- 1Centre of Imaging Diagnosis, Affiliated Tumor Hospital of Guangxi Medical University, Nanning, China
- 2Department of Clinical Medicine, Medical College of Shihezi University, Shihezi, China
Background: FAM46C is a common mutated gene in tumours. A comprehensive understanding of the relationship between FAM46C expression and pan-cancer can guide clinical prognosis and broaden the immunotherapeutic targets.
Methods: Data from The Cancer Genome Atlas and Genotype-Tissue Expression (GTEx) databases were obtained, and gene expression of different tumour types and stages was analysed. Immunohistochemical analysis was performed to detect differences in the FAM46C protein levels in normal and cancerous tissues. The genetic variation of FAM46C was characterised using cBioPortal. The clinical prognostic value of FAM46C and the impact of FAM46C expression levels on the prognosis of patients with different types of cancer were assessed based on Kaplan–Meier and Cox regression analyses. Gene set enrichment analysis (GSEA) was used to analyse the pathways associated with FAM46C. Correlations between FAM46C expression levels and immune infiltration were assessed using the TIMER2 database and CIBERSORT algorithm, and correlations between FAM46C expression and the ESTIMATE, immune and stromal scores were analysed using the ESTIMATE algorithm. In addition, we also analysed the correlation between FAM46C expression and immune activation, suppression genes and immune chemokines.
Results: The expression level of FAM46C was correlated with the prognosis of most tumours, and low expression levels often suggested a poor prognosis. FAM46C was positively correlated with the abundance of CD4+ T cells, CD8+ T cells and plasma B lymphocytes in the tumour microenvironment. FAM46C exhibited a strong correlation with immunomodulatory pathways, immunomodulatory factors and immune markers. In addition, high FAM46C expression correlated with tumour mutational burden in acute myeloid leukaemia and microsatellite instability in endometrial cancer.
Conclusion: Our study suggests that FAM46C can be a potential prognostic marker for pan-cancer, is closely associated with immune regulation and may be an immune checkpoint to guide future clinical immunotherapy.
Introduction
Cancer is one of the major diseases threatening human health, and the incidence and mortality rate of cancer are increasing every year (Sung et al., 2021). The World Health Organization estimates that the number of cancer cases worldwide may increase by 60% in the next 20 years. Although significant progress has been made in cancer diagnosis and treatment in recent years, the overall survival (OS) rate of cancer patients has not improved significantly owing to limited research on the mechanisms of cancer occurrence and development (Siegel et al., 2021).
Some studies have demonstrated that mutations and deletions of cancer-related genes are the main cause of cancer development and progression (Clinton et al., 2020). Therefore, more in-depth studies are required, which may provide relevant clues and ideas for new therapeutic approaches. The Cancer Genome Atlas (TCGA) has characterised genomic and epigenomic variations in multiple classes of tumour cells. Based on such large data samples, multiple epigenomes can be sequenced and integrated for pan-cancer analysis, and the results can be correlated with clinical and imaging manifestations for comparison. Therefore, the origin and occurrence of tumours and the underlying mechanisms of tumour cell signalling pathways can be analysed. Previous studies have suggested that tumour cells originating from different organs may exhibit the same or similar histomorphological characteristics. For example, squamous cell carcinoma can originate from the skin, head and neck, oesophagus, cervix and vagina. Moreover, tumour cells originating from the same organ can also exhibit different histomorphological characteristics; the examples include thyroid and kidney cancers. Pan-cancer analysis facilitates the study of commonality and heterogeneity of tumours based on cellular and molecular characteristics and helps to assess molecular relationships among multiple tumours and understand their underlying regulatory mechanisms. It also provides new clues to further develop the clinical treatment of cancer.
The FAM46 protein is a non-canonical poly(A) polymerase and a member of the nucleotidyltransferase (NTase) fold superfamily. It relies on the poly(A) polymerase activity to induce polyadenylation of the mRNA of target proteins, which may regulate antibody production, gene expression and development of malignant tumours in the body and induce apoptosis (Kuchta et al., 2016). It is mainly found in vertebrates, and four proteins (FAM46A/B/C/D) are mainly expressed in humans. Among these proteins, FAM46C was first identified as one of the most common mutated genes in patients with multiple myeloma (MM) and is associated with decreased OS in patients with MM (Zhu et al., 2017). Furthermore, recent studies have found a functional association between FAM46C and hepatocellular carcinoma (Zhang et al., 2017), gastric cancer (Tanaka et al., 2017), colorectal cancer (Zhang et al., 2020), oral squamous cell carcinoma (Zhuang and Lu, 2018) and prostate cancer (Ma et al., 2020). FAM46C may also inhibit tumour proliferation and metastasis and promote tumour cell apoptosis through several pathways (Kazazian et al., 2020). The absence of FAM46C is reported to be associated with recurrence and decreased OS after radical surgery for gastric cancer (Tanaka et al., 2017). In patients with pancreatic ductal adenocarcinoma, FAM46C may also mediate the development of autoimmune diseases by participating in the epidermal growth factor receptor (EFGR) and interferon (IFN) signalling pathways (Crow, 2010). However, the prognostic accuracy of FAM46C in various tumours and its role in immune regulation remain unclear.
The present study provides a comprehensive analysis of FAM46C expression in pan-cancer based on the data from TCGA, Oncomine and other databases. We analysed differential gene expressions, regulatory pathways, the prognostic accuracy of FAM46C and the correlation of FAM46C expression with immune cell infiltration and immune regulation. Our study revealed that FAM46C might be a potential clinical prognostic marker, which is closely related to immune regulation, thus providing relevant ideas for developing future tumour immunotherapy.
Methods
Clinical Specimens
A total of 20 histological specimens from patients with pathologically confirmed bladder cancer were collected. The specimens comprised 20 pairs of bladder cancer and precancerous tissues. All patients had a confirmed pathological diagnosis before tissue collection and were not treated with chemotherapy or radiotherapy. RNA-sequencing was performed for all specimens. All patients signed an informed consent form. The Medical Ethics Committee of the Affiliated Tumor Hospital of Guangxi Medical University approved the cancer specimen collection protocol. All experiments and methods were performed following relevant guidelines and regulations.
Data Collection
The RNA expression and clinical data of TCGA and GTEx cohorts were obtained from the UCSC Xena database (https://xenabrowser.net/datapages/). DNA copy numbers were downloaded from the cBioPortal database (https://www.cbioportal.org/).
Immunohistochemical Staining
All collected cancer and paraneoplastic tissues were fixed in 10% formalin for 48 h, dehydrated in ethanol gradient for 2 h and embedded in paraffin. The tissue specimens were cut into sections of 4–7 µm on a microtome and transferred to slides for drying and maintaining the morphological features of tissue specimens. The slides were dewaxed and hydrated, and the antigen was extracted. The slides were quenched in 3% hydrogen peroxide (H2O2) solution diluted in distilled water for 10 min at room temperature and incubated with anti-FAM46C(Beijing Bioss Biotechnology Co., Ltd. Antibody No.: bs-8198)for 1 h at room temperature. After incubation, the sections were stained with horseradish peroxidase (HRP)-labelled secondary antibody (Shanghai Long Island Biotechnology Co., Ltd.) for 30 min at room temperature. Subsequently, the sections were stained with diaminobenzidine (DAB) for 1 min at room temperature and haematoxylin for 3 min and rinsed with tap water for 10 min. According to the positive rate of tumour cells, the samples were divided into the high (positive rate > 25%) and low expression (positive rate < 25%) groups.
Differential Expression of FAM46C
Oncomine is the largest tumour gene microarray database to date and allows comprehensive data mining. We used the Oncomine database to analyse the differential expression of FAM46C mRNA between different tumours and normal tissues. The SangerBox software was used to compare the differences in FAM46C expression between cancerous versus paraneoplastic tissues integrated data of TCGA and GTEx databases.
Genetic Alteration Analysis
The varying genetic characteristics of FAM46C were obtained through the cBioPortal website (https://www.cbioportal.org/). We analysed the mutation frequency, copy number variation and mutation type using the “Cancer Type Summary” module of TCGA database and the “Quick Selection” and “TCGA Pan-Cancer Atlas Study” options.
Survival Analysis
We divided the tissue samples into high and low expression groups based on FAM46C mRNA expression values in the dataset, and the prognostic value of FAM46C was assessed using Kaplan–Meier survival analysis in both groups. Cox regression analysis was performed using the survive, glmnet, surviveROC and survminer R packages to analyse the correlation of FAM46C expression with OS, disease-specific survival (DSS) and progression-free survival (PFS) in different cancer types using TCGA data.
