- 1Sharda School of Agricultural Sciences, Sharda University, Greater Noida, India
- 2Plant Molecular Biology Laboratory, Department of Botany, Dayanand Anglo-Vedic (PG) College, Chhatrapati Shahu Ji Maharaj University,, Kanpur, India
- 3Molecular Biology and Biotechnology Division, CSIR-National Botanical Research Institute, Rana Pratap Marg, Lucknow, India
- 4Department of Agriculture, Integral Institute of Agricultural Science and Technology, Integral University, Lucknow, Uttar Pradesh, India
- 5Division of Genetics, ICAR-Indian Agricultural Research Institute, New Delhi, India
Nutrient deficiency has resulted in impaired growth and development of the population globally. Microgreens are considered immature greens (required light for photosynthesis and growing medium) and developed from the seeds of vegetables, legumes, herbs, and cereals. These are considered “living superfood/functional food” due to the presence of chlorophyll, beta carotene, lutein, and minerals like magnesium (Mg), Potassium (K), Phosphorus (P), and Calcium (Ca). Microgreens are rich at the nutritional level and contain several phytoactive compounds (carotenoids, phenols, glucosinolates, polysterols) that are helpful for human health on Earth and in space due to their anti-microbial, anti-inflammatory, antioxidant, and anti-carcinogenic properties. Microgreens can be used as plant-based nutritive vegetarian foods that will be fruitful as a nourishing constituent in the food industryfor garnish purposes, complement flavor, texture, and color to salads, soups, flat-breads, pizzas, and sandwiches (substitute to lettuce in tacos, sandwich, burger). Good handling practices may enhance microgreens’stability, storage, and shelf-life under appropriate conditions, including light, temperature, nutrients, humidity, and substrate. Moreover, the substrate may be a nutritive liquid solution (hydroponic system) or solid medium (coco peat, coconut fiber, coir dust and husks, sand, vermicompost, sugarcane filter cake, etc.) based on a variety of microgreens. However integrated multiomics approaches alongwith nutriomics and foodomics may be explored and utilized to identify and breed most potential microgreen genotypes, biofortify including increasing the nutritional content (macro-elements:K, Ca and Mg; oligo-elements: Fe and Zn and antioxidant activity) and microgreens related other traits viz., fast growth, good nutritional values, high germination percentage, and appropriate shelf-life through the implementation of integrated approaches includes genomics, transcriptomics, sequencing-based approaches, molecular breeding, machine learning, nanoparticles, and seed priming strategiesetc.
1 Introduction
In the past few decades, interest in organic and nutritional vegetables has gained momentum among people. That has increased the demand for sprouts and microgreens. Sprouts are considered germinated seeds that can be harvested before the growth of true leaves and consumed whole with the seed (Di Gioia et al., 2017). On the other hand, microgreens are defined as tender, immature greens that need light for photosynthesis, growing medium (soil or nutrient solution medium), and represent a 7–28 days growth cycle. Microgreens can be developed from the seeds of cereals, vegetables, legumes, and herbs, comprising two completely expanded cotyledon leaves with or without the appearance of a rudimentary pair of first true leaves (Xiao et al., 2012). The history of microgreen production can be traced back to the 1980s when it first seemed on the menu of chefs in San Francisco, California (United States Department of Agriculture, 2014). Following its popularity further, its cultivation began in the southern part of California in the 1990s, such that now microgreens are regarded as “functional foods” or “Superfoods.”
Some cereals (rice, corn, oats, wheat, and barley) and legumes (chickpeas, beans, and lentils) can also be exploited for microgreens cultivation and production due to their nutritional values. Microgreens may be used for sweet and savory dishes as a garnish. They can complement the flavor, texture, and color of salads, soups, flatbreads, pizzas, and sandwiches (alternative to lettuce in tacos/burgers/sandwiches) due to a good proportion of nutritional values and some specific metabolites. People can also supplement microgreens to prepare smoothies, juices, and health drinks. Several value-added products can be synthesized by using fresh and dry microgreens in food science laboratories as one ingredient, for example, cookies, noodles, snacks, and chips are developed. Microgreens products would be an up-and-coming resource for vegetarians as a healthy lifestyle change and generation of employment for the population engaged in agriculture and food processing sectors.
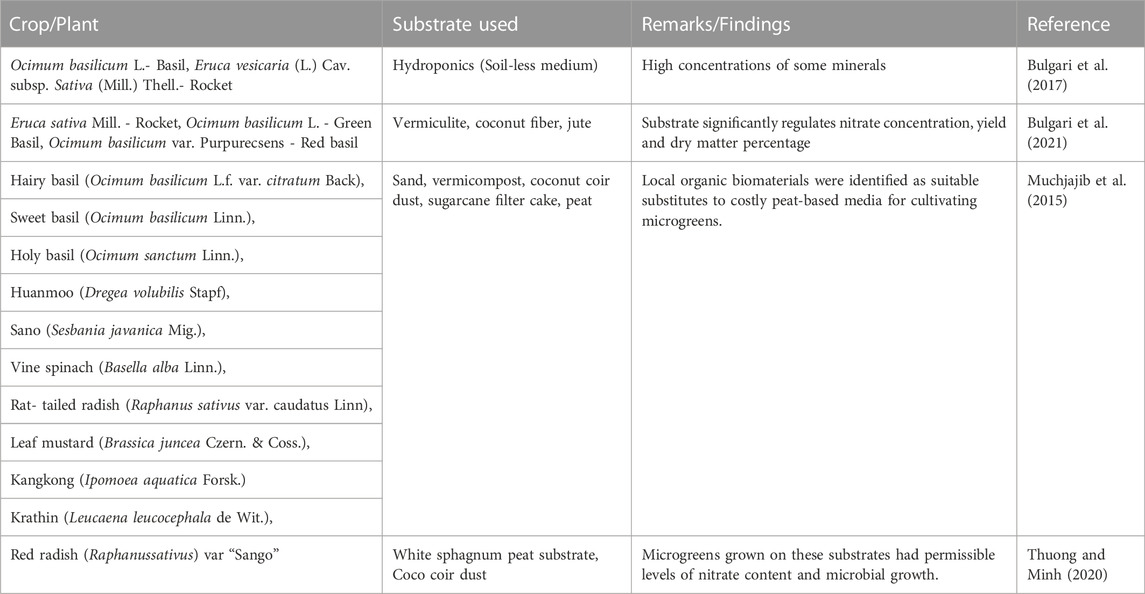
Very few reports are available on the cultivation of legumes microgreens, for example, chickpea micro-greens (Sreenivasan, 2020). The small-seeded legumes promise unmarked resources of potential ingredients for the fortification of traditional or staple foods with bioactive compounds, nutrients, and minerals (Butkutė et al., 2018). Erosion of preserved seed nutrients was observed in chickpea during the process to stimulate seedling growth that improved nutritional value with digestible protein levels ranging from 18.96% to 28.69% in 6-day-old sprouts identifying the genotypes BG-1092, ICC-11378, JG-74 as potential resources for accumulating proteins and other nutritional components at the seedling stage (Kumar et al., 2022a). Therefore, the cultivation of microgreens can be easily practiced, and cheaper sources of budding superfoods in terms of nutrition and antioxidant properties with minimum food wastage (only root) during consumption may be obtained. Thus, in the present report, we have discussed the prospects of nutraceutical aspects, health benefits, growth, and cultivation practices for microgreen production and mentioned related strategies for the first time to overcome the limitations through the utilization/integration of various OMICS approaches.
2 Nutraceutical and health benefits of microgreens: The superfoods/functional foods
Nutrients deficiency may cause serious diseases and health related issues and appropriate proportions of micronutrients, macronutrients, flavonoids and polyphenols regulate immunity and prevent from several health threats viz; osteoporosis, pharmacotherapy, COVID-19 etc (Batiha et al., 2022; Martiniakova et al., 2022; Bansal et al., 2022a; Bansal et al., 2022b). Microgreens are partially mature greens that elicit their intense flavour, aroma, texture, and nutrient properties with sensory attributes and acceptance are discussed in this review. Desirable sensory qualities and intense flavours of these superfoods have gained acclamation to be consumed as salads, garnishes, etc. The presence of minerals, vitamins, and their precursors - ascorbic acid and several other bioactive compounds tocopherols, carotenoids, betaine, phenols, glucosinolates, phytosterolsetc in microgreens add to their health and nutrition-related functional aspects (Xiao et al., 2012; Kyriacou et al., 2019a)as presented in Table 1 and Table 2. The sensory qualities of microgreens are influenced by their chemical composition (Xiao et al., 2015). This study highlights the correlation between the concentration of total phenols and with overall acceptability of sensory attributes and acceptance in terms of sweetness, sourness, bitterness, and astringency. The sensory and nutritional qualities of microgreens also vary with the growing methods. The sensory attributes and nutritional content of microgreens grown hydroponically and in soil procured from a commercial and local farm were compared (Tan et al., 2020). Nutritionally microgreens grown by either method were comparable. However, a significant difference in vitamin C content was reported. Variations in both micro-minerals (iron, zinc, copper, and manganese) and macro-mineral content (calcium, phosphorus, sodium, magnesium, chloride, potassium, and sulfur),phytochemical profile, and antioxidant capacities have been found to vary with genotype rather than growth stage as studied in microgreens of four Brassicaceae genotypes-Komatsuna, Mizuna, Pak Choi and Mibuna (Kyriacou et al., 2021a). In addition, delaying harvest from the arrival of the first to second true leaf does not seem relevant for improving bioactive compounds in microgreens (Kyriacou et al., 2021a). Regulated feeding of nutrient solution (NS) to microgreens and fertigation treatment can also influence the composition of phytochemicals and antioxidant activity apart from growth and yield (Petropoulos et al., 2021). Moreover, spinach microgreens were assessed for the effect of nutrient deprivation (0, 5, 10, and 20 days) and fertigation treatment before harvesting (Petropoulos et al., 2021). This study reported that NS feeding for a longer duration of 20 days resulted in enhanced fresh yield and content of photosynthetic pigments, including chlorophyll, beta carotene, and lutein. In contrast, the concentration of minerals like Calcium (Ca), Potassium(K), magnesium (Mg), and Phosphorus (P) were found to be lowest after 20 days in contrast to being maximum in control and 5 days of NS feeding (Petropoulos et al., 2021). Feeding the spinach microgreens for 10 days with NS yielded the best combination of yield, minerals (high), and nitrate (low) while maintaining the concentrations of bioactive compounds (Petropoulos et al., 2021). Simultaneous to the accumulation of secondary metabolites, microgreens are also known to accumulate anti-nutritive compounds like nitrate. Therefore, nutrient deprivation before harvest (DBH) was employed to reduce nitrate levels by substituting NS with osmotic water for 6 and 12 days in a garden rocket, lettuce, and mustard microgreens grown on a peat-based substrate (Kyriacou et al., 2021b). Nutrient deprivation proved a good strategy for lowering nitrate content with effective treatment duration varying from species to species. Even nitrate hyper-accumulating species like garden rockets showed an abrupt decline in nitrate concentration (Kyriacou et al., 2021b). Further, abundant secondary metabolites like flavanol glycosides, quercetin, and kaempferol glycosides were detected in Brassicaceae, and caffeoyl quinic acid in lettuce microgreens. However, total phenols increased in lettuce, reduced in the garden rocket, and unaffected in mustard microgreens in response to nutrient deprivation (Kyriacou et al., 2021b).
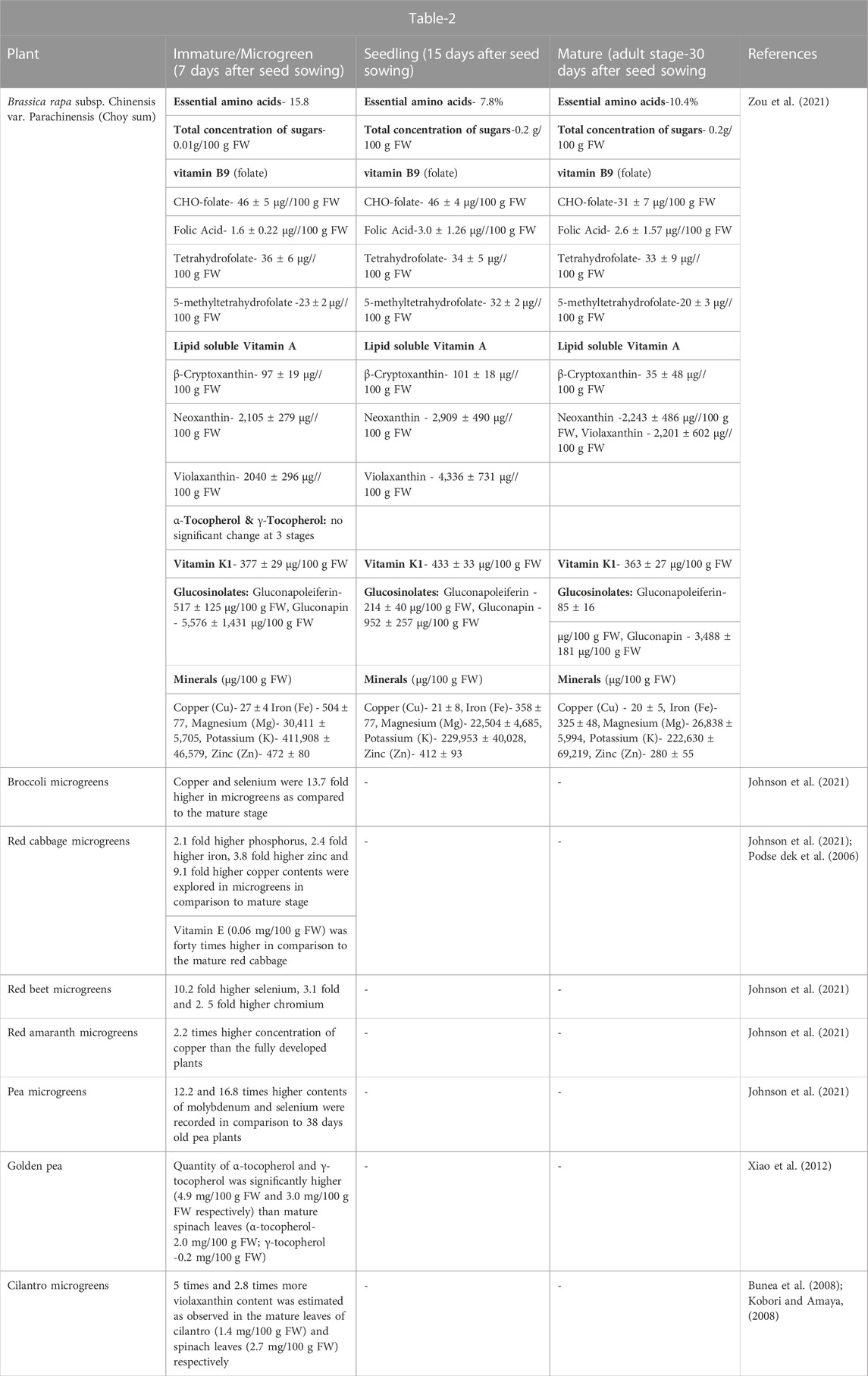
TABLE 2. Comparative analysis of nutritional components in microgreeens, mature part and different stages of plants.
The presence of bioactive compounds renders microgreens as a health beneficiary, antioxidant,anti-microbial, anti-inflammatory, and anti-carcinogenic. The health-related properties of bioactive compounds in food and herbs depend not only on their content and the amount consumed but also on their bioavailability (de la Fuente et al., 2019). Quantity and bio-accessibility of bioactive antioxidant compounds (total anthocyanins, total soluble polyphenols, ascorbic acid, total isothiocyanates), antioxidant capacity (Trolox Equivalent Antioxidant Capacity, and Oxygen Radical Absorbance capacity), macro-elements (K, Ca and Mg) and oligo elements (Fe and Zn) have been evaluated in four hydroponic Brassicaceae microgreens-broccoli, radish, kale and mustard (de la Fuente et al., 2019). The optimum amount of nutrients (macro and oligo-elements) is required for proper growth metabolism, and in contrast, deficiency may lead to life-threatening diseases in extreme circumstances. The essential nutrients are widely distributed in foods, and most people can obtain sufficient amounts by consuming a varied diet. Moreover,macro-elements (K, Ca, and Mg) and oligo-elements (Fe,Se, Cu, and Zn) remarkably stimulate the function of the immune system and are helpful in cardiopulmonary bypass (Al-Bader et al., 1998). Microgreens of soybean, green pea, garden rocket, radish, and red Rambo radish were cultivated under fluorescent and LED light conditions. The variation in anti-proliferative/pro-oxidant efficiencies of these microgreens was studied using Ewing sarcoma lines RD-ES and A673 (Truzzi et al., 2021). It was observed that all microgreen extracts could reduce cell proliferation in 2-dimensional cell cultures, while extracts from pea microgreens grown under LED light showed anti-proliferative and pro-apoptotic activity on 3-dimensional A673 and RD-ES spheroids without showing cytotoxicity on healthy L929 fibroblasts (Truzzi et al., 2021). LED and fluorescent light illuminated Red Rambo radish also exhibited anti-tumor effects on RD-ES spheroids. Further, the effects of UV-A, B, and C on inducing polyphenol content and anti-tumor activities of UV-illuminated microgreens can be an area of exploration in the future. In a study, the effect of salinity in combination with different wavelengths of light in Brassica carinata extracts from microgreens grown under different treatments of salinity and light were checked for their ability to stimulate antioxidant enzymes, including catalase (CAT), superoxide dismutase (SOD) and expressions of Nrf2 (nuclear transcription factor-erythroid 2 related factor) and HO-1 proteins (heme-oxygenase -1)on human colorectal carcinoma cells-HCT116 (Maina et al., 2021). Activation of antioxidant enzymes (SOD and CAT) and stimulation of HO-1 and Nrf2 make them preferable for the prevention and treatment of oxidative stress and inflammatory disorders (Maina et al., 2021). Microgreens can be a powerful source of nutrients owing to higher concentrations of several phytochemicals than their matured counterparts. These can be used as supplements to overcome deficiencies of several nutrients. Being a rich source of bioactive compounds like carotenoids, phenols, glucosinolates, polysterols, and many others, microgreens possess health benefits due to anti-microbial, antioxidant, anti-inflammatory, and anti-carcinogenic properties (Choe et al., 2018; Le et al., 2020).
