- 1Department of Clinical Laboratory, The First Affiliated Hospital of Guilin Medical University, Guilin, Guangxi, China
- 2Research Center of Clinical Laboratory Science, Bengbu Medical University, Bengbu, Anhui, China
- 3Department of Geriatrics, The First Affiliated Hospital of Guilin Medical University, Guilin, Guangxi, China
Purpose: To investigate the correlation between single-nucleotide polymorphisms (SNPs) of the Epidermal growth factor receptor (EGFR) gene and its protein expression with susceptibility and survival prognosis of lung cancer (LC) patients.
Methods: Using SNP-scan high-throughput technology, the EGFR gene’s rs2227983, rs2293347, and rs884225 locations were analyzed in 300 LC patients and 150 healthy individuals. And small cell lung cancer (SCLC), lung adenocarcinoma (LUAD), and lung squamous carcinoma (LUSC) were subdivided into groups for lung cancer patients. Chi-square test and logistic regression analysis were used to assess the susceptibility of LC. The correlation between SNP haplotypes and LC risk was analyzed using the SHEsis website. KM curves and Cox regression were used to analyse the association between polymorphisms and survival prognosis of LC patients. Expression differences in protein levels were analyzed using immunohistochemistry.
Results: EGFR rs2293347 was associated with LUAD, LUSC, and SCLC susceptibility, and rs884225 was associated with LUAD susceptibility. Haplotype ATT was associated with LC and histological type LUAD and SCLC susceptibility. Meanwhile, rs2293347-TT and rs884225-TT were associated with worse prognosis, and rs2293347-TT was an independent risk factor for prognosis in patients with LC. Furthermore, tumor tissue EGFR protein levels were elevated in patients with both genotypes.
Conclusion: EGFR rs2293347 (pan-subtype) and rs884225 (LUAD-specific) polymorphisms increase LC risk through elevated protein expression, with rs2293347-TT conferring worse survival. These genotype-protein correlations highlight their dual role as susceptibility markers and prognostic predictors in precision oncology.
1 Introduction
The latest annual global cancer report, released in 2022, ranked lung cancer (LC) as the second most prevalent cancer in the world and the first in terms of mortality (Siegel et al., 2022). At present, the pathogenesis of LC has not been clearly defined. Existing studies have shown that it is not only related to smoking, radiation and other factors, but also related to family genetics, immune function, endocrine metabolism, gene mutation and other factors, which is the product of the interaction between environmental factors and genetic factors (Huang et al., 2021; Malhotra et al., 2016; Krabbe et al., 2024).
Epidermal growth factor receptor (EGFR; also known as HER-1 or ErbB-1) is a transmembrane glycoprotein receptor for cell proliferation and signal transduction (Voldborg et al., 1997). Upon binding to ligands such as epidermal growth factor (EGF) (Wang et al., 2018) and transforming growth factor alpha (TGF-α) (Lin et al., 2020), it induces the formation of a dimer between two adjacent EGFR monomers, leading to inhibite EGFR tyrosine kinase structural domain activation and autophosphorylating EGFR residues, triggering a variety of downstream signaling pathways essential for the regulation of cell proliferation, differentiation, migration, invasion, metastasis and survival, including protein kinase B (PKB), stress-activated protein kinase (SAPK), and mitogen-activated Protein Kinase (MAPK). Activated protein kinase could affect normal cellular physiological processes or induce the generation and exacerbation of malignant biological behaviours (Wu and Shih, 2018; Lim et al., 2018; Oxnard et al., 2011).
Currently, more studies have highlighted that high expression of EGFR is associated with worse prognosis in various cancers, such as cervical cancer (Kim et al., 2002), bladder cancer (Colquhoun and Mellon, 2002). It remains controversial whether the EGFR pathway plays a role in the prognosis of non-small cell lung cancer (NSCLC) (Liu et al., 2017), while combined Retinoblastoma 1 (RB1) and Tumor Protein 53 (P53) mutations lead to poorer clinical outcomes in small cell lung cancer (SCLC) (Solta et al., 2024). EGFR polymorphisms, as a form of genetic variation in addition to EGFR mutations, have been reported to correlate with the prognosis of survival in patients with EGFR mutation-positive LC treated with tyrosine kinase inhibitors (TKIs) (Oxnard et al., 2011; Leonetti et al., 2019; He et al., 2021), and some studies (Bashir et al., 2019; Liu et al., 2015; Feng et al., 2014; Li et al., 2023; Bashir et al., 2018) have shown an association with susceptibility in LC patients. Of these, rs2227983 is thought to be associated with TKIs toxicity (Obradovic et al., 2023) and is associated with NSCLC risk in multiple populations (Bashir et al., 2019; Winther-Larsen et al., 2019; Zhang et al., 2013; Winther-Larsen et al., 2015; Ma et al., 2009), but it has also been shown to be unrelated to the susceptibility and prognosis of NSCLC treated with TKIs (Jurisic et al., 2020). Therefore, the results of these studies have not been harmonised and do not take into account the histological types and clinical characteristics of LC patients and the association with prognosis. This study aimed to investigate the potential correlation of EGFR gene polymorphisms and their protein levels interaction with the susceptibility, prognosis, and various clinical indicators of LC patients.
2 Materials and methods
2.1 Subjects and ethics of study
This research included 300 individuals identified with LC at The First Affiliated Hospital of Guilin Medical University’s Oncology Department between July 2021 and March 2023, categorized into LC groups based on histopathological types, with 160 in LUAD, 77 in LUSC, and 63 in SCLC. The diagnostic and pathological classification criteria of LC were based on the Chinese Society of Clinical Oncology (CSCO) 2022 Guidelines for the Diagnosis and Treatment of Primary Lung Cancer, and pathological examination (including cytology, histology, immunohistochemistry, lymph node, bronchoscopy, and lung puncture biopsy, etc.) was taken as the gold standard for the diagnosis of LC. Pathological classification of LC was based on the WHO Histological Classification of Lung Tumor (5th edition, 2021) issued by the World Health Organization (WHO). At the same time, 150 healthy volunteers were randomly recruited as healthy controls (HC group) from the health check-up centre based on age and gender of inclusion. Individuals suffering from acute liver or kidney issues, past or present autoimmune conditions, and those who were pregnant were not included. The participants were independent persons in Guangxi and had no familial ties to one another. Guilin Medical University’s Affiliated Hospital’s Medical Ethics Committee examined and sanctioned the study’s protocol (No. 2022YJSLL-78), ensuring compliance with the principles of the Declaration of Helsinki. Every participant offered to take part in this research and provided their signed informed consent. Overall Survival (OS) The outcome indicator is time to death; this death is any death from any cause and is counted. Progression Free Interval (PFI) is the period of survival without further deterioration of the disease after treatment. The outcome indicator is the occurrence of deterioration (re-admission to hospital).
2.2 DNA extraction and genotyping of the EGFR polymorphism
The workflow commenced with locus-specific probe design for SNP analysis. Subsequent steps included DNA sample preparation (30–50 ng/μL) via thermal lysis (98°C for 5 min) and probe hybridization under denaturation-renaturation conditions. Sequential reactions were then executed: (1) ligation (94°C/1°min→58°C/4 h) with optimized reagent ratios (0.5 μL ligase: 1 μL probe), (2) multiplex fluorescent PCR amplification (touchdown cycles: 62→57°C, 25 standard cycles) using precise primer-mix stoichiometry (10:1 PCR mix: primer), and (3) ABI3730XL sequencing of diluted amplicons (×10) with Liz600 size standard. Allelic discrimination was ultimately achieved through GeneMapper 4.1 analysis of raw electrophoretic data. Supplementary Table S1 provides the PCR primer sequences for the EGFR gene SNPs rs2227983, rs2293347, and rs884225. These SNPs with suballele mutation rates >5% in Asian populations from the NCBI SNP database.
