Abstract
Iron (Fe) and zinc (Zn) micronutrient deficiencies are significant health concerns, particularly among the underprivileged and resource-poor people in the semi-arid tropics globally. Pearl millet is regarded as a climate-smart crop with low water and energy footprints. It thrives well under adverse agro-ecologies such as high temperatures and limited rainfall. Pearl millet is regarded as a nutri-cereal owing to health-promoting traits such as high grain Fe and Zn content, metabolizable energy, high antioxidant and polyphenols, high proportion of slowly digestible starches, dietary fibers, and favorable essential amino acid profile compared to many cereals. Higher genetic variability for grain Fe and Zn content has facilitated considerable progress in mapping and mining QTLs, alleles and genes underlying micronutrient metabolism. This has been made possible by developing efficient genetic and genomic resources in pearl millet over the last decade. These include genetic stocks such as bi-parental RIL mapping populations, association mapping panels, chromosome segment substitution lines (CSSLs) and TILLING populations. On the genomics side, considerable progress has been made in generating genomic markers, such as SSR marker repository development. This was followed by the development of a next-generation sequencing-based genome-wide SNP repository. The circa 1,000 genomes re-sequencing project played a significant role. A high-quality reference genome was made available by re-sequencing of world diversity panel, mapping population parents and hybrid parental lines. This mini-review attempts to provide information on the current developments on mapping Fe and Zn content in pearl millet and future outlook.
Introduction
One-sixth of the global population suffers from hunger which is an unacceptable burden for humankind. A report on “The State of Food Insecurity in the World 2014” created by various organizations suggested that approximately 800 million persons were affected by undernourishment during 2012–2014, a sizable fraction of which are dwelling in the emergent nations (Bohra et al., 2016). A large fraction of the world’s population is consuming rice and wheat as a principal staple. These cereals are relatively not rich in mineral micronutrients which are essential for the human body to maintain basic metabolic rates (White and Broadley, 2009). Worldwide, South Asia and the Indian sub-continent contain 38.9% of the world’s malnourished children, impacting significantly on these economies. The order of decreasing global Fe/Zn malnutrition is as follow; India (38%), Nigeria (7%), Pakistan (6%), China (5%), Indonesia (5%), Bangladesh (4%), Ethiopia (3%), the Democratic Republic of the Congo (3%), Philippines (2%), United Republic of Tanzania (2%), Egypt (2%), Kenya (1%), Uganda (1%) and Sudan (1%) (De Onis et al., 2012).
Pearl Millet as a Nutrient-Dense Cereal
Pearl millet is a nutrient-rich staple cereal grain with a high amount of carbohydrates, proteins, fats, polyphenols, dietary fibers, vitamins, and micronutrients. Pearl millet grains are a cost-efficient energy source (360 Kcal/100 g) for humans to meet their dietary requirements than sorghum, wheat, rice and maize. Pearl millet has high levels of slowly available starch, high fiber (1.2 g/100 g), and 8–15 times greater α-amylase activity. Pearl millet flour is gluten-free with a low glycemic index and significantly rich in resistant starch, hence the ideal candidate grain for use in the functional-food market worldwide (Ragaee et al., 2006; Saleh et al., 2013). The amino acid profiling indicated that pearl millet grain is rich in histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine that are relatively higher than the other staple cereals. Pearl millet is rich in unsaturated fatty acids with a higher content of nutritionally important omega-3 fatty acids than other cereal grains.
Grain Fe/Zn Profile of Pearl Millet Relative to Other Cereals
Pearl millet grains are highly nutritious having inorganic nutrients including Fe (22–154 ppm) and Zn (19–121 ppm) (Mahendrakar et al., 2019) with relatively higher amounts of the Fe and Zn than other staple cereal such as wheat (10 ppm), rice (28 ppm), and maize (22 ppm). Genetic biofortification is a way of enhancing nutrition in staple crops in the modern period, and improved food processing and markets will be the backbone of future nutritional security (Govindaraj et al., 2019). Hence breeding for micronutrient-rich pearl millet varieties and their utilization would fulfill the suggested recommended dietary allowance (RDA) of Fe and Zn and also would encourage producing biofortified pearl millet lines (Munyaradzi, 2013). According to a recent study, having 250 g of biofortified pearl millet per day can supply 84% of the RDA for Fe and 100% of the RDA for Zn (Satyavathi et al., 2021).
Biofortification Options for Alleviating Fe/Zn Malnutrition
Crop biofortification is a viable and economical approach to handle micronutrient malnutrition, specifically in developing countries (Stein et al., 2007; Bouis et al., 2011). Biofortification is a method to improve the micronutrient content of farm harvest. This strategy includes targeting and altering the movement pathways of minerals, i.e., “pulling” nutrients from the earth and “pushing” them into edible plant parts in their bioavailable forms. This pulling and pushing approach can alleviate the malnutrition issues which impact human potential. Compared to currently available tactics namely fortification, supplementation, or dietary diversification, biofortification is a viable means of making available nutrition in remote and rural areas for delivery to the impoverished community having inadequate access to balanced and nutrient-rich food (Bouis et al., 2011).
Challenges in Biofortification and Phenotyping for grain Fe and Zn
Some of the major challenges in pearl millet biofortification include lack of access to high throughput phenotyping platforms and higher phenotyping costs. Dust contamination in the grain samples, improper/uneven seed set and soil fertility variation introduces error in measurements. The Fe/Zn trait is not simply inherited, it is largely governed by additive gene action, and shows genotype-by-environment interaction especially for small effect alleles. These pose challenges in breeding Fe-Zn dense cultivars. Also, there is a weak negative correlation between grain yield and grain Fe and Zn content, which might become significant in narrow/correlated breeding populations. Further, bioavailability-related issues such as for low phytates in the grains and absence of specific vitamins such as vitamin C in the diet need to be addressed for Fe.
