- 1Pomology Institute, Shanxi Agricultural University, Shanxi Key Laboratory of Germplasm Improvement and Utilization in Pomology, Taiyuan, Shanxi, China
- 2College of Horticulture, Shanxi Agricultural University, Jinzhong, Shanxi, China
- 3Integrative Omics and Molecular Modeling Laboratory, Department of Bioinformatics and Biotechnology, Government College University Faisalabad (GCUF), Faisalabad, Pakistan
In the genomics of plants and the phytoecosystem, Pyrus (pear) is among the most nutritious fruits and contains fiber that has great health benefits to humans. It is mostly cultivated in temperate regions and is one of the most cultivated pome fruits globally. Pears are highly subjected to biotic and abiotic stresses that affect their yield. TIR1/AFB proteins act as auxin co-receptors during the signaling of nuclear auxins and play a primary role in development-related regulatory processes and responses to biotic and abiotic stresses. However, this gene family and its members have not been explored in Pyrus genomes, and understanding these genes will help obtain useful insights into stress tolerance and ultimately help maintain a high yield of pears. This study reports a pangenome-wide investigation of TIR1/AFB genes from eight Pyrus genomes: Cuiguan (Pyrus pyrifolia), Shanxi Duli (P. betulifolia), Zhongai 1 [(P. ussuriensis × communis) × spp.], Nijisseiki (P. pyrifolia), Yunhong No.1 (P. pyrifolia), d’Anjou (P. communis), Bartlett v2.0 (P. communis), and Dangshansuli v.1.1 (P. bretschneideri). These genes were randomly distributed on 17 chromosomes in each genome. Based on phylogenetics, the identified TIR1/AFB genes were divided into six groups. Their gene structure and motif pattern showed the intraspecific structural conservation as well as evolutionary patterns of Pyrus TIR1/AFBs. The expansion of this gene family in Pyrus is mainly caused by segmental duplication; however, a few genes showed tandem duplication. Moreover, positive and negative selection pressure equally directed the gene’s duplication process. The GO and PPI analysis showed that Pyrus TIR1/AFB genes are associated with abiotic stress- and development-related signaling pathways. The promoter regions of Pyrus TIR1/AFB genes were enriched in hormone-, light-, development-, and stress-related cis elements. Furthermore, publicly available RNA-seq data analysis showed that DaTIR1/AFBs have varied levels of expression in various tissues and developmental stages, fruit hardening disease conditions, and drought stress conditions. This indicated that DaTIR1/AFB genes might play critical roles in response to biotic and abiotic stresses. The DaTIR1/AFBs have similar protein structures, which show that they are involved in the same function. Hence, this study will broaden our knowledge of the TIR1/AFB gene family in Pyrus, elucidating their contribution to conferring resistance against various environmental stresses, and will also provide valuable insights for future researchers.
Introduction
Indole-3-acetic acid (IAA) is the most common and first auxin to be discovered in plants. The auxin hormone is widely involved in plant growth, development, and morphogenesis (Kepinski and Leyser, 2005a; Zhao, 2010). The functions of auxins in plants are mainly controlled by modulating auxin synthesis metabolism, signal transduction (Petra et al., 2009), and polar transport (Mockaitis and Estelle, 2008). The classical pathway that mediates auxin signaling is known as transport inhibitor response1/auxin signaling F-box (TIR1/AFB)-mediated auxin regulation mechanism. The first auxin receptor protein to be reported was TIR1. Later, AFBs were also identified as the members of this family. The TIR1 genes belong to the F-box family of proteins whose members have highly conserved N-terminus F-box and C-terminus leucine-rich repeat (LRR) domains. The F-box domain of this family comprises the important component of the E3 ubiquitin ligase complex, which is involved in the degradation of auxin/indoleacetic acid (AUX/IAA) proteins (Hu et al., 2012).
AUX/IAA is one of the key gene families that are involved in quick response to fluctuations in auxin concentration. These gene family members are activated when the auxin level declines, form a dimer by binding to the auxin response factor (ARF), and regulate the auxin-induced gene expression. When auxin levels are high, the AUX/IAA proteins bind to the TIR1/AFBs, thus becoming degraded through ubiquitination that eliminates the inhibitory effect of AUX/IAAs. Auxin plays a major role in the growth, development, and morphogenesis of plants. Auxin response and signal transduction are strongly associated with the TIR1/AFB and AUX/IAA proteins (Paponov et al., 2008; Su et al., 2023a). Thus, the TIR1/AFB-AUX/IAA pathway is involved in the plant’s mechanism of the auxin signal’s perception, transduction, and response. It has also been demonstrated that auxin binding to TIR1/AFBs requires partial involvement of AUX/IAAs (Calderón Villalobos et al., 2012). Arabidopsis thaliana contains six TIR1/AFBs as well as 29 AUX/IAAs that are involved in diverse functions of auxin in this plant (Salehin et al., 2015).
Certain growth-related processes primarily regulated by auxin, including the elongation of the hypocotyl and lateral root development, are impaired in the mutants of tir1, having their functions lost (Ruegger et al., 1998). Overexpression of AtTIR1 leads to primary root growth inhibition, formation of a lateral root, and agravitropic root tips (Gray et al., 1999). TIR1 is the substrate receptor for the SKP-Cullin (SCF) E3 complex, which uses the ubiquitin-26S proteasome machinery to specifically break down Aux/IAA proteins; thus, SCF plays important roles in regulating cellular processes, including signal transduction (Xie et al., 2019). By directly binding to SCFTIR1, auxin molecules have the potential to enhance the interaction between the Aux/IAA and SCFTIR1 complex; TIR1 has been identified as an auxin receptor (Kepinski and Leyser, 2005b; Dharmasiri et al., 2005). Shortly after the identification of TIR1, the three genes with the names AFB1, AFB2, and AFB3 were discovered; these genes function similarly to TIR1 (Tahir ul Qamar et al., 2020). It is important to note that all four genes—TFR1, AFB1, AFB2m, and AFB3—have involvement in the way plants respond to auxin in the root, but AFB1 does not form an SCF complex as effectively as TIR1, AFB2, and AFB3 (Yu et al., 2015). Both AFB4 and AFB5 have been shown to have auxin receptor functions. The afb4 loss-of-function mutant seedlings show no discernible growth defects (Prigge et al., 2016).
Overexpression of AtTIR1 in A. thaliana led to the overproduction of lateral roots (Chen et al., 2011). Moreover, the AtAFB3 gene played a significant role in the lateral root development in response to nitrate treatment (Vidal et al., 2010). In the afb4 loss-of-function mutant seedlings, the number of lateral roots developed is lower than in the wild-type seedlings, suggesting that AFB4 is also involved in the development of lateral roots in Arabidopsis. The afb4 mutant also showed other defects in development-related processes, such as delayed flowering, lower height, and hypocotyls (Hu et al., 2012). ABA treatment generally inhibits the development of lateral roots in Arabidopsis; however, the overexpression of AtAFB2 has been demonstrated to counteract this inhibition, suggesting a response pathway from abiotic stress to this specific gene (Chen et al., 2012). Furthermore, the Arabidopsis TIR1 and AFB2 are required to inhibit the lateral root development by ABA or osmotic stress under drought stress (Chen et al., 2012). In rice, TIR1 and AFB2 were significantly down-expressed in spikelets under drought stress (Sharma et al., 2018). In maize, tomato, and potato crops, the expression of TIR1 was increased in seedlings when exposed to drought stress (Benny et al., 2019). The knockdown of soybean GmTIR1 and GmAFB3 genes resulted in fewer nodule numbers (Cai et al., 2017). Pears are also affected by fruit hardening disorder in which the top region of the pear is hardened, thus affecting the fruit quality and its commercial value. Because the pear is among the most highly cultivated and commercially important fruits, this disease and its related mechanism have been investigated in several studies (Liu et al., 2021).
Pears are one of the most significant temperate fruit trees globally and belong to the Rosaceae family and the Amygdaloideae subfamily. Pears have been cultivated for over 3,000 years, with 39 billion tons being delivered worldwide yearly (Wu et al., 2014; Ferradini et al., 2017; Wu et al., 2018). Presently, 22 species of pear with 5000 accessions have been reported. These five species are majorly cultivated for fruit production, including Pyrus bretschneideri, P. pyrifolia, P. communis, P. ussuriensis, and P. sinkiangensis (Li et al., 2022). Most cultivated pears have a diploid genome (2n = 34) that is extremely heterozygous and has several repeating sequences (Chen et al., 2023). Pear is an economical fruit with a sweet taste and great nutritional value. Abiotic stresses such as drought and biotic stresses limit the cultivation area, affecting their growth and yield. Therefore, there is an urgent need to address this situation.
TIR1/AFB genes have been shown to have important regulatory roles in auxin-regulated responses, making these major regulators of plant growth and development that are often halted by environmental factors. In this project, TIR1/AFB gene family members have been identified from eight pear genomes: Cuiguan, Shanxi Duli, Zhongai 1, Nijisseiki, Yunhong No.1, d’Anjou, Bartlett v2.0, and Dangshansuli’ v.1.1 (Chen et al., 2023). Characterization and phylogenetic analysis were performed to determine the intraspecific evolutionary relationships among these genes. The results of the analysis of protein profiles and intron/exon structures supported the classification of the Pyrus TIR1/AFB genes. In addition, the expression profiles of Pyrus TIR1/AFB genes showed that these genes had different expression patterns under abiotic (drought) and biotic stresses (disease conditions). The results of this study might help better understand the role of Pyrus TIR1/AFBs in the biotic as well as abiotic stress response of pears and provide a foundation for identifying candidate genes involved in drought response and fruit hardening disease.