2.3 Baseline information and testing indicators
The liver and kidney function indexes of each study subject were completed in this laboratory, and every testing method adhered rigorously to the guidelines of the reagent kit and the manual for instrument operation, utilizing either the Roche Cobas E701 or E801 analyzer, which was certified by ISO15189 (NO.ML00036). Blood samples analyzed in this study were collected concurrently with SNP sequencing to ensure batch consistency.
2.4 IHC (immunohistochemistry)
Human tissue experiments received approval from the First Affiliated Hospital of Guilin Medical University’s Institutional Review Board (No. 2022YJSLL-78), adhering to the Helsinki Declaration’s guidelines. Utilized protocols for primary antibody and antigen recovery included: anti-EGFR (CER-0032, MXB, Fuzhou, China). Three patients with each histological type of lung cancer were randomized to each genotype at each of the three SNPs, for a total of 81 patients. Two full senior pathologists assessed positive indicators and degree interpretation. The staining results were classified into four grades from 0 to 3+, with the following criteria: 0 as no staining; 1+ as light yellow staining of tumour cells without obvious granules or no more than 10% of tumour cells with yellow staining with obvious granules; 2+ as more than 10% of tumour cells with yellow staining with obvious granules or no more than 10% of tumour cells with brown staining with obvious granules; and 3+ as more than 10% of tumour cells with brown staining with obvious granules. The IOD/area ratio was calculated using ImageJ.
2.5 Statistical analysis
Statistical analyses for this study were completed using IBM SPSS 27.0 and R (4.2.1). Count data were described by frequency (n) or frequency [n (%)], and comparisons of count data and identification of Hardy-Weinberg equilibrium (HWE) law were carried out using Pearson’s chi-square test and chi-square test for goodness of fit in that order. Measurement data were validated for normality using the Kolmogorov-Sminov test; conformity to normal distribution was described as mean ± standard deviation, comparisons between two groups were performed using the independent samples t-test, and one-way ANOVA analysis of variance was used between multiple groups; non-conformity to normal distribution was described as median (quartile) [M (P25∼P75)], comparisons between two groups were performed using the Mann-Whitney U-test, and between multiple groups Kruskal–Wallis test was used. The relationship between biochemical markers, genetic SNP models, and LC susceptibility was evaluated using binary logistic regression, leading to the computation of odds ratio (OR) and 95% confidence interval (CI). One-way multifactor cox regression analyses and survival curves (also known as Kaplan-Meier curves) were performed using R software and the survival package [3.3.1], with the survminer package [0.4.9] and ggplot2 [3.3.6] used for visualization (variables with P-value < 0.1 in univariate analyses were enrolled in multivariate Cox models). The Tukey post hoc test was used for the analysis of protein expression differences between genotypes. P < 0.05 was considered statistically significant.
3 Results
3.1 Comparison of clinical and biochemical characteristics
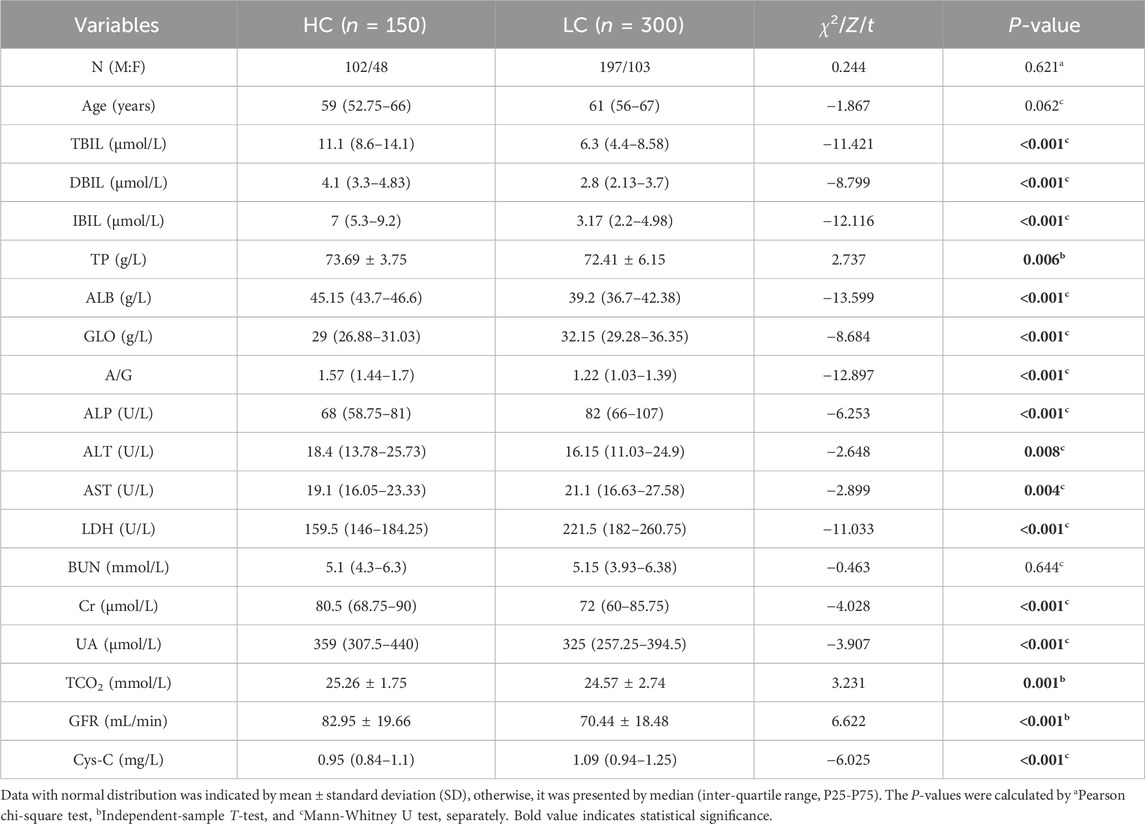
Table 1 summarizes the clinical and biochemical characteristics of HC and LC. Results indicated a notable deviation in liver and kidney function indices in LC patients relative to HC (P < 0.001), highlighting a substantial impairment in these functions in LC patients.