Two highly precise tools namely Atomic Absorption Spectrometry (AAS) and Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES) are widely used to analyze grain minerals. A detailed methodology of Fe and Zn analysis in pearl millet grain using AAS and ICP-OES is provided by Kumar et al. (2016) and Pujar et al. (2020), respectively. Both AAS and ICP-OES based methodologies are destructive and require acid digested sample preparation which is expensive, time-demanding and low throughput. Therefore, there is a demand for an alternative device for the rapid analysis of massive grain samples. The Energy-dispersive X-ray fluorescence (ED-XRF), an improved version of the XRF method, is a non-destructive technology for speedy analysis of iron (Fe) and zinc (Zn) in grains. In pearl millet, this system has been developed and validated by Govindaraj et al. (2016). This system can handle circa 250–300 samples per day at the cost of US $ < 1.0 per sample compared to the ICP-OES where the analysis cost is about US $18 per sample. The results of earlier studies show that correlations between the ICP-OES and ED-XRF values are significantly higher for both Fe and Zn content (Paltridge et al., 2012; Govindaraj et al., 2016). This can considerably reduce the cost and time for the analysis of Fe and Zn density.
Development of Genetic and Genomic Resources
Genetic Resources and Historic Breeding Efforts for Fe and Zn Rich Grains
Large variability has been reported for grain Fe and Zn in germplasm, breeding lines and cultivars. The gene bank of ICRISAT is a promising source of Fe and Zn rich genotypes. A total of 41 and 42 genotypes were richer in Fe (>80 ppm) and Zn (>60 ppm), respectively. While 33 genotypes were found to be a rich source of both minerals (Yadav et al., 2017). The analysis of 297 Iniadi germplasm from Western Africa (Togo, Eastern Ghana, Southern Burkina Faso and Western Benin) has shown wide variability for grain Fe and Zn content (Rai et al., 2008). Several studies at ICRISAT showed a wide range of variability for grain Fe and Zn densities in diverse breeding materials such as Iniadi germplasm accessions (51–121 ppm Fe; 46–87 ppm Zn), population progenies (18.0–135.0 ppm Fe; 22.0–92.0 Zn), inbred parents (30.3–102.0 ppm Fe; 27.4–84.0 ppm Zn), hybrids derived from diverse inbreds (25.8–80.0 ppm Fe; 22.0–70 ppm Zn) and commercial hybrids (31.0–61.0 ppm Fe, 32.0–54.0 ppm Zn) (Govindaraj et al., 2019). Analysis of 281 advanced breeding lines exhibited substantial variability for Fe (35–116 ppm) and Zn (21–80 ppm) (Pujar et al., 2020). ICTP 8203, an open-pollinated variety (OPV) released in India in 1988 had been found to have the highest level of Fe and Zn density (Rai et al., 2013). The efforts in biofortification have yielded nutrient-rich and high-yield cultivars in India (Rai et al., 2014). A world-first high-Fe pearl millet variety “Dhanashakti” (71 ppm Fe) was developed by utilizing the intra-population variability within ICTP 8203. Hybrid ICMH 1201 is also rich in Fe (75 ppm) and under cultivation by the name of Shakti 1201 in Maharashtra and Rajasthan. In India, the first wave of biofortified hybrids are AHB 1200 Fe (ICMH 1202), HHB 299 (ICMH 1203), and Phule Maha Shakti (ICMH 1301) officially released at the National level in India. These biofortified hybrids contain more than 70 ppm Fe and 35 ppm Zn. In general, all the commercial hybrids had 42 and 32 ppm mean Fe and Zn content, respectively (Govindaraj et al., 2019). A mega pearl millet cultivar HHB 67 Improved 2 developed through genomics-assisted breeding has been recently released and notified for cultivation in India. This hybrid is expected to contribute immensely towards food and nutritional security.
Genomic Resources
Molecular Markers
DNA markers like AFLP (Devos et al., 1995) and SSRs (Senthilvel et al., 2008; Sehgal et al., 2012) were applied to detect genetic polymorphism in pearl millet germplasm. SSR markers were also valuable for anchoring molecular linkage maps (Gonzalo et al., 2005) that were densely saturated with DArT (Supriya et al., 2011). EST-SSRs were markers of choice for linkage and association mapping since they are frequently transferable to other related plant species (Varshney et al., 2005; Singh et al., 2011; Singh et al., 2019; Srivastava et al., 2020a). Thousands of functional SSRs have been developed in pearl millet genetics and breeding (Senthilvel et al., 2008; Rajaram et al., 2013), in addition to SSCP-SNP (Bertin et al., 2005), DArT (Supriya et al., 2011; Kumar et al., 2016), CISP and SNP (Sehgal et al., 2012) markers. These molecular tools have successfully been deployed in marker-assisted backcrossing (MABC) to improve “HHB 67 Improved 2” and GHB 538 Improved hybrids from India. Pearl millet genome sequence information of circa 1,000 lines has resulted in genome-wide SNPs (low/mid/high-density panel) for trait mapping, deployment and heterotic gene pools (Varshney et al., 2017; Srivastava et al., 2020b). All these genomic resources have been made available at various public repository platforms. Some key sources include https://cegsb.icrisat.org/ipmgsc/genome.html; http://gigadb.org/dataset/100192; https://www.ncbi.nlm.nih.gov/genome/?term=pearl+millet; https://www.ncbi.nlm.nih.gov/assembly/GCA_002174835.1.
Genetic Linkage Mapping and Consensus Maps
In pearl millet, specific genetic resources were developed to map grain Fe and Zn content. This included forward genetic stocks such as bi-parental mapping populations, association mapping panels, and reverse genetic stocks as the TILLING (Targeting Induced Local Lesions in Genomes) populations. Using bi-parental mapping populations. Kumar et al. (2016) developed a linkage map of 1749 cM that was generated with 305 markers (96 codominant SSRs and 208 dominant DArTs) with inter-marker distance of 5.73 cM based on a previously reported consensus map (Rajaram et al., 2013). Kumar et al. (2018) used another population to construct a genetic linkage map with DArT and SSR markers using 317 RIL progenies derived from the (ICMS 8511-S1-17-2-1-1-B-P03 × AIMP 92901-S1-183-2-2-B-08) cross. Recently, Mahendrakar et al. (2020) reported unique genes on seven linkage groups. Figure 1 summarizes the latest mapping results for GFeC and GZnC in pearl millet from different studies. In addition, reverse genetic stocks as TILLING populations are being developed at ICRISAT in the three genetic backgrounds including that of world reference germplasm Tift 23DB1-P1-P5 for functional genomics related to Fe and Zn and other important traits.
