- 1State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai’an, Shandong, China
- 2College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an, Shandong, China
AGC protein kinases play important roles in regulating plant growth, immunity, and cell death. However, the function of AGC in Brassica rapa has not yet been clarified. In this study, 62 BrAGC genes were identified, and these genes were distributed on 10 chromosomes and divided into six subfamilies. Analysis of gene structure and conserved motifs showed that the activation segment of BrAGC genes was highly conserved, and genes of the same subfamily showed higher sequence and structural similarity. Collinearity analysis revealed that BrAGCs were more closely related to AtAGCs than to OsAGCs. Expression profile analysis revealed that BrAGCs were preferentially expressed in flowers and BrAGC26, BrAGC33, and BrAGC04 were preferentially expressed in the stigma; the expression of these genes was significantly upregulated after self-incompatibility pollination, and the expression of BrAGC13 and BrAGC32 was significantly upregulated after cross-pollination. In addition, several typical cis-elements involved in the stress response were identified in BrAGC promoters. The expression levels of BrAGC37 and BrAGC44 significantly varied under different types of abiotic stress. Collectively, we identified that BrAGC26, BrAGC33, and BrAGC44 have the greatest potential in regulating pollen-pistil interaction and abiotic stress tolerance, respectively. Our findings will aid future functional investigations of BrAGCs in B. rapa.
Introduction
Protein phosphorylation is a ubiquitous and significant post-translational modification that plays an important role in intracellular signal transduction. The activity of protein kinases is turned on or off by phosphorylation and dephosphorylation, and this mediates the responses to the internal and external environment. AGC protein kinases, which were described for the first time by Steven Hanks and Tony Hunter (Hanks and Hunter, 1995), are an important kinase family and have attracted much research attention. The activation of most AGC kinases involves the phosphorylation of two highly conserved motifs, the activation segment (T-loop) in the catalytic domain, and the hydrophobic motif located behind the catalytic domain (Pearce et al., 2010). In mammals and yeast, AGC protein kinase is an important downstream effector that senses intracellular second messengers such as cAMP, cGMP, phospholipids, and Ca2+, and it is activated by second messengers (Bögre et al., 2003). In plants, AGC protein kinases have been shown to be involved in regulating plant growth, immunity, and cell death. In 2003, 39 AGC protein kinases in Arabidopsis thaliana (AtAGC) were identified for the first time, and these were divided into six subfamilies: AGCVI, AGCVII, AGCVIIIa, AGCVIIIb, other AGCs, and PDK1 (Bögre et al., 2003).
Previous studies have shown that plant AGC protein kinases play an important role in responses to various environmental stresses and in regulating plant immunity. Concentrations of reactive oxygen species (ROS) increase when plants are exposed to stress. Oxidative signal-inducible kinase (OXI), a member of the AGC protein kinase family, plays a role in ROS-initiated downstream signaling when plants are exposed to stress. In A. thaliana, AtOXI1 mediates the transduction of oxidative stress signals under abiotic stress or during pathogen invasion as well as responses to stress (Rentel et al., 2004; Petersen et al., 2009). When plants are exposed to high-light stress, OXI can induce programmed cell death (PCD) to prevent damage induced by high light (Shumbe et al., 2016). In rice, ROS and chitin elicitors can induce the transient phosphorylation of OsOxi1, which plays a positive role in regulating plant disease resistance (Matsui et al., 2010). In tomato, AvrPto-dependent Pto-interacting protein 3 (Adi3) is a negative regulator of PCD caused by Pseudomonas syringae pv. Tomato (Pst), and it plays a role in regulating plant immunity (Devarenne et al., 2006). In wheat, TaAGC1 plays a positive regulatory role in host resistance by regulating the expression of ROS and resistance-related genes (Zhu et al., 2015). Previous studies have also shown that AGCs play a role in plant reproduction. In A. thaliana, AGC1.5 and AGC1.7 regulate the ROP/Rac small GTPase (ROP) signaling pathway through phosphorylation positive feedback to regulate the apical growth of pollen tubes (Zhang et al., 2009; Li et al., 2018). In addition, AGC protein kinases also regulate the process of plant growth and development by regulating auxin polar transport (Friml et al., 2004), sensing blue light signals, mediating plant phototropism, regulating the distribution of chloroplasts and stomatal opening and closing (Sakai et al., 2001; Inada et al., 2004), and modifying cell proliferation and embryonic development to regulate seed size (Zhang et al., 2020). The published plant AGCs are listed in Supplementary Table S1. These studies have shown that AGCs play multiple roles and regulate divergent biological processes in plants.
Chinese cabbage (Brassica rapa) is widely grown in Asia and is exposed to various types of stress during its growth. However, no studies to date have examined AGC protein kinases in Chinese cabbage. In this study, BrAGC genes were identified at the whole-genome level, and the physicochemical properties, structure of genes and proteins, and expression profiles were analyzed. The results of this study provide basic information that will aid future studies of BrAGC protein kinases.
Materials and methods
Identification of AGC gene family members in B. rapa
Whole-genome sequences, gff3 genome annotation data, and AGC amino acid (aa) sequences of B. rapa, A. thaliana, and Oryza sativa were downloaded from EnsemblPlants (http://plants.ensembl.org/). BrAGC genes were screened from the B. rapa genome using the BLASTP program with A. thaliana AGC (AtAGC) aa sequences as input to predict B. rapa AGC (BrAGC) genes. Pfam (http://pfam.xfam.org/) (Mistry et al., 2021) was used to predict the structure of the proteins of the predicted genes; candidate genes were screened based on the conserved domains, and BrAGC genes were determined. The physicochemical properties of BrAGC genes, including their isoelectric points (pI) and relative molecular masses, were analyzed using the ExPASy website (http://www.expasy.org/) (Artimo et al., 2012).
Phylogenetic tree construction, conserved motif, gene structure, and cis-element analysis
The aa sequences of BrAGCs, AtAGCs, and OsAGCs were aligned using the MUSCLE algorithm in MEGA X (Kumar et al., 2018), and an un-rooted phylogenetic tree was constructed using the maximum likelihood method in MEGA X with 1,000 bootstrap replicates. The aa sequences of the BrAGC genes were uploaded to the MEME online website (http://meme-suite.org/) (Bailey and Elkan, 1994), and the number of motifs was set to 10 to analyze the conserved motifs of BrAGC. The upstream 2000-bp sequence of each BrAGC gene was analyzed using the online PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Rombauts et al., 1999) with default parameters. BrAGC gene structure and conserved motifs were visualized using TBtools (Chen et al., 2020).
Chromosomal localization, gene replication, and collinearity analysis
The location of the BrAGC gene on each chromosome was extracted from the B. rapa gff3 genome annotation information using TBtools (Chen et al., 2020). The MCScanX (Multiple Collinearity Scan toolkit) plug-in in TBtools was used to analyze the relationships among duplicate genes within B. rapa and inter-species collinearity, and the Circos plug-in was used to visualize the results of the analysis. Non-synonymous substitutions (Ka) and synonymous substitutions (Ks) values were calculated for duplicated gene pairs using TBtools. Gene selection pressure analysis was performed using the Ka/Ks ratio. If Ka/Ks > 1, the gene has experienced positive selection; if Ka/Ks = 1, the gene has undergone neutral evolution; and if Ka/Ks < 1, the gene has experienced purifying selection (Hurst, 2002).
Protein–protein interaction networks and gene ontology analysis
STRING online website (http://cn.string-db.org) was used to predict protein–protein interaction (PPI) relationships with default parameters, and Cytoscape v3.9.1 was used to construct the interaction network (Shannon et al., 2003). Gene Ontology (GO) analysis of BrAGC genes with default parameters was conducted using the online website DAVID (http://david.ncifcrf.gov) (Dennis et al., 2003).
