- 1Neurology Department, Faculty of Medicine, Ain Shams University, Cairo, Egypt
- 2Biology Department, The American University in Cairo, Cairo, Egypt
- 3Institute of Global Health and Human Ecology, The American University in Cairo, Cairo, Egypt
Background: Vascular parkinsonism (VaP) is a subtype of parkinsonism which needs better characterization of its risks and determinants.
Objective: The aim of this report is to present an understanding of genetic risks of vascular parkinsonism.
Methods: Five participants diagnosed with VaP were recruited and Whole Exome Sequencing (WES) was performed to analyze deleterious variants in relevant genes associated with vascular and parkinsonian diseases.
Results: We identified several candidate risk variants for VaP in our patients, particularly in LRRK2, PLA2G6, TGM6, BSN, UBR4, CD36 and NOTCH3, that are different from the classical Parkinson’s disease -associated variants.
Conclusion: In this case series we highlighted the complexity of genetic contributions to VaP through predicted deleterious variants in genes associated with parkinsonism, cerebrovascular disease as well as collagen-related genes.
Introduction
Vascular Parkinsonism (VaP) is a form of parkinsonism associated with cerebrovascular disease. It predominantly affects the elderly accounting for 4.4%–12% of all parkinsonism cases worldwide (George et al., 2024), This percentage was reported as high as 28.6% in an Egyptian study (El-Tallawy et al., 2013).
Clinically, VaP presents with lower body parkinsonism, impaired gait and cognitive dysfunction, accompanied by white matter lesions (WMLs) or lacunes found in brain neuroimaging (George et al., 2024). Three main subtypes of VaP exist: acute (post-stroke), insidious (more debatable and common with gradual onset and extensive WMLs), and mixed (pathologies from both vascular and neurodegenerative diseases) (Mostile et al., 2018).
Historically, the diagnosis of insidious VaP has been controversial, lacking a distinct structural imaging pattern and having poorly established correlations between neurological symptoms and imaging findings (George et al., 2024). The syndrome’s loose criteria lead to a highly variable clinical picture, often overlapping with neurodegenerative diseases like Parkinson’s Disease (PD) and Progressive Supranuclear Palsy (PSP) as well as non-neurodegenerative conditions such as cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) and Normal Pressure Hydrocephalus (NPH) (Mostile et al., 2018).
Genetic factors, particularly mutations associated with leukoencephalopathies, such as CSF1R, COL4A1/2, NOTCH3, NOTCH2NLC and COL22A1 have emerged as potential contributors to VaP clinical picture, proposing an alternative diagnosis and warranting investigation to clarify the genetic etiology of VaP (Marsili et al., 2023). “Adult leukoencephalopathy-associated parkinsonism” has been previously suggested as alternative term to the debatable term “insidious VaP” (George et al., 2024).
This study aims to explore the genetic pathophysiology of VaP through whole exome sequencing (WES), analyzing deleterious variants in relevant genes associated with vascular and parkinsonian diseases.
Methods
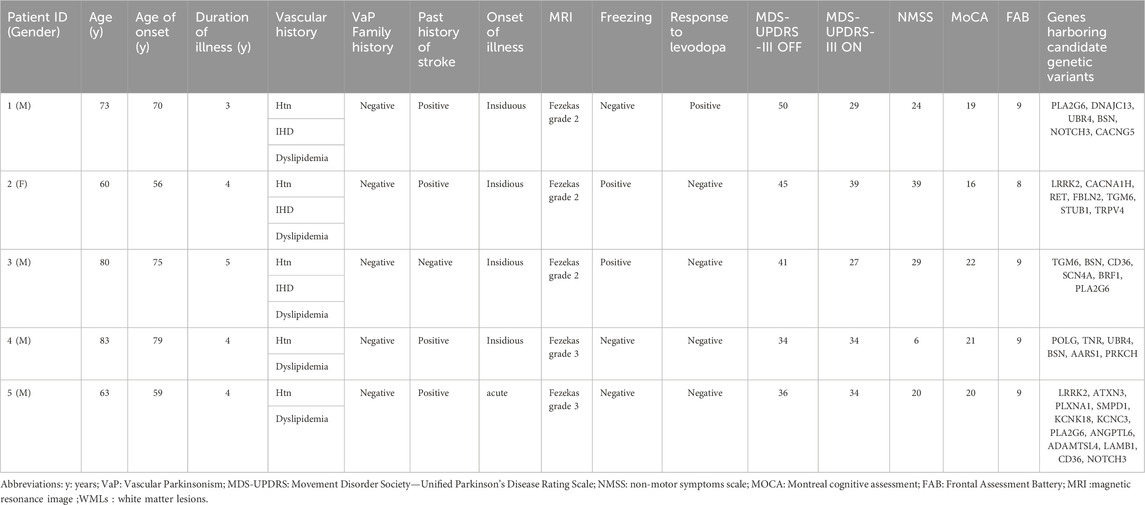
The study involved five participants diagnosed with VaP according to Zijlmans criteria (Zijlmans et al., 2004). Patients with other diagnoses such as Parkinson’s disease (PD) and atypical parkinsonism were not included. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the ethical committee of the Faculty of Medicine, Ain Shams University (George et al., 2024). All subjects gave their informed consent for inclusion before they participated in the study.
