- 1Labcorp, Durham, NC, United States
- 2Illumina, San Diego, CA, United States
- 3Labcorp, Burlington, NC, United States
- 4Wake Forest Comprehensive Cancer Center, Wake Forest School of Medicine, Department of Pathology, Winston-Salem, NC, United States
Introduction: NTRK1, NTRK2, and NTRK3 gene fusions are rare oncogenic driver alterations found in diverse tumor types of adults and children. They are clinically important biomarkers as tumors harboring these genomic alterations have high response rates to targeted therapy. Routine testing for NTRK fusions and treatment with TRK inhibitors has been recommended in multiple tumor types; however, differences between testing technologies used for detecting NTRK fusions can result in variable likelihoods of identification.
Methods: To assess the prevalence of NTRK fusions in a real-world standard-of-care setting, we analyzed data from 19,591 FFPE samples encompassing 35 solid tumor types submitted for comprehensive genomic profiling (CGP) as part of routine clinical care. CGP testing included DNA hybrid-capture sequencing to detect small variants, copy number alterations, microsatellite instability (MSI), and tumor mutational burden (TMB). RNA hybrid-capture sequencing was concurrently performed to detect fusions and splice variants. Detected NTRK fusions were categorized as oncogenic, likely oncogenic, or variant of unknown significance (VUS) based on the fusion partner, orientation, and breakpoint position.
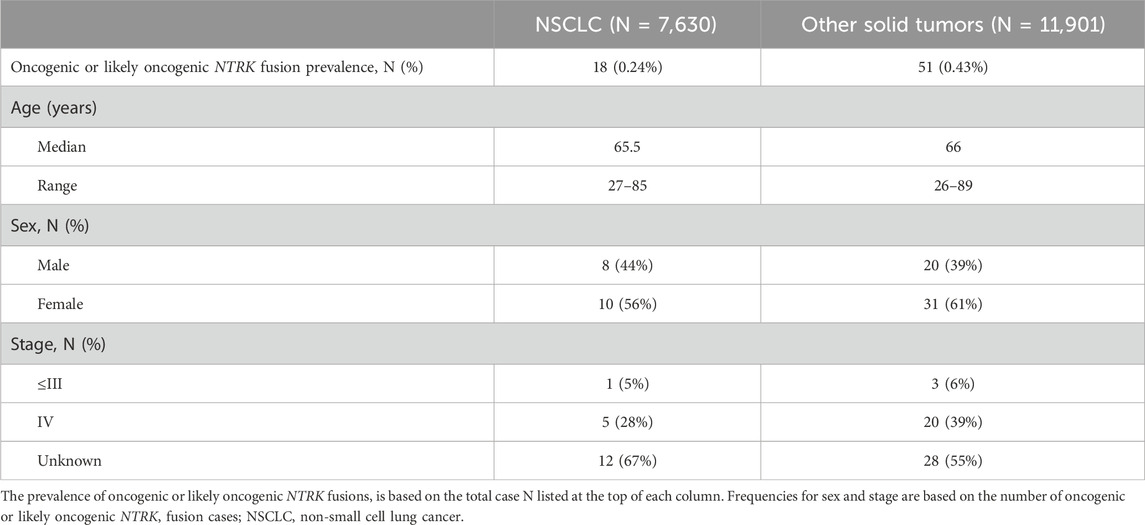
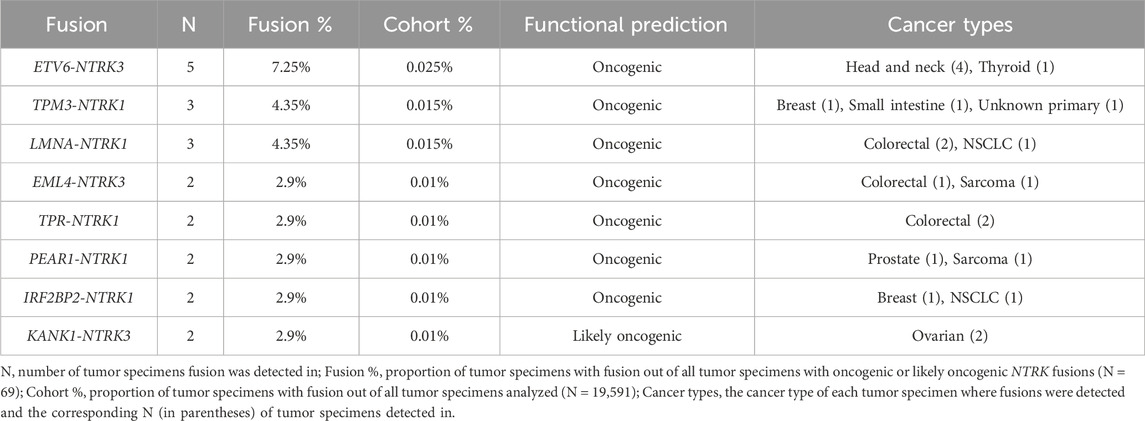
Results: 73 oncogenic or likely oncogenic NTRK fusions were identified in 69 unique tumor specimens across 19 tumor types for a total cohort prevalence of 0.35%. Tumor types with the highest NTRK fusion prevalence included glioblastoma (1.91%), small intestine (1.32%), and head and neck (0.95%) tumors with other solid tumor types ranging from 0.19% (uterine) to 0.63% (breast). We identified diverse intra and inter-chromosomal partner genes for NTRK fusions across all tumor types. Most NTRK fusions were detected in only one tumor specimen, though some recurrent fusions were noted with ETV6, TPM3, LMNA, EML4, TPR, PEAR1, IRF2BP2, and KANK1 fusion partners. Most NTRK fusions were mutually exclusive from other genomic driver alterations, however, almost a third of tumor specimens (29%) contained at least one co-occurring genomic driver, which may affect treatment decisions.
Discussion: The high prevalence of oncogenic and likely oncogenic NTRK fusions detected in our analysis suggests that RNA hybrid-capture-based sequencing for fusion detection is a highly sensitive method for identifying clinically meaningful known and novel NTRK fusions, which may be missed with other detection methods, directly impacting therapeutic options and patient outcomes.
1 Introduction
The tropomyosin receptor kinase (TRK) family of receptor tyrosine kinases are encoded by the NTRK1, NTRK2, and NTRK3 (NTRK) genes and play a critical role in neuronal development and differentiation (Amatu et al., 2019; Cocco et al., 2018). The common ligands of TRK receptors are neurotrophins, a family of growth factors critical to the functioning of the nervous system. The activation of TRK receptors by neurotrophin leads to the activation of signal cascades, including the RAS and PI3K pathways, promoting survival and other regulatory pathways in cells. The most common mechanism of oncogenic activation of the TRK receptors is via fusions involving the NTRK genes (Cocco et al., 2018). NTRK fusions are the consequence of large-scale chromosomal events, including inversions, deletions, and translocations, and result in the fusion of the C-terminal tyrosine kinase domain (TKD) of NTRK with an N-terminal fusion partner (Amatu et al., 2019; Wong et al., 2020). NTRK genes have >60 known fusion partners across multiple tumor types and the number is growing as novel fusion partners are identified (Kummar and Lassen, 2018). NTRK gene fusions are found at a low frequency (<1%) across multiple common pediatric and adult solid tumor types but are enriched (>90%) in a few rare tumors (secretory breast carcinoma, mammary analogue secretory carcinoma, cellular or mixed congenital mesoblastic nephroma, and infantile fibrosarcoma) (Amatu et al., 2019; Cocco et al., 2018; Okamura et al., 2018). In-frame NTRK fusions result in constitutive activation of the TRK receptor and downstream signaling pathways (Amatu et al., 2019; Cocco et al., 2018). Genomic characterization of NTRK fusion-positive cancers has demonstrated these tumors are typically devoid of other oncogenic drivers, with the notable exception of microsatellite instability-high (MSI-H) colorectal cancer (CRC) (Westphalen et al., 2021; Gatalica et al., 2019; Wang H. et al., 2022).
Routine testing for NTRK fusions or treatment with TRK inhibitors has been recommended in >25 different tumor types (Klink et al., 2022). Variations in testing technologies can result in significant differences in the likelihood of fusion identification (Marchio et al., 2019). Single gene tests, hotspot panels, and inadequately baited DNA-based next-generation sequencing (NGS) have technical limitations precluding accurate detection of fusions (Penault-Llorca et al., 2019; Solomon et al., 2019). Highly sensitive RNA sequencing methods such as hybrid capture are optimal for comprehensively identifying actionable fusions, including NTRK fusions in solid tumors. In non-small cell lung cancer (NSCLC), the National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology recommend biomarker testing be performed using broad NGS panels to detect druggable rearrangements or fusions involving ALK, NTRK, ROS1, and RET with consideration of RNA sequencing if not previously performed (Ettinger et al., 2022).