Most of the microgreens viz., broccoli, red beet, red amaranth, red cabbage and pea microgreens exhibit high proportion of minerals and nutritionally rich components (copper, selenium, phosphorus, iron, zinc, molybdenum and chromium) as compared to mature and other counterparts (Johnson et al., 2021) as presented in Table 2 and are being narrated as given further.
3 Comparison for nutrients and phytochemicals in microgreens and different stages of crop plants
The comparative nutrient profiling was explored in Brassica rapa subsp. Chinensis var. Parachinensis (Choy sum) at three different growth stages viz. Microgreen, seedling and adult/mature stages (7, 15 and 30 days after seed sowing) respectively (Zou et al., 2021). The content of essential amino acid was high at microgreen stage (15.8%) as compared to seedling (7.8%) and mature (10.4%) stages. Moreover, the contents of metabolites and minerals were estimated along with their changes at maturity in six crops microgreens belonging to three distinct families Brassicaceae (Eruca sativa L.) Cav. - Arugula, Brassica oleracea L. Italica Group-Broccoli and Brassica oleracea L. Capitata Group - Red cabbage), Fabaceae (Pisum sativum - Pea) and Amaranthaceae—(Amaranthus tricolor L. -Red amaranth and Beta vulgaris L. Crassa Group- Red beet) (Johnson et al., 2021). Moreover, metabolites showed significant difference (p < 0.05) and were ≥2- times greater in microgreens as compared to their mature counterparts (Johnson et al., 2021). There are 95 such metabolites in broccoli, 110 in red cabbage, 87 in arugula, 80 in red beet, 93 in pea and 101 in red amaranth microgreens, respectively. These metabolites were majorly classified as peptides, saccharides, nucleotides, amines, phenolics, lipids, organo-sulfurs, alkaloids and vitamins with prevalence of lipids and phenolics (Johnson et al., 2021). In another study, good quantities of α-tocopherol and γ-tocopherol were estimated in golden pea tendrils as 4.9 mg/100 g FW and 3.0 mg/100 g FW respectively (Xiao et al., 2012). Interestingly, cilantro microgreens and red cabbage microgreens have been reported to have more violaxanthin and vitamin E contents as compared to their mature counterparts, respectively (Podse dek et al., 2006; Bunea et al., 2008; Kobori and Amaya, 2008).
Further, now it is a proven fact that the red cabbage microgreen regulates the levels of lipids, cholesterols in human blood vessels and protects against cardiovascular diseases (Huang et al., 2016). The mustard and coriander microgreens have been reported to be very rich sources for their antioxidant, antimicrobial, anti-cancerous, anti-obesity, anti-inflammatory and antidiabetic activities (de la Fuente et al., 2020; Le et al., 2020; Saengha et al., 2021; Truzzi al., 2021; Dhakshayani and Alias 2022). Hence, appropriate daily consumption of microgreens would be a potential protective nutritional strategy to manage health, nutrition and chronic degenerative diseases (de la Fuente et al., 2020).
4 Microgreens: A novel, live, super functional food
Microgreens’ demand and preference are attributed to their aroma, tender texture, vivid colour, flavour, sensory attributes, and quick production or cultivation. Several microgreen species have peculiar colours; for example, microgreens of broccoli, spinach, and celery have been reported to be green in colour, microgreens of radish, red basil, and red cabbage are crimson, while multicolour microgreens have been developed in mustard and beet (Di Gioia et al., 2015). Some of the previous studies related to microgreens have been summarized in Table 1. Similarly, microgreens are also known to possess a distinct flavour viz; spinach and rapini taste-neutral, arugula, radish, and watercress spicy and Cucurbitaceae microgreens as bitter have been reported (Di Gioia et al., 2017).
Most of the species and varieties used in current microgreen production come from the Brassicaceae and Amaranthaceae families (Xiao et al., 2015; Kyriacou et al., 2016). In the Amaranthaceae family, some of the more popular species, subspecies, and varieties include beet, chard, and amaranth; in the Brassicaceae family, radish, broccoli, kale, cabbage, tatsoi, pakchoi, mizuna, arugula, and mustard. Microgreens of grain crops such as buckwheat, wheat, and rye have also been grown. Various medicinal and culinary herbs have also been used for microgreen production, including borage (or starflower), parsley, basil, and fenugreek, among many others (Verlinden, 2020). Microgreens’ taste can fluctuate significantly based on the variety. The widely used varieties have been explored for microgreens production from seeds of diverse plant families as mentioned further;Brassicaceae (Cauliflower, broccoli, cabbage, watercress, radish, and arugula), Apiaceae (Dill, carrot, fennel, and celery), Asteraceae (Lettuce, endive, chicory, and radicchio), Amaranthaceae (Amaranth, quinoa swiss chard, beet, and spinach), Amaryllidaceae (Garlic, onion, and leek) and Cucurbitaceae (Melon, cucumber, and squash).
Increasing interest in the production and consumption of microgreens is also due to their high nutritional content, high yield, rapid production, aroma, and other qualities. Their high nutritional qualities are mainly due to the presence of phytochemicals and other bioactive compounds, along with their antioxidant capacities. They are considered highly nutritious food because of the presence of nutrients that include proteins, minerals, vitamins, carotenoids, phenols, and glucosinolates (Ebert, 2013; Di Gioia et al., 2017). The concentrations of bioactive compounds found in microgreens and even sprouts are reported to be much higher than their mature counterparts (Kyriacou et al., 2016). For example, Broccoli microgreens grown hydroponically and in compost were found to have more nutrient content (Mn, Cu, P, K, Na, Mg, and Fe) than mature broccoli vegetables (Weber, 2017). Contrary to this, hydroponically grown fenugreek (Trigonella foenum-graecum L.), broccoli, and garden rocket (Eruca vesicaria subsp. Sativa) microgreens were reported to have lower mineral contents than their mature plants to be eaten as vegetables. Among the three, only fenugreek microgreens efficiently uptake iron in caco-2 cells (Khoja et al., 2020). Chickpea microgreens also contains a good amount of zinc, calcium, iron, antioxidants, vitamins, carbohydrates, fiber,fat, and high protein content. Moreover, enriched nutrient contents containing plants may produce microgreens with high biomass in a limited time in a cost-effective manner that would improve the cultivation and yield of microgreens.
5 Growth and cultivation practices for the production of microgreens
The basic requirements for microgreen cultivation are the availability of substances and the effect of light as narrated below.
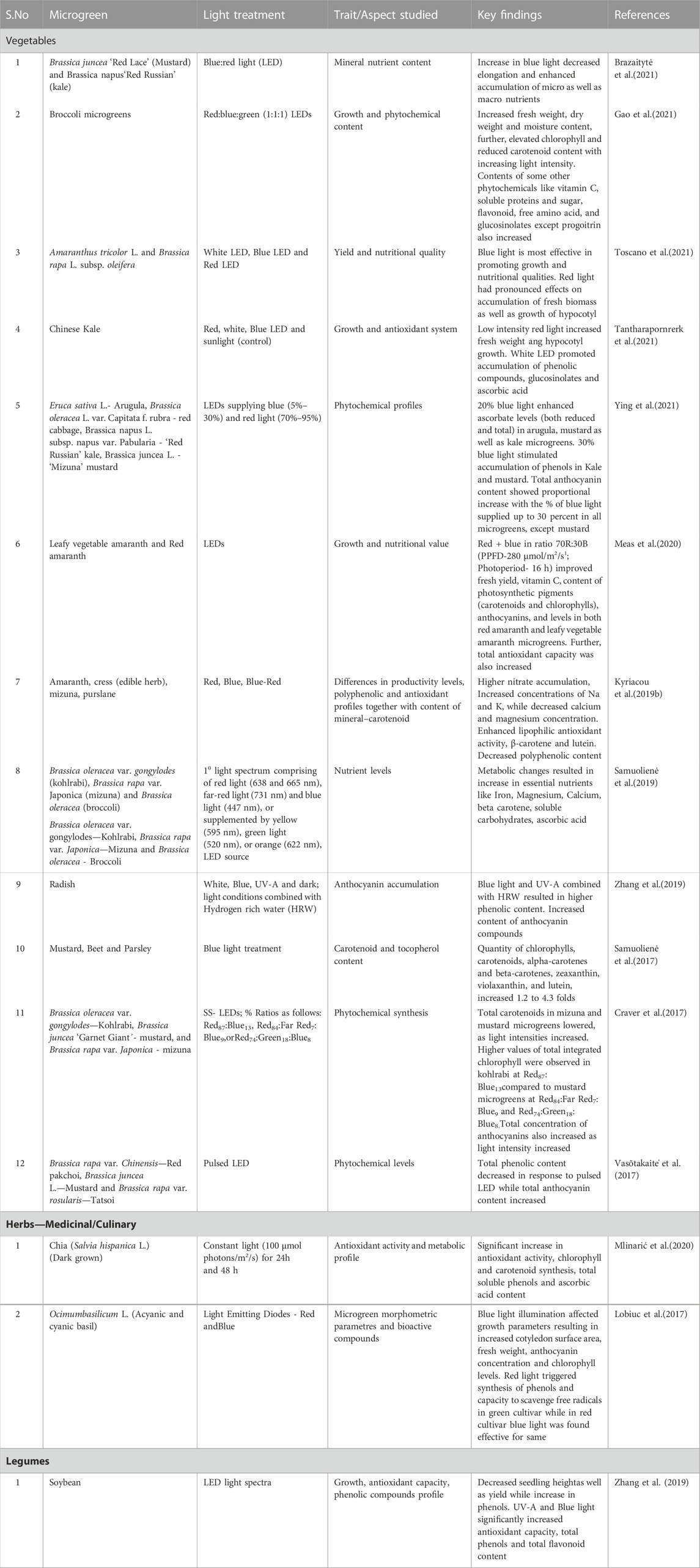
5.1 Availability of substrate for cultivation of microgreens
Various substrates have been used to grow microgreens, and their influence on yield and nutritional quality has been studied. In a study, three different substrates - vermiculite, cotton, and jute fiber were used to grow microgreens of green basil—Ocimum basilicum L., Red basil—Ocimum basilicum var. Purpurecsens and garden rocket Eruca sativa Mill in a Micro Experimental Growing System (MEG) fitted with LED lamps for light supply. In addition to that, several other substrates are also available to use further as primary medium or in combinations, for example, coco peat, coconut fiber, coconut coir dust, coconut husks, sand, jute fiber, vermicompost, sugarcane filter cake, peat and white sphagnum peat substrates presented in Table 3. A high yield of 2–3 kg/m2 was obtained. The three microgreens varied in nutritional quality, with red basil accounting for high antioxidant compounds on vermiculite and jute fiber media. At the same time, the qualitative parameters were found to be species-dependent (Bulgari et al., 2021).
5.2 Effect of light on nutritional quality and growth of microgreens
Essential growth factors like light (wavelength, intensity, and photoperiod) also influence microgreens’ biosynthesis and accumulation of phytochemicals (Delian et al., 2015). Recently, many studies have been carried out on the effect of artificial light sources like halogen lamps, high-pressure sodium lamps, fluorescent lamps, and LED lights on the growth, yield, nutrient quality, and phytochemical content of microgreens. Many studies have focused on the benefit of illumination with LED light on plant growth, quality, and accumulation of phytonutrients. Some of these studies have been tabulated (Table 4). The effect of different wavelengths of light in combination with salinity was assessed on quality in terms of antioxidant capacity, the content of phenolics and glucosinolates, as well as yield of microgreens of Brassica carinata (Maina et al., 2021). Stable cultivation of microgreens was achieved under fluorescent and blue plus red (B1R1) light conditions, which resulted in a high accumulation of biomass and glucobrassicin. Under saline conditions, blue LEDs and fluorescent light promoted antioxidant activity together with the accumulation of phenols, sinigrin and glucosinolate (Maina et al., 2021).
6 Limitations of microgreen production
The consumption and storage of microgreens pose several limitations. Poor shelf life and postharvest management of microgreens are major challenges for the concerning researchers. It is important to know if a correlation exists between produced spoilage and contamination by the human pathogen. Studies have reported seed-to-harvest pathogen infection in several microgreens like basil, lettuce, parsley, melons, and spinach (Alegbeleye et al., 2018). Growing conditions that promote microorganism growth or transfer, processing practices that expose the commodity to contaminants from animals or humans, and physiological characteristics of the plant that allow contact and binding with microorganisms, all of these factors put crops at risk (Maluin et al., 2021). As microgreens are grown in a controlled environment, they are unexposed to external agents like pests and insects. Also, there is minimal or no contamination due to almost no external application of fertilizers or manures. Strategies concerning postharvest management and enhancing the shelf life of microgreens need to be particularly developed.
7 Strategies to overcome limitations of microgreen production
There are various strategies to encounter the limitations related to microgreen cultivation and production as presented through Figure 2 and discussed below.
7.1 Selection and validation of potential crops/genotypes for microgreen traits
Identifying a diverse collection of genotypes is very important to explore the most promising genotypes containing microgreens-related traits viz., high nutrition, shelf life, sensory attributes, acceptable taste, and yield. However, microgreens’ phytoactive compounds, antioxidant capacity, shelf life, and nutrient content depend on genotypes’ genetic makeup and environmental conditions. For instance, twenty diverse genotypes of lentil and mungbean were grown as microgreens in plain and high altitude two regions (Delhi and Leh Ladakh). The investigation for profiling of phytochemical, macro and micronutrients content along with antioxidant capacity was accomplished (Mishra et al., 2021). Based on phytochemical profiles, lentil genotype L830 and mungbean genotype MH810 were identified as superior to other genotypes for contents of total flavonoids, carotenoids, ascorbic acid, antioxidant parameters, and phenols. The difference in nutritional profiles of identical genotypes under different environmental conditions was observed, probably due to variable gene expression. However, the genes and pathways governing the variation in response to different environmental conditions are yet to be elucidated. This study has provided new insights into the potential application of microgreens in harbouring the nutritional security of inhabitants of harsh environments such as the high altitudes of Leh-Ladakh (Mishra et al., 2021). Some other legume microgreens viz., sainfoin, red clover, alfalfa, chickpea, lentils, maize, cowpea and mung bean were tested for phenolic, antioxidant activity, flavonoid, carotenoid, ascorbic acid, total chlorophyll, chlorophyll a, and chlorophyll b concentrations The highest total antioxidant activity (TAA: 4,789.373 mg TE g-1), total phenolic contents (TPC: 791.770 mg GAE 100–1 g-1) in red clover and highest total flavonoid content (672.177 mg QE 100 g-1) in maize were estimated (Altuner et al., 2022). Thus, it was concluded that total phenolic contents (TPC) was considerable good in red clover (46%) and maize (73%) as compared to cowpea and other studied legumes (Altuner et al., 2022). Although, biochemical parameters have not been correlated in legumes but pigment parameters were positively associated in legumes and cereals (Altuner, 2021). Further, positive correlation was found for total antioxidant activity, total phenolic content and total ascorbic acid in cereals (Altuner, 2021).
Variation in phytochemical and antioxidant profile and macronutrient composition of red and green butterhead lettuce cultivars (Lactuca sativa L. var. Capitata–green and red Salanova) has been reported with varying stages of development at harvest (El-Nakhel et al., 2020). Though microgreens of both cultivars were rich in calcium and magnesium, the red Salanova microgreens were concluded to be highly nutrient enriched. The nutritional profiles of Chicory and lettuce microgreens of the Asteraceae family and two genotypes of Brassica (broccoli) were compared. However, the Brassica microgreens were the richest source of phenols and vitamin E; the Asteraceae microgreens were rich in carotenoids and alpha-tocopherol (Paradiso et al., 2018).
These aforesaid studies suggest that microgreens with desirable nutritional contents can be obtained by exploring and manipulating the available genetic diversity.