Materials and methods
Identification and physiochemical characterization of Pyrus TIR1/AFBs
The six A. thaliana TIR1/AFB protein sequences were retrieved using the Arabidopsis Information Resource website (https://www.arabidopsis.org/). The protein sequence FASTA files of Cuiguan, Shanxi Duli, Zhongai 1, Nijisseiki, Yunhong No.1, d’Anjou, Bartlett v2.0, and Dangshansuli’ v.1.1 were used as subject sequences to run the BLAST+ command-line tool. First, local databases for the eight Pyrus proteomes were created, and then the BLASTp search was conducted against each Pyrus proteome sequence database using TIR1/AFB protein sequences as the queries. Then, the resulting BLAST hits were filtered by steps including isoforms and duplicate removal.
The candidate Pyrus TIR1/AFB sequences were searched for the F-box protein family domains using the NCBI conserved domain database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al., 2011) and the InterPro (https://www.ebi.ac.uk/interpro/) (Hunter et al., 2009) database to identify the final protein family sequences. Physicochemical characteristics, including molecular weight, their isoelectric point (pI), aliphatic index (AI), instability index II), and the grand average of hydropathicity (GRAVY) values were predicted by using the ExPASy ProtParam tool (https://web.expasy.org/protparam/) (Gasteiger et al., 2005). The subcellular localization of these TIR1/AFB proteins was predicted by using the WoLF PSORT tool (https://wolfpsort.hgc.jp/) (Horton et al., 2007).
Phylogenetic analysis, conserved motifs, and gene structure analysis of Pyrus TIR1/AFBs
Phylogenetic analysis was conducted to evaluate the intraspecific evolutionary links among Pyrus TIR1/AFBs. A multiple sequence alignment of seven Cuiguan, six Shanxi Duli, six Zhongai 1, seven Nijisseiki, six Yunhong No.1, nine d’Anjou, five Bartlett v2.0, four Dangshansuli’ v.1.1, 6 A. thaliana (Kepinski and Leyser, 2005b), 18 Brassica juncea (Cai et al., 2000), and eight Populus (Shu et al., 2015) protein sequences was performed using ClustalW (Thompson et al., 2003). A phylogenetic tree was constructed using the IQTREE Web Server (http://iqtree.cibiv.univie.ac.at/) (Trifinopoulos et al., 2016) using the maximum likelihood (ML) method performing model selection via ModelFinder and bootstrap replicates of 1000. The tree was visualized and edited using the online Interactive Tree of Life server, iTOL (https://itol.embl.de/) (Letunic and Bork, 2007).
To evaluate the conserved common motifs present in all eight Pyrus TIR1/AFB sequences, the multiple Expectation Maximization for Motif Elicitation (MEME, https://meme-suite.org/meme/) (Bailey et al., 2009) tool was used. The conserved motif number was set to 20 to search for each sequence. The gene structures were constructed by using CDS and genomic sequences through the Gene Structure Display Server (GSDS 2.0, https://gsds.gao-lab.org/) (Hu et al., 2015). The identified motifs and gene structures were visualized using TBtools (Chen et al., 2018).
Chromosomal distribution, Ka/Ks rate, and gene duplication analysis
The chromosomal positions for each Pyrus TIR1/AFB were retrieved from the GFF/GFF3 files and mapped to chromosomes by using the gene location visualization tool of the TBtools software (Chen et al., 2018). Depending on whether the shorter gene’s length covered 70% of the longer gene and whether the two aligned genes’ similarity was equal to or greater than 70%, TIR1/AFB gene duplication occurrences were identified (Tahir ul Qamar et al., 2023). The genes were also checked to determine whether the duplication pattern was segmental or tandem. To predict the selection pressure for the duplicated genes, Ka/Ks values were also predicted using DnaSP v.6 software (Le et al., 2011; Rozas et al., 2017). Depending on whether the Ka/Ks ratio was more than, equal to, or less than one, purifying, neutral, or positive selection was analyzed (Zia et al., 2022). Furthermore, the divergence time for the duplicated gene pairs was also calculated by using the formula t = Ks/2λ×10−6, where the λ value for dicots, 1.5 × 10−8, calculates the time of duplication in million-year units (Fatima et al., 2023a).
PPI and GO enrichment analysis
The STRING database (von Mering et al., 2003) was used to analyze the PPIs among the Pyrus TIR1/AFB proteins using the amino acid sequences. The top 10 interactions were set to perform the prediction, and the interactions threshold was kept at 0.4. Furthermore, the PPI network was visualized by using Cytoscape software (Shannon et al., 2003). The GO enrichment analysis components, biological processes (BPs), cellular components (CCs), and molecular functions (MFs) were predicted using the PANNZER database (pannzer2 (helsinki.fi)) (Törönen et al., 2018).
Cis-regulatory elements prediction and expression profiling of Pyrus TIR1/AFBs
To predict the cis-regulatory elements, the 2-kb sequences upstream of the translation start site of Pyrus TIR1/AFB genes were retrieved and used to search the PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Rombauts et al., 1999).
To gain insights into the expression pattern of Pyrus Dangshansuli TIR1/AFBs, transcriptomic RNA-seq data of different developmental stages of pear fruit (BioProject: PRJNA309745), under drought stress (BioProject: PRJNA655255), and fruit hardening disease (BioProject: PRJNA763913) were obtained from the SRA-NCBI database (https://www.ncbi.nlm.nih.gov/sra). The Dangshansuli genome and annotation (GFF) files were downloaded from the Pear Genomics Database (PGDB) (http://pyrusgdb.sdau.edu.cn/) (Chen et al., 2023). The quality of the reads was evaluated by using the FastQC tool (Wingett and Andrews, 2018). The indexes of the Pyrus genome used were built by using HISAT (Kim et al., 2019), and the high-quality paired-clean reads were then mapped onto the indexed genome. The abundance estimation of gene family members was performed by using StringTie (Pertea et al., 2016). Finally, the heatmap was generated by using the fragments per kilobase of transcript per million mapped reads (FPKM) values.
3D structure prediction of TIR1/AFB proteins
A protein requires a three-dimensional (3D) structure to perform its functions properly. The 3D structures of four Dangshansuli TIR1/AFBs (DaAFB1, DaAFB3a, DaAFB3b, and DaAFB5) were predicted by using the AlphaFold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb) (Jumper et al., 2021). The predicted 3D structures were evaluated by using the SAVES (https://saves.mbi.ucla.edu/) (Zameer et al., 2022) server and MolProbity (http://molprobity.biochem.duke.edu/) (Davis et al., 2007). Finally, the UCSF ChimeraX software (Goddard et al., 2018) was used to visualize these structures (Neupane et al., 2018; Zia et al., 2024).
Results
Identification and physiochemical characteristics of TIR1/AFB genes in Pyrus genomes
A total of seven genes from the Cuiguan genome (CuTIR1/AFB), six from Shanxi Duli (ShTIR1/AFB), six from Zhongai1 (ZhTIR1/AFB), seven from Nijisseiki (NiTIR1/AFB), six from Yunhong No.1 (YuTIR1/AFB), nine from d’Anjou (AnTIR1/AFB), five from Bartlett v2.0 (BrTIR1/AFB), and four from the Dangshansuli’ v.1.1 genome (DaTIR1/AFB) were identified. All of these members were confirmed for the presence of the F-box_5 superfamily domain (Supplementary Table S3). The protein names of each member were given according to their phylogenetic relationships (Table 1).
