- 1The Suqian Clinical College of Xuzhou Medical University, Suqian, China
- 2Department of Clinical Laboratory, The Affiliated Suqian First People’s Hospital of Nanjing Medical University, Suqian, China
- 3Zhejiang University Kunshan Biotechnology Laboratory, Zhejiang University Kunshan Innovation Institute, Kunshan, China
- 4Department of R&D, Suzhou VersaBio Technologies Co Ltd., Kunshan, China
- 5Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou, China
Background: Esophageal cancer (EC) is a leading cause of cancer-related deaths in China, with the 5-year survival rate reaching less than 30%, because most cases were diagnosed and treated at the advanced stage. However, there is still a lack of low-cost, efficient, and accurate non-invasive methods for the early detection of EC at present.
Methods: A total of 48 EC plasma and 101 control plasma samples were collected in a training cohort from 1 January 2021 to 31 December 2021, and seven cancer-related DNA methylation markers (ELMO1, ZNF582, FAM19A4, PAX1, C13orf18, JAM3 and TERT) were tested in these samples to select potential markers. In total, 20 EC, 10 gastric cancer (GC), 10 colorectal cancer (CRC), and 20 control plasma samples were collected in a validation cohort to evaluate the two-gene panel.
Results: ZNF582, FAM19A4, JAM3, or TERT methylation in plasma was shown to significantly distinguish EC and control subjects (p < 0.05), and the combination of ZNF582 and FAM19A4 methylation was the two-gene panel that exhibited the best performance for the detection of EC with 60.4% sensitivity (95% CI: 45.3%–73.9%) and 83.2% specificity (95% CI: 74.1%–89.6%) in the training cohort. The performance of this two-gene panel showed no significant difference between different age and gender groups. When the two-gene panel was combined with CEA, the sensitivity for EC detection was further improved to 71.1%. In the validation cohort, the sensitivity of the two-gene panel for detecting EC, GC, and CRC was 60.0%, 30.0%, and 30.0%, respectively, with a specificity of 90.0%.
Conclusion: The identified methylation marker panel provided a potential non-invasive strategy for EC detection, but further validation should be performed in more clinical centers.
1 Introduction
Esophageal cancer (EC) is one of the most common malignant tumors worldwide, with approximately 604,100 new cases and 544,076 deaths in 2020 (Sung et al., 2021). In China, EC ranked fifth in incidence and fourth in mortality among all cancer types according to the latest nationwide statistics (Zheng et al., 2022). China is the country with the heaviest burden of EC in the world. The data from the World Health Organization indicated that EC led to approximately 324,000 new cases and 301,000 deaths in China in 2020, accounting for 53.70% and 55.35% of the global incidence and mortality of EC, respectively (He et al., 2022). Although the incidence rate and 5-year survival rate have showed a trend of gradual improvement in the last decade (Li et al., 2021; He et al., 2022), the 5-year survival rate still remained below 30% (He et al., 2020; Salta et al., 2020) likely because most EC cases were diagnosed and treated at the advanced stage (Early Diagnosis and Treatment Group of the Chinese Medical Association Oncology Branch, 2022). However, if EC can be diagnosed at an early stage, the 5-year survival rate can reach approximately 90% (Early Diagnosis and Treatment Group of the Chinese Medical Association Oncology Branch, 2022). Currently, the gold standard for EC screening is white light endoscopy, which is limited due to its high cost and invasiveness with a low acceptance rate in the Chinese population (Li et al., 2021). Therefore, a new, accurate, and non-invasive early detection method is urgently needed to reduce the risk of EC.
Blood is a widely used clinical sample type, which is easy to collect and process with high throughput. Numerous blood-based new biomarkers for early cancer detection are being studied, and some of them have been applied in clinical practice (Rosado et al., 2019). However, the traditional blood tumor markers, such as CEA and SCC-Ag, lack sufficient sensitivity and specificity, due to which their clinical application has been incompetent (Miyoshi et al., 2022). DNA methylation is an epigenetic modification associated with cell proliferation, apoptosis, and differentiation (Kulis and Esteller, 2010; Pan et al., 2018), and the changes in DNA methylation in cancer have been extensively studied and successfully developed as a powerful diagnostic tool for the early detection of cancer (Koch et al., 2018). Compared with other biomarkers, DNA methylation is a more stable and specific biomarker in blood (Chen et al., 2021; Jamshidi et al., 2022), which can significantly improve the sensitivity in early-stage cancer if multiple DNA methylation markers combined together (Cao et al., 2020; Chen et al., 2021). Our group has also developed several blood-based DNA methylation tests for the early detection of colorectal cancer (Zhao et al., 2019; Zhao et al., 2020) and gastric cancer (Miao et al., 2020), suggesting that blood-based DNA methylation markers may also be applied for the early detection of EC.
In this study, we selected seven cancer-related DNA methylation markers: engulfment and cell motility 1 (ELMO1), zinc finger protein 582 (ZNF582), family with sequence similarity 19 member A4 (FAM19A4), paired box 1 (PAX1), chromosome 13 open reading frame 18 (C13orf18), junctional adhesion molecule 3 (JAM3), and telomerase reverse transcriptase (TERT). Among them, ZNF582, PAX1, and ELMO1 have been found highly methylated in EC tissues but lack validation in plasma samples (Hu et al., 2017; Qin et al., 2019), while the methylation of FAM19A4, C13orf18, JAM3, and TERT has been reported to be associated with squamous cell carcinoma (De Strooper et al., 2016). As 90% of EC cases are esophageal squamous cell carcinoma (Henry et al., 2014), these squamous cell carcinoma-related markers have the potential to serve as early diagnostic markers for EC. Similarly, the performance of most of these markers in the plasma of EC patients has not been reported yet. Based on these principles, we evaluated their performance for EC detection in plasma samples to identify the combination of DNA methylation markers showing the best performance.