7.2 Good agricultural practices (GAP) and optimized storage conditions for pre/post-harvest management to prolong the shelf life/quality of microgreens
Microgreens are preferred to be consumed afresh, either wholesome or as garnishes or seasonings and its cultivation at home can be an excellent practical approach concerning price and sustainability that also provides fresh life and functional food on the table for growing kids and families. Maintaining food safety standards with minimal or no microbial contamination that can cause potential health hazards without compromising the sensory qualities is of utmost importance.
Microbial contamination can cause spoilage of food and products, rendering them unfit for sale and consumption. This requires adopting hygienic and healthy practices throughout the cycle, beginning from cultivation until reaching the end user. Traditional methods like chilling, freezing, pasteurization and antimicrobial compounds (chemical or biological) compromise the sensory attributes (Tropea et al., 2021). Research is now focused on improving the quality and safety of food while maintaining its nutritional and organoleptic properties. One such technology is nisin-containing nano-carriers that can be applied safely as antimicrobial agents on food products (Bahrami et al., 2019).
Microgreens have a high respiration rate at the time of harvest that affects their shelf-life and storage (Chandra et al., 2012). Hence, application of 10 mM calcium chloride to microgreens prior to harvest is effective in delaying senescence, enhancing the visual appearance, and diminishing the growth of microorganisms during storage in broccoli microgreens (Kou et al., 2014). Further, Lu et al. (2018) studied the effect of applications of CaCl2 as pre-harvest and UV-B as post-harvest on levels of Glucosinolates (GLS) and glucoerucin (GLE) for assessing and enhancing the storage quality of microgreens. It was found that the treatments with 10 mM CaCl2 followed by UV-B enhanced GLS levels andcontent of total aliphatic glucosinolates in microgreens was four times as compared to mature counterparts. These microgreens had increased biomass, calcium content, and activities of antioxidant enzymes superoxide dismutase and peroxidase. Overall shelf life, productivity, and post-harvest of microgreens were improved.
The shelf life of microgreens is a very important concern that varies from 10–15 days after harvesting, depending on the category of microgreens. Microgreens are potential plant-based food/diet full of nutrition, fibers, and antioxidants, which diminish the risk of cardiovascular disease and numerous types of cancer. The microgreens may be packed in polypropylene bags and stored at 5°C in a climate chamber or incubator for 10 days with controlled temperature and humidity. However, the 1°C storage temperature was optimum due to no chilling injury (Xiao et al., 2014). A combination of pre-harvest and post-harvest treatments, different packaging materials, and modified atmosphere packaging (MAP) regulates the shelf-life of fresh-cut microgreens and diverse sensorial characteristics. Moreover, macro-perforated packaging, including PET clamshell and LDPE self-seal bags, was also assessed for longer shelf life in radish and roselle microgreens (Ghoora and Srividya, 2020).
After harvesting, packets of microgreens should be kept at a 4–5°C and consumed within 8–10 days. Storage conditions and maintenance of shelf-life are very important to preserve the microgreens in good quality with stable nutrition. Several factors, viz., storage temperature, atmospheric composition, post-harvest light exposure, and packaging technologies, are associated with conserving fresh-cut microgreens. Further, during value-added product development, processing avenues (freezing, drying, waving, microwaving, frying, toasting, and boiling)are equally required to maintain the bio-availability of bioactive and phytochemical components of microgreens.
Different technologies and methods have been explored to maintain microgreens’ shelf life and postharvest quality for preparing ready-to-eat products through wash steps and foliage spray. Aloe vera gel-based pre-harvest spray treatment and postharvest dip coatings were tested in radish and roselle microgreens for extended shelf life due to regulation of stomata closure (Ghoora and Srividya, 2020). These procedures regulate lower physiological weight loss, respiration rate, electrolyte leakage, microbial counts, and good overall acceptability. Further, researchers concluded that aloe vera gel-based-coated microgreens exhibit minimum deteriorative postharvest changes and higher ascorbic acid content than the uncoated control. Preharvest 10 mmolL−1 CaCl2 spray without postharvest dip displayed good yield, visual quality, and extended storage life (Kou et al., 2015). The optimum quality and highest shelf life of buckwheat microgreens can be maintained and stored at 5°C through moderately high O2 (14.0–16.5 kPa) and low CO2 (1.0–1.5 kPa) content with the treatment of chlorinated wash to reduce microbial counts (Kou et al., 2013). A very interesting plant regulator is 1-methyl cyclopropane (1-MCP) which binds competitively to ethylene receptors and delays senescence resulting in active treatment to prolong the shelf life of fruits, vegetables, and edible flowers (Turner et al., 2020). However, washing treatment is equally important in maintaining prolonged shelf life and the least microbial load. Comparative analysis of treated (chlorine wash) and controlled (unwashed) Ruby radish microgreens determines that 100 ppm chlorine wash enhances the visual quality and reduces electrolyte leakage (Turner et al., 2020). In addition, 0.25%–0.50% citric acid wash followed by 50% ethanol spray and 0.25% ascorbic acid is also effective in augmenting quality score. Further, to enhance the shelf life, a potential application of “nano packaging” technology concerning microgreens can also be explored for effective postharvest management. Thus, in coherence with the farm-to-fork tradition, good agricultural practices and handling practices are crucial.
7.3 Fortification through agronomic approaches, nano-technology and seed priming for enhancing preferred qualities of microgreens
Microgreens are considered potential nutrient sources that can help overcome the deficiency of many nutrients which are not met up with the seeds or mature parts of the plant. Effective fortification strategies for producing microgreens with desired nutritional traits and shelf life can be effective tools. Insufficient availability of iron and zinc in the human diet has posed a risk of malnutrition in young children and women. To address the deficiency of iron and zinc micronutrients, microgreen produce fortification can serve as an effective but short-term approach. Fortification can be done through several approaches such as agronomic practices, application of nanotechnology, etc.
Agronomic practices are cheap and simple but non-heritable and must be done with great care due to the application method, kind and environmental considerations. This strategy emphasizes improved nutrient accessibility to plants, efficient usage of nutrients, plant mobility, and increased microbial activity. Microbes like Bacillus, Rhizobium, Azotobacter, Actinomycete, and some fungal strains, i.e., Pseudomonas indica, are used to increase nutrient availability and their uptake. Mineral nutrients show great potential for fortification when applied to the soil and the leaves. The most popular fertilizer is based on nitrogen, phosphorus, and potassium (NPK), which is vital for the health of both plants and mankind. Crops also require other micro-minerals such as iodine, zinc, copper, iron, nickel, molybdenum, manganese, etc.
In a recent study, the fortification of Brassicaceae microgreens was attempted for iron and zinc enrichment (Di Gioia et al., 2019). It involved growing red cabbage, red mustard, and arugula microgreens in nutrient solution supplemented with sulfate salts of iron and zinc at 0, 10, 20, and 40 mg L−1 and 0, 5, 10, and 20 mg L−1 concentrations, respectively. Further, investigations on the growth, yield, and mineral composition of these microgreens grown in these media composition exhibited accumulation of both iron and zinc minerals in microgreens of all three Brassicaceae members in a genotype-specific manner. Thus, this study also indicated that soil-less cultivation systems could be exploited for the production of fortification of microgreens by altering the composition of the nutrient medium. Pannico et al. (2020) have fortified selenium in green basil, purple basil, coriander, and tatsoi microgreens through a modified quarter-strength Hoagland nutrient solution with sodium selenite compound.
Nanofortification is the approach to fortify the plants or microgreens through nanoparticle application of some essential nutrients (Cu, Se, Fe, and Zn) in the form of liquid treatment as foliar and nano-fertilizers in soil or water medium (El-Ramady et al., 2021). Nanoparticles (NPs) are small materials ranging from 1 to 100 nm in size or dimension (Laurent et al., 2008), with a large surface area that allows its application in diverse fields, including fortification in plant systems. Their size and large surface area: volume ratio also contributes to their physical and chemical properties. Due to their size, the optical properties of these particles impart unique characteristic colours. Their size, property, and shape are categorized into different groups: metal NPs, ceramic NPs, polymeric NPs, and fullerenes. They find applications in several fields, including environmental engineering, biotechnology, textiles, food processing/packaging, cosmetics, plant sciences, and agriculture. Several methods are employed for the synthesis and detection of nanoparticles. The techniques used for synthesis include chemical synthesis, thermal decomposition, photo-reduction, and green synthesis. Further, characterization of synthesized NPs may be performed using UV-Vis spectroscopy, X-ray diffraction assay, Scanning electron microscopy (SEM), transmission electron microscopy (TEM), and energy dispersive analysis. The conventional chemical methods of NP synthesis are costly, and toxic chemicals used for synthesis also pose a hazard to the environment. These pave the way for the need to synthesize NPs from biological methods using plants, microorganisms, and enzymes. These methods are not only cost-effective and rapid but are also environment-friendly and safe. Several metal NPs have been synthesized using the green synthesis approach, like silver NPs, gold NPs, copper, zinc oxide, and iron oxide (Bibi et al., 2022; Eltaweil et al., 2022; Nguyen et al., 2022). These may be utilized for seed priming to improve germination, seedling growth, and nutrition. Several plant parts like roots, leaves, flowers, fruits, and stems have been used to synthesize NPs via green methods. For example, zinc oxide nanoparticles are stable oxides of metal, eco-friendly, and have no harmful effects on humans and animals. These are most interesting to researchers due to their magnetic, optical, thermal, and chemical properties. ZnO NPs also exhibit adsorption ability which increases the catalytic efficiency. Nanoparticles have been applied extensively in agriculture research and may be used as nano-fertilizers to promote plant growth.
Seed priming is an innovative and user-friendly approach to fortify seeds by treating with an appropriate number of desirable nanoparticles. Sundaria et al. (2019) investigated the effect of iron oxide nanoparticles (25–600 ppm) on wheat genotypes (WL711 and IITR2). They observed increased germination percentage, shoot length, growth parameters, and accumulation of grain iron in WL711 and IITR2 at 200ppm and 400ppm, respectively. In addition, cold plasma (CP) treatment is a pollution-free way to improve seed germination, water use efficiency, nutrient uptake, photo- and thermo-dormancy, and plant yield (Mahanta et al., 2022). Moreover, CP treatment would be a potential approach to improve microgreens performance as it plays a critical role in numerous physiological, biological, and developmental processes in plants that improve seed performance, bacterial load on seeds, altering seed coat structures, enhance seedlings growth and its association with machine learning is a sustainable approach for seed priming (Shelar et al., 2022).
7.4 OMICS and breeding approaches for microgreen biofortification
Microgreens contain various favourable attributes like a pleasing palette of colours, quality, textures, and flavours (Aroma volatiles associated with flavour) but limit their commercial use due to short shelf life. NASA scientists have also explored microgreens in space due to dynamic properties like the availability of oxygen generation, nitrogen, essential nutrients, and photoactive compounds to enhance the morale of astronauts during stretched stays away from Earth (Kyriacou et al., 2017). Several breeding approaches and multiOmics have been explored to augment the shelf life, developmental rate and nutrient content of vegetables and fruits (Kalia and Singh, 2018; Mathiazhagan et al., 2021; Reda et al., 2021; Valdes et al., 2021; AieseCigliano et al., 2022; Chakravorty et al., 2022; Dutta et al., 2022; Kumari et al., 2022; Parmar et al., 2022; Sahoo et al., 2022; Sharma et al., 2022). However, it has been observed in tomatoes that the dominance component was higher than the additive component for shelf life (Pavan and Gangaprasad, 2022). To maintain the shelf life with natural colour and flavour, an antisense gene was introduced in tomatoes by a Californian company Calgene in the 1980s and developed improved shelf-life tomatoes, i.e., popularly known as FlavrSavr tomatoes (Bruening and Lyons, 2000). In addition to that natural variant of cucumber fruit (DC-48:high shelf life) was also explored through qRT-PCR for fresh green colour and shelf life and found that Expansin (EXP), Polygalacturunase (PG), and xyloglucan endotransglucosylase linked with cell wall degradation process and regulates to maintain the fruit firmness (Pradeepkumara et al., 2022). Several such efforts are underway in several vegetable and fruit crops.
The term “biofortification” refers to the process of enhancing the nutritional value of a plant’s edible parts. It provides a long-term and sustainable alternative for supplying people with micronutrient-rich crops (Garg et al., 2018). Biofortification offers an effective, economically feasible, and sustainable means of enhancing nutritional content in crops that contribute to staple diets. This strategy typically involves interventions for improving the nutritional content, including vitamins, essential amino acids, minerals, and fatty acids, while simultaneously reducing anti-nutritive factors that hinder the bioavailability of nutrients in crop plants (Garcia-Casal et al., 2017). It has the potential to overcome malnutrition prevalent as ‘hidden hunger. In the current climate change scenario, where crop yield and nutritional quality are adversely affected, biofortification can be a successful and game-changing strategy for overcoming nutrient deficiencies, especially in developing nations. Biofortification can be done by understanding the genes, pathways and regulatory networks responsible for absorbing and transporting nutrients and adopting genetic means that involve using the natural germplasm during conventional breeding, genetic engineering (through genetic engineering or production of transgenics) and other OMICS approaches. The significance of multiomics, nutriomics and foodomics have been explained for development of desirable genotypes with potential microgreen related traits (shelf life and nutrient content) those will be also helpful for improvement of breeding cycles (Kalia and Singh, 2018; Mathiazhagan et al., 2021; Reda et al., 2021; Valdes et al., 2021; AieseCigliano et al., 2022; Bansal et al., 2022a; Chakravorty et al., 2022; Dutta et al., 2022; Kumari et al., 2022; Parmar et al., 2022; Sahoo et al., 2022; Sharma et al., 2022).
Conventional breeding depends on the genetic diversity of the gene pool for the trait of interest (TOI). The desired genes are pyramided using traditional crossing approaches, and then segregation populations are thoroughly screened. Biofortification via genetic means offers a cost-effective and relatively efficient strategy with the pre-requisite of the availability of inbred lines with high nutrient content in several crops. For millions of under privileged rural residents, sorghum is one of the most essential basic foods. It can flourish in challenging conditions. Moreover, HT12 protein increased the lysine content in sorghum (Zhao et al., 2003; Lipkie et al., 2013). The fact that sorghum is less easily digested than other main staple crops is one of the problems with eating it. Its kafirin seed storage protein is immune to protease digestion. The RNA silencing of kafirin increases the digestibility index of sorghum in combination with the suppression of kafirin-1, kafirin-2, and kafirin A1 genes (Grootboom et al., 2014; Elkonin et al., 2016). Accordingly, in soybean the globally preferred crop due to its vegetable oil and high-quality protein, the expression of the bacterial PSY gene (beta-carotene) increases the level of provitamin-A, oleic acid, and other protein contents of seed (Schmidt et al., 2015). The fruit color and freshness in strawberry were regulated by specific anthocyanin, anthocyanin related transcription factors and biosynthesis-associated gene expression (Lee et al., 2022). Thus, utilization of such nutrition-enriched developed varieties may be explored for microgreens production.
New breeding approaches, including transgenic breeding, RNA interference (RNAi), and genome editing etc. are crucial for the biofortification of crops because they provide new opportunities for developing unique genetic varieties and are being discussed under following sub heads.
7.5 Genomics and transcriptomics for microgreen traits
A number of DNA-based molecular markers viz; SSR (Simple sequence repeats),microRNA-based SSR, AFLP (Amplified fragment length polymorphism), SNP (Single Nucleotide Polymorphisms), etc. are available for diversity analysis and identification of most potential genotypes for desirable traits in different plants (Gupta et al., 2013; Maurya et al., 2015; Liu et al., 2019; Gupta et al., 2020a; Pradeepkumara et al., 2022) that would be useful to detect most potential genotypes based on phylogeny and genetics studies for particular genotype-specific microgreens. Nutritional profiling of the most diverse genotypes (range: 300–1,000 genotypes) for microgreen-related traits may be performed to detect major phytonutrient components. Evaluation of the most promising genotypes of potential plants (legumes, cereals, herbs, and vegetables) would be helpful for microgreen production based on performance for microgreen-related traits (aroma, tender texture, vivid colour, flavour, and rapid production)at different locations (high/low altitude). Genome-wide association mapping and quantitative trait loci (QTL) mapping will be helpful in detecting potential genotypes and for candidate gene identification related to microgreen related desirable traitsformolecular breeding programs.
Specific QTLs may be identified for desirable microgreens related traits (shelf life and nutrients content: Fe, Zn) in particular crop on the basis of contrasting parents and corresponding data of Genomics. The Quantitative Trait Loci have been identified for phytoactive compounds, iron, zinc and shelf-life related microgreen traits incabbage (Wu et al., 2008), broccoli (Gardner et al., 2016), wheat (Krishnappa et al., 2022), lettuce (Hayashi et al., 2012), melon (Dai et al., 2022)and chickpea (Mahto et al., 2022).