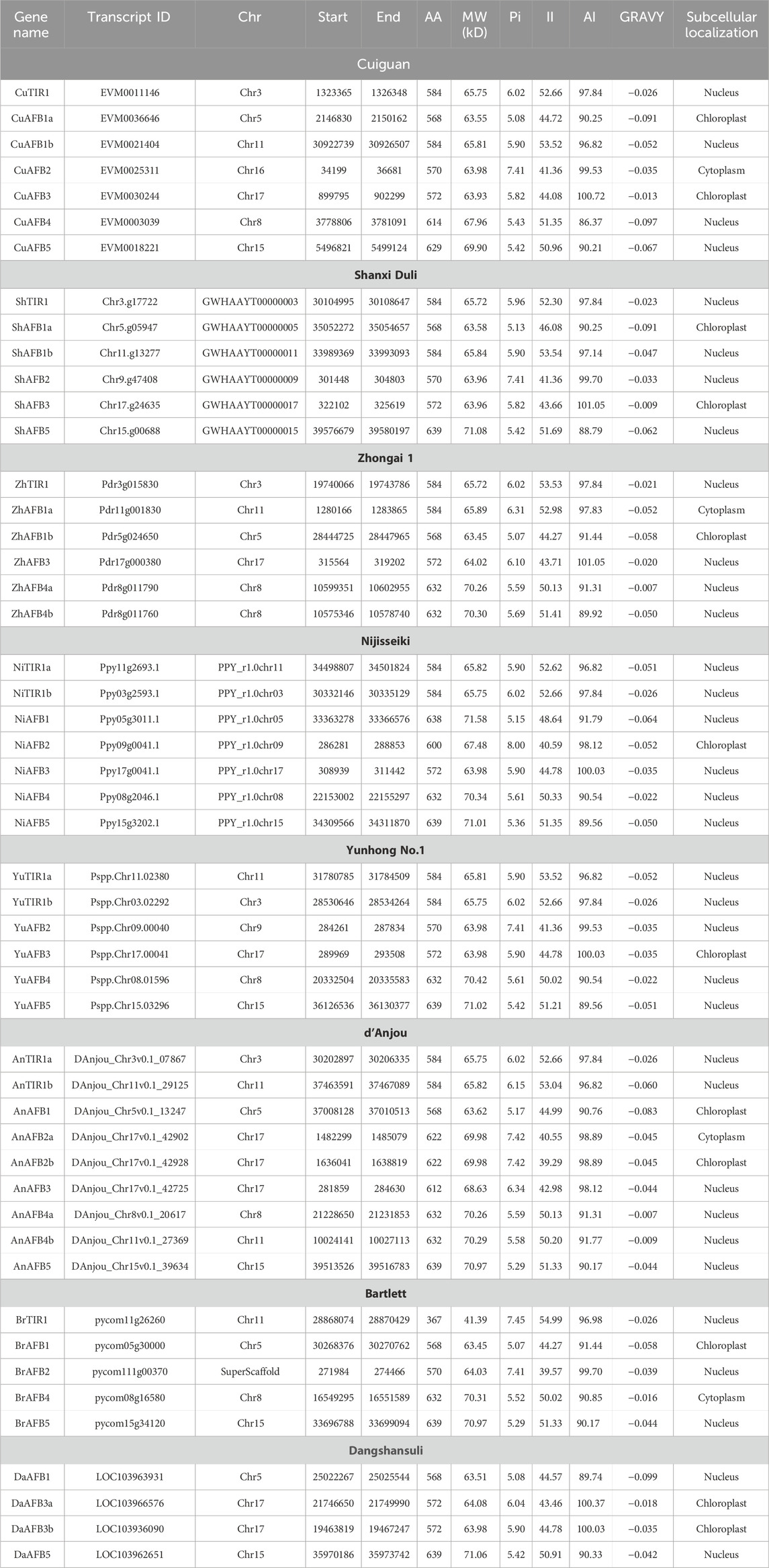
Table 1. TIR1/AFB gene family members identified in eight Pyrus genomes, their physicochemical characteristics including protein length (AA), molecular weight (MW), isoelectric point (pI), insatiability index (II), aliphatic index (AI), grand average of hydropathicity (GRAVY), and subcellular localization.
The length of the Cuiguan TIR1/AFB proteins ranged from 584aa–629aa and MW: 63 kDa–69 kDa. The CuTIR1/AFBs were located in the nucleus, chloroplast, and cytoplasm. For Shanxi Duli, the length of ShTIR1/AFB proteins ranged from 568aa–639aa and MW: 62 kDaa–71 kDa. The ShTIR1/AFBs were located in the nucleus and chloroplast. For Zhongai 1, the length of ZhTIR1/AFB proteins ranged from 568aa–632aa, MW: 63 kDa–70 kDa. The ZhTIR1/AFBs were located in the nucleus, chloroplast, and cytoplasm. For Nijisseiki, the length of NiTIR1/AFB proteins ranged from 572aa–639aa, MW: 63 kDa–71 kDa. The NiTIR1/AFBs were located in the nucleus and chloroplast. For Yunhong No. 1, the length of YuTIR1/AFB proteins ranged from 570aa–639aa and MW: 63 kDa–71 kDa. The YuTIR1/AFBs were located in the nucleus and chloroplast. For d’Anjou, the length of AnTIR1/AFB proteins ranged from 568aa–639aa and MW: 65 kDa–70 kDa. The AnTIR1/AFBs were located in the nucleus, cytoplasm, and chloroplast. For Bartlett, the length of BrTIR1/AFB proteins ranged from 568aa–639aa and MW: 41 kDa–70 kDa. The BrTIR1/AFBs were located in the nucleus, cytoplasm, and chloroplast. For Dangshansuli, the length of DaTIR1/AFB proteins ranged from 568aa–639aa and MW: 63 kDa–71 kDa. The DaTIR1/AFBs were located in the nucleus and chloroplast.
These results show that the members did not show significant differences in aa and MW values except for some proteins that showed deviant behavior. The Pyrus TIR1/AFB proteins had pI values ranging from 5–8, indicating their acidic as well as basic behavior. The II values of all these proteins were above 40, which indicates that these are unstable in the test tube. The AI values of all the Pyrus TIR1/AFB proteins indicated that these proteins are thermally stable. Furthermore, the negative GRAVY values of all these proteins indicated that these proteins are hydrophilic (Table 1).
Phylogenetic relationships of Pyrus TIR1/AFB family members
To analyze the evolutionary relationships among the TIR1/AFB members from eight Pyrus genomes, a phylogenetic tree was constructed by using 78 amino acid sequences from 11 species: eight Pyrus, A. thaliana, B. juncea, and Populus. All TIR1/AFB proteins were clustered into six groups: TIR1, AFB1, AFB2, AFB3, AFB4, and AFB5. TIR1 comprised the largest clade containing one AtTIR1, and the remaining members belonged to Pyrus genomes. AFB1 contained members from all eight Pyrus genomes. The AFB2 clade contained members from seven genomes but no members from the Zhongai1 genome. The AFB3 clade contained members from all seven genomes and no members from the Bartlett genome. The AFB4 clade contained members from six genomes, except the Shanxi Duli and Dangshansuli genomes. The AFB5 clade contained members from seven Pyrus genomes with no members from the Zhongai1 genome (Figure 1).
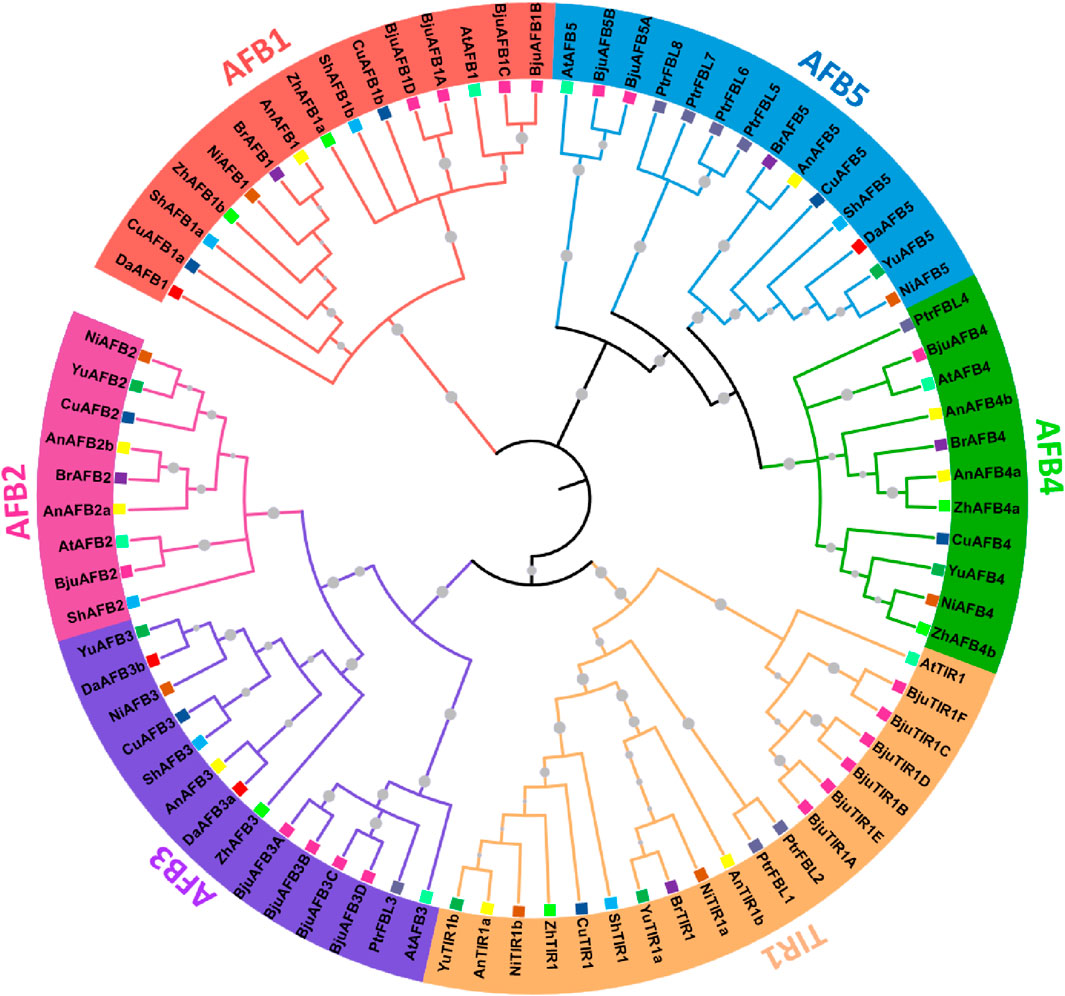
Figure 1. A maximum likelihood (ML) phylogenetic tree with 1000 bootstrap replicates (denoted by gray circle symbols) was generated using TIR1/AFB members from eight Pyrus, A. thaliana, B. juncea, and Populus genomes. Different branch colors represent different groups. Furthermore, each species is given a particular-colored square symbol at the leaf end.
Each group is represented by a particular color, and specific colors are used for each species’ genome.
