- 1Laboratório de Citogenética, Centro de Estudos Avançados da Biodiversidade, Instituto de Ciências Biológicas, Universidade Federal Do Pará (UFPA), Belém, Brazil
- 2Departamento de Biologia Estrutural, Molecular e Genética, Universidade Estadual de Ponta Grossa, Ponta Grossa, Brazil
- 3Laboratório de Genética Animal, Coordenação de Biodiversidade, Instituto Nacional de Pesquisas da Amazônia, Manaus, Brazil
- 4Department of Veterinary Medicine, Cambridge Resource Centre for Comparative Genomics, University of Cambridge, Cambridge, United Kingdom
The genus Gymnotus is a large monophyletic group of freshwater weakly-electric fishes, with wide distribution in Central and South America. It has 46 valid species divided into six subgenera (Gymnotus, Tijax, Tigre, Lamontianus, Tigrinus and Pantherus) with large chromosome plasticity and diploid numbers (2n) ranging from 34 to 54. Within this rich diversity, there is controversy about whether Gymnotus (Gymnotus) carapo species is a single widespread species or a complex of cryptic species. Cytogenetic studies show different diploid numbers for G. carapo species, ranging from 40 to 54 chromosomes with varied karyotypes found even between populations sharing the same 2n. Whole chromosome painting has been used in studies on fish species and recently has been used for tracking the chromosomal evolution of Gymnotus and assisting in its cytotaxonomy. Comparative genomic mapping using chromosome painting has shown more complex rearrangements in Gymnotus carapo than shown in previous studies by classical cytogenetics. These studies demonstrate that multiple chromosome pairs are involved in its chromosomal reorganization, suggesting the presence of a complex of cryptic species due to a post zygotic barrier. In the present study, metaphase chromosomes of G. carapo occidentalis “catalão” (GCC, 2n = 40, 30m/sm+10st/a) from the Catalão Lake, Amazonas, Brazil, were hybridized with whole chromosome probes derived from the chromosomes of G. carapo (GCA, 2n = 42, 30m/sm+12st/a). The results reveal chromosome rearrangements and a high number of repetitive DNA sites. Of the 12 pairs of G. carapo chromosomes that could be individually identified (GCA 1–3, 6, 7, 9, 14, 16 and 18–21), 8 pairs (GCA 1, 2, 6, 7, 9, 14, 20, 21) had homeology conserved in GCC. Of the GCA pairs that are grouped (GCA [4, 8], [5, 17], [10, 11] and [12, 13, 15]), most kept the number of signals in GCC (GCA [5, 17], [10, 11] and [12, 13, 15]). The remaining chromosomes are rearranged in the GCC karyotype. Analysis of both populations of the G. carapo cytotypes shows extensive karyotype reorganization. Along with previous studies, this suggests that the different cytotypes analyzed here may represent different species and supports the hypothesis that G. carapo is not a single widespread species, but a group of cryptic species.
Introduction
The genus Gymnotus (Gymnotiformes, Teleostei) is a large monophyletic group of weakly-electric freshwater fish. It is the most speciose genus of the order, with 46 validated species (Craig et al., 2019; Kim et al., 2020; Fricke et al., 2021), widely distributed in the Neotropical region (Central and South America). The highest diversity is found in the Amazon-Orinoco-Guiana basins (Mago-Leccia, 1994; Albert, 2001; Albert and Crampton, 2005).
Gymnotus is divided into six subgenera: Gymnotus, Tijax, Tigre, Lamontianus, Tigrinus and Pantherus (Craig et al., 2019) with substantial chromosome plasticity. The diploid number (2n) varies from 2n = 34 in Gymnotus capanema (Milhomem et al., 2012a) to 2n = 54 in G. carapo (Foresti et al., 1984), G. mamiraua (Milhomem et al., 2007), G. paraguensis (Margarido et al., 2007) and G. inaequilabiatus (Scacchetti et al., 2011). The growing number of studies based not only on the karyotypic formula but also on different kinds of repetitive DNA sequences such as rDNAs, satellites, microsatellites and transposable elements (Milhomem et al., 2007; Claro, 2008; Milhomem et al., 2008; Scacchetti et al., 2011; Milhomem et al., 2012a; Milhomem et al., 2012b; da Silva et al., 2014; Utsunomia et al., 2014; Almeida et al., 2015; da Silva, 2015; da Silva et al., 2016; Machado et al., 2017; Utsunomia et al., 2018), have shown many different species-specific karyotypes and even population variants.
This large variation in 2n, however, is mostly found in the G. carapo subgenus, the previous Gymnotus carapo clade (Craig et al., 2019). In all the other subgenera, the diploid number varies from 48 to 54 chromosomes, many sharing the 2n = 52 that is found in the basal subgenus, the species G. (pantherus) pantherinus (Scacchetti et al., 2011; da Silva et al., 2011; da Silva et al., 2014; Utsunomia et al., 2014; Almeida et al., 2015; da Silva et al., 2016; Machado et al., 2017; da Silva et al., 2019), and also found in Electrophorus, the sister genus of Gymnotus (Fonteles et al., 2008; Cardoso et al., 2015).
In the subgenus Gymnotus, the G. carapo species has huge karyotype diversity among populations, with 2n ranging from 40 to 54, and many karyotypic formulas within the 2n described (Table 1). The species G. carapo, based on morphology and distribution data, was divided into subspecies Gymnotus c. australis, Gymnotus c. caatingaensis, Gymnotus c. carapo, Gymnotus c. madeirensis, Gymnotus c. occidentalis, Gymnotus c. orientalis, and Gymnotus c. septentrionalis (Craig et al., 2017). While most Neotropical fish species of South America have restricted geographic distributions, these species are distributed widely (Albert and Reis, 2011; Lehmberg, 2015; Craig et al., 2017).
