparation, and Quantitative RT-PCR
Isolation of RNA from the leaves and seeds (S1–S5 stages) of different rice genotypes (PB1, IR64, Sonasal, LGR, and Nipponbare) as well as transgenic plants was carried out using TRIzol® reagent (InvitrogenTM Life Technologies, USA) as detailed previously (Singh et al., 2003; Mathew et al., 2016, 2020; Das et al., 2019). The quantification of RNA samples was done by NANODROP 2000c Spectrophotometer (Thermo Scientific, USA). For the amplification of the OsGW2 gene (LOC_Os02g14720), full-length cDNAs of five rice genotypes, mentioned above, were used. For full-length cDNA synthesis, 2 μg of purified RNA and Superscript III® first-strand cDNA synthesis kit (InvitrogenTM Life Technologies, USA) was used. Reverse transcription of purified RNA was performed with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) with random primers. Gene-specific primers were designed by using Primer Express Version 3.0 (Applied Biosystems, USA). For expression analysis of OsGW2, qRT-PCR was performed in three biological replicates using gene-specific primers (Supplementary Table 1), with Fast SYBR® Green Master Mix (Applied Biosystems, USA). Real-time PCR was done in 7500 Fast Real-Time System (Applied Biosystems, USA) using KAPA SYBR® FAST qPCR kit and 7500 software v2.0.1 for data analysis. OsACTIN1 gene was the endogenous control for leaf and seed tissue throughout the study. Relative gene expression level was calculated by the comparative Ct (2−ΔΔCt) method.
Sequence Analysis of OsGW2 in Different Rice Genotypes
The OsGW2 gene coding region was amplified from different rice genotypes (IR64, Sonasal, LGR, and PB1) using full-length cDNA for each (Supplementary Table 1) and cloned in pJET1.2 vector (Thermo Scientific, USA). Cloning was confirmed by restriction digestion by BglII and confirmed clones were sequenced on ABI 3730xl DNA Analyzer (Applied Biosystems, USA) in three replicates. The obtained sequences were analyzed by Clustal Omega tool (Sievers et al., 2011) for aligning the insert sequence with template sequence extracted from the Rice Genome Annotation Project (RGAP, http://rice.plantbiology.msu.edu/). Any difference in either nucleotide or subsequent protein sequence was noted.
Preparations of RNAi Construct and Rice Plant Transformation
To generate the RNAi construct, a 510-bp unique region of OsGW2 was delineated by Rational siRNA Design program (Reynolds et al., 2004). This unique region of OsGW2 was amplified along with CACC sequence added to the 5′ end of the forward primer (Supplementary Table 1), from the OsGW2_pJET clone. The amplified fragment was purified using GeneJET gel extraction kit (Thermo Scientific, USA), followed by cloning into Gateway® entry vector pENTRTM/D-TOPO® (InvitrogenTM, USA) containing recombination sites (attL1 and attL2) using pENTRTM/Directional TOPO® cloning kit as per the manufacturer's protocol. Cloning was confirmed by restriction digestion with AscI and NotI restriction enzymes and sequencing of two positive clones. Plants with reduced expression or knockdown of OsGW2 were made using RNAi vector pANDA (Miki and Shimamoto, 2004), in which transgene expression was under the control of the maize UBIQUITIN promoter. The insert was transferred to pANDA having kanamycin and hygromycin selection marker genes using GatewayTM LR ClonaseTM II Enzyme Mix (InvitrogenTM, USA) through Gateway® cloning technology. Target sequence was incorporated in opposite orientation in two attR sites located at both sides of the GUS linker sequence, confirmed by restriction digestion by using KpnI and SacI enzymes. The confirmed clone was introduced into the Agrobacterium tumefaciens strain EHA105. Transgenic plants were generated by transformation of the RNAi construct into rice calli (PB1 genotype IET-10364) through the Agrobacterium-mediated transformation method (Toki et al., 2006; Das et al., 2019; Mathew et al., 2020).
Phenotypic Characterization of Rice Transgenic Plants
Vegetative phenotypic characters (plant height, flag leaf length and width, number of tillers per plant, number of panicles per plant, and number of grains per panicle) were measured manually and compared with the respective wild-type plant in T0, T1, and T2 generations. Six biological replicates were taken to quantitate each vegetative character for each line. Grain parameters (length and width) were measured after the grains were harvested and dried, by using WinSEEDLETM (Regent Instruments Inc., Canada). Photographs of grain length and width were taken using a Nikon D5200 48 MP DSLR camera.
Scanning Electron Microscopy
For scanning electron microscopy analysis, the mature grains of homozygous T2 transgenic line OsGW2_RNAi_7AP9 and wild-type plants were harvested. To study cell size and number in the outer epidermis, a scanning electron microscope (SEM, Zeiss, Germany) was used to observe the central parts of the lemma surface of mature grains, under different magnifications (500×, 1,000×, and 2,500×). For analyzing starch granules, mature rice grains were dehusked manually. These were sectioned longitudinally with a scalpel and the central parts of the endosperm were photographed by SEM. To analyze cell length and cell area of the outer epidermal cells, ImageJ software was used (Schneider et al., 2012).
Subcellular Localization of OsGW2
The full-length coding sequence of OsGW2 from PB1, lacking the stop codon, was amplified (Supplementary Table 1) and cloned downstream of the YFP coding region in the pSITE-3CA-YFP vector (Chakrabarty et al., 2007) under the control of CaMV 35S promoter to generate the recombinant plasmid YFP-OsGW2. To study the subcellular localization of OsGW2, YFP-OsGW2 recombinant plasmid and control YFP vector were bombarded separately into onion peel cells by particle bombardment using Biolistic®-PDS-1000/HeTM system (Bio-Rad, USA). After incubation for 16 h in the dark at 28°C, YFP fluorescence was observed with a confocal laser microscope (TCS SP2, Leica, Germany).
Transcriptome Analysis of OsGW2_RNAi Plants
To study the functional relevance of OsGW2, RNA-seq analysis of T3 grains, from S4 stage, of OsGW2_RNAi_7AP9 transgenic plants, along with the wild-type lines, was performed in two biological replicates. RNA isolation and quantification was done as mentioned above and RNA integrity was checked on Bioanalyzer (2100 Agilent Technologies, USA). RNA having RIN ≥8.0 were used for library preparation. Total RNA (~5 μg) for each sample was used for cDNA library preparation and sequencing. Transcriptome sequencing was performed on Illumina HiseqTM 2000 platform with two independent biological replicates, each for WT and OsGW2_RNAi_7AP9. After obtaining sequencing data, different tools were used for performing preprocessing of data, alignment with reference, expression estimation, and comparison analysis, as detailed previously (Mathew et al., 2020). The preprocessing tools used were Adapter Removal v2 (version 2.2.0) and bowtie2 (version 2.2.9). Processed reads were aligned in STAR (version 2.5.3a) with the MSU rice genome database (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/all.dir). Cufflinks was used for estimating gene expression (version 2.2.1) and output was in terms of fragments per kilobase million reads (FPKM). To identify differentially expressed genes (DEGs), Cuffdiff utility provided in Cufflinks package was used. The log2 fold change cutoff was ≥1.5 (upregulated genes) and ≤−1.5 (downregulated genes) with a P-value cutoff of ≤ 0.05.
Functional annotation of DEGs in metabolic pathways/processes was studied by using databases including the KEGG pathway analysis and MSU-RGAP 7 (Ouyang et al., 2006). Characterized differentially expressed genes were separated and analyzed using RICENCODE, and seed-related genes were extracted by comparing DEGs with the data available from previous studies in the lab and online sources, as done previously (Mathew et al., 2020). Heat maps were constructed using MeV Version 4.9.0 (Saeed et al., 2003). Gene ontology enrichment for various DEGs was performed using the AgriGO software (v1.2; Du et al., 2010) with MSU-RGAP 7 as the reference annotation set. Rice GO information for biological process, cellular component, and molecular function was used for gene ontology enrichment analysis. For pathway analysis of differentially expressed genes, the MapMan software (version 3.5.1; http://mapman.gabipd.org/web/guest) was used with a P-value cutoff of ≤0.05 and mapping files from MSU-RGAP 7.
Results
OsGW2 Sequence Variation in Rice Genotypes
A natural variation (single-nucleotide polymorphism) was present in large grain-sized japonica rice genotypes in comparison with small grain-sized genotypes which was responsible for truncation of encoded OsGW2 protein functional domain in the large grain-sized genotypes (Song et al., 2007). No such variation was observed in indica genotypes (Dixit et al., 2013). In our analysis, the coding region of OsGW2 (1,278 bp) from four indica rice genotypes, selected on the basis of different grain sizes as extra large, large, medium, and small (LGR, PB1, IR64, and Sonasal), was sequenced and compared with japonica cultivar Nipponbare (Figure 1A). The coding region showed 100% similarity amongst all four indica rice genotypes (LGR, PB1, IR64, and Sonasal). However, the japonica cultivar, Nipponbare, showed variation with respect to indica sequence. Comparison of the nucleotide sequences of the alleles of OsGW2 from both indica and japonica cultivars revealed three nucleotide changes, namely two single-nucleotide polymorphism (SNP) variations and one 6-bp addition in the coding region of indica genotypes. SNP variation (G to A) at position 572 bp resulted in an amino acid change (Gly to Glu), six base pair addition at positions 714–719 resulted in the addition of two amino acids (Gln and Glu), whereas (A–G) SNP variation at 1,116 bp did not result in any amino acid variation (Figure 1A). Despite these sequence variations, the conserved domain structure of the OsGW2 protein remained the same, and it did not show truncation as reported for japonica rice genotypes with increased grain size (Song et al., 2007).
