- 1Department of Ultrasound, The 2nd Affiliated Hospital of Harbin Medical University, Harbin, China
- 2Department of Ultrasound, Harbin Red Cross Central Hospital, Harbin, China
- 3Department of Ultrasound, People’s Hospital of Zhejiang Province, Hangzhou, China
Many epidemiological studies have confirmed that ICAM-1 gene single-nucleotide polymorphisms (SNPs) are associated with susceptibility of various cancers, but there are relatively few studies on the relationship between ICAM-1 gene polymorphisms and the risk of cervical cancer. Therefore, we aimed to explore the potential role of ICAM-1 gene polymorphisms and the combined effect of SNPs in the pathogenesis of cervical cancer in Han women in northern China. This case–control group includes 488 cases of cervical cancer, 684 cases of cervical precancerous lesions, and 510 healthy females. Multiplex polymerase chain reaction (PCR) combined with the next-generation sequencing method was used for the determination of gene polymorphisms (rs5498, rs3093030, and rs281432). In our study, we divide cervical cancer into two subgroups: cervical squamous cell carcinoma (CSCC) group and cervical adenocarcinoma (CAC) group. We analyzed the alleles and genotypes of all research subjects using multivariate logistic regression analysis combined with 10,000 permutation tests. In addition, we also analyzed the distribution of haplotypes of the three SNPs in cervical cancer and cervical precancerous lesions. We found that the T allele and the dominant model of rs3093030 were associated with the susceptibility of cervical cancer (p = 0.042, p = 0.040, respectively). However, the significance disappeared after the Bonferroni correction for multiple testing (p > 0.05). For rs5498, its mutant gene G, the codominant model, and the dominant model could reduce the risk of CAC (p = 0.009, p = 0.028, p = 0.011, respectively). Significant differences remained after Bonferroni correction (p < 0.05, all). In addition, the frequency of haplotype “CTG” was significantly lower in the CAC group than in the controls. In conclusion, the study suggested that ICAM-1 gene polymorphisms may have a potential role in the pathogenesis of cervical cancer in the northern Chinese Han population.
Introduction
Cervical cancer, a serious public health concern, is the fourth most commonly diagnosed cancer and also the fourth leading cause of cancer deaths in women worldwide (Torre et al., 2015). Eighty-three percent of these deaths are reported in developing countries (Berumen-Campos, 2006). Despite that cervical cytology screening has been common in recent decades, cervical cancer is still one of the most common cancers in women (Chen et al., 2016c; Zheng et al., 2016; Siegel et al., 2017). Early cervical cancer can be treated by surgery or radiotherapy and chemotherapy with good outcomes, but metastatic cervical cancer is still incurable (Uyar and Rader, 2014). Therefore, it is particularly important to find an effective diagnosis method in the early stage. Since single-nucleotide polymorphisms (SNPs) are important genetic markers for the localization of complex diseases such as human cancer (Shastry, 2009; Tan, 2017), the study of certain SNPs related to cervical cancer will contribute to the early diagnosis and treatment of cervical cancer.
Intercellular adhesion molecule-1 (ICAM-1), also known as CD54, is a member of the immunoglobulin superfamily (IGSF) of adhesion molecules and plays an important role in mediating adhesion reactions. The ICAM-1 gene is located on chromosome 19p13.2 and has five extracellular immunoglobulin-like domains, a single transmembrane, and a short cytoplasmic tail (Springer, 1990; Staunton et al., 1990). There are two forms of ICAM-1 in vivo, namely, membrane ICAM-1 (mICAM-1) and soluble ICAM-1 (sICAM-1). ICAM-1 exists in the extracellular form of sICAM-1, which is mainly produced by mICAM-1 entering the blood after lysis by protease. ICAM-1 plays an important role in promoting adhesion in the inflammation site, controlling tumor progression and metastasis, and regulating the body’s immune response, and is widely distributed in a variety of cell types, including fibroblasts, keratinocytes, vascular endothelial cells, macrophages, and glandular epithelial cells and stromal cells of the uterus (Dustin et al., 1986; Larson and Springer, 1990; Tabibzadeh and Poubouridis, 1990).
Interestingly, several studies have shown that ICAM-1 gene polymorphisms were associated with susceptibility to breast cancer (Kammerer et al., 2004), gastric cancer (Tian et al., 2012), non-small cell lung cancer (Thanopoulou et al., 2012), hepatocellular carcinoma (Chen et al., 2016b), prostate cancer (Chen et al., 2006), melanoma (Natali et al., 1990), and ovarian cancer (Gardner et al., 1995), suggesting that the SNPs of the ICAM-1 gene may be common genetic factors that affect individual cancer susceptibility. To prove this hypothesis, we conducted a replication study to assess the relationship between ICAM-1 gene polymorphisms and the risk of cervical cancer in the northern Chinese Han population, which was different from the population studied by Sun et al. (2016).
