- 1Department of Stem Cell and Regenerative Biotechnology, Konkuk University, Seoul, South Korea
- 2College of Veterinary Medicine, Chonbuk National University, Iksan, South Korea
- 3Gift of Hope Organ & Tissue Donor Network, Itasca, IL, United States
Immortalized cell lines are valuable resources to expand the molecular characterization of major histocompatibility complex genes and their presented antigens. We generated a panel of immortalized cell lines by transfecting human telomerase reverse transcriptase (hTERT) into primary fibroblast cells prepared from ear, fetal, and lung tissues of 10 pigs from five breeds and successfully cultured them for 30–45 passages. The cell growth characteristic of the immortalized fibroblasts was similar to that of primary fibroblast, which was unable to form colonies on soft agar. The genotypes of major swine leukocyte antigen (SLA) genes, including three classical class I (SLA-1, -2, and -3) and three class II genes (DQB1, DRB1, and DQA), were determined using high-resolution typing. A total of 58 alleles, including a novel allele for SLA-2, were identified. Each cell line was unique. A cell line derived from a National Institutes of Health miniature pig was homozygous across the six major SLA genes. The expression levels of SLA classical class I genes varied among the cell lines and were slightly upregulated in the immortalized compared to the primary cells based on semiquantitative reverse transcription polymerase chain reaction. The immortalized porcine fibroblast cell lines with diverse SLA haplotypes that were developed in this study have potential to be applied in studies regarding the molecular characteristics and genetic structure of SLA genes and epitope–major histocompatibility complex interactions in pigs.
Introduction
Antigen presentation by major histocompatibility complex (MHC) class I and II molecules is an essential process in adaptive immune interactions that triggers T cell proliferation and antigen-specific cellular immune responses (Wieczorek et al., 2017). MHC class I molecules are expressed on the surface of all nucleated cells and interact with cytotoxic T cells (CTL) to trigger immune responses against intracellular pathogenic peptides, derived from viral proteins and tumor cells (Kaufman et al., 1990; Sommer, 2005). In contrast, MHC class II molecules are expressed only by professional antigen-presenting cells, including dendritic cells, mononuclear phagocytes, B cells, among others, and peptide/MHC class II complexes interact with T helper cells to mediate immune responses against extracellular pathogens (Lechler et al., 1991; Ting and Trowsdale, 2002). Therefore, the engagement of pathogen-derived antigenic peptides with MHC molecules, before recognition by surveillance T lymphocytes, plays a critical role in adaptive immunity.
MHC genes are extremely polymorphic and generating distinctive structural variations, especially in the antigen binding pockets that interact with antigenic peptides by anchoring amino acids (Rammensee, 1995). A single MHC molecule can bind to multiple peptides and a peptide could be presented by several different MHC molecules (Van Hateren et al., 2010; Eisen et al., 2012). This increases the potential diversity of antigenic peptides repertoire presented to T cells from endogenous and exogenous environments.
The activation of primary T cells requires the integration of three distinct signals, namely the three-signal theory of activation including antigen recognition, costimulation, and cytokine-mediated differentiation and expression (Sckisel et al., 2015). In these sequential processes, only antigenic peptides with stable and strong bonds to MHC molecules can be presented to T cells and induce antigen-specific immune responses (Wooldridge et al., 2012). Therefore, a detailed understanding of binding kinetics between MHC molecules and antigenic peptides is necessary to improve the efficiency of epitope screening in the development of new generation vaccines and accurate predictive algorithms (Stienekemeier et al., 2001; Lazoura and Apostolopoulos, 2005; Reche et al., 2006). In addition, the determination of MHC alleles strongly interacting with antigenic peptides of major pathogens could be exploited as a strategy to increase the resistance of animals by selecting individuals with resistance haplotypes in population (Bosch-Camós et al., 2021).
Determination of MHC alleles is important to predict adaptive immune responses of an individual. The genomic sequence-based high-resolution typing methods for SLA genes have been developed (Park et al., 2010; Thong et al., 2011; Le et al., 2012; Choi H. et al., 2015; Le et al., 2015). However, all available methods are limited to genotype a single locus. Thus, the development of high throughput typing methods suitable for parallel typing of multiple SLA genes are in need.
Several methods have been developed to estimate interaction kinetics between antigens and MHC molecules using cell-free or animal cell systems (Yaneva et al., 2010). Compared to artificial cell-free assays in bacteria and yeasts, the method using live animal cells does not require the cloning and expression of MHC or antigenic molecules. When screening against a large number of MHC alleles, the live animal cell system requires reduced labor investment for greater reproducibility (Townsend et al., 1990; Cerundolo et al., 1991; Elvin et al., 1991; Sette et al., 1994; Ottenhoff et al., 1997; Tan et al., 1997; Buus et al., 2001; Sidney et al., 2001; Narzi et al., 2008). The native structure of MHC molecules on the surface of live mammalian cells allows for a more representative characterization of antigen biding interactions in vivo compared to the cell-free system (Bodmer et al., 1989; Ljunggren et al., 1990; Storkus et al., 1993; Zeh et al., 1994; Bremers et al., 1995; van der Burg et al., 1995; Kessler et al., 2001; Turvy and Blum, 2001; Palankar et al., 2009).
Many cell lines with well characterized MHC genotypes are available for molecular characterization of human lymphocyte antigens (Sckisel et al., 2015) and epitope binding and elution assays (Townsend et al., 1990; Tan et al., 1997; Kessler et al., 2001). However, the availability of these cell lines is limited for livestock species, and only eight cell lines with genotype information of major SLA genes are available in pigs (Schook et al., 2005; Vodicka et al., 2005; Ho et al., 2009; Meurens et al., 2012). Also, the alleles of available cell lines often overlap, further limiting the coverage of analyses using major swine leukocyte antigens (SLA) alleles (Ho et al., 2009).
In immunopeptidome studies, the number of cells that are evaluated per experiment is one of the most important limitations (Melief and Kessler, 2017), and immortalization could solve that condition. The objective of this study was to establish and characterize a cell panel consisting of 10 immortalized fibroblast cell lines containing 58 SLA alleles. The availability of cell lines covering diverse alleles of SLA could serve as a valuable resource for immunogenetic analysis in pigs and the development of high-throughput SLA typing methods.
Materials and Methods
Tissues and Cell Preparation
Tissue samples (ear notch or lung tissue) were obtained from five purebred landraces on local farms, two NIH miniature pigs, one PWG micropig (PWG Genetics, Korea), and one Seoul National University miniature pig (SNU, Seoul, Korea) (Choi H. et al., 2015; Choi M. et al., 2015; Le et al., 2020). All animals were 9 weeks old. A Yorkshire uterus, with fetus, was obtained from a local slaughterhouse (Seoul, South Korea). PK15, a pig kidney fibroblast cell line (ATCC CCL-33), was purchased from American Type Culture Collection (ATCC, Manassas, VA, United States). Primary pig fibroblast cells were isolated as previously described (Denning et al., 2001). Briefly, tissue samples were rinsed in 1x phosphate buffered saline (PBS) solution containing 1,000 U/ml penicillin and 500 μg/ml streptomycin (Gibco, NY, United States), treated with 1x Antibiotic-Antimycotic (Gibco), minced with a surgical blade in a culture dish, and disaggregated in 0.25% Trypsin-EDTA (Gibco) at 37°C for 1 h in a 5% CO2 incubator (ASTEC, Fukuoka, Japan). The disaggregated cells were washed in PBS and cultured in six-well plates using Dulbecco’s Modified Eagle’s Medium (DMEM) (HyClone, UT, United States) with 10% fetal bovine serum (FBS) (Thermo Scientific, MA, United States) and 20 μg/ml gentamycin (Sigma-Aldrich, MO, United States). Subsequently, the cells were cultured in DMEM supplemented with 10% FBS and a 1% penicillin-streptomycin antibiotic mixture (Gibco). All experimental procedures were approved and performed accordance with the guidelines and regulations set by the Institute of Animal Care and Use Committee (IACUC) and the Center for Research Ethincs of Konkuk University (KU20229).
