- 1Clinical Lab, Yantai Affiliated Hospital of Binzhou Medical University, Yantai, China
- 2Department of Thoracic Cardiovascular Surgery, Hunan Province Directly Affiliated TCM Hospital, Zhuzhou, China
- 3Geneis (Beijing) Co., Ltd., Beijing, China
- 4School of Computer Science, Hunan Institute of Technology, Hengyang, China
- 5Faculty of Pediatrics, The Chinese PLA General Hospital, Beijing, China
- 6Department of Pediatric Surgery, The Seventh Medical Center of PLA General Hospital, Beijing, China
- 7National Engineering Laboratory for Birth Defects Prevention and Control of Key Technology, Beijing, China
- 8Beijing Key Laboratory of Pediatric Organ Failure, Beijing, China
Introduction: Lung cancer is one of the most frequent neoplasms worldwide with approximately 2.2 million new cases and 1.8 million deaths each year. The expression levels of programmed death ligand-1 (PDL1) demonstrate a complex association with lung cancer. Neuroblastoma is a high-risk malignant tumor and is mainly involved in childhood patients. Identification of new biomarkers for these two diseases can significantly promote their diagnosis and therapy. However, in vivo experiments to discover potential biomarkers are costly and laborious. Consequently, artificial intelligence technologies, especially machine learning methods, provide a powerful avenue to find new biomarkers for various diseases.
Methods: We developed a machine learning-based method named LDAenDL to detect potential long noncoding RNA (lncRNA) biomarkers for lung cancer and neuroblastoma using an ensemble of a deep neural network and LightGBM. LDAenDL first computes the Gaussian kernel similarity and functional similarity of lncRNAs and the Gaussian kernel similarity and semantic similarity of diseases to obtain their similar networks. Next, LDAenDL combines a graph convolutional network, graph attention network, and convolutional neural network to learn the biological features of the lncRNAs and diseases based on their similarity networks. Third, these features are concatenated and fed to an ensemble model composed of a deep neural network and LightGBM to find new lncRNA–disease associations (LDAs). Finally, the proposed LDAenDL method is applied to identify possible lncRNA biomarkers associated with lung cancer and neuroblastoma.
Results: The experimental results show that LDAenDL computed the best AUCs of 0.8701, 107 0.8953, and 0.9110 under cross-validation on lncRNAs, diseases, and lncRNA-disease pairs on Dataset 1, respectively, and 0.9490, 0.9157, and 0.9708 on Dataset 2, respectively. Furthermore, AUPRs of 0.8903, 0.9061, and 0.9166 under three cross-validations were obtained on Dataset 1, and 0.9582, 0.9122, and 0.9743 on Dataset 2. The results demonstrate that LDAenDL significantly outperformed the other four classical LDA prediction methods (i.e., SDLDA, LDNFSGB, IPCAF, and LDASR). Case studies demonstrate that CCDC26 and IFNG-AS1 may be new biomarkers of lung cancer, SNHG3 may associate with PDL1 for lung cancer, and HOTAIR and BDNF-AS may be potential biomarkers of neuroblastoma.
Conclusion: We hope that the proposed LDAenDL method can help the development of targeted therapies for these two diseases.
1 Introduction
Long non-coding RNAs (lncRNAs) are non-coding RNAs with more than 200 nucleotides (Bertone et al., 2004; Peng et al., 2022a; Peng et al., 2022b). LncRNAs play an important role in the development and progression of various diseases (Lanjanian et al., 2021; Meng et al., 2021; Yang and Li 2021; Peng et al., 2022c). LncRNAs have dense associations with many diseases, for example, lung cancer, colorectal cancer, prostate cancer, and Alzheimer’s disease (Klattenhoff et al., 2013; Tan et al., 2013; Chakravarty et al., 2014; He et al., 2014; Zhang et al., 2014). LncRNA H19 is associated with the under-regulation of renal carcinoma cells (Wang et al., 2015). The expression of EGOT in breast cancer is much lower than one in adjacent noncancerous tissues (Broadbent et al., 2008). NEAT1 is overexpressed in prostate cancer cells (Pasmant et al., 2011). The identification of lncRNA-disease associations (LDAs) helps us to further understand the biological processes and the molecular mechanisms of various complex diseases. However, the number of known and experimentally validated LDAs is very small. Thus, it is important to identify potential LDAs. Determining LDAs through in vivo experiments is costly and time-consuming, therefore, it is necessary to design efficient computational approaches for identifying potential LDAs (Meng et al., 2021; Peng et al., 2022d). Computational LDA prediction methods are categorized as biological network-based methods and machine learning-based methods.
Biological network-based methods use network algorithms for association prediction (Liu et al., 2023a). This type of method first constructs heterogeneous networks of lncRNAs and diseases and then identifies LDAs via matrix decomposition, random walk, and so on. To predict potential LDAs, LRWRHLDA combined Laplace normalized random walk with restart (Wang et al., 2022), LDGRNMF used graph regularized nonnegative matrix factorization (Wang et al., 2021), DSCMF developed a dual sparse collaborative matrix factorization approach (Liu et al., 2021a), RWSF-BLP added random walk-based multi-similarity fusion to bidirectional label propagation (Xie et al., 2021), HBRWRLDA utilized bi-random walk on hypergraphs (Xie et al., 2022), and MHRWRLDA exploited a random walk model with restart through multiplex and heterogeneous networks (Yao et al., 2021).
With the fast advance of RNA sequencing technologies, artificial intelligence has obtained wide applications in biomedical data analysis (Peng et al., 2023a; Peng et al., 2023b; Xu et al., 2023). Notably, artificial intelligence technologies, especially machine learning methods, have been widely applied to predict miRNA-disease associations (Liu et al., 2022) and circRNA-disease associations (Liu et al., 2023b). To find new LDAs, HGATLDA developed a novel heterogeneous graph attention network model (Zhao et al., 2022), DeepMNE extracted multi-omics data and designed a deep multi-network embedding model (Ma, 2022), iLncDA-LTR is a rank-based method (Wu et al., 2022), MAGCNSE utilized a graph convolutional network (Liang et al., 2022), LDAformer extracted topological features and used a transformer encoder for LDA classification (Zhou et al., 2022), BiGAN explored a bidirectional generative adversarial network (Yang et al., 2021), and SVDNVLDA extracted linear and non-linear features and used an XGBoost for LDA prediction (Li et al., 2021).
Computational methods have found many potential LDAs, however, network-based methods were more likely to favor well-investigated lncRNAs or diseases and can not predict LDAs for new lncRNAs or new diseases. Machine learning-based methods failed to effectively integrate different kernels from multiple data sources. Thus, in this study, we developed a machine learning-based method named LDAenDL to detect potential lncRNA biomarkers for lung cancer and neuroblastoma based on an ensemble of a deep neural network and LightGBM.