Materials and Methods
Participants
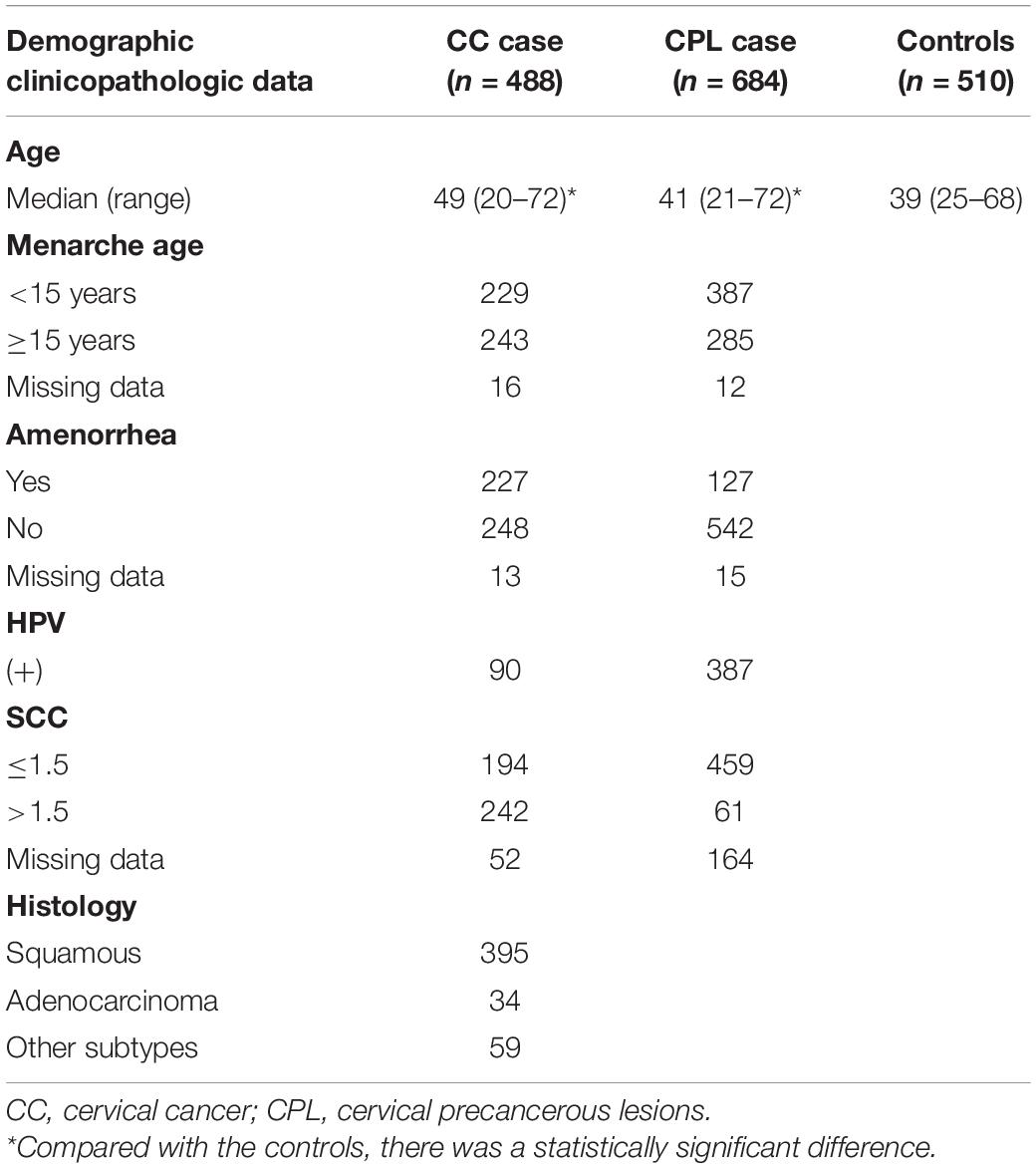
A total of 488 cervical cancer patients, 684 cervical precancerous lesion patients, and 510 healthy females were recruited from the second affiliated hospital of Harbin Medical University in Heilongjiang Province, China, from September 2014 to October 2018 in this study. The cervical cancer group consists of 395 cases of cervical squamous cell carcinoma (CSCC), 34 cases of cervical adenocarcinoma (CAC), and 59 cases of other pathological types of cancer. On this basis, we have further analyzed the relationship between common pathological types of cervical cancer, CSCC and CAC, and ICAM-1 gene polymorphisms. In addition, for cervical precancerous lesions, only patients with high-grade intraepithelial lesions, which included moderate and severe dysplasia and carcinoma in situ, were recruited. All the patients obtained their tissue specimens by surgery or biopsy and were further diagnosed by the pathologists of the Second Affiliated Hospital of Harbin Medical University. At the same time, the exclusion criteria were as follows: cervical benign lesions, cervical benign tumors, other cervical malignant tumors, and patients with cervical lesions who have undergone preoperative radiotherapy and chemotherapy. These healthy women were selected from a population of TCT-negative, cancer-free, or cancer-free family history who had undergone regular physical examinations at the Second Affiliated Hospital of Harbin Medical University or who volunteered to participate in epidemiological investigations. The cases and controls were non-probabilistic continuous samples. All participants were biologically unrelated Han Chinese female living in northern China.
The research was approved by the ethics committee of the Second Affiliated Hospital of Harbin Medical University, and informed consent for participation in this study was obtained from all subjects.
Selection of SNPs
Based on Sun et al.’s (2016) study of cervical carcinogenesis in Taiwanese women, as well as published candidate gene-association studies, and combined with the characteristics of the East Asian population in HapMap, we selected three ICAM-1 gene SNPs, rs5498, rs3093030, and rs281432. The minor allele frequency of the three SNPs is shown as follows: rs5498 PG = 0.381, rs3093030 PT = 0.317, and rs281432 PG = 0.360. Of the SNPs, rs5498 (A > G) was located in the exon region, rs3093030 (C > T) was SNP between the ICAM-1 and ICAM-4 genes, and rs281432 (C > G) was located in the intron region.
Extraction of DNA
Peripheral venous blood was collected from all subjects on admission and placed in a 2% EDTA-Na2 anticoagulant tube, which was then refrigerated at −80°C until DNA extraction. The genomic DNA of all subjects was extracted in strict accordance with the standard steps of the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China). DNA samples were amplified by PCR. All genotyping experiments were completed by Shanghai Biowing Applied Biotechnology Company1 using the multiplex PCR method combined with next-generation sequencing methods (Chen et al., 2016a). Primer 3 online software (Version 0.4.0)2 was used to design amplification primers for rs5498, rs3093030, and rs281432 of the ICAM-1 gene. The amplification primers are as follows: for rs5498 sense primer 5′-TGCCCATCGGGGAATCAG-3′ and anti-sense primer 5′-CAGTGATGATGACAATCTCATACC-3′, for rs3093030 sense primer 5′-TAATAAAGCTTTCTCAACTGCCTC-3′ and anti-sense primer 5′-TCTGGAGAGGAGTCTTTAGTTTTC-3′, and for rs281432 sense primer 5′-GATTGATGGGAGGAAGGGTG-3′ and anti-sense primer 5′-TGTCCTTCCCTTCTTGAATACG-3′. After PCR amplification, the PCR products were purified by TIANgel Midi Purification Kit (Tiangen Biotech, China). The purified PCR products were then paired-end sequenced (2 × 150 bp) by Illumina HiSeq X Ten platform according to the SOP. The readings were aligned to the human reference genome using Burrows–Wheeler Aligner (BWA, v0.7.12) (Li, 2011), and Samtools (v0.1.19) was used for SNP calling and genotyping.