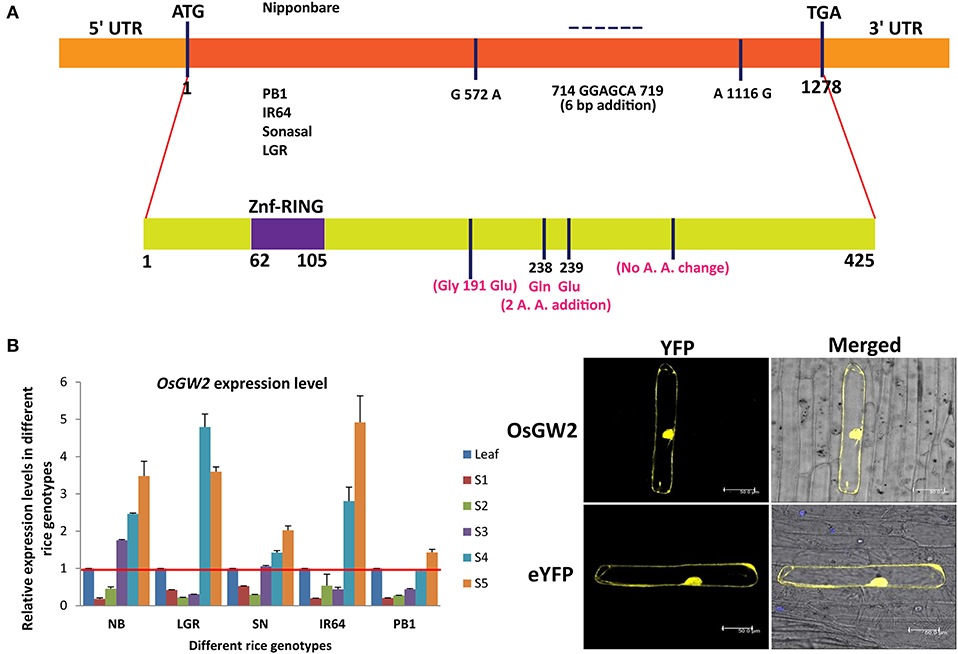
Figure 1. Structure, expression, and subcellular localization of OsGW2. (A) OsGW2 nucleotide (upper panel) and amino acid (lower panel, marked in pink color) sequence variation in indica rice genotypes (IR64, PB1, Sonasal, and LGR) including substitutions and additions in comparison with the japonica rice genotype (Nipponbare) and protein structure variation. A. A. represents amino acid. (B) Expression levels of OsGW2 in seed tissues (S1–S5) of five different rice genotypes determined by qRT-PCR. The rice OsACTIN1 gene was used as an internal control. Data show fold changes with respect to the flag leaf of the same genotype, and bar represents mean ± SD (n = 3). (C) Subcellular localization of YFP-tagged OsGW2 in onion peel cells (top panel). Lower panel represents empty vector control. Scale bar, 50 μm.
OsGW2 Expresses Mainly in Seed Stages and Its Protein Localizes to the Nucleus and Cytoplasm
To determine the expression patterns of OsGW2, transcript levels were examined by qRT-PCR in S1–S5 seed developmental stages of four indica and one japonica rice genotype in comparison with their respective flag leaf. These rice genotypes show variation in grain size parameters, viz. grain width, length, and weight (Supplementary Figure 1). Previous data showed that OsGW2 expressed constitutively in vegetative as well as reproductive tissues (Song et al., 2007). qRT-PCR data revealed that OsGW2 expressed at lower levels in S1–S3 stages and had higher expression in S4–S5 stages of seed development, which indicates its essential role in rice seed development (Figure 1B). To study the subcellular localization of OsGW2, OsGW2 yellow fluorescent protein (YFP-OsGW2) fusion construct was designed where expression was controlled by the CaMV 35S promoter. In vivo protein targeting in onion epidermal cells by bombarding with the YFP-OsGW2 fusion construct showed that it localized to both the nucleus and cytoplasm. This was observed in six independent cells (Figure 1C).
Suppressed Expression of OsGW2 Alters Grain Morphology
To functionally validate OsGW2, we generated OsGW2 RNA interference (RNAi) lines in indica PB1 (IET-10364) background, and a 510-bp unique region was targeted to suppress the expression of OsGW2. A total of 10 OsGW2_RNAi transgenic lines (RNAi_2, 4, 7, 9, 13, 15, 17, 21, 23, 24) were obtained through tissue culture. Each plant of all these transgenic lines in T0 generation was found positive by PCR amplification of hygromycin resistance gene (Hygromycin phosphotransferase II) resulting in an amplicon of 850 bp (Supplementary Figure 2). Vegetative parameters (plant height, flag leaf length, flag leaf width, number of tillers per plant) did not show any morphological difference between RNAi and wild-type PB1 plants. However, transgenic plants in T0 generation showed significantly higher grain width (23–38%), grain length (5–15%), and grain weight (19–58%) when compared with the wild-type lines (Figures 2A–C; Supplementary Figure 2). In T1 generation, four lines (RNAi 2A, 7A, 9B, and 24P2) were selected out of 10 lines which showed 3:1 segregation ratio (Supplementary Table 2). These OsGW2 RNAi plants had significantly higher grain width, length, and weight in comparison with PB1 seeds, in T1 generation (Figures 2D–F). Other phenotypic parameters did not show significant difference in comparison with wild-type plants as before. The four lines were taken forward, and in T2 generation, suppressed expression of OsGW2 (RNAi 2AP9, 7AP9, 9BP3, and 24P2P5) was screened and used for further analysis. Two lines 2AP9 and 7AP9 had homozygous plants. Positive OsGW2_RNAi transgenic plants along with the wild-type lines were grown and analyzed (Figure 3A; Supplementary Figure 3). Expression levels were significantly reduced (53.88–84.84%) in the third leaf (as the gene expresses ubiquitously) of OsGW2_RNAi T2 transgenic plants. T3 transgenic seeds from S4 stage of the two homozygous lines also showed a significant decrease (66–88%) in the expression of GW2 (Figure 3B). The grains of T2 generation plants were significantly larger than those of the wild-type lines (Figure 3E), showing increased grain width (4.0–18.98%) and grain length (4.09–6.19%) (Figures 3C,D,F,G). GW2 is known to affect seed width in japonica rice (Song et al., 2007). It has also been shown to affect width and, hence, weight in wheat (Sestili et al., 2019). In our experiments, T1 seeds showed a significant increase in length. T2 seeds did show an increase in length but it was not significant (Figure 2E). Again, T3 seeds from two homozygous lines (Figure 3G) showed a significant increase in length. This may be possible because the t-test was performed on 10 T1 seeds from one plant, in triplicates. Further, 10 T2 seeds were taken from three plants and 100 T3 seeds were taken from three plants to calculate seed parameters. All T1 seeds generated by each plant and T3 seeds from two homozygous plants showed a significant increase in seed length, probably because of less variation in the sample taken. Furthermore, the 30-grain weight at the mature stage of OsGW2_RNAi T3 seeds was 12.94–42.96% higher than that of the wild-type grains (Figure 3H). These results indicated that reduced expression of OsGW2 could significantly increase grain width and weight even in indica rice, where a natural polymorphism was absent.
