- 1College of Landscape Architecture, Sichuan Agricultural University, Chengdu, China
- 2College of Horticultural Biotechnology, South China Agricultural University, Guangzhou, China
Reverse transcription quantitative real-time PCR (RT-qPCR) is a common way to study gene regulation at the transcriptional level due to its sensibility and specificity, but it needs appropriate reference genes to normalize data. Ananas comosus var. bracteatus, with white-green chimeric leaves, is an important pantropical ornamental plant. Up to date, no reference genes have been evaluated in Ananas comosus var. bracteatus. In this work, we used five common statistics tools (geNorm, NormFinder, BestKeeper, ΔCt method, RefFinder) to evaluate 10 candidate reference genes. The results showed that Unigene.16454 and Unigene.16459 were the optimal reference genes for different tissues, Unigene.16454 and zinc finger ran-binding domain-containing protein 2 (ZRANB2) for chimeric leaf at different developmental stages, isocitrate dehydrogenase [NADP] (IDH) and triacylglycerol lipase SDP1-like (SDP) for seedlings under different hormone treatments. The comprehensive results showed IDH, pentatricopeptide repeat-containing protein (PPRC), Unigene.16454, and caffeoyl-CoA O methyltransferase 5-like (CCOAOMT) are the top-ranked stable genes across all the samples. The stability of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was the least during all experiments. Furthermore, the reliability of recommended reference gene was validated by the detection of porphobilinogen deaminase (HEMC) expression levels in chimeric leaves. Overall, this study provides appropriate reference genes under three specific experimental conditions and will be useful for future research on spatial and temporal regulation of gene expression and multiple hormone regulation pathways in Ananas comosus var. bracteatus.
Introduction
Ananas comosus var. bracteatus is a monocotyledon from the Bromeliaceae family that originates from tropical and subtropical areas of South America (Baker and Collins, 1939; Duval et al., 1997). Historically, it has been cultivated for fiber and fruit juice and is now a popular pantropical ornamental plant (Baker and Collins, 1939; Duval et al., 1997; Ma et al., 2015). In addition, Ananas comosus var. bracteatus is an excellent material for studying leaf albino mechanisms due to its white-green chimeric leaf (Mao et al., 2020). The formation of leaf albino mutants involves multiple biological processes, including photosynthesis, chlorophyll biosynthesis, chloroplast development, and hormone signaling pathways (Córdoba et al., 2016; Gao et al., 2016; Zhang et al., 2018; Zuo et al., 2019; Liu et al., 2020). Thus, complicated molecular regulatory networks of genetics, signaling, and metabolic mechanisms may exist in Ananas comosus var. bracteatus. Gene expression analysis is a popular and powerful tool to elucidate these complicated molecular regulatory networks (Bustin et al., 2005).
Reverse transcription-quantitative real-time PCR (RT-qPCR) is the most common method for analyzing gene expression level and validating the results obtained from other methods such as RNA-Seq (Bustin, 2002; Gachon et al., 2004; Bustin et al., 2005; Erickson et al., 2009; Kozera and Rapacz, 2013; Takamori et al., 2017). However, the prerequisite for avoiding inaccurate results by RT-qPCR is to select reliable reference genes for standard correction (Derveaux et al., 2010). Generally speaking, an ideal reference gene should be stably expressed independently of any experimental conditions (Kozera and Rapacz, 2013). Housekeeping genes, such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin (ACT), elongation factor-1a (EF1α), and 18S ribosomal RNA (18S), play essential roles in basic cellular progress, suggesting that they are stably expressed (Nicot et al., 2005; Garg et al., 2010; Maroufi et al., 2010). Based on the assumption that housekeeping genes are expressed at constant levels under all conditions, they are usually used as reference genes without verifying their stability. However, several studies indicated that the transcription stability of housekeeping genes is affected by internal and external conditions (Czechowski et al., 2005; Gutierrez et al., 2008; Sgamma et al., 2016; Wan et al., 2017). Therefore, no single reference gene, including housekeeping genes, is appropriate in all experimental conditions. As a result, reference genes must be systematically screened prior to specific experimental conditions (Cheng et al., 2017).
Previous studies on Ananas comosus var. comosus recommended several reference genes for gene expression analysis in pineapple plants, and mainly assayed reproductive organ development and abiotic/biotic stress conditions (Ma et al., 2012; Chen et al., 2019; Jin et al., 2020). For instance, housekeeping genes GAPDH and 18S are considered reference genes during pineapple organ development. In addition, several novel reference genes have been found based on pineapple genome and transcriptome databases. This method has been successfully applied to several plants, such as Arabidopsis thaliana, Oryza sativa, and Solanum lycopersicum (Czechowski et al., 2005; Narsai et al., 2010; Müller et al., 2015). Given this genetic background and biological process, the reference genes in Ananas comosus var. comosus may not be directly applied in Ananas comosus var. bracteatus (Galli et al., 2015; Jian et al., 2016; Wang and Lu, 2016). Therefore, systematic selection of reliable reference genes should be performed in Ananas comosus var. bracteatus.
In this study, we evaluated the expression stability of 10 candidate reference genes in different tissues (root, leaf, stem, and flesh), chimeric leaf at different developmental stages, and seedlings under different hormone treatments [Indole-3-acetic Acid (IAA), Trans-zeatin (T-z), Gibberellin A3 (GA3), Abscisic Acid (ABA), Brassinolide (BR)]. Gene expression stability was assayed by four common statistics tools including geNorm, NormFinder, BestKeeper, and ΔCt method (Vandesompele et al., 2002; Andersen et al., 2004; Pfaffl et al., 2004; Silver et al., 2006), then the comprehensive rank was obtained by RefFinder (Xie et al., 2012). Finally, porphobilinogen deaminase (HEMC) was analyzed to compare stable and unstable reference genes. Our results provide appropriate reference genes under three specific experimental conditions and will be useful for future research on the spatial and temporal regulation of gene expression and multiple hormone regulation pathways in Ananas comosus var. bracteatus.
Materials and Methods
Plant Materials
Ananas comosus var. bracteatus was obtained from a garden in Zhanjiang (Guangdong, China) and cultivated in a greenhouse at Sichuan Agricultural University (Sichuan, China). Green tissue culture plantlets were derived from stem explants of Ananas comosus var. bracteatus. The stem explants were sterilized and then transplanted on MS medium containing 6-BA (3 mg/L) and NAA (2 mg/L). After 1 month, callus was transplanted on MS medium containing 6-BA (0.5 mg/L) and NAA (3 mg/L). Then, green tissue culture plantlets were planted in plastic pots.