FIGURE 1
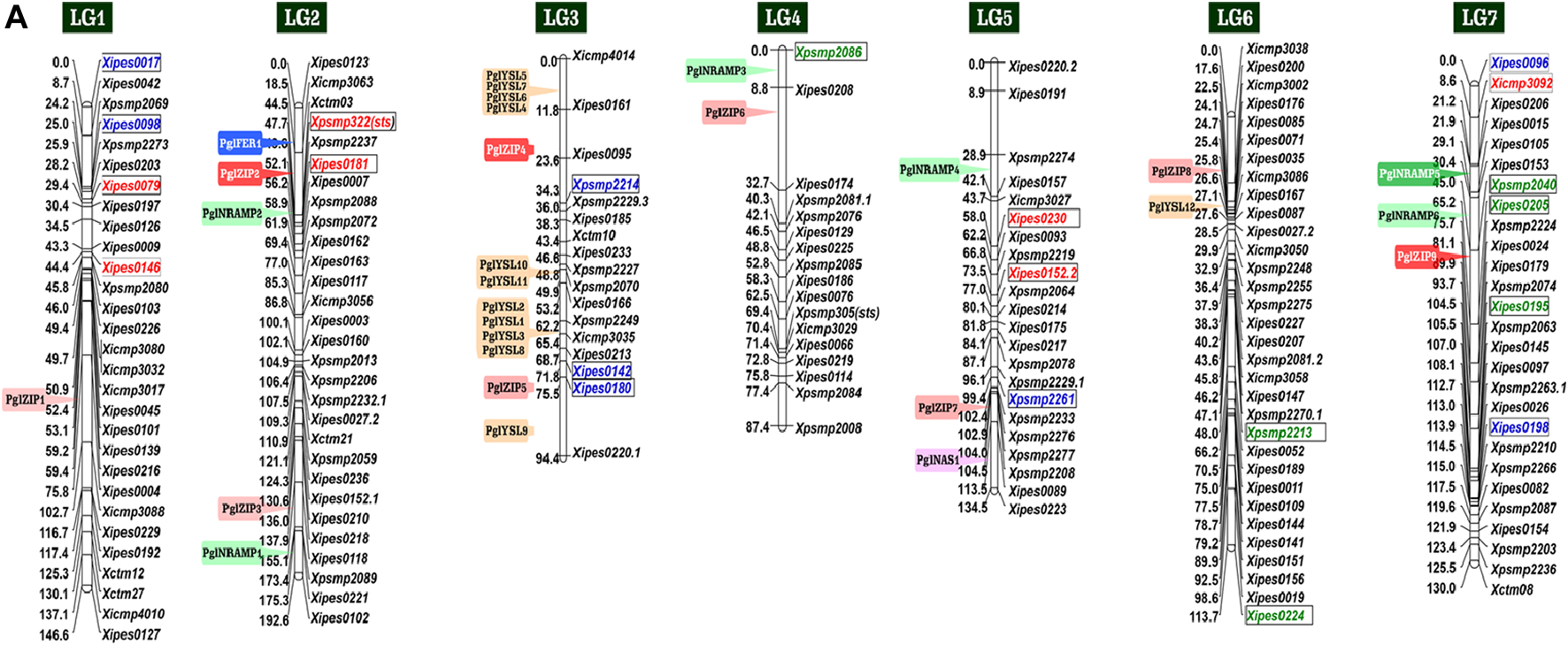
List of the reported linked markers for QTLs, alleles and candidate genes for high grain Fe and Zn content in pearl millet on the consensus map of Rajaram et al. (2013)Note: Red font color indicates favorable allele for grain Fe content, green for grain Zn content and blue for both grain Fe and Zn content. The 29 genes for grain Fe and Zn homeostasis belonging to five broad families are in solid color boxes. The five candidate genes for grain Fe and Zn are in solid boxes with white font (Data source: Mahendrakar et al., 2020).
Marker-Trait Association for Grain Iron and Zinc Density
Significant efforts have been made to develop a fine linkage map and Fe-Zn QTLs in pearl millet (Kumar et al., 2016). Hundreds of markers were employed to generate a linkage map for mapping QTLs for GFeC and GZnC using 106 RILs (F6) derived from ICMB 841-P3 × 863B-P2 cross in pearl millet (Kumar et al., 2016). Using phenotypic data from two environments, two co-localized QTLs for Fe and Zn content on LG3 were detected. Similarly, for open-pollinated (OP) seeds, the QTL analysis detected two Fe-QTLs on LG3 and LG5, and two Zn-QTLs on LG3 and LG7. The total phenotypic variance for Fe-QTL and Zn-QTLs in OP seeds was 16 and 42%, respectively (Kumar et al., 2016). In another analogous study, 11 Fe-QTLs and 8 Zn-QTLs have been identified, among them three major QTLs on LG1 and two on LG7 for both Fe and Zn were co-mapped in pearl millet (Kumar et al., 2018). The phenotypic variance explained by different Fe-QTLs and Zn-QTLs ranged between 9.0–31.9% and 9.4–30.4%, respectively (Table 1). A total of 46 QTLs in pearl millet were detected with phenotypic variance ranged from 6.7 to 20.5% and 5.7–26.4% for GFeC and GZnC QTLs, respectively (Kumar et al., 2016; Kumar et al., 2018). Although, linkage analysis in most of the investigations indicates the existence of genes/QTLs at a distance of 5.73–23.0 cM from the flanking markers for MABC programs and functional validation of candidate genes (Vengadessan et al., 2013). In addition to SSR markers, genome-wide SNPs are more suited for genome-wide association studies (GWAS), and genomic selection (GS) and QTL mapping for Fe/Zn traits in pearl millet (Srivastava et al., 2020a). A panel of pearl millet germplasm lines was evaluated under the different agro-climates using SSRs for identification of strong maker trait associations (MTAs) using GWAS, in which Xicmp3092 marker exhibited a significant association with GFeC on LG7 and Xpsmp2086, Xpsmp2213 and Xipes0224 markers were showed association with GZnC on LG4 and LG6, respectively (Anuradha et al., 2017). Conserved association for GFeC and GZnC was demonstrated by Xipes0180, Xpsmp2261, and Xipes0096 markers on LG3, LG5, and LG7, respectively (Anuradha et al., 2017). Using 144 multi-parent advance generation inter-cross (MAGIC) plus population, Descalsota et al. (2018) identified SNPs for grain Fe and Zn. They identified few Fe and Zn homeostasis genes such as OsMTP6, OsNAS3, OsMT2D, OsVIT1 and OsNRAMP7 which were localized with QTLs. This information may be helpful in the biofortification of Fe and Zn in pearl millet. The LG3 grain Fe and Zn QTL (Kumar et al., 2016) interval was sequenced for the development of SNP markers. A 4-SNP panel is being used in the breeding programs for rejecting lines with poor grain Fe and Zn content.