2 Materials and methods
2.1 Sample collection
From 1 January 2021 to 31 December 2021, 59 EC plasma and 102 control plasma samples were enrolled in the training cohort, and from 1 January 2023 to 31 August 2023, 20 EC, 10 gastric cancer (GC), 10 colorectal cancer (CRC), and 20 control plasma samples were collected in the validation cohort. All participants were examined via endoscopy. EC patients were confirmed by pathological diagnoses, and the control subjects were people who underwent physical examination with no evidence of disease. The inclusion criteria for all participants were as follows: patients with the age of 18 years and above, no history of esophageal cancer, no pregnancy, having undergone complete endoscopy, and the results of EC patients were confirmed via pathology. The exclusion criteria for all samples were as follows: hemolysis, insufficient plasma volume, and insufficient cell-free DNA determined using an internal control (ACTB). The blood sample was collected from each subject using a 4-mL K2EDTA tube and stored at room temperature (20°C ± 5°C) for no more than 24 h. The blood sample was centrifuged two times at 1,350 ± 100 g to separate plasma, and the obtained plasma was stored at −80°C for long-term storage. Finally, 11 EC plasma and one control plasma samples were excluded according to the aforementioned criteria, and 48 EC and 101 control subjects were included for further analysis.
2.2 cfDNA extraction and bisulfite treatment
Cell-free DNA (cfDNA) from 0.5–1.0 mL plasma of EC patients and control subjects was purified and bisulfite-treated with Blood Sample Pretreatment Kit for DNA Methylation Detection (Suzhou VersaBio Technologies Co., Ltd., Kunshan, China). The purification and conversion steps were performed according to Miao et al. (2020). Briefly, 1.0 mL lysis buffer and 20 μL of protease K solution were added to each plasma sample and incubated at room temperature for 10 min. Subsequently, 750 μL of ethanol and 20 μL of magnetic bead solution were added to each sample and allowed to incubate at room temperature for 25 min. Following this, 100 μL of elution buffer was introduced to obtain the cfDNA solution. Afterward, 100 μL cfDNA solution, 150 μL of conversion buffer, and 25 μL of protectant were added, and the mixture was incubated at 80°C for 45 min. Then, 1 mL of wash buffer A and 20 μL of magnetic beads were added, followed by a 25-min incubation period. After washing two times in wash buffer B, the purified and converted product was eluted in 100 μL of elution buffer.
2.3 Quantitative methylation-specific PCR
The methylation of ELMO1, ZNF582, FAM19A4, PAX1, C13orf18, JAM3, and TERT in plasma was analyzed using quantitative methylation-specific PCR (qMSP) assay kits from Suzhou VersaBio Technologies Co., Ltd. (Kunshan, China). The qMSP assay for each methylation marker contained two methylation-specific primers and one methylation-specific probe. The qMSP reaction was performed in a total reaction volume of 30 μL, including 15 μL DNA and 15 μL pre-master mix on an ABI 7500 instrument (Applied Biosystems, Foster City, CA, United States). The concentration of each primer was 0.4 μM, and the concentration of each probe was 0.2 μM. The qMSP reaction was initially denatured at 95°C for 20–30 min, followed by 50 cycles at 95°C for 10 s, 56°C–58°C for 30 s, and 72°C for 0–15 s, and a final cooling to 40°C for 30 s. For each qMSP reaction, an internal control, ACTB, was simultaneously detected in the same tube to monitor the reaction and normalize the methylation level of each marker. Positive and negative controls were run in parallel with samples each time. The sequences of primers and probes used in this study are listed in Supplementary Table S1. Samples were analyzed using Applied Biosystems Real-Time PCR Software v2.4.
2.4 Blood CEA level measurement
CEA levels were measured using a Roche Cobas 8000 electrochemiluminescence instrument at the Department of Clinical Laboratory of The Affiliated Suqian First People’s Hospital of Nanjing Medical University. The normal reference value was CEA≤5 ng/mL.
2.5 Data analysis
The cfDNA concentration was considered insufficient if the Ct value of ACTB was more than 40.0; thus, the reaction was considered “valid.” Receiver operating characteristic (ROC) curves were plotted with Ct values or ∆Ct values, and the area under the curve (AUC) values were calculated. Ct values were set to the maximal PCR cycle numbers of 50 for samples with no amplification signals in the qMSP reaction (Supplementary Tables S2, S3) (He et al., 2020). A ∆Ct value was defined as the difference between the Ct values of the methylation marker and ACTB. A multinomial logistic regression was performed to calculate the probability, which was used as the test variable to run an ROC curve for two or three biomarker combination. GraphPad Prism 8.0 was used for all statistical analyses, Pearson’s chi-squared test for sensitivity comparison among groups, and the Mann–Whitney U-test for the differences in methylation levels.