2 Materials and methods
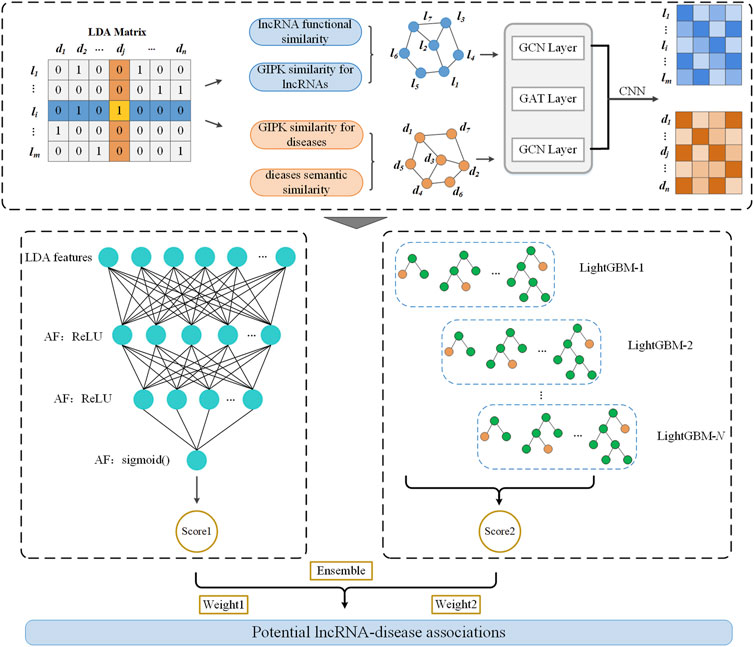
As shown in Figure 1, LDAenDL first computes the Gaussian kernel similarity and functional similarity of lncRNAs and the Gaussian kernel similarity and semantic similarity of diseases to obtain their similar networks. Next, LDAenDL combines a graph convolutional network (GCN) (Kipf and Welling, 2016), graph attention network (GAT) (Velickovic et al., 2017), and convolutional neural network (Gu et al., 2018) to learn the biological features of lncRNAs and diseases based on their similarity networks. Third, these features are concatenated and fed to an ensemble model composed of a deep neural network (DNN) and LightGBM to find new LDAs. Finally, LDAenDL was applied to identify possible lncRNA biomarkers associated with lung cancer and neuroblastoma.
2.1 Data preparation
We used two human LDA datasets that were provided by Chen et al. (2012) and Cui et al. (2018). Dataset 1 contains 605 LDAs between 157 diseases and 82 lncRNAs. Dataset 2 contains 1,529 LDAs between 190 diseases and 89 lncRNAs. An LDA network can be denoted as
2.2 Similarity computation
Inspired by the LDA-DLPU method (Peng et al., 2022a), we computed the Gaussian kernel similarity and functional similarity of lncRNAs and the Gaussian kernel similarity and semantic similarity of diseases. Based on the computed lncRNA similarity and disease similarity matrices, we learned the features of lncRNAs and diseases by combining a GCN, GAT, and CNN.
2.3 Feature learning
Dai et al. (2022) designed a hybrid graph representation learning model (GraphCDA) to represent the features of circRNAs and diseases and obtained better circRNA-disease association prediction performance. Inspired by GraphCDA proposed by Dai et al. (2022), we exploit a GraphCDA-based LDA feature learning model.
2.3.1 Graph convolutional network
A GCN was applied to obtain the feature representations of lncRNAs and diseases based on their similarity networks. For a GCN G, it is denoted as an adjacency matrix
where
2.3.2 Graph attention network
A GAT (Veličković et al., 2017) uses multi-head attention to set weights for all adjacent nodes based on their importance. LDAenDL introduces a GAT layer between two GCN layers to help the GCN to extract high-level features of lncRNAs and diseases.
For the GCN G, a GAT layer outputs node representations
For
where
where || denotes a concatenation operation,
2.3.3 Feature representation of lncRNAs and diseases
For a lncRNA similarity network
Thus, a 1D CNN is used to produce the lncRNA feature representation matrix
Similarly, the graph feature representations of diseases at different levels are denoted by Eq. 7:
A 1D CNN is used to produce the disease feature representation matrix
2.3.4 Preference matrix construction
The preference matrix