High Resolution SLA Typing
Genomic DNA was extracted from ∼0.5 g of tissue using the standard protocol (Sambrook et al., 1989). Briefly, tissues were incubated with lysis buffer [10 mM Tris-HCl (pH 8.0) and 0.1 M EDTA] containing 0.5% sodium dodecyl sulfate and 1 mg/ml proteinase K (Promega, Madison, WI, United States) at 55°C for 6 h, followed by phenol extraction. For cells, DNA and RNA were isolated simultaneously from 5×106 cells using an AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Genomic DNA-based high-resolution typing of SLA-1, SLA-2, SLA-3, SLA-DQA, SLA-DQB1, and SLA-DRB1 was conducted for each gene as previously described (Park et al., 2010; Thong et al., 2011; Le et al., 2012; Choi H. et al., 2015; Le et al., 2015; Le et al., 2020). Briefly, locus-specific amplicons of each SLA gene were generated in a 20 μL reaction containing 50–100 ng of genomic DNA, 0.5 μM primers (SLA1-e1F1 and SLA-e4R4, SLA2-e1F2 and SLA-e4R4, SLA3-spF5 and SLA-e4R4, DQAi1F3 and DQAe3R1, DQB1F-119 and DQB1R+295, and DRB1F-22 and DRB1R+284 for SLA-1, SLA-2, SLA-3, SLA-DQA, SLA-DQB1, and SLA-DRB1, respectively, Supplementary Table S1), 200 μM dNTPs, 1x PCR buffer, and 0.5 U of Supertherm™ DNA polymerase (JMR Holdings, Kent, United Kingdom) using an ABI 9700 Thermocycler (Applied Biosystems, Foster City, CA, United States). The cycling profiles consisted of the following steps: initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 1 min, annealing at a specific temperature for each locus (Supplementary Table S1) for 1 min, extension at 72°C for 2 min, and a final extension at 72°C for 5 min. PCR products were analyzed by 1.5% agarose gel electrophoresis. Amplicons were then subjected to direct sequencing using locus-specific sequencing primers (SLA1-Seq. 2-F and SLA1-Seq. 3-F, SLA-i1F and SLA-i2F, SLA3-seq2-F6, SLA3-seq2-R7, SLA3-seq3-F4 and SLA3-seq3-R5, DQAi1F4, Sq-mul-DQB1 and Sq-mul-DRB1 for SLA-1, SLA-2, SLA-3, SLA-DQA, SLA-DQB1, and SLA-DRB1, respectively, Supplementary Table S1) using an ABI PRISM BigDye™ Terminator Cycle Sequencing Kit (Applied Biosystems) following the manufacturer’s protocol. Allelic determination of each SLA gene was conducted by aligning the sequence-based typing results with the annotated reference sequences from a local SLA database (containing all SLA sequences uploaded to GenBank) using a multiple alignment tool, CLC Main Workbench 7 (Qiagen CLC bio, Hilden, Germany). To separate and identify each allele in the heterozygotes, the PCR products were cloned when the pattern was observed for the first time, and then the individual clones of each allele were sequenced as described previously (Thong et al., 2011). At least two clones for each allele were sequenced from both the 3′ and 5′ directions. Observed heterozygosity was calculated as the number of heterozygotes divided by the total number of individuals.
Transfection and Selection of Cells
Cells were transfected with pBABE-hTERT-Puro (Counter et al., 1998) using LipofectaminTM 3000 (Thermo Scientific) following the manufacturer’s protocol. Cell selection occurred after 48 h when transfected cells were placed under selective pressure by adding 2 μg/ml puromycin (Sigma-Aldrich, Germany) for 4 weeks. The cells were transferred and cultured in DMEM supplemented with 10% FBS and a 1% penicillin-streptomycin antibiotic mixture (Gibco) at 37°C and 5% CO2. Cells were frozen overnight at −80°C in DMEM containing 20% FBS and 10% dimethyl sulfoxide, without antibiotics, and then stored in liquid nitrogen. Before use, the cells were rapidly thawed in a water bath at 37°C.
Characterization of Cell Growth and Soft Agar Assay
Cells (2 × 105 cells/well) were seeded in a six-well plate and cultured in the same conditions as described for cell selection. Cells were trypsinized at 24, 48, and 72 h post-seeding, counted using the trypan blue exclusion method (Strober, 2001), and the number of cells were plotted against each time point. To measure cell anchorage-independent proliferation, 1 × 104 cells/well of randomly selected immortalized fibroblasts were cultured for 2 weeks in a six-well soft agar dish with 0.8% agar and 0.7% agarose in the bottom and upper layers, respectively. Cells were fed with cell culture media every 2 days. Colony formation was monitored using a light microscope (Model BX51 TF, Olympus, Tokyo, Japan).
Expression Analysis of hTERT
Total RNA from 2 × 105 cells was extracted using an RNA/DNA mini kit (Qiagen, Hilden, Germany) and subjected to cDNA synthesis by SuperiorScript III Reverse Transcriptase (Enzynomics, Korea) according to the manufacturer’s protocol. PCR was performed in a 20 µL reaction mixture containing 2 µL cDNA, 0.5 µM primers, 0.5 U of Supertherm DNA polymerase (JMR Holdings) in 1.2x PCR buffer (1.5 nM MgCl2), and 0.1 mM dNTPs using the ABI 9700 Thermocycler (Applied Biosystems). The amplification primers for hTERT and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were hTERT-F (5′- GCCGAGACCAAGCACTTCCTCTACT-3′) and hTERT-R (5′-GCAACTTGCTCCAGACACTCTTCCG-3′), and GAPDH-SP-F (5′-GCTACACTGAGGACCAGGTTG-3′) and GAPDH-SP-R (5′- AGGAGATGCTCGGTGTGTTG-3′), respectively. The PCR profile consisted of an initial denaturation cycle of 5 min at 95°C, 35 cycles for 30 s at 95°C, 30 s at 63°C, 45 s at 72°C, and a final extension of 5 min at 72°C for hTERT. The cycling conditions for GAPDH was similar to that of hTERT except that the denaturation cycle was 3 min (at the same temperature), the number of annealing cycles was 25 at 58°C, and extension was performed for 30 s at 72°C. Subsequently, PCR products were electrophoresed on 1% agarose gel and band intensity was compared between samples.