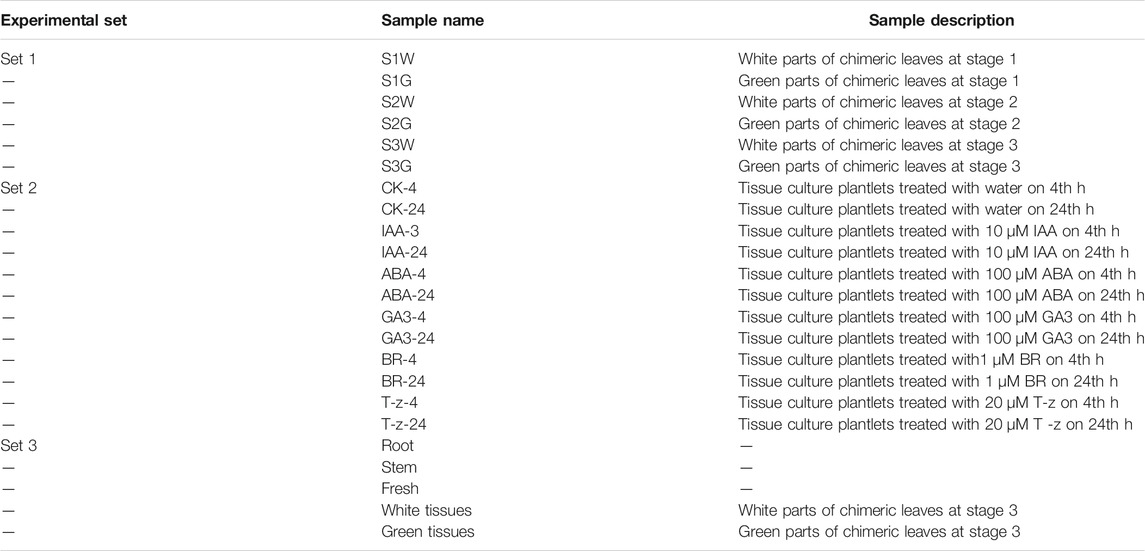
There are three experimental sets in this study (Table 1). In experimental set 1, chimeric leaves at different developmental stages were collected. Chimeric leaf development stages (S) were as follows: stage 1 (S1, no chlorophyll is visible on the leaf); stage 2 (S2, chlorophyll becomes visible on leaf center); stage 3 (S3, two thirds of the leaf are fully pigmented by chlorophyll) (Additional File 1: Supplementary Figure S1). In experimental set 2, green tissue culture plantlets with 10–12 expanded leaves were treated with different hormones. Then, leaves were collected after 4 and 24 h hormone treatments, respectively. In experimental set 3, roots, stem, flesh, white tissues, and green tissues were collected separately. All samples were stored at −80°C until RNA extraction.
RNA Extraction and cDNA Synthesis
Total RNA was extracted using LABGENETM Plant RNA Isolation Kit (Cat. No. LB1111, LABGENE Biotechnology Co., Ltd., Chengdu, China). The concentration and purity of extracted RNA were measured using NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, United States), and 1.2% (w/v) agarose gel electrophoresis was performed to assess the RNA integrity. When A260/A280 ratio was between 1.8 and 2.2, and A260/A230 ratio was higher than 1.8, RNA can be used for further analysis. cDNA was synthesized from 1.0 μg RNA using Evo M-MLV RT Kit whit gDNA Clean for qPCR II (Code No. AG11711, Accurate Biotechnology Co., Ltd., Wuhan, China), then stored at −20°C until use.
Candidate Gene Selection and Primer Design
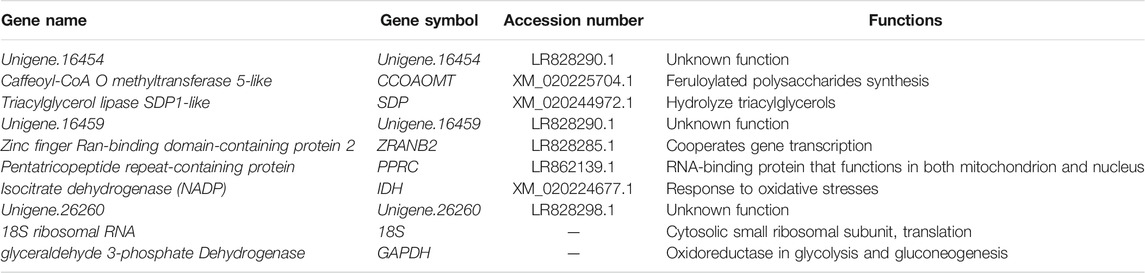
The 10 candidate reference genes included two previously assessed pineapple reference genes, GAPDH, 18S, as well as 8 novel candidate reference genes chosen from the transcriptome of Ananas comosus var. bracteatus (NCBI accessions: PRJNA564223), Unigene.16454, caffeoyl-CoA O methyltransferase 5-like (CCOAOMT), triacylglycerol lipase SDP1-like (SDP), Unigene.16459, zinc finger ran-binding domain-containing protein 2 (ZRANB2), pentatricopeptide repeat-containing protein (PPRC), and isocitrate dehydrogenase (NADP) (IDH), Unigene.26260 (Table 2, Additional file 2: Supplementary Table S1). Primer specificity was judged by gel electrophoresis and melting-curve analyses. In addition, the amplification efficiency (E) of the primer was calculated by standard curve.
RT-qPCR Analysis
The RT-qPCR was performed with CFX96 Real-time PCR Detection System (Bio-Rad, US) using the SYBR® Green Premix Pro Taq HS qPCR Kit (Cat No. AG11701, Accurate Biotechnology Co., Ltd., Wuhan, China). 25 μL reaction consisted 12.5 μL 2X SYBR® Green Pro Taq HS Premix, 0.5 μL each primer (10 μM), 1 μL of cDNA, and 10.5 μL dH2O. The following RT-qPCR amplification program was 95°C for 30 s, and 40 cycles of 95°C for 5 s, the optimal temperature for each primer for 15 and 15 s for 72°C. The dissociation curve was then generated by melting the amplicon from 60 to 95°C. All RT-qPCR experiments were performed in three biological replicates and three technical replicates. A no-template control and a reverse transcription negative control were also performed.
Gene Expression Stability Analysis
First of all, the mean Cq value was calculated by three biological replicates to analyze expression levels. Secondly, geNorm, NormFinder, BestKeeper, and ΔCt method were applied to evaluate expression stability (Vandesompele et al., 2002; Andersen et al., 2004; Pfaffl et al., 2004; Silver et al., 2006). Finally, RefFinder online (http://blooge.cn/RefFinder/) was used to obtain comprehensive ranks (Xie et al., 2012).
Validation of Identified Reference Genes
HEMC is a key gene involved in chlorophyll biosynthesis and was used as a target gene to verify the results of our experiment. The most and least stable reference gene, as well as the combination of the two best reference genes, were used respectively to normalize the expression profile of HEMC in the green and white parts of the chimeric leaf at different developmental stages. The relative expression was conducted with the comparative 2−ΔΔCT method (Livak and Schmittgen, 2001).
Gene Expression Analysis of AbGLK1
Golden 2-like, or GLK, transcription factors are known to be involved in chloroplast development and have also been implicated in various hormone signaling pathways. (Fitter et al., 2002; Savitch et al., 2007; Gutiérrez et al., 2008; Waters et al., 2009). The expression profiles of AbGLK1 during chimeric leaf development and response to hormone stimuli were obtained according to the comparative 2−ΔΔCT method (Livak and Schmittgen, 2001). Sample collections and experiments were performed as described above.