Accordingly, transcriptomics based specific mRNA expression quantitative trait loci (eQTLs)and splicing quantitative trait loci (sQTL) may also be identified for desirable microgreens related traits (shelf life, Fe, Zn) in particular crop on the basis of contrasting parents, standard population size and corresponding nutrition data (Agarwal et al., 2014; Zhu et al., 2022) as identified for several complex traits (Khokhar et al., 2019; Qi et al., 2022). Based on the desirable trait evaluation, a set of candidate genotypes may be selected for transcriptomics study to identify the differentially expressed genes (DEGs) for nutrition and related traits (Figure 2). The candidate genotypes should differ for traits, including bioactive compounds, phytochemicals, antioxidant capacities, mineral composition, yield, and biomass-related traits. The transcriptomic analysis will help to identify the differentially expressed transcripts, biological processes, and molecular pathways for all the contrasting traits, including nutritional and shelf-life-related traits. Once identified, the differential transcripts/genes will be converted into user-friendly markersto be utilized through breeding approaches for future microgreens production.
7.6 Proteomics and metabolomics for microgreen traits
Proteomics and metabolomics have been explored in several microgreens viz; broccoli (Sun et al., 2015), brassica (Castellaneta et al., 2022) and other leafy vegetables also (Sahoo et al., 2022). Novel datasets for microgreens may be generated through research activities, and prospects of encroachment in OMICS approaches. The improvements will consequence through the combined relationship of proteomics and metabolomics for nutritionally rich microgreen development. Protein specific quantitative trait loci (pQTLs), metabolic quantitative trait loci (mQTLs) and micronutrient quantitative trait loci (nutriQTL) play dynamic role in Physiological processes and molecular pathways that may be further identified for desirable microgreens related traits (shelf life, Fe, Zn) as investigated earlier (Engelken et al., 2016; Zhou et al., 2021). Furthermore, proteomics and metabolomics approaches will be a major advancement for microgreen improvement in terms of proteins, metabolites, and bioactive compounds. They will be helpful from plant breeding to OMICS-assisted plant molecular breeding (Langridge and Fleury, 2011).
Variations in the quality and quantity of microgreen proteins and metabolites can be investigated through analysis of proteome composition and changes to developmental stages, including stress-response mechanisms for the enhancement of proteome coverage data and further improvement of protein quality and shelf-life. The establishment of novel approaches related to proteomic and metabolites pipelines would be useful for data analysis associated with different kinds of growth and stress conditions for microgreen-related traits as already explored in several crops through proteome mapping, comparative analysis of proteomics, post-translational modifications, and protein-protein interaction networks, 2D gel electrophoresis coupled with MALDI-TOF (Vanderschuren et al., 2013; Katam et al., 2015). These approaches would be helpful for the purposeful annotation of desirable proteins that participate in metabolism (nitrogen, amino acid, carbon and energy, and Reactive Oxygen Species), stress response, secondary metabolism, and signal transduction (Gupta et al., 2019) that will be further regulated and improve nutrition quality and extended shelf-life.
Good accuracy, speed improvements, sensitivity perfections in mass spectrometry (MS) applications, and software tool improvements have all benefitted high-throughput protein quantification that will be further useful for comparative analysis of proteomics profile in association with differential expression analysis as explored related to stress responses in legume crops (Pandey et al., 2008; Abdallah et al., 2012; Hu et al., 2015).
Metabolite profiling provides the appropriate data and depth information on metabolic networks responsible for a diverse range of desirable phenotypic traits and undesirable traits that can be regulated through plant metabolic engineering (Fernie and Schauer, 2009). The literature highlights two important nuclear magnetic resonance (NMR) and mass spectrometry (MS)-based metabolomics profiling techniques. It was usually necessary to combine several analytical methods to extract a wider variety of multiple plant metabolites from a single MS (Arbona et al., 2013). Additional methods include Fourier Transform Infrared spectroscopy and MS (FIA/MS), and flow injection-based analysis.
Integrating metabolomics, transcriptomics, bioinformatics platforms, and phenomics to evaluate genetically diverse individuals and improve gene identification accuracy enables the detection of unique metabolic QTLs and candidate genes for the targeted trait that will be cooperative for microgreens improvements. Moreover, a combination of metabolomics screening and genomic-assisted selection strategy has been identified to increase yields, reducing the time spent discovering novel traits and allelic mutations (Fernie and Schauer, 2009).
7.7 Pan genomics for microgreen traits
A species’ pan-genome refers to all of its genes collectively. Due to the variety in genomic sequences, it has been determined that a single organism cannot have all of a species’ genes. Completeness (i.e., the presence of all functioning genes), stability (i.e., the presence of distinctive catechistic properties), comprehensibility (i.e., the presence of all genomic data for all species or individuals), and effectiveness are the desired characteristics of an ideal pan-genome (i.e., organized data structure). Recently, a 592.58 Mb chickpea pangenome with 29,870 genes was created (Varshney et al., 2021). In order to create the pan-genome, 3,366 accessions totalling 3,171 farmed and 195 wild ones were used in whole genome sequencing. This comprehensive genome analysis provided important details on the genomic regions frequently chosen during domestication, the best haplotypes, and the locations of harmful allele targets. The newly discovered genes that encode reactions to oxidative stress, stimuli, heat shock proteins, cellular (acidic pH) and cold responses may help to modify microgreen cultivation.
7.8 Transgenic approaches for microgreen traits
In order to introduce tolerance or resistance to diverse abiotic and biotic problems, genetic manipulation has been extensively used to identify and transfer resistant gene(s) from a variety of resources to desirable plants for a targeted trait. Today, different genes are used in plants, and transgenic plants have been created through Agrobacterium-mediated transformation (Sharma et al., 2006), electroporation of intact axillary buds (Chowrira et al., 1996), particle gun bombardment (Indurker et al., 2007). Agrobacterium-mediated explant transformation is the most frequently employed technique to create transgenic pulse crops by inserting transgenes from diverse sources to produce transgenic plants.
Various transgenic plants have been developed for several desired traits. Further, several genes have also been identified for insect pest (protease inhibitor genes, α-amylase inhibitor genes, lectin genes, Cry genes from Bacillus thuringiensis, chitinase gene) and disease resistance for example fungal (antifungal protein genes, stilbene synthase gene), viral (coat protein genes of viruses) and bacterial (T4 lysozyme gene) (Dita et al., 2006; Eapen, 2008). The impact of endogenous genes could be regulated by modifying biological processes and metabolic pathways to boost carotenoids and flavanoids using various abiotic stimuli such as drought, salinity, mineral toxins, cold, temperature and RNA interference technologies (Eapen, 2008). Interestingly, desirable multi-trait transgenic plant can be developed through in vitro gene stacking system: GuanNan Stacking (Qin et al., 2022). However, transgenic rice have been developed by construction of binary vector and insertion of five desirable foreign genes that would be helpful for regulation of metabolic engineering and trait improvements through breeding and multiomics approaches in future (Qin et al., 2022). Moreover, improvement of chickpea and pigeonpea have been explored through transgenic and molecular approaches (Arya and Mishra 2022).
7.9 Genome editing for microgreen traits
In plant genome editing, sequence-specific nucleases modify specific genes in the selected crop to construct transgene free plants. Moreover, different sequence-specific nucleases including ZFNs, TALENs, and the CRISPR-Cas9 systems have been explored to alter the genome of the targeted plant, fruits and vegetables (Upadhyaya et al., 2009; Mathiazhagan et al., 2021; Chakravorty et al., 2022; Kumari et al., 2022). CRISPR genome editing uses RNA-guided DNA endonucleases (Cas9/13), but these complexes form at the specific target site to execute targeted gene editing (Ducreux et al., 2004; Dancs et al., 2008). Enhancing shelf life and plant storage features by genome editing would be a valuable venture (Kuzmina, 2020). Although, biofortification for cytokinin using gene editing for improving nutrition in chickpeas has recently been projected (Mahto et al., 2022).
7.10 Sequencing-based approaches for microgreen traits
With the advances in the NGS based technologies, trait mapping has become an easy job to do. Not only are these technologies time saving but also reduces the cost at basal levels. The genetic mapping is based on recombination (the exchange of DNA sequence between sister chromatids during meiosis) and the centimorgan (cM) distance measured between the markers by representing approximately 1% of the recombination frequency, while the physical map is based on the alignment of the DNA sequences with distance between markers measured in base pairs. However, the high-resolution physical maps serve as the scaffold for genome sequence assembly to identify the most accurate distance between the markers and the genes linked in addition to exploring of the potential candidate gene(s) linked to desired traits. The trait mapping through sequencing approaches may be categorized into two classes i) Sequencing of complete populations for trait mapping and ii) Sequencing of pooled samples for trait mapping (Singh et al., 2022). Researchers have great interest in its genomic properties, which provides a valuable marker for crop improvement. Wu et al. (2020) identified multiple high-quality SNPs that would serve as an important resource for the mungbean’s nutritional improvement and cultivation. Due to the advancement in sequencing techniques, Dasgupta et al. (2021) conducted the bulk-RNASeq-based gene expression analysis across mungbean genotypes to identify disease-resistance genes. Bhardwaj et al. (2015) did an RNAseq-based analysis to identify drought stress-regulated genes in Brassica juncea. Guo et al. (2021) emphasized the genomic-based structural variant that indicated the diversification of different morphotypes of Brassica oleracea. These approaches are equally applicable and may be explored for improving microgreen traits (Mishra et al., 2021).
7.12 Epigenomics for microgreen traits
The increasing world population and changing climate increase the demand for greater crop productivity. The selection of appropriate genetic techniques and desirable heritable DNA sequences have led to notable genetic advancements in many crop species. Specific methylation quantitative trait loci (meQTLs) may be identified for desirable microgreens related traits (shelf life, Fe, Zn) (Kumar et al., 2016; Kumar et al., 2022b). Further, correlation between transcriptome (eQTL) and methylome (meQTLs) have been established for genetic regulation of complex traits (Oliva et al., 2022). In addition, a better comprehension of and capacity to choose advantageous epigenomic modifications is suggested to incorporate a more effective and comprehensive approach to crop improvement (AieseCigliano et al., 2022; Chandana et al., 2022). This is because many plant stress responses are governed by epigenomic processes, notably through cell-autonomous epigenetic switching. This makes it possible to register and remember random genetic signals. According to a report, the memory-directed alteration may result in an increased ability to resist stress in the future (Berr et al., 2011).
The mechanisms governing plant-stress interactions and conditions are revealed by studying the roles of epigenetics causing stressors, such as histone changes and DNA methylations (Chinnusamy and Zhu, 2009). Numerous heritable modifications originating from mitotic and meiotic divisions (variations in the heredity of epigenetic markers) were observed during gene expression studies and are steadily transmitted from one generation to the next that were not encoded in the DNA sequence itself (Tsaftaris and Polidors, 2000; Berger et al., 2009; Chen et al., 2010).
Stout epigenetic alterations are mitotically transmitted through genomic imprinting, but transient epigenetic alterations are not heritable (Spillane et al., 2001). Until they are lost or removed, epigenetic alterations produced during meiosis are always transmissible from one generation to the next without the need for initial stimulation. The loss could be from a genetic mutation, unintentional (for unknown reasons), or result from environmental factors. These are distinct from the ones that brought about the original epigenetic changes. In plants, heterosis is exhibited in hybrids for high biomass (Gupta et al., 2020b), a straightforward epigenetic assumption.
7.13 Genomics to artificial intelligence for microgreen traits
Machine learning (ML) and Deep Learning (DL) is the component of artificial intelligence (AI) that includes mathematical models of data to improve plant performance based on statistics, predictive modeling, and data analysis. It may be considered artificial intelligence based on plant breeding or crop improvement. Precision agriculture and crop improvement through artificial intelligence are new for plants and are untouched areas for microgreens. Prediction of shelf-life is also possible through exact measurement of plant characters (accurate interpretation of high-throughput phenotypic data) by good quality imaging techniques (greater than 7,000 high-resolution images of 300 GBytes) and proficient analysis of refined extracted data using artificial intelligence.
Microgreens-specific genomics and machine learning-based innovative approaches may be employed in other crop plants that have been proposed worldwide. Genomics approaches help to identify the “SNP” and “SSR '' markers and annotate the genes. Furthermore, a detailed analysis of these markers could help identify the nearest genes (QTLs), as shown in Figure 1. Another important genomic layer of information is “Bulk-RNASeq,” which can be implemented for accurate data analysis and prediction. ML aims at providing innovative approaches for prediction-based model development; Girma (2019) discussed the ML approach in mungbean to classify the raw quality of samples by analyzing digital imaging data; Jung et al. (2021) applied the deep learning algorithm to correctly identify the Brassica napa varieties, followed by a cross-validation approach. Another important microgreen vegetable crop is Broccoli (Brassica oleracea L. var. italica). The importance lies in the broccoli head portion, which helps to assess plant quality and different biotic and abiotic stress. Zhou et al. (2020) used the “Improved ResNet” to extract the broccoli pixels from the background data. In another study based on broccoli head estimation, Kusumam et al. (2017) applied the deep learning approach for 3D-vision-based detection. DL based semantic segmentation models have been applied in another microgreen plant cabbage for crop estimation (Jo et al., 2021). One of the studies by Wolanin et al. (2020) used the DL method for the time series data to estimate the wheat yield in the Indian wheat belt. AI has been applied to the barley seeds too. Various studies have been published where machine learning has been used for the identification of lentil-based rust disease identification (Singh et al., 2019). Applying machine learning models and techniques predicted the shelf life of Okra (Iorliam et al., 2021) and muskmelons (Albert-Weiss and Osman, 2022). Thus, active learning would be helpful to predict the shelf life of different types of microgreens through Support Vector Machine, Logistic Regression K-Nearest Neighbour algorithms,Naïve Bayes and Decision Tree.
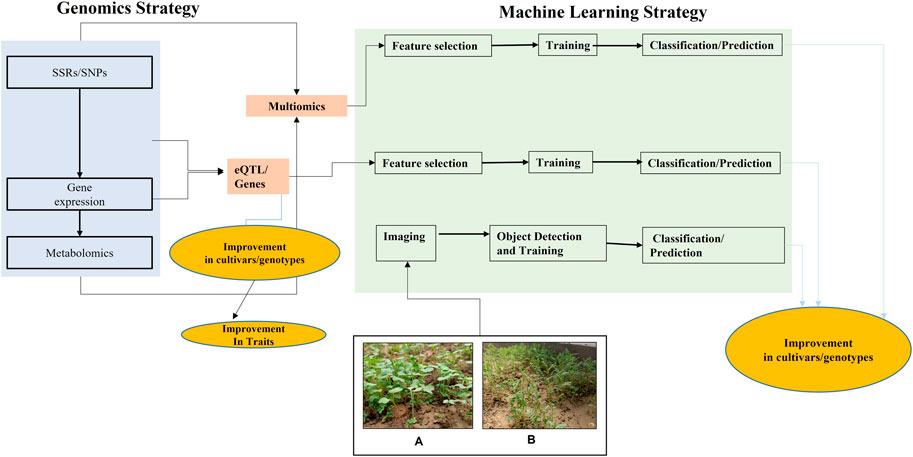
FIGURE 1. Systematic workflow of OMICS analysis and its association with machine learning for microgreens improvement and shelf-life prediction: Genomic data could be used to explore the markers in the form of SSRs, SNPs. Bulk-RNASeq and metabolomics data provides another layer of information that would enable Multi-OMICS analysis for cultivation improvement. Machine learning strategies that make the use of both “numeric” and “imaging” data would enable the development of prediction and classification models.
7.14 Bioinformatics and molecular databases for microgreen traits
The plant research group requires efficient bioinformatics pipelines and a system to support efforts to analyse microgreen-related targeted plant genomes through functional genomics due to the rapid progress of publicly available databases from different kinds of tissues, development, environments, and stress treatment. The comprehensive model plant genomics, transcriptomics, and proteomics databases can be used to identify appropriate microgreen genotypes. The genome sequences of several plants (Medicago truncatula, Glycine max, and Lotus japonicus) and reference plant species (Arabidopsis thaliana and Populus trichocarpa) are already available to investigate gene function, biological processes, metabolic pathways, and genome evolution (Li et al., 2012). The available data bases viz., The Legume Information System (LIS; https://legumeinfo.org) and KnowPulse (https://knowpulse.usask.ca)are very informative and useful computational genomics platforms to evaluate molecular markers, diversity analysis, comparative genomics, gene annotation, novel transcription factors, sequence variants, phenotypic traits informationand to map SNPs, QTLs, long non-coding RNAs and to identify candidate genesfor selection of microgreen oriented suitable chickpea, faba bean, common bean, lentil, andfield pea germplasm (Doddamani et al., 2015; Verma et al., 2015; Dash et al., 2016; Gayaliet al., 2016; Sanderson et al., 2019; Lee et al., 2022).
8 Integrating various OMICS approaches for microgreen traits
Prospective OMICS approaches have been investigated in many plants to improve the desirable traits and elucidated earlier (AieseCigliano et al., 2022; Chakravorty et al., 2022; Dutta et al., 2022; Gupta, 2022; Gupta and Tyagi, 2022; Parmar et al., 2022; Sharma and Gupta, 2022; Singh et al., 2022). Re-sequencing activities of whole-genome employing genetic diversity, domestication patterns, evolutionary analysis, population structure, and linkage disequilibrium for chickpea improvement as a result of the technical advancements that upgraded chickpea (an orphan crop) to a potential geneticcrop (Varshney et al., 2019).