NTRK gene fusions were the first gene-specific alterations to receive US Food and Drug Administration (FDA) approval in a histology-agnostic manner across all solid tumors (Drilon, 2019). Larotrectinib (inhibitor of TRKA, TRKB, and TRKC) and entrectinib (inhibitor of TRKA, B and C, ROS1, and ALK) are both FDA-approved for adult and pediatric patients with metastatic, unresectable solid tumors harboring NTRK fusions that have progressed on prior therapies or when no suitable treatment is available. In phase I and II trials of larotrectinib, an objective response was observed in 79% of evaluable patients, with 16% having complete responses (Hong et al., 2020). At a median follow-up of 25.8 months, entrectinib demonstrated a complete or partial response in 61.2% of patients (Demetri et al., 2022). In 2024, the FDA granted accelerated approval to repotrectinib as the third pan-tumor approval for the treatment of adult and pediatric patients (12 years or older) with unresectable solid tumors harboring an NTRK gene fusion with locally advanced or metastatic disease after treatment progression or those with no satisfactory alternative therapy options, based on efficacy data from the phase 1/2 TRIDENT-1 trial in TRK inhibitor naïve patients and those who previously received a TRK inhibitor (Solomon et al., 2023). Entrectinib, larotrectinib, and repotrectinib are effective across pediatric and adult solid tumors and responses are independent of the NTRK fusion gene and fusion partner. Despite the efficacy of TRK inhibitors, acquired resistance to TRK inhibitors, including the emergence of on-target NTRK kinase domain mutations or off-target bypass mechanisms, remains a clinical unmet need. Second-generation TRK inhibitors are in development for on-target NTRK kinase domain mutations (Drilon, 2019; Cocco et al., 2019a).
The immunotherapy biomarker landscape in patients with NTRK fusions has been explored, and the association of NTRK fusions with tumor mutational burden (TMB) is discrepant between studies, except for in microsatellite instability-high (MSI-H) CRC (Gatalica et al., 2019; Wang H. et al., 2022; Rosen et al., 2020; Bazhenova et al., 2021). Programmed death-ligand 1 (PD-L1) expression has been detected in around 25% of NTRK fusion-positive cases, including MSI-H cases (Gatalica et al., 2019). NCCN guidelines emphasize that for patients with advanced NSCLC, testing for all recommended biomarkers should be performed before initiating single-agent immunotherapy due to the lack of response in tumors harboring actionable driver alterations (Ettinger et al., 2022; Mazieres et al., 2019). However, there has been minimal investigation of treatment sequencing in patients with NTRK fusion-positive tumors concerning targeted therapy versus immunotherapy and the role of immunotherapy after targeted therapy resistance.
To assess the prevalence of NTRK fusions in a real-world standard-of-care setting, we (Amatu et al., 2019) describe the landscape of oncogenic and likely oncogenic NTRK fusions detected across solid tumors, including novel fusion partners, by hybrid-capture next-generation RNA sequencing performed in a reference laboratory as part of patients’ routine clinical care and (Cocco et al., 2018) characterize the co-occurring alterations and immunotherapy-related signatures detected in NTRK fusion-positive solid tumors.
2 Methods
2.1 Patient cohort
Approval for this study, including waiver of informed consent, was obtained from the Western Institutional Review Board Copernicus Group (WCG protocol # 1340120).
We retrospectively analyzed comprehensive genomic profiling (CGP) and PD-L1 22C3 immunohistochemistry (IHC) data from 19,591 FFPE solid tumor samples submitted for NGS testing at a reference laboratory (OmniSeq/Labcorp, Buffalo, NY, United States) as part of patients’ routine clinical care between June 2021 and August 2024 (Supplementary Table S1).
2.2 Comprehensive genomic profiling (CGP)
DNA and RNA were co-extracted from FFPE tissue specimens and submitted for library preparation and sequencing using the hybrid-capture-based TruSight® Oncology (TSO) 500 assay (Illumina, San Diego, CA, United States) as part of OmniSeq® INSIGHT (OmniSeq/Labcorp, Buffalo, NY, United States). OmniSeq® INSIGHT is a comprehensive genomic and immune profiling assay performed in a laboratory accredited by the College of American Pathologists and certified by the Clinical Laboratory Improvement Amendments (Conroy et al., 2021). Within the genomic profiling framework of the assay, DNA sequencing with hybrid capture detects small nucleotide variants (SNVs) in exonic regions of 523 genes, copy number variants (CNVs) in 59 genes, and genomic signatures including MSI and TMB. RNA sequencing with hybrid capture detects fusions and splice variants in 55 genes. Variant annotation is performed using the GenomeOncology Precision Oncology Platform (GenomeOncology, Cleveland, OH, United States). Only genomic alterations annotated as known pathogenic were analyzed in the current study.
2.3 NTRK fusion analysis and classification
NTRK fusions were called and assembled using the Manta fusion caller (Chen et al., 2016), and candidate fusions were annotated as [5′ gene]-[3′ gene] when ≥5 unique candidate gene fusion reads were detected (Conroy et al., 2021). Detected fusions were labeled oncogenic or likely oncogenic based on published NTRK fusion oncogenicity classification guidelines outlined by the ClinGen NTRK Fusions Somatic Cancer Variant Curation Expert Panel (Saliba et al., 2022). Based on these guidelines, NTRK fusions were classified as oncogenic if (Amatu et al., 2019) the NTRK gene component was the 3′ partner of the fusion and contained the TKD region and (Cocco et al., 2018) the 5′ gene component was a known fusion partner of NTRK genes. Known fusion partners included canonical partners or partners with clinical evidence of validity. Canonical fusion partners were defined as those reported in Catalogue Of Somatic Mutations In Cancer (COSMIC) (Sondka et al., 2024) and GENIE (AACR Project GENIE Consortium, 2017) databases or >3 studies in the scientific literature as curated by Salida et al., 2022 (Saliba et al., 2022). Partners with clinical evidence of validity were those identified in clinical studies that supported the FDA approvals of larotrectinib or entrectinib (Hong et al., 2020; Doebele et al., 2020; Drilon et al., 2017). NTRK fusions were classified as likely oncogenic if (Amatu et al., 2019) the NTRK gene component was the 3′ partner of the fusion and contained the TKD region and (Cocco et al., 2018) the 5′ gene component was a novel fusion partner where the reading frame was preserved (Saliba et al., 2022). An NTRK fusion that did not meet the oncogenic or likely oncogenic classification criteria was labeled as a variant of unknown significance (VUS).
2.4 Immunohistochemical studies
PD-L1 protein expression on the surface of tumor cells was measured by Dako PD-L1 IHC 22C3 pharmDx (Agilent, Santa Clara, CA, United States). A board-certified anatomical pathologist scored cell surface PD-L1 expression according to published guidelines as tumor proportion score (TPS) or combined positive score (CPS) depending on the tumor type (Patel and Kurzrock, 2015). TPS represents the percentage of tumor cells with positive PD-L1 out of all viable tumor cells. CPS represents the ratio of total PD-L1 positive cells (tumor and non-tumor cells) in a tumor specimen relative to all viable tumor cells. Both are scored numerically with values from 0 to 100.
2.5 Statistical analysis
Statistical analysis and plot generation were performed in R v 4.4.2 (https://www.r-project.org/). Basic plotting was performed using the ggplot2 v 3.5.1 (https://cran.r-project.org/web/packages/ggplot2) and ggpubr v 0.6.0 (https://cran.r-project.org/web/packages/ggpubr). Circos plots were generated using the R package circlize v 0.4.16 (https://cran.r-project.org/web/packages/circlize). The Wilcoxon rank-sum test was used to test for differences in TMB and PD-L1 IHC between NTRK-positive and negative tumors. TMB was given a pseudo-count of 1 and then log-transformed before statistical testing.
3 Results
3.1 NTRK fusions are present across solid tumor types and histologies
We retrospectively analyzed data from 19,591 FFPE samples encompassing 35 solid tumor types submitted for CGP as part of routine clinical care (Supplementary Table S1). The largest tumor type group was NSCLC making up more than a third of cases included in the analysis (39%) (Figure 1A; Table 1). Overall, 73 oncogenic or likely oncogenic NTRK fusions were identified in 69 unique tumor specimens across 19 tumor types for a total cohort prevalence of 0.35% (Figure 1B, first column; Supplementary Table S2). The prevalence of NTRK fusions in NSCLC was below that of the total cohort prevalence (0.24%) and above that in other solid tumor types (0.43%) (Table 1). Tumor types with the highest NTRK fusion prevalence included glioblastoma (1.91%, N = 3 out of 157), small intestine (1.32%, N = 1 out of 76), and head and neck (0.95%, N = 4 out of 423) tumors with other solid tumor types ranging from 0.19% (uterine, N = 1 out of 514) to 0.63% (breast, N = 9 out of 1,423) (Figure 1B). The median age of patients with NTRK fusions was 66 years (range 26–89 years) with a slight overrepresentation of females in NSCLC (56%) and other solid tumors (61%) (Table 1; Supplementary Table S2). For patients with staging information, almost all were advanced stage (86%) (Table 1; Supplementary Table S2).