Results
Primer Specificity and Amplification Efficiency of the Candidate Reference Genes
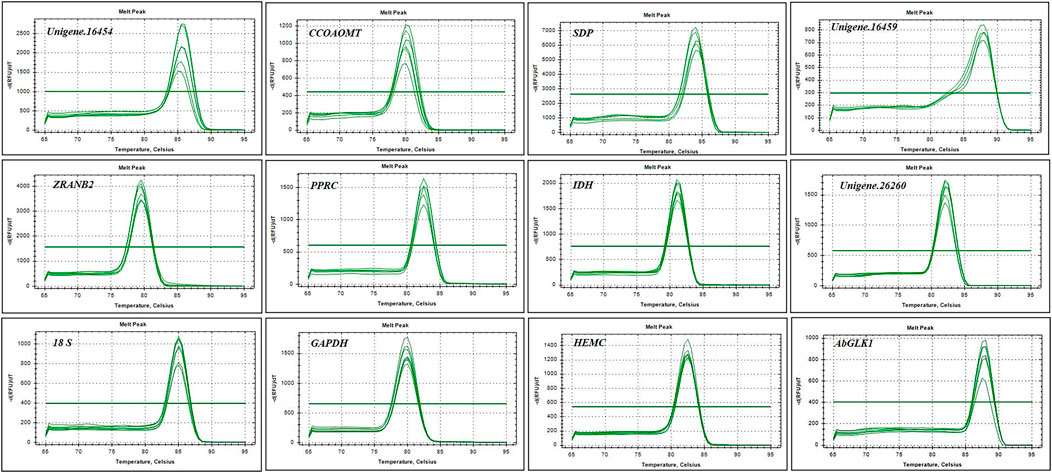
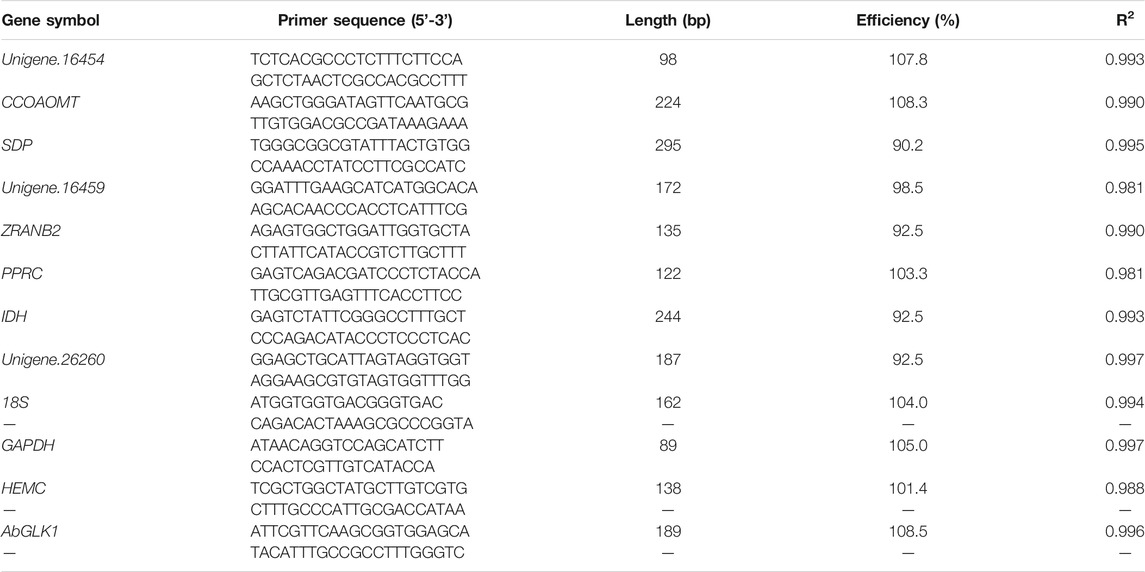
Gel electrophoresis and melting curve analysis showed that all primer pairs present a single band and a single peak (Figures 1,2), which validates the specificity of primers. No amplification appeared in the negative control. In addition, the amplification efficiency for each primer pair varied from 90.2% for SDP to 108.3% for CCOAOMT, and the correlation coefficients (R2) were all higher than 0.98 (Table 3). Therefore, these primers were used in further RT-qPCR analysis.

FIGURE 1. Amplified fragment of 10 candidate genes on 1.2% agarose gel. M represents the DL2000 DNA marker.
Expression Levels of Candidate Reference Genes
Raw quantification cycle (Cq) values can directly reflect gene expression level, and therefore provide basic data support to evaluate candidate reference genes in tested samples (Yan et al., 2011; Chen et al., 2015). In this study, three specific experimental sets were analyzed individually (Table 1). The raw Cq values of the 10 genes are shown in Additional file 3: Supplementary Table S2 and were plotted directly by Boxplot (Figure 3). In all experimental conditions, 18S and PPRC genes showed the highest and lowest expression levels, respectively. In detail, the Cq means values ranged from 19.74 (18S) to 25.96 (PPRC) in different tissues, 19.57 (18S) to 25.68 (PPRC) in chimeric leaf at different developmental stages, 19.83 (18S) to 26.03 (PPRC) in seedlings under different hormone treatments, and 20.14 (18S) to 26.23 (PPRC) in all samples. There was no gene to express stably in all experimental conditions, so it was necessary to rank the expression stabilities by using statistical algorithms.
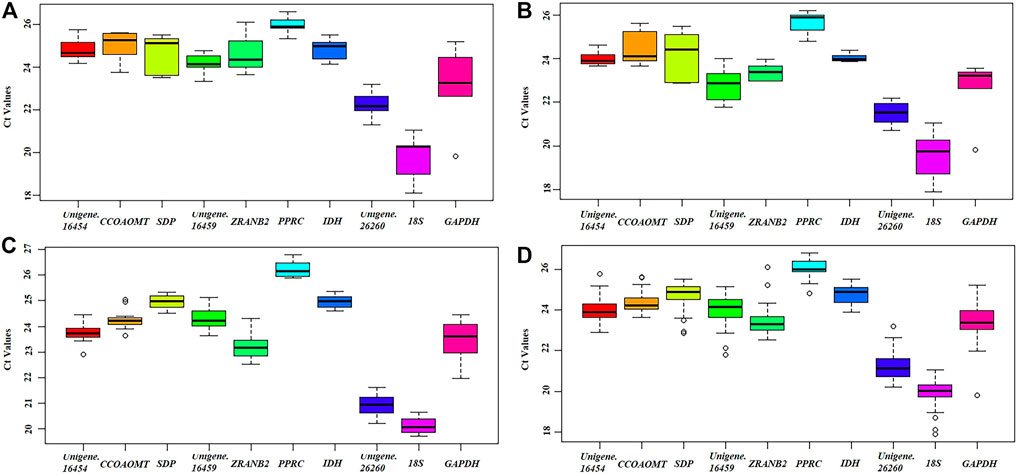
FIGURE 3. Distribution of Cq values of candidate reference genes in all samples. (A) Tissue, (B) Developmental stage, (C) Hormone treatment, (D) Global analysis. Boxplots show the 25th and 75th percentiles, mean, and outliers.
Expression Stability Analysis by geNorm
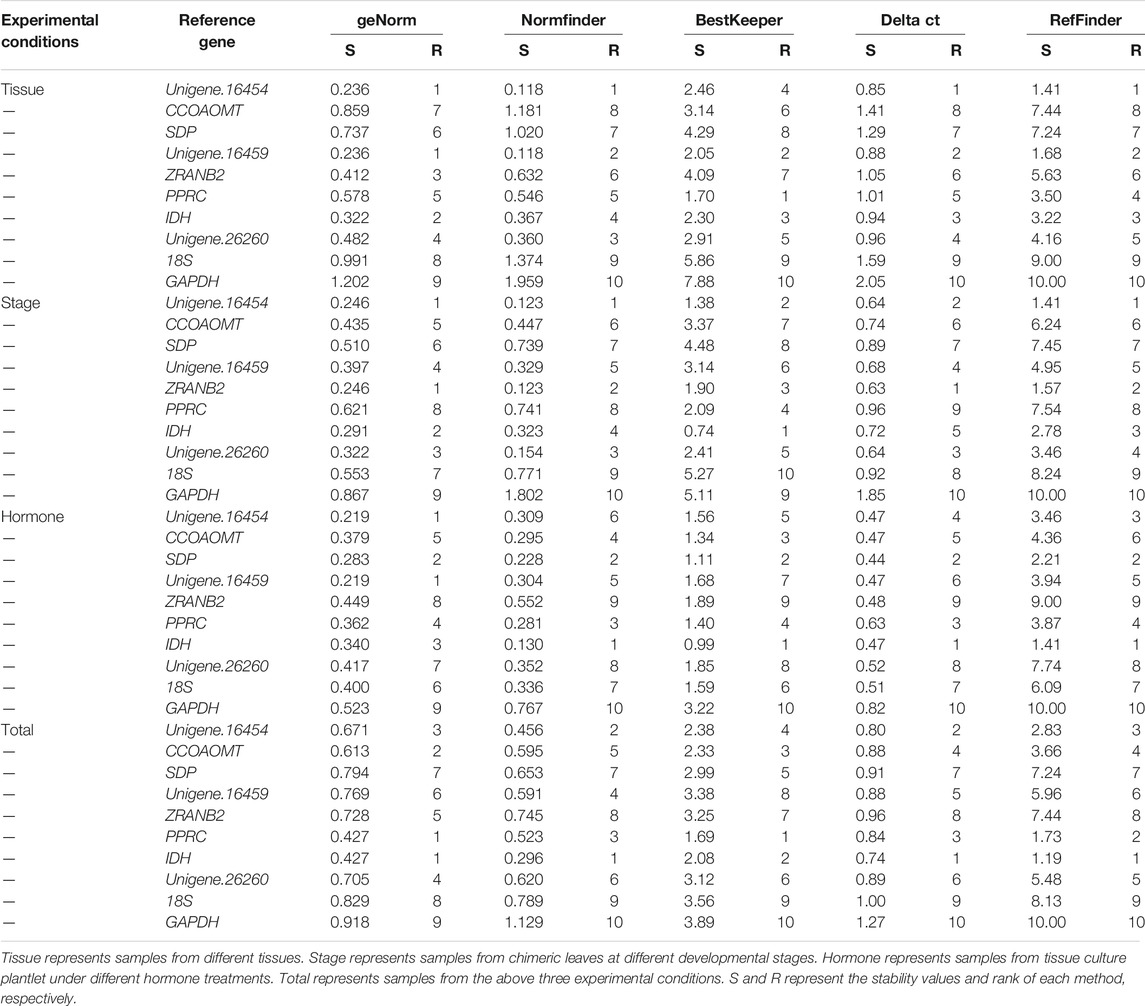
GeNorm calculates the average pairwise expression ratio to evaluate expression stability, and genes with lower stability measure (M) values were more stable (Vandesompele et al., 2002). For tissue samples, Unigene.16454 and Unigene.16459 (0.236) had the lowest M values. In addition, Unigene.16454 and ZRANB2 (0.241) were the most stably expressed genes in the chimeric leaf during different developmental stages. Under hormone treatments, the two most stably expressed genes were Unigene.16454 and Unigene.16459 (0.219). Concerning all samples tested, PPRC and IDH (0.421) were the best reference genes. Notably, GAPDH was considered the least stable gene in all experimental conditions (Table 4).