Immune Infiltration Analysis
We comprehensively analysed the correlation between FAM46C expression and immune infiltration using the TIMER2 database and CIBERSORT algorithm. In addition, we analysed the relationship between FAM46C and immune cells such as CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells (DCs) at B-cell abundance. The immune, matrix and ESTIMATE scores of FAM46C expression were evaluated using the ESTIMATE algorithm.
Gene Set Enrichment Analysis
We used the clusterProfiler R package for performing gene set enrichment analysis (GSEA) to identify signalling pathways affected by FAM46C. Gene sequences obtained based on the fold change of mean gene expression between patients in the high and low FAM46C expression groups were entered into a file. Biological processes were evaluated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and HALLMARK pathway analyses.
Statistical Analysis
Based on the mRNA expression of FAM46C, the samples were divided into a high and low expression group. The Spearman’s correlation was used to assess the correlation between FAM46C expression and relevant targets, including tumour mutational burden (TMB) and microsatellite instability (MSI). We used Cox regression analysis to examine hazard ratios (HRs) and log-rank p values of survival analyses, and p < 0.05 was considered statistically significant.
Results
Analysis of FAM46C Expression in Pan-Cancer
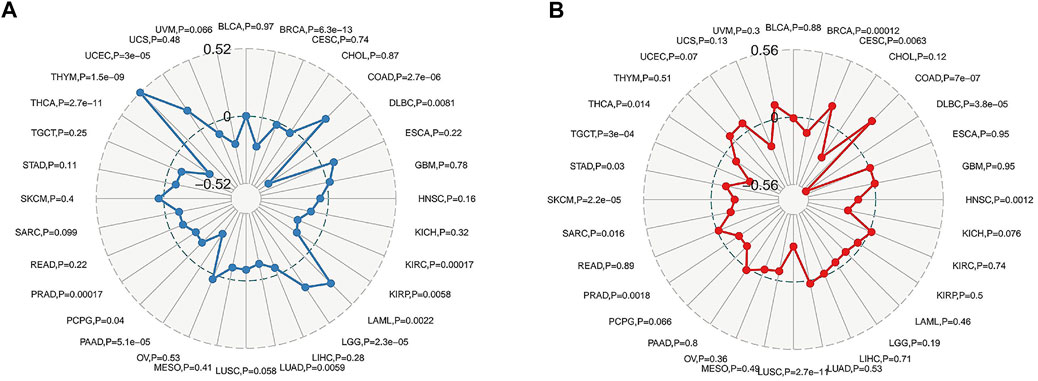
The Oncomine data revealed that the mRNA expression levels of FAM46C were significantly decreased in most cancers, such as bladder, brain and central nervous system, colorectal, stomach, kidney and pancreatic cancers; leukaemia and sarcoma (SARC). However, the mRNA expression of FAM46C was high in hepatocellular carcinoma and lung cancer (Figure 1A). In addition, considering the small number of normal samples in TCGA, we integrated data from normal tissues in the GTEx database and tumour tissues in TCGA database to analyse the differential expression of FAM46C in 20 cancers. Consequently, low FAM46C expression was observed in eight tumours: bladder uroepithelial carcinoma (BLCA), colon adenocarcinoma (COAD), oesophageal cancer (ESCA), kidney chromophobe (KICH), prostate adenocarcinoma (PRAD), rectal adenocarcinoma (READ), stomach adenocarcinoma (STAD) and thyroid carcinoma (THCA) (Figure 1B). These results revealed that the mRNA expression of FAM46C was low in various tumours, with FAM46C playing a protective role in tumour development.
FIGURE 1. Different levels of FAM46C expression in pan-cancer. (A) FAM46C expression in different cancer and paired normal tissues in the Oncomine dataset. The number in each cell is the number of datasets. (B) FAM46C expression in tumour tissues from TCGA cohort and normal tissues from TCGA and GTEx cohorts (*p < 0.05, **p < 0.01, ***p < 0.001).
Relationship Between FAM46C Expression and Tumour Stages
We further evaluated the expression levels of FAM46C in different tumour stages. As shown in Figure 2, the FAM46C expression levels are significantly higher in patients with stage I–II disease than in patients with stage III–IV disease in renal papillary cell carcinoma (KIRP), lung adenocarcinoma (LUAD) and testicular germ cell tumour (TGCT) (all p < 0.01). However, a decreasing trend in the FAM46C expression levels was observed in patients with stage III–IV tumours such as THCA, liver hepatocellular carcinoma (LIHC), lung squamous carcinoma (LUSC), mesothelioma (MESO) and READ; however, the difference was not statistically significant (Supplementary Figure 1).
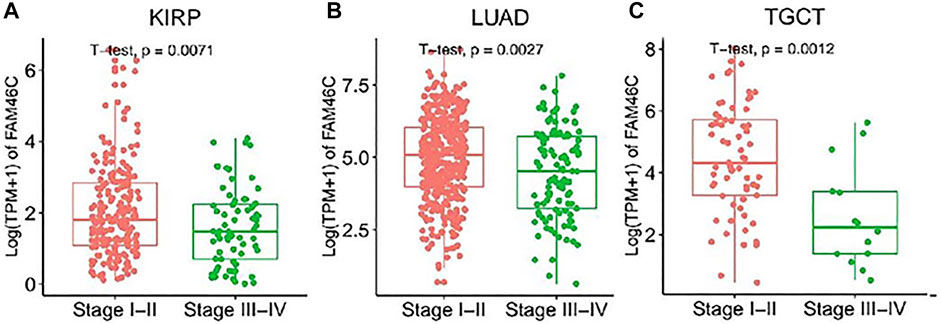
FIGURE 2. FAM46C expression in pan-cancer with different World Health Organizatio stages. (A–C) Comparison of early and late expression of FAM46C in different tumours.
Genetic Alterations in FAM46C
We extracted FAM46C mutation data from TCGA dataset and used the cBioPortal tool to analyse the extracted data and correlation between FAM46C mutations and tumours. As shown in Figure 3A, FAM46C has the highest mutation frequency (>6%) in patients with cutaneous melanoma. However, it was less frequently mutated in LIHC, KIRP and low-grade glioma (LGG) of the brain (approximately <1%). Furthermore, the main types of “amplified” copy number alterations (CNAs) were observed in ovarian plasmacytoid cystic adenocarcinoma (OV) and pancreatic adenocarcinoma (PAAD), and FAM46C was amplified in all patients with PAAD. The loci and types of FAM46C mutation and the number of patients harbouring the mutation are shown in Figure 3B. The main type of genetic alteration in FAM46C was missense mutation. As demonstrated in Figure 3C, GSEA reveals that FAM46C is positively correlated with DNA copy number in OV, breast invasive carcinoma (BRCA), ESCA, pheochromocytoma and paraganglioma (PCPG), LIHC, acute myeloid leukaemia (LAML), STAD, LUAD, LUSC and head and neck squamous cell carcinoma (HNSC).
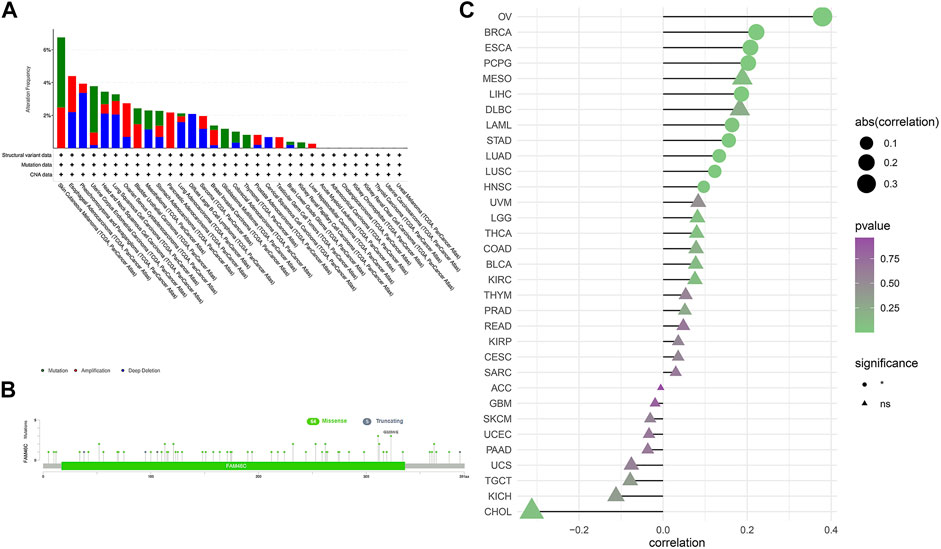
FIGURE 3. Characterisation of FAM46C gene mutations in different TCGA tumour samples as analysed using the cBioPortal tool. (A) The type and frequency of FAM46C gene mutations in different cancers. (B) The mutation sites of FAM46C. (C) The correlation between copy number changes and FAM46C expression in each tumour sample.