Allele, Genotype, and Haplotype Association Analysis
In order to find the relationship between ICAM-1 gene polymorphisms and cervical cancer and cervical precancerous lesion susceptibility, we used the following models: codominant, dominant, and recessive models. Allele frequencies and genotype distribution (under 2-df codominant, 1-df dominant, and 1-df recessive models) were compared between groups using the χ2 test of independence with a 2 × 2 contingency. Haplotype analysis between controls and cases was implemented using the SHEsis software3 (Shi and He, 2005; Li et al., 2009). Where appropriate, the odds ratios (OR) with corresponding 95% confidence intervals (CI) were calculated. Empirical p values were calculated by 10,000 permutation tests using the Max(T) permutation procedure implemented in PLINK 1.9 software4 (Purcell et al., 2007). Moreover, multiple comparisons were counteracted by using the Bonferroni correction in PLINK 1.9 software. P < 0.05 was considered statistically significant.
Multivariable Logistic Regression Analysis
Multivariable logistic regression analysis (using SPSS V21.0 for Windows) was used to assess the association of the allele and genotype of each SNP with cervical cancer and cervical precancerous lesion after adjustment for age.
Results
SNP Genotype and Quality
Linkage disequilibrium among the SNPs was tested using SHEsis software. All SNPs were found to be in Hardy–Weinberg equilibrium in the case group and controls (Table 1). In order to ensure the accuracy of the study, we set the quality control for 73 samples with an accuracy rate of 98.5%, providing evidence for the reliability of subsequent studies.
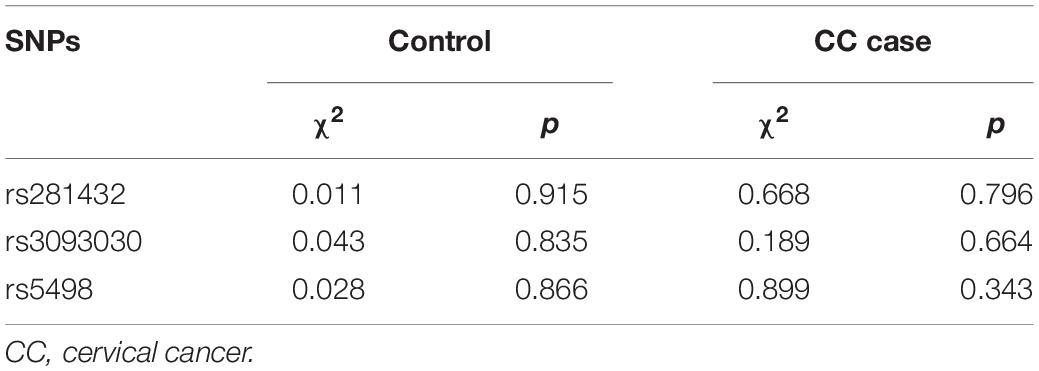
Table 1. Hardy–Weinberg equilibrium test results of ICAM-1 SNPs in the cervical cancer and control groups.
Clinical Characteristics of the Study Population
The characteristics of the individuals enrolled in our study are listed in Table 2. The ages of the subjects with cervical precancerous lesions and cervical cancer and the controls were significantly different (p < 0.05). In the cervical cancer group, there were 395 patients with squamous cell carcinoma, 34 patients with adenocarcinoma, and 59 patients with other pathological types.
ICAM-1 SNPs and Cervical Cancer
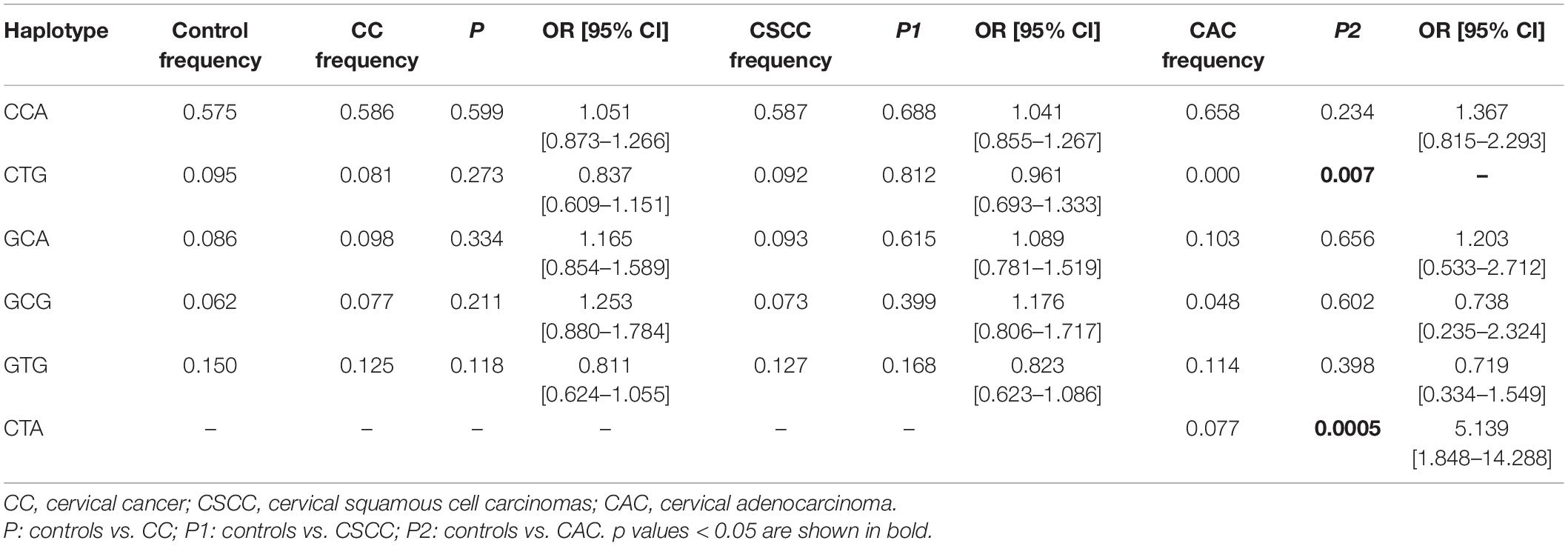
We genotyped three ICAM-1 gene polymorphisms as follows: rs5498, rs3093030, and rs281432. The genotype and allele frequencies of each SNP between the controls and cervical cancer patients and subgroups, CSCC and CAC, are summarized in Table 3. The results showed that the frequency of the rs3093030 C/T allele was significantly different between the controls and cervical cancer patients (p = 0.042). However, logistic regression analysis-adjusted age factor and Bonferroni correction showed no significant statistical significance (p = 0.060, p = 0.125, respectively). The frequency of the rs5498 A/G allele was significantly different between the controls and CAC patients (p = 0.009). The results of p after logistic regression analysis-adjusted age factors showed that the frequency of the rs5498 A/G allele in CAC patients was also significantly different from controls (p = 0.016). This difference remained statistically significant after Bonferroni correction (p = 0.028). For the rs281432 C/G allele, compared with the controls, there was no significant correlation in either the cervical cancer patients or the subgroups, CSCC and CAC (p > 0.05).