TABLE 1
| Sl. no. | Objective | Plant accessions/population used | Range of GFeC (ppm) | Range of GZnC (ppm) | DNA markers used | Number of identified QTLs/genes/MTAs linked with GFeC and GZnC | Associated markers/candidate genes | Phenotypic variance (R2) | References |
| 1. | Identification and validation of candidate genes underlying GFeC and GZnC metabolism | Two contrast parents of RILs population AIMP 92,901 (high grain Fe and Zn), and ICMS 8511 (low grain Fe and Zn) | 22.93–154.5 | 19.31–121.00 | N/A | 5 Candidate gene includes; PglFER1, PglZIP2, PglZIP4, PglNramp5, and PglZIP9 controlling with GFeC and GZnC | Ferritin-like gene, PglFER1 (Fe), PglZIP and PglNRAMP (Fe and Zn) | N/A | Mahendrakar et al. (2020) |
| 2. | To map QTLs linked with GFeC and GZnC in an iniadi-derived immortal pearl millet mapping population | A panel of 317 F6 recombinant inbred lines (RILs) derived from the (ICMS 8511-S1-17-2-1-1-B-P03 × AIMP 92901-S1-183-2-2-B-08) cross | 20–131 | 18–110 | 196 (177 DArTs & 19 SSRs) | Total 11 QTLs detected for GFeC and 8 for GZnC. 3 QTLs co-mapped for both GFeC-GZnC observed, 1 on LG1 and 2 on LG7 | Fe-Zn: pgpb10531-pgpb9130, Fe: pgpb8427-pgpb13221 pgpb11938-pgpb8987 Zn: Xipes198-pgpb8427 pgpb12329-pgpb9721 |
GFeC: 9.0–31.9% (cumulative 74%). GZnC: 9.4–30.4% (cumulative 65%) | Kumar et al. (2018) |
| 3. | Genome-wide association mapping of QTLs for Fe & Zn | An association mapping panel comprised of 130 diverse lines (B-, R- & advanced breeding lines) | 32.3–111.9 | 26.6–73.7 | 267 markers (250 SSRs & 17 genic markers) | SSRs associated with both grain GZnC and GZnC on LG3, LG5 and LG7 | Xipes 0810 (aspartic proteinase gene) Xpsmp 2261 (intergenic region) Xipes 0096 |
11.4, 13.3 and 11.3% R2, respectively | Anuradha et al. (2017) |
| 4. | Mapping of QTLs controlling high GFeC and GZnC in self and open-pollinated grains of pearl millet | A set of 144 RILs derived from the (ICMB 841-P3 × 863B-P2) cross | a) Fe-Self 28.4–124 | a) Zn-Self 28.7–119.8 | 305 (96 SSRs & 208 DArTs) | In selfed seed, a 2 co-localized QTL for GFeC and GZnC on LG3. b) In open pollinated seeds 2 QTLs for GFeC on LG 3 & LG5, and 2 QTLs for GZnC on LG 3 & LG7. | Fe & Zn: Xpsmp2214-Xipes0142. OP Fe: Xpsmp2214-Xipes0142 Pgbp5908-Pgpb6674 OP Zn: Xpsmp2214-Xipes0142. Xpsmp2040-Pgpb10727 |
Fe: 19.0%; Zn: 36.0% Fe: 16.0%; Zn: 42.0% | Kumar et al. (2016) |
| b) Fe-OP 22.4–77.4 | b) Zn- OP 21.9–73.7 |
Summary of various studies for high grain Fe (GFeC) and Zn (GZnC) content in pearl millet.
N/A = not applicable.
Evolution of Genotyping Platforms
With the advent and recent advancement of next-generation sequencing (NGS) technology with affordable cost, high-throughput genotyping platforms (HTGP) have emerged. These HTGPs revolutionized genotyping assays in a cost-efficient manner and rapid protocols. Using NGS-based genotyping platforms, assaying for genomic variations, high-resolution linkage mapping, identification of candidate gene(s) and natural allelic variants underlying for QTLs controlling yield-related traits have become regular practices for crop species. In pearl millet, various next-generation sequencing-based genotyping platforms have been developed and optimized. These can help generate thousands of SNPs in various genetic stocks, breeding populations, and landraces. Genotyping-by-sequencing (GbS) is a speedy, economical and reduced representation sequencing method that is frequently deployed for genome-wide genetic variability profiling in cereal crops (Elshire et al., 2011). Over singe (ApeK1) enzyme GbS, two enzyme combinations (SphI and MluC1) double digest restriction associated DNA (ddRAD) was found to be superior (Srivastava et al., 2016) and was used to fine map the LG2 drought tolerance QTL (Srivastava et al., 2017). The other genotyping platforms that are available include tunable-GbS (tGbS) (Ott et al., 2017; Liang et al., 2018), DArT-seq (Allan et al., 2020); and RNase H2 enzyme-dependent amplicon sequencing (rhAmpSeq) (Dobosy et al., 2011).