Analysis of the Protein Expression Levels of FAM46C
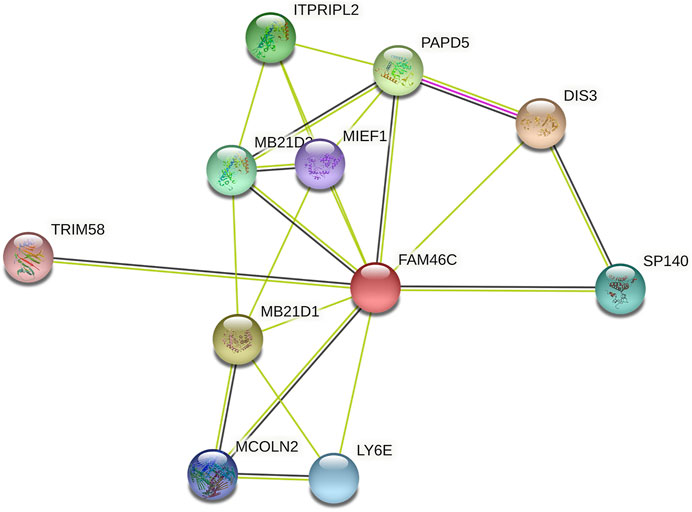
After analysing the mRNA expression, mutations and CNAs of FAM46C, we assessed the protein expression level of FAM46C in tumours. Using the Human Protein Atlas (HPA) database, we found that the expression level of FAM46C was significantly decreased in skin and kidney cancer tissues compared with normal tissues (Figures 4A–D). In addition, we performed immunohistochemical and statistical analyses on paired tissue specimens from patients with bladder cancer, and the results revealed that the expression level of FAM46C was significantly decreased in bladder cancer (p = 0.014), which was consistent with the analysis of the HPA database (Figures 5A–C). Furthermore, to assess the relationship between FAM46C and protein interactions, we constructed a protein–protein interaction (PPI) network and found that FAM46C was closely associated with ITPRIPL2, PAPD5, DIS3, SP140, MB21D2, MIEF1, TRIM58, MB21D1, MCOLN2, LY6E and other proteins (Figure 6). Some previous studies have suggested that the SP140 protein may play a role in chromogranin-mediated regulation of gene expression involved in the pathogenesis of LAML (Hoang et al., 2019). However, DIS3 and PAPD5 may act as RNA ectodomain complexes involved in the degradation or transcription of various cellular RNAs (Hu et al., 2020; Liu et al., 2020). Moreover, MIEF1 and LY6E may mediate different signalling pathways to induce apoptosis in the corresponding cancer cells (Yeom et al., 2016; Hou et al., 2019; Zhou et al., 2019). The expression level of TRIM58 protein may be related to the prognosis of patients with LUAD and COAD (Liu et al., 2018; Chen et al., 2021). In addition, the MCOLN2 protein expression level may regulate the development of paediatric acute lymphoblastic leukaemia (Almamun et al., 2015).
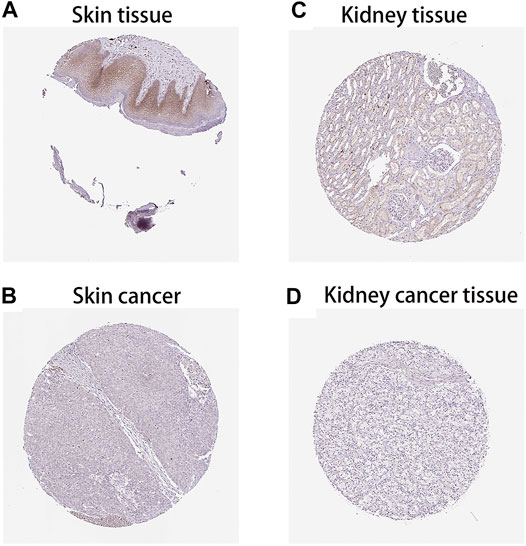
FIGURE 4. Protein levels of FAM46C in tumour and normal tissues from the Human Protein Atlas (HPA) database. (A,C) Normal skin and kidney tissues, respectively. (B,D) Skin and kidney cancer tissues, respectively.
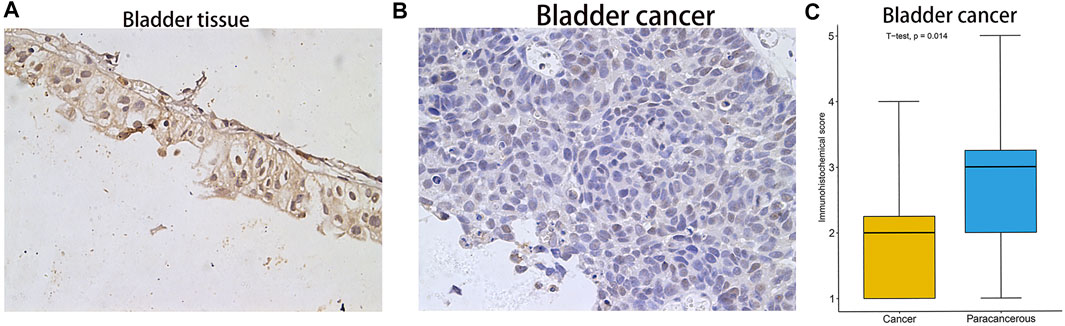
FIGURE 5. Immunohistochemical staining results and statistical plots of FAM46C protein expression in normal bladder and bladder cancer tissues. (A) Represent 20× immunohistochemical image of bladder tissue. (B) Represent 20× immunohistochemical image of bladder cancer. (C) Statistical plots of FAM46C protein expression in bladder cancer and normal tissues.
Prognostic Significance of FAM46C
We analysed the clinical value of FAM46C using univariate Cox regression and Kaplan–Meier analyses. Univariate Cox regression analysis revealed that high expression of FAM46C was associated with a better prognosis. The results of OS analysis revealed that FAM46C was associated with improved prognosis in adrenocortical carcinoma (ACC) (p = 0.01,HR = 0.28), BLCA (p = 0.01, HR = 0.49), BRCA (p = 0.02, R = 0.59), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (p = 0.01, HR = 0.59), COAD (p = 0.03, HR = 0.59), KICH (p = 0.01, HR = 0.21), PAAD (p = 0.02, HR = 0.38) and SARC (p = 0.01, HR = 0.58) (Figure 7A).The DSS analysis revealed that high expression of FAM46C was significantly associated with improved prognosis in 11 out of 32 tumours (Figure 7B). According to data on progression-free interval (PFI), high expression of FAM46C was significantly associated with prognosis in 12 out of 32 tumours (Figure 7C).
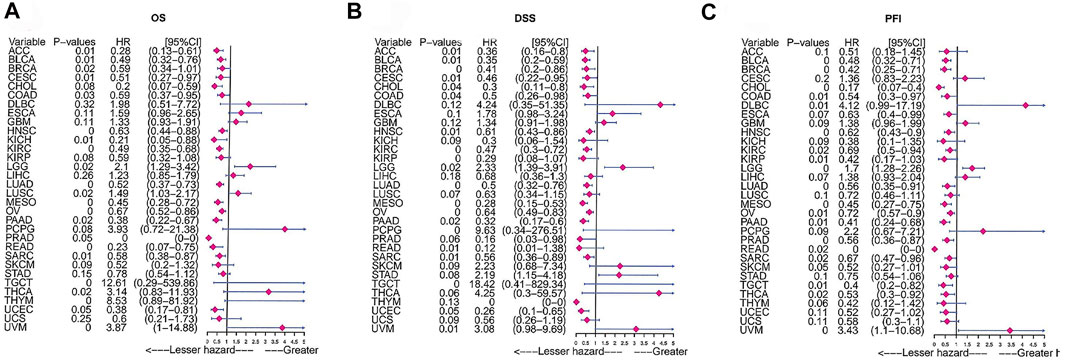
FIGURE 7. Prognostic analysis of FAM46C via univariate Cox regression. The results of (A) OS analysis, (B) DSS analysis and (C) PFI analysis in pan-cancer.