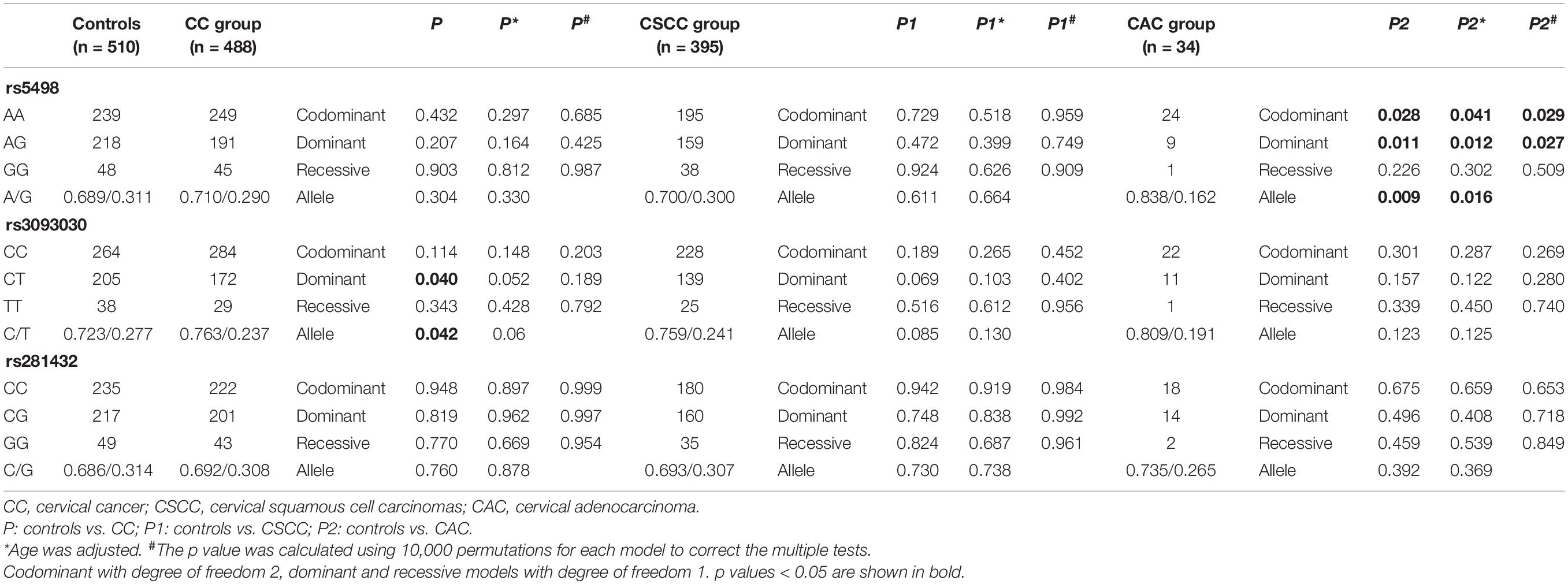
Table 3. Distribution of genotypes and alleles of three SNPs in the cervical cancer and control groups.
For the rs5498 SNP, the codominant model and dominant model distributions were significantly different in controls and CAC patients (p = 0.028, p = 0.011, respectively). Moreover, after performing 10,000 permutations, the p value was marginally significant (codominant model p = 0.029, dominant model p = 0.027). After Bonferroni correction, we found that there were marginal differences under the codominant model and dominant model (p = 0.049, p = 0.042, respectively). For the rs3093030 SNP, compared with the controls, dominant model distribution in cervical cancer patients was significantly different (p = 0.040). After 10,000 permutations, the p value of the dominant model was 0.189, and the significance disappeared after Bonferroni correction for multiple testing, indicating that the controls have no significant association with cervical cancer patients. Neither the genotype nor the allele frequencies of the rs281432 SNP were significantly different compared controls with cervical cancer patients or subgroups, CSCC and CAC.