3.2 Relationship between EGFR gene polymorphism and LC susceptibility
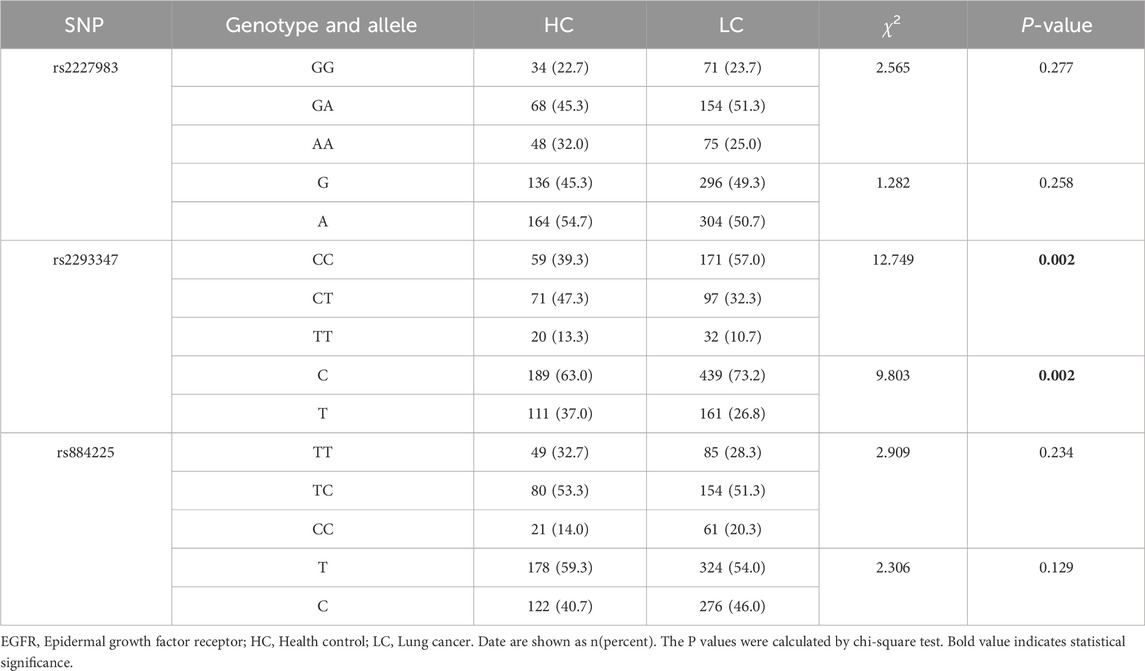
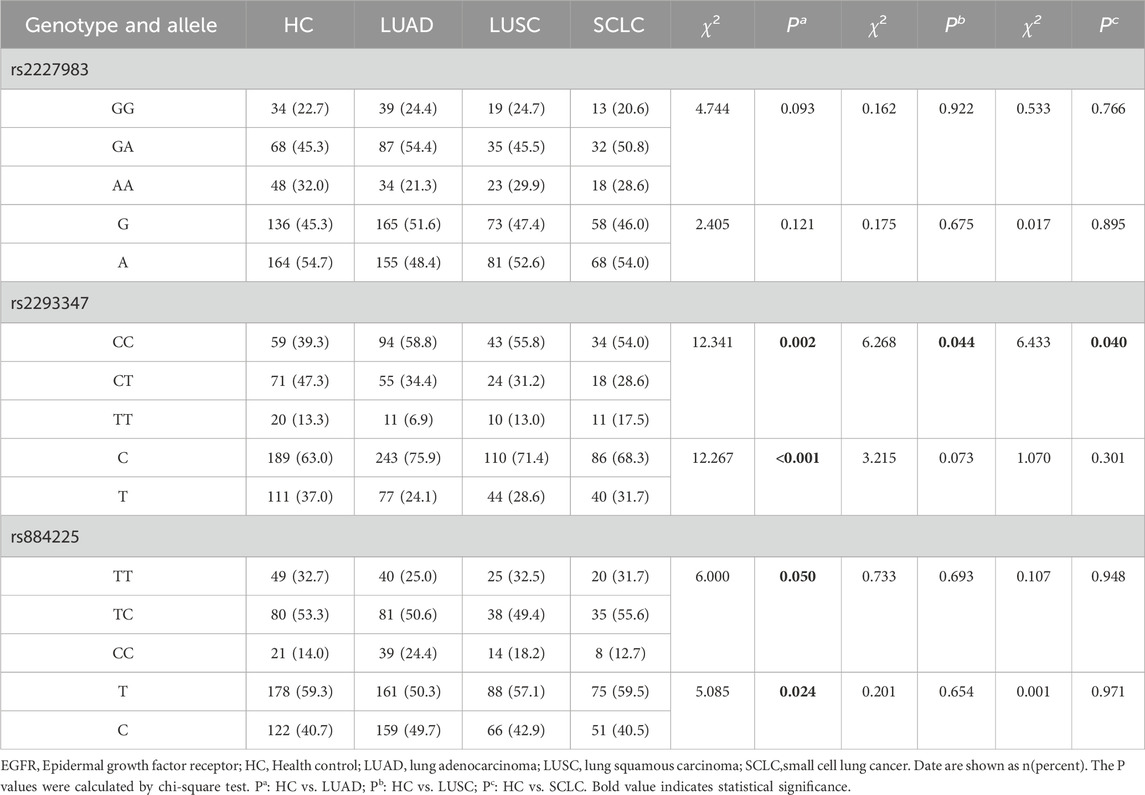
The results of multiple fluorescence PCR typing of the EGFR gene of rs2227983, rs2293347, and rs884225 are shown in Supplementary Figure S1, in which the pure genotypes are single peaks and the heterozygous genotypes are double peaks. Hardy-Weinberg’s law of genetic equilibrium of HC, LC, and population size (P > 0.05) was in agreement with these three SNPs, indicating that selected subjects were well represented in the population. The rs2293347 CC genotype and C allele frequencies were higher in the LC patients than in the HC population, and the CT and TT genotypes and T allele frequencies were lower than in the HC population (χ2 = 12.794 and 9.803, both P = 0.002, Table 2). Notable variances were observed in the rs2293347 genotype and allele frequency among the LUAD, LUSC, and SCLC groups versus the HC group (all P < 0.05, Table 3), along with a marked disparity in allele frequency at the rs884225 in the LUAD group versus the HC group (P = 0.024, Table 3).
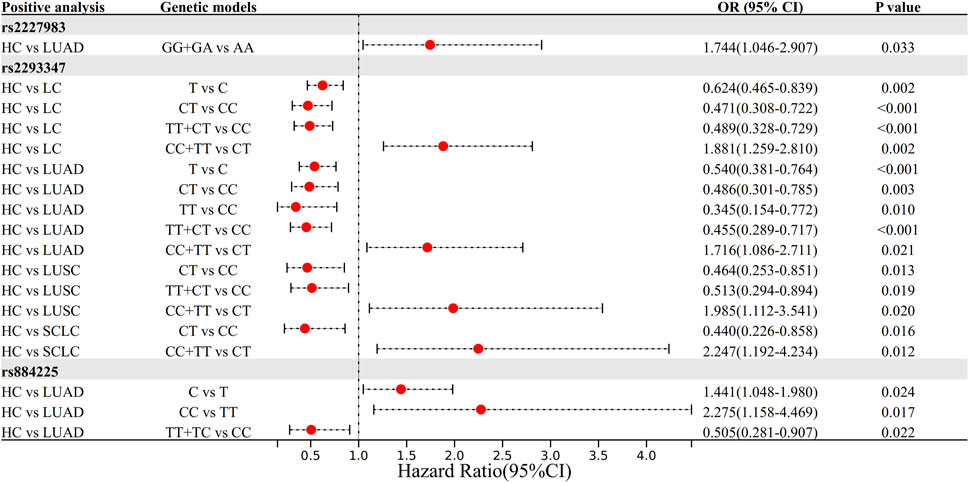
Genetic regression models showed that rs2293347CT and TT + CT genotypes were 0.471 and 0.489 times more susceptible to the risk of LC than the CC genotype, respectively (OR = 0.471 and 0.489, 95% CI = 0.308–0.722 and 0.328–0.729, both P < 0.001), the T allele was 0.624 times more likely to be susceptible to LC than the C allele (OR = 0.624, 95% CI = 0.465–0.839, P = 0.002), and the CC + TT genotype was 1.881 times more likely to be susceptible to LC than the CT genotype (OR = 1.881, 95% CI = 1.259–2.810, P = 0.002).