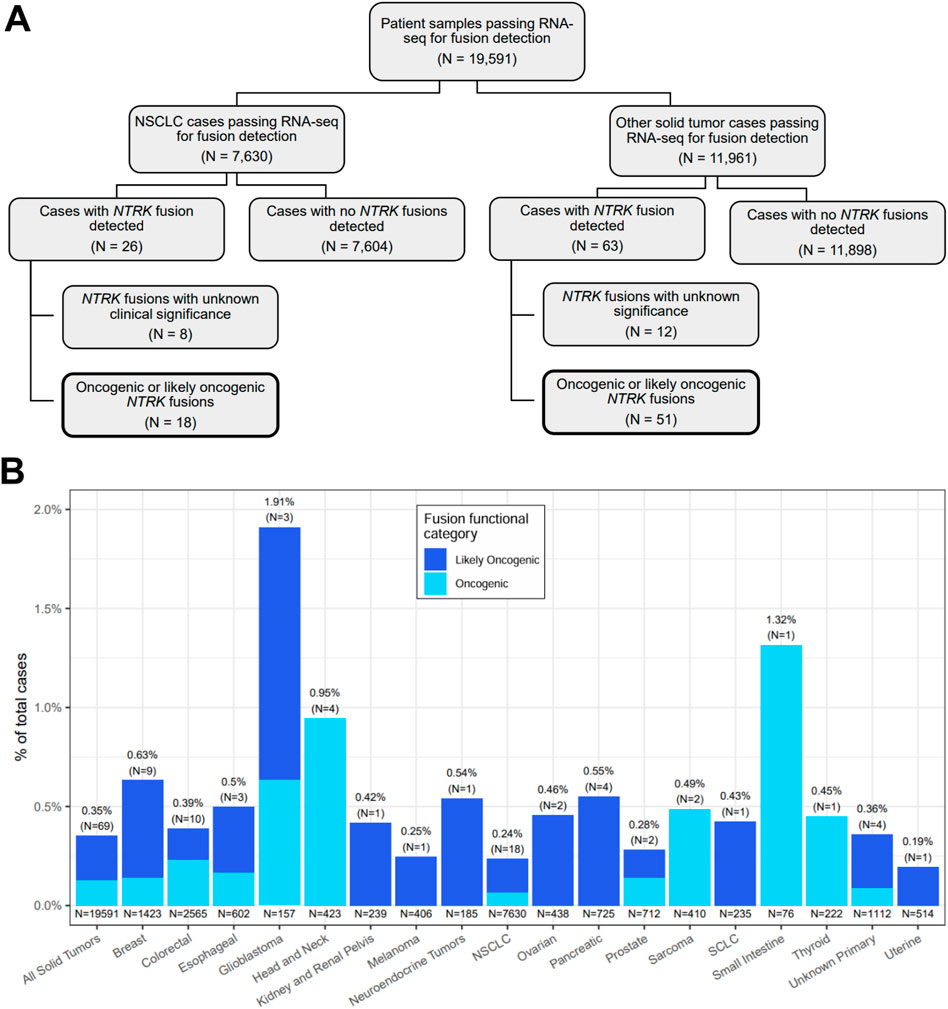
Figure 1. Overview of NTRK fusion case selection and detection. (A) A flow chart showing the breakdown of NTRK fusion cases detected out of 19,591 patient tumor samples tested with and passing the RNA-seq fusion detection component of the TruSight Oncology 500 assay. The remaining figures in the current study focused on cases where oncogenic or likely oncogenic NTRK fusions were detected (boxes with bold borders). (B) Distributions of oncogenic and likely oncogenic NTRK fusions stratified by cancer type. Each bar represents the prevalence of cases with NTRK fusions estimated to be oncogenic or likely oncogenic out of the total number of cases that passed RNA-seq for fusion detection. The N in parentheses represents the number of cases with an oncogenic or likely oncogenic NTRK fusion. The N under each bar represents the total number of cases included in a group. Each bar is colored by the fraction of cases with NTRK fusions predicted to be likely oncogenic or oncogenic via manual data inspection by two independent investigators. NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer.
NTRK fusions in NSCLC tumors were identified across NSCLC histologic types, including poorly differentiated adenocarcinoma (Figure 2A), lung adenocarcinoma (Figure 2B), and lung adenocarcinoma with enteric differentiation (Figure 2C). For comparison, Figure 2D highlights an ovarian carcinoma with a KANK-NTRK3 fusion.
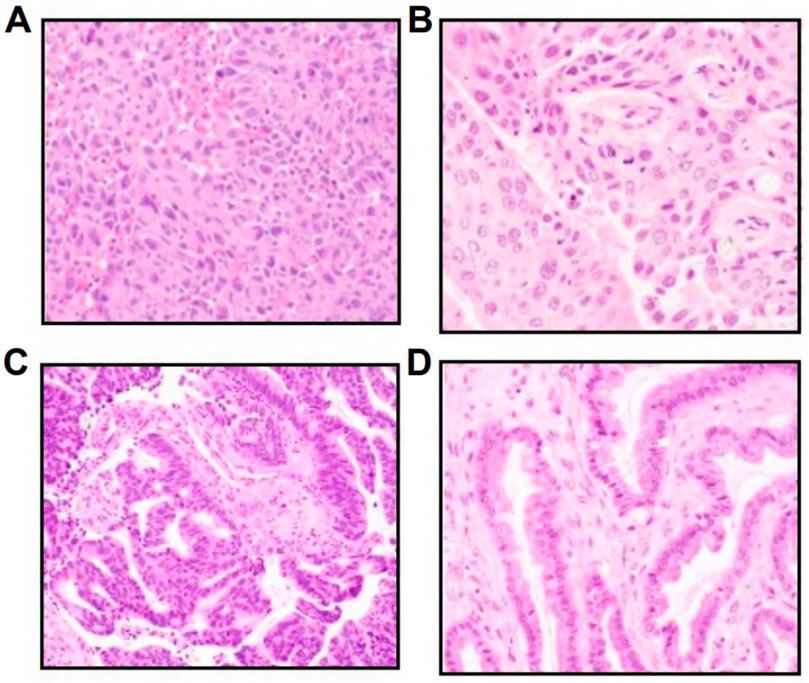
Figure 2. Hematoxylin and eosin images of diverse NTRK-positive NSCLC histologies, including (A) a poorly differentiated adenocarcinoma with a RABGAP1L-NTRK1 fusion, (B) a lung adenocarcinoma with a TP53-NTRK1 fusion, (C) a lung adenocarcinoma with enteric differentiation with a PRKACA-NTRK2 fusion, and (D) an ovarian carcinoma with a KANK-NTRK3 fusion.
3.2 Genomic structure of NTRK fusions
Fusion of the NTRK 3′ TKD with a 5′ upstream gene results in a chimeric oncoprotein having ligand-independent constitutive TRK kinase activity (Figure 3A). There are currently multiple generations of NTRK-targeted therapies, some approved and others in development, that bind the TRK domain and block downstream growth signaling pathways (Figure 3A). A specific example of a fusion and its structure that was detected in the present study is shown in Figure 3B, where we detail the fusion locations for the known LMNA-NTRK1 fusion, with breakpoints in intron 3 (LMNA) and intron 11 (NTRK1), and the resulting LMNA-NTRK1 fused mRNA.