Patients were subjected to comprehensive clinical evaluations, including motor and cognitive assessments (George et al., 2024).
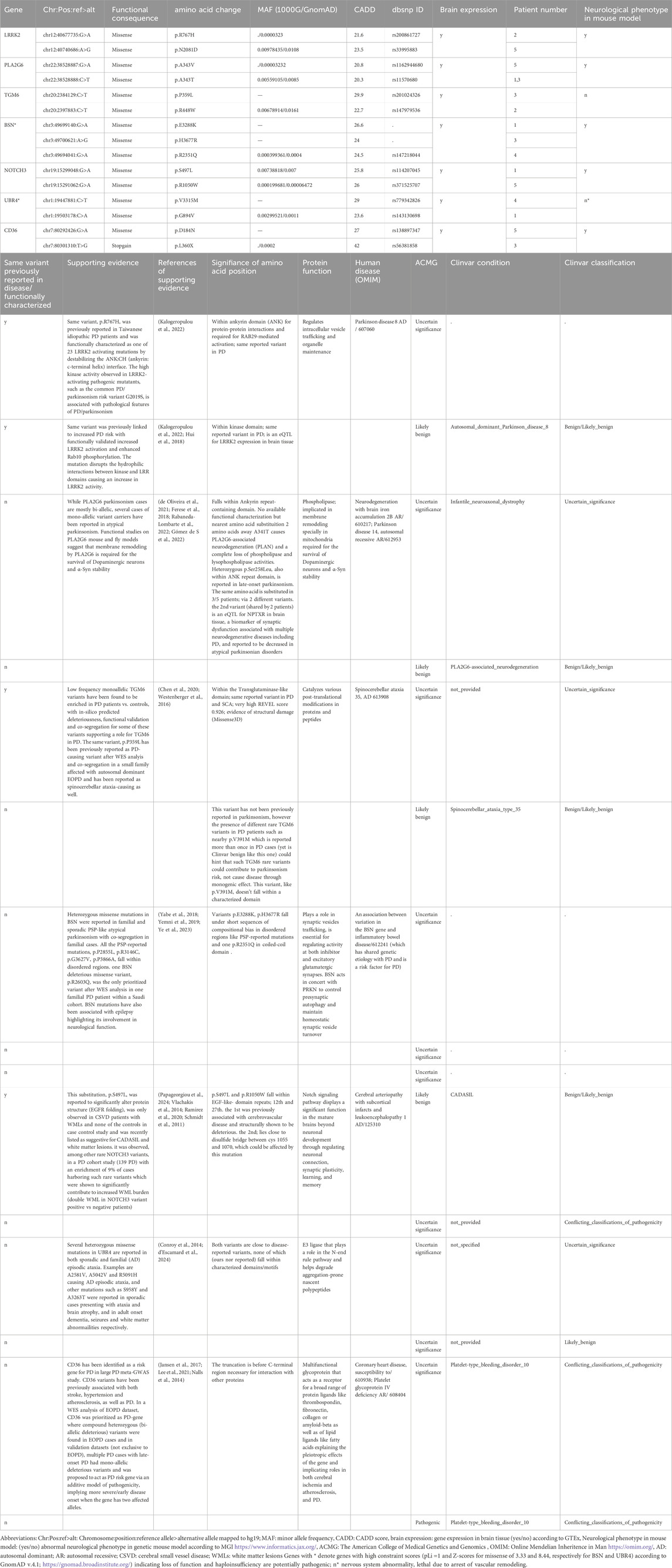
Whole Exome Sequencing (WES) was performed on DNA isolated from peripheral blood using Illumina Novaseq 6000 platform with a coverage of 100x. Variants with read depth <10 were filtered out. ANNOVAR was used for variant annotation (Wang, 2010) for filtering variants with a minor allele frequency (MAF ≤1%) according to 1000 Genomes or GnomAD and loss-of-function (LoF) or missense functional consequence according to RefSeq annotation with CADD score >20 (representing the most deleterious 1% of all amino acid substitutions) (Rentzsch et al., 2019) and consensus of deleteriousness by 2 or more tools provided by ANNOVAR dbnsfp47a database (for functional prediction of variants in whole-exome data). Variants within genes associated with parkinsonian or cerebrovascular disease were prioritized. For further assessment of the clinical relevance of the prioritized genes, the genes harboring the prioritized variants were checked for expression in brain tissue in GTEx (https://gtexportal.org/home/) and the Mouse Genome Informatics (MGI; https://www.informatics.jax.org/) to identify genes whose knockout demonstrated an abnormal neurological phenotype in mice. The use of the American College of Medical Genetics ACMG or Clinvar classification of pathogenicity was not assumed appropriate for genetic analysis of parkinsonism as it is intended for Mendelian single-gene disorders while mostly parkinsonism is sporadic and possibly multiple genetic risk factors are involved, however both annotations were reported for further insight. From this prioritized gene list, we chose to highlight genes, harboring deleterious variants, that were observed in 2 or more patients using a voting approach, allowing for allelic heterogeneity where different deleterious variants within same gene may cause disease or increase risk of disease (Chen et al., 2013).