Conserved motifs and gene structure analysis of Pyrus TIR1/AFBs
The conserved motifs and gene structures were analyzed to reveal the evolutionary pattern among Pyrus TIR1/AFBs. All members of each subfamily shared highly conserved motifs. Members of TIR1, AFB1, AFB2, and AFB3 have exactly similar motif patterns (conserved motifs 3–16). Meanwhile, only one member of the Bartlett genome, BrTIR1, had a different conservation pattern, with only motifs 3–10 being conserved. Members belonging to groups AFB4 and AFB5 showed a similar pattern as the groups mentioned earlier with some other motifs (motifs 1 and 2) (Figure 2A,C). This high conservation of motifs implies no major differences in the structure and functions of Pyrus TIR1/AFBs.
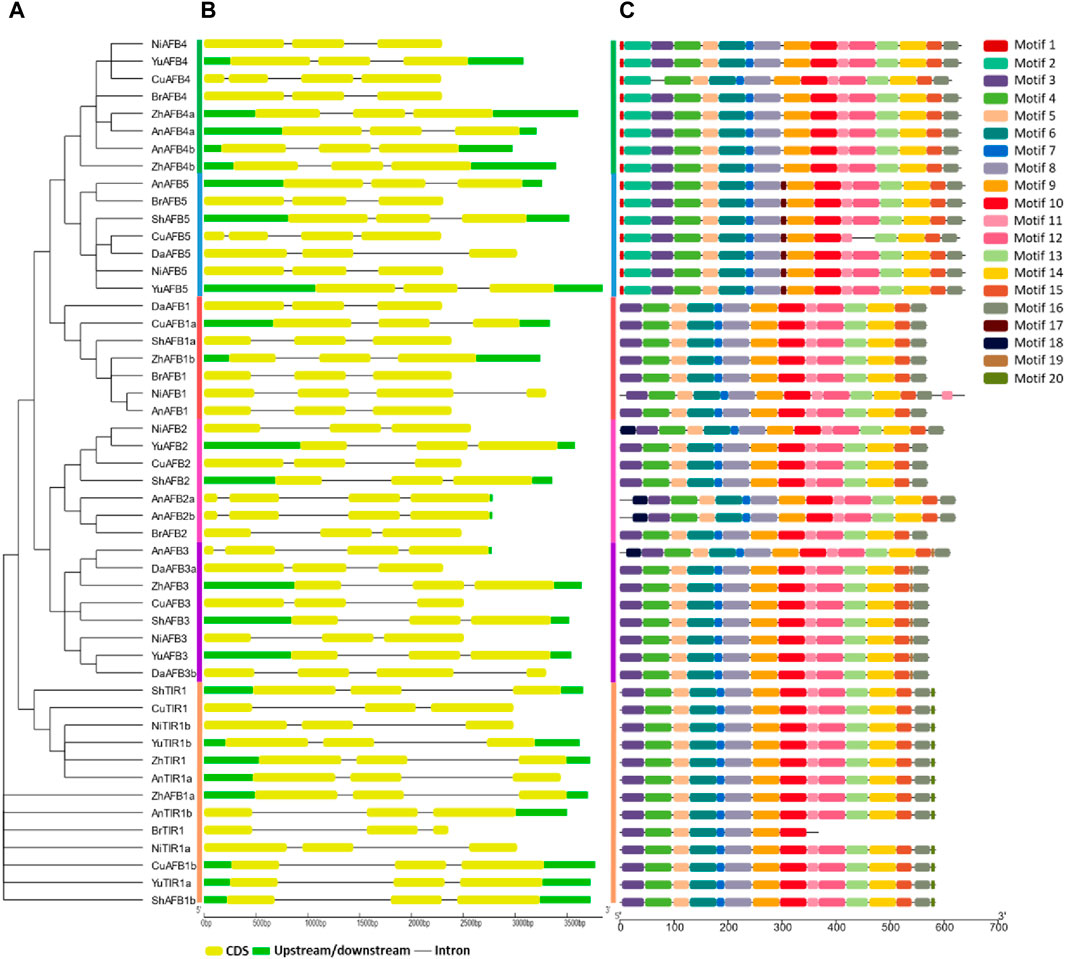
Figure 2. (A) Phylogenetic tree, motif pattern, and gene structures of Pyrus TIR1/AFBs, (B) gene structure showing conservation pattern of exons and introns, and (C) conserved motifs determined using the MEME suits.
Similarly, the gene structure was also found to be highly conserved among the members of the same subfamily. Almost all members contained at least three exons and two introns. A few members of the AFB2 subfamily (AnAFB2a, AnAFB2b, and AnAFB3) contained four conserved exons with three introns (Figure 2B). This conservation of motifs and exons among the Pyrus TIR1/AFBs indicated that these genes are conserved across eight Pyrus genomes.
Chromosomal mapping and gene duplication analysis
The chromosomal gene localization was identified to evaluate the gene distribution pattern of Pyrus TIR1/AFBs on 17 chromosomes of each Pyrus genome. This analysis showed that all genes were unevenly distributed on chromosomes. In the Cuiguan genome, seven CuTIR1/AFBs were found to be localized on seven of seventeen chromosomes (Chr3, 5, 8, 11, 15, 16, and 17). The remaining chromosomes had no CuTIR1/AFB genes present on them. In Shanxi Duli, six ShTIR1/AFBs were scattered on six of seventeen chromosomes. In the Zhongai1 genome, six ZhTIR1/AFBs were distributed on five of seventeen chromosomes. In the Nijisseiki genome, seven NiTIR1/AFBs were distributed on seven chromosomes. In the Yunhong No.1 genome, six YuTIR1/AFBs were unevenly distributed on six chromosomes. Seven AnTIR1/AFBs in d’Anjou genomes were distributed on six chromosomes. Five Bartlett v2.0 BrTIR1/AFBs were distributed on four chromosomes (Supplementary Figures S1–S7). Four DaTIR1/AFB members from the Dangshansuli’ v.1.1 genome were distributed on 3 of 17 chromosomes (Figure 3).
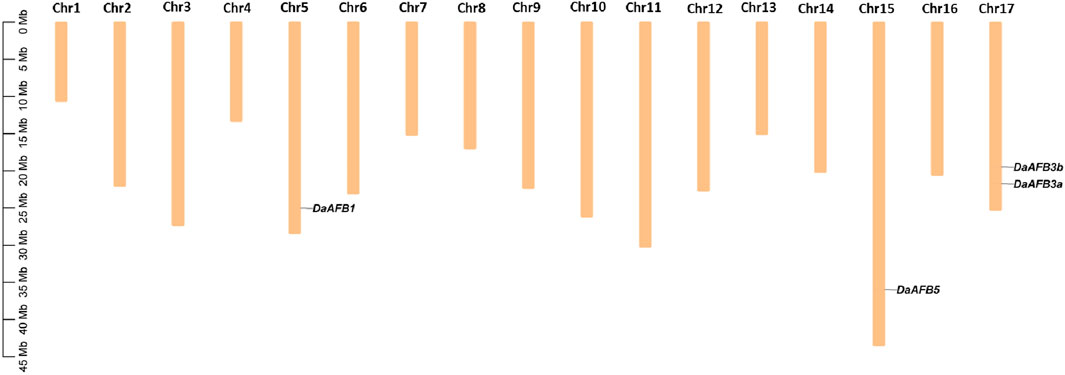
Figure 3. Chromosomal map showing four DaTIR1/AFBs distributed on 3 of 17 Dangshansuli’ v.1.1 chromosomes.
Gene duplication events were also analyzed among members of each Pyrus TIR1/AFBs (Table 2). The Cuiguan genome contained four duplicated pairs of genes, with all of them being segmentally duplicated. The genome of Shanxi Duli contained five segmentally duplicated pairs of ShTIR1/AFBs. The Zhongai1 genome contained four duplicated pairs of ZhTIR1/AFBs. Of these four pairs, one (ZhAFB4a/ZhAFB4b) exhibited tandem duplication, while the rest showed segmental duplication. The Nijisseiki genome contained five pairs of segmentally duplicated NiTIR1/AFBs. The Yunhong No.1 genome contained four pairs of YuTIR1/AFB genes that originated through segmental duplication. The d’Anjou genome exhibited eight pairs of duplicated genes. Of these two pairs, AnAFB2a/AnAFB3 and AnAFB2b/AnAFB3 had tandem duplication, while the rest were segmentally duplicated. The Bartlett genome contained two pairs of segmentally duplicated genes, BrTIR1/BrAFB2 and BrAFB4/BrAFB5. The Dungshanxuli genome exhibited only one segmentally duplicated gene pair, DaAFB1/DaAFB3a.