We analysed the relationship between FAM46C expression and OS, DSS and PFI using Kaplan–Meier analysis. The results revealed that high FAM46C expression was associated with OS and was a protective factor in ACC (p = 0.012, HR = 0.28), BRCA (p = 0.017, HR = 0.59), BLCA (p = 0.015, HR = 0.49), CESC (p = 0.011, HR = 0.51), COAD (p = 0.028, HR = 0.59), KICH (p = 0.015, HR = 0.21), HNSC (p = 0.002, HR = 0.63), renal clear cell carcinoma (KIRC) (p < 0.001, HR = 0.49), LUAD (p < 0.001, HR = 0.52), MESO (p = 0.003, HR = 0.45), OV (p = 0.002, HR = 0.67), READ (p = 0.002, HR = 0.23), PAAD (p = 0.016, HR = 0.38) and SARC (p = 0.006, HR = 0.58) (Figure 8A). The DSS analysis revealed that FAM46C was the most important protective factor in ACC (p = 0.013, HR = 0.36), BRCA (p < 0.001, HR = 0.41), cholangiocarcinoma (CHOL) (p = 0.043, HR = 0.3), BLCA (p = 0.008, HR = 0.35), CESC (p = 0.008, HR = 0.46), COAD (p = 0.039, HR = 0.5), HNSC (p = 0.006, HR = 0.61), KIRC (p < 0.001, HR = 0.47), KIRP (p = 0.003, HR = 0.29), LUAD (p < 0.001, HR = 0.5), MESO (p = 0.003, HR = 0.28), OV (p = 0.001, HR = 0.64), READ (p = 0.007, HR = 0.12), PAAD (p = 0.018, HR = 0.32), SARC (p = 0.008, HR = 0.56), uterine corpus endometrial carcinoma (UCEC) (p = 0.047, HR = 0.26) (Figure 8B). The PFI analysis revealed that FAM46C was a protective factor in BRCA (p < 0.001, HR = 0.42), CHOL (p < 0.001, HR = 0.17), BLCA (p = 0.005, HR = 0.48), COAD (p = 0.013, HR = 0.54), HNSC (p = 0.003, HR = 0.62), KIRC (p = 0.017, HR = 0.69), KIRP (p = 0.008, HR = 0.42), MESO (p = 0.004, HR = 0.45), OV (p = 0.005, HR = 0.75), PAAD (p = 0.011, HR = 0.41), PRAD (p = 0.005, HR = 0.56), SARC (p = 0.021, HR = 0.67), TGCT (p = 0.007, HR = 0.4) and THCA (p = 0.019, HR = 0.53) (Figure 8C).

FIGURE 8. Prognostic analysis of FAM46C using Kaplan–Meier curves for (A) OS, (B) DSS and (C) PFI in different tumours.
Gene Set Enrichment Analysis of FAM46C
To further explore the possible signalling pathways involving FAM46C, we evaluated 33 tumour types in TCGA database using GSEA, and gene enrichment pathways were analysed using KEGG and HALLMARK. According to KEGG analysis, cell adhesion, intestinal immunity and aldosterone–sodium ion regulation pathways were enriched in high FAM46C expression, suggesting that FAM46C had regulatory effects on these pathways (Figure 9A). However, biosynthetic pathways such as pentose phosphate and pyrimidine metabolism and pathogenic mechanisms such as those in Alzheimer’s and Huntington’s diseases were enriched in low FAM46C expression, suggesting that FAM46C might play an inhibitory role in these pathways (Figure 9B). According to the HALLMARK pathway analysis, ultraviolet (UV) receptor signalling pathways were enriched in high FAM46C expression, suggesting that FAM46C expression was positively correlated with these pathways (Figure 9C). However, oxidative phosphorylation, reactive oxygen metabolism and pathways such as MYC genes and glycolysis were enriched in low FAM46C expression, suggesting that FAM46C expression was negatively correlated with these pathways owing to its inhibitory effects (Figure 9D).
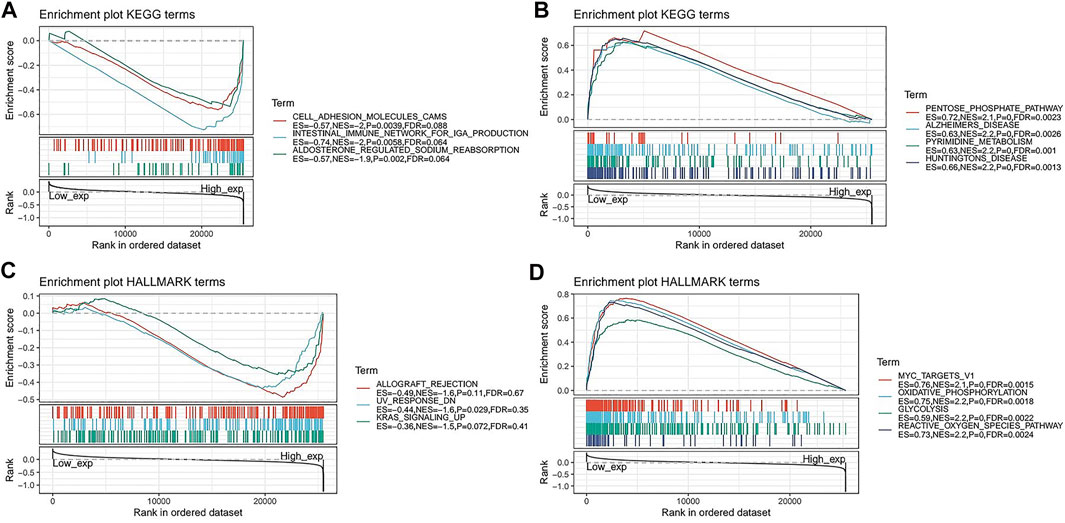
FIGURE 9. Gene set enrichment analysis of FAM46C. Functional terms linked with FAM46C enrichment through (A,B) KEGG pathway analysis and (C,D) HALLMARK pathway analysis.
Immune Cell Infiltration Analyses
To assess the relationship between FAM46C expression and immune cell infiltration, we used two different sources of immune cell infiltration data for correlation analysis. The TIMER2 database results revealed that in most tumours, FAM46C was positively correlated with CD4+ and CD8+ T-cell infiltration levels and negatively correlated with CD4+ Th1-cell infiltration levels (p < 0.05) (Figures 10A,B). We performed correlation analysis on published data using the CIBERSORT algorithm and evaluated 26 immune cells. The results revealed that FAM46C was positively correlated with the infiltration levels of plasma B cells, regulatory memory CD4+ T cells and CD8+T cells but was negatively correlated with the infiltration levels of bone marrow DCs and naive CD4+ T cells (p < 0.05) (Figure 10C). These results suggested that FAM46C promoted T-cell infiltration, thus explaining the protective role of FAM46C in most tumours.
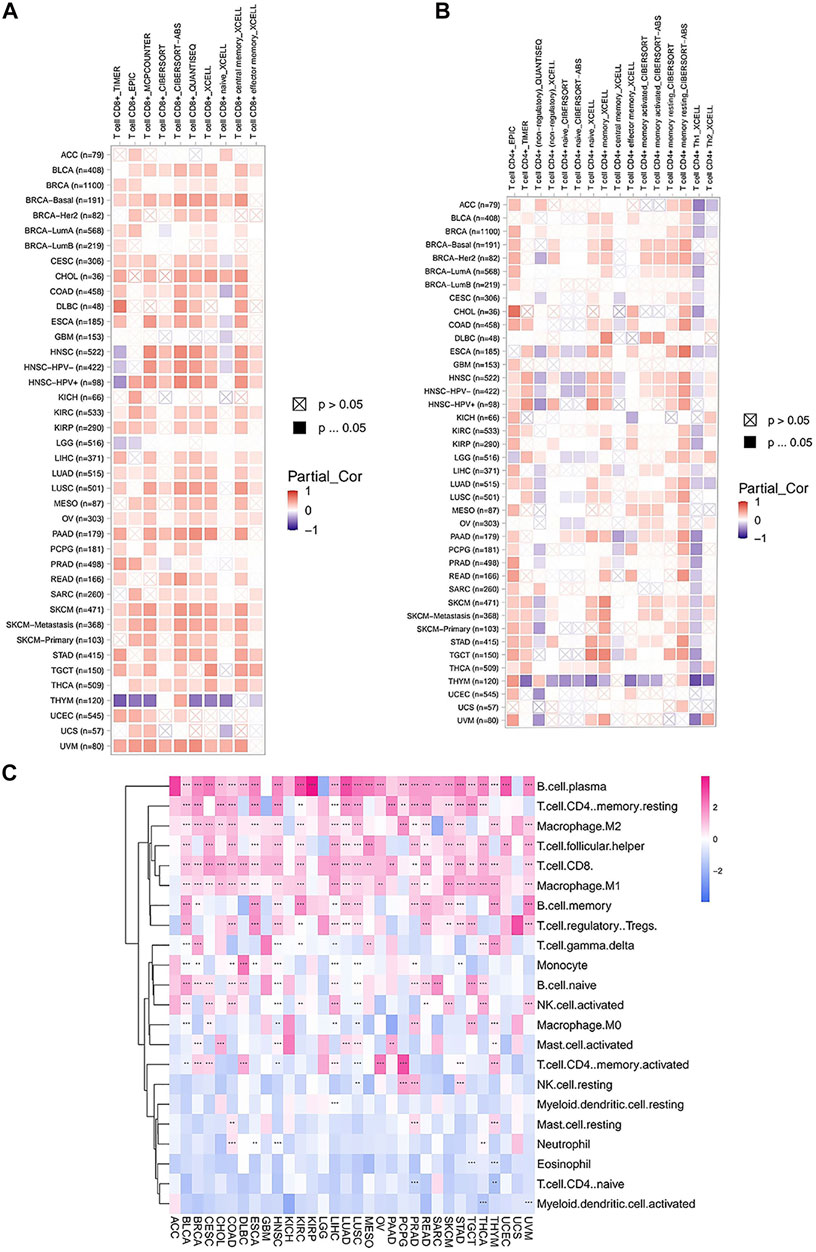
FIGURE 10. Correlation analysis between FAM46C expression and immune infiltration of cells. Correlation of FAM46C expression with (A) CD8+ T-cell infiltration, (B) CD4+ T-cell infiltration and (C) different types of immune infiltrating cells in pan-cancer.