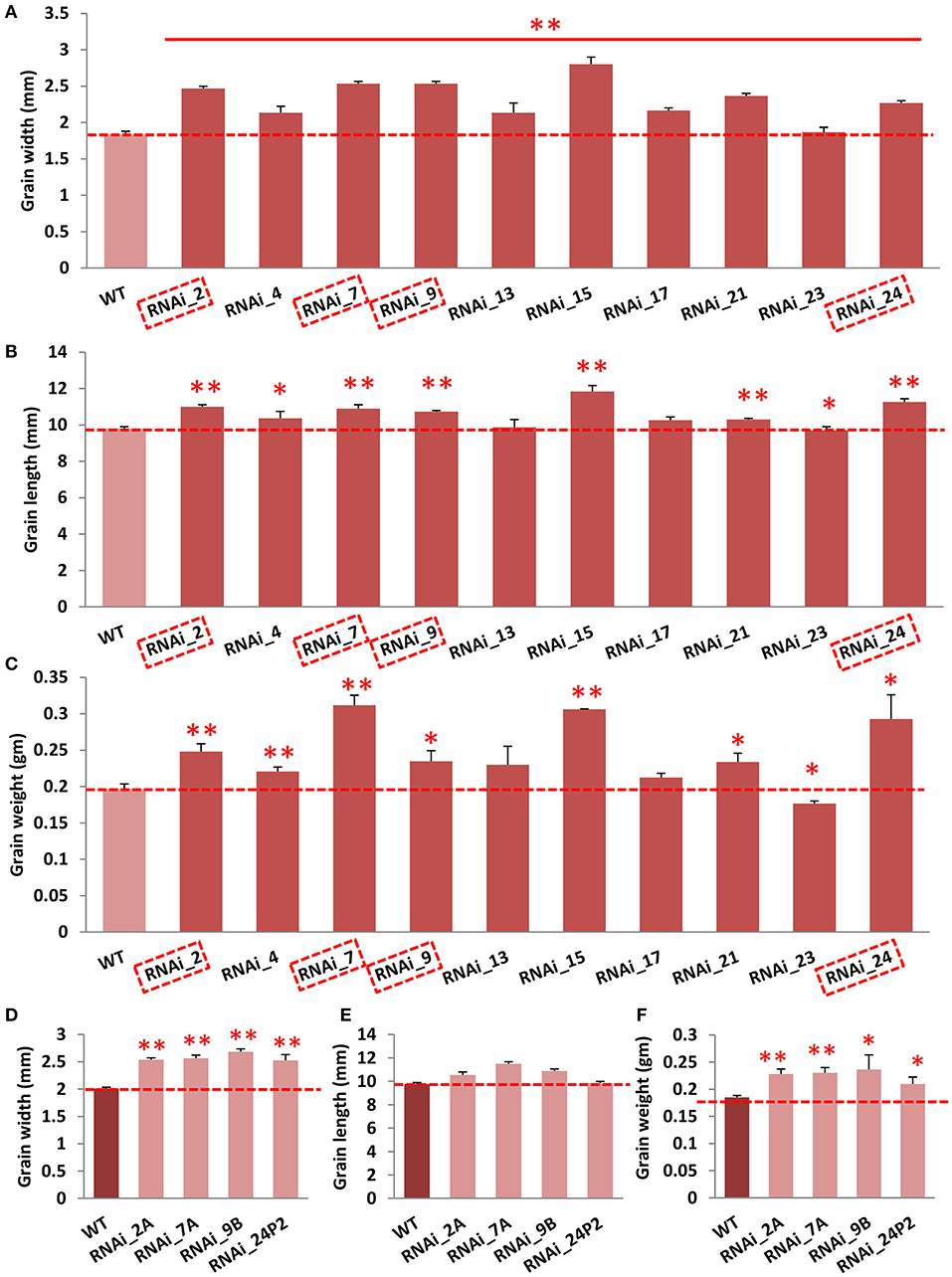
Figure 2. Effect of OsGW2 knockdown on width, length, and weight of T1 and T2 rice grains. (A–C) Grain width, length, and 10-grain weight, respectively, of OsGW2_RNAi T1 seeds. (D–F) Grain width, length, and 10-grain weight, respectively, of OsGW2_RNAi T2 seeds. WT represents wild-type PB1 grains; grain length and width were measured with WINSEEDLETM. RNAi_2, 4, 7, 9, 13, 15, 17, 21, 23, and 24 are 10 different transgenic lines in T0 generation. Dashed boxes in (A–C) represent selected transgenic lines for further analyses which follow 3:1 segregation in T1 generation. Hence, RNAi_2A, 7A, 9B, and 24P2 are four different transgenic lines in T1 generation, selected from T0 generation. Bar represents mean ± SD [n = 10 in (A–E); n = 3 in (C,F)]. **P < 0.01; *P < 0.05, determined by Student's t-test.
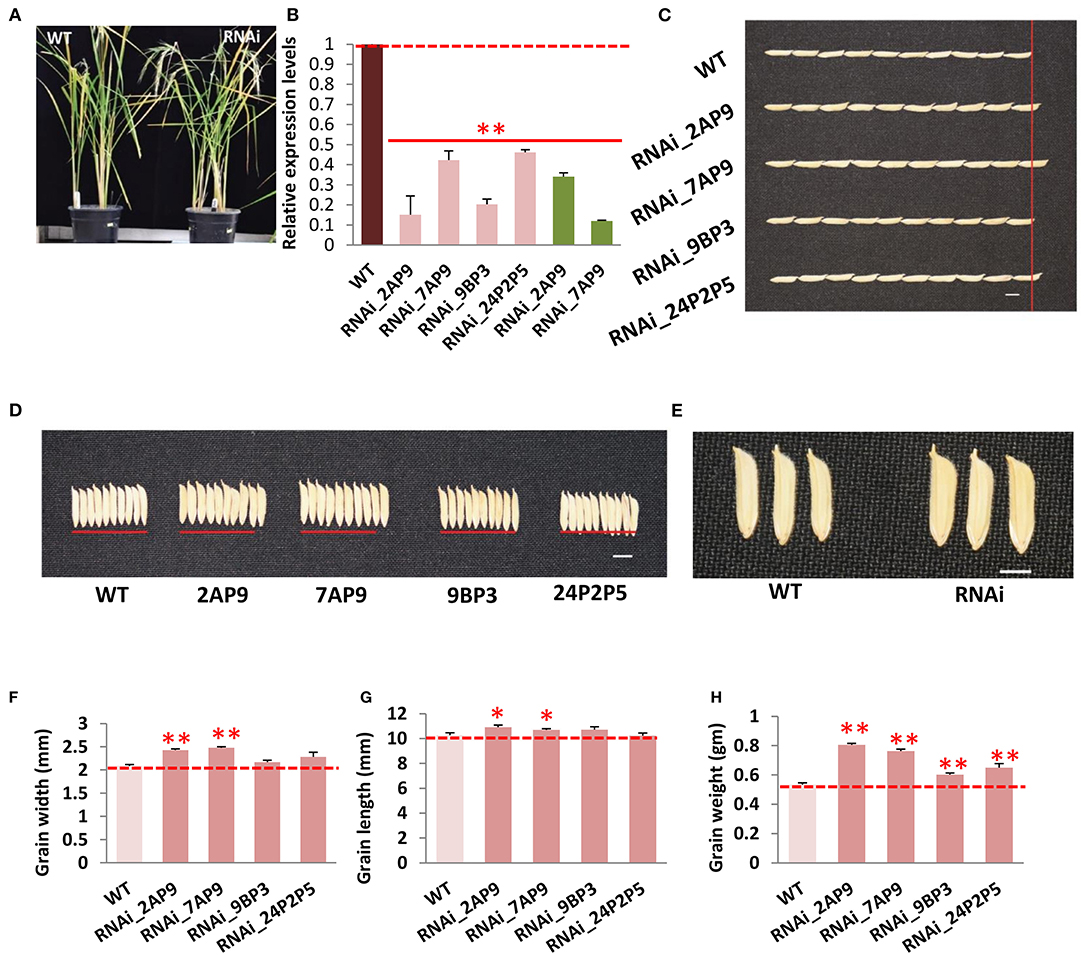
Figure 3. Phenotypic characterization of OsGW2_RNAi T2 generation transgenic plants. (A) Gross morphology of wild-type (WT) and OsGW2_RNAi plants at the mature stage. (B) Relative expression levels, by qRT-PCR, of GW2 in OsGW2_RNAi plants (T2 flag leaf, pink bars and T3 grain, green bars) as compared with the respective wild-type tissues. Data show fold changes in expression level with respect to the wild-type lines; values represent mean ± SD (n = 3), **P < 0.01, Student's t-test. (C) Grain length of OsGW2_RNAi T3 seeds. (D) Grain width of OsGW2 RNAi T3 seeds. (E) Zoomed in image of grains of WT and OsGW2_RNAi_7AP9 T3 grains. Bar = 1 cm. Comparison of (F) grain width, (G) grain length, and (H) 30-grain weight of OsGW2_RNAi T3 seeds with wild-type PB1 seeds. Error bar represents mean ± SD [n = 100*3 in (F,G), n = 3 in (H)]. **P ≤ 0.01, and *P ≤ 0.05, calculated by Student's t-test. RNAi (2AP9, 7AP9, 9BP3, 24P2P5) are four different lines in T2 generation.
OsGW2 Regulates Rice Grain Size by Influencing Cell Division and Expansion
Spikelet hull size is delimiting to the final grain size. Since cell proliferation and/or cell expansion is responsible for alteration in grain size, the cell number of the OsGW2_RNAi_7AP9 line was calculated in the central part of the lemma epidermis using scanning electron microscopy images (Figure 4A). The cell length and cell area of the outer lemma epidermis of the wild-type and OsGW2_RNAi_7AP9 T3 grains were compared by SEM (Figures 4B,C). The images revealed that total lemma cell number in OsGW2_RNAi was increased by 12.94% compared with that of the wild-type lines (Figure 4D), resulting from a significant decrease in cell length and cell area. This suggested that decreased expression level of OsGW2 contributes to higher grain width by affecting both cell division and cell expansion to regulate the number and size of cells of the spikelet hull during the process of development. To check whether or not the suppressed expression of OsGW2 influences the structure of starch granules in the endosperm, we observed seed cross-sections by a scanning electron microscope. In the WT endosperm, starch granules were tightly packed. The spherical starch granules (Dong et al., 2015) were more in comparison with the polyhedron-shaped ones. However, in the OsGW2_RNAi_7AP9 line, starch granules were loosely packed and were larger in size, and the distribution of spherical and polyhedron granules was similar (Figure 4E). Thus, suppression of OsGW2 additionally resulted in modifications in starch granule size.