Plant material and stress treatments
The first-generation hybrid cultivar “848” of B. rapa with stable self-incompatibility was used for stress treatments. The plump seeds were sown in nursery pots filled with substrate and cultivated in a plant incubator (16-h light/8-h dark photoperiod at 25°C, light intensity 2000 lx), When the seedlings had six leaves, seedlings with similar growth status were subjected to stress treatments. The seedlings were placed in a hydroponic system with 150 mM NaCl to simulate salt stress and 15% PEG 6000 to simulate drought conditions. For the cold stress treatment, plants were exposed to 4°C. The duration of all stress treatments was 2, 4, 6, and 12 h. The stigma of ZY15 was pollinated using the pollen of ZY15 and 14CR, which corresponded to self-incompatibility (SI) and cross-pollination (CP), respectively. There were three biological replicates for each treatment group, and samples were stored at −80°C for subsequent RNA extraction.
Total RNA extraction and qRT-PCR
Total RNA was extracted using the SteadyPure Plant RNA Extraction Kit (Code No. AG21017, Accurate Biotechnology, China); samples were reverse-transcribed using TransScript® Uni All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (TransGen, AU341-02, Beijing, China) for qRT-PCR analysis. qRT-PCR was performed on a qTOWER3 qPCR machine (Analytikjena, Germany) using the ChamQ SYBR qPCR Master Mix (Q711-03, Vazyme, China), and BrActin2 was used as the reference gene, analysis of the relative expression levels of each gene was conducted using the 2−ΔΔCt method. The specific primer sequences are listed in Supplementary Table S2.
Expression analysis
The transcriptome sequences of B. rapa of different tissues from NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) (Barrett and Edgar, 2006) with the accession number GSE43245 (Tong et al., 2013) were downloaded, and the data were normalized using the transcripts per million (TPM) method. The unpollinated stigmas and stigmas at 5, 10, and 20 min after SI and CP of B. rapa were collected and stored in liquid nitrogen. Three biological replicates were collected for each sample. The samples were subjected to transcriptome sequencing on the Illumina NovaSeq 6000 platform (unpublished) by BioMarker Technologies (Beijing, China). The number of mapped reads and transcript lengths in the samples were normalized after sequencing. The abundance of transcripts was measured using fragments per kilobase of transcript per million fragments mapped (Trapnell et al., 2010). Gene expression profile heatmaps were built using TBtools (Chen et al., 2020).
Result
Genome-wide identification and physicochemical characterization of B. rapa AGC genes
A total of 62 BrAGC genes were screened from the B. rapa genome using the BLAST program with the AtAGC gene sequences as input. The genes were renamed as BrAGC01–BrAGC62 and divided into six subfamilies. The physicochemical properties of AtAGC genes and BrAGC genes were analyzed and listed in Table 1; the lengths of the BrAGC proteins ranged from 76 aa (BrAGC59 and BrAGC61) to 1281 aa (BrAGC27), the molecular weight ranged from 8.30 kDa (BrAGC59) to 141.65 kDa (BrAGC27), and the isoelectric point (pI) ranged from 4.85 (BrAGC61) to 9.52 (BrAGC28 and BrAGC62).
Chromosomal distribution, gene duplication, and phylogenetic analysis
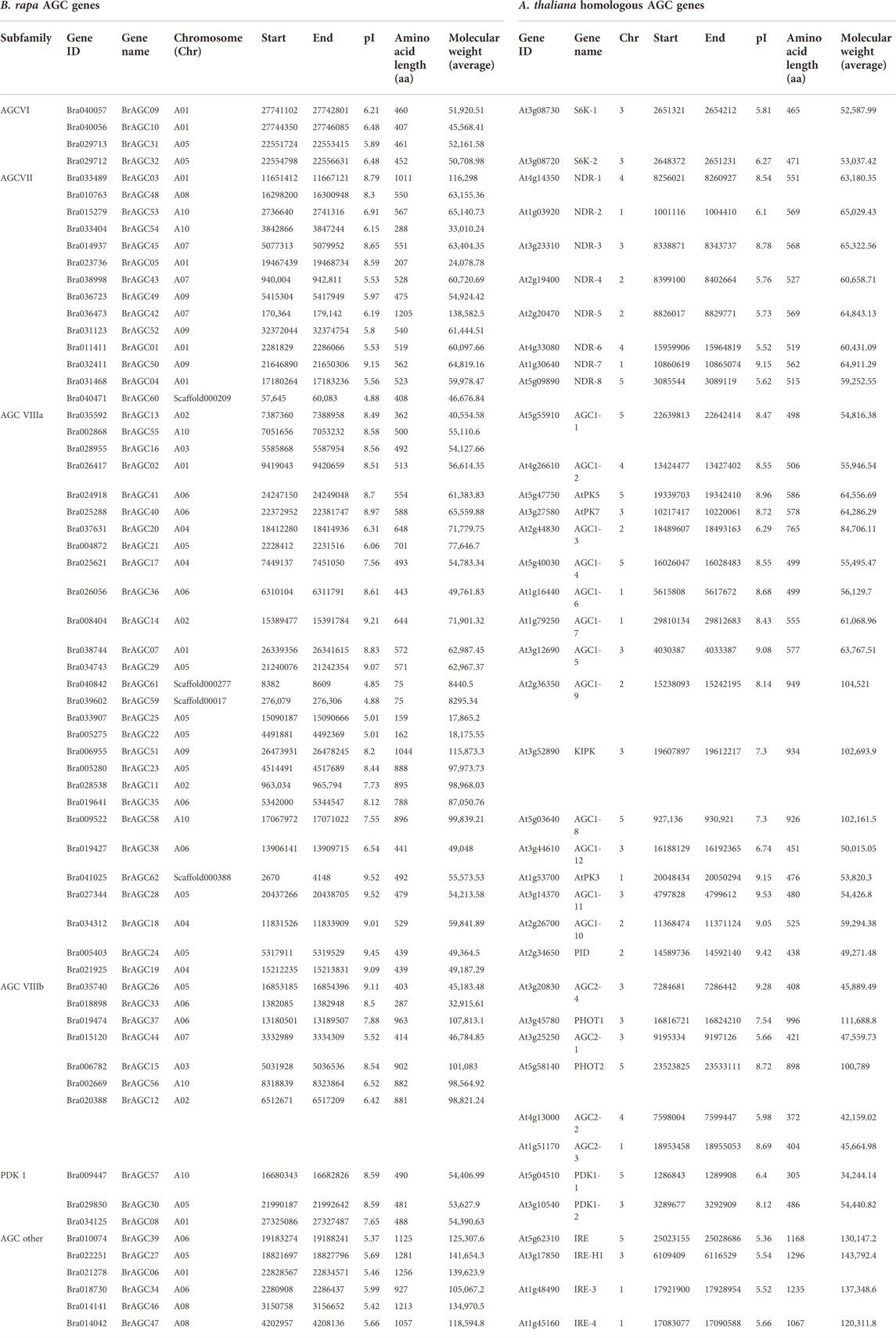
According to the gff3 genome annotations, 58 BrAGCs were mapped to the chromosomes of B. rapa, and the remaining four genes were mapped to the chromosome scaffold (Figure 1A). Among them, 12 BrAGCs were found on chromosome A05, accounting for 19% of all BrAGC genes. This was followed by A01 (10 genes); A06 (9 genes); A10 (6 genes); A02, A04, A07, and A09 (4 genes); A08 (3 genes); and A03 (2 genes). Gene duplication is considered a major cause of gene family expansion in the genome (Cannon et al., 2004). To clarify the associations among BrAGCs, we performed intraspecific collinearity analysis and detected two tandem duplications and 17 chromosome segmental duplications in BrAGCs (Figure 1A; Supplementary Table S3). The number of segmental duplications was 8.5 times the number of tandem duplications; this might be a major reason for the expansion of BrAGC genes in the B. rapa genome. We analyzed selection on gene pairs in BrAGC using the Ka/Ks ratio. Ka was much less than Ks (Ka/Ks < 1) for all tandem repeat gene pairs and segmentally duplicated gene pairs; this indicates that each replicated gene pair has experienced strong purifying selection; the Ka/Ks ratio was highest for BrAGC34/BrAGC46 (0.61) (Supplementary Table S3).