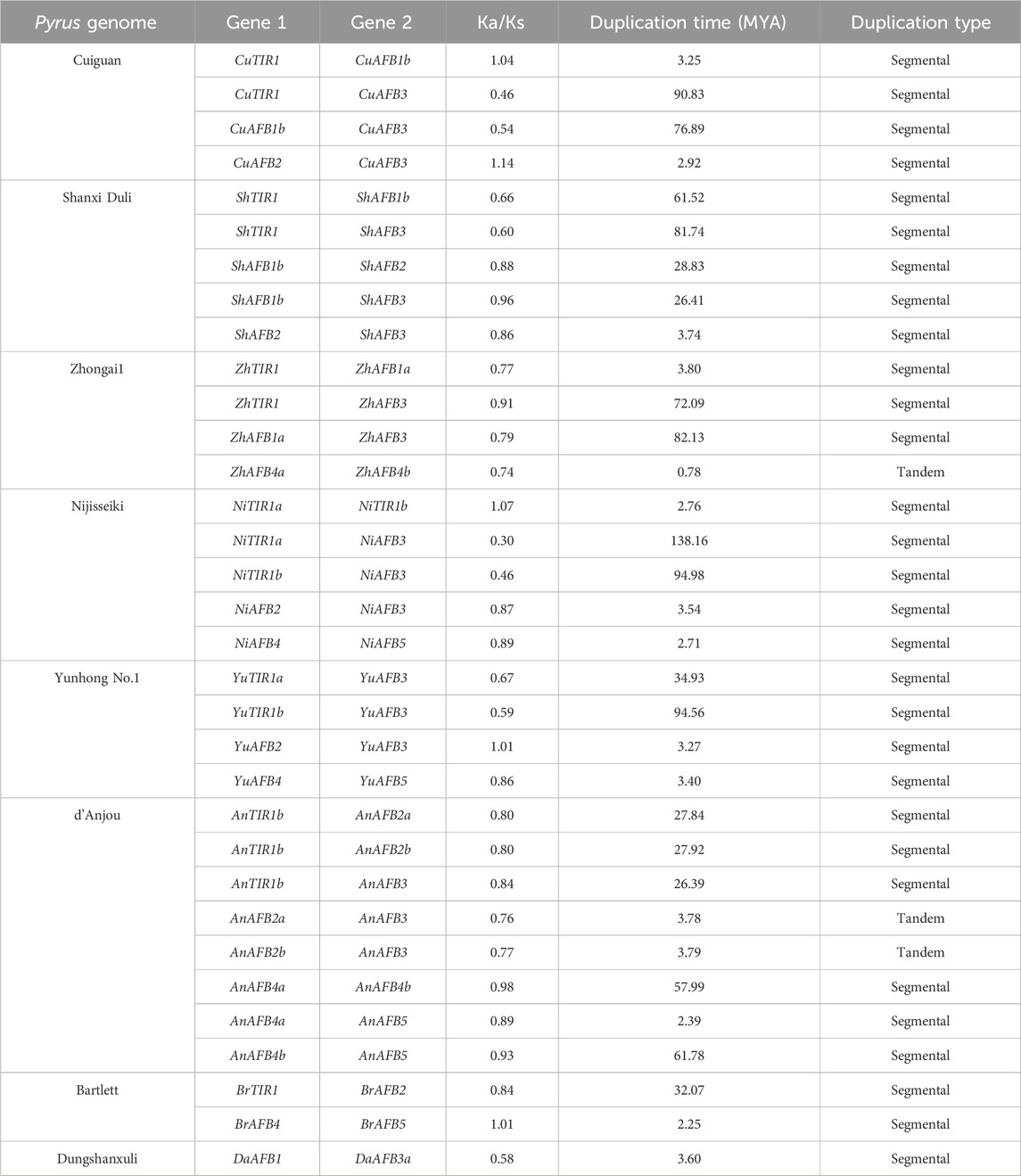
Table 2. Duplication data on Pyrus TIR1/AFBs, rate of synonymous (Ka) and non-synonymous mutations (Ks), duplication time (MYA), and the type of duplication.
To analyze the evolutionary constraints of the repeated Pyrus TIR1/AFB genes, the Ka, Ks, and Ka/Ks ratios of all para-homologous gene pairs were also calculated. The Ka/Ks ratio ranged from 0.5 to 1.1 in the eight genomes, which shows both positive and negative selection events. The time of divergence of all 30 duplicated gene pairs of Pyrus TIR1/AFBs was between 0.7 and 90.83 million years ago (MYA), which suggests a significant period of evolutionary divergence (Table 2).
PPI and GO enrichment analyses
A PPI network of the Pyrus TIR1/AFBs was generated to understand the functional diversity among members. The three Dangshansuli members, DaAFB1, DaAFB3b, and DaAFB5, interacted with several other proteins. Most of the interactions were identified with 1AA and AUX proteins, which shows the potential roles of these Pyrus members in auxin regulation, thus their involvement in the growth and development of plants (Figure 4A).
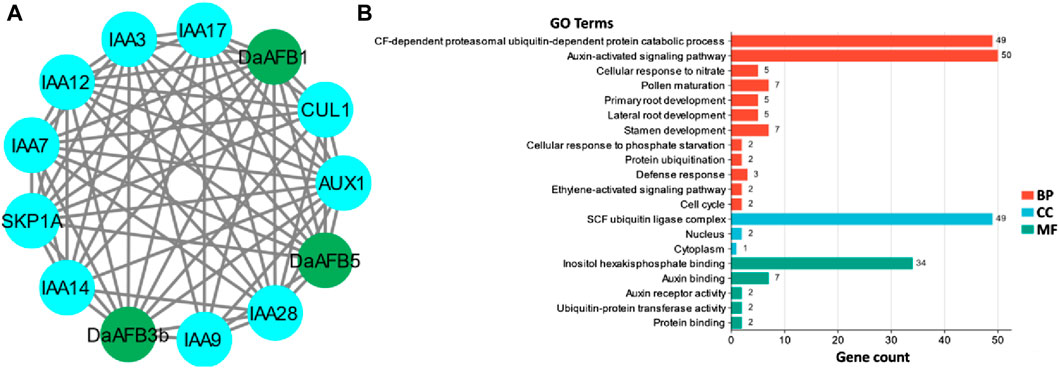
Figure 4. (A) Interactions among Pyrus TIR1/AFBs (shown in dark green nodes) and other homologous proteins (shown in blue color nodes). Gray lines indicate the interactions. (B) Predicted biological processes (BP), cellular components (CC), and molecular functions (MF) associated with Pyrus TIR1/AFBs.
GO enrichment analysis was carried out to ascertain the molecular roles of Pyrus TIR1/AFBs in a dynamic manner. Based on this GO analysis, Pyrus TIR1/AFB genes were classified into three different major categories: BPs, CCs, and MFs. Major BPs predicted included auxin-activated signaling pathways, defense responses, and other developmental and signaling pathways. These genes were found in CCs, including the nucleus, cytoplasm, and SCF ubiquitin ligase complex. The MFs associated with these genes included auxin and protein binding activity (Figure 4B; Supplementary Table S2). All these terms clearly indicate the functional involvement of Pyrus TIR1/AFBs in auxin signaling and drought stress responsiveness, thus resulting in growth.
Cis-regulatory element analysis of Pyrus TIR1/AFBs
To gain better insights into the diverse stress responses of Pyrus TIR1/AFBs, cis-regulatory elements in their promoter sequences were analyzed. In all genomes, cis elements associated with stress, such as light, hormones, and development-related responsiveness, were abundantly found. In relation to these elements, it was discovered that G-box, GT1-motif, and GATA-motif—cis elements Box 4—were implicated in the regulation of light stress. Hormone responsiveness was linked to five cis elements: P-box, TGA-element, ABRE, CGTCA-motif, and TCA-element. Furthermore, it was discovered that the GC-motif, LTR, TC-rich repeats, and MBS are the four cis elements associated with stress responsiveness. Developmental processes involved five elements: CAT-box, MBSI, circadian, HD-Zip 1, and o2-site. In Pyrus TIR1/AFBs, the presence of these elements shows their hormones, stress, and development-related responses (Figure 5; Supplementary Table S1).
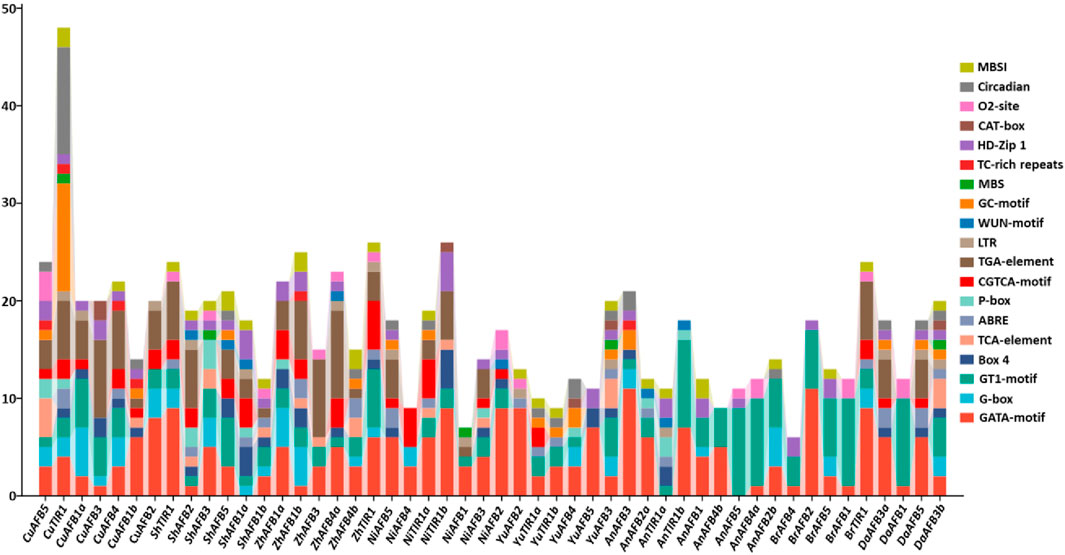
Figure 5. The Pyrus TIR1/AFB genes’ upstream promoter regions contain cis-regulatory elements. Every bar represents a distinct element found in a given gene.