ICAM-1 SNPs and Cervical Precancerous Lesions
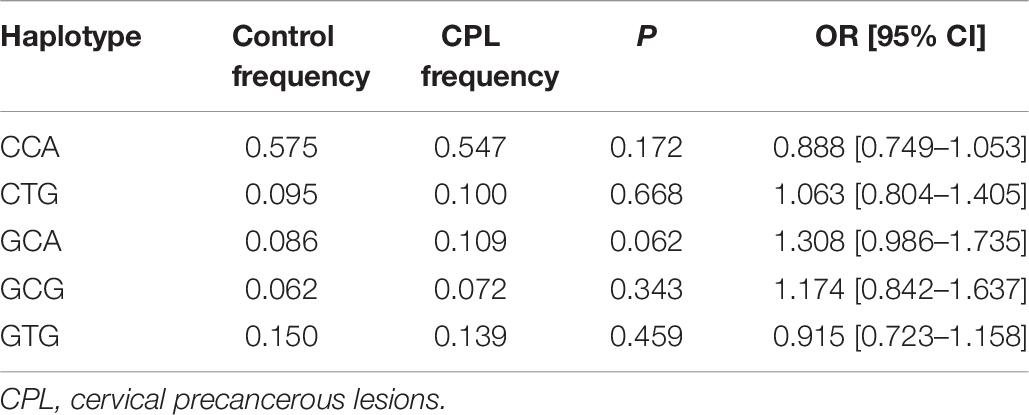
Table 4 shows the distribution of the genotype and allele frequencies of the three SNPs of the ICAM-1 gene in the healthy control group and the cervical precancerous lesion group. With healthy women as a control, there was no significant difference between the three SNPs and the genetic susceptibility of patients with cervical precancerous lesions (p > 0.05, all). We further analyzed the relationship between cervical precancerous lesions and cervical cancer (Table 4); neither the genotype nor the allele frequencies of the polymorphisms were significantly different (p > 0.05, all). After using logistic regression analysis to adjust for the age factors, there was no difference with cervical precancerous lesions (p > 0.05, all). There was no statistical significance in the results obtained by 10,000 permutations. Similarly, the consistency of the results of 10,000 permutations provides evidence for the accuracy of our research.
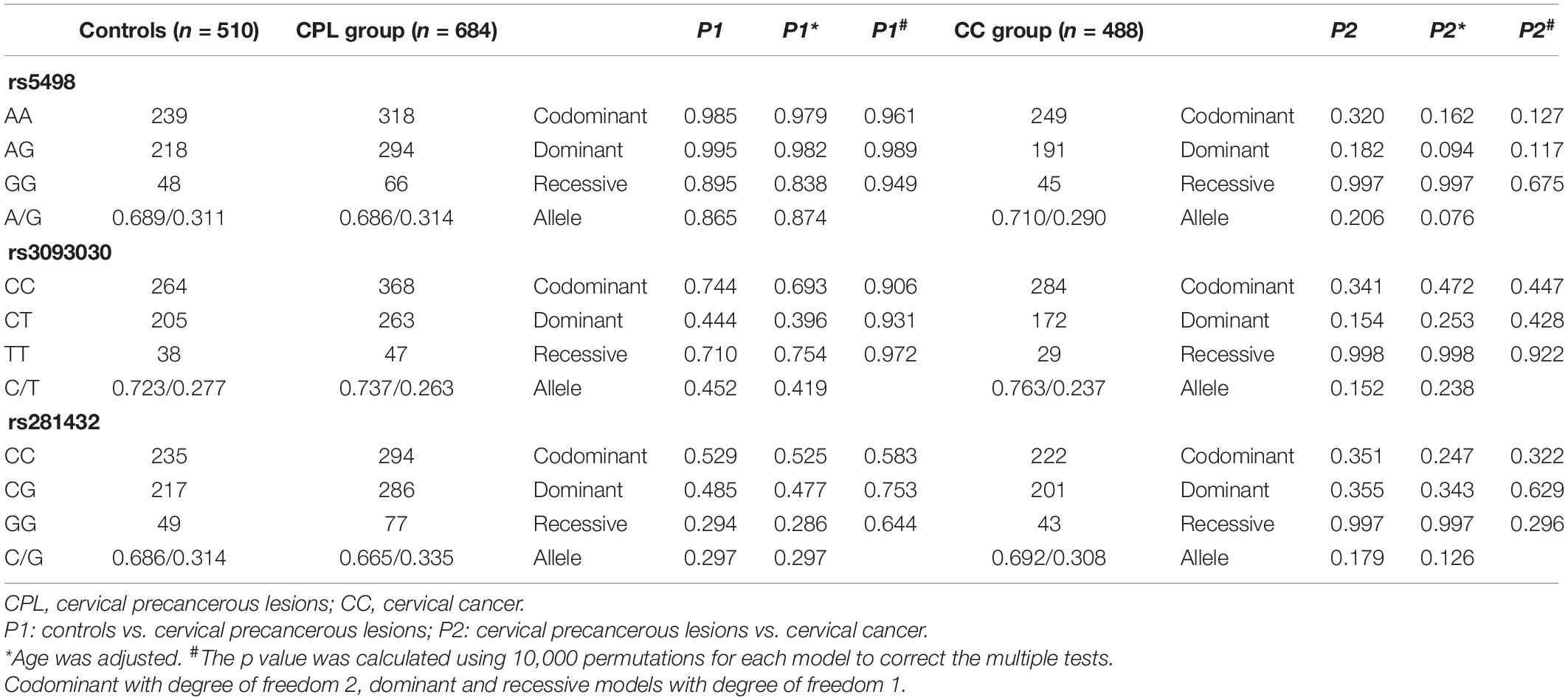
Table 4. The distribution of the three SNP genotypes and alleles of the ICAM-1 gene in the cervical precancerous lesion group and the control group.
Haplotype Analysis
The relationship between the haplotype distribution of the ICAM-1 gene and cervical cancer and cervical precancerous lesions is shown in Tables 5, 6. For haplotype analyses, we found the “CTA” haplotype only in the CAC group. Furthermore, the frequency of “CTG” was significantly lower in the CAC group than in the controls (p = 0.007).
Discussion
Cervical cancer is a gynecological malignant tumor with high mortality (Dongol et al., 2014). Its occurrence and development are a long-term, continuous, multifactor, and multistep complex process. Persistent infection of high-risk HPV and chronic inflammation are currently recognized causes of cervical cancer (Feng et al., 2016; Yang et al., 2016; Li et al., 2017; Wang and Luo, 2018). More and more studies show that genetic variation plays an indispensable role in the pathogenesis of cervical cancer. SNPs are one of the most common types of genetic variation in humans. It is widely present in the human genome and is considered to be the third-generation genetic marker, which is closely related to the susceptibility to many diseases (Shastry, 2009; Tan, 2017). Based on the findings of previous studies, there was no research on the relationship between the SNPs of the ICAM-1 gene and the risk of cervical cancer in the northern Chinese population. Therefore, we reported for the first time the association between ICAM-1 gene polymorphisms and the risk of cervical cancer in the northern Chinese population.