Expression Analysis of SLA Genes and Vimentin
Isolated total RNA was subjected to RNase-free DNaseI (Qiagen Sciences Inc., Germantown, MD, United States) treatment, and RNA quality was analyzed on 2% formaldehyde agarose gel. Reverse transcription was performed in a 20 μL reaction mixture using 1 μg of total RNA with oligo-(dT)15 and SuperScript® III Reverse Transcriptase (Invitrogen, MA, United States), for 50 min at 50°C with inactivation for 15 min at 72°C. In contrast to the materials used in SLA typing, the target cDNA was amplified using 2 μL of cDNA with Pyrobest polymerase (TaKaRa Bio, Shiga, Japan). The primers used to amplify the entire coding region of SLA-1, SLA-2, SLA-3, and SLA-DRB1 were SLA1-e1F1 and SLA-e4R4, SLA2-e1F and SLA-e4R4, SLA1/3f#92 and SLA3r#121, CpDRB5a and CpDRB3b, respectively (Ho et al., 2006; Thong et al., 2011; Choi H. et al., 2015; Le et al., 2020) (Supplementary Table S1). The thermal cycling profile included a 3 min denaturation step at 95°C, 32 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 65°C, 61°C, 50°C, and 63°C for SLA-1, -2, -3, and -DRB1, respectively, 5 min extension at 72°C, and a final extension of 30 min at 72 °C. The amplification of vimentin was carried out using a primer set, Vimentin-F and Vimentin-R (Supplementary Table S1, Shi et al., 2013), using 2 μL of cDNA in the same way as above under the PCR profile consisting of an initial denaturation cycle of 3 min at 95°C, 35 cycles for 30 s at 95°C, 30 s at 58°C, 30 s at 72°C, and a final extension of 5 min at 72°C. GAPDH was used as a control. The level of expression was estimated as the photo density ratio of the SLA-1, -2, -3, and -DRB1 amplicons relative to GAPDH.
SLA Class I Immuno-Cytochemical Analysis
Cells were seeded on glass cover slips in 6-well plates at a density of 1.2 × 105 cells/well in DMEM in the same conditions described above. After 2°days, cover slips with attached cells were washed three times with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. Cells were washed three times with cold PBS before blocking with 5% bovine serum albumin (BSA) in PBS for 30 min at room temperature. The cells were incubated with a mouse monoclonal antibody specific to pig SLA Class I (Bio-Rad, CA, United States) at 1:250 dilution in PBS with 2.5% BSA at room temperature for 2 h. After washing in PBS, cells were incubated with goat anti-mouse antibody conjugated with Alexa Fluor 568 (Invitrogen) at a dilution of 1:500 in PBS containing 3% BSA for 1 h at room temperature in the dark. After washing in cold PBS, cover slips were mounted with Vectashield (Vector, Burlingame, CA, United States). The cells were observed under a fluorescence microscope (BX51TF, Olympus).
Results
Genetic Diversity of the Three SLA Classical Class I and Three Class II Genes
To establish a panel of cell lines with diverse SLA haplotypes, we conducted high-resolution typing of three SLA classical class I genes, SLA-1, SLA-2, and SLA-3, and three SLA class II genes, SLA-DQA, SLA-DQB1, and SLA-DRB1, from 10 individuals of five different breeds (Table 1). A total of 16, 13, 8, 6, 8, and 7 alleles were identified for SLA-1, SLA-2, SLA-3, SLA-DQA, SLA-DQB1, and SLA-DRB1, respectively. A new SLA-2 allele was identified showing 27 nucleotide differences from SLA-2*13:02. The sequence of the new allele was confirmed by cloning, submitted to NCBI (accession number MZ700355), by the ISAG SLA nomenclature committee.
The mean heterozygosity of SLA-1, SLA-2, SLA-3, SLA-DQA, SLA-DQB1, and SLA-DRB1 was 90%, 80%, 60%, 50%, 70%, and 60%, respectively, with SLA-1 and DQA showing the highest and the lowest heterozygosity, respectively. The typing results for individuals corresponding to iKUF cells 02, 04, 06, and 09 showed the presence of three alleles in SLA-1, indicating the presence of the duplication haplotypes. The allelic constitution for each SLA gene was consistent to the reported allelic frequencies of the corresponding breeds (Park et al., 2010; Thong et al., 2011; Le et al., 2012; Choi H. et al., 2015; Le et al., 2015; Le et al., 2020).
In our cell panel, iKUF04, which originated from an inbred NIH miniature pig, was homozygous across all six loci (SLA-1: 04:01; SLA-2: 04:01; SLA-3: 04:01; DQA: 02:01; DQB1: 04:01:01; DRB1: 02:01) (Table 1). Based on this, we were able to determine the haplotype of iKUF03 (SLA-1: 02:01-07:01; SLA-2: 02:01; SLA-3: 03:03; DQA: 02:01; DQB1: 02:01; DRB1: 02:01) by subtracting the identified haplotype of iKUF04 from the genotyping results of iKUF03. For the rest of the cell lines, we were unable to separate the combined genotyping results of the six SLA genes into individual haplotypes due to the unavailability of family typing results.
Establishment of 10 Immortalized Porcine Fibroblast Cell Lines With High Resolution SLA Genotypes
Primary fibroblast cells obtained from different tissues of 10 pigs were transfected with the hTERT expression construct. After culturing for 4 weeks in the selection medium containing puromycin, ∼10–50% of cells survived. A small number of cells were found to grow rapidly from enlarged and flattened senescent cells when they were relaxed in a puromycin-free culture. These cells were isolated and cultured until passage number 20 to make a homogeneous population. The passage numbers of the established fibroblast cell lines ranged from 30 to 45 (Table 1). We considered the cells with uniform morphology after 30 passages to be immortalized (Figure 1). To characterize the growth of the immortalized cells, we randomly selected three cell lines, iKUF08, iKUF09, and iKUF10, and compared their proliferation rates together with that of PK15, a commercially available porcine fibroblast cell line (Figure 2). Cell proliferation rates were much higher in PK15 and iKUF09 than in iKUF08 and iKUF10, showing variations in cell growth among them. The difference in proliferation rates among iKUF08, iKUF09, and iKUF10 was not associated to passage number (Figure 2 and Table 1).
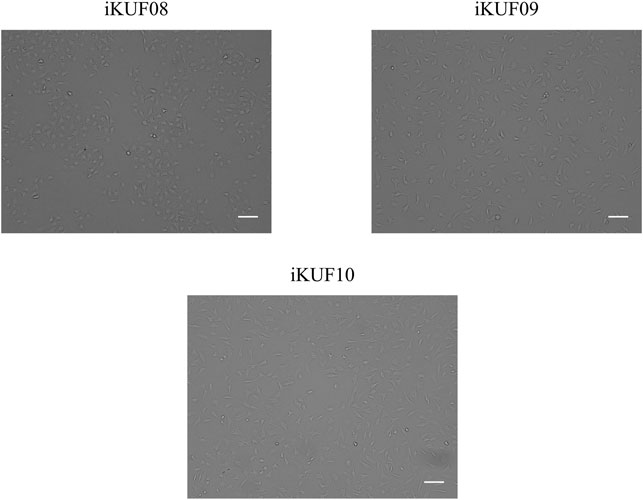
FIGURE 1. Morphological comparison of immortalized porcine fibroblast cell lines. Three cell lines, iKUF08, iKUF09, and iKUF10, were randomly selected from 10 developed cell lines. The names of the cell lines are indicated on top. Scale bar: 100 µm. Magnification: × 10.
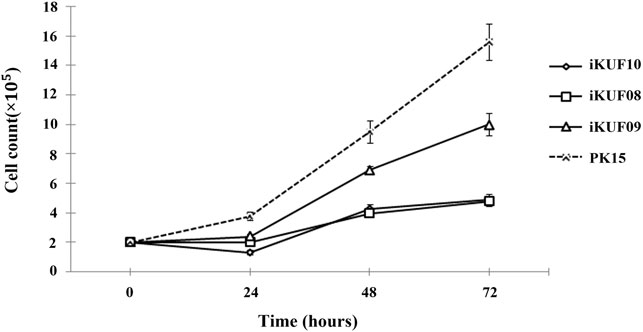
FIGURE 2. Growth curves of immortalized fibroblast cells. Three randomly selected cell lines, iKUF08, iKUF09, and iKUF10, and a commercially available PK15 were used. The culture duration is indicated on the x-axis.