Expression profiling of Pyrus TIR1/AFBs
Transcriptome expression data were used to determine the expression level of four DaTIR1/AFBs in tissues, including fruit, leaves, petals, sepals, ovaries, stems, and buds. DaAFB1 showed a decline in expression in ovary tissues. However, it was highly expressed in stem tissues. DaAFB5 exhibited a higher expression in leaves and bud tissues (Figure 6A).
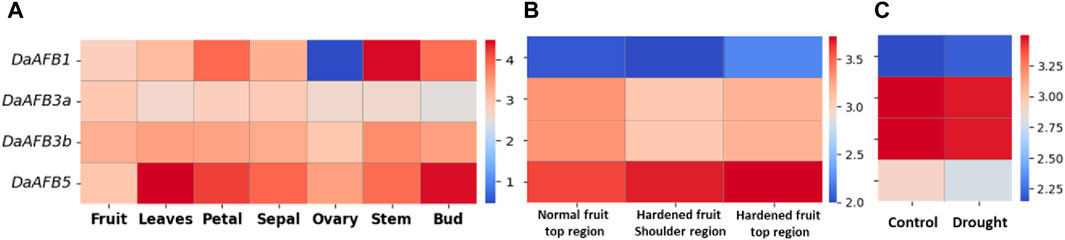
Figure 6. Heatmap showing the expression pattern of DaTIR1/AFBs in (A) different tissues where blue shows a decreased expression downregulation and red shows an increased expression, (B) disease condition where the normal fruit region is compared with hardened fruit, and (C) drought stress condition.
Expression profiles of DaTIR1/AFBs in fruit hardening disease conditions were also performed. DaAFB1 was highly downregulated under disease conditions. DaAFB5 showed an increased expression. However, DaAFB3a/3b showed no significant change in expression (Figure 6B). Under drought stress, DaAFB1 showed a large decrease in expression. DaAFB3a/3b showed overexpression in drought stress, and DaAFB5 exhibited no specific change in expression (Figure 6C). These results show the differential expression pattern at various stages and biotic and abiotic stress conditions.
3D structure prediction of DaTIR1/AFB proteins
To gain further insights into the structural and functional diversity, the 3D structures of the four DaTIR1/AFB proteins were modeled. The four proteins showed high conservation in 3D structures. All these four proteins contained similar structures of helices and turns. All the proteins shared a similar helical structure at the C and N termini. This conservation of structure suggests the potential similar functions of DaTIR1/AFB proteins as the DaAFB1 showed similar expression results in ovary tissues, fruit hardening, and drought stress conditions. Similarly, DaAFB5 also showed conservation in expression levels in various tissues and disease conditions (Figure 7).
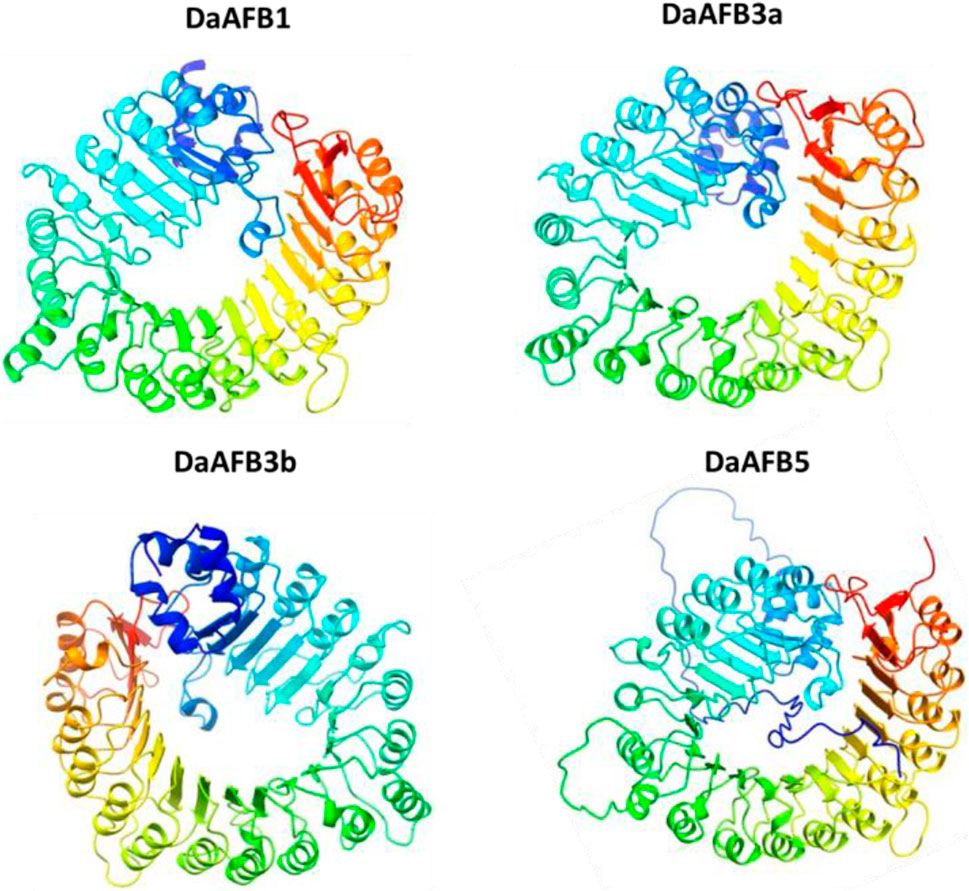
Figure 7. Predicted 3D structures of four DaTIR1/AFB proteins. In structures, blue and cyan colors represent the helices; yellow-green color patterns represent the turns.
Discussion
TIR1/AFB proteins have critical roles in auxin signaling. In Arabidopsis, these gene family members comprise a group of related proteins that include TIR1 and AFB1/2/3/4/5. Previous studies have reported that TIR1, along with AFBs, functions to regulate auxin signaling, which ultimately impacts plant growth and development (Dharmasiri et al., 2005). The present study reports a pangenome-wide identification of the TIR1/AFB gene family in eight Pyrus genomes. The TIR1/AFB gene family has already been reported in Arabidopsis, where the TIR1/AFB gene showed a significant role in lateral root development and hypocotyl elongation (Ruegger et al., 1998). In B. juncea var. tumida (Cai et al., 2000), 18 members showed differential expression in different tissues and salt stress. Similarly, eight members of this gene family have been reported in the Populus genome, where TIR1s showed differential expression in stem growth and drought stress (Shu et al., 2015). Moreover, this gene family has also been analyzed in Physcomitrium patens, Fragaria vesca (Fatima et al., 2023b), and Selaginella moellendorffii genomes (Su et al., 2023b), where these genes showed the tissue-specific expression patterns. These results suggest that this gene family contributes to plant development and responses to abiotic stresses.
The phylogenetic tree revealed that Pyrus TIR1/AFB proteins are divided into six subfamilies, namely, TRI1, AFB1, AFB2, AFB3, AFB4, and AFB5. This division of clades is consistent with other species, including Arabidopsis and Populus. In B. juncea, the BjuTIR1/AFBs are also divided into the same six clusters of subfamilies. The specific functions of these gene family members vary across and within the clades. For instance, the AFB4 and AFB5 shared the same clade but showed distinct specificities for auxin (Prigge et al., 2020). The tree showed that AFB1s and TIR1s clustered on the individual clades, while AFB2 and AFB3 originated from the same clade and diverged into two subclades, thus expanding independently. Similarly, AFB4 and AFB5 also arose from the same clade and then evolved separately (Dharmasiri et al., 2005). Most of the Pyrus TIR1/AFB proteins contained three conserved exons with two introns except for BjuAFB1D and BjuAFB1D, which contained four exons with three introns. This pattern of exon-intron conservation aligns with the Pyrus members that also contain three exons. Similarly, members of this gene family also showed comparable results in rice (Wang et al., 2007) and maize (Xing et al., 2011) genomes. However, members belonging to AFB4 and AFB5 clades had motifs 2 and 3 conserved. Similarly, Arabidopsis AFB2, AFB3, AFB4, and AFB5 showed certain conserved motifs (Du et al., 2022). This presence and absence of certain motifs indicates that TIR1/AFBs may have different functions.
The evolution of the members of this gene family was examined by comparing Pyrus TIR1/AFBs. It was found that almost all genes evolved through segmental duplication. Only one member from the Zhongai1 genome and two from the d’Anjou genome showed tandem duplication. The four Populus gene pairs (PtrFBL1/PtrFBL2, PtrFBL3/PtrFBL4, PtrFBL5/PtrFBL6, and PtrFBL7/PtrFBL8) were found to be originated from the genome duplication. These members also exhibited a similar pattern of intensive segmental recombination (Shu et al., 2015). Similarly, the multiple auxin receptor homologs in B. juncea also originated due to genome duplication (Cai et al., 2000). However, TIR1/AFBs from land plants exhibited tandem duplication to be more dominant during genome evolution (Su et al., 2023b).