Transcriptome Assemblies and Candidate Genes
To discover underlying mineral homeostasis, transcriptomics may pave the way to gene discovery and increased biosynthesis of useful nutritional products (Hirai et al., 2004). Knowledge of the genes controlling specific stages in the Fe and Zn metabolic pathways is relatively meagre in pearl millet. Recently, using transcriptomic studies, the key candidate genes were discovered for grain Fe and Zn metabolism in pearl millet (Mahendrakar et al., 2020). A total of 29 unique genes were used for spatio-temporal characterization using expression studies for different developmental stages in contrasting genotypes for GFeC and GZnC. Tissue and stage-specific expressions of Fe and Zn genes in contrasting genotypes for GFeC and GZnC were recorded (Mahendrakar et al., 2020). Results revealed that PglZIP, PglNRAMP, PglYSL, and PglFER family genes were candidates for GFeC and GZnC. The evolutionary relationship of the 29 unique genes showed ortholog events with other related cereals studied (Figure 2). To further support the reported candidate genes, protein-protein interaction (PPI) was studied. The PglFER and PglZIP gene families interacted with various other proteins associated with Fe and Zn metabolism along with metabolism of carbohydrates, hexokinase, protein kinase, ATP binding protein and phosphotransferase activity (Figure 3). Ferritin-like gene (PglFER-1) was found to be the most potent candidate gene for GFeC. For further validation, genomic regions underlying GFeC and GZnC were deciphered by annotating QTL regions for grain Fe and Zn densities. Expressed genes were correlated with major QTL co-localized on LG7 for GFeC and GZnC (Mahendrakar et al., 2020). The study provided a useful understanding of different Fe and Zn metabolism gene homologs and laid a foundation for functional dissection. Ogo et al. (2011) reported overexpression of OsIRO2 resulted in enhanced accumulation of Fe in brown rice grain even in calcareous soil and further recorded high Zn in grains. By overexpressing the soybean-ferritin gene, a high iron rice variety called IR68144 was developed in which iron concentration was increased by 3.7 fold in polished rice grain (Vasconcelos et al., 2003). Another group overexpressed soybean ferritin gene (soyfer1) in the same rice variety and interbred with Swarna rice variety resulting in a new variety with 2.54 fold and 1.54 fold more Fe and Zn content in rice grain, respectively (Paul et al., 2014).
FIGURE 2

Phylogeny tree of reported 29 pearl millet candidate genes (prefix Pgl) for Fe and Zn metabolism with that of other cereal crops showing evolutionary relationship and ortholog events with other cereal crops (Data source: Mahendrakar et al., 2020).
FIGURE 3
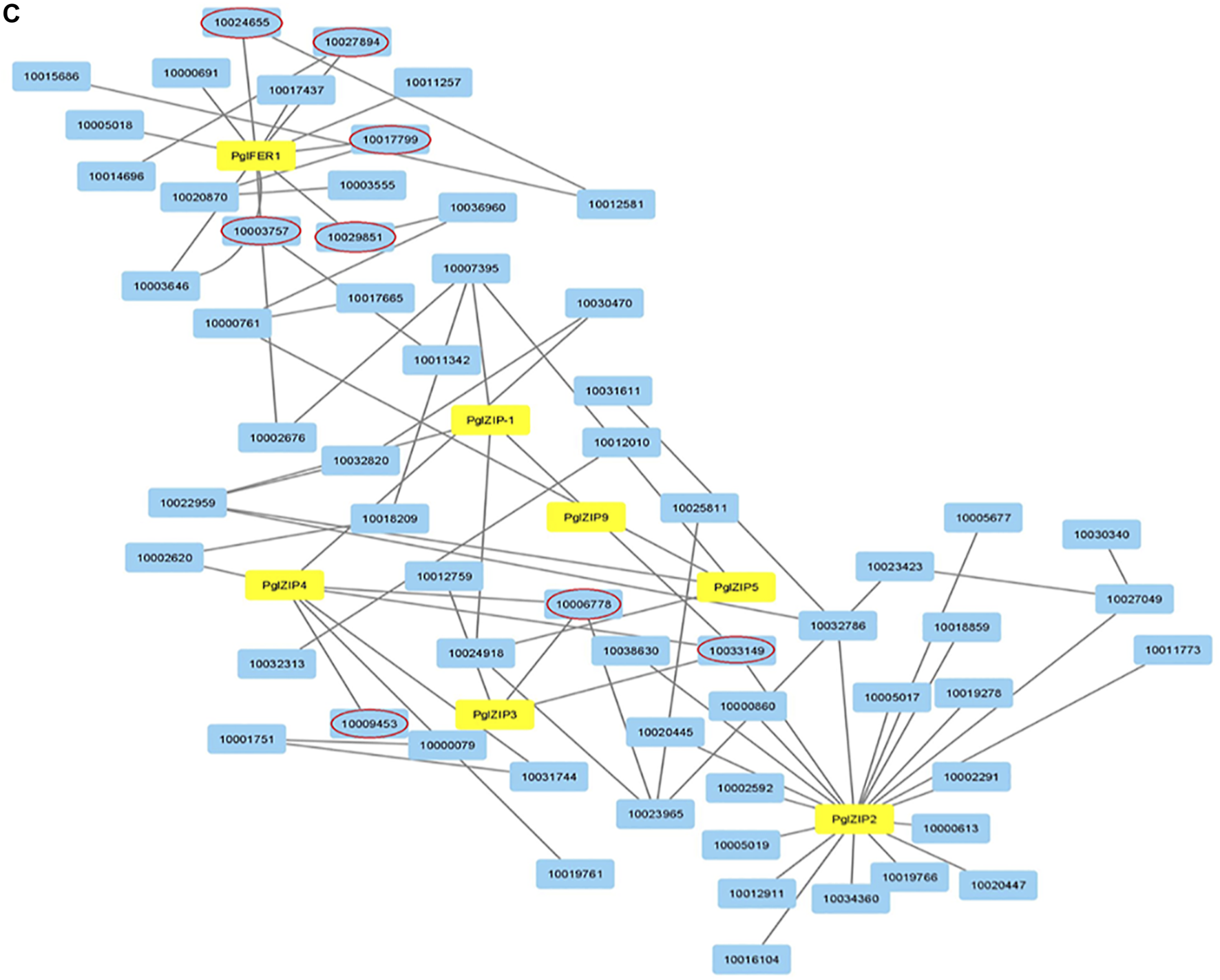
Protein-protein interaction (PPI) network for 7 putative proteins. Yellow colored nodes have been short listed from 29 unique proteins (Mahendrakar et al., 2020) and other interacted nodes (red circled) involved in Fe and Zn metabolism and transport and other functions like ATP binding, phosphotransferase activity, carbohydrate metabolism and hexokinase activity (Data source: Mahendrakar et al., 2020).