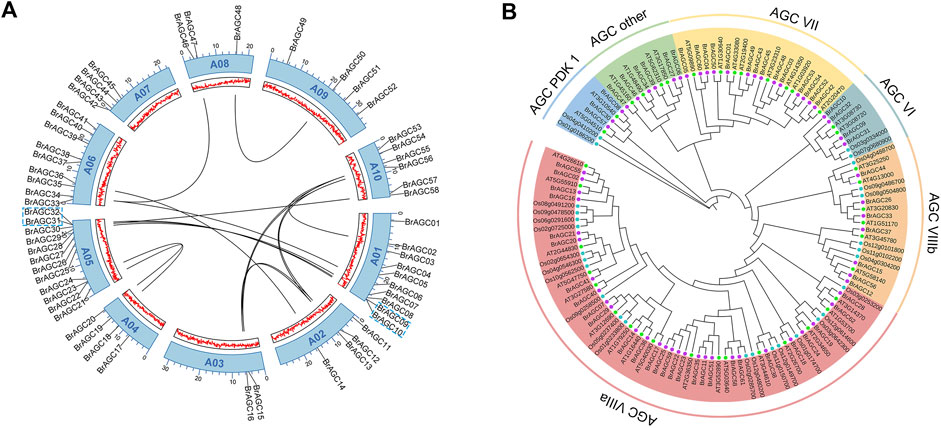
FIGURE 1. Chromosome localization and phylogenetic analysis of BrAGC genes (A) Gene distribution and duplication events of BrAGC genes. The gray line is the segmentally duplicated gene pair, and the tandemly duplicated gene is indicated by blue dotted lines. (B) Phylogenetic tree from the maximum likelihood analysis of the AGC gene family in B. rapa, A. thaliana, and O. sativa.
To further clarify the evolutionary relationships among BrAGC genes, we performed phylogenetic analysis on the AGC genes in B. rapa, A. thaliana, and O. sativa and constructed an ML tree (Figure 1B). We divided BrAGC genes into six subfamilies: AGCVI, AGCVII, AGCVIIIa, AGCVIIIb, other AGCs, and AGCPDK1. The largest and smallest subfamilies were AGCVIIIa and AGCPDK1, which contained 28 and three genes, respectively.
Gene structure and conserved motif analysis
To clarify the composition and function of BrAGC genes, we visualized and analyzed the exon–intron structure of BrAGC genes (Figure 2A). The length and distribution of exons and introns of BrAGC genes within subfamilies were similar, and the degree of similarity was positively correlated with clustering relationships. However, the structure of the coding regions of BrAGC genes varied among subfamilies. Among all BrAGC genes, 96% of genes in the AGCVIIIa subfamily and BrAGC44, BrAGC26, and BrAGC33 in the AGCVIIIb subfamily lack or contain only 1 to 2 introns, whereas the remaining genes have more introns (e.g., up to 25 in BrAGC03).
FIGURE 2. Gene structure and conserved motifs of BrAGC genes. (A) Composition and distribution of conserved motifs of BrAGC genes. (B) Exon–intron structure of BrAGC genes. Green box represents exons, and lines connecting exons are introns. (C) Ten conserved motif sequences of BrAGC genes. (D) Motif logo of motif2, motif3, and motif5.
We predicted 10 conserved motifs of BrAGC genes (Figures 2B,C). The number and distribution of motifs of different genes varied and ranged from 1 to 10. Motif6, 9, 4, 7, 2, 5, 1, 8, and 3 are highly conserved in BrAGC genes and exist in 77.4% of BrAGC genes in the order of 5′–3′. Motif2 contains the Mg2+-binding loop, and motif5 is located at its C-terminal and contains a T-loop and P+1 loop (Figure 2D), which constitutes the activation segment of the AGC protein kinase. These two motifs are highly conserved in BrAGCs, with 88.7% and 93.5% of the genes containing motif2 and motif5, respectively. In addition, motif3 contains the hydrophobic motif FxxxW, which is located at the C-terminus of the BrAGC genes (Figure 2D), and it is the most widely distributed motif. In AGCVII, 64.3% of the genes contain all 10 motifs, and motif10 only exists in this subfamily. We speculated that motif10 might contribute to the functional diversity of the AGCVII subfamily. The distribution of motifs of genes in the same subfamily gene was similar, and the degree of similarity was positively correlated with clustering relationships. These results are consistent with the findings of the gene structure analysis and confirm the subfamily division of BrAGC genes.
Gene ontology analysis
We performed GO analysis on BrAGC genes to determine their functions (Supplementary Table S4; Figure 3). GO is divided into three parts: biological process, molecular function, and cellular component. BrAGC genes is mainly involved in phosphorylation-related biological processes, with a total of 38 genes enriched in phosphorylation, followed by 16 genes enriched in intracellular signal transduction, 4 genes enriched in response to stimulus, and 4 genes enriched in protein-chromophore linkage. In the molecular function category, 38 genes were enriched in protein serine/threonine kinase activity, 36 genes were enriched in ATP binding, and 4 genes were enriched in photoreceptor activity.
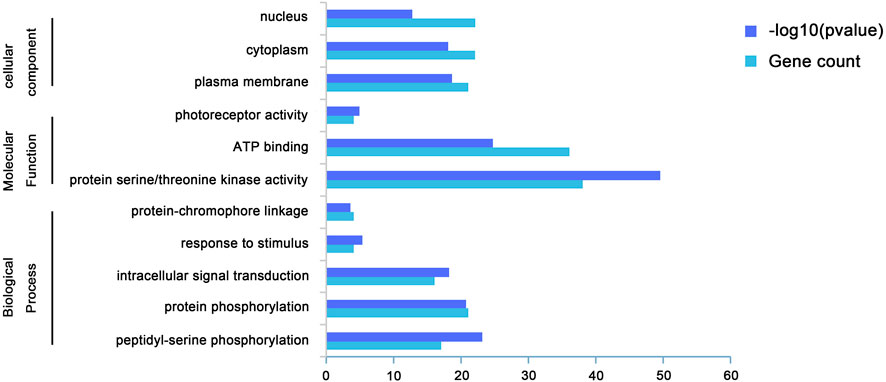
FIGURE 3. Gene Ontology of BrAGC genes in molecular function, biological process, and cellular component categories.
Collinearity analysis of B. rapa AGC
To further clarify the origin and evolution of BrAGC genes, comparative syntenic relationships between B. rapa and two typical model plants, A. thaliana and O. sativa, were analyzed (Figure 4). There were 76 collinear pairs of BrAGC genes and AtAGC genes. There were only eight collinear pairs of BrAGC genes and OsAGC genes. These findings indicate that B. rapa and A. thaliana are closely related and functionally similar.
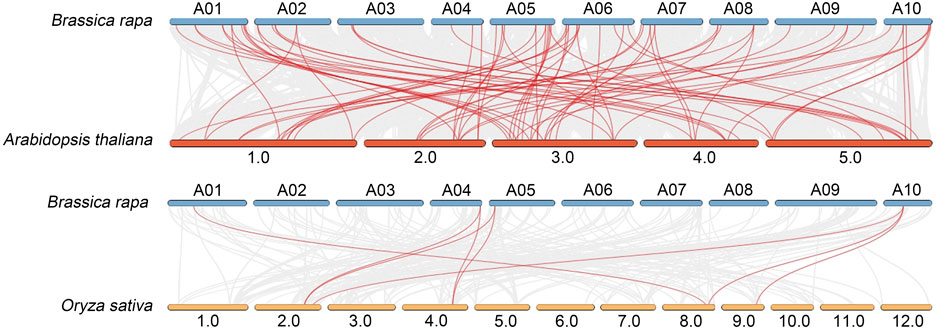
FIGURE 4. Collinearity analysis of B. rapa, A. thaliana, and O. sativa AGC genes. Homologous gene pairs of AGC protein kinases between species are linked by red lines.
