- 1Institute of Genetics and Physiology, Almaty, Kazakhstan
- 2The University of Winnipeg, Winnipeg, MB, Canada
- 3Al-Farabi Kazakh National University, Almaty, Kazakhstan
- 4Oskemen Bilim-Innovation Lyceum, Ust’-Kamenogorsk, Kazakhstan
Ethnogenesis of Kazakhs took place in Central Asia, a region of high genetic and cultural diversity. Even though archaeological and historical studies have shed some light on the formation of modern Kazakhs, the process of establishment of hierarchical socioeconomic structure in the Steppe remains contentious. In this study, we analyzed haplotype variation at 15 Y-chromosomal short-tandem-repeats obtained from 1171 individuals from 24 tribes representing the three socio-territorial subdivisions (Senior, Middle and Junior zhuz) in Kazakhstan to comprehensively characterize the patrilineal genetic architecture of the Kazakh Steppe. In total, 577 distinct haplotypes were identified belonging to one of 20 haplogroups; 16 predominant haplogroups were confirmed by SNP-genotyping. The haplogroup distribution was skewed towards C2-M217, present in all tribes at a global frequency of 51.9%. Despite signatures of spatial differences in haplotype frequencies, a Mantel test failed to detect a statistically significant correlation between genetic and geographic distance between individuals. An analysis of molecular variance found that ∼8.9% of the genetic variance among individuals was attributable to differences among zhuzes and ∼20% to differences among tribes within zhuzes. The STRUCTURE analysis of the 1164 individuals indicated the presence of 20 ancestral groups and a complex three-subclade organization of the C2-M217 haplogroup in Kazakhs, a result supported by the multidimensional scaling analysis. Additionally, while the majority of the haplotypes and tribes overlapped, a distinct cluster of the O2 haplogroup, mostly of the Naiman tribe, was observed. Thus, firstly, our analysis indicated that the majority of Kazakh tribes share deep heterogeneous patrilineal ancestries, while a smaller fraction of them are descendants of a founder paternal ancestor. Secondly, we observed a high frequency of the C2-M217 haplogroups along the southern border of Kazakhstan, broadly corresponding to both the path of the Mongolian invasion and the ancient Silk Road. Interestingly, we detected three subclades of the C2-M217 haplogroup that broadly exhibits zhuz-specific clustering. Further study of Kazakh haplotypes variation within a Central Asian context is required to untwist this complex process of ethnogenesis.
Introduction
Central Asia is a region populated by a wide range of ethnicities and characterized by heterogeneous economic and linguistic landscapes. Being located along the Silk Road, Central Asian populations have been genetically and culturally influenced by a millennia-long interplay between East and West that underpins its highly diverse genetic landscape. Genetic studies of Bronze Age (3100–1300 BC) remains from Central Asia show substantial temporal changes in the genetic composition of populations, indicating extensive migrations and west-to-east expansions of sedentary herders from the western steppe that formed a homogeneous gene pool by the end of the second millennium BCE (Allentoft et al., 2015; Narasimhan et al., 2019; Lalueza-Fox et al., 2004). In the Iron Age (1300–900 BC), nomadic pastoralists spread through the Eurasian steppe, dispersing the Scythian culture. Analysis of ancient DNA from Sakas and Sarmatians burials, belonging to the Scythian culture, demonstrates an increase of Iranian and eastern Eurasian genetic influx in southern and eastern samples, respectively (Gnecchi-Ruscone et al., 2021; Unterländer et al., 2017). During the first millennium CE, multiple confederations and empires were formed on the territory of modern Kazakhstan that were associated with substantial gene flow. For instance, the male-biased westward expansion of the Xiongnu nomads from the eastern steppe led to significant admixture of east Eurasian lineages into Central Sakas and displacement of the Indo-European Kangju and Wusun people (Damgaard et al., 2018). Subsequently, diverse Turkic nomadic states formed and blended into each other, resulting in gene flows between heterogeneous populations of the former Hunnic empire (Damgaard et al., 2018; Gnecchi-Ruscone et al., 2021). Following the Mongol invasion of the territory in 1211, the Golden Horde was established in the 13th century, that underwent series of fragmentation in the following centuries, resulting in the establishment of the Kazakh Khanate (1465–1847). During this time, nomadic tribes of different origins lived throughout the territory of present-day Kazakhstan, and eventually they were organized into three socio-territorial groups (zhuzes) based largely on geographical origin: Senior zhuz, Middle zhuz, and Junior zhuz (Figure 1) (Akishev et al., 1996).
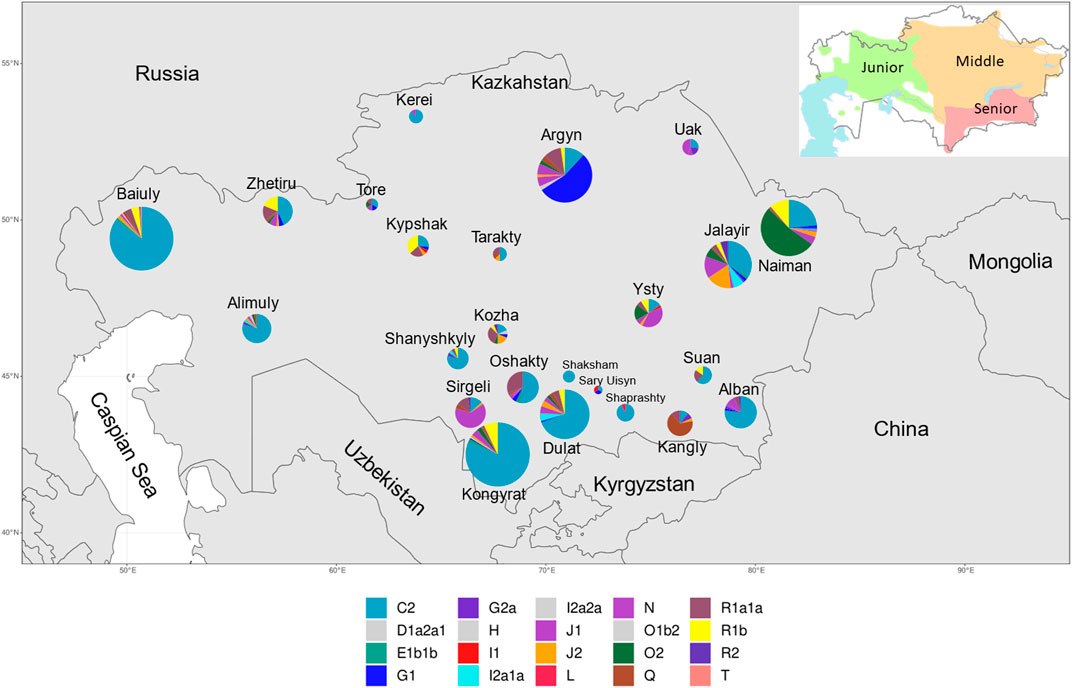
FIGURE 1. Frequency of Y-STR haplogroups by approximate geographic center of 22 Kazakh tribes, plus the Kozha and Tore in Kazakhstan. Size of pie chart is proportional to the total number of individuals sampled. The relative proportion of each haplogroup (see legend for colouration) is depicted in each pie chart. The smaller map in the top right corner shows approximate territories of Kazakh zhuzes in early 20th century. Reproduced from Wassily 2014.
The nomadic society of the Kazakh Steppe was organized based on a hierarchical patrilineal clan system of genealogical lineages. Individuals of the same genealogical lineage claim to share a common ancestor, and multiple genealogical lineages combine into clans that, collectively, form tribes. The 12 tribes of the Senior zhuz primarily occupied Southern and South-Eastern Kazakhstan, the seven tribes of the Middle zhuz reside in Eastern, Northern and Central Kazakhstan, while the three tribes in the Junior zhuz traditionally lived in Western Kazakhstan (Figure 1). Some of the steppe clans were not affiliated to the zhuzes, notably the clergy (Kozha and Sunak) and aristocracy (Tore). Representatives of the Kozha and Sunak clans link their ancestry to Islamic missionaries who originated from paternal-line relatives of the Prophet Muhammad. Tore people claim to be direct descendants of Genghis Khan. In contrast to sedentary farmer populations of Central Asia, Kazakhs have practiced exogamous marriages: a partner must be chosen from a different clan, and women integrate into the clan of their husband.