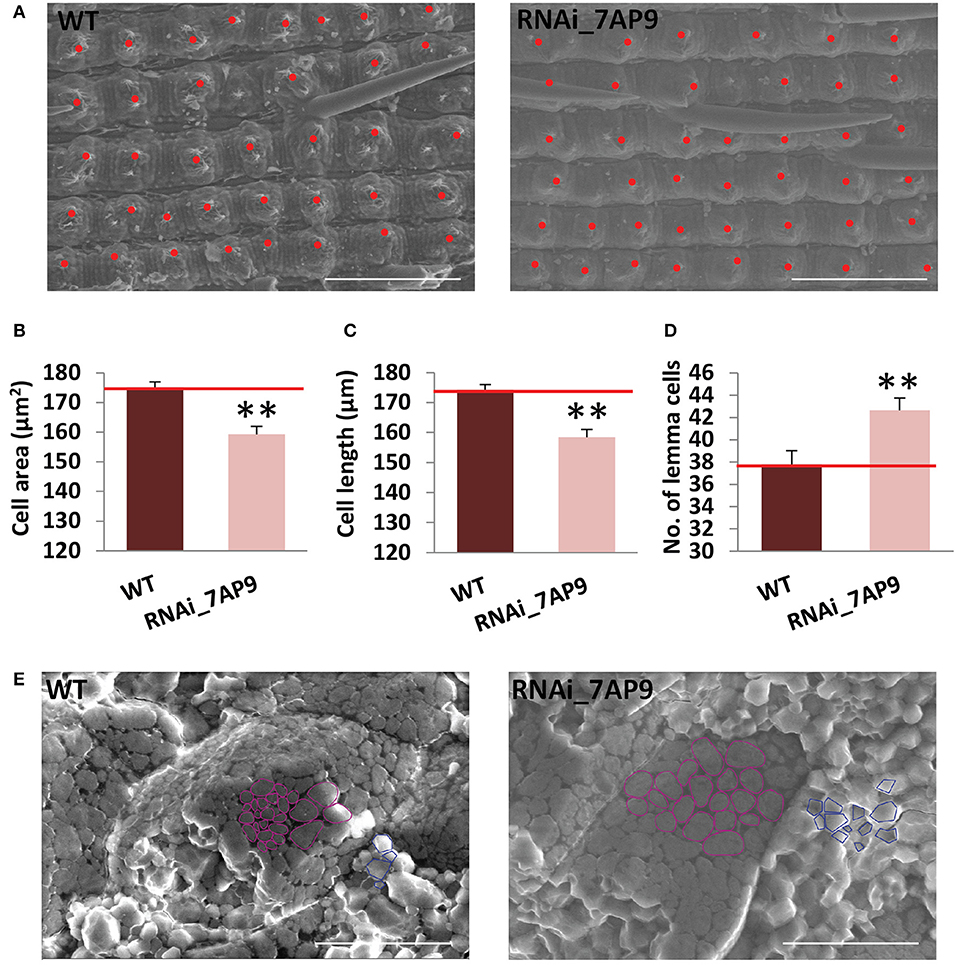
Figure 4. OsGW2 influences cell proliferation and expansion in T3 spikelet hulls. (A) SEM photographs of the outer surface of lemma from wild-type and OsGW2_RNAi_7AP9 T3 seeds. Each red dot represents a cell. (B–D) Cell area, cell length, and number of cells on the epidermis of lemma surface as seen in the area under the microscope. Bar represents mean ± SD; n = 3. **P ≤ 0.01, calculated by Student's t-test. (E) Scanning electron microscopy (SEM) analysis of starch granules of endosperm in the wild-type and OsGW2_RNAi_7AP9 T3 seeds at the mature stage. As a representation, spherical granules are outlined in magenta, while the polyhedron ones are by a blue outline. WT represents wild-type; RNAi_7AP9 represents the knockdown line of OsGW2. Scale bar in (A) and (E) = 20 μm.
Transcriptome Analysis
The transcriptome of the developing seed was compared at S4 stage between WT and OsGW2_RNAi_7AP9 (a homozygous line) to elucidate genes and pathways which could be altering the grain size. Two biological replicates of each sample were used for sequencing and four libraries were generated in total. Each library had more than 50–78 million clean reads after quality control and 89–93% reads mapped uniquely to the rice reference sequences (Supplementary Table 3). Pearson correlation coefficient (R2) was calculated between biological replicates. Correlation was higher (0.98) within the biological replicates than that between the WT and OsGW2_RNAi_7AP9 line. Published data shows that in rice, using RNA sequencing data from two biological replicates, ~22–61 million clean reads have been generated and 62–92% of these reads map to the reference genome (Ding et al., 2017; Yuenyong et al., 2018; Jung et al., 2019; Zha et al., 2019). The transcripts with log2 fold change ≥1.5 (upregulated genes) and ≤(−1.5) (downregulated genes) with a P-value cutoff of ≤0.05, in OsGW2_RNAi_7AP9 transgenic line in comparison with WT, were considered as DEGs. Thus, we could identify 1,426 DEGs, of which 636 (44.60%) DEGs had higher expression in the OsGW2_RNAi_7AP9 line compared with the WT (upregulated genes), while 790 (55.40%) DEGs showed lower expression in the OsGW2_RNAi_7AP9 line compared with the WT (downregulated genes) (Figure 5A; Supplementary Table 4). The expression data obtained through RNA sequencing was validated by comparing the expression of OsGW2 in RNAi_7AP9 T3 seeds generated by qRT-PCR (Figure 3B). The expression patterns were found consistent for OsGW2 in both analyses (Figure 5B).
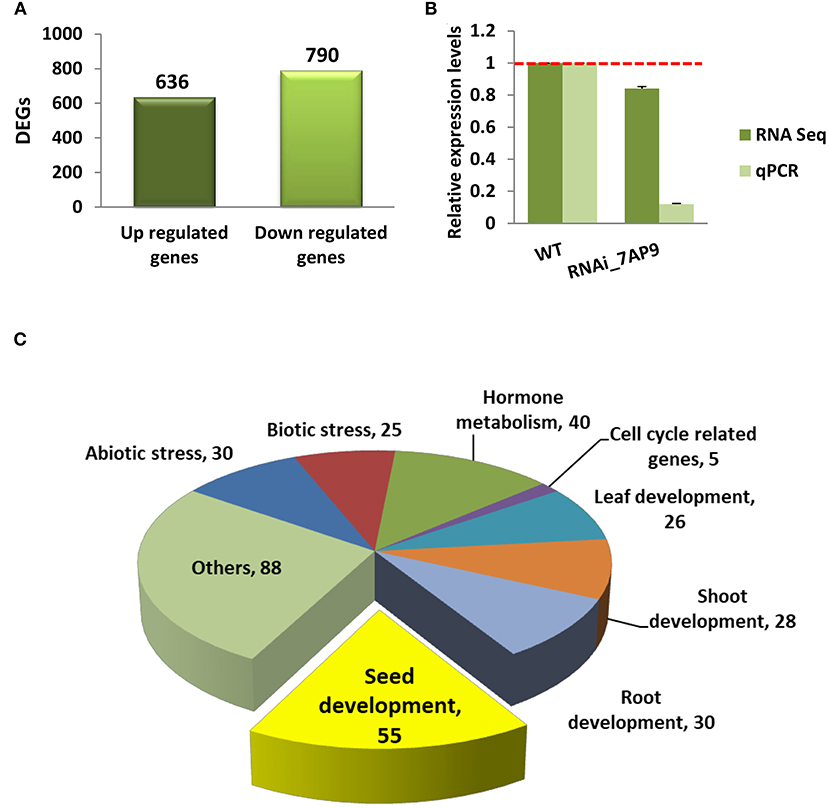
Figure 5. Transcriptome analysis of OsGW2_RNAi T3 generation S4 stage seeds. (A) The number of up- and downregulated DEGs between WT and OsGW2_RNAi_7AP9 seeds is represented as bars. The DEGs with log2 fold change ≥1.5 (upregulated genes) and ≤-1.5 (downregulated genes) with a P-value cutoff of ≤0.05 were considered as significant DEGs. (B) qRT-PCR validation of RNA-seq-based expression levels of OsGW2. qRT-PCR values are the same as in Figure 3B. Data are mean ± SD for two biological replicates. (C) Categorization of functionally characterized DEGs with their overlapping functional terms through RICENCODE. Yellow color in the pie chart represents DEGs specifically involved in seed development. Colors in the pie chart represent different plant development and metabolism-related processes.