Chromosome painting is as an important tool in comparative cytogenetics studies of fish species. The results have helped to solve various issues in this field and have giving insights into several evolutionary issues (Barby et al., 2019). It has helped to bring insights into the origin of B chromosomes (Vicari et al., 2011; Scudeler et al., 2015; Utsunomia et al., 2016), the origin and evolution of sex chromosomes (Carvalho et al., 2017; Cioffi et al., 2017; de Oliveira et al., 2017; de Moraes et al., 2017; Yano et al., 2017; de Freitas et al., 2018) and helped in the understanding of chromosomal evolution and relationships between closely related species (Ráb et al., 2008; Nagamachi et al., 2010; Nagamachi et al., 2013; Milhomem et al., 2013; Machado et al., 2018; Cioffi et al., 2019). Nagamachi et al. (2010) produced whole chromosome probes (WCP) from G. c. orientalis (GCA42, 2n = 42, 30m/sm+12st/a) by flow sorting, and hybridized these probes to the cytotypes of G. c. orientalis with 2n = 40 (GCA40, 34m/sm+6st/a). Two other studies with WCP from GCA42 mapped the karyotypes of G. capanema (GCP 2n = 34, Nagamachi et al., 2013) and G. arapaima (GAR 2n = 44, Machado et al., 2018). Those studies showed a higher level of chromosomal rearrangement than previously thought between these species.
In this study we used GCA42 WCP (Nagamachi et al., 2010) for mapping the karyotype of G. c. occidentalis “Catalão” (GCC 2n = 40), a distinctive population which has been proposed as a new species (da Silva et al., 2014). The results were compared with those obtained from GCA40 (Nagamachi et al., 2010), GCP34 (Nagamachi et al., 2013) and GAR44 (Machado et al., 2018).
Material and Methods
Sampling
Samples of G. carapo “Catalão” (GCC) were collected in Amazonas, Brazil (Figure 1). The Cytogenetics Laboratory from Centro de Estudos Avançados da Biodiversidade (UFPA) has permit number 19/2003 from the Ministry of Environment for sample transport and permit 52/2003 for using the samples for research. The Ethics Committee from Para Federal University (Comitê de Ética Animal da Universidade Federal do Pará) approved this research (Permit 68/2015). Sample collections were authorized by Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) and Secretaria de Estado de Meio Ambiente do Pará (SEMA-PA) under permit 020/2005 (Registration: 207419).
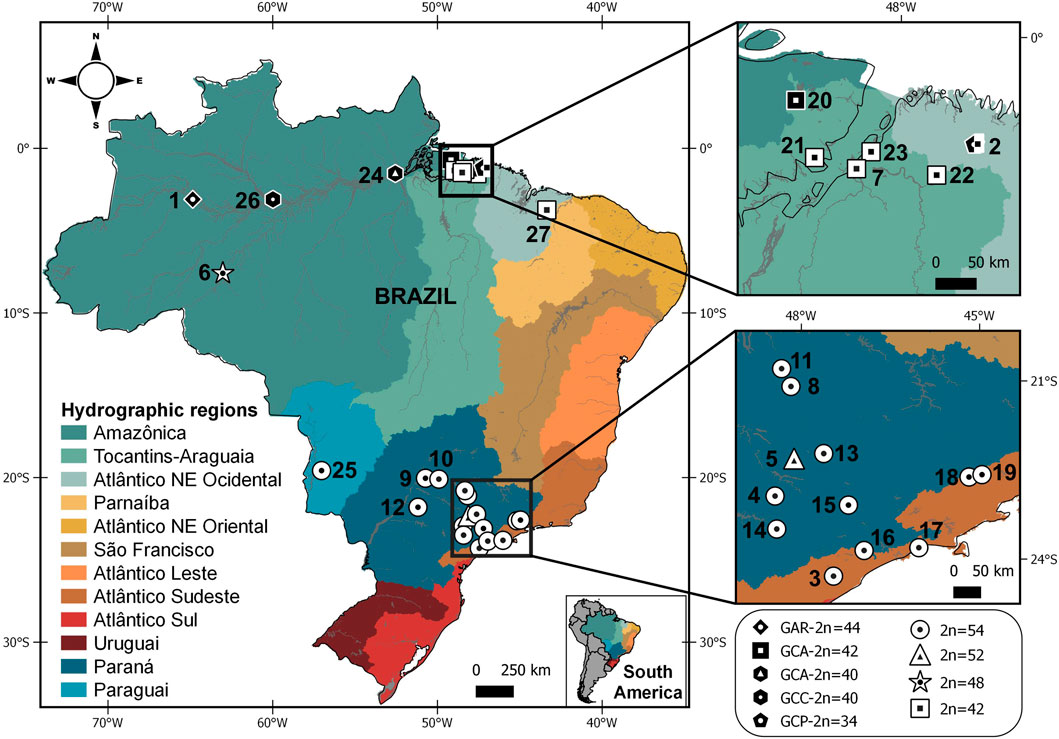
FIGURE 1. Map showing the Brazilian hydrographic regions and sample points for Gymnotus arapaima, G. capanema, and G. carapo, with karyotypic information (2n = diploid number) from the present study and from the literature. Each symbol indicates the karyotype obtained from the locality; more than one symbol indicates that distinct karyotypes were collected at the same locality. Symbols in white denote specimens analyzed by classic cytogenetics; symbols in black denote specimens analyzed by chromosome painting with G. carapo (2n = 42) whole-chromosome probes (Table 1; Nagamachi et al., 2010; Nagamachi et al., 2013; Machado et al., 2018, and present study). The numbers refer to localities detailed on Table 1.
Map
A distribution map was made using QGIS v.3.10.7. The shapefiles containing country limits were obtained from DIVA-GIS (Hijmans et al., 2004). We used the hydrographic regions limits provided by Braga et al. (2008) and we created the shapefiles on QGIS v.3.10.7. The localities numbered are shown on Table 1.
Whole Chromosome Painting
Whole Chromosome Probes (WCP) from G. carapo (2n = 42; 30m/sm+12st/a; Nagamachi et al., 2010) were hybridized onto metaphases of G. carapo “Catalão”. Chromosome painting techniques followed Yang et al. (1995) with the modifications proposed by Nagamachi et al. (2010). Chromosomes were classified morphologically according to Levan et al. (1964) with modifications. The karyotype was organized following da Silva et al. (2014).
Results
Chromosome Painting in Gymnotus carapo Occidentalis “Catalão”
Gymnotus carapo occidentalis “Catalão” (GCC) has 2n = 40 with 30m/sm+10st/a chromosomes (Figure 2A) without differentiated sex chromosomes in male and female specimens.