Despite the globalization of the last centuries and the move to a sedentary lifestyle, the tribal-clan structure of the Kazakh people has persisted, and many modern Kazakhs know the tribal affiliation and history of their clan. Being a patrilineal custom, the transgenerational transmission of tribal-clan affiliation resembles inheritance of the non-recombining part of Y chromosome, even though the former is a social entity. The analysis of genetic markers of the Y chromosome has been successfully employed in many studies of human populations to reconstruct migration routes; the combination of extended Y-haplotypes in patrilocal communities with genealogical data can enhance our understanding of the fine-scale demographic dynamics of a population (Wells et al., 2001).
To date, a multitude of studies has employed genetic markers to investigate the genetic diversity and differentiation of the Kazakh population in global (Wells et al., 2001; Underhill et al., 2010; Underhill et al., 2015; Unterländer et al., 2017), regional (Karafet et al., 2002; Lalueza-Fox et al., 2004; M.; Zhabagin et al., 2017) and local (Gokcumen et al., 2008; Balmukhanov et al., 2013; Tarlykov et al., 2013; Wen et al., 2020; M.; Zhabagin et al., 2021) contexts. The accumulated data provide preliminary insights into the demographic history of Kazakhs. For instance, Central Asian populations possess high levels of mtDNA and Y chromosomal haplotype diversity (Underhill et al., 1997; Comas et al., 2004), although paternal genetic markers are less polymorphic than maternal ones (Gokcumen et al., 2008; Tarlykov et al., 2013; Shan et al., 2014a). An earlier study also indicated potential discrepancies between the present-day geographic distribution of the Kazakh tribes and their ancestral relationships to neighboring populations (Tarlykov et al., 2013). Overall, however, prior studies have been hampered by one or more weaknesses such as small sample size, disregard for tribal affiliation, genealogical information, and/or insufficient geographical coverage. Here, we performed one of the largest study to date, N = 1,171 of Y-chromosomal haplotype diversity among all extant Kazakh tribes including the Tore and Kozha, with the primary goal of assessing the relationship of tribes and zhuz among the Kazakh people of modern-day Kazakhstan.
Materials and Methods
Samples and DNA extraction: A total of 1171 Kazakh males were included in this study. Blood or saliva was sampled from unrelated males from all-known Kazakh clans throughout five geographic regions in Kazakhstan (Supplementary Tables S1, S2). Individual and ethnological information, such as ethnicity and tribal-clan affiliation were self-declared and collected using an approved interview form from all individuals for which blood/saliva samples were obtained. Individuals with admixed ethnicity in their paternal lineage were removed from the study. Written informed consent was obtained from all participants to perform genetic analyses. The study was approved by the local ethics committee for biological research at the National Center for Biotechnology. Genomic DNA was extracted from all samples using the QIAamp DNA Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol and quantified spectrophotometrically (BioPhotometer Plus, Eppendorf, Germany) and fluorometrically (Qubit 2.0, Thermo Fisher Scientific, United States).
Y-STR genotyping: Samples were genotyped with the AmpFLSTR Y-filer PCR Amplification Kit (Thermo Fisher Scientific, United States) generating STR profiles at 17-loci (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a, DYS385b, DYS438, DYS439, DYS437, DYS448, DYS458, DYS456, DYS635, and Y-GATA-H4) (Supplementary Table S2). Fragments were separated and visualized on the ABI PRISM 310 Genetic Analyzer (Applied Biosystems, United States) and alleles called using GeneMapper IDX V1.4 (Applied Biosystems, United States). Due to difficulties in correctly assigning alleles to the duplicated DYS385a/b loci, these loci were removed and the haplotype of all individuals at 15 loci were used in further analyses. The raw data were submitted to the Y-Chromosome Haplotype Reference Database (YHRD) and is under the accession number YA004686.
Haplogroup prediction and Y-SNP genotyping: Y-haplogroups were predicted using the online Y-DNA Haplogroup Predictors NevGen (http://www.nevgen.org) as well as Whit-Athey’s (http://www.hprg.com). Putative Y-haplogroups for 16 of the 20 predicted haplogroups were then definitively determined through the analysis of 16 Y-SNPs, genotyped by RFLP or allele specific PCR using nine primers designed from previous studies and seven primer pairs designed for this study (Supplementary Table S3). In Y-STR haplotypes confirmed by Y-SNP analyses were confirmed for 1164 men (Supplementary Table S2). The nomenclature of Karafet et al., 2008, Underhill et al., 2010, and Myres et al., 2011 was used for SNP-analyses as they incorporate the latest information from the International Society of Genetic Genealogy (ISOGG) regarding Y-haplogroup confirmation (https://isogg.org/tree/index.html).
Data Analyses
Haplogroups: To visualize the clustering of Y-haplogroups in the region, the Y-haplogroups of all individuals were plotted with respect to their birthplace and sampling location (Supplementary Figures S1, S2, respectively). Haplogroup frequencies were calculated by direct counting and the frequency of Y-haplogroups by tribe calculated and plotted with respect to the approximate geographic center of the tribes.
Haplotypes: The frequency of alleles (repeat lengths) for each Y-STR marker was calculated by direct counting, and the genetic diversity (GD) of single-markers was calculated using Nei’s formula GD = n(1-Σpi2)/(n-1), where Pi is the relative frequency of the ith allele and n is the sample size (Nei, 1987). The number of unique haplotypes and their frequencies were obtained with the help of the R package Pegas (Paradis, 2010). Haplotype diversity was calculated in an analogous way to GD except by replacing the allele frequencies (Pi) by the relative frequencies of each haplotype. Haplotype discrimination capacity (DC) was calculated as the ratio of unique haplotypes in the sample (i.e. (n haplotypes)/N) ∗100).
Genetic distance and differentiation among tribes: The pairwise genetic distances between individuals in each of the 24 tribes was estimated using Weir and Cockerham pairwise FST for haploid data (Weir and Cockerham 1984). An analysis of molecular variance was performed to assess the components of molecular variance as explained by both tribe (model: ∼tribe), and tribe nested in zhuz (∼zhuz/tribe). The significance of the covariance components was assessed using a permutation test following Excoffier (Excoffier et al., 1992). A Mantel test was performed to test for a linear relationship between the Euclidean geographic distance between all pairs of individuals’ birthplace and the corresponding genetic distance of their Y-haplotypes estimated by Edward’s distance, and the significance of the regression was tested by a randomization procedure as implemented in the R-package Ade4 (v 1.7–15).
Structure and Principal Coordinate Analysis: An analysis of the inferred number of ancestral Y-STR groups, K was performed using the program Structure 2.3.4 (Pritchard et al., 2000). Given the nearly complete linkage and haploid nature of the Y-STR, we assessed the haplotype structure within haplogroups and tribes using a “no admixture” model, which assumes that each individual originate from one of the K ancestral groups. The structure analyses was performed on 1163 individuals (all of those that were SNP-genotyped and excluding haplogroup T present in a single individual) using a person’s tribal affiliation and haplogroup as priors. To assess the best estimate of K, the number of ancestral Y-STR groups in the total population, simulations were carried out for K = 1 to 25 with three replicates for each value of K. To select the best estimate of K, the second-order rate of change of the likelihood function with respect to K was estimated, using the program Structure Harvester (Earl and vonHoldt 2012), and the value of K with the highest likelihood selected. We also examined the inferred posterior probability that individual i is from the kth group assuming a prior probability of 1/K. To further explore the relationship among tribes, a principal coordinate analysis (PcoA aka, multidimensional scaling, MDS) was performed by using the pairwise genetic distance between individuals based on Meirmans PT, as above, performing the PcoA and then extracting the first two dimensions for visualization with the help of the R package Ape (Paradis et al., 2004). The individual coordinates of all 1171 individuals were plotted with respect to 1) Y-haplogroups and 2) the tribal affiliations of individuals.
