- 1Innovative Institute of Animal Healthy breeding, College of Animal Sciences and Technology, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 2Yingde Yingxin Agriculture Co., Ltd., Qingyuan, Guangdong, China
- 3Laboratory of Animal Disease Detection, Qingyuan Animal Disease Prevention Control Center, Qingyuan, Guangdong, China
Introduction: Toll-like receptors (TLRs) are pattern recognition receptors essential for immune defense against pathogens, activating the host’s immune response by recognizing conserved pathogen structures. The Chinese spiny frog (Quasipaa spinosa), an amphibian native to southern China and northern Vietnam, has been severely impacted by recent infectious disease outbreaks caused by bacterial, viral, and parasitic infections, which threaten the sustainable development of the Q. spinosa farming industry. However, the roles of Q. spinosa TLRs (QsTLRs) in combating these exogenous pathogens have not yet been explored.
Methods: In the study, using the whole genome data of Q. spinosa, bioinformatics tools were employed to identify and analyze the TLR gene family. The bacteria Elizabethkingia miricola, a common pathogen, which causes the cataract disease and can lead to serious death of the frog. Here, we selected the bacteria to conduct the challenge experiment in order to characterize the immune responses of the TLR genes of Q. spinosa against bacterial infection.
Results: The analysis identified 17 members of the TLR gene family in Q. spinosa. Phylogenetic analysis revealed that QsTLRs can be classified into seven subfamilies: TLR1, TLR3, TLR4, TLR5, TLR7, TLR11, and TLR13. Conserved synteny analysis indicated that Q. spinosa is more closely related to Rana temporaria than to Xenopus laevis. Protein structure prediction and motif analysis demonstrated that all QsTLRs are relatively conserved in both structure and function. mRNA expression levels of QsTLRs in spleen tissues were measured following stimulation with Elizabethkingia miricola, which revealing that 15 QsTLR genes exhibited up-regulation at various time points post-stimulation.
Discussion: These findings provide a comprehensive understanding of the QsTLR gene family and lay the groundwork for future studies exploring the functional evolution of the amphibian TLR gene family.
1 Introduction
Toll-like receptors (TLRs) are pattern recognition receptors that detect conserved pathogen-associated molecular patterns (PAMPs), playing a critical role in immune defense against pathogen invasion (Akira et al., 2006). As transmembrane (TM) proteins, TLRs initiate the production of immune effector molecules by recognizing conserved pathogen structures. Each member of this extensive family has the function of specifically distinguishes between pathogen classes, coordinating appropriate adaptive immune responses (Takeda and Akira, 2015). TLRs are defined as type I transmembrane receptors characterized by leucine-rich repeats (LRRs) in their extracellular domain and a Toll/interleukin-1 (IL-1) receptor domain (TIR domain) in the C-terminal (Mills et al., 2000). Upon LRR recognition of a specific pathogen ligand, the TIR domain recruits downstream signaling proteins, forming a cascade that promotes the production of pro-inflammatory cytokines and interferons (IFNs), ultimately aiding in pathogen clearance (Lim and Staudt, 2013). Through different TLRs, the body recognizes potential pathogens and activates the immune response. TLR3, TLR7, TLR8, TLR9, TLR10, TLR11, TLR12, and TLR13 are localized in endosomal membranes, where they detect nucleic acids or proteins. Specifically, TLR3 recognizes double-stranded RNA (dsRNA) and mRNA, TLR7 detects single-stranded RNA (ssRNA), immunoadjuvants, and guanosine, TLR8 responds to immunoadjuvants, ssRNA, and uridine, TLR9 identifies single-stranded unmethylated 5′-C-phosphate-G-3′ (CpG)-DNA and 5′-xCx DNA sequences, TLR10 recognizes dsRNA, TLR11/12 complex detects profilin, and TLR13 targets bacterial 23S ribosomal RNA (Alexopoulou et al., 2001; Jurk et al., 2002; Diebold et al., 2004; Hidmark et al., 2012; Raetz et al., 2013; Lee et al., 2018; Ohto et al., 2018). Previous studies have identified 13 TLR members in mammals, each functioning as sensors for distinct PAMPs. A total of 27 TLR family members have been identified in vertebrates (Zhang et al., 2017), with 10 members (TLR1-TLR10) found in humans (Homo sapiens) and 13 members (TLR1-TLR9, TLR11-TLR13) in mice (Mus musculus) (Holcombe et al., 2011). At least 21 TLRs have been identified in teleosts, including several “non-mammalian” TLRs such as TLR18-26. In amphibians, TLR family investigations have been reported for Xenopus (Xenopus laevis) and two salamander species (Lissotriton montandoni and Lissotriton vulgaris), with 19 TLR genes identified in Xenopus (Ishii et al., 2007) and 16 in salamanders (Babik et al., 2014). These species also possess non-mammalian TLRs such as TLR19, TLR21, and TLR22, although their numbers are fewer than those found in fish. Amphibians, living both in aquatic and terrestrial environments, may have evolved a unique TLR family to adapt to their complex habitats (Ishii et al., 2007). To test this hypothesis, further studies on a broader range of amphibian species are needed to investigate their TLR gene families.
The Chinese spiny frog (Quasipaa spinosa), also known as the stone frog or rock frog, belongs to the Dicroglossidae family within the order Anura. Primarily found in southern China and northern Vietnam, it inhabits rocky streams in evergreen forests and open fields at altitudes ranging from 500 m to 1,500 m above sea level (Yu et al., 2016b). Due to its significant medicinal and nutritional value, there is a growing demand for its meat, which has driven the expansion of the frog farming industry in China (Shu, 2000). However, high-density farming practices have led to outbreaks of infectious diseases, including “rotting skin” disease (Liu et al., 2024), cataract disease, ascites disease, and meningoseptica bacteremia (Lei et al., 2019), caused by bacteria, viruses, and parasites. These diseases result in substantial economic losses and hinder the development of the Q. spinosa farming industry. Despite the critical role of TLRs in innate immunity, the composition and immune functions of the TLR family in Q. spinosa have yet to be characterized. Given the importance of TLRs in pathogen recognition, it is essential to explore the TLR family in this species further.
In this study, 17 TLRs were identified from the Q. spinosa genome database, and bioinformatic analyses were performed to investigate their gene structures and phylogenetic relationships. Additionally, TLR gene expression was analyzed at various time points following pathogen challenges in spleen tissue. The results provide essential genomic data to understand the potential functions of the TLR gene family in Q. spinosa and offer preliminary insights into the evolutionary mechanisms of TLRs in amphibian innate immunity.
2 Materials and methods
2.1 Identification of members of the TLR gene family
The complete whole genome data of Q. spinosa was downloaded from the DRYAD data platform (https://datadryad.org/stash) in order to identify members of the TLR gene family of species Q. spinosa (Hu et al., 2022). In addition, homologous TLR protein sequences of H. sapiens, X. laevis, Nanorana parkeri, Rana temporaria and Danio rerio were downloaded from the National Center for Biotechnology Information (NCBI) databases. They were used as query sequences for searching against the whole genome of Q. spinosa to identify candidate TLR family members via the TBLASTN of local Blast2.2-26 (Camacho et al., 2009), with an e-value of 1 × 10−5. The sequences of candidate TLR family members of Q. spinosa (QsTLRs) were obtained and further confirmed by performing a comparison between them and the NCBI protein sequence database.
2.2 Gene structure characterisation and protein-conserved domain prediction
The Expasy ProtParam tool was used to calculate the amino acid sequences of QsTLRs (Wilkins et al., 1999). The subcellular localisation prediction was performed using the WOLF PSORTY (Horton et al., 2007). Exon-intron structure of QsTLRs were analyzed using the online gene structure visualisation server GSDS (Hu et al., 2015). Conservative motifs were evaluated using MEME suite 5.5.5 online tools (Bailey et al., 2015) and the final genetic structures were visualised using TBtools local visual software. Protein conserved domains were identified and annotated using the normal mode of the online Simple Modular Architecture Research Tool (SMART) (Schultz et al., 1998) with all parameters at default levels. The TIR domains were compared using GeneDoc multi-sequence alignment software.
2.3 Phylogenetic and syntenic analysis of the TLR gene family in Q. spinosa
Molecular phylogenetic analysis was constructed based on the predicted amino acid sequences of TLR genes in Q. spinosa and the orthologous sequences in other representative vertebrates, which included Anolis carolinensis, Chrysemys picta, D. rerio, Lateolabrax maculatus, H. sapiens, M. musculus, X. laevis and N. parkeri (the corresponding TLR sequences that were used to create phylogenetic tree can be seen in Supplementary Table S1). Multiple sequences were aligned using the MUSCLE program in MEGA 11 with default parameters (Tamura et al., 2021). The tree was constructed using the neighbour-joining (NJ) method with a bootstrapping value setting of 1,000 replications (Saitou and Nei, 1987).
2.4 Analyses of conserved synteny
The reported genomic data of representative Anurans of X. laevis (ID: GCF_017654675) and R. temporaria (ID: GCF_905171775) were downloaded from the NCBI database by MCScanX (Wang et al., 2012) for homozygosity analysis in order to further investigate the conservation of the QsTLR genes in Anurans. The TLRs were also further conserved analysis with protein sequences of 50 KP neighbouring genes upstream and downstream of them and finally visualised using IBS2.0 software (Xie et al., 2022).
2.5 Challenge experiment and sample collection
The bacteria Elizabethkingia miricola is a common pathogen found in Q. spinosa, which causes the cataract disease and serious death of the frog. Here we selected the bacteria for conducting the challenge experiment (E. miricola was isolated from diseased Q. spinosa and stored in the laboratory) to characterize the immune responses of the TLR genes of Q. spinosa against bacterial infection.