LUAD subgroup analysis showed that the rs2227983 GG + GA genotype was 1.744 times more susceptible to LUAD risk than the AA type (OR = 1.744, 95% CI = 1.046–2.907, P = 0.033); and that the rs2293347 CT, TT, and TT + CT genotypes were 0.486, 0.345, and 0.455 times (OR = 0.486, 0.345, 0.455, 95% CI = 0.301–0.785, 0.154–0.772, 0.289–0.717, P = 0.003, 0.010, P < 0.001), and the T allele was 0.540 times more common than the C allele (OR = 0.540, 95% CI = 0.381–0.764, P < 0.001), the CC + TT genotype was 1.716 times more common than the CT type (OR = 1.716, 95% CI = 1.086–2.711, P = 0.021); rs884225 CC genotype was 2.275 times more common than the TT type (OR = 2.275, 95% CI = 1.158–4.469, P = 0.017), the C allele was 1.441 times more common than the T allele (OR = 1.441, 95% CI = 1.048–1.980, P = 0.024), and the TT + TC genotype was 0.505 times more common than the CC phenotype (OR = 0.505, 95% CI = 0.281–0.907, P = 0.022).
LUSC subgroup analysis showed that rs2293347 CT and TT + CT genotypes were 0.464 and 0.513 times more likely to be susceptible to LUSC than the CC genotype (OR = 0.464 and 0.513, 95% CI = 0.253–0.851 and 0.294–0.894, P = 0.013 and 0.019 respectively), and the CC + TT genotype was 1.985 times more likely to be susceptible than the CT genotype (OR = 1.985, 95% CI = 1.112–3.541, P = 0.020).
SCLC subgroup analysis showed that the rs2293347 CT genotype was 0.440 times more likely to be susceptible to SCLC than the CC genotype (OR = 0.440, 95% CI = 0.226–0.858, P = 0.016), and the CC + TT genotype was 2.247 times more likely to be susceptible than the CT genotype (OR = 2.247, 95% CI = 1.192–4.234, P = 0.012). All positive results were visualized as Figure 1, and all results are shown in Supplementary Tables S2–S5.
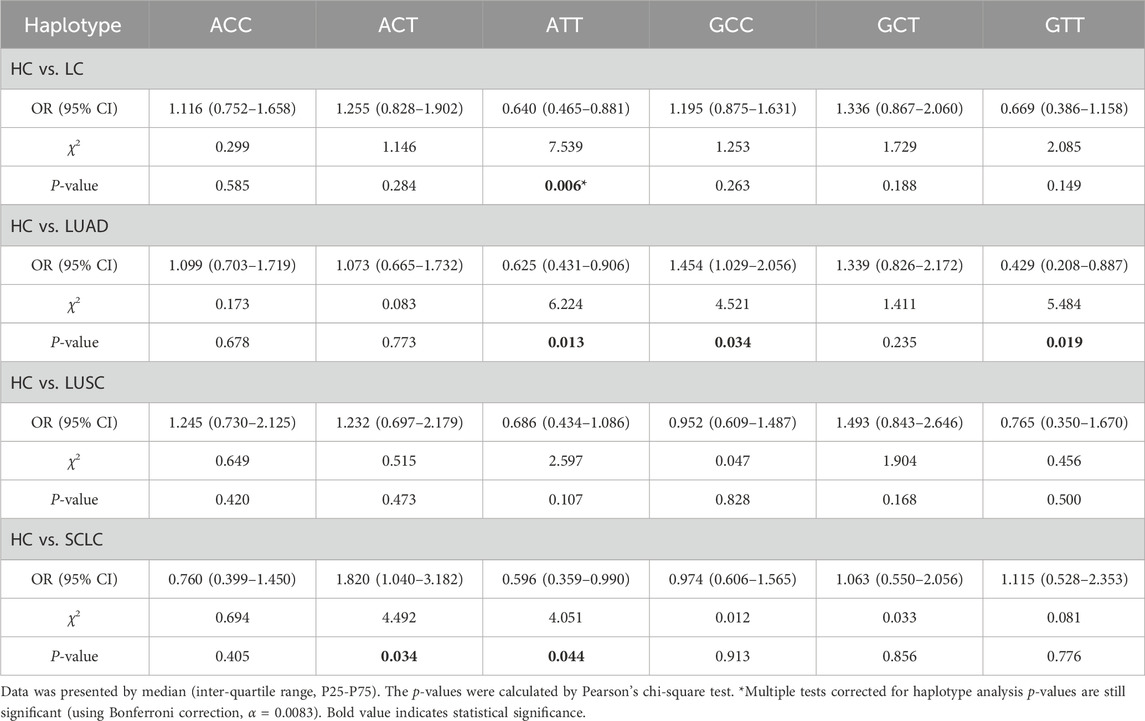
3.3 Haplotype analysis of EGFR
Possible haplotypes at the rs2227983, rs2293347, and rs884225 SNPs of the EGFR gene were identified by SHEsis online software (Shi and He, 2023), including ACC, ACT, ATT, GCC, GCT, and GTT (Table 4). The GTC and ATC haplotypes were excluded from analysis due to low frequency, which precluded reliable conformation assessment. The ATT haplotype may act as a protective factor, reducing LC susceptibility risk (OR = 0.640, 95% CI = 0.465–0.881, P = 0.006); ATT and GTT haplotypes may be related to reduce the risk of LUAD susceptibility as protective factors (OR = 0.625 and 0.429, 95% CI = 0.431–0.906 and 0.208–0.887, P = 0.013 and 0.019, respectively); and GCC haplotype may be associated with a reduced risk of LUAD susceptibility as a risk factor (OR = 1.454, 95% CI = 1.029–2.056, P = 0.034); ATT haplotype may be associated with decreased risk of susceptibility to SCLC (OR = 0.596, 95% CI = 0.359–0.990, P = 0.044); ACT haplotype may be associated with associated with increased risk of SCLC susceptibility (OR = 1.820, 95% CI = 1.040–3.182, P = 0.034); all haplotypes were not associated with risk of LUSC susceptibility (all P > 0.05). However, after correction for multiple testing, only the ATT haplotype association in the HC vs. LC group remained significant (Bonferroni correction, α = 0.0083), and the associations of the other haplotypes may be false positives or need to be verified with larger samples.