Results
Y-chromosome haplogroups in Kazakh population: 1171 Kazakh males were included in the study: 433 total individuals representing the 12 tribes in the Senior zhuz, 475 from seven tribes in the Middle zhuz, 241 from the Junior zhuz, and 22 samples from the Kozha (n = 16) and Tore (n = 6) tribes (Supplementary Table S1). Although the analysis performed here is based on the highest hierarchical affiliation of a person to their tribe/zhuz, all but 142 individuals reported their family lineage/clan and this data could be used in future analyses (given in Supplementary Table S2). All individuals included in the study were living in Kazakhstan at the time of the study, 61 were born outside of Kazakhstan (Supplementary Table S2, Supplementary Figure S1 for map of birth locations and Supplementary Figure S2 for sampling locations). Gene diversity of the single locus markers for each of the 15 Y-STR’s ranged from 0.3305 (DYS438) to 0.7699 (DYS635) (Supplementary Table S4). The locus with the highest diversity, DYS635, harbored eight allelic classes, while the least diverse loci DYS393, DYS391 and DYS437 each had five alleles, with 129 alleles scored at all 15 loci (Supplementary Table S4). Haplotypes were submitted to two online Y-DNA haplogroup predictors (NevGen and Whit-Athey’s) to assign a tentative haplogroup to each individual (given in Supplementary Table S2). Sixteen of the 20 predicted haplogroups were confirmed by SNP genotyping using primers obtained from the literature or developed for this study (Supplementary Table S3). Of the 1171 males for which a Y-haplotype was obtained, 1164 were assigned to one of 16 Y-haplogroups based on the combined Y-STR - SNP data, while seven individuals were assigned to one of four additional Y-haplogroups (D1a2a1, H, I2a2a, O1b2) based on the NevGen prediction alone (Supplementary Table S2). These seven individuals were included in some analyses (those based purely on haplotypes for which only the allele sizes/locus are used) but not all analyses; these four haplogroups require Y-SNP genotyping to be confirmed.
The most frequent haplogroup in the Kazakh population was C2-M217 (51.9% - 608 men). Haplogroup C2-M217 was present in all examined tribes and its frequency ranged from 11% in Kangly and Argyn to 100% in Shaksham (n = 6) (Figure 1 and Supplementary Table S5). Another important component of the Kazakh gene pool is represented by the haplogroup R (12.8%), which has three subclades: R1a1a-M17 (6.5%, 76 individuals), R1b-M343 (5.6%, 65 individuals) and R2-M479 (0.8%, nine individuals). The R1a1a-M17 haplogroup was observed in 18 of the 24 tribes, and was most frequent in the Kozha clan (31.3%, five individuals) and Oshakty tribe (31%, 13 individuals). Subclade R1b-M343 was found in 12 Kazakh tribes and had the highest frequency (36.8%, seven individuals) in the Kypshak tribe. Lastly, the subclade R2-M479 was observed in five tribes at low frequencies, and was most prevalent in the Kozha clan (6.3%, one person) and Jalayir tribe (5.4%, five individuals) (Figure 1 and Supplementary Table S5).
Haplogroups O (represented by O2-M122 and O1b2, the latter predicted by NevGen), G (G1-M285 and G2a-P15), N-M231, J (J1-M267 and J2-M172), and Q-M242 were observed at frequencies <10% across tribes, but were found at higher frequencies in one or a few tribes. For example, haplogroup O2-M122, had a frequency of 8.03% in the sampled population, but was found in 52.3% of the individuals from the Naiman tribe. Haplogroup G had a global frequency of 7.9%, with the majority (7.1%) belonging to subclade G1-M285 and 0.8% to subclade G2a-P15, but the frequency varied between tribes, and ∼54% of the males from the Argyn tribe (n = 126) harbored haplogroup G1-M285. The highest frequency of the haplogroup G2a-P15 was observed in the Uak tribe (18.2%, 11 individuals). The N-M231 haplogroup had a global frequency of ∼6.9%, but was prevalent in the Sirgeli (64.1% of 39 individuals) and Uak (45.5% of 11 individuals) tribes, Tore tribe (16.7% from six individuals) and Jalayir tribe (15.1% from 93 individuals). Haplogroup J, represented by J1-M267 and J2-M172, had a global frequency of ∼6.2%, being frequent in the Ysty tribe (J1-M267 (39.4%, 13 individuals) and J2-M172 (3%, one person)). Additionally, J2-M172 was also observed in the Kozha clan (18.8%, three individuals). Lastly, haplogroup Q had a low overall frequency of ∼3.1%, but was highly represented in the Kangly tribe (66.7%, 27 individuals), while its frequency was <5.5% in all other tribes (Supplementary Table S5). The other haplogroups show frequencies in the sampled population lower than 2% (Supplementary Figure S3).
The assigned Y-haplogroup of each individual was plotted with respect to the location of their birth and sampling location (Supplementary Figures S1, S2) and the frequency of the haplogroups by tribe was plotted with respect to the approximate geographic center of the territory occupied by a tribe in the past (Figure 1). Overlaying the Y-haplogroup assignments on the map of Kazakhstan with the approximate route of the Mongolian invaders who rampaged through Central Asia in the 13th century (Zerjal et al., 2003), shows the historical mark of this invasion since haplogroup C2 is more frequent in the southern and western portions of the country where the Mongols passed (Supplementary Figure S1). For example, tribes in the southern and western regions of Kazakhstan, such as the Alban, Kongyrat, Dulat, Baiuly and Alimuly, have frequencies of C2-M217 ≥ 70%, while tribes located in the center and northeast of the country, such as the Argyn, Uak, Naiman, Kangly, Ysty and Kypshak, have lower frequencies (<30% in most cases) (see Figure 1 and Supplementary Table S5).
Haplotype Diversity: From the 1171 individuals, 577 distinct haplotypes were found, of which 429 were observed once and the remaining 148 were observed between 2 and 51 times and 15 haplotypes observed ≥10 times (Supplementary Table S6). Overall, this resulted in a Y chromosome haplotype diversity of 0.9938 ± 0.0006, reflecting the deep paternal lineages of different origins in the sample. However, the discriminatory capacity of the samples based on the haplotype frequency distribution was 55.17%, reflecting the high frequency of a few haplotypes. Seven of the top nine most frequent haplotypes belonged to the C2 haplogroup (Supplementary Table S6). The two most common haplotypes (Ht1, Ht2, Supplementary Table S6) belonged to the C2 haplogroup and were both observed in 51 individuals while a third C2 haplotype (Ht3) was observed in 37 individuals. Ht1 was present in 38 individuals from the Baiuly tribe and six individuals from the Alimuly tribe (Supplementary Table S7) both of which belong to the Junior zhuz, located in western Kazakhstan (Figure 1). Ht2 was identified in 32 individuals in the Dulat tribe and 10 individuals in Alban, both belonging to the Senior zhuz in the southeastern region (Figure 1 and Supplementary Table S7). Finally, Ht3 was observed in 37 individuals from the Kongyrat, a member of the Middle zhuz (Figure 1 and Supplementary Table S7). The most frequent non-C2 haplotype (Ht4) belonged to haplogroup G1 and was found in 34 individuals, 26 of which were in the Argyn tribe belonging to the Middle zhuz, located in the center-north of the country (Figure 1 and Supplementary Table S7).
Population Structure: The analysis of molecular variance revealed that between ∼73% (Model: ∼tribe) and ∼71% (Model: ∼zhuz/tribe) of the variation in Y-STR diversity is attributable to variation within tribes. Variation in Y-STR diversity among zhuz, explained ∼9% of the variation in YSTR-diversity, a value that was lower than expected if the diversity was distributed randomly among the highest hierarchical level using a Monte Carlo permutation test, while the amount of variation within tribes was greater than expected (Table 1 and Supplementary Figure S4). Estimates of the fixation index ΦST, were significant and between 0.273 (model ∼tribe) and 0.2923 (model ∼ zhus/tribe), such that between 27.3 and 29.3% of the total variation is due to inter-tribal differentiation (Table 1). Pairwise Weir and Cockerham’s FST values between tribes varied between ∼0 (−0.0003, Baiuly vs Alimuly) and 0.19 (Sirgeli vs Shaksham) (Supplementary Table S8).