3 Results
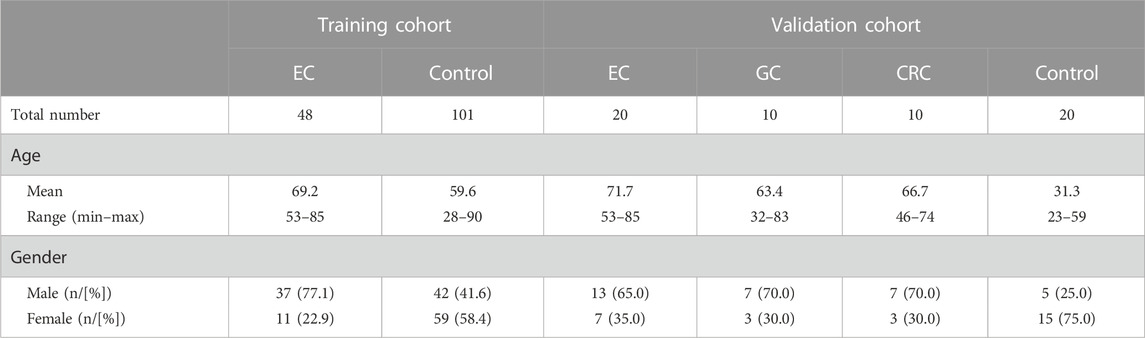
This study included 48 qualified EC samples and 101 qualified control subjects in the training cohort for data analysis (Table 1). The age of EC patients ranged from 53 to 85 years, with a mean of 69.2. The percentage of male patients was 77.1% in the EC group. In the control group, the age of the subjects ranged from 28 to 90 years, with a mean of 59.6, and 41.6% were males (Table 1). In the validation cohort, the mean age for EC, GC, CRC, and control subjects was 71.7, 63.4, 66.7, and 31.3, respectively. The proportion of male patients for EC, GC, CRC, and control subjects was 65.0%, 70.0%, 70.0, and 25.0%, respectively (Table 1).
To identify potential DNA methylation markers for EC, the Ct values and △Ct values for each marker were used to plot ROC curves, and potential markers were selected based on AUC values and p-values. As shown in Table 2, for ROC curves plotted with Ct values in the training cohort, only ZNF582 could significantly discriminate EC from control subjects, with an AUC value of 0.660. If ROC curves were plotted with △Ct values, ZNF582, FAM19A4, JAM3, and TERT all showed a significant difference between EC and control subjects, and all of their AUC values were greater than or equal to 0.600. Particularly for ZNF582, the AUC value obtained with △Ct values was larger than that obtained with Ct values.
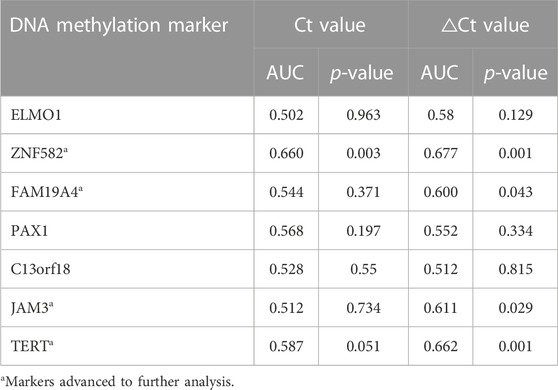
TABLE 2. Discrimination of each DNA methylation marker in the training cohort for esophageal cancer detection in plasma samples.
Each DNA methylation marker was further evaluated for its ability to discriminate between EC and control plasma samples in the training cohort with both Ct and △Ct values as they represent absolute and normalized methylation levels, respectively. Only the Ct values of ZNF582 and TERT showed a significant difference between the EC and control groups (Figures 1C,M). In comparison, the △Ct values of ZNF582, FAM19A4, JAM3, and TERT displayed significant differences between the EC and control groups (Figures 1D,F,L,M). Comprehensive analysis of the aforementioned results showed the same trend between AUC values and methylation levels. Therefore, the Ct value of ZNF582 and △Ct values of ZNF582, FAM19A4, JAM3, and TERT were chosen for combination biomarker analysis.
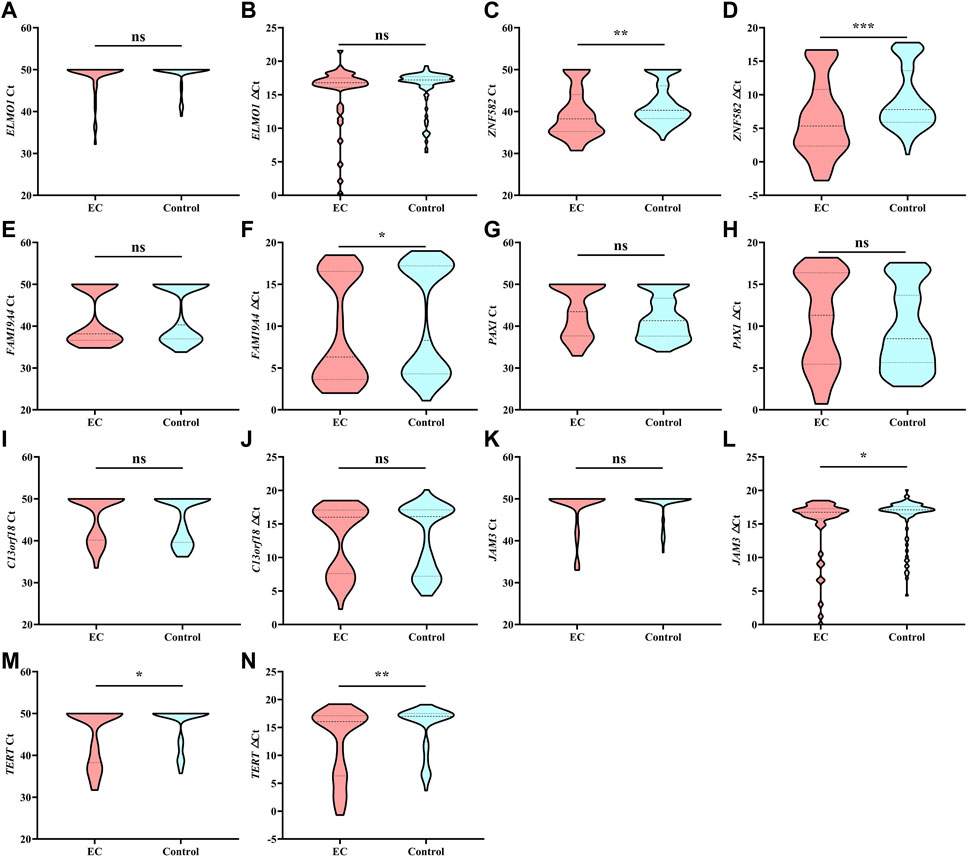
FIGURE 1. Methylation level of each DNA methylation marker in the training cohort analyzed by Ct values and △Ct values. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, no significant difference.