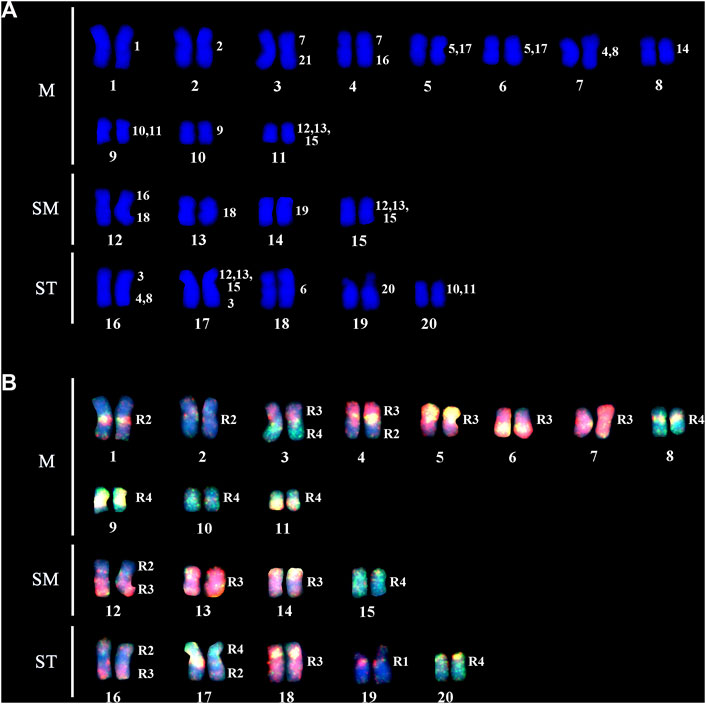
FIGURE 2. (A) A DAPI stained karyotype of GCC; the numbers on the right represent the G. carapo (GCA42) equivalent chromosomes. (B) Dual color fish with the probes of R3 (pairs 4–8 and 17–19; red) and R4 (pairs 9–15 and 21; green). Chromosome segments hybridizing with 2 colors indicate repetitive DNA sequences. The chromosomes or segments in blue (DAPI) represent the NOR-bearing chromosome of GCA42 (pair 20) and the chromosomes corresponding to R2 of GCA42 (pairs 1–3 and 16).
The regions of homology with GCA42 are indicated on the karyotype of GCC arranged from DAPI-stained chromosomes (Figure 2A).
Dual color FISH with the probes of GCA42 from R3 (pairs 4–8 and 17–19; red) and R4 (pairs 9–15 and 21; green) define the chromosome groups in GCC40 corresponding to the four groups in Figure 2B. Chromosome segments hybridizing with 2 colors indicate repetitive DNA sequences. The chromosomes or segments in blue (DAPI) represent the GCA42 NOR-bearing chromosome (pair 20) and the chromosomes corresponding to R2 (pairs 1–3 and 16).
From the 12 chromosome pairs of GCA42 that can be individually differentiated (pairs 1–3, 6, 7, 9, 14, 16 and 18–21), 8 pairs (1, 2, 6, 9, 14, 19, 20, 21) conserve homeology within GCC40 (pairs 1, 2, 3, 8, 10, 14, 18, 19). GCA42 pair 20 hybridizes one whole chromosome in GCC40, pair 19. Four chromosome pairs of GCA42 (3, 7, 16, and 18) show 2 signals on GCC40 (Figure 2).
The GCA42 probes that represent two chromosome pairs (4,8), (10,11) and (5,17) all reveal 2 signals, and the one that represents three pairs (12, 13, 15) reveals 3 signals on the GCC40 chromosomes.
The following chromosome associations of GCA42 are present in GCC40 pairs: 3 (7/C/21); 4 (7/C/16); 12 (16/C/16/18); 16 (3/C/3/[4, 8]); 17 ([12, 13, 15]/C/[12, 13, 15]/3), where C = centromere.
Comparative Chromosome Painting Analysis Among Different Gymnotus Karyotypes
From the 12 chromosome pairs in GCA42 that can be identified individually (1–3, 6, 7, 9, 14, 16, 18–21), GCC shows conserved synteny in 8 pairs (1, 2, 6, 9, 14, 19, 20, 21); GCC shares the same 8 pairs with GCA40, and also the same eight pairs shared between GCA40 and GCA42, grouping them together and closer to each other than the other analyzed karyotypes. From the pairs that are grouped in GCA42 [(4, 8), (10,11), (5,17), (12, 13, 15)], all keep the same number of signals in GCC40, while GCA40 has an extra signal for (5,17) and for (12, 14, 15) (Figure 3). The 16/18 syntenic associations are shared between GCC40 (16/C/16/18) and GCA40 (18/C/16) indicating pericentric inversions (Figures 3, 4).
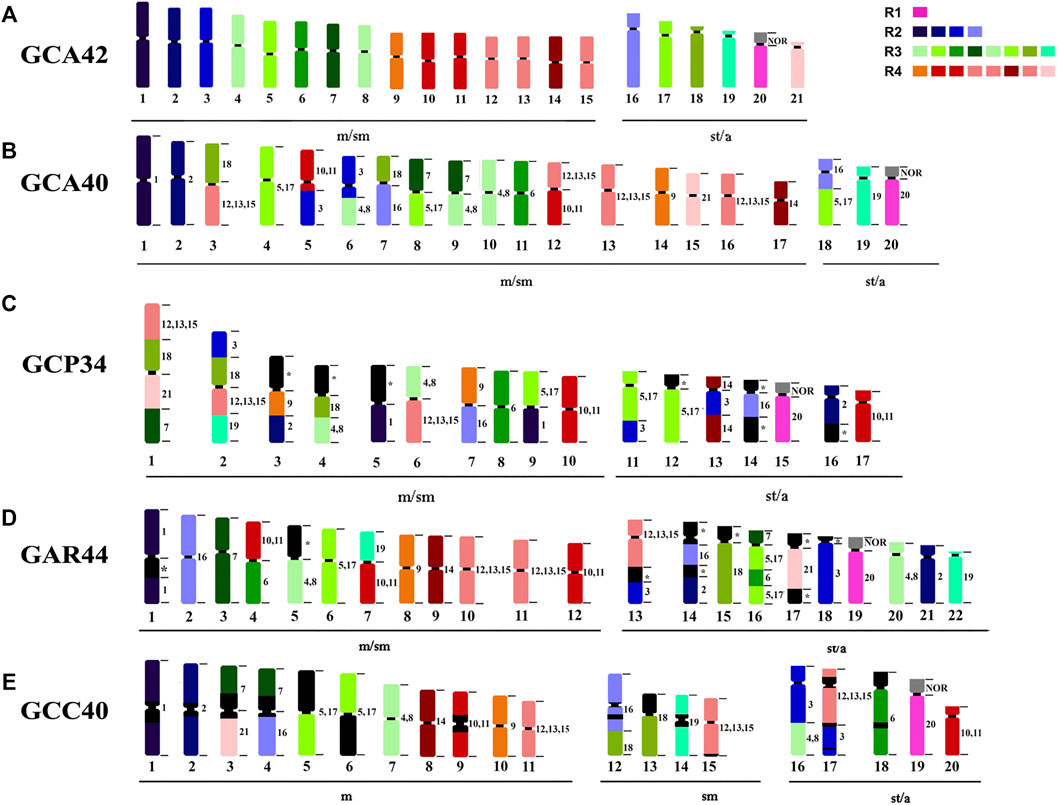
FIGURE 3. Ideogram showing chromosome rearrangements obtained by WCP. R1, R2, R3 and R4 representing the colors of the four major regions of the G. carapo 2n = 42 probes. (A) G. carapo 2n = 42 (GCA42) utilized in the production of the probes, each different color represents a chromosome or chromosome group; (B) G. carapo 2n = 40 (GCA40) (Nagamachi et al., 2010); (C) G. capanema 2n = 34 (GCP34) (Nagamachi et al., 2013); (D) G. arapaima 2n = 44 (GAR44) (Machado et al., 2018); (E) G. carapo “catalão” 2n = 40 (GCC40) (present study).