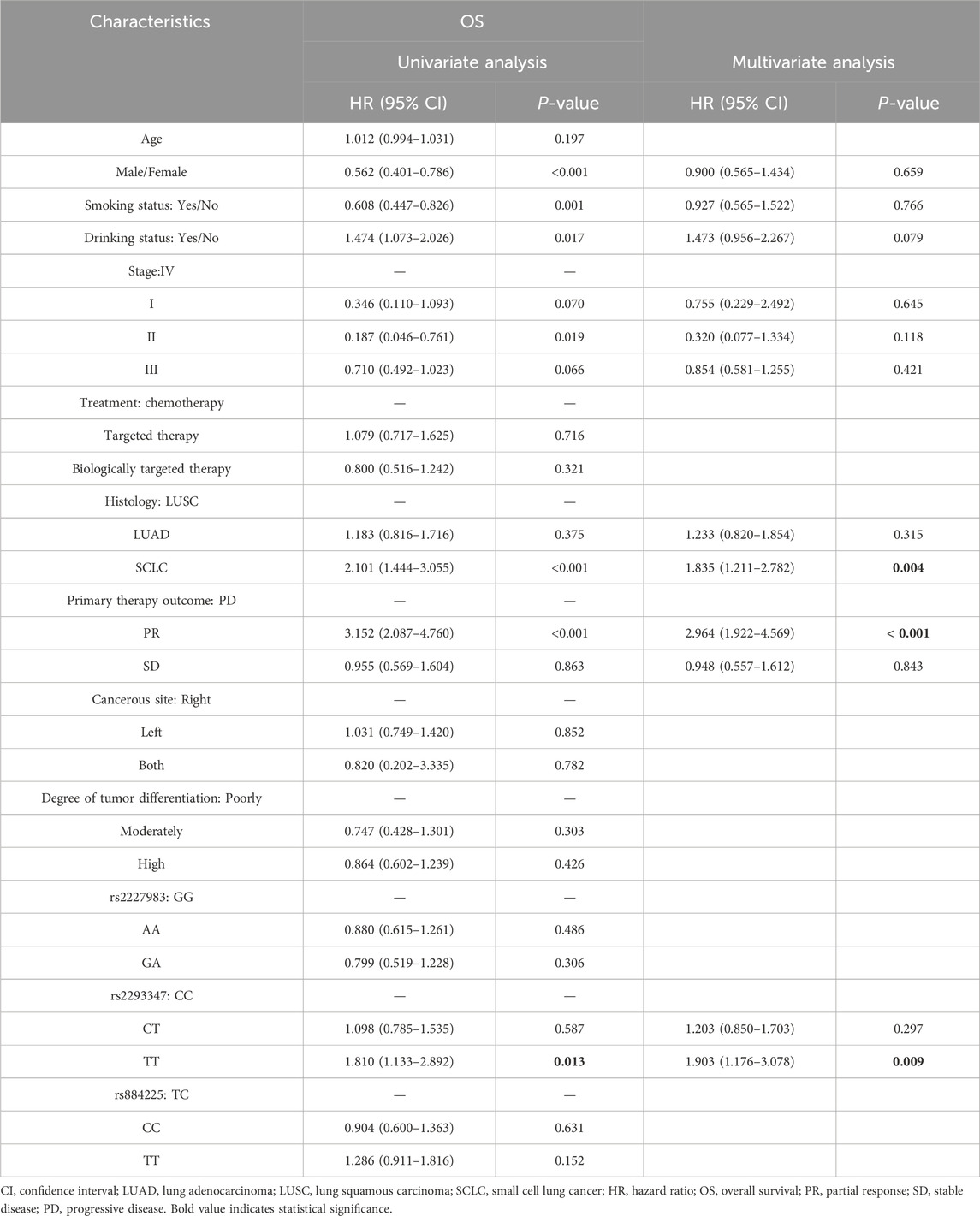
3.4 Relationship between EGFR gene polymorphism and prognosis of LC
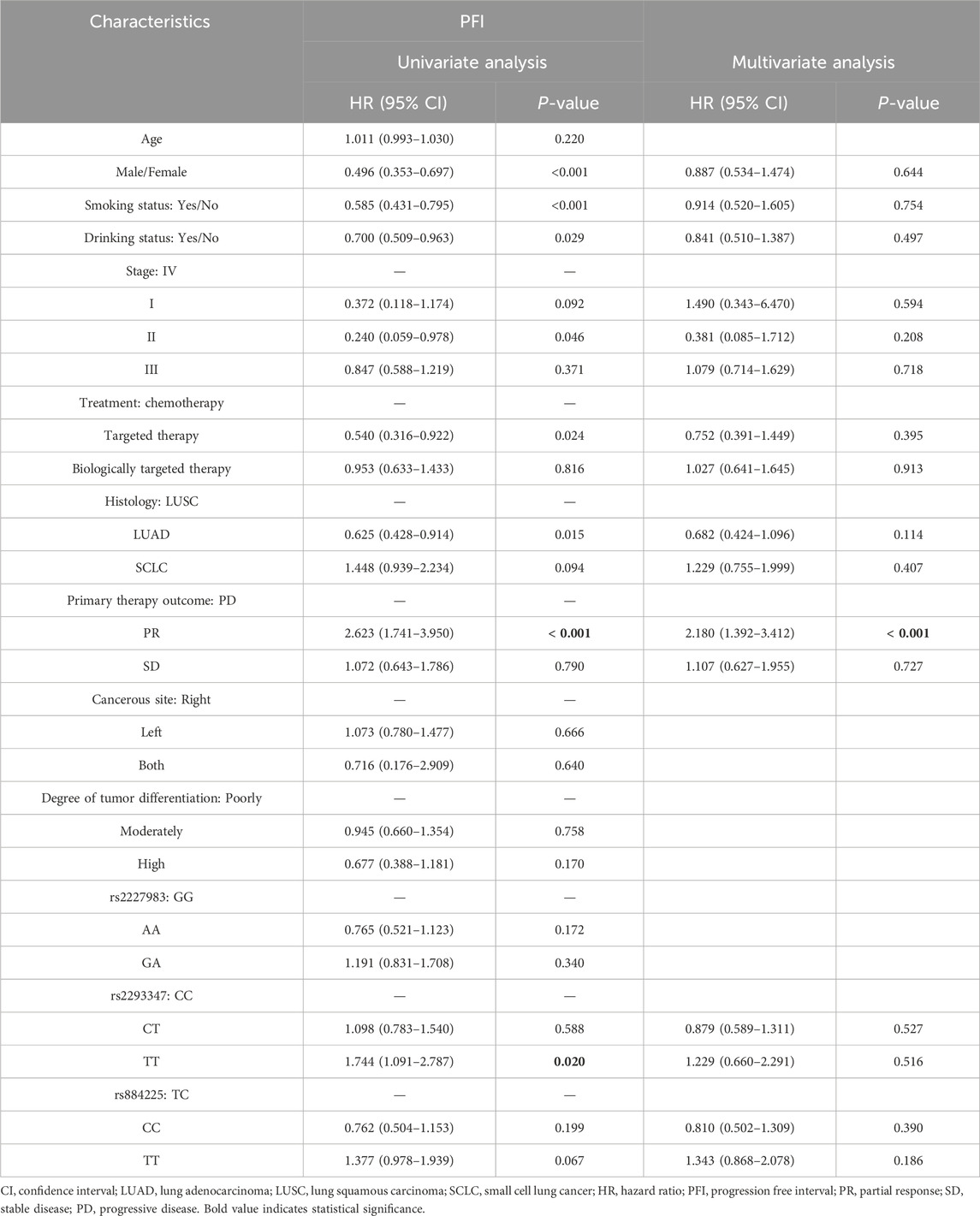
The median overall survival (OS) of all LC patients was 938 days, and rs2293347-TT had a significantly shorter OS compared to CC (710 days vs. 1,014 days, P = 0.047) (Figure 2A). The median Progression Free Interval (PFI) for all LC patients was 41 days, and rs2293347-TT had a shortened PFI compared to CC (25 days vs. 47 days, P = 0.090) but the difference was not statistically significant (Figure 2B), rs884225-TT had a significantly shorter PFI compared to CC (28 days vs. 93 days, P = 0.022) (Figure 2C).