All possible combinations of multiple methylation markers in the training cohort are displayed in Table 3, and their AUC values and p-values were calculated. The combination of △Ct values for ZNF582 and FAM19A4 showed the largest AUC value. Meanwhile, the △Ct value of ZNF582 alone showed a sensitivity of 43.8% (95% CI: 29.8%–58.7%), with a specificity of 93.1% (95% CI: 85.6%–96.9%), and the sensitivity and specificity of FAM19A4 △Ct value alone were 25.0% (95% CI: 14.1%–39.9%) and 90.1% (95% CI: 82.1%–94.9%), respectively. When ZNF582 and FAM19A4 were combined, the sensitivity was improved to 60.4% (95% CI: 45.3%–73.9%), while the specificity decreased to 83.2% (95% CI: 74.1%–89.6%). Nonetheless, the combination of ZNF582 and FAM19A4 still achieved the largest Youden index compared to single markers (Table 4). As for the positive predictive value (PPV), the combination of ZNF582 and FAM19A4 also achieved a compromise result compared with the single gene; however, it has the highest negative predictive value (NPV), which represents that the assay can detect more EC cases with higher sensitivity (Table 4). Therefore, the combination of ZNF582 and FAM19A4 △Ct values was the most discriminant two-gene panel for the detection of EC. Sensitivities of this two-gene panel showed no significant difference between different age and gender groups (Figure 2). Based on the aforementioned results, the cut-off values for this two-gene panel are as follows: ZNF582 △Ct < 4.30, FAM19A4 △Ct < 3.65.
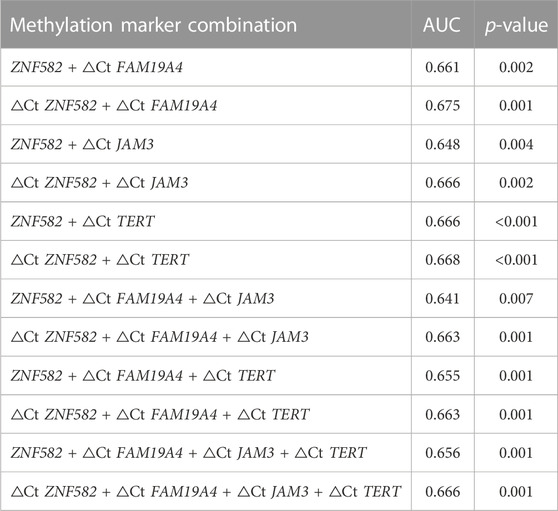
TABLE 3. Discrimination of several methylation marker combinations in the training cohort for esophageal cancer detection in plasma samples.
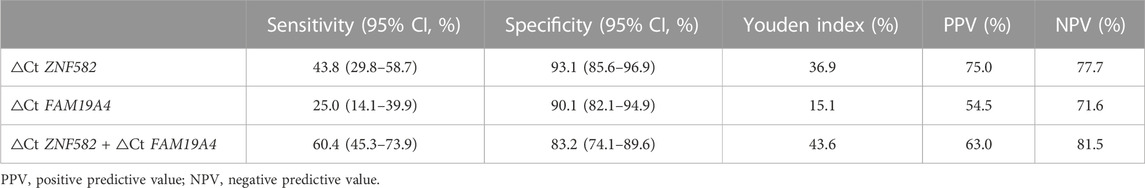
TABLE 4. Sensitivities and specificities of potential DNA methylation markers for the detection of esophageal cancer in the training cohort.
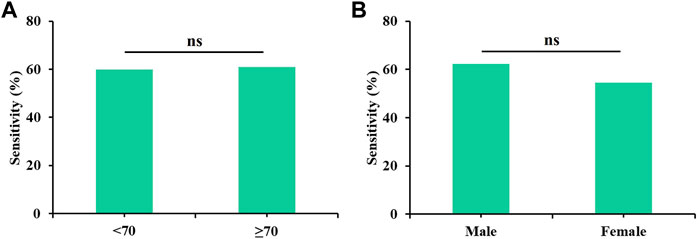
FIGURE 2. Sensitivities of the two-gene panel containing ZNF582 and FAM19A4 in the training cohort for the detection of esophageal cancer in different groups. (A) age, (B) gender.
CEA is a commonly used blood-based protein marker for cancer detection in clinics. The diagnostic efficacies of CEA and two-gene DNA methylation panel for EC detection were compared. In 45 EC patients, the sensitivity of CEA was only 22.2%, which was less than the sensitivities of ZNF582 and FAM19A4 (46.7% and 26.7%, respectively) (Figure 3A). In contrast, CEA showed the highest specificity of 93.7% among the three markers (Figure 3B). When ZNF582 or FAM19A4 was combined with CEA, their sensitivities increased to 57.8% and 44.4% (Figure 3A), while their specificities were 86.3% and 83.2%, respectively (Figure 3B). However, if the two-gene panel was combined with CEA, sensitivity was improved from 62.2% to 71.1% and specificity decreased from 83.2% to 75.8%. When Youden indexes were compared, the combination of two-gene panel and CEA showed the largest Youden index of 46.9%, whereas Youden indexes of two-gene panel and CEA alone were 43.6% and 15.9%, respectively (Figure 3C). The AUC value of CEA for discriminating EC from control subjects was 0.661 (95% CI: 0.563–0.759), and the AUC value of two-gene panel was 0.673 (95% CI: 0.566–0.779). In comparison, the combination of two-gene panel and CEA improved the AUC value to 0.677 (95% CI: 0.571–0.784) (Figure 4).
