- 1Department of Medical Genetics/Prenatal Diagnostic Center, West China Second University Hospital, Sichuan University, Chengdu, China
- 2Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Chengdu, China
F-box and leucine-rich repeat protein 4 (FBXL4) plays a crucial role in mitochondrial bioenergetics, mitochondrial DNA (mtDNA) maintenance, and mitochondrial dynamics. The variations in the FBXL4 gene can give rise to encephalomyopathy mitochondrial DNA depletion syndrome-13 (MTDPS13) characterized by the reduction of mtDNA copy number, leading to deficiencies in mitochondrial functions, which is a serious and rare autosomal recessive genetic disorder. Patients with FBXL4 variations are usually diagnosed due to the emergence of symptoms in the early stages of life. Commonly observed are lactic acidemia, developmental retardation, and hypotonia. A portion of patients may be accompanied by comorbidities such as cardiovascular diseases, epilepsy, ophthalmopathy, hearing impairment, and movement disorders. Currently, there have been no reported cases of prenatal diagnosis for FBXL4 gene variations. Here, we report for the first time the prenatal diagnosis of a fetus with a compound heterozygous mutation in the FBXL4 gene (NM_012160.5: c.1288C>T, p. Arg430* and c.518_523del, p. Glu173_Leu175delinsVal) by trio-WES, the nonsense mutation (c.1288C>T) was reported only once in an unrelated individual and no detailed clinical phenotype; the deletion mutation (c.518_523del) has not been reported yet. Additionally, we monitor prenatal phenotypes of fetus at different stages of pregnancy using ultrasound and magnetic resonance imaging (MRI), present prenatally with nuchal translucency (NT) thickening and progressive brain developmental abnormalities. Our report indicates that the application of trio whole exome sequencing (trio-WES) and imaging monitoring can facilitate prenatal diagnosis of FBXL4 gene-related MTDPS13, and this will modify the decision-making process for couples with FBXL4 variations.
1 Introduction
Mitochondria are the energy-producing structures in cells and the primary site for aerobic respiration. Mitochondrial autophagy is a process by which damaged, dysfunctional, or excessive mitochondria are selectively processed through autophagy, playing a vital role in maintaining cellular equilibrium (Aman et al., 2021). F-box and leucine-rich repeat protein 4 (FBXL4) is a member of the F-box protein family, and a protein located on the outer mitochondrial membrane that participates in regulating mitochondrial autophagy (Cao et al., 2023). The human genome encodes 69 F-box proteins, which are regarded as substrate receptors to facilitate the recognition of the Skp1-Cul1-F-box (SCF) E3 ubiquitin ligase complex. The SCF-FBXL4 E3 ubiquitin ligase complex is situated on the outer mitochondrial membrane, constitutively mediates the ubiquitination and degradation of BNIP3L/NIX and BNIP3 mitochondrial autophagy receptors to suppress mitochondrial autophagy. The post-translational regulation of BNIP3L and BNIP3 is disrupted in mitochondrial DNA depletion syndrome 13 (MTDPS13), a multisystem disorder disease caused by variations in the FBXL4 gene (Cao et al., 2023). MTDPS13 is a rare autosomal recessive disease induced by biallelic mutations in the F-box and leucine-rich repeat (LRR) protein 4 gene (FBXL4) (Gai et al., 2013), characterized by increased mitochondrial autophagy and mitochondrial DNA (mtDNA) depletion in FBXL4 knockout mice and patient fibroblasts (Alsina et al., 2020).
Since its first description in 2013 (Bonnen et al., 2013; Gai et al., 2013), a total of 59 pathogenic variations in FBXL4 have been identified in 112 patients with autosomal recessive inheritance. These variations occur through homozygosity or compound heterozygosity (Gao et al., 2024). Our case will be the 113rd reported case and the first prenatal diagnosis. Among them, the deletion mutation (c.518_523del) is a novel variation in FBXL4 gene. Patients with FBXL4 variations are typically diagnosed due to the manifestation of symptoms in the early stages of life. The associated symptoms of MTDPS13 can vary considerably among different patients, FBXL4 variations tend to exert more severe influences on energy-demanding organs such as the brain, heart, muscles, and the renal system. Commonly reported symptoms encompass hypotonia, neurodevelopmental delay, lactic acidemia, microcephaly, hyperammonemia, and growth failure. Additionally, a series of less frequent manifestations have been identified, including extensive neurological, ophthalmological, cardiac, gastrointestinal, urogenital, and immunological manifestations. Special facial features have been observed in some affected individuals. Abnormalities detected in patients undergoing brain magnetic resonance imaging (MRI) include white matter abnormalities, diffuse cerebral atrophy, basal ganglia abnormalities, ventricular dilation, periventricular cysts, brainstem abnormalities, thinning of the corpus callosum, and arachnoid cysts (El-Hattab et al., 2017; Gao et al., 2024). The prognosis of MTDPS13 patients is extremely unfavorable, with a high mortality rate. The causes of death include sepsis, severe acidosis, pneumonia, and cardiopulmonary failure (El-Hattab et al., 2017).