All identified DEGs (1,426 in total) in OsGW2_RNAi_7AP9 plants were analyzed by comparison with functionally characterized rice genes from the RICENCODE database. A total of 115 genes were well characterized with specific function (Supplementary Table 5). These genes were involved in the regulation of development of the seed, leaf, root, and shoot, apart from the genes related to the cell cycle, hormone signaling, and biotic and abiotic stress tolerance. Overlapping functions of these 115 characterized genes were represented a total of 327 times with respect to different functional terms (leaf, root, shoot, flower, and seed development, etc.). Of these, 55 genes were known to be involved in seed development in rice (Figure 5C, Supplementary Table 6).
To gain insight into the function of OsGW2 in seeds, DEGs in OsGW2_RNAi_7AP9 T3 seeds were compared with previously generated DEGs in five seed developmental stages (S1–S5) in the rice genotype IR64 (Sharma et al., 2012). The same analysis has also been successfully done for rice transgenic plants with altered expression of another seed-specific gene ONAC025. DEGs common to both transgenic plant and seed are the ones downstream to the transgene (Mathew et al., 2020). This analysis unveiled a correlation of DEGs in the OsGW2_RNAi_7AP9 transgenic line with seed-related genes. Of the 636 genes upregulated as a result of OsGW2 knockdown, 262 genes were also differentially expressed in the different seed developmental stages in comparison with the Y-leaf stage (Figure 6A). Of these, 131 genes were upregulated in both OsGW2_RNAi_7AP9 plants and S1–S5 seed developmental stages, of which 23 were seed-specific (Figure 6B). The upregulated seed-specific genes in OsGW2_RNAi_7AP9 plants were involved in various seed development and related processes/pathways. One such seed-specific transcription factor, OsMADS29, has been shown to affect many processes. Knockdown and overexpression of OsMADS29 revealed its function in hormone homeostasis, starch biosynthesis in the endosperm, and embryo development (Yang et al., 2012; Nayar et al., 2013). Also, 81 genes showed downregulation in various seed stages compared with the Y-leaf stage (Supplementary Figure 6A). Similarly, among the 50 downregulated genes in OsGW2_RNAi_7AP9 plants, 42 showed upregulation in the seed stages and eight of them showed downregulation in the seed stages compared with the Y-leaf stage (Figure 6A; Supplementary Figures 6B,C).
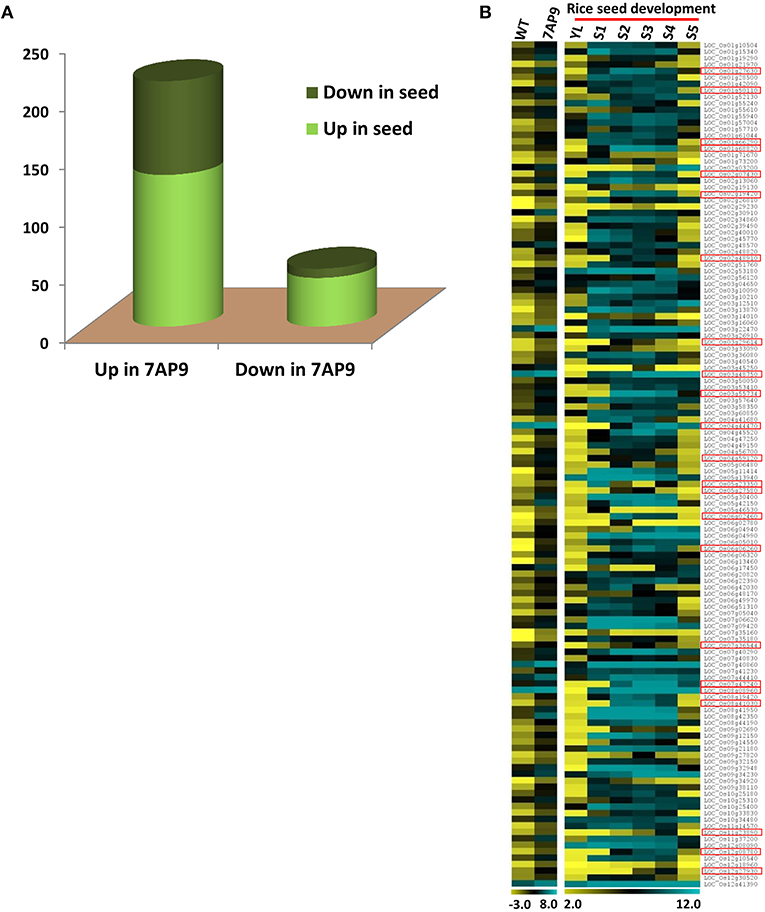
Figure 6. Comparison of transcriptomes of OsGW2_RNAi seed and five rice seed development stages. (A) Graph represents DEGs in both IR64 seed development stages and in OsGW2_RNAi_7AP9 seeds. The first and second bars represent gene numbers up- and downregulated, respectively, in OsGW2_RNAi_7AP9 seeds. Gene numbers up- and downregulated in rice seed are marked in light and dark green, respectively. (B) Heat map shows log2 expression values of genes upregulated in both OsGW2_RNAi_7AP9 seeds and in five stages of rice seed development. Upregulated seed-specific genes have been demarcated with red boxes in the figure. Color bar below the heat map shows the range of log2 GCRMA or FPKM values, for microarray and RNA-seq, respectively. YL represents Y-leaf and S1-S5 represent five stages of rice seed development.
Furthermore, the total DEGs were analyzed for their roles in metabolic pathways by MapMan. There was enrichment of secondary metabolites, starch and sucrose biosynthesis, and lipid metabolism, besides amino acid metabolism and cell wall biosynthetic pathways in the OsGW2_RNAi_7AP9 line (Figure 7A). A parallel level of pathway enrichment was also observed by KEGG. KEGG analysis of all DEGs revealed that photosynthesis, sucrose, and starch metabolism DEGs were mostly upregulated (Supplementary Figure 4). We found that genes involved in signaling pathways, including components of ABA, cytokinin, ethylene, GA, IAA, jasmonic acid, and transcription regulatory elements (including TFs), were enriched. Similarly, genes involved in protein modification and protein degradation were found to be enriched in OsGW2_RNAi_7AP9 (Figure 7B).
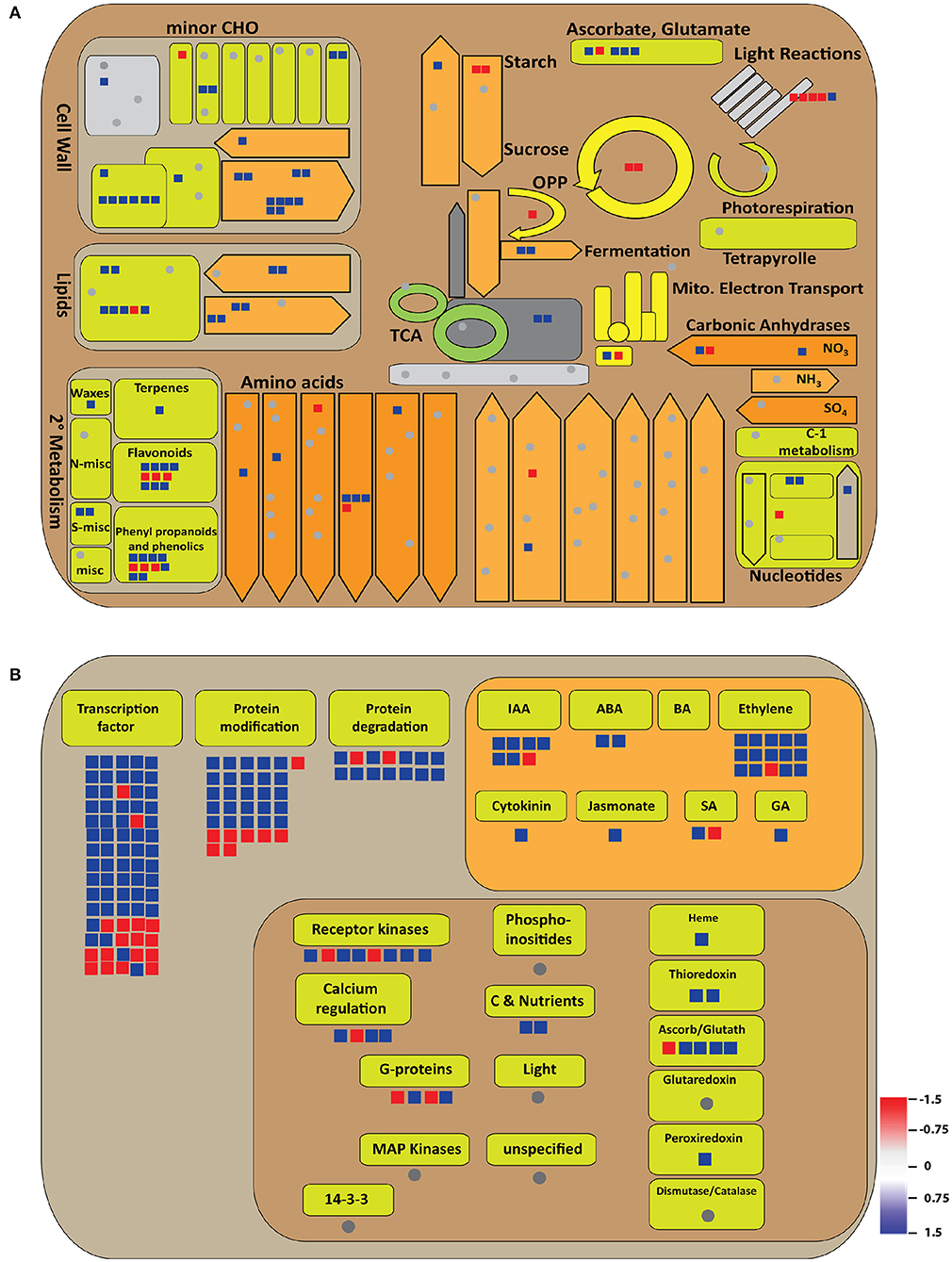
Figure 7. Illustration of metabolic processes involving significant DEGs (log2 fold change ≥1.5 or ≤−1.5) as visualized by the MapMan tool. (A) Metabolism and (B) regulation overview have been shown. Blue and red colors signify up- and downregulated genes, respectively, as shown in the color bar in (B).