In recent years, a large amount of evidence has shown that ICAM-1 has a dual role in the occurrence and development of tumors (Reina and Espel, 2017). ICAM-1 not only participates in the recognition and killing of tumor cells by immune cells but also plays a certain role in the process of tumor evading the body’s immunity. ICAM-1 can not only mediate the nonspecific adhesion between tumor cells and T cells and facilitate the recognition of tumor cells by T cells but also bind to the LFA-1 ligand on the surface of T cells to form a complex ICAM-1/LFA-1. Tumor necrosis factor-α (TNF-α) and interleukin-1 (IL-1) secreted by ICAM-1/LFA-1 can not only kill tumor cells directly but also significantly upregulate the expression of ICAM-1 on the surface of tumor cells, to promote the recognition of immune cells to tumor cells and further expand its killing effect (Nakayama et al., 2011). Therefore, some scholars believe that the formation of the ICAM-1/LFA-1 complex is a positive feedback mechanism in the immune process of tumor cells, which helps to eliminate tumor cells in the body (Ren et al., 2010). However, sICAM-1, as a competitive inhibitor of ICAM-1, can competitively bind to the LFA-1 ligand, inhibit the formation of the ICAM-1/LFA-1 complex, weaken its killing effect, and cause tumor cells to evade the body’s immune surveillance (Dustin et al., 1986; Nasu et al., 1997; Li and Gao, 2008), resulting in tumor development and metastasis.
This study focused on the relationship between the rs5498, rs3093030, and rs281432 polymorphisms of the ICAM-1 gene and the susceptibility of cervical cancer in the northern Chinese population. Our results indicated that rs3093030 was significantly associated with the susceptibility of cervical cancer, that is, carrying the T allele and the dominant model has a protective effect on the susceptibility of cervical cancer (p = 0.042, OR = 0.811, 95% CI: 0.663∼0.993; p = 0.040, OR = 0.769, 95% CI: 0.598–0.988). It was speculated that the protective effect may be related to the increase of sICAM-1 concentration by rs3093030 (Bielinski et al., 2011; Lin et al., 2013; Ghasemian et al., 2015). The rs3093030 located at the 3′ end of the ICMA-1 gene has no known function (Bielinski et al., 2008). Therefore, rs3093030 may affect the concentration level of sICAM-1 through a special mechanism, thereby affecting the competitive effect with ICAM-1. On the one hand, ICAM-1 can bind more to the ligand LAF-1 and exert its killing effect in tumor cells. On the other hand, it also reduces the immune evasion of tumor cells, inhibits the occurrence and development of tumors, and protects the susceptibility of tumors. However, after Bonferroni correction, we did not find a significant association between the rs3093030 polymorphisms and cervical cancer. Therefore, it is necessary to further study whether there is an association between rs3093030 and the susceptibility to cervical cancer. In addition, our study did not find that rs5498 was associated with the risk of cervical cancer, but subgroup analysis showed that rs5498 was associated with the risk of CAC. In the codominant model and dominant model, it has a protective effect on the susceptibility of CAC (codominant p = 0.028; dominant p = 0.011, OR = 0.374, 95% CI: 0.175–0.799), and its mutant gene G can also be used as a protective factor to reduce the risk of CAC (p = 0.009, OR = 0.045, 95% CI: 0.221–0.827). The logistic regression analysis showed that rs5498 was significantly associated with CAC (codominant p = 0.041; dominant p = 0.012, OR = 0.362, 95% CI: 0.164–0.799; allele p = 0.016, OR = 0.431, 95% CI: 0.218–0.853). Meanwhile, the correlation remained after 10,000 permutation tests and after Bonferroni correction. rs5498 may be closely related to the pathological classification of cervical cancer. Rs5498 was located 3 bp upstream of the splice donor site, Bielinski et al. (2011) confirmed that when this site was mutated, it will cause the destruction of the splice site, causing the G mutant gene vector to produce a higher concentration of sICAM-1. This process will help tumor cells to escape from the body’s immune killing and immune escape occurs. However, in our study, false-positive results due to the small sample size of the CAC group were not ruled out. Therefore, the impact of rs5498 on cervical cancer was controversial, and further classification and repeated studies with large sample sizes were needed. Bielinski et al. confirmed that rs281432 was associated with sICAM-1 (Bielinski et al., 2011). In addition, it has been reported that elevated serum sICAM-1 levels may contribute to the diagnosis of malignancy in suspected breast neoplastic diseases (Altomonte et al., 1999). However, no significant association between rs281432 SNP and cervical cancer was found in this study, or in the subgroup.
This study also analyzed the relationship between the SNPs of the ICAM-1 gene and cervical precancerous lesions in the Han population in northern China. The results showed that the rs5498, rs3093030, and rs281432 of the ICAM-1 gene were not significantly associated with the susceptibility of cervical precancerous lesions. It was inconsistent with the research results reported by Sun et al. (2016) that rs281432 and rs5498 can increase the risk of cervical precancerous lesions. The reasons for the two different research results may be attributed to the following points. Firstly, limited sample sizes, living environment (such as lifestyle, education methods, and socioeconomic factors), and ethnic difference may change the genetic effects of ICAM-1 gene mutations. Secondly, other genes or factors may be involved in the development of cervical cancer. Last but not least, the relationship between the ICAM-1 gene and the risk of cervical cancer may not be achieved by affecting the structure and function of encoding protein.
To assess the combined effect of SNPs on the risk of cervical cancer and to identify the possible risk haplotypes in the population, we performed multilocus haplotype analysis. The haplotype analysis showed that “CTG” was significantly associated with CAC risk (p = 0.007). The result of the above haplotype frequency distribution was significantly lower in the CAC cases than in the controls. The haplotype “CTG” was found to significantly reduce the risk of CAC.