Conclusion and Future Outlook
To enhance the genetic gains for grain micronutrient contents and yield, it is essential to understand the germplasm, breeding lines, and functional and translational genomics. The availability of cost-efficient NGS-based technologies have revolutionized whole-genome sequencing and transcriptome analysis. This has led to genome-wide genetic dissections of untapped valuable alleles. The identified genomic regions/QTLs controlling micronutrient accumulation and homeostasis into elite hybrid parental lines and OPVs may pave the way to breed high grain Fe/Zn cultivars using low density markers such as the available 4-SNP panel and pan-genome marker-assisted selection schemes. The promising lines with favourable alleles bred by using such technologies may be useful for generating new cultivars which accumulate all major alleles regulating grain iron and zinc metabolism. The Indian Council of Agricultural Research (ICAR) minimum standards for Fe (42 ppm) and Zn (32 ppm) in pearl millet cultivar release policy and the product profiles support use of molecular markers in mainstreaming these traits. National and international gene banks should sequentially genotype the germplasm to map all essential nutritional traits to explore traits specific germplasm and its novel genes. Thus, with the help of nutrigenomic advances, the next generation of nutrient-dense farmer preferred cultivars may be bred with precision and efficiency in pearl millet.
Statements
Author contributions
RKS planned and coordinated this study. RKS, CS, MDM, RBS, SK, MG, and IG contributed to this work and drafted the manuscript.
Funding
Funding support from the Department of Biotechnology (DBT) and the CGIAR Research Program on Dryland Cereals is gratefully acknowledged. This work has been published as part of the CGIAR Research Program on Dryland Cereals.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer (SP) declared a past co-authorship/collaboration with one of the authors (RKS) to the handling Editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Allan V. Vetriventhan M. Senthil R. Geetha S. Deshpande S. Rathore A. et al (2020). Genome-Wide DArTSeq Genotyping and Phenotypic Based Assessment of within and Among Accessions Diversity and Effective Sample Size in the Diverse Sorghum, Pearl Millet, and Pigeonpea Landraces. Front. Plant Sci.11, 587426. 10.3389/fpls.2020.587426
2
Anuradha N. Satyavathi C. T. Bharadwaj C. Nepolean T. Sankar S. M. Singh S. P. et al (2017). Deciphering Genomic Regions for High Grain Iron and Zinc Content Using Association Mapping in Pearl Millet. Front. Plant Sci.8, 412. 10.3389/fpls.2017.00412
3
Bertin I. Zhu J. H. Gale M. D. (2005). SSCP-SNP in Pearl Millet-A New Marker System for Comparative Genetics. Theor. Appl. Genet.110, 1467–1472. 10.1007/s00122-005-1981-0
4
Bohra A. Jha U. C. Kumar S. (2016). “Enriching Nutrient Density in Staple Crops Using Modern "-Omics" Tools,” in Biofortification of Food Crops. Editors SinghU.PraharajC. S.SinghS. S.SinghN. P . (New Delhi: Springer), 85–103. 10.1007/978-81-322-2716-8_8
5
Bouis H. E. Hotz C. McClafferty B. Meenakshi J. V. Pfeiffer W. H. (2011). Biofortification: a New Tool to Reduce Micronutrient Malnutrition. Food Nutr. Bull.32 (1_Suppl. 1), S31–S40. 10.1177/15648265110321s105
6
De Onis M. Blössner M. Borghi E. (2012). Prevalence and Trends of Stunting Among Pre-school Children, 1990-2020. Public Health Nutr.15 (1), 142–148. 10.1017/s1368980011001315
7
Descalsota G. I. L. Swamy B. P. M. Zaw H. Inabangan-Asilo M. A. Amparado A. Mauleon R. et al (2018). Genome-wide Association Mapping in a rice MAGIC Plus Population Detects QTLs and Genes Useful for Biofortification. Front. Plant Sci.9, 1347. 10.3389/fpls.2018.01347
8
Devos K. M. Pittaway T. S. Busso C. S. Gale M. D. Witcombe J. R. Hash C. T. (1995). Molecular Tools for the Pearl Millet Nuclear Genome. Int. Sorghum Millets Newslett.36, 64–66.
