- Department of Endocrinology, Shanxi Provincial People’s Hospital, Taiyuan, China
Objective: To screen for possible pathogenic mutations in polycystic ovary syndrome (PCOS) patients with diabetes and preliminarily explore the relationship between genotype and phenotype to offer a research basis for PCOS pathogenesis with diabetes.
Methods: Four patients with PCOS and diabetes were recruited and their demographic and clinical data were collected. Genomic DNA was extracted from peripheral blood leukocytes of the study subjects. High-throughput whole-exome sequencing was conducted to identify candidate genes that could play a pathogenic role in PCOS with diabetes in Aiji Taikang. The sequencing data obtained were evaluated using a variety of bioinformatics tools. Verification of candidate sites was done by Sanger sequencing.
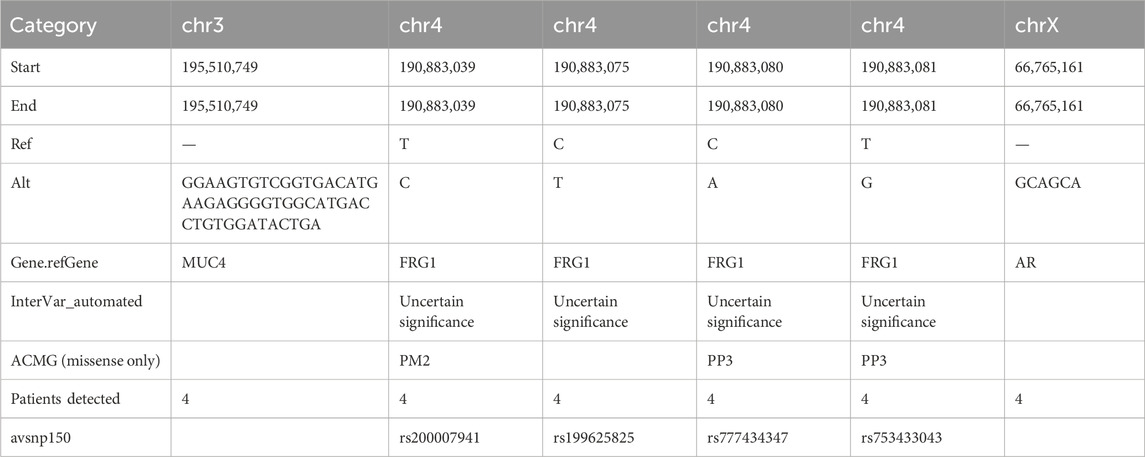
Results: Based on whole-exome sequencing, six mutations residing in three genes were detected in these four patients: (1) MUC4 located at Chr 3q29, (2) FSHD region gene 1 (FRG1)gene located at Chr 4q35.2, and (3) androgen receptor (AR) located at Chr Xq11-q12 were detected in these four patients (every patients had the 6 mutations). Of the six genetic mutations, an insertion/deletion (indel) mutation was found in the mucin 4 (MUC4) gene [MUC4:NM_018406.6:2/25:c.7701_7702insTCAGTATCCACAGGTCATGCCACCCCTCTTCATGTCACCGACACTTCC:p.(Ser2567_Ala2568insSerValSerThrGlyHisAlaThrProLeuHisValThrAspThrSer)], and an indel mutation in the AR gene (AR:NM_000044:exon1:c.173_174insGCAGCA:p. Q58delinsQQQ), while the other four were missense single-nucleotide polymorphisms (SNPs) located in FRG1 of uncertain significance (FRG1:NM_004477:exon8:c.T692C:p. L231P, FRG1:NM_004477:exon8:c.C728T:p.T243M, FRG1:NM_004477:exon8:c.C733A:p.L245M, FRG1:NM_004477:exon8:c.T734G:p.L245R). A Mucin 4 (MUC4) gene indel mutation was detected at the same site in four patients, which could be associated with endometriosis-related infertility. The AR gene indel mutation, AR:NM_000044:exon1:c.173_174insGCAGCA: p. Q58delinsQQQ was detected simultaneously in four patients.
Conclusion: Whole exome sequencing can quickly identify candidate genes for genes. Gaining an in-depth understanding of the AR mutations underlying PCOS with diabetes will deepen our understanding of the endocrine factors involved in the disease etiology, and provide potential targets for treatment.
Introduction
Polycystic ovary syndrome (PCOS) is a highly prevalent, heterogeneous disease in women of childbearing age. It is characterized by anovulation, irregular menstruation, amenorrhea, hirsutism, and infertility. Additionally, diverse metabolic symptoms have been reported, including insulin resistance (IR) diabetes, obesity, extensive coronary artery disease, hypertension, endometrial hyperplasia, ovarian cancer, and breast cancer (Al-Saleh, 2022; Chen et al., 2021; Smith et al., 2021; Falcetta et al., 2021). Currently, the Rotterdam consensus criteria have been used for PCOS diagnosis, and at least two of the following items must be met: hyperandrogenemia, oligomenorrhea or amenorrhea, and polycystic ovarian changes (unilateral or bilateral ovaries 2–9 mm, number of follicles ≥ 12, or ovarian volume ≥10 mL, ovarian volume = 0.5 × length × width × thickness) (Lauritsen et al., 2014). Treatment options and interventions include lifestyle adjustments, hyperandrogenemia, insulin resistance, recommended metformin use, and ovulation induction therapy in patients with fertility requirements.