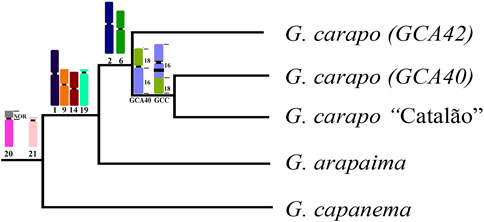
FIGURE 4. Representative phylogeny based on da Silva et al. (2019), with the syntenic blocks shared by the nodes. Chromosome numbers refer to the G. carapo 2n = 42 chromosomes (see Figure 3).
Compared to GAR, GCC shares five of the individually identified chromosomes (1, 9, 14, 20, 21), as well as the same number of signals in the groups (4, 8) and (12, 13, 15). There is also a similar chromosome rearrangement in GCC 17 and GAR 13 that is not shared with the other G. carapo cytotypes. However, as it is not possible to differentiate between GCA (2n = 42) 12,13, and 15, we cannot infer that the involved chromosome is the same or is different (Figure 3; Table 2).
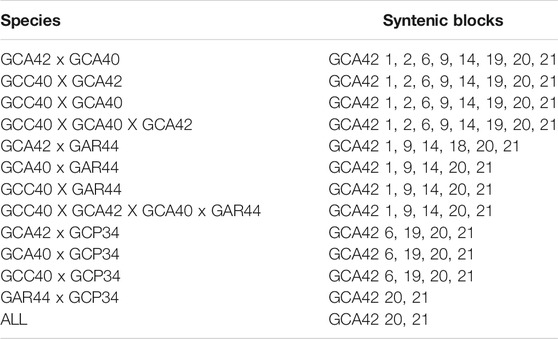
TABLE 2. Syntenic blocks shared among analyzed species with WCP. GCA42—Gymnotus carapo 2n = 42; GCA40—G. carapo 2n = 40; GCC40—G. carapo “Catalão” 2n = 40; GAR44 - G. arapaima 2n = 44; GCP34—G. capanema 2n = 34.
Compared to GCP, GCC shares three individual pairs (GCA 1,20,21) and the same number of signals as GCA (4,8), (10,11) and (12, 13, 15). All species share homeology to GCA 1,20,21 (Figure 3; Table 2).
The syntenic block of GCA42 6 is conserved in four of the five analyzed karyotypes by painting, except for GAR (Figure 3; Table 2), in which it is divided into two signals in pairs 4 and 16, while the syntenic block 18 of GCA 42 is shared with GAR, but not with GCC or GCA 40 (Figure 3; Table 2).
Discussion
Gymnotus carapo occidentalis “Catalão” has 2n = 40 (GCC, 30m/sm+10st/a), the same as G. carapo orientalis (GCA) 2n = 40 (28m/sm+12st/a), but with a different karyotype. It is hypothesized that the basal diploid number for Gymnotidae is 2n = 52 (da Silva et al., 2019), as the sister species Electrophorus electricus and G. pantherinus (sister species to all Gymnotus, Craig et al., 2019) both have 2n = 52.
This variation in karyotype can be consistently observed along the hydrographic regions. The 2n = 42 is found in the G. c. orientalis locations in the “Tocantins-Araguaia” region, while the 2n = 40 is observed in the G. c. orientalis (GCA40) located in the “Amazonica” region. While sharing the same 2n GCA40, GCC was sampled in a lake in an area close Negro river, in G. c. occidentalis occurrence area and also has significant karyotype differences. The 2n = 48 is found only in G. c. madeirensis (Table 1; Figure 1). The higher 2n = 52 and 54 is found only in G. c. australis, distributed along the Paraguai, “Paraná” and “Atlântico Sudeste” hydrographic regions.
When compared to all cytotypes of G. carapo in the literature, there is a tendency to a reduction in diploid number in the “Amazonica”, “Tocantins-Araguaia” and “Atlântico Nordeste Ocidental” hydrographic regions, a trait also shared with G. arapaima (Figure 1). Whereas the cytotypes in the “Paraguai”, “Paraná” and “Atlântico Sudeste” hydrographic regions have a higher chromosome number, with all locations having 2n = 54 except in one population that has 2n = 52 (Table 1; Figure 1) similar to more basal 2n in the genus. This suggests that the reduction in diploid number in the amazon region happened after colonization of the area.
Whole chromosome probes of GCA42 have been used in previous studies comparing two cytotypes of G. carapo (GCA42 and GCA40), G. capanema (GCP34) and G. arapaima (GAR44). The results demonstrate highly rearranged karyotypes, more than found by classical cytogenetics alone (Nagamachi et al., 2010; Nagamachi et al., 2013; Machado et al., 2018). The same results are observed in G. carapo “Catalão” (present study), confirming that the chromosomal evolution in this group is quite complex.