Although Sun et al. (2016) confirmed that ICAM-1 gene polymorphism is significantly associated with cervical cancer, the sample size is relatively small, including only 91 cases of cervical cancer, 63 cases of cervical precancerous lesions, and 290 healthy controls. A large sample size is very important in exploring and verifying the potential mechanism of cervical cancer in Chinese women. This study analyzes and elaborates from multiple angles such as cervical cancer subgroups and cervical precancerous lesions, which will play an important role in future research on cervical cancer with different pathological types. This study confirmed for the first time the correlation between ICAM-1 gene polymorphisms and cervical cancer in the northern Chinese Han population. This discovery can guide us to develop new understanding and insights into the mechanism and pathological process of cervical cancer. In addition, this study also has some unavoidable limitations. First of all, this study uses a case–control study, and selection bias cannot be avoided. Second, the sample size of the CAC group in this study is small, so it is necessary to conduct a larger sample size replication study to confirm the results. Finally, our study population is limited to Han women in northern China. In order to improve this study, people from different regions and ethnic backgrounds should be included.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) are: FigShare, doi:106084/m9.figshare.14039183.
Ethics Statement
Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YF: data collection, data analysis, and manuscript writing. XL and QM: data analysis. SZ, MZ, SL, and LF: data collection. JT and LS: project development. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82071929).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are deeply grateful to all the study participants. We are also grateful to the National Natural Science Foundation of China (No. 82071929).
Footnotes
- ^ http://www.biowing.com.cn
- ^ http://frodo.wi.mit.edu/
- ^ http://analysis.bio-x.cn/myAnalysis.php
- ^ http://www.Cog-genomics.org/plink/1.9/
References
Altomonte, M., Fonsatti, E., Lamaj, E., Cattarossi, I., Cattelan, A., and Maio, M. (1999). Differential levels of soluble intercellular adhesion molecule-1 (sICAM-1) in early breast cancer and benign breast lesions. Breast Cancer Res. Treat. 58, 19–23. doi: 10.1023/a:1006280729252
Berumen-Campos, J. (2006). [Human papilloma virus and cervical cancer]. Gac. Med. Mex. 142(Suppl. 2) 51–59.
Bielinski, S. J., Pankow, J. S., Li, N., Hsu, F. C., Adar, S. D., Jenny, N. S., et al. (2008). ICAM1 and VCAM1 polymorphisms, coronary artery calcium, and circulating levels of soluble ICAM-1: the multi-ethnic study of atherosclerosis (MESA). Atherosclerosis 201, 339–344. doi: 10.1016/j.atherosclerosis.2008.02.031
Bielinski, S. J., Reiner, A. P., Nickerson, D., Carlson, C., Bailey, K. R., Thyagarajan, B., et al. (2011). Polymorphisms in the ICAM1 gene predict circulating soluble intercellular adhesion molecule-1(sICAM-1). Atherosclerosis 216, 390–394. doi: 10.1016/j.atherosclerosis.2011.02.018
Chen, H., Hernandez, W., Shriver, M. D., Ahaghotu, C. A., and Kittles, R. A. (2006). ICAM gene cluster SNPs and prostate cancer risk in African Americans. Hum. Genet. 120, 69–76. doi: 10.1007/s00439-006-0184-3
Chen, K., Zhou, Y. X., Li, K., Qi, L. X., Zhang, Q. F., Wang, M. C., et al. (2016a). A novel three-round multiplex PCR for SNP genotyping with next generation sequencing. Anal. Bioanal. Chem. 408, 4371–4377. doi: 10.1007/s00216-016-9536-6
Chen, T. P., Lee, H. L., Huang, Y. H., Hsieh, M. J., Chiang, W. L., Kuo, W. H., et al. (2016b). Association of intercellular adhesion molecule-1 single nucleotide polymorphisms with hepatocellular carcinoma susceptibility and clinicopathologic development. Tumour Biol. 37, 2067–2074. doi: 10.1007/s13277-015-3992-z
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016c). Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132. doi: 10.3322/caac.21338
Dongol, S., Tai, Y., Shao, Y., Jiang, J., and Kong, B. (2014). A retrospective clinicopathological analysis of small-cell carcinoma of the uterine cervix. Mol. Clin. Oncol. 2, 71–75. doi: 10.3892/mco.2013.193
Dustin, M. L., Rothlein, R., Bhan, A. K., Dinarello, C. A., and Springer, T. A. (1986). Induction by IL 1 and interferon-gamma: tissue distribution, biochemistry, and function of a natural adherence molecule (ICAM-1). J. Immunol. 137, 245–254.
Feng, D., Yan, K., Zhou, Y., Liang, H., Liang, J., Zhao, W., et al. (2016). Piwil2 is reactivated by HPV oncoproteins and initiates cell reprogramming via epigenetic regulation during cervical cancer tumorigenesis. Oncotarget 7, 64575–64588. doi: 10.18632/oncotarget.11810
Gardner, M. J., Jones, L. M., Catterall, J. B., and Turner, G. A. (1995). Expression of cell adhesion molecules on ovarian tumour cell lines and mesothelial cells, in relation to ovarian cancer metastasis. Cancer Lett. 91, 229–234. doi: 10.1016/0304-3835(95)03743-g
Ghasemian, A., Najar Peerayeh, S., Bakhshi, B., and Mirzaee, M. (2015). The microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) genes among clinical isolates of Staphylococcus aureus from hospitalized children. Iran. J. Pathol. 10, 258–264.
Kammerer, S., Roth, R. B., Reneland, R., Marnellos, G., Hoyal, C. R., Markward, N. J., et al. (2004). Large-scale association study identifies ICAM gene region as breast and prostate cancer susceptibility locus. Cancer Res. 64, 8906–8910. doi: 10.1158/0008-5472.CAN-04-1788
Larson, R. S., and Springer, T. A. (1990). Structure and function of leukocyte integrins. Immunol. Rev. 114, 181–217. doi: 10.1111/j.1600-065x.1990.tb00565.x
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, M., and Gao, C. J. (2008). [An update on beta2 integrin LFA-1 and ligand ICAM-1 signaling]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 16, 213–216.