Diverse risk factors are associated with PCOS in adults, including obesity, low exercise, high serum glucose levels, insulin resistance, type 1 or 2 diabetes, gestational diabetes, and mental health disorders. In recent years, research on PCOS pathogenesis has focused predominantly on genome-wide association studies, androgen biosynthesis and metabolism, and oxidative stress cycle markers. However, PCOS is a common female endocrine disorder that affects women of reproductive age and is often associated with metabolic diseases, including diabetes. Despite rigorous efforts, the genetic variants underlying PCOS associated with diabetes remain largely unknown.
Polycystic ovary syndrome (PCOS) is a multifactorial and polygenic disease involving a spectrum of environmental and lifestyle factors, and multiple genetic mutations. For example, genes, such as androgen receptor (AR) and sex hormone-binding globulin(SHBG), are involved in steroid hormone changes (Khan et al., 2019; Muccee et al., 2022; Samant et al., 2025; Villuendas et al., 2003). Several researchers have reported AR mutations in PCOS patients (Hickey et al., 2002; Wang et al., 2015; Shah et al., 2008; Schüring et al., 2012). Several studies have demonstrated that PCOS incidence is familial and inherited predominantly in an autosomal dominant manner. PCOS is a complex polygenic disease with an X-linked dominant inheritance or autosomal dominant inheritance pattern that does not conform to the Mendelian inheritance law. Most studies suggest that PCOS is a syndrome of endocrine and metabolic abnormalities caused by heredity, environment, and lifestyle, wherein genetic factors play an important role.
Exome sequencing is a high-throughput sequencing method that can simultaneously detect all exon mutations in the human genome at one time, which is of significant value for exploring disease-related genes (Azad et al., 2024; Dauda et al., 2022; Aqeel et al., 2022). Several studies have identified disease-modifying genes using exonic approaches. Several studies on PCOS have been conducted using exome sequencing. Herein, PCOS-associated genes were investigated using exome sequencing. The aim of this study was to identify potential pathogenic mutations in patients with polycystic ovary syndrome (PCOS) and diabetes and to investigate the genotype-phenotype relationship, thereby offering a research foundation for understanding PCOS pathogenesis associated with diabetes.
Materials and methods
In this case-control study, four women were recruited from Shanxi Provincial People’s Hospital. Among these subjects who were all Han Chinese women, were diagnosed as PCOS according to the Rotterdam diagnostic criteria. None of the subjects had any other diseases, and none had undergone chemotherapy or radiotherapy. All were unrelated individuals of reproductive age who had not received hormonal therapy for at least 3 months prior to the study.
The clinical examinations conducted on the patients
Patients 1 and 2 were diagnosed with diabetes using a 75 g Oral Glucose Tolerance Test (OGTT) at 4 weeks postpartum, fasting (7.5 mM), and 2-h blood glucose 11.5 mmol/L. Patient 2, her blood sugar was elevated, her fasting blood sugar was 8.5 mmol/L, and her 2-h postprandial blood sugar was 15.6 mmol/L. The third patient had normal blood sugar. Patient 4 measured 12.5 mmol/L of blood glucose 2 h post meals,sex hormone test demonstrated that her testosterone level was elevated.
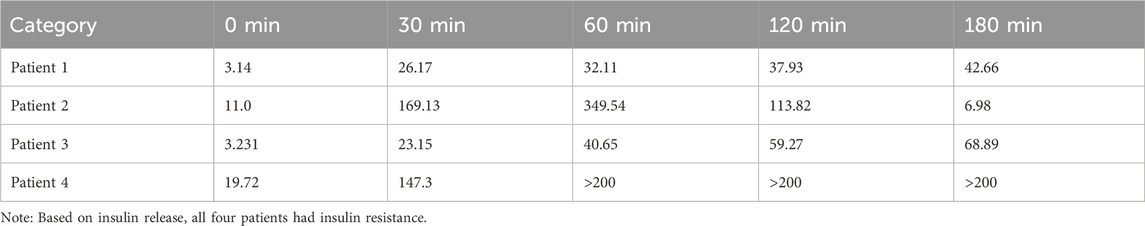
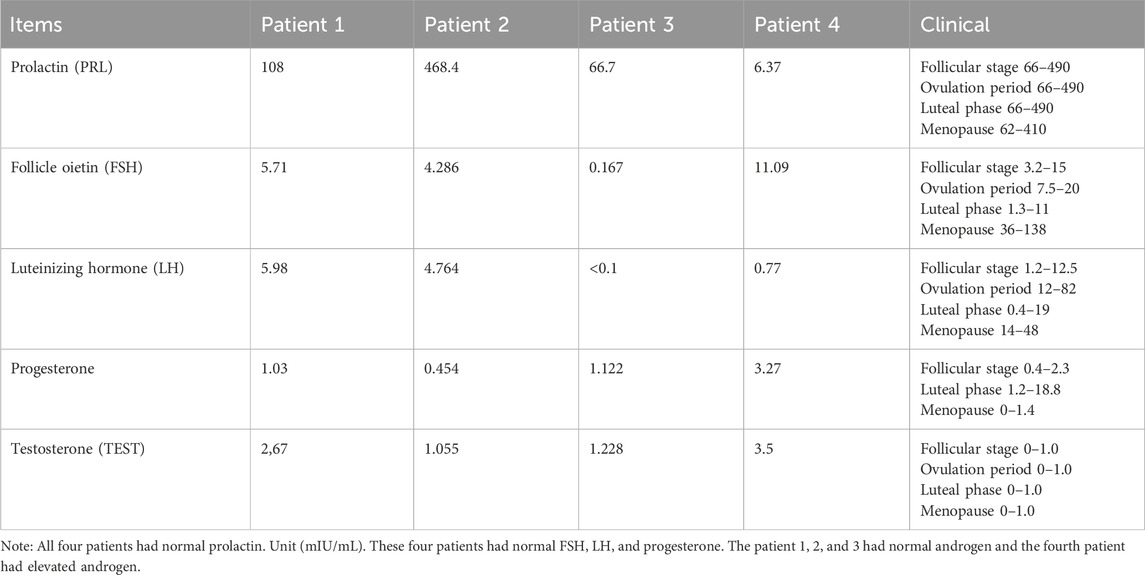
Based on insulin release, all four patients had insulin resistance. All four patients had normal prolactin.