Q. spinosa (average weight of 100 ± 10 g) were cultivated in a raising farm in Qingyuan City, Guangdong Province. Prior to the experiments, the frogs were acclimatised in circulating water with a temperature of 25°C for 2°weeks and fed daily with a common diet of yellow mealworm. The frogs were then divided into treated and PBS control groups, with 60 frogs in each group (54 frogs were used as experimental and the rest were supplemental). The treated group were all injected intraperitoneally with 0.1 mL (4.0 × 106 CFU/mL) of E. miricola and frogs in the control group were injected with an equal amount of sterile PBS. To reduce the impact of random errors, improve the reliability and statistical validity of the experimental results, after infection, 5 frogs (N = 5) were chosen at random from both the control and treated groups after 0, 6, 9, 12 and 24 h and euthanised after being anaesthetized with MS-222 (Sigma, United States) before tissue dissection. Spleen, kidney, and liver tissues were sampled and immediately frozen in liquid nitrogen and stored in −80°C refrigerator for subsequent RNA extraction. All animal experiments were conducted in accordance with the guidelines for the care and use of laboratory animals that have been approved by the Institutional Animal Care and Use Committee in Zhongkai University of Agriculture and Engineering, Guangdong, China.
2.6 RNA extraction, cDNA synthesis and qPCR analysis
The total RNA was extracted from the spleen, kidney, and liver tissue using TRIzol reagent (Invitrogen, United States) and digested with RNase free DNase I (Thermo Scientific, United States) according to the instructions of the manufacturer. The quality of extracted RNA was examined by 1% agarose gel electrophoresis and NanoDrop One microvolume-uv-spectrophotometer (Thermo Fisher Scientific, United States) with an A260/280 ratio of between 1.8 and 2.1. The first strand cDNA was synthesised using HiScript II-RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China) following the protocol and the cDNA products were stored at −20°C for further experiments.
Quantitative real-time PCR (qRT-PCR) was used for detecting the mRNA level of TLR genes using a CFX Connect Real-Time PCR Detection Systems (Bio-Rad, California, United States). The gene-specific primers for qRT-PCR were designed based on each TLR gene sequence using primer 5 software (Supplementary Table S2). The total volume of the qRT-PCR reaction was 20 μL, which included 10 µL of ChamQ SYBR qPCR Master Mix (2×), 1 µL of cDNA (3 times dilution of template), 0.4 µL of each primer (10 µM) and 8.2 µL of RNase-free water. The qRT-PCR of each sample was performed in triplicate according to the following conditions: denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Melting curve analyses were performed at the end of each amplification to check the specificity of the reaction. The β-actin gene was amplified in parallel for normalisation.
2.7 Statistical analysis
All data were first subjected to Shapiro-Wilk normality test and Levene’s chi-square test. The number of threshold cycles (CT values) was collected and the 2-△△CT method was used to calculate the expression levels of each gene. Significant differences between samples were assessed using one-way ANOVA followed by Tukey’s HSD post hoc correction. Expression patterns of differentially expressed genes were visualized using the R package pheatmap (version 1.2.12) (FDR-adjusted). After Z-score normalization, clustered heat maps were constructed using Euclidean distance and Ward.D2 clustering algorithm to elucidate the underlying patterns of gene expression.
3 Results
3.1 Identification and characterisation of QsTLRs in Q. spinosa
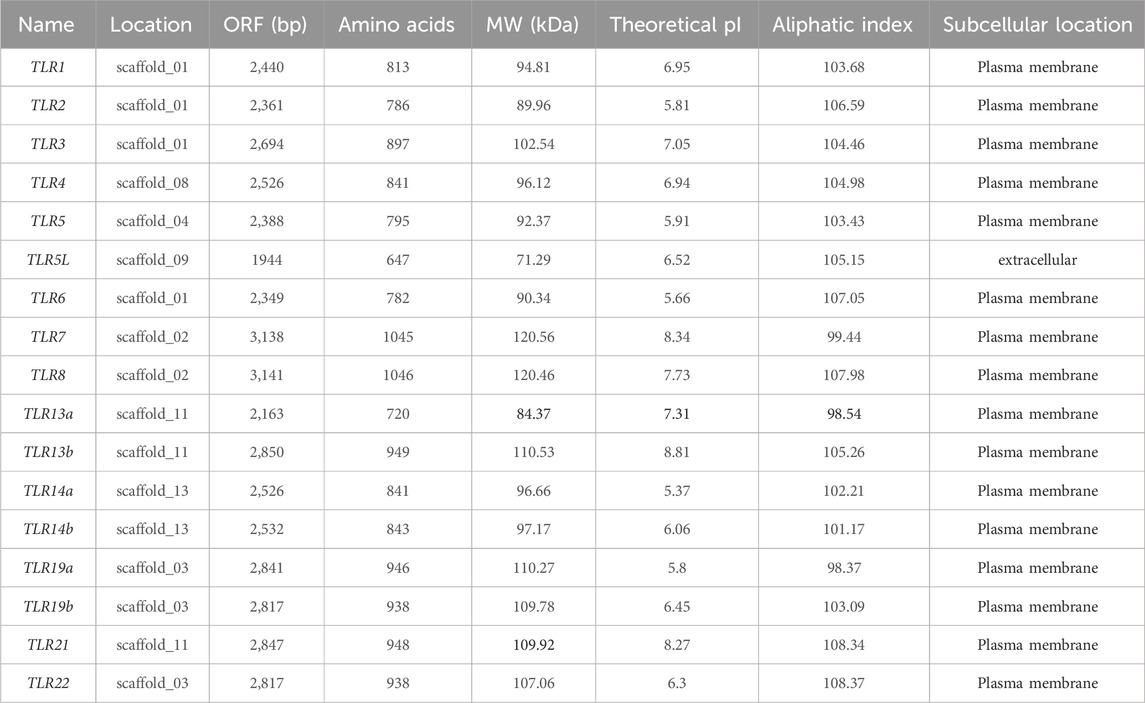
Following local identification and bioinformatic analysis, 17 different TLR genes were identified from the Q. spinosa genome database (Table 1): QsTLR1, QsTLR2, QsTLR3, QsTLR4, QsTLR5, QsTLR5L, QsTLR6, QsTLR7, QsTLR8, QsTLR13a, QsTLR13b, QsTLR14a, QsTLR14b, QsTLR19a, QsTLR19b, QsTLR21, QsTLR22. 17 QsTLR genes were located on 9 of the 13 largest scaffolds in Q. spinosa genome. The open reading frame (ORF) length of the 17 genes ranged from 1944 to 3138 bp, encoding 720 to 1046 amino acids. The physicochemical property analysis found the predicted molecular weights of QsTLRs to range from 71.29 to 120.56 kDa, and the theoretical pI values were between 5.66 and 8.81. Aliphatic analysis revealed most of the QsTLR proteins to be hydrophobic proteins, excluding QsTLR7, QsTLR13a and QsTLR19a. The predicted subcellular location suggested that most QsTLR proteins were targeted to the plasma membrane, with the exception of QsTLR5L which was extracellular proteins.
3.2 Phylogenetic relation of the TLR gene family among several vertebrates
A neighbour-joining (NJ) phylogenetic tree was constructed based on full-length amino acid sequences of QsTLRs and 18 other vertebrates to investigate the phylogenetic relationships of TLRs between Q. spinosa and other vertebrates (Figure 1). The phylogenetic tree revealed that all vertebrate TLRs were mainly clustered into seven major subfamilies and named TLR1-subfamily (TLR1/1L/2/6/14a/14b), TLR3-subfamily, TLR4-subfamily, TLR5-subfamily (5/5M/5L), TLR7-subfamily (TLR7/8/9), TLR11-subfamily (TLR12/19) and TLR13-subfamily (TLR13/21/22). The TLR1 subfamily contained the maximum number of TLR members, including TLR1, TLR1L, TLR2, TLR6, TLR14a and TLR14b. The TLR3 and TLR4 subfamilies both contained only 1 TLR member, while the TLR5 subfamily contained TLR5, TLR5M, TLR5S and TLR5L members. Q. spinosa had several representative TLR genes from major vertebrates and homologues to other vertebrate TLRs that were found to be highly conserved and clustered in the same subfamilies. However, no homologues of TLR9 and TLR12 were found in Q. spinosa genome in comparison to the TLRs of Xenopus tropicalis (Ishii et al., 2007). TLR4, which was missing in some fish such as Siniperca chuatsi (Wang et al., 2021), L. maculatus (Fan et al., 2019) and Lethenteron japonicum (Kasamatsu et al., 2010), in addition to the predicted TLR4 of X. tropicalis (Ishii et al., 2007), was identified in Q. spinosa. The results show that QsTLRs clustered in the same branch as homologues of other vertebrate TLRs. High support rates among the seven TLR subfamilies suggest that they were both evolutionarily and functionally related to each other.