We used binary cross-entropy as the activation function to evaluate the difference between the preference matrix
2.4 LDA prediction
2.4.1 DNN
We built a DNN to predict new LDAs based on known LDAs and the learned LDA features. The DNN contains an input layer, an output layer, and multiple hidden layers. In the input layer, there are F neurons that are the same as the number of LDA features.
Given an LDA sample
where
The hidden layer is represented by Eq. 10:
where
The output in the
where
2.4.2 LightGBM
In this section, we built a LightGBM (Ke et al., 2017) to identify new LDAs. For a training set
LightGBM integrates
The regression trees are expressed as
At step
The objective function (15) is rapidly approximated with Newton’s method (Sun et al., 2020).
To solve the objective function of LightGBM, we removed the constant term for simplicity, and model (15) can be represented as Eq. 16:
where
Given a certain tree structure
where
where
2.4.3 Ensemble learning
Through the solution of models (12) and (15), we can identify potential LDAs based on a DNN and LightGBM. Ensemble learning has better prediction accuracy than a single model. To further improve LDA prediction accuracy, we combined a DNN and LightGBM and developed an ensemble model for LDA identification through soft voting in Eq. 16:
where
3 Results
3.1 Evaluation metrics
In this article, we compared our proposed LDAenDL method with four LDA prediction methods, SDLDA, LDNFSGB, IPCAF, and LDASR. Precision, recall, accuracy, F1-score, AUC, and AUPR were used to compare the performance of LDAenDL with the four methods. The six metrics have been defined by Peng et al. (2022b) (Shen et al., 2022).
3.2 Comparison of LDAenDL with the other four methods
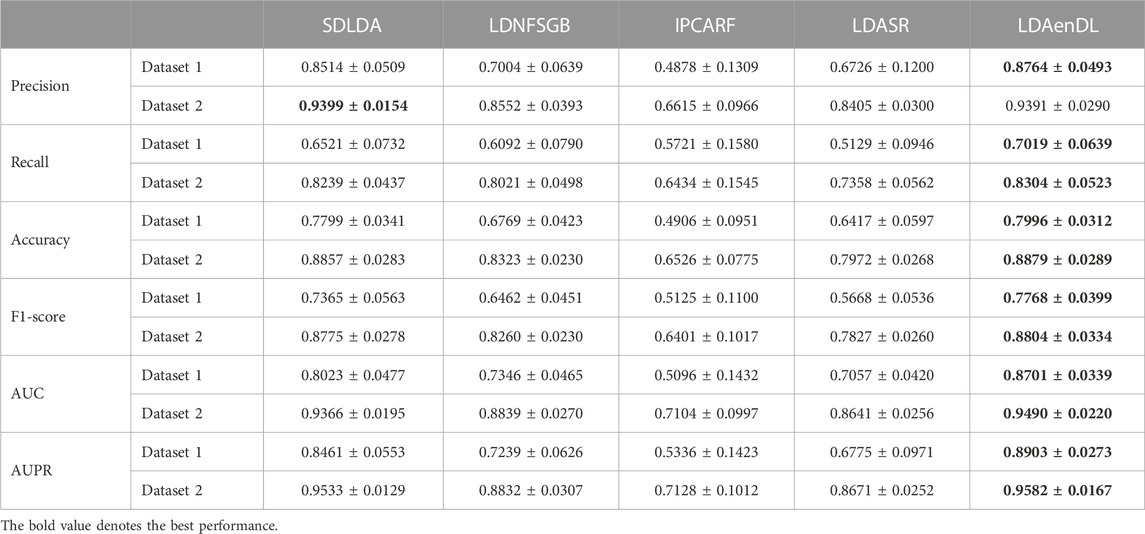
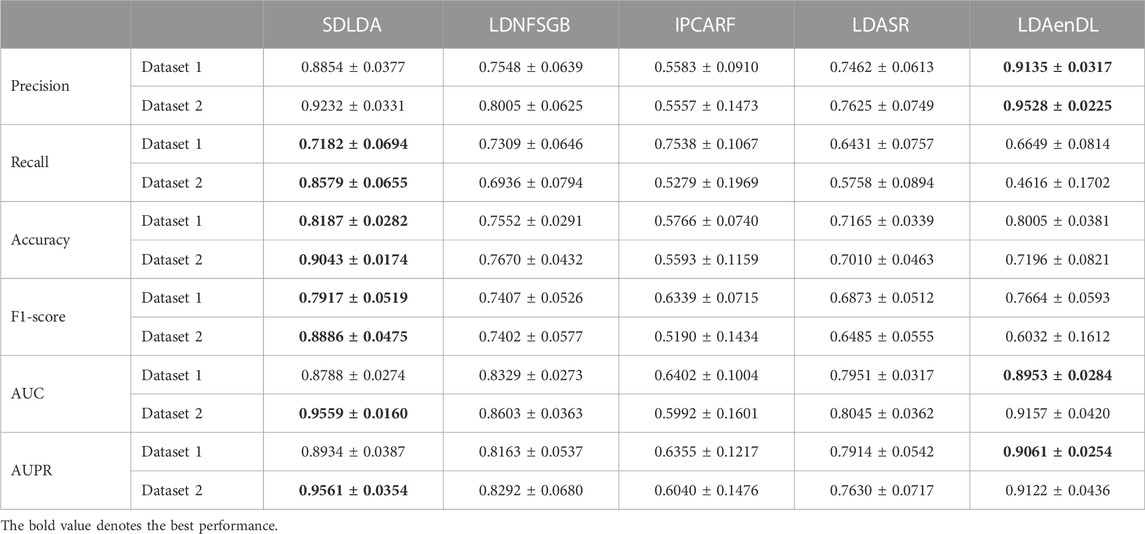
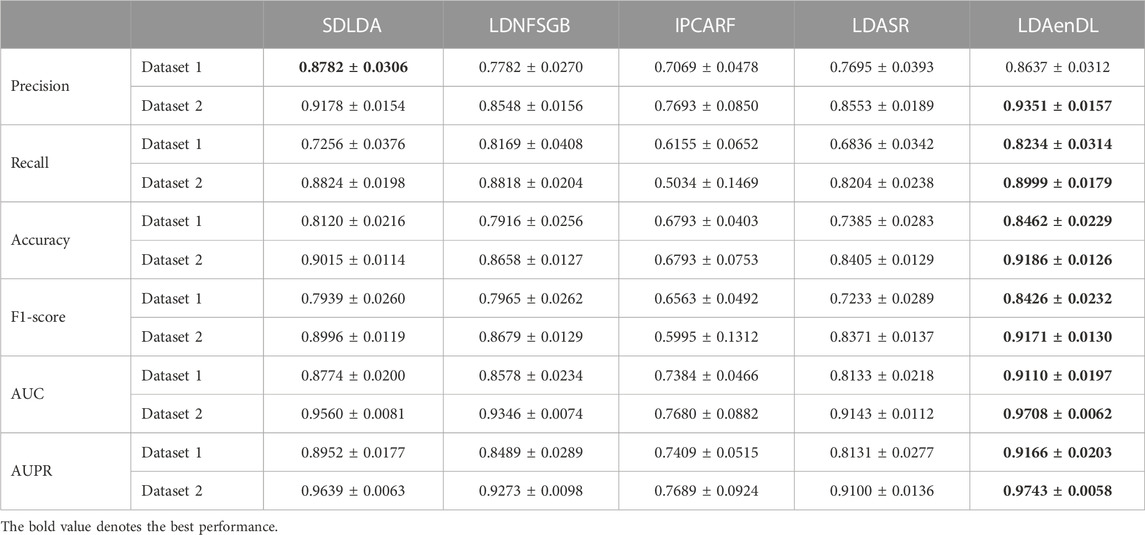
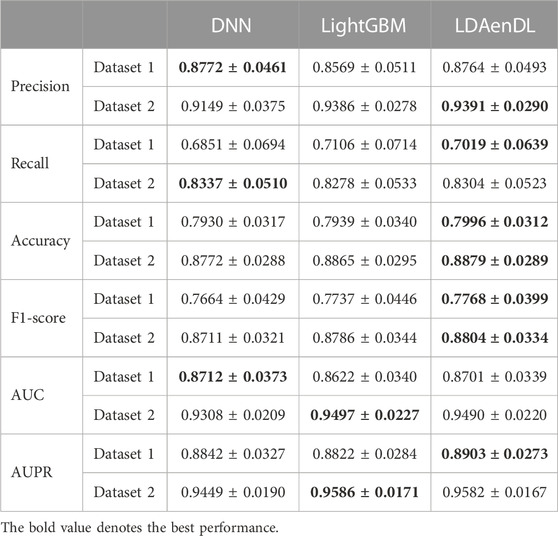
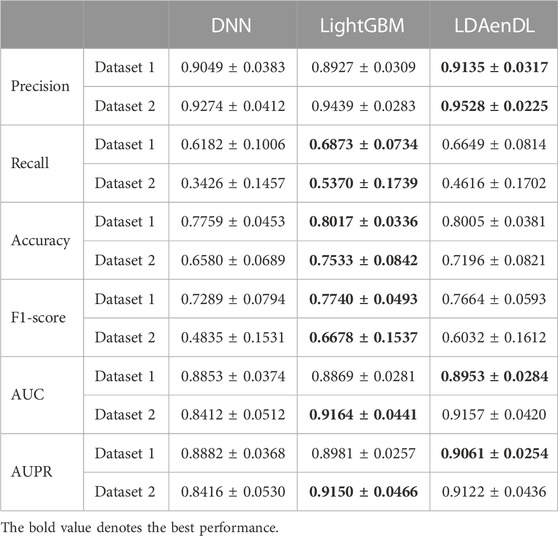
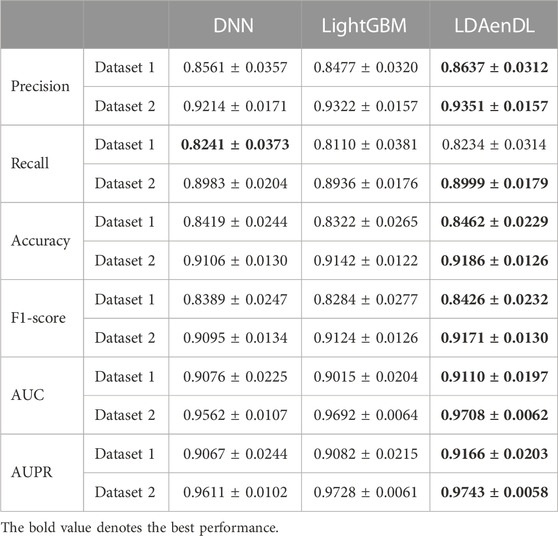
To implement the performance evaluation, inspired by the three cross-validations proposed by Zhou et al. (2021), we conducted cross-validations on lncRNAs (CV1), diseases (CV2), and lncRNA-disease pairs (CV3). Tables 1–3 give the precision, recall, accuracy, F1-score, AUC, and AUPR under CV1, CV2, and CV3 on two LDA datasets. In Tables 1–6, the bold font in each row denotes the best performance.
Under CV1, LDAenDL randomly took 80% of lncRNAs as training samples, and the rest were taken as test samples to investigate the LDA prediction ability for new lncRNAs. The results from Table 1 show that our proposed LDAenDL approach obtained the best precision, recall, accuracy, F1-score, AUC, and AUPR on two datasets under CV1 except that it computed slightly lower precision on Dataset 2 (0.9391 vs. 0.9399). It computed the highest AUPRs of 0.8903 and 0.9582, and far exceeded the AUPR values computed by SDLDA (i.e., 0.8461 and 0.9533).