Cis-elements in promoters
The transcription of genes is mainly controlled by the recognition and binding of the DNA sequence motif of the cis-regulatory region by transcription factors (TFs), which activate or inhibit transcription to mediate the response to changes in the external environment (Qiu, 2003). To explore the mechanism by which BrAGC genes mediate the response to abiotic stress, the 2,000-bp upstream region of the coding sequences of BrAGC genes was used for cis-element prediction. A total of 1,532 cis-elements were predicted in the promoter region of BrAGC genes (Figure 5), and these were involved in growth and development response, phytohormone response, and stress response. There were 752 light-responsive elements in 62 BrAGC genes, and these were the most abundant cis-elements. In addition, the promoter region of BrAGC genes contains several phytohormone response elements; 79% of BrAGC genes contain abscisic acid-responsive elements (ABRE, total of 154); 60% of BrAGC genes contain MeJA-responsive elements (CGTCA-motif and TGACG-motif, total of 146); and 85% of BrAGC genes contain anaerobic induction elements (ARE, total of 141). The above data indicate that these elements might be involved in abiotic stress and the hormone response. Differences in the cis-elements of genes indicate that BrAGC genes might play different roles in plant growth and development.
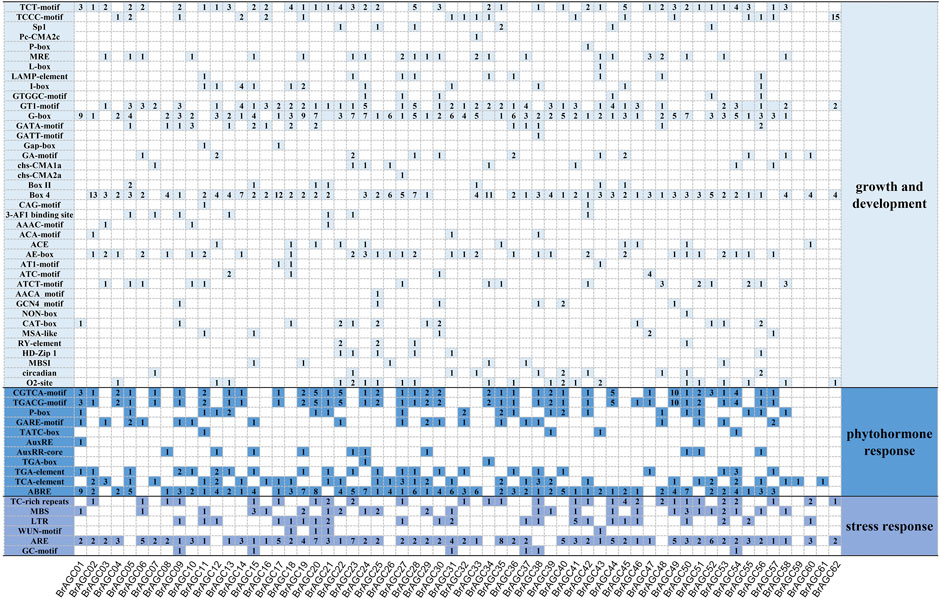
FIGURE 5. Cis-element analysis in the promoter region of BrAGC genes. The number of cis-elements in each gene is indicated by numbers.
Expression patterns in different tissues
To explore the potential functions of BrAGC genes, we analyzed the expression levels of BrAGC genes in the scallus, flower, leaf, root, silique, and stem of B. rapa (Figure 6; Supplementary Table S5). A total of 59 genes (except BrAGC59, BrAGC05, and BrAGC60) were expressed in at least one tissue, and 46 genes were expressed in all six tested tissues. Most BrAGC genes showed obvious expression differences among tissues, and genes preferentially expressed in flowers accounted for 35.5% (N = 22) of all genes; only 3.2% (N = 2) of genes were most highly expressed in the leaves. Some genes showed tissue-specific expression. For example, the expression level of BrAGC12 in the stem was more than 10 times that in other tissues, and the expression level of BrAGC33 in silique was nearly five times that in other tissues.
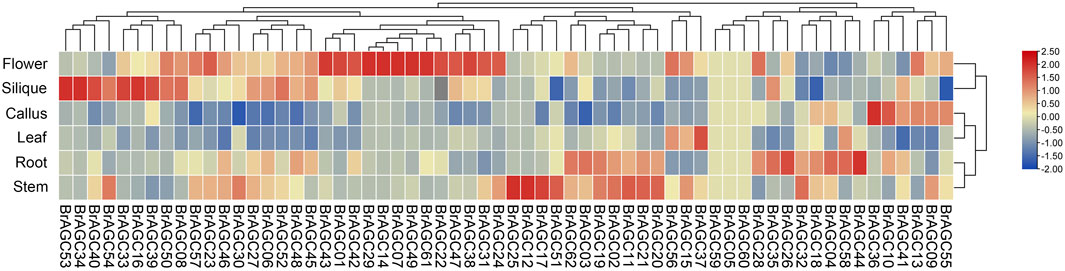
FIGURE 6. Expression profiles of BrAGC genes in different tissues of B. rapa. The abundance of each gene was measured using TPM.
Expression patterns during the self-incompatibility and cross-pollination response
B. rapa is a typical self-incompatible vegetable. Some protein kinases have been shown to be involved in the regulation of SI and self-compatibility processes, such as FERONIA receptor kinase, which plays an important role in regulating the SI response (Li et al., 2016; Duan et al., 2020; Zhang et al., 2021). AGC protein kinases in A. thaliana have been shown to regulate the polar growth of pollen tubes (McCormick, 1993; Li et al., 2018). To study the function of BrAGC genes in the SI and CP response of B. rapa, the expression levels of BrAGC genes in unpollinated (UP) stigma and the stigma after CP and SI pollination at different times were detected by RNA-seq (Figure 7A; Supplementary Table S6), and 11 genes with high expression in the stigma were selected for qRT-PCR to verify the reliability of the RNA-seq results (Figure 7B). The relative expression levels of BrAGC genes in the stigma varied under different pollination conditions. After CP pollination, the relative expression levels of BrAGC55, BrAGC13, BrAGC32, and BrAGC33 increased, and the most significant increases were observed for BrAGC13 and BrAGC32; the expression levels of these genes were increased by more than five times compared with that of the control. In addition, the expression levels of BrAGC23, BrAGC37, BrAGC12, BrAGC31, and BrAGC04 decreased. After SI pollination, the relative expression levels of BrAGC23, BrAGC37, BrAGC26, BrAGC33, BrAGC32, BrAGC41, and BrAGC04 increased, and the most significant increases were observed for BrAGC26, BrAGC33, and BrAGC04; the expression levels of these genes were increased by more than five times compared with the expression of these genes in the control. The qRT-PCR results were consistent with the RNA-seq results; thus, the qRT-PCR results were used in subsequent analyses.