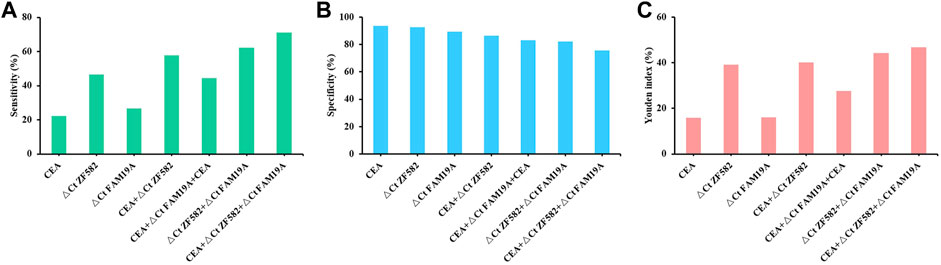
FIGURE 3. Sensitivity (A), specificity (B), and Youden index (C) of methylation markers combined with CEA for the detection of esophageal cancer.
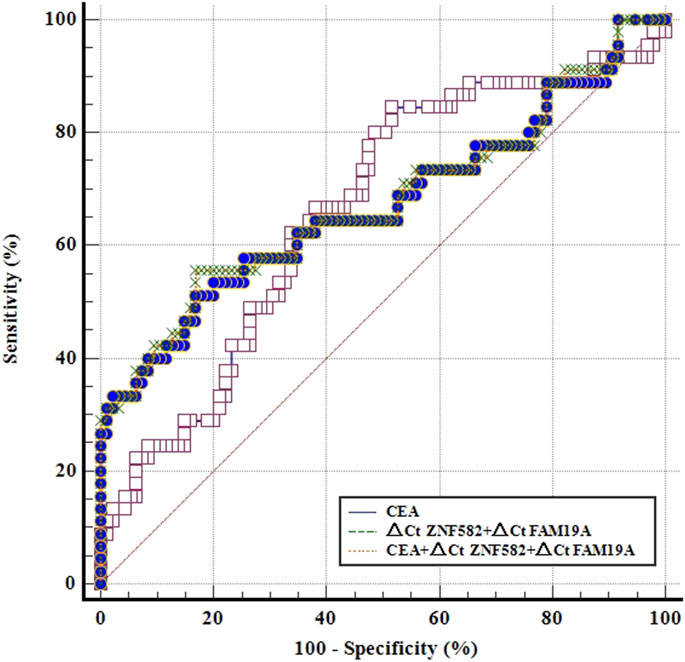
FIGURE 4. ROC curves for methylation markers combined with CEA for the detection of esophageal cancer in the training cohort.
In order to further evaluate the performance of the two-gene panel for the detection of EC, we included 20 EC, 10 GC, 10 CRC, and 20 control subjects in a validation cohort and compared its sensitivity and specificity in each group. As shown in Figure 5A, the two-gene panel displayed higher sensitivity (60.0%) than single genes (50.0% and 15.0%) for the detection of EC, with a specificity of 90.0%. In contrast, the sensitivity of the two-gene panel in detecting GC and CRC was only 30.0%, demonstrating its higher specificity for EC. Additionally, as shown in Figure 5B, the two-gene panel exhibited an AUC value of 0.845 when distinguishing EC from control subjects. The AUC value for distinguishing EC from the combination of GC and CRC is 0.710, while the AUC value for distinguishing the combination of GC and CRC from control subjects was 0.682.
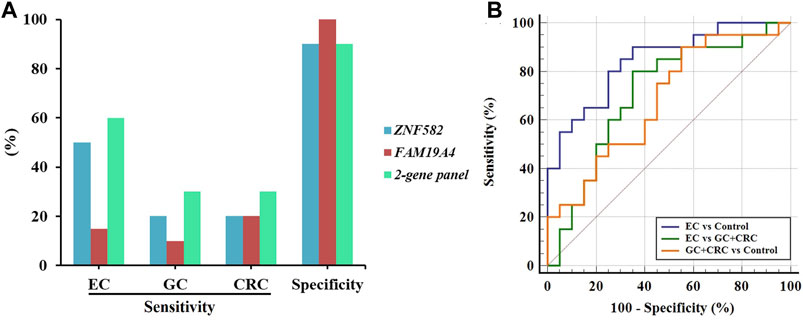
FIGURE 5. Performance of the two-gene panel for the detection of EC in the validation cohort. (A) Sensitivities and specificities of the two-gene panel for the detection of EC, GC, and CRC. (B) Comparison of ROC curves of the two-gene panel for the detection of EC, GC, and CRC.
4 Discussion
EC is a leading cause of cancer-related deaths in China, and early detection and screening is one of the most effective strategies for reducing its incidence and mortality. DNA methylation is the most reliable biomarker at present, with high stability and specificity, and several blood-, stool-, or cell-based commercial kits have been developed and approved by regulatory authorities for the early detection of colorectal cancer, lung cancer, and cervical cancer (Potter et al., 2014; Zhang et al., 2017; Dippmann et al., 2020; Wang et al., 2022). These positive outcomes suggested that DNA methylation markers might be developed into non-invasive diagnostic methods for the early detection of EC.