Correlation of FAM46C With Immunomodulation-Related Genes and Chemokines
The association between FAM46C and immune-regulated genes in pan-cancer was observed using the Spearman’s correlation based on the stromal, immune and ESTIMATE scores (p < 0.05 and |R| > 0.3). FAM46C expression was specifically correlated with stromal fraction in HNSC (R = 0.4, p < 0.05), LGG (R = 0.395, p < 0.05) and LUSC (R = 0.592, p < 0.05) (Supplementary Figure 2). In addition, a specific correlation was observed between immune scores and FAM46C in BLCA (R = 0.443, p < 0.05), COAD (R = 0.539, p < 0.05), diffuse large B-cell lymphoma (DLBC) (R = 0.486, p < 0.05), HNSC (R = 0.577, p < 0.05), LGG (R = 0.415, p < 0.05), LUSC (R = 0.671, p < 0.05), skin cutaneous melanoma (SKCM) (R = 0.595, p < 0.05), TGCT (R = 0.559, p < 0.05) and uterine carcinosarcoma (UCS) (R = 0.359, p < 0.05) in which there was (Supplementary Figure 3). In addition, a higher correlation was observed between FAM46C and ESTIMATE scores in BLCA (R = 0.44, p < 0.05), COAD (R = 0.463, p < 0.05), DLBC (R = 0.386, p < 0.05), HNSC (R = 0.548, p < 0.05), LGG (R = 0.42, p < 0.05), LUSC (R = 0.666, p < 0.05), MESO (R = 0.33, p < 0.05) and SKCM (R = 0.589, p < 0.05), which was used as a combined ratio to assess the immune and stromal components in tumour tissues (Supplementary Figure 4).
Furthermore, the correlation between FAM46C and immune activation genes in pan-cancer was analysed. FAM46C expression was found to be positively correlated with TNFRSF17 and TNFRSF13B, except in DLBC, thymoma (THYM) and UCS. In most human tumours, FAM46C was negatively correlated with CD70. In addition, we also found that FAM46C expression exhibited a more significant negative correlation with CD276 and TNFSF9 in LUAD and LUSC (both p < 0.01) (Figure 11A). Immunosuppression also plays an important role in the immune regulation of tumours. The results revealed that FAM46C expression was positively correlated with TIGIT and negatively correlated with PVRL2 in most tumours, especially in LUAD, SARC and TGCT (all p < 0.01) (Figure 11B). We also investigated the specific association between FAM46C expression and chemokines and their receptors in pan-cancer and found that FAM46C expression was positively correlated with immune chemokines (CXCL9 and CXCL13) and immune chemokine receptors (CCR2 and CCR4) in most tumours (all p < 0.01) (Figures 11C,D).
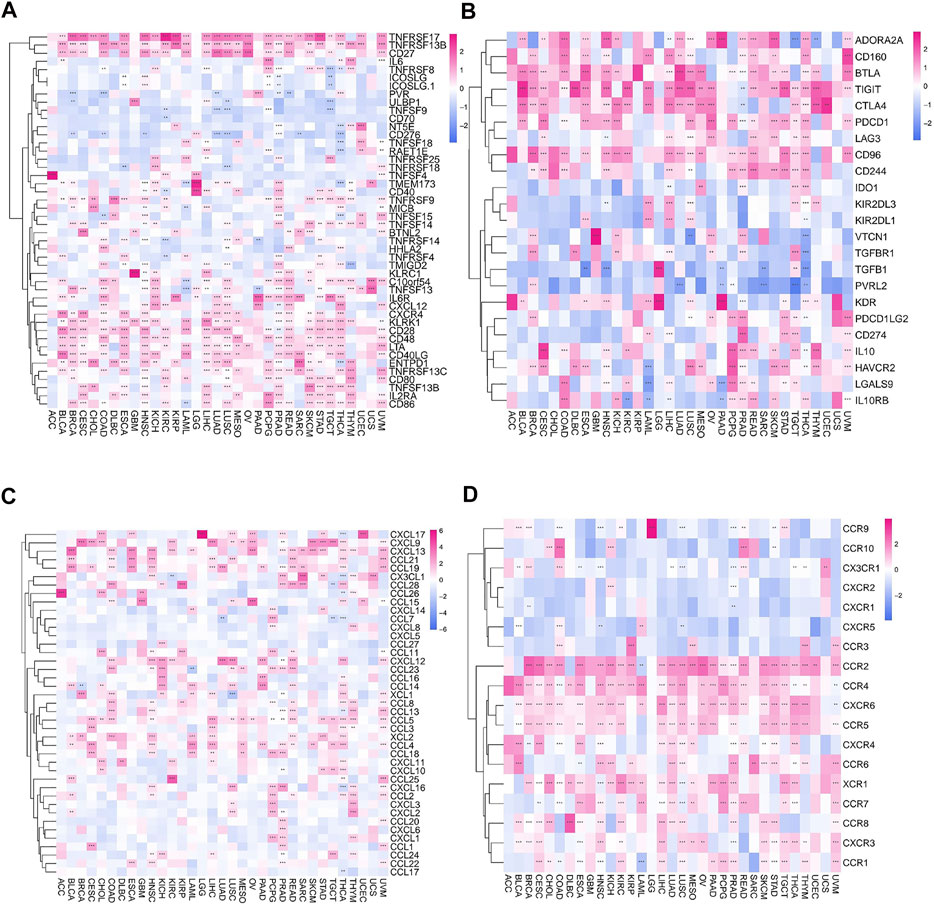
FIGURE 11. Relationship between FAM46C expression and immune-related factors. Relationship between FAM46C expression and (A) immune activating genes, (B) immunosuppressive genes, (C) immune chemokines and (D) immune chemokine receptors (*p < 0.05, **p < 0.01, ***p < 0.001).
FAM46C Expression Is Related to Immune Checkpoint Genes in Human Cancers
As mentioned above, we explored the role of FAM46C in immunomodulation. Currently, immunotherapy is one of the important tumour treatment approaches. Immunotherapy controls tumour progression by restarting and maintaining the tumour–immunity cycle and restoring the normal anti-tumour immune response of the body. Previous studies have demonstrated the significant effect of immune checkpoint (ICP) genes on immunotherapy (Zerfas et al., 2020). To further explore the potential of FAM46C in immunotherapy, we analysed the relationship between FAM46C expression and ICP genes. As demonstrated in Figure 12, FAM46C expression is positively correlated with 47 ICP genes in BLCA, BRCA, CESC, COAD, ESCA, HNSC, KIRC, LAML, LIHC, LUAD, LUSC, PAAD, PCPG, PRAD, READ, SKCM, STAD, TGCT, THCA, THYM and uveal melanoma (UVM). In particular, more than 30 of the 47 ICP genes were associated with FAM46C expression in COAD, SKCM, HNSC and UVM. In addition, FAM46C expression exhibited a positive correlation with several tumour types, suggesting that FAM46C promotes ICP gene activation by participating in several different signalling pathways and is a potential immunotherapeutic target (Figure 12).
