- 1Department of Medical Genetics, Hunan Provincial Maternal and Child Healthcare Hospital, Changsha, China
- 2Department of General Surgery, Changsha Central Hospital Affiliated to University of South China, Changsha, China
- 3Department of Ultrasonography, Hunan Provincial Maternal and Child Healthcare Hospital, Changsha, China
- 4Center for Medical Genetics and Hunan Key Laboratory of Medical Genetics School of Life Sciences, School of Life Sciences, Central South University, Changsha, China
- 5Department of Medical Genetics, Hunan Children’s Hospital, Changsha, China
Chromosomal mosaicism remains a perpetual diagnostic and clinical dilemma. In the present study, we detected two prenatal trisomy 9 mosaic syndrome cases by using multiple genetic testing methods. The non-invasive prenatal testing (NIPT) results suggested trisomy 9 in two fetuses. Karyotype analysis of amniocytes showed a high level (42%–50%) of mosaicism, and chromosomal microarray analysis (CMA) of uncultured amniocytes showed no copy number variation (CNV) except for large fragment loss of heterozygosity. Ultrasound findings were unmarkable except for small for gestational age. In Case 1, further umbilical blood puncture confirmed 22.4% and 34% trisomy 9 mosaicism by CMA and fluorescent in situ hybridization (FISH) respectively. After comprehensive consideration of the genetic and ultrasound results, the two gravidas decided to receive elective termination and molecular investigations of multiple tissue samples from the aborted fetus and the placenta. The results confirmed the presence of true fetoplacental mosaicism with levels of trisomy 9 mosaicism from 76% to normal in various tissues. These two cases highlight the necessity of genetic counseling for gravidas whose NIPT results highly suggest the risk of chromosome 9 to ascertain the occurrence of mosaicism. In addition, the comprehensive use of multiple genetic techniques and biological samples is recommended for prenatal diagnosis to avoid false-negative results. It should also be noted that ultrasound results of organs with true trisomy 9 mosaicism can be free of structural abnormalities during pregnancy.
Introduction
Trisomy 9 is an uncommon chromosomal abnormality that can occur in a mosaic or non-mosaic state (Cantú et al., 1996). Full trisomy 9 syndrome can be lethal with an incidence of 2.2%–2.7% in first-trimester spontaneous abortions (Ferreres et al., 2008; Benn and Grati., 2021), but trisomy 9 mosaicism syndrome has been reported compatible with life (Bruns and Campbell. 2015). More than 100 cases of mosaic trisomy 9 have been reported in the literature (Li et al., 2021). Most individuals with trisomy 9 and trisomy 9 mosaicism have prenatal and perinatal issues, including intrauterine growth retardation or “small size”, oligohydramnios, placental insufficiency, premature rupture of membranes, and skeletal abnormalities (Chen et al., 2010; Bruns and Campbell. 2015). Although there is no specific intrauterine ultrasound phenotype, more uncommon mosaic chromosomal abnormalities have been detected prenatally by invasive genetic tests following the sonographic diagnosis of fetal anomalies or high risk of the non-invasive prenatal testing (NIPT) (Wang et al., 2020; Lee et al., 2018). The diagnosis of chromosomal mosaicism in the prenatal stage is fraught with uncertainty and multiple factors need to be considered in order to gauge the likely impact. In genetic counseling for mosaicism, multiple influencing factors need to be considered comprehensively, including the chromosome position, type of mosaicism, distribution of the abnormal cell line in the fetus, assay noise, and culture artifacts. Herein, we present two cases of fetoplacental mosaic trisomy 9 based on a series of results from prenatal screening to diagnosis and follow-up observations on abnormal cell distributions in various tissues within the fetus. It should be addressed that ultrasound results of organs with true trisomy 9 mosaicism can be free of structural abnormalities during pregnancy.
Case report
Case 1
The gravida was a 26-year-old woman (gravida 2, para 0) with no family history of chromosomal abnormalities and a sign of spontaneous abortion during early pregnancy. She underwent maternal serum screening at 12 weeks of gestation, which revealed a risk for Down syndrome of 1 in 200. Due to concerns about the unavoidable risks of invasive prenatal diagnosis methods, she chose NIPT for fetal autosomal aneuploidies screening at 16+1 weeks of gestation after genetic counseling. Sample preparation, maternal plasma DNA sequencing, and bioinformatics analysis for NIPT were carried out using BGI platform (MGISEQ-2000, Shenzhen, China) as previously described (Xiang et al., 2023). NIPT analysis showed that the Z-scores of other chromosomes were all in the normal range (−3 < Z < 3) except for chromosome 9 (Z = 8.9898) with a cell-free fetal DNA fraction at 11.08% (3.5% is the least reliable cell-free fetal DNA level), suggesting a possibility of trisomy 9 or trisomy 9 mosaicism.