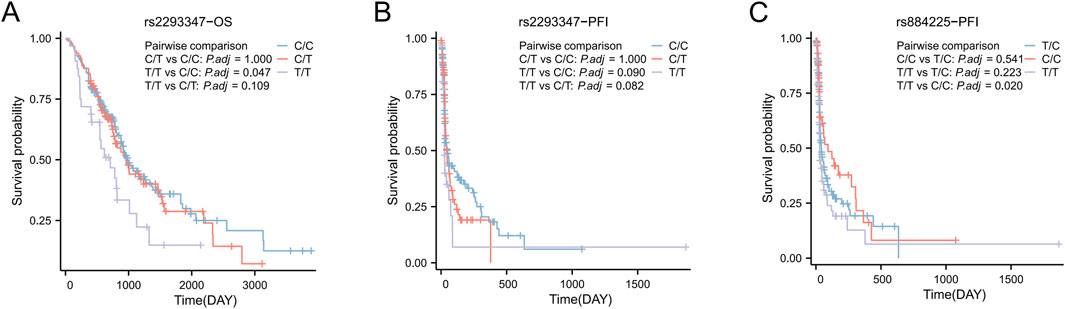
Figure 2. Kaplan–Meier survival curves of Progression Free Interval (PFI) and overall survival (OS) for all LC patients. (A,B) OS and PFI for LC patients with rs2293347 polymorphism (C) PFI for LC patients with rs884225 polymorphism.
In univariate Cox analysis, rs2293347-TT was related to lower OS and PFI (HR = 1.810, 95% CI = 1.133–2.892, P = 0.013; HR = 1.744, 95% CI = 1.091–2.787, P = 0.020). Multivariate Cox analysis showed that rs2293347-TT was an independent adverse factor for OS (HR = 1.903, 95% CI = 1.176–3.078, P = 0.009), but not an independent adverse factor for PFI (HR = 1.229, 95% CI = 0.660–2.291, P = 0.516) (Tables 5, 6).
3.5 Association between EGFR gene polymorphisms and LC tissue protein levels
Upon categorizing and examining the immunohistochemical outcomes by genotype, it was discovered that rs2293347-TT protein’s expression peaked across all LC histological types, with TT and CT surpassing CC (Figures 3A, 4A–C), and that rs884225-TT had the highest protein level in the LUAD population, with both TT and CT exceeding CC (Figures 3B, 4D). In addition, rs2227983 in all histological types and rs88425 in LUSC and SCLC histological types did not differ in genotype protein levels.

Figure 3. Relationship between protein expression and genotype of EGFR in LC patients. (A) rs2293347 (B) rs884225.
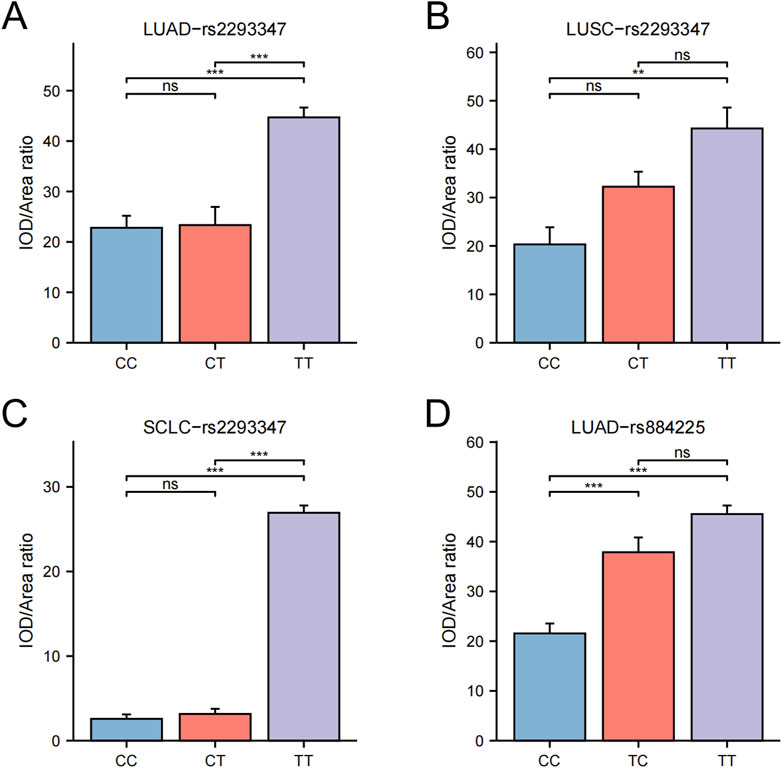
Figure 4. Differential statistics of EGFR genotypes and protein levels in LC patients. (A) rs2293347-LUAD (B) rs2293347-LUSC (C) rs2293347-SCLC (D) rs884225-LUAD.
4 Discussion
To our knowledge, this is the first clinical research to check into the susceptibility to EGFR gene polymorphisms in Chinese patients with LC and to explore the relationship between EGFR protein levels in their lung cancer tissues. We found that EGFR gene polymorphisms were significantly associated with susceptibility and prognosis of LC and its histological types and were responsible for influencing the expression of protein levels.
With extensive research in molecular genetics, a variety of genetic and epigenetic changes have been potentially associated with the risk of developing LC (Zhu et al., 2015). The rs2293347 resides in exon 25, the C-terminal structural segment of the EGFR gene’s regulatory area (Mitsudomi and Yatabe, 2010). This genetic variation, A→G, leads to the substitution of aspartic acid in codon 994 with an identical amino acid in exon 25 s coding region, a synonymous SNP. Such synonymous SNPs do not result in coding sequence changes and may not be involved in affecting the inherent biological function of the protein (Kimchi-Sarfaty et al., 2007), but may lead to changes in protein number, structure, activity and function by affecting mRNA stability, selective splicing and translation kinetics (Sauna et al., 2007). In the present study, there was a statistically significant difference in the frequency of distribution of the rs2293347 SNP of the EGFR gene between the HC population and LC patients, which was significantly correlated with LC susceptibility, and this correlation persisted in subsequent subgroup analysis of pathology types. This is consistent with the results of a Jordanian population-based study (Bashir et al., 2019). Anne’s research (Winther-Larsen et al., 2019) revealed a notable extension in both progression-free survival (PFS) and overall survival (OS) among NSCLC patients with the CT genotype, in contrast to those with the 181946C>T (rs2293347) CC genotype, with numerous prior studies indicating a link between this particular SNP and LC susceptibility. And several previous studies (Zhang et al., 2013; Winther-Larsen et al., 2015; Ma et al., 2009) have reported that this polymorphism is significantly associated with efficacy response in NSCLC patients treated with TKI. Our KM curves and Cox regression results showed that compared to CC and CT, TT was an independent prognostic risk factor, with a significant reduction in both PFI and OS. Also in this study, carrying the CC genotype at this SNP has the potential to increase the risk of susceptibility to LUAD, LUSC and SCLC, carrying the CT or TT genotypes may be protective against the risk of susceptibility to LUAD, and carrying the CT genotype may be protective against the risk of susceptibility to LUSC and SCLC. Our research also revealed a link between rs2293347 and the expression of EGFR protein in LUAD, LUSC, and SCLC tumors, as identified through immunohistochemical staining. Additionally, lung tumor tissues from LUAD, LUSC, and SCLC patients with the TT genotype exhibited notably elevated EGFR levels compared to those with CC and CT genotypes, and that EGFR over expression has been demonstrated to be involved in numerous EGFR over expression is involved in the formation and progression of many malignant solid tumors, which effectively validates the TT genotype as an independent prognostic risk factor in this study. In conclusion, SNP at the rs2293347 of the EGFR gene may have the potential to predict LC susceptibility and prognostic risk, and its TT genotype is a protective factor against susceptibility to LC, but it may contribute to the increased worse prognosis risk of LC by inducing the upregulation of EGFR protein expression. Furthermore, considering the proximity of the rs2293347 SNP to the tyrosine kinase structural domain (Mitsudomi and Yatabe, 2010) and its correlation with the efficacy response of TKIs, this SNP may be a good candidate for future clinical trials targeting the EGFR gene to improve the clinical outcomes of LC patients.