The more comprehensive filtered gene list, not restricted to parkinsonism or cerebrovascular-associated genes, was used for enrichment analysis to investigate overrepresented pathways using enrichr (Chen et al., 2013). This analysis was done with the hypothesis in mind that pathways important for cerebrovascular function or PD/parkinsonism pathogenesis could be overrepresented by deleterious variants in their corresponding genes where the polygenic defects contribute to the disease. This was done for the whole filtered gene list as well as a subset of genes harboring the most deleterious variants, predicted using REVEL cut-off of 0.7, recommended for prioritizing pathogenic variants (Ioannidis et al., 2016; Hopkins et al., 2023).
Results
Participants had a mean age of 71.8 ± 11.2 years. Notable vascular risk factors were present and there was no family history of PD. Approximately 80% did not significantly respond to the levodopa challenge test, and 80% showed insidious onset of symptoms. Cognitive impairment was observed across the cohort (Table 1).
No pathogenic mutations, according to ACMG or Clinvar which are intended classifications for monogenic diseases, were found in known PD genes. Variants of uncertain significance, however, were identified in several PD/parkinsonism genes like LRRK2, PLA2G6, EIF4G1, POLG and DNAJC13. Using the voting approach, a total of seven genes were highlighted (Table 2), including monogenic PD genes (LRRK2, PLA2G6), monogenic parkinsonism genes (TGM6, BSN), genes associated with both vascular function and PD (UBR4 and CD36) and monogenic cerebrovascular disease genes (NOTCH3). The whole list of variants in 53 prioritized genes associated with either parkinsonian (39 genes) or cerebrovascular disease (14 genes) categories are provided (Supplementary Table S1).
Pathway enrichment analysis revealed a significant over-representation of pathways related to collagen and extracellular matrix organization, as well as vascular transport and glycogen catabolism (Supplementary Tables S2, S3).
Discussion
Parkinsonism can arise from various neurodegenerative conditions, with increasing evidence that mutations in monogenic PD genes may act as susceptibility factors for sporadic forms of parkinsonism (Lesage and Brice, 2012). In VaP, a causal link between clinical strokes and parkinsonism remains elusive. Studies have shown only 10% of cases with striatal or cerebral infarcts, and a third of cases of all types of ischemic strokes develop parkinsonism (Voss and Dorsey, 2010). This possibly suggests that genetic or environmental risk factors, present in this subgroup, are necessary for developing parkinsonism. Relevantly, an association between prior strokes and developing PD was reported by a large retrospective study (Kummer et al., 2019). PD is predominantly sporadic where polygenic and environmental components are assumed to cause disease. These results support a possible interaction between PD/parkinsonism polygenic risk factors and vascular risk factors for developing VaP.
This study investigated genetic risk factors for parkinsonism and cerebrovascular disease in a series of VaP patients, prioritizing variants in 7 relevant genes using a voting approach. These genes include LRRK2, PLA2G6, TGM6, BSN, UBR4, CD36 and NOTCH3 (Table 2).
LRRK2 mutations are a major cause to autosomal dominant inherited late-onset PD as well as sporadic parkinsonism by increasing disease susceptibility (Blauwendraat et al., 2020). In PD recessive genes such as PLA2G6, the prevalence of heterozygous rare variants is significantly higher in familial and sporadic parkinsonism cases than in controls highlighting possible contribution to disease susceptibility, as has been reported for PINK1 and PRKN (Hayashida et al., 2021). The polygenic interaction of such heterozygous mutations in PD genes in PD pathogenesis is also exemplified in familial PD cases with digenic heterozygous mutations in PINK1/DJ1, PINK1/PRKN or PRKN/LRRK2 (Hayashida et al., 2021).