These four patients had normal FSH, LH, and progesterone. The patient 1, 2, and 3 had normal androgen and the fourth patient had elevated androgen.
Whole-exome sequencing and bioinformatics analysis
DNA extraction
A 5 mL venous blood sample was collected from each patient. Heparin was used as the anticoagulant. Human peripheral blood leukocyte genomic DNA was extracted using the OMEGA SE Blood DNA Kit and sent to Aiji Taikang for whole-exome sequencing (Heald et al., 2020).
High-throughput whole-sequencing sequencing
Genomic DNA samples from the four patients were sent to Aiji Taikang for whole sequencing using the Illumina NovaSeq 6000 sequencing platform and paired-end sequencing.
Bioinformatics analysis of exome sequencing
The sequencing quality of the raw sequence reads was evaluated by FastQC (De Sena Brandine and Smith, 2019), and low-quality reads were removed by Trimmomatic to obtain high-quality sequences, which were aligned with the human reference genome (hg19) by Burrows-Wheeler Aligner (BWA) (Li and Durbin, 2009). Duplicates were marked by samblaster (Faust and Hall, 2014) by comparing read sequences with control sequences. GATK HaplotypeCaller (McKenna et al., 2010) was used for mutation analysis, and the National Center for Biotechnology Information (NCBI) dbSNP and ClinVar, 1000 Genomes database, dbNSFP (Liu et al., 2016), and other databases were used to annotate the population frequencies of the variant sites.
Analysis of mutations and candidate genes
Populations with mutation frequencies greater than 1% and mutations with no biological significance (intron regions, synonymous mutations, and other mutations that did not affect protein function) were filtered out. The intersection of the mutations in the four patients was evaluated.
Verification of candidate sites
The AR gene mutation site was verified by first-generation sequencing using primers with the sequences 5′ AAGACCTACGAGGAGCTTTCC 3′ and 5′ TTGGGGAGAACCATCCTCACC 3′. PCR was conducted using a Bio-Rad T100 for amplification. PCR system: 1 µL DNA template, 1 µL each of upstream and downstream primers, 0.5 µL Taq enzyme (TAKARA), 5 µL dNTP, 4 µL buffer, and the remaining volume adjusted to 50 μL with double-distilled water. The reaction conditions were as follows: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 30 s; annealing temperature at 56°C for 30 s; extension at 72°C for 45 s; and extension at 72°C for 5 min post 30 cycles. The resulting PCR products were stored at 4°C. Post the reaction, the cells were electrophoresed on a 1% agarose gel, stained with ethidium bromide, and the bands were observed under UV light. The PCR products were sent to Shanghai Shenggong Biological Company for sequencing. The obtained sequence was aligned with each exon sequence offered by GenBank and further searches were conducted to determine whether these mutations were present in the NCBI dbSNP or Human Gene Mutation Database (HGMD).
Results
All four patients were Asian. All patients exhibited insulin resistance, hyperandrogenemia, and normal prolactin levels.
History of present illness
Patient one was a 32 years old. In 2003, the patient experienced menstrual disorders for no obvious reason. In February 2018, she became pregnant post ovulation induction. Her blood glucose levels increased during the pregnancy. The patient was diagnosed with “gestational diabetes.” Type 2 diabetes was diagnosed a 75 g Oral Glucose Tolerance Test (OGTT) at 4 weeks postpartum, fasting (7.5 mM), and 2-h blood glucose 11.5 mmol/L according to the 2020 Chinese Diabetes Prevention and Treatment Guidelines. The sequencing data were obtained during pregnancy.
Patient 2 is female, 31 years old. In June 2019, she visited our hospital due to amenorrhea for 3 months. Simultaneously, her blood sugar level was elevated; her fasting blood sugar was 8.5 mmol/L, and her 2-h postprandial blood sugar was 15.6 mmol/L.
Patient 3, female 21 years old. She visited a doctor for menstrual disorders with symptoms of dry mouth, polydipsia, polyuria, and weight loss.
Patient 4 is female, 32 years old. She visited the dermatology department of our hospital with acne. Sex hormone tests revealed elevated testosterone levels. The patient was transferred to our department. The patient denied menstrual disorders and claimed to have normal menstrual flow. She had been experiencing dry mouth recently and measured 12.5 mmol/L blood glucose 2 h post meals at home.
Patient 1 was 168 cm tall, weighed 85 kg, had a 124/70 mmHg blood pressure, and body mass index (BMI) of 30 kg/m2. She was obese, hirsute, and had visible acne on her face, acanthosis nigricans and no edema in the lower limbs. Physical examination of the remaining three patients demonstrated no obvious abnormalities. All four patients had a family history of polycystic ovaries and diabetes. General clinical data included routine blood tests, liver function, renal function, glycosylated hemoglobin, cortisol rhythm, gynecological color Doppler ultrasonography, thyroid function, androgen and prolactin levels, adrenal CT), and electrocardiography. This study was approved by the Research Ethics Commission of the Shanxi Provincial People’s Hospital (201992).