There is no reported prenatal diagnostic of MTDPS13 linked to FBXL4. For the neonates died with FBXL4 variations, the review of prenatal phenotypes show periventricular cysts, periventricular echogenicity, ventriculomegaly, thin corpus callosum, mega cisterna magna, and large cavum (Saini et al., 2022), polyhydramnios and cerebellar atrophy (van Rij et al., 2016a). Therefore, prenatal diagnostic for FBXL4 variants is important, which will help pregnant’ women making decision. This case will report the prenatal diagnostic of MTDPS13 linked to FBXL4 detected by MRI and trio Whole Exome Sequencing (trio-WES) and its detailed prenatal phenotypes.
2 Fetal phenotype
The prenatal diagnostic center received a referral for a 32-year-old pregnant woman. The obstetric history of pregnant woman includes two pregnancies and one birth. The couple is not relatives. Besides, the family history was non-contributory. The pregnant’s previous ultrasound had no significant findings except increased nuchal translucency (NT: 3.7 mm). At 22+3 weeks of gestation, the pregnant woman underwent amniocentesis for copy number variation sequencing (CNV-seq) of the fetal amniotic fluid, which did not reveal any CNVs or chromosome aneuploidies that may lead to NT thickening. After receiving thorough consultation from a geneticist, the pregnant woman agreed to perform trio-WES on the genomic DNA extracted from the fetal amniotic fluid and parental peripheral blood, which identified a compound heterozygous variant in the FBXL4 gene. After reviewing literature and databases, no prenatal report of NT thickening related to FBXL4 gene was found, which may be associated with prenatal phenotypes such as abnormal brain development (including ventricular dilatation, thin corpus callosum, cerebellar hypoplasia, etc.) and polyhydramnios (Saini et al., 2022; van Rij et al., 2016b).
In order to further evaluate the prognosis of the fetus and the pathogenicity of FBXL4 gene variations, the geneticist recommended conducting fetal brain MRI. The targeted fetal brain MRI at 26+4 weeks of gestation showed a subependymal cyst located on the left side of the anterior horn of the lateral ventricle (Figure 1A). Reexamination MRI performed at 35 weeks of gestation showed the subependymal cyst had enlarged compared to the previous examination (Figure 1B), along with bilateral lateral ventricle dilatation (Figure 1C) and the enlarged posterior fossa (Figure 1D). The targeted ultrasound at 35+6 weeks of gestation also indicated a subependymal cyst (Figure 1E), as well as a fetal posterior fossa measuring approximately 1.0 cm (Figure 1F).
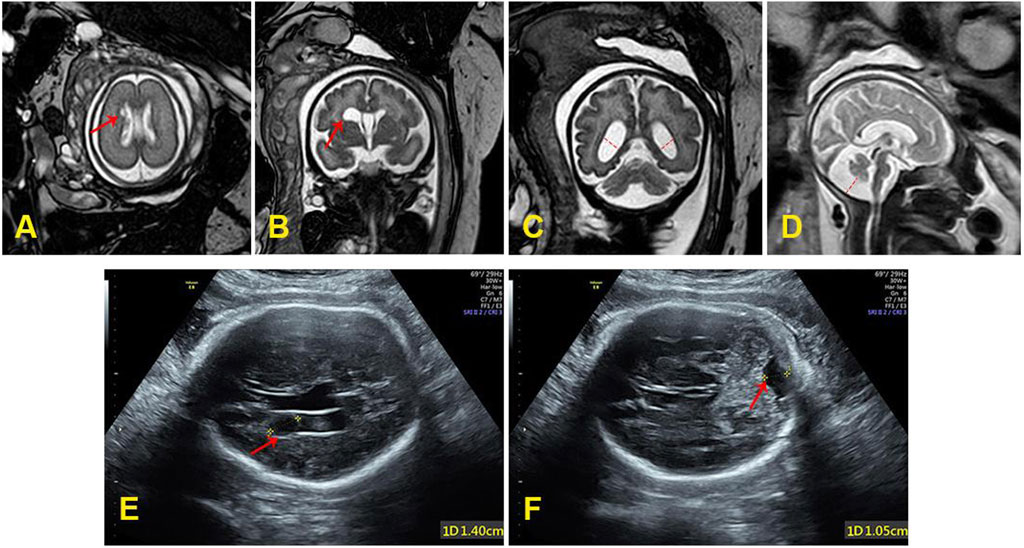
Figure 1. (A) The subependymal cyst (0.5 cm × 0.3 cm) at 26+4 weeks of gestation. (B) The subependymal cyst (1.62 cm × 1.26 cm × 0.88 cm) at 35 weeks of gestation. (C) The bilateral lateral ventricle dilatation (left: 1.19 cm, right: 1.01 cm) at 35 weeks of gestation. (D) The enlarged posterior fossa (1.37 cm) at 35 weeks of gestation. (E) The subependymal cyst (1.4 cm) at 35+6 weeks of gestation. (F) The posterior fossa was close to 1.0 cm at 35+6 weeks of gestation.