TGM6 and BSN mutations are monogenic causes for late-onset atypical parkinsonism such as spinocerebellar ataxia and progressive supranuclear palsy (PSP)- like syndromes and are associated with PD (Chen et al., 2020; Yabe et al., 2018) PSP-like syndrome with multi-infarct and poor/no L-dopa response has been described as one of the clinical syndromes of VaP (Vizcarra et al., 2015).
CD36 and UBR4 are implicated in both vascular functions and parkinsonism. CD36 was identified as a PD risk gene in PD meta-GWAS study and large-scale PD case control WES study (Jansen et al., 2017) and is also implicated in cerebral ischemia (Lee et al., 2021). UBR4 was identified as a monogenic cause for familial adult-onset episodic ataxia, WMLs and brain atrophy via AD, is associated with PD and early onset dementia, and its role in vascular physiology and functionality was recently unraveled through driving collagen/matrix production necessary for vascular development (Conroy et al., 2014; d’Escamard et al., 2024). Worth mentioning UBR4 and BSN have high constraint scores (GnomAD; https://gnomad.broadinstitute.org/) and a probability of being loss-of-function intolerant (pLI) 1.0, indicating loss of function and haploinsufficiency are potentially pathogenic (Table 2).
NOTCH3 mutations are causative of CADASIL, characterized by recurrent strokes, migraines and not uncommonly late-onset parkinsonism (Rowley, 2013). Characteristic cysteine-altering mutations distributed throughout the 34 epidermal growth factor-like repeats (EGFRs) that comprise the extracellular domain of the NOTCH3 receptor cause the disease with a highly heterogenous clinical phenotype with stroke onset ranging from the fourth to eighth decade (Cho et al., 2022). EGFr 1-6 pathogenic variants are associated with a more severe phenotype compared with EGFr 7–34 22,which was not supported in a recent study reporting severe phenotype in EGFRs 18–34 (Cho et al., 2022). Furthermore, a recent study demonstrated that the disease is not always fully penetrant, by studying the carriers of NOTCH3 cysteine-altering mutations in EGF repeats 1 to 34 in a large exome database (the exome aggregation consortium) reporting a 100-fold higher frequency of CADASIL (Coupland et al., 2018; Papageorgiou et al., 2024).
Recently, some NOTCH3 cysteine-sparing mutations such as (p.R61W, p.R75P, p.D80G, p.R213K, p.L1515P, A1604T) were reported to cause milder vascular disease, referred to as CADASIL-like (Coupland et al., 2018), meeting several but not all clinical criteria for CADASIL, with support from co-segregation genetic analysis from multiple families, structural or functional analysis of the mutations (Coupland et al., 2018). Such cys-sparing mutations may produce an altered Notch signaling output resulting in subtler aspects of vascular disease without causing the entire spectrum of classical CADASIL symptoms Interestingly, a report on 7 cases with a clinical profile of VaP secondary to CADASIL showed that all NOTCH3 mutations were cysteine sparing (Guo et al., 2022). This draws attention to the relevance of NOTCH3 genetic testing in VaP and functional characterization of cys-sparing mutations Furthermore, a link between NOTCH3 genetic variants, PD and WMLs has been previously noted, where PD patients showed an enrichment of rare missense NOTCH3 variants compared to controls, and a doubling of WML burden between NOTCH3 variant positive vs. negative patients with a large effect size for periventricular WMLs and lacunes, as well as a significant negative correlation between WML and global cognition were reported (Ramirez et al., 2020). Interestingly, no association between NOTCH3 genetic variants and early-onset or familial PD was found in a Chinese study (Zeng et al., 2022). The difference in significance of the association between NOTCH3 genetic variants and PD cases not of early age of onset and with WMLs and lacunes, which greatly overlaps with sporadic VaP, but not early onset or familial PD might indicate a possible contribution of NOTCH3 genetic risk variants to VaP or sporadic parkinsonism in general. Relevantly, and in addition to parkinsonism (Vascular parkinsonism) being reported as a late but not rare feature of CADASIL in NOTCH3 genetic variant carriers, parkinsonism has been reported as a novel onset symptom in CADASIL patients with NOTCH3 cys mutations, with similar VaP clinical picture of progressive gait instability, rigidty and WMLs (Wang et al., 2023; Arbeu et al., 2024). Finally, co-occurrence of CADASIL and parkinsonian syndromes (progressive parkinsonism in one case and autopsy-confirmed PD in another), have been reported, further establishing NOTCH3 as a PD and parkinsonism gene (Murray et al., 2017). Taken together, as WES analysis gains broader application, attention has shifted toward the contribution of deleterious mutations in NOTCH3 in PD and parkinsonism, complementing their established association with CADASIL and further underscoring the intersection between parkinsonism and vascular dysfunction (Lesage and Brice, 2012).