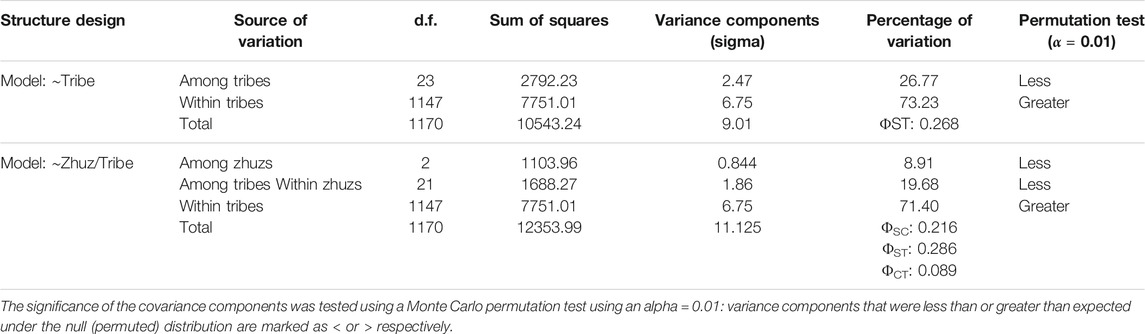
TABLE 1. Analysis of Molecular Variance (AMOVA) among Y-STR haplotypes of 1171 men in Kazakhstan using two models 1) tribal affiliations (model: ∼tribe), and 2) tribal affiliation nested in Zhus (model: ∼zhus/tribe).
Mantel test: Despite the apparent higher frequency of the C2-M217 haplogroup along the southern and western borders of Kazakhstan, tracking the Mongolian invasion (Supplementary Figure S1), there was not a significant correlation between the genetic and geographic distance among individuals based on a linear regression between all individuals pairwise. The regression line explained only 0.12% of the variation in the data and the permutation test failed to reject the hypothesis of no spatial structure (Supplementary Figure S5).
Structure analyses: Further analysis of the haplotype structure using the program Structure implementing a “no admixture” model and using both the tribe and haplogroup as priors, found that the best estimate of K was 20 (Supplementary Table S9). Visualization of the structure of the Y-STR’s by tribe (Figure 2A) and haplogroup (Figure 2B), confirms the presence of at least three subgroups within C2-M217: lime green (Senior zhuz), teal blue (Middle zhuz) and pink (Junior zhuz). Similarly, it identified the presence of some homogenous haplogroups (e.g. G1, O2 and R1a1a) that are strongly associated with some tribes (e.g. G1- Argyn, O2 – Naiman).
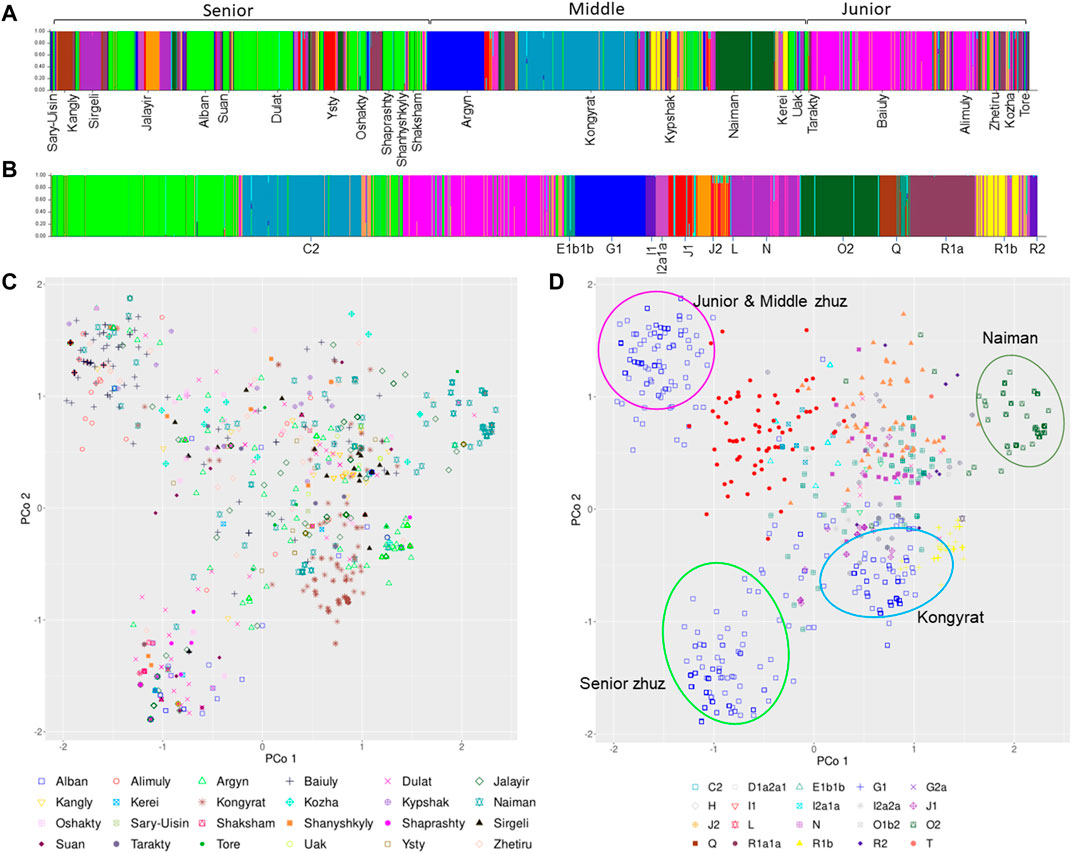
FIGURE 2. Posterior Probability that an individual is assigned to a specific tribe (A) or haplogroup (B) based on a no admixture model in the program Structure. Principal Coordinate Analysis (also known as Multidimensional Scaling Plot) showing the relationship among 1171 Kazakh men genotyped at 17 Y-STR’s by tribe (C) or and haplogroup (D).
MultiDimensional Scaling: The principal coordinate analysis (PCoA) provided a closer examination of the relationship among A) tribes and B) haplogroups. This revealed that while many of the haplogroups fall in the middle of the coordinate with somewhat distinct clustering (Figure 2D), the haplogroups are diffusely distributed among tribes (Figure 2C). On the other hand, there are three distinct clusters of haplogroup C2-M217 that fall in the upper and lower left or middle bottom (Figure 2B), which broadly correspond to the three subgroups found in the Structure analyses. Haplogroup C2-M217 cluster 1 is found within numerous tribes in the Junior and Middle zhuz, notably the Baiuly, Argyn, Alimuly and Kypshak and corresponds to the pink haplotypes in the Structure analyses (Figures 2B,D). On the other hand, the second C2-M217 haplogroup is found among diverse members of the Senior zhuz, including the Alban, Shaprashty, Oshakty, Dulat and Suan, all found in south-east Kazakhstan and corresponds to the lime-green Structure haplotypes (Figures 2B,D). Lastly, the third cluster of C2 haplogroups is found predominantly among members of the Kongyrat tribe corresponding to the teal blue Structure haplotype (Figures 2B,D). Other distinct clusters in the MDS analyses pertain to haplotype O2 (upper right, Figure 2D), which is most frequent in the Naiman tribe, but is also found in the Jalayir tribe (Figure 2C) and for haplotype G1 (brown crosses middle right, Figure 2D), which is found in many tribes, particularly the Argyn (Figure 2C). The remaining haplotypes have overlapping ranges on the pCoA plot (Figures 2A,B). This indicates that there are further subclade differences among individuals within the C2-M217 haplogroup that require further subtyping. Thus, by combining the results of the pCoA and Structure analyses indicate that are broad differences among the haplogroups among the three zhuzes, in particular differences in the C2-M217, haplogroup among zhuz, as well as large differences in the frequency of certain Y-haplogroups among tribes.
Discussion
In this study, we present the most comprehensive study of Y-STR diversity in Kazakhstan, with 1171 samples representing all of the extant tribes living within the territory of Kazakhstan. Haplotype diversity of Y-STR in Kazakhs reached a value of 0.9929, reflecting the deep paternal lineages of different origins in the sample. Our results agree with another recent study from Kazakhstan (0.9936) (Zhabagin et al., 2019), while the haplotype diversity of Kazakhs from Xinjiang (Shan et al., 2014a; Shan et al., 2014b) was found to be lower, possibly due to a founder effect of the Kazakhs that migrated to China. Most of the gene diversity (GD) estimates derived from the Y-profiler Y-STRs were consistent between Kazakhs from Kazakhstan and China (Shan et al., 2014a; Shan et al., 2014b), however, GD of DYS448 was two-fold higher in Xinjiang Kazakhs. Interestingly, the most frequent haplotype in Kazakhs from China is also one the most common haplotypes among Kazakhs from Kazakhstan. The Ht8 haplotype is associated with O2 haplogroup that accounted for 52.2% of individuals from the Naiman tribe, historically situated in Eastern Kazakhstan. Moreover, Kazakh populations in the Altai Region, Russia, are also characterized by a significant fraction of O2 individuals (31–40%) (Dulik et al., 2011; Kharkov 2012). Surprisingly, despite the high frequency of O2 haplotype in a Kazakh population studied by Shan et al. (Shan et al., 2014a; Shan et al., 2014b), no individuals with the O2 haplogroup were found in Kazakhs from Northwest China (Shou et al., 2010).