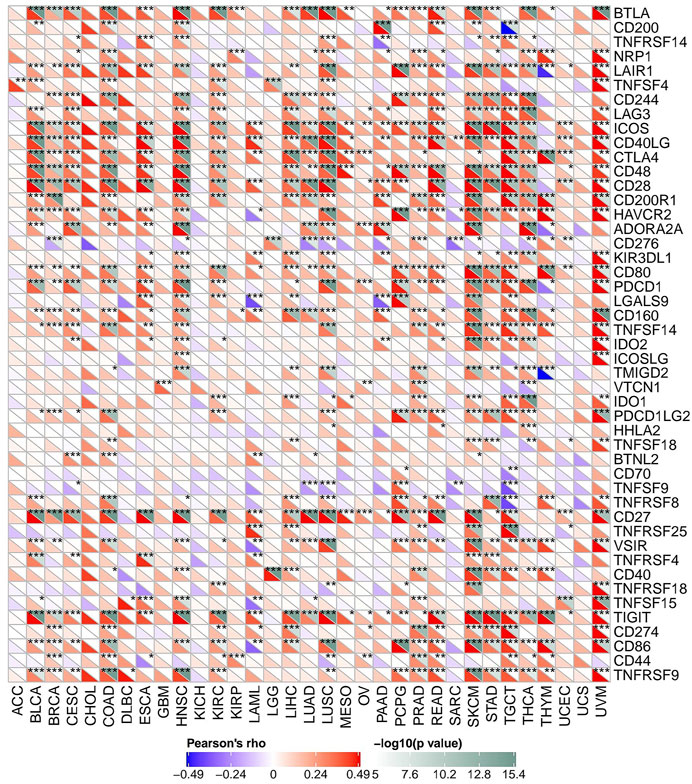
FIGURE 12. The association between FAM46C expression and immune checkpoint genes in pan-cancer (*p < 0.05, **p < 0.01, ***p < 0.001).
Relationship Between FAM46C Expression and TMB and MSI
TMB is an indicator of the extent of mutations in tumour cell species (Nikanjam et al., 2018). MSI, caused by defects in mismatch repair (MMR) genes, is closely associated with tumorigenesis and is an important molecular marker for the prognosis of solid tumours and the development of adjuvant immunotherapy regimens (Srinivas et al., 2001). The relationship between FAM46C expression and TMB and MSI can be evaluated for determining the potential clinical feasibility of immunisation with FAM46C. We used the Spearman’s correlation to assess the correlation between FAM46C expression and TMB and MSI. As demonstrated in Figures 13A,B, FAM46C is positively correlated with high TMB in LAML (p = 0.0022) and high MSI in CESC (p = 0.0063).
Discussion
The FAM46C gene is widely expressed in animal genomes, and its regulation encodes a non-canonical polymerase. Previously, mutations in the FAM46C gene were reported to be associated with the development and invasion of myeloma, with frequent mutations in FAM46C affecting the outcome and clinical prognosis of patients with myeloma. However, in recent years, abnormal expression levels of FAM46C have been found in several cancers. FAM46C overexpression inhibits invasion and metastasis and induces cell cycle arrest in hepatocellular carcinoma, whereas FAM46C deletion is associated with tumour progression and poor prognosis. In a study, FAM46C expression was significantly reduced in colorectal cancer tissues and increased after norethindrone (NCTD) treatment (Wan et al., 2017). These studies suggest that FAM46C is functionally associated with various human tumours, which guided us to explore the correlation of FAM46C expression levels with immunity, tumour staging and prognosis in various cancers to determine the potential of FAM46C in predicting clinical prognosis and immunotherapy. Therefore, we conducted a comprehensive study on FAM46 expression levels in different cancers using the Oncomine, TCGA and TIMER2 data.
First, we assessed the mRNA expression levels of FAM46C in tumours in the Oncomine and TCGA datasets. The results revealed that the expression level of FAM46C was significantly decreased in most cancers. Furthermore, we evaluated the mRNA expression levels of FAM46C in different stages of the same tumour based on TCGA clinical data. The results suggested a decreasing trend of FAM46C expression levels in patients with stage III–IV disease in most tumours. In addition, we used univariate Cox regression and Kaplan–Meier analyses to investigate the clinical prognostic value of FAM46C expression levels in tumours. It has been reported that reduced FAM46C expression levels are associated with shorter OS in patients with gastric cancer (Tanaka et al., 2017). We considered that the study endpoint of OS includes death from non-cancerous causes, which has limitations in reflecting tumour biological behaviour and treatment. To more effectively reflect the clinical factors affecting patients, we analysed the data related to PFI and DSS, and the combined results revealed that the prognosis of most tumours correlated with FAM46C expression levels. In particular, in BRCA, BLCA, KIRC, LUAD, MESO, OV, PAAD, SARC and other tumours, the prognosis of patients in the high FAM46C expression group was significantly better than that in the low expression group. Therefore, it can be tentatively concluded that FAM46C plays a protective role in most tumours.
In previous studies, mutations and changes in the protein expression levels of FAM46C have been mostly reported in multiple myeloma. However, the mutations and protein expression levels of FAM46C were associated with various tumours in our study. Recent studies have demonstrated that gene mutations alter gene expression levels in tumours and induce changes in the tumour microenvironment (TME). Genes may also bind to new loci to adapt to the new microenvironment via genetic recombination (Li et al., 2019). We found that mutations in the FAM46C gene affected its expression levels, which was further associated with altered biological behaviour in various tumours, indicating that FAM46C has the potential to predict prognosis and treatment outcomes in patients with tumours. In addition to significantly decreased FAM46C mRNA expression levels in most tumours, the expression levels of FAM46C and related proteins exhibited a decreasing trend in most tumours. It has been demonstrated that deletion of some genes in tumour cells results in reduced mRNA and protein abundance, which leads to increased tumour cell migration (Herrero et al., 2020), suggesting that the expression levels of related genes and proteins are related to tumorigenesis and invasion. On this basis, we compared our paired immunohistochemical and statistical results in cancerous and paracancerous tissues in the HPA database, and the paired results showed consistency. Immunohistochemical comparison validated the potential of FAM46C as a prognostic marker for tumours. In addition, the PPI network indicated that FAM46C was involved in regulating the functions of various proteins that contribute to apoptosis and immune recognition of the corresponding cancer cells by mediating different signalling pathways.
The reciprocal protein relationships and differential expression of mRNA reflect the potential value of FAM46C in predicting prognosis and modulating immunity. To assess the clinical application of FAM46C and the practicality and feasibility of tumour treatment, we analysed the relationship between FAM46C and immune pathways, cellular infiltration, immunomodulatory factors and immune checkpoints. According to the KEGG and HALLMARK pathway analyses, FAM46C was enriched in different immune pathways, and the expression level of FAM46C was closely related to the regulation of different immune and metabolic pathways. A study elucidated that the intestinal immune system is a signalling pathway that produces IgA and inhibits the proliferation and migration of hepatocellular carcinoma (Yang et al., 2018), which is consistent with our findings that the signalling pathway was enriched in high FAM46C expression, suggesting that FAM46C contributes to this pathway. Previous studies have reported that over-activation of the pentose phosphate pathway plays an important role in promoting the growth of cancer cells (Jiang et al., 2014; Ge et al., 2020). Therapeutic strategies to modulate and balance the pentose phosphate pathway are valuable for tumour treatment. Furthermore, MYC migration and overexpression may lead to increased aggressiveness of related tumours (Walker et al., 2014). In our study, the pentose phosphate and MYC pathways were enriched in low FAM46C expression, which further demonstrated the tumour suppressive role of FAM46C. This finding indicates that FAM46C can positively regulate immune responses and related metabolic pathways to exert a protective role in tumours.
Our study also highlights that FAM46C is closely associated with immune cell infiltration. Our results revealed that FAM46C was positively correlated with CD4+ T cell-, CD8+ T cell- and plasma B-cell infiltration levels and negatively correlated with naive CD4+ T-cell infiltration levels. A study demonstrated that FAM46C expression was decreased and the threshold of T-cell activation was increased in mice infected with Andes virus (ANDV), and FAM46C was one of the immune regulatory genes involved in the functioning of the immune system (Campbell et al., 2015). Seweryn Mroczek et al. found that B lymphocyte proliferation was significantly faster after strong induction of FAM46C protein in FAM46C-FLAG knockout mice, and FAM46C functioned as a tumour suppressor in multiple myeloma (Mroczek et al., 2017). Our findings are consistent with their findings, which suggest that FAM46C is associated with the production of B and T cells.