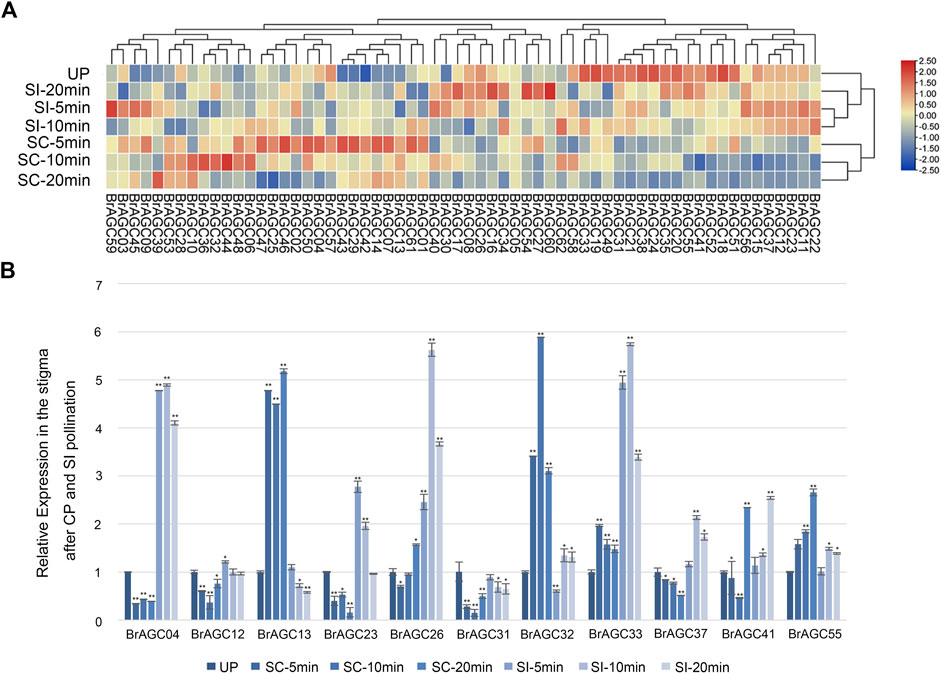
FIGURE 7. Expression profiles of BrAGC genes during the SI and CP response. (A) Heatmap showing the expression patterns of BrAGC genes in the UP stigma and under CP and SI pollination after 5, 10, and 20 min. (B) Transcriptional expression patterns of 11 BrAGC genes in the stigma after CP and SI pollination at 5, 10, and 20 min. The above experiments were performed using UP as the control; each group was subjected to three biological replicates, and error bars indicate standard errors. Asterisks above data bars indicate statistical significance (two-tailed t-test; *p < 0.05; **p < 0.01).
Expression patterns in response to abiotic stress
Cold stress, salt stress, and drought stress are common threats in B. rapa production. To identify the key genes involved in responses to stress, 10 genes with high expression levels in B. rapa were selected for qRT-PCR detection after various stress treatments. BrAGC genes could respond to abiotic stress. Under the three stresses, the relative expression of BrAGC37 and BrAGC44 was significantly upregulated and downregulated, respectively. The expression patterns of the other genes varied under different types of stress.
Under salt stress (Figure 8A), the relative expression levels of BrAGC41, BrAGC37, and BrAGC21 were significantly upregulated at each time point after treatment. The expression of BrAGC44 and BrAGC26 was downregulated at each time point after treatment. The expression of BrAGC33 and BrAGC56 first increased and then decreased, and the expression of these genes peaked after 2 h of salt treatment.
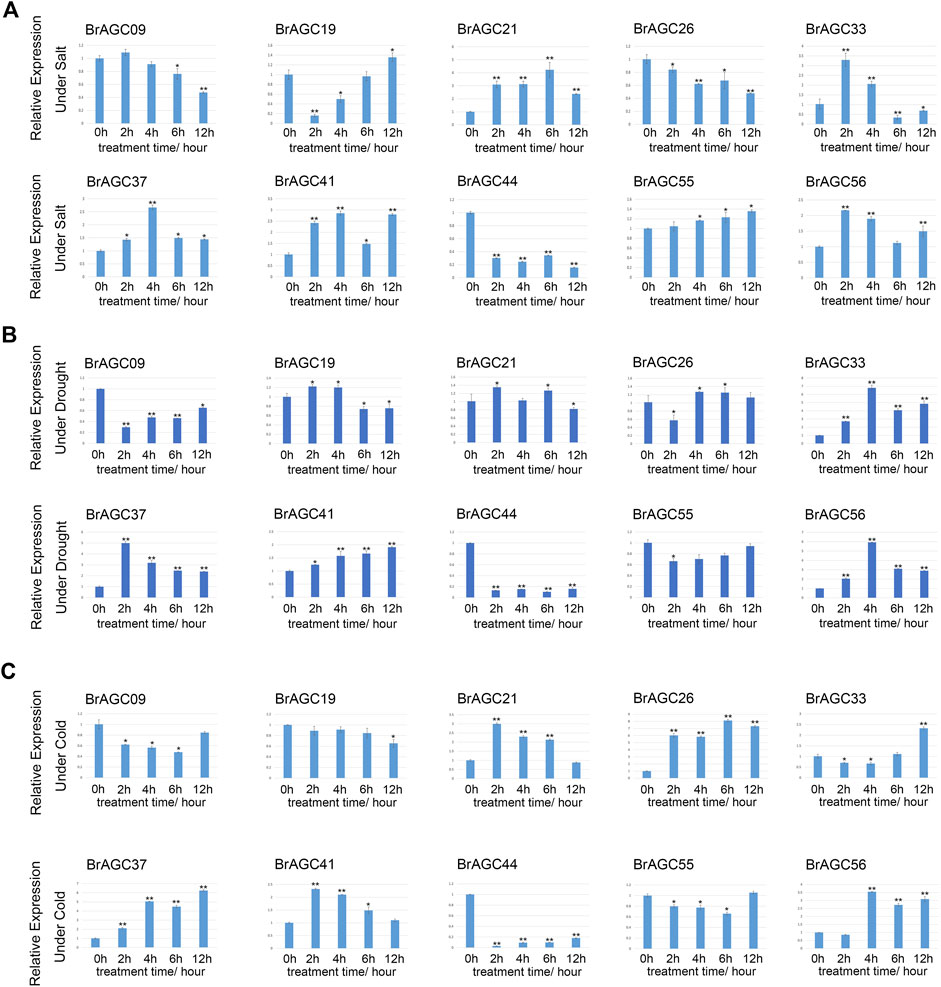
FIGURE 8. Expression profiles of 10 BrAGC genes under abiotic stress were analyzed by qRT-PCR. (A) Salt treatment. (B) Drought treatment. (C) Cold treatment. The above experiments were performed using 0 h as the control, and the treatment time was set to 2, 4, 6, and 12 h. Each group was subjected to three biological replicates, and error bars indicate standard errors. Asterisks above data bars indicate statistical significance (two-tailed t-test; *p < 0.05; **p < 0.01).
Under drought stress (Figure 8B), the relative expression levels of BrAGC37, BrAGC56, and BrAGC33 were significantly upregulated, and the expression levels peaked at 2 or 4 h. The expression levels of BrAGC44, BrAGC55, and BrAGC09 were significantly downregulated, and the expression of BrAGC44 showed the most significant downregulation among all genes.
The relative expression levels of BrAGC21, BrAGC26, BrAGC37, BrAGC41, and BrAGC56 were higher under cold stress compared with under control conditions (Figure 8C). The relative expression levels of BrAGC26 and BrAGC37 were positively correlated with the treatment time. The relative expression levels of BrAGC21 and BrAGC41 peaked after 2 h of cold stress and then gradually decreased. The relative expression of BrAGC44 decreased significantly after cold stress treatment.