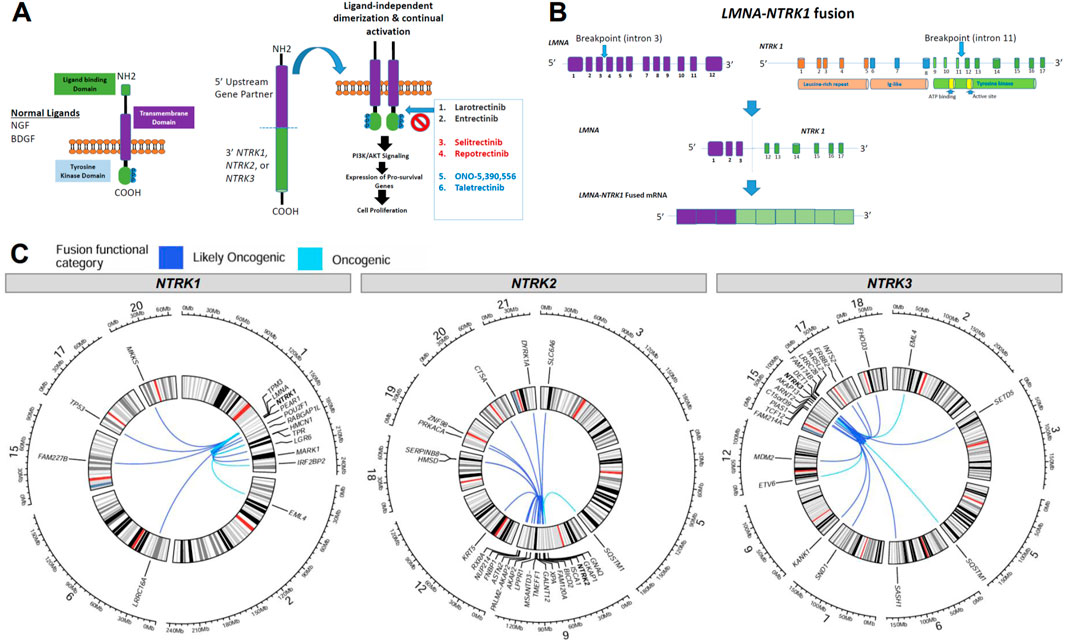
Figure 3. Overview of NTRK fusion mechanisms and fusion partners. (A) Schematic of the normal neurotrophic tropomyosin receptor kinase (NTRK) structure and oncogenic NTRK fusion structure with downstream pathways and TRK inhibitor therapy options, with next-generation drugs in red and blue. (B) An example of breakpoint locations in an NTRK fusion (denoted with arrows) in an LMNA-NTRK1 fusion identified in this analysis, resulted in fused mRNA. (C) Circos plots demonstrating the locations of NTRK genes and fusion partners for oncogenic or likely oncogenic fusions identified in the present study. Lines connect the 5′ and 3′ genes in each gene fusion identified. The color of the lines denotes if a fusion was inferred to be oncogenic or likely oncogenic.
We identified other, diverse partner genes for oncogenic and likely oncogenic NTRK fusions across all tumor types including intra and inter-chromosomal rearrangements with intra-chromosomal fusions identified more frequently (Figure 3C). For NTRK1-detected fusions, 10 included partners within chromosome 1 (e.g., RABGAP1L, PEAR1, LMNA, HMCN1) while 5 inter-chromosomal rearrangements were identified with genes in chromosomes 2 (EML4), 6 (LRRC16A), 15 (FAM227B), 17 (TP53), and 20 (MKKS) (Figure 3C, first panel). NTRK2-detected fusions had the highest number of unique fusion partners, 15 included partners within chromosome 9 (e.g., GNAQ) while 9 inter-chromosomal rearrangements were identified with genes in chromosomes 3 (SLAC6A6), 5 (SQSTM1), 12 (KRT5), 18 (HMSD, SERPINB8), 19 (PRKACA, ZNF98), 20 (CTSA), and 21 (DYRK1A) (Figure 3C, second panel). For NTRK3-detected fusions, 10 included partners within chromosome 15 (e.g., ARNT2, AKAP13) while 11 inter-chromosomal rearrangements were identified with genes in chromosomes 2 (EML4), 3 (SETD5), 5 (SQSTM1), 6 (SASH1), 7 (SND1), 9 (KANK1), 12 (ETV6, MDM2), 17 (ERBB2, INTS2), and 18 (FHOD3) (Figure 3C, third panel). Most NTRK fusions were detected in only one tumor specimen, though some recurrent fusions were noted with ETV6, TPM3, LMNA, EML4, TPR, PEAR1, IRF2BP2, and KANK1 fusion partners (Table 2; Supplementary Table S2).
We identified fusion breakpoint locations in introns 1, 6, 9, 11, and exon 10 in NTRK1 (Figure 4A; Supplementary Table S2). In NTRK2, breakpoints were found more ubiquitously across the NTRK2 gene including introns 1, 4, 6, 10, 11, 15, 16, and in exons 5, 12, 15, and 17 (Figure 4B; Supplementary Table S2). In NTRK3, we identified focalized breakpoints associated with 4–6 gene partners in exons 4, 14, and 15 while other breakpoints were associated with fewer gene partners including intron 3 and exons 2, 3, 6, and 12 (Figure 4C; Supplementary Table S2). We identified 41 novel fusion partners across the NTRK genes including HMCN, ASTN2, MSANTD3-TMEFF1, PRKACA, FAM174B, PIAS1, and others. We identified fewer known fusion partners (N = 18), which suggests the potential for further investigation into novel fusion partners to increase this number and bring more clinical evidence. Detected known fusion partners included LMNA, PEAR1, RABGAP1L, TP53, EML4, TMP3, LGR6, TPR, and IRF2BP2 for NTRK1 (Figure 4A), GKAP1 and SQSTM1 for NTRK2 (Figure 4B), and KANK1, SASH1, ETV6, EML4, ARNT2, SQSTM1, and TARSL2 for NTRK3 (Figure 4C).
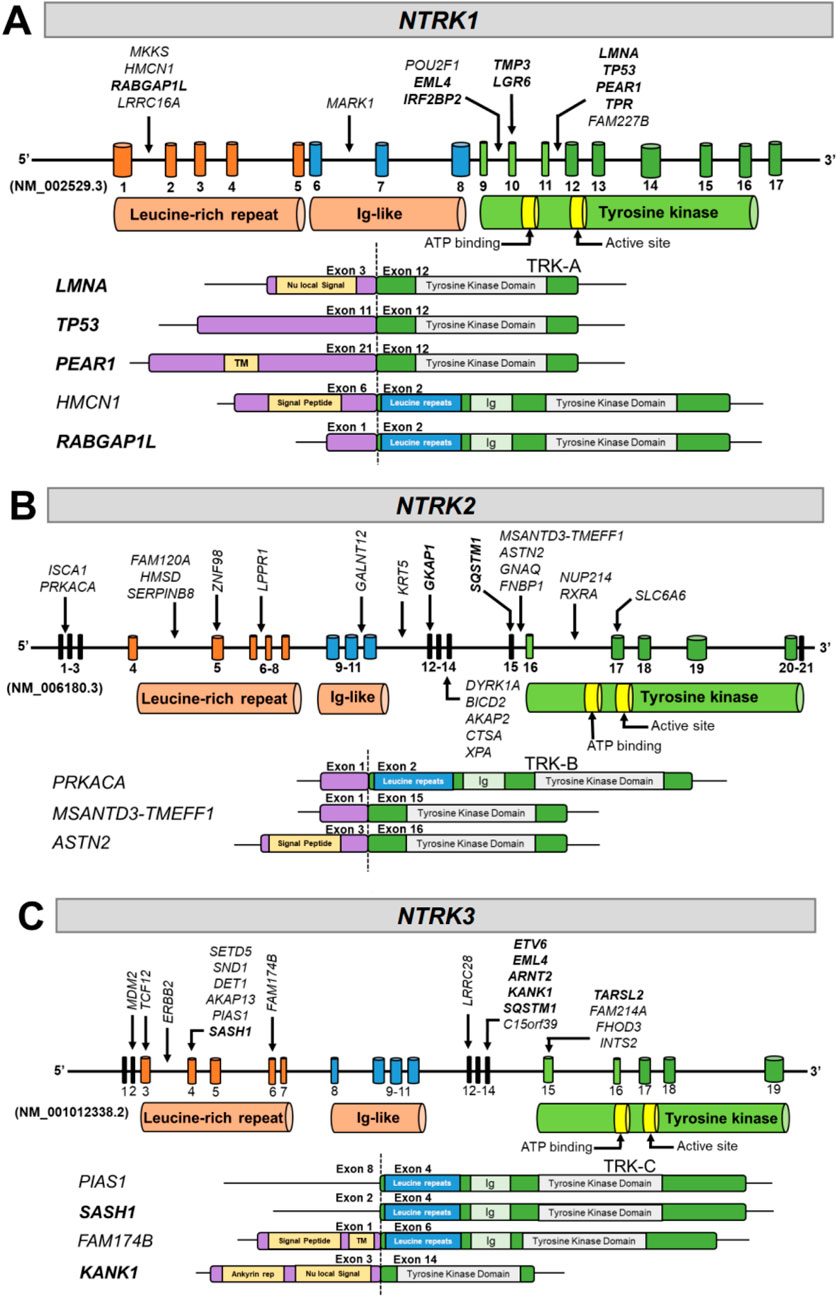
Figure 4. Diagrams of NTRK fusions identified in the present study. The resulting exonic structure, including retained exons and functional domains for each partner gene and NTRK gene, are represented for (A) NTRK1, (B) NTRK2, and (C) NTRK3. Arrows next to gene partner names denote breakpoints involved in each fusion. Known fusion partners have been bolded. TM, transmembrane; Ig, immunoglobulin.