3 Diagnostic method
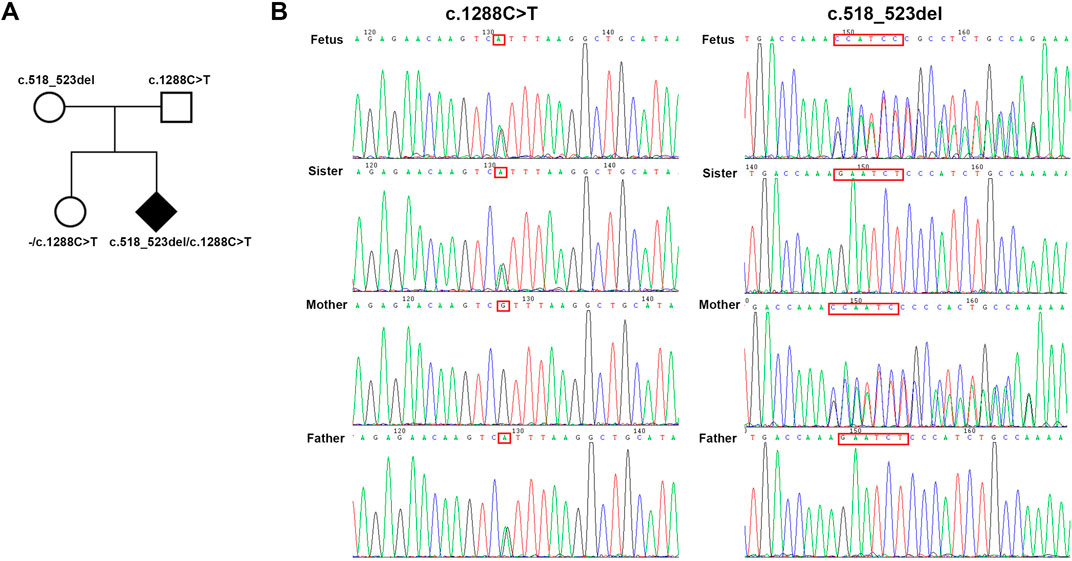
The trio-WES analysis was performed using an Illumina NovaSeq6000 platform. Sequencing reads were aligned to the reference human genome GRCh38/hg38 with Burrows-Wheeler Aligner (v0.7.17). Functional annotation was performed using the ENLIVEN variants interpretation system. Suspected variants identified through Trio-WES were confirmed by Sanger sequencing (c.1288C>T-Forward sequence: TCAGAGTAGTCAGAAGGCATCA; c.1288C>T-Reversed sequence: TGAATTGTCTTGCAGCCACTT, c.518_523del-Forward sequence: GCACTGGCTTGTCCTTCAC; c.518_523del-Reversed sequence: AGGTCTGTGGCATTTGGTTT).
4 Genetic results and pregnancy outcomes
A compound heterozygous variation in the FBXL4 gene (NM_012160.5: c.1288C>T, p. Arg430* and c.518_523del, p. Glu173_Leu175delinsVal) was detected by trio-WES (Table 1). The c.1288C>T variation was inherited from unaffected father. This variation has been documented in the HGMD database and was identified in patient diagnosed with MTDPS13. However, no specific phenotypes description was provided in the article (Smedley et al., 2021).
