- 1Plant Genomics and Molecular Breeding Laboratory, National Institute for Biotechnology and Genetic Engineering, Faisalabad, Pakistan
- 2Department of Biotechnology, Pakistan Institute of Engineering and Applied Sciences (PIEAS), Islamabad, Pakistan
- 3Department of Biosciences, COMSATS University Islamabad, Islamabad, Pakistan
- 4State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai'an, Shandong, China
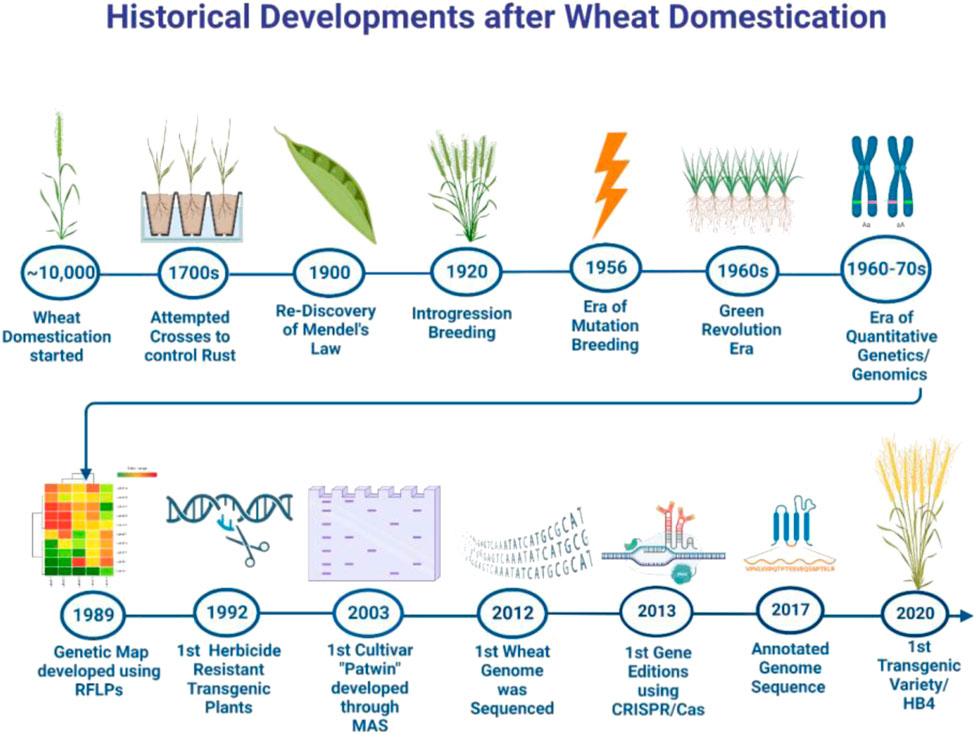
Domestication of wheat started with the dawn of human civilization. Since then, improvement in various traits including resistance to diseases, insect pests, saline and drought stresses, grain yield, and quality were improved through selections by early farmers and then planned hybridization after the discovery of Mendel’s laws. In the 1950s, genetic variability was created using mutagens followed by the selection of superior mutants. Over the last 3 decades, research was focused on developing superior hybrids, initiating marker-assisted selection and targeted breeding, and developing genetically modified wheat to improve the grain yield, tolerance to drought, salinity, terminal heat and herbicide, and nutritive quality. Acceptability of genetically modified wheat by the end-user remained a major hurdle in releasing into the environment. Since the beginning of the 21st century, changing environmental conditions proved detrimental to achieving sustainability in wheat production particularly in developing countries. It is suggested that high-tech phenotyping assays and genomic procedures together with speed breeding procedures will be instrumental in achieving food security beyond 2050.
Introduction
Wheat (Triticum aestivum L.) is cultivated in 89 countries to feed around 2.5 billion people—one-fifth of the total world population. Bread wheat is grown on about 95% of the total wheat cropped area while the remaining 5% area is covered by the durum wheat (Mastrangelo and Cattivelli, 2021). The contribution of durum wheat in total wheat production is also around 5%.
Wheat domestication and human civilization evolved simultaneously in the history of mankind. Among the cereals, it is one of the most important crops with relatively more potential to get adapted in challenging environments. With the increasing food demand and depleting agricultural land, it is pivotal to enhance the grain yield in a sustainable way to feed the increasing human population beyond 2025. Nearly 100% increase in wheat production is inevitable to meet the global food requirements by the end of 2050 (Senker, 2011). Concerning the advancements in research and development, all the major events and technologies that paced up wheat research and have a plethora of contributions towards wheat improvement after its domestication as a cereal crop have been described in this article.
Origin and domestication of wheat
The hexaploid wheat (AABBDD) contains three different genomes each derived from different diploid species viz., Triticum urartu (AA genome), Aegilops speltoides (controversial, BB genome), and Aegilops tauschii (DD genome) (Feldman et al., 1995; Nesbitt and Samuel, 1996). According to the archeological records, wheat originated in Southeast Turkey. Initially, the progenitor species containing AA and BB subgenomes were discovered (Aaronsohn, 1910) and these were hybridized followed by a doubling of chromosomes which resulted in tetraploid fertile wheat, T. turgidum (AABB) (v. Tschermak and Von, 1914). Then the T. turgidum, wild emmer, was domesticated in Fertile Crescent. Afterward, T. turgidum hybridized with a diploid specie A. tauschii (Kihara, 1944; McFadden, 1944; McFadden and Sears, 1946) which resulted in the formation of hexaploid wheat (AABBDD). The hexaploid bread wheat evolved in the Fertile Crescent (Figure 1). It is worth mentioning that tetraploid ancestors spread into the natural range of diploid species Ae. tauschii. Because of its high acceptance as an ultimate source of calories, it was spread into different parts of the world via different routes (Figure 1). After domestication, hexaploid wheat was cultivated and selected in diverse geographical regions for centuries which resulted in present-day cultivated bread wheat (McFadden and Sears, 1946; Dubcovsky and Dvorak, 2007). Among diploids, einkorn wheat, Triticum monococcum, is considered the first domesticated hulled wheat. The historical record shows that it was domesticated 12,000—c. 8,500 years ago in the Pre-Pottery Neolithic period (Zaharieva and Monneveux, 2014). However, cultivated tetraploids Triticum dicoccum (wheat emmer) and Triticum durum (tetraploid durum), both arose from wild ancestors.
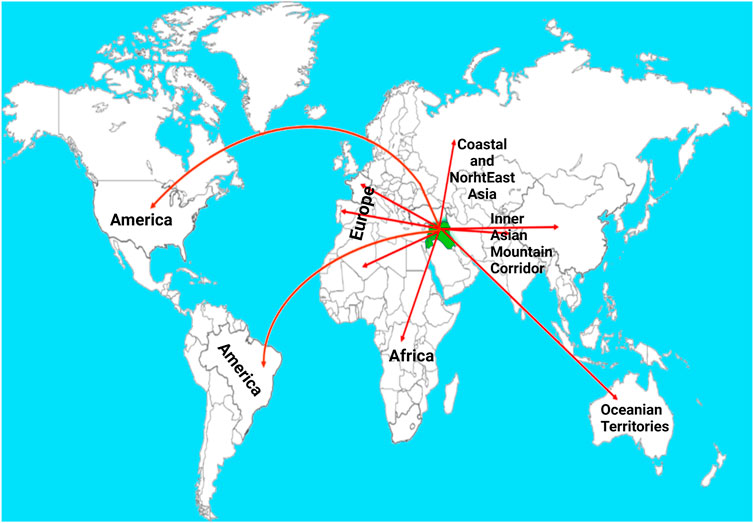
FIGURE 1. Routes showing the migration of wheat from the center of origin-Fertile Crescent-to other continents of the world. The green color indicates Fertile Crescent and the red lines indicate different known routes (Pont et al., 2019). Post-Domestication to Mendel’s Era.
Initially, the wheat was spread to Greece, Cyprus, India, and Egypt followed by other countries around the world (Cooper, 2015). Domesticated wheat had large-sized hulled seeds attached to the ear as compared to its wild species. The wheat grain in primitively cultivated species was long, thin, and small in size. The first naturally mutated traits in the wild germplasm were non-brittle rachis and naked grain that were responsible for the domestication of wheat (Pourkheirandish et al., 2018). The process of natural selection played a significant role in shaping the present-day cultivated wheat. Selections were made by the early farmers on the basis of phenotypic traits such as grain size, grain color and non-shattering type (Eckardt, 2010). Seeds from the best plants were used to grow the next generation while inferior quality seeds were discarded (Charmet, 2011).
During the 1800s, several wheat varieties were developed by selecting superior lines by the wheat breeders. Wheat growers or distributors were used to select different wheat spikes with desirable quality followed by assigning a new fancy name to the selected spikes such as ‘Thomas Rust Resistant’ for wheat variety resistant to rust and ‘Landretti’s Hard Winter’ for resistance to changing climate (Spennemann, 2001).
In the 18th century, wheat ‘rust’ was scientifically described for the first time (Eriksson and Henning, 1896). Efforts were made to improve wheat varieties mainly for high yield and resistance to diseases. Similarly, wheat breeding for protein content and baking quality was initiated by William James Farrer (Wrigley et al., 1981). Then in 1873, interspecific hybridization between rye and wheat was successfully made, and thereafter enormous valuable hybrids of wheat showed significant improvements in yield and other traits such as early maturity, rust-resistance with stiff straws, gluten content, and non-shattering traits were developed (Beach, 1923).
Wheat genetics in post-mendelian era
In the late 19th century, Mendel published data pertaining to his historic experiments conducted on pea plants. In the early 1900s, his work was re-discovered which provided him recognition as the “Founder of Genetics” (Wrigley et al., 1981). Since then, breeding based upon scientific knowledge started and the whole research perspective was shifted towards improving desirable traits such as plant height, seed color, and seed shape instead of the plant as a whole (Biffen, 1905). In 1916, the first hard wheat variety ‘Yeoman’ having low protein content was developed through hybridization. It was discovered that for achieving genetic stability, more than 10 generations were required for fixing the traits (Bajaj, 1990). In 1920, the stem rust gene, Sr2 was incorporated into wheat from tetraploid emmer wheat (Singh and McIntosh, 1984). Following several hybridization experiments, Italian landraces and inbred lines were crossed with the Japanese variety ‘Akakomugi’. Resultantly, new varieties harboring improved resistance to rust diseases, early flowering, and early maturing were developed. These varieties were used in other wheat breeding programs, and laid down a firm foundation for achieving green revolution (Salvi et al., 2013). During the early period of hybridization-based breeding programs, varieties depicting high yield potential were developed without focusing on improving resistance to biotic stresses. Another winter wheat variety ‘Turkey red’ was also developed and cultivated in the United States (Olmstead and Rhode, 2002). Drought resistance was incorporated from landraces to ‘Aragon 03’ (Royo and Briceño-Félix, 2011). Later on, wheat varieties with high Zn and protein content as well as biotic and abiotic stress tolerance were also developed through conventional breeding.
The era of mutagenesis
By the process of mutagenesis, novel genetic variability in plants was induced by exposing them to physical or chemical mutagens (de Oliveira Camargo et al., 2000). Over the last century, physical mutagens, for example, gamma rays, UV rays, fast neutrons, and the chemical mutagens such as sodium azide, N-methyl-N-nitrosourea, ethyl methansulfonate and hydrogen fluoride have been widely used. Biological mutagens like Agrobacterium are also being used (Krishnan et al., 2009). Recently, the mutant population was developed by exposing the seeds of a wheat cultivar ‘Punjab-11’ to gamma-rays. The developed mutants were found to be resistant to either leaf rust, yellow rust or, stem rust (Hussain et al., 2021). Few of these mutants also demonstrated high grain quality traits as compared to wild type (Zulfiqar et al., 2021).
Mutation breeding techniques were resurrected during early years of the 21st century due to a better understanding of mutagens, their use, the process of mutagenesis, and its application in related disciplines. Nowadays, traditional approaches being used for the selection of mutants in second and third generations have provided high yielding as well as better quality varieties (Singh and Balyan, 2009; Albokari, 2014). Consequently, a huge number of varieties with improved traits have been released through mutation induction which reveals the economic impact of this technology (Micke et al., 1990; Jankowicz-Cieslak et al., 2017). To date, ∼ 3400 mutant varieties have been produced through mutagenesis directly or indirectly, including 265 varieties of bread wheat (https://nucleus.iaea.org/sites/mvd/SitePages/Search.aspx) (Figure 2). However, the majority of the varieties (∼85%) are the result of mutation inductions through gamma rays. All these varieties released through mutation breeding are high yielding, with better tolerance to pests, diseases, and biotic and abiotic stresses.