The TRK TKD is encoded by different exons in each gene NTRK gene. The TKD is encoded by exons 9-17 for NTRK1, 16-20 in NTRK2, and 15-19 in NTRK3 (Figure 4). The TKD was present in all gene fusions identified with NTRK as the 3′ fusion partner (Figures 4A–C). Transmembrane domains were present in the partner genes PEAR1 and FAM174B (Figures 4A,C), and 6 of 12 NTRK fusions contained the transmembrane domain in the TRK protein; however, a transmembrane domain is not required for a fusion to be functional.
3.3 Genomic driver alterations that co-occur with NTRK fusions
We evaluated co-occurring genomic alterations across all samples with an oncogenic or likely oncogenic NTRK fusion (Figure 5; Supplementary Table S3). The most common co-occurring alterations were small SNVs, or insertions/deletions found in 86% (N = 59) of oncogenic or likely oncogenic NTRK-positive tumors. CNVs and non-NTRK fusions co-occurred at similar frequencies in 29% (N = 20) and 26% (N = 18) of NTRK-positive tumors, respectively (Supplementary Table S3). The gene most commonly co-mutated was TP53, which was altered in 50.7% of cases, while other commonly co-mutated genes included ARID1A (13%), KRAS (13%), and NOTCH1 (10.1%) (Figure 5; Supplementary Table S3). Co-occurring CNVs typically involved amplification of MDM2 or loss of PTEN (∼4% of NTRK-positive tumors respectively) (Figure 5; Supplementary Table S3). Most NTRK fusions were mutually exclusive from other genomic driver alterations, however, almost a third of tumor specimens (29%) contained at least one co-occurring genomic driver (Supplementary Table S3). In NSCLC, co-occurring oncogenic driver mutations were identified in 61% of cases (N = 11 out of 18) including ALK, KRAS, BRAF, and EGFR (Supplementary Table S3). A breakdown of co-occurring alteration frequencies in patient tumors with oncogenic and likely oncogenic NTRK fusions and NTRK fusions of unknown significance, along with AMP/CAP/ASCO Tier associations, can be found in Supplementary Table S4.
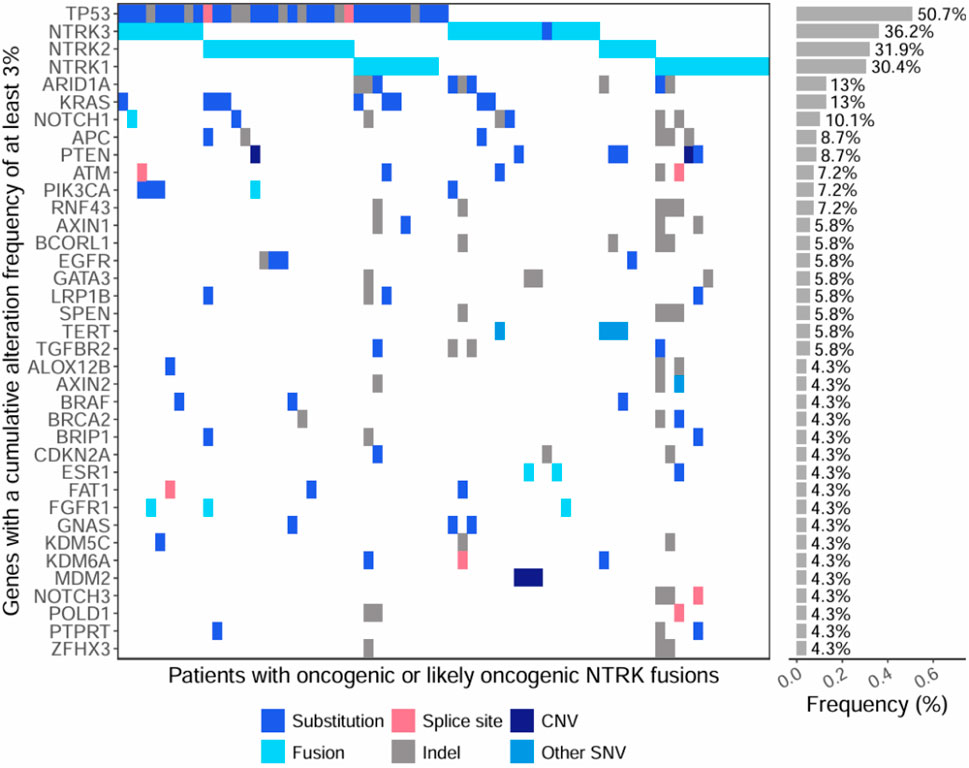
Figure 5. Oncoprint of co-occurring mutations within genes with a cumulative alteration frequency of at least 3%. The first section of the plot shows individual mutations detected within tumors of patients with oncogenic or likely oncogenic NTRK fusions (x-axis). The second section of the plot shows the cumulative alteration frequency of each gene (y-axis). Colors represent the type of genomic alteration identified in the genes.
3.4 Immunotherapy biomarkers: TMB, MSI, and PD-L1 IHC
We next sought to characterize distributional differences in immunotherapy biomarkers (TMB, PD-L1 IHC, MSI) between tumors harboring oncogenic or likely oncogenic NTRK fusions and NTRK-negative tumors (Figure 6). We found TMB and PD-L1 IHC scores were significantly lower in NTRK-positive head and neck tumors compared to NTRK-negative (1.1 vs. 5.4 mutations/Mb, 0 vs. 10 CPS, P < 0.03) (Figures 6A,B). An opposite trend was observed in CRC tumors where NTRK-positive tumors trended toward higher TMB scores (7.8 vs. 6.2 mutations/Mb, P = 0.06) and had significantly higher PD-L1 IHC scores (20 vs. 0 CPS, P < 0.0001) (Figures 6A,B). No significant differences in TMB or PD-L1 IHC were observed when analyzing all solid tumors at once (P > 0.8) (Figures 6A,B). MSI-H was seen at a higher frequency in NTRK-positive tumors in CRC and small intestine cases (50% vs. 6.6%) while MSI-H cases were only observed in NTRK-negative cases in other solid tumors (Figure 6C).
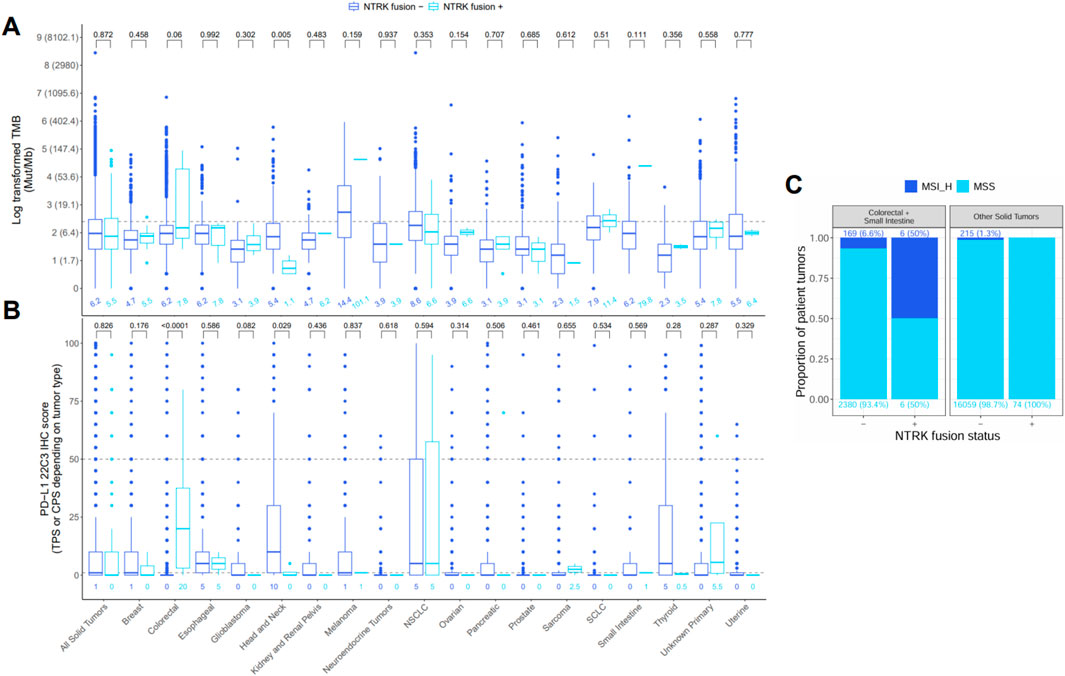
Figure 6. Distribution of (A) tumor mutational burden (TMB), (B) PD-L1 22C3 immunohistochemistry (IHC), and (C) microsatellite instability (MSI) in tumors negative (−) or positive (+) for oncogenic or likely oncogenic NTRK fusions. The bottom, middle, and top horizontal boundaries of each box in the box plots represent the first, second (median), and third quartiles of the data for a particular age group. The lines extending from the two ends of each box represent 1.5x outside the interquartile range. Points beyond the lines are considered outliers. Values above each pair of box plots represent the uncorrected P-values from testing differences between NTRK–or + tumors using the Wilcoxon rank-sum test. Values below each box plot represent the median value for a group. The dashed line represents the threshold for high (≥10) and low (<10) values for TMB and the high (≥50), low (1–49), and negative (<1) values for PD-L1 IHC. PD-L1 IHC was calculated using the tumor proportion score (TPS) or combined positive score (CPS) depending on tumor type. Values for TMB were log-transformed for testing and plotting, but the corresponding untransformed values are provided in parentheses next to each log-transformed value on the y-axis. Bar plots of MSI represent the proportion of cases that were MSI high or microsatellite stable (MSS) in NTRK - or + tumors. Differences in MSI status were not tested due to low sample size in the MSI-high (MSI_H) groups.