The karyotypes of the three G. carapo cytotypes analyzed by chromosome painting (GCA42, GCA40 and GCC40) are more similar to each other than to those of GAR44 or GCP34 and share a uniform amount of synteny. However, they have multiple species-specific rearrangements, which probably constitute a post zygotic barrier that would result in an inviable or infertile hybrid (Figure 4). We observe the same pattern when compared to G. arapaima, explained by the fact that they are sister species with recent divergence (Brochu, 2011) in relation to G. capanema (Craig et al., 2019).
This large number of chromosomal rearrangements in Gymnotus carapo, demonstrated by chromosome painting, indicates that the different cytotypes constitute a complex of cryptic species as already suggested (Milhomem et al., 2008; Nagamachi et al., 2010). The chromosomal speciation must have played a key role in this process that, if associated with small-inbred demes, could have facilitated the fixation of chromosomal rearrangements (King, 1993).
Currently these cryptic species (with the same morphology but different cytotypes) are in allopatry, which corroborates the pattern found in many Neotropical freshwater fish groups. This could be due to the dynamics of the river networks, including the fragmentation and merging of adjacent rivers, that led to increased species richness closer to the core region of the Amazon basin (Albert and Reis, 2011; Tagliacollo et al., 2016; Albert et al., 2018a; Albert et al., 2018b; Albert et al., 2020).
The taxonomy of G. carapo is quite complex and has been discussed in many studies. Some authors report it as a single generalized species (Craig et al., 2017), other studies show a paraphyletic group within a monophyletic complex of related species (Brochu, 2011; da Silva et al., 2019) and still others suggest it to be a complex of cryptic species (Milhomem et al., 2008; Nagamachi et al., 2010). Within this context, some phylogenetic studies with molecular data show polytomy of the G. carapo complex, consisting of G. carapo, G. arapaima, and G. ucamara (Lovejoy et al., 2010; Brochu, 2011) and other studies show species of the subgenus Gymnotus nested within G. carapo lineages (Lehmberg et al., 2018; Craig et al., 2019; da Silva et al., 2019).
In conclusion, the results presented here support that these populations with different cytotypes of G. carapo analysed (G. carapo occidentalis “Catalão”, G. c. orientalis GCA42 and GCA 40, along with the geographic-specific 2n = 48 and 2n = 54) may be a cryptic species complex. Analyses by chromosome painting of more cytotypes of G. carapo as well as other species of this genus coupled with molecular studies of those samples could help elucidate the chromosomal evolution and pattern of speciation in the group and help identify same-species populations from endemic species that have recently diverged.
Data Availability Statement
All data presented in this study are found in the article.
Ethics Statement
The animal study was reviewed and approved by The Ethics Committee from Para Federal University (Comitê de Ética Animal da Universidade Federal do Pará) approved this research (Permit 68/2015).
Author Contributions
MM: Conceptualization; Data Curation; Formal analysis; Investigation; Methodology; Visualization; Writing original draft; Writing review and editing. MS: Investigation; Methodology; Visualization; Writing review and editing. EF: Investigation; Methodology; Funding acquisition; Visualization; Writing review and editing. PM: Investigation; Methodology; Visualization; Writing review and editing. MF-S: Investigation; Methodology; Resources; Visualization; Writing review and editing. JP: Data Curation; Formal analysis; Funding acquisition; Resources; Visualization; Writing review and editing. CN: Data Curation; Formal analysis; Funding acquisition; Project administration; Resources; Supervision; Visualization; Writing review and editing.
Funding
The authors thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support on a project coordinated by CN (Edital Pró-Amazônia Proc 047/2012); the FAPESPA for financial support (Edital Vale–Proc 2010/110447) and Banco Nacional de Desenvolvimento Econômico e Social–BNDES (2.318.697.0001) on a project coordinated by JP; the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for financial support on project coordinated by EF (Edital Universal Proc 473812/2011-8). CYN (305880/2017-9), JP (305876/2017-1) and EF (301886/2019-9), are grateful to CNPq for Productivity Grants and MS (160155/2018-5) for a scholarship from CNPq. This study is part of the Doctoral Thesis in Genetic and Molecular Biology of MM who is recipient of a CNPq Doctor Scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to members of the team of the cytogenetics laboratory UFPA for the fieldwork and chromosomal preparations. To MSc. Jorge Rissino, to MSc. Shirley Nascimento and Maria da Conceição for assistance in laboratory work.
References
Albert, J. S., Craig, J. M., Tagliacollo, V. A., and Petry, P. (2018a). “Upland and lowland Fishes: a Test of the River Capture Hypothesis,” in Mountains, Climate and Biodiversity. Editors C. Hoorn, A. Perrigo, and A. Antonelli (Chichester: Wiley Blackwell), 273–294.
Albert, J. S., and Crampton, W. G. R. (2005). “Diversity and Phylogeny of Neotropical Electric Fishes (Gymnotiformes),” in Electroreception (New York: Springer), 360–409. doi:10.1017/s1477200005001726
Albert, J. S. (2001). Species Diversity and Phylogenetic Systematics of American Knifefishes (Gymnotiformes, Teleostei), 190. Ann Arbor, MI: Miscellaneous Publications. University of Michigan, 1–129.
Albert, J. S., Tagliacollo, V. A., and Dagosta, F. (2020). Diversification of Neotropical Freshwater Fishes. Annu. Rev. Ecol. Evol. Syst. 51 (1), 27–53. doi:10.1146/annurev-ecolsys-011620-031032
Albert, J. S., Val, P., and Hoorn, C. (2018b). The Changing Course of the Amazon River in the Neogene: center Stage for Neotropical Diversification. Neotrop. Ichthyol. 16, e180033. doi:10.1590/1982-0224-20180033
Almeida, J. S., Migues, V. H., Diniz, D., and Affonso, P. R. A. M. (2015). A Unique Sex Chromosome System in the Knifefish Gymnotus Bahianus with Inferences about Chromosomal Evolution of Gymnotidae. J. Hered. 106, 177–183. doi:10.1093/jhered/esu087
Barby, F. F., Bertollo, L. A. C., de Oliveira, E. A., Yano, C. F., Hatanaka, T., Ráb, P., et al. (2019). Emerging Patterns of Genome Organization in Notopteridae Species (Teleostei, Osteoglossiformes) as Revealed by Zoo-FISH and Comparative Genomic Hybridization (CGH). Sci. Rep. 9 (1), 1112. doi:10.1038/s41598-019-38617-4
Braga, B. P. F., Flecha, R., Pena, D. S., and Kelman, J. (2008). Pacto federativo e gestão de águas. Estud. Av. 22 (63), 17–42. doi:10.1590/S0103-40142008000200003
Brochu, K. (2011). Molecular Phylogenetics of the Neotropical Electric Knifefish Genus Gymnotus (Gymnotidae, Teleostei): Biogeography and Signal Evolution of the Trans-andean Species. [Master dissertation]. Toronto (ON): University of Toronto.