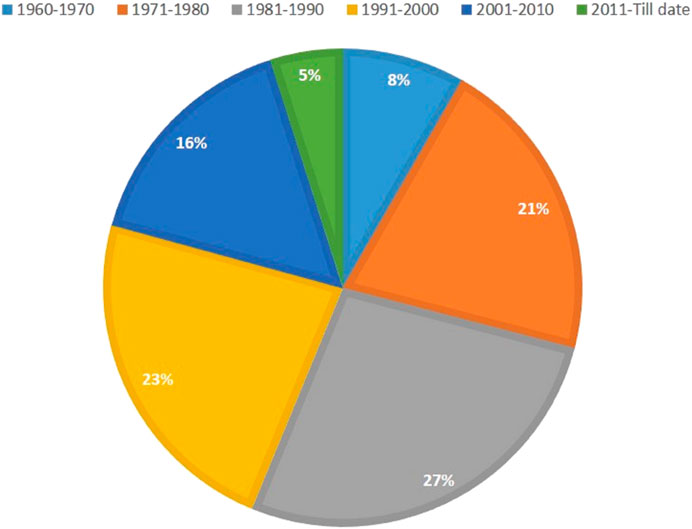
FIGURE 2. Percentage of mutant wheat varieties developed through mutagenesis in various decades (Data source: https://nucleus.iaea.org/sites/mvd/SitePages/Search.aspx).
The use of chemical mutagens on a large scale has been started in 2000. Ethyl methanesulphonate (EMS) –the most commonly used mutagen-generates random point mutations in the entire genome (Brini and Masmoudi, 2014). For example, a single nucleotide polymorphism (SNP) in Lr21 was identified in one of the resistant mutants (to leaf rust) of ‘NN-Gandum-1’. This mutation caused a substitution of glutamic acid with alanine (Hussain et al., 2018). Few of these mutants also demonstrated better tolerance to drought stress as compared to wild type (Zahra et al., 2021). It is suggested that mutagenesis experiments are effective in inducing useful mutations in wheat which can be used in forward and reverse genetic studies for gaining insights into the important biological traits of the complex wheat genome.
In the beginning of the 21st century, an advanced method, Targeting Induced Local Lesions IN Genomes (TILLING) was used for identifying point mutations in targeted genes of mutants and or genetic stocks (Henikoff and Comai, 2003). In 2009, through TILLING, complete waxy wheat was bred by crossing Wx-A1 and Wx-D1 truncation mutant whereas Wx-B1was naturally null in both of them (Dong et al., 2009). Similarly, a waxy gene GBSS-I (granule-bound starch synthase I) in near null waxy mutant was identified (Slade et al., 2012). In 2012, the wheat mutant gene MNR220 was identified that carried resistance locus to powdery mildew and other types of rust. The mutant population resulted after TILLING with many novel alleles that could be a good genetic resource for improvement of wheat (Chen et al., 2012). Novel genetic variations in the SBEIIa gene for amylose content, TaAGP gene for starch biosynthesis and, TaMlo gene for durable resistance against powdery mildew were also identified through TILLING (Slade et al., 2012; Acevedo-Garcia et al., 2017; Guo et al., 2017).
Introgression wheat breeding
Among cereals, a lot of introgression work was done on wheat (Dempewolf et al., 2017), resulting in significant improvement in the genetic diversity of wheat (Lu and Ellstrand, 2014). The genetic sources of wheat, consisting of wild relatives, landraces, and close relatives have contributed significantly to adding novel genetic variations to modern wheat cultivars (Molnar-Lang et al., 2016). In 1930, stem rust resistance Sr2 gene was introgressed into cultivated wheat from its wild relative emmer wheat cultivar “Yaroslav” (McFadden and Sears, 1946). Several genes against biotic stresses such as Ug99 were found in Aegilops. Genes for stem rust including Sr33, Sr45, Sr45, Sr46, and SrTA1662 were introgressed and localized into the genome of cultivated wheat (Olson et al., 2013).
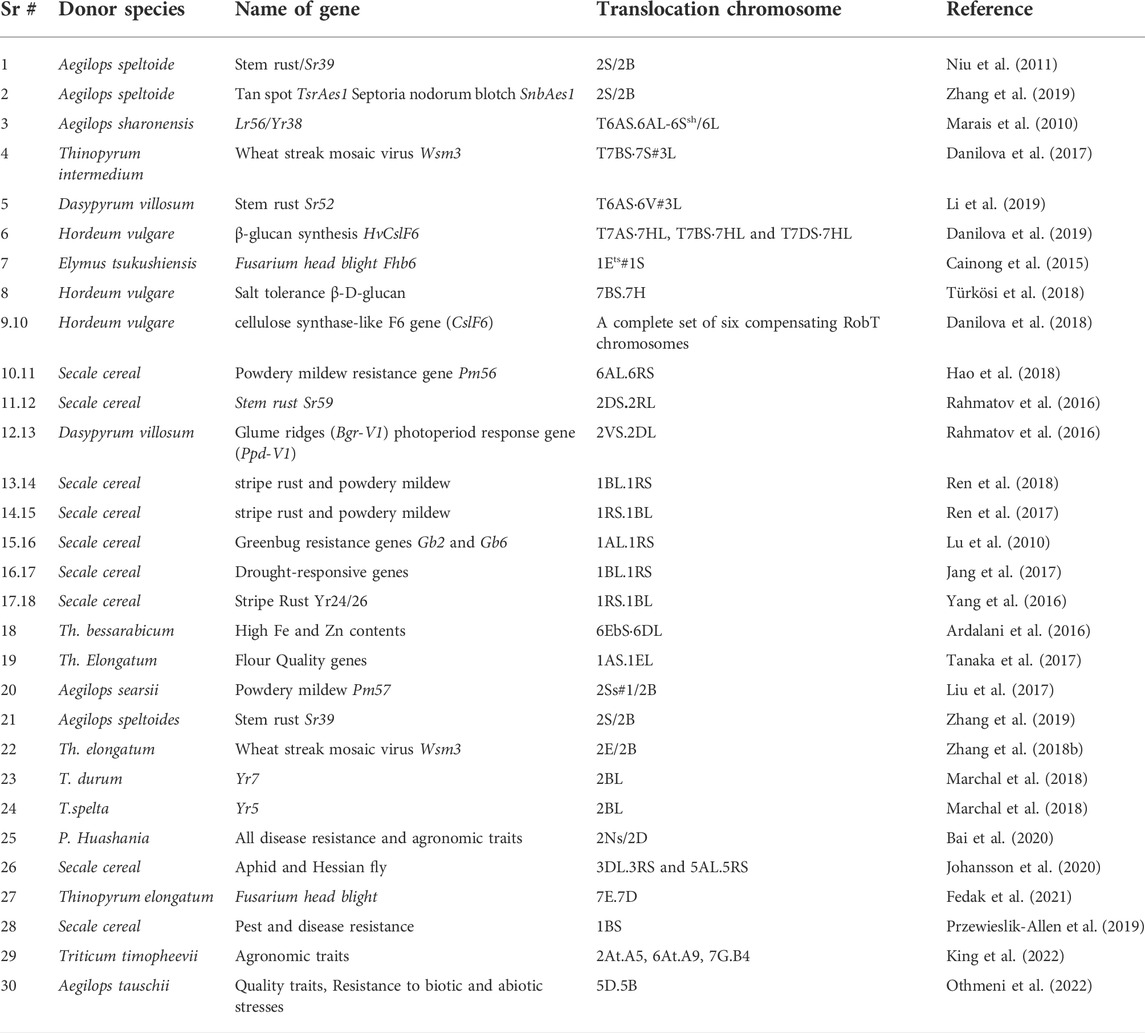
Secale cereale, commonly known as rye, is one of the most important wheat relatives which was used for incorporating several genes into the cultivated wheat. After hybridization with rye, translocations and substitutions played a role in transferring genes responsible for high yield and disease resistance (Rabinovich, 1998). The most important non-Triticum introgressions in the wheat genome were 1BL/1RS, 1DL/1RS and 1AL/1RS translocations that contained biotic and abiotic stress resistance genes (Rabinovich, 1998; Mago et al., 2015). The 1BL/1RS translocation between wheat chromosome ‘1B’ and rye chromosome ‘1R’ carrying genes for leaf rust (Lr26), stem rust (Sr31), stripe rust (Yr9), and powdery mildew (Pm8) improved the resistance to fungal diseases in wheat (Singh et al., 1990; Friebe et al., 1996; Friebe et al., 1999; Baffes, 2005). The wheat lines containing the 1RS chromosomal arm exhibited a substantial increase in the root length and spike length (Liu et al., 2020a). Likewise, introgression of 4R and 6R chromosomes from rye cultivar ‘Kriszta’ resulted in a significant increase in protein content (Schneider et al., 2016). A novel stem rust resistant gene Sr59, yellow rust resistant gene Yr83, and powdery mildew resistant gene Pm56 from rye were introgressed into wheat as a 2DS:2RL and 6AL:6RS Robertsonian translocations, respectively (Table 1) (Rahmatov et al., 2016; Hao et al., 2018)
Deficiency of essential micronutrients also called as hidden hunger has affected around two billion people in the world. As a major staple crop, wheat provides almost 20% protein and energy to mankind. Hence, wheat is an ideal candidate for biofortification. Improvement of protein content in wheat grain has been remained a major breeding objective of several wheat groups around the globe. In wild emmer wheat, a Gpc-B1 locus was discovered which can enhance the content of protein, Zn and Fe; hence can be incorporated into cultivated wheat to increase its nutritional value (Uauy et al., 2006). This gene was introgressed into two cultivars HUW468 and HUW234 through marker assisted backcrossing (Vishwakarma et al., 2014; Mishra et al., 2015). The introgression of Gpc-B1 gene for increasing grain protein content has also been achieved in 10 elite wheat cultivars (Kumar et al., 2011). Moreover, introgression of Gpc-B1 gene was also performed in different spring wheat cultivars (Carter et al., 2012; Tabbita et al., 2012; Eagles et al., 2014), where grain protein contents were substantially increased. Tolerance to salt, and (1,3;1,4))-β-D-glucan content were also transferred in wheat as a 7BS.7HS wheat/barley Robertsonian translocation (Türkösi et al., 2018). Recombinant inbred wheat lines were developed by introgression with A. caudate; newly developed lines showed improved disease resistance. Also, resistance to the take-all disease was incorporated in the 2NS/2D substitution line of bread wheat by making a cross with Psathyrostachys huashania Keng (Table 1) (Bai et al., 2020). Recent technological advancements in genomics and cytogenetics offer new avenues for transferring alien genes to wheat, avoiding issues like linkage drag.
The era of the green revolution in wheat production
In 1950, photoperiod insensitive (ppd1 and ppd2) genes were transferred into wheat for expanding germplasm usage globally (Rajaram, 2001). After the wheat rust epidemic (1951–1954) in North America, research for the development of rust-resistant modern cultivars was initiated by the CIMMYT (International Maize and Wheat Improvement Center, Mexico). Another initiative that prevented the outbreak of the famine in 1970s was the introduction of genes which resulted in a significant reduction of plant height in wheat. These genes were derived from a wheat genotype ‘Norin-10’. The semi-dwarf “Norin-10” was about 60 cm in height, more responsive to nitrogen fertilizer, resistant to rust, and had lodging resistance. Norin-10 was estimated to be cultivated on 15–18 million acres worldwide (Reitz and Salmon, 1968). The genes responsible for conferring short height were named Rht-B1 and Rht-D1 (Khush, 1999). The introduction of these genes in wheat paved the way for wheat breeding aimed at enhancing yield potential which ultimately helped in alleviating hunger and poverty across the globe. These varieties brought a green revolution in several developing countries like Pakistan, India, Turkey, Afghanistan, etc. Dr. Norman Borlaug was awarded Nobel Peace Prize for his brilliant work (Borlaug, 2007).
After the Green revolution, the wheat yield increased many folds (Figure 3), however, the nutritional quality was compromised (Ortiz-Monasterio et al., 2007). Increasing the nutritional quality of cultivated wheat became another challenge for wheat breeders. Also, the other disadvantage of the green revolution was that many old varieties disappeared, and many of these led to extinction (Eliazer Nelson et al., 2019). Almost 63% of wheat varieties released in the 21st century contain “Green revolution” alleles (Würschum et al., 2017). But the performance of varieties having these alleles was not satisfactory in dry and warm regions owing to arrested growth of coleoptile and seedling emergence (Rebetzke et al., 2014). An alternative dwarfing allele, Rht18 was identified that has no impact on coleoptile length, and hence can be used to replace previous dwarfing genes in target environments (Pearce, 2021). In the post-green evolution era, stem rust had also threatened these short-statured varieties. So, during this period, Sr2, Sr5, Sr6, Sr7a, Sr7b, Sr8a, Sr9b, Sr9d, Sr9e, Sr9g, Sr10, Sr11, Sr12, Sr17, Sr24, Sr26, Sr30, Sr31 and Sr36 genes were incorporated into wheat (Knott, 1988). However, later on, new devastating rust races evolved that became a serious threat to wheat germplasm, globally. Stem rust race Ug99 and its variants caused 80–100% yield losses in different countries of the world (Khan et al., 2013). These fast-evolving races of rust put the attention of breeders toward pyramiding two or three major genes to induce durable resistance in wheat. Later on, different varieties having minor genes for stem rust, leaf rust, and yellow rust were developed to avoid the issue of resistance breakdown (Figure 4).