Following post-test genetic counseling for the NIPT results, the gravida agreed to receive amniocentesis for further analysis at 22 weeks of gestation. Genetic amniocentesis test revealed a karyotype of mos 47,XX,+9 [25]/46,XX [25], indicating a level of 50% trisomy 9 mosaicism (Figure 1A). Parental karyotypes of peripheral blood were normal. Chromosomal microarray analysis (CMA) of uncultured amniocytes with the Affymetrix CytoScan®750 K Array (Affymetrix Inc., CA, United States) revealed an 18.54 Mb loss of heterozygosity at 9p24.3p22.1, arr [GRCh37] 9p24.3p22.1 (216,123_18,758,836) × 2 hmz (Figure 1B). As there was no copy number variation (CNV) in the CMA results, and the prenatal ultrasound findings at 23+3 weeks of gestation were unremarkable, further invasive prenatal diagnosis by percutaneous umbilical blood sampling was performed at 27 weeks of gestation to confirm the trisomy 9 mosaicism. Karyotype analysis of umbilical blood identified only 1 cell with an abnormal 47,XX,+9 karyotype among 100 metaphase cells. According to the International System for Human Cytogenomic Nomenclature (McGowan-Jordanet al., 2020) guidelines and recommendations (McGowan-Jordanet al., 2020), this karyotype should be normally reported as 46,XX. Interphase fluorescent in situ hybridization (FISH) analysis on uncultured fetal cord blood showed 22.4% (35/156) trisomy 9 mosaicism (Figure 1C). CMA analysis on the DNA extracted from uncultured umbilical blood detected a gene dosage increase of chromosome 9 and loss of heterozygosity for a large fragment of chromosome 9: arr [GRCh37] 9p24.3q34.3 (208,454_141,018,648) × 3 [0.34]; arr [GRCh37] 9p24.3p22.1 (216,123_18,782,021) × 2 hmz (Figure 1D). Ultrasound findings at 28+4 weeks of gestation showed no structural abnormalities except for those slightly smaller than the gestational age, with a biparietal diameter of 7.8 cm (97.5th), a head circumference of 26.1 cm (14.7th), an abdominal circumference of 21.8 cm (2nd), a humerus length of 4.7 cm (14.7th), a femur length of 5.2 cm (14.7th), and a fetal weight assessment of 1040 g (5.5th). The gravida chose to terminate the pregnancy at 29+4 weeks of gestation.
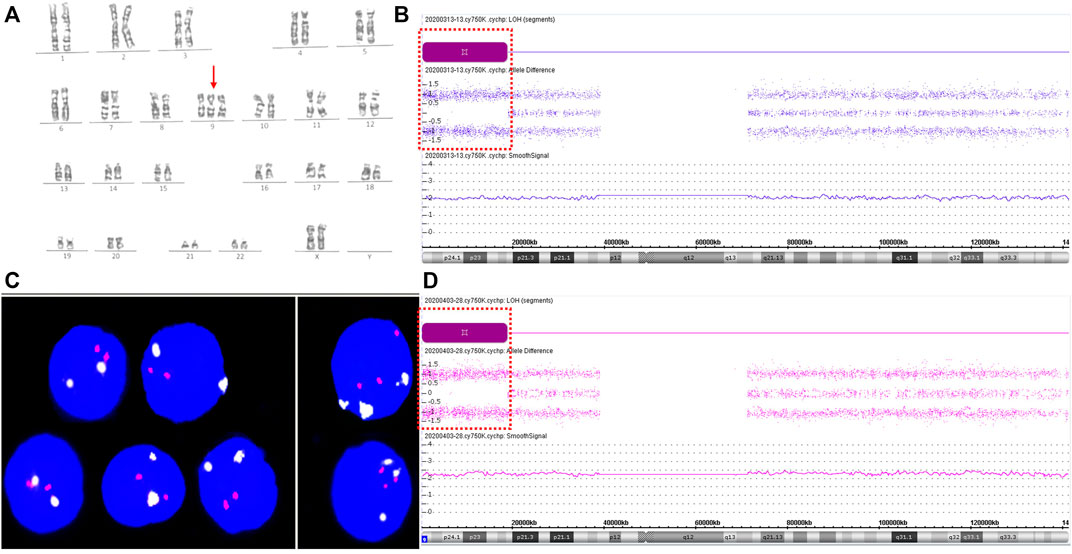
FIGURE 1. Results for trisomy 9 mosaicism in Case1. (A) Karyotype profiles indicating trisomy 9 (red arrows indicate the chromosome 9). (B) CMA assay results of the uncultured amniotic fluid sample for Case 1. Loss-of-heterozygosity (LOH) at 9p24.3p22.1 detected by CMA (positions of LOH are indicated by the dashed boxes). (C) FISH profiles of 22.4% trisomy 9 mosaicism in uncultured interphase umbilical blood cells using chromosome 9 centromere-specific (white) and 9qter telomeres-specific probe (red). (D) CMA assay results of uncultured umbilical blood sample for Case 1. LOH at 9p24.3p22.1 detected by CMA (positions of LOH are indicated by the dashed boxes). SmoothSignal showed 34% trisomy 9 mosaicism (x-axis: chromosomes 9; y-axis: copy number).