In addition, GeNorm can determine the optimal number of reference genes by calculating pairwise variation value (Vn/Vn +1, V-value) (Vandesompele et al., 2002). The number of reference genes increased to n+1 when the V-value was higher than 0.15 (Vandesompele et al., 2002). As shown in Figure 4, two reference genes were sufficient for data normalization in tissue samples, developmental stage samples, and hormone treatment samples. For all samples tested, however, the V-value of V2/V3 and V3/V4 were both above 0.15, and thus four reference genes should be selected to normalize gene expression results.
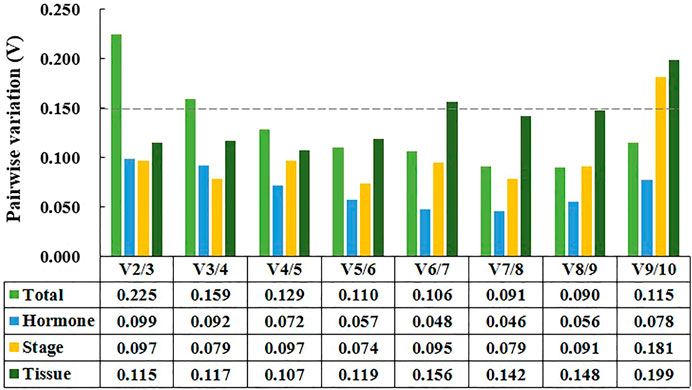
FIGURE 4. Pairwise variation (Vn/Vn+1) calculated by for geNorm. Tissue represents samples from different tissues. Stage represents samples from chimeric leaves at different developmental stages. Hormone represents samples from tissue culture plantlet under different hormone treatments. Total represents samples from the above three experimental conditions.
Expression Stability Analysis by NormFinder
NormFinder has a similar mathematical model to GeNorm, and genes with higher stability values (SV) were considered as less stable (Pfaffl et al., 2004). The most stably expressed genes were Unigene.16459 (SV = 0.118) and Unigene.16459 (SV = 0.118) for tissue samples, Unigene.16454 (SV = 0.123) and ZRANB2 (SV = 0.123) for developmental stage samples, IDH (SV = 0.130) for hormone treatment samples, IDH (SV = 0.296) for all samples (Table 4). Like geNorm, GAPDH was ranked as the worst reference gene in all experimental conditions (Table 4).
Expression Stability Analysis by BestKeeper
BestKeeper program evaluates the expression stabilities by the standard deviation (SD) and coefficient variance (CV) (Andersen et al., 2004). The lower CV and SD (CV ± SD) value, the higher stability. As shown in Table 4, PPRC (1.35 ± 0.5) and IDH (0.60 ± 0.14) were the most stably expressed genes for tissue samples and developmental stage samples, respectively. IDH (0.79 ± 0.20) and SDP (0.89 ± 0.22) were the two most reliable genes for hormone treatment samples. In addition, PPRC (1.34 ± 0.35) and IDH (1.67 ± 0.41) were the two most stable reference genes for all tested samples. The SD value of stable expression genes should be lower than the threshold of 1 (Andersen et al., 2004). As shown in Table 4, the SD value of GAPDH was higher than 1 in all experimental conditions, suggesting it was an unstable gene.
Expression Stability Analysis by ΔCt Method
ΔCt method ranks the candidate reference genes based on average standard deviation (SD), which compared the relative expression of “pairs of genes” within each sample (Silver et al., 2006). Unigene.16454 (0.85) was the most stable reference gene for tissue samples, ZRANB2 (0.63) for developmental stage samples, and IDH (0.47) for hormones treatments samples (Table 4). Concerning all samples tested, IDH (0.74) was the best reference gene (Table 4).
Comprehensive Stability Analysis of Candidate Reference Genes
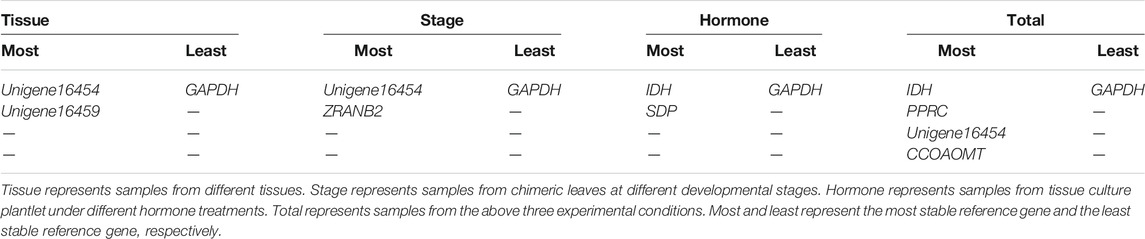
The four statistical algorithms created different ranking patterns. To obtain a consensus result, a comprehensive analysis based on RefFinder was thus carried out (Xie et al., 2012). Given the optimal number of reference genes, reference genes in three specific experimental conditions were determined in this study. We recommend Unigene.16454 and Unigene.16459 as reference genes for tissue samples, Unigenes.16454 and ZRANB2 for developmental stage samples, IDH and SDP for hormone treatment samples, IDH, PPRC, Unigene.16454, and CCOAOMT for all samples tested (Table 5).
Validation of the Recommended Reference Genes
In Ananas comosus var. bracteatus the chlorophyll content of green parts was significantly higher than white parts, which results in leaf albino phenotype (Xue et al., 2019). The HEMC gene encodes enzymes for the first rate-limiting step in chlorophyll biosynthesis. Therefore, we chose HEMC as the target gene to verify the optimal reference genes in this study. As shown in Figure 5, HEMC is expressed in chimeric leaves under all development stages. In detail, the highest expression was in white parts at stage S3, followed by green parts at stage S2. The relative expression levels of HEMC were found to be unbiased when Unigene.16454 and/or ZRANB2 (Figure 5) were used. However, the expression pattern of HEMC changed when the least stable gene, GAPDH, was used (Figure 5). These results verified the reliability of the recommended reference gene and provided theoretical support for future insight on gene expression patterns in Ananas comosus var. bracteatus.