For immune stimulators and suppressors, our findings indicate that FAM46C is associated with multiple immune drivers in most tumours. Previous studies have reported that CD70 is overexpressed in various tumours. Elevated levels of CD70 expression enhance the suppressive effect of TME on the immune system (Jacobs et al., 2015). Anti-CD70 therapy has emerged as a new tumour immunotherapy approach. A study validated the efficacy of anti-CD70 in glioblastoma, wherein anti-CD70 treatment contributed to the regression of glioblastoma (Jin et al., 2018). However, other tumours that we analysed in this study, such as LUAD and THCA, have not yet been experimentally validated, which will be a target for our future study. However, the correlation between FAM46C expression and immunomodulators suggests that FAM46C has the potential to predict tumour progression and guide immunotherapy.
In addition, our study found that FAM46C expression was closely associated with ICP genes in most tumours, especially in COAD, SKCM, HNSC and UVM. The abnormal expression level of ICP genes in tumour cells is an important reason for the incomplete activation of T cells, which may explain the suppressive effect of TME on the immune response (Zerfas et al., 2020). In previous clinical trials, cancer vaccines have been effective in the treatment of prostate cancer (Jafari et al., 2020), although they could not overcome the suppressive effect of TME on the immune system owing to the weak sensitivity of monoclonal antibodies to immune loci. However, a considerable number of studies have confirmed that ICP genes play a key role in promoting the initiation and progression of tumours and inhibition of dominant immune cells in TME. However, most patients are resistant to a single ICP inhibitor. Therefore, identification of more promoters related to ICP therapy is the future direction of tumour treatment. Based on the findings of previous studies and our study, FAM46C can be considered a factor associated with ICP immunotherapy, which may be an ideal treatment strategy in the future.
According to TMB and MSI analyses, high expression of FAM46C was associated with LAML and CESC. Higher levels of TMB were found in patients with LAML, with the effectiveness of immunotherapy being higher in this group of patients (Galanina et al., 2018). This finding indicates that TMB is one of the methods to evaluate immunotherapy and also highlights the need to monitor gene mutations in patients with haematological tumours. It has also been reported in the literature that endometrial and cervical cancers in the presence of MSI are better treated with PD-1 immunotherapy (Bonneville et al., 2017), and treatment with immune checkpoint inhibitor is a promising therapeutic approach for future individualised testing. Therefore, the regulation of FAM46C is effective in tumours with high MSI.
However, despite the comprehensive and systematic analysis of FAM46C using different databases and statistical methods for cross-validation, this study has some limitations. First, bias and discrepancies between different databases may lead to systematic bias. Second, in vivo/in vitro experiments are required to validate the potential function of FAM46C and improve the credibility of our conclusions. Third, although our findings support that FAM46C expression is associated with immune cell infiltration and clinical prognosis in pan-cancer, we lack direct evidence for demonstrating the prognostic effect of FAM46C on immune infiltration. Therefore, the exact underlying immunomodulatory mechanisms of FAM46C remain to be further investigated. Fourth, the effect of FAM46C expression levels on prognosis and targeted therapies should be validated in clinical trials. In conclusion, the current role of FAM46C in immune infiltration in pan-cancer requires further validation by prospective studies to further guide the clinical treatment of tumours.
Conclusion
FAM46C can be used as a potential prognostic marker for pan-cancer, is closely related to immune regulation and may become an immune checkpoint to guide future clinical immunotherapy.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by The Medical Ethics Committee of the Affiliated Tumor Hospital of Guangxi Medical University. The ethics committee waived the requirement of written informed consent for participation.
Author Contributions
Conceived and designed the experiments:ZW, JD, WX; Performed the data collection: ZW, JD; Analyzed the data: ZW, JD, WX; Contributed reagents/materials/analysis tools: ZW, JD, WX; Contributed to the writing of the manuscript: ZW, JD, WX; All authors reviewed the manuscript.
Funding
This work was supported by the Guangxi Medical and health appropriate technology and popularization and application project (Grant No. S2020093).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.810252/full#supplementary-material
Supplementary Figure 1 | FAM46C expression in pan-cancer with different World Health Organization stages. The differential expression of FAM46C is not statistically significant (p > 0.05) in the WHO staging of different cancer types in TCGA database.
Supplementary Figure 2 | Analysis of the association between FAM46C expression and stromal scores using the Spearman’s rho test. The data with p < 0.05 are plotted.
Supplementary Figure 3 | Analysis of the association between FAM46C expression and immune scores using the Spearman’s rho test. The data with p < 0.05 are plotted.
Supplementary Figure 4 | Analysis of the association between FAM46C expression and the ESTIMATE scores using the Spearman’s rho test. The data with p < 0.05 are plotted.
References
Almamun, M., Levinson, B. T., van Swaay, A. C., Johnson, N. T., McKay, S. D., Arthur, G. L., et al. (2015). Integrated Methylome and Transcriptome Analysis Reveals Novel Regulatory Elements in Pediatric Acute Lymphoblastic Leukemia. Epigenetics 10 (9), 882–890. doi:10.1080/15592294.2015.1078050
Bonneville, R., Krook, M. A., Kautto, E. A., Miya, J., Wing, M. R., Chen, H.-Z., et al. (2017). Landscape of Microsatellite Instability across 39 Cancer Types. JCO precision Oncol. 2017 (1), 1–15. doi:10.1200/PO.17.00073
Campbell, C. L., Torres-Perez, F., Acuna-Retamar, M., and Schountz, T. (2015). Transcriptome Markers of Viral Persistence in Naturally-Infected andes Virus (Bunyaviridae) Seropositive Long-Tailed Pygmy rice Rats. PloS one 10 (4), e0122935. doi:10.1371/journal.pone.0122935
Chen, X., Wang, Y., Qu, X., Bie, F., Wang, Y., and Du, J. (2021). TRIM58 Is a Prognostic Biomarker Remodeling the Tumor Microenvironment in KRAS-Driven Lung Adenocarcinoma. Future Oncol. 17 (5), 565–579. doi:10.2217/fon-2020-0645
Clinton, S. K., Giovannucci, E. L., and Hursting, S. D. (2020). The World Cancer Research Fund/American Institute for Cancer Research Third Expert Report on Diet, Nutrition, Physical Activity, and Cancer: Impact and Future Directions. J. Nutr. 150 (4), 663–671. doi:10.1093/jn/nxz268
Crow, M. K. (2010). Type I Interferon in Organ-Targeted Autoimmune and Inflammatory Diseases. Arthritis Res. Ther. 12 (Suppl. 1), S5. doi:10.1186/ar2886
Galanina, N., Bejar, R., Choi, M., Goodman, A., Wieduwilt, M., Mulroney, C., et al. (2018). Comprehensive Genomic Profiling Reveals Diverse but Actionable Molecular Portfolios across Hematologic Malignancies: Implications for Next Generation Clinical Trials. Cancers 11 (1), 11. doi:10.3390/cancers11010011
Ge, T., Yang, J., Zhou, S., Wang, Y., Li, Y., and Tong, X. (2020). The Role of the Pentose Phosphate Pathway in Diabetes and Cancer. Front. Endocrinol. 11, 365. doi:10.3389/fendo.2020.00365