4 Discussion
NTRK gene fusions are oncogenic drivers found in diverse pediatric and adult tumors. Intra- and inter-chromosomal rearrangements involving the 3′ regions of NTRK genes and the 5′ region of a partner gene can produce a constitutively active TRK fusion protein that drives uncontrolled downstream signaling (Penault-Llorca et al., 2019). Though these gene fusions are rare, they are clinically important targets due to high response rates to targeted therapy and durable responses in patients with TRK fusion-driven advanced and metastatic tumors (Hong et al., 2020; Demetri et al., 2022). It is recommended that all patients with TRK fusion-positive tumors be identified so they can receive approved targeted therapies. In our analysis, 19,531 FFPE solid tumor samples underwent successful RNA hybrid-capture-based NGS and functional characterization of detected NTRK fusions, revealing 73 oncogenic or likely oncogenic NTRK fusions identified in 69 (0.35%) adult patients (Figure 1; Supplementary Table S2). Among these fusions, 41 novel 5′ partners were identified (Figure 4; Supplementary Table S2).
Though there are several techniques available to identify NTRK gene fusions, the wide range of gene partners and the presence of large intronic regions where breakpoints often occur can make identification of these fusions challenging (Marchio et al., 2019). Techniques such as IHC and fluorescence in situ hybridization (FISH) are often readily available; however, these methods can produce high rates of false negatives and false positives. Additionally, they do not identify the fusion partner as they only interrogate one target at a time. Reverse transcriptase polymerase chain reaction (RT-PCR) has the advantage of high sensitivity and specificity; however, target sequences must be known ahead of time, which decreases the likelihood of detection of novel fusion partners. Though clinical guidelines acknowledge multiple techniques for NTRK gene fusion detection, NGS is the recommended method (Ettinger et al., 2022).
The advantages of using NGS for gene fusion detection include the ability to evaluate multiple actionable targets at once and the potential for detecting novel fusion partners (Marchio et al., 2019). Several commercially available DNA-based hybrid-capture NGS assays exist that sequence full exons or hotspot regions and a select number of introns for detecting common rearrangement and fusion breakpoints (Marchio et al., 2019). Though many breakpoints occur within introns, the number of intronic regions and complete coverage of select introns in targeted panels is often limited to avoid compromising the sequencing capacity of desired exons in key genes of interest (Solomon et al., 2019). Therefore, adequate sequencing of all introns with a DNA-only hybrid capture approach is not feasible when large or complex intronic regions are present. This is especially true for NTRK genes that contain large introns that cannot be included in a targeted DNA-only NGS approach. These panels are often designed to sequence a limited number of select introns within NTRK1 and NTRK2. With this approach, breakpoints occurring in introns that are not sequenced in these targeted panels will be missed. Additionally, it is not feasible to sequence the large NTRK3 introns with targeted DNA panels. These panels are restricted to identifying only NTRK3 gene fusions with its most common partner, ETV6, based on select ETV6 introns. Because of these limitations, DNA-only targeted NGS platforms will not identify all NTRK gene fusions.
RNA-based NGS assays for gene fusion detection eliminate the need for sequencing introns, since they are spliced out in mRNA, and can target known fusion exons within multiple oncogenes. In addition, the detection of RNA fusions indicates successful transcription of the gene fusion and provides insight into possible translation to a functional oncogenic fusion protein. The advantages of using PCR amplicon-based RNA panels for fusion detection include shorter and simpler workflows and compatibility with lower quantities of RNA; however, this technique is limited to a small number of gene targets and detection of novel fusion partners is not possible with classical amplicon approaches. Hybrid capture-based RNA panels typically require high quantities of RNA with more complex and longer workflows but also capture large numbers of gene targets and novel fusions with increased sensitivity and specificity as only one fusion partner needs to be known (Solomon et al., 2019; Singh, 2022; Bruno and Fontanini, 2020).
We utilized an RNA hybrid capture-based NGS approach designed to detect fusions and splice variants in 55 genes including NTRK1, NTRK2, and NTRK3 fusions, resulting in a detection rate of 0.35% across a real-world, pan-tumor cohort. The prevalence of NTRK fusions in this study was higher than in previous studies using DNA hybrid capture NGS with or without RNA sequencing using targeted multiplex PCR for fusion detection (Westphalen et al., 2021; Rosen et al., 2020). We also used a stringent approach to characterize each NTRK fusion as functionally oncogenic or likely oncogenic (Saliba et al., 2022). This resulted in 28 additional NTRK fusions detected in this study being classified as VUS and not carried forward in downstream characterizations (Supplementary Table S2). Of the 41 novel fusion partners, 51% (N = 21) were NTRK2 fusions with the majority having intronic breakpoints (Figure 4B). Detection of novel NTRK2 fusions is limited using DNA-only hybrid capture approaches due to limited intronic baiting, which would miss most of the novel NTRK2 fusions identified in this analysis (Westphalen et al., 2021). An oncogenic, canonical GNAQ-NTRK2 fusion identified in a patient with esophageal adenocarcinoma patient would have likely been missed as breakpoints within both genes were in intronic regions (intron 1 of GNAQ, intron 15 of NTRK2) that are not typically covered in DNA-only hybrid capture panels. Considering this patient did not have any other actionable co-mutations (CHEK1 deletion (S343fs) and TP53 substitution (R248Q), Supplementary Table S3), the identification of this GNAQ-NTRK2 fusion would be critical for treatment decisions. Additionally, novel NTRK3 fusions would not have been detected by DNA NGS targeted panels if breakpoints occurred in NTRK3 introns and did not have ETV6 as the 5′ fusion partner. The identification of these 41 novel NTRK fusion partners in our analysis provides additional evidence that RNA hybrid-capture NGS testing is a superior method for NTRK fusion detection over DNA NGS targeted panels and speaks to the need for a well-designed RNA NGS panel to capture all potential NTRK fusions.
We assessed TMB and MSI genomic signatures by DNA NGS and PD-L1 expression by IHC in NTRK fusion-positive and negative patient tumors. Consistent with a previous report, TMB and PD-L1 expression were found to be significantly lower in NTRK fusion-positive head and neck cancer specimens compared to fusion-negative cases (Figures 6A,B), suggesting these NTRK fusions are the dominant targetable oncogenic drivers (Xu et al., 2021). DNA NGS was also used to detect co-occurring genomic alterations. Of the 12 NTRK fusion-positive CRC and small intestine cases, 50% (N = 6) were MSI-H (Figure 6C) and 92% (N = 11) were KRAS, NRAS, and BRAF wild-type, consistent with published data (Cocco et al., 2019b). One NTRK fusion-positive CRC patient’s tumor was MSI-H, TMB-H (91.6 mut/Mb) and PD-L1 IHC positive (CPS score = 80%) with a POLD1 co-mutation, and another case with metastatic small intestine cancer was also MSI-H and TMB-H (80 mut/Mb) with a POLD1 co-mutation (Supplementary Tables S2, S3). This suggests these patients are more likely to benefit from immune checkpoint inhibitor (ICI) therapy. ICIs such as pembrolizumab are effective treatment options for patients with MSI-H or TMB-H metastatic CRC (Benson et al., 2021; Andre et al., 2020; Samstein et al., 2019). However, there is currently no evidence to suggest the timing of immunotherapy treatment for NTRK-positive CRC patients. Of the 18 NTRK fusion-positive patients with NSCLC, 6 had PD-L1 IHC expression with TPS ≥1–49%, and 6 had TPS ≥50% for consideration of pembrolizumab (Figure 1B; Supplementary Table S2).