Recent genomics methods offer the potential to accelerate gene discovery, marker creation, molecular breeding, trait mapping, and productivity advances in microgreens, among other processes (Figure 2). Integration of precise phenotypic variation, low-frequency variants, and sequence information approach would be helpful for the selection of the most appropriate accessions with desirable key traits like biomass components, biotic and abiotic stress tolerance, and nutritional traits (Roorkiwal et al., 2020). A broad range of molecular markers (SSR, SNP, and DArT) have been discovered in chickpea that has been facilitated by NGS technology (Whole-genome re-sequencing, genotyping by sequencing, skim sequencing, RAD-Seq, and lower-depth sequencing). They can also be used for the development of chickpea microgreens (Kale et al., 2015; Varshney, 2016; Varshney et al., 2018).
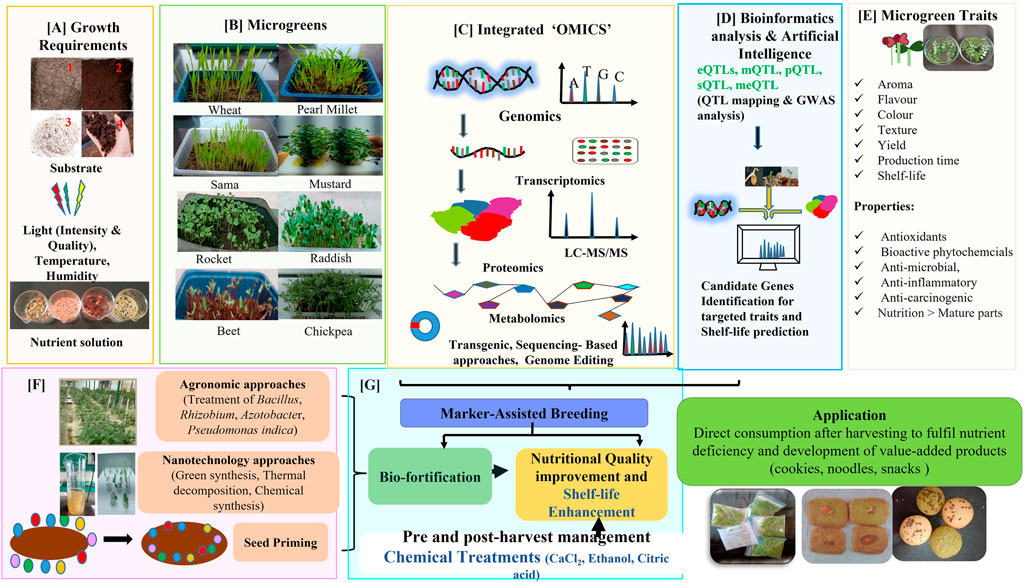
FIGURE 2. Summarizes prospects of microgreens as budding live functional food. (A) Growth conditions required for microgreens cultivation which includes a variety of substrates like vermiculite (1), cocopeat (2), perlite (3) and vermicompost (4), light (quality, intensity and duration), temperature, humidity and nutrient solution. (B) Microgreens of different plant species: Wheat, Pearl millet, Sama, Mustard, Rocket, Raddish, Beet and Chickpea. (C) Numerous “OMICS’ approaches such as Genomics, Transcriptomics, Proteomics, Metabolomics, Epigenomics, along with Transgenics, Gene editing and Sequencing based approaches can be integrated with bioinformatics tools and artificial intelligence (D) to tag Quantitative Trait Loci (eQTLs: mRNA expression Quantitative Trait Loci; meQTLs: Methylation Quantitative Trait Loci; PQTL: Protein Quantitative Trait Loci; sQTL: splicing Quantitative Trait Loci; mQTLs: Metabolic Quantitative Trait Loci) and candidate genes identification for microgreen related desirable traits (E) like nutrients, flavour, colour, early germination, yield and shelf life. The data generated from Integrated “OMICS’ approaches can be further utilized in molecular breeding to produce nutritionally rich varieties with improved shelf life through Marker-Assisted Breeding and producing biofortified microgreens with targeted micro/macro nutrients (like iron, zinc, magnesium, calcium) enrichments. (F) Biofortification of target microgreen is also possible by agronomic approaches (incubation with microorganisms like Bacillus, Rhizobium, Azotobacter, Pseudomonas indica), nanotechnology (nano-biofortification) and seed priming. Bioavailability of nutrients and minerals of microgreens can be stabilized through pre and post-harvest management strategy (G). Improved microgreens with desired nutrients can be either consumed fresh as garnishes in soups, sandwiches, salads or processed to develop value-added products (like noodles, breads, drinks, cookies etc.) to overcome nutrients deficiency.
There is a huge gap from genome to phenome in agricultural plants to identify the particular phenotype based on their DNA sequence information and genetics. Thus, it is crucial to integrate multi-OMICS information in one place from several branches of OMICs platforms, including phenomics, genomics transcriptomics, proteomics, epigenomics, and metabolomics. Using all the OMICS technology, the genotype-phenotype divide in any microgreens can be closed with precision phenotyping.
To learn new things about the potential genes and biological processes involved, analysis at genomics, transcriptomics, proteomics, epigenomics, and metabolomics levels can be done depending on the study’s goal. Moreover, it has been reported by Mannur et al. (2019) that Fusarium wilt resistance loci (foc 4) from WR 315 Annigeri 1 has been made available as “Super Annigeri one″ for commercial production in India using a genomics strategy. Thus, it is evident that various other studies have also proposed OMICS-based integration methods (Argelaguet et al., 2018; Bhardwaj and Steen, 2020; Mahto et al., 2022). Cai et al., 2021 discussed applicability of OMICS data (transcriptome and proteomics) for barley and identified the connection between sugar metabolism and wild barley. Li et al. (2019) used a single omics layer of information, i.e., transcriptome, and revealed the gene expression patterns of sulforaphane metabolism in Broccoli florets. For microgreens, this field is still lagging (Figure 1). There is a need to exploit the information from different resources to identify various biological factors that would help microgreen plants’ cultivation and breeding processes.
Candidate genes have been identified and incorporated in desirable plant through Genomics (QTL mapping), transcriptomics and transgenic approaches for microgreens related traits (shelf life and nutrient content: Fe, Zn) in different plant systems viz cabbage (Wu et al., 2008), broccoli (Gardner et al., 2016), wheat (Krishnappa et al., 2022), lettuce (Hayashi et al., 2012), melon (Dai et al., 2022) and chickpea (Mahto et al., 2022). Metabolomics and proteomics have been explored in several microgreens viz., broccoli (Sun et al., 2015), brassica (Castellaneta et al., 2022) and other leafy vegetables also (Sahoo et al., 2022). Thus, it is evident that we may identify and predict candidate genes associated with microgreens related traits (shelf life, desirable nutrients content, developmental rate and phytochemicals) may be incorporated in targeted crop varieties of fruits and vegetables utilizing the crop specific data bases of genomics, transcriptomics and metabolomics, marker-assisted selection, GWAS, bioinformatics, AI approaches utilizing the databases of specific crop transgenic CRISPR/Cas9 and gene editing approaches (Mathiazhagan et al., 2021; Valdes et al., 2021; Chakravorty et al., 2022; Parmar et al., 2022; Sahoo et al., 2022). Thus combination of multiomics, foodomics in association with nutriomics would be fruitful for regulation of nutritional balance, health management and treatment of diseases (Bansal et al., 2022a).
9 Conclusion and future perspective
Optimization of light, substrate, and temperature would be helpful for good quality microgreen cultivation containing desirable aroma traits, tender texture, vivid colour, flavour, sensory attributes, and rapid production. Further evaluation of genotypes would be helpful for the selection of the most potential genotypes for the mass production of microgreens. Genomics and transcriptomics approaches may be explored for candidate gene identification for microgreens and important nutritional traits. Explored genes or associated SNPs may be developed and explored for user-friendly markers for marker-assisted selection and metabolic pathways with the integration of multi-OMICS approaches. Characterization of microgreens can be performed by combining the most standardized condition for the growth of microgreens with enhanced nutrient quality and bioavailability. Profiling of phytochemicals, nutrients, and minerals should be studied for good quality microgreens and biofortify further using traditional and novel biofortification approaches. For commercialization and popularization of the microgreen’s cultivation and harvest, shelf-life should be focused. The development of post-harvest technology for enhanced storability of microgreens is very important for synthesizing value-added, tasty, and nutritional products.
Due to the nutritional content inclusion of microgreens in the diet have several health benefits, as evident from the literature. However, microgreens’ growth, yield, and nutritional content can vary with the growing method (soil, compost, or hydroponic), intensity and quality of illumination, and composition of plant nutrient solution. Establishing hydroponics and vertical farming systems will enhance the cultivation of nutritionally rich microgreens with an easy harvest. The development of technologies to preserve microgreens is needed for a more extended period with minimal changes in their phytochemicals and nutrients. Novel value-added products (for example, drink, juices, cookies, noodles, etc.) may be developed with the integration of microgreens as one of the ingredients having wider acceptability and enhanced nutrition, especially for elderly persons, infants, young growing children, and sick persons due to excellent digestibility. Microgreens cultivation is a very easy and promising strategy to initiate at home or as a start-up for beginners/poor farmers in their respective localities, including people with malnutrition in India. The persons engaged in microgreens cultivation, production, and utilization may explore the food processing and packaging industry to enhance the market regarding agriculture and nutritionally rich products. Thus, microgreens will occupy a central place in the future food industry.\
Author contributions
AG and RK conceptualized and supervised the manuscript writing. TS, DS, AB and SS collected the related literature and contributed to the original writing. RK extended their help in inference, review and AG did editing of the manuscript. All authors went through the final manuscript draft and approved it.
Funding
The study was supported by School of Agricultural Sciences, Sharda University, and ICAR-Indian Agricultural Research Institute, New Delhi.
Acknowledgments
The authors gratefully acknowledge the Sharda School of Agricultural Sciences, Sharda University, Greater Noida, Uttar Pradesh, India; ICAR-Indian Agricultural Research Institute, New Delhi, India; Integral Institute of Agricultural Sharda School of Agricultural Sciences, Sharda University, Greater Noida Uttar Pradesh, India; Institute of Clinical Molecular Biology (IKMB), Kiel, Germany; Dayanand Anglo-Vedic (PG) College, Chhatrapati Shahu Ji Maharaj University, Kanpur, Uttar Pradesh, India. All authors are grateful to Dolly Wattal Dhar, Dean, Sharda School of Agricultural Sciences, Sharda University, Greater Noida, Uttar Pradesh, India for professional editing. The lead author RK gratefully acknowledges the DST-Science and Engineering Research Board for providing the financial support to carry out the manuscript preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdallah, C., Dumas-Gaudot, E., Renaut, J., and Sergeant, K. (2012). Gel-based and gel-free quantitative proteomics approaches at a glance. Int. J. Plant Genomics 2012, 1–17. doi:10.1155/2012/494572
Agarwal, S., Vgn, T. V., Kotla, A., Mangrauthia, S. K., and Neelamraju, S. (2014). Expression patterns of QTL based and other candidate genes in Madhukar× Swarna RILs with contrasting levels of iron and zinc in unpolished rice grains. Gene 546 (2), 430–436. doi:10.1016/j.gene.2014.05.069
AieseCigliano, R., Aversano, R., Di Matteo, A., Palombieri, S., Termolino, P., Angelini, C., et al. (2022). Multi-omics data integration provides insights into the post-harvest biology of a long shelf-life tomato landrace. Hortic. Res. 9 uhab042, doi:10.1093/hr/uhab042
Al-Bader, A., Christenson, J. T., Simonet, F., Abul, H., Dashti, H., and Schmuziger, M. (1998). Inflammatory response and oligo-element alterations following cardiopulmonary bypass in patients undergoing coronary artery bypass grafting. Cardiovasc. Surg. 6 (4), 406–414. doi:10.1016/s0967-2109(98)00022-2
Albert-Weiss, D., and Osman, A. (2022). Interactive deep learning for shelf life prediction of muskmelons based on an active learning approach. Sensors 22 (2), 414. doi:10.3390/s22020414
Alegbeleye, O. O., Singleton, I., and Sant’Ana, A. S. (2018). Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: A review. Food Microbiol. 73, 177–208. doi:10.1016/j.fm.2018.01.003
Altuner, F. (2021). Determination of biochemical composition and pigment content in legume and cereal microgreens. Legume Res-An Int. J. 1, 8. doi:10.18805/LR-635
Altuner, F., Tunçtürk, R., Oral, E., and Tunçtürk, M. (2022). Determınatıon of the content of antıoxıdants and bıochemıcal composition of legume mıcrogreens. J. Elem. 1 (2022), 165–180. doi:10.5601/jelem.2022.27.1.2178
Arbona, V., Manzi, M., de Ollas, C., and Gómez-Cadenas, A. (2013). Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 14 (3), 4885–4911. doi:10.3390/ijms14034885
Argelaguet, R., Velten, B., Arnol, D., Dietrich, S., Zenz, T., Marioni, J. C., et al. (2018). Multi-omics factor analysis—A framework for unsupervised integration of multi-omics data sets. Mol. Sys. Biol. 14 (6), 8124. doi:10.15252/msb.20178124
Arya, M., and Mishra, S. B. (2022). “Transgenic and molecular approaches for pigeonpea and chick pea improvement,” in Technologies in plant biotechnology and breeding of field crops (Singapore: Springer), 239–272.
Bahrami, A., Delshadi, R., Jafari, S. M., and Williams, L. (2019). Nano encapsulated nisin: An engineered natural antimicrobial system for the food industry. Trends Food Sci. Technol. 94, 20–31. doi:10.1016/j.tifs.2019.10.002
Bansal, P., Bansal, R., and Arora, M. (2022a). “Traditional nutritional approaches and nutriomics evidence: Nutrition from inception to evidence,” in Nutriomics (United States: CRC Press), 23–47.
Bansal, T., Kawatra, A., and Sangwan, V. (2022b). Sensorial, nutritional and shelf life evaluation of bio-fortified millet based cookies supplemented with carrot powder and sesame. Asian JDairy Food Res. 41 (1), 83–88. doi:10.18805/ajdfr.DR-1696
Batiha, G. E. S., Al-Gareeb, A. I., Qusti, S., Alshammari, E. M., Kaushik, D., Verma, R., et al. (2022). Deciphering the immunoboosting potential of macro and micronutrients in COVID support therapy. Environ Sci Pollut. Res 1, 43516–43531. doi:10.1007/s11356-022-20075-7
Berger, S. L., Kouzarides, T., Shiekhattar, R., and Shilatifard, A. (2009). An operational definition of epigenetics. Genes & Dev. 23 (7), 781–783. doi:10.1101/gad.1787609
Berr, A., Shafiq, S., and Shen, W. H. (2011). Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta. 1809 (10), 567–576. doi:10.1016/j.bbagrm.2011.07.001
Bhardwaj, A. R., Joshi, G., Kukreja, B., Malik, V., Arora, P., Pandey, R., et al. (2015). Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 15 (1), 9–15. doi:10.1186/s12870-014-0405-1
Bhardwaj, A., and Steen, K. V. (2020). “Multi-omics data and analytics integration in ovarian cancer,” in IFIP International Conference on Artificial Intelligence Applications and Innovations, Neos Marmaras, Greece, 5-7 June, 347–435.