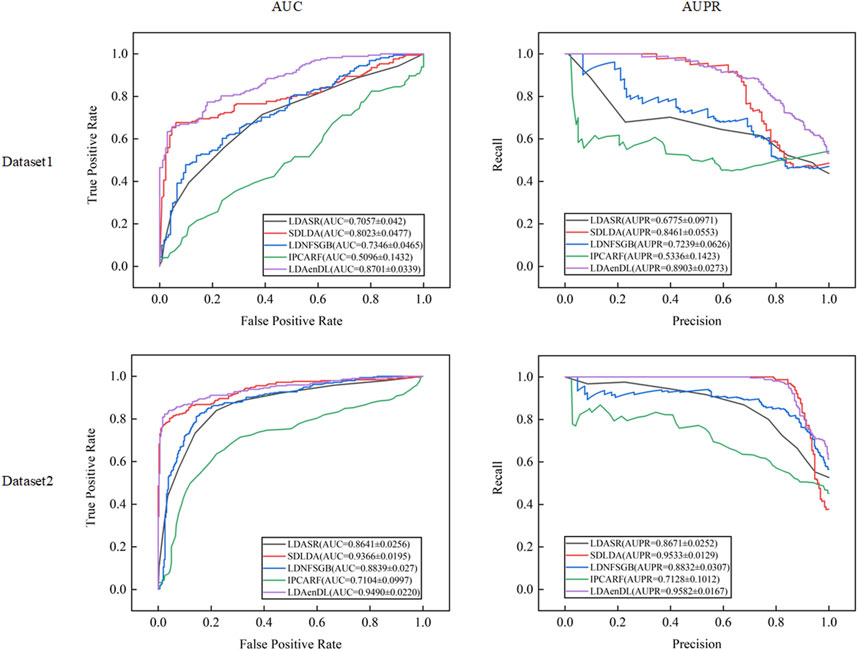
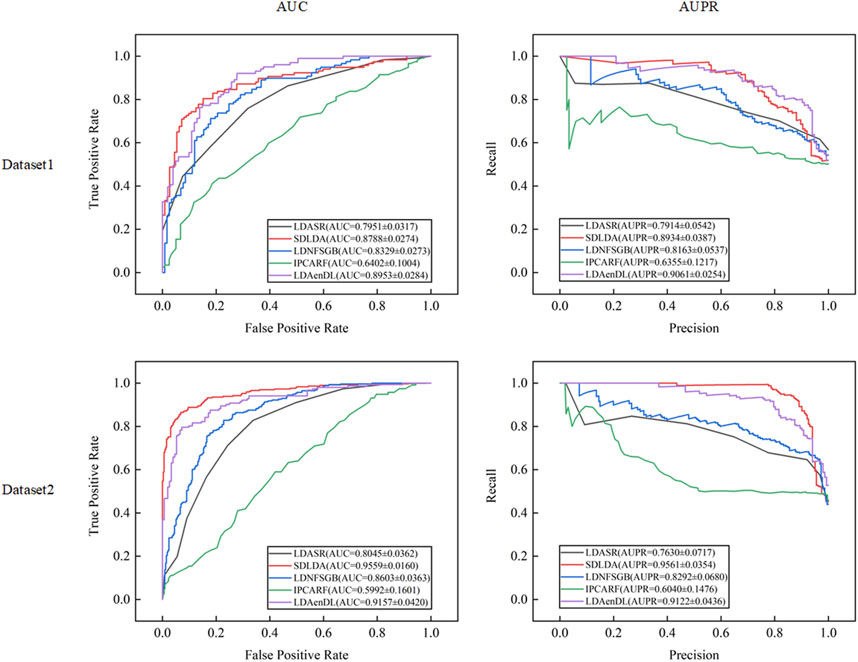
Figure 2 shows the AUC and AUPR values computed by LDAenDL and the other four methods on two datasets under CV1. The results demonstrated that LDAenDL can discover possible diseases associated with a new lncRNA.
Under CV2, LDAenDL randomly took 80% of diseases as training samples, and the rest were taken as test samples to investigate the LDA prediction ability for new diseases. The results from Table 2 show that our proposed LDAenDL approach obtained better precision, AUC, and AUPR on two datasets under CV2. However, SDLDA computed higher recall, accuracy, and F1-score than LDAenDL, which may be caused by smaller disease samples.
Figure 3 shows the AUC and AUPR values computed by LDAenDL and the other four methods on two datasets under CV2. The results show that LDAenDL can be applied to screen possible lncRNAs associated with a new disease.
Under CV3, LDAenDL randomly took 80% of lncRNA-disease pairs as training samples, and the rest were taken as test samples to investigate the LDA prediction ability. The results from Table 3 show that our proposed LDAenDL approach obtained the best precision, recall, accuracy, F1-score, AUC, and AUPR on two datasets under CV3. It computed the highest AUCs of 0.9110 and 0.9708 and far exceeded those computed by SDLDA (i.e., 0.8774 and 0.9560). Furthermore, our LDAenDL approach computed the highest AUPRs of 0.9166 and 0.9743 and far exceeded those computed by SDLDA (i.e., 0.8952, and 0.9639).
Figure 4 shows the AUC and AUPR values computed by LDAenDL and the other four methods on two datasets under CV3. The results demonstrated that LDAenDL could find potential LDAs based on known LDAs.
3.3 Comparison of LDAenDL with individual models
To measure the effect of the ensemble algorithm on LDA prediction performance, we compared LDAenDL with two individual models, DNN, and LightGBM. Tables 4–6 show the precision, recall, accuracy, F1-score, AUC, and AUPR of the DNN, LightGBM, and LDAenDL under CV1, CV2, and CV3, respectively.
Under CV1, as shown in Table 4, LDAenDL outperformed the DNN and LightGBM on two LDA datasets for the majority of conditions. LDAenDL computed the best accuracy and F1-score on the two datasets. Although LDAenDL computed slightly lower AUC value than the DNN on dataset 1, and still slightly lower AUC than LightGBM on dataset 2, their differences were very small. For example, the DNN computed an AUC of 0.8712 while LDAenDL computed 0.8701 on dataset 1, and the DNN calculated an AUC of 0.9497 while LDAenDL calculated 0.9490 on dataset 2. LDAenDL obtained the best AUPR on dataset 1, and LightGBM obtained an AUPR of 0.9586 while LDAenDL obtained an AUPR of 0.9582.
Under CV2, as shown in Table 5, LDAenDL outperformed the DNN under all conditions on two LDA datasets. Recall, accuracy, and F1-score computed by LightGBM were slightly better than LDAenDL on the two datasets. But it calculated the best AUC and AUPR on dataset 1.
Under CV3, as shown in Table 6, LDAenDL computed the highest precision, recall, accuracy, F1-score, AUC, and AUPR on the two LDA datasets except that it computed a slightly lower recall on dataset 1. The results demonstrate that LDAenDL is appropriate to predict possible LDAs from unknown lncRNA-disease pairs.