The novel c.518_523del variation was inherited from the unaffected mother. Following sanger sequencing verification, the full sister of the fetus, who does not exhibit any abnormal phenotype, only inherits the father’s nonsense variation (Figure 2B). The remaining verification results are consistent with trio-WES (Figure 2A).
Finally, the family opted for pregnancy termination following a comprehensive multidisciplinary consultation.
5 Disscusion
The NT thickening of the fetus in the early stage of pregnancy is a common phenotypic manifestation of chromosomal abnormalities. Besides, it is associated with various fetal malformations, dysplasias, deformations, disruptions, and an increased risk of adverse perinatal outcomes caused by genetic syndromes (Souka et al., 2005). Our case represents the first report of a compound heterozygous variation in the FBXL4 gene being identified in cases of NT thickening during early pregnancy. Notably, we detected subarachnoid cysts in the fetus at mid-pregnancy through targeted brain MRI, with cyst enlargement, ventriculomegaly, and enlargement of the posterior fossa cistern in the late pregnancy. The prenatal phenotypes of fetus brain verified the trio-WES result, FBXL4 variants can lead to severe influence on brain (El-Hattab et al., 2017), which suggests that FBXL4 is the pathogenic gene for the fetus. Besides, this indicates that the phenotypes of most patients with severe encephalopathy due to MTDPS13 have already emerged prenatally but have been overlooked as they were not detected during routine prenatal examinations. Therefore, in the limited prenatal diagnostic window period, diagnostic methods should be fast and comprehensive. With the informed consent of the pregnant woman and her family, for fetuses with prenatal phenotypes such as NT thickening (NT > 3.5 mm, especially), it is recommended to consider simultaneous chromosome and genetic testing (trio-WES) and strengthen follow-up imaging monitoring. Additionally, detailed family history queries might be necessary. The comprehensive testing can aid in decision-making for the pregnant women, offering more evidence.
FBXL4 is 621 amino acids in length and comprises an N-terminal mitochondrial targeting sequence (MTS) (amino acids 1–26), an F-box domain (amino acids 277–332), and 9 leucine-rich repeat (LRR) domain (amino acids 340–609) (Figure 3A) (Bonnen et al., 2013; Gai et al., 2013). Two mutations of FBXL4 were detected in this fetus: a nonsense variation c.1288C>T (p. Arg430*) and a deletion variation c.518_523del (p. Glu173_Leu175delinsVal). The nonsense variation resulted in a reduction of 191 amino acids in FBXL4 and disrupted the LRR sequence (Figure 3B). The deletion variation caused change near F-BOX in the protein structure (Figure 3C). These structural changes may have severe influence on the function of FBXL4 protein. Additionally, relevant studies have indicated that the phenotypes of nonsense and deletion variations are more severe compared to missense variations (El-Hattab et al., 2017). The average lifespan of patients is 4 years, with a median of 2 years (El-Hattab et al., 2017). The fetus in this case also exhibited severe brain structural developmental abnormalities prenatally. Therefore, the pregnant couple chose to terminate the pregnancy.

Figure 3. The schematic of FBXL4 and FBXL4’s variants predicted structure by AlphaFold2. (A) Schematic of FBXL4 domain organization and predicted structure. (B) The comparison between the structures of FBXL4 with its nonsense variation (c.1288C>T, p. R430*). (C) The comparison between the structures of FBXL4 with its deletion variation (c.518_523del, p. E173_L175delinsV).