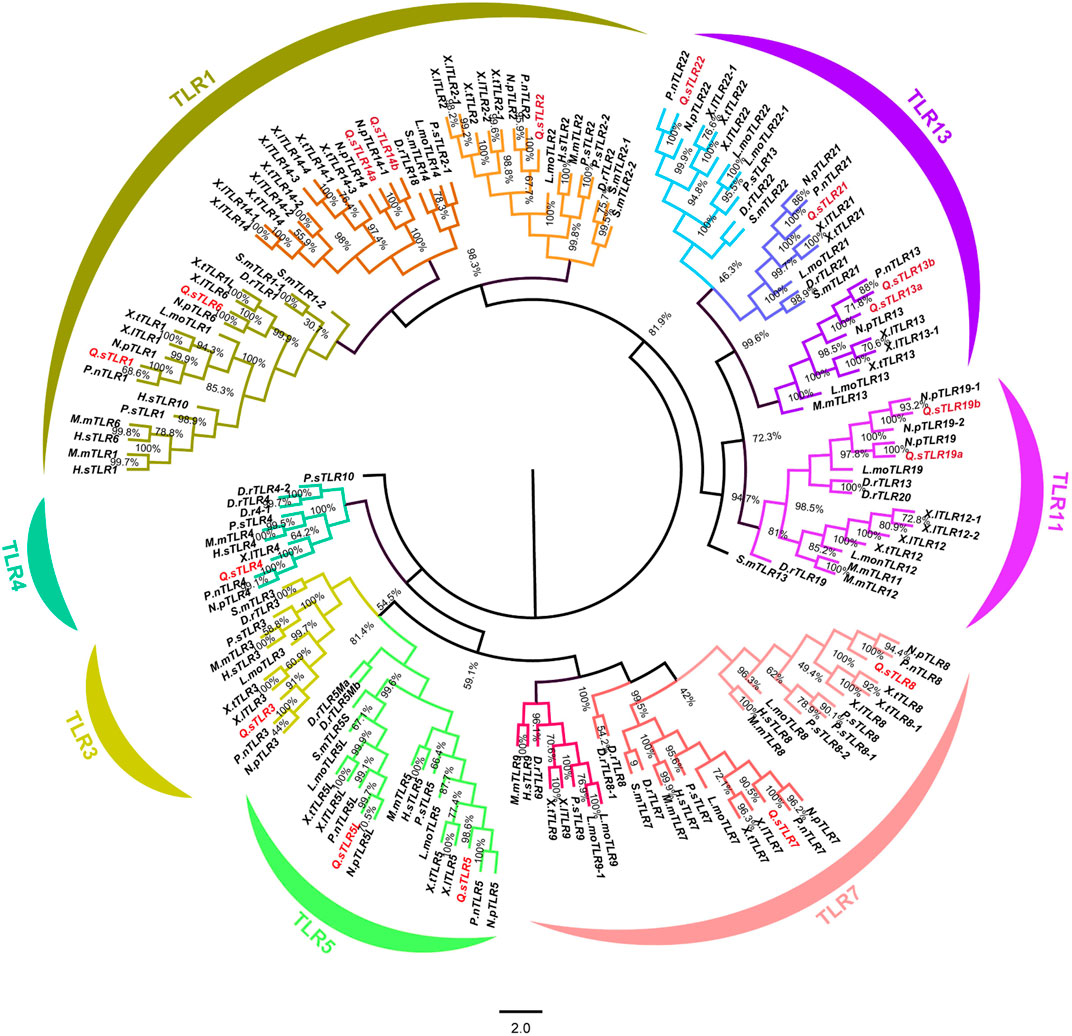
Figure 1. Phylogenetic tree of TLR gene families of 18 selected representative species. TLR genes of Q. spinosa were marked in red. The phylogenetic tree was divided into seven different subfamilies and each subfamily was marked in different colour.
3.3 Analyses of conserved synteny
Chromosomal homology analysis was conducted using Xenopus laevis and Rana temporaria as representative species (Figure 2) to investigate the homology of TLR genes in Q. spinosa within the Anuran order. The results revealed high chromosomal homology among the three species, with Q. spinosa showing chromosomal breakage and fusion events compared to X. laevis. Notably, a closer genetic relationship was observed between Q. spinosa and R. temporaria. To explore the conservation of the TLR gene family in these species, upstream and downstream genes of the TLR loci were analyzed (Figure 2; Supplementary Figure S1). The positions of TLR genes in Anurans appeared to be conserved, as the surrounding genes exhibited similar arrangements. However, some differences in the copy number and location of TLRs were observed across family members. For example, in X. laevis, TLR22 and TLR19 were located on different chromosomes, whereas in both Q. spinosa and R. temporaria, these two TLRs were located on the same chromosome, likely due to genetic variation and adaptation to different habitats.
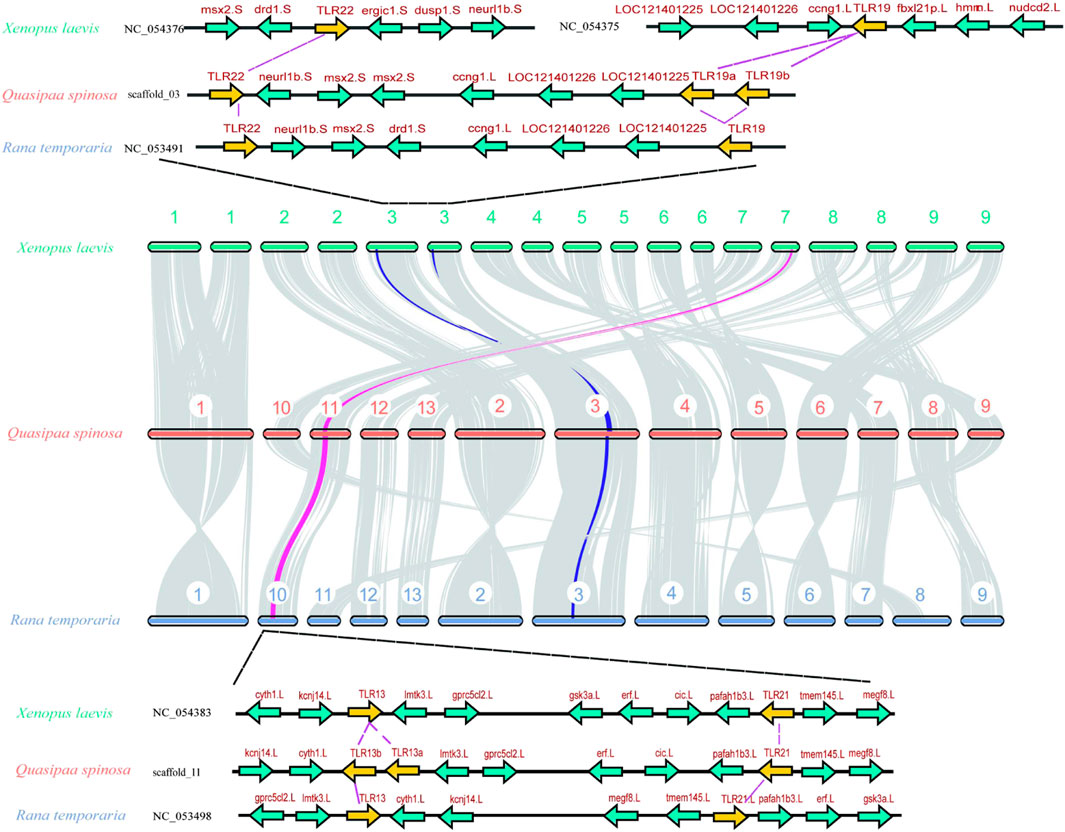
Figure 2. A Syntenic analysis of the chromosomes between Q. spinosa, R. temporaria and X. laevis. The conserved commonality manifested by some gene loci near representative TLR members.
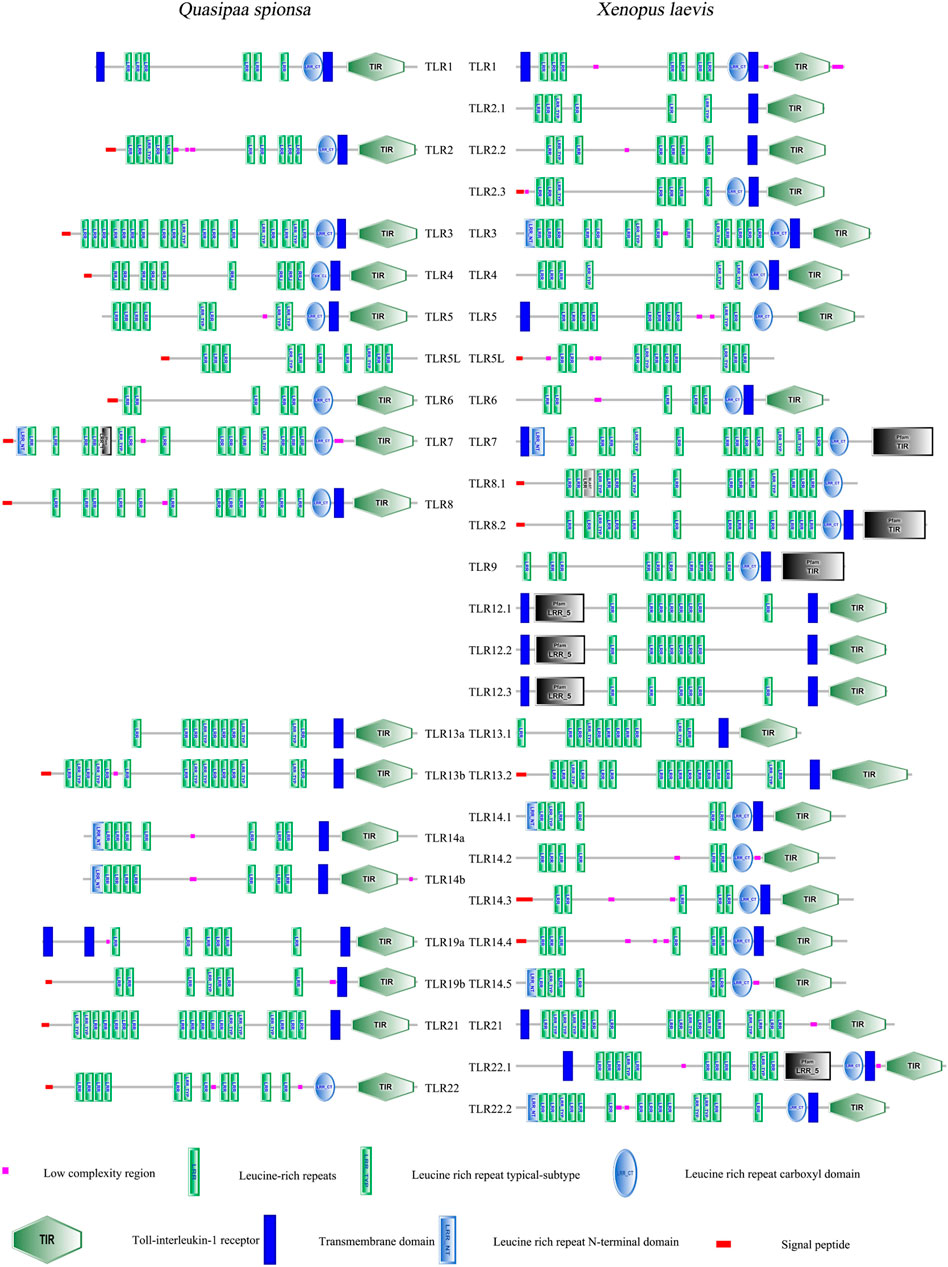
3.4 Gene structure characterisation and protein domain prediction
To further investigate the structural features of QsTLRs, protein models were predicted based on known TLR sequences from X. laevis using SMART software. The results (Figure 3) indicated that the proteins encoded by QsTLRs primarily consist of three functional domains: the LRR domain, the TM domain, and the intracellular Toll/interleukin-1 receptor (TIR) domain. All QsTLRs contained these three domains, with exceptions such as QsTLR5L, which lacked both the TM and TIR domains, and QsTLR6, QsTLR7, and QsTLR22, which lacked the TM domain.