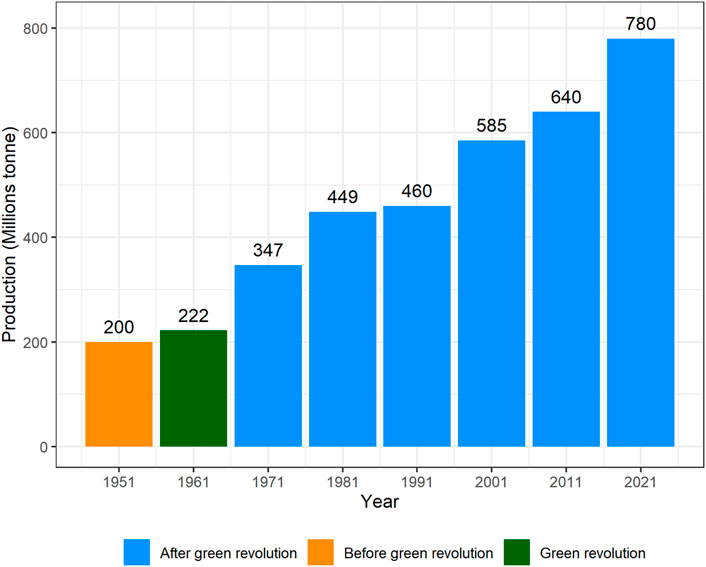
FIGURE 3. Worldwide wheat production after the green revolution (FAOSTAT, 2022).
Hybrid wheat: Future prospects
Breeding hybrids remains instrumental in uplifting the production of several crop species including corn. There is a dire need to breed for adding resilience to changing environments and high-yielding wheat varieties to address the pressing issue of global food security. The exploitation of hybrid vigor may entail the desired increase in wheat yield (Longin et al., 2013), biotic/abiotic stress tolerance as well as grain quality (Gowda et al., 2012; Longin et al., 2013; Longin et al., 2014; Mühleisen et al., 2014; Longin et al., 2015; Jiang et al., 2017; Thorwarth et al., 2018);
The development of wheat hybrid is entirely dependent upon manual crossing between the two inbred lines. Efforts to develop hybrid wheat have a long history, however, the success rate is very slow because of its autogamous nature and tedious procedure of making crosses. Heterosis was first time reported in wheat in 1919, since then several efforts have been attempted to establish a well-defined hybrid production system in wheat (Freeman, 1919; Pickett, 1993). During the 1950s and 1980s, hybrid wheat breeding showed 10% mid-parent heterosis for grain yield. For hybrid development, cytoplasmic male sterility (CMS) and chemical hybridizing agents (CHA) were two methods practiced for removing pollens of the lines used as female (Kihara, 1951). The CMS and restorer genes were derived from T. timopheevii (Wilson, 1962). The research efforts being done in hybrid breeding could not be continued in real spirit owing to the maintenance of three lines, and some genetic issues related to complete restoration of fertility as well as the undesirable effect of alien cytoplasm (Singh et al., 2010).
Recent years witnessed the cloning of several male sterility genes which provided new tools for producing hybrid seeds in wheat. The dominant male-sterility gene Ms2 present in common bread wheat facilitated the evolution of several lines and cultivars, and could further be utilized in developing a high-throughput hybrid production system (Ni et al., 2017). The cloning of a nuclear recessive male sterility gene Ms1 also provides a new resource for large-scale commercial hybrid seed production (Wang et al., 2017). In addition, orthologous male sterile genes identified in grasses could help to understand the male sterile trait in wheat. In total, three homoeoalleles of OsNP1 showed similar expression to OsNP1 and ZmIPE1 in wheat. The optimized CRISPR/Cas9 mediated triple homozygous Tanp1 mutant displayed complete male sterility and only one wild type of TaNP1 gene was sufficient for the maintenance of male fertility (Li et al., 2022). This work provided an optimized CRISPR/Cas9 vector system in wheat, elucidated the highly conserved function of TaNP1 genes, and produced complete male sterile mutants which can be used in hybrid seed production. This genetic male sterile line will lead to two-line hybrid seed production in wheat. The two-line system will be free of negative effects associated with alloplasms, cytoplasm, and restorer line. This two-line system is relatively simple and potentially more efficient as compared to the existing 3-lines hybrid system; which demands a high cost of maintenance with a limited level of success in restoring fertility in F1 hybrids (Li et al., 2020). Despite several limitations, different attempts to develop wheat hybrids were made but still, hybrid breeding is not completely established.
During the 1990s, hybrid wheat programs were re-initiated by the CIMMYT-Monsanto joint project. Over the last few decades, global area under hybrid wheat is increasing; in Europe hybrid wheat cultivation impressively increased from 100,000 ha to 560,000 ha during 2017–2018. The success of hybrid wheat production can be witnessed in winter wheat variety ‘Hystar’ produced by the joint collaboration of Germany and Portugal. Moreover, several public and private companies have launched programs for the development hybrid wheat such as CROPCO’s program in the United Kingdom. Syngenta claims for the release first wheat hybrid in India in near future. In China, more than 50 wheat hybrids having 10–20% increased yield potential have been developed; out of these, seven hybrids (Yunza 6, Yunza 3, Jingmai 6, Mianyang 32, Jingmai 7, Yunza 5, Mianzamai 168) were approved. In another study, 3000 wheat lines were evaluated for hybrid wheat production which highlighted the importance of hybrid wheat production targeted at multiple traits (Baenziger et al., 2019). The recent studies suggest that recurrent genomic selection may help in achieving long-term selection gain of hybrid breeding (Rembe et al., 2019). By reducing the cost of hybrid seeds production, a better understanding of heterosis, developing heterotic groups, and incorporating novel technologies for fertility restoration, wheat hybrids can be developed in the future.
Use of DNA markers in wheat breeding
Research on the development of DNA markers started in the early 1980s (Figure 4). In 1984, the restriction fragment length polymorphism (RFLP) assay was deployed for developing linkage and physical maps and studying the extent of genetic diversity among the wheat genotypes (Chao et al., 1989; Lagudah et al., 1991). Subsequently, PCR-based fingerprinting assays including random amplified polymorphic DNA (RAPD), sequence characterized amplified regions (SCAR), and simple sequence repeats (SSRs) were used for the amplification of desired DNA fragments. These DNA markers were associated with several traits including disease resistance, kernel traits, lodging, vernalization response, etc. (Galiba et al., 1995; Keller et al., 1999a; Keller et al., 1999b; Shahid et al., 2002; Campbell et al., 1999; Rahman et al., 2004; Marza et al., 2006). Consequently, several quantitative trait loci (QTLs) associated with heat stress and drought stress were identified which were located on different chromosomes (Malik and Malik, 2015; Sarkar et al., 2021). The use of DNA markers in marker-assisted breeding paved the way for the identification and selection of desirable genotypes/cultivars. Vrn, Ppd, and Rht were frequently used in marker-assisted selection to develop wheat cultivars/lines. By using RAPD and SCAR molecular markers, Dn 2 genes linked to Russian aphid resistance were identified which were used in developing new resistant wheat cultivars. The SSRs for Fusarium head blight resistance (FHB), pre-harvest sprouting tolerance, and mildew resistance were identified and used in marker-assisted selection in wheat (Del Blanco et al., 2003; Kottearachchi et al., 2006; Tucker et al., 2006). Similarly, SSR markers were also used in marker-assisted backcross breeding (MABB) for introducing traits such as grain protein content and drought tolerance in wheat (Davies et al., 2006; Rai et al., 2018).
Single nucleotide polymorphism (SNPs) is an efficient marker system (Gupta et al., 1999) that is found in abundance throughout the genome. Recently, technological advancements have resulted in the identification of important SNPs linked to traits such as tiller number, spikelets per spike, plant height, spike length, protein content, and grain yield per spike (Marcotuli et al., 2017; Chai et al., 2018; Wang et al., 2018).
Development and utilization of SNP chips in wheat breeding
The SNPs-based platforms, containing informative SNPs are attractive and powerful tools used for studying the genetic diversity in wheat. The diversity array technology (DArT) was among the initial array-based technologies that generated hundreds of anonymous markers in wheat (Allen et al., 2011). A 9K SNP chip was developed for genotyping 2994 wheat lines. The generated information was used in genome-wide association studies (Cavanagh et al., 2013). The development of Illumina 90K gene-associated SNPs array designed to identify polymorphism in wheat, and KASP assays accelerated the use of SNP markers in wheat breeding (Allen et al., 2011; Wang S. et al., 2014). For detecting polymorphism in primary, secondary, and tertiary pools, an 820 K Affymetrix Axiom SNP array was developed (Winfield et al., 2016). The Wheat Breeders’ 35K Axiom array was later derived from the Wheat 820K SNP array (Allen et al., 2017). Another genome-specific and widely used 660K SNP array was fabricated for studying the polymorphism among the wheat genotypes (Sun et al., 2020). Moreover, some other SNP chips including 15K SNP (Boeven et al., 2016; Qaseem et al., 2019), and 55K SNP developed from 660K has been widely used for GWAS in wheat (Ye et al., 2019; Jin et al., 2020).
Whole genome sequencing of wheat
The huge size of the wheat genome (∼17.6 Gb) contains approximately 90% repetitive DNA which was the main hurdle in sequencing the whole genome (Li et al., 2004; Wanjugi et al., 2009). The International Wheat Genome Sequencing Consortium (IWGSC) was initiated to sequence the complex genome of wheat (Gill et al., 2004). Finally, in 2012, the first whole genome sequencing information of a Chinese Spring wheat variety “CS42” was released by deploying the short gun whole genome sequencing technique (Brenchley et al., 2012). Afterward, one chromosome at a time was sequenced and the genome assembly of 10.2 billion bases was constructed (IWGSC: The International Wheat Genome Sequencing Consortium, 2014). In the third attempt, 12.7 billion bases were assembled (Clavijo et al., 2017). Eventually, in the year, 2017–2018, the wheat genome of Chinese spring wheat was sequenced and released as the first reference wheat genome. With the rapid advancements in sequencing and assembly tools, many wheat cultivars were resequenced including the Chinese wheat-rye 1RS.1BL translocation cultivar “Aikang 58” (Ru et al., 2020), French bread wheat cultivar “Renan” (Aury et al., 2022), transformation-amenable common wheat cultivar “Fielder” (Sato et al., 2021), etc.
Genome editing as an emerging technique
Genome editing can specifically modify the genome by inducing insertions, deletions, substitutions, and or targeted mutations (Zhang J. et al., 2018). Earlier genome editing tools such as zinc finger nucleases (ZNFs) and transcription activator-like effector nucleases (TALENs) have been replaced with Clustered Regularly Interspaced Short Palindromic Repeats and associated protein 9 (CRISPR Cas/9). CRISPR/Cas9, a highly precise genome editing tool is used to induce specific double-stranded breaks in the genome (Azhar et al., 2021). This assay is relatively more economical and user-friendly than other editing assays including ZNFs and TALENs (Razzaq et al., 2021). Previously, CRISPR/Cas9 and transcription activator-like effector nucleases (TALEN) were used to knock out Mildew Locus O (Mlo) gene for enhancing resistance to powdery mildew (Wang C. et al., 2014).
Due to allohexaploid nature of wheat, it is very difficult to target three or more genes simultaneously. After successful development of stable plants in wheat, α-gliadin genes were targeted to decrease the gluten content (Sánchez-León et al., 2018). Similarly, TaGW2 homeologs were knocked out to develop mutant lines containing high protein content and grain weight (Zhang Y. et al., 2018). Furthermore, EDR1 homeologs and TaEDR1 lines showed resistance to powdery mildew (Zhang et al., 2017). To understand meiotic crossover, TaZIP4-B2 was targeted (Rey et al., 2018). Likewise, through Agrobacterium-mediated CRISPR/Cas9, each of the Qsd1 homeo alleles was targeted to suppress pre-harvest sprouting (Abe et al., 2019). Haploid wheat was developed by editing matrilineal (TaMTL) that triggers haploid production and centromere-specific histone H3 (CENH3I) gene that plays a vital role in the segregation of chromosomes during cell division (Liu et al., 2020b).