In contrast to high haplotype diversity, the overall discrimination capacity of the 15 Y-STR loci was only 0.5517. This suggests that despite including only unrelated males in the study, individuals from the same or different tribes may have identical haplotypes presumably reflecting the deep patrilineal descent among Kazakhs. For example, the discrimination capacity in Chinese Han from Shanxi Province, Northern China was 0.9865, which indicates a high potential for differentiating between male individuals in this population (Bai et al., 2013). But in Kazakh populations from Xinjiang, Northwest China the discrimination capacity was 0.5950 (Shan et al., 2014a). The discrimination capacity in our Kazakh sample is also lower compared to some data from European and Asian populations for the same set of 15 Y-STR loci (Turrina et al., 2006; Roewer et al., 2007; Robino et al., 2008; Lacau et al., 2011; Liu et al., 2020). It should be borne in mind that diversity indices vary depending on the Y-STR genotyping systems used. As the number of marker sets increases, diversity indices also increase (Purps et al., 2014; Khubrani et al., 2018; Zhabagin et al., 2019; Liu et al., 2020).
The results of the AMOVA and Mantel tests in our study confirmed that there is significantly less genetic variation among zhuzes than expected under a hierarchical model of genetic structure. This suggests that the zhuz structure is not the primary influence on genetic relationships among Kazakh tribes, and that the division into zhuzes was conditional rather than socio-territorial as suggested by other authors (Ashirbekov et al., 2018; Zhabagin et al., 2018). Nevertheless, approximately, 10% of the genetic variation among individuals was accounted for by variation among zhuzes. Furthermore, the MDS analyses indicated some differences in haplogroup structure among tribes/zhuzes, particularly for the C2-M217 haplogroup (see below). In the AMOVA analyses, partitioning the genetic variance within and among tribes (model: ∼tribe), revealed that ∼27% of the genetic variance is found between tribes and ∼73% within tribes. This is similar to a recent study by Ashirbekov et al. (2017) who surveyed Y-STR polymorphism, including more detailed analyses of haplogroup subclades based on SNP polymorphism, for 1269 Kazakh men sampled from 10 tribes in Southern Kazakhstan (Ashirbekov et al., 2017). Overall, they find ∼22% of the genetic variance between tribes and 78% within tribes. We did not find evidence of a relationship between genetic and geographic (birthplace) distances of individuals, despite the apparent higher frequency of the C-M217 haplogroup along the southern and western borders of Kazakhstan. We suggest that a more sophisticated spatial analysis is required to show that the C haplogroup exhibits a higher frequency in the southern part of the country.
The MDS and Structure analysis showed that there is considerable diversity in some haplogroups. Particularly interesting is the haplogroup C-M217. It is the most common haplogroup in modern Kazakhs, but the analysis shown here reveal that there at least three distinct sub-clusters of this haplogroup. Оne of the C2-M217 subgroups is dominant in tribes of the Junior zhuz (mostly in tribes Baiuly, Alimuly), one in the middle (Kongyrat tribe) and one in the Senior zhuz (in almost all tribes). We assume that these clusters represent different daughter branches of the C-M217 haplogroup. A Y-STR study by Ashirbekov et al. sampled 564 individuals (Ashirbekov et al., 2018) from ten tribes in the Senior zhuz, five tribes in the Middle zhuz and three tribes in Junior zhuz, and identified three daughter branches of the haplogroup C-M217: C-M401, C-M86 and C-M407. The authors note the predominance of the С-М401 subgroup in the tribes of the Senior zhuz, the С-М86 subgroup in the Junior zhuz tribes, and the С-М407 subgroup in the tribe of the Middle zhuz - the Kongyrat. This result is consistent with our observation. Further in-depth analysis of a number of single nucleotide markers of the C-M217 haplogroup will make it possible to determine which subgroups of this haplogroup are precisely present in the population of modern Kazakhs.
In conclusion, although several papers have described genetic polymorphism at Y-STR’s among Kazakh tribes, this is the largest study, published in English, and represents individuals from tribes in all three zhuzes as well as individuals from the Kozha and Tore tribes. Overall, we find evidence of genetic differentiation between zhuz (∼10%) and between tribes within zhuz (∼20%) suggesting that there are differences in haplogroup structure among Kazakh tribes. Although we did not find evidence of a linear relationship between genetic and geographic distance among paired individuals (i.e. non-significant Mantel test), we observed the imprint of higher frequencies of C2 haplogroups along the southern and western border of Kazakhstan, which corresponds to both the path of the Mongolian invasion and the approximate route of the ancient Silk Road. A broader spatial and temporal analysis of the Y-STR diversity among Kazakh tribes within the context of other groups in Central Asia is needed to further elucidate this dynamic history Abilev et al., 2012, Akerov, 2016, Artykbaev, 2020, Balaganskaya et al., 2011, Balanovsky et al., 2015, Barinova, 2016, Beisenov et al., 2015, Cai et al., 2011, Damba et al., 2018, Derenko et al., 2007, Ding et al., 2020, Herrera and Garcia-Bertrand, 2018, Hollard et al., 2014, Ilumä e et al., 2016, Ismagulov, 1970, Ismagulov, 1982, Jeong et al., 2019, Keyser et al., 2009, Kozhanuly, 2018, Malyarchuk et al., 2010, Meirmans, 2006, Roewer et al., 2013, Sabitov, 2013, Shi et al., 2005, Shi et al., 2013, Wei et al., 2018, Huang et al., 2018, Zegura et al., 2004, Zhabagin et al., 2016, Zhabagin et al., 2014, Zhabagin et al., 2020, Zhong et al., 2010.
Data Availability Statement
The raw Y-STR data were submitted to the Y-Chromosome Haplotype Reference Database (YHRD) under the accession number YA004686. Other datasets for this study can be found in the Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Local ethics committee for biological research at the National Center for Biotechnology, Nur-Sultan, Kazakhstan. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceived and designed the experiments: EKh, SG, and LD Performed the experiments: EKh, IK, OI, BB, LS, AG, EKh, GZ, LM, NK, AA, MB, KB, AP, GA, AF, and AS Analyzed the data: EK, ZZ, IK, and SG Contributed reagents/materials/analysis tools: BB and AA Wrote the paper: EKh, SG, and IK.
Funding
This research was funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (Programs No. OR11465435 and BR05233709).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the donors for their contributions to this work and all those who helped with sample collection. We are grateful to Oraz Sapargali, Saltanat Abdikerim and other our colleagues for laboratory assistance. We are grateful to Nursaule Rsalieva for verification the correct spelling of the names of the Kazakh tribes. We also thank Talgat Yechshzhanov and Sergey Yegorov for assistance in scientific collaboration.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.801295/full#supplementary-material
References
Abilev, S., Malyarchuk, B., Derenko, M., Wozniak, M., Grzybowski, T., and Zakharov, I. (2012). The Y-Chromosome C3* Star-Cluster Attributed to Genghis Khan's Descendants Is Present at High Frequency in the Kerey Clan from Kazakhstan. Hum. Biol. 84 (1), 79–89. doi:10.3378/027.084.0106
Akerov, T. A. (2016). On the Origin of the Naiman. J. Sib. Fed. Univ. Humanit. Soc. Sci. 9 (9), 2071–2081. doi:10.17516/1997-1370-2016-9-9-2071-2081
Akishev, K., Baipakov, K., and Kumekov, B. (1996). History of Kazakhstan in 4 Volumes, 1. Almaty: Atamura. Available at: https://www.twirpx.com/file/988541/.
Allentoft, M. E., Sikora, M., Sjögren, K.-G., Rasmussen, S., Rasmussen, M., Stenderup, J., et al. (2015). Population Genomics of Bronze Age Eurasia. Nature 522 (7555), 167–172. doi:10.1038/nature14507
Artykbaev, Z. (2020). O Kazahskom Plemeni Sirgeli: Problemy Proishozhdenija [Kazakh Tribe Sirgeli: Problems of Origin]. North-Eastern humanitarian Bull. 2, 34–39. doi:10.25693/SVGV.2020.31.2.004
Ashirbekov, Y., Botbaev, D., Belkozhaev, A., Abaildayev, A., and Aitkhozhina, N. (2017). Raspredelenie Gaplogrupp Y-Hromosomy Kazahov Juzhno-Kazahstanskoj, Zhambylskoj I Almatinskoj Oblasti [Distribution of Y-Chromosome Haplogroups of Kazakhs in South Kazakhstan, Zhambyl and Almaty Regions]. Almaty: Reports of National Academy of Sciences of the Republic of Kazakhstan, 6, 25–30.