The TLR proteins of Q. spinosa exhibited varying numbers of LRR domains, ranging from 5 to 18. This variation in LRR count may be linked to their distinct mechanisms for recognizing PAMPs. The intracellular TIR domain was identified as a crucial functional region for signal transduction (Oda and Kitano, 2006; Kawai and Akira, 2010). Comparative analysis of the TIR domains of QsTLRs (excluding QsTLR5L) revealed three highly conserved regions, named Supplementary Boxes 1–3. These conserved motifs suggest that the signal transduction mechanism is preserved across these TLRs, as highlighted in Supplementary Figure S2.
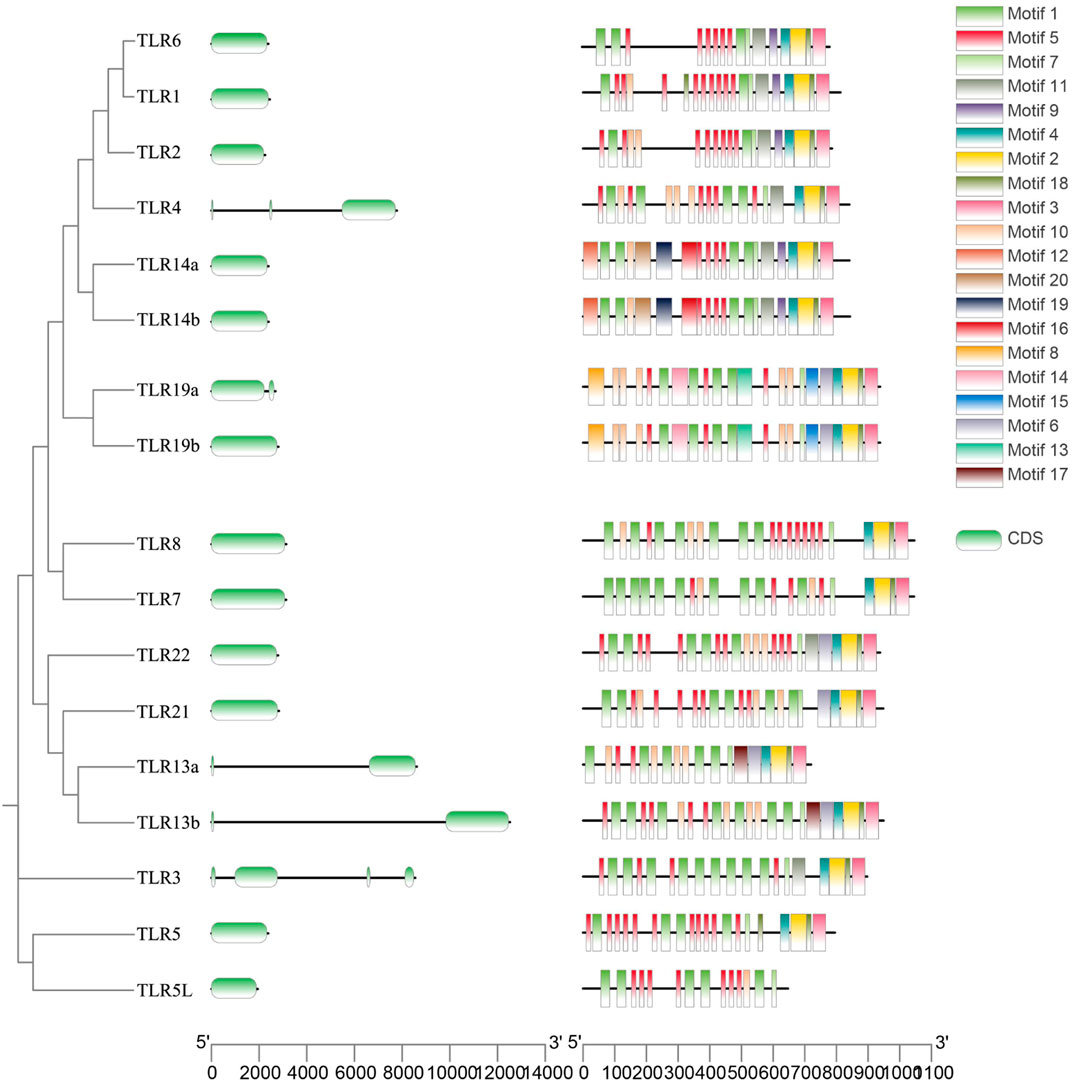
A comparative analysis of the 17 QsTLRs identified 20 conserved motifs, which were organized according to their frequency of occurrence (Figure 4). Motifs 1, 5, and 7 were widely distributed across the TLR protein sequences of Q. spinosa. The distribution of motifs within the same subfamily, such as QsTLR1/QsTLR6, QsTLR7/QsTLR8, QsTLR14a/QsTLR14b, and QsTLR19a/QsTLR19b, was highly similar, suggesting functional conservation within subfamilies. Most QsTLRs contained a single exon, while QsTLR13a, QsTLR13b, and QsTLR19a had two exons, QsTLR4 contained three exons, and QsTLR3 had four.
3.5 Expression of TLR genes at mRNA level in Q. spinosa following the stimulation of E. miricola
To assess the potential function of TLR genes in Q. spinosa in response to bacterial infection, the expression profiles of 17 TLR genes were evaluated in spleen, kidney, and liver tissues following challenge with E. miricola. The results showed a significant upregulation of 15 TLR genes in spleen tissue at varying time points. However, no consistent patterns were observed in liver and kidney tissues (Supplementary Figure S3), likely because these tissues are not the primary immune organs in Q. spinosa. Consequently, the analysis focused on spleen tissue expression (Figure 5). QsTLR1, QsTLR4, QsTLR8, QsTLR21, and QsTLR22 were significantly upregulated at 12 h (P < 0.001) and subsequently downregulated at 24 h (P < 0.001). QsTLR6 was upregulated at 6 h (P < 0.05), while QsTLR7 showed upregulation at 9 h (P < 0.001), both briefly downregulated at 12 h and then significantly upregulated again at 24 h. QsTLR3 exhibited continuous upregulation at all time points tested. For TLR genes with two subtypes, such as QsTLR5/QsTLR5L, QsTLR13a/QsTLR13b, and QsTLR14a/QsTLR14b, similar expression patterns were observed. QsTLR5 and QsTLR5L were both significantly upregulated at 12 h (P < 0.01), with QsTLR5 downregulated at 24 h (P < 0.001), while QsTLR5L remained continuously upregulated at 24 h. QsTLR13a and QsTLR13b were both significantly downregulated at 6 h (P < 0.001) and then gradually upregulated at subsequent time points. QsTLR14a and QsTLR14b were significantly upregulated at 6 h (P < 0.05), followed by a slow downregulation and then upregulation at 24 h. Expression patterns for QsTLR19a and QsTLR19b differed: QsTLR19a was downregulated after infection but significantly upregulated at 12 h, while QsTLR19b showed continuous upregulation from 6 to 24 h.