Genomic selection
Genomic selection (GS) helps to select superior genotypes by integrating genotypic and phenotypic data of a training population (TP) to predict breeding values (GEBVs) of a breeding population (BP) (Meuwissen et al., 2001). On the basis of these GEBVs, better performing plants can be selected for use as parent genotypes in the next breeding programs. The similarity index of the molecular marker profile of these individuals with TP allows us to predict the best performing plants. Over time, GS has been efficiently used in wheat breeding due to its high accuracy. In the wheat breeding program, genotype-by-sequencing (GBS) was used to detect polymorphisms followed by the estimation of prediction accuracy (Poland et al., 2012). Relatively, low to moderate prediction accuracy (0.28 and 0.42) was recorded that was higher than the established marker platforms. Another study showed that Fusarium head blight resistance showed moderate to high prediction accuracies (0.67–0.82) making it a promising approach for improving resistance to Fusarium (Arruda et al., 2015). In GS, the prediction accuracy for grain quality ranged from 0.27 to 0.81, thus showed its potential for deploying in a wheat breeding program (Sandhu et al., 2021).
Speed breeding—A way to accelerate breeding cycles
Conventional wheat breeding systems take several years to release a stable variety which hinders the breeding progress. However, speed breeding made it possible to harvest up to 4–6 generations of spring/winter wheat per year (Watson et al., 2018). The photoperiod of wheat has been extended to accelerate the developmental stages of plants. Plants grown under controlled environment chambers (speed breeding) reach the anthesis and heading stage almost half the time earlier than the plants grown under natural field conditions. The plant grown under light supplemented glasshouses have been evaluated on the basis of their germination rate.
Speed breeding is instrumental in advancing 5–6 generations per year. Thus, speed breeding can help in lessening the time required for each breeding cycle (Watson et al., 2018). Several protocols have been proposed for undertaking phenotypic characterization of wheat plants which can add synergism to speed breeding. For instance, grain dormancy to tolerate sprouting after harvesting (Hickey et al., 2009, 2010), resistance to stripe rust in wheat (Hickey et al., 2011), screening of root traits for improving adaptation to drought stress (Richard et al., 2015), resistance to yellow spot disease (Dinglasan et al., 2016), and leaf rust resistance traits (Riaz et al., 2016) were targeted through speed breeding in wheat. Several other traits including disease related traits, plant height, root traits, and flowering time have also been screened through speed breeding in wheat (Alahmad et al., 2018; Ghosh et al., 2018). Genomic selection together with speed breeding was used to increase genetic gain (Watson et al., 2019). Recent advancements in high-throughput phenotyping further reduced the obstacles in the progress of plant breeding and genetics. Hence, speed breeding coupled with high-throughput phenotyping can help in discovery of novel and desirable traits in a more sustainable way (Al-Tamimi et al., 2016). For instance, selection of some root and seedling traits through speed breeding helped in rapid selection of mature plants with improved root architecture (Richard et al., 2015).
High throughput phenotyping
High throughput phenotyping assays have been used to monitor and measure several traits in a large number of plants, simultaneously. These techniques take advantage of the latest automated sensors and imaging tools. Through HTP approaches, non-destructive data can be collected more accurately from trials. The unmanned aerial vehicle (UAV)-based RGB imagery HTP approach was used to estimate wheat plant height (Volpato et al., 2021). The UAV was used to select wheat genotypes for grain yield in early selection cycles (Hu et al., 2020). In addition, complex traits such as lodging were also assessed using a UAV system which proved that HTP can be used to study complex traits (Singh et al., 2019).
Development of genetically modified wheat
In 1992, the first transgenic wheat conferring tolerance to herbicide was developed (Vasil et al., 1992). The complex nature of the wheat genome together with its acceptability by the public remained a major challenge in extending research on the development of transgenic wheat (Shewry Jones, 2005). Initially, transgenic wheat was developed using the biolistic method. Later on, Agrobacterium-mediated transformation approach was used. In 2004, the first genetically modified wheat round-up ready (MON- 71800) was developed by Monsanto through the introduction of the CP4 Esps gene—conferring resistance to glyphosate (herbicide). Several transgenic lines were developed containing avidin gene conferring resistance to insect pests (Abouseadaa et al., 2015). Moreover, transgenic wheat lines containing DREB1A, HDG11, WRKY2, TaSHN1, NAC, and bZIP2 were produced which had a high tolerance to drought and increased yield potential (Pellegrineschi et al., 2004; Xue et al., 2011; Li et al., 2016; Bi et al., 2018; Gao et al., 2018; Luang et al., 2018) Similarly, heat resistant transgenic wheat was produced which were overexpressing TaHsfA6f, TaFER-5B, TaHsfC2a genes (Xue et al., 2015; Zang et al., 2017; Hu et al., 2018). Some other researchers produced wheat lines conferring resistance to viruses, showing high nutritional quality and improved yield (Sivamani et al., 2000; Xue et al., 2004; Tamás et al., 2009). In 2020, Argentina was the first country to approve drought-resistant GM wheat Bioceres HB4 (Sheridan, 2021). Recently, Brazil also approved HB4 developed by Argentina for consumption as flour.
Contribution of wheat towards global food security in changing climates
Wheat is consumed by 2.5 billion people as a staple crop and it contributes to global food security by providing 20% calories and proteins. Escalating temperature can severely affect the average wheat yield across the globe. It was projected that a 1°C rise in temperature can suppress yield by 10%. In the coming years, the impact of changing environmental conditions and wheat production to feed extra 3 billion people will be the major challenges for wheat breeders (Curtis and Halford, 2014). The annual yield should be increased to 1.6% to meet the increasing demand of the human population under the scenario of changing environments (Ray et al., 2013; Wheeler and Von Braun, 2013; FAO, 2017). Another major menace is the limited or non-availability of irrigation water which is required for maintaining normal wheat growth. It has been reported that drought alone can reduce yield by up to 86%, and the condition can be worsened under the changing environmental conditions (Shafeeq and Zafar, 2006; Joshi et al., 2007; Prasad et al., 2011). Development of resilient wheat varieties that can demonstrate high yield potential is required for sustaining wheat production worldwide. Also, the changing climate can foster the evolution of new strains of pathogens and diseases. For example, new races/strains of rust disease can overcome the available resistance in cultivated wheat varieties. Rust diseases can cause 15%–20% wheat yield losses worldwide (Figueroa et al., 2018). The incorporation of rust-resistant genes against evolving pathogens into high-yielding wheat varieties is very important for sustainable wheat production. Thus, new resilient wheat varieties under the changing environmental conditions can ensure global food security.
Increasing grain yield to ensure food security
Grain yield (Y)-a complex quantitative trait-is affected significantly by biotic and abiotic stresses. Grain yield is dependent on biomass (B) and grain harvest index (HI) (Yield = B × HI). During the green revolution, HI was improved by about 60% by the incorporation of height-related genes in old wheat varieties. Yield can be improved by increasing photosynthetic area or capacity (Parry et al., 2011). Canopy architecture, large-spike, and spike fertility can also contribute to high yield (Gaju et al., 2009; Murchie et al., 2009; Reynolds et al., 2009). For pyramiding all these traits in one cultivar, DNA markers can be used for monitoring the introgression of these traits. Likewise, wheat hybrid breeding can also enhance wheat production in the future.
Eradication of malnutrition by quality improvement
Micronutrient deficiency is also a major challenge, almost three billion people are affected by these deficiencies globally (Welch and Graham, 2004). Children and females in developing countries are more prone to Zinc (17%) and Iron (33%) deficiencies worldwide (Wessells and Brown, 2012; Kassebaum et al., 2014). To address micronutrient deficiencies, wheat is the best candidate crop as it is consumed by a large population globally. It has been reported that Zn and Fe concentration is relatively high in closely related wild wheat species (Çakmak et al., 2004). Provitamin content has been increased by expressing bacterial CrtB and CrtI gene through transgenic methods. Similarly, protein content has been enhanced by expressing Amaranthus albumin gene and Fe content by the soybean ferritin gene in wheat (Cong et al., 2009; Wang C. et al., 2014). However, varieties expressing high-quality traits have relatively low yield potential. For wider acceptability, support price (premium) for such varieties should be announced by the regulators for encouraging their cultivation. Alternatively, some transgenic wheat lines expressing high Zn and Fe contents should be allowed for cultivation in restricted parts of wheat-growing countries that can be mixed in flour of non-transgenic wheat varieties.
Enhancing resilience to stresses
Development of varieties having high yield potential and resilience to stresses is the need of the hour (Juliana et al., 2019). Wild relative of wheat such as Aegilops tauschii (DD) is a good source for climate resilience because it can easily be crossed with durum (AABB) or bread wheat (AABBDD) to generate synthetic wheat (Elbashir et al., 2017). Hybrid wheat is another promising approach as it has higher yield stability and tolerance to stresses. Hybrids in wheat have been produced with resistance to Fusarium head blight, frost resistant, leaf rust resistance and, Septoria tritici blotch resistance showing hybrid wheat potential with context to climate change (Longin et al., 2013; Miedaner et al., 2017).
Conclusion
Wheat is one of the ancient crops and it is the crop of the future. Since its domestication, breeders and farmers have modified wheat continuously through selections and by incorporating different genes for short plant height, and biotic and abiotic stresses. Despite the extensive research, still there is a gap between the total wheat production and consumption, particularly in developing countries. The current rate of genetic gain is alarming which would not help in meeting the food demand of the growing human population in 2050. Changes in climatic conditions may further worsen the situation by inviting new pests and diseases, reducing yield due to terminal heat, and altered rainfall patterns may reduce the cultivated area of wheat in several countries. Under such circumstances, genes conferring resilience to rust diseases, terminal heat, drought, and salinity are required to be introduced into wheat cultivars. For increasing yield potential by 30%, it is extremely important to find new genetic solutions for tackling the issue of male sterility and restoration in hybrid wheat. For example, the adoption of new technologies including high throughput phenotyping, gene editing, speed breeding, molecular breeding, and selection strategies can accelerate the magnitude of genetic gains of the newly developed varieties. Thus, wheat production can be sustained beyond 2050.
Author contributions
SG and SZ prepared a preliminary draft; MS edited gave critical inputs; JL helped in editing the MS; JW conceived and edited the MS. MR prepared the outlines and edited extensively primary and subsequent draft of the article; All authors read and approved the article.
Funding
This work was supported by the Shandong Provincial Natural Science Foundation project (ZR2021ZD31).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aaronsohn, A. (1910). Agricultural and botanical explorations in Palestine. Washington, DC: US Government Printing Office.
Abe, F., Haque, E., Hisano, H., Tanaka, T., Kamiya, Y., Mikami, M., et al. (2019). Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell. Rep. 28, 1362–1369. doi:10.1016/j.celrep.2019.06.090
Abouseadaa, H. H., Osman, G. H., Ramadan, A. M., Hassanein, S. E., Abdelsattar, M. T., Morsy, Y. B., et al. (2015). Development of transgenic wheat (Triticum aestivum L.) expressing avidin gene conferring resistance to stored product insects. BMC Plant Biol. 15, 183–188. doi:10.1186/s12870-015-0570-x
Acevedo‐Garcia, J., Spencer, D., Thieron, H., Reinstädler, A., Hammond‐Kosack, K., Phillips, A. L., et al. (2017). mlo‐based powdery mildew resistance in hexaploid bread wheat generated by a non‐transgenic TILLING approach. Plant Biotechnol. J. 15, 367–378. doi:10.1111/pbi.12631
Al-Tamimi, N., Brien, C., Oakey, H., Berger, B., Saade, S., Ho, Y. S., et al. (2016). Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat. Commun. 7 (1), 1–11. doi:10.1038/ncomms13342
Alahmad, S., Dinglasan, E., Leung, K. M., Riaz, A., Derbal, N., Voss-Fels, K. P., et al. (2018). Speed breeding for multiple quantitative traits in durum wheat. Plant methods 14 (1), 36–15. doi:10.1186/s13007-018-0302-y
Albokari, M. (2014). Induction of mutants in durum wheat (Triticum durum desf cv. samra) using gamma irradiation. Pak. J. Bot. 46, 317–324.