9
Dobosy J. R. Rose S. D. Beltz K. R. Rupp S. M. Powers K. M. Behlke M. A. et al (2011). RNase H-dependent PCR (rhPCR): Improved Specificity and Single Nucleotide Polymorphism Detection Using Blocked Cleavable Primers. BMC Biotechnol.11, 80. 10.1186/1472-6750-11-80
10
Elshire R. J. Glaubitz J. C. Sun Q. Poland J. A. Kawamoto K. Buckler E. S. et al (2011). A Robust, Simple Genotyping-By-Sequencing (GBS) Approach for High Diversity Species. PLoS One6, e19379. 10.1371/journal.pone.0019379
11
Gonzalo M. J. Oliver M. Garcia-Mas J. Monfort A. Dolcet-Sanjuan R. Katzir N. et al (2005). Simple-sequence Repeat Markers Used in Merging Linkage Maps of Melon (Cucumis Melo L.). Theor. Appl. Genet.110, 802–811. 10.1007/s00122-004-1814-6
12
Govindaraj M. Rai K. N. Cherian B. Pfeiffer W. H. Kanatti A. Shivade H. (2019). Breeding Biofortified Pearl Millet Varieties and Hybrids to Enhance Millet Markets for Human Nutrition. Agriculture9 (5), 106. 10.3390/agriculture9050106
13
Govindaraj M. Rai K. N. Pfeiffer W. H. Kanatti A. Shivade H. H. (2016). Energy-dispersive X-ray Fluorescence Spectrometry for Cost-Effective and Rapid Screening of Pearl Millet Germplasm and Breeding Lines for Grain Iron and Zinc Density. Commun. Soil Sci. Plant Anal.47 (18), 2126–2134. 10.1080/00103624.2016.1228938
14
Hirai M. Y. Yano M. Goodenowe D. B. Kanaya S. Kimura T. Awazuhara M. et al (2004). From the Cover: Integration of Transcriptomics and Metabolomics for Understanding of Global Responses to Nutritional Stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci.101 (27), 10205–10210. 10.1073/pnas.0403218101
15
K. Srivastava R. Bollam S. Pujarula V. Pusuluri M. Singh R. B. Potupureddi G. et al (2020b). Exploitation of Heterosis in Pearl Millet: A Review. Plants9 (7), 807. 10.3390/plants9070807
16
Kumar S. Hash C. Nepolean T. Mahendrakar M. Satyavathi C. Singh G. et al (2018). Mapping Grain Iron and Zinc Content Quantitative Trait Loci in an Iniadi-Derived Immortal Population of Pearl Millet. Genes9 (5), 248. 10.3390/genes9050248
17
Kumar S. Hash C. T. Thirunavukkarasu N. Singh G. Rajaram V. Rathore A. et al (2016). Mapping Quantitative Trait Loci Controlling High Iron and Zinc Content in Self and Open Pollinated Grains of Pearl Millet [Pennisetum Glaucum (L.) R. Br.]. Front. Plant Sci.7, 1636. 10.3389/fpls.2016.01636
18
Liang Z. Gupta S. K. Yeh C.-T. Zhang Y. Ngu D. W. Kumar R. et al (2018). Phenotypic Data from Inbred Parents Can Improve Genomic Prediction in Pearl Millet Hybrids. G3: Genes Genomes Genet.8, 2513–2522. 10.1534/g3.118.200242
19
Mahendrakar M. D. Kumar S. Singh R. B. Rathore A. Potupureddi G. Kavi Kishor P. B. et al (2019). Genetic Variability, Genotype × Environment Interaction and Correlation Analysis for Grain Iron and Zinc Contents in Recombinant Inbred Line Population of Pearl Millet [Pennisetum Glaucum (L). R. Br.]. Indian J. Genet.79 (3), 545–551. 10.31742/ijgpb.79.3.3
20
Mahendrakar M. D. Parveda M. Kishor P. K. Srivastava R. K. (2020). Discovery and Validation of Candidate Genes for Grain Iron and Zinc Metabolism in Pearl Millet [Pennisetum Glaucum (L.) R. Br.].. Scientific Rep.10 (1), 1–16. 10.1038/s41598-020-73241-7
21
Munyaradzi M. (2013). Biofortified Pearl Millet 'can Combat Iron Deficiency. Available at: SciDev.Net (Accessed August 29, 2013).
22
Ogo Y. Itai R. N. Kobayashi T. Aung M. S. Nakanishi H. Nishizawa N. K. (2011). OsIRO2 Is Responsible for Iron Utilization in rice and Improves Growth and Yield in Calcareous Soil. Plant Mol. Biol.75, 593–605. 10.1007/s11103-011-9752-6
23
Ott A. Liu S. Schnable J. C. Yeh C.-T. E. Wang K.-S. Schnable P. S. (2017). tGBS Genotyping-By-Sequencing Enables Reliable Genotyping of Heterozygous Loci. Nucleic Acids Res.45 (21), e178. 10.1093/nar/gkx853
24
Paltridge N. G. Palmer L. J. Milham P. J. Guild G. E. Stangoulis J. C. R. (2012). Energy-dispersive X-ray Fluorescence Analysis of Zinc and Iron Concentration in rice and Pearl Millet Grain. Plant Soil361, 251–260. 10.1007/s11104-011-1104-4
25
Paul S. Ali N. Datta S. K. Datta K. (2014). Development of an Iron-Enriched High-Yieldings Indica rice Cultivar by Introgression of a High-Iron Trait from Transgenic Iron-Biofortified rice. Plant Foods Hum. Nutr.69, 203–208. 10.1007/s11130-014-0431-z
26
Pujar M. Govindaraj M. Gangaprasad S. Kanatti A. Shivade H. (2020). Genetic Variation and Diversity for Grain Iron, Zinc, Protein and Agronomic Traits in Advanced Breeding Lines of Pearl Millet [Pennisetum Glaucum (L.) R. Br.] for Biofortification Breeding. Genet. Resour. Crop Evol.67, 2009–2022. 10.1007/s10722-020-00956-x
27
Rai K. N. Hash C. T. Singh A. K. Velu G. (2008). Adaptation and Quality Traits of a Germplasm-Derived Commercial Seed Parent of Pearl Millet. Plant Genetic Resour. Newslett.154, 20–24.
28
Rai K. N. Yadav O. P. Rajpurohit B. S. Patil H. T. Govindaraj M. Khairwal I. S. et al (2013). Breeding Pearl Millet Cultivars for High Iron Density With Zinc Density as an Associated Trait. J. SAT Agric. Res.11, 1–7.
29
Rai K. N. Patil H. T. Yadav O. P. Govindaraj M. Khairwal I. S. Cherian B. et al (2014). Dhanashakti: A High-Iron Pearl Millet Variety. Ind. Farm.64, 32–34.
30
Ragaee S. Abdelaal E. Noaman M. (2006). Antioxidant Activity and Nutrient Composition of Selected Cereals for Food Use. Food Chem.98, 32–38. 10.1016/j.foodchem.2005.04.039
31
Rajaram V. Nepolean T. Senthilvel S. Varshney R. K. Vadez V. Srivastava R. K. et al (2013). Pearl Millet [Pennisetum glaucum(L.) R. Br.] Consensus Linkage Map Constructed Using Four RIL Mapping Populations and Newly Developed EST-SSRs. BMC Genom.14, 159. 10.1186/1471-2164-14-159
32
Saleh A. S. M. Zhang Q. Chen J. Shen Q. (2013). Millet Grains: Nutritional Quality, Processing, and Potential Health Benefits. Compr. Rev. Food Sci. Food Saf.12, 281–295. 10.1111/1541-4337.12012
33
Satyavathi C. T. Ambawat S. Khandelwal V. Govindaraj M. Neeraja C. N. (2021). Micronutrient Rich Pearl Millet for Nutritionally Secure IndiaICAR-AII India Coordinated Research Project on Pearl Millet. Rajasthan, India: Jodhpur, 18.