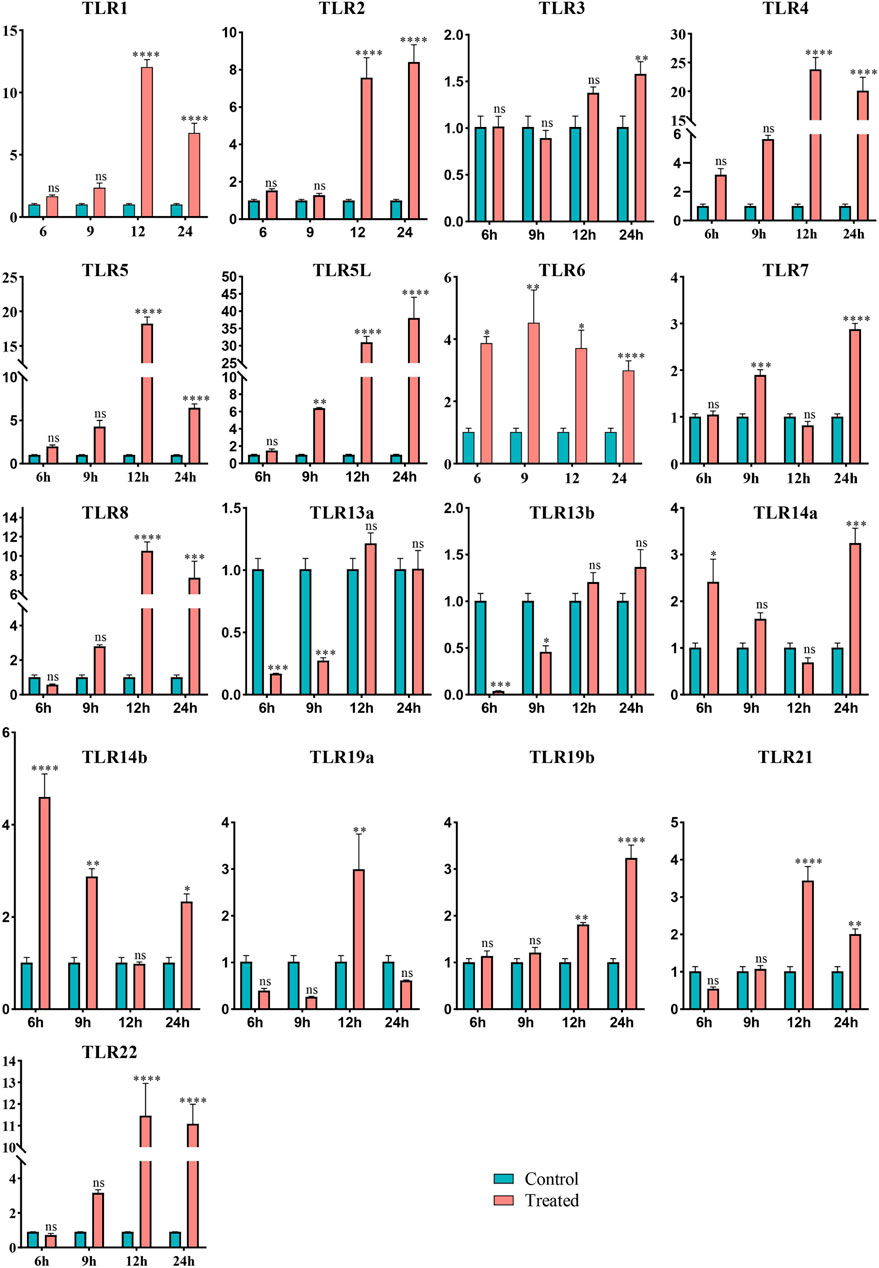
Figure 5. Expression profiles of TLR genes in Q. spinosa spleen tissue after E. miricola injection. Relative gene expression levels of QsTLRs were normalized to β-actin. Asterisks indicate statistically significant differences in upregulation/downregulation at different time points (*: P < 0.05, **: P < 0.01, ***: P < 0.001, ****: P < 0.0001).
Clustering analysis of the expression profiles categorized the QsTLR genes into four groups (Supplementary Figure S4). Type III, the largest group, included QsTLR3, QsTLR7, QsTLR13a, QsTLR13b, QsTLR14a, QsTLR14b, QsTLR19a, QsTLR19b, and QsTLR21. Type II consisted of QsTLR1, QsTLR2, QsTLR6, QsTLR8, and QsTLR22. QsTLR4 and QsTLR5 were classified as Type I, while QsTLR5L was separated as a distinct group, Type IV.
4 Discussion
TLRs have been identified in several vertebrates, including bovines (Fisher et al., 2011), birds (Velová et al., 2018), D. rerio (Chen et al., 2021), Megalobrama amblycephala (Lai et al., 2017), N. parkeri (Zhang L. et al., 2022), X. tropicalis (Ishii et al., 2007), Pelophylax nigromaculatus (Zhang L. et al., 2022) and L. montandoni (Babik et al., 2014). However, the nucleotide sequences of TLRs from Q. spinosa are currently unavailable in the NCBI database. In this study, 17 QsTLR sequences were identified from the whole genome of Q. spinosa, and their nucleotide and protein sequences were analyzed. Additionally, the expression profile of these TLRs was examined through qRT-PCR following E. miricola infection, aiming to elucidate the role of QsTLRs in pathogen resistance.
4.1 Potential reasons for variation of TLRs in amphibians
TLR4 is present across most vertebrates, from fish to mammals (Velová et al., 2018). In fish, TLR4 has been observed in Cypriniformes species with varying numbers of subtypes, such as three subtypes (TLR4ba/bb/al) in D. rerio (Chen et al., 2021), three subtypes (TLR4-1/2/3) in Cyprinus carpio(Gong et al., 2017) and one subtype in M. amblycephala (Lai et al., 2017), However, TLR4 is absent in many other bony fish, such as Perciformes (Martínez-López et al., 2023). In reptiles and mammals, a single copy of TLR4 is present (Zhou et al., 2016). In amphibians, TLR4 has been detected in the genomes of X. laevis and X. tropicalis (Fisher et al., 2023), as well as in Bombina maxima transcriptomic data (Zhao et al., 2014). Similarly, one TLR4 was identified in the Q. spinosa genome in this study. In contrast, no TLR4 was found in the Caudata L. montandoni genome, suggesting that TLR4 may be exclusive to Anuran amphibians (Babik et al., 2014). TLR5 is present in various vertebrates and replicates across multiple species. Two types of TLR5 are found in bony fish: soluble TLR5S and membrane-embedded TLR5M (Oshiumi et al., 2003). In mammals, including humans and mice, only the membrane-embedded TLR5M has been identified. However, amphibians (Babik et al., 2014) and some reptiles (such as turtles and the anole lizard (Abdullayev et al., 2013)), possess a variant known as TLR5L, which shares a similar protein structure to TLR5S. In this study, TLR5L was also found in Q. spinosa. Phylogenetic analysis showed that TLR5L clustered with TLR5S as a sister group, suggesting that TLR5S and TLR5L likely resulted from a duplication event of TLR5 following the divergence of fish and amphibians. It is speculated that TLR5S and TLR5L may be functionally homologous. Furthermore, TLR5 appears to be expanded in aquatic animals, such as fish and amphibians, compared to terrestrial animals, indicating its significant role in mediating resistance to the complex aquatic environments that these organisms inhabit.
The TLR11 subfamily is both unique and complex, comprising four TLR genes: TLR11, TLR12, TLR19, and TLR20. In vertebrates, the existence of this subfamily spans from fish to mammals, although not all members are present in every group (Zhang L. et al., 2022). For instance, reptiles such as the Chinese soft-shelled turtle (Liu et al., 2019) and anole lizard lack members of the TLR11 subfamily (Abdullayev et al., 2013). In other vertebrates, TLR11 has only been identified in mammals, such as M. musculus. TLR12 has been found in both mammals and amphibians, while TLR20 exists only in fish, with multiple copies identified in species like D. rerio (six copies of TLR20) (Pietretti et al., 2014; Lv et al., 2023). TLR19 has been observed in both fish and amphibians but is absent in amniotes (Wang et al., 2015; Gong et al., 2017; Zhang L. et al., 2022). In Q. spinosa, TLR11 and TLR12 were not identified, but two copies of TLR19 (a and b) were found within the TLR11 subfamily. Phylogenetic analysis showed that the two TLR19 genes clustered with TLR19 genes from other species, and TLR19 was closely related to TLR12, suggesting that these genes may share homologous roles in host defense against pathogenic microorganisms.
Amphibians, as ancient vertebrates, occupy a critical evolutionary position, acting as a “bridge” between aquatic and terrestrial vertebrates (Inger et al., 1986). Compared to other animals, amphibians exhibit increased copy numbers of certain TLR genes, and some TLR genes are unique to fish and amphibians, being absent in other terrestrial vertebrates. For instance, TLR13 appears in multiple copies in both fish and amphibians (Ishii et al., 2007; Liu et al., 2019; Wang et al., 2021; Zhang L. et al., 2022), while only a single copy is retained in reptiles and mammals (Song et al., 2015; Yu et al., 2016a). Similarly, TLR14 exists in one or two copies in amphibians (Zhang L. et al., 2022), but only one copy is found in most reptiles. Interestingly, TLR14 genes are absent in fish, birds, and mammals. TLR19, previously thought to be unique to bony fish with one copy, has been shown to also exist and expand in amphibians (Wang et al., 2015; Altmann et al., 2016; Qi et al., 2017; Liu et al., 2019). Multiple copies of TLR22 are found in fish, with species such as Gymnocypris eckloni having two copies (TLR22a and TLR22b) (Qi et al., 2017), Boleophthalmus pectinrostris having four copies (TLR22a to d) (Qiu et al., 2019) and Coregonus maraena possessing as many as ten copies (TLR22a to j) (Altmann et al., 2016). In amphibians, one or two copies of TLR22 remain, but it is completely absent in birds and mammals (Babik et al., 2014). The expansion and diversity of TLRs in cephalochordates and echinoderms are believed to reflect the evolutionary response to a variety of microorganisms and pathogens in aquatic environments (Rast et al., 2006; Huang et al., 2008; Messier-Solek et al., 2010). Similarly, the diversity of TLRs in amphibians plays a pivotal role in their ability to resist pathogenic bacteria during life domain migration.
4.2 Characterization of TLRs in Q. spinosa
Similar to other vertebrates, the proteins encoded by QsTLRs possess three typical functional regions: the extracellular region, the TM region, and the C-terminal intracellular region (TIR) (Figure 3). The extracellular region consists of 2–25 LRR domains, each containing 20 to 30 amino acids. These LRRs are essential for recognizing PAMPs in pathogenic organisms, such as bacteria, parasites, and fungi (Uematsu and Akira, 2006). The variation in the number of LRRs among species and TLR family members appears to be an evolutionary adaptation, enabling hosts to better recognize and respond to diverse PAMPs. Consequently, there is considerable variability in the number of LRRs across different TLRs. The TIR domain, which plays a vital role in TLR signaling, is highly conserved in Q. spinosa (O'Neill and Bowie, 2007). The functional conservation of the TIR domain across TLR members is mainly concentrated in three critical motifs (Supplementary Figure S2): Supplementary Box 1 ((F/Y) DAFISY), Supplementary Box 2 (LC---RD---PG), and Supplementary Box 3 (a conserved (FW) surrounded by basic residues). Notably, phenylalanine (F) in Supplementary Box 1 can be substituted with tyrosine (Y). This study found that proteins encoded by QsTLRs retain the important motifs of the TIR structural domain. Interestingly, proline (P) in Supplementary Box 2—an essential residue with auxiliary recognition functions—was found to be conserved in various fish species, such as yellowtail leucocytes (Reyes-Becerril et al., 2016), Sebastiscus marmoratus (Zhang Y. et al., 2022), Larimichthys crocea (Sun et al., 2016) and Seriola lalandi (Reyes-Becerril et al., 2016). This suggests that variations in conserved residue sites between amphibians and fish are not coincidental.