Clinical guidelines emphasize testing for actionable biomarkers (including ALK, BRAF, EGFR, MET exon 14 skipping, NTRK, RET, and ROS) should be performed before administering first-line ICIs due to decreased efficacy when tumors harbor co-occurring driver mutations (Ettinger et al., 2022; Mazieres et al., 2019). Two NTRK fusion-positive NSCLC patients had co-occurring KRAS G12C driver mutations (Supplementary Table S3). Four NTRK fusion-positive patients had co-occurring EGFR driver mutations (3 with EGFR L858R and 1 with EGFR exon 19 deletion), where first-line immunotherapy may have decreased efficacy (Supplementary Table S3) and could also lead to severe immune-related adverse events when followed by osimertinib (Lee et al., 2017; Schoenfeld et al., 2019). Five of these lung cancer patients had novel NTRK2 fusions with intronic breakpoints that would likely have been missed if using a DNA NGS targeted panel (Supplementary Table S2), potentially leading to first-line immune checkpoint inhibitor therapy with decreased efficacy in the presence of a co-occurring oncogenic driver.
Of the 10 NTRK fusion-positive breast cancer patients, only 1 patient had an additional actionable PIK3CA E545K mutation (Supplementary Table S3). A novel ERBB2-NTRK3 fusion was identified from a breast mammary adenocarcinoma specimen that was also confirmed to have an ERBB2 copy number gain (estimated to be a 7-fold change) and a co-occurring TP53 mutation (Supplementary Tables S2, S3). The pathology report confirmed this tumor to be HER2 positive (HER2 IHC 3+). In previous NGS genomic profiling studies of HER2+ breast cancer patients, fusion events are common, with ERBB2 being the most frequently involved gene (Chen et al., 2020) with the most frequent co-mutations found in TP53 (Schubert et al., 2023; Wang T. et al., 2022). ERBB2 fusions most commonly occur in chromosome 17, with the most frequent breakpoint identified within exon 27, corresponding to the chromosome and breakpoint identified in this ERBB2-NTRK3 fusion (Supplementary Table S2). HER2+ patients with tumors harboring ERBB2 fusions have been shown to achieve a pathological complete response with chemotherapy plus trastuzumab, indicating these fusions do not lead to resistance to therapy (Lesurf et al., 2017). This novel ERBB2-NTRK3 fusion provides an additional targeted therapy option after progression on available anti-HER2 agents.
Additional co-occurring actionable genomic alterations were identified in 5 NTRK fusion-positive patients: CRC (CCDC6-RET fusion), pancreatic (ITSN2-ALK fusion), prostate (PTEN loss, TBL1XR1-PIK3CA fusion), ovarian (PIK3CA mutation) (Supplementary Table S3). A patient originally diagnosed with uterine endometrioid adenocarcinoma had additional clinically relevant PIK3CA and TP53 mutations that further characterize this patient as having pleomorphic rhabdomyosarcoma (Pinto et al., 2018). In summary, most NTRK fusion-positive patients did not have an actionable co-occurring alteration where additional targeted therapies would have been available, highlighting the need for accurate NTRK fusion testing.
5 Conclusion
NTRK fusions are clinically relevant driver alterations across solid tumor types. These fusions are difficult to detect, as breakpoints can occur within introns, and they have a wide range of partner genes. RNA hybrid capture-based sequencing in a real-world standard-of-care setting revealed the highest reportable oncogenic and likely oncogenic NTRK fusion prevalence (0.35%) across solid tumors with 41 previously unreported novel NTRK fusion partners. These data emphasize the importance of CGP using a well-designed RNA hybrid-capture sequencing assay to identify all NTRK fusions across malignancies from a broad range of histologic subtypes for optimal patient treatment management.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Western Institutional Review Board Copernicus Group. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
ZW: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review and editing. MT: Conceptualization, Data curation, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review and editing. ES: Conceptualization, Data curation, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review and editing. AR: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review and editing. MG: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review and editing. KA: Data curation, Formal Analysis, Investigation, Methodology, Writing – review and editing. RP: Writing – review and editing. SH: Project administration, Supervision, Writing – review and editing. SP: Data curation, Resources, Writing – review and editing. TJ: Resources, Supervision, Writing – review and editing. BC: Resources, Supervision, Writing – review and editing. ME: Resources, Supervision, Writing – review and editing. PS: Conceptualization, Resources, Supervision, Writing – review and editing. SR: Resources, Supervision, Writing – review and editing. ES: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
Authors MT, ES, AR, and PS are employed by and have an equity interest in Illumina. ZDW, MG, KA, RAP, SH, SP, TJJ, BC, ME, SHR, and EAS are employed by and have an equity interest in Labcorp.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1550706/full#supplementary-material
References
AACR Project GENIE Consortium, André, F., Arnedos, M., Baras, A. S., Baselga, J., Bedard, P. L., et al. (2017). AACR Project GENIE: powering precision medicine through an international Consortium. Cancer Discov. 7 (8), 818–831. doi:10.1158/2159-8290.CD-17-0151
Amatu, A., Sartore-Bianchi, A., Bencardino, K., Pizzutilo, E. G., Tosi, F., and Siena, S. (2019). Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann. Oncol. 30 (Suppl. 8), viii5–viii15. doi:10.1093/annonc/mdz383
Andre, T., Shiu, K. K., Kim, T. W., Jensen, B. V., Jensen, L. H., Punt, C., et al. (2020). Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 383 (23), 2207–2218. doi:10.1056/NEJMoa2017699
Bazhenova, L., Lokker, A., Snider, J., Castellanos, E., Fisher, V., Fellous, M., et al. (2021). TRK fusion cancer: patient characteristics and survival analysis in the real-world setting. Target Oncol. 16 (3), 389–399. doi:10.1007/s11523-021-00815-4
Benson, A. B., Venook, A. P., Al-Hawary, M. M., Arain, M. A., Chen, Y. J., Ciombor, K. K., et al. (2021). Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 19 (3), 329–359. doi:10.6004/jnccn.2021.0012
Bruno, R., and Fontanini, G. (2020). Next generation sequencing for gene fusion analysis in lung cancer: a literature review. Diagn. (Basel) 10 (8), 521. doi:10.3390/diagnostics10080521
Chen, B., Zhang, G., Wei, G., Wang, Y., Guo, L., Lin, J., et al. (2020). Heterogeneity of genomic profile in patients with HER2-positive breast cancer. Endocr. Relat. Cancer 27 (3), 153–162. doi:10.1530/ERC-19-0414
Chen, X., Schulz-Trieglaff, O., Shaw, R., Barnes, B., Schlesinger, F., Kallberg, M., et al. (2016). Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32 (8), 1220–1222. doi:10.1093/bioinformatics/btv710
Cocco, E., Benhamida, J., Middha, S., Zehir, A., Mullaney, K., Shia, J., et al. (2019b). Colorectal carcinomas containing hypermethylated MLH1 promoter and wild-type BRAF/KRAS are enriched for targetable kinase fusions. Cancer Res. 79 (6), 1047–1053. doi:10.1158/0008-5472.CAN-18-3126
Cocco, E., Scaltriti, M., and Drilon, A. (2018). NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 15 (12), 731–747. doi:10.1038/s41571-018-0113-0
Cocco, E., Schram, A. M., Kulick, A., Misale, S., Won, H. H., Yaeger, R., et al. (2019a). Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat. Med. 25 (9), 1422–1427. doi:10.1038/s41591-019-0542-z
Conroy, J. M., Pabla, S., Glenn, S. T., Seager, R. J., Van Roey, E., Gao, S., et al. (2021). A scalable high-throughput targeted next-generation sequencing assay for comprehensive genomic profiling of solid tumors. PLoS One 16 (12), e0260089. doi:10.1371/journal.pone.0260089