In addition to the 7 highlighted genes by voting, other VaP-relevant candidate variants in our prioritized list of vascular and parkinsonian associated genes are reported in supplementary data with their supporting evidence. These genes include: monogenic parkinsonism genes (POLG), monogenic PD genes such as (PARK7, DNAJC13), PD risk genes (SMPD1), PD drug transporter and response gene (SLC22A1), genes harboring the same pathogenic variants previously reported in familial cases of PD (TNR), familial migraine with aura (KCNK18), a risk factor for both strokes and parkinsonism, or genes harboring variants falling in important domains and directly neighboring pathogenic variants previously reported in parkinsonism and familial intracranial aneurysms (PLXNA1 and ANGPTL6).
Enrichment of deleterious variants in collagen and ECM genes, which play a crucial role in blood vessel integrity and elasticity, in our VaP sample supports their reported contribution to both vascular and neurological dysfunction. This significant enrichment was evident in our filtered gene list harboring LoFs and in silico predicted deleterious variants, and also evident when using only the most deleterious subset of that list, filtered by REVEL score (Supplementary Tables S2, S3). Genetic susceptibility to periventricular venous collagenosis; i.e., increased collagen deposition in vessels leading to wall-thickening and stenosis, has been proposed as a key factor in WML development, which is common and extensively found in the subcortex in VaP (Lin et al., 2017). Genetic studies consistently implicate different collagen genes, which are core components of the basement membrane in blood vessels, such as in COL4A1/2, COL3A1, COL5A2, in CSVD and stroke through predisposing to arterial wall weakness. Furthermore, collagen genes, COL6A1-6 have been associated with movement disorders such as muscular dystrophies, dystonia, restless leg syndrome, epilepsy and defects in dopaminergic signaling (Gregorio et al., 2022). A new implication for COL22A1, previously associated with muscular dystrophy, in intracranial aneurysms was reported where a heterozygous mutation functionally characterized to impair vessel integrity and increase cerebral hemorrhages in Zebrafish was reported in a patient diagnosed and presenting with VaP, who was later suggested by the authors to be probably misdiagnosed and that the patients’ WMLs were due to adult-onset leukoencephalopathy due to the collagenopathy (Marsili et al., 2023). Collectively, this study highlights the complexity of genetic contributions to VaP through predicted deleterious variants in genes associated with parkinsonism, cerebrovascular disease as well as collagen-related genes. These findings support the notion of a polygenic interplay of risk factors contributing to VaP, rather than it being solely a feature of monogenic leukoencephalopathies due to single mutations in genes such as CSF1R, COL4A1/2, NOTCH3, and COL22A1 as recently proposed (Marsili et al., 2023; Rowley, 2013; Guo et al., 2022). This polygenic model is also more consistent with the sporadic nature of the disease.
Importantly, the presence of variants previously linked to classical Parkinson’s disease (PD) such as LRRK2 p.R767H and p.N2081D, TGM6 p.P359, and TNR p.T166A does not confound the diagnosis of VaP in our cohort. These findings align with current understanding that mutations in monogenic PD-associated genes may serve as risk modifiers for both familial and sporadic forms of parkinsonism (Lesage and Brice, 2012).
Moreover, the identification of variants like NOTCH3 p.S497L, previously reported in CADASIL and PD, and KCNK18 p.F139Wfs*25, associated with familial migraine with aura and proposed as a risk factor for both CADASIL and VaP, reinforces the overlapping phenotypic spectrum among these conditions. Our data support previous observations that parkinsonism may be an initial or co-occurring manifestation in carriers of NOTCH3 variants, especially considering the incomplete penetrance and variable expression of CADASIL.
Given the clinical overlap between VaP, PD, CADASIL and other diseases, and the ongoing debate regarding VaP as a distinct entity, further genetic studies involving larger cohorts are necessary to validate these associations and explore whether these variants represent common genetic risk factors. Future research should also focus on functional characterization of the identified variants and their clinical relevance in the pathogenesis of VaP.
Data availability statement
The datasets presented in this article are not readily available because of ethical and privacy restrictions. Requests to access the datasets should be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Ain Shams Faculty of medicine ethical approval. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
AS: Conceptualization, Formal Analysis, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing. SE-S: Data curation, Formal Analysis, Writing – original draft. PG: Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. TR: Methodology, Supervision, Writing – review and editing. MF: Methodology, Supervision, Writing – review and editing. MY: Data curation, Formal Analysis, Writing – review and editing. ME-B: Methodology, Supervision, Writing – review and editing. MA: Conceptualization, Methodology, Supervision, Writing – review and editing. MS: Methodology, Supervision, Writing – review and editing, Conceptualization, Formal Analysis, Funding acquisition, Resources, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The American University in Cairo, Faculty Support grant (MS).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1579454/full#supplementary-material
References
Arbeu, R. M., Madariaga, C. B., and Castillo Torres, S. (2024). “Early-onset parkinsonism as presenting manifestation of CADASIL in a Mexican woman with NOTCH3 c.1732C>T pathogenic variant,” in International parkinson and movement disorder society.