4.3 Immune response of QsTLRs against E. miricola infection
Previous research has demonstrated the functional similarity of factors involved in resistance to viral or non-viral exogenous attacks and in signaling cascade transduction, despite being derived from different species (Liu et al., 2020). This functional similarity is particularly evident among fish, reptiles, and mammals. However, the role of the TLR family in amphibians remains unexplored, and the functional comparison of TLR genes between amphibians and other vertebrates is not yet understood.
E. miricola is a common pathogen in Q. spinosa as well as other frogs, including P. nigromaculatus (Hu et al., 2017; Li et al., 2023), Lithobates pipiens (Trimpert et al., 2021), and Rana catesbeiana (Wei et al., 2023). Infected Q. spinosa exhibits typical symptoms such as cataracts with white, cloudy eyes, and reduced movement (Lei et al., 2019). The spleen, kidney, and liver often become enlarged or hemorrhagic (Li et al., 2023; Wei et al., 2023). This disease outbreak leads to high mortality rates in frog farming. Therefore, investigating the roles of QsTLRs in the immune response following E. miricola infection could provide valuable insights into enhancing innate immunity and developing immune adjuvants. In this study, E. miricola was selected as the pathogen for challenge experiments, and the expression levels of 17 TLR genes were assessed in the spleen, kidney, and liver tissue, to explore the potential anti-pathogenic immune responses of TLRs in Q. spinosa.
The results revealed significant temporal variation in the expression of most genes, suggesting their involvement in the immune response to E. miricola infection. TLR2 and TLR4 in fish and reptiles have been shown to recognize and respond to Gram-negative bacterial invasions, such as Aeromonas hydrophila, with activation occurring in the spleen (Zhang et al., 2013; Lai et al., 2017; Samanta et al., 2017; Liu et al., 2019). In this study, the expressions of QsTLR2 and QsTLR4 followed an upregulation trend, peaking at 12 h post-infection, similar to the expression patterns observed in Chinese soft-shelled turtle after A. hydrophila infection (Liu et al., 2019), indicating their active participation in the immune response to E. miricola in Q. spinosa. TLR5 in fish and reptiles, such as Anolis carolinensis, is known to recognize bacterial flagellin and LPS, initiating immune responses in the spleen (Voogdt et al., 2016; Wang et al., 2021; Zhang Y. et al., 2022). In Q. spinosa, both QsTLR5 and QsTLR5L exhibited a significant and sustained upregulation until 12 h post-infection, indicating that TLR5 functions similarly across vertebrates in immune responses triggered by bacterial pathogens. However, while QsTLR5 expression decreased at 24 h, QsTLR5L remained upregulated, suggesting that the two TLR5 genes in Q. spinosa play critical roles in the immune response to E. miricola infection, with QsTLR5L contributing functional diversity and complexity.
Genes undergoing whole-genome duplication (WGD) or local replication events typically avoid gene loss through subfunctionalization or neofunctionalization (Jaillon et al., 2004). Fish TLR13 is upregulated in response to poly (I:C), LPS, and PNG stimulation (Wang et al., 2021), with a similar response detected in the spleen of Pelodiscus sinensis (Liu et al., 2019). In mammals, such as mice, TLR13 is activated by bacterial 23S rRNA and viral ssRNA, suggesting that TLR13 in mammals, reptiles, and fish may share functional similarities (Song et al., 2015). However, antibiotic-resistant bacterial 23S rRNA and synthetic oligonucleotides containing methylated adenosine or guanosine have been shown to inhibit proper activation of TLR13 (Oldenburg et al., 2012). In this study, QsTLR13a and QsTLR13b were significantly downregulated following infection, returning to baseline levels at 12 h, a response distinct from that observed in fish or mammals. This may indicate that E. miricola employs immune evasion strategies that prevent normal immune recognition by TLR13. In fish, TLR19 is known to recognize dsRNAs (e.g., poly (I:C)) (Zhang Y. et al., 2022) and bacteria stimuli (Zhang et al., 2013). In this study, QsTLR19b exhibited a continuous upregulation at 24 h after infection, similar to the expression patterns seen in Ictalurus punctatus (Zhang et al., 2013). However, QsTLR19a displayed a distinct expression pattern, with upregulation only observed at 12 h, suggesting that TLR19a may function in a temporally regulated manner or be subject to more complex regulatory mechanisms. Although subtypes resulting from gene replication display some functional divergence, these differences may reflect evolutionary adaptations in response to the challenges posed by complex habitats.
Gene clustering analysis was performed to investigate the expression patterns of different subfamily members. The results showed that QsTLR5 and QsTLR4 exhibited highly similar expression profiles, suggesting they may work synergistically to recognize different receptor molecules. This complementary function could enable the detection of multiple signaling pathways (like downstream signaling), thereby preventing immune escape. QsTLR5L also responded to E. miricola infection and triggered an immune response; however, QsTLR5L did not cluster with QsTLR5 but instead formed a distinct branch with consistently upregulated expression. This indicates that QsTLR5L functions independently of QsTLR5 and may represent a fish-like variant, possibly an evolutionarily adapted TLR5S in frogs, tailored to meet the immunological demands of their complex aquatic and terrestrial environments. Further studies on TLR5S in fish and TLR5L in amphibians are needed to confirm this hypothesis.
In terms of functional analysis, the expression of QsTLR genes following E. miricola stimulation was initially examined. However, due to the absence of viral pathogens in the study, immune responses in QsTLRs to viral stimulation could not be assessed. Additionally, the expression profiles of QsTLRs were only analyzed in spleen tissues post-infection, which constitutes a limitation of this study. Although the expression of QsTLRs in liver and kidney tissues was also analyzed, the results were inconsistent, lacking any clear trends. This suggests that the kidney and liver may not be primary immune tissues in frogs, failing to generate immune responses after infection. A similar observation was made in P. sinensis, where immune responses to A. hydrophila were only detected in the spleen, not in the liver or kidney (Zhou et al., 2016; Liu et al., 2019). Therefore, only the spleen expression results are presented here. Despite the lack of further functional characterization, this study provides the first expression profile of Q. spinosa following E. miricola infection and identifies key TLR members (TLR4, TLR5, and TLR5L) likely involved in the immune response. This lays the groundwork for future studies exploring the role of TLRs in pathogen recognition in other frog species and investigates the complementary roles of different TLR members in downstream signaling pathways.
In conclusion, 17 QsTLRs were identified in Q. spinosa, offering a preliminary understanding of the TLR gene family in this species. The expression changes of these genes in response to E. miricola infection were analyzed, providing a basis for further research into the role of Q. spinosa in defending against exogenous pathogens.
5 Conclusion
In this study, 17 members of the TLR gene family were identified from the whole genome sequences of Q. spinosa. Phylogenetic analysis showed that the QsTLRs were highly homologous with their homologs in other vertebrates. Analysis of protein structural domains and motifs showed that the QsTLRs proteins were highly structurally conserved. qRT-PCR results showed that 17 QsTLRs responded positively to the attack by E. miricola, with different regulatory tendencies. 15 QsTLR genes displayed upregulation trends at different time intervals. Overall, these results provided a comprehensive understanding of the QsTLRs and provided a theoretical basis for further investigation on the immunological function as well as vaccine development in Chinese spiny frog.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by laboratory animals that have been approved by the Institutional Animal Care and Use Committee in Zhongkai University of Agriculture and Engineering, Guangdong, China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RL: Data curation, Funding acquisition, Methodology, Visualization, Writing – review and editing. ZL: Investigation, Methodology, Validation, Visualization, Writing – original draft. ZG: Data curation, Investigation, Methodology, Writing – original draft. DM: Data curation, Software, Visualization, Writing – original draft. ZZ: Software, Visualization, Writing – original draft. JZ: Data curation, Software, Visualization, Writing – original draft. ML: Funding acquisition, Project administration, Writing – review and editing. HX: Methodology, Project administration, Supervision, Writing – review and editing. MZ: Methodology, Writing – review and editing. TG: Data curation, Methodology, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The study was supported by the Science and Technology Plan Project of Qingyuan City (NO. 2022KJJH064, NO. 2023KJJ005) and the Guangdong Provincial Department of Science and Technology-Rural Science and Technology Commissioner Project (NO. KTP20210367). We are grateful to Innnovative Institute of Animal Healthy Breeding, Zhongkai University of Agriculture and Engineering for providing the facilities in the laboratory that make this study happen.