When the hTERT expression of immortalized cell lines was evaluated using semiquantitative reverse transcription PCR, the 778 bp hTERT specific amplicon was observed in all the cell lines (Supplementary Figure S1). The level of hTERT expression varied among the cell lines with iKUF01, iKUF05, iKUF08, iKUF09, and iKUF10 showing the highest levels of expression. The level of hTERT expression was not directly related to proliferation rate (Figure 2 and Supplementary Figure S1).
Anchorage-independent Growth of Immortalized Fibroblast Cells
Anchorage-independent growth is the ability of transformed cells to grow without requiring a solid surface. We analyzed the anchorage dependency of cell growth for PK15 as well as three randomly selected cell lines, iKUF08, iKUF09, and iKUF10 (with passage numbers 32, 42, and 45, respectively). All four cell lines grew well on soft agar, with PK15 forming numerous colonies. In contrast, the hTERT transfected cells did not form any colonies, even after 2 weeks of culture (Figure 3).
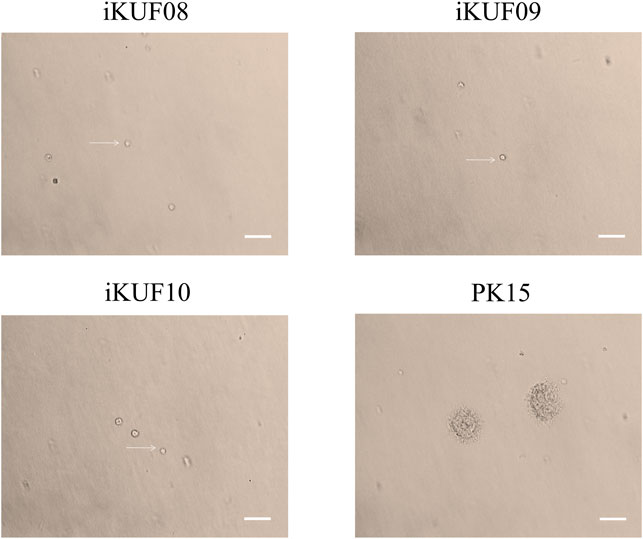
FIGURE 3. Anchorage-independent growth of immortalized porcine fibroblast cells in soft agar. Three randomly selected cell lines, iKUF08, iKUF09, and iKUF10, and a commercially available PK15 were used. The cell names are indicated on top. Arrows indicate single cells. Scale bar: 100 µm. Magnification: × 10.
Variations of SLA Classical Class I Gene Expression Among Immortalized Fibroblast Cell Lines
Given that the immortalized fibroblast cells in this study were sampled from different breeds and tissue types (ear, fetus, and lung), we analyzed the expression of three SLA class I genes, SLA-1, SLA-2, and SLA-3, and a class II gene, SLA-DRB1, using semiquantitative RT-PCR (Figure 4). Seven of the 10 cell lines, iKUF01, 04, 05, 06, 07, 08, and 09, expressed all three classical class I genes, though at varying levels. iKUF01 and iKUF05 showed higher SLA-2 expression than the other cell lines. SLA-1 expression was higher in iKUF04 and iKUF08 than in other cell lines. iKUF02 and iKUF10 lacked the expression of SLA-3 and SLA-1, respectively. iKUF03 showed only a low level of SLA-1 expression with no or undetectable levels of SLA-2 and SLA-3 expression. Interestingly, the levels and patterns of SLA class I gene expression varied among iKUF cell lines 01, 03, 04, 05, 06, 09, and 10 that were derived from the ear of different individuals, indicating, presumably, the presence of individual or haplotype-based differences in the expression of SLA class I genes. However, additional experiments should be conducted to prove this more clearly. In general, the expression of SLA-3 was lower than that of SLA-1 and SLA-2. The expression of SLA-DRB1 was not observed in any of the cell lines, owing to the fact that fibroblasts do not express MHC class II molecules without cytokine stimulation (Umetsu et al., 1986; Kim et al., 2012).
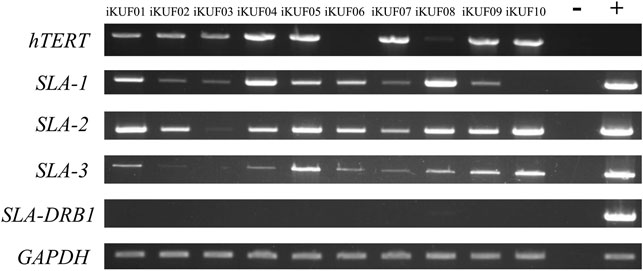
FIGURE 4. Comparison of the expression of SLA-1, -2, -3, and -DRB1 in immortalized fibroblast cell lines. Semi quantitative reverse transcription PCRs were carried out for SLA-1, -2, -3, and -DRB1. GAPDH was used as a control. The numbers of the cell lines are on top. “–” and “+” indicate negative and positive controls, respectively. The representative image from the triplicate analysis is shown.
Increased Expression of SLA Class I Genes in Immortalized Fibroblast Cells
We compared the expression levels of SLA-1, -2, and -3 between iKUF08, iKUF09, and iKUF10 cells before and after immortalization (Figure 5). In both phases, all three genes were expressed in iKUF08 and iKUF09, but iKUF10 cells lacked SLA-1 expression. The expression levels of SLA classical class I genes varied among the cell lines and were slightly upregulated in the immortalized compared to the primary cells based on semiquantitative reverse transcription polymerase chain reaction. The expression of SLA classical class I gene was also analyzed at the protein level by immunostaining with Pan-SLA classical class I-specific antibodies (Figure 6). Although the signal intensity of SLA-1 appeared to be consistent to that of the mRNA expression, clear conclusion on the protein expression level could not be drawn due to the quantitative limitations of immunohistochemical analysis.
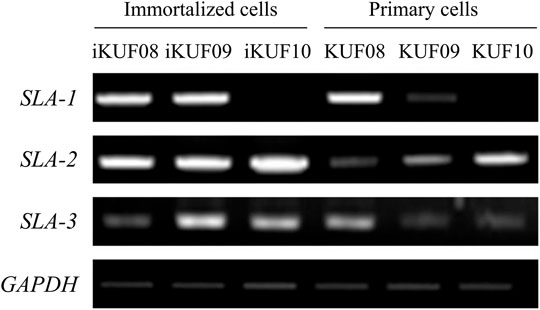
FIGURE 5. Comparison of SLA classical class I gene expression between primary and immortalized cells. Three immortalized cell lines, iKUF08, iKUF09, and iKUF10, are analyzed. KUF08, KUF09, and KUF10 are primary cells before immortalized. The names of the cells are on top. The expression of SLA-1, -2, and -3 are being compared. GAPDH is used as a control. The representative image from the triplicate analysis is shown.