3.4 Case study
3.4.1 Identifying possible lncRNA biomarkers for lung cancer
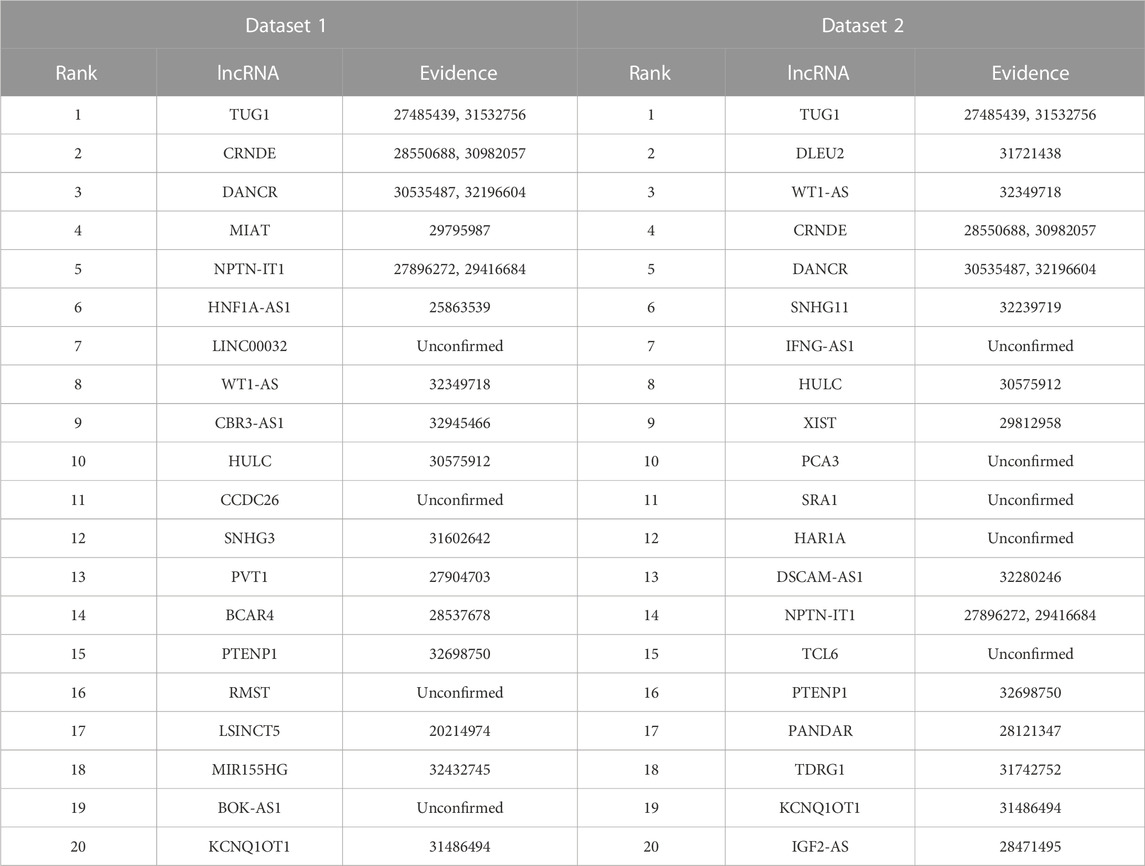
Lung cancer is one of the most prevalent causes of mortality globally. It mainly contains small cell lung cancer and non-small cell lung cancer. Targeted drug therapy is its one therapeutic option (Lahiri et al., 2023). We used the proposed LDAenDL method to predict possible lncRNA biomarkers for lung cancer. Table 7 shows the predicted top 20 lncRNA biomarkers for lung cancer. The 20 lncRNA biomarkers associated with lung cancer have no known association information with lung cancer in the two datasets.
In dataset 1, LDAenDL predicted that CCDC26 could be associated with lung cancer. CCDC26 can enhance thyroid cancer malignant progression (Ma et al., 2021). It promotes imatinib resistance in human gastrointestinal stromal tumors (Yan et al., 2019). Its inhibition could increase the sensitivity of doxorubicin in MDR-CML cells (Liu et al., 2021b). In this study, we predicted that CCDC26 could be associated with lung cancer in dataset 1.
In dataset 2, LDAenDL predicted that IFNG-AS1 could be associated with lung cancer. IFNG-AS1 has been reported in long-lasting memory T cells (Castellucci et al., 2021). It can boost interferon gamma generation in human natural killer cells (Stein et al., 2019). We identified that IFNG-AS1 could be associated with lung cancer in Dataset 2.
Figure 5 shows the top 20 predicted lncRNAs associated with lung cancer in each of the two datasets. Yellow solid lines and blue solid lines denote lncRNA-lung cancer associations confirmed by the literatures among the predicted top 20 associations on datasets 1 and 2, respectively. Grey solid lines denote the predicted and co-occurring lncRNA-lung cancer associations that can be confirmed by the literatures in the two datasets, and grey dashed lines denote the predicted and unconfirmed lncRNA-lung cancer associations in the two datasets. The repeated lncRNAs in the two datasets have been removed.
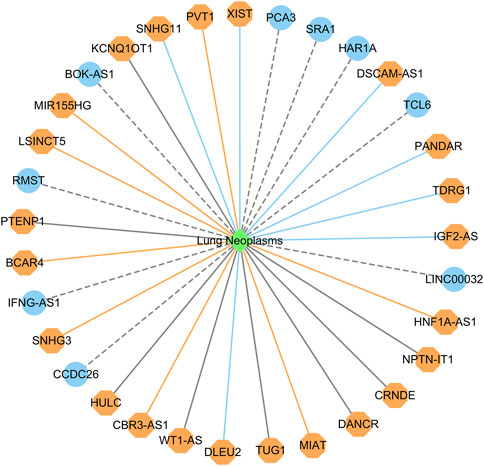
FIGURE 5. The top 20 predicted lncRNA biomarkers for lung cancer in each of the two datasets (The repeated lncRNAs in the two datasets have been removed). This figure was drawn using Cytoscape (Shannon et al., 2003).
3.4.2 Identifying possible lncRNAs associated with PDL1 for lung cancer
Recent advances in lung cancer treatment have demonstrated significant responses in patients when they were treated with programmed death-1/programmed death-ligand 1 (PD-1/PD-L1) checkpoint blockade immunotherapies (Lahiri et al., 2023). To find possible lncRNAs associated with PDL1 for lung cancer, inspired by LPI-DLDN proposed by Peng et al. (2022a), we first downloaded the sequence of PDL1 from the UniProt database. Next, we extracted the biological features of PDL1 and depicted PDL1 as a 10,029-dimensional vector using BioTriangle. Finally, we used cosine similarity to compute the similarities between PDL1 and the other proteins in a lncRNA-protein interaction dataset (Li et al., 2015) and found the top 3 proteins with the highest interaction probabilities with PDL1. The results show that SNHG3 has a higher interaction probability with PDL1 and has been reported to be associated with lung cancer.