Cardoso, A. L., Ready, J. S., Pieczarka, J. C., Milhomem, S. S. R., Figueiredo-Ready, W. M. B. d., Silva, F. H. R., et al. (2015). Chromosomal Variability between Populations of Electrophorus Electricus Gill, 1864 (Pisces: Gymnotiformes: Gymnotidae). Zebrafish 12 (6), 440–447. doi:10.1089/zeb.2014.1059
Carvalho, P. C., de Oliveira, E. A., Bertollo, L. A. C., Yano, C. F., Oliveira, C., Decru, E., et al. (2017). First Chromosomal Analysis in Hepsetidae (Actinopterygii, Characiformes): Insights into Relationship between African and Neotropical Fish Groups. Front. Genet. 8, 203. doi:10.3389/fgene.2017.00203
Cioffi, M. B., Ráb, P., Ezaz, T., Bertollo, L. A. C., Lavoué, S., Oliveira, E. A., et al. (2019). Deciphering the Evolutionary History of Arowana Fishes (Teleostei, Osteoglossiformes, Osteoglossidae): Insight from Comparative Cytogenomics. Ijms 20, 4296. doi:10.3390/ijms20174296
Cioffi, M. d. B., Yano, C. F., Sember, A., and Bertollo, L. A. C. (2017). Chromosomal Evolution in Lower Vertebrates: Sex Chromosomes in Neotropical Fishes. Genes 8, 258. doi:10.3390/genes8100258
Claro, F. L. (2008). Gymnotus Carapo and Gymnotus Sylvius (Teleostei: Gymnotidae): Uma Abordagem Citogenético-Molecular. Master’s Dissertation. São Paulo, SP: Universidade de São Paulo.
Craig, J. M., Crampton, W. G. R., and Albert, J. S. (2017). Revision of the Polytypic Electric Fish Gymnotus carapo (Gymnotiformes, Teleostei), with Descriptions of Seven Subspecies. Zootaxa 4318, 401–438. doi:10.11646/zootaxa.4318.3.1
Craig, J. M., Kim, L. Y., Tagliacollo, V. A., and Albert, J. S. (2019). Phylogenetic Revision of Gymnotidae (Teleostei: Gymnotiformes), with Descriptions of Six Subgenera. Plos One 14 (11), e0224599. doi:10.1371/journal.pone.0224599
da Silva, M. (2015).Análise Biogeográfica do Gênero Gymnotus (Gymnotidae, Gymnotiformes), Por Meio de Marcadores Cariotípicos e Moleculares. Dissertation. Manaus, AM: Instituto Nacional de Pesquisas da Amazônia.
da Silva, M., Barbosa, P., Artoni, R. F., and Feldberg, E. (2016). Evolutionary Dynamics of 5S rDNA and Recurrent Association of Transposable Elements in Electric Fish of the Family Gymnotidae (Gymnotiformes): the Case of Gymnotus Mamiraua. Cytogenet. Genome Res. 149, 297–303. doi:10.1159/000449431
da Silva, M., Matoso, D. A., Artoni, R. F., and Feldberg, E. (2019). Karyotypic Diversity and Evolutionary Trends in Neotropical Electric Fish of the Genus Gymnotus (Gymnotiformes: Gymnotidae). Zebrafish 16 (3), 308–320. doi:10.1089/zeb.2018.1716
da Silva, M., Matoso, D. A., Artoni, R. F., and Feldberg, E. (2014). New Approach Data in Electric Fish (Teleostei: Gymnotus): Sex Chromosome Evolution and Repetitive DNA. Zebrafish 11, 528–535. doi:10.1089/zeb.2013.0966
da Silva, M., Matoso, D. A., Vicari, M. R., de Almeida, M. C., Margarido, V. P., and Artoni, R. F. (2011). Physical Mapping of 5S rDNA in Two Species of Knifefishes: Gymnotus Pantanal and Gymnotus Paraguensis (Gymnotiformes). Cytogenet. Genome Res. 134, 303–307. doi:10.1159/000328998
de Freitas, N. L., Al-Rikabi, A. B. H., Bertollo, L. A. C., Ezaz, T., Yano, C. F., de Oliveira, E. A., et al. (2018). Early Stages of XY Sex Chromosomes Differentiation in the Fish Hoplias malabaricus (Characiformes, Erythrinidae) Revealed by DNA Repeats Accumulation. Cg 19 (3), 216–226. doi:10.2174/1389202918666170711160528
de Moraes, R. L. R., Bertollo, L. A. C., Marinho, M. M. F., Yano, C. F., Hatanaka, T., Barby, F. F., et al. (2017). Evolutionary Relationships and Cytotaxonomy Considerations in the GenusPyrrhulina(Characiformes, Lebiasinidae). Zebrafish 14 (6), 536–546. doi:10.1089/zeb.2017.1465
de Oliveira, E. A., Sember, A., Bertollo, L. A. C., Yano, C. F., Ezaz, T., Moreira-Filho, O., et al. (2017). Tracking the Evolutionary Pathway of Sex Chromosomes Among Fishes: Characterizing the Unique XX/XY1Y2 System in Hoplias malabaricus (Teleostei, Characiformes). Chromosoma 127 (1), 115–128. doi:10.1007/s00412-017-0648-3
Fernandes-Matioli, F. M. C., Marchetto, M. C. N., Almeida-Toledo, L. F., and Toledo-Filho, S. A. (1998). High Intraspecific Karyological Conservation in Four Species ofGymnotus(Pisces: Gymnotiformes) from Southeastern Brazilian Basins. Caryologia 51, 221–234. doi:10.1080/00087114.1998.10797414
Fonteles, S. B. A., Lopes, C. E., Akama, A., Fernandes, F. M. C., Porto-Foresti, F., Senhorini, J. A., et al. (2008). Cytogenetic Characterization of the Strongly Electric Amazonian Eel, Electrophorus Electricus (Teleostei, Gymnotiformes), from the Brazilian Rivers Amazon and Araguaia. Genet. Mol. Biol. 31, 227–230. doi:10.1590/S1415-47572008000200010
Foresti, F., de Almeida Toledo, L. F., and de Almeida Toledo, S. (1984). Chromosome Studies inGymnotus CarapoandGymnotus Sp.(Pisces, Gymnotidae). Caryologia 37, 141–146. doi:10.1080/00087114.1984.10797693
Hijmans, R. J., Guarino, L., Bussink, C., Mathur, P., Cruz, M., Barrentes, I., et al. DIVA-GIS. Vsn 5.0. A Geographic Information System for the Analysis of Species Distribution Data. 2004. Available at: http://www.diva-gis.org.