Previous studies of the rs884225 SNP in exon 28 of the EGFR gene have shown that the rs884225 SNP is associated with the risk of developing Adverse reaction (ADR) in TKIs-treated patients with advanced NSCLC in the Chinese population (Winther-Larsen et al., 2019). In addition, Fan et al. (2019) showed that rs884225 was significantly related to EGFR expression levels and contributed to the risk of susceptible NSCLC, which is consistent with our results. However, a study from a Jordanian population reported that the rs884225 SNP was not significantly related to LC susceptibility (Bashir et al., 2019). Our study showed that the rs884225 SNP was significantly associated with LUAD susceptibility and EGFR protein expression levels in LUAD tumor tissues. We hypothesize that the rs884225 SNP could act as a possible genetic indicator for forecasting the likelihood of developing LUAD. Its T allele and TT genotype may serve as effective protective factors for susceptibility, but its resulting elevated EGFR protein levels may be responsible for a worse prognosis.
EGFR gene mutations are particularly important in guiding clinical therapeutic regimens, but their detection is often limited by factors such as difficulty in sourcing tumour tissues, technical complexity, and high cost. EGFR SNP testing offers practical advantages as a prognostic/diagnostic marker due to its cost-effectiveness and minimal sample requirements. Meanwhile, Ma’s study (Ma et al., 2011) showed that rs2293347 was associated with the efficacy of gefitinib in advanced NSCLC patients, and Zhang et al. (2013) found that the rs2293347 affects the OS of patients with LC in a population in southern China, which are further evidences of the potential of EGFR polymorphisms to guide targeted therapeutic strategies and personalized medicine. The inclusion of testing for EGFR gene polymorphisms in lung cancer screening programmes, especially for high-risk populations, can help in the early identification of individuals carrying unfavourable variants, and the adoption of targeted lifestyle modifications and regular surveillance to reduce the risk of morbidity. Individualised treatment plans are developed based on the patient’s EGFR gene variant status, including initial targeted drug selection, monitoring drug responsiveness, predicting recurrence risk, and adjusting treatment strategies to maximise therapeutic benefits. However, the above results are only speculative based on our results and the current study, and it is important to study how changes in EGFR expression in Chinese patients affect LC susceptibility and prognosis. Regardless, this study provides valuable insight into the role of EGFR in LC development and prognosis. Additionally, it proposes new strategies to assess risk and tailor interventions for patients with different histological types of LC in China.
Our study and analysis have certain limitations that need to be clarified. Firstly, our study focused on populations and patient tissues, and PDX mice and lung cancer cells should be investigated in future studies to uncover EGFR SNPs. Second, population segmentation needs to be strengthened, e.g., by differentiating between specific drugs and modalities under different treatment modalities, as well as independent analyses for small cell lung cancer. Meanwhile, the number of cases for certain indicators has decreased due to the lack of basic clinical data. Finally, as a cross-sectional, single-center study, we need to conduct multicenter longitudinal studies for further validation.
5 Conclusion
EGFR rs2293347 (pan-subtype) and rs884225 (LUAD-specific) polymorphisms increase LC risk through elevated protein expression, with rs2293347-TT conferring worse survival. These genotype-protein correlations highlight their dual role as susceptibility markers and prognostic predictors in precision oncology.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the Guilin Medical University's Affiliated Hospital’s Medical Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
CZ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Software, Writing – original draft. ZW: Conceptualization, Methodology, Project administration, Supervision, Validation, Writing – review and editing. YL: Conceptualization, Formal Analysis, Methodology, Software, Writing – review and editing. JC: Data curation, Investigation, Project administration, Supervision, Validation, Writing – review and editing. DY: Conceptualization, Formal Analysis, Methodology, Validation, Writing – review and editing. YW: Conceptualization, Project administration, Validation, Visualization, Writing – review and editing. YQ: Conceptualization, Data curation, Funding acquisition, Resources, Visualization, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the general program of Guangxi Natural Science Foundation (2025GXNSFHA069198), the Guangxi Medical and Health Appropriate Technology Development and Application Project (S2024041), the self-funded project of Guangxi Traditional Chinese Medicine (GXZYC20230356), the Innovation Project of Guangxi Graduate Education (No. YCSW2024441) and Guangxi Medical and Health Key Cultivation Discipline Construction Project (No. 12023).
Acknowledgments
The author of this article expresses gratitude for the collaborative efforts and contributions made by all members involved.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1591539/full#supplementary-material
SUPPLEMENT FIGURE 1 | Capillary electrophoresis peak map of EGFR gene polymorphism. (A) rs222798 (B) rs2293347 (C) rs884225.
References
Bashir, N. A., Morad, F. R., Khabour, O. F., AlFaqih, M. A., and Khassawneh, B. Y. (2019). The EGFR rs2233947 polymorphism is associated with lung cancer risk: a study from Jordan. Acta Biochim. Pol. 66, 315–319. doi:10.18388/abp.2019_2781
Bashir, N. A., Ragab, E. S., Khabour, O. F., Khassawneh, B. Y., Alfaqih, M. A., and Momani, J. A. (2018). The association between epidermal growth factor receptor (EGFR) gene polymorphisms and lung cancer risk. Biomolecules 8, 53. doi:10.3390/biom8030053
Colquhoun, A. J., and Mellon, J. K. (2002). Epidermal growth factor receptor and bladder cancer. Postgrad. Med. J. 78, 584–589. doi:10.1136/pmj.78.924.584
Fan, Z., Yang, J., Zhang, D., Zhang, X., Ma, X., Kang, L., et al. (2019). The risk variant rs884225 within EGFR impairs miR-103a-3p’s anti-tumourigenic function in non-small cell lung cancer. Oncogene 38, 2291–2304. doi:10.1038/s41388-018-0576-6
Feng, X., Qin, J.-J., Zheng, B.-S., Huang, L.-L., Xie, X.-Y., and Zhou, H.-F. (2014). Association of epidermal growth factor receptor (EGFR) gene polymorphism with lung cancer risk: a systematic review. J. Recept. Signal Transduct. Res. 34, 333–334. doi:10.3109/10799893.2014.885052
He, J., Huang, Z., Han, L., Gong, Y., and Xie, C. (2021). Mechanisms and management of 3rd‑generation EGFR‑TKI resistance in advanced non‑small cell lung cancer (Review). Int. J. Oncol. 59, 90. doi:10.3892/ijo.2021.5270
Huang, Y., Zhu, M., Ji, M., Fan, J., Xie, J., Wei, X., et al. (2021). Air pollution, genetic factors, and the risk of lung cancer: a prospective study in the UK biobank. Am. J. Respir. Crit. Care Med. 204, 817–825. doi:10.1164/rccm.202011-4063OC
Jurisic, V., Vukovic, V., Obradovic, J., Gulyaeva, L. F., Kushlinskii, N. E., and Djordjević, N. (2020). EGFR polymorphism and survival of NSCLC patients treated with TKIs: a systematic review and meta-analysis. J. Oncol. 2020, 1973241. doi:10.1155/2020/1973241
Kim, Y. B., Kim, G. E., Cho, N. H., Pyo, H. R., Shim, S. J., Chang, S. K., et al. (2002). Overexpression of cyclooxygenase-2 is associated with a poor prognosis in patients with squamous cell carcinoma of the uterine cervix treated with radiation and concurrent chemotherapy. Cancer 95, 531–539. doi:10.1002/cncr.10684
Kimchi-Sarfaty, C., Oh, J. M., Kim, I.-W., Sauna, Z. E., Calcagno, A. M., Ambudkar, S. V., et al. (2007). A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528. doi:10.1126/science.1135308
Krabbe, J., Steffens, K. M., Drießen, S., and Kraus, T. (2024). Lung cancer risk and occupational pulmonary fibrosis: systematic review and meta-analysis. Eur. Respir. Rev. Off. J. Eur. Respir. Soc. 33, 230224. doi:10.1183/16000617.0224-2023
Leonetti, A., Sharma, S., Minari, R., Perego, P., Giovannetti, E., and Tiseo, M. (2019). Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 121, 725–737. doi:10.1038/s41416-019-0573-8
Li, Z.-X., Wang, F., Sun, Z.-Y., Xu, C., Zou, Z.-X., Zhao, Z.-X., et al. (2023). Prognostic implications of the EGFR polymorphism rs763317 and clinical variables among young Chinese lung cancer population. Neoplasma 70, 443–450. doi:10.4149/neo_2023_230305N115
Lim, S. M., Syn, N. L., Cho, B. C., and Soo, R. A. (2018). Acquired resistance to EGFR targeted therapy in non-small cell lung cancer: mechanisms and therapeutic strategies. Cancer Treat. Rev. 65, 1–10. doi:10.1016/j.ctrv.2018.02.006
Lin, Y.-T., Tsai, T.-H., Wu, S.-G., Liu, Y.-N., Yu, C.-J., and Shih, J.-Y. (2020). Complex EGFR mutations with secondary T790M mutation confer shorter osimertinib progression-free survival and overall survival in advanced non-small cell lung cancer. Lung Cancer Amst. Neth. 145, 1–9. doi:10.1016/j.lungcan.2020.04.022
Liu, C., Xu, X., and Zhou, Y. (2015). Association between EGFR polymorphisms and the risk of lung cancer. Int. J. Clin. Exp. Pathol. 8, 15245–15249.