Bibi, I., Ghulam, T., Kamal, S., Jilani, K., Alwadai, N., and Iqbal, M. (2022). Green synthesis of iron nanoparticles and photocatalytic activity evaluation for the degradation of methylene blue dye. Z. Phys. Chem. 236, 1191–1201. doi:10.1515/zpch-2021-3128
Brazaitytė, A., Miliauskienė, J., Vaštakaitė-Kairienė, V., Sutulienė, R., Laužikė, K., Duchovskis, P., et al. (2021). Effect of different ratios of blue and red LED light on Brassicaceae microgreens under a controlled environment. Plants 10 (4), 801. doi:10.3390/plants10040801
Bruening, G., and Lyons, J. (2000). The case of the FLAVR SAVR tomato. Calif. Agri 54 (4), 6–7. doi:10.3733/ca.v054n04p6
Bulgari, R., Baldi, A., Ferrante, A., and Lenzi, A. (2017). Yield and quality of basil, Swiss chard, and rocket microgreens grown in a hydroponic system. N. Z. J. Crop Hortic. Sci. 45 (2), 119–129. doi:10.1080/01140671.2016.1259642
Bulgari, R., Negri, M., Santoro, P., and Ferrante, A. (2021). Quality evaluation of indoor-grown microgreens cultivated on three different substrates. Horticulturae 7 (5), 96. doi:10.3390/horticulturae7050096
Bunea, A., Andjelkovic, M., Socaciu, C., Bobis, O., Neacsu, M., Verhé, R., et al. (2008). Total and individual carotenoids and phenolic acids content in fresh, refrigerated and processed spinach (Spinacia oleracea L.). Food Chem. 108 (2), 649–656. doi:10.1016/j.foodchem.2007.11.056
Butkutė, B., Taujenis, L., and Norkevičienė, E. (2018). Small-seeded legumes as a novel food source. Variation of nutritional, mineral and phytochemical profiles in the chain: Raw seeds-sprouted seeds-microgreens. Molecules 24 (1), 133. doi:10.3390/molecules24010133
Cai, S., Shen, Q., Huang, Y., Han, Z., Wu, D., Chen, Z. H., et al. (2021). Multi-omics analysis reveals the mechanism underlying the edaphic adaptation in wild barley at evolution slope (tabigha). Adv. Sci. 8 (20), 2101374. doi:10.1002/advs.202101374
Castellaneta, A., Losito, I., Cisternino, G., Leoni, B., Santamaria, P., Calvano, C. D., et al. (2022). All ion fragmentation analysis enhances the untargeted profiling of glucosinolates in Brassica microgreens by liquid chromatography and high-resolution mass spectrometry. J. AmSoc Mass Spectrom. 33 (11), 2108–2119. doi:10.1021/jasms.2c00208
Chakravorty, M., Nanda, M., Arora, N., Singh, S., Kumar, V., and Deshwal, S. (2022). CRISPR-Cas9 mediated genome tailoring to improve nutritional quality and shelf life in crops: A review. Plant 31, 100369. doi:10.1016/j.plgene.2022.100369
Chandana, B. S., Mahto, R. K., Singh, R. K., Ford, R., Vaghefi, N., Gupta, S. K., et al. (2022). Epigenomics as potential tools for enhancing magnitude of breeding approaches for developing climate resilient chickpea. Front. Genet. 13, 900253. doi:10.3389/fgene.2022.900253
Chandra, D., Kim, J. G., and Kim, Y. P. (2012). Changes in microbial population and quality of microgreens treated with different sanitizers and packaging films. Hortic. Environ. Biotech. 53 (1), 32–40. doi:10.1007/s13580-012-00756
Chen, H. M., Chen, L. T., Patel, K., Li, Y. H., Baulcombe, D. C., and Wu, S. H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U. S. A. 107 (34), 15269–15274. doi:10.1073/pnas.1001738107
Chinnusamy, V., and Zhu, J. K. (2009). Epigenetic regulation of stress responses in plants. Curr.Opin. Plant Biol. 12 (2), 133–139. doi:10.1016/j.pbi.2008.12.006
Choe, U., Yu, L. L., and Wang, T. T. (2018). The science behind microgreens as an exciting new food for the 21st century. J. Agric. Food Chem. 66 (44), 11519–11530. doi:10.1021/acs.jafc.8b03096
Chowrira, G. M., Akella, V., Fuerst, P. E., and Lurquin, P. F. (1996). Transgenic grain legumes obtained by in planta electroporation-mediated gene transfer. Mol. Biotech. 5 (2), 85–96. doi:10.1007/BF02789058
Craver, J. K., Gerovac, J. R., Lopez, R. G., and Kopsell, D. A. (2017). Light intensity and light quality from sole-source light-emitting diodes impact phytochemical concentrations within Brassica microgreens. J. Am. Soc. Hortic. Sci. 142 (1), 3–12. doi:10.21273/JASHS03830-16
Dai, D., Zeng, S., Wang, L., Li, J., Ji, P., Liu, H., et al. (2022). Identification of fruit firmness QTL ff2. 1 by SLAF-BSA and QTL mapping in melon. Euphytica 218 (5), 1–15. doi:10.21203/rs.3.rs-1006787/v1
Dancs, G., Kondrak, M., and Banfalvi, Z. (2008). The effects of enhanced methionine synthesis on amino acid and anthocyanin content of potato tubers. BMC Plant Biol. 8 (1), 65. doi:10.1186/1471-2229-8-65
Dasgupta, U., Mishra, G. P., Dikshit, H. K., Mishra, D. C., Bosamia, T., Roy, A., et al. (2021). Comparative RNA-Seq analysis unfolds a complex regulatory network imparting yellow mosaic disease resistance in mungbean [Vigna radiata (L.) R. Wilczek]. Plos one 16 (1), 0244593. doi:10.1371/journal.pone.0244593
Dash, S., Campbell, J. D., Cannon, E. K., Cleary, A. M., Huang, W., Kalberer, S. R., et al. (2016). Legume information system (legume info. Org): A key component of a set of federated data resources for the legume family. Nucl. Acids Res. 44 (D1), D1181–D1188. doi:10.1093/nar/gkv1159
De la Fuente, B., López-García, G., Máñez, V., Alegría, A., Barberá, R., and Cilla, A. (2020). Antiproliferative effect of bioaccessible fractions of four Brassicaceae microgreens on human colon cancer cells linked to their phytochemical composition. Antioxidants 9 (5), 368–383. doi:10.3390/antiox9050368
de la Fuente, B., López-García, G., Máñez, V., Alegría, A., Barberá, R., and Cilla, A. (2019). Evaluation of the bioaccessibility of antioxidant bioactive compounds and minerals of four genotypes of Brassicaceae microgreens. Foods 8 (7), 250. doi:10.3390/foods8070250
Delian, E., Chira, A., Bădulescu, L., and Chira, L. (2015). Insights into microgreens physiology. Sci. Pap. Ser. B Hortic. 59, 447–454.
Dhakshayani, G. M., and Alias, P. S. J. (2022). A comparative study of phytochemical, antioxidant, anticarcinogenic, and antidiabetic potential of coriander (Coriandrum sativum L.): Microgreen and mature plant. Foods Raw Materi 10 (2), 283–294. doi:10.21603/2308-4057-2022-2-539
Di Gioia, F., Leoni, B., and Santamaria, P. (2015). “The selection of the species to grow,” in Eco-logicaeditore. Microgreens(Bari) (Bari: Eco-logica).
Di Gioia, F., Petropoulos, S. A., Ozores-Hampton, M., Morgan, K., and Rosskopf, E. N. (2019). Zinc and iron agronomic biofortification of Brassicaceae microgreens. Agronomy 9 (11), 677. doi:10.3390/agronomy9110677
Di Gioia, F., Renna, M., and Santamaria, P. (2017). “Sprouts, microgreens and “baby leaf” vegetables”,” in Minimally processed refrigerated fruits and vegetables (Boston: Springer), 403–432.
Dita, M. A., Rispail, N., Prats, E., Rubiales, D., and Singh, K. B. (2006). Biotechnology approaches to overcome biotic and abiotic stress constraints in legumes. Euphytica 147 (1), 1–24. doi:10.1007/s10681-006-6156-9
Doddamani, D., Khan, A. W., Katta, M. A. V. S. K., Agarwal, G., Thudi, M., Ruperao, P., et al. (2015). CicArVarDB: SNP and InDel database for advancing genetics research and breeding applications in chickpea. Database 19, bav078–7. doi:10.1093/database/bav078
Ducreux, L. J. M., Morris, W. L., Hedley, P. E., Shepherd, T., Davies, H. V., Millam, S., et al. (2004). Metabolic engineering of high carotenoid potato tubers containing enhanced levels of β-carotene and lutein. J. Exp. Bot. 56 (409), 81–89. doi:10.1093/jxb/eri016
Dutta, B., Lahiri, D., Nag, M., Abukhader, R., Sarkar, T., Pati, S., et al. (2022). Multi-omics approach in amelioration of food products. FrontMicrobiol 13, 955683. doi:10.3389/fmicb.2022.955683
Eapen, S. (2008). Advances in development of transgenic pulse crops. Biotechnol. Adv. 26 (2), 162–168. doi:10.1016/j.biotechadv.2007.11.001
Ebert, A. W. (2013). “Sprouts, microgreens, and edible flowers: The potential for high value specialty produce in asia,” in SEAVEG 2012: High Value Vegetables in Southeast Asia: Production, Supply and Demand, Chiang Mai, Thailand, 24-26 January 2012, 216–227.
El-Nakhel, C., Pannico, A., Graziani, G., Kyriacou, M. C., Giordano, M., Ritieni, A., et al. (2020). Variation in macronutrient content, phytochemical constitution and in vitro antioxidant capacity of green and red butterhead lettuce dictated by different developmental stages of harvest maturity. Antioxidants 9 (4), 300. doi:10.3390/antiox9040300
El-Ramady, H., Abdalla, N., Elbasiouny, H., Elbehiry, F., Elsakhawy, T., Omara, A. E. D., et al. (2021). Nano-biofortification of different crops to immune against COVID-19: A review. Ecotoxicol. Environ. Saf. 222, 112500. doi:10.1016/j.ecoenv.2021.112500
Elkonin, L. A., ItalianskayaI, J. V., Domanina, V. N., Selivanov, N. Y., Rakitin, A. L., and Ravi, N. V. (2016). Transgenic Sorghum with improved digestibility of storage proteins obtained by Agrobacterium-mediated transformation. Russ. J. Plant Physiol. 63 (5), 678–689. doi:10.1134/S1021443716050046
Eltaweil, A. S., Fawzy, M., Hosny, M., Abd El-Monaem, E. M., Tamer, T. M., and Omer, A. M. (2022). Green synthesis of platinum nanoparticles using Atriplex halimus leaves for potential antimicrobial, antioxidant, and catalytic applications. Arab. J. Chem. 15, 103517. doi:10.1016/j.arabjc.2021.103517
Engelken, J., Espadas, G., Mancuso, F. M., Bonet, N., Scherr, A. L., Jímenez-Álvarez, V., et al. (2016). Signatures of evolutionary adaptation in quantitative trait loci influencing trace element homeostasis in liver. Mol. BiolEvol 33 (3), 738–754. doi:10.1093/molbev/msv267
Fernie, A. R., and Schauer, N. (2009). Metabolomics-assisted breeding: A viable option for crop improvement? Trends Genet. 25 (1), 39–48. doi:10.1016/j.tig.2008.10.010
Gao, M., He, R., Shi, R., Zhang, Y., Song, S., Su, W., et al. (2021). Differential effects of low light intensity on broccoli microgreens growth and phytochemicals. Agronomy 11 (3), 537. doi:10.3390/agronomy11030537
Garcia-Casal, M. N., Peña-Rosas, J. P., and Giyose, B.Consultation Working Groups (2017). Staple crops biofortified with increased vitamins and minerals: Considerations for a public health strategy. Ann. N. Y. Acad. Sci. 1390 (1), 3–13. doi:10.1111/nyas.13293
Gardner, A. M., Brown, A. F., and Juvik, J. A. (2016). QTL analysis for the identification of candidate genes controlling phenolic compound accumulation in broccoli (Brassica oleracea L. var. italica). Mol. Breed. 36 (6), 81–12. doi:10.1007/s11032-016-0497-4
Garg, M., Sharma, N., Sharma, S., Kapoor, P., Kumar, A., Chunduri, V., et al. (2018). Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 5, 12. doi:10.3389/fnut.2018.00012
Gayali, S., Acharya, S., Lande, N. V., Pandey, A., Chakraborty, S., and Chakraborty, N. (2016). CicerTransDB 1.0: A resource for expression and functional study of chickpea transcription factors. BMC Plant Biol. 16 (1), 169. doi:10.1186/s12870-016-0860-y
Ghoora, M. D., and Srividya, N. (2020). Effect of packaging and coating technique on postharvest quality and shelf life of Raphanus sativus L. and Hibiscus sabdariffa L. microgreens. Foods 9 (5), 653. doi:10.3390/foods9050653
Girma, T. (2019). Feature extraction and classification of green mung bean using machine learning techniques. Ethiopia: Diss. ADDIS ABABA Science and Technology University.
Grootboom, A. W., Mkhonza, N. L., Mbambo, Z., O’Kennedy, M. M., Da Silva, L. S., Taylor, J., et al. (2014). Co-suppression of synthesis of major α-kafirin sub-class together with γ-kafirin-1 and γ-kafirin-2 required for substantially improved protein digestibility in transgenic Sorghum. Plant Cell Rep. 33 (3), 521–537. doi:10.1007/s00299-013-1556-5
Guo, N., Wang, S., Gao, L., Liu, Y., Wang, X., Lai, E., et al. (2021). Genome sequencing sheds light on the contribution of structural variants to Brassica oleracea diversification. BMC Biol. 19 (1), 93–15. doi:10.1186/s12915-021-01031-2
Gupta, A., Bhardwaj, A., Sawant, S. V., and Yadav, H. K. (2019). Utilization and characterization of genome-wide SNP markers for assessment of ecotypic differentiation in Arabidopsis thaliana. Int. J. Curr. Microbiol. App. Sci. 8 (6), 158–173. doi:10.20546/ijcmas.2019.806.020
Gupta, A., Jaiswal, V., Sawant, S. V., and Yadav, H. K. (2020a). Mapping QTLs for 15 morphometric traits in Arabidopsis thaliana using Col-0 × Don-0 population. Physiol. Mol. Biol. Plants 26 (5), 1021–1034. doi:10.1007/s12298-020-00800-7
Gupta, A. (2022). “Mainstreaming of underutilized oilseed safflower crop through biotechnological approaches for improving economic and environmental sustainability,” in Biotechnological innovations for environmental bioremediation (Singapore: Springer), 397–418.
Gupta, A., Maurya, R., Roy, R. K., Sawant, S. V., and Yadav, H. K. (2013). AFLP based genetic relationship and population structure analysis of Canna - an ornamental plant. Sci. Hortic. 154, 1–7. doi:10.1016/j.scienta.2013.02.005
Gupta, A., Sawant, S. V., and Yadav, H. K. (2020b). Assessment of phenotypic developmental traits and hybrid vigour in Arabidopsis thaliana. Indian J. Biotechnol. 19 (2), 102–112.
Gupta, A., and Tyagi, T. (2022). “Phytoremediation and therapeutic potential of neglected plants: An invasive aquatic weeds and ornamental plant,” in Biotechnological innovations for environmental bioremediation (Singapore: Springer), 259–290.
Hayashi, E., You, Y., Lewis, R., Calderon, M. C., Wan, G., and Still, D. W. (2012). Mapping QTL, epistasis and genotype× environment interaction of antioxidant activity, chlorophyll content and head formation in domesticated lettuce (Lactuca sativa). TheorApplGenet 124 (8), 1487–1502. doi:10.1007/s00122-012-1803-0
Hu, J., Rampitsch, C., and Bykova, N. V. (2015). Advances in plant proteomics toward improvement of crop productivity and stress resistancex. Front. Plant Sci. 6, 209. doi:10.3389/fpls.2015.00209
Huang, H., Jiang, X., Xiao, Z., Yu, L., Pham, Q., Sun, J., et al. (2016). Red cabbage microgreens lower circulating low-density lipoprotein (LDL), liver cholesterol, and inflammatory cytokines in mice fed a high-fat diet. J. Agric. Food Chem. 64 (48), 9161–9171. doi:10.1021/acs.jafc.6b03805
Indurker, S., Misra, H. S., and Eapen, S. (2007). Genetic transformation of chickpea (Cicerarietinum L.) with insecticidal crystal protein gene using particle gun bombardment. Plant Cell Rep. 26 (6), 755–763. doi:10.1007/s00299-006-0283-6
Iorliam, I. B., Ikyo, B. A., Iorliam, A., Okube, E. O., Kwaghtyo, K. D., and Shehu, Y. I. (2021). Application of machine learning techniques for Okra shelf-life prediction. J. Data Anal. Info Process. 9 (3), 136–150. doi:10.4236/jdaip.2021.93009
Jo, Y., Lee, S., Lee, Y., Kahng, H., Park, S., Bae, S., et al. (2021). Semantic segmentation of cabbage in the South Korea highlands with images by unmanned aerial vehicles. Appl. Sci. 11 (10), 4493. doi:10.3390/app11104493
Johnson, S. A., Prenni, J. E., Heuberger, A. L., Isweiri, H., Chaparro, J. M., Newman, S. E., et al. (2021). Comprehensive evaluation of metabolites and minerals in 6 microgreen species and the influence of maturity. Curr. Dev. Nutr. 5 (2), nzaa180–12. doi:10.1093/cdn/nzaa180
Jones-Baumgardt, C., Llewellyn, D., Ying, Q., and Zheng, Y. (2019). Intensity of sole-source light-emitting diodes affects growth, yield, and quality of Brassicaceae microgreens. Hort. Sci. 54 (7), 1168–1174. doi:10.21273/HORTSCI13788-18
Jung, M., Song, J. S., Hong, S., Kim, S., Go, S., Lim, Y. P., et al. (2021). Deep learning algorithms correctly classify Brassica rapa varieties using digital images. Front. Plant Sci. 12, 738685. doi:10.3389/fpls.2021.738685
Kale, S. M., Jaganathan, D., Ruperao, P., Chen, C., Punna, R., Kudapa, H., et al. (2015). Prioritization of candidate genes in “QTL-hotspot” region for drought tolerance in chickpea (Cicer arietinum L.). Sci. Rep. 5 (1), 15296–15314. doi:10.1038/srep15296
Kalia, P., and Singh, S. (2018). “Breeding for improving nutritional qualities and shelf life in vegetable crops,” in Advances in postharvest technologies of vegetable crops (Canada: Apple Academic Press), 127–169.