Protein-protein interaction networks
AGC protein kinases are highly conserved in eukaryotes, and BrAGC genes are closely related to AtAGC genes. The functions of the AtAGC genes have been thoroughly studied. We performed PPI network analysis on AtAGC protein kinases to further determine their functions. A total of 44 nodes and 52 edges were predicted in the PPI networks (Figure 9A; Supplementary Table S7), and five functional partners of AtAGC genes, TOR, PDK, EIF3A, RPT3, and NPH3, were predicted. These genes play a role in stress responses (Ahn et al., 2011), auxin signal transduction (Deng et al., 2016; Tan et al., 2020), and plant phototropism response (Motchoulski and Liscum, 1999). This indicates that AGC protein kinases might regulate associated biological processes through its close relationship with these proteins. We also found that At3g25250 (BrAGC44 homologous gene) interacted with zinc-finger (C2H2-type) protein (At5G59820, AtZat12) (Figure 9B). AtZat12 has been shown to play a key role in ROS and abiotic stress signal transduction, and an interaction was detected between At3g20830 (BrAGC26 and BrAGC33 homologous gene) and At4g16340, a gene encoding guanine nucleotide exchange factor (GEF) (Figure 9C; Supplementary Table S7). These results further indicate that BrAGC genes have important and diverse function.
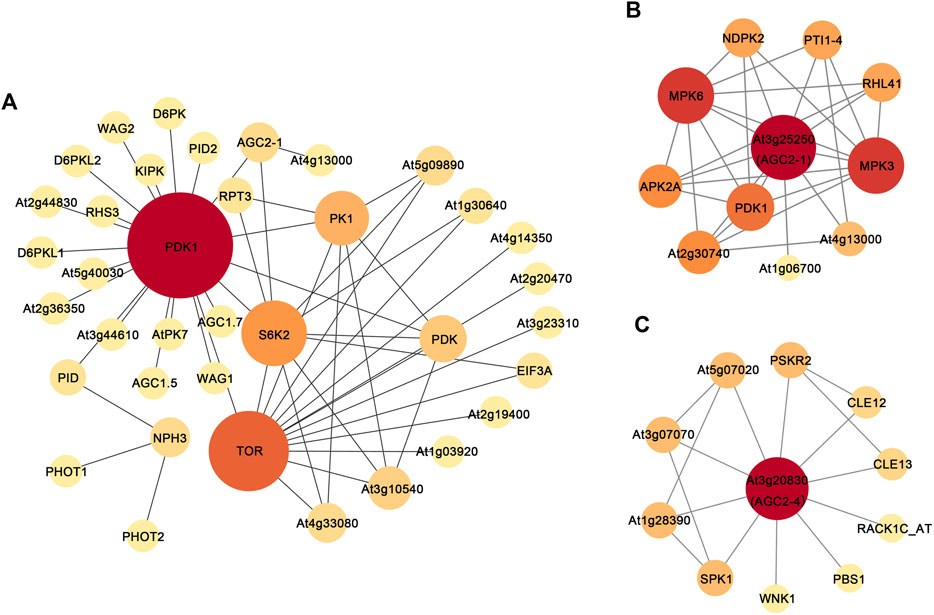
FIGURE 9. Protein-protein interaction networks of AGC protein kinase in A. thaliana. (A) All AtAGCs PPIs. (B) At3g25250 (BrAGC44 homologous gene) PPIs. (C) At3g20830 (BrAGC26 and BrAGC33 homologous gene) PPIs. Each node represents a protein, and interacting protein connected by line. Node size and fill color shade are positively correlated with degree centrality.
Discussion
AGC protein kinases are a subgroup of serine/threonine protein kinases that are widespread in eukaryotes, and they play important roles in regulating plant growth and development, as well as resistance, immunity, and programmed cell death. No studies to date have identified the AGC protein kinases in B. rapa, and the functions of BrAGC genes have not yet been reported. We identified 62 BrAGC genes in B. rapa and analyzed their physicochemical properties, structural characteristics, and expression patterns. The results of this study will aid future studies of the functions of BrAGC genes.
Whole-genome duplication (WGD), segmental duplication, and tandem duplication are the main drivers of gene family expansion in species, and these processes can enrich the functional diversity of gene family members (Li et al., 2020). A total of 17 segmentally duplicated gene pairs were identified among BrAGC genes, and the number of segmentally duplicated gene pairs was 8.5 times that of the number of tandemly duplicated gene pairs. This indicates that segmental duplication is an important driver of the expansion of BrAGC genes in the B. rapa genome and that it has promoted the emergence of new functional genes in the BrAGC gene family (Cannon et al., 2004). The Ka/Ks ratio of duplicated gene pairs in BrAGCs was substantially less than 1, which suggests that BrAGC genes have experienced purifying selection. The number of BrAGC genes (N = 62) was higher than that of AtAGC genes (N = 39), which might reflect the three WGDs that B. rapa has experienced (common to all Brassicaceae members), as well as a Brassica-specific whole-genome triplication (WGT) event (Wang et al., 2011; Cheng et al., 2013). The diploid B. rapa genome has been rediploidized through chromosomal rearrangements and gene loss (Cai et al., 2021). Collinearity analysis between BrAGCs and AGCs in the model plants A. thaliana and O. sativa has shown that the number of collinear gene pairs of BrAGCs and AtAGCs was more than 9 times higher than that between BrAGCs and OsAGCs, indicating that the evolutionary relationships between BrAGCs and AtAGCs are closer than those between BrAGCs and OsAGCs. This allows us to speculate on the functions of AtAGC genes orthologous to BrAGCs. We also found that one AtAGC gene could be mapped to up to five BrAGC genes, which reflects the WGT event in B. rapa.
The activation segment of protein kinases usually includes the Mg2+-binding loop (DFG), T-loop, and the P+1 loop (Nolen et al., 2004; Taylor and Kornev, 2011), and the C-terminus of AGC protein kinases has a hydrophobic motif (FxxF) that regulates its interaction with PDK1, which is called the PDK1-interacting fragment (PIF). In plant AGC kinases, glycine (G) in the Mg2+-binding loop (DFG) is replaced by aspartic acid (D), and there are 36–118 aa insertion domains between the Mg2+-binding loop (DFDLS) and the T-loop (SFVGT), which are typical features of plant AGC kinases (Zegzouti et al., 2006). Analysis of the structural characteristics of BrAGC genes revealed that the characteristic motifs of plant AGC protein kinases are conserved in BrAGCs, such as motif3 containing the hydrophobic motif FxxxW, which is present in 98.4% of BrAGC genes. It can interact with the T-loop in motif5 to promote phosphorylation (Frödin et al., 2002). Special motifs also exist in different subfamilies. For example, motif10 exists at the N-terminus of 85.7% AGCVII subfamily genes, which is a motif specific to this subfamily. The conserved domain (NCBI Accession number: cl45907) corresponding to motif10 plays a role in the regulation of cellular functions such as cell morphogenesis, cell division, proliferation, and apoptosis (Lu et al., 2020), and the functions of the homologous genes of the subfamily in regulating cell division and cell morphology have been verified in yeast and other eukaryotes (Tamaskovic et al., 2003). Thus, the genes in the AGCVII subfamily might play a role in cellular metabolism.
Most plants in Brassicaceae have evolved SI to resist SI pollination and promote CP, which can help maintain the hybrid vigor and genetic diversity of plants (Takayama and Isogai, 2005). BrAGCs were preferentially expressed in flowers (Figure 5), and AGC protein kinases in A. thaliana have been shown to regulate the polar growth of pollen tubes (McCormick, 1993; Li et al., 2018). Suggesting that AGC protein kinases play an important role in sexual reproduction in plants. The expression levels of BrAGC genes in the stigma were detected at different times following SI and CP. The upregulated genes BrAGC13 and BrAGC32 after CP and the upregulated genes BrAGC26, BrAGC33, and BrAGC04 after SI pollination might be involved in the interaction between pollen and the stigma in B. rapa. In addition, PPI network analysis showed that the homologous gene At3g20830 of BrAGC26 and BrAGC33 interacts with At4g16340. At4g16340 encodes SPIKE1 (SPK1), the guanine nucleotide exchange factor (GEF) in Arabidopsis. AGC1.5 in A. thaliana can directly interact with GEFs to control the polar growth of pollen tubes (Li et al., 2018). Therefore, BrAGC26 and BrAGC33 might play a role in regulating the pollen–pistil interaction; additional studies are needed to confirm this possibility.