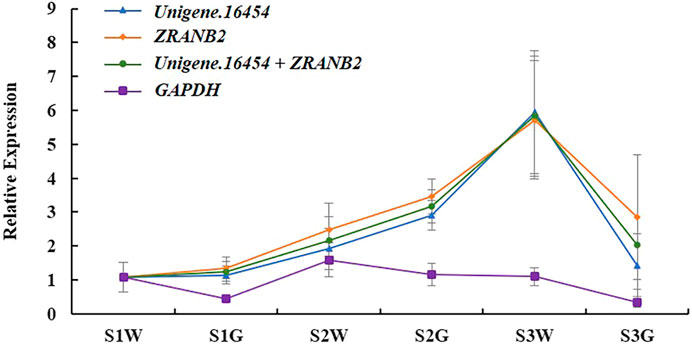
FIGURE 5. Relative expression patterns of HEMC. S1W, S2W, and S3W mean white parts of chimeric leaves at S1, S2, and S3, respectively. S1G, S2G, and S3G mean green parts of chimeric leaves at S1, S2, and S3, respectively. Error bars for RT-qPCR show the standard deviation of three replicates.
Expression Profile of AbGLK1 During Chimeric Leaf Development and Response to Hormone Stimuli
In Ananas comosus var. bracteatus, few normal chloroplasts were observed in the white parts of the chimeric leaf (Mao et al., 2020). GLK transcription factors are known to be involved in chloroplast development, therefore, we investigated the expression of AbGLK1 during chimeric leaf development. As shown in Figure 6A, the transcript abundance of AbGLK1 in chimeric leaf development stage S2 was relatively lower than that of stage S3, suggesting that the expression level of AbGLK1 was affected by the stage of development. Furthermore, the expression level of AbGLK1 in the white parts at stage S3 was about 0.5 times higher than that of stage S2. While the expression level of AbGLK1 in the green parts at stage S3 was about 3.5 times higher than that of stage S2. These results showed that the expression profile of AbGLK1 in the white parts differed from the green parts during chimeric leaf development. Therefore, the regulation of AbGLK1 should be the focus of the investigation. As shown in Figure 6B, the hormone signaling was activated after hormone treatments in Ananas comosus var. bracteatus. The fluctuating expression level of AbGLK1 was in response to various hormone stimuli. In detail, IAA and GA3 caused significant upregulation of AbGLK1 in response to 4 h treatment (p < 0.05). BR caused slight upregulation of AbGLK1 in response to 4 h treatment (p > 0.05). T-z and ABA caused slight downregulation of AbGLK1 in response to 4 h treatment (p > 0.05).
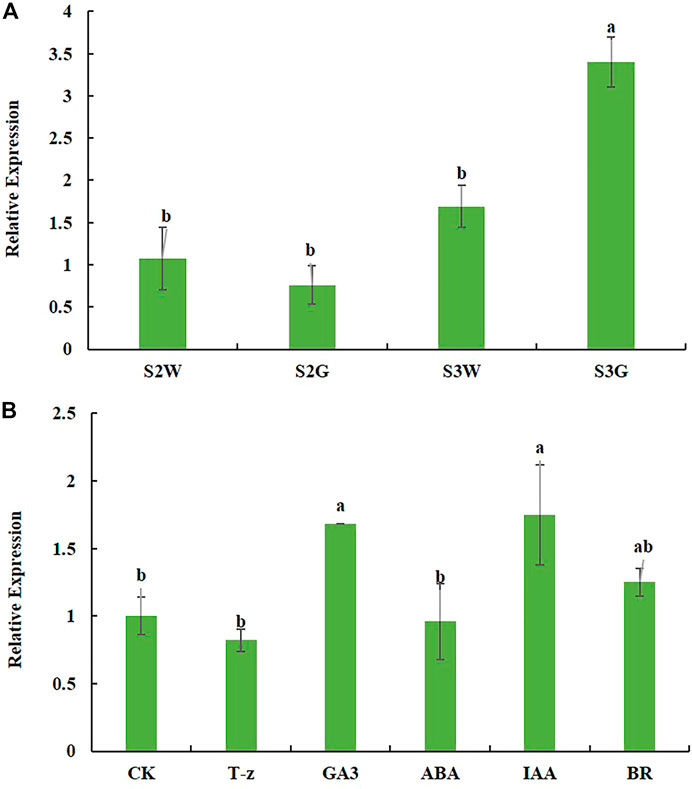
FIGURE 6. Expression profile of AbGLK1 during chimeric leaf development (A) and response to hormone stimuli (B). S2W and S3W mean white parts of chimeric leaves at S2 and S3, respectively. S2G and S3G mean green parts of chimeric leaves at S2 and S3, respectively. CK, T-z, ABA, IAA, BR mean tissue culture plantlets treated hormone treatments, respectively. Error bars for RT-qPCR show the standard deviation of three replicates. Different lowercase letters indicate significant differences at the 0.05 level.
Discussion
Ananas comosus var. bracteatus is one of the most important pantropical ornamental plants because of its bright pink-to-red colored fruit and white-green chimeric leaves (Ma et al., 2015). Chimeric leaves are optimal materials for the study of plant tissue formation and development as well as the interaction between cells (Burge et al., 2002; Stegemann and Bock, 2009). When breeding ornamental plants, white-green chimeric leaves are a valuable trait for their aesthetic and commercial value. Research into leaf albino mechanisms is beneficial for leaf color artificial regulation of Ananas comosus var. bracteatus. Gene expression analysis is crucial for obtaining insight into complex molecular mechanisms (Bustin et al., 2005). RT-qPCR is a highly sensitive and specific technique for gene expression analysis, but its accuracy requires appropriate reference genes for data normalization (Bustin, 2002; Gachon et al., 2004; Bustin et al., 2005; Erickson et al., 2009; Kozera and Rapacz, 2013; Takamori et al., 2017). However, reference genes focused on Ananas comosus var. bracteatus have never been investigated. In this study, we identified reference genes for three specific experimental conditions. Tissue samples were mainly used for gene spatial expression, developmental stage samples for gene temporal expression. And hormone treatment samples are mainly used to study multiple hormone signaling pathways in Ananas comosus var. bracteatus.
In the present study, transcriptome sequencing data was fully utilized to select 8 novel candidate reference genes in Ananas comosus var. bracteatus. Compared to novel candidate reference genes, traditional housekeeping genes 18S and GAPDH were identified and fluctuated considerably in Ananas comosus var. bracteatus. Studies in Camellia sinensis and Brassica napus have also revealed that the expression of traditional housekeeping genes varies considerably under certain conditions (Hao et al., 2014; Yang et al., 2014). These results suggest that sequencing generates large-scale gene segments to provide a resource for screening superior reference genes (He et al., 2016; Liang et al., 2018; Pombo et al., 2019). In this study, novel stably expressed genes mostly differ from those previously found in other plants. The novel reference genes standing out in the present study are involved in various basic physiological processes, thus providing the basis for their normalization reliability and expression stability. For instance, ZRANB2 influences the alternative splicing of multiple genes in the transcriptome (Yang et al., 2013). IDH is involved in nitrogen metabolism, redox regulation, senescence, and responses to oxidative stress (Leterrier et al., 2016).