Laboratory information
Patient 1: 8 points adrenocorticotropic hormone (ACTH) 38.66 (6.0–40) pg/mL, fasting blood glucose 11.59 mmol/L, glycosylated hemoglobin 10.5%.
Normal thyroid function, 8 points of cortisol 159.7 ng/mL (72.6–233.8), 4 points of 98.87 ng/mL, 0 points of cortisol 27.33 ng/mL, 24-h urine free cortisol 305.24 µg/24 h, follicle-stimulating hormone (FSH) 6.711 mIU/mL, luteinizing hormone (LH) 8.088 mIU/mL, testosterone 0.342 (0–1) ng/mL.
Patient 1 was diagnosed with diabetes based on fasting blood glucose and A1C. Patient 1’s cortisol rhythm and ACTH levels were normal, except for hypercortisolism. Patient 1 had normal levels of sex hormones.
Patient 2: 8 points of cortisol 167.43 ng/mL (72.6–233.8) 4 points of 86.54, 0 points of cortisol 30.00, 24-h urine free cortisol 220.00 µg/24 h, glycosylated hemoglobin 8.5%.
Patient 2’s cortisol rhythm and ACTH levels were normal, except for hypercortisolism. The second patient had diabetes mellitus.
Patient 3: 8 points cortisol 233.0 ng/mL (72.6–233.8) 4 points 142.5 ng/mL, 0 points 60.00 ng/mL, 24 h urinary free cortisol 125.554 µg/24 h, 8 points ACTH 37.17 (6.0–40) pg/mL, glycosylated hemoglobin 7.1%, thin-slice computed tomography (CT) of the adrenal glands demonstrated no abnormalities.
Patient 3’s cortisol rhythm and ACTH levels were normal, except for hypercortisolism. Patient 3 had a history of diabetes.
Patient 4: Gynecological color Doppler ultrasound: The uterine size was 42.7 × 41.0 × 34.8 mm. The left ovary was 35.0 × 16.0 mm; the right ovary was 36.5 × 16.6 mm, all demonstrated more than 12 follicles. Results: Several follicles were observed in ovaries. see Figure 1.
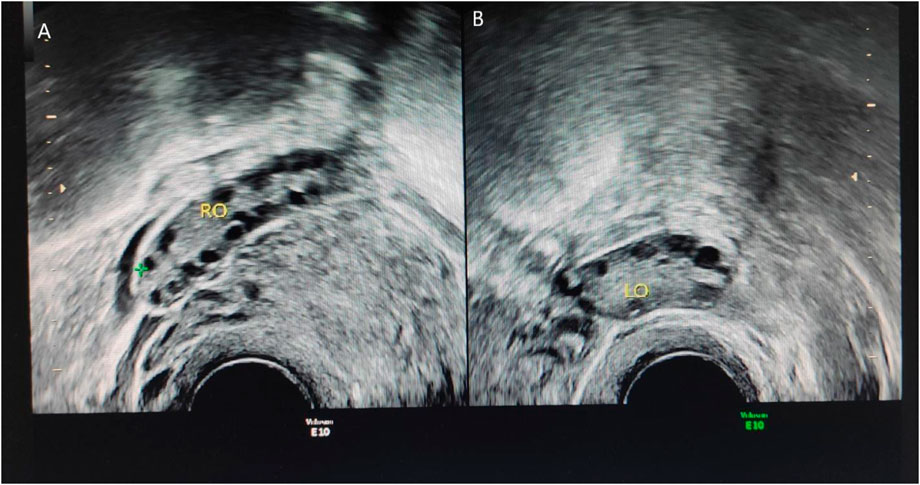
Figure 1. Ultrasound results of patient 4. Thin-slice CT of the adrenal glands demonstrates no abnormalities. (A) represents the right ovary of the patient. The right ovary is 22.4 × 9.9 mm, and approximately five follicles can be seen in the right ovary. The larger diameter is approximately 8.6 mm, and the smaller diameter is approximately 4.0 mm. (B) represents the left ovary of the patient, the size of the left ovary is 24.0 × 9.7 mm, and approximately four follicles can be seen in the left ovary, the larger diameter is approximately 7 mm, and the smaller diameter is approximately 4.6 mm indicates large volume of both ovaries accompanied by more follicles.
Data on insulin function in the four patients are depicted in Table 1, and data on related indicators of sex hormone parameters of the four patients are depicted in Table 2.
These four patients had normal FSH, LH, and progesterone.
The patient 1, 2, and 3 had normal androgen and the fourth patient had elevated androgen.
Gynecological ultrasound (ovulation)
More than 12 follicles were observed in the left ovary, all of which were less than 8 mm in diameter, and several anechoic follicles were observed in the right ovary, which were approximately 19.5 × 19.0 mm in diameter. Results: The right ovary was large, with multiple follicular echoes. Several follicles were observed in the left ovary. Pelvic fluid was noted see Figure 2.
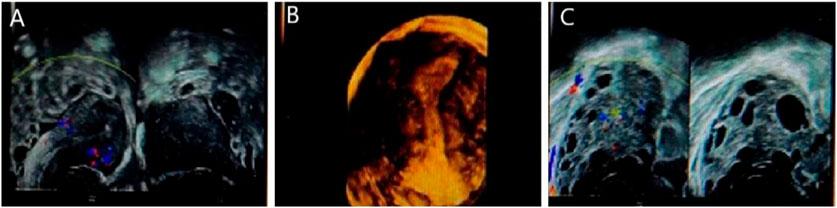
Figure 2. Gynecological ultrasound of patient 1. (A) represents the right ovary of the patient, with several echoless are as visible, approximately 19.5 × 19.0 mm. (B) represents uterus of the patient, and (C) represents more than 12 follicles in the left ovary, all less than 8 mm in diameter. Indicates large volume of the right ovary with multiple follicle-like echoes. More follicles in the left ovary, accompanied by pelvic effusion.