Demetri, G. D., De Braud, F., Drilon, A., Siena, S., Patel, M. R., Cho, B. C., et al. (2022). Updated integrated analysis of the efficacy and safety of entrectinib in patients with NTRK fusion-positive solid tumors. Clin. Cancer Res. 28 (7), 1302–1312. doi:10.1158/1078-0432.CCR-21-3597
Doebele, R. C., Drilon, A., Paz-Ares, L., Siena, S., Shaw, A. T., Farago, A. F., et al. (2020). Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 21 (2), 271–282. doi:10.1016/S1470-2045(19)30691-6
Drilon, A. (2019). TRK inhibitors in TRK fusion-positive cancers. Ann. Oncol. 30 (Suppl. 8), viii23–viii30. doi:10.1093/annonc/mdz282
Drilon, A., Siena, S., Ou, S. I., Patel, M., Ahn, M. J., Lee, J., et al. (2017). Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 7 (4), 400–409. doi:10.1158/2159-8290.CD-16-1237
Ettinger, D. S., Wood, D. E., Aisner, D. L., Akerley, W., Bauman, J. R., Bharat, A., et al. (2022). Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 20 (5), 497–530. doi:10.6004/jnccn.2022.0025
Gatalica, Z., Xiu, J., Swensen, J., and Vranic, S. (2019). Molecular characterization of cancers with NTRK gene fusions. Mod. Pathol. 32 (1), 147–153. doi:10.1038/s41379-018-0118-3
Hong, D. S., DuBois, S. G., Kummar, S., Farago, A. F., Albert, C. M., Rohrberg, K. S., et al. (2020). Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 21 (4), 531–540. doi:10.1016/S1470-2045(19)30856-3
Klink, A. J., Kavati, A., Gassama, A. T., Kozlek, T., Gajra, A., and Antoine, R. (2022). Timing of NTRK gene fusion testing and treatment modifications following TRK fusion status among US oncologists treating TRK fusion cancer. Target Oncol. 17 (3), 321–328. doi:10.1007/s11523-022-00887-w
Kummar, S., and Lassen, U. N. (2018). TRK inhibition: a new tumor-agnostic treatment strategy. Target Oncol. 13 (5), 545–556. doi:10.1007/s11523-018-0590-1
Lee, C. K., Man, J., Lord, S., Links, M., Gebski, V., Mok, T., et al. (2017). Checkpoint inhibitors in metastatic EGFR-mutated non-small cell lung cancer-A meta-analysis. J. Thorac. Oncol. 12 (2), 403–407. doi:10.1016/j.jtho.2016.10.007
Lesurf, R., Griffith, O. L., Griffith, M., Hundal, J., Trani, L., Watson, M. A., et al. (2017). Genomic characterization of HER2-positive breast cancer and response to neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041 (Alliance) trial. Ann. Oncol. 28 (5), 1070–1077. doi:10.1093/annonc/mdx048
Marchio, C., Scaltriti, M., Ladanyi, M., Iafrate, A. J., Bibeau, F., Dietel, M., et al. (2019). ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann. Oncol. 30 (9), 1417–1427. doi:10.1093/annonc/mdz204
Mazieres, J., Drilon, A., Lusque, A., Mhanna, L., Cortot, A. B., Mezquita, L., et al. (2019). Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann. Oncol. 30 (8), 1321–1328. doi:10.1093/annonc/mdz167
Okamura, R., Boichard, A., Kato, S., Sicklick, J. K., Bazhenova, L., and Kurzrock, R. (2018). Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis. Oncol. 2018, 1–20. doi:10.1200/PO.18.00183
Patel, S. P., and Kurzrock, R. (2015). PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther. 14 (4), 847–856. doi:10.1158/1535-7163.MCT-14-0983
Penault-Llorca, F., Rudzinski, E. R., and Sepulveda, A. R. (2019). Testing algorithm for identification of patients with TRK fusion cancer. J. Clin. Pathol. 72 (7), 460–467. doi:10.1136/jclinpath-2018-205679
Pinto, A., Kahn, R. M., Rosenberg, A. E., Slomovitz, B., Quick, C. M., Whisman, M. K., et al. (2018). Uterine rhabdomyosarcoma in adults. Hum. Pathol. 74, 122–128. doi:10.1016/j.humpath.2018.01.007
Rosen, E. Y., Goldman, D. A., Hechtman, J. F., Benayed, R., Schram, A. M., Cocco, E., et al. (2020). TRK fusions are enriched in cancers with uncommon histologies and the absence of canonical driver mutations. Clin. Cancer Res. 26 (7), 1624–1632. doi:10.1158/1078-0432.CCR-19-3165
Saliba, J., Church, A. J., Rao, S., Danos, A., Furtado, L. V., Laetsch, T., et al. (2022). Standardized evidence-based approach for assessment of oncogenic and clinical significance of NTRK fusions. Cancer Genet. 264-265, 50–59. doi:10.1016/j.cancergen.2022.03.001
Samstein, R. M., Lee, C. H., Shoushtari, A. N., Hellmann, M. D., Shen, R., Janjigian, Y. Y., et al. (2019). Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51 (2), 202–206. doi:10.1038/s41588-018-0312-8
Schoenfeld, A. J., Arbour, K. C., Rizvi, H., Iqbal, A. N., Gadgeel, S. M., Girshman, J., et al. (2019). Severe immune-related adverse events are common with sequential PD-(L)1 blockade and osimertinib. Ann. Oncol. 30 (5), 839–844. doi:10.1093/annonc/mdz077
Schubert, L., Elliott, A., Le, A. T., Estrada-Bernal, A., Doebele, R. C., Lou, E., et al. (2023). ERBB family fusions are recurrent and actionable oncogenic targets across cancer types. Front. Oncol. 13, 1115405. doi:10.3389/fonc.2023.1115405
Singh, R. R. (2022). Target enrichment approaches for next-generation sequencing applications in oncology. Diagn. (Basel) 12 (7), 1539. doi:10.3390/diagnostics12071539
Solomon, B. J., Drilon, A., Lin, J. J., Bazhenova, L., Goto, K., De Langen, J., et al. (2023). 1372P Repotrectinib in patients (pts) with NTRK fusion-positive (NTRK+) advanced solid tumors, including NSCLC: update from the phase I/II TRIDENT-1 trial. Ann. Oncol. 34, S787–S788. doi:10.1016/j.annonc.2023.09.2405
Solomon, J. P., Benayed, R., Hechtman, J. F., and Ladanyi, M. (2019). Identifying patients with NTRK fusion cancer. Ann. Oncol. 30 (Suppl. 8), viii16–viii22. viii16-viii22. doi:10.1093/annonc/mdz384
Sondka, Z., Dhir, N. B., Carvalho-Silva, D., Jupe, S., Madhumita, M. L. K., et al. (2024). COSMIC: a curated database of somatic variants and clinical data for cancer. Nucleic Acids Res. 52 (D1), D1210–D1217. doi:10.1093/nar/gkad986
Wang, H., Li, Z. W., Ou, Q., Wu, X., Nagasaka, M., Shao, Y., et al. (2022a). NTRK fusion positive colorectal cancer is a unique subset of CRC with high TMB and microsatellite instability. Cancer Med. 11 (13), 2541–2549. doi:10.1002/cam4.4561
Wang, T., Wei, L., Lu, Q., Shao, Y., You, S., Yin, J. C., et al. (2022b). Landscape of potentially targetable receptor tyrosine kinase fusions in diverse cancers by DNA-based profiling. NPJ Precis. Oncol. 6 (1), 84. doi:10.1038/s41698-022-00325-0
Westphalen, C. B., Krebs, M. G., Le Tourneau, C., Sokol, E. S., Maund, S. L., Wilson, T. R., et al. (2021). Genomic context of NTRK1/2/3 fusion-positive tumours from a large real-world population. NPJ Precis. Oncol. 5 (1), 69. doi:10.1038/s41698-021-00206-y
Wong, D., Yip, S., and Sorensen, P. H. (2020). Methods for identifying patients with tropomyosin receptor kinase (TRK) fusion cancer. Pathol. Oncol. Res. 26 (3), 1385–1399. doi:10.1007/s12253-019-00685-2
Keywords: NTRK, fusions, clinical utility, targeted therapy, genomics, next-generation sequencing, diagnostics, oncology
Citation: Wallen ZD, Tierno M, Schnettler E, Roos A, Green M, Amoah K, Previs RA, Hastings S, Pabla S, Jensen TJ, Caveney B, Eisenberg M, Sathyan P, Ramkissoon SH and Severson EA (2025) RNA hybrid-capture next-generation sequencing has high sensitivity in identifying known and less characterized oncogenic and likely oncogenic NTRK fusions in a real-world standard-of-care setting. Front. Genet. 16:1550706. doi: 10.3389/fgene.2025.1550706
Received: 16 January 2025; Accepted: 23 April 2025;
Published: 02 May 2025.
Edited by:
Ricardo Fujita, Universidad de San Martin de Porres, PeruReviewed by:
Bruce Frederic Smith, Auburn University, United StatesYosuke Seto, Japanese Foundation For Cancer Research, Japan
Copyright © 2025 Wallen, Tierno, Schnettler, Roos, Green, Amoah, Previs, Hastings, Pabla, Jensen, Caveney, Eisenberg, Sathyan, Ramkissoon and Severson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eric A. Severson, ZXJpYy5zZXZlcnNvbkBsYWJjb3JwLmNvbQ==
†These authors have contributed equally to this work
 Zachary D. Wallen
Zachary D. Wallen Marni Tierno
Marni Tierno Erica Schnettler2†
Erica Schnettler2†