Katam, K., Jones, K. A., and Sakata, K. (2015). Advances in proteomics and bioinformatics in agriculture research and crop improvement. J. Proteomics Bioinform 8 (3), 39. doi:10.4172/2Fjpb.1000372
Kaur, N., Singh, B., Kaur, A., and Yadav, M. P. (2022). Impact of growing conditions on proximate, mineral, phenolic composition, amino acid profile, and antioxidant properties of black gram, mung bean, and chickpea microgreens. J. Food Process Preserv 46, e16655. doi:10.1111/jfpp.16655
Khoja, K., Buckley, A., Aslam, F. M., Sharp, A. P., and Latunde-Dada, G. O. (2020). In vitro bioaccessibility and bioavailability of iron from mature and microgreen Fenugreek, Rocket and Broccoli. Nutrients 12 (4), 1057. doi:10.3390/nu12041057
Khokhar, W., Hassan, M. A., Reddy, A. S., Chaudhary, S., Jabre, I., Byrne, L. J., et al. (2019). Genome-wide identification of splicing quantitative trait loci (sQTLs) in diverse ecotypes of Arabidopsis thaliana. Front plant sci 10, 1160. doi:10.3389/fpls.2019.01160
Kobori, C. N., and Amaya, D. B. R. (2008). Uncultivated Brazilian green leaves are richer sources of carotenoids than are commercially produced leafy vegetables. Food Nutr. Bull. 29 (4), 320–328. doi:10.1177/156482650802900408
Kong, Y., Kamath, D., and Zheng, Y. (2019). Blue versus red light can promote elongation growth independent of photoperiod: A study in four Brassica microgreens species. HortSci 54 (11), 1955–1961. doi:10.21273/HORTSCI14286-19
Kou, L., Luo, Y., Yang, T., Xiao, Z., Turner, E. R., Lester, G. E., et al. (2013). Postharvest biology, quality and shelf life of buckwheat microgreens. LWT-Food Sci. Tech. 51 (1), 73–78. doi:10.1016/j.lwt.2012.11.017
Kou, L., Yang, T., Liu, X., and Luo, Y. (2015). Effects of pre-and postharvest calcium treatments on shelf life and postharvest quality of broccoli microgreens. HortSci 50 (12), 1801–1808. doi:10.21273/HORTSCI.50.12.1801
Kou, L., Yang, T., Luo, Y., Liu, X., Huang, L., and Codling, E. (2014). Pre-harvest calcium application increases biomass and delays senescence of broccoli microgreens. Postharvest Biol. Technol. 87, 70–78. doi:10.1016/j.postharvbio.2013.08.004
KraljCigić, I., Rupnik, S., Rijavec, T., PoklarUlrih, N., and Cigić, B. (2020). Accumulation of agmatine, spermidine, and spermine in sprouts and microgreens of alfalfa, fenugreek, lentil, and daikon radish. Foods 9 (5), 547–567. doi:10.3390/foods9050547
Krishnappa, G., Khan, H., Krishna, H., Kumar, S., Mishra, C. N., Parkash, O., et al. (2022). Genetic dissection of grain iron and zinc, and thousand kernel weight in wheat (Triticum aestivum L.) using genome-wide association study. Sci. Rep. 12 (1), 12444–12514. doi:10.1038/s41598-022-15992-z
Kumar, S., Hash, C. T., Thirunavukkarasu, N., Singh, G., Rajaram, V., Rathore, A., et al. (2016). Mapping quantitative trait loci controlling high iron and zinc content in self and open pollinated grains of pearl millet [Pennisetum glaucum (L.) R. Br.]. Front Plant Sci 7, 1636. doi:10.3389/fpls.2016.01636
Kumar, R., Singh, R. K., Misra, J., Yadav, A., Kumar, A., Yadav, R., et al. (2022a). Dissecting proteomic estimates for enhanced bioavailable nutrition during varied stages of germination and identification of potential genotypes in chickpea. Legume Res. 45 (9), 1082–1087 doi:10.18805/LR-4531
Kumar, S., Seem, K., Kumar, S., Vinod, K. K., Chinnusamy, V., and Mohapatra, T. (2022b). Pup1 QTL regulates gene expression through epigenetic modification of DNA under phosphate starvation stress in rice. Front. Plant Sci. 13, 871890. doi:10.3389/fpls.2022.871890
Kumari, C., Sharma, M., Kumar, V., Sharma, R., Kumar, V., Sharma, P., et al. (2022). Genome editing technology for genetic amelioration of fruits and vegetables for alleviating post-harvest loss. Bioengineering 9 (4), 176. doi:10.3390/bioengineering9040176
Kusumam, K., Krajník, T., Pearson, S., Duckett, T., and Cielniak, G. (2017). 3D-vision based detection, localization, and sizing of broccoli heads in the field. J. Field Robot. 34 (8), 1505–1518. doi:10.1002/rob.21726
Kuzmina, Y. (2020). Methods of genome editing for increasing the shelf life of tomato fruit. Plant Biotechnol. Breed. 3, 31–39. doi:10.30901/2658-6266-2020-1-o6
Kyriacou, M. C., De Pascale, S., Kyratzis, A., and Rouphael, Y. (2017). Microgreens as a component of space life support systems: A cornucopia of functional food. Front. Plant Sci. 1587, 1587. doi:10.3389/fpls.2017.01587
Kyriacou, M. C., El-Nakhel, C., Graziani, G., Pannico, A., Soteriou, G. A., Giordano, M., et al. (2019a). Functional quality in novel food sources: Genotypic variation in the nutritive and phytochemical composition of thirteen microgreens species. Food Chem. 277, 107–118. doi:10.1016/j.foodchem.2018.10.098
Kyriacou, M. C., El-Nakhel, C., Pannico, A., Graziani, G., Soteriou, G. A., Giordano, M., et al. (2019b). Genotype-specific modulatory effects of select spectral bandwidths on the nutritive and phytochemical composition of microgreens. Front. Plant Sci. 10, 1501. doi:10.3389/fpls.2019.01501
Kyriacou, M. C., El-Nakhel, C., Pannico, A., Graziani, G., Zarrelli, A., Soteriou, G. A., et al. (2021a). Ontogenetic variation in the mineral, phytochemical and yield attributes of brassicaceous microgreens. Foods 10 (5), 1032. doi:10.3390/foods10051032
Kyriacou, M. C., El-Nakhel, C., Soteriou, G. A., Graziani, G., Kyratzis, A., Antoniou, C., et al. (2021b). Pre harvest nutrient deprivation reconfigures nitrate, mineral, and phytochemical content of microgreens. Foods 10 (6), 1333. doi:10.3390/foods10061333
Kyriacou, M. C., Rouphael, Y., Di Gioia, F., Kyratzis, A., Serio, F., Renna, M., et al. (2016). Micro-scale vegetable production and the rise of microgreens. Trends Food Sci. Technol. 57, 103–115. doi:10.1016/j.tifs.2016.09.005
Langridge, P., and Fleury, D. (2011). Making the most of ‘omics’ for crop breeding. Trends Biotech. 29 (1), 33–40. doi:10.1016/j.tibtech.2010.09.006
Laurent, S., Forge, D., Port, M., Roch, A., Robic, C., Vander Elst, L., et al. (2008). Magnetic iron oxide nanoparticles: Synthesis, stabilization, vectorization, physicochemical characterizations, and biological applications. Chem. Rev. 108 (6), 2064–2110. doi:10.1021/cr068445e
Le, T. N., Chiu, C. H., and Hsieh, P. C. (2020). Bioactive compounds and bioactivities of Brassicaoleracea L. var. italica sprouts and microgreens: An updated overview from a nutraceutical perspective. Plants 9 (8), 946–969. doi:10.3390/plants9080946
Lee, C., Lee, J., and Lee, J. (2022). Relationship of fruit color and anthocyanin content with related gene expression differ in strawberry cultivars during shelf life. SciHortic 301, 111109. doi:10.1016/j.scienta.2022.111109
Li, J., Dai, X., Liu, T., and Zhao, P. X. (2012). Legume IP: An integrative database for comparative genomics and transcriptomics of model legumes. Nucl. Acids Res. 40 (D1), D1221–D1229. doi:10.1093/nar/gkr939
Li, Z., Liu, Y., Li, L., Fang, Z., Yang, L., Zhuang, M., et al. (2019). Transcriptome reveals the gene expression patterns of sulforaphane metabolism in broccoli florets. PLoS One 14 (3), 0213902. doi:10.1371/journal.pone.0213902
Lipkie, T. E., De Moura, F. F., Zhao, Z-Y., Albertsen, M. C., Che, P., Glassman, K., et al. (2013). Bioaccessibility of carotenoids from transgenic provitamin A biofortified Sorghum. J. Agric. Food Chem. 61 (24), 5764–5771. doi:10.1021/jf305361s
Liu, W., Min, X., and Wang, Y. (2019). Genome-wide development of microRNA-based SSR markers in Medicago truncatula with their transferability analysis and utilization in related legume species. Model Legume Medicago truncatula 2019, 936–945. doi:10.1002/9781119409144.ch121
Lobiuc, A., Vasilache, V., Oroian, M., Stoleru, T., Burducea, M., Pintilie, O., et al. (2017). Blue and red LED illumination improves growth and bioactive compounds contents in acyanic and cyanic Ocimumbasilicum L. microgreens. Molecules 22 (12), 2111. doi:10.3390/molecules22122111
Lu, Y., Dong, W., Alcazar, J., Yang, T., Luo, Y., Wang, Q., et al. (2018). Effect of pre harvest CaCl2 spray and postharvest UV-B radiation on storage quality of Broccoli microgreens, a richer source of glucosinolates. J. Food Compost Anal. 67, 55–62. doi:10.1016/j.jfca.2017.12.035
Mahanta, S., Habib, M. R., and Moore, J. M. (2022). Effect of high-voltage atmospheric cold plasma treatment on germination and heavy metal uptake by soybeans (Glycine max). Int. J. Mol. Sci. 23 (3), 1611. doi:10.3390/ijms23031611
Mahto, R. K., Singh, C., Chandana, B. S., Singh, R. K., Verma, S., Gahlaut, V., et al. (2022). Chickpea biofortification for cytokinin dehydrogenase via genome editing to enhance abiotic-biotic stress tolerance and food security. Front. Genet. 13, 900324. doi:10.3389/fgene.2022.900324
Maina, S., Ryu, D. H., Cho, J. Y., Jung, D. S., Park, J. E., Nho, C. W., et al. (2021). Exposure to salinity and light spectra regulates glucosinolates, phenolics, and antioxidant capacity of Brassica carinata L. Microgreens. Antioxidants. 10 (8), 1183. doi:10.3390/antiox10081183
Maluin, F. N., Hussein, M. Z., Nik Ibrahim, N. N. L., Wayayok, A., and Hashim, N. (2021). Some emerging opportunities of nanotechnology development for soilless and microgreen farming. Agronomy 11 (6), 1213. doi:10.3390/agronomy11061213
Mannur, D. M., Babbar, A., Thudi, M., Sabbavarapu, M. M., Roorkiwal, M., Yeri, S. B., et al. (2019). Super Annigeri 1 and improved JG 74: Two Fusarium wilt-resistant introgression lines developed using marker-assisted backcrossing approach in chickpea (Cicer arietinum L.). Mol. Breed. 39 (1), 2–13. doi:10.1007/s11032-018-0908-9
Martiniakova, M., Babikova, M., Mondockova, V., Blahova, J., Kovacova, V., and Omelka, R. (2022). The role of macronutrients, micronutrients and flavonoid polyphenols in the prevention and treatment of osteoporosis. Nutrients 14 (3), 523. doi:10.3390/nu14030523
Mathiazhagan, M., Chidambara, B., Hunashikatti, L. R., and Ravishankar, K. V. (2021). Genomic approaches for improvement of tropical fruits: Fruit quality, shelf life and nutrient content. Genes 12 (12), 1881. doi:10.3390/genes12121881
Maurya, R., Gupta, A., Singh, S. K., Rai, K. M., Katiyar, R., Sawant, S. V., et al. (2015). Genomic-derived microsatellite markers for diversity analysis in Jatropha curcas. Trees 29 (3), 849–858. doi:10.1007/s00468-015-1166-7
Meas, S., Luengwilai, K., and Thongket, T. (2020). Enhancing growth and phytochemicals of two amaranth microgreens by LEDs light irradiation. Sci. Hortic. 265, 109204. doi:10.1016/j.scienta.2020.109204
Mishra, G. P., Dikshit, H. K., Tontang, M. T., Stobdan, T., Sangwan, S., Aski, M. S., et al. (2021). Diversity in phytochemical composition, antioxidant capacities, and nutrient contents among Mungbean and Lentil microgreens when grown at plain-altitude region (Delhi) and high-altitude region (Leh-Ladakh), India. Front. Plant Sci. 12, 710812. doi:10.3389/fpls.2021.710812
Mlinarić, S., Gvozdić, V., Vuković, A., Varga, M., Vlašiček, I., Cesar, V., et al. (2020). The effect of light on antioxidant properties and metabolic profile of chia Microgreens. Appl. Sci. 10 (17), 5731. doi:10.3390/app10175731
Muchjajib, U., Muchjajib, S., Suknikom, S., and Butsai, J. (2015). Evaluation of organic media alternatives for the production of microgreens in Thailand. Acta Hortic. 1102, 157–162. doi:10.17660/ActaHortic.2015.1102.19
Nguyen, T. H. A., Nguyen, V. C., Phan, T. N. H., Vasseghian, Y., Trubitsyn, M. A., Nguyen, A. T., et al. (2022). Novel biogenic silver and gold nanoparticles for multifunctional applications: Green synthesis, catalytic and antibacterial activity, and colorimetric detection of Fe (III) ions. Chemosphere 287, 132271. doi:10.1016/j.chemosphere.2021.132271
Niroula, A., Khatri, S., Timilsina, R., Khadka, D., Khadka, A., and Ojha, P. (2019). Profile of chlorophylls and carotenoids of wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) microgreens. J. Food Sci. Tech. 56 (5), 2758–2763. doi:10.1007/s13197-019-03768-9
Oliva, M., Demanelis, K., Lu, Y., Chernoff, M., Jasmine, F., Ahsan, H., et al. (2022). DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nat. Genet. 2022, 1–11. doi:10.1038/s41588-022-01248-z
Pandey, A., Chakraborty, S., Datta, A., and Chakraborty, N. (2008). Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Mol. Cell Proteom 7 (1), 88–107. doi:10.1074/mcp.M700314-MCP200
Pannico, A., El-Nakhel, C., Graziani, G., Kyriacou, M. C., Giordano, M., Soteriou, G. A., et al. (2020). Selenium biofortification impacts the nutritive value, polyphenolic content, and bioactive constitution of variable microgreens genotypes. Antioxidants 9 (4), 272. doi:10.3390/antiox9040272
Paradiso, V. M., Castellino, M., Renna, M., Gattullo, C. E., Calasso, M., Terzano, R., et al. (2018). Nutritional characterization and shelf-life of packaged microgreens. Food Funct. 9 (11), 5629–5640. doi:10.1039/C8FO01182F
Parmar, S., Sharma, V., Bomireddy, D., Soni, P., Joshi, P., Gangurde, S. S., et al. (2022). Recent advances in genetics, genomics, and breeding for nutritional quality in groundnut, Accel. Plant Breed. 4, 111–137. doi:10.1007/978-3-030-81107-5_4
Pavan, M. P., and Gangaprasad, S. (2022). Studies on mode of gene action for fruit quality characteristics governing shelf life in tomato (Solanum lycopersicum L.). Sci. Hortic. 293, 110687. doi:10.1016/j.scienta.2021.110687
Petropoulos, S. A., El-Nakhel, C., Graziani, G., Kyriacou, M. C., and Rouphael, Y. (2021). The effects of nutrient solution feeding regime on yield, mineral profile, and phytochemical composition of spinach microgreens. Horticulturae 7 (7), 162. doi:10.3390/horticulturae7070162
Podse dek, A., Sosnowska, D., Redzynia, M., and Anders, B. (2006). Antioxidant capacity and content of Brassica oleracea dietary antioxidants. Int. J. Food Sci. Technol. 41 (1), 49–58. doi:10.1111/j.1365-2621.2006.01260.x
Pradeepkumara, N., Sharma, P. K., Munshi, A. D., Behera, T. K., Bhatia, R., Kumari, K., et al. (2022). Fruit transcriptional profiling of the contrasting genotypes for shelf life reveals the key candidate genes and molecular pathways regulating post-harvest biology in cucumber. Genomics 114 (2), 110273. doi:10.1016/j.ygeno.2022.110273
Qi, T., Wu, Y., Fang, H., Zhang, F., Liu, S., Zeng, J., et al. (2022). Genetic control of RNA splicing and its distinct role in complex trait variation. Nat. Genet. 54 (9), 1355–1363. doi:10.1038/s41588-022-01154-4
Qin, G., Wu, S., Zhang, L., Li, Y., Liu, C., Yu, J., et al. (2022). An efficient modular gateway recombinase-based gene stacking system for generating multi-trait transgenic plants. Plants 11 (4), 488. doi:10.3390/plants11040488
Reda, T., Thavarajah, P., Polomski, R., Bridges, W., Shipe, E., and Thavarajah, D. (2021). Reaching the highest shelf: A review of organic production, nutritional quality, and shelf life of kale (Brassica oleracea var. acephala). Plants, People, Planet 3 (4), 308–318. doi:10.1002/ppp3.10183
Roorkiwal, M., Bharadwaj, C., Barmukh, R., Dixit, G. P., Thudi, M., Gaur, P. M., et al. (2020). Integrating genomics for chickpea improvement: Achievements and opportunities. Theor. Appl. Genet. 133 (5), 1703–1720. doi:10.1007/s00122-020-03584-2
Saengha, W., Karirat, T., Buranrat, B., Matra, K., Deeseenthum, S., Katisart, T., et al. (2021). Cold plasma treatment on mustard green seeds and its effect on growth, isothiocyanates, antioxidant activity and anticancer activity of microgreens. Int. J. Agric. Biol. 25 (33), 667–676. doi:10.17957/IJAB/15.1715
Sahoo, M. R., Naresh, P., Kumari, M., and Acharya, G. C. (2022). “Omics in leafy vegetables: Genomics, transcriptomics, proteomics, metabolomics, and multiomics approaches,” in Omics in horticultural crops (United States: Academic Press), 281–302.