Allen, A. M., Barker, G. L., Berry, S. T., Coghill, J. A., Gwilliam, R., Kirby, S., et al. (2011). Transcript‐specific, single‐nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol. J. 9, 1086–1099. doi:10.1111/j.1467-7652.2011.00628.x
Allen, A. M., Winfield, M. O., Burridge, A. J., Downie, R. C., Benbow, H. R., Barker, G. L., et al. (2017). Characterization of a Wheat Breeders’ Array suitable for high‐throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol. J. 15, 390–401. doi:10.1111/pbi.12635
Ardalani, S., Mirzaghaderi, G., and Badakhshan, H. (2016). A Robertsonian translocation from Thinopyrum bessarabicum into bread wheat confers high iron and zinc contents. Plant Breed. 135, 286–290. doi:10.1111/pbr.12359
Arruda, M. P., Brown, P. J., Lipka, A. E., Krill, A. M., Thurber, C., and Kolb, F. L. (2015). Genomic selection for predicting Fusarium head blight resistance in a wheat breeding program. Plant Genome 8. doi:10.3835/plantgenome2015.01.0003
Aury, J.-M., Engelen, S., Istace, B., Monat, C., Lasserre-Zuber, P., Belser, C., et al. (2022). Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding. GigaScience 11, giac034. doi:10.1093/gigascience/giac034
Azhar, M. T., Atif, R. M., Israr, M., Khan, A. I., Khalid, S., and Rana, I. A. (2021). A discussion on cotton transformation during the last decade (2010–2021); an update on present trends and future prospects. J. Cotton Res. 4 (1), 1–14.
Baenziger, P., Belamkar, V., Easterly, A., Garst, N., Stoll, H., Ibrahim, A., et al. (2019). Developing the tools for hybrid wheat: American perspective. EWG Breed. methods-workshop hybrid wheat held Febr. 19, 2019.
Baffes, J. (2005). The “cotton problem”. World Bank Res. Observer 20, 109–144. doi:10.1093/wbro/lki004
Bai, S., Yuan, F., Zhang, H., Zhang, Z., Zhao, J., Yang, Q., et al. (2020). Characterization of the wheat-psathyrostachys huashania Keng 2ns/2D substitution line H139: A novel germplasm with enhanced resistance to wheat take-all. Front. Plant Sci. 11, 233. doi:10.3389/fpls.2020.00233
Bi, H., Shi, J., Kovalchuk, N., Luang, S., Bazanova, N., Chirkova, L., et al. (2018). Overexpression of the TaSHN1 transcription factor in bread wheat leads to leaf surface modifications, improved drought tolerance, and no yield penalty under controlled growth conditions. Plant Cell. Environ. 41, 2549–2566. doi:10.1111/pce.13339
Biffen, R. H. (1905). Mendel's laws of inheritance and wheat breeding. J. Agric. Sci. 1, 4–48. doi:10.1017/s0021859600000137
Boeven, P. H., Longin, C. F. H., Leiser, W. L., Kollers, S., Ebmeyer, E., and Würschum, T. (2016). Genetic architecture of male floral traits required for hybrid wheat breeding. Theor. Appl. Genet. 129 (12), 2343–2357. doi:10.1007/s00122-016-2771-6
Brenchley, R., Spannagl, M., Pfeifer, M., Barker, G. L., D’Amore, R., Allen, A. M., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710. doi:10.1038/nature11650
Brini, F., and Masmoudi, K. (2014). “Biotechnology for drought and salinity tolerance of crops,” in Physiological mechanisms and adaptation strategies in plants under changing environment (Springer), 97–113.
Cainong, J. C., Bockus, W. W., Feng, Y., Chen, P., Qi, L., Sehgal, S. K., et al. (2015). Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor. Appl. Genet. 128, 1019–1027. doi:10.1007/s00122-015-2485-1
Çakmak, İ., Torun, A., Millet, E., Feldman, M., Fahima, T., Korol, A., et al. (2004). Triticum dicoccoides: An important genetic resource for increasing zinc and iron concentration in modern cultivated wheat. Soil Sci. plant Nutr. 50, 1047–1054. doi:10.1080/00380768.2004.10408573
Campbell, K. G., Bergman, C. J., Gualberto, D. G., Anderson, J. A., Giroux, M. J., Hareland, G., et al. (1999). Quantitative trait loci associated with kernel traits in a soft× hard wheat cross. Crop Sci. 39, 1184–1195. doi:10.2135/cropsci1999.0011183x003900040039x
Carter, A. H., Santra, D. K., and Kidwell, K. K. (2012). Assessment of the effects of the Gpc‐B1 allele on senescence rate, grain protein concentration and mineral content in hard red spring wheat (Triticum aestivum L.) from the Pacific Northwest Region of the USA. Plant Breed. 131 (1), 62–68. doi:10.1111/j.1439-0523.2011.01900.x
Cavanagh, C. R., Chao, S., Wang, S., Huang, B. E., Stephen, S., Kiani, S., et al. (2013). Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. U. S. A. 110, 8057–8062. doi:10.1073/pnas.1217133110
Chai, L., Chen, Z., Bian, R., Zhai, H., Cheng, X., Peng, H., et al. (2018). Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.). Theor. Appl. Genet. 131, 2621–2637. doi:10.1007/s00122-018-3177-4
Chao, S., Sharp, P., Worland, A., Warham, E., Koebner, R., and Gale, M. (1989). RFLP-based genetic maps of wheat homoeologous group 7 chromosomes. Theor. Appl. Genet. 78, 495–504. doi:10.1007/BF00290833
Charmet, G. (2011). Wheat domestication: Lessons for the future. C. R. Biol. 334, 212–220. doi:10.1016/j.crvi.2010.12.013
Chen, L., Huang, L., Min, D., Phillips, A., Wang, S., Madgwick, P. J., et al. (2012). Development and characterization of a new TILLING population of common bread wheat (Triticum aestivum L.). PLoS One 7, e41570. doi:10.1371/journal.pone.0041570
Clavijo, B. J., Venturini, L., Schudoma, C., Accinelli, G. G., Kaithakottil, G., Wright, J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27, 885–896. doi:10.1101/gr.217117.116
Cong, L., Wang, C., Chen, L., Liu, H., Yang, G., and He, G. (2009). Expression of phytoene synthase1 and carotene desaturase crtI genes result in an increase in the total carotenoids content in transgenic elite wheat (Triticum aestivum L.). J. Agric. Food Chem. 57, 8652–8660. doi:10.1021/jf9012218
Cooper, R. (2015). Re-discovering ancient wheat varieties as functional foods. J. Tradit. Complement. Med. 5, 138–143. doi:10.1016/j.jtcme.2015.02.004
Curtis, T., and Halford, N. (2014). Food security: The challenge of increasing wheat yield and the importance of not compromising food safety. Ann. Appl. Biol. 164, 354–372. doi:10.1111/aab.12108
Danilova, T. V., Friebe, B., Gill, B. S., Poland, J., and Jackson, E. (2018). Development of a complete set of wheat–barley group-7 Robertsonian translocation chromosomes conferring an increased content of β-glucan. Theor. Appl. Genet. 131, 377–388. doi:10.1007/s00122-017-3008-z
Danilova, T. V., Poland, J., and Friebe, B. (2019). Production of a complete set of wheat–barley group-7 chromosome recombinants with increased grain β-glucan content. Theor. Appl. Genet. 132, 3129–3141. doi:10.1007/s00122-019-03411-3
Danilova, T. V., Zhang, G., Liu, W., Friebe, B., and Gill, B. S. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of a wheat streak mosaic virus and Triticum mosaic virus resistance gene Wsm3 from Thinopyrum intermedium to wheat. Theor. Appl. Genet. 130, 549–556. doi:10.1007/s00122-016-2834-8
Davies, J., Berzonsky, W. A., and Leach, G. D. (2006). A comparison of marker-assisted and phenotypic selection for high grain protein content in spring wheat. Euphytica 152, 117–134. doi:10.1007/s10681-006-9185-5
de Oliveira Camargo, C. E., Neto, A. T., Ferreira Filho, A. W., and Felicio, J. C. (2000). Genetic control of aluminum tolerance in mutant lines of the wheat cultivar Anahuac. Euphytica 114, 47–53. doi:10.1023/a:1003993320432
Del Blanco, I., Frohberg, R., Stack, R., Berzonsky, W., and Kianian, S. (2003). Detection of QTL linked to Fusarium head blight resistance in Sumai 3-derived North Dakota bread wheat lines. Theor. Appl. Genet. 106, 1027–1031. doi:10.1007/s00122-002-1137-4
Dempewolf, H., Baute, G., Anderson, J., Kilian, B., Smith, C., and Guarino, L. (2017). Past and future use of wild relatives in crop breeding. Crop Sci. 57, 1070–1082. doi:10.2135/cropsci2016.10.0885
Dinglasan, E., Godwin, I. D., Mortlock, M. Y., and Hickey, L. T. (2016). Resistance to yellow spot in wheat grown under accelerated growth conditions. Euphytica 209, 693–707. doi:10.1007/s10681-016-1660-z
Dong, C., Dalton‐Morgan, J., Vincent, K., and Sharp, P. (2009). A modified TILLING method for wheat breeding. Plant Genome 2. doi:10.3835/plantgenome2008.10.0012
Dubcovsky, J., and Dvorak, J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866. doi:10.1126/science.1143986
Eagles, H. A., McLean, R., Eastwood, R. F., Appelbee, M. J., Cane, K., Martin, P. J., et al. (2014). High-yielding lines of wheat carrying Gpc-B1 adapted to Mediterranean-type environments of the south and west of Australia. Crop Pasture Sci. 65 (9), 854–861.
Elbashir, A. A. E., Gorafi, Y. S. A., Tahir, I. S. A., Kim, J.-S., and Tsujimoto, H. (2017). Wheat multiple synthetic derivatives: A new source for heat stress tolerance adaptive traits. Breed. Sci. 67, 248–256. doi:10.1270/jsbbs.16204
Eliazer Nelson, A. R. L., Ravichandran, K., and Antony, U. (2019). The impact of the Green Revolution on indigenous crops of India. J. Ethn. Food. 6, 8–10. doi:10.1186/s42779-019-0011-9
Eriksson, J., and Henning, E. J. (1896). Die getreideroste, ihre geschichte und natur sowie massregeln genen dieselben: Bericht über die am experimentalfelde der Kgl. schwedischen landbau-akademie in den jahren 1890-93 mit staatsunten stützung ausgeführte untersuchung. Stockholm: PA Norstedt & söner.
Fedak, G., Chi, D., Wolfe, D., Ouellet, T., Cao, W., Han, F., et al. (2021). Transfer of fusarium head blight resistance from Thinopyrum elongatum to bread wheat cultivar Chinese Spring. Genome 64, 997–1008. doi:10.1139/gen-2020-0151
Feldman, M., Lupton, F., and Miller, T. (1995). “Wheats,” in Evolution of crop plants. Editors J. Smartt, and N. w. Simmonds. 2nd (London: Longman Scientific), 184–192.