A subsequent autopsy of the aborted fetus confirmed the prenatal ultrasound finding of no structural anomalies. The copy number variation sequencing (CNV-seq) (NextSeq CN500 platform, Berry Genomics, Beijing, China) analysis of the maternal and fetal center of the placenta verified trisomy 9 with the level of 80% and 81% mosaicism respectively. The levels of trisomy 9 mosaicism were 6% in the DNA of uncultured amniocytes following CMA testing, 23% in uncultured umbilical blood, and 21% in the umbilical cord (Supplementary Figure S1). The trisomy 9 mosaicism levels within various tissues were 60% in the uterus, 35% in the lung, 20% in the kidney, 15% in the skeletal muscle, 15% in the small intestine, 11% in the ovaries, and 10% in the heart. No trisomy 9 mosaicism was detected in the skin, thymus, adrenal glands, and brain (Supplementary Figure S2).
Case 2
A 31-year-old woman (gravid 2, para 0) was referred to the Hunan Provincial Maternal and Child Health Care Hospital because of the high risk of NIPT results. NIPT using the MGISEQ-2000 platform (BGI, Shenzhen, China) at 14 gestational weeks showed that the Z-score of chromosome 9 was outside the normal range (14.8715, 11.931% cell-free fetal DNA fraction), suggesting a high risk of trisomy 9. There was no signs of miscarriage during early pregnancy, and all common laboratory parameters were within normal reference ranges. The gravida underwent amniocentesis at 18 weeks of gestation. Genetic amniocentesis analysis revealed the existence of trisomy 9 mosaicism, mos 47,XY,+9 [21]/46,XY [29] with a 42% level of trisomy 9 mosaicism (Figure 2A). Parental karyotypes of peripheral blood were normal. CMA of uncultured amniocytes with the Affymetrix CytoScan®750 K Array showed no CNV on chromosome 9 except for 24.96 Mb and 19.22 Mb loss of heterozygosity at 9p23p13.1 and 9q33.1q34.3: arr [GRCh37] 9p23p13.1 (13,814,169_38,771,831) × 2 hmz and arr [GRCh37] 9q33.1q34.3 (121,790,571_141,011,581) × 2 hmz (Figure 2B). Prenatal ultrasound at 20+5 weeks of gestation showed no structural anomalies except for a slightly wider bilateral renal pelvis (0.5–0.52 cm for the left side and 0.6–0.63 cm for the right side). The pregnancy was terminated at 21+5 weeks of gestation upon the request of the parents.