Liu, X., Wang, P., Zhang, C., and Ma, Z. (2017). Epidermal growth factor receptor (EGFR): a rising star in the era of precision medicine of lung cancer. Oncotarget 8, 50209–50220. doi:10.18632/oncotarget.16854
Ma, F., Sun, T., Shi, Y., Yu, D., Tan, W., Yang, M., et al. (2009). Polymorphisms of EGFR predict clinical outcome in advanced non-small-cell lung cancer patients treated with Gefitinib. Lung Cancer Amst. Neth 66, 114–119. doi:10.1016/j.lungcan.2008.12.025
Ma, F., Xu, B., Lin, D., Sun, T., and Shi, Y. (2011). Effect of rs2293347 polymorphism in EGFR on the clinical efficacy of gefitinib in patients with non-small cell lung cancer. Zhongguo Fei Ai Za Zhi Chin. J. Lung Cancer 14, 642–645. doi:10.3779/j.issn.1009-3419.2011.08.02
Malhotra, J., Malvezzi, M., Negri, E., La Vecchia, C., and Boffetta, P. (2016). Risk factors for lung cancer worldwide. Eur. Respir. J. 48, 889–902. doi:10.1183/13993003.00359-2016
Mitsudomi, T., and Yatabe, Y. (2010). Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 277, 301–308. doi:10.1111/j.1742-4658.2009.07448.x
Obradovic, J., Todosijevic, J., and Jurisic, V. (2023). Side effects of tyrosine kinase inhibitors therapy in patients with non-small cell lung cancer and associations with EGFR polymorphisms: a systematic review and meta-analysis. Oncol. Lett. 25, 62. doi:10.3892/ol.2022.13649
Oxnard, G. R., Arcila, M. E., Sima, C. S., Riely, G. J., Chmielecki, J., Kris, M. G., et al. (2011). Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 17, 1616–1622. doi:10.1158/1078-0432.CCR-10-2692
Sauna, Z. E., Kimchi-Sarfaty, C., Ambudkar, S. V., and Gottesman, M. M. (2007). Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res. 67, 9609–9612. doi:10.1158/0008-5472.CAN-07-2377
Shi, Y. Y., and He, L. (2023). Publisher Correction: SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 15, 97–98. doi:10.1038/sj.cr.7290272
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33. doi:10.3322/caac.21708
Solta, A., Ernhofer, B., Boettiger, K., Megyesfalvi, Z., Heeke, S., Hoda, M. A., et al. (2024). Small cells - big issues: biological implications and preclinical advancements in small cell lung cancer. Mol. Cancer 23, 41. doi:10.1186/s12943-024-01953-9
Voldborg, B. R., Damstrup, L., Spang-Thomsen, M., and Poulsen, H. S. (1997). Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 8, 1197–1206. doi:10.1023/a:1008209720526
Wang, Z.-F., Ren, S.-X., Li, W., and Gao, G.-H. (2018). Frequency of the acquired resistant mutation T790 M in non-small cell lung cancer patients with active exon 19Del and exon 21 L858R: a systematic review and meta-analysis. BMC Cancer 18, 148. doi:10.1186/s12885-018-4075-5
Winther-Larsen, A., Fynboe Ebert, E. B., Meldgaard, P., and Sorensen, B. S. (2019). EGFR gene polymorphism predicts improved outcome in patients with EGFR mutation-positive non-small cell lung cancer treated with erlotinib. Clin. Lung Cancer 20, 161–166. doi:10.1016/j.cllc.2019.02.011
Winther-Larsen, A., Nissen, P. H., Jakobsen, K. R., Demuth, C., Sorensen, B. S., and Meldgaard, P. (2015). Genetic polymorphism in the epidermal growth factor receptor gene predicts outcome in advanced non-small cell lung cancer patients treated with erlotinib. Lung Cancer Amst. Neth. 90, 314–320. doi:10.1016/j.lungcan.2015.09.003
Wu, S.-G., and Shih, J.-Y. (2018). Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol. Cancer 17, 38. doi:10.1186/s12943-018-0777-1
Zhang, L., Yuan, X., Chen, Y., Du, X.-J., Yu, S., and Yang, M. (2013). Role of EGFR SNPs in survival of advanced lung adenocarcinoma patients treated with Gefitinib. Gene 517, 60–64. doi:10.1016/j.gene.2012.12.087
Keywords: EGFR, polymorphism, lung cancer, susceptibility, prognosis
Citation: Zuo C, Wang Z, Liu Y, Cheng J, Yang D, Wang Y and Qiao Y (2025) EGFR polymorphisms drive lung cancer risk and survival disparities: a genotype-expression-outcome cohort study. Front. Genet. 16:1591539. doi: 10.3389/fgene.2025.1591539
Received: 11 March 2025; Accepted: 30 April 2025;
Published: 14 May 2025.
Edited by:
Elisa Frullanti, University of Siena, ItalyCopyright © 2025 Zuo, Wang, Liu, Cheng, Yang, Wang and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongchao Qiao, cWlhb3ljQGdsbWMuZWR1LmNu; Yu Wang, d2FuZ3l1MTAxMTAxQDE2My5jb20=
 Chao Zuo
Chao Zuo Ziqiang Wang
Ziqiang Wang Yi Liu1
Yi Liu1 Yongchao Qiao
Yongchao Qiao