Ashirbekov, Y. Y., Khrunin, A. V., Botbayev, D. M., Belkozhaev, A. M., Abaildayev, A. O., Rakhimgozhin, M. B., et al. (2018). Molecular Genetic Analysis of Population Structure of the Great Zhuz Kazakh Tribal Union Based on Y-Chromosome Polymorphism. Mol. Genet. Microbiol. Virol. 33 (2), 91–96. doi:10.3103/S0891416818020040
Bai, R., Zhang, Z., Liang, Q., Lu, D., Yuan, L., Yang, X., et al. (2013). Haplotype Diversity of 17 Y-STR Loci in a Chinese Han Population Sample from Shanxi Province, Northern China. Forensic Sci. Int. Genet. 7 (1), 214–216. doi:10.1016/j.fsigen.2012.10.004
Balaganskaya, O., Lavryashina, M., Kuznetchova, M. A., Romanov, A. G., Dibirova, Kh. D., Frolova, S. A., et al. (2011). Gene Pool of the Altay Ethnic Groups (From Russia, Kazakhstan, and Mongolia) Analyzed by the Y Chromosomal Markers. Moscow: Moscow University Anthropology Bulletin (Vestnik Moskovskogo Universiteta. Seria XXIII. Antropologia), 2, 25–36.
Balanovsky, O., Zhabagin, M., Agdzhoyan, A., Chukhryaeva, M., Zaporozhchenko, V., Utevska, O., et al. (2015). Deep Phylogenetic Analysis of Haplogroup G1 Provides Estimates of SNP and STR Mutation Rates on the Human Y-Chromosome and Reveals Migrations of Iranic Speakers. PLoS ONE 10 (4), e0122968. doi:10.1371/journal.pone.0122968
Balmukhanov, T., Bekseitov, E., Akhmetollaev, I., Khanseitova, A., Botbaev, D., Belkozhaev, A., et al. (2013). Investigation of Y-Chromosome Microsatellite STR Loci in Kazakh Population. Proc. Natl. Acad. Sci. Republic Kazakhstan 4, 91–95.
Barinova, E. B. (2016). Some Aspects of Forming the Population of East and Central Asia in Ancient Times. RUDN J. World Hist. 3 (December), 40–51.
Beisenov, A. Z., Ismagulova, A. O., Kitov, E. P., and Kitova, A. O. (2015). Naselenie Central’nogo Kazahstana V I Tys. Do n.Je. Almaty [The Population of Central Kazakhstan in the 1st Millennium BC].
Cai, X., Qin, Z., Wen, B., Xu, S., Wang, Y., Lu, Y., et al. (2011). Human Migration Through Bottlenecks from Southeast Asia into East Asia During Last Glacial Maximum Revealed by Y Chromosomes. PLoS ONE 6 (8), e24282. doi:10.1371/journal.pone.0024282
Comas, D., Plaza, S., Wells, R. S., Yuldaseva, N., Lao, O., Calafell, F., et al. (2004). Admixture, Migrations, and Dispersals in Central Asia: Evidence from Maternal DNA Lineages. Eur. J. Hum. Genet. 12 (6), 495–504. doi:10.1038/sj.ejhg.5201160
Damba, L. D., Balanovskaya, Е. V., Zhabagin, M. K., Yusupov, Y. М., Bogunov, Y. V., Sabitov, Z. M., et al. (2018). Estimating the Impact of the Mongol Expansion Upon the Gene Pool of Tuvans. Vestn. Vogis 22 (August), 611–619. doi:10.18699/VJ18.402
Damgaard, P. d. B., Marchi, N., Rasmussen, S., Peyrot, M., Renaud, G., Korneliussen, T., et al. (2018). 137 Ancient Human Genomes from Across the Eurasian Steppes. Nature 557 (7705), 369–374. doi:10.1038/s41586-018-0094-2
Derenko, M. V., Malyarchuk, B. A., Wozniak, M., Denisova, G. A., Dambueva, I. K., Dorzhu, C. M., et al. (2007). Distribution of the Male Lineages of Genghis Khan's Descendants in Northern Eurasian Populations. Russ. J. Genet. 43 (3), 334–337. doi:10.1134/S1022795407030179
Ding, J., Fan, H., Zhou, Y., Wang, Z., Wang, X., Song, X., et al. (2020). Genetic Polymorphisms and Phylogenetic Analyses of the Ü-Tsang Tibetan from Lhasa Based on 30 Slowly and Moderately Mutated Y-STR Loci. Forensic Sci. Res. 0 (0), 1–8. doi:10.1080/20961790.2020.1810882
Dulik, M. C., Osipova, L. P., and Schurr, T. G. (2011). Y-chromosome Variation in Altaian Kazakhs Reveals a Common Paternal Gene Pool for Kazakhs and the Influence of Mongolian Expansions. PLoS ONE 6 (3), e17548. doi:10.1371/journal.pone.0017548
Earl, D. A., and vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conservation Genet. Resour. 4 (2), 359–361. doi:10.1007/s12686-011-9548-7
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of Molecular Variance Inferred from Metric Distances Among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 131 (2), 479–491. doi:10.1093/genetics/131.2.479
Gnecchi-Ruscone, G. A., Khussainova, E., Kahbatkyzy, N., Musralina, L., Spyrou, M. A., Bianco, R. A., et al. (2021). Ancient Genomic Time Transect from the Central Asian Steppe Unravels the History of the Scythians. Sci. Adv. 7 (13), eabe4414. doi:10.1126/sciadv.abe4414
Gokcumen, O., Dulik, M. C., Pai, A. A., Zhadanov, S. I., Rubinstein, S., Osipova, L. P., et al. (2008). Genetic Variation in the Enigmatic Altaian Kazakhs of South-Central Russia: Insights into Turkic Population History. Am. J. Phys. Anthropol. 136 (3), 278–293. doi:10.1002/ajpa.20802
Herrera, R. J., and Garcia-Bertrand, R. (2018). Ancestral DNA, Human Origins, and Migrations. Elsevier Science.
Hollard, C., Keyser, C., Giscard, P.-H., Tsagaan, T., Bayarkhuu, N., Bemmann, J., et al. (2014). Strong Genetic Admixture in the Altai at the Middle Bronze Age Revealed by Uniparental and Ancestry Informative Markers. Forensic Sci. Int. Genet. 12 (September), 199–207. doi:10.1016/j.fsigen.2014.05.012
Huang, Y.-Z., Pamjav, H., Flegontov, P., Stenzl, V., Wen, S.-Q., Tong, X.-Z., et al. (2018). Dispersals of the Siberian Y-Chromosome Haplogroup Q in Eurasia. Mol. Genet. Genomics 293 (1), 107–117. doi:10.1007/s00438-017-1363-8
Ilumäe, A.-M., Reidla, M., Chukhryaeva, M., Järve, M., Post, H., Karmin, M., et al. (2016). Human Y Chromosome Haplogroup N: A Non-trivial Time-Resolved Phylogeography that Cuts Across Language Families. Am. J. Hum. Genet. 99 (1), 163–173. doi:10.1016/j.ajhg.2016.05.025
Ismagulov, O. (1982). Jetnicheskaja Antropologija Kazahstana: Somatologicheskoe Issledovanie [Ethnic Anthropology of Kazakhstan: Somatological Study]. Nauka.
Ismagulov, O. (1970). Naselenie Kazahstana Ot Jepohi Bronzy Do Sovremennosti: (Paleoantropologicheskoe Issledovanie) [ Population of Kazakhstan from the Bronze Age to the Present: (Paleoanthropological Study)]. Nauka.