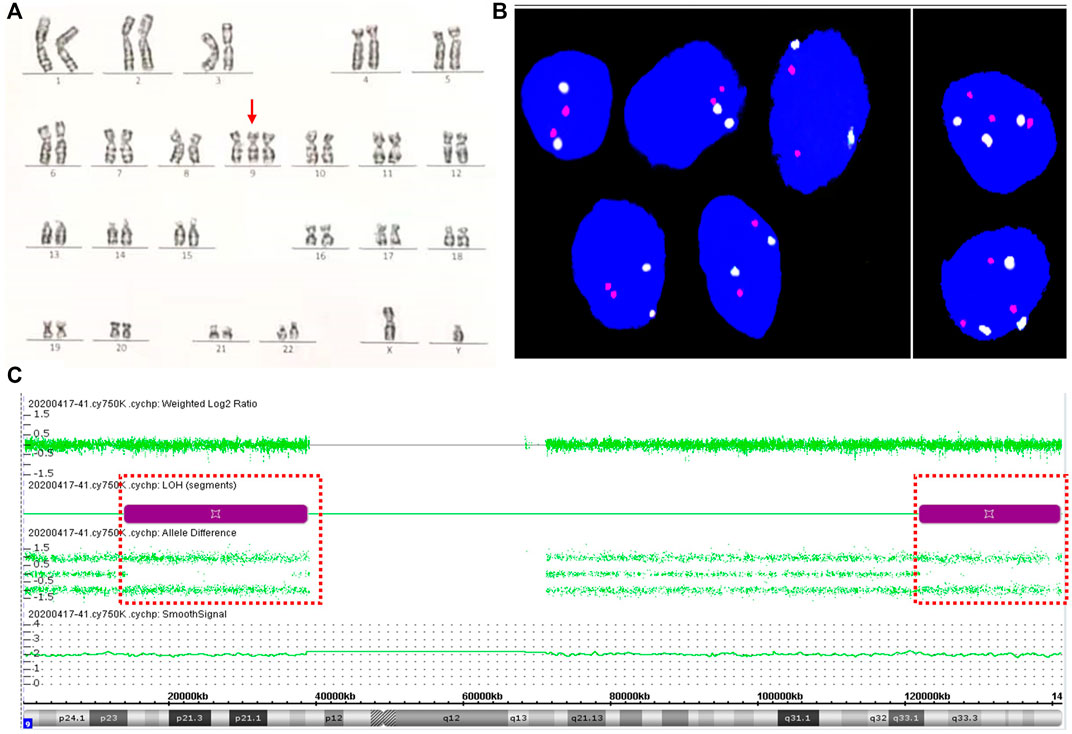
FIGURE 2. Results for trisomy 9 mosaicism in Case 2. (A) Karyotype profiles indicating trisomy 9 (red arrows indicated the chromosome 9). (B) FISH profiles of 8.2% trisomy 9 mosaicism in interphase uncultured oral mucosal cells using chromosome 9 centromere-specific (white) and 9qter telomeres-specific probe (red). (C) CMA assay results of uncultured amniocytes for Case 2. LOH at 9p23p13.1 and 9q33.1q34.3 detected by CMA (positions of LOH are indicated by the dashed boxes).
Upon approval from the parents, an autopsy of the aborted fetus was performed, and the result showed no significant structural anomalies. FISH analysis of uncultured oral mucosal cells with chromosome 9p-ter and 9q-ter FISH probes detected trisomy 9 in 16 (8.2%) of 196 cells examined (Figure 2C). CNV-seq analysis of the maternal and fetal center of the placenta confirmed placental mosaicism with chromosome 9 of 2.76 triploid equivalents (Supplementary Figure S3). CNV-seq of the multiple tissue samples of the aborted fetus confirmed true fetal mosaicism. The levels of trisomy 9 mosaicism within various tissues were variable: 10% in the cord portion close to the fetus, 15% in the cord portion close to the placenta (Supplementary Figure S3), 20% in the heart, 12% in the skeletal muscle, 10% in the brain, 8% in the stomach, and 6% in the adrenal glands. Trisomy 9 was not detected in cutaneous cells, small intestine, kidney, and the DNA of uncultured amniocytes following CMA testing (Supplementary Figure S4).
Discussion
Owing to the advantage of NIPT in detecting genome-wide chromosomal anomalies (Bianchi and Wilkins-Haug., 2014), more studies have investigated the use of NIPT in detecting rare autosomal trisomies, including trisomy 9 (Lee et al., 2018; Wang et al., 2020; Li et al., 2022). However the positive predictive value of NIPT for rare autosomal trisomies is low (4%–6%) in the general obstetrical population (van der Meij et al., 2019; Van Den Bogaert et al., 2021), and approximately 40% of all rare autosomal trisomy cases culminate in adverse perinatal outcomes (Xiang et al., 2023). Since most of the circulating fetal DNA in maternal plasma is derived primarily from the placental trophoblasts (Flori et al., 2004), and trisomy 9 is usually miscarried in the first trimester (López-Félix et al., 2017), invasive prenatal testing is strongly recommended for all gravidas with positive NIPT results of trisomy 9 to exclude placental or fetal mosaicism.