3.4.3 Identifying possible lncRNA biomarkers for neuroblastoma
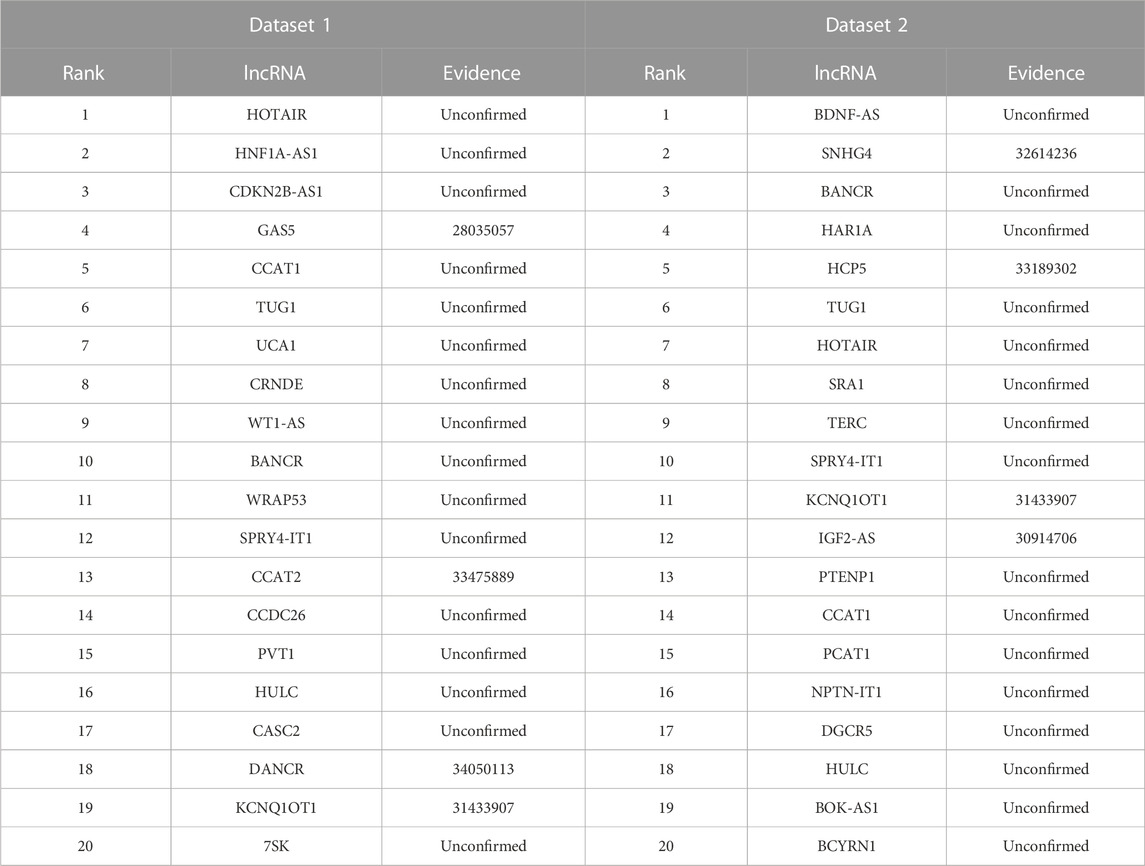
Neuroblastoma is the most frequent pediatric solid tumor and accounts for approximately 15% of childhood cancer-related mortality (Zafar et al., 2021). We used the proposed LDAenDL method to identify possible lncRNA biomarkers for neuroblastoma. Table 8 shows the top 20 predicted lncRNA biomarkers for neuroblastoma in each of the two datasets. The repeated lncRNAs in the two datasets have been removed.
In dataset 1, we predicted that HOTAIR could be associated with neuroblastoma with the highest probability. HOTAIR is a novel oncogenic biomarker in human cancer (Rajagopal et al., 2020). Its knockdown can promote radiosensitivity in colorectal cancer (Liu et al., 2020). It also can enhance the carcinogenesis of gastric (Zhang et al., 2020). We identified that HOTAIR may be one biomarker of neuroblastoma in dataset 1.
In dataset 2, we predicted that BDNF-AS could be associated with neuroblastoma with the highest probability. PABPC1-induced stabilization of BDNF-AS helps the inhibition of malignant progression in glioblastoma cells (Su et al., 2020). It can regulate the miR-9-5p/BACE1 pathway that affects neurotoxicity in Alzheimer’s disease (Ding et al., 2022). We identified that BDNF-AS is a possible biomarker of neuroblastoma in dataset 2.
Figure 6 shows the top 20 predicted lncRNAs associated with neuroblastoma in each of the two datasets. Yellow solid lines and blue solid lines denote lncRNA-neuroblastoma associations confirmed by the literatures among the predicted top 20 associations on datasets 1 and 2, respectively. Grey solid lines denote the predicted and co-occurring lncRNA-neuroblastoma associations that can be confirmed by the literatures in the two datasets, and grey dashed lines denote the predicted and unconfirmed lncRNA-neuroblastoma associations in the two datasets. The repeated lncRNAs in the two datasets have been removed.
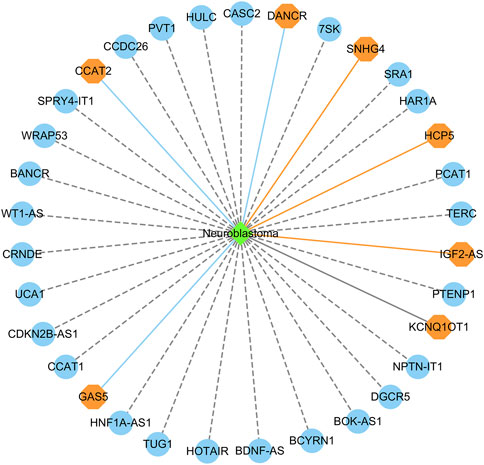
FIGURE 6. The top 20 predicted lncRNA biomarkers for neuroblastoma in each of the two datasets. (The repeated lncRNAs in the two datasets have been removed). This figure was drawn using Cytoscape (Shannon et al., 2003).
4 Conclusion
Lung cancer and neuroblastoma are two human diseases that severely affect the human body. Detecting new biomarkers for them contributes to their diagnosis and therapy. Experimental biomarker identification methods are costly and laborious. Thus, we developed a machine learning-based method named LDAenDL to predict possible lncRNA biomarkers for the two diseases based on an ensemble of a deep neural network and LightGBM. LDAenDL first computed lncRNA similarity and disease similarity and then combined a GCN, GAT, and CNN to learn the biological features of lncRNAs and diseases. Finally, these features were fed to a DNN and LightGBM to find new LDAs.
LDAenDL was compared with the other four classical LDA prediction methods (i.e., SDLDA, LDNFSGB, IPCAF, and LDASR). The results showed that LDAenDL computed the best AUCs and AUPRs under three cross-validations on two LDA datasets, demonstrating the optimal LDA prediction performance of LDAenDL. We further identified possible lncRNA biomarkers for lung cancer and neuroblastoma. The results demonstrated that CCDC26 and IFNG-AS1 may be new biomarkers for lung cancer, SNHG3 may be associated with PDL1 for lung cancer, and HOTAIR and BDNF-AS may be potential biomarkers for neuroblastoma.
In the future, we will combine data from multiple sources, for example, miRNA, circRNA, and drugs, to improve LDA identification performance. We will also design a new deep-learning model to efficiently extract the biological features of lncRNAs and diseases for LDA prediction. We hope that the proposed LDAenDL can help the development of targeted therapies for these two diseases.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
Conceptualization: ZS, HL, ZL, and LD; Investigation: ZS and HL; Methodology: ZS, HL, ZL, and LD; Project administration: YW and LD; Software: ZS and ZL; Writing-original draft: ZS and HL; Writing-review and editing: ZS, HL, ZL, and LD. All authors contributed to the article and approved the submitted version.