Discussion
GRAIN WIDTH 2 codes for an E3 ubiquitin ligase. A natural variation in GW2 causes truncation of the protein-interacting domain in a long grain japonica rice cultivar, WY3, resulting in increased grain width and weight (Song et al., 2007). Recent steps toward the identification of the molecular mechanism of GW2 show that it interacts with EXPLA1, a cell wall-loosening protein (Choi et al., 2018), and chitinase 14 (CHT14) and phosphoglycerate kinase (PGK) which are involved in carbohydrate metabolism (Lee et al., 2018). Oochikara (a natural japonica rice gw2 mutant) has loosely packed spherical starch granules (Choi et al., 2018), also observed in knockdown indica rice in our experiments (Figure 4E). Since indica rice lacks the SNP leading to a stop codon (Dixit et al., 2013), the role of such a GW2 allele has hitherto not been explored in these varieties. This is the first report of functional relevance of GW2 in indica rice, which shows that its downregulation promotes seed width, length and weight, even in indica rice, and elucidates the genes downstream to GW2 responsible for this phenotype.
In the present study, four indica rice genotypes, with variation in seed size, lacked the natural variation of japonica genotypes, though GW2 expressed in all. In order to test if the GW2 protein has a functional role in indica rice, we generated a knockdown of OsGW2 in indica rice. The aim was to examine if a knockdown would generate a phenotype similar to a knockout phenotype observed in japonica genotypes. A similar RNAi-based knockdown approach has been adopted to explore the GW2 function in wheat which resulted in higher grain width and starch content (Sestili et al., 2019). While knockout (KO) is increasingly being used to study gene functions, the methodology has certain limitations, such as the unavailability of PAM sites, off-target effects, and most importantly a “fitness cost” (Ahmad et al., 2018). Partial knockdown of genes allows us to study the function of essential genes, while complete knockout of genes may be lethal to the plants (Boettcher and McManus, 2017; Moreira et al., 2020). Hence, RNAi-mediated knockdown of genes has its own utility. It has also been suggested to first characterize a gene by RNAi methodology and, subsequently, edit it by the CRISPR/Cas technique (Ansari et al., 2020). We did not generate KO in this attempt as the gene expresses ubiquitously, and a KO may hamper plant growth.
Grain weight, a key determinant of grain yield in rice, is positively associated with grain size parameters (grain length, width, and thickness; Shi et al., 2019). Majority of the genes (GW5, qGL3, UBP15/LG1, GL4, SPL16, etc.) impact grain size by affecting the rate of cell division/proliferation. Few genes (SPL13, KNAT7) were reported to control grain size by influencing cell expansion (Weng et al., 2008; Qi et al., 2012; Zhang et al., 2012; Wang et al., 2015, 2019; Si et al., 2016; Liu et al., 2017; Wu et al., 2017; Gao et al., 2019; Shi et al., 2019). In the last decade, some genes were reported that regulate grain size by controlling both cell proliferation as well as expansion (GS2, GS5; Li et al., 2011; Duan et al., 2015; Xu et al., 2015). In our study, SEM analysis of the central portion of the hull of mature grains of the knockdown line showed that an increase in grain size resulted from increased cell number. Previously, in Arabidopsis, it was observed that E3 ubiquitin ligase, DA2, and EOD1 also regulate grain size by affecting cell proliferation (Li et al., 2008; Xia et al., 2013). GW2 controls grain size in rice in a similar way as DA2 and EOD1 in Arabidopsis. Downregulation of OsGW2 led to a wider and larger grain. Collectively, OsGW2 negatively regulates grain size by affecting cell proliferation in the spikelet hull even in indica rice.
In dicot plants, endosperm disappears with embryo maturation, whereas in monocots, the endosperm covers the major proportion of the seed after maturation and stores starch in amyloplasts (Olsen et al., 1999). Sugars translocate to the endosperm from vegetative tissues and are converted to amylose and amylopectin at the time of grain filling and stored in the form of starch granules in amyloplasts (Nayar et al., 2013; Sonnewald and Kossmann, 2013). The starch granules in the endosperm of the OsGW2 knockdown lines were larger and more loosely packed. OsGW2 knockdown affected the size, shape, and number of starch granules. Transcriptome data suggested that transcript levels of starch metabolism/synthesis-related genes (OsIAA9, OsMADS6, OsMADS29) are upregulated in the silenced OsGW2_RNAi plants (Supplementary Table 6).
Hormone signal transduction, cell cycle, and sucrose metabolism-related genes are well known to be involved in grain size regulation through various metabolic processes (Li and Li, 2016; Lee et al., 2017). In our transcriptome study, KEGG, MapMan, and GO analyses revealed that DEGs that were significantly enriched pertained to biological processes (cellular process, primary and secondary metabolic processes), metabolism (carbohydrate, lipid, amino acid, and nucleotide), hormone signal transduction, starch metabolism, stress response, photosynthesis, etc. (Figure 7; Supplementary Figures 4, 5). Plant hormones are important grain size regulators. Auxins, cytokinins, and brassinosteroids play an important role in determining rice grain size. Auxin-responsive genes (OsARF4, Gnp4/LAX2, BG1, qTGW3, TGW6), a cytokinin-related gene (OsCKX2/Gn1a), and brassinosteroid-related genes (GS5, qGL3, GS2/OsGRF4) regulate grain size by being involved in signaling, biosynthesis, and transport (Azizi et al., 2019; Li and Li, 2019). In our transcriptome data, majority of auxin-responsive genes, including OsMGH3, OsMADS29, OsRAA1, and OsIAA9, showed upregulation in OsGW2 knockdown plants (Supplementary Table 6). In addition to this, cytokinin- and brassinosteroid-related genes were also upregulated in the OsGW2_RNAi_7AP9 line. These results provide evidence for hormone-responsive genes being important regulators of grain size in rice. Several genes (GS3, GS5, GW5, qGL3, TGW6, GIF1, GW8, etc.) are preferentially or specifically expressed in seed tissues and regulate grain size in rice by being part of various pathways like the ubiquitin–proteasome pathway, G-protein signaling, plant hormone signaling, mitogen-activated protein kinase (MAPK) signaling, and some transcription factors (Zuo and Li, 2014). Upregulation of starch metabolism and cell cycle-related genes (OsMADS6, OsMADS29, OsYUC9, and OsKRP3) in OsGW2 knockdown plants also indicates the pathways through which GW2 regulates seed development in rice. The DEGs generated by transcriptome analysis will in all probability be downstream of OsGW2 substrates, which regulate grain size in rice. We can speculate that upregulated genes in OsGW2_RNAi plants that are also seed-specific might be involved in direct grain size regulation.
Conclusion
Here, we described some important facets of OsGW2-mediated regulation of grain width and weight in indica rice. Both cell division and expansion in the hull of spikelet are responsible for higher grain width as well as weight in OsGW2 knockdown plants in indica rice. Additionally, OsGW2 also affects starch granule morphology and their packing. We further conducted RNA sequencing to elucidate the active molecular mechanisms in OsGW2 knockdown plants and identified enriched DEGs involved in hormone signal transduction and starch metabolism. Transcriptome analyses also suggested that OsGW2 plays a crucial role in the regulation of cellular and metabolic processes relevant to grain size regulation.
Data Availability Statement
RNA-seq was performed for one wild type line and one transgenic line (RNAi_7AP9), in biological replicates, using Illumina® HiseqTM. RNA sample preparation kit v.2 (Illumina; USA). The sequences generated were submitted to NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra/PRJNA659415) with SRA accession number PRJNA659415. Biosample accession numbers for individual samples are SAMN15905438_WT_Rep1, SAMN15905439_WT_Rep2, SAMN15905440_7AP9_Rep1, SAMN15905441_7AP9_Rep2.