Different genetic techniques have respective advantages in detecting mosaicism, and attention should be paid to excluding culture artifacts in detecting the mosaicism of cultured samples. For the flask method of karyotyping, we followed the current international practice and analyzed at least 50 cells from a minimum of two different culture vessels. Cytogenetic abnormalities confined to two or more flasks are considered level III true mosaicism (Hsu and Benn., 1999). Nevertheless, with the developments in molecular genetics, more studies have demonstrated inconsistent mosaic ratios between the results of cultured karyotype and uncultured CMA or FISH (Chen et al., 2016, Chen et al., 2012, Ma et al., 2015). Inconsistent results between CNV-seq/CMA (uncultured samples) and karyotyping (cultured samples) in the prenatal diagnosis of mosaic trisomy 9 may be attributed to the variable proliferation of cells with different karyotypes under in vitro cell culture, which is especially common in prenatal diagnosis of mosaic trisomy 9 (Chen et al., 2010; Tang et al., 2019). The growth advantages of trisomy 9 amniotic fluid cells is entirely useless in cord blood cell culture in our cases as previously reported (Miryounesi et al., 2016), which may be the reason why trisomy 9 is less frequently diagnosed in children by karyotyping (Li et al., 2021). Therefore, attention should be paid to culture artifacts and technical limitations of karyotyping in prenatal diagnosis of mosaicism. Besides, umbilical blood sampling for rapid confirmation of trisomy 9 mosaicism by karyotyping is not practical and extensive use of multiple techniques is strongly suggested in prenatal diagnosis of mosaicism suggested by NIPT.
The clinical significance of uniparental disomy (UPD) lies in its ability of producing either aberrant patterns of imprinting or homozygosity for recessive mutations. GLIS3 (MIM: 610199) is only paternally imprinted gene on chromosome 9, which is implicated in neonatal diabetes and pancreatic development by autosomal recessive inheritance instead of imprinting (Yang et al., 2011; Kang et al., 2009). According to the molecular results performed in the present cases, the large block(s) of homozygosity of chromosome 9 detected by SNP-array in both amniocytes and umbilical blood is most probably the result of postzygotic trisomy rescue combined with mitotic recombination. Among the 5135 cases included in ChromosOmics Database (Liehr, 2023, https://cs-tl.de/DB/CA/UPD/0-Start.html), only 53 cases have been referred to chromosome 9 and limited prenatal clinical significance of UPD 9 has been reported in the literature (Slater et al., 2000; Chen et al., 2022, Chen et al., 2010). Besides, the UPDs of most cases were reported after the discovery of trisomy 9 mosaicism at cytogenetic prenatal diagnosis (Liehr, 2023, https://cs-tl.de/DB/CA/UPD/0-Start.html), which makes the clinical phenotype analysis of UPD 9 more difficult.
Chromosomal mosaicism presents a major interpretative dilemma in prenatal genetic counseling. Genetic counseling for clinical outcomes of chromosomal mosaicism in pregnant women needs to be assessed on the case-by-case basis and comprehensively consider multiple factors, including the timing of the initial event, gene-phenotype associations of referred chromosome, the ratio and distribution of the normal/abnormal cells in tissues. The placental findings from NIPT and the trisomic rescue observed by SNP-array and karyotyping, suggest that our cases are fetoplacental trisomy 9 mosaicism, which was further confirmed by CNV-seq testing of the aborted placental and fetal tissues. Prenatal clinical features of mosaic trisomy 9 are often complicated with intrauterine growth retardation and/or “small” size, which is not specific. Live births with trisomy 9 mosaicism may present with characteristic phenotypic features, such as craniofacial abnormalities (small palpebral fissures, bulbous nose, micrognathia, abnormal ears, scoliosis, low-set ears and micrognathia), cardiac abnormalities, feeding (gastroesophageal reflux) and breathing difficulties, cryptorchidism, hip dysplasia, seizures, and developmental delay (Li et al., 2021; Bruns and Campbell. 2015). The incidence and severity of malformations and intellectual disability correlate with the percentage of trisomic cells in different tissues (Lee et al., 2018), while the types of tissue sources that can be obtained for prenatal diagnosis are limited. Furthermore, the fetus in this report exhibited no structural abnormality in prenatal ultrasound, suggesting that fetal organs with trisomy 9 mosaicism may not always present with abnormal clinical manifestations during the prenatal period. Therefore, genetic counseling for evaluating the likely phenotype of mosaic trisomy 9 in prenatal diagnosis is very difficult and challenging.