Two or more reference genes can avoid misinterpretation for normalization (Xu et al., 2017; Wang et al., 2019). Based on a pairwise variation analysis, in this study, two reference genes were required to present accurate expression patterns in tissue samples, developmental stage samples, and hormone treatment samples. In all samples tested, however, the four reference genes do not necessarily guarantee data accuracy. Similarly, the numbers of reference genes also increased under all tested samples in Sapium sebiferum and Lilium spp. (Chen et al., 2017; Xu et al., 2017). For the results obtained with all samples set, it should be considered that such samples may be subject to diverse physiological states. Besides the accuracy of data, however, cost-effectiveness should be taken into consideration (Ferradás et al., 2016). These results indicated that it is better to identify reference genes in specific experimental sets rather than simultaneously analyzing them in all tested samples, which was consistent with previous reports.
The use of different algorithms results in potentially contradictory results from the same data. In this study, four popular statistical algorithms (geNorm, NormFinder, BestKeeper, and ΔCt method) were applied to avoid inaccurate results, respectively. Results showed that the stability rank varied with statistical algorithms. For example, geNorm ranked Unigene.16454 and Unigene.16459 as the most stable gene under hormone treatments, but the other three programs ranked IDH as the most stable. The different results among different programs have also been reported in diverse species, such as Cichorium intybus, Actinidia deliciosa, Cannabis Sativa, Sorghum bicolor, and Vigna sinensis (Delporte et al., 2015; Petriccione et al., 2015; Mangeot-Peter et al., 2016; Sudhakar et al., 2016; Amorim et al., 2018). However, there is a lack of consensus on which type of statistical algorithms should be selected to rank gene expression stability. To obtain an entirely satisfactory solution, RefFinder was applied in this study. RefFinder calculates the geometric mean (GM) of the ranking values from geNorm, NormFinder, BestKeeper, and the ΔCt method then ranks overall stability (Xie et al., 2004). To verify the reliability of recommended reference genes, the expression patterns of HEMC were detected in chimeric leaves at different development stages, using the most and least stable reference genes for normalization. The HEMC showed consistent expression profiles when the two most stable genes were used for normalization either alone or in combination. However, the expression pattern of HEMC changed when the least stable gene was used. These results further confirm the reliability of the recommended reference gene and demonstrated that the use of inappropriate reference genes may lead to inaccurate results.
Expression analysis of a given gene plays vital a role in further revealing its function. In the present study, the expression level AbGLK1 was increased with the stage of development. However, the increased rate was inconsistent in the green and white parts of the chimeric leaf. The higher expression level was in green parts at stage S3. A previous study showed that a significant downregulation of CsGLKs likely caused the chloroplast defect (Hao et al., 2019). Therefore, the expression silencing of AbGLK1 should be responsible for the chloroplast developmental defects in the white parts of the chimeric leaf. Consistent with the roles of hormone and GLK genes in photosynthetic development, GLK1 and GLK2 were both found to be regulated by various hormone signaling in Arabidopsis thaliana (Ponomareva, 2012). In this study, hormone IAA, GA3, and BR caused upregulation of AbGLK1, while T-z and ABA caused downregulation, suggesting that hormone signaling was activated in Ananas comosus var. bracteatus. In conclusion, the expression silencing of AbGLK1 may therefore be due to various hormone signaling pathways in the regulation of AbGLK1. The mechanisms of AbGLK1 regulation should be further verified, which would help reveal its function in chimeric leaf formation.
Conclusion
The present study focused on Ananas comosus var. bracteatus, a pantropical ornamental with white-green chimeric leaf. To obtain greater insight into Ananas comosus var. bracteatus, a pilot study was conducted to identify reference genes to ensure the accurate presentation of transcript levels by RT-qPCR. 10 candidate reference genes were evaluated to identify the appropriate reference genes in three specific experimental conditions, including tissue types, developmental stage samples, and hormone treatment samples. In general, Unigene.16454 and Unigene.16459 were the best reference genes for tissue samples, Unigene.16454 and ZRANB2 for developmental stages samples, IDH and SDP for hormone treatments samples, and IDH, PPRC, Unigene.16454 and CCOAOMT for all samples tested. These results are useful for future research on spatial and temporal regulation of gene expression and multiple hormone regulation pathways in Ananas comosus var. bracteatus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JM and YH conceived and designed the research. MM, XZ, HH, JL, LF, and WY performed the research. MM, YX, XL, JL, and HZ analyzed the data. MM and JM drafted and edited the paper. All authors contributed, read, and approved the final version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 31971704; No. 31770743). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.716137/full#supplementary-material
References
Amorim, L. L. B., Ferreira-Neto, J. R. C., Bezerra-Neto, J. P., Pandolfi, V., de Araújo, F. T., da Silva Matos, M. K., et al. (2018). Cowpea and Abiotic Stresses: Identification of Reference Genes for Transcriptional Profiling by qPCR. Plant Methods 14, 88. doi:10.1186/s13007-018-0354-z
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: a Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and colon Cancer Data Sets. Cancer Res. 64, 5245–5250. doi:10.1158/0008-5472.CAN-04-0496
Baker, K. F., and Collins, J. L. (1939). Notes on the Distribution and Ecology of Ananas and Pseudananas in South America. Am. J. Bot. 26, 697–702. doi:10.1002/j.1537-2197.1939.tb09339.x
Burge, G. K., Morgan, E. R., and Seelye, J. F. (2002). Opportunities for Synthetic Plant Chimera Breeding: Past and Future. Plant Cell 70, 13–21. doi:10.1023/A:1016088621471
Bustin, S. A., Benes, V., Nolan, T., and Pfaffl, M. W. (2005). Quantitative Real-Time RT-PCR - a Perspective. J. Mol. Endocrinol. 34, 597–601. doi:10.1677/jme.1.01755
Bustin, S. (2002). Quantification of mRNA Using Real-Time Reverse Transcription PCR (RT-PCR): Trends and Problems. J. Mol. Endocrinol. 29, 23–39. doi:10.1677/jme.0.0290023
Chen, H., Hu, B., Zhao, L., Shi, D., She, Z., Huang, X., et al. (2019). Differential Expression Analysis of Reference Genes in Pineapple (Ananas Comosus L.) during Reproductive Development and Response to Abiotic Stress, Hormonal Stimuli. Trop. Plant Biol. 12 (2), 67–77. doi:10.1007/s12042-019-09218-2
Chen, Y., Tan, Z., Hu, B., Yang, Z., Xu, B., Zhuang, L., et al. (2015). Selection and Validation of Reference Genes for Target Gene Analysis with Quantitative RT-PCR in Leaves and Roots of Bermudagrass under Four Different Abiotic Stresses. Physiol. Plantarum 155 (2), 138–148. doi:10.1111/ppl.12302
Cheng, Y., Bian, W., Pang, X., Yu, J., Ahammed, G. J., Zhou, G., et al. (2017). Genome-wide Identification and Evaluation of Reference Genes for Quantitative RT-PCR Analysis during Tomato Fruit Development. Front. Plant Sci. 8, 1440. doi:10.3389/fpls.2017.01440
Córdoba, J., Molina-Cano, J.-L., Martínez-Carrasco, R., Morcuende, R., and Pérez, P. (2016). Functional and Transcriptional Characterization of a Barley Mutant with Impaired Photosynthesis. Plant Sci. 244, 19–30. doi:10.1016/j.plantsci.2015.12.006
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W.-R. (2005). Genome-wide Identification and Testing of superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi:10.1104/pp.105.063743