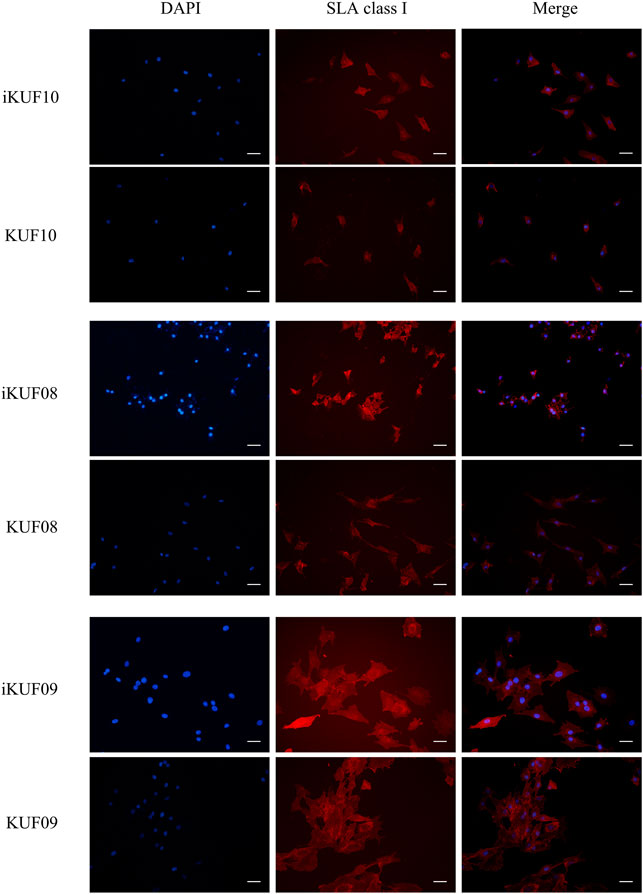
FIGURE 6. Immunohistochemical comparison of SLA classical class I gene expression between primary and immortalized cells. Three immortalized cell lines, iKUF10, iKUF08, and iKUF09 and corresponding primary cells, KUF10, KUF08 and KUF09 were used. The first and second columns show the results of DAPI staining and immunostaining by anti-pan SLA classical class I antibodies, respectively. The merged images of the two are shown in the third column. The first and second rows correspond to immortalized and primary cells, respectively. Scale bar: 50 µm. Magnification: × 20. The representative image from the triplicate analysis is shown.
Discussion
Epstein–Barr virus-transformed B-lymphoblastoid cell lines and extensive curation have been important developments in the molecular characterization of MHC genes and MHC-antigen interactions related to HLA (Yang et al., 1989). Similar to B-lymphoblastoid cell lines, hTERT-transformed cells have extended cellular lifespans and are effectively used to prepare cells in sufficient number while maintaining their biological properties (Saldanha et al., 2003). In this study, we created a panel of 10 hTERT-transformed porcine fibroblast cell lines, conducted high resolution analysis of the major classical class I and class II SLA genes, including SLA-1, SLA-2, SLA-3, SLA-DQB1, SLA-DRB1, and SLA-DQA, and characterized the expression patterns of three class I genes and DRB1 in each cell line. However, owing to the unavailability of MHC haplotype reference information, we were unable to reliably characterize the haplotypes of most of the six SLA genes in pigs. Together with other publicly available pig cell lines (Ho et al., 2009; Le et al., 2018), immortalized porcine cells could be used to establish a reference cell line panel for immunogenetic studies in pigs.
Except for nucleotide substitutions, structural variations in the SLA gene region, including SLA-1 duplication in certain haplotypes, are not well studied in pigs (Le et al., 2020). To collectively improve our scientific understanding of haplotype diversity, gene duplication, pseudogenization, and conflicts in sequence assembly at the SLA region require systematic analysis using a common and reliable resource. Homozygous cell lines across the entire MHC region would be particularly useful. For HLA, the DNA fragments of the ∼5 Mbp MHC region of 95 MHC homozygous B-lymphoblastoid cell lines were captured and analyzed together with six alternative MHC reference sequences (Norman et al., 2017). In pigs, only three cell lines PK13, PK15, and 3D4/21 were reported as possibly homozygous across the entire SLA region, although the haplotype was identical for PK13 and PK15 (Ho et al., 2009; Le et al., 2018). Limited progress has been made with regard to porcine B-lymphoblastoid cell lines (Kaeffer, 1994).
The use of different typing methods between laboratories can contribute to heterogeneity in typing results (Erlich, 2012). The use of reference cell lines with diverse SLA haplotypes could resolve this artefact of methodological differences (Ho et al., 2009). Typing methods for SLA genes employing high throughput and parallel typing of multiple loci from genomic DNA have not been available. A gene panel of well characterized cell lines could greatly contribute to the development of the typing technology and the evaluation of its accuracy (Creary et al., 2019).
Understanding the kinetics of epitope binding to MHC molecules is important for the development of effective vaccines. However, resources for the systematic analysis of interactions between antigenic epitopes and SLA molecules are limited in pigs. We previously reported that the affinity of PCV2 ORF2 as an epitope to MHC class II molecules varied depending on the MHC haplotype (Le et al., 2018) and showed that the use of immortalized PAM cells can instead be used to evaluate the MHC binding affinity to antigenic peptides. The cell lines reported in this study could be applied in determining the binding affinity of epitopes to MHC class I proteins, a critical factor for CTL activation, without the complication of peptides binding to class II SLA molecules in the cell-based assay.
Our immortalized fibroblast cell lines showed lower growth rates than PK15 and did not form colonies under the anchorage-independent culture condition, suggesting greater similarity in phenotype to primary cells than available ATCC fibroblast cell lines. This is consistent with the results of a previous study on hTERT transfected porcine alveolar macrophages (Le et al., 2018). These characteristics may be more suitable in molecular biology studies that require primary fibroblast cells with relatively short passage numbers and showing lower accumulation of genetic mutations that could alter their original characteristics.
Resilience after pathogenic infection is likely to be affected by an individual’s immune capacity. Considering the functionality of SLA molecules, their expression should affect the consequence of adaptive immune responses and general animal health. The expression of MHC class I molecules varies depending on cell types and tissue origins (Fanbo et al., 2005). Our results showed variation in the expression level of SLA classical class I genes among the cell lines, independent of tissue type or individual breed. However, these results are only based on in vitro analysis. In vivo studies are required to describe the natural variation of SLA class I gene expression among individual pigs of different SLA haplotypes. Consistent to our observation, haplotype-specific expression patterns of SLA class I genes have been previously reported (Kita et al., 2012). Our fibroblast cell panel could be applied to investigate the regulatory mechanisms or genetic factors affecting the expression of SLA genes, which could inform animal breeding programs and improve the immune capacity within animal breeds. The cell lines also could be useful to study molecular biological mechanisms influencing MHC class I expression including autophagy and ER stress in pigs (Crotzer and Blum, 2009; Granados et al., 2009).
In this study, the expression levels of SLA class I genes were slightly higher in immortalized cells than in primary cells (Figure 5). Modo et al. (2003) found higher levels of MHC expression in the proliferative than in the non-proliferative or differentiating conditions in conditionally immortalized murine neural stem cells. Redondo et al. (1998) also showed that the expression level of MHC class II genes varied in lung tumor cell lines and observed a positive correlation between the expression of HLA-DR and proto-oncogene c-myc. The expression of MHC genes could be influenced by the rate of cell proliferation and may explain the increased expression of SLA class I genes in the immortalized fibroblast cell lines.
The development of diverse cell lines with various MHC haplotypes could be useful for immunogenetic analyses in pigs but the availability of these cell lines has been limited. Fibroblasts are relatively easy to isolate, culture, and immortalize compared to other livestock cell types and, therefore, appropriate to develop stable cell lines with confirmed MHC genotypes. Studies on variations in the genetic structures of SLA and their expression using reference cell lines have great potential to enhance our knowledge regarding the adaptive capacity and regulatory elements of the porcine immune system. Although MHC genes of mammalian species are evolutionary conserved, the organization of the MHC region in pigs differs from others in that MHC class II and I genes are separated by a centromere (Lunney et al., 2009). The immortalized fibroblast cell lines in this study also could be useful to understand the influence of the centromere on generating haplotype diversity of the MHC region through genetic recombination.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institute of Animal Care and Use Committee of Konkuk University.