Blauwendraat, C., Nalls, M. A., and Singleton, A. B. (2020). The genetic architecture of Parkinson’s disease. Lancet Neurol. 19 (2), 170–178. doi:10.1016/S1474-4422(19)30287-X
Chen, E. Y., Tan, C. M., Kou, Y., Duan, Q., Wang, Z., Meirelles, G. V., et al. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 14, 128. doi:10.1186/1471-2105-14-128
Chen, K., Lu, Y., Peng, F., Yu, H. L., Wu, J. Y., Tan, Y., et al. (2020). TGM6 variants in Parkinson’s disease: clinical findings and functional evidence. J. Integr. Neurosci. 19 (1), 51–64. doi:10.31083/J.JIN.2020.01.1203
Cho, B. P. H., Jolly, A. A., Nannoni, S., Tozer, D., Bell, S., and Markus, H. S. (2022). Association of NOTCH3 variant position with stroke onset and other clinical features among patients with CADASIL. Neurology 99 (5), E430–E439. doi:10.1212/WNL.0000000000200744
Conroy, J., McGettigan, P., Murphy, R., Webb, D., Murphy, S. M., McCoy, B., et al. (2014). A novel locus for episodic ataxia: UBR4 the likely candidate. Eur. J. Hum. Genet. 22 (4), 505–510. doi:10.1038/ejhg.2013.173
Coupland, K., Lendahl, U., and Karlström, H. (2018). Role of NOTCH3 mutations in the cerebral small vessel disease cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Stroke 49 (11), 2793–2800. doi:10.1161/STROKEAHA.118.021560
de Oliveira, P., Montanaro, V., Carvalho, D., Martins, B., Ferreira, A., and Cardoso, F. (2021). Severe early-onset parkinsonian syndrome caused by PLA2G6 heterozygous variants. Mov. Disord. Clin. Pract. 8 (5), 794–796. doi:10.1002/mdc3.13230
d’Escamard, V., Kadian-Dodov, D., Ma, L., Lu, S., King, A., Xu, Y., et al. (2024). Integrative gene regulatory network analysis discloses key driver genes of fibromuscular dysplasia. Nat. Cardiovasc. Res. 3 (9), 1098–1122. doi:10.1038/s44161-024-00533-w
El-Tallawy, H. N., Farghaly, W. M., Shehata, G. A., Rageh, T. A., Hakeem, N. M. A., Hamed, M. A. A., et al. (2013). Prevalence of Parkinson’s disease and other types of parkinsonism in Al kharga district, Egypt. Neuropsychiatr. Dis. Treat. 9, 1821–1826. doi:10.2147/NDT.S48318
Ferese, R., Scala, S., Biagioni, F., Giardina, E., Zampatti, S., Modugno, N., et al. (2018). Heterozygous PLA2G6 mutation leads to iron accumulation within basal ganglia and Parkinson’s disease. Front. Neurol. 9 (JUL), 536. doi:10.3389/fneur.2018.00536
George, P., Roushdy, T., Fathy, M., Hamid, E., Ibrahim, Y. A., El-Belkimy, M., et al. (2024). The clinical and neuroimaging differences between vascular parkinsonism and Parkinson’s disease: a case-control study. BMC Neurol. 24 (1), 56. doi:10.1186/s12883-024-03556-9
Gómez de San José, N., Massa, F., Halbgebauer, S., Oeckl, P., Steinacker, P., and Otto, M. (2022). Neuronal pentraxins as biomarkers of synaptic activity: from physiological functions to pathological changes in neurodegeneration. J. Neural Transm. 129 (2), 207–230. doi:10.1007/s00702-021-02411-2
Gregorio, I., Mereu, M., Contarini, G., Bello, L., Semplicini, C., Burgio, F., et al. (2022). Collagen VI deficiency causes behavioral abnormalities and cortical dopaminergic dysfunction. DMM Dis. Models Mech. 15 (9), dmm049481. doi:10.1242/dmm.049481
Guo, W., Xu, B., Sun, H., Ma, J., Mei, S., Zeng, J., et al. (2022). Case Report: progressive asymmetric parkinsonism secondary to CADASIL without dementia. Front. Neurol. 12, 760164. doi:10.3389/fneur.2021.760164
Hayashida, A., Li, Y., Yoshino, H., Daida, K., Ikeda, A., Ogaki, K., et al. (2021). The identified clinical features of Parkinson’s disease in homo-heterozygous and digenic variants of PINK1. Neurobiol. Aging 97, 146.e1–146.e13. doi:10.1016/j.neurobiolaging.2020.06.017