J. S. Albert, and R. Reis (Editors) (2011). Historical Biogeography of Neotropical Freshwater Fishes (Berkeley, USA: Univ of California Press). doi:10.1525/california/9780520268685.001.0001
Kim, L. Y., Crampton, W. G. R., and Albert, J. S. (2020). Two New Species of Gymnotus (Gymnotiformes: Gymnotidae) from Brazil and Historical Biogeography of the Subgenus Lamontianus. Copeia 108 (3), 468–484. doi:10.1643/ci-19-205
King, M. (1993). Species Evolution - the Role of Chromosome Change. Cambridge, England: Cambridge University Press, 335.
Lehmberg, E. (2015). Biogeography of the Weakly Electric Knifefish Gymnotus carapo (Teleostei: Gymnotidae) in the Guianas. Toronto: University of Toronto. Available at: https://tspace.library.utoronto.ca/bitstream/1807/69686/3/Lehmberg_Emma_S_201506_MSc_thesis.pdf (Accessed January 19, 2022).
Lehmberg, E. S., Elbassiouny, A. A., Bloom, D. D., López‐Fernández, H., Crampton, W. G. R., and Lovejoy, N. R. (2018). Fish Biogeography in the “Lost World” of the Guiana Shield: Phylogeography of the Weakly Electric Knifefish Gymnotus carapo (Teleostei: Gymnotidae). J. Biogeogr. 45, 815–825. doi:10.1111/jbi.13177
Levan, A., Fredga, K., and Sandberg, A. A. (1964). Nomenclature for Centromeric Position on Chromosomes. Hereditas 52, 201–220. doi:10.1111/j.1601-5223.1964.tb01953.x
Lovejoy, N. R., Lester, K., Crampton, W. G. R., Marques, F. P. L., and Albert, J. S. (2010). Phylogeny, Biogeography, and Electric Signal Evolution of Neotropical Knifefishes of the Genus Gymnotus (Osteichthyes: Gymnotidae). Mol. Phylogenet. Evol. 54, 278–290. doi:10.1016/j.ympev.2009.09.017
Machado, M. A., Pieczarka, J. C., Silva, F. H. R., O'Brien, P. C. M. O., Ferguson-Smith, M. A., and Nagamachi, C. Y. (2018). Extensive Karyotype Reorganization in the Fish Gymnotus arapaima (Gymnotiformes, Gymnotidae) Highlighted by Zoo-FISH Analysis. Front. Genet. 9 (8), 1–19. doi:10.3389/fgene.2018.00008
Machado, M. d. A., Cardoso, A. L., Milhomem-Paixão, S. S. R., Pieczarka, J. C., and Nagamachi, C. Y. (2017). Gymnotus Coatesi (Gymnotiformes): a Case of Colocation of Multiple Sites of 18S rDNA with Telomeric Sequences. Zebrafish 14, 459–463. doi:10.1089/zeb.2017.1435
Mago-Leccia, F. (1994). Electric Fishes of the continental Waters of America: Classification and Catalogue of the Electric Fishes of the Order Gymnotiformes (Teleostei: Ostariophysi), with Descriptions of New Genera and Species. Caracas, Venezuela: Fundacion para Desarrollo de las Ciencias Fiscias, Matematicas y Naturales.
Margarido, V. P., Bellafronte, E., and Moreira-Filho, O. (2007). Cytogenetic Analysis of Three sympatricGymnotus(Gymnotiformes, Gymnotidae) Species Verifies Invasive Species in the Upper Paraná River basin, Brazil. J. Fish. Biol. 70, 155–164. doi:10.1111/j.1095-8649.2007.01365.x
Milhomem, S. S., Pieczarka, J. C., Crampton, W. G., Silva, D. S., De Souza, A. C., Carvalho, J. R., et al. (2008). Chromosomal Evidence for a Putative Cryptic Species in the Gymnotus carapo Species-Complex (Gymnotiformes, Gymnotidae). BMC Genet. 9, 75. doi:10.1186/1471-2156-9-75
Milhomem, S. S. R., Crampton, W. G. R., Pieczarka, J. C., dos Santos Silva, D., Cardoso, A. L., da Silva, P. C., et al. (2012b). Chromosomal and Electric Signal Diversity in Three Sympatric Electric Knifefish Species (Gymnotus, Gymnotidae) from the central Amazon Floodplain. Rev. Fish. Biol. Fish. 22, 485–497. doi:10.1007/s11160-011-9239-1
Milhomem, S. S. R., Crampton, W. G. R., Pieczarka, J. C., Shetka, G. H., Silva, D. S., and Nagamachi, C. Y. (2012a). Gymnotus Capanema, a New Species of Electric Knife Fish (Gymnotiformes, Gymnotidae) from Eastern Amazonia, with Comments on an Unusual Karyotype. J. Fish. Biol. 80, 802–815. doi:10.1111/j.1095-8649.2012.03219.x
Milhomem, S. S. R., Pieczarka, J. C., Crampton, W. G. R., De Souza, A. C. P., Carvalho Jr, J. R., and Nagamachi, C. Y. (2007). Differences in Karyotype between Two Sympatric Species of Gymnotus (Gymnotiformes: Gymnotidae) from the Eastern Amazon of Brazil. Zootaxa 1397, 55–62. doi:10.11646/zootaxa.1397.710.11646/zootaxa.1397.1.7
Milhomem, S. S. R., Scacchetti, P. C., Pieczarka, J. C., Ferguson-Smith, M. A., Pansonato-Alves, J. C., O’Brien, P. C. M., et al. (2013). Are NORs Always Located on Homeologous Chromosomes? A FISH Investigation with rDNA and Whole Chromosome Probes in Gymnotus Fishes (Gymnotiformes). PLoS ONE 8, e55608. doi:10.1371/journal.pone.0055608