QC of raw data
In the raw whole-exome sequencing data, the four patients had an average of approximately 70 M reads and the Q30 average was greater than 90%. Post quality control, an average of approximately 68 M high-quality reads was obtained, and the Q30 average was greater than 93%. The quality of the sequencing reads was high, and the high-quality sequencing data satisfied the downstream analysis. Summaries of high-quality sequences are presented in Supplementary Table S1; Supplementary Figure S1.
Alignment to reference genome
High-quality sequence reads were aligned with the reference genome (hg19). Approximately 99% of the sequence reads obtained from the four patients aligned to the reference genome. Approximately 10% of duplicate reads were found in four patients. The whole exome region was sequenced with an average coverage of 99% for all four patients. The average coverage depth of the four patients was 110X, and the average 50X coverage region was 79%. The comparison was normal and met the requirements for identifying mutations. Quality control of the alignment of the four samples is depicted in Supplementary Table S2.
Mutation identification
Genome Analysis Toolkit (GATK) was used to identify mutations in the four patients. On an average, 83,566 single-nucleotide polymorphisms (SNPs) were identified in each patient. Approximately 98% of the SNPs were annotated to the NCBI dbSNP database, and approximately 98% were annotated to the Genome Aggregation Database (gnomAD). The proportion of SNPs in exons was approximately 27%, and more than half of the SNPs were in the intronic region. The number of SNPs and annotations of the four patients are depicted in Supplementary Table S3.
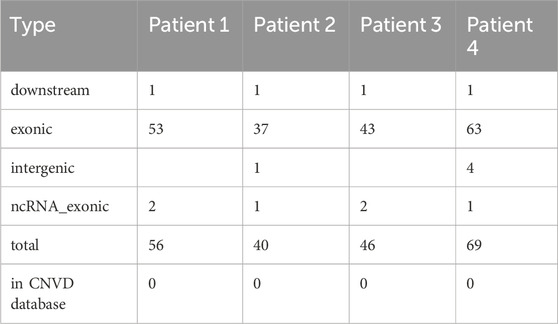
On average, 13,333 indels were identified for each patient; approximately 90% of indels could be annotated to the NCBI dbSNP database, and approximately 92% of indels could be annotated to the gnomAD database. The proportion of indels in the exons was approximately 5%, and more than 78% of the indels were in the intronic region. The number of SNPs and annotations for the four patients are depicted in Supplementary Table S4. On average, 52 CNVs were identified in each patient, approximately 90% of which were located in the exonic region. Additionally, none of the CNVs were annotated in the CNVD database. The number of CNVs and annotations for the four patients are depicted in Table 3.
Identification of candidate causative genes and location
Mutations were filtered based on the mutation frequency in the population database. Considering the distribution of the four patients, we enumerated the intersections of gene mutations across the four patients. Three genes (mucin 4 (MUC4), FSHD region gene 1 (FRG1), and AR) were mutated in four patients. We found that FRG1 had multiple mutations, and four missense mutations were annotated in the NCBI dbSNP database. MUC4 and AR harbored indel mutations. Based on a literature search, we believe that AR (AR:NM_000044:exon1:c.173_174insGCAGCA:p. Q58delinsQQQ) is likely to be a genetic variant that could serve as a potential therapeutic target. To the best of our knowledge, this variant has not been reported previously. An overview of these three gene mutations is presented in Table 4.
First-generation sequencing verification of the AR mutation sites
First-generation sequencing verification of the AR indel mutation.
Through first-generation sequencing (Sanger sequencing), we verified the AR indel mutations in these four patients (Figure 3).
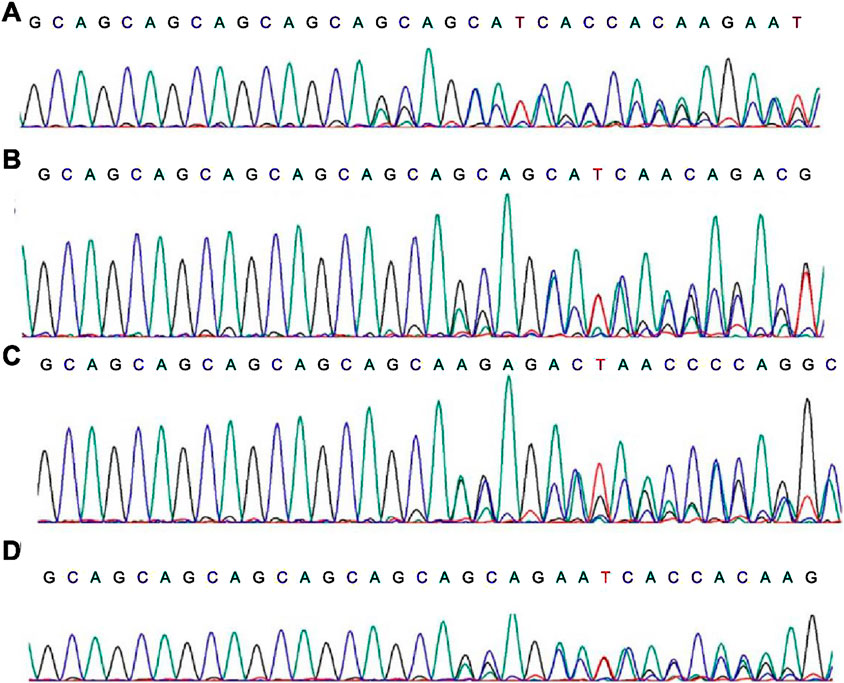
Figure 3. First-generation sequencing results of the AR mutation sites. (A–D) stands for patient 1, 2, 3, 4
Discussion
PCOS is a common reproductive, endocrine, and metabolic disease with high genetic heterogeneity. The clinical manifestations can be accompanied by metabolic disorders, including glucose metabolism, obesity, and insulin resistance (Bell et al., 2021). Among all clinical manifestations, most PCOS patients have varying degrees of hyperandrogenemia. The high androgen level of the ovarian tissue in patients with polycystic ovaries can lead to infertility. Currently, PCOS incidence is increasing annually; however, the disease etiology and influencing factors have not been completely elucidated. This lack of understanding has increased the difficulty of clinical research and slowed progress in clinical diagnosis and treatment strategies. Therefore, it is vital to conduct further genetic studies.