Conflict of interest
Authors ML, HX were employed by Yingde Yingxin Agriculture Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1569669/full#supplementary-material
References
Abdullayev, I., Kirkham, M., Björklund Å, K., Simon, A., and Sandberg, R. (2013). A reference transcriptome and inferred proteome for the salamander Notophthalmus viridescens. Exp. Cell Res. 319 (8), 1187–1197. doi:10.1016/j.yexcr.2013.02.013
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124 (4), 783–801. doi:10.1016/j.cell.2006.02.015
Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001). Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413 (6857), 732–738. doi:10.1038/35099560
Altmann, S., Korytář, T., Kaczmarzyk, D., Nipkow, M., Kühn, C., Goldammer, T., et al. (2016). Toll-like receptors in maraena whitefish: evolutionary relationship among salmonid fishes and patterns of response to Aeromonas salmonicida. Fish andand Shellfish Immunol. 54, 391–401. doi:10.1016/j.fsi.2016.04.125
Babik, W., Dudek, K., Fijarczyk, A., Pabijan, M., Stuglik, M., Szkotak, R., et al. (2014). Constraint and adaptation in newt toll-like receptor genes. Genome Biol. Evol. 7 (1), 81–95. doi:10.1093/gbe/evu266
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43 (W1), W39–W49. doi:10.1093/nar/gkv416
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinforma. 10, 421. doi:10.1186/1471-2105-10-421
Chen, H., Liang, Y., Han, Y., Liu, T., and Chen, S. (2021). Genome-wide analysis of Toll-like receptors in zebrafish and the effect of rearing temperature on the receptors in response to stimulated pathogen infection. J. Fish. Dis. 44 (3), 337–349. doi:10.1111/jfd.13287
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S., and Reis e Sousa, C. (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303 (5663), 1529–1531. doi:10.1126/science.1093616
Fan, H., Wang, L., Wen, H., Wang, K., Qi, X., Li, J., et al. (2019). Genome-wide identification and characterization of toll-like receptor genes in spotted sea bass (Lateolabrax maculatus) and their involvement in the host immune response to Vibrio harveyi infection. Fish and Shellfish Immunol. 92, 782–791. doi:10.1016/j.fsi.2019.07.010
Fisher, C. A., Bhattarai, E. K., Osterstock, J. B., Dowd, S. E., Seabury, P. M., Vikram, M., et al. (2011). Evolution of the bovine TLR gene family and member associations with Mycobacterium avium subspecies paratuberculosis infection. PLOS ONE 6 (11), e27744. doi:10.1371/journal.pone.0027744
Fisher, M., James-Zorn, C., Ponferrada, V., Bell, A. J., Sundararaj, N., Segerdell, E., et al. (2023). Xenbase: key features and resources of the Xenopus model organism knowledgebase. Genetics 224 (1), iyad018. doi:10.1093/genetics/iyad018
Gong, Y., Feng, S., Li, S., Zhang, Y., Zhao, Z., Hu, M., et al. (2017). Genome-wide characterization of Toll-like receptor gene family in common carp (Cyprinus carpio) and their involvement in host immune response to Aeromonas hydrophila infection. Comp. Biochem. Physiol. Part D. Genomics Proteomics 24, 89–98. doi:10.1016/j.cbd.2017.08.003
Hidmark, A., von Saint Paul, A., and Dalpke, A. H. (2012). Cutting edge: TLR13 is a receptor for bacterial RNA. J. Immunol. 189 (6), 2717–2721. doi:10.4049/jimmunol.1200898
Holcombe, A. O., Linares, D., and Vaziri-Pashkam, M. (2011). Perceiving spatial relations via attentional tracking and shifting. Curr. Biol. 21 (13), 1135–1139. doi:10.1016/j.cub.2011.05.031
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 (Web Server issue), W585–W587. doi:10.1093/nar/gkm259
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi:10.1093/bioinformatics/btu817
Hu, R., Yuan, J., Meng, Y., Wang, Z., and Gu, Z. (2017). Pathogenic Elizabethkingia miricola infection in cultured black-spotted frogs, China, 2016. Emerg. Infect. Dis. 23 (12), 2055–2059. doi:10.3201/eid2312.170942
Hu, X., Jiang, Z., Ming, Y., Jian, J., Jiang, S., Zhang, D., et al. (2022). A chromosomal level genome sequence for Quasipaa spinosa (Dicroglossidae) reveals chromosomal evolution and population diversity. Mol. Ecol. Resour. 22 (4), 1545–1558. doi:10.1111/1755-0998.13560
Huang, S., Yuan, S., Guo, L., Yu, Y., Li, J., Wu, T., et al. (2008). Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 18 (7), 1112–1126. doi:10.1101/gr.069674.107
Inger, R. F., Szarski, H., Kollros, J. J., Duellman, W. E., and Trueb, L. (1986). Biology of Amphibians. Copeia 1986 (2), 549–553. doi:10.2307/1445022
Ishii, A., Kawasaki, M., Matsumoto, M., Tochinai, S., and Seya, T. (2007). Phylogenetic and expression analysis of amphibian Xenopus Toll-like receptors. Immunogenetics 59 (4), 281–293. doi:10.1007/s00251-007-0193-y
Jaillon, O., Aury, J. M., Brunet, F., Petit, J. L., Stange-Thomann, N., Mauceli, E., et al. (2004). Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431 (7011), 946–957. doi:10.1038/nature03025
Jurk, M., Heil, F., Vollmer, J., Schetter, C., Krieg, A. M., Wagner, H., et al. (2002). Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol. 3 (6), 499. doi:10.1038/ni0602-499
Kasamatsu, J., Oshiumi, H., Matsumoto, M., Kasahara, M., and Seya, T. (2010). Phylogenetic and expression analysis of lamprey toll-like receptors. Dev. and Comp. Immunol. 34 (8), 855–865. doi:10.1016/j.dci.2010.03.004
Kawai, T., and Akira, S. (2010). The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11 (5), 373–384. doi:10.1038/ni.1863
Lai, R. F., Jakovlić, I., Liu, H., Zhan, F. B., Wei, J., and Wang, W. M. (2017). Molecular characterization and immunological response analysis of toll-like receptors from the blunt snout bream (Megalobrama amblycephala). Dev. Comp. Immunol. 67, 471–475. doi:10.1016/j.dci.2016.09.005
Lee, S. M., Yip, T. F., Yan, S., Jin, D. Y., Wei, H. L., Guo, R. T., et al. (2018). Recognition of double-stranded RNA and regulation of interferon pathway by toll-like receptor 10. Front. Immunol. 9, 516. doi:10.3389/fimmu.2018.00516
Lei, X. P., Yi, G., Wang, K. Y., OuYang, P., Chen, F., Huang, X. L., et al. (2019). Elizabethkingia miricola infection in Chinese spiny frog (Quasipaa spinosa). Transbound. Emerg. Dis. 66 (2), 1049–1053. doi:10.1111/tbed.13101
Li, S., Wang, X., Lu, Y., Wang, J., Yu, D., Zhou, Z., et al. (2023). Co-infections of Klebsiella pneumoniae and Elizabethkingia miricola in black-spotted frogs (Pelophylax nigromaculatus). Microb. Pathog. 180, 106150. doi:10.1016/j.micpath.2023.106150
Lim, K. H., and Staudt, L. M. (2013). Toll-like receptor signaling. Cold Spring Harb. Perspect. Biol. 5 (1), a011247. doi:10.1101/cshperspect.a011247
Liu, G., Zhang, H., Zhao, C., and Zhang, H. (2020). Evolutionary history of the toll-like receptor gene family across vertebrates. Genome Biol. Evol. 12 (1), 3615–3634. doi:10.1093/gbe/evz266
Liu, T., Han, Y., Chen, S., and Zhao, H. (2019). Genome-wide identification of Toll-like receptors in the Chinese soft-shelled turtle Pelodiscus sinensis and expression analysis responding to Aeromonas hydrophila infection. Fish. Shellfish Immunol. 87, 478–489. doi:10.1016/j.fsi.2019.01.052
Liu, W., Tao, Y.-H., Lu, C.-P., Zhang, L., Chen, J., and Lin, Z.-H. (2024). Transcriptomic analysis of skin immunity genes in the Chinese spiny frog (Quasipaa spinosa) after Proteus mirabilis infection. Comp. Biochem. Physiology Part D Genomics Proteomics 49, 101172. doi:10.1016/j.cbd.2023.101172
Lv, M., Zhang, J., Wang, W., Jiang, R., and Su, J. (2023). Re-identification and characterization of grass carp Ctenopharyngodon idella TLR20. Fish. Shellfish Immunol. Rep. 5, 100119. doi:10.1016/j.fsirep.2023.100119
Martínez-López, A., Tyrkalska, S. D., Alcaraz-Pérez, F., Cabas, I., Candel, S., Martínez Morcillo, F. J., et al. (2023). Evolution of LPS recognition and signaling: the bony fish perspective. Dev. Comp. Immunol. 145, 104710. doi:10.1016/j.dci.2023.104710
Messier-Solek, C., Buckley, K. M., and Rast, J. P. (2010). Highly diversified innate receptor systems and new forms of animal immunity. Semin. Immunol. 22 (1), 39–47. doi:10.1016/j.smim.2009.11.007
Mills, C. D., Kincaid, K., Alt, J. M., Heilman, M. J., and Hill, A. M. (2000). M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 164 (12), 6166–6173. doi:10.4049/jimmunol.164.12.6166
Oda, K., and Kitano, H. (2006). A comprehensive map of the toll-like receptor signaling network. Mol. Syst. Biol. 2 (1), 0015. doi:10.1038/msb4100057
Ohto, U., Ishida, H., Shibata, T., Sato, R., Miyake, K., and Shimizu, T. (2018). Toll-like receptor 9 contains two DNA binding sites that function cooperatively to promote receptor dimerization and activation. Immunity 48 (4), 649–658. doi:10.1016/j.immuni.2018.03.013