In this study, we tested the methylation of ELMO1, ZNF582, FAM19A4, PAX1, C13orf18, JAM3, and TERT in plasma for their abilities to discriminate EC from control subjects. The results of ELMO1, PAX1, and C13orf18 methylation showed no significant difference between the EC and control groups. In Tang et al. (2019), PAX1 methylation detected EC with 96.0% sensitivity and 51.4% specificity in tissue samples, and Qin et al. (2019) showed that ELMO1 methylation significantly discriminated EC from control subjects in tissue and plasma samples. The inconsistency between our results and the previous studies may be due to the following reasons: a) the quantity of methylated genomic DNA in tissue samples is far more than that in a small volume of plasma; thus, PAX1 methylation in plasma may not be detected due to low cfDNA amount. b) The plasma volume used in this study is only approximately 1/4 of that used in the previous study, which may have affected ELMO1 sensitivity, as our previous study demonstrated that decreasing plasma volume significantly decreased sensitivities of methylation markers in plasma (Zhang et al., 2021).
The methylation of FAM19A4, JAM3, and TERT has been identified as diagnostic biomarkers for cervical neoplasia (Jiang et al., 2012; Yin et al., 2015; Bonde et al., 2021). Cervical cancer includes two histologic subtypes, cervical squamous cell carcinoma and cervical adenocarcinoma (Kang et al., 2006), while EC includes the same two histologic subtypes, esophageal adenocarcinoma and esophageal squamous carcinoma (Testa et al., 2017). Therefore, cervical cancer and EC may have similar pathological processes. In addition, several cervical cancer-related methylation markers have also demonstrated aberrant DNA methylation in EC (Rad et al., 2016). ZNF582 methylation is another confirmed marker associated with cervical cancer (Li et al., 2019), and ZNF582 methylation has also been reported as a potential marker for EC detection in tissue samples (Tang et al., 2019), although the study of ZNF582 methylation in plasma samples is insufficient. After three rounds of selection, ZNF582 and FAM19A4 methylation markers were found to be the best combination for the detection of EC in plasma. The results in Table 2 and Figure 1 indicated that while ZNF582 methylation was the best single marker for EC detection in plasma, the combination of ZNF582 and FAM19A4 methylation could further improve the diagnostic capability.
CEA is a commonly used traditional cancer marker for cancer treatment monitoring and detection (Leja and Linē, 2021), but its sensitivity for EC detection is no more than 30% such that CEA is always combined with other markers to improve its sensitivity (Bagaria et al., 2013). In this study, CEA showed 22.2% sensitivity and 93.7% specificity for EC detection, while the two-gene panel showed a significantly higher sensitivity of 62.2%, almost 2.8-fold of CEA. Furthermore, if the two-gene panel and CEA were combined, sensitivity further increased to 71.1%. CEA and the two-gene panel are both blood-based markers; thus, only one tube of blood needs to be drawn for the simultaneous detection of these two markers. Therefore, it can not only improve the sensitivity of EC detection but also increase the screening throughput, allowing more people to participate in the early detection of EC.
There are some limitations to this study. The number of EC cases was relatively small, and more subjects need to be enrolled in multiple centers in future studies to validate the performance of the two-gene panel for EC detection. In addition, the detailed pathological information of EC patients, such as cancer stage, tumor size and differentiation, and more characteristics of the patients (smoking, cancer type, and medication), was not collected and analyzed. Such information can help us evaluate the performance of DNA methylation markers more comprehensively.
5 Conclusion
In this study, we demonstrated that four methylation markers can significantly distinguish EC from control subjects in plasma, and two methylation markers, ZNF582 and FAM19A4, were selected as the combination with the most potential for the early detection of EC. Therefore, non-invasive detection of EC based on DNA methylation markers in plasma can be an effective strategy to reduce the incidence and mortality of EC.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics statement
All participants have acknowledged and signed the informed consent. This study was performed according to the principles of the Declaration of Helsinki and approved by the Institutional Review Board of The Affiliated Suqian First People’s Hospital of Nanjing Medical University (Ethics Committee Reference Number: 2021-SL-0072). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
BP and GZ performed the statistical analyses and drafted the manuscript. ZG, YW, MW, XW, and SX participated in sample collection and data analysis. BP, GZ, and MZ conceived the study and participated in the design and coordination of the study. All authors read and approved the final manuscript.
Funding
This research was supported by the Suqian Key Laboratory of Science and Technology Project (Suqian Clinical Medical Laboratory) (Grant No. M201902), Maternal and Child Scientific Research Project of Jiangsu Province (Grant No. F201868), Jiangsu Province Six Talents Project (Grant No. YY-230), and Kunshan Association for Science and Technology Youth Talent Promotion Project.