In the promoter regions, cis elements were predicted to gain a better understanding of the role of Pyrus TIR1/AFBs under various environmental conditions. Similar to land plants P. patens, S. moellendorfii, A. thaliana, and F. vesca, these gene family members contained a large number of hormone-responsive elements (auxin, salicylic acid, gibberellin, and abscisic acid). Moreover, elements related to growth and drought stress were also found (Su et al., 2023a). The promoter of BjuTIR1/AFB genes also contained elements related to plant responses to biotic as well as abiotic stress (Cai et al., 2000). Furthermore, the functional prediction through PPI and GO analysis also showed the conservation and involvement of these genes in auxin responsiveness and stress tolerance mechanisms. PPI analysis revealed that DaAFB1, DaAFB5, and DaAFB3b showed high interactions with IAA proteins. Studies have shown that the IAA proteins help in mediating tolerance to stresses (such as drought) in plants (Salehin et al., 2019). These DaAFBs have been shown to be involved in BPs such as auxin-activated signaling pathways and lateral root development processes. The Arabidopsis AFB1 has been shown to have a specialized function in rapid auxin-dependent inhibition of root growth and the early phase of root gravitropism (Prigge et al., 2020). Thus, the Pyrus TIR1/AFBs can be deemed to be potentially involved in abiotic and biotic stress responses mediated by auxin.
RNA-seq expression data analysis revealed that Pyrus TIR1/AFB genes express differently in different tissues, fruit hardening disease, and drought stress conditions. Drought stress affects the expression of multiple TIR1/AFB genes, indicating a potential role for the TIR1/AFB family in the drought tolerance pathway (Du et al., 2022). In Arabidopsis, the TIR1 showed an increased expression under drought stress (Benny et al., 2019). The rice TIR1 and AFB2 showed decreased expression under drought stress (Sharma et al., 2018). In wheat, the roots exposed to drought stress showed an increase in expression of AFB2, suggesting its key role in drought stress responsiveness (Dalal et al., 2018). Due to the targeting and reduction of AsAFB2 and AsTIR1 expression, creeping bentgrass (Agrostis stolonifera L.) overexpressing the rice pri-miR393a showed increased tolerance to drought stress. DaAFB5 was highly expressed in tissues, while DaAFB1 exhibited low expression. The BjuTIR1/AFBs also showed differential expression patterns in different tissues, including root, stem, leaf, flower, pod, and swollen stem. In contrast, these gene family members had halted expression in salt stress (Cai et al., 2000). Promoter, PPI, GO, and expression analyses revealed that DaTIR1/AFBs are involved in auxin-mediated stress responsiveness to both abiotic and biotic stresses. Therefore, these genes can be used in future breeding studies to improve Pyrus stress resilience to various environmental stresses.
Conclusion
The present study provides a systematic as well as comparative analysis of TIR1/AFB genes in eight economically important and nutritious Pyrus genomes. A total of seven genes from the Cuiguan genome (CuTIR1/AFB), six from Shanxi Duli (ShTIR1/AFB), six from Zhongai1 (ZhTIR1/AFB), seven from Nijisseiki (NiTIR1/AFB), six from Yunhong No.1 (YuTIR1/AFB), nine from d’Anjou (AnTIR1/AFB), five from Bartlett v2.0 (BrTIR1/AFB), and four from the Dangshansuli’ v.1.1 genome (DaTIR1/AFB) were identified. These genes were classified into six groups. All the members from the same group shared conservation in motif and gene structure. Most of the genes originated through segmental duplication, and a few originated through tandem duplication. A large number of cis elements related to hormones, stress, and development were found in the promoter region of these Pyrus TIR1/AFBs. PPI and GO also revealed the involvement of these genes in auxin-mediated stress-related and development-related processes. Structure prediction showed high conservation among members of DaTIR1/AFBs. DaTIR1, DaTIR3a, DaTIR3b, and DaTIR5 showed differential expression in various tissues, fruit hardening disease, and drought stress. Our results provide a solid foundation to further investigate the function of TIR1/AFBs in regulating various abiotic and environmental stress responses in Pyrus.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author contributions
SY: writing–original draft. XY: writing–review and editing. XG: writing–review and editing. KF: writing–original draft. MT: writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was partially supported by the Natural Science Foundation of Shanxi Province (202103021224122), the earmarked fund for CARS (CARS-28-29), the Shanxi Fruit Industry Technology System Construction Project (SXFRS-2023), subtasks of national key R&D plan projects (2021YFD1901105-X-Y), the Central Leading Local Science and Technology Development Fund Project: research, development, and demonstration of key technologies for cold shed planting of Yulu xiangli pear in Northern Xinjiang, the Special Project for Guiding the Transformation of Scientific and Technological Achievements in Shanxi Province (202204021301037), the XPCC Science and Technology Planning Project (2023AB004-01, the Shanxi Province Science and Technology Major Special Plan (202201140601027-2), and Shanxi Agricultural University (YZGC036).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1393487/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | CuTIR1/AFBs distributed on Cuiguan chromosomes.
SUPPLEMENTARY FIGURE S2 | ShTIR1/AFBs distributed on Shanxi Duli chromosomes.
SUPPLEMENTARY FIGURE S3 | ZhTIR1/AFBs distributed on Zhongai1 chromosomes.
SUPPLEMENTARY FIGURE S4 | NiTIR1/AFBs distributed on Nijisseiki chromosomes.
SUPPLEMENTARY FIGURE S5 | YuTIR1/AFBs distributed on Yunhong No.1 chromosomes.
SUPPLEMENTARY FIGURE S6 | AnTIR1/AFBs distributed on d’Anjou chromosomes.
SUPPLEMENTARY FIGURE S7 | BrTIR1/AFBs distributed on Bartlett chromosomes.
SUPPLEMENTARY TABLE S1 | Identified cis elements in Pyrus TIR1/AFB genes.
SUPPLEMENTARY TABLE S2 | Gene ids, their GO enrichment classes, and description of predicted GO terms.
SUPPLEMENTARY TABLE S3 | Domains identified in Pyrus TIR1/AFB members.
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Benny, J., Pisciotta, A., Caruso, T., and Martinelli, F. (2019). Identification of key genes and its chromosome regions linked to drought responses in leaves across different crops through meta-analysis of RNA-Seq data. BMC Plant Biol. 19, 194–218. doi:10.1186/s12870-019-1794-y
Cai, Z., Wang, Y., Zhu, L., Tian, Y., Chen, L., Sun, Z., et al. (2017). GmTIR1/GmAFB3-based auxin perception regulated by miR393 modulates soybean nodulation. New Phytol. 215, 672–686. doi:10.1111/nph.14632
Cai, Z., Zeng, D., Liao, J., Cheng, C., Sahito, Z. A., Xiang, M., et al. (2000). Genome-wide analysis of auxin receptor family genes in Brassica juncea var. tumida. Genes. (Basel). 10, 165–215. doi:10.3390/genes10020165
Calderón Villalobos, L. I. A., Lee, S., De Oliveira, C., Ivetac, A., Brandt, W., Armitage, L., et al. (2012). A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485. doi:10.1038/nchembio.926
Chen, C., Xia, R., Chen, H., and He, Y. (2018). TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. TBtools, a Toolkit Biol. Integr. Var. HTS-Data Handl. Tools a User-Friendly Interface, 289660. doi:10.1101/289660
Chen, H., Li, Z., and Xiong, L. (2012). A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett. 586, 1742–1747. doi:10.1016/j.febslet.2012.05.013
Chen, S., Sun, M., Xu, S., Xue, C., Wei, S., Zheng, P., et al. (2023). The pear genomics database (PGDB): a comprehensive multi-omics research platform for Pyrus spp. BMC Plant Biol. 23, 430. doi:10.1186/s12870-023-04406-5
Chen, Z.-H., Bao, M.-L., Sun, Y.-Z., Yang, Y.-J., Xu, X.-H., Wang, J.-H., et al. (2011). Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 77, 619–629. doi:10.1007/s11103-011-9838-1
Dalal, M., Sahu, S., Tiwari, S., Rao, A. R., and Gaikwad, K. (2018). Transcriptome analysis reveals interplay between hormones, ROS metabolism and cell wall biosynthesis for drought-induced root growth in wheat. Plant Physiol. biochem. 130, 482–492. doi:10.1016/j.plaphy.2018.07.035
Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., et al. (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, 375–383. doi:10.1093/nar/gkm216
Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., et al. (2005). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 9, 109–119. doi:10.1016/j.devcel.2005.05.014
Du, W., Lu, Y., Li, Q., Luo, S., Shen, S., Li, N., et al. (2022). TIR1/AFB proteins: active players in abiotic and biotic stress signaling. Front. Plant Sci. 13, 1083409. doi:10.3389/fpls.2022.1083409
Fatima, K., Sadaqat, M., Azeem, F., Rao, M. J., Albekairi, N. A., Alshammari, A., et al. (2023a). Integrated omics and machine learning-assisted profiling of cysteine-rich-receptor-like kinases from three peanut spp. revealed their role in multiple stresses. Front. Genet. 14, 1–17. doi:10.3389/fgene.2023.1252020
Fatima, K., Sadaqat, M., Azeem, F., and Tahir ul Qamar, M., Chapter 7 - role of integrative omics and bioinformatics approaches in berries research and genetic improvement, in: I. Khalifa, and A. B. T.-B. B. C. B.-P. Nawaz (Eds.), Germany, Academic Press, 2023b: pp. 159–192. doi:10.1016/B978-0-323-95600-0.00005-5
Ferradini, N., Lancioni, H., Torricelli, R., Russi, L., Dalla Ragione, I., Cardinali, I., et al. (2017). Characterization and phylogenetic analysis of ancient Italian landraces of pear. Front. Plant Sci. 8, 751. doi:10.3389/fpls.2017.00751
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., Bairoch, A., et al. (2005). Protein identification and analysis tools on the ExPASy server. Proteomics Protoc. Handb., 571–607. doi:10.1385/1-59259-890-0:571
Goddard, T. D., Huang, C. C., Meng, E. C., Pettersen, E. F., Couch, G. S., Morris, J. H., et al. (2018). UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. doi:10.1002/pro.3235
Gray, W. M., Del Pozo, J. C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., et al. (1999). Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes. Dev. 13, 1678–1691. doi:10.1101/gad.13.13.1678
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi:10.1093/nar/gkm259
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi:10.1093/bioinformatics/btu817
Hu, Z., Keçeli, M. A., Piisilä, M., Li, J., Survila, M., Heino, P., et al. (2012). F-box protein AFB4 plays a crucial role in plant growth, development and innate immunity. Cell. Res. 22, 777–781. doi:10.1038/cr.2012.12
Hunter, S., Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Binns, D., et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res. 37, 211–215. doi:10.1093/nar/gkn785
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kepinski, S., and Leyser, O., Plant development: auxin in loops, Curr. Biol. 15 (2005a) R208–R210. doi:10.1016/j.cub.2005.03.012
Kepinski, S., and Leyser, O. (2005b). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. doi:10.1038/nature03542
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi:10.1038/s41587-019-0201-4
Le, D. T., Nishiyama, R., Watanabe, Y., Mochida, K., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2011). Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 18, 17–29. doi:10.1093/dnares/dsq032
Letunic, I., and Bork, P. (2007). Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi:10.1093/bioinformatics/btl529
Li, J., Zhang, M., Li, X., Khan, A., Kumar, S., Allan, A. C., et al. (2022). Pear genetics: recent advances, new prospects, and a roadmap for the future.