Figueroa, M., Hammond‐Kosack, K. E., and Solomon, P. S. (2018). A review of wheat diseases—A field perspective. Mol. Plant Pathol. 19, 1523–1536. doi:10.1111/mpp.12618
Freeman, G. F. (1919). The heredity of quantitative characters in wheat. Genetics 4, 1–93. doi:10.1093/genetics/4.1.1
Friebe, B., Jiang, J., Raupp, W., McIntosh, R., and Gill, B. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica 91, 59–87. doi:10.1007/bf00035277
Friebe, B., Kynast, R., Hatchett, J., Sears, R., Wilson, D., and Gill, B. (1999). Transfer of wheat-rye translocation chromosomes conferring resistance to hessian fly from bread wheat into durum wheat. Crop Sci. 39, 1692–1696. doi:10.2135/cropsci1999.3961692x
Gaju, O., Reynolds, M., Sparkes, D., and Foulkes, M. (2009). Relationships between large‐spike phenotype, grain number, and yield potential in spring wheat. Crop Sci. 49, 961–973. doi:10.2135/cropsci2008.05.0285
Galiba, G., Quarrie, S. A., Sutka, J., Morgounov, A., and Snape, J. W. (1995). RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor. Appl. Genet. 90, 1174–1179. doi:10.1007/BF00222940
Gao, H., Wang, Y., Xu, P., and Zhang, Z. (2018). Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 9, 997. doi:10.3389/fpls.2018.00997
Ghosh, S., Watson, A., Gonzalez-Navarro, O. E., Ramirez-Gonzalez, R. H., Yanes, L., Mendoza-Suárez, M., et al. (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 13 (12), 2944–2963. doi:10.1038/s41596-018-0072-z
Gill, B. S., Appels, R., Botha-Oberholster, A.-M., Buell, C. R., Bennetzen, J. L., Chalhoub, B., et al. (2004). A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics 168, 1087–1096. doi:10.1534/genetics.104.034769
Gowda, M., Longin, C. F. H., Lein, V., and Reif, J. C. (2012). Relevance of specific versus general combining ability in winter wheat. Crop Sci. 52, 2494–2500. doi:10.2135/cropsci2012.04.0245
Guo, H., Yan, Z., Li, X., Xie, Y., Xiong, H., Liu, Y., et al. (2017). Development of a high-efficient mutation resource with phenotypic variation in hexaploid winter wheat and identification of novel alleles in the TaAGP. L-B1 gene. Front. Plant Sci. 8, 1404. doi:10.3389/fpls.2017.01404
Gupta, P., Varshney, R., Sharma, P., and Ramesh, B. (1999). ReviewMolecular markers and their applications in wheat breeding. Plant Breed. 118, 369–390. doi:10.1046/j.1439-0523.1999.00401.x
Hao, M., Liu, M., Luo, J., Fan, C., Yi, Y., Zhang, L., et al. (2018). Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat. Front. Plant Sci. 9, 1040. doi:10.3389/fpls.2018.01040
Henikoff, S., and Comai, L. (2003). Single-nucleotide mutations for plant functional genomics. Annu. Rev. Plant Biol. 54, 375–401. doi:10.1146/annurev.arplant.54.031902.135009
Hickey, L. T., Dieters, M. J., DeLacy, I. H., Christopher, M. J., Kravchuk, O. Y., and Banks, P. M. (2010). Screening for grain dormancy in segregating generations of dormant × non-dormant crosses in white-grained wheat (Triticum aestivum L.). Euphytica 172, 183–195. doi:10.1007/s10681-009-0028-z
Hickey, L. T., Dieters, M. J., DeLacy, I. H., Kravchuk, O. Y., Mares, D. J., and Banks, P. M. (2009). Grain dormancy in fixed lines of white-grained wheat (Triticum aestivum L.) grown under controlled environmental conditions. Euphytica 168, 303–310. doi:10.1007/s10681-009-9929-0
Hickey, L. T., Lawson, W., Platz, G. J., Dieters, M., Arief, V. N., German, S., et al. (2011). Mapping Rph20: A gene conferring adult plant resistance to Puccinia hordei in barley. Theor. Appl. Genet. 123, 55–68. doi:10.1007/s00122-011-1566-z
Hu, X. J., Chen, D., Lynne Mclntyre, C., Fernanda Dreccer, M., Zhang, Z. B., Drenth, J., et al. (2018). Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA‐mediated regulatory pathway. Plant Cell. Environ. 41, 79–98. doi:10.1111/pce.12957
Hu, Y., Knapp, S., and Schmidhalter, U. (2020). Advancing high-throughput phenotyping of wheat in early selection cycles. Remote Sens. 12, 574. doi:10.3390/rs12030574
Hussain, M., Gul, M., Kamal, R., Iqbal, M. A., Zulfiqar, S., Abbas, A., et al. (2021). Prospects of developing novel genetic resources by chemical and physical mutagenesis to enlarge the genetic window in bread wheat varieties. Agriculture 11, 621. doi:10.3390/agriculture11070621
Hussain, M., Iqbal, M. A., Till, B. J., and Rahman, M.-u.-. (2018). Identification of induced mutations in hexaploid wheat genome using exome capture assay. PLoS One 13, e0201918. doi:10.1371/journal.pone.0201918
Jang, J. H., Jung, W. J., Kim, D. Y., and Seo, Y. W. (2017). cDNA-AFLP analysis of 1BL. 1RS under water-deficit stress and development of wheat-rye translocation-specific markers. N. Z. J. Crop Hortic. Sci. 45, 150–164. doi:10.1080/01140671.2016.1269018
Jankowicz-Cieslak, J., Tai, T. H., Kumlehn, J., and Till, B. J. (2017). Biotechnologies for plant mutation breeding: Protocols. Springer Nature.
Jiang, Y., Schmidt, R. H., Zhao, Y., and Reif, J. C. (2017). A quantitative genetic framework highlights the role of epistatic effects for grain-yield heterosis in bread wheat. Nat. Genet. 49, 1741–1746. doi:10.1038/ng.3974
Jin, J., Duan, S., Qi, Y., Yan, S., Li, W., Li, B., et al. (2020). Identification of a novel genomic region associated with resistance to Fusarium crown rot in wheat. Theor. Appl. Genet. 133 (7), 2063–2073. doi:10.1007/s00122-020-03577-1
Johansson, E., Henriksson, T., Prieto-Linde, M. L., Andersson, S., Ashraf, R., and Rahmatov, M. (2020). Diverse wheat-alien introgression lines as a basis for durable resistance and quality characteristics in bread wheat. Front. Plant Sci. 11, 1067. doi:10.3389/fpls.2020.01067
Joshi, A., Chand, R., Arun, B., Singh, R., and Ortiz, R. (2007). Breeding crops for reduced-tillage management in the intensive, rice–wheat systems of South Asia. Euphytica 153, 135–151. doi:10.1007/s10681-006-9249-6
Juliana, P., Poland, J., Huerta-Espino, J., Shrestha, S., Crossa, J., Crespo-Herrera, L., et al. (2019). Improving grain yield, stress resilience and quality of bread wheat using large-scale genomics. Nat. Genet. 51, 1530–1539. doi:10.1038/s41588-019-0496-6
Kassebaum, N. J., Jasrasaria, R., Naghavi, M., Wulf, S. K., Johns, N., Lozano, R., et al. (2014). A systematic analysis of global anemia burden from 1990 to 2010. Blood 123, 615–624. doi:10.1182/blood-2013-06-508325
Keller, M., Karutz, C., Schmid, J., Stamp, P., Winzeler, M., Keller, B., et al. (1999a). Quantitative trait loci for lodging resistance in a segregating wheat× spelt population. Theor. Appl. Genet. 98, 1171–1182. doi:10.1007/s001220051182
Keller, M., Keller, B., Schachermayr, G., Winzeler, M., Schmid, J., Stamp, P., et al. (1999b). Quantitative trait loci for resistance against powdery mildew in a segregating wheat× spelt population. Theor. Appl. Genet. 98, 903–912. doi:10.1007/s001220051149
Khan, M. H., Bukhari, A., Dar, Z. A., and Rizvi, S. M. (2013). Status and strategies in breeding for rust resistance in wheat. Agric. Sci. 4, 292–301. doi:10.4236/as.2013.46042
Khush, G. S. (1999). Green revolution: Preparing for the 21st century. Genome 42, 646–655. doi:10.1139/g99-044
Kihara, H. (1944). Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric. Hort. 19, 13–14.
Kihara, H. (1951). Substitution of nucleus and its effects on genome manifestations. Cytologia 16, 177–193. doi:10.1508/cytologia.16.177
King, J., Grewal, S., Othmeni, M., Coombes, B., Yang, C.-y., Walter, N., et al. (2022). Introgression of the Triticum timopheevii genome into wheat detected by chromosome-specific kompetitive allele specific PCR markers. Front. Plant Sci. 13, 919519. doi:10.3389/fpls.2022.919519
Knott, D. (1988). “Using polygenic resistance to breed for stem rust resistance in wheat,” in Breeding strategies for resistance to the rusts of wheat (Mexico: El Batan). Chapter 4.
Kottearachchi, N., Uchino, N., Kato, K., and Miura, H. (2006). Increased grain dormancy in white-grained wheat by introgression of preharvest sprouting tolerance QTLs. Euphytica 152, 421–428. doi:10.1007/s10681-006-9231-3
Krishnan, A., Guiderdoni, E., An, G., Hsing, Y.-i. C., Han, C.-d., Lee, M. C., et al. (2009). Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 149, 165–170. doi:10.1104/pp.108.128918
Kumar, J., Jaiswal, V., Kumar, A., Kumar, N., Mir, R. R., Kumar, S., et al. (2011). Introgression of a major gene for high grain protein content in some Indian bread wheat cultivars. Field Crops Res. 123 (3), 226–233. doi:10.1016/j.fcr.2011.05.013
Lagudah, E., Appels, R., Brown, A., and McNeil, D. (1991). The molecular–genetic analysis of Triticum tauschii, the D-genome donor to hexaploid wheat. Genome 34, 375–386. doi:10.1139/g91-059
Li, H., Dong, Z., Ma, C., Tian, X., Qi, Z., Wu, N., et al. (2019). Physical mapping of stem rust resistance gene Sr52 from Dasypyrum villosum based on ph1b-induced homoeologous recombination. Int. J. Mol. Sci. 20, 4887. doi:10.3390/ijms20194887
Li, J., Wang, Z., He, G., Ma, L., and Deng, X. W. (2020). CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J. Genet. Genomics 47 (5), 263–272. doi:10.1016/j.jgg.2020.05.004
Li, L., Zheng, M., Deng, G., Liang, J., Zhang, H., Pan, Z., et al. (2016). Overexpression of AtHDG11 enhanced drought tolerance in wheat (Triticum aestivum L.). Mol. Breed. 36, 23. doi:10.1007/s11032-016-0447-1
Li, S., Lin, D., Zhang, Y., Deng, M., Chen, Y., Lv, B., et al. (2022). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460. doi:10.1038/s41586-022-04395-9
Li, W., Zhang, P., Fellers, J. P., Friebe, B., and Gill, B. S. (2004). Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 40, 500–511. doi:10.1111/j.1365-313X.2004.02228.x
Liu, H., Tang, H., Ding, P., Mu, Y., Habib, A., Liu, Y., et al. (2020a). Effects of the 1BL/1RS translocation on 24 traits in a recombinant inbred line population. Cereal Res. Commun. 48, 225–232. doi:10.1007/s42976-020-00027-y
Liu, H., Wang, K., Jia, Z., Gong, Q., Lin, Z., Du, L., et al. (2020b). Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 71, 1337–1349. doi:10.1093/jxb/erz529
Liu, W., Koo, D.-H., Xia, Q., Li, C., Bai, F., Song, Y., et al. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 130, 841–848. doi:10.1007/s00122-017-2855-y
Longin, C. F. H., Gowda, M., Mühleisen, J., Ebmeyer, E., Kazman, E., Schachschneider, R., et al. (2013). Hybrid wheat: Quantitative genetic parameters and consequences for the design of breeding programs. Theor. Appl. Genet. 126, 2791–2801. doi:10.1007/s00122-013-2172-z
Longin, C. F. H., Mi, X., Melchinger, A. E., Reif, J. C., and Würschum, T. (2014). Optimum allocation of test resources and comparison of breeding strategies for hybrid wheat. Theor. Appl. Genet. 127, 2117–2126. doi:10.1007/s00122-014-2365-0
Longin, C. F. H., Mi, X., and Würschum, T. (2015). Genomic selection in wheat: Optimum allocation of test resources and comparison of breeding strategies for line and hybrid breeding. Theor. Appl. Genet. 128, 1297–1306. doi:10.1007/s00122-015-2505-1
Lu, B. R., and Ellstrand, N. (2014). World food security and the tribe Triticeae (Poaceae): genetic resources of cultivated, wild, and weedy taxa for crop improvement, 52. Wiley Online Library, 661–666.
Lu, H., Rudd, J. C., Burd, J., and Weng, Y. (2010). Molecular mapping of greenbug resistance genes Gb2 and Gb6 in T1AL. 1RS wheat‐rye translocations. Plant Breed. 129, 472–476.
Luang, S., Sornaraj, P., Bazanova, N., Jia, W., Eini, O., Hussain, S. S., et al. (2018). The wheat TabZIP2 transcription factor is activated by the nutrient starvation-responsive SnRK3/CIPK protein kinase. Plant Mol. Biol. 96, 543–561. doi:10.1007/s11103-018-0713-1
Mago, R., Zhang, P., Vautrin, S., Šimková, H., Bansal, U., Luo, M.-C., et al. (2015). The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat. Plants 1, 1–13. doi:10.1038/nplants.2015.186
Malik, S., and Malik, T. A. (2015). Genetic mapping of potential QTLs associated with drought tolerance in wheat. JAPS J. Animal Plant Sci. 25 (4).
Marais, G., Badenhorst, P., Eksteen, A., and Pretorius, Z. (2010). Reduction of Aegilops sharonensis chromatin associated with resistance genes Lr56 and Yr38 in wheat. Euphytica 171, 15–22. doi:10.1007/s10681-009-9973-9
Marchal, C., Zhang, J., Zhang, P., Fenwick, P., Steuernagel, B., Adamski, N. M., et al. (2018). BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants 4, 662–668. doi:10.1038/s41477-018-0236-4
Marcotuli, I., Gadaleta, A., Mangini, G., Signorile, A. M., Zacheo, S. A., Blanco, A., et al. (2017). Development of a high-density SNP-based linkage map and detection of QTL for β-glucans, protein content, grain yield per spike and heading time in durum wheat. Int. J. Mol. Sci. 18, 1329. doi:10.3390/ijms18061329
Marza, F., Bai, G.-H., Carver, B., and Zhou, W.-C. (2006). Quantitative trait loci for yield and related traits in the wheat population Ning7840 x Clark. Theor. Appl. Genet. 112, 688–698. doi:10.1007/s00122-005-0172-3
Mastrangelo, A. M., and Cattivelli, L. (2021). What makes bread and durum wheat different? Crop Pasture Sci. 26, 677–684. doi:10.1071/cp14106
McFadden, E. S., and Sears, E. R. (1946). The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37, 81 107–116. doi:10.1093/oxfordjournals.jhered.a105590
Meuwissen, T. H., Hayes, B. J., and Goddard, M. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829. doi:10.1093/genetics/157.4.1819
Miedaner, T., Schulthess, A. W., Gowda, M., Reif, J. C., and Longin, C. F. H. (2017). High accuracy of predicting hybrid performance of Fusarium head blight resistance by mid-parent values in wheat. Theor. Appl. Genet. 130, 461–470. doi:10.1007/s00122-016-2826-8
Mishra, V. K., Gupta, P. K., Arun, B., Chand, R., Vasistha, N. K., Vishwakarma, M. K., and Joshi, A. K. (2015). Introgression of a gene for high grain protein content (Gpc-B1) into two leading cultivars of wheat in Eastern Gangetic Plains of India through marker assisted backcross breeding.