Delporte, M., Legrand, G., Hilbert, J.-L., and Gagneul, D. (2015). Selection and Validation of Reference Genes for Quantitative Real-Time PCR Analysis of Gene Expression in Cichorium Intybus. Front. Plant Sci. 6, 651. doi:10.3389/fpls.2015.00651
Derveaux, S., Vandesompele, J., and Hellemans, J. (2010). How to Do Successful Gene Expression Analysis Using Real-Time PCR. Methods 50 (4), 227–230. doi:10.1016/j.ymeth.2009.11.001
Duval, M. F., Coppens d'Eeckenbrugge, G., Ferreira, F. R., Bianchetti, L. D. B., and Cabral, J. R. S. (1997). First Results from Joint EMBRAPA-CIRAD Ananas Germplasm Collecting in Brazil and French Guyana. Acta Hortic. 425, 137–144. doi:10.17660/ActaHortic.1997.425.14
Erickson, H. S., Albert, P. S., Gillespie, J. W., Rodriguez-Canales, J., Marston Linehan, W., Pinto, P. A., et al. (2009). Quantitative RT-PCR Gene Expression Analysis of Laser Microdissected Tissue Samples. Nat. Protoc. 4, 902–922. doi:10.1038/nprot.2009.61
Ferradás, Y., Rey, L., Martínez, Ó., Rey, M., and González, M. V. (2016). Identification and Validation of Reference Genes for Accurate Normalization of Real-Time Quantitative PCR Data in Kiwifruit. Plant Physiol. Biochem. 102, 27–36. doi:10.1016/j.plaphy.2016.02.011
Fitter, D. W., Martin, D. J., Copley, M. J., Scotland, R. W., and Langdale, J. A. (2002). GLKgene Pairs Regulate Chloroplast Development in Diverse Plant Species. Plant J. 31, 713–727. doi:10.1046/j.1365-313X.2002.01390.x
Gachon, C., Mingam, A., and Charrier, B. (2004). Real-time PCR: what Relevance to Plant Studies? J. Exp. Bot. 55, 1445–1454. doi:10.1093/jxb/erh181
Galli, V., Borowski, J. M., Perin, E. C., Messias, R. d. S., Labonde, J., Pereira, I. d. S., et al. (2015). Validation of Reference Genes for Accurate Normalization of Gene Expression for Real Time-Quantitative PCR in Strawberry Fruits Using Different Cultivars and Osmotic Stresses. Gene 554, 205–214. doi:10.1016/j.gene.2014.10.049
Gao, M., Hu, L., Li, Y., and Weng, Y. (2016). The Chlorophyll-Deficient golden Leaf Mutation in Cucumber Is Due to a Single Nucleotide Substitution in CsChlI for Magnesium Chelatase I Subunit. Theor. Appl. Genet. 129, 1961–1973. doi:10.1007/s00122-016-2752-9
Garg, R., Sahoo, A., Tyagi, A. K., and Jain, M. (2010). Validation of Internal Control Genes for Quantitative Gene Expression Studies in Chickpea (Cicer Arietinum L.). Biochem. Biophysical Res. Commun. 396, 283–288. doi:10.1016/j.bbrc.2010.04.079
Gutierrez, L., Mauriat, M., Gunin, S., Pelloux, J., Lefebvre, J.-F., Louvet, R., et al. (2008). The Lack of a Systematic Validation of Reference Genes: a Serious Pitfall Undervalued in Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis in Plants. Plant Biotechnol. J. 6, 609–618. doi:10.1111/j.1467-7652.2008.00346.x
Gutiérrez, R. A., Stokes, T. L., Thum, K., Xu, X., Obertello, M., Katari, M. S., et al. (2008). Systems Approach Identifies an Organic Nitrogen-Responsive Gene Network that Is Regulated by the Master Clock Control Gene CCA1. Proc. Natl. Acad. Sci. 105, 4939–4944. doi:10.1073/pnas.0800211105
Hao, X., Horvath, D., Chao, W., Yang, Y., Wang, X., and Xiao, B. (2014). Identification and Evaluation of Reliable Reference Genes for Quantitative Real-Time PCR Analysis in Tea Plant (Camellia Sinensis (L.) O. Kuntze). Ijms 15 (12), 22155–22172. doi:10.3390/ijms151222155
Hao, X., Zhang, W., Liu, Y., Zhang, H., Ren, H., Chen, Y., et al. (2020). Pale green Mutant Analyses Reveal the Importance of CsGLKs in Chloroplast Developmental Regulation and Their Effects on Flavonoid Biosynthesis in tea Plant. Plant Physiol. Biochem. 146, 392–402. doi:10.1016/j.plaphy.2019.11.036
He, Y., Yan, H., Hua, W., Huang, Y., and Wang, Z. (2016). Selection and Validation of Reference Genes for Quantitative Real-Time PCR in Gentiana Macrophylla. Front. Plant Sci. 07, 945. doi:10.3389/fpls.2016.00945
Jin, X., Hou, Z., Zhao, L., Liu, L., Priyadarshani, S. V. G. N., Wang, L., et al. (2020). Genome-Wide Identification and Evaluation of New Reference Genes in Pineapple (Ananas Comosus L.) during Stamen and Ovule Development. Trop. Plant Biol. 13, 371–381. doi:10.1007/s12042-020-09269-w
Kozera, B., and Rapacz, M. (2013). Reference Genes in Real-Time PCR. J. Appl. Genet. 54 (4), 391–406. doi:10.1007/s13353-013-0173-x
Leterrier, M., Barroso, J. B., Valderrama, R., Begara-Morales, J. C., Sánchez-Calvo, B., Chaki, M., et al. (2016). Peroxisomal NADP-Isocitrate Dehydrogenase Is Required for Arabidopsis Stomatal Movement. Protoplasma 253 (2), 403–415. doi:10.1007/s00709-015-0819-0
Li, J., Han, J., Hu, Y., and Yang, J. (2016). Selection of Reference Genes for Quantitative Real-Time PCR during Flower Development in Tree Peony (Paeonia Suffruticosa Andr.). Front. Plant Sci. 7, 516. doi:10.3389/fpls.2016.00516
Liang, W., Zou, X., Carballar-Lejarazú, R., Wu, L., Sun, W., Yuan, X., et al. (2018). Selection and Evaluation of Reference Genes for qRT-PCR Analysis in Euscaphis Konishii Hayata Based on Transcriptome Data. Plant Methods 14, 42. doi:10.1186/s13007-018-0311-x
Liu, L., Lin, N., Liu, X., Yang, S., Wang, W., and Wan, X. (2020). From Chloroplast Biogenesis to Chlorophyll Accumulation: The Interplay of Light and Hormones on Gene Expression in Camellia Sinensis Cv. Shuchazao Leaves. Front. Plant Sci. 11, 256. doi:10.3389/fpls.2020.00256
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Ma, J., He, Y., Wu, C., Liu, H., and Hui, Z. (2012). Cloning and Selection of Reference Genes for Gene Expression Studies in Ananas Comosus. Afr. J. Biotechnol. 11 (29), 7424–7433. doi:10.5897/AJB12.1110
Ma, J., Kanakala, S., He, Y., Zhang, J., and Zhong, X. (2015). Transcriptome Sequence Analysis of an Ornamental Plant, Ananas Comosus Var. Bracteatus, Revealed the Potential Unigenes Involved in Terpenoid and Phenylpropanoid Biosynthesis. Plos One 10 (3), e0119153. doi:10.1371/journal.pone.0119153
Mangeot-Peter, L., Legay, S., Hausman, J.-F., Esposito, S., and Guerriero, G. (2016). Identification of Reference Genes for RT-qPCR Data Normalization in Cannabis Sativa Stem Tissues. Ijms 17 (9), 1556. doi:10.3390/ijms17091556
Mao, M., Xue, Y., He, Y., Zhou, X., Rafique, F., Hu, H., et al. (2020). Systematic Identification and Comparative Analysis of Lysine Succinylation between the green and white Parts of Chimeric Leaves of Ananas Comosus Var. Bracteatus. BMC Genomics 21, 383. doi:10.1186/s12864-020-6750-6
Maroufi, A., Van Bockstaele, E., and De Loose, M. (2010). Validation of Reference Genes for Gene Expression Analysis in Chicory (Cichorium Intybus) Using Quantitative Real-Time PCR. BMC Mol. Biol 11, 15–27. doi:10.1186/1471-2199-11-15
Müller, O. A., Grau, J., Thieme, S., Prochaska, H., Adlung, N., Sorgatz, A., et al. (2015). Genome-wide Identification and Validation of Reference Genes in Infected Tomato Leaves for Quantitative RT-PCR Analyses. PLoS One 10, e0136499. doi:10.1186/1471-2199-11-15
Narsai, R., Ivanova, A., Ng, S., and Whelan, J. (2010). Defining Reference Genes in Oryza Sativausing Organ, Development, Biotic and Abiotic Transcriptome Datasets. BMC Plant Biol. 10, 56. doi:10.1186/1471-2229-10-56
Nicot, N., Hausman, J.-F., Hoffmann, L., and Evers, D. (2005). Housekeeping Gene Selection for Real-Time RT-PCR Normalization in Potato during Biotic and Abiotic Stress. J. Exp. Bot. 56, 2907–2914. doi:10.1093/jxb/eri285
Petriccione, M., Mastrobuoni, F., Zampella, L., and Scortichini, M. (2015). Reference Gene Selection for Normalization of RT-qPCR Gene Expression Data from Actinidia Deliciosa Leaves Infected with Pseudomonas syringae Pv. Actinidiae. Sci. Rep. 5, 16961. doi:10.1038/srep16961
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper - Excel-based Tool Using Pair-wise Correlations. Biotechnol. Lett. 26, 509–515. doi:10.1023/b:bile.0000019559.84305.47
Pombo, M. A., Ramos, R. N., Zheng, Y., Fei, Z., Martin, G. B., Rosli, H. G., et al. (2019). Transcriptome-based Identification and Validation of Reference Genes for Plant-Bacteria Interaction Studies Using Nicotiana Benthamiana. Sci. Rep. 9, 1632. doi:10.1038/s41598-018-38247-2
Ponomareva, E. (2012). Effects of Different Signaling Pathways on Regulation of GLK GARP Transcription Factors in Arabidopsis thaliana. [dissertation/master’s Thesis]. Ottawa (IL): University of Ottawa.