B. rapa is often threatened by abiotic stress, and this has favored the evolution of mechanisms to resist abiotic stress (Nakashima and Yamaguchi-Shinozaki, 2013). More than 96% of AGCVIIIa genes and BrAGC44, BrAGC26, and BrAGC33 in AGCVIIIb lack introns or only contain 1 to 2 introns; by contrast, other subfamilies have more introns. Previous studies have demonstrated that the expression levels of genes with fewer introns can rapidly change in response to stress (Jeffares et al., 2008). Fewer introns are conducive to the transcriptional regulation of genes under stress and facilitate stress responses. In other plants, AGCVIII kinase is involved in the regulation of plant growth and development and in responses to stress. We speculate that genes in the AGCVIII subfamily might play an important role in stress responses (Rentel et al., 2004; Devarenne et al., 2006; Petersen et al., 2009; Matsui et al., 2010; Zhu et al., 2015). Therefore, AGCVIII subfamily members might play important roles in the stress responses of B. rapa. To further predict the genes that might be involved in responses to abiotic stress, we analyzed the PPI networks of AtAGC genes and the cis-elements of BrAGC genes and characterized the expression patterns of some genes under low-temperature stress, salt stress, and drought stress. We found that TOR, a predicted functional partner of AtAGC genes, is closely related to several AtAGC protein kinases and controls plant growth under environmental stress (Ahn et al., 2011). AGC protein kinases can affect the kinase activity of TOR through phosphorylation (Hálová et al., 2013). Therefore, AGC protein kinases may regulate the response to external stress through cooperation with TOR. In addition, we found that the relative expression levels of BrAGC21, BrAGC26, BrAGC37, BrAGC41, and BrAGC56 were upregulated under cold stress, and the promoter regions of BrAGC21, BrAGC37, and BrAGC41 contained low temperature-responsive elements (LTRs). Under drought stress, the relative expression levels of BrAGC33, BrAGC56, and BrAGC37 were upregulated, and BrAGC56, BrAGC21, and BrAGC41 contained drought-inducible elements (MBS). Under salt stress, the relative expression levels of BrAGC41, BrAGC37, and BrAGC21 were upregulated, and BrAGC21 and BrAGC41 contained defense and stress-responsive elements (TC-rich repeats). Phytohormone response elements are widely distributed in BrAGC genes, including those for gibberellin (P-box, GARE-motif, and TATC-box), auxin (AuxRE, AuxRR-core, TGA-box, and TGA-element), salicylic acid (TCA-element), and MeJA (CGTCA-motif and TGACG-motif), and AtAGC protein kinase can interact with PDK and TOR to regulate auxin signal transduction (Deng et al., 2016; Tan et al., 2020). Therefore, BrAGC genes may affect plant growth and development and environmental stress responses through the hormone signal regulation network. The promoter regions containing these elements have been shown to regulate the expression of related genes, which allows plants to respond to changes in the external environment. In addition, the expression of BrAGC37 was upregulated under three types of abiotic stress, and the relative expression of BrAGC44 was significantly decreased under these three abiotic stresses. Plants can enhance their resistance to other stresses after experiencing certain stresses; this might contribute to the cross-adaptation process of B. rapa and have a conserved function in the response to abiotic stress. ROS are important signaling molecules that play a role in the responses of plants to biotic and abiotic stresses, resistance to pathogen invasion, and the regulation of programmed cell death (Apel and Hirt, 2004). In many plants, the expression of AGC protein kinase member OXI is induced by oxidative stress; this protein plays key roles in plant defense and immunity. In addition, PPI network analysis predicted that AtZat12 is a functional partner of AtOxi1. AtZat12; it was also predicted to be involved in ROS metabolism and play a role in various abiotic stresses (Rizhsky et al., 2004; Davletova et al., 2005). BrAGC44 is orthologous to AtOxi1, and the relative expression of BrAGC44 is significantly downregulated under the three abiotic stresses. Based on the above conclusions, BrAGC44 might mediate the resistance of plants to stress by regulating ROS.
Conclusion
We identified 62 BrAGCs in B. rapa. Based on the comprehensive analysis of sequence features, cis-elements, expression profiles of different tissues, abiotic stress tolerance, sexual reproductive processes, and the published data, we found that BrAGC26, BrAGC33, and BrAGC44 likely regulate pollen–pistil interactions and abiotic stress tolerance, respectively.
Data availability statement
The data presented in the study are deposited in the NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE43245.
Author contributions
JH conceived the project; JH conceptualized and designed the experiments; XW performed the bioinformatic analysis and experiments with help from LP, XG, TL, and JL; XW wrote the manuscript. JH and QD revised the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (32272745).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1044853/full#supplementary-material
References
Ahn, C. S., Han, J. A., Lee, H. S., Lee, S., and Pai, H. S. (2011). The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell. 23 (1), 185–209. doi:10.1105/tpc.110.074005
Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi:10.1146/annurev.arplant.55.031903.141701
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40 (W1), W597–W603. doi:10.1093/nar/gks400
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Barrett, T., and Edgar, R. (2006). Gene expression omnibus: Microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 411, 352–369. doi:10.1016/S0076-6879(06)11019-8
Bögre, L., Ökrész, L., Henriques, R., and Anthony, R. G. (2003). Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 8 (9), 424–431. doi:10.1016/S1360-1385(03)00188-2
Cai, X., Chang, L., Zhang, T., Chen, H., Zhang, L., Lin, R., et al. (2021). Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 22 (1), 166–224. doi:10.1186/s13059-021-02383-2
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4 (1), 10–21. doi:10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Cheng, F., Mandáková, T., Wu, J., Xie, Q., Lysak, M. A., and Wang, X. (2013). Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell. 25 (5), 1541–1554. doi:10.1105/tpc.113.110486
Davletova, S., Schlauch, K., Coutu, J., and Mittler, R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139 (2), 847–856. doi:10.1104/pp.105.068254
Deng, K., Yu, L., Zheng, X., Zhang, K., Wang, W., Dong, P., et al. (2016). Target of rapamycin is a key player for auxin signaling transduction in Arabidopsis. Front. Plant Sci. 7, 291. doi:10.3389/fpls.2016.00291
Dennis, G., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., et al. (2003). David: Database for annotation, visualization, and integrated discovery. Genome Biol. 4 (5), P3. doi:10.1186/gb-2003-4-5-p3
Devarenne, T. P., Ekengren, S. K., Pedley, K. F., and Martin, G. B. (2006). Adi3 is a Pdk1‐interacting AGC kinase that negatively regulates plant cell death. EMBO J. 25 (1), 255–265. doi:10.1038/sj.emboj.7600910
Duan, Q., Liu, M-C. J., Kita, D., Jordan, S. S., Yeh, F-L. J., Yvon, R., et al. (2020). FERONIA controls pectin-and nitric oxide-mediated male–female interaction. Nature 579 (7800), 561–566. doi:10.1038/s41586-020-2106-2