Li, P., Tan, Y., Zhu, L. X., Zhou, L. N., Zeng, P., Liu, Q., et al. (2017). Prognostic value of HPV DNA status in cervical cancer before treatment: a systematic review and meta-analysis. Oncotarget 8, 66352–66359. doi: 10.18632/oncotarget.18558
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z., et al. (2009). A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 19, 519–523. doi: 10.1038/cr.2009.33
Lin, C. W., Chuang, C. Y., Tang, C. H., Chang, J. L., Lee, L. M., Lee, W. J., et al. (2013). Combined effects of icam-1 single-nucleotide polymorphisms and environmental carcinogens on oral cancer susceptibility and clinicopathologic development. PLoS One 8:e72940. doi: 10.1371/journal.pone.0072940
Nakayama, Y., Ishikawa, C., Tamaki, K., Senba, M., Fujita, J., and Mori, N. (2011). Interleukin-1 alpha produced by human T-cell leukaemia virus type I-infected T cells induces intercellular adhesion molecule-1 expression on lung epithelial cells. J. Med. Microbiol. 60, 1750–1761. doi: 10.1099/jmm.0.033456-0
Nasu, K., Narahara, H., Etoh, Y., Kawano, Y., Hirota, Y., and Miyakawa, I. (1997). Serum levels of soluble intercellular adhesion molecule-1 (ICAM-1) and the expression of ICAM-1 mRNA in uterine cervical cancer. Gynecol. Oncol. 65, 304–308. doi: 10.1006/gyno.1997.4636
Natali, P., Nicotra, M. R., Cavaliere, R., Bigotti, A., Romano, G., Temponi, M., et al. (1990). Differential expression of intercellular adhesion molecule 1 in primary and metastatic melanoma lesions. Cancer Res. 50, 1271–1278.
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Reina, M., and Espel, E. (2017). Role of LFA-1 and ICAM-1 in cancer. Cancers 9:153. doi: 10.3390/cancers9110153
Ren, G., Zhao, X., Zhang, L., Zhang, J., L’Huillier, A., Ling, W., et al. (2010). Inflammatory cytokine-induced intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 in mesenchymal stem cells are critical for immunosuppression. J. Immunol. 184, 2321–2328. doi: 10.4049/jimmunol.0902023
Shastry, B. S. (2009). SNPs: impact on gene function and phenotype. Methods Mol. Biol. 578, 3–22. doi: 10.1007/978-1-60327-411-1_1
Shi, Y. Y., and He, L. (2005). SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 15, 97–98. doi: 10.1038/sj.cr.7290272
Siegel, R. L., Miller, K. D., and Jemal, A. (2017). Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30. doi: 10.3322/caac.21387
Springer, T. A. (1990). Adhesion receptors of the immune system. Nature 346, 425–434. doi: 10.1038/346425a0
Staunton, D. E., Dustin, M. L., Erickson, H. P., and Springer, T. A. (1990). The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell 61, 243–254. doi: 10.1016/0092-8674(90)90805-o
Sun, Y. H., Yang, S. F., Liu, Y. F., Ko, J. L., Wu, C. H., Wu, T. F., et al. (2016). Single-nucleotide polymorphisms and haplotypes of intercellular adhesion molecule-1 in uterine cervical carcinogenesis in Taiwanese women. Reprod. Sci. 23, 401–408. doi: 10.1177/1933719115604731
Tabibzadeh, S. S., and Poubouridis, D. (1990). Expression of leukocyte adhesion molecules in human endometrium. Am. J. Clin. Pathol. 93, 183–189. doi: 10.1093/ajcp/93.2.183
Tan, H. (2017). The association between gene SNPs and cancer predisposition: correlation or causality? EBioMedicine 16, 8–9. doi: 10.1016/j.ebiom.2017.01.047
Thanopoulou, E., Kotzamanis, G., Pateras, I. S., Ziras, N., Papalambros, A., Mariolis-Sapsakos, T., et al. (2012). The single nucleotide polymorphism g.1548A >G (K469E) of the ICAM-1 gene is associated with worse prognosis in non-small cell lung cancer. Tumour Biol. 33, 1429–1436. doi: 10.1007/s13277-012-0393-4
Tian, M. M., Sun, Y., Li, Z. W., Wu, Y., Zhao, A. L., and Li, J. Y. (2012). Polymorphisms of ICAM-1 are associated with gastric cancer risk and prognosis. World J. Gastroenterol. 18, 368–374. doi: 10.3748/wjg.v18.i4.368
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Uyar, D., and Rader, J. (2014). Genomics of cervical cancer and the role of human papillomavirus pathobiology. Clin. Chem. 60, 144–146. doi: 10.1373/clinchem.2013.212985
Wang, Y., and Luo, T. (2018). LINC00673 rs11655237 polymorphism is associated with increased risk of cervical cancer in a Chinese population. Cancer Control 25:1073274818803942. doi: 10.1177/1073274818803942
Yang, L., Yi, K., Wang, H., Zhao, Y., and Xi, M. (2016). Comprehensive analysis of lncRNAs microarray profile and mRNA-lncRNA co-expression in oncogenic HPV-positive cervical cancer cell lines. Oncotarget 7, 49917–49929. doi: 10.18632/oncotarget.10232
Keywords: cervical cancer, ICAM-1, SNPs, genetic association, multiplex polymerase chain reaction
Citation: Feng Y, Li X, Ma Q, Zhang S, Zhu M, Li S, Fang L, Tian J and Sun L (2021) Intercellular Adhesion Molecule-1 Gene Polymorphisms and Susceptibility to Cervical Cancer in the Northern Chinese Han Population. Front. Genet. 12:668539. doi: 10.3389/fgene.2021.668539
Received: 16 February 2021; Accepted: 15 June 2021;
Published: 27 July 2021.
Edited by:
Yufang Pei, Soochow University Medical College, ChinaReviewed by:
Tomas Drgon, United States Food and Drug Administration, United StatesApostolos Papachristos, University of Chicago, United States
Copyright © 2021 Feng, Li, Ma, Zhang, Zhu, Li, Fang, Tian and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Litao Sun, bGl0YW9zdW4xOTcxQGFsaXl1bi5jb20=; Jiawei Tian, and0aWFuMjAwNEAxNjMuY29t
 Yanan Feng1
Yanan Feng1 Litao Sun
Litao Sun