In conclusion, the findings in the present study suggest that attention should be paid to the possibility of mosaicism and placental mosaicism in gravidas with positive NIPT results of trisomy 9, and the comprehensive use of multiple genetic techniques and biological samples is strongly suggested for the diagnosis of trisomy 9 mosaicism. Genetic counseling for clinical outcomes of trisomy 9 mosaicism during pregnancy should be provided on the case-by-case basis by comprehensive consideration of the prenatal results and the fact that true fetal trisomy 9 mosaicism can be free of structural anomalies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The study was approved by the Ethics Committee of Hunan Provincial Maternal and Child Health Care Hospital (EC20190101). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
Author contributions
NM, ZZ, YP, and HX had major roles in the design of the study. NM and HX drafted the manuscript. ZZ and HK performed the fetal autopsy examination. NM, JH, and JP performed the molecular genetic experiments and analyzed the data. SY, JL, and HW analyzed the clinical data. JC, WT, and ZL performed data collection and follow-up outcomes. NM and RH performed the cytogenetic experiments and analyzed the data. YP and HX are corresponding authors of this manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Science and Technology Innovation Program of Hunan Province (2021SK50609, 2019SK1014), the National Key R&D Program of China (2019YFC1005100, 2021YFC1005300).
Acknowledgments
We thank all the families for their contributions. We also thank the laboratory staff at the Prenatal Diagnosis Center of Hunan Province, Hunan Provincial Maternal and Child Health Care Hospital, and the staff of Berry Genomics Corporation, Wang, who has offered professional help of language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1121121/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | CNV-seq profiles of Case 1 for multiple samplings showed various levels of trisomy 9. CNV-seq profiles are shown for uncultured amniotic fluid, maternal center of the placenta, fetal center of the placenta, umbilical cord, and umbilical blood of Case 1 with 6%, 81%, 80%, 21% and 23% trisomy 9 mosaicism, respectively. The blue line represents the mean copy number and the black box represents the centromere (x-axis: chromosomes 9; y-axis: copy number).
SUPPLEMENTARY FIGURE S2 | CNV-seq profiles of Case 1 for multiple samplings showed various levels of trisomy 9. CNV-seq profiles are shown for uterus, lung, kidney, heart and skin of Case1 with 60%, 35%, 20% and 10% trisomy 9 mosaicism and normal result, respectively. The blue line represents the mean copy number and the black box represents the centromere (x-axis: chromosomes 9; y-axis: copy number).
SUPPLEMENTARY FIGURE S3 | CNV-seq profiles of Case 2 for multiple samplings showed various levels of trisomy 9. CNV-seq profiles are shown for uncultured amniotic fluid, maternal center of the placenta, fetal center of the placenta, cord portion close to the placenta and cord portion close to the fetus of Case 2 with normal result, 76%, 76%, 15% and 10% trisomy 9 mosaicism, respectively. The blue line represents the mean copy number and the black box represents the centromere (x-axis: chromosomes 9; y-axis: copy number).
SUPPLEMENTARY FIGURE S4 | CNV-seq profiles of Case 2 for multiple samplings showed various levels of trisomy 9. CNV-Seq profiles are shown for the heart, brain, stomach, adrenal glands, and skin of Case 2 with 20%, 10%, 8%, 6% trisomy 9 mosaicism and normal result, respectively. The blue line represents the mean copy number and the black box represents the centromere (x-axis: chromosomes 9; y-axis: copy number).