Herein, four PCOS patients had abnormal blood sugar levels. Post treatment, such as hypoglycemia and weight loss, the patient’s symptoms significantly improved, and the same gene mutation was found. Patient 1 was administered oral metformin 0.25 g thrice a day, fasting blood glucose fluctuated between 5–6 mmol/L, and blood glucose fluctuated between 7–8 mmol/L 2 h post meals, and menstruation returned to normal. Patient 2 was administered oral metformin (0.5 g) thrice a day, acarbose 50 mg, thrice a day, fasting blood glucose fluctuated between 5–6 mmol/L, and blood glucose fluctuated between 7–8 mmol/L 2 h post meals, and menstruation returned to normal. Patient 3 had slightly higher blood sugar levels. After guiding diet and exercise, blood sugar levels and menstruation returned to normal. Patient 4: After a subcutaneous injection of a GLP-1 receptor agonist, the blood glucose and body weight of the patient reached standard levels, and menstruation was normal.
The detection of AR gene mutations in diabetic patients with PCOS indicates that mutations in the AR gene can lead to not only IR (Gao et al., 2020; Luo et al., 2021; Yu et al., 2020; Echiburú et al., 2020) but also PCOS. The influence of CAG repeat mutations of impacting testosterone AR gene on insulin resistance in women with PCOS has been previously reported (Lin et al., 2008). This offers a basis for early PCOS prevention and treatment.
The AR gene comprises 11 exons located at q11-q12 on the X chromosome. Exon 1 of the AR gene contains a polymorphic CAG repeat sequence, the number of which is typically between 10 and 35, which encodes the polyglutamine fragment of the AR transactivation domain. There is evidence that the CAG number is negatively correlated with AR transcriptional activity, and decreased AR sensitivity is associated with increased mortality and decreased blood glucose levels in patients with type 2 diabetes (Yu et al., 2014).
The expression of AR in PCOS is different in PCOS patients. The correlation between AR expression and serum FSH LH may be related to the number of follicles in PCOS (Lin et al., 2005). Previous studies have demonstrated that serine/arginine-rich splicing factors (SRSFs) mediate the role of AR in ovarian granulosa cells in patients with polycystic ovaries and it has been demonstrated that SRSFs are regulated by the corresponding microRNAs (miRNAs) (Rana et al., 2011). Alteration of SRSF expression interferes with the alternative splicing process of AR, which ultimately leads to a decline in AR function and ovarian androgens accumulation. Previous studies have demonstrated that ARs in PCOS patients is closely associated with hyperandrogenemia and abnormal follicular development. This study demonstrated that CK2A plays a role in PCOS development by phosphorylating and stabilizing AR, increasing AR expression, and increasing ovulation- and androgen synthesis-related gene expression (Supplementary Figure S2).
Using targeted next-generation sequencing (NGS), researchers studied DNA methylation patterns in the promoter regions of several genes, including anti-Müllerian hormone (AMH) and AR, in children born to 24 women with PCOS (12 of whom received metformin during pregnancy) and 24 women without PCOS in early infancy (2–3 months) (Möhlig et al., 2006). Female children demonstrated differences between groups at one CpG site of LEPR, two CpG sites of LEP, one CpG site of ADIPOR2, and two CpG sites of AR, whereas male children demonstrated differences in five CpG sites of LEP, three CpG sites of AMH, and nine CpG sites of AR, indicating that intrauterine PCOS environment can influence the sex-dependent acquisition of DNA methylation of certain genes.
Women with polycystic ovary syndrome have higher rates of miscarriage, gestational diabetes mellitus, gestational hypertension, preeclampsia, and cesarean section. Given the high prevalence of PCOS, exploring new biomarkers, perfecting diagnostic criteria, and improving treatment methods are essential to improve the accuracy and effectiveness of interventions, which will have a positive impact on patients’ lives (Stener-Victorin et al., 2024; Salari et al., 2024; Bahri Khomami et al., 2024). PCOS is associated with diverse risk factors in adults, including insulin resistance, type 1 or 2 diabetes, gestational diabetes, and mental health disorders. The limitations of this study include the lack of access to the genotype information of parents of the patients. Because this study aimed to investigate PCOS in a specific population, we had access to a limited sample size, which may have affected the generalizability of this study. Future studies with larger sample sizes are warranted.
Conclusion
An indel mutation was identified in the AR (AR:NM_000044:exon1:c.173_174insGCAGCA:p. Q58delinsQQQ) was detected simultaneously in four patients, which provides new potential pathogenic sites and therapeutic targets for PCOS Patients with diabetes.