Author Contributions
QL and CP designed and coordinated the study. QL, SY, MC, and HJ performed the experiments. QL, SY, MC and CP analyzed the data and interpreted the results. QL, SY, and CP wrote the manuscript. QL, MC, W-IK and C-SH provided technical and material support. The final version of the manuscript has been reviewed by all authors.
Funding
This work was supported by the Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ016221), Rural Development Administration, South Korea.
Conflict of Interest
Author C-SH was employed by the company Gift of Hope Organ & Tissue Donor Network.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.815328/full#supplementary-material
References
Bodmer, H. C., Bastin, J. M., Askonas, B. A., and Townsend, A. R. (1989). Influenza-specific Cytotoxic T-Cell Recognition Is Inhibited by Peptides Unrelated in Both Sequence and MHC Restriction. Immunology 66 (2), 163–169.
Bosch-Camós, L., López, E., Navas, M. J., Pina-Pedrero, S., Accensi, F., Correa-Fiz, F., et al. (2021). Identification of Promiscuous African Swine Fever Virus T-Cell Determinants Using a Multiple Technical Approach. Vaccines 9 (1), 29. doi:10.3390/vaccines9010029
Bremers, A. J. A., van der Burg, S. H., Kuppen, P. J. K., Kast, W. M., van de Velde, C. J. H., and Melief, C. J. M. (1995). The Use of Epstein-Barr Virus-Transformed B Lymphocyte Cell Lines in a Peptide-Reconstitution Assay. J. Immunother. 18 (2), 77–85. doi:10.1097/00002371-199508000-00001
Buus, S., Lise Lauemøller, S., Stryhn, A., and Østergaard Pedersen, L. (2001). “Measurement of Peptide-MHC Interactions in Solution Using the Spin Column Filtration Assay,” in Current Protocols in Immunology. Editors J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, W. Strober, and E. M. Shevach (Hoboken, New Jersey, United States: John Wiley & Sons).
Cerundolo, V., Elliott, T., Elvin, J., Bastin, J., Rammensee, H.-G., and Townsend, A. (1991). The Binding Affinity and Dissociation Rates of Peptides for Class I Major Histocompatibility Complex Molecules. Eur. J. Immunol. 21 (9), 2069–2075. doi:10.1002/eji.1830210915
Choi, H., Le, M. T., Lee, H., Choi, M.-K., Cho, H.-S., Nagasundarapandian, S., et al. (2015a). Sequence Variations of the Locus-specific 5′ Untranslated Regions of SLA Class I Genes and the Development of a Comprehensive Genomic DNA-Based High-Resolution Typing Method for SLA-2. Tissue Antigens 86 (4), 255–266. doi:10.1111/tan.12648
Choi, M., Lee, J., Le, M. T., Nguyen, D. T., Park, S., Soundrarajan, N., et al. (2015b). Genome-wide Analysis of DNA Methylation in Pigs Using Reduced Representation Bisulfite Sequencing. DNA Res. 22 (5), 343–355. doi:10.1093/dnares/dsv017
Counter, C. M., Hahn, W. C., Wei, W., Caddle, S. D., Beijersbergen, R. L., Lansdorp, P. M., et al. (1998). Dissociation Among In Vitro Telomerase Activity, Telomere Maintenance, and Cellular Immortalization. Proc. Natl. Acad. Sci. 95 (25), 14723–14728. doi:10.1073/pnas.95.25.14723
Creary, L. E., Guerra, S. G., Chong, W., Brown, C. J., Turner, T. R., Robinson, J., et al. (2019). Next-generation HLA Typing of 382 International Histocompatibility Working Group Reference B-Lymphoblastoid Cell Lines: Report from the 17th International HLA and Immunogenetics Workshop. Hum. Immunol. 80 (7), 449–460. doi:10.1016/j.humimm.2019.03.001
Crotzer, V. L., and Blum, J. S. (2009). Autophagy and its Role in MHC-Mediated Antigen Presentation. J. Immunol. 182 (6), 3335–3341. doi:10.4049/jimmunol.0803458
Denning, C., Dickinson, P., Burl, S., Wylie, D., Fletcher, J., and Clark, A. J. (2001). Gene Targeting in Primary Fetal Fibroblasts from Sheep and Pig. Cloning and Stem Cells 3 (4), 221–231. doi:10.1089/15362300152725945
Eisen, H. N., Hou, X. H., Shen, C., Wang, K., Tanguturi, V. K., Smith, C., et al. (2012). Promiscuous Binding of Extracellular Peptides to Cell Surface Class I MHC Protein. Proc. Natl. Acad. Sci. 109 (12), 4580–4585. doi:10.1073/pnas.1201586109
Elvin, J., Cerundolo, V., Elliott, T., and Townsend, A. (1991). A Quantitative Assay of Peptide-dependent Class I Assembly. Eur. J. Immunol. 21 (9), 2025–2031. doi:10.1002/eji.1830210909
Erlich, H. (2012). HLA DNA Typing: Past, Present, and Future. Tissue Antigens 80 (1), 1–11. doi:10.1111/j.1399-0039.2012.01881.x
Fanbo, J., Chen, C., Yajun, D., Jun, Y., and Songnian, H. (2005). Analysis of Porcine MHC Expression Profile. Chin. Sci. Bull. 50 (9), 880–890. doi:10.1007/bf02897382
Granados, D. P., Tanguay, P.-L., Hardy, M.-P., Caron, É., de Verteuil, D., Meloche, S., et al. (2009). ER Stress Affects Processing of MHC Class I-Associated Peptides. BMC Immunol. 10 (1), 10. doi:10.1186/1471-2172-10-10
Ho, C.-S., Rochelle, E. S., Martens, G. W., Schook, L. B., and Smith, D. M. (2006). Characterization of Swine Leukocyte Antigen Polymorphism by Sequence-Based and PCR-SSP Methods in Meishan Pigs. Immunogenetics 58 (11), 873–882. doi:10.1007/s00251-006-0145-y
Ho, C. S., Franzo-Romain, M. H., Lee, Y. J., Lee, J. H., and Smith, D. M. (2009). Sequence-based Characterization of Swine Leucocyte Antigen Alleles in Commercially Available Porcine Cell Lines. Int. J. Immunogenet. 36 (4), 231–234. doi:10.1111/j.1744-313X.2009.00853.x
Kaeffer, B. (1994). Immortal Porcine Lymphoblastoid Cell Lines: Interest for Veterinary and Medical Research. Vet. Res. 25 (5), 425–441.
Kaufman, J., Skjoedt, K., and Salomonsen, J. (1990). The MHC Molecules of Nonmammalian Vertebrates. Immunol. Rev. 113, 83–117. doi:10.1111/j.1600-065x.1990.tb00038.x
Kessler, J. H., Benckhuijsen, W. E., Mutis, T., Melief, C. J. M., van der Burg, S. H., and Drijfhout, J. W. (2001). “Competition-Based Cellular Peptide Binding Assay for HLA Class I,” in Current Protocols in Immunology. Editors J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, W. Strober, and E. M. Shevach (Hoboken, New Jersey, United States: John Wiley & Sons).