Conflict of interest
Author YW was employed by Geneis (Beijing) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bertone, P., Stolc, V., Royce, T. E., Rozowsky, J. S., Urban, A. E., Zhu, X., et al. (2004). Global identification of human transcribed sequences with genome tiling arrays. Science 306 (5705), 2242–2246. doi:10.1126/science.1103388
Broadbent, H. M., Peden, J. F., Lorkowski, S., Goel, A., Ongen, H., Green, F., et al. (2008). Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 17 (6), 806–814. doi:10.1093/hmg/ddm352
Castellucci, L. C., Almeida, L., Cherlin, S., Fakiola, M., Francis, R. W., Carvalho, E. M., et al. (2021). A genome-wide association study identifies SERPINB10, CRLF3, STX7, LAMP3, IFNG-AS1, and KRT80 as risk loci contributing to cutaneous leishmaniasis in Brazil. Clin. Infect. Dis. 72 (10), e515–e525. doi:10.1093/cid/ciaa1230
Chakravarty, D., Sboner, A., Nair, S. S., Giannopoulou, E., Li, R., Hennig, S., et al. (2014). The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5 (1), 5383. doi:10.1038/ncomms6383
Chen, G., Wang, Z., Wang, D., Qiu, C., Liu, M., Chen, X., et al. (2012). LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic acids Res. 41 (D1), D983–D986. doi:10.1093/nar/gks1099
Cui, T., Zhang, L., Huang, Y., Yi, Y., Tan, P., Zhao, Y., et al. (2018). MNDR v2. 0: an updated resource of ncRNA–disease associations in mammals. Nucleic acids Res. 46 (D1), D371–D374. doi:10.1093/nar/gkx1025
Dai, Q., Liu, Z., Wang, Z., Duan, X., and Guo, M. (2022). GraphCDA: a hybrid graph representation learning framework based on GCN and GAT for predicting disease associated circRNAs. Briefings in Bioinformatics 23 (5), bbac379. doi:10.1093/bib/bbac379
Ding, Y., Luan, W., Wang, Z., and Cao, Y. (2022). LncRNA BDNF-AS as ceRNA regulates the miR-9-5p/BACE1 pathway affecting neurotoxicity in Alzheimer's disease. Archives Gerontology Geriatrics 99, 104614. doi:10.1016/j.archger.2021.104614
Gu, J., Wang, Z., Kuen, J., Ma, L., Shahroudy, A., Shuai, B., et al. (2018). Recent advances in convolutional neural networks. Pattern Recognit. 77, 354–377. doi:10.1016/j.patcog.2017.10.013
He, X., Tan, X., Wang, X., Jin, H., Liu, L., Ma, L., et al. (2014). C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumor Biol. 35, 12181–12188. doi:10.1007/s13277-014-2526-4
Ke, G., Meng, Q., Finley, T., Wang, T., Chen, W., Ma, W., et al. (2017). Lightgbm: a highly efficient gradient boosting decision tree. Adv. neural Inf. Process. Syst. 30. doi:10.5555/3294996.3295074
Kipf, T. N., and Welling, M. (2016). Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907.
Klattenhoff, C. A., Scheuermann, J. C., Surface, L. E., Bradley, R. K., Fields, P. A., Steinhauser, M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 152 (3), 570–583. doi:10.1016/j.cell.2013.01.003
Lahiri, A., Maji, A., Potdar, P. D., Singh, N., Parikh, P., Bisht, B., et al. (2023). Lung cancer immunotherapy: progress, pitfalls, and promises. Mol. Cancer 22 (1), 40–37. doi:10.1186/s12943-023-01740-y
Lanjanian, H., Nematzadeh, S., Hosseini, S., Torkamanian-Afshar, M., Kiani, F., Moazzam-Jazi, M., et al. (2021). High-throughput analysis of the interactions between viral proteins and host cell RNAs. Comput. Biol. Med. 135, 104611. doi:10.1016/j.compbiomed.2021.104611
Li, A., Ge, M., Zhang, Y., Peng, C., and Wang, M. (2015). Predicting long noncoding RNA and protein interactions using heterogeneous network model. BioMed Res. Int. 2015, 671950. doi:10.1155/2015/671950
Li, J., Li, J., Kong, M., Wang, D., Fu, K., and Shi, J. (2021). Svdnvlda: predicting lncRNA-disease associations by singular value decomposition and node2vec. BMC Bioinforma. 22, 538. doi:10.1186/s12859-021-04457-1
Liang, Y., Zhang, Z. Q., Liu, N. N., Wu, Y. N., Gu, C. L., and Wang, Y. L. (2022). Magcnse: predicting lncRNA-disease associations using multi-view attention graph convolutional network and stacking ensemble model. BMC Bioinforma. 23 (1), 189. doi:10.1186/s12859-022-04715-w
Liu, Y., Chen, X., Chen, X., Liu, J., Gu, H., Fan, R., et al. (2020). Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell. Death Dis. 11 (3), 175. doi:10.1038/s41419-020-2268-8
Liu, J. X., Gao, M. M., Cui, Z., Gao, Y. L., and Li, F. (2021a). Dscmf: prediction of LncRNA-disease associations based on dual sparse collaborative matrix factorization. BMC Bioinforma. 22 (3), 241. doi:10.1186/s12859-020-03868-w
Liu, Z., Wang, Y., Xu, Z., Yuan, S., Ou, Y., Luo, Z., et al. (2021b). Analysis of ceRNA networks and identification of potential drug targets for drug-resistant leukemia cell K562/ADR. PeerJ 9, e11429. doi:10.7717/peerj.11429
Liu, W., Lin, H., Huang, L., Peng, L., Tang, T., Zhao, Q., et al. (2022). Identification of miRNA–disease associations via deep forest ensemble learning based on autoencoder. Briefings Bioinforma. 23 (3), bbac104. doi:10.1093/bib/bbac104
Liu, W., Yang, Y., Lu, X., Fu, X., Sun, R., Yang, L., et al. (2023a). Nsrgrn: a network structure refinement method for gene regulatory network inference. Briefings Bioinforma. 24 (3), bbad129. doi:10.1093/bib/bbad129
Liu, W., Tang, T., Lu, X., Fu, X., Yang, Y., and Peng, L. (2023b). Mpclcda: predicting circRNA–disease associations by using automatically selected meta-path and contrastive learning. Briefings Bioinforma. 24, bbad227. doi:10.1093/bib/bbad227
Ma, X., Li, Y., Song, Y., and Xu, G. (2021). Long noncoding RNA CCDC26 promotes thyroid cancer malignant progression via miR-422a/EZH2/Sirt6 axis. OncoTargets Ther. 14, 3083–3094. doi:10.2147/OTT.S282011
Ma, Y. (2022). Deepmne: deep multi-network embedding for lncRNA-disease association prediction. IEEE J. Biomed. Health Inf. 26 (7), 3539–3549. doi:10.1109/JBHI.2022.3152619
Meng, J., Kang, Q., Chang, Z., and Luan, Y. (2021). PlncRNA-HDeep: plant long noncoding RNA prediction using hybrid deep learning based on two encoding styles. BMC Bioinforma. 22 (3), 242. doi:10.1186/s12859-020-03870-2
Pasmant, E., Sabbagh, A., Vidaud, M., and Bièche, I. (2011). ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 25 (2), 444–448. doi:10.1096/fj.10-172452
Peng, L., Huang, L., Lu, Y., Liu, G., Chen, M., and Han, G. (2022a). “Identifying possible lncRNA-disease associations based on deep learning and positive-unlabeled learning,” in 2022 IEEE international conference on bioinformatics and biomedicine (BIBM) (IEEE), 168–173.