Jeong, C., Balanovsky, O., Lukianova, E., Kahbatkyzy, N., Flegontov, P., Zaporozhchenko, V., et al. (2019). The Genetic History of Admixture Across Inner Eurasia. Nat. Ecol. Evol. 3 (6), 966–976. doi:10.1038/s41559-019-0878-2
Karafet, T. M., Mendez, F. L., Meilerman, M. B., Underhill, P. A., Zegura, S. L., and Hammer, M. F. (2008). New Binary Polymorphisms Reshape and Increase Resolution of the Human Y Chromosomal Haplogroup Tree. Genome Res. 18 (5), 830–838. doi:10.1101/gr.7172008
Karafet, T. M., Osipova, L. P., Gubina, M. A., Posukh, O. L., Zegura, S. L., and Hammer, M. F. (2002). High Levels of Y-Chromosome Differentiation Among Native Siberian Populations and the Genetic Signature of a Boreal Hunter-Gatherer Way of Life. Hum. Biol. 74 (6), 761–789. doi:10.1353/hub.2003.0006
Keyser, C., Bouakaze, C., Crubézy, E., Nikolaev, V. G., Montagnon, D., Reis, T., et al. (2009). Ancient DNA Provides New Insights into the History of South Siberian Kurgan People. Hum. Genet. 126 (3), 395–410. doi:10.1007/s00439-009-0683-0
Kharkov, V. N. (2012). Structure and Phylogeography of Gene Pools of Aboriginal Peoples of Siberia Based on Y-Chromosomal Markers. Tomsk: Dr Sci Biol thesis (Research Institute of Medical Genetics).
Khubrani, Y. M., Wetton, J. H., and Jobling, M. A. (2018). Extensive Geographical and Social Structure in the Paternal Lineages of Saudi Arabia Revealed by Analysis of 27 Y-STRs. Forensic Sci. Int. Genet. 33 (March), 98–105. doi:10.1016/j.fsigen.2017.11.015
Kozhanuly, M. N. (2018). Nekotorye Drevnie Jetnotoponimy Mangistauskogo Regiona [Some Ancient Ethnotonyms of the Mangystau Region]. Int. Res. J. 6 (72), 106–109. doi:10.23670/IRJ.2018.72.6.044
Lacau, H., Bukhari, A., Gayden, T., La Salvia, J., Regueiro, M., Stojkovic, O., et al. (2011). Y-STR Profiling in Two Afghanistan Populations. Leg. Med. 13 (2), 103–108. doi:10.1016/j.legalmed.2010.11.004
Lalueza-Fox, C., Sampietro, M. L., Gilbert, M. T. P., Castri, L., Facchini, F., Pettener, D., et al. (2004). Unravelling Migrations in the Steppe: Mitochondrial DNA Sequences from Ancient Central Asians. Proc. R. Soc. Lond. B 271 (1542), 941–947. doi:10.1098/rspb.2004.2698
Liu, J., Wang, R., Shi, J., Cheng, X., Hao, T., Guo, J., et al. (2020). The Construction and Application of a New 17-Plex Y-STR System Using Universal Fluorescent PCR. Int. J. Leg. Med 134 (6), 2015–2027. doi:10.1007/s00414-020-02291-3
Malyarchuk, B., Derenko, M., Denisova, G., Wozniak, M., Grzybowski, T., Dambueva, I., et al. (2010). Phylogeography of the Y-Chromosome Haplogroup C in Northern Eurasia. Ann. Hum. Genet. 74 (6), 539–546. doi:10.1111/j.1469-1809.2010.00601.x
Meirmans, P. G. (2006). Using the Amova Framework to Estimate a Standardized Genetic Differentiation Measure. Evolution 60 (11), 2399–2402. doi:10.1111/j.0014-3820.2006.tb01874.x
Myres, N. M., Rootsi, S., Lin, A. A., Järve, M., King, R. J., Kutuev, I., et al. (2011). A Major Y-Chromosome Haplogroup R1b Holocene Era Founder Effect in Central and Western Europe. Eur. J. Hum. Genet. 19 (1), 95–101. doi:10.1038/ejhg.2010.146
Narasimhan, V. M., Patterson, N., Moorjani, P., Rohland, N., Bernardos, R., Mallick, S., et al. (2019). The Formation of Human Populations in South and Central Asia. Science 365 (6457), eaat7487. doi:10.1126/science.aat7487
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: Analyses of Phylogenetics and Evolution in R Language. Bioinformatics 20 (2), 289–290. doi:10.1093/bioinformatics/btg412
Paradis, E. (2010). Pegas: An R Package for Population Genetics with an Integrated-Modular Approach. Bioinformatics 26, 419–420. doi:10.1093/bioinformatics/btp696
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of Population Structure Using Multilocus Genotype Data. Genetics 155 (2), 945–959. doi:10.1093/genetics/155.2.945
Purps, J., Siegert, S., Willuweit, S., Nagy, M., Alves, C., Salazar, R., et al. (2014). A Global Analysis of Y-Chromosomal Haplotype Diversity for 23 STR Loci. Forensic Sci. Int. Genet. 12 (September), 12–23. doi:10.1016/j.fsigen.2014.04.008
Robino, C., Crobu, F., Di Gaetano, C., Bekada, A., Benhamamouch, S., Cerutti, N., et al. (2008). Analysis of Y-Chromosomal SNP Haplogroups and STR Haplotypes in an Algerian Population Sample. Int. J. Leg. Med. 122 (3), 251–255. doi:10.1007/s00414-007-0203-5
Roewer, L., Krüger, C., Willuweit, S., Nagy, M., Rodig, H., Kokshunova, L., et al. (2007). Y-chromosomal STR Haplotypes in Kalmyk Population Samples. Forensic Sci. Int. 173 (2–3), 204–209. doi:10.1016/j.forsciint.2006.11.013
Roewer, L., Nothnagel, M., Gusmão, L., Gomes, V., González, M., Corach, D., et al. (2013). Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans. Plos Genet. 9 (4), e1003460. doi:10.1371/journal.pgen.1003460
Sabitov, Z. (2013). Jetnogenez Kazahov S Tochki Zrenija Populjacionnoj Genetiki [Ethogenesis of Kazakhs: Population Genetics Perspective]. The Russ. J. Genet. Genealogy 5 (1), 29–47.
Shan, W., Ablimit, A., Zhou, W., Zhang, F., Ma, Z., and Zheng, X. (2014a). Genetic Polymorphism of 17 Y Chromosomal STRs in Kazakh and Uighur Populations from Xinjiang, China. Int. J. Leg. Med. 128 (5), 743–744. doi:10.1007/s00414-013-0948-y
Shan, W., Ren, Z., Wu, W., Hao, H., Abulimiti, A., Chen, K., et al. (2014b). Maternal and Paternal Diversity in Xinjiang Kazakh Population from China. Russ. J. Genet. 50 (11), 1218–1229. doi:10.1134/S1022795414110143
Shi, H., Dong, Y.-l., Wen, B., Xiao, C.-J., Underhill, P. A., Shen, P.-d., et al. (2005). Y-chromosome Evidence of Southern Origin of the East Asian-specific Haplogroup O3-M122. Am. J. Hum. Genet. 77 (3), 408–419. doi:10.1086/444436
Shi, H., Qi, X., Zhong, H., Peng, Y., Zhang, X., Ma, R. Z., et al. (2013). Genetic Evidence of an East Asian Origin and Paleolithic Northward Migration of Y-Chromosome Haplogroup N. PLoS ONE 8 (6), e66102. doi:10.1371/journal.pone.0066102
Shou, W.-H., Qiao, E.-F., Wei, C.-Y., Dong, Y.-L., Tan, S.-J., Shi, H., et al. (2010). Y-chromosome Distributions Among Populations in Northwest China Identify Significant Contribution from Central Asian Pastoralists and Lesser Influence of Western Eurasians. J. Hum. Genet. 55 (5), 314–322. doi:10.1038/jhg.2010.30
Tarlykov, P. V., Zholdybayeva, E. V., Akilzhanova, A. R., Nurkina, Z. M., Sabitov, Z. M., Rakhypbekov, T. K., et al. (2013). Mitochondrial and Y-Chromosomal Profile of the Kazakh Population from East Kazakhstan. Croat. Med. J. 54 (1), 17–24. doi:10.3325/cmj.2013.54.17
Turrina, S., Atzei, R., and De Leo, D. (2006). Y-chromosomal STR Haplotypes in a Northeast Italian Population Sample Using 17plex Loci PCR Assay. Int. J. Leg. Med 120 (1), 56–59. doi:10.1007/s00414-005-0054-x
Underhill, P. A., Jin, L., Lin, A. A., Mehdi, S. Q., Jenkins, T., Vollrath, D., et al. (1997). Detection of Numerous Y Chromosome Biallelic Polymorphisms by Denaturing High-Performance Liquid Chromatography. Genome Res. 7 (10), 996–1005. doi:10.1101/gr.7.10.996
Underhill, P. A., Myres, N. M., Rootsi, S., Metspalu, M., Zhivotovsky, L. A., King, R. J., et al. (2010). Separating the Post-Glacial Coancestry of European and Asian Y Chromosomes within Haplogroup R1a. Eur. J. Hum. Genet. 18 (4), 479–484. doi:10.1038/ejhg.2009.194
Underhill, P. A., Poznik, G. D., Rootsi, S., Järve, M., Lin, A. A., Wang, J., et al. (2015). The Phylogenetic and Geographic Structure of Y-Chromosome Haplogroup R1a. Eur. J. Hum. Genet. 23 (1), 124–131. doi:10.1038/ejhg.2014.50
Unterländer, M., Palstra, F., Lazaridis, I., Pilipenko, A., Hofmanová, Z., Groß, M., et al. (2017). Ancestry and Demography and Descendants of Iron Age Nomads of the Eurasian Steppe. Nat. Commun. 8 (March), 14615. doi:10.1038/ncomms14615
Wassily, (2014). Approximate Areas Occupied by the Three Kazakh Zhuzes in the Early 20th century. Image. Available at: https://upload.wikimedia.org/wikipedia/commons/e/ef/%D0%96%D1%83%D0%B7.svg.