Friml, J., Yang, X., Michniewicz, M., Weijers, D., Quint, A., Tietz, O., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 (5697), 862–865. doi:10.1126/science.1100618
Frödin, M., Antal, T. L., Dümmler, B. A., Jensen, C. J., Deak, M., Gammeltoft, S., et al. (2002). A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. Embo J. 21 (20), 5396–5407. doi:10.1093/emboj/cdf551
Hálová, L., Du, W., Kirkham, S., Smith, D. L., and Petersen, J. (2013). Phosphorylation of the TOR ATP binding domain by AGC kinase constitutes a novel mode of TOR inhibition. J. Cell. Biol. 203 (4), 595–604. doi:10.1083/jcb.201305103
Hanks, S. K., and Hunter, T. (1995). The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification 1. FASEB J. 9 (8), 576–596. doi:10.1096/fasebj.9.8.7768349
Hurst, L. D. (2002). The ka/ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 18 (9), 486. doi:10.1016/s0168-9525(02)02722-1
Inada, S., Ohgishi, M., Mayama, T., Okada, K., and Sakai, T. (2004). RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell. 16 (4), 887–896. doi:10.1105/tpc.019901
Jeffares, D. C., Penkett, C. J., and Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24 (8), 375–378. doi:10.1016/j.tig.2008.05.006
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Li, C., Wu, H-M., and Cheung, A. Y. (2016). FERONIA and her pals: Functions and mechanisms. Plant Physiol. 171 (4), 2379–2392. doi:10.1104/pp.16.00667
Li, E., Cui, Y., Ge, F. R., Chai, S., Zhang, W. T., Feng, Q. N., et al. (2018). AGC1.5 kinase phosphorylates RopGEFs to control pollen tube growth. Mol. Plant 11 (9), 1198–1209. doi:10.1016/j.molp.2018.07.004
Li, Y., Lin-Wang, K., Liu, Z., Allan, A. C., Qin, S., Zhang, J., et al. (2020). Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 148, 817–832. doi:10.1016/j.ijbiomac.2020.01.167
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48 (D1), D265–D268. doi:10.1093/nar/gkz991
Matsui, H., Yamazaki, M., Kishi-Kaboshi, M., Takahashi, A., and Hirochika, H. (2010). AGC kinase OsOxi1 positively regulates basal resistance through suppression of OsPti1a-mediated negative regulation. Plant Cell. Physiol. 51 (10), 1731–1744. doi:10.1093/pcp/pcq132
Mccormick, S. (1993). Male gametophyte development. Plant Cell. 5 (10), 1265–1275. doi:10.1105/tpc.5.10.1265
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49 (D1), D412–D419. doi:10.1093/nar/gkaa913
Motchoulski, A., and Liscum, E. (1999). Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286 (5441), 961–964. doi:10.1126/science.286.5441.961
Nakashima, K., and Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell. Rep. 32 (7), 959–970. doi:10.1007/s00299-013-1418-1
Nolen, B., Taylor, S., and Ghosh, G. (2004). Regulation of protein kinases: Controlling activity through activation segment conformation. Mol. Cell. 15 (5), 661–675. doi:10.1016/j.molcel.2004.08.024
Pearce, L. R., Komander, D., and Alessi, D. R. (2010). The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell. Biol. 11 (1), 9–22. doi:10.1038/nrm2822
Petersen, L. N., Ingle, R. A., Knight, M. R., and Denby, K. J. (2009). OXI1 protein kinase is required for plant immunity against Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 60 (13), 3727–3735. doi:10.1093/jxb/erp219
Qiu, P. (2003). Recent advances in computational promoter analysis in understanding the transcriptional regulatory network. Biochem. Biophys. Res. Commun. 309 (3), 495–501. doi:10.1016/j.bbrc.2003.08.052
Rentel, M. C., Lecourieux, D., Ouaked, F., Usher, S. L., Petersen, L., Okamoto, H., et al. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427 (6977), 858–861. doi:10.1038/nature02353
Rizhsky, L., Davletova, S., Liang, H., and Mittler, R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 279 (12), 11736–11743. doi:10.1074/jbc.M313350200
Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27 (1), 295–296. doi:10.1093/nar/27.1.295
Sakai, T., Kagawa, T., Kasahara, M., Swartz, T. E., Christie, J. M., Briggs, W. R., et al. (2001). Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. U. S. A. 98 (12), 6969–6974. doi:10.1073/pnas.101137598
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shumbe, L., Chevalier, A., Legeret, B., Taconnat, L., Monnet, F., and Havaux, M. (2016). Singlet oxygen-induced cell death in Arabidopsis under high-light stress is controlled by OXI1 kinase. Plant Physiol. 170 (3), 1757–1771. doi:10.1104/pp.15.01546
Takayama, S., and Isogai, A. (2005). Self-incompatibility in plants. Annu. Rev. Plant Biol. 56, 467–489. doi:10.1146/annurev.arplant.56.032604.144249
Tamaskovic, R., Bichsel, S. J., and Hemmings, B. A. (2003). NDR family of AGC kinases--essential regulators of the cell cycle and morphogenesis. FEBS Lett. 546 (1), 73–80. doi:10.1016/s0014-5793(03)00474-5
Tan, S., Zhang, X., Kong, W., Yang, X. L., Molnár, G., Vondráková, Z., et al. (2020). The lipid code-dependent phosphoswitch PDK1-D6PK activates PIN-mediated auxin efflux in Arabidopsis. Nat. Plants 6 (5), 556–569. doi:10.1038/s41477-020-0648-9
Taylor, S. S., and Kornev, A. P. (2011). Protein kinases: Evolution of dynamic regulatory proteins. Trends biochem. Sci. 36 (2), 65–77. doi:10.1016/j.tibs.2010.09.006
Tong, C., Wang, X., Yu, J., Wu, J., Li, W., Huang, J., et al. (2013). Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC genomics 14 (1), 689–710. doi:10.1186/1471-2164-14-689
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28 (5), 511–515. doi:10.1038/nbt.1621
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43 (10), 1035–1039. doi:10.1038/ng.919
Zegzouti, H., Li, W., Lorenz, T. C., Xie, M., Payne, C. T., Smith, K., et al. (2006). Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J. Biol. Chem. 281 (46), 35520–35530. doi:10.1074/jbc.M605167200
Zhang, L., Huang, J., Su, S., Wei, X., Yang, L., Zhao, H., et al. (2021). FERONIA receptor kinase-regulated reactive oxygen species mediate self-incompatibility in Brassica rapa. Curr. Biol. 31 (14), 3004–3016.e4. doi:10.1016/j.cub.2021.04.060
Zhang, Y., He, J., and Mccormick, S. (2009). Two Arabidopsis AGC kinases are critical for the polarized growth of pollen tubes. Plant J. 58 (3), 474–484. doi:10.1111/j.1365-313X.2009.03792.x
Zhang, Y., Yao, W., Wang, F., Su, Y., Zhang, D., Hu, S., et al. (2020). AGC protein kinase AGC1-4 mediates seed size in Arabidopsis. Plant Cell. Rep. 39 (6), 825–837. doi:10.1007/s00299-020-02533-z
Keywords: Brassica rapa, AGC protein kinase, gene family, bioinformatics, expression profile
Citation: Wu X, Pan L, Guo X, Li T, Li J, Duan Q and Huang J (2022) Genome-wide identification and expression analysis of BrAGC genes in Brassica rapa reveal their potential roles in sexual reproduction and abiotic stress tolerance. Front. Genet. 13:1044853. doi: 10.3389/fgene.2022.1044853
Received: 15 September 2022; Accepted: 14 October 2022;
Published: 26 October 2022.
Edited by:
Romit Seth, North Carolina State University, United StatesReviewed by:
Vishal Sharma, Institute of Himalayan Bioresource Technology (CSIR), IndiaGopal Singh, Institute of Bioorganic Chemistry (Polska Akademia Nauk—PAN), Poland
Copyright © 2022 Wu, Pan, Guo, Li, Li, Duan and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiabao Huang, amJodWFuZzIwMThAc2RhdS5lZHUuY24=
 Xiaoyu Wu
Xiaoyu Wu Lianhui Pan1,2
Lianhui Pan1,2 Qiaohong Duan
Qiaohong Duan Jiabao Huang
Jiabao Huang