Liu, X., Li, S., Feng, X., and Li, L. (2021). Study on cell wall composition, fruit quality and tissue structure of hardened ‘suli’ pears (Pyrus bretschneideri rehd). J. Plant Growth Regul. 40, 2007–2016. doi:10.1007/s00344-020-10248-4
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, 225–229. doi:10.1093/nar/gkq1189
Mockaitis, K., and Estelle, M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell. Dev. Biol. 24, 55–80. doi:10.1146/annurev.cellbio.23.090506.123214
Neupane, S., Andersen, E. J., Neupane, A., and Nepal, M. P. (2018). Genome-wide identification of NBS-encoding resistance genes in sunflower (Helianthus annuus L.). Genes. (Basel) 9, 384. doi:10.3390/genes9080384
Paponov, I. A., Paponov, M., Teale, W., Menges, M., Chakrabortee, S., Murray, J. A. H., et al. Comprehensive transcriptome analysis of auxin responses in Arabidopsis, Mol. Plant. 1 (2008) 321–337. doi:10.1093/mp/ssm021
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi:10.1038/nprot.2016.095
Petrasek, J., and Friml, J. (2009). Auxin transport routes in plant development. Development 136, 2675–2688. doi:10.1242/dev.030353
Prigge, M. J., Greenham, K., Zhang, Y., Santner, A., Castillejo, C., Mutka, A. M., et al. (2016). The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 Genes., Genomes, Genet. 6, 1383–1390. doi:10.1534/g3.115.025585
Prigge, M. J., Platre, M., Kadakia, N., Zhang, Y., Greenham, K., Szutu, W., et al. (2020). Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. Elife 9, e54740. doi:10.7554/eLife.54740
Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi:10.1093/nar/27.1.295
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi:10.1093/molbev/msx248
Ruegger, M., Dewey, E., Gray, W. M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes. Dev. 12, 198–207. doi:10.1101/gad.12.2.198
Salehin, M., Bagchi, R., and Estelle, M. (2015). SCFTIR1/AFB-Based auxin perception: mechanism and role in plant growth and development. Plant Cell. 27, 9–19. doi:10.1105/tpc.114.133744
Salehin, M., Li, B., Tang, M., Katz, E., Song, L., Ecker, J. R., et al. (2019). Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 10, 4021. doi:10.1038/s41467-019-12002-1
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Sharma, L., Dalal, M., Verma, R. K., V Kumar, S. V., Yadav, S. K., Pushkar, S., et al. (2018). Auxin protects spikelet fertility and grain yield under drought and heat stresses in rice. Environ. Exp. Bot. 150, 9–24. doi:10.1016/j.envexpbot.2018.02.013
Shu, W., Liu, Y., Guo, Y., Zhou, H., Zhang, J., Zhao, S., et al. (2015). A Populus TIR1 gene family survey reveals differential expression patterns and responses to 1-naphthaleneacetic acid and stress treatments. Front. Plant Sci. 6, 719–810. doi:10.3389/fpls.2015.00719
Su, L., Zhang, T., Yang, B., Dong, T., Liu, X., Bai, Y., et al. AUX/IAAs and their implications for the morphogenesis of land plants, (2023b) 1–10.
Su, L., Zhang, T., Yang, B., Dong, T., Liu, X., Bai, Y., et al. (2023a). Different evolutionary patterns of TIR1/AFBs and AUX/IAAs and their implications for the morphogenesis of land plants. BMC Plant Biol. 23, 265. doi:10.1186/s12870-023-04253-4
Tahir ul Qamar, M., Zhu, X., Khan, M. S., Xing, F., and Chen, L.-L., Pan-genome: a promising resource for noncoding RNA discovery in plants, Plant Genome. 13 (2020) e20046. doi:10.1002/tpg2.20046
Tahir ul Qamar, M., Sadaqat, M., Zhu, X.-T., Li, H., Huang, X., Fatima, K., et al. (2023). Comparative genomics profiling revealed multi-stress responsive roles of the CC-NBS-LRR genes in three mango cultivars. Front. Plant Sci. 14, 1285547. doi:10.3389/fpls.2023.1285547
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2003). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinforma. 00, Unit 2.3–22. doi:10.1002/0471250953.bi0203s00
Törönen, P., Medlar, A., and Holm, L. (2018). PANNZER2: a rapid functional annotation web server. Nucleic Acids Res. 46, W84-W88–W88. doi:10.1093/nar/gky350
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi:10.1093/nar/gkw256
Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 107, 4477–4482. doi:10.1073/pnas.0909571107
von Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., and Snel, B. (2003). STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261. doi:10.1093/nar/gkg034
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi:10.1016/j.gene.2007.01.006
Wingett, S. W., and Andrews, S. (2018). FastQ Screen: a tool for multi-genome mapping and quality control. F1000Research 7, 1338. doi:10.12688/f1000research.15931.2
Wu, J., Li, L.-T., Li, M., Khan, M. A., Li, X.-G., Chen, H., et al. (2014). High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J. Exp. Bot. 65, 5771–5781. doi:10.1093/jxb/eru311
Wu, J., Wang, Y., Xu, J., Korban, S. S., Fei, Z., Tao, S., et al. (2018). Diversification and independent domestication of Asian and European pears. Genome Biol. 19, 77. doi:10.1186/s13059-018-1452-y
Xie, J., Jin, Y., and Wang, G. (2019). The role of SCF ubiquitin-ligase complex at the beginning of life. Reprod. Biol. Endocrinol. 17, 101. doi:10.1186/s12958-019-0547-y
Xing, H., Pudake, R. N., Guo, G., Xing, G., Hu, Z., and Zhang, Y. (2011). Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize.
Yu, H., Zhang, Y., Moss, B. L., Bargmann, B. O. R., Wang, R., Prigge, M., et al. (2015). Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat. Plants 1, 14030–14038. doi:10.1038/nplants.2014.30
Zameer, R., Fatima, K., Azeem, F., Algwaiz, H. I. M., Sadaqat, M., Rasheed, A., et al. (2022). Genome-wide characterization of superoxide dismutase (SOD) genes in daucus carota: novel insights into structure, expression, and binding interaction with hydrogen peroxide (H2O2) under abiotic stress condition. Front. Plant Sci. 13, 870241–870315. doi:10.3389/fpls.2022.870241
Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. doi:10.1146/annurev-arplant-042809-112308
Zia, K., Rao, M. J., Sadaqat, M., Azeem, F., Fatima, K., Tahir ul Qamar, M., et al. (2022). Pangenome-wide analysis of cyclic nucleotide-gated channel (CNGC) gene family in citrus Spp. Revealed their intraspecies diversity and potential roles in abiotic stress tolerance. Front. Genet. 13, 1034921. doi:10.3389/fgene.2022.1034921
Keywords: Pyrus, pangenome-wide, TIR1/AFBs, evolutionary pattern, gene ontology, fruit hardening disease, abiotic stress, drought stress
Citation: Yang S, Yu X, Gao X, Fatima K and Tahir Ul Qamar M (2024) Comparative genomic profiling of transport inhibitor Response1/Auxin signaling F-box (TIR1/AFB) genes in eight Pyrus genomes revealed the intraspecies diversity and stress responsiveness patterns. Front. Genet. 15:1393487. doi: 10.3389/fgene.2024.1393487
Received: 29 February 2024; Accepted: 09 April 2024;
Published: 10 May 2024.
Edited by:
Sajid Shokat, International Atomic Energy Agency, AustriaReviewed by:
Lihu Wang, Hebei University of Engineering, ChinaMuhammad Amjad Nawaz, Tomsk State University, Russia
Copyright © 2024 Yang, Yu, Gao, Fatima and Tahir Ul Qamar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Yang, eXMyMDA4MDgwOEAxNjMuY29t; Muhammad Tahir Ul Qamar, dGFoaXJ1bHFhbWFyQGdjdWYuZWR1LnBr
 Sheng Yang1*
Sheng Yang1* Kinza Fatima
Kinza Fatima Muhammad Tahir Ul Qamar
Muhammad Tahir Ul Qamar