Author Contributions
GP made the constructs, and RR and GP generated the rice transgenic plants. AV conducted all the experiments and analyzed the data. AKT and PA supervised the study and guided the experimental design and data analysis. AV and PA wrote the manuscript. All authors read/corrected the manuscript and approved it.
Funding
This work was supported by research grant no. BT/AB/NIPGR/SEED BIOLOGY/2012 to PA and BT/PR12394/AGIII/103/891/2014 to AKT from the Department of Biotechnology, Ministry of Science and Technology, India, and NIPGR core grant to PA; junior and senior research fellowships from the Department of Biotechnology, India, to AV; and junior and senior research fellowships from the Council of Scientific and Industrial Research (CSIR) to RR.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the support provided by the central instrumentation, microscopy, and plant growth facilities of NIPGR and the DBT e-library consortium for providing online access to the articles.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.600378/full#supplementary-material
Supplementary Figure 1. Comparison of grain parameters in five different rice genotypes. (A) Grain width, (B) grain length, and (C) grain weight. Error bar represents mean ± SD; n = 100. Grain width and length were measured with WINSEEDLETM. Nipponbare, LGR, Sonasal, IR64, and PB1 represent different rice genotypes.
Supplementary Figure 2. Screening of OsGW2_RNAi transgenic plants and effect of GW2 on grain size parameters. (A) Screening of OsGW2_RNAi T0 transgenic plants by PCR amplification of hygromycin resistance gene (Hygromycin phosphotransferase II). L represents Gene Ruler 100 bp DNA Ladder Mix. 1–35 are different transgenic lines in T0 generation. Ten positive T0 plants marked in red (2, 4, 7, 9, 13, 15, 17, 21, 23, 24) were grown further and analyzed. Effect of GW2 on grain size parameters; grain length (B) and width (C) of OsGW2_RNAi T1 seeds and; grain length (D) and width (E) of GW2 RNAi T2 seeds. In T1 generation four lines (RNAi_2A, 7A, 9B, and 24P2) were selected out of ten lines which showed 3:1 segregation ratio. Bar = 1 cm; WT represents wild-type PB-1 grains; grain length and width measured with 30 cm scale by aligning 10 seeds of each line length wise and breadth wise. Red line in (B–E) represents WT measurements for comparison.
Supplementary Figure 3. Screening of OsGW2_RNAi plants in T2 generation by PCR amplification of hygromycin resistance gene (Hygromycin phosphotransferase II). L represents Gene Ruler 1 kb DNA Ladder Mix. RNAi (2AP3, 2AP9, 7AP8, 7AP9, 9BP3, 9BP4, 24P2P5, 24P2P6) are different lines in T2 generation. N and P represent negative and positive control, respectively.
Supplementary Figure 4. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of DEGs. KEGG pathway analysis of (A) up-regulated and (B) down-regulated genes. The KEGG pathways with FDR value ≤0.05 were significantly enriched. Numbers represent DEGs enriched in each pathway.
Supplementary Figure 5. GO term enrichment analysis through AgriGO of DEGs in OsGW2_RNAi transgenic plants. The terms enriched under (A) biological process, (B) cellular component, and (C) molecular function have been shown. Each box contains GO terms, GO IDs, and statistical information. GO terms with FDR value ≤0.05 show significant enrichment. Yellow, orange, and red colors represent significant GO terms and white color represents non-significant terms as shown in color legend in (B). Arrows represent relationship between GO terms as shown in (B).
Supplementary Figure 6. Heat maps showing comparison of OsGW2_RNAi_7AP9 seeds RNAseq data with rice seed development microarray data. (A) DEGs down-regulated in IR64 seeds (S1–S5 stages) and up-regulated in OsGW2_RNAi_7AP9 transgenic seeds. (B) DEGs up-regulated in IR64 seeds and down-regulated in OsGW2_RNAi_7AP9 transgenic seeds. (C) DEGs down-regulated in both IR64 seeds as well as OsGW2_RNAi_7AP9 transgenic seeds. S1–S5 represent different seed developmental stages, YL represents Y leaf. WT and 7AP9 represent wild-type and OsGW2_RNAi transgenic line, respectively. The DEGs with log2 fold change ≥1.5 (up-regulated genes) and ≤−1.5 (down-regulated genes) with p-value cut off of ≤0.05 were considered as significant DEGs.
References
Agarwal, P., Arora, R., Ray, S., Singh, A. K., Singh, V. P., Takatsuji, H., et al. (2007). Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485. doi: 10.1007/s11103-007-9199-y
Agarwal, P., Kapoor, S., and Tyagi, A. K. (2011). Transcription factors regulating the progression of monocot and dicot seed development. Bioessays 33, 189–202. doi: 10.1002/bies.201000107
Ahmad, S., Wei, X., Sheng, Z., Hu, P., and Tang, S. (2018). CRISPR/Cas9 for development of disease resistance in plants: recent progress, limitations, and future prospects. Brief. Funct. Genomics 19, 26–39. doi: 10.1093/bfgp/elz041
Ansari, W. A., Chandanshive, S. U., Bhatt, V., Nadaf, A. B., Vats, S., Katara, J. L., et al. (2020). Genome editing in cereals: approaches, applications, and challenges. Int. J. Mol. Sci. 21, 1–32. doi: 10.3390/ijms21114040
Azizi, P., Osman, M., Hanafi, M. M., Sahebi, M., Rafii, M. Y., Taheri, S., et al. (2019). Molecular insights into the regulation of rice kernel elongation. Crit. Rev. Biotechnol. 39, 904–923. doi: 10.1080/07388551.2019.1632257
Boettcher, M., and McManus, M. T. (2017). Choosing the right tool for the job: RNAi, TALEN or CRISPR. Physiol. Behav. 176, 139–148. doi: 10.1016/j.molcel.2015.04.028
Chakrabarty, R., Banerjee, R., Chung, S., Farman, M., Citovsky, V., Hogenhout, S. A., et al. (2007). pSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana—virus interactions. Mol. Plant Microbe Interact. 20, 740–750. doi: 10.1094/MPMI-20-7-0740
Choi, B. S., Kim, Y. J., Markkandan, K., Koo, Y. J., Song, J. T., and Seo, H. S. (2018). GW2 functions as an E3 ubiquitin ligase for rice expansin-like 1. Int. J. Mol. Sci. 19, 1–12. doi: 10.3390/ijms19071904
Das, S., Parida, S. K., Agarwal, P., and Tyagi, A. K. (2019). Transcription factor OsNF-YB9 regulates reproductive growth and development in rice. Planta 250, 1849–1865. doi: 10.1007/s00425-019-03268-2
Ding, W., Tong, H., Zheng, W., Ye, J., Pan, Z., Zhang, B., et al. (2017). Isolation, characterization, and transcriptome analysis of a cytokinin receptor mutant osckt1 in rice. Front. Plant Sci. 8:88. doi: 10.3389/fpls.2017.00088
Dixit, N., Dokku, P., Amitha Mithra, S. V., Parida, S. K., Singh, A. K., Singh, N. K., et al. (2013). Haplotype structure in grain weight gene GW2 and its association with grain characteristics in rice. Euphytica 192, 55–61. doi: 10.1007/s10681-012-0852-4
Dong, X., Zhang, D., Liu, J., Liu, Q. Q., Liu, H., Tian, L., et al. (2015). Plastidial disproportionating enzyme participates in starch synthesis in rice endosperm by transferring maltooligosyl groups from amylose and amylopectin to amylopectin. Plant Physiol. 169, 2496–2512. doi: 10.1104/pp.15.01411
Du, Z., Zhou, X., Ling, Y., Zhang, Z., and Su, Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. doi: 10.1093/nar/gkq310
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2:15203. doi: 10.1038/nplants.2015.203
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Gao, X., Zhang, J.-Q., Zhang, X., Zhou, J., Jiang, Z., Huang, P., et al. (2019). Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 31, 1077–1093. doi: 10.1105/tpc.18.00836
Huang, K., Wang, D., Duan, P., Zhang, B., Xu, R., Li, N., et al. (2017). WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size, and shape in rice. Plant J. 91, 849–860. doi: 10.1111/tpj.13613
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Jung, I. J., Ahn, J. W., Jung, S., Hwang, J. E., Hong, M. J., Choi, H. I. I., et al. (2019). Overexpression of rice jacalin-related mannose-binding lectin (OsJAC1) enhances resistance to ionizing radiation in Arabidopsis. BMC Plant Biol. 19:561. doi: 10.1186/s12870-019-2056-8
Kubo, M., and Purevdorj, M. (2004). The future of rice production and consumption. J. Food Distrib. Res. 35, 128–142. doi: 10.22004/ag.econ.27145