Conflict of interest
Authors GZ, YW, XW, and SX were employed by Suzhou VersaBio Technologies Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1222617/full#supplementary-material
References
Bagaria, B., Sood, S., Sharma, R., and Lalwani, S. (2013). Comparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis). Cancer Biol. Med. 10, 148–157. doi:10.7497/j.issn.2095-3941.2013.03.005
Bonde, J., Floore, A., Ejegod, D., Vink, F.J., Hesselink, A., van de Ven, P.M., et al. (2021). Methylation markers FAM19A4 and miR124-2 as triage strategy for primary human papillomavirus screen positive women: A large European multicenter study. Int. J. cancer 148, 396–405. doi:10.1002/ijc.33320
Cao, Y., Zhao, G., Yuan, M., Liu, X., Ma, Y., Cao, Y., et al. (2020). KCNQ5 and C9orf50 methylation in stool DNA for early detection of colorectal cancer. Front. Oncol. 10, 621295. doi:10.3389/fonc.2020.621295
Chen, Z., Zhao, G., Wang, K., Wang, X., Ma, Y., Xiong, S., et al. (2021). Blood leukocytes methylation levels analysis indicate methylated plasma test is a promising tool for colorectal cancer early detection. J. Cancer 12, 3678–3685. doi:10.7150/jca.57114
De Strooper, L.M.A., Verhoef, V.M.J., Berkhof, J., Hesselink, A.T., de Bruin, H.M.E., van Kemenade, F.J., et al. (2016). Validation of the FAM19A4/mir124-2 DNA methylation test for both lavage- and brush-based self-samples to detect cervical (pre)cancer in HPV-positive women. Gynecol. Oncol. 141, 341–347. doi:10.1016/j.ygyno.2016.02.012
Dippmann, C., Schmitz, M., Wunsch, K., Schütze, S., Beer, K., Greinke, C., et al. (2020). Triage of hrHPV-positive women: comparison of two commercial methylation-specific PCR assays. Clin. epigenetics 12, 171. doi:10.1186/s13148-020-00963-w
Early Diagnosis and Treatment Group of the Chinese Medical Association Oncology Branch (2022). Chinese expert consensus on early diagnosis and treatment of esophageal cancer. Zhonghua zhong liu za zhi Chin. J. Oncol. 44, 1066–1075. doi:10.3760/cma.j.cn112152-20220220-00114
He, J., Chen, W.Q., Li, Z.S., Li, N., Ren, J.S., Tian, J.H., et al. (2022). China guideline for the screening, early detection and early treatment of esophageal cancer (2022, Beijing). Zhonghua zhong liu za zhi Chin. J. Oncol. 44, 491–522. doi:10.3760/cma.j.cn112152-20220517-00348
He, Y., Liang, D., Du, L., Guo, T., Liu, Y., Sun, X., et al. (2020). Clinical characteristics and survival of 5283 esophageal cancer patients: A multicenter study from eighteen hospitals across six regions in China. Cancer Commun. Lond. Engl. 40, 531–544. doi:10.1002/cac2.12087
Henry, M.A., Lerco, M.M., Ribeiro, P.W., and Rodrigues, M.A. (2014). Epidemiological features of esophageal cancer. Squamous cell carcinoma versus adenocarcinoma. Acta cir. bras. 29, 389–393. doi:10.1590/s0102-86502014000600007
Huang, J., Wang, G., Tang, J., Zhuang, W., Wang, L.P., Liou, Y.L., et al. (2017). DNA methylation status of PAX1 and ZNF582 in esophageal squamous cell carcinoma. Int. J. Environ. Res. public health 14, 216. doi:10.3390/ijerph14020216
Jamshidi, A., Liu, M.C., Klein, E.A., Venn, O., Hubbell, E., Beausang, J.F., et al. (2022). Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell 40, 1537–1549.e12. doi:10.1016/j.ccell.2022.10.022
Jiang, J., Zhao, L.J., Zhao, C., Zhang, G., Zhao, Y., Li, J.R., et al. (2012). Hypomethylated CpG around the transcription start site enables TERT expression and HPV16 E6 regulates TERT methylation in cervical cancer cells. Gynecol. Oncol. 124, 534–541. doi:10.1016/j.ygyno.2011.11.023
Kang, S., Kim, J.W., Kang, G.H., Lee, S., Park, N.H., Song, Y.S., et al. (2006). Comparison of DNA hypermethylation patterns in different types of uterine cancer: cervical squamous cell carcinoma, cervical adenocarcinoma and endometrial adenocarcinoma. Int. J. cancer 118, 2168–2171. doi:10.1002/ijc.21609
Koch, A., Joosten, S.C., Feng, Z., de Ruijter, T.C., Draht, M.X., Melotte, V., et al. (2018). Analysis of DNA methylation in cancer: location revisited. Nat. Rev. Clin. Oncol. 15, 459–466. doi:10.1038/s41571-018-0004-4
Kulis, M., and Esteller, M. (2010). DNA methylation and cancer. Adv. Genet. 70, 27–56. doi:10.1016/b978-0-12-380866-0.60002-2
Leja, M., and Linē, A. (2021). Early detection of gastric cancer beyond endoscopy - new methods. Best Pract. Res. Clin. gastroenterology 50-51, 101731–101751. doi:10.1016/j.bpg.2021.101731
Li, J., Xu, J., Zheng, Y., Gao, Y., He, S., Li, H., et al. (2021). Esophageal cancer: epidemiology, risk factors and screening. Chin. J. cancer Res. = Chung-kuo yen cheng yen chiu 33, 535–547. doi:10.21147/j.issn.1000-9604.2021.05.01
Li, N., He, Y., Mi, P., and Hu, Y. (2019). ZNF582 methylation as a potential biomarker to predict cervical intraepithelial neoplasia type III/worse: A meta-analysis of related studies in Chinese population. Medicine 98, e14297. doi:10.1097/md.0000000000014297