34
Sehgal D. Rajaram V. Armstead I. P. Vadez V. Yadav Y. P. Hash C. T. et al (2012). Integration of Gene-Based Markers in a Pearl Millet Genetic Map for Identification of Candidate Genes Underlying Drought Tolerance Quantitative Trait Loci. BMC Plant Biol.12, 9. 10.1186/1471-2229-12-9
35
Senthilvel S. Jayashree B. Mahalakshmi V. Kumar P. S. Nakka S. Nepolean T. et al (2008). Development and Mapping of Simple Sequence Repeat Markers for Pearl Millet from Data Mining of Expressed Sequence Tags. BMC Plant Biol.8, 119. 10.1186/1471-2229-8-119
36
Singh R. B. Singh B. Singh R. K. (2019). Cross-taxon Transferability of Sugarcane Expressed Sequence Tags Derived Microsatellite (EST-SSR) Markers across the Related Cereal Grasses. J. Plant Biochem. Biotechnol.28, 176–188. 10.1007/s13562-019-00502-6
37
Singh R. K. Singh R. B. Singh S. P. Sharma M. L. (2011). Identification of Sugarcane Microsatellites Associated to Sugar Content in Sugarcane and Transferability to Other Cereal Genomes. Euphytica182, 335–354. 10.1007/s10681-011-0484-0
38
Srivastava R. K. Lachagari V. B. R. Vadez V. Kholova J. Lekkala S. P. Blumwald E. (2016). “Genotyping-by-Sequencing Using Double Digest Restriction Associated DNA (ddRAD) Approach for Fine Mapping LG2 Drought Tolerance QTL in Pearl Millet,” in Plant and Animal Genome Conference XXIV, San Diego, CA, January 08-13, 2016.
39
Srivastava R. K. Singh R. B. Pujarula V. L. Bollam S. Pusuluri M. Chellapilla T. S. et al (2020a). Genome-Wide Association Studies and Genomic Selection in Pearl Millet: Advances and Prospects. Front. Genet.10, 1389. 10.3389/fgene.2019.01389
40
Srivastava R. K. Vadez V. Kholova J. Lachagari V. B. R. Mahendrakar M. D. Katiyar P. et al (2017). “Fine Mapping of the Linkage Group 2 Drought Tolerance QTL in Pearl Millet [Pennisetum Glaucum (L.) R. Br.],” in InterDrought-V, Hyderabad (India), Feb 21-25, 025.
41
Stein A. J. Qaim M. Meenakshi J. V. Nestel P. Sachdev H. P. S. Bhutta Z. A. (2007). “Biofortification, an Agricultural Micronutrient Intervention: its Potential Impact and Cost-Effectiveness,” in Consequences and Control of Micronutrient Deficiencies: Science, Policy, and Programs, Istanbul, April 16–18.
42
Supriya A. Senthilvel S. Nepolean T. Eshwar K. Rajaram V. Shaw R. et al (2011). Development of a Molecular Linkage Map of Pearl Millet Integrating DArT and SSR Markers. Theor. Appl. Genet.123, 239–250. 10.1007/s00122-011-1580-1
43
Varshney R. K. Graner A. Sorrells M. E. (2005). Genic Microsatellite Markers in Plants: Features and Applications. Trends Biotechnol.23, 48–55. 10.1016/j.tibtech.2004.11.005
44
Varshney R. K. Shi C. Thudi M. Mariac C. Wallace J. Qi P. et al (2017). Pearl Millet Genome Sequence Provides a Resource to Improve Agronomic Traits in Arid Environments. Nat. Biotechnol.35, 969–976. 10.1038/nbt.3943
45
Vasconcelos M. Datta K. Oliva N. Khalekuzzaman M. Torrizo L. Krishnan S. et al (2003). Enhanced Iron and Zinc Accumulation in Transgenic rice with the Ferritin Gene. Plant Sci.164, 371–378. 10.1016/s0168-9452(02)00421-1
46
Vengadessan V. Rai K. N. Kannan Bapu J. R. Hash C. T. Bhattacharjee R. Senthilvel S. et al (2013). Construction of Genetic Linkage Map and QTL Analysis of Sink-Size Traits in Pearl Millet (Pennisetum Glaucum). Inter. Schol. Res. Not.10.5402/2013/471632
47
White P. J. Broadley M. R. (2009). Biofortification of Crops with Seven mineral Elements Often Lacking in Human Diets - Iron, Zinc, Copper, Calcium, Magnesium, Selenium and Iodine. New Phytol.182 (1), 49–84. 10.1111/j.1469-8137.2008.02738.x
48
Yadav O. P. Upadhyaya H. D. Reddy K. N. Jukanti A. K. Pandey S. Tyagi R. K. et al (2017). Genetic Resources of Pearl Millet: Status and Utilization. Indian J. Plant Genet Resour.30, 31–47. 10.5958/0976-1926.2017.00004.3
Summary
Keywords
micronutrient malnutrition, iron and zinc deficiency, Pearl millet [Pennisetum glaucum (L.) R. Br.], next-generation sequencing (NGS), forward genetics, reverse genetics
Citation
Srivastava RK, Satyavathi CT, Mahendrakar MD, Singh RB, Kumar S, Govindaraj M and Ghazi IA (2021) Addressing Iron and Zinc Micronutrient Malnutrition Through Nutrigenomics in Pearl Millet: Advances and Prospects. Front. Genet. 12:723472. doi: 10.3389/fgene.2021.723472
Received
10 June 2021
Accepted
23 September 2021
Published
18 November 2021
Volume
12 - 2021
Edited by
Christian Meyer, INRA UMR1318 Institut Jean Pierre Bourgin, France
Reviewed by
Muhammad Azhar Nadeem, Sivas University of Science and Technology, Turkey
Swati Puranik, Aberystwyth University, United Kingdom
Updates
Copyright
© 2021 Srivastava, Satyavathi, Mahendrakar, Singh, Kumar, Govindaraj and Ghazi.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rakesh K. Srivastava, r.k.srivastava@cgiar.org
This article was submitted to Plant Genomics, a section of the journal Frontiers in Genetics
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.