Molnar-Lang, M., Ceoloni, C., and Dolezel, J. (2016). Alien introgression in wheat cytogenetics, molecular biology, and genomics. Cereal Res. Commun. 44, 535–536.
Mühleisen, J., Piepho, H.-P., Maurer, H. P., Longin, C. F. H., and Reif, J. C. (2014). Yield stability of hybrids versus lines in wheat, barley, and triticale. Theor. Appl. Genet. 127, 309–316. doi:10.1007/s00122-013-2219-1
Mukhtar, S., Rahman, M., and Zafar, Y. (2002). Assessment of genetic diversity among wheat (Triticum aestivum L.) cultivars from a range of localities across Pakistan using random amplified polymorphic DNA (RAPD) analysis. Euphytica 128 (3), 417–425.
Murchie, E., Pinto, M., and Horton, P. (2009). Agriculture and the new challenges for photosynthesis research. New Phytol. 181, 532–552. doi:10.1111/j.1469-8137.2008.02705.x
Nesbitt, M., and Samuel, D. (1996). “From staple crop to extinction? The archaeology and history of the hulled wheat,” in Hulled Wheats, Promoting the conservation and used of underutilised and neglected crops. Editors S. Padulosi, K. Hammer, and J. Heller (Rome: IPGRI), 40–99.
Ni, F., Qi, J., Hao, Q., Lyu, B., Luo, M.-C., Wang, Y., et al. (2017). Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat. Commun. 8, 1–12. doi:10.1038/ncomms15121
Niu, Z., Klindworth, D. L., Friesen, T. L., Chao, S., Jin, Y., Cai, X., et al. (2011). Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187, 1011–1021. doi:10.1534/genetics.110.123588
Olmstead, A. L., and Rhode, P. W. (2002). The red queen and the hard reds: Productivity growth in American wheat, 1800–1940. J. Econ. Hist. 62, 929–966. doi:10.1017/s0022050702001602
Olson, E. L., Rouse, M. N., Pumphrey, M. O., Bowden, R. L., Gill, B. S., and Poland, J. A. (2013). Simultaneous transfer, introgression, and genomic localization of genes for resistance to stem rust race TTKSK (Ug99) from Aegilops tauschii to wheat. Theor. Appl. Genet. 126, 1179–1188. doi:10.1007/s00122-013-2045-5
Ortiz-Monasterio, J. I., Palacios-Rojas, N., Meng, E., Pixley, K., Trethowan, R., and Pena, R. (2007). Enhancing the mineral and vitamin content of wheat and maize through plant breeding. J. Cereal Sci. 46, 293–307. doi:10.1016/j.jcs.2007.06.005
Othmeni, M., Grewal, S., Walker, J., Yang, C.-y., King, I. P., and King, J. (2022). Assessing the potential of using the Langdon 5D (5B) substitution line for the introgression of Aegilops tauschii into durum wheat. Front. Plant Sci. 13, 927728. doi:10.3389/fpls.2022.927728
Parry, M. A., Reynolds, M., Salvucci, M. E., Raines, C., Andralojc, P. J., Zhu, X.-G., et al. (2011). Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J. Exp. Bot. 62, 453–467. doi:10.1093/jxb/erq304
Pearce, S. (2021). Towards the replacement of wheat ‘Green Revolution’genes. J. Exp. Bot. 72, 157–160. doi:10.1093/jxb/eraa494
Pellegrineschi, A., Reynolds, M., Pacheco, M., Brito, R. M., Almeraya, R., Yamaguchi-Shinozaki, K., et al. (2004). Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 47, 493–500. doi:10.1139/g03-140
Pickett, A. A. (1993). Hybrid wheat-results and problems. Germany: Fortschritte der Pflanzenzüchtung.
Poland, J. A., Endelman, J., Dawson, J., Rutkoski, J., Wu, S., Manes, Y., et al. (2012). Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 5, 113. doi:10.3835/plantgenome2012.06.0006
Pont, C., Leroy, T., Seidel, M., Tondelli, A., Duchemin, W., Armisen, D., et al. (2019). Tracing the ancestry of modern bread wheats. Nat. Genet. 51, 905–911. doi:10.1038/s41588-019-0393-z
Pourkheirandish, M., Dai, F., Sakuma, S., Kanamori, H., Distelfeld, A., Willcox, G., et al. (2018). On the origin of the non-brittle rachis trait of domesticated einkorn wheat. Front. Plant Sci. 8, 2031. doi:10.3389/fpls.2017.02031
Prasad, P., Pisipati, S., Momčilović, I., and Ristic, Z. (2011). Independent and combined effects of high temperature and drought stress during grain filling on plant yield and chloroplast EF‐Tu expression in spring wheat. J. Agron. Crop Sci. 197, 430–441. doi:10.1111/j.1439-037x.2011.00477.x
Przewieslik-Allen, A. M., Burridge, A. J., Wilkinson, P. A., Winfield, M. O., Shaw, D. S., McAusland, L., et al. (2019). Developing a high-throughput SNP-based marker system to facilitate the introgression of traits from Aegilops species into bread wheat (Triticum aestivum). Front. Plant Sci. 9, 1993. doi:10.3389/fpls.2018.01993
Qaseem, M. F., Qureshi, R., Shaheen, H., and Shafqat, N. (2019). Genome-wide association analyses for yield and yield-related traits in bread wheat (Triticum aestivum L.) under pre-anthesis combined heat and drought stress in field conditions. PLoS One 14 (3), e0213407. doi:10.1371/journal.pone.0213407
Rabinovich, S. V. (1998). Importance of wheat-rye translocations for breeding modern cultivar of Triticum aestivum L. Euphytica 100, 323–340. doi:10.1023/a:1018361819215
Rahman, M., Chowdhary, M. A., Iqbal, M. J., and Zafar, Y. (2004). Application of random amplified polymorphic DNA (RAPD) technique for the identification of markers linked to salinity tolerance in wheat (Triticum aestivum l.). Pak. J. Bot. 36 (3), 595–602.
Rahmatov, M., Rouse, M. N., Nirmala, J., Danilova, T., Friebe, B., Steffenson, B. J., et al. (2016). A new 2DS· 2RL Robertsonian translocation transfers stem rust resistance gene Sr59 into wheat. Theor. Appl. Genet. 129, 1383–1392. doi:10.1007/s00122-016-2710-6
Rai, N., Bellundagi, A., Kumar, P. K., Kalasapura Thimmappa, R., Rani, S., Sinha, N., et al. (2018). Marker‐assisted backcross breeding for improvement of drought tolerance in bread wheat (Triticum aestivum L. em Thell). Plant Breed. 137, 514–526. doi:10.1111/pbr.12605
Rajaram, S. (2001). “Prospects and promise of wheat breeding in the 21 st century,” in Wheat in a global environment (Springer), 37–52.
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PloS one 8, e66428. doi:10.1371/journal.pone.0066428
Razzaq, A., Zafar, M. M., Ali, A., Hafeez, A., Batool, W., Shi, Y., et al. (2021). Cotton germplasm improvement and progress in Pakistan. J. Cotton Res. 4 (1), 1–14. doi:10.1186/s42397-020-00077-x
Rebetzke, G. J., Verbyla, A. P., Verbyla, K. L., Morell, M. K., and Cavanagh, C. R. (2014). Use of a large multiparent wheat mapping population in genomic dissection of coleoptile and seedling growth. Plant Biotechnol. J. 12, 219–230. doi:10.1111/pbi.12130
Reitz, L., and Salmon, S. (1968). Origin, history, and use of Norin 10 wheat. Crop Sci. 8, 686–689. doi:10.2135/cropsci1968.0011183x000800060014x
Rembe, M., Zhao, Y., Jiang, Y., and Reif, J. C. (2019). Reciprocal recurrent genomic selection: An attractive tool to leverage hybrid wheat breeding. Theor. Appl. Genet. 132, 687–698. doi:10.1007/s00122-018-3244-x
Ren, T., Ren, Z., Yang, M., Yan, B., Tan, F., Fu, S., et al. (2018). Novel source of 1RS from Baili rye conferred high resistance to diseases and enhanced yield traits to common wheat. Mol. Breed. 38, 101–109. doi:10.1007/s11032-018-0856-4
Ren, T., Tang, Z., Fu, S., Yan, B., Tan, F., Ren, Z., et al. (2017). Molecular cytogenetic characterization of novel wheat-rye T1RS. 1BL translocation lines with high resistance to diseases and great agronomic traits. Front. Plant Sci. 8, 799. doi:10.3389/fpls.2017.00799
Rey, M.-D., Martín, A. C., Smedley, M., Hayta, S., Harwood, W., Shaw, P., et al. (2018). Magnesium increases homoeologous crossover frequency during meiosis in ZIP4 (Ph1 gene) mutant wheat-wild relative hybrids. Front. Plant Sci. 9, 509. doi:10.3389/fpls.2018.00509
Reynolds, M., Foulkes, M. J., Slafer, G. A., Berry, P., Parry, M. A., Snape, J. W., et al. (2009). Raising yield potential in wheat. J. Exp. Bot. 60, 1899–1918. doi:10.1093/jxb/erp016
Riaz, A., Periyannan, S., Aitken, E., and Hickey, L. (2016). A rapid phenotyping method for adult plant resistance to leaf rust in wheat. Plant Methods 12, 17. doi:10.1186/s13007-016-0117-7
Richard, C., Hickey, L., Fletcher, S., Chenu, K., Borrell, A., and Christopher, J. (2015). High-throughput phenotyping of wheat seminal root traits in a breeding context. Procedia Environ. Sci. 29, 102–103. doi:10.1016/j.proenv.2015.07.179
Ru, Z., Juhasz, A., Li, D., Deng, P., Zhao, J., Gao, L., et al. (2020). 1RS. 1BL molecular resolution provides novel contributions to wheat improvement. New York: bioRxiv.
Sheridan, C. (2021). Questions swirl around failures of disease-modifying Huntington’s drugs. Nat. Biotechnol. 39, 650. doi:10.1038/s41587-021-00955-y
Salvi, S., Porfiri, O., and Ceccarelli, S. (2013). Nazareno strampelli, the ‘prophet’of the green revolution. J. Agric. Sci. 151, 1–5. doi:10.1017/s0021859612000214
Sánchez‐León, S., Gil‐Humanes, J., Ozuna, C. V., Giménez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low‐gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. doi:10.1111/pbi.12837
Sandhu, K. S., Aoun, M., Morris, C. F., and Carter, A. H. (2021). Genomic selection for end-use quality and processing traits in soft white winter wheat breeding program with machine and deep learning models. Biology 10, 689. doi:10.3390/biology10070689
Sarkar, S., Islam, A. A., Barma, N., and Ahmed, J. (2021). Tolerance mechanisms for breeding wheat against heat stress: A review. South Afr. J. Bot. 138, 262–277. doi:10.1016/j.sajb.2021.01.003
Sato, K., Abe, F., Mascher, M., Haberer, G., Gundlach, H., Spannagl, M., et al. (2021). Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder. DNA Res. 28, dsab008. doi:10.1093/dnares/dsab008
Schneider, A., Rakszegi, M., Molnár-Láng, M., and Szakács, É. (2016). Production and cytomolecular identification of new wheat-perennial rye (Secale cereanum) disomic addition lines with yellow rust resistance (6R) and increased arabinoxylan and protein content (1R, 4R, 6R). Theor. Appl. Genet. 129, 1045–1059. doi:10.1007/s00122-016-2682-6
Senker, P. (2011). Foresight: The future of food and farming, final project report. Taylor & Francis.
Shafeeq, S., and Zafar, Y. (2006). Genetic variability of different wheat (Triticum aestivum L.) genotypes/cultivars under induced water stress. Pak. J. Bot. 38, 1671–1678.