In conclusion, our case is the first case of prenatal diagnosis of MTDPS13, provides a detailed description of the prenatal phenotype during pregnancy, which will improve decision-making for couples in ongoing pregnancies with FBXL4 variation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The studies involving human participants were reviewed and approved by West China Second University Hospital, Sichuan University, Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
YZ: Writing – original draft. JC: Writing – original draft. SY: Writing – original draft. HW: Writing – review and editing. YX: Writing – review and editing. SL: Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2022YFC2703300 and 2022YFC2703302).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alsina, D., Lytovchenko, O., Schab, A., Atanassov, I., Schober, F. A., Jiang, M., et al. (2020). FBXL4 deficiency increases mitochondrial removal by autophagy. EMBO Mol. Med. 12 (7), e11659. doi:10.15252/emmm.201911659
Aman, Y., Schmauck-Medina, T., Hansen, M., Morimoto, R. I., Simon, A. K., Bjedov, I., et al. (2021). Autophagy in healthy aging and disease. Nat. Aging 1 (8), 634–650. doi:10.1038/s43587-021-00098-4
Bonnen, P. E., Yarham, J. W., Besse, A., Wu, P., Faqeih, E. A., Al-Asmari, A. M., et al. (2013). Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am. J. Hum. Genet. 93 (3), 471–481. doi:10.1016/j.ajhg.2013.07.017
Cao, Y., Zheng, J., Wan, H., Sun, Y., Fu, S., Liu, S., et al. (2023). A mitochondrial SCF-FBXL4 ubiquitin E3 ligase complex degrades BNIP3 and NIX to restrain mitophagy and prevent mitochondrial disease. EMBO J. 42 (13), e113033. doi:10.15252/embj.2022113033
El-Hattab, A. W., Dai, H., Almannai, M., Wang, J., Faqeih, E. A., Al Asmari, A., et al. (2017). Molecular and clinical spectra of FBXL4 deficiency. Hum. Mutat. 38 (12), 1649–1659. doi:10.1002/humu.23341
Gai, X., Ghezzi, D., Johnson, M. A., Biagosch, C. A., Shamseldin, H. E., Haack, T. B., et al. (2013). Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. Am. J. Hum. Genet. 93 (3), 482–495. doi:10.1016/j.ajhg.2013.07.016
Gao, K., Xu, X., and Wang, C. (2024). FBXL4 mutation-caused mitochondrial DNA depletion syndrome is driven by BNIP3/BNIP3L-dependent excessive mitophagy. Trends Mol. Med. 30 (2), 113–116. doi:10.1016/j.molmed.2023.11.017
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17 (5), 405–424. doi:10.1038/gim.2015.30
Saini, N., Vijayasree, V., Nandury, E. C., Dalal, A., and Aggarwal, S. (2022). Prenatal phenotype of FBXL4-associated encephalomyopathic mitochondrial DNA depletion syndrome-13. Prenat. Diagn. 42 (13), 1682–1685. doi:10.1002/pd.6272
Smedley, D., Smith, K. R., Martin, A., Thomas, E. A., McDonagh, E. M., Cipriani, V., et al. (2021). 100,000 genomes pilot on rare-disease diagnosis in health care - preliminary report. N. Engl. J. Med. 385 (20), 1868–1880. doi:10.1056/NEJMoa2035790
Souka, A. P., Von Kaisenberg, C. S., Hyett, J. A., Sonek, J. D., and Nicolaides, K. H. (2005). Increased nuchal translucency with normal karyotype. Am. J. Obstetrics Gynecol. 192 (4), 1005–1021. doi:10.1016/j.ajog.2004.12.093
van Rij, M. C., Jansen, F. A., Hellebrekers, D. M., Onkenhout, W., Smeets, H. J., Hendrickx, A. T., et al. (2016a). Polyhydramnios and cerebellar atrophy: a prenatal presentation of mitochondrial encephalomyopathy caused by mutations in the FBXL4 gene. Clin. Case Rep. 4 (4), 425–428. doi:10.1002/ccr3.511
van Rij, M. C., Jansen, F. A. R., Hellebrekers, D. M. E. I., Onkenhout, W., Smeets, H. J. M., Hendrickx, A. T., et al. (2016b). Polyhydramnios and cerebellar atrophy: a prenatal presentation of mitochondrial encephalomyopathy caused by mutations in the FBXL4 gene. Clin. Case Rep. 4 (4), 425–428. doi:10.1002/ccr3.511
Keywords: prenatal diagnosis, FBXL4, NT thickening, MTDPS13, trio-WES
Citation: Zhai Y, Chen J, Yang S, Wang H, Xiao Y and Liu S (2025) Prenatal diagnosis of a compound heterozygous variation in the FBXL4 gene by trio-WES and imaging monitoring: a case report. Front. Genet. 16:1539288. doi: 10.3389/fgene.2025.1539288
Received: 04 December 2024; Accepted: 08 April 2025;
Published: 25 April 2025.
Edited by:
Michela Grosso, University of Naples Federico II, ItalyReviewed by:
María Pilar Bayona-Bafaluy, University of Zaragoza, SpainMichele Migliavacca, Albert Einstein Israelite Hospital, Brazil
Copyright © 2025 Zhai, Chen, Yang, Wang, Xiao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyuan Xiao, eXl4aWFvMjAxMEAxNjMuY29t; Shanling Liu, c3Vubnk2MzBAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Yujia Zhai
Yujia Zhai Jing Chen
Jing Chen Shuo Yang
Shuo Yang He Wang1,2
He Wang1,2 Yuanyuan Xiao
Yuanyuan Xiao Shanling Liu
Shanling Liu