Nagamachi, C. Y., Pieczarka, J. C., Milhomem, S. S., O'Brien, P. C., de Souza, A. C., and Ferguson-Smith, M. A. (2010). Multiple Rearrangements in Cryptic Species of Electric Knifefish, Gymnotus carapo (Gymnotidae, Gymnotiformes) Revealed by Chromosome Painting. BMC Genet. 11, 1. doi:10.1186/1471-2156-11-28
Nagamachi, C. Y., Pieczarka, J. C., Milhomem, S. S. R., Batista, J. A., O'Brien, P. C. M., and Ferguson-Smith, M. A. (2013). Chromosome Painting Reveals Multiple Rearrangements betweenGymnotus capanemaandGymnotus carapo(Gymnotidae, Gymnotiformes). Cytogenet. Genome Res. 141, 163–168. doi:10.1159/000354988
Ráb, P., Rábová, M., Pereira, C. S., Collares-Pereira, M. J., and Pelikánová, Š. (2008). Chromosome Studies of European Cyprinid Fishes: Interspecific Homology of Leuciscine Cytotaxonomic Marker-The Largest Subtelocentric Chromosome Pair as Revealed by Cross-Species Painting. Chromosome Res. 16, 863–873. doi:10.1007/s10577-008-1245-3
R. Fricke, W. N. Eschmeyer, and R. van der Laan (Editors) (2021). Eschmeyer's Catalog of Fishes: Genera, Species, References. (San Francisco, United States: California Academy of Sciences). Available at: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
Scacchetti, P., Pansonato-Alves, J., Utsunomia, R., Oliveira, C., and Foresti, F. (2011). Karyotypic Diversity in Four Species of the Genus Gymnotus Linnaeus, 1758 (Teleostei, Gymnotiformes, Gymnotidae): Physical Mapping of Ribosomal Genes and Telomeric Sequences. Ccg 5, 223–235. doi:10.3897/compcytogen.v5i3.1375
Scudeler, P. E. S., Diniz, D., Wasko, A. P., Oliveira, C., and Foresti, F. (2015). Whole Chromosome Painting of B Chromosomes of the Red-Eye Tetra Moenkhausia sanctaefilomenae (Teleostei, Characidae). Ccg 9 (4), 661–669. doi:10.3897/CompCytogen.v9i4.5460
Tagliacollo, V. A., Bernt, M. J., Craig, J. M., Oliveira, C., and Albert, J. S. (2016). Model-based Total Evidence Phylogeny of Neotropical Electric Knifefishes (Teleostei, Gymnotiformes). Mol. Phylogenet. Evol. 95, 20–33. doi:10.1016/j.ympev.2015.11.007
Utsunomia, R., Melo, S., Scacchetti, P. C., Oliveira, C., Machado, M. d. A., Pieczarka, J. C., et al. (2018). Particular Chromosomal Distribution of Microsatellites in Five Species of the Genus Gymnotus (Teleostei, Gymnotiformes). Zebrafish 15 (4), 398–403. doi:10.1089/zeb.2018.1570
Utsunomia, R., Scacchetti, P. C., Pansonato-Alves, J. C., Oliveira, C., and Foresti, F. (2014). Comparative Chromosome Mapping of U2 snRNA and 5S rRNA Genes in Gymnotus Species (Gymnotiformes, Gymnotidae): Evolutionary Dynamics and Sex Chromosome Linkage in G. Pantanal. Cytogenet. Genome Res. 142, 286–292. doi:10.1159/000362258
Utsunomia, R., Silva, D. M. Z. d. A., Ruiz-Ruano, F. J., Araya-Jaime, C., Pansonato-Alves, J. C., Scacchetti, P. C., et al. (2016). Uncovering the Ancestry of B Chromosomes in Moenkhausia sanctaefilomenae (Teleostei, Characidae). PLoS ONE 11 (3), e0150573. doi:10.1371/journal.pone.0150573
Vicari, M. R., de Mello Pistune, H. F., Castro, J. P., de Almeida, M. C., Bertollo, L. A. C., Moreira-Filho, O., et al. (2011). New Insights on the Origin of B Chromosomes in Astyanax scabripinnis Obtained by Chromosome Painting and FISH. Genetica 139, 1073–1081. doi:10.1007/s10709-011-9611-z
Yang, F., Carter, N. P., Shi, L., and Ferguson-Smith, M. A. (1995). A Comparative Study of Karyotypes of Muntjacs by Chromosome Painting. Chromosoma 103, 642–652. doi:10.1007/BF00357691
Keywords: amazon, banded knife-fish, whole chromosome painting, FISH, complex of species
Citation: Machado MA, da Silva M, Feldberg E, O’Brien PCM, Ferguson-Smith MA, Pieczarka JC and Nagamachi CY (2022) Chromosome Painting in Gymnotus carapo “Catalão” (Gymnotiformes, Teleostei): Dynamics of Chromosomal Rearrangements in Cryptic Species. Front. Genet. 13:832495. doi: 10.3389/fgene.2022.832495
Received: 09 December 2021; Accepted: 10 February 2022;
Published: 24 March 2022.
Edited by:
Tony Silveira, Federal University of Rio Grande, BrazilReviewed by:
Jack Craig, Temple University, United StatesDenisa Šimoníková, Academy of Sciences of the Czech Republic, Czechia
Copyright © 2022 Machado, da Silva, Feldberg, O’Brien, Ferguson-Smith, Pieczarka and Nagamachi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cleusa Yoshiko Nagamachi, Y2xldXNhQHVmcGEuYnI=, Y2xldXNhbmFnYW1hY2hpQGdtYWlsLmNvbQ==
 Milla de Andrade Machado
Milla de Andrade Machado Maelin da Silva
Maelin da Silva Eliana Feldberg
Eliana Feldberg Patricia Caroline Mary O’Brien
Patricia Caroline Mary O’Brien Malcolm Andrew Ferguson-Smith
Malcolm Andrew Ferguson-Smith Julio Cesar Pieczarka
Julio Cesar Pieczarka Cleusa Yoshiko Nagamachi
Cleusa Yoshiko Nagamachi