Shewry Jones, P. R. (2005). Transgenic wheat: Where do we stand after the first 12 years? Ann. Appl. Biol. 147, 1–14. doi:10.1111/j.1744-7348.2005.00009.x
Singh, D., Wang, X., Kumar, U., Gao, L., Noor, M., Imtiaz, M., et al. (2019). High-throughput phenotyping enabled genetic dissection of crop lodging in wheat. Front. Plant Sci. 10, 394. doi:10.3389/fpls.2019.00394
Singh, N., and Balyan, H. (2009). Induced mutations in bread wheat (Triticum aestivum L.) CV.” Kharchia 65” for reduced plant height and improve grain quality traits. Adv. Biol. Res. 3, 215–221.
Singh, N., Shepherd, K., and McIntosh, R. (1990). Linkage mapping of genes for resistance to leaf, stem and stripe rusts and ω-secalins on the short arm of rye chromosome 1R. Theor. Appl. Genet. 80, 609–616. doi:10.1007/BF00224219
Singh, R., and McIntosh, R. (1984). Complementary genes for reaction to Puccinia recondita tritici in Triticum aestivum. I. Genetic and linkage studies. Can. J. Genet. Cytol. 26, 723–735. doi:10.1139/g84-115
Singh, S., Chatrath, R., and Mishra, B. (2010). Perspective of hybrid wheat research: A review. Indian J. Agric. Sci. 80, 1013–1027.
Sivamani, E., Brey, C. W., Dyer, W. E., Talbert, L. E., and Qu, R. (2000). Resistance to wheat streak mosaic virus in transgenic wheat expressing the viral replicase (NIb) gene. Mol. Breed. 6, 469–477. doi:10.1023/a:1026576124482
Slade, A. J., McGuire, C., Loeffler, D., Mullenberg, J., Skinner, W., Fazio, G., et al. (2012). Development of high amylose wheat through TILLING. BMC Plant Biol. 12, 69–17. doi:10.1186/1471-2229-12-69
Sun, C., Dong, Z., Zhao, L., Ren, Y., Zhang, N., and Chen, F. (2020). The Wheat 660K SNP array demonstrates great potential for marker‐assisted selection in polyploid wheat. Plant Biotechnol. J. 18, 1354–1360. doi:10.1111/pbi.13361
Tabbita, F., Lewis, S. M., Vouilloz, J. P., Ortega, M. D. L. Á. H., Kade, M., Abbate, P. E., et al. (2012). The effects of GpcB1 locus on high grain protein concentration introgressed into Argentinean wheat germplasm.
Tamás, C., Kisgyörgy, B. N., Rakszegi, M., Wilkinson, M. D., Yang, M.-S., Láng, L., et al. (2009). Transgenic approach to improve wheat (Triticum aestivum L.) nutritional quality. Plant Cell. Rep. 28, 1085–1094. doi:10.1007/s00299-009-0716-0
Tanaka, H., Nabeuchi, C., Kurogaki, M., Garg, M., Saito, M., Ishikawa, G., et al. (2017). A novel compensating wheat–Thinopyrum elongatum Robertsonian translocation line with a positive effect on flour quality. Breed. Sci. 67, 509–517. doi:10.1270/jsbbs.17058
The International Wheat Genome Sequencing Consortium (2014). International wheat genome sequencing Consortium.
Thorwarth, P., Piepho, H. P., Zhao, Y., Ebmeyer, E., Schacht, J., Schachschneider, R., et al. (2018). Higher grain yield and higher grain protein deviation underline the potential of hybrid wheat for a sustainable agriculture. Plant Breed. 137, 326–337. doi:10.1111/pbr.12588
Tschermak, V., and Von, E. (1914). Die Verwertung der Bastardierung für phylogenetische Fragen in der Getreidegruppe. Zeitschr. F. Pflanzenzüchtung. 2, 291–312.
Tucker, D., Griffey, C., Liu, S., and Saghai Maroof, M. (2006). Potential for effective marker‐assisted selection of three quantitative trait loci conferring adult plant resistance to powdery mildew in elite wheat breeding populations. Plant Breed. 125, 430–436. doi:10.1111/j.1439-0523.2006.01233.x
Türkösi, E., Darko, E., Rakszegi, M., Molnár, I., Molnár-Láng, M., and Cseh, A. (2018). Development of a new 7BS. 7HL winter wheat-winter barley robertsonian translocation line conferring increased salt tolerance and (1, 3; 1, 4)-β-D-glucan content. PLoS One 13, e0206248. doi:10.1371/journal.pone.0206248
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., and Dubcovsky, J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301. doi:10.1126/science.1133649
Vasil, V., Castillo, A. M., Fromm, M. E., and Vasil, I. K. (1992). Herbicide resistant fertile transgenic wheat plants obtained by microprojectile bombardment of regenerable embryogenic callus. Nat. Biotechnol. 10, 667–674. doi:10.1038/nbt0692-667
Vishwakarma, M. K., Mishra, V. K., Gupta, P. K., Yadav, P. S., Kumar, H., and Joshi, A. K. (2014). Introgression of the high grain protein gene Gpc-B1 in an elite wheat variety of Indo-Gangetic Plains through marker assisted backcross breeding. Curr. Plant Biol. 1, 60–67. doi:10.1016/j.cpb.2014.09.003
Volpato, L., Pinto, F., González-Pérez, L., Thompson, I. G., Borém, A., Reynolds, M., et al. (2021). High throughput field phenotyping for plant height using UAV-based RGB imagery in wheat breeding lines: Feasibility and validation. Front. Plant Sci. 12, 591587. doi:10.3389/fpls.2021.591587
Wang, C., Zeng, J., Li, Y., Hu, W., Chen, L., Miao, Y., et al. (2014a). Enrichment of provitamin A content in wheat (Triticum aestivum L.) by introduction of the bacterial carotenoid biosynthetic genes CrtB and CrtI. J. Exp. Bot. 65, 2545–2556. doi:10.1093/jxb/eru138
Wang, R., Liu, Y., Isham, K., Zhao, W., Wheeler, J., Klassen, N., et al. (2018). QTL identification and KASP marker development for productive tiller and fertile spikelet numbers in two high-yielding hard white spring wheat cultivars. Mol. Breed. 38, 135. doi:10.1007/s11032-018-0894-y
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014b). Characterization of polyploid wheat genomic diversity using a high‐density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi:10.1111/pbi.12183
Wang, Z., Li, J., Chen, S., Heng, Y., Chen, Z., Yang, J., et al. (2017). Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. U. S. A. 114, 12614–12619. doi:10.1073/pnas.1715570114
Wanjugi, H., Coleman-Derr, D., Huo, N., Kianian, S. F., Luo, M.-C., Wu, J., et al. (2009). Rapid development of PCR-based genome-specific repetitive DNA junction markers in wheat. Genome 52, 576–587. doi:10.1139/g09-033
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M.-D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29. doi:10.1038/s41477-017-0083-8
Watson, A., Hickey, L. T., Christopher, J., Rutkoski, J., Poland, J., and Hayes, B. J. (2019). Multivariate genomic selection and potential of rapid indirect selection with speed breeding in spring wheat. Crop Sci. 59 (5), 1945–1959. doi:10.2135/cropsci2018.12.0757
Welch, R. M., and Graham, R. D. (2004). Breeding for micronutrients in staple food crops from a human nutrition perspective. J. Exp. Bot. 55, 353–364. doi:10.1093/jxb/erh064
Wessells, K. R., and Brown, K. H. (2012). Estimating the global prevalence of zinc deficiency: Results based on zinc availability in national food supplies and the prevalence of stunting. PloS one 7, e50568. doi:10.1371/journal.pone.0050568
Wheeler, T., and Von Braun, J. (2013). Climate change impacts on global food security. Science 341, 508–513. doi:10.1126/science.1239402
Wilson, J. (1962). Male sterility interaction of the Triticum aestivum nucleus and Triticum timopheevi cytoplasm. Wheat Int. Serv. 14, 29–30.
Winfield, M. O., Allen, A. M., Burridge, A. J., Barker, G. L., Benbow, H. R., Wilkinson, P. A., et al. (2016). High‐density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 14, 1195–1206. doi:10.1111/pbi.12485
Wrigley, C. W., Robinson, P. J., and Williams, W. T. (1981). Association between electrophoretic patterns of gliadin proteins and quality characteristics of wheat cultivars. J. Sci. Food Agric. 32, 433–442. doi:10.1002/jsfa.2740320503
Würschum, T., Langer, S. M., Longin, C. F. H., Tucker, M. R., and Leiser, W. L. (2017). A modern Green Revolution gene for reduced height in wheat. Plant J. 92, 892–903. doi:10.1111/tpj.13726
Xue, G.-P., Drenth, J., and McIntyre, C. L. (2015). TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 66, 1025–1039. doi:10.1093/jxb/eru462
Xue, G.-P., Way, H. M., Richardson, T., Drenth, J., Joyce, P. A., and McIntyre, C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4, 697–712. doi:10.1093/mp/ssr013
Xue, Z.-Y., Zhi, D.-Y., Xue, G.-P., Zhang, H., Zhao, Y.-X., and Xia, G.-M. (2004). Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci. 167, 849–859. doi:10.1016/j.plantsci.2004.05.034
Yang, E., Li, G., Li, L., Zhang, Z., Yang, W., Peng, Y., et al. (2016). Characterization of stripe rust resistance genes in the wheat cultivar Chuanmai45. Int. J. Mol. Sci. 17, 601. doi:10.3390/ijms17040601
Ye, X., Li, J., Cheng, Y., Yao, F., Long, L., Wang, Y., et al. (2019). Genome-wide association study reveals new loci for yield-related traits in Sichuan wheat germplasm under stripe rust stress. BMC genomics 20 (1), 640. doi:10.1186/s12864-019-6005-6
Zaharieva, M., and Monneveux, P. (2014). Cultivated einkorn wheat (Triticum monococcum L. subsp. monococcum): The long life of a founder crop of agriculture. Genet. Resour. Crop Evol. 61, 677–706. doi:10.1007/s10722-014-0084-7
Zahra, S., Shaheen, T., Hussain, M., Zulfiqar, S., and Rahman, M.-u. (2021). Multivariate analysis of mutant wheat (Triticum aestivum L.) lines under drought stress. Turk. J. Agric. For. 45, 617–633. doi:10.3906/tar-2106-73
Zang, X., Geng, X., Wang, F., Liu, Z., Zhang, L., Zhao, Y., et al. (2017). Overexpression of wheat ferritin gene TaFER-5B enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging. BMC Plant Biol. 17, 14–13. doi:10.1186/s12870-016-0958-2
Zhang, J., Zhang, H., Botella, J. R., and Zhu, J. K. (2018a). Generation of new glutinous rice by CRISPR/Cas9‐targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 60, 369–375. doi:10.1111/jipb.12620
Zhang, W., Zhu, X., Zhang, M., Chao, S., Xu, S., and Cai, X. (2018b). Meiotic homoeologous recombination-based mapping of wheat chromosome 2B and its homoeologues in Aegilops speltoides and Thinopyrum elongatum. Theor. Appl. Genet. 131, 2381–2395. doi:10.1007/s00122-018-3160-0
Zhang, W., Zhu, X., Zhang, M., Shi, G., Liu, Z., and Cai, X. (2019). Chromosome engineering-mediated introgression and molecular mapping of novel Aegilops speltoides-derived resistance genes for tan spot and Septoria nodorum blotch diseases in wheat. Theor. Appl. Genet. 132, 2605–2614. doi:10.1007/s00122-019-03374-5
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi:10.1111/tpj.13599
Zhang, Y., Li, D., Zhang, D., Zhao, X., Cao, X., Dong, L., et al. (2018c). Analysis of the functions of Ta GW 2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. doi:10.1111/tpj.13903
Keywords: domestication, hybrid wheat, NGS, CRISPR, genomic selection (GS), climate change, food security
Citation: Gohar S, Sajjad M, Zulfiqar S, Liu J, Wu J and Rahman M-u- (2022) Domestication of newly evolved hexaploid wheat—A journey of wild grass to cultivated wheat. Front. Genet. 13:1022931. doi: 10.3389/fgene.2022.1022931
Received: 19 August 2022; Accepted: 12 September 2022;
Published: 03 October 2022.
Edited by:
Karansher Singh Sandhu, Bayer Crop Science (United States), United StatesReviewed by:
Dinesh Kumar Saini, South Dakota State University, United StatesNastaran Mehri, Ardebil Agriculture and Natural Resources Research Centere (AREEO), Iran
Copyright © 2022 Gohar, Sajjad, Zulfiqar, Liu, Wu and Rahman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiajie Wu, amlhamlld3VAc2RhdS5lZHUuY24=; Mehboob-ur- Rahman, bWVoYm9vYl9wYmRAeWFob28uY29t
 Sasha Gohar
Sasha Gohar Muhammad Sajjad
Muhammad Sajjad Sana Zulfiqar
Sana Zulfiqar Jiajun Liu4
Jiajun Liu4 Jiajie Wu
Jiajie Wu Mehboob-ur- Rahman
Mehboob-ur- Rahman