Oldenburg, M., Krüger, A., Ferstl, R., Kaufmann, A., Nees, G., Sigmund, A., et al. (2012). TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337 (6098), 1111–1115. doi:10.1126/science.1220363
O'Neill, L. A., and Bowie, A. G. (2007). The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7 (5), 353–364. doi:10.1038/nri2079
Oshiumi, H., Tsujita, T., Shida, K., Matsumoto, M., Ikeo, K., and Seya, T. (2003). Prediction of the prototype of the human Toll-like receptor gene family from the pufferfish, Fugu rubripes, genome. Immunogenetics 54 (11), 791–800. doi:10.1007/s00251-002-0519-8
Pietretti, D., Scheer, M., Fink, I. R., Taverne, N., Savelkoul, H. F., Spaink, H. P., et al. (2014). Identification and functional characterization of nonmammalian Toll-like receptor 20. Immunogenetics 66 (2), 123–141. doi:10.1007/s00251-013-0751-4
Qi, D., Xia, M., Chao, Y., Zhao, Y., and Wu, R. (2017). Identification, molecular evolution of toll-like receptors in a Tibetan schizothoracine fish (Gymnocypris eckloni) and their expression profiles in response to acute hypoxia. Fish. Shellfish Immunol. 68, 102–113. doi:10.1016/j.fsi.2017.07.014
Qiu, H. T., Fernandes, J. M. O., Hong, W. S., Wu, H. X., Zhang, Y. T., Huang, S., et al. (2019). Paralogues from the expanded Tlr11 gene family in mudskipper (Boleophthalmus pectinirostris) are under positive selection and respond differently to LPS/poly(I:C) challenge. Front. Immunol. 10, 343. doi:10.3389/fimmu.2019.00343
Raetz, M., Kibardin, A., Sturge, C. R., Pifer, R., Li, H., Burstein, E., et al. (2013). Cooperation of TLR12 and TLR11 in the IRF8-dependent IL-12 response to Toxoplasma gondii profilin. J. Immunol. 191 (9), 4818–4827. doi:10.4049/jimmunol.1301301
Rast, J. P., Smith, L. C., Loza-Coll, M., Hibino, T., and Litman, G. W. (2006). Genomic insights into the immune system of the sea urchin. Science 314 (5801), 952–956. doi:10.1126/science.1134301
Reyes-Becerril, M., Ascencio-Valle, F., Hirono, I., Kondo, H., Jirapongpairoj, W., Esteban, M. A., et al. (2016). TLR21's agonists in combination with Aeromonas antigens synergistically up-regulate functional TLR21 and cytokine gene expression in yellowtail leucocytes. Dev. Comp. Immunol. 61, 107–115. doi:10.1016/j.dci.2016.03.012
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 (4), 406–425. doi:10.1093/oxfordjournals.molbev.a040454
Samanta, M., Basu, M., Swain, B., Paichha, M., Lenka, S. S., Das, S., et al. (2017). Molecular cloning and characterization of LrTLR4, analysis of its inductive expression and associated down-stream signaling molecules following lipopolysaccharide stimulation and Gram-negative bacterial infection. Fish and Shellfish Immunol. 60, 164–176. doi:10.1016/j.fsi.2016.11.028
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95 (11), 5857–5864. doi:10.1073/pnas.95.11.5857
Shu, M. (2000). An analysis of the nutritive compositions in muscle of Rana spinosa II. Compositions of amino acids and mineral elements. J. Zhejiang Univ. 27 (5), 553–559. doi:10.3785/j.issn.1008-9497.2000.05.0553
Song, W., Wang, J., Han, Z., Zhang, Y., Zhang, H., Wang, W., et al. (2015). Structural basis for specific recognition of single-stranded RNA by Toll-like receptor 13. Nat. Struct. Mol. Biol. 22 (10), 782–787. doi:10.1038/nsmb.3080
Sun, M., Mu, Y., Ding, Y., Ao, J., and Chen, X. (2016). Molecular and functional characterization of Toll-like receptor 21 in large yellow croaker (Larimichthys crocea). Fish. Shellfish Immunol. 59, 179–188. doi:10.1016/j.fsi.2016.10.024
Takeda, K., and Akira, S. (2015). Toll-like receptors. Curr. Protoc. Immunol. 109, 14.12.1–14.12.10. doi:10.1002/0471142735.im1412s109
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi:10.1093/molbev/msab120
Trimpert, J., Eichhorn, I., Vladimirova, D., Haake, A., Schink, A. K., Klopfleisch, R., et al. (2021). Elizabethkingia miricola infection in multiple anuran species. Transbound. Emerg. Dis. 68 (2), 931–940. doi:10.1111/tbed.13761
Uematsu, S., and Akira, S. (2006). Toll-like receptors and innate immunity. J. Mol. Med. 84 (9), 712–725. doi:10.1007/s00109-006-0084-y
Velová, H., Gutowska-Ding, M. W., Burt, D. W., and Vinkler, M. (2018). Toll-like receptor evolution in birds: gene duplication, pseudogenization, and diversifying selection. Mol. Biol. Evol. 35 (9), 2170–2184. doi:10.1093/molbev/msy119
Voogdt, C. G., Bouwman, L. I., Kik, M. J., Wagenaar, J. A., and van Putten, J. P. (2016). Reptile Toll-like receptor 5 unveils adaptive evolution of bacterial flagellin recognition. Sci. Rep. 6, 19046. doi:10.1038/srep19046
Wang, J., Zhang, Z., Fu, H., Zhang, S., Liu, J., Chang, F., et al. (2015). Structural and evolutionary characteristics of fish-specific TLR19. Fish. Shellfish Immunol. 47 (1), 271–279. doi:10.1016/j.fsi.2015.09.005
Wang, K. L., Chen, S. N., Huo, H. J., and Nie, P. (2021). Identification and expression analysis of sixteen Toll-like receptor genes, TLR1, TLR2a, TLR2b, TLR3, TLR5M, TLR5S, TLR7−9, TLR13a−c, TLR14, TLR21−23 in Mandarin fish Siniperca chuatsi. Dev. and Comp. Immunol. 121, 104100. doi:10.1016/j.dci.2021.104100
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wei, D., Cheng, Y., Xiao, S., Liao, W., Yu, Q., Han, S., et al. (2023). Natural occurrences and characterization of Elizabethkingia miricola infection in cultured bullfrogs (Rana catesbeiana). Front. Cell Infect. Microbiol. 13, 1094050. doi:10.3389/fcimb.2023.1094050
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi:10.1385/1-59259-584-7:531
Xie, Y., Li, H., Luo, X., Li, H., Gao, Q., Zhang, L., et al. (2022). IBS 2.0: an upgraded illustrator for the visualization of biological sequences. Nucleic Acids Res. 50 (W1), W420–W426. doi:10.1093/nar/gkac373
Yu, D., Wu, Y., Xu, L., Fan, Y., Peng, L., Xu, M., et al. (2016a). Identification and characterization of toll-like receptors (TLRs) in the Chinese tree shrew (Tupaia belangeri chinensis). Dev. and Comp. Immunol. 60, 127–138. doi:10.1016/j.dci.2016.02.025
Yu, D., Zheng, R., Lu, Q., Yang, G., Fu, Y., and Zhang, Y. (2016b). Genetic diversity and population structure for the conservation of giant spiny frog (Quasipaa spinosa) using microsatellite loci and mitochondrial DNA. Asian Herpetological Res. 7 (2), 75–86. doi:10.16373/j.cnki.ahr.150040
Zhang, J., Liu, S., Rajendran, K. V., Sun, L., Zhang, Y., Sun, F., et al. (2013). Pathogen recognition receptors in channel catfish: III Phylogeny and expression analysis of Toll-like receptors. Dev. and Comp. Immunol. 40 (2), 185–194. doi:10.1016/j.dci.2013.01.009
Zhang, L., Liu, G., Xia, T., Yang, X., Sun, G., Zhao, C., et al. (2022). Evolution of toll-like receptor gene family in amphibians. Int. J. Biol. Macromol. 208, 463–474. doi:10.1016/j.ijbiomac.2022.03.112
Zhang, X. T., Zhang, G. R., Shi, Z. C., Yuan, Y. J., Zheng, H., Lin, L., et al. (2017). Expression analysis of nine Toll-like receptors in yellow catfish (Pelteobagrus fulvidraco) responding to Aeromonas hydrophila challenge. Fish. Shellfish Immunol. 63, 384–393. doi:10.1016/j.fsi.2017.02.021
Zhang, Y., Wang, X., Han, F., and Gao, T. (2022). Genome-wide identification, characterization and expression analysis of toll-like receptors in marbled rockfish (Sebastiscus marmoratus). Int. J. Mol. Sci. 23 (19), 11357. doi:10.3390/ijms231911357
Zhao, F., Yan, C., Wang, X., Yang, Y., Wang, G., Lee, W., et al. (2014). Comprehensive transcriptome profiling and functional analysis of the frog (Bombina maxima) immune system. DNA Res. 21 (1), 1–13. doi:10.1093/dnares/dst035
Keywords: Quasipaa spinosa, genome-wide identification, Toll-like receptor, Elizabethkingia miricola, immune response
Citation: Li Z, Gao Z, Mo D, Zhu Z, Zhang J, Liu M, Xiao H, Zhou M, Gao T and Liang R (2025) Genome-wide identification and characterisation of Toll-like receptors in Chinese spiny frog (Quasipaa spinosa). Front. Genet. 16:1569669. doi: 10.3389/fgene.2025.1569669
Received: 01 February 2025; Accepted: 23 May 2025;
Published: 06 June 2025.
Edited by:
Yonggang Niu, Dezhou University, ChinaReviewed by:
Wael Kamel Elfeil, Suez Canal University, EgyptEleni Voukali, Charles University, Czechia
Copyright © 2025 Li, Gao, Mo, Zhu, Zhang, Liu, Xiao, Zhou, Gao and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teng Gao, Z2FvdGVuZzIwMjRAMTI2LmNvbQ==; Rishen Liang, bGlhbmdyaXNoZW4wMUAxNjMuY29t
†These authors have contributed equally to this work
 Zehong Li1†
Zehong Li1† Meng Zhou
Meng Zhou Rishen Liang
Rishen Liang