Miao, J., Liu, Y., Zhao, G., Liu, X., Ma, Y., Li, H., et al. (2020). Feasibility of plasma-methylated SFRP2 for early detection of gastric cancer. Cancer controlJ. Moffitt Cancer Cent. 27, 1073274820922559. doi:10.1177/1073274820922559
Miyoshi, J., Zhu, Z., Luo, A., Toden, S., Zhou, X., Izumi, D., et al. (2022). A microRNA-based liquid biopsy signature for the early detection of esophageal squamous cell carcinoma: A retrospective, prospective and multicenter study. Mol. cancer 21, 44. doi:10.1186/s12943-022-01507-x
Pan, Y., Liu, G., Zhou, F., Su, B., and Li, Y. (2018). DNA methylation profiles in cancer diagnosis and therapeutics. Clin. Exp. Med. 18, 1–14. doi:10.1007/s10238-017-0467-0
Potter, N.T., Hurban, P., White, M.N., Whitlock, K.D., Lofton-Day, C.E., Tetzner, R., et al. (2014). Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin. Chem. 60, 1183–1191. doi:10.1373/clinchem.2013.221044
Qin, Y., Wu, C.W., Taylor, W.R., Sawas, T., Burger, K.N., Mahoney, D.W., et al. (2019). Discovery, validation, and application of novel methylated DNA markers for detection of esophageal cancer in plasma. Clin. cancer Res.official J. Am. Assoc. Cancer Res. 25, 7396–7404. doi:10.1158/1078-0432.Ccr-19-0740
Rad, A., Esmaeili Dizghandi, S., Abbaszadegan, M.R., Taghechian, N., Najafi, M., and Forghanifard, M.M. (2016). SOX1 is correlated to stemness state regulator SALL4 through progression and invasiveness of esophageal squamous cell carcinoma. Gene 594, 171–175. doi:10.1016/j.gene.2016.08.045
Rosado, M., Silva, R., Marian, G.B., Jhon, G.J., Manadas, B., and Anjo, S.I. (2019). Advances in biomarker detection: alternative approaches for blood-based biomarker detection. Adv. Clin. Chem. 92, 141–199. doi:10.1016/bs.acc.2019.04.003
Salta, S., Macedo-Silva, C., Miranda-Gonçalves, V., Lopes, N., Gigliano, D., Guimarães, R., et al. (2020). A DNA methylation-based test for esophageal cancer detection. Biomark. Res. 8, 68. doi:10.1186/s40364-020-00248-7
Sung, H., Ferlay, J., Siegel, R.L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA a cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tang, L., Liou, Y.L., Wan, Z.R., Tang, J., Zhou, Y., Zhuang, W., et al. (2019). Aberrant DNA methylation of PAX1, SOX1 and ZNF582 genes as potential biomarkers for esophageal squamous cell carcinoma. Biomed. Pharmacother. = Biomedecine Pharmacother. 120, 109488. doi:10.1016/j.biopha.2019.109488
Testa, U., Castelli, G., and Pelosi, E. (2017). Esophageal cancer: genomic and molecular characterization, stem cell compartment and clonal evolution. Med. (Basel, Switz. 4, 67. doi:10.3390/medicines4030067
Wang, Z., Shang, J., Zhang, G., Kong, L., Zhang, F., Guo, Y., et al. (2022). Evaluating the clinical performance of a dual-target stool DNA test for colorectal cancer detection. J. Mol. diagnosticsJMD 24, 131–143. doi:10.1016/j.jmoldx.2021.10.012
Yin, A., Zhang, Q., Kong, X., Jia, L., Yang, Z., Meng, L., et al. (2015). JAM3 methylation status as a biomarker for diagnosis of preneoplastic and neoplastic lesions of the cervix. Oncotarget 6, 44373–44387. doi:10.18632/oncotarget.6250
Zhang, C., Yu, W., Wang, L., Zhao, M., Guo, Q., Lv, S., et al. (2017). DNA methylation analysis of the SHOX2 and RASSF1A panel in bronchoalveolar lavage fluid for lung cancer diagnosis. J. Cancer 8, 3585–3591. doi:10.7150/jca.21368
Zhang, G., He, F., Zhao, G., Huang, Z., Li, X., Xia, X., et al. (2021). Combining serum DNA methylation biomarkers and protein tumor markers improved clinical sensitivity for early detection of colorectal cancer. Int. J. genomics 2021, 6613987. doi:10.1155/2021/6613987
Zhao, G., Li, H., Yang, Z., Wang, Z., Xu, M., Xiong, S., et al. (2019). Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med. 8, 5619–5628. doi:10.1002/cam4.2475
Zhao, G., Ma, Y., Li, H., Li, S., Zhu, Y., Liu, X., et al. (2020). A novel plasma based early colorectal cancer screening assay base on methylated SDC2 and SFRP2. Clin. chimica acta; Int. J. Clin. Chem. 503, 84–89. doi:10.1016/j.cca.2020.01.010
Keywords: esophageal cancer, DNA methylation, non-invasive, early detection, plasma
Citation: Pei B, Zhao G, Geng Z, Wang Y, Wang M, Wang X, Xiong S and Zheng M (2023) Identifying potential DNA methylation markers for the detection of esophageal cancer in plasma. Front. Genet. 14:1222617. doi: 10.3389/fgene.2023.1222617
Received: 14 May 2023; Accepted: 21 September 2023;
Published: 04 October 2023.
Edited by:
Rodrigo Alexandre Panepucci, University of Sao Paulo, BrazilReviewed by:
Weijuan Cai, Central People’s Hospital of Zhanjiang, ChinaDahmane Oukrif, University College London, United Kingdom
Copyright © 2023 Pei, Zhao, Geng, Wang, Wang, Wang, Xiong and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Pei, c3FwZWliaW5nQG5qbXUuZWR1LmNu; Guodong Zhao, Z2Rkbjg5NDI5QGhvdG1haWwuY29t
†These authors have contributed equally to this work
 Bing Pei1,2*†
Bing Pei1,2*† Guodong Zhao
Guodong Zhao Shangmin Xiong
Shangmin Xiong