References
Benn, P., and Grati, F. R. (2021). Aneuploidy in first trimester chorionic villi and spontaneous abortions: Windows into the origin and fate of aneuploidy through embryonic and fetal development. Prenat. Diagn 41 (5), 519–524. doi:10.1002/pd.5795
Bianchi, D. W., and Wilkins-Haug, L. (2014). Integration of noninvasive DNA testing for aneuploidy into prenatal care: What has happened since the rubber met the road? Clin. Chem. 60 (1), 78–87. doi:10.1373/clinchem.2013.202663
Bruns, D. A., and Campbell, E. (2015). Twenty-five additional cases of trisomy 9 mosaic: Birth information, medical conditions, and developmental status. Am. J. Med. Genet. A 167A(5), 997–1007. doi:10.1002/ajmg.a.36977
Cantú, E. S., Eicher, D. J., Pai, G. S., Donahue, C. J., and Harley, R. A. (1996). Mosaic vs. Nonmosaic trisomy 9: Report of a liveborn infant evaluated by fluorescence in situ hybridization and review of the literature. Am. J. Med. Genet. 62 (4), 330–335. doi:10.1002/(SICI)1096-8628(19960424)62:4<330:AID-AJMG1>3.0.CO;2-V
Chen, C. P., Chern, S. R., Wu, P. S., Chen, S. W., Wu, F. T., Chen, L. F., et al. (2022). Detection of maternal uniparental disomy 9 in association with low-level mosaic trisomy 9 at amniocentesis in a pregnancy associated with intrauterine growth restriction, abnormal first-trimester screening result (low PAPP-A and low PlGF), maternal preeclampsia and a favorable outcome. Taiwan J. Obstet. Gynecol. 61 (1), 141–145. doi:10.1016/j.tjog.2021.11.024
Chen, C. P., Lin, H. M., Su, Y. N., Chern, S. R., Tsai, F. J., Wu, P. C., et al. (2010). Mosaic trisomy 9 at amniocentesis: Prenatal diagnosis and molecular genetic analyses. Taiwan J. Obstet. Gynecol. 49 (3), 341–350. doi:10.1016/S1028-4559(10)60071-X
Chen, C. P., Su, Y. N., Chern, S. R., Chen, Y. T., Wu, P. S., Su, J. W., et al. (2012). Mosaic trisomy 2 at amniocentesis: Prenatal diagnosis and molecular genetic analysis. Taiwan J. Obstet. Gynecol. 51 (4), 603–611. doi:10.1016/j.tjog.2012.09.016
Chen, C. P., Wang, L. K., Chern, S. R., Chen, Y. N., Chen, S. W., Wu, P. S., et al. (2016). Mosaic trisomy 17 at amniocentesis: Prenatal diagnosis, molecular genetic analysis, and literature review. Taiwan J. Obstet. Gynecol. 55 (5), 712–717. doi:10.1016/j.tjog.2016.07.006
Ferreres, J. C., Planas, S., Martínez-Sáez, E. A., Vendrell, T., Peg, V., Salcedo, M. T., et al. (2008). Pathological findings in the complete trisomy 9 syndrome: Three case reports and review of the literature. Pediatr. Dev. Pathol. 11 (1), 23–29. doi:10.2350/06-08-0143.1
Flori, E., Doray, B., Gautier, E., Kohler, M., Ernault, P., Flori, J., et al. (2004). Circulating cell-free fetal DNA in maternal serum appears to originate from cyto- and syncytio-trophoblastic cells. Case report. Hum. Reprod. 19 (3), 723–724. doi:10.1093/humrep/deh117
Hsu, L. Y., and Benn, P. A. (1999). Revised guidelines for the diagnosis of mosaicism in amniocytes. Prenat. Diagn 19 (11), 1081–1090. doi:10.1002/(sici)1097-0223(199911)19:11<1081:aid-pd682>3.0.co;2-z
Kang, H. S., Kim, Y. S., ZeRuth, G., Beak, J. Y., Gerrish, K., Kilic, G., et al. (2009). Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol. Cell. Biol. 29 (24), 6366–6379. doi:10.1128/MCB.01259-09
Lee, C. Y., Su, H. J., Cheng, Y. T., Ku, Y. L., Ngo, Y. G., Chen, C. M., et al. (2018). Detection of fetal trisomy 9 mosaicism by noninvasive prenatal testing through maternal plasma DNA sequencing. Taiwan J. Obstet. Gynecol. 57(4), 594–597. doi:10.1016/j.tjog.2018.06.021
Li, H., Lu, L., Yao, Y., Gao, T., Jiang, Y., Zhang, C., et al. (2022). Perinatal outcomes of prenatal cases testing positive for trisomy 9 by noninvasive prenatal testing. Taiwan J. Obstet. Gynecol. 61 (6), 965–970. doi:10.1016/j.tjog.2022.07.006
Li, M., Glass, J., Du, X., Dubbs, H., Harr, M. H., Falk, M., et al. (2021). Trisomy 9 mosaic syndrome: Sixteen additional patients with new and/or less commonly reported features, literature review, and suggested clinical guidelines. Am. J. Med. Genet. A 185 (8), 2374–2383. doi:10.1002/ajmg.a.62251
Liehr, T.(2009). Cases with uniparental disomy. [DATASET]. Available at: https://cs-tl.de/DB/CA/UPD/0-Start.html.