Kim, M., Seo, H., Choi, Y., Shim, J., Bazer, F. W., and Ka, H. (2012). Swine Leukocyte Antigen-DQ Expression and its Regulation by Interferon-Gamma at the Maternal-Fetal Interface in Pigs. Biol. Reprod. 8643 (2), 43–11. doi:10.1095/biolreprod.111.094011
Kita, Y. F., Ando, A., Tanaka, K., Suzuki, S., Ozaki, Y., Uenishi, H., et al. (2012). Application of High-Resolution, Massively Parallel Pyrosequencing for Estimation of Haplotypes and Gene Expression Levels of Swine Leukocyte Antigen (SLA) Class I Genes. Immunogenetics 64 (3), 187–199. doi:10.1007/s00251-011-0572-2
Lazoura, E., and Apostolopoulos, V. (2005). Insights into Peptide-Based Vaccine Design for Cancer Immunotherapy. Cmc 12 (13), 1481–1494. doi:10.2174/0929867054039017
Le, M., Choi, H., Choi, M.-K., Cho, H., Kim, J.-H., Seo, H. G., et al. (2015). Development of a Simultaneous High Resolution Typing Method for Three SLA Class II Genes, SLA-DQA, SLA-DQB1, and SLA-DRB1 and the Analysis of SLA Class II Haplotypes. Gene 564 (2), 228–232. doi:10.1016/j.gene.2015.03.049
Le, M. T., Choi, H., Choi, M.-K., Nguyen, D. T., Kim, J.-H., Seo, H. G., et al. (2012). Comprehensive and High-Resolution Typing of Swine Leukocyte Antigen DQA from Genomic DNA and Determination of 25 New SLA Class II Haplotypes. Tissue Antigens 80 (6), 528–535. doi:10.1111/tan.12017
Le, M. T., Choi, H., Lee, H., Le, V. C. Q., Ahn, B., Ho, C.-S., et al. (2020). SLA-1 Genetic Diversity in Pigs: Extensive Analysis of Copy Number Variation, Heterozygosity, Expression, and Breed Specificity. Sci. Rep. 10 (1), 743. doi:10.1038/s41598-020-57712-5
Lechler, R., Batchelor, R., and Lombardi, G. (1991). The Relationship between MHC Restricted and Allospecific T Cell Recognition. Immunol. Lett. 29 (1-2), 41–50. doi:10.1016/0165-2478(91)90197-i
Ljunggren, H.-G., Stam, N. J., Öhlén, C., Neefjes, J. J., Höglund, P., Heemels, M.-T., et al. (1990). Empty MHC Class I Molecules Come Out in the Cold. Nature 346 (6283), 476–480. doi:10.1038/346476a0
Lunney, J. K., Ho, C.-S., Wysocki, M., and Smith, D. M. (2009). Molecular Genetics of the Swine Major Histocompatibility Complex, the SLA Complex. Develop. Comp. Immunol. 33 (3), 362–374. doi:10.1016/j.dci.2008.07.002
Melief, C. J. M., and Kessler, J. H. (2017). Novel Insights into the HLA Class I Immunopeptidome and T-Cell Immunosurveillance. Genome Med. 9 (1), 1–3. doi:10.1186/s13073-017-0439-8
Meurens, F., Summerfield, A., Nauwynck, H., Saif, L., and Gerdts, V. (2012). The Pig: a Model for Human Infectious Diseases. Trends Microbiol. 20 (1), 50–57. doi:10.1016/j.tim.2011.11.002
Modo, M., Mellodew, K., and Rezaie, P. (2003). In Vitro expression of Major Histocompatibility Class I and Class II Antigens by Conditionally Immortalized Murine Neural Stem Cells. Neurosci. Lett. 337 (2), 85–88. doi:10.1016/s0304-3940(02)01301-0
Narzi, D., Winkler, K., Saidowsky, J., Misselwitz, R., Ziegler, A., Böckmann, R. A., et al. (2008). Molecular Determinants of Major Histocompatibility Complex Class I Complex Stability. J. Biol. Chem. 283 (34), 23093–23103. doi:10.1074/jbc.M710234200
Norman, P. J., Norberg, S. J., Guethlein, L. A., Nemat-Gorgani, N., Royce, T., Wroblewski, E. E., et al. (2017). Sequences of 95 Human MHC Haplotypes Reveal Extreme Coding Variation in Genes Other Than Highly Polymorphic HLA Class I and II. Genome Res. 27 (5), 813–823. doi:10.1101/gr.213538.116
Ottenhoff, T. H., Geluk, A., Toebes, M., Benckhuijsen, W. E., van Meijgaarden, K. E., and Drijfhout, J. W. (1997). A Sensitive Fluorometric Assay for Quantitatively Measuring Specific Peptide Binding to HLA Class I and Class II Molecules. J. Immunol. Methods 200 (1-2), 89–97. doi:10.1016/s0022-1759(96)00190-1
Palankar, R., Skirtach, A. G., Kreft, O., Bédard, M., Garstka, M., Gould, K., et al. (2009). Controlled Intracellular Release of Peptides from Microcapsules Enhances Antigen Presentation on MHC Class I Molecules. Small 5 (19), 2168–2176. doi:10.1002/smll.200900809
Park, K., Choi, H., Thong, L. M., Kwon, O.-J., Kim, J.-H., Lee, H.-T., et al. (2010). Simple and Comprehensive SLA-DQB1 Genotyping Using Genomic PCR and Direct Sequencing. Tissue Antigens 76 (4), 301–310. doi:10.1111/j.1399-0039.2010.01522.x
Rammensee, H.-G. (1995). Chemistry of Peptides Associated with MHC Class I and Class II Molecules. Curr. Opin. Immunol. 7 (1), 85–96. doi:10.1016/0952-7915(95)80033-6
Reche, P. A., Keskin, D. B., Hussey, R. E., Ancuta, P., Gabuzda, D., and Reinherz, E. L. (2006). Elicitation from Virus-Naive Individuals of Cytotoxic T Lymphocytes Directed against Conserved HIV-1 Epitopes. Med. Immunol. 5 (1), 1. doi:10.1186/1476-9433-5-1
Redondo, M., Ruiz‐Cabello, F., Concha, A., Hortas, M. L., Serrano, A., Morell, M., et al. (1998). Differential Expression of MHC Class II Genes in Lung Tumour Cell Lines. Eur. J. Immunogenet. 25 (6), 385–391. doi:10.1046/j.1365-2370.1998.00116.x
Saldanha, S. N., Andrews, L. G., and Tollefsbol, T. O. (2003). Analysis of Telomerase Activity and Detection of its Catalytic Subunit, hTERT. Anal. Biochem. 315 (1), 1–21. doi:10.1016/s0003-2697(02)00663-2
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York, United States: Cold Spring Harbor Laboratory Press.