Hopkins, J. J., Wakeling, M. N., Johnson, M. B., Flanagan, S. E., and Laver, T. W. (2023). REVEL is better at predicting pathogenicity of loss-of-function than gain-of-function variants. Hum. Mutat. 2023, 8857940. doi:10.1155/2023/8857940
Hui, K. Y., Fernandez-Hernandez, H., Hu, J., Schaffner, A., Pankratz, N., Hsu, N. Y., et al. (2018). Functional variants in the LRRK2 gene confer shared effects on risk for Crohn’s disease and Parkinson’s disease. Sci. Transl. Med. 10 (423), eaai7795. doi:10.1126/scitranslmed.aai7795
Ioannidis, N. M., Rothstein, J. H., Pejaver, V., Middha, S., McDonnell, S. K., Baheti, S., et al. (2016). REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99 (4), 877–885. doi:10.1016/j.ajhg.2016.08.016
Jansen, I. E., Ye, H., Heetveld, S., Lechler, M. C., Michels, H., Seinstra, R. I., et al. (2017). Discovery and functional prioritization of Parkinson’s disease candidate genes from large-scale whole exome sequencing. Genome Biol. 18 (1), 22. doi:10.1186/s13059-017-1147-9
Kalogeropulou, A. F., Purlyte, E., Tonelli, F., Lange, S. M., Wightman, M., Prescott, A. R., et al. (2022). Impact of 100 LRRK2 variants linked to Parkinson’s disease on kinase activity and microtubule binding. Biochem. J. 479 (17), 1759–1783. doi:10.1042/BCJ20220161
Kummer, B. R., Diaz, I., Wu, X., Aaroe, A. E., Chen, M. L., Iadecola, C., et al. (2019). Associations between cerebrovascular risk factors and parkinson disease. Ann. Neurol. 86 (4), 572–581. doi:10.1002/ana.25564
Lee, D. H., Won, G. W., Lee, Y. H., Shin, J. S., Ku, E. J., Oh, T. K., et al. (2021). Association between rs1761667 CD36 polymorphism and risk of stroke in Korean patients with type 2 diabetes. Chin. Med. J. Engl. 134 (19), 2385–2387. doi:10.1097/CM9.0000000000001501
Lesage, S., and Brice, A. (2012). Role of Mendelian genes in “sporadic” parkinson’s disease, Park. Relat. Disord., 18; S66, S70. doi:10.1016/S1353-8020(11)70022-0
Lin, J., Wang, D., Lan, L., and Fan, Y. (2017). Multiple factors involved in the pathogenesis of white matter lesions. Biomed. Res. Int. 2017, 9372050. doi:10.1155/2017/9372050
Marsili, L., Kauffman, M. A., Rufin Florat, D., Zaidi, A., Botsford, V., Sharma, J., et al. (2023). Resolving ‘vascular parkinsonism’ –COL22A1 as a genetic adult-onset leukoencephalopathy. Park. Relat. Disord. 117, 105898. doi:10.1016/j.parkreldis.2023.105898
Mostile, G., Nicoletti, A., and Zappia, M. (2018). Vascular parkinsonism: still looking for a diagnosis. Front. Neurol. 9 (JUN), 411. doi:10.3389/fneur.2018.00411
Murray, N., Pepper, E., Blackie, J., Ronan, A., and Sugo, E. (2017). Co-occurrence of cadasil and a parkinsonian syndrome: a report of two cases and review of the literature. J. Neurol. Neurosurg. Psychiatry 88 (5), e1.35–e1. doi:10.1136/jnnp-2017-316074.38
Nalls, M. A., Pankratz, N., Lill, C. M., Do, C. B., Hernandez, D. G., Saad, M., et al. (2014). Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46 (9), 989–993. doi:10.1038/ng.3043
Papageorgiou, L., Papa, L., Papakonstantinou, E., Mataragka, A., Dragoumani, K., Chaniotis, D., et al. (2024). SNP and structural study of the notch superfamily provides insights and novel pharmacological targets against the CADASIL syndrome and neurodegenerative diseases. Genes (Basel) 15 (5), 529. doi:10.3390/genes15050529
Rabaneda-Lombarte, L., Beyer, K., Ispierto, L., and Vilas-Rolan, D. (2022). Atypical parkinsonism related to a rare variant in the PLA2G6 gene. Mov. Disord. 37. Available online at: https://www.mdsabstracts.org/abstract/do-mutations-in-the-tgm6-sca35-gene-cause-early-onset-parkinsons-disease.