Wei, T., Liao, F., Wang, Y., Pan, C., Xiao, C., and Huang, D. (2018). A Novel Multiplex Assay of SNP-STR Markers for Forensic Purpose. PLoS ONE 13 (7), e0200700. doi:10.1371/journal.pone.0200700
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-Statistics for the Analysis of Population Structure. Evolution 38 (6), 1358–1370. doi:10.2307/2408641
Wells, R. S., Yuldasheva, N., Ruzibakiev, R., Underhill, P. A., Evseeva, I., Blue-Smith, J., et al. (2001). The Eurasian Heartland: A Continental Perspective on Y-Chromosome Diversity. Proc. Natl. Acad. Sci. 98 (18), 10244–10249. doi:10.1073/pnas.171305098
Wen, S.-Q., Sun, C., Song, D.-L., Huang, Y.-Z., Tong, X.-Z., Meng, H.-L., et al. (2020). Y-chromosome Evidence Confirmed the Kerei-Abakh Origin of Aksay Kazakhs. J. Hum. Genet. 65 (9), 797–803. doi:10.1038/s10038-020-0759-1
Zegura, S. L., Karafet, T. M., Zhivotovsky, L. A., and Hammer, M. F. (2004). High-Resolution SNPs and Microsatellite Haplotypes Point to a Single, Recent Entry of Native American Y Chromosomes into the Americas. Mol. Biol. Evol. 21 (1), 164–175. doi:10.1093/molbev/msh009
Zerjal, T., Xue, Y., Bertorelle, G., Wells, R. S., Bao, W., Zhu, S., et al. (2003). The Genetic Legacy of the Mongols. Am. J. Hum. Genet. 72 (3), 717–721. doi:10.1086/367774
Zhabagin, M., Balanovska, E., Sabitov, Z., Kuznetsova, M., Agdzhoyan, A., Balaganskaya, O., et al. (2017). The Connection of the Genetic, Cultural and Geographic Landscapes of Transoxiana. Sci. Rep. 7 (1), 3085. doi:10.1038/s41598-017-03176-z
Zhabagin, M., Dibirova, H. D., Frolova, S. A., Sabitov, Z., Yusupov, Y. M., Utevska, O., et al. (2014). The Relation between the Y-Chromosomal Variation and the Clan Structure: The Gene Pool of the Steppe Aristocracy and the Steppe Clergy of the Kazakhs. Mosc. Univ. Anthropol. Bull. 1, 96–101.
Zhabagin, M. K., Balanovsky, О. Е., Sabitov, Z. M., Temirgaliyev, A. Z., Agdzhoyan, A. T., Koshel, S. M., et al. (2018). Reconstructing the Genetic Structure of the Kazakh from Clan Distribution Data. Vestn. Vogis 22 (November), 895–904. doi:10.18699/VJ18.431
Zhabagin, M. K., Sabitov, Z., Agdzhoyan, A., Yusupov, Y. M., Bogunov, Y., Lavryashina, M. B., et al. (2016). Genezis Krupnejshej Rodoplemennoj Gruppy Kazahov – Argynov – V Kontkste Populjacionnoj Genetiki [Genesis of the Largest Tribal Group of Kazakhs - the Argyns - in the Context of Population Genetics]. Moscow: Vestnik Moskovskogo Universiteta. Seria XXIII. Antropologia [Moscow University Anthropology Bulletin] 4, 59–68.
Zhabagin, M., Sabitov, Z., Tarlykov, P., Tazhigulova, I., Junissova, Z., Yerezhepov, D., et al. (2020). The Medieval Mongolian Roots of Y-Chromosomal Lineages from South Kazakhstan. BMC Genet. 21 (S1), 87. doi:10.1186/s12863-020-00897-5
Zhabagin, M., Sabitov, Z., Tazhigulova, I., Alborova, I., Agdzhoyan, A., Wei, L.-H., et al. (2021). Medieval Super-grandfather Founder of Western Kazakh Clans from Haplogroup C2a1a2-M48. J. Hum. Genet. 66, 707–716. doi:10.1038/s10038-021-00901-5
Zhabagin, M., Sarkytbayeva, A., Tazhigulova, I., Yerezhepov, D., Li, S., Akilzhanov, R., et al. (2019). Development of the Kazakhstan Y-Chromosome Haplotype Reference Database: Analysis of 27 Y-STR in Kazakh Population. Int. J. Leg. Med 133 (4), 1029–1032. doi:10.1007/s00414-018-1859-8
Keywords: Y-chromosome, Y-STR, haplotypes, haplogroups, Kazakhstan, MDS plot, Kazakh tribes
Citation: Khussainova E, Kisselev I, Iksan O, Bekmanov B, Skvortsova L, Garshin A, Kuzovleva E, Zhaniyazov Z, Zhunussova G, Musralina L, Kahbatkyzy N, Amirgaliyeva A, Begmanova M, Seisenbayeva A, Bespalova K, Perfilyeva A, Abylkassymova G, Farkhatuly A, Good SV and Djansugurova L (2022) Genetic Relationship Among the Kazakh People Based on Y-STR Markers Reveals Evidence of Genetic Variation Among Tribes and Zhuz. Front. Genet. 12:801295. doi: 10.3389/fgene.2021.801295
Received: 25 October 2021; Accepted: 10 December 2021;
Published: 07 January 2022.
Edited by:
Maxat Zhabagin, National Center for Biotechnology, KazakhstanReviewed by:
Riga Wu, Sun Yat-sen University, ChinaMiriam Baeta, University of Basque Country UPV/EHU, Spain
Atif Adnan, Naif Arab University for Security Sciences (NAUSS), Saudi Arabia
Copyright © 2022 Khussainova, Kisselev, Iksan, Bekmanov, Skvortsova, Garshin, Kuzovleva, Zhaniyazov, Zhunussova, Musralina, Kahbatkyzy, Amirgaliyeva, Begmanova, Seisenbayeva, Bespalova, Perfilyeva, Abylkassymova, Farkhatuly, Good and Djansugurova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elmira Khussainova, a2h1c3NhaW5vdmFAbWFpbC5ydQ==; Sara V. Good, cy5nb29kQHV3aW5uaXBlZy5jYQ==
 Elmira Khussainova
Elmira Khussainova Ilya Kisselev
Ilya Kisselev Olzhas Iksan1,3
Olzhas Iksan1,3 Bakhytzhan Bekmanov
Bakhytzhan Bekmanov Liliya Skvortsova
Liliya Skvortsova Alexander Garshin
Alexander Garshin Zhassulan Zhaniyazov
Zhassulan Zhaniyazov Gulnur Zhunussova
Gulnur Zhunussova Lyazzat Musralina
Lyazzat Musralina Mamura Begmanova
Mamura Begmanova Anastasia Perfilyeva
Anastasia Perfilyeva Sara V. Good
Sara V. Good Leyla Djansugurova
Leyla Djansugurova