Herrero, A. B., Quwaider, D., Corchete, L. A., Mateos, M. V., García‐Sanz, R., and Gutiérrez, N. C. (2020). FAM46C Controls Antibody Production by the Polyadenylation of Immunoglobulin mRNAs and Inhibits Cell Migration in Multiple Myeloma. J. Cel Mol Med 24 (7), 4171–4182. doi:10.1111/jcmm.15078
Hoang, P. H., Cornish, A. J., Dobbins, S. E., Kaiser, M., and Houlston, R. S. (2019). Mutational Processes Contributing to the Development of Multiple Myeloma. Blood Cancer J. 9 (8), 60. doi:10.1038/s41408-019-0221-9
Hou, Y., Lan, C., Kong, Y., Zhu, C., Peng, W., Huang, Z., et al. (2019). Genetic Ablation of TAZ Induces HepG2 Liver Cancer Cell Apoptosis through Activating the CaMKII/MIEF1 Signaling Pathway. Onco Targets Ther. 12, 1765–1779. doi:10.2147/OTT.S196142
Hu, J.-L., Liang, H., Zhang, H., Yang, M.-Z., Sun, W., Zhang, P., et al. (2020). FAM46B Is a Prokaryotic-like Cytoplasmic Poly(A) Polymerase Essential in Human Embryonic Stem Cells. Nucleic Acids Res. 48 (5), 2733–2748. doi:10.1093/nar/gkaa049
Jacobs, J., Deschoolmeester, V., Zwaenepoel, K., Rolfo, C., Silence, K., Rottey, S., et al. (2015). CD70: An Emerging Target in Cancer Immunotherapy. Pharmacol. Ther. 155, 1–10. doi:10.1016/j.pharmthera.2015.07.007
Jafari, S., Molavi, O., Kahroba, H., Hejazi, M. S., Maleki-Dizaji, N., Barghi, S., et al. (2020). Clinical Application of Immune Checkpoints in Targeted Immunotherapy of Prostate Cancer. Cell. Mol. Life Sci. 77 (19), 3693–3710. doi:10.1007/s00018-020-03459-1
Jiang, P., Du, W., and Wu, M. (2014). Regulation of the Pentose Phosphate Pathway in Cancer. Protein Cell 5 (8), 592–602. doi:10.1007/s13238-014-0082-8
Jin, L., Ge, H., Long, Y., Yang, C., Chang, Y., Mu, L., et al. (2018). CD70, a Novel Target of CAR T-Cell Therapy for Gliomas. Neuro. oncol 20 (1), 55–65. doi:10.1093/neuonc/nox116
Kazazian, K., Haffani, Y., Ng, D., Lee, C. M. M., Johnston, W., Kim, M., et al. (2020). FAM46C/TENT5C Functions as a Tumor Suppressor through Inhibition of Plk4 Activity. Commun. Biol. 3 (1), 448. doi:10.1038/s42003-020-01161-3
Kuchta, K., Muszewska, A., Knizewski, L., Steczkiewicz, K., Wyrwicz, L. S., Pawlowski, K., et al. (2016). FAM46 Proteins Are Novel Eukaryotic Non-canonical Poly(A) Polymerases. Nucleic Acids Res. 44 (8), 3534–3548. doi:10.1093/nar/gkw222
Li, F., Kitajima, S., Kohno, S., Yoshida, A., Tange, S., Sasaki, S., et al. (2019). Retinoblastoma Inactivation Induces a Protumoral Microenvironment via Enhanced CCL2 Secretion. Cancer Res. 79 (15), 3903–3915. doi:10.1158/0008-5472.CAN-18-3604
Liu, M., Zhang, X., Cai, J., Li, Y., Luo, Q., Wu, H., et al. (2018). Downregulation of TRIM58 Expression Is Associated with a Poor Patient Outcome and Enhances Colorectal Cancer Cell Invasion. Oncol. Rep. 40 (3), 1251–1260. doi:10.3892/or.2018.6525
Liu, X., Tang, H., Liu, J., and Wang, X. (2020). hsa_circRNA_101237: A Novel Diagnostic and Prognostic Biomarker and Potential Therapeutic Target for Multiple Myeloma. Cancer Manag. Res. 12, 2109–2118. doi:10.2147/CMAR.S241089
Ma, L., He, H., Jiang, K., Jiang, P., He, H., Feng, S., et al. (2020). FAM46C Inhibits Cell Proliferation and Cell Cycle Progression and Promotes Apoptosis through PTEN/AKT Signaling Pathway and Is Associated with Chemosensitivity in Prostate Cancer. Aging 12 (7), 6352–6369. doi:10.18632/aging.103030
Mroczek, S., Chlebowska, J., Kuliński, T. M., Gewartowska, O., Gruchota, J., Cysewski, D., et al. (2017). The Non-canonical Poly(A) Polymerase FAM46C Acts as an Onco-Suppressor in Multiple Myeloma. Nat. Commun. 8 (1), 619. doi:10.1038/s41467-017-00578-5
Nikanjam, M., Cohen, P. R., Kato, S., Sicklick, J. K., and Kurzrock, R. (2018). Advanced Basal Cell Cancer: Concise Review of Molecular Characteristics and Novel Targeted and Immune Therapeutics. Ann. Oncol. 29 (11), 2192–2199. doi:10.1093/annonc/mdy412
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer Statistics, 2021. CA A. Cancer J. Clin. 71 (1), 7–33. doi:10.3322/caac.21654
Srinivas, P. R., Kramer, B. S., and Srivastava, S. (2001). Trends in Biomarker Research for Cancer Detection. Lancet Oncol. 2 (11), 698–704. doi:10.1016/s1470-2045(01)00560-5
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tanaka, H., Kanda, M., Shimizu, D., Tanaka, C., Kobayashi, D., Hayashi, M., et al. (2017). FAM46C Serves as a Predictor of Hepatic Recurrence in Patients with Resectable Gastric Cancer. Ann. Surg. Oncol. 24 (11), 3438–3445. doi:10.1245/s10434-016-5636-y
Walker, B. A., Wardell, C. P., Brioli, A., Boyle, E., Kaiser, M. F., Begum, D. B., et al. (2014). Translocations at 8q24 Juxtapose MYC with Genes that Harbor Superenhancers Resulting in Overexpression and Poor Prognosis in Myeloma Patients. Blood Cancer J. 4, e191. doi:10.1038/bcj.2014.13
Wan, X. Y., Zhai, X. F., Jiang, Y. P., Han, T., Zhang, Q. Y., and Xin, H. L. (2017). Antimetastatic Effects of Norcantharidin on Hepatocellular Carcinoma Cells by Up-Regulating FAM46C Expression. Am. J. Transl Res. 9 (1), 155–166.
Yang, Z., Tao, Y., Xu, X., Cai, F., Yu, Y., and Ma, L. (2018). Bufalin Inhibits Cell Proliferation and Migration of Hepatocellular Carcinoma Cells via APOBEC3F Induced Intestinal Immune Network for IgA Production Signaling Pathway. Biochem. biophys. Res. Commun. 503 (3), 2124–2131. doi:10.1016/j.bbrc.2018.07.169
Yeom, C. J., Zeng, L., Goto, Y., Morinibu, A., Zhu, Y., Shinomiya, K., et al. (2016). LY6E: a Conductor of Malignant Tumor Growth through Modulation of the PTEN/PI3K/Akt/HIF-1 axis. Oncotarget 7 (40), 65837–65848. doi:10.18632/oncotarget.11670
Zerfas, B. L., Maresh, M. E., and Trader, D. J. (2020). The Immunoproteasome: An Emerging Target in Cancer and Autoimmune and Neurological Disorders. J. Med. Chem. 63 (5), 1841–1858. doi:10.1021/acs.jmedchem.9b01226
Zhang, Q.-Y., Yue, X.-Q., Jiang, Y.-P., Han, T., and Xin, H.-L. (2017). FAM46C Is Critical for the Anti-proliferation and Pro-apoptotic Effects of Norcantharidin in Hepatocellular Carcinoma Cells. Sci. Rep. 7 (1), 396. doi:10.1038/s41598-017-00313-6
Zhang, S., Yang, Y., Hua, Y., Hu, C., and Zhong, Y. (2020). NCTD Elicits Proapoptotic and Antiglycolytic Effects on Colorectal Cancer Cells via Modulation of Fam46c Expression and Inhibition of ERK1/2 Signaling. Mol. Med. Rep. 22 (2), 774–782. doi:10.3892/mmr.2020.11151
Zhou, J., Zhang, S., Li, Z., Chen, Z., Xu, Y., Ye, W., et al. (2019). Yap-Hippo Promotes A549 Lung Cancer Cell Death via Modulating MIEF1-Related Mitochondrial Stress and Activating JNK Pathway. Biomed. Pharmacother. 113, 108754. doi:10.1016/j.biopha.2019.108754
Zhu, Y. X., Shi, C.-X., Bruins, L. A., Jedlowski, P., Wang, X., Kortüm, K. M., et al. (2017). Loss of FAM46C Promotes Cell Survival in Myeloma. Cancer Res. 77 (16), 4317–4327. doi:10.1158/0008-5472.CAN-16-3011
Keywords: human cancer, immunomodulation, immunotherapy, prognosis, tumour microenvironment
Citation: Deng J, Xiao W and Wang Z (2022) FAM46C as a Potential Marker for Pan-Cancer Prognosis and Predicting Immunotherapeutic Efficacy. Front. Genet. 13:810252. doi: 10.3389/fgene.2022.810252
Received: 11 November 2021; Accepted: 18 January 2022;
Published: 09 February 2022.
Edited by:
Je-Keun Rhee, Soongsil University, South KoreaReviewed by:
Opeyemi Soremekun, MRC/UVRI and MRC/UVRI at London School of Health and Tropical Medicine, UgandaRui Vitorino, University of Aveiro, Portugal
Copyright © 2022 Deng, Xiao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng Wang, Mjg1NDg0NzQzQHFxLmNvbQ==
 Jiehua Deng
Jiehua Deng Wei Xiao2
Wei Xiao2