Ramirez, J., Dilliott, A. A., Binns, M. A., Breen, D. P., Evans, E. C., Beaton, D., et al. (2020). Parkinson’s disease, NOTCH3 genetic variants, and white matter hyperintensities. Mov. Disord. 35 (11), 2090–2095. doi:10.1002/mds.28171
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J., and Kircher, M. (2019). CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47 (D1), D886–D894. doi:10.1093/nar/gky1016
Rowley, H. A. (2013). Parkinsonism is a late, not rare, feature of CADASIL A study on Italian patients carrying the R1006C mutation. In: Stroke, 44. S53. S54. doi:10.1161/STROKEAHA.113.001939
Schmidt, H., Zeginigg, M., Wiltgen, M., Freudenberger, P., Petrovic, K., Cavalieri, M., et al. (2011). Genetic variants of the NOTCH3 gene in the elderly and magnetic resonance imaging correlates of age-related cerebral small vessel disease. Brain 134 (11), 3384–3397. doi:10.1093/brain/awr252
Vizcarra, J. A., Lang, A. E., Sethi, K. D., and Espay, A. J. (2015). Vascular parkinsonism: deconstructing a syndrome. Mov. Disord. 30 (7), 886–894. doi:10.1002/mds.26263
Vlachakis, D., Tsaniras, S. C., Loannidou, K., Papageorgiou, L., Baumann, M., and Kossida, S. (2014). A series of Notch3 mutations in CADASIL; insights from 3D molecular modelling and evolutionary analyses. Available online at: http://meme.sdsc.edu/meme4_6_1/.
Voss, T. S., and Dorsey, E. R. (2010). Parkinsonism: vascular. Encycl. Mov. Disord. Three-Volume Set, 416–420. doi:10.1016/B978-0-12-374105-9.00092-7
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from next-generation sequencing data nucleic acids research.
Wang, X., Ke, M., Fan, P., Ding, Y., and Zhang, Y. (2023). Parkinsonism is a new pattern onset of CADASIL patients carrying with R544Cmutation: a case Report. doi:10.21203/rs.3.rs-2679681/v1
Westenberger, A., Svetel, M., Dragaševic, N., Brænne, I., Dobricic, V., Hicks, A. A., et al. (2016). Do mutations in the TGM6 (SCA35) gene cause early-onset Parkinson’s disease? Mov. Disord. 31. Available online at: https://www.mdsabstracts.org/abstract/atypical-parkinsonism-related-to-a-rare-variant-in-the-pla2g6-gene/.
Yabe, I., Yaguchi, H., Kato, Y., Miki, Y., Takahashi, H., Tanikawa, S., et al. (2018). Mutations in bassoon in individuals with familial and sporadic progressive supranuclear palsy-like syndrome. Sci. Rep. 8 (1), 819. doi:10.1038/s41598-018-19198-0
Yemni, E. Al, Monies, D., Alkhairallah, T., Bohlega, S., Abouelhoda, M., Magrashi, A., et al. (2019). Integrated analysis of whole exome sequencing and copy number evaluation in parkinson’s disease. Sci. Rep. 9 (1), 3344. doi:10.1038/s41598-019-40102-x
Zeng, Q., Pan, H., Zhao, Y., Wang, Y., Xu, Q., Tan, J., et al. (2022). Association between NOTCH3 gene and Parkinson’s disease based on whole-exome sequencing. Front. Aging Neurosci. 14, 995330. doi:10.3389/fnagi.2022.995330
Keywords: vascular, parkinsonism, leukoencephalopathy, genetics, Egyptian
Citation: Shalash A, El-Shafie S, George P, Roushdy T, Fathy M, Yousef MH, El-Belkimy M, Abdulghani MO and Salama M (2025) Case Report: Decoding genetic risks of vascular parkinsonism: a case series. Front. Genet. 16:1579454. doi: 10.3389/fgene.2025.1579454
Received: 20 February 2025; Accepted: 27 June 2025;
Published: 25 July 2025.
Edited by:
Jordi Pérez-Tur, Spanish National Research Council (CSIC), SpainCopyright © 2025 Shalash, El-Shafie, George, Roushdy, Fathy, Yousef, El-Belkimy, Abdulghani and Salama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali Shalash, YWxpX25ldXJvQHlhaG9vLmNvbQ==; Mohamed Salama, TW9oYW1lZC1TYWxhbWFAYXVjZWd5cHQuZWR1
†These authors have contributed equally to this work
 Ali Shalash
Ali Shalash Salma El-Shafie
Salma El-Shafie Peter George1
Peter George1 Tamer Roushdy
Tamer Roushdy Mai Fathy
Mai Fathy Mohamed H. Yousef
Mohamed H. Yousef Mohamed Salama
Mohamed Salama