Savitch, L. V., Subramaniam, R., Allard, G. C., and Singh, J. (2007). The GLK1 'regulon' Encodes Disease Defense Related Proteins and Confers Resistance to Fusarium Graminearum in Arabidopsis. Biochem. Biophysical Res. Commun. 359, 234–238. doi:10.1016/j.bbrc.2007.05.084
Sgamma, T., Pape, J., Massiah, A., and Jackson, S. (2016). Selection of Reference Genes for Diurnal and Developmental Time-Course Real-Time PCR Expression Analyses in Lettuce. Plant Methods 12, 21. doi:10.1186/s13007-016-0121-y
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol 7, 33. doi:10.1186/1471-2199-7-33
Stegemann, S., and Bock, R. (2009). Exchange of Genetic Material between Cells in Plant Tissue Grafts. Science 324, 649–651. doi:10.1126/science.1170397
Sudhakar Reddy, P., Srinivas Reddy, D., Sivasakthi, K., Bhatnagar-Mathur, P., Vadez, V., and Sharma, K. K. (2016). Evaluation of Sorghum [Sorghum Bicolor (L.)] Reference Genes in Various Tissues and under Abiotic Stress Conditions for Quantitative Real-Time PCR Data Normalization. Front. Plant Sci. 7, 529. doi:10.3389/fpls.2016.00529
Takamori, L. M., Pereira, A. V. C., Maia Souza, G., Vieira, L. G. E., and Ferreira Ribas, A. (2017). Identification of Endogenous Reference Genes for RT-qPCR Expression Analysis in Urochloa Brizantha under Abiotic Stresses. Sci. Rep. 7 (1), 8502. doi:10.1038/s41598-017-09156-7
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 3 (7). research0034.1. doi:10.1186/gb-2002-3-7-research0034
Wan, Q., Chen, S., Shan, Z., Yang, Z., Chen, L., Zhang, C., et al. (2017). Stability Evaluation of Reference Genes for Gene Expression Analysis by RT-qPCR in Soybean under Different Conditions. Plos One 12 (12), e0189405. doi:10.1371/journal.pone.0189405
Wang, M., and Lu, S. (2016). Validation of Suitable Reference Genes for Quantitative Gene Expression Analysis in Panax Ginseng. Front. Plant Sci. 6, 1259. doi:10.3389/fpls.2015.01259
Wang, X., Wu, Z., Bao, W., Hu, H., Chen, M., Chai, T., et al. (2019). Identification and Evaluation of Reference Genes for Quantitative Real-Time PCR Analysis in Polygonum Cuspidatum Based on Transcriptome Data. BMC Plant Biol. 19, 498. doi:10.1186/s12870-019-2108-0
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., and Langdale, J. A. (2009). GLK Transcription Factors Coordinate Expression of the Photosynthetic Apparatus in Arabidopsis. Plant Cell 21, 1109–1128. doi:10.1105/tpc.108.065250
Xie, F., Xiao, P., Chen, D., Xu, L., and Zhang, B. (2012). miRDeepFinder: a miRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 80, 75–84. doi:10.1007/s11103-012-9885-2
Xu, L., Xu, H., Cao, Y., Yang, P., Feng, Y., Tang, Y., et al. (2017). Validation of Reference Genes for Quantitative Real-Time PCR during Bicolor Tepal Development in Asiatic Hybrid Lilies (Lilium spp.). Front. Plant Sci. 8, 669. doi:10.3389/fpls.2017.00669
Yan, J., Yuan, F., Long, G., Qin, L., and Deng, Z. (2011). Selection of Reference Genes for Quantitative Real-Time RT-PCR Analysis in Citrus. Mol. Biol. Rep. 39, 1831–1838. doi:10.1007/s11033-011-0925-9
Yang, H., Liu, J., Huang, S., Guo, T., Deng, L., and Hua, W. (2014). Selection and Evaluation of Novel Reference Genes for Quantitative Reverse Transcription PCR (qRT-PCR) Based on Genome and Transcriptome Data in Brassica Napus L. Gene 538 (1), 113–122. doi:10.1016/j.gene.2013.12.057
Yang, Y. H. J., Markus, M. A., Mangs, A. H., Raitskin, O., Sperling, R., and Morris, B. J. (2013). ZRANB2 Localizes to Supraspliceosomes and Influences the Alternative Splicing of Multiple Genes in the Transcriptome. Mol. Biol. Rep. 40 (9), 5381–5395. doi:10.1007/s11033-013-2637-9
Zhang, T., Feng, P., Li, Y., Yu, P., Yu, G., Sang, X., et al. (2018). VIRESCENT-ALBINO LEAF 1 Regulates Leaf Colour Development and Cell Division in rice. J. Exp. Bot. 69 (20), 4791–4804. doi:10.1093/jxb/ery250
Keywords: Ananas comosus var bracteatus, RT-qPCR, reference genes, tissue, chimeric leaf development, hormone stimuli, gene expression
Citation: Mao M, Xue Y, He Y, Zhou X, Hu H, Liu J, Feng L, Yang W, Luo J, Zhang H, Li X and Ma J (2021) Validation of Reference Genes for Quantitative Real-Time PCR Normalization in Ananas comosus var. bracteatus During Chimeric Leaf Development and Response to Hormone Stimuli. Front. Genet. 12:716137. doi: 10.3389/fgene.2021.716137
Received: 28 May 2021; Accepted: 28 September 2021;
Published: 21 October 2021.
Edited by:
Deepmala Sehgal, International Maize and Wheat Improvement Center, MexicoReviewed by:
Yulong Ren, Institute of Crop Sciences (CAAS), ChinaSudhakar Reddy Palakolanu, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India
Copyright © 2021 Mao, Xue, He, Zhou, Hu, Liu, Feng, Yang, Luo, Zhang, Li and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ma, anVubWEzNjVAaG90bWFpbC5jb20=
 Meiqin Mao1
Meiqin Mao1 Yanbin Xue
Yanbin Xue Yehua He
Yehua He Jun Ma
Jun Ma