López-Félix, J., Flores-Gallegos, L., Garduño-Zarazúa, L., Leis-Márquez, T., Juárez-García, L., Meléndez-Hernández, R., et al. (2017). Partial trisomy 9: Prenatal diagnosis and recurrence within same family. Clin. Case Rep. 5 (6), 986–992. doi:10.1002/ccr3.970
Ma, J., Cram, D. S., Zhang, J., Shang, L., Yang, H., and Pan, H. (2015). Birth of a child with trisomy 9 mosaicism syndrome associated with paternal isodisomy 9: Case of a positive noninvasive prenatal test result unconfirmed by invasive prenatal diagnosis. Mol. Cytogenet. 8, 44. doi:10.1186/s13039-015-0145-4
McGowan-Jordan, J., Hastings, R. J., and Moore, S. (2020). ISCN 2020: An International System for Human Cytogenomic Nomenclature. Basel, Switzerland: Karger.
Miryounesi, M., Dianatpour, M., Shadmani, Z., and Ghafouri-Fard, S. (2016). Report of a case with trisomy 9 mosaicism. Iran. J. Med. Sci. 41 (3), 249–252.
Slater, H. R., Ralph, A., Daniel, A., Worthington, S., and Roberts, C. (2000). A case of maternal uniparental disomy of chromosome 9 diagnosed prenatally and the related problem of residual trisomy. Prenat. Diagn 20 (11), 930–932. doi:10.1002/1097-0223(200011)20:11<930:aid-pd955>3.0.co;2-e
Tang, H. S., Wang, D. G., Huang, L. Y., and Li, D. Z. (2019). Chromosomal microarray analysis detects trisomy 9 mosaicism in a prenatal case not revealed by conventional cytogenetic analysis of cord blood. J. Obstet. Gynaecol. 39 (1), 123–125. doi:10.1080/01443615.2018.1439905
Van Den Bogaert, K., Lannoo, L., Brison, N., Gatinois, V., Baetens, M., Blaumeiser, B., et al. (2021). Outcome of publicly funded nationwide first-tier noninvasive prenatal screening. Genet. Med. 23 (6), 1137–1142. doi:10.1038/s41436-021-01101-4
van der Meij, K., Sistermans, E. A., Macville, M., Stevens, S., Bax, C. J., Bekker, M. N., et al. (2019). TRIDENT-2: National implementation of genome-wide non-invasive prenatal testing as a First-Tier screening test in The Netherlands. Am. J. Hum. Genet. 105 (6), 1091–1101. doi:10.1016/j.ajhg.2019.10.005
Wang, C., Chen, Y., Zhao, J., and Xia, Y. (2020). Prenatal diagnosis and genetic counseling of low-level trisomy 9 mosaicism with a favorable outcome. Taiwan J. Obstet. Gynecol. 59 (5), 786–787. doi:10.1016/j.tjog.2020.07.032
Xiang, J., Li, R., He, J., Wang, X., Yao, L., Song, N., et al. (2023). Clinical impacts of genome-wide noninvasive prenatal testing for rare autosomal trisomy. Am. J. Obstet. Gynecol. MFM. 5(1), 100790. doi:10.1016/j.ajogmf.2022.100790
Keywords: karyotype, copy number variation sequencing, non-invasive prenatal testing, chromosomal microarray analysis, mosaicism, trisomy 9
Citation: Ma N, Zhu Z, Hu J, Pang J, Yang S, Liu J, Chen J, Tang W, Kuang H, Hu R, Li Z, Wang H, Peng Y and Xi H (2023) Case report: Detection of fetal trisomy 9 mosaicism by multiple genetic testing methods: Report of two cases. Front. Genet. 14:1121121. doi: 10.3389/fgene.2023.1121121
Received: 13 December 2022; Accepted: 27 February 2023;
Published: 10 March 2023.
Edited by:
Bettina Blaumeiser, University of Antwerp, BelgiumReviewed by:
Thomas Liehr, Friedrich Schiller University Jena, GermanyLinda Randolph, Children’s Hospital of Los Angeles, United States
Copyright © 2023 Ma, Zhu, Hu, Pang, Yang, Liu, Chen, Tang, Kuang, Hu, Li, Wang, Peng and Xi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Peng, cGVuZ3lpbmdweUBob3RtYWlsLmNvbQ==; Hui Xi, NTQ1NDg2NjBAcXEuY29t
†These authors have contributed equally to this work
 Na Ma
Na Ma Zhenhua Zhu2
Zhenhua Zhu2 Jing Liu
Jing Liu Zhuo Li
Zhuo Li Ying Peng
Ying Peng Hui Xi
Hui Xi