Lee, K. H., Park, S. W., Kim, Y. J., Koo, Y. J., Song, J. T., and Seo, H. S. (2018). Grain width 2 (GW2) and its interacting proteins regulate seed development in rice (Oryza sativa L.). Bot. Stud. 59:23. doi: 10.1186/s40529-018-0240-z
Lee, Y., Choi, M. S., Lee, G., Jang, S., Yoon, M. R., Kim, B., et al. (2017). Sugary endosperm is modulated by starch branching enzyme IIa in rice (Oryza sativa L.). Rice 10:33. doi: 10.1186/s12284-017-0172-3
Li, N., and Li, Y. (2014). Ubiquitin-mediated control of seed size in plants. Front. Plant Sci. 5:332. doi: 10.3389/fpls.2014.00332
Li, N., and Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, N., and Li, Y. (2019). Molecular networks of seed size control in plants. J. Exp. Bot. 66, 1087–1097. doi: 10.1093/jxb/eru549
Li, N., Xu, R., Duan, P., and Li, Y. (2018). Control of grain size in rice. Plant Reprod. 31, 1–15. doi: 10.1007/s00497-018-0333-6
Li, Q., Li, L., Yang, X., Warburton, M. L., Bai, G., Dai, J., et al. (2010). Relationship, evolutionary fate, and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10:143. doi: 10.1186/1471-2229-10-143
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3:17043. doi: 10.1038/nplants.2017.43
Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U.S.A. 107, 19579–19584. doi: 10.1073/pnas.1014419107
Mathew, I. E., Das, S., Mahto, A., and Agarwal, P. (2016). Three rice NAC transcription factors heteromerize and are associated with seed size. Front. Plant Sci. 7:1638. doi: 10.3389/fpls.2016.01638
Mathew, I. E., Priyadarshini, R., Mahto, A., Jaiswal, P., Parida, S. K., and Agarwal, P. (2020). SUPER STARCHY1/ONAC025 participates in rice grain filling. Plant Direct 4, 1–25. doi: 10.1002/pld3.249
Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495. doi: 10.1093/pcp/pch048
Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16, 3181–3195. doi: 10.1105/tpc.104.161220
Moreira, D., Pereira, A. M., Lopes, A. L., and Coimbra, S. (2020). The best CRISPR/Cas9 versus RNA interference approaches for Arabinogalactan proteins' study. Mol. Biol. Rep. 47, 2315–2325. doi: 10.1007/s11033-020-05258-0
Nayar, S., Sharma, R., Tyagi, A. K., and Kapoor, S. (2013). Functional delineation of rice MADS29 reveals its role in embryo and endosperm development by affecting hormone homeostasis. J. Exp. Bot. 64, 4239–4253. doi: 10.1093/jxb/ert231
Olsen, O.-A., Linnestad, C., and Nichols, S. E. (1999). Developmental biology of the cereal endosperm. Trends Plant Sci. 4, 253–257. doi: 10.1016/S1360-1385(99)01431-4
Ouyang, S., Zhu, W., Hamilton, J., Lin, H., Campbell, M., Childs, K., et al. (2006). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 35, D883–D887. doi: 10.1093/nar/gkl976
Qi, P., Lin, Y.-S., Song, X.-J., Shen, J.-B., Huang, W., Shan, J.-X., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22, 1666–1680. doi: 10.1038/cr.2012.151
Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W. S., and Khvorova, A. (2004). Rational siRNA design for RNA interference. Nat. Biotechnol. 22, 326–330. doi: 10.1038/nbt936
Rosegrant, M. W., and Cline, S. A. (2003). Global food security: challenges and policies. Science 302, 1917–1919. doi: 10.1126/science.1092958
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. doi: 10.2144/03342mt01
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Sestili, F., Pagliarello, R., Zega, A., Saletti, R., Pucci, A., Botticella, E., et al. (2019). Enhancing grain size in durum wheat using RNAi to knockdown GW2 genes. Theor. Appl. Genet. 132, 419–429. doi: 10.1007/s00122-018-3229-9
Sharma, R., Agarwal, P., Ray, S., Deveshwar, P., Sharma, P., Sharma, N., et al. (2012). Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Funct. Integr. Genomics 12, 229–248. doi: 10.1007/s10142-012-0274-3
Shi, C., Ren, Y., Liu, L., Wang, F., Zhang, H., Tian, P., et al. (2019). Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol. 180, 381–391. doi: 10.1104/pp.19.00065
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Singh, G., Kumar, S., and Singh, P. (2003). A quick method to isolate RNA from wheat and other carbohydrate-rich seeds. Plant Mol. Biol. Report. 21, 93–93. doi: 10.1007/BF02773401
Smalle, J., and Vierstra, R. D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. doi: 10.1146/annurev.arplant.55.031903.141801
Song, X.-J., Huang, W., Shi, M., Zhu, M.-Z., and Lin, H.-X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Sonnewald, U., and Kossmann, J. (2013). Starches-from current models to genetic engineering. Plant Biotechnol. J. 11, 223–232. doi: 10.1111/pbi.12029
Toki, S., Hara, N., Ono, K., Onodera, H., Tagiri, A., Oka, S., et al. (2006). Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47, 969–976. doi: 10.1111/j.1365-313X.2006.02836.x
Vierstra, R. D. (2009). The ubiquitin−26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. doi: 10.1038/nrm2688
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, S., Yang, H., Mei, J., Liu, X., Wen, Z., Zhang, L., et al. (2019). Rice homeobox protein KNAT7 integrates the pathways regulating cell expansion and wall stiffness. Plant Physiol. 181, 669–682. doi: 10.1104/pp.19.00639
Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209. doi: 10.1038/cr.2008.307
Wu, W., Liu, X., Wang, M., Meyer, R. S., Luo, X., Ndjiondjop, M.-N., et al. (2017). A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 3:17064. doi: 10.1038/nplants.2017.64
Xia, T., Li, N., Dumenil, J., Li, J., Kamenski, A., Bevan, M. W., et al. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25, 3347–3359. doi: 10.1105/tpc.113.115063
Xing, Y., and Zhang, Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. doi: 10.1146/annurev-arplant-042809-112209
Xu, C., Liu, Y., Li, Y., Xu, X., Xu, C., Li, X., et al. (2015). Differential expression of GS5 regulates grain size in rice. J. Exp. Bot. 66, 2611–2623. doi: 10.1093/jxb/erv058
Yang, X., Wu, F., Lin, X., Du, X., Chong, K., Gramzow, L., et al. (2012). Live and let die–The Bsister MADS-Box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS ONE 7:e51435. doi: 10.1371/journal.pone.0051435
Ying, J. Z., Gao, J. P., Shan, J. X., Zhu, M. Z., Shi, M., and Lin, H. X. (2012). Dissecting the genetic basis of extremely large grain shape in rice cultivar “JZ1560.” J. Genet. Genomics 39, 325–333. doi: 10.1016/j.jgg.2012.03.001
Yuenyong, W., Chinpongpanich, A., Comai, L., Chadchawan, S., and Buaboocha, T. (2018). Downstream components of the calmodulin signaling pathway in the rice salt stress response revealed by transcriptome profiling and target identification. BMC Plant Biol. 18:335. doi: 10.1186/s12870-018-1538-4
Zha, K., Xie, H., Ge, M., Wang, Z., Wang, Y., Si, W., et al. (2019). Expression of maize MADS transcription factor ZmES22 negatively modulates starch accumulation in rice endosperm. Int. J. Mol. Sci. 20:483. doi: 10.3390/ijms20030483
Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U.S.A. 109, 21534–21539. doi: 10.1073/pnas.1219776110
Zhao, M., Zhao, M., Gu, S., Sun, J., Ma, Z., Wang, L., et al. (2019). DEP1 is involved in regulating the carbon–nitrogen metabolic balance to affect grain yield and quality in rice (Oryza sativa L.). PLoS ONE 14:e0213504. doi: 10.1371/journal.pone.0213504
Keywords: E3 ubiquitin ligase, grain width, GW2, Oryza sativa, seed development
Citation: Verma A, Prakash G, Ranjan R, Tyagi AK and Agarwal P (2021) Silencing of an Ubiquitin Ligase Increases Grain Width and Weight in indica Rice. Front. Genet. 11:600378. doi: 10.3389/fgene.2020.600378
Received: 29 August 2020; Accepted: 27 November 2020;
Published: 12 January 2021.
Edited by:
Christian Meyer, INRA UMR1318 Institut Jean Pierre Bourgin, FranceReviewed by:
Longbiao Guo, Chinese Academy of Agricultural Sciences, ChinaHimanshu Sharma, National Agri-Food Biotechnology Institute, India
Copyright © 2021 Verma, Prakash, Ranjan, Tyagi and Agarwal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pinky Agarwal, cGlua3kuYWdhcndhbEBuaXBnci5hYy5pbg==; cGlua3kuYWdhcndhbEBnbWFpbC5jb20=
 Ankit Verma
Ankit Verma Geeta Prakash1,2
Geeta Prakash1,2 Rajeev Ranjan
Rajeev Ranjan Akhilesh K. Tyagi
Akhilesh K. Tyagi Pinky Agarwal
Pinky Agarwal