Samuolienė, G., Brazaitytė, A., Viršilė, A., Miliauskienė, J., Vaštakaitė-Kairienė, V., and Duchovskis, P. (2019). Nutrient levels in Brassicaceae microgreens increase under tailored light-emitting diode spectra. Front. Plant Sci. 10, 1475. doi:10.3389/fpls.2019.01475
Samuolienė, G., Viršilė, A., Brazaitytė, A., Jankauskienė, J., Sakalauskienė, S., Vaštakaitė, V., et al. (2017). Blue light dosage affects carotenoids and tocopherols in microgreens. Food Chem. 228, 50–56. doi:10.1016/j.foodchem.2017.01.144
Sanderson, L. A., Caron, C. T., Tan, R., Shen, Y., Liu, R., and Bett, K. E. (2019). KnowPulse: A web-resource focused on diversity data for pulse crop improvement. Front. Plant Sci. 10, 965. doi:10.3389/fpls.2019.00965
Sangwan, S., Kukreja, B., Mishra, G. P., Dikshit, H. K., Singh, A., Aski, M., et al. (2022). Yield optimization, microbial load analysis, and sensory evaluation of mungbean (Vigna radiata L.), lentil (Lens culinaris subsp. culinaris), and Indian mustard (Brassica juncea L.) microgreens grown under greenhouse conditions. Plos one 17 (5), 0268085. doi:10.1371/journal.pone.0268085
Schmidt, M. A., Parrott, W. A., Hildebrand, D. F., Berg, R. H., Cooksey, A., Pendarvis, K., et al. (2015). Transgenic soyabean seeds accumulating β-carotene exhibit the collateral enhancements of oleate and protein content traits. Plant Biotech. J. 13 (4), 590–600. doi:10.1111/pbi.12286
Sharma, K. K., Lavanya, M., and Anjaiah, V. (2006). Agrobacterium-mediated production of transgenic pigeon pea (Cajanus cajan L. Mill sp.) expressing the synthetic Bt cry1Ab gene. Vitro Cell Dev. Biol. Plant 42 (2), 165–173. doi:10.1079/IVP2005730
Sharma, S., Shree, B., Sharma, D., Kumar, S., Kumar, V., Sharma, R., et al. (2022). Vegetable microgreens: The gleam of next generation super foods, their genetic enhancement, health benefits and processing approaches. Food Res. Int. 155, 111038. doi:10.1016/j.foodres.2022.111038
Sharma, T., and Gupta, A. (2022). “Integrated OMICS approaches to ameliorate the abiotic stress in,” in Plant stress: Challenges and management in the new decade (Cham: Springer), 361–373.
Shelar, A., Singh, A. V., Dietrich, P., Maharjan, R. S., Thissen, A., Didwal, P. N., et al. (2022). Emerging cold plasma treatment and machine learning prospects for seed priming: A step towards sustainable food production. RSC Adv. 12 (17), 10467–10488. doi:10.1039/D2RA00809B
Singh, K., Kumar, S., and Kaur, P. (2019). Automatic detection of rust disease of Lentil by machine learning system using microscopic images. Int. J. Elect. Com. Eng. 9 (1), 660–666. doi:10.11591/ijece.v9i1.pp660-666
Singh, R. K., Singh, C., Chandana, B. S., Mahto, R. K., Patial, R., Gupta, A., et al. (2022). Exploring chickpea germplasm diversity for broadening the genetic base utilizing genomic resourses. Front. Genet. 1836, 905771. doi:10.3389/fgene.2022.905771
Spillane, C., Steimer, A., Grossniklaus, U., and GrossniklaUs, U. (2001). Apomixis in agriculture: The quest for clonal seeds. Sex. Plant Reprod. 14 (4), 179–187. doi:10.1007/s00497-001-0117-1
Sreenivasan, E. (2020). Preliminary report on multiple harvests of microgreens from Chickpea (Cicer arietinum) seeds. Intl J. Trend Sci. Res. Dev. 4, 306–308. IJTSRD33337.
Sun, J., Kou, L., Geng, P., Huang, H., Yang, T., Luo, Y., et al. (2015). Metabolomic assessment reveals an elevated level of glucosinolate content in CaCl2 treated broccoli microgreens. J. Agric. Food Chem. 63 (6), 1863–1868. doi:10.1021/jf504710r
Sundaria, N., Singh, M., Upreti, P., Chauhan, R. P., Jaiswal, J. P., and Kumar, A. (2019). Seed priming with iron oxide nanoparticles triggers iron acquisition and biofortification in wheat (Triticum aestivum L.) grains. J. Plant Growth Reg. 38 (1), 122–131. doi:10.1007/s00344-018-9818-7
Tan, L., Nuffer, H., Feng, J., Kwan, S. H., Chen, H., Tong, X., et al. (2020). Antioxidant properties and sensory evaluation of microgreens from commercial and local farms. Food Sci. Hum. Wellness. 9 (1), 45–51. doi:10.1016/j.fshw.2019.12.002
Tantharapornrerk, N., Vichitsoonthonkul, T., Techavuthiporn, C., and Photchanachai, S. (2021). Growth and antioxidant system of Chinese kale microgreens in response to different illumination of light sources. N. Z. J. Crop Hort. Sci. 2021, 1–15. doi:10.1080/01140671.2021.1958876
Thuong, V. T., and Minh, H. G. (2020). Effects of growing substrates and seed density on yield and quality of radish (Raphanus sativus) microgreens. Res. Crops 21 (3), 579–586. doi:10.31830/2348-7542.2020.091
Toscano, S., Cavallaro, V., Ferrante, A., Romano, D., and Patané, C. (2021). Effects of different light spectra on final biomass production and nutritional quality of two microgreens. Plants 10 (8), 1584. doi:10.3390/plants10081584
Tropea, A., Potortì, A. G., Lo Turco, V., Russo, E., Vadalà, R., Rando, R., et al. (2021). Aqua feed production from fermented fish waste and lemon peel. Fermentation 7 (4), 272. doi:10.3390/fermentation7040272
Truzzi, F., Whittaker, A., Roncuzzi, C., Saltari, A., Levesque, M. P., and Dinelli, G. (2021). Microgreens: Functional food with anti proliferative cancer properties influenced by light. Foods 10 (8), 1690. doi:10.3390/foods10081690
Tsaftaris, A. S., and Polidoros, A. N. (2000). DNA methylation and plant breeding. Plant Breed. Rev. 18, 87–176. doi:10.1002/9780470650158.ch3
Turner, E. R., Luo, Y., and Buchanan, R. L. (2020). Microgreen nutrition, food safety, and shelf life: A review. J. Food Sci. 85 (4), 870–882. doi:10.1111/1750-3841.15049
Upadhyaya, C. P., Young, K. E., Akula, N., Soon Kim, H., Heung, J. J., Oh, O., et al. (2009). Over-expression of strawberry d-galacturonic acid reductase in potato leads to accumulation of vitamin C with enhanced abiotic stress tolerance. Plant Sci. 177 (6), 659–667. doi:10.1016/j.plantsci.2009.08.004
United States Department of Agriculture (2014). Specialty greens pack a nutritionalpunch, AgResearch Magazine. Available at: http://agresearchmag.ars.usda.gov/2014/jan/greens.
Valdes, A., Álvarez-Rivera, G., Socas-Rodríguez, B., Herrero, M., Ibáñez, E., and Cifuentes, A. (2021). Foodomics: Analytical opportunities and challenges. Anal. Chem. 94 (1), 366–381. doi:10.1021/acs.analchem.1c04678
Vanderschuren, H., Lentz, E., Zainuddin, I., and Gruissem, W. (2013). Proteomics of model and crop plant species: Status, current limitations and strategic advances for crop improvement. J. Proteomics 93, 5–19. doi:10.1016/j.jprot.2013.05.036
Varshney, R. K. (2016). Exciting journey of 10 years from genomes to fields and markets: Some success stories of genomics-assisted breeding in chickpea, pigeonpea and groundnut. Plant Sci. 242, 98–107. doi:10.1016/j.plantsci.2015.09.009
Varshney, R. K., Pandey, M. K., Bohra, A., Singh, V. K., Thudi, M., and Saxena, R. K. (2019). Toward the sequence-based breeding in legumes in the post-genome sequencing era. TheorAppl Genet. 132 (3), 797–816. doi:10.1007/s00122-018-3252-x
Varshney, R. K., Roorkiwal, M., Sun, S., Bajaj, P., Chitikineni, A., Thudi, M., et al. (2021). A chickpea genetic variation map based on the sequencing of 3,366 genomes. Nature 599 (7886), 622–627. doi:10.1038/s41586-021-04066-1
Varshney, R. K., Thudi, M., Pandey, M. K., Tardieu, F., Ojiewo, C., Vadez, V., et al. (2018). Accelerating genetic gains in legumes for the development of prosperous smallholder agriculture: Integrating genomics, phenotyping, systems modelling and agronomy. J. Exp. Bot. 69 (13), 3293–3312. doi:10.1093/jxb/ery088
Vaštakaitė, V., Viršilė, A., Brazaitytė, A., Samuolienė, G., Jankauskienė, J., Novičkovas, A., et al. (2017). Pulsed light-emitting diodes for a higher phytochemical level in microgreens. J. Agri Food Chem. 65 (31), 6529–6534. doi:10.1021/acs.jafc.7b01214
Verlinden, S. (2020). Microgreens: Definitions, product types, and production practices. Hortic. Rev. 47, 85–124. doi:10.1002/9781119625407.ch3
Verma, M., Kumar, V., Patel, R. K., Garg, R., and Jain, M. (2015). CTDB: An integrated chickpea transcriptome database for functional and applied genomics. PLoS ONE 10 (8), e0136880. doi:10.1371/journal.pone.0136880
Weber, C. F. (2017). Broccoli microgreens: A mineral-rich crop that can diversify food systems. Front. Nutr. 4, 7. doi:10.3389/fnut.2017.00007
Wolanin, A., Mateo-García, G., Camps-Valls, G., Gómez-Chova, L., Meroni, M., Duveiller, G., et al. (2020). Estimating and understanding crop yields with explainable deep learning in the Indian Wheat Belt. Environ. Res. Lett. 15 (2), 024019. doi:10.1088/1748-9326/ab68ac
Wu, J., Yuan, Y. X., Zhang, X. W., Zhao, J., Song, X., Li, Y., et al. (2008). Mapping QTLs for mineral accumulation and shoot dry biomass under different Zn nutritional conditions in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Soil 310 (1), 25–40. doi:10.1007/s11104-008-9625-1
Wu, X., Islam, A. F., Limpot, N., Mackasmiel, L., Mierzwa, J., Cortés, A. J., et al. (2020). Genome-wide Snp identification and association mapping for seed mineral concentration in mung bean (Vigna Radiata L.). Front. Genet. 11, 656. doi:10.3389/fgene.2020.00656
Xiao, Z., Lester, G. E., Luo, Y., and Wang, Q. (2012). Assessment of vitamin and carotenoid concentrations of emerging food products: Edible microgreens. J. Agri Food Chem. 60 (31), 7644–7651. doi:10.1021/jf300459b
Xiao, Z., Lester, G. E., Park, E., Saftner, R. A., Luo, Y., and Wang, Q. (2015). Evaluation and correlation of sensory attributes and chemical compositions of emerging fresh produce: Microgreens. Postharvest Biol. Tech. 110, 140–148. doi:10.1016/j.postharvbio.2015.07.021
Xiao, Z., Luo, Y., Lester, G. E., Kou, L., Yang, T., and Wang, Q. (2014). Postharvest quality and shelf life of radish microgreens as impacted by storage temperature, packaging film, and chlorine wash treatment. LWT-Food Sci. Tech. 55 (2), 551–558. doi:10.1016/j.lwt.2013.09.009
Ying, Q., Jones-Baumgardt, C., Zheng, Y., and Bozzo, G. (2021). The Proportion of blue light from light-emitting diodes alters microgreen phytochemical profiles in a species-specific manner. HortSci 56 (1), 13–20. doi:10.21273/HORTSCI15371-20
Ying, Q., Kong, Y., Jones-Baumgardt, C., and Zheng, Y. (2020a). Responses of yield and appearance quality of four Brassicaceae microgreens to varied blue light proportion in red and blue light-emitting diodes lighting. Sci. Hortic. 259, 108857. doi:10.1016/j.scienta.2019.108857
Ying, Q., Kong, Y., and Zheng, Y. (2020b). Applying blue light alone, or in combination with far-red light, during nighttime increases elongation without compromising yield and quality of indoor-grown microgreens. HortSci 55 (6), 876–881. doi:10.21273/hortsci14899-20
Zhang, X., Bian, Z., Li, S., Chen, X., and Lu, C. (2019). Comparative analysis of phenolic compound profiles, antioxidant capacities, and expressions of phenolic biosynthesis-related genes in soybean microgreens grown under different light spectra. J Agri Food Chem 67 (49), 13577–13588. doi:10.1021/acs.jafc.9b05594
Zhang, X., Wei, J., Tian, J., Li, N., Jia, L., Shen, W., et al. (2019). Enhanced anthocyanin accumulation of immature radish microgreens by hydrogen-rich water under short wavelength light. Sci. Horti 247, 75–85. doi:10.1016/j.scienta.2018.11.060
Zhao, Z. Y., Glassman, K., Sewalt, V., Wang, N., Miller, M., Chang, S., et al. (2003). “Nutritionally improved transgenic sorghum”,” in Plant Biotechnology2002 and beyond (Netherlands: Springer), 413–416.
Zhou, C., Hu, J., Xu, Z., Yue, J., Ye, H., and Yang, G. (2020). A monitoring system for the segmentation and grading of broccoli head based on deep learning and neural networks. Front. Plant Sci. 11, 402. doi:10.3389/fpls.2020.00402
Zhou, Q., Fu, Z., Liu, H., Wang, J., Guo, Z., Zhang, X., et al. (2021). Mining novel kernel size-related genes by pQTL mapping and multi-omics integrative analysis in developing maize kernels. Plant Biotechnol. J. 19 (8), 1489–1491. doi:10.1111/pbi.13634
Zhu, F., Jadhav, S. S., Tohge, T., Salem, M. A., Lee, J. M., Giovannoni, J. J., et al. (2022). A comparative transcriptomics and eQTL approach identifies SlWD40 as a tomato fruit ripening regulator. Plant Physiol. 190 (1), 250–266. doi:10.1093/plphys/kiac200
Keywords: biofortification, functional food, microgreen, nutrition, OMICS, shelf life
Citation: Gupta A, Sharma T, Singh SP, Bhardwaj A, Srivastava D and Kumar R (2023) Prospects of microgreens as budding living functional food: Breeding and biofortification through OMICS and other approaches for nutritional security. Front. Genet. 14:1053810. doi: 10.3389/fgene.2023.1053810
Received: 26 September 2022; Accepted: 05 January 2023;
Published: 25 January 2023.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Rutwik Barmukh, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaSandhya Tyagi, United Arab Emirates University, United Arab Emirates
Copyright © 2023 Gupta, Sharma, Singh, Bhardwaj, Srivastava and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Astha Gupta, YXN0aGFuYnJpQGdtYWlsLmNvbQ==; Rajendra Kumar, cmFqZW5kcmFrNjRAeWFob28uY28uaW4=
†These authors have contributed equally to this work
 Astha Gupta
Astha Gupta Tripti Sharma
Tripti Sharma Surendra Pratap Singh
Surendra Pratap Singh Archana Bhardwaj
Archana Bhardwaj Deepti Srivastava
Deepti Srivastava Rajendra Kumar
Rajendra Kumar