Data availability statement
The datasets presented in this article are not readily available because restrictions outlined in Regulations on the Management of Human Genetic Resources of the People's Republic of China. Requests to access the datasets should be directed to Corresponding author.
Ethics statement
The studies involving humans were approved by Institutional Review Board of Shanxi Provincial People’s Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
CW: Data curation, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1541946/full#supplementary-material
References
Al-Saleh, I. (2022). The relationship between urinary phthalate metabolites and polycystic ovary syndrome in women undergoing in vitro fertilization: nested case-control study. Chemosphere 286 (Pt 1), 131495. doi:10.1016/j.chemosphere.2021.131495
Aqeel, M., Ajmal, W., Mujahid, Q., Murtaza, M., Almuqbil, M., Ghazanfar, S., et al. (2022). Genome-wide identification of potential mRNAs in drought response in wheat (Triticum aestivum L.). Genes 13(10): 1906, doi:10.3390/genes13101906
Azad, H., Akbar, M. Y., Sarfraz, J., Haider, W., Riaz, M. N., Ali, G. M., et al. (2024). G-acp: a machine learning approach to the prediction of therapeutic peptides for gastric cancer. J. Biomol. Struct. Dyn. 1–14. doi:10.1080/07391102.2024.2323141
Bahri Khomami, M., Shorakae, S., Hashemi, S., Harrison, C. L., Piltonen, T. T., Romualdi, D., et al. (2024). Systematic review and meta-analysis of pregnancy outcomes in women with polycystic ovary syndrome. Nat. Commun. 15(1): 5591, doi:10.1038/s41467-024-49749-1
Bell, R. J., Islam, R. M., Skiba, M. A., Herbert, D., Martinez Garcia, A., and Davis, S. R. (2021). Substituting serum anti-Müllerian hormone for polycystic ovary morphology increases the number of women diagnosed with polycystic ovary syndrome: a community-based cross-sectional study. Hum. Reprod. 37, 109–118. doi:10.1093/humrep/deab232
Chen, J., Zhou, Q., Zhang, Y., Tan, W., Gao, H., Zhou, L., et al. (2021). Discovery of novel serum metabolic biomarkers in patients with polycystic ovarian syndrome and premature ovarian failure. Bioengineered 12 (1), 8778–8792. doi:10.1080/21655979.2021.1982312
Dauda, W. P., Morumda, D., Abraham, P., Adetunji, C. O., Ghazanfar, S., Glen, E., et al. (2022). Genome-wide analysis of cytochrome P450s of Alternaria species: evolutionary origin, family expansion and putative functions. J. Fungi 8(4): 324, doi:10.3390/jof8040324
De Sena Brandine, G., and Smith, A. D. (2019). Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res 8, 1874. doi:10.12688/f1000research.21142.2
Echiburú, B., Milagro, F., Crisosto, N., Pérez-Bravo, F., Flores, C., Arpón, A., et al. (2020). DNA methylation in promoter regions of genes involved in the reproductive and metabolic function of children born to women with PCOS. Epigenetics 15 (11), 1178–1194. doi:10.1080/15592294.2020.1754674
Falcetta, P., Benelli, E., Molinaro, A., Di Cosmo, C., Bagattini, B., Del Ghianda, S., et al. (2021). Effect of aging on clinical features and metabolic complications of women with polycystic ovary syndrome. J. Endocrinol. Invest 44 (12), 2725–2733. doi:10.1007/s40618-021-01594-5
Faust, G. G., and Hall, I. M. (2014). SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505. doi:10.1093/bioinformatics/btu314
Gao, X. Y., Liu, Y., Lv, Y., Huang, T., Lu, G., Liu, H. B., et al. (2020). Role of androgen receptor for reconsidering the “True” polycystic ovarian morphology in PCOS. Sci. Rep. 10, 8993. doi:10.1038/s41598-020-65890-5
Heald, A. H., Yadegar, F. G., Livingston, M., Fachim, H., Lunt, M., Narayanan, R. P., et al. (2020). Androgen receptor-reduced sensitivity is associated with increased mortality and poorer glycaemia in men with type 2 diabetes mellitus: a prospective cohort study. Cardiovasc Endocrinol. Metab. 10 (1), 37–44. doi:10.1097/XCE.0000000000000230
Hickey, T., Chandy, A., and Norman, R. J. (2002). The androgen receptor CAG repeat polymorphism and X-chromosome inactivation in Australian Caucasian women with infertility related to polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 87 (1), 161–165. doi:10.1210/jcem.87.1.8137
Khan, M. J., Ullah, A., and Basit, S. (2019). Genetic basis of polycystic ovary syndrome (PCOS): current perspectives. Appl. Clin. Genet. 12, 249–260. doi:10.2147/TACG.S200341
Lauritsen, M. P., Bentzen, J. G., Pinborg, A., Loft, A., Forman, J. L., Thuesen, L. L., et al. (2014). The prevalence of polycystic ovary syndrome in a normal population according to the Rotterdam criteria versus revised criteria including anti-Mullerian hormone. Hum. Reprod. 29 (4), 791–801. doi:10.1093/humrep/det469
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi:10.1093/bioinformatics/btp324