Schook, L., Beattie, C., Beever, J., Donovan, S., Jamison, R., Zuckermann, F., et al. (2005). Swine in Biomedical Research: Creating the Building Blocks of Animal Models. Anim. Biotechnol. 16 (2), 183–190. doi:10.1080/10495390500265034
Sckisel, G. D., Bouchlaka, M. N., Monjazeb, A. M., Crittenden, M., Curti, B. D., Wilkins, D. E. C., et al. (2015). Out-of-Sequence Signal 3 Paralyzes Primary CD4+ T-cell-dependent Immunity. Immunity 43 (2), 240–250. doi:10.1016/j.immuni.2015.06.023
Sette, A., Sidney, J., del Guercio, M.-F., Southwood, S., Ruppert, J., Dahlberg, C., et al. (1994). Peptide Binding to the Most Frequent HLA-A Class I Alleles Measured by Quantitative Molecular Binding Assays. Mol. Immunol. 31 (11), 813–822. doi:10.1016/0161-5890(94)90019-1
Shi, J. W., Liu, W., Zhang, T. T., Wang, S. C., Lin, X. L., Li, J., et al. (2013). The Enforced Expression of c-Myc in Pig Fibroblasts Triggers Mesenchymal-Epithelial Transition (MET) via F-Actin Reorganization and RhoA/Rock Pathway Inactivation. Cell Cycle 12 (7), 1119–1127. doi:10.4161/cc.24164
Sidney, J., Southwood, S., Oseroff, C., del Guercio, M.-F., Sette, A., and Grey, H. M. (2001). “Measurement of MHC/Peptide Interactions by Gel Filtration,” in Current Protocols in Immunology. Editors J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, W. Strober, and E. M. Shevach (Hoboken, New Jersey, United States: John Wiley & Sons). doi:10.1002/0471142735.im1803s31
Sommer, S. (2005). The Importance of Immune Gene Variability (MHC) in Evolutionary Ecology and Conservation. Front. Zool 2 (1), 16. doi:10.1186/1742-9994-2-16
Stienekemeier, M., Falk, K., Rötzschke, O., Weishaupt, A., Schneider, C., Toyka, K. V., et al. (2001). Vaccination, Prevention, and Treatment of Experimental Autoimmune Neuritis (EAN) by an Oligomerized T Cell Epitope. Proc. Natl. Acad. Sci. 98 (24), 13872–13877. doi:10.1073/pnas.241504598
Storkus, W. J., Zeh, H. J., Salter, R. D., and Lotze, M. T. (1993). Identification of T-Cell Epitopes. J. Immunother. 14 (2), 94–103. doi:10.1097/00002371-199308000-00003
Strober, W. (2001). Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. Appendix 3B. doi:10.1002/0471142735.ima03bs21
Tan, T. L. R., Geluk, A., Toebes, M., Ottenhoff, T. H. M., and Drijfhout, J. W. (1997). A Novel, Highly Efficient Peptide-HLA Class I Binding Assay Using Unfolded Heavy Chain Molecules: Identification of HIV-1 Derived Peptides that Bind to HLA-A*0201 and HLA-A*0301. J. Immunological Methods 205 (2), 201–209. doi:10.1016/S0022-1759(97)00086-0
Thong, L. M., Choi, H., Kwon, O.-J., Kim, J.-H., Kim, Y.-B., Oh, J.-W., et al. (2011). Systematic Analysis of Swine Leukocyte Antigen-DRB1 Nucleotide Polymorphisms Using Genomic DNA-Based High-Resolution Genotyping and Identification of New Alleles. Tissue Antigens 77 (6), 572–583. doi:10.1111/j.1399-0039.2011.01662.x
Ting, J. P., and Trowsdale, J. (2002). Genetic Control of MHC Class II Expression. Cell 109 (Suppl. 1), S21–S33. doi:10.1016/s0092-8674(02)00696-7
Townsend, A., Elliott, T., Cerundolo, V., Foster, L., Barber, B., and Tse, A. (1990). Assembly of MHC Class I Molecules Analyzed In Vitro. Cell 62 (2), 285–295. doi:10.1016/0092-8674(90)90366-m
Turvy, D. N., and Blum, J. S. (2001). “Biotin Labeling and Quantitation of Cell-Surface Proteins,” in Current Protocols in Immunology. Editors J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, W. Strober, and E. M. Shevach (Hoboken, New Jersey, United States: John Wiley & Sons).
Umetsu, D. T., Katzen, D., Jabara, H. H., and Geha, R. S. (1986). Antigen Presentation by Human Dermal Fibroblasts: Activation of Resting T Lymphocytes. J. Immunol. 136 (2), 440–445.
Van Chanh Le, Q., Le, T. M., Cho, H. S., Kim, W. I., Hong, K., Song, H., et al. (2018). Analysis of Peptide-SLA Binding by Establishing Immortalized Porcine Alveolar Macrophage Cells with Different SLA Class II Haplotypes. Vet. Res. 49 (1), 96–10. doi:10.1186/s13567-018-0590-2
van der Burg, S. H., Ras, E., Drijfhout, J. W., Benckhuijsen, W. E., Bremers, A. J. A., Melief, C. J. M., et al. (1995). An HLA Class I Peptide-Binding Assay Based on Competition for Binding to Class I Molecules on Intact Human B Cells. Hum. Immunol. 44 (4), 189–198. doi:10.1016/0198-8859(95)00105-0
Van Hateren, A., James, E., Bailey, A., Phillips, A., Dalchau, N., and Elliott, T. (2010). The Cell Biology of Major Histocompatibility Complex Class I Assembly: towards a Molecular Understanding. Tissue Antigens 76 (4), 259–275. doi:10.1111/j.1399-0039.2010.01550.x
Vodička, P., Smetana, K., Dvořánková, B., Emerick, T., Xu, Y. Z., Ourednik, J., et al. (2005). The Miniature Pig as an Animal Model in Biomedical Research. Ann. N.Y Acad. Sci. 1049, 161–171. doi:10.1196/annals.1334.015
Wieczorek, M., Abualrous, E. T., Sticht, J., Álvaro-Benito, M., Stolzenberg, S., Noé, F., et al. (2017). Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol. 8, 292. doi:10.3389/fimmu.2017.00292
Wooldridge, L., Ekeruche-Makinde, J., van den Berg, H. A., Skowera, A., Miles, J. J., Tan, M. P., et al. (2012). A Single Autoimmune T Cell Receptor Recognizes More Than a Million Different Peptides. J. Biol. Chem. 287 (2), 1168–1177. doi:10.1074/jbc.M111.289488
Yaneva, R., Schneeweiss, C., Zacharias, M., and Springer, S. (2010). Peptide Binding to MHC Class I and II Proteins: New Avenues from New Methods. Mol. Immunol. 47 (4), 649–657. doi:10.1016/j.molimm.2009.10.008
Yang, S. Y., Milford, E., Hämmerling, U., and Dupont, B. (1989). “Description of the Reference Panel of B-Lymphoblastoid Cell Lines for Factors of the HLA System: the B-Cell Line Panel Designed for the Tenth International Histocompatibility Workshop,” in Immunobiology of HLA. Editor B. Dupont (Berlin/Heidelberg, Germany: Springer), 11–19. doi:10.1007/978-1-4612-3552-1_2
Keywords: fibroblast, immortalization, swine leukocyte, pig, major histocompatibility complex, hTERT
Citation: Le QVC, Youk S, Choi M, Jeon H, Kim W-, Ho C- and Park C (2022) Development of an Immortalized Porcine Fibroblast Cell Panel With Different Swine Leukocyte Antigen Genotypes. Front. Genet. 13:815328. doi: 10.3389/fgene.2022.815328
Received: 15 November 2021; Accepted: 14 January 2022;
Published: 07 February 2022.
Edited by:
Manuel Alfonso Patarroyo, Colombian Institute of Immunology Foundation, ColombiaReviewed by:
Hua-Ji Qiu, Harbin Veterinary Research Institute (CAAS), ChinaDiego Ordoñez, Universidad de Ciencias Aplicadas y Ambientales, Colombia
Carmen Celis, Universidad de Ciencias Aplicadas y Ambientales, Colombia
Copyright © 2022 Le, Youk, Choi, Jeon, Kim, Ho and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chankyu Park, Y2hhbmt5dUBrb25rdWsuYWMua3I=
†These authors have contributed equally to this work and share first authorship
 Quy Van Chanh Le
Quy Van Chanh Le SeungYeon Youk1†
SeungYeon Youk1† Hyoim Jeon
Hyoim Jeon Chak-Sum Ho
Chak-Sum Ho Chankyu Park
Chankyu Park