Peng, L., Tan, J., Tian, X., and Zhou, L. (2022b). EnANNDeep: an ensemble-based lncRNA–protein interaction prediction framework with adaptive k-nearest neighbor classifier and deep models. Interdiscip. Sci. Comput. Life Sci. 14 (1), 209–232. doi:10.1007/s12539-021-00483-y
Peng, L., Wang, C., Tian, X., Zhou, L., and Li, K. (2022c). Finding lncrna-protein interactions based on deep learning with dual-net neural architecture. IEEE/ACM Trans. Comput. Biol. Bioinforma. 19 (6), 3456–3468. doi:10.1109/TCBB.2021.3116232
Peng, L., Wang, F., Wang, Z., Tan, J., Huang, L., Tian, X., et al. (2022d). Cell–cell communication inference and analysis in the tumour microenvironments from single-cell transcriptomics: data resources and computational strategies. Briefings Bioinforma. 23 (4), bbac234. doi:10.1093/bib/bbac234
Peng, L., Tan, J., Xiong, W., Zhang, L., Wang, Z., Yuan, R., et al. (2023a). Deciphering ligand–receptor-mediated intercellular communication based on ensemble deep learning and the joint scoring strategy from single-cell transcriptomic data. Comput. Biol. Med. 16 (2023), 107137. doi:10.1016/j.compbiomed.2023.107137
Peng, L., Yuan, R., Han, C., Han, G., Tan, J., Wang, Z., et al. (2023b). CellEnBoost: a boosting-based ligand-receptor interaction identification model for cell-to-cell communication inference. IEEE Trans. NanoBioscience, 1–11. doi:10.1109/TNB.2023.3278685
Rajagopal, T., Talluri, S., Akshaya, R. L., and Dunna, N. R. (2020). HOTAIR LncRNA: a novel oncogenic propellant in human cancer. Clin. Chim. acta 503, 1–18. doi:10.1016/j.cca.2019.12.028
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shen, L., Liu, F., Huang, L., Liu, G., Zhou, L., and Peng, L. (2022). VDA-RWLRLS: an anti-SARS-CoV-2 drug prioritizing framework combining an unbalanced bi-random walk and Laplacian regularized least squares. Comput. Biol. Med. 140, 105119. doi:10.1016/j.compbiomed.2021.105119
Stein, N., Berhani, O., Schmiedel, D., Duev-Cohen, A., Seidel, E., Kol, I., et al. (2019). IFNG-AS1 enhances interferon gamma production in human natural killer cells. Iscience 11, 466–473. doi:10.1016/j.isci.2018.12.034
Su, R., Ma, J., Zheng, J., Liu, X., Liu, Y., Ruan, X., et al. (2020). PABPC1-induced stabilization of BDNF-AS inhibits malignant progression of glioblastoma cells through STAU1-mediated decay. Cell. Death Dis. 11 (2), 81. doi:10.1038/s41419-020-2267-9
Sun, X., Liu, M., and Sima, Z. (2020). A novel cryptocurrency price trend forecasting model based on LightGBM. Finance Res. Lett. 32, 101084. doi:10.1016/j.frl.2018.12.032
Tan, L., Yu, J. T., Hu, N., and Tan, L. (2013). Non-coding RNAs in Alzheimer's disease. Mol. Neurobiol. 47, 382–393. doi:10.1007/s12035-012-8359-5
Veličković, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., and Bengio, Y. (2017). Graph attention networks. arXiv preprint arXiv:1710.10903.
Velickovic, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., and Bengio, Y. (2017). Graph attention networks. stat 1050 (20), 10–48550. doi:10.48550/arXiv.1710.10903
Wang, L., Cai, Y., Zhao, X., Jia, X., Zhang, J., Liu, J., et al. (2015). Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma 62 (3), 412–418. doi:10.4149/neo_2015_049
Wang, M. N., You, Z. H., Wang, L., Li, L. P., and Zheng, K. (2021). Ldgrnmf: lncRNA-disease associations prediction based on graph regularized non-negative matrix factorization. Neurocomputing 424, 236–245. doi:10.1016/j.neucom.2020.02.062
Wang, L., Shang, M., Dai, Q., and He, P. A. (2022). Prediction of lncRNA-disease association based on a Laplace normalized random walk with restart algorithm on heterogeneous networks. BMC Bioinforma. 23 (1), 5–20. doi:10.1186/s12859-021-04538-1
Wu, H., Liang, Q., Zhang, W., Zou, Q., Hesham, A. E. L., and Liu, B. (2022). iLncDA-LTR: identification of lncRNA-disease associations by learning to rank. Comput. Biol. Med. 146, 105605. doi:10.1016/j.compbiomed.2022.105605
Xie, G., Huang, B., Sun, Y., Wu, C., and Han, Y. (2021). RWSF-BLP: a novel lncRNA-disease association prediction model using random walk-based multi-similarity fusion and bidirectional label propagation. Mol. Genet. Genomics 296, 473–483. doi:10.1007/s00438-021-01764-3
Xie, G., Zhu, Y., Lin, Z., Sun, Y., Gu, G., Li, J., et al. (2022). Hbrwrlda: predicting potential lncRNA–disease associations based on hypergraph bi-random walk with restart. Mol. Genet. Genomics 297 (5), 1215–1228. doi:10.1007/s00438-022-01909-y
Xu, J., Xu, J., Meng, Y., Lu, C., Cai, L., Zeng, X., et al. (2023). Graph embedding and Gaussian mixture variational autoencoder network for end-to-end analysis of single-cell RNA sequencing data. Cell. Rep. Methods 3, 100382. doi:10.1016/j.crmeth.2022.100382
Yan, J., Chen, D., Chen, X., Sun, X., Dong, Q., Hu, C., et al. (2019). Downregulation of lncRNA CCDC26 contributes to imatinib resistance in human gastrointestinal stromal tumors through IGF-1R upregulation. Braz. J. Med. Biol. Res. 52, e8399. doi:10.1590/1414-431x20198399
Yang, Q., and Li, X. (2021). BiGAN: lncRNA-disease association prediction based on bidirectional generative adversarial network. BMC Bioinforma. 22, 357. doi:10.1186/s12859-021-04273-7
Yang, M., Zhao, L., Hu, X., Feng, H., and Kang, X. (2021). Identification of key mRNAs and lncRNAs associated with the effects of anti-TWEAK on osteosarcoma. Curr. Bioinforma. 16 (1), 154–161. doi:10.2174/1574893615999200626191405
Yao, Y., Ji, B., Lv, Y., Li, L., Xiang, J., Liao, B., et al. (2021). Predicting LncRNA–disease association by a random walk with restart on multiplex and heterogeneous networks. Front. Genet. 12, 712170. doi:10.3389/fgene.2021.712170
Zafar, A., Wang, W., Liu, G., Wang, X., Xian, W., McKeon, F., et al. (2021). Molecular targeting therapies for neuroblastoma: progress and challenges. Med. Res. Rev. 41 (2), 961–1021. doi:10.1002/med.21750
Zhang, E. B., Yin, D. D., Sun, M., Kong, R., Liu, X. H., You, L. H., et al. (2014). P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell. death Dis. 5 (5), e1243. doi:10.1038/cddis.2014.201
Zhang, J., Qiu, W. Q., Zhu, H., Liu, H., Sun, J. H., Chen, Y., et al. (2020). HOTAIR contributes to the carcinogenesis of gastric cancer via modulating cellular and exosomal miRNAs level. Cell. death Dis. 11 (9), 780. doi:10.1038/s41419-020-02946-4
Zhao, X., Zhao, X., and Yin, M. (2022). Heterogeneous graph attention network based on meta-paths for lncrna–disease association prediction. Briefings Bioinforma. 23 (1), bbab407. doi:10.1093/bib/bbab407
Zhou, L., Wang, Z., Tian, X., and Peng, L. (2021). LPI-deepGBDT: a multiple-layer deep framework based on gradient boosting decision trees for lncRNA–protein interaction identification. BMC Bioinforma. 22 (1), 479. doi:10.1186/s12859-021-04399-8
Keywords: lncRNA, biomarker, lung cancer, neuroblastoma, deep neural network, LightGBM
Citation: Su Z, Lu H, Wu Y, Li Z and Duan L (2023) Predicting potential lncRNA biomarkers for lung cancer and neuroblastoma based on an ensemble of a deep neural network and LightGBM. Front. Genet. 14:1238095. doi: 10.3389/fgene.2023.1238095
Received: 10 June 2023; Accepted: 19 July 2023;
Published: 16 August 2023.
Edited by:
Junlin Xu, Hunan University, ChinaReviewed by:
XianFang Tang, Wuhan Textile University, ChinaWenyan Wang, Anhui University of Technology, China
Copyright © 2023 Su, Lu, Wu, Li and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zejun Li, bHpqZm94QGhuaXQuZWR1LmNu; Lian Duan, ZHVhbmxpYW4zMDFAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Zhenguo Su1†
Zhenguo Su1† Yan Wu
Yan Wu Zejun Li
Zejun Li Lian Duan
Lian Duan