Lin, H. Y., Xu, Q., Yeh, S., Wang, R. S., Sparks, J. D., and Chang, C. (2005). Insulin and leptin resistance with hyperleptinemia in mice lacking androgen receptor. Diabetes 54 (6), 1717–1725. doi:10.2337/diabetes.54.6.1717
Lin, H. Y., Yu, I. C., Wang, R. S., Chen, Y. T., Liu, N. C., Altuwaijri, S., et al. (2008). Increased hepatic steatosis and insulin resistance in mice lacking hepatic androgen receptor. Hepatology 47 (6), 1924–1935. doi:10.1002/hep.22252
Liu, X., Wu, C., Li, C., and Boerwinkle, E. (2016). dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 37, 235–241. doi:10.1002/humu.22932
Luo, J., Ye, H., Hao, L., Sun, Y., Li, R., Li, Y., et al. (2021). SRSFs mediate the function of AR in the ovarian granulosa cells of patients with PCOS. Genes Dis. 8, 94–109. doi:10.1016/j.gendis.2019.09.005
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi:10.1101/gr.107524.110
Möhlig, M., Jürgens, A., Spranger, J., Hoffmann, K., Weickert, M. O., Schlösser, H. W., et al. (2006). The androgen receptor CAG repeat modifies the impact of testosterone on insulin resistance in women with polycystic ovary syndrome. Eur. J. Endocrinol. 155 (1), 127–130. doi:10.1530/eje.1.02195
Muccee, F., Bijou, O., Harakeh, S., Adawiyah, R., Sayyed, R. Z., Haghshenas, L., et al. (2022). In-Silico investigation of effects of single-nucleotide polymorphisms in PCOS-associated CYP11A1 gene on mutated proteins. Genes 13(7): 1231, doi:10.3390/genes13071231
Rana, K., Fam, B. C., Clarke, M. V., Pang, T. P., Zajac, J. D., and MacLean, H. E. (2011). Increased adiposity in DNA binding-dependent androgen receptor knockout male mice associated with decreased voluntary activity and not insulin resistance. Am. J. Physiol. Endocrinol. Metab. 301 (5), E767–E778. doi:10.1152/ajpendo.00584.2010
Salari, N., Nankali, A., Ghanbari, A., Jafarpour, S., Ghasemi, H., Dokaneheifard, S., et al. (2024). Global prevalence of polycystic ovary syndrome in women worldwide: a comprehensive systematic review and meta-analysis. Archives Gynecol. obstetrics 310(3): 1303–1314. doi:10.1007/s00404-024-07607-x
Samant, M., Bhat, M., Dadachanji, R., Sudhakar, D. V. S., Patil, A., and Mukherjee, S. (2022). Whole exome sequencing uncovers rare variants associated with PCOS susceptibility in Indian women. Syst. Biol. Reproductive Med. 71(1): 76–89. doi:10.1080/19396368.2025.2471418
Schüring, A. N., Welp, A., Gromoll, J., Zitzmann, M., Sonntag, B., Nieschlag, E., et al. (2012). Role of the CAG repeat polymorphism of the androgen receptor gene in polycystic ovary syndrome (PCOS). Exp. Clin. Endocrinol. Diabetes 120 (2), 73–79. doi:10.1055/s-0031-1291343
Shah, N. A., Antoine, H. J., Pall, M., Taylor, K. D., Azziz, R., and Goodarzi, M. O. (2008). Association of androgen receptor CAG repeat polymorphism and polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 93 (5), 1939–1945. doi:10.1210/jc.2008-0038
Smith, J., Ayre, J., Jansen, J., Cvejic, E., McCaffery, K. J., Doust, J., et al. (2021). Impact of diagnostic labels and causal explanations for weight gain on diet intentions, cognitions and emotions: an experimental online study. Appetite 167, 105612. doi:10.1016/j.appet.2021.105612
Stener-Victorin, E., Teede, H., Norman, R. J., Legro, R., Goodarzi, M. O., Dokras, A., et al. (2024). Polycystic ovary syndrome. Nat. Rev. Dis. Prim. 10(1): 27, doi:10.1038/s41572-024-00511-3
Villuendas, G., Tosi, F., Sancho, J., Moghetti, P., and San Millán, J. L. (2003). Association between the D19S884 marker at the insulin receptor gene locus and polycystic ovary syndrome. Fertil. Steril. 79(1): 219–220. doi:10.1016/s0015-0282(02)04570-3
Wang, F., Pan, J., Liu, Y., Meng, Q., Lv, P., Qu, F., et al. (2015). Alternative splicing of the androgen receptor in polycystic ovary syndrome. Proc. Natl. Acad. Sci. U S A. 112 (15), 4743–4748. doi:10.1073/pnas.1418216112
Yu, C. J., Liu, X., Zhou, Z. Y., Chen, X. J., Meng, Y. C., Gu, H. C., et al. (2020). The casein kinase 2α promotes the occurrence polycystic ovary syndrome. Biochem. Biophys. Res. Commun. 525, 121–128. doi:10.1016/j.bbrc.2020.02.065
Keywords: polycystic ovary syndrome, diabetes, whole-exome sequencing, bioinformatics analysis, AR gene, insulin resistance
Citation: Wang C (2025) Molecular variants of multiple genes were revealed by whole-exome sequencing in PCOS patients with diabetes. Front. Genet. 16:1541946. doi: 10.3389/fgene.2025.1541946
Received: 22 January 2025; Accepted: 25 April 2025;
Published: 23 May 2025.
Edited by:
Ahmed Rebai, Centre of Biotechnology of Sfax, TunisiaReviewed by:
Shakira Ghazanfar, National Institute for Genomics and Advanced Biotechnology (NIGAB), PakistanAkeem Babatunde Sikiru, Federal University of Agriculture Zuru, Nigeria
Copyright © 2025 Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenglin Wang, d2NsNzY1NDMxMUAxNjMuY29t
 Chenglin Wang
Chenglin Wang